Abstract
Background:
Rearrangements involving the MYC protooncogene are common in newly diagnosed multiple myeloma (MM), but their prognostic significance is still unclear. The purpose of this study was to assess the impact of MYC rearrangement on clinical characteristics, treatment response, and survival in newly diagnosed MM.
Patients and Methods:
This is a retrospective study including 1342 patients seen in Mayo Clinic in Rochester, MN, from January 2006 to January 2018, and who had cytogenetic testing by fluorescence in-situ hybridization at diagnosis, including MYC testing using the break apart FISH probe (8q24.1)
Results:
A rearrangement involving MYC was found in 8% of patients, and was associated with elevated β2-microglobulin, ≥50% bone marrow plasma cells, IgA MM, and the co-occurrence of trisomies. There were no differences in overall response rates between patients with and without MYC rearrangement when induction chemotherapy was proteasome inhibitor (PI)-based, immunomodulatory drug (IMiD)-based or PI+IMiD-based. Overall survival was shorter in patients with MYC rearrangement compared to patients without MYC rearrangement (5.3 vs. 8.0 years, P<0.001). MYC rearrangement was associated with increased risk of death on multivariate analysis when high-risk cytogenetic abnormalities, ISS stage III, and ≥70 years of age were included (risk ratio: 1.5, P=0.007)
Conclusion:
MYC rearrangement is associated with high disease burden and is an independent adverse prognostic factor in newly diagnosed MM.
Keywords: MYC rearrangement, MYC translocation, survival, multiple myeloma, FISH
INTRODUCTION
Multiple myeloma (MM) is a clonal plasma cell disorder accounting for ~20% of hematologic malignancies in the United States.1 Patients with MM exhibit a wide range of clinical presentations, diverse cytogenetic profiles, and heterogeneous outcomes. The utilization of Interphase fluorescence in-situ hybridization (FISH) for cytogenetic analysis in newly diagnosed patients with MM has identified recurrent primary and secondary cytogenetic abnormalities in the majority of MM patients.2, 3 Among those, immunoglobulin gene translocations t(4;14), t(14;16), t(14;20), and deletion in the short arm of chromosome 17 (17q13) have been identified as adverse risk factors.5 While the prognostic significance of these abnormalities has been established, the prognostic significance of other recurring cytogenetic abnormalities remains equivocal. Rearrangements involving the MYC proto-oncogene on the long arm of chromosome 8 (8q24.1) are secondary cytogenetic abnormalities detected by FISH in ~15% of newly diagnosed patients with MM.4 These arrangements, which include insertions, inversions and translocations,5 result in overexpression of MYC, which encodes a transcription factor involved in various cellular functions like growth6 and proliferation,7 metabolism,6 protein synthesis (translation)8 and apoptosis.9 MYC rearrangements result in the juxtaposition of super enhancers of immunoglobulin and other gene loci with MYC,10, 11 leading to MYC overexpression and malignant transformation.12, 13 The activation of MYC is a key event in the progression from MGUS and smoldering myeloma (SMM) to symptomatic myeloma.14 While some studies have found MYC rearrangements to be associated with inferior outcomes,13, 15 other studies failed to show prognostic significance.16 The objective of this study was to assess the impact of MYC translocation on clinical characteristics, treatment response, and survival in newly diagnosed MM patients.
METHODS
Patients and study design:
This is a retrospective study including patients with MM seen in Mayo Clinic in Rochester, Minnesota within 90 days from diagnosis in the period from January, 2006 to January 2018. All patients were identified using a prospectively maintained database; additional clinical and laboratory data was obtained by review of electronic medical records. All patients had authorized the use of their electronic medical record data for research. We included 1342 patients who had cytogenetic analysis by FISH performed within 1 year from diagnosis and less than 6 months from the start of first-line treatment, and in whom FISH analysis including the probe for MYC translocation. The study was approved by the Mayo Clinic Institutional Review Board. FISH analysis was performed as previously described,17 using unsorted plasma cells, identified using cytoplasmic immunoglobulin stain. The FISH panel used to detect primary and secondary abnormalities included the following probes 5: 1q/1p (1q22/TP73) (In house, custom developed), 3 centromere (D3Z1) (Abbott Molecular), 7 centromere (D7Z1) (Abbott Molecular), 9 centromere (D9Z1) (Abbott Molecular), 15 centromere (D15Z4) (Abbott Molecular), 13q (RB1/LAMP1) (Abbott Molecular) and chromosome 17 (TP53/D17Z1) (Abbott Molecular) enumeration probes. Dual-color, dual-fusion probes targeting t(11;14) CCND1/IgH (Abbott Molecular), and break apart probe targeting IgH (in house, custom developed) and 8q24.1 (MYC) (Abbott Molecular) were used. If an IgH rearrangement other than t(11;14) was found by the IgH break apart probe, reflex testing was done using dual-color, dual-fusion probes to identify the translocation partner: t(4;14)(p16.3;q32) FGFR3/IgH, t(14;16)(q32;q23) IgH/MAF, t(14;20)(q32;q12) IgH/MAFB, and t(6;14)(p21;q32) CCND3/IgH (Abbott Molecular). The MYC break apart probe was introduced for clinical use as part of the Myeloma FISH panel in Mayo Clinic starting August 2014. For samples obtained before this date, testing for MYC rearrangement was performed as an add-on test on by scoring a total of 200 cells from whole marrow samples not subjected to plasma cell enrichment. The threshold for MYC abnormality using this technique was 6.5%. After this date, MYC rearrangement testing was performed as part of the myeloma FISH panel by scoring a total of 50 cells from samples enriched with plasma cells using the cytoplasmic immunoglobulin stain. A sample was considered positive for a MYC rearrangement if this abnormality was detected in at least 5 of 50 of cells scored.
Statistical analysis:
Baseline clinical characteristics were compared between patients with a MYC rearrangement and those without a MYC rearrangement using Fisher’s exact test for categorical variables and Wilcoxon Rank Sum test for continuous variables. Based on initial therapy, patients were grouped into one of 3 groups: 1) PI-based (proteasome inhibitor only), 2) IMiD-based (Immunomodulatory drug only) and 3) PI + IMiD-based treatment. We compared treatment responses to first-line induction therapy, and the time to next treatment (TTNT) between the 2 groups. Fisher’s exact test was used to compare treatment responses. The impact of MYC translocation on overall survival (OS) was evaluated using univariate and multivariate cox proportional hazards model. High-risk (HR) translocations were defined by the presence of any of: t(4;14), t(14;16) or t(14;20);3, 18 all other translocations were considered standard-risk (SR) translocations. OS was calculated from the time of diagnosis. TTNT was defined as time of start of first-line treatment to time of start of second-line treatment. OS and TTNT curves were estimated using the Kaplan-Meier method and compared using the log-rank test. For all tests, 2-sided P values <0.05 were considered statistically significant. All statistical analyses were performed using the JMP software (SAS, Cary, NC).
RESULTS
Association with clinical characteristics:
Overall, a MYC rearrangement was found in 111 patients (8%); the rate was similar with both techniques used (7.7% using plasma cell-enriched samples and 8.7% using non-enriched samples). Compared to patients without MYC rearrangement, patients with MYC rearrangement were more likely to have elevated β2-microglobulin (>3.5 μg/ml) (71% vs. 58%, P=0.01), ≥50% bone marrow plasma cells (70% vs. 54%, P=0.003), lytic lesions (78% vs. 68%, P=0.04) and IgA MM (35% vs. 24%, P=0.04). In addition, MYC rearrangement was associated with trisomies (71% vs. 58%, P=0.006). In contrast, a MYC rearrangement was less likely to be present with t(11;14) rearrangement (10% vs. 21%, P=0.004). Otherwise, there were no differences in baseline characteristics or co-occurrence of cytogenetic abnormalities between patients with and without MYC rearrangement (Table 1).
Table 1: Clinical characteristics.
Comparison of clinical characteristics, prevalence of cytogenetic abnormalities and first-line treatments in patients with +1q and those without +1q. The median (interquartile range) is presented for continuous variables and number (percentage) for categorical variables. Abbreviations: B2M: beta2microglobulin, BMPCs: bone marrow plasma cells, del: deletion, Hb: hemoglobin, HR: high-risk, IMiD: immunomodulatory drug, ISS: international staging system, LC: light chain, LDH: lactate dehydrogenase, MM: multiple myeloma, PCLI: plasma cell labeling index, PI: proteasome inhibitor, PS: performance status, SR: standard-risk, Abn: abnormal, WBC: white blood cell.
| All (N=1342) |
No MYC Abn (N=1231) |
MYC Abn (N=111) |
P value | |
|---|---|---|---|---|
| Age (years) | ||||
| Median | 64 (57-71) | 64 (57-71) | 65 (58-71) | 0.49 |
| ≥70 (vs <70) | 373 (28) | 339 (28) | 34 (31) | 0.51 |
| Male | 819 (61) | 757 (61) | 62 (56) | 0.26 |
| ECOG PS | ||||
| ≥ 2 (vs 0-1) | 94 (19) | 87 (20) | 7 (17) | 0.84 |
| Hb (g/dL) | ||||
| Median | 10.9 (9.4-12.4) | 11.0 (9.4-12.5) | 10.5 (9.4-12.1) | 0.25 |
| < 10 (vs ≥ 10) | 397 (33) | 360 (33) | 37 (36) | 0.59 |
| Platelets (x 10 9 /L) | ||||
| Median | 210 (163-259) | 210 (163-259) | 213 (149-258) | 0.78 |
| < 150 (vs ≥ 150) | 167 (20) | 146 (19) | 21 (25) | 0.19 |
| WBC (x 10 9 /L) | 5.3 (4.0-7.0) | 5.4 (4.0-7.1) | 5.2 (3.9-6.2) | 0.14 |
| Creatinine (mg/dL) | 1.0 (0.9-1.5) | 1.0 (0.9-1.5) | 1.0 (0.9-1.4) | 0.98 |
| Creatinine ≥2 | 189 (16) | 175 (16) | 14 (14) | 0.45 |
| LDH (units/L) | ||||
| Median | 165 (138-201) | 164 (137-198) | 172 (150-215) | 0.034 |
| >222 vs (≤222) | 145 (16) | 131 (16) | 14 (20) | 0.40 |
| B2M (μg/ml) | ||||
| Median | 4.1 (2.8-7.4) | 4.1 (2.8-7.4) | 4.9 (3.4-8.2) | 0.009 |
| >3.5 vs (≤ 3.5) | 622 (59) | 557 (58) | 65 (71) | 0.01 |
| >5.5 vs (≤ 5.5) | 377 (36) | 339 (35) | 38 (42) | 0.25 |
| Albumin (g/dL) | ||||
| Median | 3.6 (3.2-3.8) | 3.6 (3.2-3.8) | 3.6 (3.2-3.9) | 0.66 |
| ≤3.5 (vs >3.5) | 502 (49) | 459 (49) | 43 (48) | 0.91 |
| Calcium (mg/dL) | ||||
| Median | 9.5 (9.1-10.1) | 9.5 (9.1-10.1) | 9.6 (9.2-10.2) | 0.16 |
| ≥ 11 (vs < 11) | 107 (9) | 99 (10) | 8 (8) | 0.72 |
| Lytic lesions | 744 (69) | 668 (68) | 76 (78) | 0.04 |
| % BMPCs | ||||
| Median | 50 (30-70) | 50 (30-70) | 60 (42-80) | <0.001 |
| ≥ 50% (vs <50%) | 690 (55) | 619 (54) | 71 (70) | 0.003 |
| Serum M spike (g/dL) | 2.5 (0.7-3.9) | 2.5 (0.6-3.9) | 3.0 (1.3-4.0) | 0.08 |
| Urine M spike (g/24 hrs.) | 0.04 (0-0.47) | 0.03 (0-0.42) | 0.13 (0-0.74) | 0.11 |
| Urine albumin (g/24 hrs.) | 0.05 (0.02-0.14) | 0.05 (0.02-0.14) | 0.06 (0.03-0.14) | 0.39 |
| Ig Isotype | ||||
| IgA | 259 (25) | 230 (24) | 29 (35) | 0.04 |
| IgG | 613 (59) | 565 (60) | 48 (58) | 0.82 |
| LC MM | 139 (13) | 133 (14) | 6 (7) | 0.09 |
| Involved LC | ||||
| Lambda | 377 (36) | 354 (37) | 23 (27) | |
| Kappa | 665 (64) | 602 (63) | 63 (73) | 0.06 |
| ISS Stage | ||||
| I | 243 (24) | 227 (25) | 16 (18) | |
| II | 379 (38) | 346 (38) | 33 (38) | |
| III | 378 (38) | 340 (37) | 38 (44) | |
| ISS III (vs I/II) | 378 (38) | 340 (37) | 38 (44) | 0.25 |
| PCLI (%) | ||||
| Median | 0.8 (0.3-1.5) | 0.8 (0.2-1.5) | 1.0 (0.6-1.6) | 0.01 |
| ≥2% (vs. <2%) | 79 (19) | 68 (18) | 11 (23) | 0.42 |
| SR FISH abnormalities | ||||
| Trisomy | 773 (59) | 694 (58) | 79 (71) | 0.006 |
| t(11;14) | 269 (20) | 258 (21) | 11 (10) | 0.004 |
| Del(13q) | 122 (9) | 114 (9) | 8 (7) | 0.61 |
| Monosomy 13 | 516 (39) | 469 (39) | 47(43) | 0.42 |
| HR FISH translocations | 193 (15) | 172 (14) | 21 (19) | 0.16 |
| t(4;14) | 126 (10) | 113 (9) | 13 (12) | 0.40 |
| t(14;16) | 54 (4) | 48 (4) | 6 (5) | 0.45 |
| t(14;20) | 13 (1) | 11 (1) | 2 (2) | 0.30 |
| Del(17p)/monosomy 17 | 168 (13) | 157 (13) | 11 (10) | 0.46 |
| First-line induction chemotherapy | ||||
| PI-based | 459 (36) | 414 (35) | 45 (42) | |
| IMiD-based | 458 (36) | 423 (36) | 35 (32) | |
| PI+IMiD | 365 (28) | 338 (29) | 27 (25) | |
| Other | 8 (1) | 7 (1) | 1 (1) | |
| First-line transplant | 570 (44) | 527 (45) | 43 (40) |
Efficacy of first-line treatment
Treatment data were available for 1290 patients, including 1190 with treatment response data; of these, 411, 429, 345, and 5 patients received treatment with PI-based, IMiD-based, PI+IMiD-based, and other treatments, respectively. PI-based regimens were bortezomib-based (373 patients) or ixazomib-based (38 patients); IMiD-based regimens were lenalidomide-based (411 patients) or thalidomide-based (18 patients); PI+IMiD combinations included bortezomib+lenalidomide (358 patients), carfilzomib+lenalidomide (27 patients), bortezomib+thalidomide (11 patients), ixazomib+lenalidomide (8 patients) and carfilzomib+thalidomide (7 patients). There was no difference in overall response rate (ORR) between patients with and without MYC rearrangement with PI-based (76% vs. 80%, P=0.53), IMiD-based (91% vs. 82%, P=0.24) or PI+IMiD-based (88 vs. 96%, P=0.13) induction chemotherapy. Patients with MYC rearrangement had lower rates of ≥very good partial response (VGPR), compared to patients without MYC rearrangement, with PI+IMiD-based treatment (35% vs. 60%, P=0.02). There was no significant difference in ≥VGPR rate between the 2 groups, with PI-based (39% vs. 44%, P=0.73) or IMiD-based (36% vs. 29%, P=0.33) treatments. Almost all patients who underwent post-induction transplant (570 patients) achieved at least a partial response to treatment; a ≥VGPR was achieved in 85% and 80% of patients with and without MYC rearrangement, respectively (P=0.54).
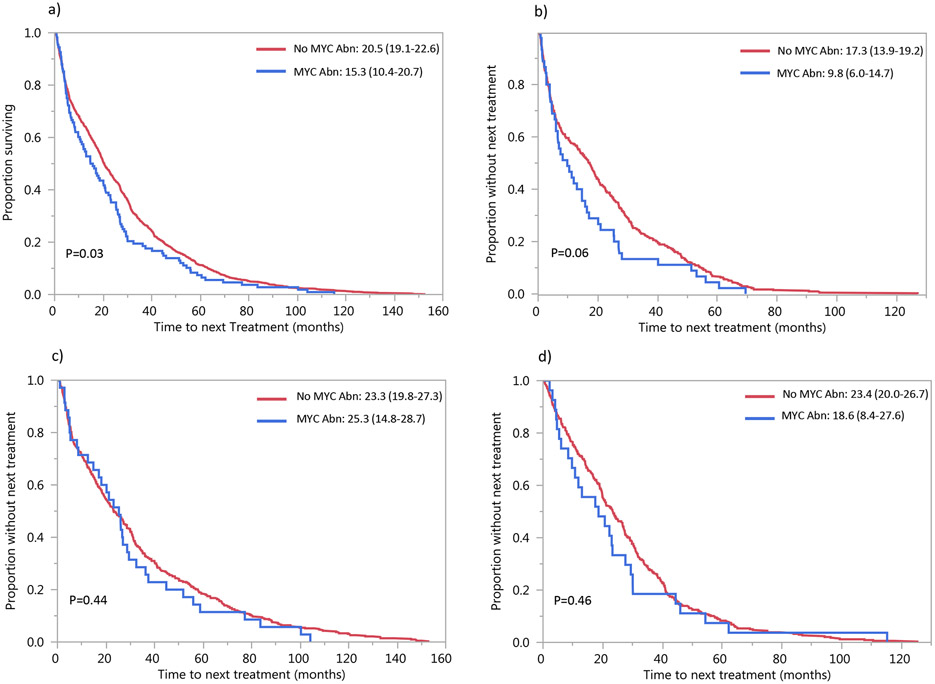
Overall, TTNT was shorter in patients with MYC rearrangement (15.3, 95%CI: 10.4-20.7) compared to those without MYC rearrangement (20.5, 95%CI: 19.1-22.6), P=0.03 (Figure 1a). TTNT based on the induction regimen is shown in Figure 1b, 1c, and 1d. The TTNT for patients with and without MYC rearrangement was 26.0 vs. 30.6 months (P=0.08) among those who underwent transplant post-first line induction therapy (n=570), and 6.5 vs. 7.9 months (P=0.30) among those who received chemotherapy only.
Figure 1: TTNT after first-line treatment.
Median TTNT (95%CI) in months in patients with MYC abnormality (blue curve) and without MYC abnormality (red curve) a) overall and among those who received b) PI-based, c) IMiD-based and d) PI+IMiD-based first-line treatment. Abbreviations: Abn: abnormality, IMiD: immunomodulatory drug, PI: proteasome inhibitor,, TTNT: time to next treatment.
Survival outcomes:
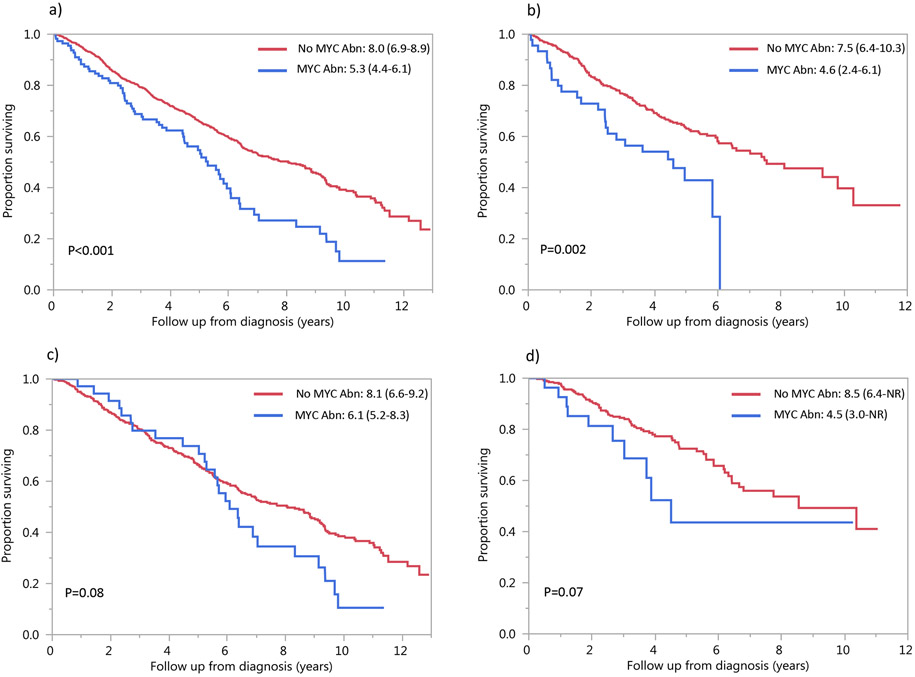
The median follow-up in the entire cohort was 4.0 (interquartile range: 2.2-6.1) years; median OS was 8.6 (95%CI: 6.5-8.6) years. OS was significantly shorter in patients with MYC rearrangement [median OS: 5.3 (95%CI: 4.4-6.1) years] compared to patients without MYC rearrangement [median OS: 8.0 (95%CI: 6.9-8.9) years, P=<0.001] (Figure 2a). OS based on induction therapy is shown in Figure 2b, 2c, 2d. OS in patients with MYC rearrangement compared to those without MYC rearrangement was 3.9 vs. 6.3 years (P<0.001) among patients receiving first-line treatment with chemotherapy only; for those who underwent transplant after first-line induction chemotherapy, OS was 6.4 vs. 10.4 years (P=0.07).
Figure 2: OS by first-line treatment:
Median OS (95% CI) in years in patients with MYC abnormality (blue curve) and without MYC abnormality (red curve) a) overall and among those who received b) PI-based, c) IMiD-based and d) PI+IMiD-based first line treatment. Abbreviations: Abn: abnormality, IMiD: immunomodulatory drug, NR: not reached, OS: overall survival, PI: proteasome inhibitor.
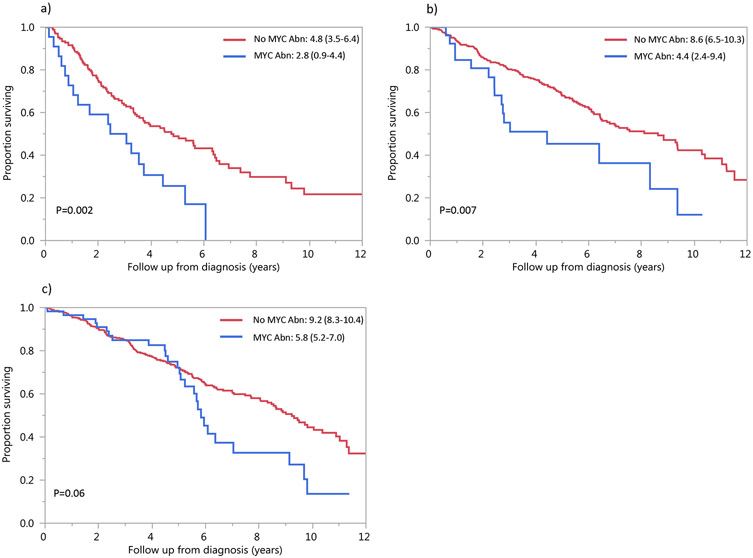
The impact of MYC rearrangement on OS was evaluated among patients with HR translocations, SR translocations, and trisomies (without IgH translocations). Among patients with HR translocations (n=193), patients with a concurrent MYC rearrangement had decreased OS compared to those without MYC rearrangement (2.8 vs. 4.8 years, P=0.002). Among patients with SR translocations (n=445), OS was also shorter in patients with MYC rearrangement (4.4 vs. 8.6 years, P=0.007). In patients with trisomies without IgH rearrangement (n=538), there was a trend towards decreased OS in patients with MYC rearrangement (5.8 vs. 9.2 years, P=0.06). The survival curves are shown in Figure 3a, 3b, 3c.
Figure 3: OS by cytogenetic group:
Median OS (95% CI) in years in patients with MYC abnormality (blue curve) and without MYC abnormality (red curve) among patients with a) HR translocations, b) SR translocations or c) trisomies. Abbreviations: Abn: abnormality.
A MYC rearrangement was associated with increased mortality on univariate analysis [Risk ratio (RR): 1.7, 95%CI: 1.3-2.2, P<0.001]. In a multivariate model including MYC rearrangement and other high risk cytogenetic abnormalities [HR translocations, del(17p) and 1q gain], MYC rearrangement was independently associated with increased risk of death [RR: 1.7 (95%CI: 1.3-2.2), P<0.001).When advanced ISS (stage III), and old age (≥ 70 years) were also included in the multivariate model, MYC rearrangement retained its prognostic value [RR: 1.5 (95%CI: 1.1-2.0), P=0.01) (table 2).
Table 2: Univariate and multivariate models.
Univariate and multivariate analysis including MYC rearrangement, HR cytogenetic abnormalities, ISS stage and age ≥70. Abbreviations: HR: high-risk, ISS: international staging system, OS: overall survival, RR: risk ratio.
| Variable | Univariate | Multivariate (HR FISH abnormalities only) |
Multivariate (All) | |||
|---|---|---|---|---|---|---|
| OS RR (95%CI) |
P Value |
OS RR (95%CI) |
P value |
OS RR (95%CI) |
P value |
|
| MYC rearrangement | 1.7 (1.3-2.2) |
<0.001 | 1.7 (1.3-2.2) |
<0.001 | 1.5 (1.1-2.0) |
0.01 |
| HR IgH translocation | 2.0 (1.6-2.5) |
<0.001 | 1.6 (1.3-2.0) |
<0.001 | 1.9 (1.5-2.5) |
<0.001 |
| Del(17p) | 2.0 (1.6-2.5) |
<0.001 | 2.0 (1.5-2.5) |
<0.001 | 1.6 (1.2-2.1) |
<0.001 |
| 1q gain/amplification | 1.9 (1.6-2.3) |
<0.001 | 1.7 (1.4-2.0) |
<0.001 | 1.4 (1.2-1.8) |
0.001 |
| ISS stage III | 1.9 (1.6-2.3) |
<0.001 | - | - | 1.7 (1.4-2.1) |
<0.001 |
| Age ≥ 70 | 2.1 (1.8-2.5) |
<0.001 | - | - | 2.3 (1.8-2.8) |
<0.001 |
We performed subgroup analysis including 72 patients where data on the proportion of cells harboring a MYC rearrangement was available. A total of 200 cells were scored from samples not enriched for plasma cells. The median plasma cell percentage was 12% (IQR: 9-19); 17 (24%) had ≥20% plasma cells with MYC rearrangement, including 4 (6%) with ≥ 50% plasma cells. The median OS was 9.4 (95%CI: 4.4-NR) years in patients with MYC rearrangements in ≥ 20% of cells, and 5.2 (95%CI: 3.0-6.4) years in with rearrangement in <20% of cells (P=0.13)
DISCUSSION
Chromosomal translocations involving the MYC protooncogene have been reported in 13-15% of newly diagnosed MM patients using FISH,4, 16 and associated with elevated β2 microglobulin,4 ISS stage II/III,13 extramedullary disease and plasmablastic morphology.15 In this study, MYC rearrangement was detected in ~8% of newly diagnosed patients who underwent cytogenetic testing by FISH, and was associated with elevated β2-microglobulin (>3.5 μg/ml), ≥50% bone marrow plasma cells, lytic lesions and IgA MM. Patients with MYC rearrangement were more likely to have trisomies. While some studies have shown MYC rearrangements to be associated with hyperdiploidy,11, 12 others have found equal prevalence in hyperdiploid and non-hyperdiploid MM.13, 19 In contrast to the study by Mikulasova et al. using 1267 samples from newly diagnosed myeloma patients, we did not find a significant association between MYC rearrangements and advanced ISS stage. However in their study, next-generation sequencing was used to detect MYC rearrangements, which were detected in 36% of samples.20 The reported prevalence of MYC rearrangements in the literature ranges from 10% to 50% in newly diagnosed MM.11-13, 21 This variability is due to differences in both the method and FISH probes used; the detection rate of MYC rearrangements by FISH is lower than that by genome sequencing techniques, where small insertions and cryptic translocations may be missed by FISH.5 In this study, the prevalence of MYC rearrangements was lower than that reported in other studies utilizing FISH, which may be attributed to the method used. Although we utilized 2 different techniques for detection of MYC rearrangements, the rate was similar with both techniques, suggesting that MYC rearrangements were not underestimated when non-plasma enriched samples were used.
The impact of MYC rearrangements on survival has not been yet established, with previous studies showing inconsistent results.13, 15, 16 This may also be attributed to variability in the methodologies used to detect MYC rearrangements, heterogeneity in patient characteristics and treatments, small sample size, and short follow up. In a previous study at MD Anderson, MM patients with rearrangements involving MYC (23 patients) had decreased progression-free and overall survival when compared to matched controls who did not have MYC rearrangements. On the other hand, their outcomes were comparable to patients with plasma cell leukemia (without MYC rearrangements).15 In another study including 55 patient samples from the MRC Myeloma IX trial, MYC rearrangements detected by targeted capture-based sequencing were associated with decreased progression-free (P=0.032) and overall (P=0.035) survival; this was retained on multivariate analysis when other adverse translocations were included. 13 On the other hand, a study including newly diagnosed patients (<66 years) from the IFM99 trials, showed that MYC translocations had no prognostic impact among patients treated with VAD induction followed by double intensive therapy.16
In this study, patients with MYC rearrangement had similar response rates to therapy with novel agents, compared to those without MYC rearrangement, but had inferior survival; the impact of MYC rearrangement on OS was retained in a multivariate model including HR translocations, del(17p), 1q gain, ISS stage III, and age ≥70 years. The presence of MYC rearrangement discriminated patients with different prognosis within the HR and SR IgH rearrangement groups. Our results, based on a large sample with long median follow up, suggest that MYC rearrangement has an independent prognostic impact in newly diagnosed MM patients and may have a role in further risk stratification if incorporated into the current model.
When we evaluated the impact of the clone size, we did not observe a statistically significant difference in OS between patients with ≥20% and <20% plasma cells harboring MYC rearrangement, but this analysis was limited by small sample size, particularly for patients with larger clone sizes; this association should be evaluated in future large studies.
This study is limited by its retrospective nature and heterogeneity of treatment regimens. It is also important to highlight that our findings are only applicable to patients MYC rearrangements detected by FISH using the MYC break apart probe. Future large prospective studies are needed to confirm our findings, and evaluate the prognostic significance of MYC rearrangements when detected by more sensitive methodologies.
CONCLUSION
A rearrangement involving MYC was found in 8% of newly diagnosed multiple myeloma patients, and was associated with high tumor burden and hyperdiploidy. A MYC rearrangement detected by FISH was associated with increased risk of death independent of age, advanced stage or co-occurrence of high-risk cytogenetic abnormalities. This abnormality may have a role in risk stratification of newly diagnosed MM patients.
Statement of translational relevance:
MYC rearrangements are commonly detected by FISH in newly diagnosed multiple myeloma (MM) patients, but their prognostic value not yet been established; at this time, they are not included in risk stratification systems for newly diagnosed MM. In this study, we show that MYC rearrangements are associated with significantly inferior outcomes, independent of the presence of other high risk cytogenetic abnormalities. The results from this large, unselected cohort reflecting real-word practice, and with long-term follow up, justify the inclusion of MYC rearrangements in future risk stratification systems, which has the potential to improve outcome prediction in newly diagnosed MM patients, and to guide treatment choices. In addition, these results have the potential to guide clinical trial designs for high risk patients, which in turn can inform treatment decisions in the future.
Acknowledgments
Financial support: None
Footnotes
Conflict of Interest Disclosures: P.K. received research funding from Takeda Pharmaceuticals, Celgene, and Amgen. A.D. received research funding from Celgene, Millennium Pharmaceuticals, Pfizer, and Janssen and received a travel grant from Pfizer. M.A.G. served as a consultant for Millennium Pharmaceuticals and received honoraria from Celgene, Millennium Pharmaceuticals, Onyx Pharmaceuticals, Novartis, GlaxoSmithKline, Prothena, Ionis Pharmaceuticals, and Amgen. M.Q.L. received research funding from Celgene. N.L. serves on an advisory board for Takeda Pharmaceuticals. S.K.K. served as a consultant for Celgene, Millennium Pharmaceuticals, Onyx Pharmaceuticals, Janssen, and Bristol-Myers Squibb and received research funding from Celgene, Millennium Pharmaceuticals, Novartis, Onyx Pharmaceuticals, AbbVie, Janssen, and Bristol-Myers Squibb. The remaining authors declare no competing financial interests.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2.Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64(4):1546–58. [DOI] [PubMed] [Google Scholar]
- 3.Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23(12):2210–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avet-Loiseau H, Gerson F, Magrangeas F, Minvielle S, Harousseau JL, Bataille R, et al. Rearrangements of the c-myc oncogene are present in 15% of primary human multiple myeloma tumors. Blood. 2001;98(10):3082–6. [DOI] [PubMed] [Google Scholar]
- 5.Smadbeck J, Peterson JF, Pearce KE, Pitel BA, Figueroa AL, Timm M, et al. Mate pair sequencing outperforms fluorescence in situ hybridization in the genomic characterization of multiple myeloma. Blood Cancer J. 2019;9(12):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, et al. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9(21):1255–8. [DOI] [PubMed] [Google Scholar]
- 7.Prochownik EV. c-Myc: linking transformation and genomic instability. Curr Mol Med. 2008;8(6):446–58. [DOI] [PubMed] [Google Scholar]
- 8.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10(4):301–9. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27(50):6462–72. [DOI] [PubMed] [Google Scholar]
- 10.Dib A, Gabrea A, Glebov OK, Bergsagel PL, Kuehl WM. Characterization of MYC translocations in multiple myeloma cell lines. J Natl Cancer Inst Monogr. 2008(39):25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misund K, Keane N, Stein CK, Asmann YW, Day G, Welsh S, et al. MYC dysregulation in the progression of multiple myeloma. Leukemia. 2020;34(1):322–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Affer M, Chesi M, Chen WG, Keats JJ, Demchenko YN, Roschke AV, et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia. 2014;28(8):1725–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker BA, Wardell CP, Brioli A, Boyle E, Kaiser MF, Begum DB, et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J. 2014;4:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011;25(6):1026–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glitza IC, Lu G, Shah R, Bashir Q, Shah N, Champlin RE, et al. Chromosome 8q24.1/c-MYC abnormality: a marker for high-risk myeloma. Leuk Lymphoma. 2015;56(3):602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007;109(8):3489–95. [DOI] [PubMed] [Google Scholar]
- 17.Fonseca R, Blood E, Rue M, Harrington D, Oken MM, Kyle RA, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101(11):4569–75. [DOI] [PubMed] [Google Scholar]
- 18.Stewart AK, Bergsagel PL, Greipp PR, Dispenzieri A, Gertz MA, Hayman SR, et al. A practical guide to defining high-risk myeloma for clinical trials, patient counseling and choice of therapy. Leukemia. 2007;21(3):529–34. [DOI] [PubMed] [Google Scholar]
- 19.Gabrea A, Martelli ML, Qi Y, Roschke A, Barlogie B, Shaughnessy JD Jr., et al. Secondary genomic rearrangements involving immunoglobulin or MYC loci show similar prevalences in hyperdiploid and nonhyperdiploid myeloma tumors. Genes Chromosomes Cancer. 2008;47(7):573–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikulasova A, Ashby C, Tytarenko RG, Qu P, Rosenthal A, Dent JA, et al. Microhomology-mediated end joining drives complex rearrangements and overexpression of MYC and PVT1 in multiple myeloma. Haematologica. 2020;105(4):1055–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szabo AG, Gang AO, Pedersen MO, Poulsen TS, Klausen TW, Norgaard P. Overexpression of c-myc is associated with adverse clinical features and worse overall survival in multiple myeloma. Leuk Lymphoma. 2016;57(11):2526–34. [DOI] [PubMed] [Google Scholar]