Abstract
Background
Primary ciliary dyskinesia (PCD) is a heterogeneous disease with a diverse clinical and genetic spectrum among populations worldwide. Few cases of pediatric PCD have been reported from China.
Research Question
What are the clinical and genotypic characteristics of children with PCD in China?
Study Design and Methods
Clinical characteristics, laboratory findings, and genetic results obtained for 75 patients with PCD were reviewed retrospectively at a single center in China. Genetic sequencing was conducted using whole-exome screening.
Results
Patient median age at diagnosis was 7.0 years (range, 2 months-14 years). Of 75 patients, 88% (66/75) had chronic wet cough, 77% (58/75) had recurrent sinusitis, 76% (57/75) had bronchiectasis, 40% (30/75) had neonatal respiratory distress, and 28% (21/75) had coexistent asthma. Notably, postinfectious bronchiolitis obliterans (PIBO) as first presentation was found in 8% of children (6/75). Genes with the highest incidence of mutations were DNAH11 (15/51), followed by DNAH5 (9/51), CCDC39 (5/51), DNAH1 (4/51), and CCNO (3/51). Four genes (DNAI1, HEATR2, RSPH9, and DNAAF3) each were respectively found in two patients, and seven genes (CCDC40, LRRC6, SPAG1, RSPH4A, ARMC4, CCDC114, and DNAH14, a novel gene) each were mutated once. No differences in classical clinical features were observed among patients with commonly observed PCD-associated genotypes. However, three of six PIBO patients carried DNAH1 mutations.
Interpretation
Besides typical clinical features, PIBO was observed as the first presentation of pediatric PCD in China. An association of the novel gene DNAH14 with PCD was observed, expanding the PCD genotypic spectrum.
Key Words: China, DNAH14, gene, postinfectious bronchiolitis obliterans, primary ciliary dyskinesia
Abbreviations: CA, central apparatus; IDA, inner dynein arm; MTD, microtubular disorganization; nNO, nasal nitric oxide; ODA, outer dynein arm; PCD, primary ciliary dyskinesia; PIBO, postinfectious bronchiolitis obliterans; TEM, transmission electron microscopy
Primary ciliary dyskinesia (PCD) is a clinically and genetically heterogeneous disorder caused by abnormal function of motile cilia. Patients typically seek treatment with chronic wet cough, sinusitis, bronchiectasis, otitis media, and neonatal respiratory distress, with approximately 50% of patients exhibiting situs inversus.1 Although more than 40 disease-causing genes have been identified, in 20% to 30% of individuals with definitive PCD diagnosis, no genetic cause has been identified.2,3 The PCD-associated genetic spectrum varies among populations of different countries4; the most frequently reported genes have been DNAH5 and DNAH11, although in some countries such as Tunisia and Egypt, CCDC39 and CCDC40 are most common.5, 6, 7 Recently, genotype-phenotype relationships have been described in the PCD population.8 Patients with certain variant alleles of CCDC39 or CCDC40 exhibit worse lung function and rapid disease progression, whereas patients with certain variants of DNAH9 often demonstrate phenotypically mild respiratory dysfunction or normal lung function.3,8,9 Nevertheless, progress has been made in the discovery of associations of genotype-phenotype relationships with PCD, although more research is needed.
The prevalence of PCD is approximately 1:10,000 to 1:15,000 in Europeans, but has been reported to be much higher in the British Asian population.10,11 Notably, China hosts the largest population in Asia, but few PCD patients have been reported there. It is likely that heterogeneity of PCD phenotypic characteristics and clinical presentation have led to the assumption that PCD is rare in China. However, this apparent lack of cases in fact may reflect the fact PCD patients have not been detected or have been misdiagnosed there because of a lack of appropriate diagnostic facilities and clinician training in China.
Herein, we summarize the clinical and genotypic characteristics of 75 children with PCD receiving care at a single treatment center in China. These findings should enhance our understanding of the clinical and genetic features of PCD. In addition, discovery of novel PCD-associated genes can facilitate disease diagnosis and guide development of therapeutic treatments for PCD.
Methods
Patient Cohort
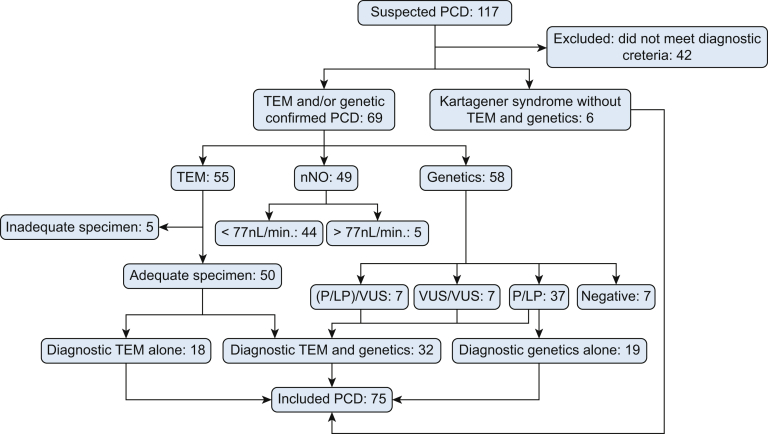
We retrospectively reviewed records of 117 children with suspected PCD treated between January 2012 and November 2019 at the Department of Respiratory Medicine, Beijing Children’s Hospital, Beijing, China. Seventy-five patients were included, with 69 diagnoses confirmed according to guidelines of the European Respiratory Society, the American Thoracic Society, or both.12,13 PCD was diagnosed based on at least one of the following criteria: recognized ciliary ultrastructural defect; biallelic pathogenic variants in a PCD-associated gene; at least two of the four key clinical features for PCD, combined with low nasal nitric oxide (nNO) level (excluding cystic fibrosis), namely, unexplained neonatal respiratory distress in a term infant, year-round daily cough beginning before 6 months of age, year-round daily nasal congestion beginning before 6 months of age, or organ laterality defect; and Kartagener syndrome, which was considered to be PCD even if the previously mentioned criteria were not fulfilled completely.14 Patients without the above diagnostic criteria were excluded.
Identification of identical pathogens derived from two cultured sputum specimens or both sputum and BAL specimens was conducted. Clinical data were obtained from electronic medical records. Clinical features of PCD patients were evaluated for associations with common PCD-associated genotypes (n ≥ 3).
Informed consent was obtained from all participants or legal guardians. This study was conducted in accordance with tenets of the amended Declaration of Helsinki and was approved by Beijing Children’s Hospital Ethics Committee (approval no. 2020-k-21).
nNO Measurement
nNO was measured during quiet exhalation using an EcoMedics CLD 88 Analyzer. Measurements of nNO in cooperative children > 5 years of age followed American Thoracic Society and European Respiratory Society recommendations.15 For younger or uncooperative children, nasal sampling was performed for 60 s during tidal breathing by measuring nitric oxide values of both nostrils, and the greater value was recorded. nNO production (in nanoliters per min) was calculated by multiplying nNO concentration (parts per billion) by sampling flow rate (0.3 L/min).
Transmission Electron Microscopy
All samples were obtained by bronchial biopsy. Bronchial mucosa biopsy specimens immediately were immersed in glutaraldehyde at 4°C, washed overnight, then fixed in 1% osmium tetroxide. After dehydration, samples were embedded in epoxy resin. After polymerization, several sections were placed onto copper grids and then stained with aqueous uranyl acetate and Reynold’s lead citrate. Ciliary ultrastructure was analyzed using a transmission electron microscope (JEM-1400; Jeol).
Immunofluorescence Analysis of DNAH14 Expression in Human Respiratory Cells
Respiratory epithelial cells were obtained by nasal brush biopsy and were suspended in cell culture medium. Samples were spread onto glass slides, air dried, and stored at –80°C or used directly. Cells were treated with 4% paraformaldehyde, washed with phosphate-buffered saline, and permeabilized for 20 min at room temperature with 0.5% Trition X-100. Next, cells were incubated with primary antibodies (monoclonal mouse antiacetylated α-tubulin [Sigma] or rabbit polyclonal anti-DNAH14 [Invitrogen]) overnight at 4°C. Alexa Fluor 594 antimouse antibody and Alexa Fluor 488 antirabbit antibody were used as secondary antibodies (Molecular Probes, Invitrogen). DNA was stained using DAPI (Beyotime). Confocal and high-resolution images were obtained using a Zeiss LSM 980 Airyscan2 (Carl Zeiss Ltd.).
Genetic Sequencing and Variant Assessment
Genomic DNA samples were prepared, and exonic DNA sequences were enriched using the NimbleGen 2.0 probe sequence capture array (Roche). Sequencing was performed using the Illumina HiSeq2500 platform (Illumina), and raw data were assessed for quality using the base-calling method according to the manufacturer’s instructions. Low-quality sequence reads were removed (quality scores < 20). Remaining sequence reads were aligned to the National Center for Biotechnology Information human reference genome (hg19) using Burrows-Wheeler alignment. SAMtools and Pindel were used to call single-nucleotide polymorphisms and insertions and deletions. Synonymous changes and single-nucleotide polymorphisms with minor allele frequencies of > 5% were removed (http://www.ncbi.nlm.nih.gov/projects/SNP). Candidate missense mutations were evaluated using SIFT (http://sift.bii.a-star.edu.sg/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), and the Mutation Taster application (http://www.mutationtaster.org/) to determine the pathogenicity. Splice-site changes were assessed using MaxEntScan (http://hollywood.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html) and dbscSNV (http://www.liulab.science/dbscsnv.html) software packages. American College of Medical Genetics and Genomics guidelines were referenced for variant evaluation.
Statistical Analyses
Statistical analyses were performed using Statistical Package for the Social Sciences version 22.0 software (SPSS, Inc.). Continuous data were expressed as mean ± SD or median. Intergroup differences were compared using Kruskal-Wallis tests for continuous dependent variable values and Fisher exact tests for comparisons of proportions. The Mann-Whitney U test was used for post-doc comparisons. Statistical tests were two sided, with statistical significance for P < .05.
Results
Demographic and Clinical Features of Patients
A total of 75 diagnosed pediatric PCD patients constituted the cohort for the current analysis (Fig 1, e-Table 1). Of all patients, 45 (60%) were males and 30 (40%) were females. No parent was diagnosed with PCD, whereas two siblings had PCD. The median age at onset was 3 months (range, 1 day-10 years), and the median age at diagnosis was 7.0 years (range, 2 months-14 years). Nasal nitric oxide testing of 49 individuals revealed a median nNO value of 19.2 nL/min (minimum, 1.3 nL/min; maximum, 196.9 nL/min), with normal nNO values observed for five patients (e-Table 1). Approximately 8% of patients (6/75) had a family history of PCD (parents or siblings with PCD or bronchiectasis). Eighty-eight percent of patients (66/75) had recurrent wet cough, 77% of patients (58/75) had sinusitis, 76% of patients (57/75) had bronchiectasis, and 61% of patients (46/75) had atelectasis of the right middle lobe. Forty percent of patients (30/75) had neonatal respiratory distress, 25% of patients (19/75) had otitis media, and 11% of patients (8/75) had hearing impairment (Fig 2A). Further examination revealed that 20% of patients (15/75) had situs inversus totalis and 7% of patients (5/75) had congenital heart defects (ventricular septal defect, atrial septal defect, mitral insufficiency, and tricuspid insufficiency); postinfectious bronchiolitis obliterans (PIBO) (Fig 2C, 2D) occurred after affliction with severe mycoplasma and adenovirus pneumonia in 8% of patients (6/75), whereas five of six PIBO patients had never experienced recurrent respiratory infection or wet cough before PIBO diagnosis.
Figure 1.
Consolidated Standards of Reporting Trials diagram outlining the included patients. LP = likely pathogenic; nNO = nasal nitric oxide; P = pathogenic; PCD = primary ciliary dyskinesia; TEM = transmission electron microscopy; VUS = variants of uncertain significance.
Figure 2.
A, Bar graph showing the main clinical findings in patients with PCD. Percentage distribution of clinical manifestations in 75 definite PCD patients. B, Bar graph showing the percentage distribution of genetic analysis in 52 individuals. C, High-resolution CT scan showing atelectasis of the right middle lobe with mosaic pattern, air trapping, bronchiectasis, and hyperlucency in one patient with PCD. D, High-resolution CT scan showing bronchiectasis and bilateral mosaic ground-glass patterns with air trapping in another patient with PCD. CHD = congenital heart disease; NRD = neonatal respiratory distress; PCD = primary ciliary dyskinesia; PIBO = postinfectious bronchiolitis obliterans.
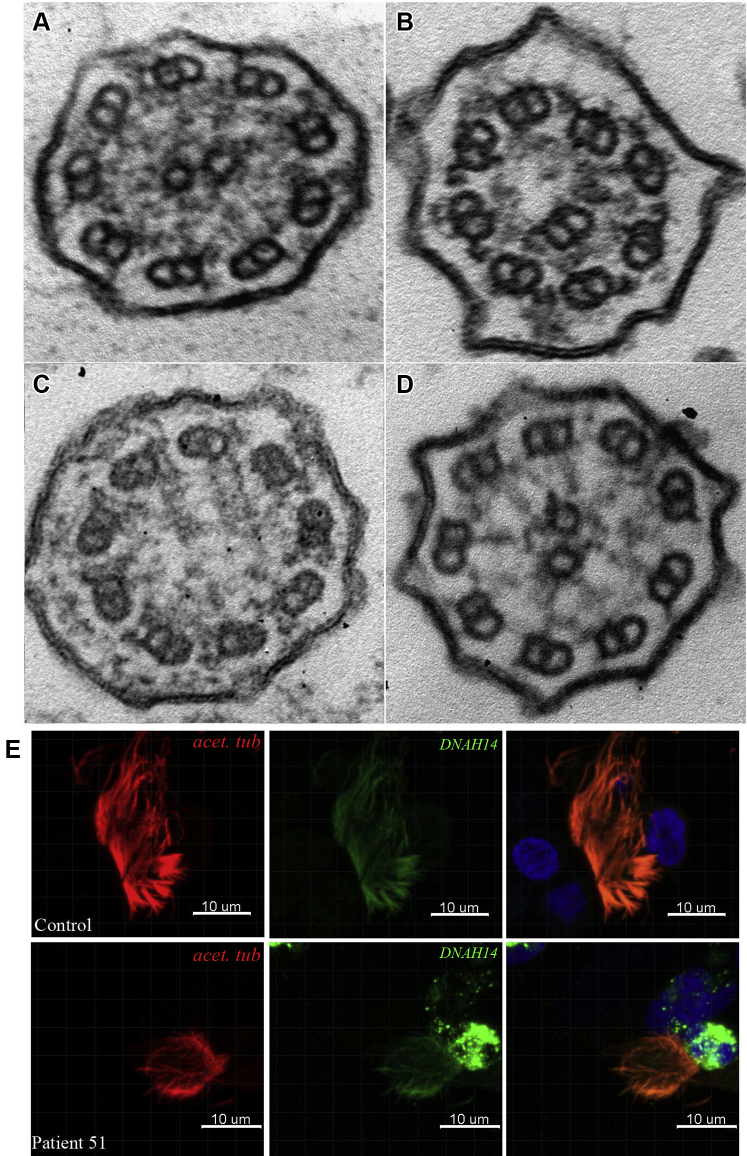
Transmission electron microscopy (TEM) analysis of bronchial mucosal biopsy specimens obtained from 52 patients revealed ciliary ultrastructural defects that are presented in Table 1, along with mutations detected during patient genetic testing. In 16% of patients (8/50), outer dynein arm (ODA) defects were detected (Fig 3). Inner dynein arm (IDA) defects in conjunction with central apparatus (CA) defects and microtubule disorganization (MTD) were classified as IDA/CA/MTD.8 The IDA defects, CA defects, and MTD were detected in 16% of patients (8/50), ODA and IDA defects were detected in 24% of patients (12/50), CA or IDA defects alone were detected in 20% of patients (10/50), with oligocilia detected in 8% of patients (4/50), normal structure detected in 10% of patients (5/50), and conical protrusions detected in 6% of patients (3/50) (Table 1).
Table 1.
Genetic and TEM Information in This Cohort
| Patient No. | Gene | Complementary DNA Change (Protein Change) | dbSNP | Type of Mutation | Source | GnomAD, All | ACMG Guideline |
Mutation State | Reference | TEM | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Evidence | ||||||||||
| 1 | CCDC39 | c.1308delG "(p.Q436Hfs∗10) | NA | Frameshift | P | None | Path | PVS1+PM2+PM3 | Compound heterozygous | Current | IDA/CA/MTD |
| c.1167+1G→A (P?02) | rs1292030955 | Splicing | M | None | Path | PVS1+PM2+PP3 | |||||
| 2 | CCDC39 | c.1363-2A→G (P?) | NA | Splicing | P | None | Path | PVS1+PM2+PP3 | Compound heterozygous | Current | IDA/CA/MTD |
| c.210+1G→C (P?) | NA | Splicing | M | None | Path | PVS1+PM2+PP3 | |||||
| 3 | CCDC39 | c.526_c.527delCT (p.L176Afs∗10) | rs780175755 | Frameshift | P | None | Path | PVS1+PS1+PM2 | Compound heterozygous | 23255504 | IDA/CA/MTD |
| c.2542G→T (p.E848X) | NA | Nonsense | M | None | Path | PVS1+PM2+PP3 | |||||
| 4/5 | CCDC39 | c.872delT (p.L291Xfs∗1) | NA | Frameshift | P | None | Path | PVS1+PM2+PP1 | Compound heterozygous | Current | Inadequate/NA |
| c.833_c.834delAG (p.E278Vfs∗2) | NA | Frameshift | M | None | Path | PVS1+PM2+PP1 | |||||
| 6 | CCDC40 | c.961C→T (p.R321X) | rs754867753 | Nonsense | M | None | Path | PVS1+PS1+PM2 | Compound heterozygous | 21131974/current | IDA/CA/MTD |
| c.2330G→A (p.W777X) | NA | Nonsense | P | None | Path | PVS1+PM2+PP3 | |||||
| 7 | CCNO | c.267_c.268insAGCCC (p.V90Sfs∗6) | rs939597472 | Frameshift | P | None | Path | PVS1 +PM2+PM5 | Compound heterozygous | 24747639/current | NA |
| c.940G→T (p.E314X) | NA | Nonsense | M | None | Path | PVS1+PM2+PM3 | |||||
| 8 | CCNO | c.262_c.263insGGCCC (p.Q88Rfs∗8) | rs587777502 | Frameshift | M/P | None | Path | PVS1+PS1+PM2 | Homozygous | 24747639 | NA |
| 9 | CCNO | c.262_c.263insGGCCCGGCCC (p.Q88Rfs∗51) | rs587777502 | Frameshift | P/M | None | Path | PVS1+PM2+PP1 | Homozygous | Current | Oligocilia |
| 10 | DNAAF3 | c.813_c.814insCGAC (p.A272Rfs∗17) | NA | Frameshift | M | None | Path | PVS1+PM2+PP1 | Compound heterozygous | Current | ODA+IDA |
| c.677G→A (p.G226E) | rs1171652733 | Missense | P | None | LP | PM2+PM3+PP2+PP3 | |||||
| 11 | DNAAF3 | c.271C→A (p.P91T) | NA | Missense | P/M | None | LP | PM1+PM2+PP3+PP4 | Homozygous | Current | ODA+IDA |
| 12 | DNAH1 | c.5356C→T (p.R1786C) | rs759042431 | Missense | M | 0.000073 | LP | PM1+PM2+PP3+PP4 | Compound heterozygous | Current | NA |
| c.1286+7C→A (P?) | rs570694571 | Splicing near | P | None | VUS | PM2+BP4 | |||||
| 13 | DNAH1 | c.2912G→A (p.R971H) | rs190420231 | Missense | P | 0.000097 | LP | PM1+PM2+PP3+PP4 | Compound heterozygous | Current | Normal |
| c.11135G→A (p.R3712Q) | rs200395837 | Missense | M | 0.000065 | VUS | PM1+PP4 | |||||
| 14 | DNAH1 | c.2610G→A (p.W870X) | NA | Nonsense | P/M | None | Path | PVS1+PM2+PP3 | Homozygous | 28577616 | NA |
| 15 | DNAH1 | c.3836A→G (p.K1279R) | rs142595873 | Missense | M | 0.0009 | LP | PM1+PM3+PP3+PP1 | Compound heterozygous | 25877616 | NA |
| c.6328_6337del (p.S2110Gfs ∗19) | NA | Frameshift | P | None | Path | PVS1+PM2+PP1 | |||||
| 16 | DNAH11 | c.4321C→T (p.R1441W) | rs183489539 | Missense | P | None | VUS | PM1+PM2+PP3 | Compound heterozygous | Current | Normal |
| c.3025C→G (p.L1009V) | rs542059067 | Missense | M | None | VUS | PM2+PP3 | |||||
| 17 | DNAH11 | c.4670G→T (p.C1557F) | NA | Missense | P | None | LP | PM1+PM2+PP3+PP4 | Compound heterozygous | Current | NA |
| c.9220C→G (p.Q3074E) | rs775542623 | Missense | M | 0.000054 | LP | PM1+PM2+PP3+PP4 | |||||
| 18 | DNAH11 | c.8005C→T (p.Q2669X) | NA | Nonsense | P | None | Path | PVS1+PM2+PP4 | Compound heterozygous | Current | Normal |
| c.13163-2_13163-1insCAAT (P?) | NA | Splicing | M | None | Path | PVS1+PM2+PP4 | |||||
| 19 | DNAH11 | c.3020T→G (p.L1007X) | rs1480698078 | Nonsense | P | None | Path | PVS1+PM2+PP4 | Compound heterozygous | Current | NA |
| c.12934-3_12934-2insG (P?) | NA | Splicing | M | None | Path | PVS1+PM2+PP4 | |||||
| 20 | DNAH11 | c.11383_11386delAGAA (p.K3797Rfs∗2) | NA | Frameshift | De novo | None | Path | PVS+PS2+PM2 | Compound heterozygous | Current | CA |
| c.1942delT (p.W649Gfs∗19) | NA | Frameshift | M | None | Path | PVS1+PM2+PP4 | |||||
| 21 | DNAH11 | c.6165C→G (p.Y2055X) | rs184241449 | Nonsense | M | None | Path | PVS1+PM2+PP3 | Compound heterozygous | Current | Oligocilia |
| c.11924C→T (p.S3975L) | NA | Missense | P | None | LP | PM1+PM2+PM3+PP3 | |||||
| 22 | DNAH11 | c.6632_6633insTGAAC (p.F2214Nfs∗24) | NA | Frameshift | M | None | Path | PVS1+PM2+PP4 | Compound heterozygous | Current | NA |
| c.10053_10054insAA (p.T3352Kfs∗19) | NA | Frameshift | P | None | Path | PVS1+PM2+PP4 | |||||
| 23 | DNAH11 | c.13373C→T (p.P4458L) | rs72658835 | Missense | P | None | Path | PS1+PM1+PM2+PP3 | Compound heterozygous | 22184204 | NA |
| exon21-66del (P?) | NA | Deletion | M | None | Path | PVS1+PM2+PM3 | |||||
| 24 | DNAH11 | c.727A→G (p.I243V) | rs189000268 | Missense | P | 0.0004 | VUS | PM1+PM2+BP4 | Compound heterozygous | Current | Oligocilia |
| c.1452T→G (p.F484L) | rs755256285 | Missense | M | 0.000046 | LP | PM1+PM2+PP3+PP4 | |||||
| 25 | DNAH11 | c.6937C→G (p.R2313G) | NA | Missense | P | None | LP | PM1+PM2+PP1+PP4 | Compound heterozygous | Current | Oligocilia |
| c.5513T→A (p.V1838D) | rs1336120886 | Missense | M | None | LP | PM&+PM2+PP3+PP4 | |||||
| 26 | DNAH11 | c.12153T→A (p.D4051E) | NA | Missense | M | None | VUS | PM2+PP3 | Compound heterozygous | Current | NA |
| c.2485C→T (p.R829C) | rs767607595 | Missense | P | None | VUS | PM2+PP1 | |||||
| 27 | DNAH11 | c.13276T→C (p.W4426R) | rs527548552 | Missense | M | 0.0001 | LP | PM1+PM2+PP1+PP3 | Compound heterozygous | Current | NA |
| c.10433G→A (p.R3478Q) | rs117729990 | Missense | P | 0.0008 | VUS | PP3+PP4 | |||||
| 28 | DNAH11 | c.10851A→T (p.R3617S) | rs373667589 | Missense | P | None | VUS | PM2+PP3+PP4 | Compound heterozygous | Current | NA |
| c.13189C→A (p.H4397N) | rs774489385 | Missense | M | None | LP | PM1+PM2+PP3+PP4 | |||||
| 29 | DNAH11 | c.971C→T (p.A324V) | rs79874320 | Missense | M | 0.0003 | LP | PM1+PM2+PM3 | Compound heterozygous | Current | Oligocilia |
| c.6165C→G (p.Y2055X) | rs184241449 | Nonsense | P | None | Path | PVS1+PM2+PP3 | |||||
| 30 | DNAH11 | c.12679A→G (p.S4227G) | rs574488123 | Missense | P | 0.0002 | VUS | PM2+PP1+PP4 | Compound heterozygous | Current | conical Protrusions |
| c.5523G→A (p.S1841S) | rs750430725 | Missense | M | None | VUS | PM2+PP3+PP4 | |||||
| 31 | DNAH5 | c.997C→T (p.R333X) | rs771956532 | Nonsense | P | 0.000032 | Path | PVS1+PM2 +PP5 | Compound heterozygous | Current | NA |
| c.7918_c.7919insA (p.M2640Nfs∗18) | NA | Frameshift | M | None | Path | PVS1+PM2+PP4 | |||||
| 32 | DNAH5 | c.12813G→A (p.W4271X) | NA | Nonsense | P | None | Path | PVS1+PM2+PP4 | Compound heterozygous | Current | ODA |
| c.5928delA (p.A1977Pfs∗77) | NA | Frameshift | M | None | Path | PVS1+PM2+PP4 | |||||
| 33 | DNAH5 | c.5563_c.5564insA (p.I1855Nfs∗6) | rs752925056 | Frameshift | P | None | Path | PVS1+PM2+PP4 | Compound heterozygous | 11788826 | Inadequate |
| c.4147_c.4148delAinsTCC (p.I1383Sfs∗22) | NA | Frameshift | M | None | Path | PVS1+PM2+PP4 | |||||
| 34 | DNAH5 | c.8029C→T (p.R2677X) | rs775946081 | Nonsense | P | None | Path | PVS1+PM2+PP4 | Compound heterozygous | 16627867 | ODA |
| c.6647delA (p. K2216Rfs∗20) | NA | Nonsense | M | None | Path | PVS1+PM2+PP4 | |||||
| 35 | DNAH5 | c.4114C→G (p.Q1372E) | NA | Missense | M | None | LP | PM1+PM2+PM5+PP3 | Compound heterozygous | Current | ODA |
| gain1 exon37 (P?) | NA | Duplication | De novo | None | Path | PVS1+PM2+PP4 | |||||
| 36 | DNAH5 | c.8383C→T (p.R2795X) | rs560398270 | Nonsense | M | None | Path | PVS1+PM2+PP3 | Compound heterozygous | 30067075/Current | ODA |
| c.8311C→T (p.R2771C) | rs769557047 | Missense | P | None | LP | PM1+PM2+PM3+PP3 | |||||
| 37 | DNAH5 | c.12472C→T (p.R4158W) | rs3756672 | Missense | P | 0.0023 | VUS | PM1+BP6 | Compound heterozygous | Current | NA |
| c.13286G→A (p.R4429Q) | rs61744047 | Missense | M | 0.0023 | VUS | PM1+PP5+BP6 | |||||
| 38 | DNAH5 | c.11023_c.11028+17delTTTAAGGTAGGTTAAACCTATGGinsC (p.F3675_K3676del) | NA | Deletion | De novo | None | LP | PS2+PM2+PM4+PP4 | Compound heterozygous | Current | ODA |
| c.8366A→G (p.H2789R) | NA | Missense | P | None | LP | PS2+PM2+PM3 | |||||
| 39 | DNAH5 | c.9502C→T (p.R3168X) | rs863224504 | Nonsense | P | None | Path | PVS1+PM2+PP4 | Compound heterozygous | Current | NA |
| c.6061+1G→A (P?) | rs772658660 | Splicing | M | None | Path | PVS1+PM2+PP4 | |||||
| 40 | DNAI1 | c.1738G→A (p.D580N) | NA | Missense | P | None | LP | PM1+PM2+PP3+PP4 | Compound heterozygous | Current | ODA |
| c.482C→T (p.T161I) | NA | Missense | M | None | LP | PM1+PM2+PP1+PP4 | |||||
| 41 | DNAI1 | c.1612G→A (p.A538T) | rs368248592 | Missense | P/M | 0.000065 | LP | PS1+PM2+PP3 | Homozygous | Current | ODA |
| 42 | HEATR2 | c.2511G→A (p.S837S) | rs143524894 | Synonymous | P | 0.000016 | VUS | PM2+PP1 | Compound heterozygous | Current | NA |
| c.2420A→G (p.D807G) | rs201177019 | Missense | M | 0.0008 | LP | PM1+PM2+PP1+PP4 | |||||
| 43 | HEATR2 | c.2011G→A (p.A671T) | rs751454661 | Missense | P | 0.0002 | LP | PS4+PM1+PM2 | Compound heterozygous | Current | ODA+IDA |
| c.788G→A (p.R263Q) | rs201059622 | Missense | M | None | VUS | PM1+PM2+PP1 | |||||
| 44 | LRRC6 | exon 8-10 del (P?) | NA | Deletion | P/M | None | Path | PVS1+PM2+PP4 | Homozygous | Current | ODA+IDA |
| 45 | RSPH4A | c.1454G→A (p.R485Q) | rs1296082790 | Missense | P/M | None | VUS | PM2+PP3+PP4 | Homozygous | Current | CA |
| 46 | RSPH9 | c.421G→A (p.V141M) | rs2295947 | Missense | M | 0.0005 | VUS | PM2+PP3+BP1 | Compound heterozygous | Current | CA |
| c.467G→A (p.R156Q) | rs201032669 | Missense | P | None | VUS | PM1+PP3+PP5 | |||||
| 47 | RSPH9 | c.467G→A (p.R156Q) | rs201032669 | Missense | P | None | VUS | PM1+PP3+PP5 | Compound heterozygous | 25789548 | Conical protrusions |
| c.-41T→C (P?) | NA | Non-coding | M | None | VUS | PM2+PP1 | |||||
| 48 | SPAG1 | c.1649_c.1650insA (p.I550Ifs∗10) | NA | Frameshift | P | None | Path | PVS1+PM2+PP4 | Compound heterozygous | Current | ODA+IDA |
| c.691delT (p.Y231Ifs∗12) | NA | Frameshift | M | None | Path | PVS1+PM2+PP4 | |||||
| 49 | CCDC114 | c.-41_2A→C (P?) | rs963154615 | Splicing | P | None | Path | PVS1+PM2+PP3 | Compound heterozygous | Current | NA |
| c.705_c.706insGCAG (p.P236Afs∗11) | rs1294704273 | Frameshift | M | None | Path | PVS1+PM2+PM3 | |||||
| 50 | ARMC4 | c.2306C→A (p.P769H) | rs117515566 | Missense | P | 0.000032 | LP | PM1+PM2+PP3 | Compound heterozygous | Current | ODA |
| c.1679C→T (p.A560V) | rs749814633 | Missense | M | 0.000097 | Path | PS4+PM1+PM2+PP3 | |||||
| 51 | DNAH14 | c.4172 _c.4173insAT (p.R1391Rfs∗26) | rs77953334 | Frameshift | M | 0.0031 | Path | PVS1+PM2+PP4 | Compound heterozygous | Current | IDA |
| c.5068delG (p.G1690Vfs∗12) | rs577893006 | Frameshift | P | 0.0002 | Path | PVS1+PM2+PP4 | |||||
ACMG = American College of Medical Genetics and Genomics; dbSNP = Single Nucleotide Polymorphism Database; GnomAD = Genome Aggregation Database; IDA = inner dynein arm; LP = likely pathogenic; M = maternal; NA = not applicable; ND = not determined; ODA = outer dynein arm; P = paternal; Path = pathogenic; TEM = transmission electron microscopy; VUS = variant of uncertain significance.
Figure 3.
A-D, Ultrastructural defects in selected patients. A, Outer dynein arm (ODA) defect. B, Absence of inner dynein arm (IDA) defect in conjunction with central apparatus (CA) defects and microtubular disorganization. C, CA defect alone. D, ODA and IDA defects. E, Expression of DNAH14 is reduced in respiratory cilia in patient 51.
At least two culture specimens were obtained successfully from 46 patients and were confirmed to harbor a pathogenic species if the same pathogens were detected in at least two cultured specimens. Streptococcus pneumoniae was the most commonly isolated pathogen (54%), followed by Haemophilus influenzae (28%), Pseudomonas aeruginosa (11%), and Staphylococcus aureus (11%).
Most children showed normal spirometry indexes or only mild obstructive impairment. However, patients with PIBO exhibited moderate to severe small airways obstruction.
Genetic Testing Findings and Genotype Associations With Phenotype
Biallelic variants of 97 autosomal recessive genes were identified. Seven were homozygous and only one ethnic minority family reported consanguineous marriage. Genetic analysis revealed frequencies of pathogenic or likely pathogenic variants (Table 1, e-Table 2): 29.4% with DNAH11 variants (15 individuals), 17.6% with DNAH5 variants (nine individuals), 9.8% with CCDC39 variants (five individuals), 7.8% with DNAH1 variants (four individuals), 5.9% with CCNO variants (three individuals), and 3.9% with variants of DNAI1, HEATR2, RSPH9, or DNAAF3 (two individuals for each). CCDC40, LRRC6, SPAG1, ARMC4, RSPH4A, CCDC114, and DNAH14 mutated in one individual each. The distribution of genetic variants is shown in Figure 2B. Patient 33 carried two compound heterozygote variants at both DNAH5 and RSPH4A loci; both DNAH5 variants were considered pathogenic, whereas both RSPH4A variants were variants of uncertain significance according to American College of Medical Genetics and Genomics scoring guidelines.
In addition, we identified a novel gene (DNAH14) in a 5-year-old patient (e-Fig 1). This boy demonstrated a recurrent wet cough that had persisted for the previous 2 years. Chest CT scan revealed bronchiectasis affecting the right middle lobe. He also exhibited chronic sinusitis, otitis media, and a low nNO level (63 nL/min). TEM findings demonstrated an absence of IDA defects and the presence of a few shortened or truncated ODA projections (e-Fig 1). Trio whole-exome sequencing was undertaken for the proband and the proband’s parents. Sanger sequencing confirmed segregation of DNAH14 variants in this family, together supporting a definitive PCD diagnosis.
Key clinical features were evaluated based on commonly observed genotypic associations, with generally no significant correlations between genotype and typical PCD phenotypic features observed (Table 2). However, PIBO was more common in patients with DNAH1 variant alleles compared with those in the CCDC39 group (P = .026). Median FEV1 % predicted values were lower in patients with CCDC39 variant alleles compared with other groups, although these results did not reach statistical significance because of the low number of patients in this group.
Table 2.
Demographics and Clinical Characteristics of Patients According to Main Genetic Analysis
| Variable | DNAH11 (n = 15) | DNAH5 (n = 9) | CCDC39 (n = 5) | CCNO (n = 3) | DNAH1 (n = 4) | P Value |
|---|---|---|---|---|---|---|
| Male sex | 8 (53) | 6 (67) | 2 (40) | 3 (100) | 2 (50) | .846a |
| Age at diagnosis, y | 6.2 ± 3.5 | 7.1 ± 3.9 | 10.5 ± 2.4 | 6.2 ± 3.2 | 3.6 ± 3.1 | .193b |
| nNO, nL/min | 9.6 (8.2-23.5), n = 10 | 12.6 (8.8-16.7), n = 6 | 11.4 (9.9-37.3), n = 4 | 20.8 (16.5-55.8), n = 3 | 37.1 (9.24-65.0), n = 2 | .363b |
| FEV1 % predicted | 94 (34-135) | 108 (98-115) | 63 (20-89) | 71 (67-81) | N/A | .056bc |
| NRD | 5 (33) | 5 (56) | 4 (80) | 2 (67) | 1(25) | .342a |
| Asthma | 4 (27) | 3 (33) | 2 (40) | 2 (67) | 0 | .481a |
| Bronchiectasis | 10 (67) | 9 (100) | 4 (80) | 1 (33) | 3 (75) | .124a |
| Atelectasis | 9 (60) | 8 (89) | 4 (80) | 3 (100) | 2 (50) | .536a |
| PIBO | 0 | 0 | 1 (20) | 0 | 3 (75) | .026a |
| SI | 0 | 2 (22.2) | 1 (20%) | 0 | 0 | 1.0a |
Data are presented as No. (%), mean ± SD, or median (interquartile range). N/A = not applicable; nNO = nasal nitric oxide; NRD = neonatal respiratory distress; PIBO = postinfectious bronchiolitis obliterans; SI = situs inversus.
Fisher exact test.
Kruskal-Wallis test.
No statistically significant on post hoc tests. Median (first quartile to third quartile [n ≥ 4], minimum-maximum value [n < 4]).
Immunofluorescence Analysis of DNAH14 in Respiratory Epithelial Cells
High-resolution immunofluorescence staining demonstrated DNAH14 localization throughout the entire length of the ciliary axoneme in respiratory epithelial cells from heathy control participants. By contrast, DNAH14 expression was reduced in ciliary axonemes of patient 51 (Fig 3E, e-Fig 1).
Discussion
Herein, to the best of our knowledge, we report results of the most comprehensive analysis to date of phenotypic, genotypic, and clinical features associated with pediatric PCD patients originating from across China. The median age of diagnosis in the pediatric population was 7.0 years, later than in European and North American pediatric patients.16,17 This diagnostic delay in China may stem from a lack of appropriate diagnostic facilities and trained diagnosticians for PCD detection. Nevertheless, the median age of diagnosis in our patient cohort has improved markedly since 2018.
In this study, classical clinical PCD features, including bronchiectasis, sinusitis, and otitis media, were observed, and these observations were consistent with results of previous studies.18, 19, 20 Meanwhile, no laterality defects were observed in patients with RSPH9 and RSPH4A mutations, as noted in previous reports. In our cohort, only 20% of patients exhibited situs inversus totalis, a lower frequency than reported previously. Interestingly, some patients demonstrated PIBO, a symptom not associated previously with PCD that was diagnosed based on clinical manifestations, chest high-resolution CT scanning, and lung function.21 These patients exhibited persistent wheezing, dyspnea, or both after experiencing pneumonia. High-resolution CT scan findings included mosaic perfusion, air trapping, and bronchiectasis, with lung function showing moderate to severe small airways obstruction. In addition, approximately 30% of patients had coexistent asthma, a greater percentage than reported previously, whereas patients with CCNO variants were more likely to have asthma (although these results were not statistically significant). When taken together, these observations highlight the variety and complexity of PCD in Chinese children.
A total of 95 gene mutations were identified in this study, of which 85 were novel and 10 had been reported previously.22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32 More than 50% of variants were loss-of-function mutations (nonsense, frameshift, and splice acceptor or donor site). Following American College of Medical Genetics and Genomics guidelines for mutation interpretation, most mutant alleles were classified as pathogenic or likely pathogenic. As for patients possessing variants of uncertain significance, TEM findings and low nNO production levels indicated that such variant of uncertain significance genes may involve in PCD patients. Notably, seven mutations found in members of our cohort families were homozygous, with only one ethnic minority family reporting consanguineous marriage, for an overall low recorded consanguinity rate.
It was reported previously that the most common PCD-causing gene was DNAH5, followed by DNAH11, DNAI1, and CCDC39.33 The distribution of DNAH11 and DNAH5 in our study is consistent with results of patient studies from North America and other regions,5,8 with the genetic basis of disease still unknown in about 20% to 30% of patients. In our PCD patient cohort, negative genetic results were revealed in seven patients (approximately 12%) who were diagnosed clinically with PCD by TEM, key clinical features combined with low level of nNO, or both.34 Initially, two compound heterozygous mutations of HYDIN were identified in four patients with central pair deficiency revealed by TEM. However, the presence of HYDIN2, a pseudogene with very high homology to exon 6-28 and 30-83 of HYDIN, interferes the detection of HYDIN variants when using next-generation sequencing analysis.35 To identify variants in HYDIN or pseudogene (HYDIN2), we designed a gene-specific primer based on the principle of amplification-refractory mutation system (which depends on a single base difference at the 3′ end of the primer) for polymerase chain reaction analysis. Results of amplification-refractory mutation system polymerase chain reaction analysis revealed that variants in three patients were mapped on HYDIN2, whereas variants in four patients were not determined, which indicated that it was difficult to distinguish all of the variants using only amplification-refractory mutation system polymerase chain reaction analysis. In future, further research is needed to make genetic verification of HYDIN mutations.
A previous study suggested that gene interactions may influence PCD phenotype.36 Herein, we observed one patients carrying two different variants (heterozygous, homozygous, or both). One patient (patient 33) carried a compound heterozygote of variant alleles of both DNAH5 and RSPH4A. Generally, DNAH5 variants caused ODA defects, whereas RSPH4A mutations lead to ciliary central pair defects. TEM results for patient 33 revealed ultrastructural dynein arm defects. Digenic mode of inheritance in PCD was mentioned in a previous study.28 Indeed, although the effects of these variants with uncertain significance on PCD phenotype are not clear, their existence may imply a molecular mechanism underlying PCD that is based on digenic inheritance. To confirm this hypothesis, functional experiments involving a larger number of patients are warranted.
We also analyzed possible genotype-phenotype relationships. Genotype analysis did not reveal significant differences in prevalence of clinical features across groups. Moreover, patients with CCDC39 variants exhibited worse lung function than those with variants of other genes studied herein, in agreement with previous studies.8 Notably, the incidence of PIBO was higher in patients possessing the DNAH1 mutation compared with patients in the CCDC39 group. In fact, although few studies have investigated the association of PCD with DNAH1 variants, DNAH1 mutations were reported in a family study of siblings who were affected by PCD. Interestingly, DNAH1 is required during spermatogenesis for ciliary IDA formation, with DNAH1 mutations often reported in infertile men.37, 38, 39 However, the pathogenesis of DNAH1 to PCD not yet clear, warranting further study.
To date, few pathogenic genetic variants that cause PCD have been identified with genes affecting only IDA complexes.40 In our cohort, DNAH14 was identified as a novel PCD-associated gene. DNAH14 encodes an axonemal dynein heavy chain that has not yet been associated with human disease. Herein, a 5-year-old boy demonstrated a classical clinical PCD phenotype and recognized TEM defect. Trio whole-exome sequencing was undertaken for the proband and the proband’s parents, and two mutations within DNAH14 were identified that may be relevant to PCD. Sanger sequencing confirmed segregation of DNAH14 variants in this family. Immunofluorescence results revealed that expression of DNAH14 was reduced in the patient’s respiratory ciliary cells, which suggests that it may be involved in causing PCD.
In conclusion, results presented herein describing the clinical and genetic spectrum of PCD in China do not agree completely with findings of previous reports. In this study, PCD patients with PIBO were reported for the first time, and a novel PCD-associated gene was identified. These results should enhance our understanding of PCD pathogenic processes for improvement of early disease detection and improved disease management and prognosis.
Acknowledgments
Author contributions: S. Z., X. N., and G. W. designed the study and secured funding. Y. G., H. Y., and S. Z. led data management and statistical analyses and drafted the manuscript. S. Z., H. Y., Y. G., H. X., X. T., X. Z., and H. L. recruited and evaluated patients. X. Y., H. Y., and S. Z. reviewed electron photomicrographs and led standardization of study TEM. C. H., S. Z., and Y. G. coordinated and validated genetic studies. X. Z. and Y. G. performed and studies analyses of nNO.
Financial/nonfinancial disclosures: None declared.
Other contributions: The authors thank the children and their families who participated in this study and all the physicians for their help in accomplishing this work.
Additional information: The e-Figure and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
Drs Guan and Yang contributed equally to this manuscript.
FUNDING/SUPPORT: This study was supported by the Pediatric Specialty Collaborative Development Center of Beijing Hospital Authority [Grant XTZD20180103].
Supplementary Data
References
- 1.Kennedy M.P., Omran H., Leigh M.W. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation. 2007;115:2814–2821. doi: 10.1161/CIRCULATIONAHA.106.649038. [DOI] [PubMed] [Google Scholar]
- 2.Horani A., Ferkol T.W. Advances in the genetics of primary ciliary dyskinesia: clinical implications. Chest. 2018;154:645–652. doi: 10.1016/j.chest.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas J.S., Davis S.D., Omran H., Shoemark A. Primary ciliary dyskinesia in the genomics age. Lancet Respir Med. 2020;8(2):202–216. doi: 10.1016/S2213-2600(19)30374-1. [DOI] [PubMed] [Google Scholar]
- 4.Fassad M.R., Patel M.P., Shoemark A. Clinical utility of NGS diagnosis and disease stratification in a multiethnic primary ciliary dyskinesia cohort. J Med Genet. 2020;57(5):322–330. doi: 10.1136/jmedgenet-2019-106501. [DOI] [PubMed] [Google Scholar]
- 5.Emiralioğlu N., Taşkıran E.Z., Koşukcu C. Genotype and phenotype evaluation of patients with primary ciliary dyskinesia: first results from Turkey. Pediatr Pulmonol. 2020;55(2):383–393. doi: 10.1002/ppul.24583. [DOI] [PubMed] [Google Scholar]
- 6.Mani R., Belkacem S., Soua Z. Primary ciliary dyskinesia gene contribution in Tunisia: identification of a major Mediterranean allele. Hum Mutat. 2020;41(1):115–121. doi: 10.1002/humu.23905. [DOI] [PubMed] [Google Scholar]
- 7.Fassad M.R., Shoman W.I., Morsy H. Clinical and genetic spectrum in 33 Egyptian families with suspected primary ciliary dyskinesia. Clin Genet. 2020;97(3):509–515. doi: 10.1111/cge.13661. [DOI] [PubMed] [Google Scholar]
- 8.Davis S.D., Ferkol T.W., Rosenfeld M. Clinical features of childhood primary ciliary dyskinesia by genotype and ultrastructural phenotype. Am J Respir Crit Care Med. 2015;191(3):316–324. doi: 10.1164/rccm.201409-1672OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loges N.T., Antony D., Maver A. Recessive DNAH9 loss-of-function mutations cause laterality defects and subtle respiratory ciliary-beating defects. Am J Hum Genet. 2018;103(6):995–1008. doi: 10.1016/j.ajhg.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damseh N., Quercia N., Rumman N., Dell S.D., Kim R.H. Primary ciliary dyskinesia: mechanisms and management. Appl Clin Genet. 2017;10:67–74. doi: 10.2147/TACG.S127129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Callaghan C., Chetcuti P., Moya E. High prevalence of primary ciliary dyskinesia in a British Asian population. Arch Dis Child. 2010;95(1):51–52. doi: 10.1136/adc.2009.158493. [DOI] [PubMed] [Google Scholar]
- 12.Lucas J.S., Barbato A., Collins S.A. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur Respir J. 2017;49(1):1601090. doi: 10.1183/13993003.01090-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro A.J., Davis S.D., Polineni D. Diagnosis of primary ciliary dyskinesia. An Official American Thoracic Society Clinical Practice Guideline. Am J Respir Crit Care Med. 2018;197(12):e24–e39. doi: 10.1164/rccm.201805-0819ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leigh M.W., Pittman J.E., Carson J.L. Clinical and genetic aspects of primary ciliary dyskinesia / Kartagener syndrome. Genet Med. 2009;11(7):473–487. doi: 10.1097/GIM.0b013e3181a53562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang Z., Xinglan W., Huimin L., Wei W., Shunying Z. The value of nasal nitric oxide measurement in the diagnosis of primary ciliary dyskinesia. Pediatr Invest. 2019;3(4):209–213. doi: 10.1002/ped4.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuehni C.E., Frischer T., Strippoli M.P. Factors influencing age at diagnosis of primary ciliary dyskinesia in European children. Eur Respir J. 2010;36(6):1248–1258. doi: 10.1183/09031936.00001010. [DOI] [PubMed] [Google Scholar]
- 17.Welch J.E., Hogan M.B., Wilson N.W. Ten-year experience using a plastic, disposable curette for the diagnosis of primary ciliary dyskinesia. Ann Allergy Asthma Immunol. 2004;93(2):189–192. doi: 10.1016/S1081-1206(10)61474-3. [DOI] [PubMed] [Google Scholar]
- 18.Goutaki M., Meier A.B., Halbeisen F.S. Clinical manifestations in primary ciliary dyskinesia: systematic review and meta-analysis. Eur Respir J. 2016;48:1081–1095. doi: 10.1183/13993003.00736-2016. [DOI] [PubMed] [Google Scholar]
- 19.Leigh M.W., Ferkol T.W., Davis S.D. Clinical features and associated likelihood of primary ciliary dyskinesia in children and adolescents. Ann Am Thorac Soc. 2016;13:1305–1313. doi: 10.1513/AnnalsATS.201511-748OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frija-Masson J., Bassinet L., Honore I. Clinical characteristics, functional respiratory decline and follow-up in adult patients with primary ciliary dyskinesia. Thorax. 2017;72:154–160. doi: 10.1136/thoraxjnl-2015-207891. [DOI] [PubMed] [Google Scholar]
- 21.Kavaliunaite E., Aurora P. Diagnosing and managing bronchiolitis obliterans in children. Expert Rev Respir Med. 2019;13:481–488. doi: 10.1080/17476348.2019.1586537. [DOI] [PubMed] [Google Scholar]
- 22.Moonnumakal S.P., Fan L.L. Bronchiolitis obliterans in children. Curr Opin Pediatr. 2008;20:272–278. doi: 10.1097/MOP.0b013e3282ff62e9. [DOI] [PubMed] [Google Scholar]
- 23.Olbrich H., Schmidts M., Werner C. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am J Hum Genet. 2012;91:672–684. doi: 10.1016/j.ajhg.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antony D., Becker-Heck A., Zariwala M.A. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum Mutat. 2013;34:462–472. doi: 10.1002/humu.22261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker-Heck A., Zohn I.E., Okabe N. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat Genet. 2011;43:79–84. doi: 10.1038/ng.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallmeier J., Al-Mutairi D.A., Chen C.T. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Genet. 2014;46:646–651. doi: 10.1038/ng.2961. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Jin H., Han F. Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in Chinese. Clin Genet. 2017;91:313–321. doi: 10.1111/cge.12857. [DOI] [PubMed] [Google Scholar]
- 28.Knowles M.R., Leigh M.W., Carson J.L. Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax. 2012;67:433–441. doi: 10.1136/thoraxjnl-2011-200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olbrich H., Häffner K., Kispert A. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 30.Hornef N., Olbrich H., Horvath J. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am J Respir Crit Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis S.D., Rosenfeld M., Lee H.S. Primary ciliary dyskinesia: longitudinal study of lung disease by ultrastructure defect and genotype. Am J Respir Crit Care Med. 2019;199:190–198. doi: 10.1164/rccm.201803-0548OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frommer A., Hjeij R., Loges N.T. Immunofluorescence analysis and diagnosis of primary ciliary dyskinesia with radial spoke defects. Am J Respir Cell Mol Biol. 2015;53:563–573. doi: 10.1165/rcmb.2014-0483OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zariwala MA, Knowles MR, Leigh MW. Primary ciliary dyskinesia. Seattle (WA): University of Washington, Seattle; 2007 Jan 24 [updated 2019 Dec 5]. https://www.ncbi.nlm.nih.gov/books/NBK1122/. Accessed March 31, 2021.
- 34.O’Callaghan C., Rutman A., Williams G., Kulkarni N., Hayes J., Hirst R.A. Ciliated conical epithelial cell protrusions point towards a diagnosis of primary ciliary dyskinesia. Respir Res. 2018;19:125. doi: 10.1186/s12931-018-0782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cindrić S., Dougherty G.W., Olbrich H. SPEF2-and HYDIN-mutant cilia lack the central pair associated protein SPEF2 aiding PCD diagnostics. Am J Respir Cell Mol Biol. 2020;62:382–396. doi: 10.1165/rcmb.2019-0086OC. [DOI] [PubMed] [Google Scholar]
- 36.Pereira R., Barbosa T., Gales L. Clinical and genetic analysis of children with Kartagener syndrome. Cells. 2019;8:E900. doi: 10.3390/cells8080900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amiri-Yekta A., Coutton C., Kherraf Z.E. Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum Reprod. 2016;31:2872–2880. doi: 10.1093/humrep/dew262. [DOI] [PubMed] [Google Scholar]
- 38.Ben Khelifa M., Coutton C., Zouari R. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imtiaz F., Allam R., Ramzan K., Al-Sayed M. Variation in DNAH1 may contribute to primary ciliary dyskinesia. BMC Med Genet. 2015;16:14. doi: 10.1186/s12881-015-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bustamante-Marin X.M., Horani A., Stoyanova M. Mutation of CFAP57, a protein required for the asymmetric targeting of a subset of inner dynein arms in chlamydomonas, causes primary ciliary dyskinesia. PLoS Genet. 2020;16(8) doi: 10.1371/journal.pgen.1008691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.