Abstract
Insects are the most diversified and species-rich group of animals and harbor an immense diversity of viruses. Several taxa in the flavi-like superfamily, such as the genus Flavivirus, are associated with insects; however, systematic studies on insect virus genetic diversity are lacking, limiting our understanding of the evolution of the flavi-like superfamily. Here, we examined the diversity of flavi-like viruses within the most complete and up-to-date insect transcriptome collection comprising 1,243 insect species by employing a Flaviviridae RdRp profile hidden Markov model search. We identified seventy-six viral sequences in sixty-one species belonging to seventeen insect, one entognathan, and one arachnidan orders. Phylogenetic analyses revealed that twenty-seven sequences fell within the Flaviviridae phylogeny but did not group with established genera. Despite the large diversity of insect hosts studied, we only detected one virus in a blood-feeding insect, which branched within the genus Flavivirus, indicating that this genus likely diversified only in hematophagous arthropods. Nine new jingmenviruses with novel host associations were identified. One of the jingmenviruses established a deep rooting lineage additional to the insect- and tick-associated clades. Segment co-segregation phylogenies support the separation of tick- and insect-associated groups within jingmenviruses, with evidence for segment reassortment. In addition, fourteen viruses grouped with unclassified flaviviruses encompassing genome length of up to 20 kb. Species-specific clades for Hymenopteran- and Orthopteran-associated viruses were identified. Forty-nine viruses populated three highly diversified clades in distant relationship to Tombusviridae, a plant-infecting virus family, suggesting the detection of three previously unknown insect-associated families that contributed to tombusvirus evolution.
Keywords: flavi-like virus, insect virus, phylogeny, positive-sense RNA, tombusvirus
1. Introduction
During the last decade, the scientific interest in arthropod-borne viruses (arboviruses) has broadened to include arthropod- and, in particular, insect-specific viruses (Bolling et al. 2015). Whereas the arthropod vector in the arboviral transmission cycle was originally noted for its function to transmit the virus to vertebrates, it is becoming more apparent that the evolutionary origins of arboviruses may lie in arthropods. Arthropod-specific viruses could thus be regarded as precursors of arboviruses, even if they appear to be phylogenetically distinct (Marklewitz et al. 2015). In major groups of viruses, arthropod-associated members moreover seem to represent a greater sample from a common ancestral diversity than vertebrate-specific members. For instance, within bunya- and mononegaviruses, known vertebrate pathogenic viruses are embedded in a diversity of novel arthropod-specific viruses discovered only in the past years (Li et al. 2015; Käfer et al. 2019). The great majority in the phylum Arthropoda are insects. Insects are the most abundant and diversified animal group with an estimated number of ∼5.5 million species representing about eighty percent of the world’s species (Stork 2018). Metagenomic studies revealed an enormous virus diversity in insects (Li et al. 2015; Shi et al. 2016a,b; Käfer et al. 2019), yet only a fraction of the tremendous diversity of insect species has been analyzed.
The genus Flavivirus (sensu stricto, relates to ICTV-classified species) within the Flaviviridae family hosts an extensive list of human-pathogenic arboviruses, transmitted by mosquitoes and ticks. On the other hand, all other established Flaviviridae genera (Pegivirus, Pestivirus, and Hepacivirus) contain only vertebrate-infecting viruses that are not associated with arthropod vectors and show a lower intra-genetic diversity in comparison to the genus Flavivirus. New hepaci- and pegi-like viruses in virus discovery studies have only been reported to occur in primates and mammals (Porter et al. 2020). Recent findings of viruses that have different genome organizations from classical flaviviruses but group within the family Flaviviridae in phylogenetic analyses based on the RNA-dependent RNA polymerase (RdRp) gene, indicate that the evolution of this family is complex and not well understood. One of these groups contains viruses that have been found in bees, flies, and aphids, as well as in plant and nematode hosts (Kobayashi et al. 2013; Bekal et al. 2014; Shi et al. 2016a; Teixeira et al. 2016; Webster et al. 2016; Remnant et al. 2017; Kondo et al. 2020; Faizah et al. 2020). This group contains viruses with genomic length of 16–23 kb as opposed to 10 kb in classical flaviviruses, albeit with a similar organization. Another unclassified virus group contains segmented viruses, tentatively named ‘jingmenviruses’, which were recently discovered in ticks and several insects, as well as in humans with reported febrile illness (Kuivanen et al. 2019; Wang et al. 2019a,b). These data suggest that non-hematophagous insect hosts played an important role in flavivirus evolution. However, systematic studies in insects are lacking.
The Flaviviridae and Tombusviridae families are grouped in the so-called flavi-like superfamily (Koonin et al. 2015); however, the evolution of host associations within and between both families remains elusive. Tombusviruses are known to infect angiosperm plants in more than fifteen different orders, mainly causing leaf mottling and deformations, and stunting. Virus spread occurs by contact between plants, pollen, or seeds, or by contact to infected soil or water (Herrera-Vásquez et al. 2009; Mehle and Ravnikar 2012). There is little knowledge if virions can also be mechanically transmitted by arthropods perhaps acting as carriers for seeds or pollen.
Current knowledge on flavi-related viruses is mainly driven by interests in human or domestic animal health, and strategies to combat their associated repercussions. The same pattern occurs similarly in the tombus-related viruses with research lines mostly drawn across mitigation strategies for plant viral disease. Despite the need for a broad understanding of flavivirus evolution (Blitvich and Firth 2017) and RNA virus evolution in general (Koonin and Dolja 2018), only few studies have previously focused on a systematic and comprehensive search in non-typical tombusvirus hosts, that is plant hosts, or non-hematophagous flavivirus hosts, other than mosquitoes or ticks (Kobayashi et al. 2013; Bekal et al. 2014; Teixeira et al. 2016; Shi et al. 2016b; Webster et al. 2016; Remnant et al. 2017; Parry and Asgari 2019; Kondo et al. 2020). Crucially too, understanding the evolution of the flavi-like superfamily requires a unified sampling strategy of large organismic host groups in higher taxonomic ranks, such as orders or classes, and in a variety of geographic locations. However, sampling is often performed on a basis of limited geographical sites or organismic groups for reasons of capacity. Also, the practice of sample pooling, though assisting in the era of next-generation sequencing, introduces doubts in ascertaining species-specific host associations as well as in genome assembly (especially for segmented viruses) when individual samples are not retained. Here, we explored the diversity of flavi-related viruses in the largest insect collection of transcriptomes sampled worldwide representing all extant insect orders (Misof et al. 2014). Our findings unveil the evolution of the flavi-like superfamily within insects, contributing new coding-complete viral genomes and previously unknown insect host associations, even at the order level.
2. Materials and methods
2.1 Virus identification
Screening of insect transcriptome data to identify and filter viral sequences was done as described previously (Käfer et al. 2019). In brief, worldwide collected samples from 1,243 insects and other arthropods were sequenced in an Illumina HiSeq2000 platform (Misof et al. 2014). RNAseq raw data were assembled using SOAPdenovo-Trans-31kmer (version 1.01) (Li et al. 2010), and checked for quality and cross contaminations with VecScreen (www.ncbi.nlm.nih.gov/tools/vecscreen/), and UniVec database build 7.0 (www.ncbi.nlm.nih.gov/tools/vecscreen/univec/). Nucleotide assemblies were translated in all six open reading frames (ORFs) with the fastatranslate program within the package EXONERATE (version 2.2.0) (Slater and Birney 2005). The ORF library was computationally screened for viral sequences with the software HMMER version 3 (Eddy 2011), utilizing an amino acid profile hidden Markov model (pHMM) built based on a T-coffee alignment (Expresso mode) (Notredame et al. 2000) of 138 Flaviviridae RdRp sequences. The transcriptome data stem from the 1KITE consortium (NCBI Umbrella BioProject accession number: PRJNA183205, ‘The 1KITE project: evolution of insects’). The detection of distant evolutionary homologs by utilizing pHMMs comes with the downside of inflating the results with redundant sequences. Filtering out redundant matches included a two-fold approach as previously described (Käfer et al. 2019). As a first step, all hits were aligned to the initial flavivirus template RdRp alignment and sequences that were too short and/or did not overlap with the core RdRp motifs were subsequently removed. Phylogenetic tree inference with PhyML v.3.2.0 (Guindon et al. 2010) assisted further in identifying erroneously matched sequences by inspecting the topologies for instances of long branch attraction. In the second step, all sequences were compared against the non-redundant NCBI protein database, which had been filtered for viral sequences beforehand, using blastp of the BLAST+ suite v.2.2.28 (Camacho et al. 2009). The combination of results from both approaches along with a cut-off of 300 nucleotides that served as the minimum sequence length yielded the final viral sequences.
2.2 Genome assembly and annotation
Search for further viral sequence information was performed using full genome sequences of related viruses downloaded from GenBank (Supplementary Table S1). In particular, for segmented viruses (jingmenviruses, the genus Dianthovirus of Tombusviridae, Hubei tombus-like virus 28, and Wuhan insect virus 35 and -21), the corresponding sequences for non-RdRp segments were downloaded from GenBank and served as the basis to create per-segment sequence databases. Insect transcriptome assemblies were matched to amino acid libraries per genomic segment using diamond version 0.9.26 (Buchfink et al. 2015). A read-mapping step using Geneious (Geneious v.9.1.8, Biomatters, 581 Auckland, New Zealand, https://www.geneious.com) was applied to all viral sequences in order to verify the assemblies and potentially elongate them at their 5′ or 3′ ends. Genome annotations were carried out using the webservice mode of InterProScan software (Jones et al. 2014) for protein domain predictions. Cleavage sites were predicted by aligning the obtained sequences to known flaviviruses, as well as by the webservice mode of SignalP-5.0 (Armenteros et al. 2019) for host signalase site prediction.
2.3 Phylogenetic analyses
Amino acid multiple sequence alignments of the RdRp gene for Flaviviridae- and Tombusviridae-related sequences were performed using the E-INS-i mode of mafft v7.407 (Katoh and Standley 2013). Alignments were 351 and 900 amino acids long for the Flaviviridae- and Tombusviridae-related sequences, respectively. Unclassified viral sequences that have previously been reported as flavi-related, as well as unclassified flaviviruses and tombusviruses were included in the alignments. The LG amino acid substitution model was selected for both alignments based on the Bayesian Information Criterion, with the options that empirically count amino acid frequencies from the data and allow a proportion of invariable sites. Maximum likelihood phylogenies were inferred in RAxML-NG version 0.7.0 BETA (Kozlov et al. 2019) using 1,000 bootstrap replicates and the transfer bootstrap expectation metric for clade credibility. Trees were visualized using the R package ggtree (Yu et al. 2017). Phylogenetic co-segregation of different jingmenvirus segments was based on maximum likelihood phylogenies of the corresponding viral proteins, following the alignment and tree inference methods as described above. Tanglegrams were visualized using the R package dendextend (Galili 2015).
3. Results
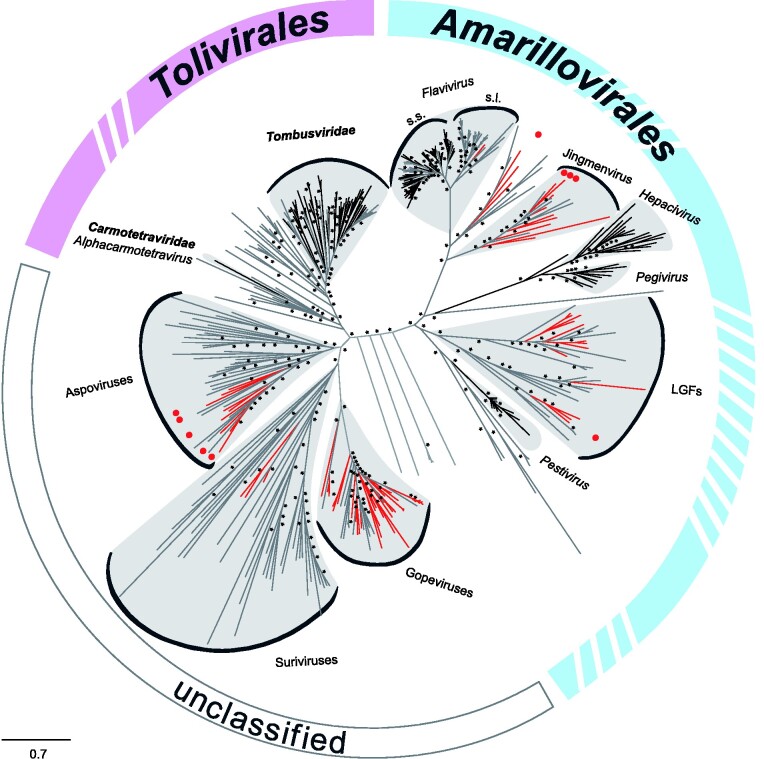
Our search, based on an amino acid profile hidden Markov model (pHMM) of the Flaviviridae RdRp gene, yielded a broad diversity of previously unknown viruses. In total, 162 putative viral sequences showing 18–61 per cent amino acid identity to flaviviruses and 22–68 per cent amino acid identity to tombusviruses were identified. The pHMM search, while based on a Flaviviridae RdRp model, delivered hits for both Flaviviridae and Tombusviridae, confirming the grouping of the two families in the flavi-like superfamily and their phylogenetic clustering within the Kitrinoviricota phylum (Wolf et al. 2018). An overview unrooted phylogeny is shown in Fig. 1 that includes Amarillovirales and Tolivirales, the two virus orders of Flaviviridae and Tombusviridae, respectively. Extensive filtering and verification of the pHMM hits based on sequence length (>300 nucleotides), taxonomic grouping, and presence of a continuous ORF containing the conserved canonical glycine–aspartate–aspartate (GDD) motif resulted in seventy-six unique viral sequences (n = twenty-seven flavi-related and n = forty-nine tombus-related) that were included in downstream phylogenetic and genome organization analyses. An average of 1.5 sequences per insect transcriptome with a maximum of six sequences per insect transcriptome were identified. Average read number per assembled sequence was 19,518 reads, with a minimum of twenty-seven and a maximum of 1,756,653 reads per assembled sequence. These viral sequences were identified in sixty-one species belonging to seventeen insect orders (such as Trichoptera, Embioptera, and Odonata), one entognathan order (Diplura), and one arachnidan order (Scorpiones), with nearly each detected virus associated with a different insect species. Transcriptomes consisted of both individual specimens (n = twenty-seven) and pools of two to sixty individuals of the same species (n = thirty-four). Details of host associations, taxonomic classification, and sampling date and location are given in Supplementary Table S1. Novel viruses were named after the host order, viral family, and the designation ‘OKIAV’ (for ‘1KITE insect-associated virus’), followed by the sample ID (e.g. Hymenopteran flavi-related virus OKIAV350) in conformity with viruses previously identified in the same sample set (Käfer et al. 2019).
Figure 1.
Maximum likelihood phylogeny of Flaviviridae (order: Amarillovirales), and Tombusviridae and Carmotetraviridae (order: Tolivirales). The phylogenetic inference was based on an amino acid alignment of the RdRp region using RAxML-NG version 0.7.0 BETA (Kozlov et al. 2019). ICTV-classified viruses are shown in black, unclassified viruses in grey, and new viruses described in this study are marked in red. Red dots represent complete genomes of new viruses described in the present study. Bootstrap values above seventy are marked with an asterisk.
3.1 Genome and phylogenetic analyses of novel flavi-related viruses
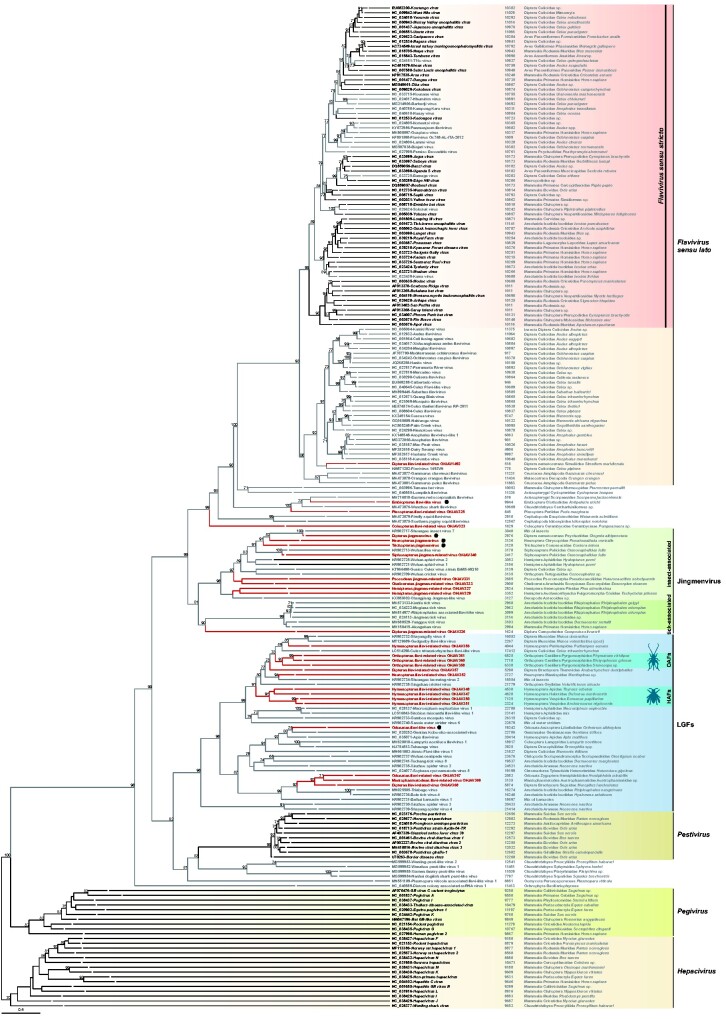
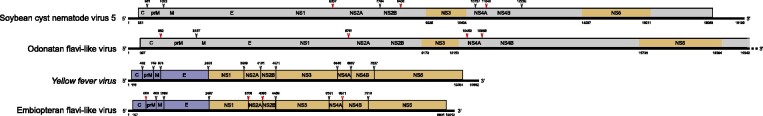
The following four viral sequences Dipteran flavi-related virus OKIAV1492, Embiopteran flavi-like virus (OKIAV324), Plecopteran flavi-related virus OKIAV325, and Coleopteran flavi-related virus OKIAV323 established four lineages in phylogenetic sister relationship to members of the genus Flavivirus (Fig. 2). Coleopteran flavi-related virus OKIAV323 branched basal to the genus Flavivirus and to the unclassified flavi-related viruses and was identified in a longhorn beetle (Pempsamacra sp.). Dipteran flavi-related virus OKIAV1492 was found in a black fly (Simulium meridionale) and shared a recent ancestor with the classical insect-specific flaviviruses. Black flies feed on vertebrates, for example birds and humans (DeFoliart and Rao 1965), supporting the notion that the genus Flavivirus (sensu lato) diversified within blood-feeding arthropods. Embiopteran flavi-like virus was found in a webspinner and grouped with Plecopteran flavi-related virus OKIAV325, detected in a stonefly, as well as with viruses from marine hosts, such as sharks and molluscs, and a bat host. Embiopteran flavi-like virus comprised a coding-complete flavivirus genome with a typical genome organization as shown in Fig. 3. Putative protein cleavage sites were similar as in other flaviviruses and are listed in Table 1. Exact size of corresponding proteins may differ from classical flaviviruses.
Figure 2.
Maximum likelihood phylogeny of flaviviruses. The phylogenetic inference was based on an amino acid alignment of the RdRp region with 1,000 bootstrap replicates, using RAxML-NG version 0.7.0 BETA (Kozlov et al. 2019). ICTV-classified viruses that belong to the Flaviviridae family are shown in black, unclassified flavi-related viruses in grey, and new flavi-related viral taxa described in this study are marked in red. Genomic sequence length is noted for every viral taxon and for segmented viruses sequence length corresponds to the RdRp-encoding segment. Viruses with a coding-complete genome described in this study are marked by a black dot. The tree is rooted to the branch leading to Hepacivirus. Bootstrap values below seventy are not shown.
Figure 3.
Genome organization of Embiopteran flavi-like virus and Odonatan flavi-like virus. Reference genomes of Yellow fever virus and Soybean cyst nematode virus 5 are shown for comparison. Nucleotide positions of ORF start/end are indicated. Arrows point to cleavage sites: black arrows for sites similar to reference sequences, red arrows for sequence length/position variation in comparison to reference sequences. A complete list of the sequence stretches around the cleavage sites can be found in Table 1.
Table 1.
Putative polyprotein cleavage sites of Embiopteran flavi-like virus, Odonatan flavi-like virus, and closely related flaviviruses.
| Cleavage site | JEV | WNV | YFV | Embiopteran flavi-like virus | SbCNV-5 | Odonatan flavi-like virus |
|---|---|---|---|---|---|---|
| C/prM | IAYAGA/MKLSNF | IASVGA/VTLSN | LLMTGG/VTLVRK | LVMVAA/AQFSAD | ILLGGG/ARFVRK | FIGKET/VKSAA |
| pr/M | SKRSRR/SVSVQT | SRRSRR/SLTVQT | SRRSRR/AIDLPT | KTRLR/VAISIP | STRGKR/AAAKSS | DRSARP/AHAGRK |
| prM/E | VAPAYS/FNCLGM | VAPAYS/FNCLGM | VGPAYS/AHCIGI | YLVVGS/KACHQV | ? | ? |
| E/NS1 | TNVHA/DTGCAI | VNVHA/DTGCAI | SLGVGA/DOGCAI | FWGVKG/DEMVLS | ? | ? |
| NS1/NS2A | QVDAF/NGEMV | QVNAY/NADMID | RSWVTA/GEIHAY | KPVYTS/GYYHDL | DSVDTA/SLRHRL | KGIDDV/YNETNK |
| NS2A/NS2B | PNKKR/GWPATE | PNRKR/GWPATE | RIFGRR/SIPVNE | IYRRKR/PKHDDP | YPFPKR/SSGWNE | ? |
| NS2B/NS3 | LKTTKR/GGVFWD | LQYTKR/GGVLWD | VRGARR/SGDVLW | YGQWGQ/RGTIMD | SGTERR/VSVAEG | ? |
| NS3/NS4A | AAGKR/SAISFI | ASGKR/SQIGLI | FAEGRR/GAAEVL | KYARLR/GKHASF | LSTGRF/GLFKTQ | HANFKR/DNVKKA |
| NS4A/NS2B | GVVAA/NEYGM | SAVAA/NEMGW | VSAVAA/NELGML | NPQIIS/ALIEVK | RSAKKE/LEGMDE | GKLKEM/LAGLKN |
| NS2B/NS5 | PSLKR/GRPGG | PGLKR/GGAKG | MKTGRR/GSANG | FETPRT/GSSHAE | SAHAKK/EGKDKA | ? |
3.2 Genome and phylogenetic analyses of novel jingmenviruses
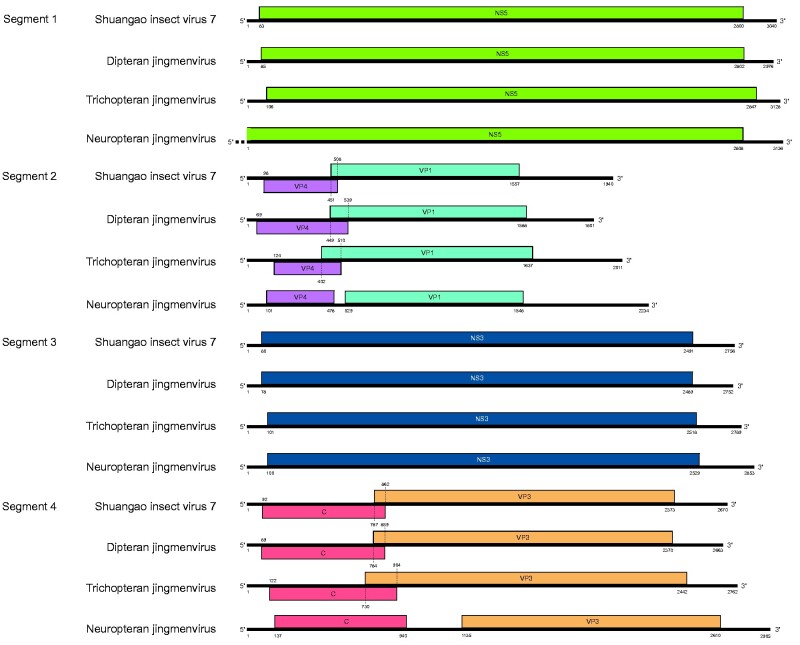
We have identified nine jingmenviruses, of which eight grouped with the insect-associated clade and one was placed on a long branch basal to the tick-associated clade based on phylogenetic analyses of RdRp proteins (Fig. 2). A search for non-RdRp genome segments yielded in total fourteen additional segments. Coding-complete genomes, each with four segments, were assembled for the following three viruses: Dipteran jingmenvirus (OKIAV332), Trichopteran jingmenvirus (OKIAV337), and Neuropteran jingmenvirus (OKIAV339). As shown in Fig. 4, these genomes have a typical jingmenvirus-like genome organization. However, segment 2 and segment 4 of Neuropteran jingmenvirus differed from those of other insect-associated jingmenviruses as the respective ORFs did not overlap (Garry and Garry 2020). Overlapping ORFs are generally encountered in insect-associated jingmenviruses but have also been observed in some tick-associated jingmenviruses (Temmam et al. 2019).
Figure 4.
Genome organization of Dipteran jingmenvirus, Trichopteran jingmenvirus, and Neuropteran jingmenvirus. Nucleotide positions of ORF start/end are indicated. The genome of Shuangao insect virus 7 is shown for comparison.
Our data on a novel jingmenvirus in bristletails (Campodea silvestrii) established a new deep branch (Dipluran jingmen-related virus OKIAV326) in addition to the insect- and tick-associated clades, and indicated that the group of jingmenviruses is more diversified than previously shown (Zhang et al. 2020) (Fig. 2). Dipluran jingmen-related virus OKIAV326 shared a most recent common ancestor with the tick-associated jingmenviruses and not as expected with the insect-associated jingmenviruses. This topology was confirmed by additional phylogenetic analyses based on an alignment containing only jingmenviruses (Supplementary Fig. S1).
Genetic markers that differ between insect- and tick-associated jingmenviruses and that are present on genomic segments other than the RdRp could not be analyzed for Dipluran jingmen-related virus OKIAV326 as only the RdRp-encoding segment 1 was identified. Siphonapteran jingmen-related virus OKIAV340 grouped with Wuhan flea virus and both viruses were found in the same host species suggesting that this cluster of jingmenviruses is associated with fleas (order Siphonaptera). Of note, Shuangao insect virus 7 was identified in a mix of two dipteran and one neuropteran insects that belong to the Psychodidae and Chrysopidae families (Shi et al. 2016a), the same families of the hosts of its sister taxa, Dipteran jingmenvirus and Neuropteran jingmenvirus. However, Dipteran jingmenvirus and Shuangao insect virus 7 were more closely related (Fig. 2), suggesting that Psychodidae species are the likely hosts for Shuangao insect virus 7.
In addition to the establishment of host-specific groups, the geographical distribution of jingmenviruses expanded significantly with the inclusion of data from this study, adding Australia, USA, Japan, and countries of Central Europe to the areas where jingmenviruses have been detected in previous studies (see Table 2 for detailed host associations and sampling sites).
Table 2.
Host associations down to the species level (wherever available) and geographic locations of jingmenviruses shown in Fig. 2.
| Virus | Order | Family | Species | Geographic location (reference) |
|---|---|---|---|---|
| Shuangao insect virus 7 | Diptera Neuroptera | Psychodidae Chrysopidae | Chrysopidae sp. Psychoda alternate Diptera sp. | China: Zhejiang (Shi et al. 2016) |
| Dipteran jingmenvirus | Diptera | Psychodidae | Clogmia albipunctata | USA: North Carolina (this study) |
| Neuropteran jingmenvirus | Neuroptera | Chrysopidae | Pseudomallada ventralis | Austria: near Vienna (this study) |
| Trichopteran jingmenvirus | Trichoptera | Conoesucidae | Costora delora | Australia: Victoria (this study) |
| Siphonapteran jingmen-related virus OKIAV340 | Siphonaptera | Pulicidae | Ctenocephalides felis | USA (this study) |
| Wuhan flea virus | Siphonaptera | Pulicidae | Ctenocephalides felis | China: Hubei (Shi et al. 2016) |
| Wuhan aphid virus 1 | Hemiptera | Aphididae | Hyalopterus pruni | China: Hubei (Shi et al. 2016) |
| Wuhan aphid virus 2 | Hemiptera | Aphididae | Mix of Hyalopterus pruni and Aulacorthum magnoliae | China: Hubei (Shi et al. 2016) |
| Cheliceratan jingmen-related virus OKIAV333 | Scorpiones | Euscorpiidae | Euscorpius sicanus | Italy: Sicily (this study) |
| Psocodean jingmen-related virus OKIAV331 | Psocoptera | Pseudocaeciliidae | Heterocaecilius solocipennis | Japan: Hokkaido (this study) |
| Guaico Culex virus | Diptera | Culicidae | Culex coronator, Culex interrogator, Culex declarator | Brazil: Nhecolandia (Pauvolid-Corrêa et al. 2016) Trinidad: Aripo (Ladner et al. 2016) Peru: Loreto (Ladner et al. 2016) Panama: Soberania, Achiotes (Ladner et al. 2016) |
| Wuhan cricket virus | Orthoptera | Tettigoniidae | Conocephalus sp. | China: Hubei (Shi et al. 2016) |
| Hemipteran jingmen-related virus OKIAV329 | Hemipteran | Cixiidae | Tachycixius pilosus | Germany: Thuringia (this study) |
| Hemipteran jingmen-related virus OKIAV327 | Hemipteran | Pleidae | Plea minutissima | Germany: Lower Saxony (this study) |
| Changjiang Jingmen-like virus | Decapoda | Cambaridae | Procambarus clarkii | China: Hubei (Shi et al. 2016) |
| Mogiana tick virus | Ixodida Artiodactyla | Ixodidae Bovidae | Rhipicephalus microplus Bos sp. | Brazil: Uberlandia (Maruyama et al. 2014;Villa et al. 2017;Souza et al. 2018;Pascoal et al. 2019) Trinidad and Tobago (Sameroff et al. 2019) |
| Kindia tick virus | Ixodida | Ixodidae | Rhipicephalus geigyi | Guinea (Ternovoi et al.) |
| Rhipicephalus associated flavi-like virus | Ixodida | Ixodidae | Rhipicephalus microplus | China: Yunnan (Xia et al. 2015) |
| Jingmen tick virus | Chiroptera Primates Primates Diptera Ixodida | Pteropodidae Hominidae Cercopithecidae Culicidae Ixodidae | Pteropus lylei Homo sapiens Piliocolobus rufomitratus Armigeres sp., Anopheles sp., Culex sp. Amblyomma testudinarium, Dermacentor nuttalli, Haemaphysalis longicornis, Haemaphysalis campanulata, Haemaphysalis flava, Hyalomma marginatum, Ixodes sinensis, Ixodes granulatus, Ixodes ricinus, Rhipicephalus microplus, Rhipicephalus sanguineus | China: Hubei, Heilongjiang (Qin et al. 2014;Jia et al. 2019;Meng et al. 2019) Lao PDR (Temmam et al. 2019) Uganda: Kibale National Park (Ladner et al. 2016) Kosovo (Emmerich et al. 2018) French Antilles (Temmam et al. 2019) Cambodia (Temmam et al. 2019) France (Temmam et al. 2019) Turkey (Dinçer et al. 2019) |
| Yanggou tick virus | Ixodida | Ixodidae | Dermacentor nuttalli | China: Xinjiang (Shen et al. unpublished data) |
| Alongshan virus | Ixodida Artiodactyla Primates Diptera | Ixodidae Bovidae Hominidae Culicidae | Ixodes ricinus, Ixodes persulcatus Bos sp., Ovis aries Homo sapiens Culex pipiens, Culex tritaeniorhynchus, Anopheles yatsushiroensis, Aedes vexans | Finland: south-eastern Finland (Kuivanen et al. 2019) Russia: Chelyabinsk, Republic of Karelia (Kholodilov et al. 2020) China: Hulunbuir, Hinggan, Qiqihar, Greater Khingan, Jilin (Wang et al. 2019a,b) |
| Dipluran jingmen-related virus OKIAV326 | Diplura | Campodeidae | Campodea silvestrii | Germany: North Rhine Westphalia (this study) |
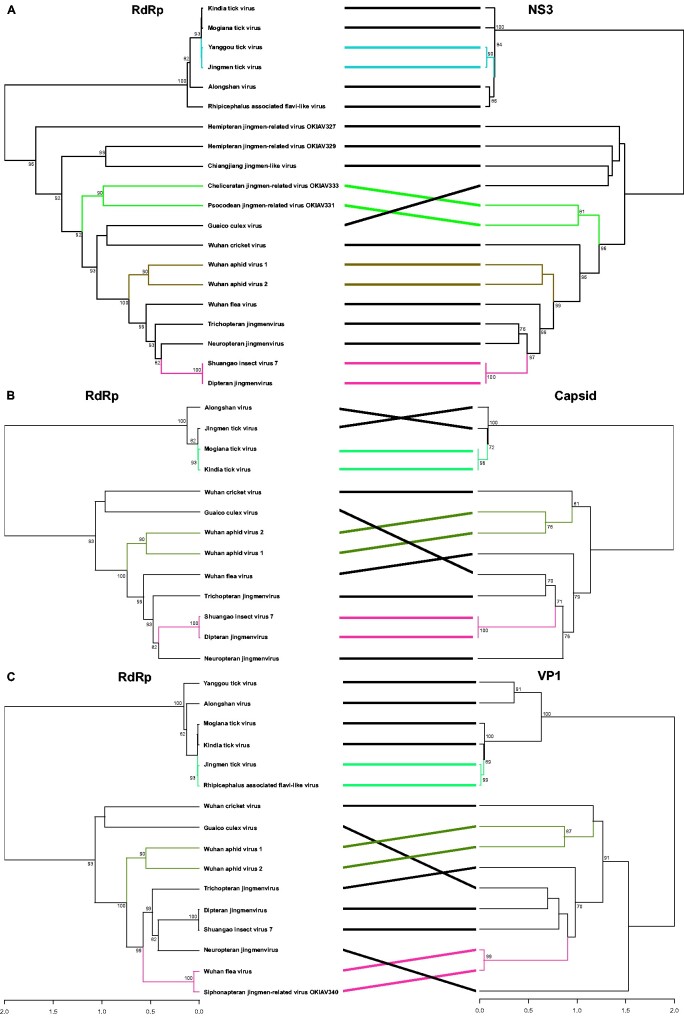
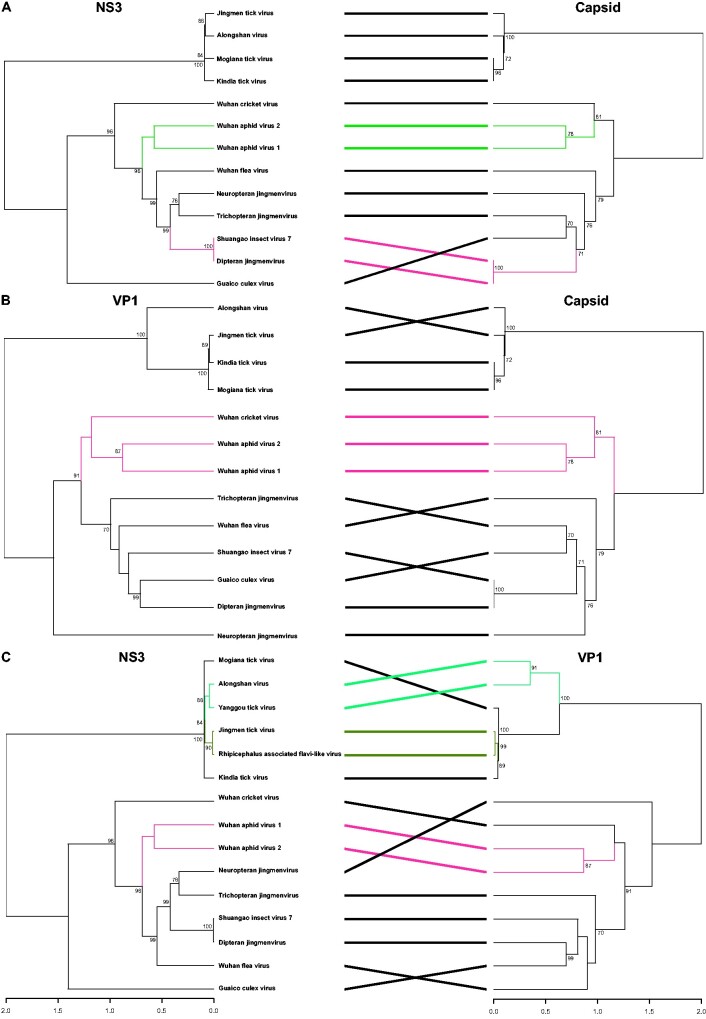
Segment co-segregation phylogenetic analyses shown in Figs. 5 and 6 reveal a consistent phylogenetic grouping among segment proteins for viruses that occur in the same host group, thus confirming the observation of intra-host adaptation for the tick-associated jingmenviruses (Temmam et al. 2019) and extending the phylogenetic congruence to some of the insect-associated jingmenviruses. All phylogenetic co-segregation topologies were identical for Wuhan aphid virus 1 and Wuhan aphid virus 2, two jingmenviruses that were both encountered in aphids. The RdRp, capsid, and NS3 phylogenies were also topologically consistent for the grouping of Shuangao insect virus 7 and Dipteran jingmenvirus, supporting the hypothesis of a dipteran host of Shuangao insect virus 7. Phylogenetic incongruence among the remaining jingmenviruses suggests an independent evolutionary scheme of reassortment events of the segments coding for the different proteins.
Figure 5.
Phylogenetic co-segregation of jingmenviruses. Analyses have been performed between RdRp and NS3, capsid, and VP1 genes. Topologically congruent clades are highlighted in color. Branches in black indicate taxa that do not share a common topological pattern in the respective tree pairs. Bootstrap values below seventy are not shown.
Figure 6.
Phylogenetic co-segregation of jingmenviruses. Analyses have been performed between NS3, capsid, and VP1 genes. Topologically congruent clades are highlighted in color. Branches in black indicate taxa that do not share a common topological pattern in the respective tree pairs. Bootstrap values below seventy are not shown.
3.3 Genome and phylogenetic analyses of a novel clade of unclassified flavi-related viruses
We have identified fourteen viruses within a clade of unclassified flaviviruses with genome length ranging from 16–26 kb, marked as LGFs for ‘large genome flaviviruses’ in Fig. 2. This clade is highly diversified, especially in comparison to pestiviruses that branch as sister clade, and associated with a wide diversity of arthropod hosts. Two novel host-specific subgroups were established based on four viruses detected in hymenopterans and three viruses detected in orthopterans, marked as HAFs and OAFs for Hymenoptera- and Orthoptera-associated flaviviruses, respectively. Hymenopteran flavi-related virus OKIAV356 did not group within the HAF clade as confirmed by tree inference analyses based on a clade-specific alignment. Whereas HAFs and OAFs formed host-specific subclades, LGFs detected in other insects did not form such groups, e.g. viruses detected in Odonata or Diptera did not group together. The nearly coding-complete genome of Odonatan flavi-like virus (OKIAV365) is comparable to the genome of Soybean cyst nematode virus 5 in Fig. 3. However, as LGFs show very limited similarity to flaviviruses and as no complete genome annotation for any LGF is available, annotations were only possible for the NS3 and NS5 genes.
3.4 Genome and phylogenetic analyses of novel tombus-related viruses
We detected forty-nine tombus-related viruses, which, together with other invertebrate viruses, established three distinct phylogenetic clades in distant relationship to the family Tombusviridae (Fig. 7). The three clades were provisionally named Gopeviruses, Suriviruses, and Aspoviruses. The clade of Gopeviruses was mostly populated by viruses identified within this study and showed several host-specific subclades. The Dipteran tombus-related viruses OKIAV386, -OKIAV387, and -OKIAV388 grouped together, and the subclade was named flower-feeding Diptera-associated tombus-related viruses (FF-DATs), stemming from three different Brachycera species that feed on nectar and pollen. The larvae of Pterodontia mellii are parasitic to spiders, which could explain the close relationship between Dipteran tombus-related virus OKIAV386 and Hubei tombus-like virus 30 that was found in spiders (Shi et al. 2016). Six viruses were identified in a wingless ectoparasite of bees, Braula coeca, and grouped in a monophyletic subclade of wingless Diptera-associated tombus-related viruses (W-DATs). The sister phylogenetic relationship of subclade W-DATs to Fig wasp tombus-like virus 1 and to the subclade of Hymenoptera-associated tombus-related viruses (HATs) indicated that the HATs subclade probably harbors a larger diversity of hymenopteran viruses, yet to be discovered. Both the W-DATs and HATs clades consist of viruses identified in monospecific insect hosts. The short genetic distance within the W-DATs and HATs subclades suggested a co-evolutionary relationship between hosts and viruses. Brachycera species host the viruses identified within the Diptera-associated tombus-related subclade (DATs), with the exception of Dipteran tombus-related virus OKIAV407 that stems from a Nematocera species.
Figure 7.
Maximum likelihood phylogeny of the families Tombusviridae and Carmotetraviridae and related viruses. The phylogenetic inference was based on an amino acid alignment of the RdRp region with 1,000 bootstrap replicates, using RAxML-NG version 0.7.0 BETA (Kozlov et al. 2019). ICTV-classified viruses of the families Tombusviridae and Carmotetraviridae are shown in black, unclassified tombus-related viruses in grey, and tombus-related viral taxa described in this study in red. Genomic sequence length is noted for every viral taxon and for segmented viruses sequence length corresponds to the RdRp-encoding segment. Viruses with a coding-complete genome described in this study are marked by a black dot. The tree is rooted to the branch leading to the lower part of the tree which includes Tombusviridae and Carmotetraviridae. Bootstrap values below seventy are not shown.
Within the clade of Suriviruses, the insect-associated virus diversity dropped significantly, as this clade contains mostly viruses with non-insect arthropod hosts, such as Crustacea and Myriapoda. Only three viruses from this study fell within this clade, each stemming from a host of a different insect order (Fig. 7). Interestingly, Leptomonas pyrrhocoris tombus-like virus 1 has a protozoan origin, found in the trypanosoma species Leptomonas pyrrhocoris (Grybchuk et al. 2018). It has been suggested that a possible transmission route for this parasite to acquire Leptomonas pyrrhocoris tombus-like virus 1 is through the parasite’s host, firebugs (order: Hemiptera, species: Pyrrhocoris apterus) (Grybchuk et al. 2018). Yet, no closely related virus of hemipteran origin was detected in our sample set.
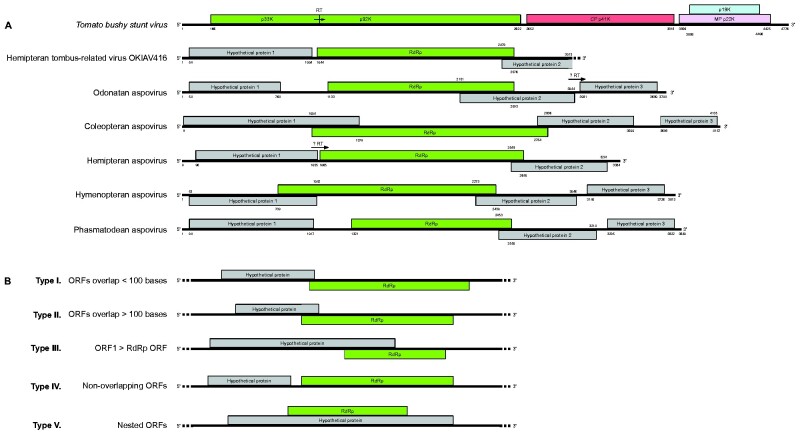
Nine viruses from the present study were found within the clade of Aspoviruses (Fig. 7). Six of them are potentially coding-complete viral genomes (genome organization shown in Fig. 8A) and each belongs to a host of a different insect order. The genomes of these viruses showed similar organizations to the ones of tombusviruses, though with longer regions of overlapping ORFs. All tombusviruses share the common characteristic of amber codon readthrough in ORF1 which results in the expression of a larger protein (reviewed in Sit and Lommel 2015). Amber codons (UAG triplets) appeared in the predicted protein 1 of Hemipteran aspovirus (OKIAV417) and in the predicted protein 2 of Odonatan aspovirus (OKIAV411). We could not verify whether those two codons undergo readthrough, but it seems a likely outcome in both cases as this is a common observation for ORF1 across all tombusviruses. Viruses of the genus Machlomovirus involve an additional readthrough event in ORF3 (Scheets 2016). Fig. 8B shows five different types of genome organizations of incomplete tombus-related sequences listed in Table 3. Because some of the tombus-related viruses (Hubei tombus-like virus 28 and Wuhan insect virus 35 and -21), as well as the genus Dianthovirus within the Tombusviridae family have a segmented genome, we searched for the presence of additional genome segments in our data. However, we only identified monopartite tombus-related viral sequences.
Figure 8.
(A) Genome organization of tombus-related viral sequences. The genome of Tomato bushy stunt virus is shown for comparison. Potential amber codon readthroughs are indicated with ‘RT’. Nucleotide positions of ORF start/end are indicated. (B) Five scheme types corresponding to the genome organization for smaller-than-genome viral sequences: I. ORF1 and RdRp ORF overlap for less than 100 bases; II. ORF1 and RdRp ORF overlap for more than 100 bases; III. ORF1 is longer than RdRp ORF; IV. ORF1 and RdRp ORF do not overlap; V. ORF1 and RdRp ORF are nested. Correspondence of genome scheme to viral sequence is listed in Table 3.
Table 3.
Genome scheme type to viral sequence correspondence from Fig. 8B.
| Type I | Coleopteran tombus-related virus OKIAV396, Hymenopteran tombus-related virus OKIAV377, -OKIAV378, -OKIAV379, -OKIAV390, -OKIAV415, Megalopteran tombus-related virus OKIAV398, Neuropteran tombus-related virus OKIAV373, Odonatan tombus-related virus OKIAV382 |
| Type II | Coleopteran tombus-related virus OKIAV372, -OKIAV397, Dipteran tombus-related virus OKIAV374, -OKIAV400, -OKIAV404, -OKIAV405, -OKIAV409, -OKIAV410, Hymenopteran tombus-related virus OKIAV383, -OKIAV384, Odonatan tombus-related virus OKIAV381, Zygentoman tombus-related virus OKIAV389 |
| Type III | Dipteran tombus-related virus OKIAV374, -OKIAV375, -OKIAV387, -OKIAV388, Hymenopteran tombus-related virus OKIAV385, Raphidiopteran tombus-related virus OKIAV395 |
| Type IV | Coleopteran tombus-related virus OKIAV419, Hymenopteran tombus-related virus OKIAV371, Odonatan tombus-related virus OKIAV420 |
| Type V | Dipluran tombus-related virus OKIAV422 |
Sequence length for all sequences below is in the range of 2–2.8 kb.
4. Discussion
In this study, we discovered sequences ascribable to seventy-six novel insect-associated viruses in seventeen insect, one entognathan, and one arachnidan host orders, related to two of the largest families within the Kitrinoviricota phylum: Flaviviridae and Tombusviridae. Our data contribute to the known flavi-related insect-specific viral diversity introducing new insect host associations of non-typical flavivirus hosts, such as the Mantophasmatodea, Trichoptera, and Neuroptera orders. Phylogenetic relationships and distances suggest the establishment of two new taxonomic genera within the Flaviviridae family (Fig. 2). In addition, our sequences populate three highly diversified phylogenetic clades grouping as sister clades to the Tombusviridae (Fig. 7), each as diverse as to signify taxonomic assignment of a new family.
Phylogenetic inference of identified sequences in relationship to flaviviruses revealed that the new sequences were widely distributed across the phylogeny and did not group with the established genera of the family Flaviviridae (Fig. 2). Interestingly, no novel virus from the present study was identified within the genus Flavivirus (sensu stricto). The same applied to the vertebrate-infecting virus genera Pestivirus, Pegivirus, and Hepacivirus. Despite the large number of diverse Diptera species (n = eighty-one) that were sampled in the course of the 1KITE project (Misof et al. 2014), the sampling did not include any members of Culicidae, the family that includes all species of mosquitoes. Ticks were also not included in the sampling, which explains the absence of viral sequences within the Flavivirus genus in traditional arthropod hosts altogether. However, the absence of viruses from this genus (s.s.) in such a comprehensive sample of insects covering all extant orders is remarkable. Taken together with the occurrence of Dipteran flavi-like virus OKIAV1492 in a blood-feeding black fly (Simuliidae, Simulium meridionale), these findings provide evidence that the evolution of the genus Flavivirus (s.l.) is likely associated with blood-feeding arthropods. Viruses of this genus mainly diversified in Culicidae and Ixodida. Whether the ancestors of this genus originated in vertebrates or in arthropods before they acquired their dual-host tropism is still unclear as insect-specific viruses as well as crustacean-, fish-, and cephalopod-infecting viruses have been found in basal phylogenetic relationship to the genus Flavivirus (s.s.) (Soto et al. 2020).
Until now, jingmenviruses have been described in a set of hosts that, apart from mammals, includes ticks, mosquitoes, fleas, and non-bloodfeeding insects (Maruyama et al. 2014; Qin et al. 2014; Ladner et al. 2016; Shi et al. 2016a,b; Pauvolid-Corrêa et al. 2016; Villa et al. 2017; Emmerich et al. 2018; Kuivanen et al. 2019; Wang et al. 2019a,b; Temmam et al. 2019; Kholodilov et al. 2020; Ternovoi et al.; Shen et al. unpublished data). This group of viruses has not been taxonomically classified yet but is phylogenetically related to the genus Flavivirus based on NS3 and NS5 protein similarities. Yet, their genomes, unlike flaviviruses, are segmented and generally consist of four segments (with the exception of Guaico Culex virus that has an additional fifth segment, however experimentally shown to be dispensable for genome replication (Ladner et al. 2016)). We identified jingmenviruses in hosts with a variety of ecological lifestyles such as drain flies, booklice, aquatic insects, scorpions, and soil-dwelling bristletails, thereby enriching the current knowledge of jingmenvirus host range.
Previous phylogenetic reconstructions separated jingmenviruses into two clades according to their host associations: the insect- and the tick-associated clades (Temmam et al. 2019). In addition to the distinct host range, the insect- and tick-associated clades were shown to differ by a number of genetic features. For example, conserved nucleotide stretches in the 5′ and 3′ untranslated regions (UTRs), and a 3′-end poly-A tail were present in all four genomic segments in tick-associated but not in insect-associated jingmenviruses underlining the differentiation of these two groups. Of note, for Dipluran jingmen-related virus OKIAV326, only a partial sequence of the RdRp genomic segment was retrieved. The 5′-end of the segment is missing but the 3′-end is available and lacks a poly-A tail. Dipluran jingmen-related virus OKIAV326 shared a most recent common ancestor with the tick-associated jingmenviruses. These findings imply that the ancestor of jingmenviruses may have existed in insects with genomic segments not containing 3′-end poly-A tails. Recently, class II viral fusion protein (VFP) domains were identified in envelope glycoproteins of tick-associated jingmenviruses, whereas the insect-associated viruses did not contain class II VFPs. Class II VFPs in tick-associated jingmenviruses stem likely from flavivirus class II VPFs and have probably been secondarily lost in the insect-associated clade when this clade separated from the tick-associated clade via divergent evolution, segment reassortment or recombination (Garry and Garry 2020). The presence of a mucin-like domain, an ectodomain of beta-sheets and alpha-helices in the tick-associated jingmenvirus glycoproteins, further differentiates them from the insect-associated jingmenvirus glycoproteins. None of the four insect-associated jingmenvirus glycoprotein segments reported here contained the fusion peptide or the mucin-like domain present in the tick-associated jingmenviruses. The phylogenetic separation of the tick-associated from the insect-associated jingmenviruses was apparent and maintained in all phylogenetic segment co-segregation analyses (Figs. 5 and 6), supporting previous suggestions on divergent evolution of the envelope glycoprotein structure in tick- and insect-associated jingmenviruses (Garry and Garry 2020).
Segmentation within the Flaviviridae-related viruses is suggested to have evolved only once because of the monophyletic nature of the jingmenvirus clade (Shi et al. 2016a). Although the non-structural proteins NS3 and NS5 share sequence similarities with flaviviruses, homologs of the jingmenvirus structural proteins, glycoprotein and capsid, are yet unknown (Qin et al. 2014). Explanations for the unrelatedness to the flavivirus structural proteins may lie within the hypothesis that two segments of an ancestral flavivirus genome were perhaps co-packaged or captured by structural ‘orphan’ proteins of a co-infecting virus (Qin et al. 2014).
A series of previously described viruses with distinct genome organization (similar to flaviviruses, but with substantially larger genome length: 16–23 kb) (Kobayashi et al. 2013; Bekal et al. 2014; Shi et al. 2016a; Teixeira et al. 2016; Webster et al. 2016; Remnant et al. 2017; Kondo et al. 2020; Faizah et al. 2020), share homologous sequence regions with flaviviruses mainly in the protease, helicase, and RNA-dependent RNA polymerase (RdRp) genes. We identified fourteen viruses that together with the previously described viruses formed the phylogenetic clade of LGFs within the Flaviviridae (Fig. 2). The distinct genomic length and organization (Fig. 3), as well as the phylogenetic distances of LGFs (Fig. 2) signify their classification as a separate genus within the Flaviviridae family. LGFs show a remarkably broad spectrum of host associations, ranging from gentian plants to nematodes and other arthropods. LGFs grouped as sister clade to the genus Pestivirus which contains vertebrate-infecting viruses only, livestock pathogenic viruses, with genomes of approximately 12 kb. The observed genetic divergence of LGFs was much greater than that of pestiviruses reflected by multiple highly diverse clades separated by long branches suggesting that the putative insect-specific LGF viruses are ancestral to the mammalian pestiviruses.
Numerous tombus-related viruses have recently been described in non-plant hosts, such as marine invertebrates and terrestrial arthropods, demonstrating both mono- and bipartite genome organizations (Shi et al. 2016b). These viruses show a large genetic diversity and branch basal to the plant-associated Tombusviridae suggesting that the RNA virome of angiosperm plants evolved via horizontal virus transfer from invertebrates (Shi et al. 2016b; Dolja and Koonin 2018; Wolf et al. 2018). Phylogenetic analyses of the forty-nine newly identified tombus-related viruses in this study showed that the sequences fell within three highly diversified major clades, each being as diverse as the family Tombusviridae, and shared a most recent common ancestor with the family Tombusviridae (Fig. 7). Our findings corroborate the horizontal virus transfer theory of plant virus evolution via multiple events of horizontal virus transfer from different invertebrate hosts (Dolja and Koonin 2018). As the origin of invertebrates precedes that of angiosperm plants, the members of Tombusviridae possibly evolved after being passed on to angiosperm plant hosts from invertebrates due to the tight biological associations formed between these two host groups (Dolja and Koonin 2018). Also, the absence of tombus-related viruses in unicellular eukaryotic hosts indicates the absence of a common viral ancestry that preceded the last eukaryotic common ancestor (Dolja and Koonin 2011). Despite the position of Leptomonas pyrrhocoris tombus-like virus 1 in the clade of Suriviruses (Fig. 7), the transmission route of this virus likely involves the trypanosomatid firebug host (Grybchuk et al. 2018). These findings together with the novel viruses identified in insect hosts within the present study suggest that insect-associated viruses played an ancestral role in plant-insect virus exchange but are not involved in current virus transmission (Dolja and Koonin 2011, 2018).
With the given ease of generating environmental and organismic metagenomic data, large projects of virus sequence discoveries will accompany us in the future years. The number of uncultivated virus genomes has surpassed that of viral genomes stemming from cultivated isolates (Roux et al. 2019). An important effort has been made to define the minimum information that should accompany uncultured viral genomes with their sequence publication (Roux et al. 2019). Crucially also, the ICTV has made an important step to align with the current flow of metagenomic sequence information, allowing the classification of viruses that stem from metagenomic sources (Simmonds et al. 2017). Nevertheless, efforts towards elaborate examinations of sequence information should extend to protect the quality of published sequence information.
Supplementary data
Supplementary data are available at Virus Evolution online.
Supplementary Material
Acknowledgements
We thank Dr. Karen Meusemann, Dr. Jeanne Wilbrandt, and Dr. Tanja Ziesmann for providing the metadata of the 1KITE transcriptomes. Computation has been performed on the HPC for Research cluster of the Berlin Institute of Health.
Funding
This work was supported by grants from the Deutsche Forschungsgemeinschaft DR 772/7-2 (grant to C.D.), the German Ministry of Research DZIF TTU 01.801 (grant to C.D.), and the Federal Ministry of Education and Research BMBF 01KI1716 (grant to S.J.) as part of the Research Network Zoonotic Infectious Diseases. Work of BM has been funded by a Leibniz competition grant ‘Graduate School on Biodiversity Genomics’.
Conflict of interest: None declared.
Data availability
Sequences are available under GenBank accession numbers MW208755-MW208806 and MW314679-MW314716 (Supplementary Table S1).
References
- Armenteros J. J. A. et al. (2019) ‘SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks’, Nature Biotechnology, 37: 420–3. [DOI] [PubMed] [Google Scholar]
- Bekal S. et al. (2014) ‘A Novel Flavivirus in the Soybean Cyst Nematode’, The Journal of General Virology, 95: 1272–80. [DOI] [PubMed] [Google Scholar]
- Blitvich B. J., Firth A. E. (2017) ‘A Review of Flaviviruses That Have No Known Arthropod Vector’, Viruses, 9: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolling B. G. et al. (2015) ‘Insect-Specific Virus Discovery: Significance for the Arbovirus Community’, Viruses, 7: 4911–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D. H. (2015) ‘Fast and Sensitive Protein Alignment Using DIAMOND’, Nature Methods, 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Camacho C. et al. (2009) ‘BLAST+: Architecture and Applications’, BMC Bioinformatics, 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFoliart G. R., Rao M. R. (1965) ‘The Ornithophilic Black Fly Simulium meridionale Riley (Diptera: Simuliidae) Feeding on Man during Autumn’, Journal of Medical Entomology, 2: 84–5. [DOI] [PubMed] [Google Scholar]
- Dinçer E. et al. (2019) ‘Survey and Characterization of Jingmen Tick Virus Variants’, Viruses, 11: 1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja V. V., Koonin E. V. (2011) ‘Common Origins and Host-Dependent Diversity of Plant and Animal Viromes’, Current Opinion in Virology, 1: 322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja V. V., Koonin E. V. (2018) ‘Metagenomics Reshapes the Concepts of RNA Virus Evolution by Revealing Extensive Horizontal Virus Transfer’, Virus Research, 244: 36–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. (2011) ‘Accelerated Profile HMM Searches’, PLoS Computational Biology, 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich P. et al. (2018) ‘Viral Metagenomics, Genetic and Evolutionary Characteristics of Crimean-Congo Hemorrhagic Fever Orthonairovirus in Humans, Kosovo’, Infection, Genetics and Evolution, 65: 6–11. [DOI] [PubMed] [Google Scholar]
- Faizah A. N. et al. (2020) ‘Deciphering the Virome of Culex vishnui Subgroup Mosquitoes, the Major Vectors of Japanese Encephalitis, in Japan’, Viruses, 12: 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili T. (2015) ‘dendextend: An R Package for Visualizing, Adjusting and Comparing Trees of Hierarchical Clustering’, Bioinformatics, 31: 3718–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry C. E., Garry R. F. (2020) ‘Proteomics Computational Analyses Suggest That the Envelope Glycoproteins of Segmented Jingmen Flavi-Like Viruses Are Class II Viral Fusion Proteins (β-Penetrenes) with Mucin-Like Domains’, Viruses, 12: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grybchuk D. et al. (2018) ‘Viral Discovery and Diversity in Trypanosomatid Protozoa with a Focus on Relatives of the Human Parasite Leishmania’, Proceedings of the National Academy of Sciences of the United States of America, 115: E506–E515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0’, Systematic Biology, 59: 307–21. [DOI] [PubMed] [Google Scholar]
- Herrera-Vásquez J. A. et al. (2009) ‘Seed Transmission of Melon Necrotic Spot Virus and Efficacy of Seed-Disinfection Treatments’, Plant Pathology, 58: 436–42. [Google Scholar]
- Jia N. et al. (2019) ‘Emergence of Human Infection with Jingmen Tick Virus in China: A Retrospective Study’, EBioMedicine, 43: 317–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. et al. (2014) ‘InterProScan 5: Genome-Scale Protein Function Classification’, Bioinformatics, 30: 1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käfer S. et al. (2019) ‘Re-Assessing the Diversity of Negative Strand RNA Viruses in Insects’, PLoS Pathogens, 15: e1008224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodilov I. S. et al. (2020) ‘Isolation and Characterisation of Alongshan Virus in Russia’, Viruses, 12: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K. et al. (2013) ‘Gentian Kobu-Sho-Associated Virus: A Tentative, Novel Double-Stranded RNA Virus That is Relevant to Gentian Kobu-Sho Syndrome’, Journal of General Plant Pathology, 79: 56–63. [Google Scholar]
- Kondo H. et al. (2020) ‘Virome Analysis of Aphid Populations That Infest the Barley Field: The Discovery of Two Novel Groups of Nege/Kita-Like Viruses and Other Novel RNA Viruses’, Frontiers in Microbiology, 11: 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Dolja V. V. (2018) ‘Metaviromics: A Tectonic Shift in Understanding Virus Evolution’, Virus Research, 246: A1–A3. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Dolja V. V., Krupovic M. (2015) ‘Origins and Evolution of Viruses of Eukaryotes: The Ultimate Modularity’, Virology, 479-480: 2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov A. M. et al. (2019) ‘RAxML-NG: A Fast, Scalable, and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference’, Bioinformatics, 35: 4453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen S. et al. (2019) ‘Detection of Novel Tick-Borne Pathogen, Alongshan Virus, in Ixodes ricinus Ticks, South-Eastern Finland, 2019’, Euro Surveill, 24: e1900394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. T. et al. (2016) ‘A Multicomponent Animal Virus Isolated from Mosquitoes’, Cell Host & Microbe, 20: 357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-X. et al. (2015) ‘Unprecedented Genomic Diversity of RNA Viruses in Arthropods Reveals the Ancestry of Negative-Sense RNA Viruses’, eLife, 4: e05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. et al. (2010) ‘De Novo Assembly of Human Genomes with Massively Parallel Short Read Sequencing’, Genome Research, 20: 265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklewitz M. et al. (2015) ‘Evolutionary and Phenotypic Analysis of Live Virus Isolates Suggests Arthropod Origin of a Pathogenic RNA Virus Family’, Proceedings of the National Academy of Sciences, 112: 7536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S. R. et al. (2014) ‘Characterisation of Divergent Flavivirus NS3 and NS5 Protein Sequences Detected in Rhipicephalus microplus Ticks from Brazil’, Memorias Do Instituto Oswaldo Cruz, 109: 38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle N., Ravnikar M. (2012) ‘Plant Viruses in Aqueous Environment–Survival, Water Mediated Transmission and Detection’, Water Research, 46: 4902–17. [DOI] [PubMed] [Google Scholar]
- Meng F. et al. (2019) ‘Virome Analysis of Tick-Borne Viruses in Heilongjiang Province, China’, Ticks and Tick-Borne Diseases, 10: 412–20. [DOI] [PubMed] [Google Scholar]
- Misof B. et al. (2014) ‘Phylogenomics Resolves the Timing and Pattern of Insect Evolution’, Science, 346: 763–7. [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Heringa J. (2000) ‘T-Coffee: A Novel Method for Fast and Accurate Multiple Sequence Alignment’, Journal of Molecular Biology, 302: 205–17. [DOI] [PubMed] [Google Scholar]
- Parry R., Asgari S. (2019) ‘Discovery of Novel Crustacean and Cephalopod Flaviviruses: Insights into the Evolution and Circulation of Flaviviruses between Marine Invertebrate and Vertebrate Hosts’, Journal of Virology, 93: e00432–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoal J. d O. et al. (2019) ‘Detection and Molecular Characterization of Mogiana Tick Virus (MGTV) in Rhipicephalus microplus Collected from Cattle in a Savannah Area, Uberlândia, Brazil’, Ticks and Tick-Borne Diseases, 10: 162–5. [DOI] [PubMed] [Google Scholar]
- Pauvolid-Corrêa A. et al. (2016) ‘Novel Viruses Isolated from Mosquitoes in Pantanal’, Genome Announcements, 4: e01195–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter A. F. et al. (2020) ‘Novel Hepaci- and Pegi-like Viruses in Native Australian Wildlife and Non-Human Primates’, Virus Evolution, 6: veaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X.-C. et al. (2014) ‘A Tick-Borne Segmented RNA Virus Contains Genome Segments Derived from Unsegmented Viral Ancestors’, Proceedings of the National Academy of Sciences of the United States of America, 111: 6744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remnant E. J. et al. (2017) ‘A Diverse Range of Novel RNA Viruses in Geographically Distinct Honey Bee Populations’, Journal of Virology, 91: e00158–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S. et al. (2019) ‘Minimum Information about an Uncultivated Virus Genome (MIUViG)’, Nature Biotechnology, 37: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameroff S. et al. (2019) ‘Viral Diversity of Tick Species Parasitizing Cattle and Dogs in Trinidad And Tobago’, Scientific Reports, 9: 10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheets K. (2016) ‘Analysis of Gene Functions in Maize Chlorotic Mottle Virus’, Virus Research, 222: 71–9. [DOI] [PubMed] [Google Scholar]
- Shi M. et al. (2016a) ‘Divergent Viruses Discovered in Arthropods and Vertebrates Revise the Evolutionary History of the Flaviviridae and Related Viruses’, Journal of Virology, 90: 659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M. et al. (2016b) ‘Redefining the Invertebrate RNA Virosphere’, Nature, 540: 539–43. [DOI] [PubMed] [Google Scholar]
- Simmonds P. et al. (2017) ‘Consensus Statement: Virus Taxonomy in the Age of Metagenomics’, Nature Reviews Microbiology, 15: 161–8. [DOI] [PubMed] [Google Scholar]
- Sit T. L., Lommel S. A. (2015) ‘Tombusviridae’, eLS, 1–9. [Google Scholar]
- Slater G. S. C., Birney E. (2005) ‘Automated Generation of Heuristics for Biological Sequence Comparison’, BMC Bioinformatics, 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto E. et al. (2020) ‘First Isolation of a Novel Aquatic Flavivirus from Chinook Salmon (Oncorhynchus tshawytscha) and Its In Vivo Replication in a Piscine Animal Model’, Journal of Virology, 94: e00337-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza W. d. et al. (2018) ‘Viral Diversity of Rhipicephalus microplus Parasitizing Cattle in Southern Brazil’, Scientific Reports, 8: 16315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork N. E. (2018) ‘How Many Species of Insects and Other Terrestrial Arthropods Are There on Earth? ’, Annual Review of Entomology, 63: 31–45. [DOI] [PubMed] [Google Scholar]
- Teixeira M. et al. (2016) ‘A Novel Virus from Macrosiphum euphorbiae with Similarities to Members of the Family Flaviviridae’, Journal of General Virology, 97: 1261–71. [DOI] [PubMed] [Google Scholar]
- Temmam S. et al. (2019) ‘Insights into the Host Range, Genetic Diversity, and Geographical Distribution of Jingmenviruses’ mSphere, 4: e00645–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternovoi V. A. et al. Complete Coding Genome Sequence for Novel Multicomponent Kindia Tick Virus Isolated from Ticks Collected in Guinea. submitted to Microbiology Resource Announcements.
- Villa E. C. et al. (2017) ‘Complete Coding Genome Sequence for Mogiana Tick Virus, a Jingmenvirus Isolated from Ticks in Brazil’, Genome Announcements, 5: e00232–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-D. et al. (2019a) ‘A New Segmented Virus Associated with Human Febrile Illness in China’, The New England Journal of Medicine, 380: 2116–25. [DOI] [PubMed] [Google Scholar]
- Wang Z.-D. et al. (2019b) ‘Prevalence of the Emerging Novel Alongshan Virus Infection in Sheep and Cattle in Inner Mongolia, Northeastern China’, Parasites & Vectors, 12: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C. L. et al. (2016) ‘Twenty-Five New Viruses Associated with the Drosophilidae (Diptera)’, Evolutionary Bioinformatics Online, 12: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y. I. et al. (2018) ‘Origins and Evolution of the Global RNA Virome’, mBio, 9: e02329–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H. et al. (2015) ‘Metagenomic Profile of the Viral Communities in Rhipicephalus spp. ticks from Yunnan, China’, PLoS One, 10: e0121609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. et al. (2017) ‘ggtree: An R Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data’, Methods in Ecology and Evolution, 8: 28–36. [Google Scholar]
- Zhang X. et al. (2020) ‘The Discovery of Segmented Flaviviruses: Implications for Viral Emergence’, Current Opinion in Virology, 40: 11–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences are available under GenBank accession numbers MW208755-MW208806 and MW314679-MW314716 (Supplementary Table S1).