Abstract
Background
Asthma exacerbations result in significant health and economic burden, but are difficult to predict.
Research Question
Can machine learning (ML) models with large-scale outpatient data predict asthma exacerbations?
Study Design and Methods
We analyzed data extracted from electronic health records (EHRs) of asthma patients treated at the Cleveland Clinic from 2010 through 2018. Demographic information, comorbidities, laboratory values, and asthma medications were included as covariates. Three different models were built with logistic regression, random forests, and a gradient boosting decision tree to predict: (1) nonsevere asthma exacerbation requiring oral glucocorticoid burst, (2) ED visits, and (3) hospitalizations.
Results
Of 60,302 patients, 19,772 (32.8%) had at least one nonsevere exacerbation requiring oral glucocorticoid burst, 1,748 (2.9%) requiring and ED visit and 902 (1.5%) requiring hospitalization. Nonsevere exacerbation, ED visit, and hospitalization were predicted best by light gradient boosting machine, an algorithm used in ML to fit predictive analytic models, and had an area under the receiver operating characteristic curve of 0.71 (95% CI, 0.70-0.72), 0.88 (95% CI, 0.86-0.89), and 0.85 (95% CI, 0.82-0.88), respectively. Risk factors for all three outcomes included age, long-acting β agonist, high-dose inhaled glucocorticoid, or chronic oral glucocorticoid therapy. In subgroup analysis of 9,448 patients with spirometry data, low FEV1 and FEV1 to FVC ratio were identified as top risk factors for asthma exacerbation, ED visits, and hospitalization. However, adding pulmonary function tests did not improve models’ prediction performance.
Interpretation
Models built with an ML algorithm from real-world outpatient EHR data accurately predicted asthma exacerbation and can be incorporated into clinical decision tools to enhance outpatient care and to prevent adverse outcomes.
Key Words: acute asthma exacerbation, asthma, machine learning
Abbreviations: AI, artificial intelligent; AUC, area under the receiver operating characteristic curve; EHR, electronic health record; HDiCS, high-dose inhaled corticosteroid; iCS, inhaled corticosteroid; LABA, long-acting β-agonist; LightGBM, light gradient boosting machine; ML, machine learning; PFT, pulmonary function testing; SHAP, shapley additive explanation
Asthma, a common disease, is associated with significant health and economic burden. In the United States, nearly half of the 25 million people with asthma report one or more asthma attacks every year.1,2 In fact, asthma resulted in about 307.8 physician office visits, 55.9 ED visits, and 5.9 hospital admissions per 10,000 civilian population in 2016,1 with an estimated direct and indirect cost of $80 billion in the United States alone in 2013.3 Despite guideline-directed therapy, more than 60% of individuals with asthma continued to report that their asthma was uncontrolled. According to the Global Initiative for Asthma guidelines,4 patients with asthma should undergo routine risk assessment and their plan of care should be adjusted based on their asthma control and risk of acute exacerbation to prevent adverse outcomes.
Recently, the US health-care system has shifted toward pay for performance, with the goal of improving patient outcome and reducing the costs of asthma care. Outcome measures such as asthma-related ED visits and hospitalizations are monitored closely.5 Therefore, identifying populations at high risk for decompensation is imperative for early individualized interventions to reinforce asthma action plans and to determine the need for escalation of therapy. Ultimately, the goal is to reduce disease burden on both individuals and the health-care system.
Unfortunately, acute asthma exacerbations can be difficult to predict. In the past, several studies developed asthma exacerbation prediction models with traditional statistical methods and known risk factors, including FEV1, smoking status, BMI, Asthma Control Questionnaire score, severity of asthma score, and history of severe acute exacerbation; however, the performances of these models were suboptimal.6, 7, 8 Machine learning (ML) is a field of artificial intelligence (AI) that uses mathematical methods to analyze the data. The prediction models built with ML attempt to learn the relationships or patterns between the input variables and specific outcome.9 ML applications in the medical field have become more popular, having been used to interpret ECG findings,10 to classify heart failure,11 and to predict diabetes outcomes.12 Many studies also have used AI for asthma diagnosis,13 severity classification,6 risk stratification, and phenotype subcategorization.7,14 AI also was used to predict risk of asthma-related hospitalization at the time of ED encounter.15,16 Compared with traditional statistics, ML offers advantages of higher accuracy and the capacity to handle large-scale data. To our knowledge, ambulatory data have not been studied with ML to predict the risk of asthma exacerbation.
To address the knowledge gap, we aimed to develop ML models with large-scale and real-world local data to predict asthma exacerbations. We also evaluated the performance and interpretability of the prediction models derived from complicated mathematical ML algorithms as compared with classic statistical methods using logistic regression.
Methods
Study Design and Setting
Cleveland Clinic is a health-care system with more than 200 outpatient sites and 15 hospitals in the United States. De-identified data of patients with asthma seen within our system from 2010 through 2018 were extracted from electronic health records (EHRs) using eResearch (Cleveland Clinic Enterprise Data Center) (e-Appendix 1). The study was approved by the Cleveland Clinic Institutional Review Board.
Study Samples
We identified all patients ever diagnosed with asthma at the Cleveland Clinic Health System using International Classification of Diseases, Clinical Modification, codes (Ninth Revision, 493.xx; or Tenth Revision, J45.xx) between 2010 and 2018. We limited our analysis to patients 18 to 80 years of age who received asthma rescue or controller medication for more than 6 months. We also excluded pregnant women, active smokers, former smokers with a remote smoking history of more than 10 pack-years, and patients with any other chronic pulmonary diseases (e-Fig 1). We then replicated our results in an independent database of 12,093 individuals seen for the first time for asthma at the Cleveland Clinic Health System between January 1, 2019, and November 15, 2020. Patients included in the replicative 2019-2020 cohort were not included previously in the 2010-2018 cohort.
Predictors
Input variables for ML models included information on demographics, clinical characteristics, laboratory test measurements, asthma medication use, and comorbidities. Covariates were selected according to theory, logic, and prior evidence (e-Table 1). Severe asthma was identified by the need for high-dose inhaled corticosteroids (HDiCS) with or without inhaled long-acting β-agonists (LABAs), chronic systemic glucocorticoid therapy with prescription of 3 months or longer, biological therapy, or the presence in the EHR of an International Classification of Diseases, Tenth Revision, Clinical Modification code of J45.5 (severe persistent asthma). Subgroup analysis in patients with pulmonary function testing (PFT) was performed separately to evaluate the independent impact of spirometry.
Outcomes Measures
Nonsevere asthma exacerbation was defined as an exacerbation requiring an oral glucocorticoid burst prescription that was limited to fewer than 28 days, whereas severe exacerbation was defined as an asthma exacerbation event requiring an ED visit or hospitalization. Three prediction models were built separately to assess different outcomes of interests: oral glucocorticoid bursts, ED visits, and hospitalization. In prediction models, we used binary outcomes defined as whether a patient had events during the study period.
Statistical Analysis
The dataset was split randomly into two groups to develop the models: 80% of the data served as a training set and 20% served as a validation set. Predictive models were built with: (1) logistic regression, (2) random forests, and (3) the light gradient boosting machine (LightGBM) algorithm.17 Logistic regression analysis and random forests were carried out or constructed with the Scikit-Learn package in Python,18 whereas LightGBM is a widely used gradient-boosting framework that uses a tree-based algorithm to perform multiclass classification or regression.17 LightGBM was designed to be accurate, efficient, and fast, which are advantages in handling large-scale data. All models were optimized to avoid overfitting with parameters tuning and restricting the decision tree’s freedom. As compared with logistic regression and random forests, LightGBM used NaN to represent missing values by default and were dealt separately than zero, because missing values were interpreted as containing information. For example, the algorithm considered no IgE data (NaN) and IgE level of 0 as two different categories during tree building.
The models were interpreted with the shapley additive explanations (SHAP) approach, which is based on classic Shapley values from game theory.19 This approach explained the models at the level of individual patients based on additive of numeric computed credit—SHAP value—of each feature. The SHAP value reflects the contribution of each input feature (eg, age, BMI, race, and asthma severity) in predicting the output results (eg, risk of asthma-related ED visit) in each patient. It also integrated all computed local credits of each feature that affected risk prediction for patients across the entire database. SHAP values were generated by the Python Shap version 0.35.0 package, and the results of model interpretation are demonstrated graphically.
The performance of prediction models was evaluated by computing the area under the receiver operating characteristic curve (AUC), sensitivity, specificity, positive predictive value, and negative predictive value. The accuracy of probabilistic prediction was measured using the Brier score (e-Table 2), which reflects the mean squared error of the forecast. A lower Brier score value implies more accurate prediction. All analyses were performed with Scikit-Learn version 0.21.3 and Python version 3.6.6.
Results
Patient Characteristics
Between 2010 and 2018, inclusion criteria were met for 113,668 patients with asthma. Of these, we excluded 19,378 patients with chronic lung diseases, 27,459 patients with age outside of 18 to 80 years, and 6,529 pregnant women. A total of 60,302 patients were included in the analysis. The baseline patient characteristics are shown in Table 1. In this cohort, the median age was 47.5 years (interquartile range, 60.7-29.4 years), 62.1% were women, 76.3% were White, and the mean follow-up duration was 3 years (interquartile range, 4.0-1.0 years). The most common comorbidities were sinusitis, hypertension, and depression. Overall, 19,772 patients (32.8%) experienced at least one nonsevere exacerbation and required oral glucocorticoid burst therapy, 1,748 (2.9%) required an ED visit but not hospitalization, and 902 (1.5%) were hospitalized. On average, the asthma exacerbation rates were: 0.23 exacerbation per patient-year for oral glucocorticoid bursts, 0.03 per patient-year for ED visits, and 0.007 per patient-year for hospitalizations. The performance of prediction models is summarized in Table 2.
Table 1.
Clinical Characteristics of the Study Patients, According to Presence or Absence of Exacerbation
| Characteristic | All Patients (N = 60,302) | No Exacerbation (n = 37,880) | Nonsevere Exacerbation (n = 19,772)a | Severe Exacerbation (n = 2,650)b | P Value |
|---|---|---|---|---|---|
| Age, y | 47.5 (60.7-29.4) | 46.3 (60.4-28.5) | 49.9 (61.7-33.2) | 41.8 (55.4-25.6) | < .001 |
| Female sex | 37,443 (62.1) | 23,335 (61.6) | 12,468 (63.1) | 1,640 (61.9) | < .001 |
| Race | |||||
| White | 45,998 (76.3) | 29,003 (76.6) | 15,580 (78.8) | 1,415 (53.4) | < .001 |
| Black | 9,259 (15.4) | 5,528 (14.6) | 2,743 (13.9) | 988 (37.3) | |
| Asian | 800 (1.3) | 528 (1.4) | 247 (1.2) | 25 (0.9) | |
| Other | 4,245 (7.0) | 2,821 (7.4) | 1,202 (6.1) | 222 (8.4) | |
| BMI, kg/m2 | 28.7 (34.3-24.4) | 28.2 (33.8-24.0) | 29.4 (35.0-25.1) | 30.2 (36.9-25.4) | < .001 |
| Treatment | |||||
| SABA | 44,615 (74.0) | 25,206 (66.5) | 16,792 (84.9) | 2,617 (98.8) | < .001 |
| LDiCS | 34,384 (57.0) | 17,910 (47.3) | 14,340 (72.5) | 2,134 (80.5) | < .001 |
| iCS + LABA or HDiCS | 33,871 (56.2) | 17,550 (46.3) | 14,201 (71.8) | 2,120 (80.0) | < .001 |
| Chronic steroid | 1,041 (1.7) | 260 (0.7) | 613 (3.1) | 189 (7.1) | < .001 |
| Theophylline | 199 (0.3) | 62 (0.2) | 99 (0.5) | 38 (1.4) | < .001 |
| Leukotriene inhibitor | 14,096 (23.4) | 6,752 (17.8) | 6,289 (31.8) | 1,055 (39.8) | < .001 |
| Anti-IgE biological therapy | 257 (0.4) | 65 (0.2) | 161 (0.8) | 31 (1.2) | < .001 |
| Nasal steroids | 35,028 (58.1) | 18,363 (48.5) | 14,521 (73.4) | 2,144 (80.9) | < .001 |
| Nasal antihistamines | 3,867 (6.4) | 1,666 (4.4) | 2,012 (10.2) | 189 (7.1) | < .001 |
| Comorbidities | |||||
| Hypertension | 18,488 (30.7) | 10,631 (28.1) | 6,932 (35.1) | 925 (34.9) | < .001 |
| Diabetes | 6,599 (10.9) | 4,002 (10.6) | 2,216 (11.2) | 381 (14.4) | < .001 |
| Liver disease | 2,962 (4.9) | 1,591 (4.2) | 1,225 (6.2) | 146 (5.5) | < .001 |
| Renal failure | 2,024 (3.4) | 1,101 (2.9) | 812 (4.1) | 111 (4.2) | < .001 |
| Sleep apnea | 8,983 (14.9) | 4,928 (13.0) | 3,548 (17.9) | 507 (19.1) | < .001 |
| Depression | 12,251 (20.3) | 6,778 (17.9) | 4,760 (24.1) | 713 (26.9) | < .001 |
| GERD | 16,296 (27.0) | 8,844 (23.3) | 6,648 (33.6) | 804 (30.3) | < .001 |
| Sinusitis | 20,827 (34.5) | 10,080 (26.6) | 9,663 (48.9) | 1,084 (40.9) | < .001 |
| Anemia | 7,940 (13.2) | 4,237 (11.2) | 3,231 (16.3) | 472 (17.8) | < .001 |
Data are presented as No. (%) or median (interquartile range), unless otherwise indicated. GERD = gastroesophageal reflux disease; HDiCS = high-dose inhaled corticosteroid; iCS= inhaled corticosteroid; LABA = long-acting β-agonist; LDiCS = low-dose inhaled corticosteroid; SABA = short-acting β-agonist.
Requiring oral prednisone bursts, but no ED visits or hospitalizations.
Requiring ED visits or hospitalizations.
Table 2.
Prediction Performance of ML Models
| Prediction Model | AUC | P Value | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| Nonsevere exacerbation | ||||||
| Logistic regression | 0.71 (0.70-0.72) | Reference | 0.60 (0.59-0.62) | 0.71 (0.70-0.72) | 0.52 (0.51-0.54) | 0.77 (0.76-0.78) |
| Random forest | 0.68 (0.67-0.69) | < .01 | 0.60 (0.58-0.61) | 0.67 (0.66-0.68) | 0.49 (0.48-0.50) | 0.76 (0.75-0.77) |
| LightGBM | 0.71 (0.70-0.72) | .44 | 0.64 (0.62-0.65) | 0.67 (0.66-0.68) | 0.51 (0.49-0.52) | 0.78 (0.77-0.78) |
| ED visit | ||||||
| Logistic regression | 0.78 (0.76-0.80) | Reference | 0.67 (0.62-0.71) | 0.77 (0.76-0.77) | 0.10 (0.09-0.11) | 0.98 (0.98-0.99) |
| Random forest | 0.84 (0.82-0.86) | .17 | 0.75 (0.71-0.79) | 0.78 (0.77-0.79) | 0.12 (0.11-0.13) | 0.99 (0.98-0.99) |
| LightGBM | 0.88 (0.86-0.89) | < .01 | 0.84 (0.81-0.88) | 0.76 (0.75-0.77) | 0.12 (0.11-0.13) | 0.99 (0.99-0.99) |
| Hospitalization | ||||||
| Logistic regression | 0.81 (0.77-0.84) | Reference | 0.76 (0.70-0.82) | 0.74 (0.73-0.74) | 0.04 (0.04-0.05) | 1 (0.99-1) |
| Random forest | 0.79 (0.76-0.83) | .47 | 0.59 (0.52-0.66) | 0.86 (0.85-0.87) | 0.06 (0.05-0.07) | 0.99 (0.99-0.99) |
| LightGBM | 0.85 (0.82-0.88) | < .01 | 0.86 (0.81-0.91) | 0.73 (0.72-0.73) | 0.05 (0.04-0.05) | 1 (1-1) |
Data are presented as No. (%) or median (interquartile range). AUC = area under the receiver operating characteristic curve; LightGBM = light gradient boosting machine; ML = machine learning; NPV = negative predictive value; PPV = positive predictive value.
Predicting Nonsevere Exacerbation
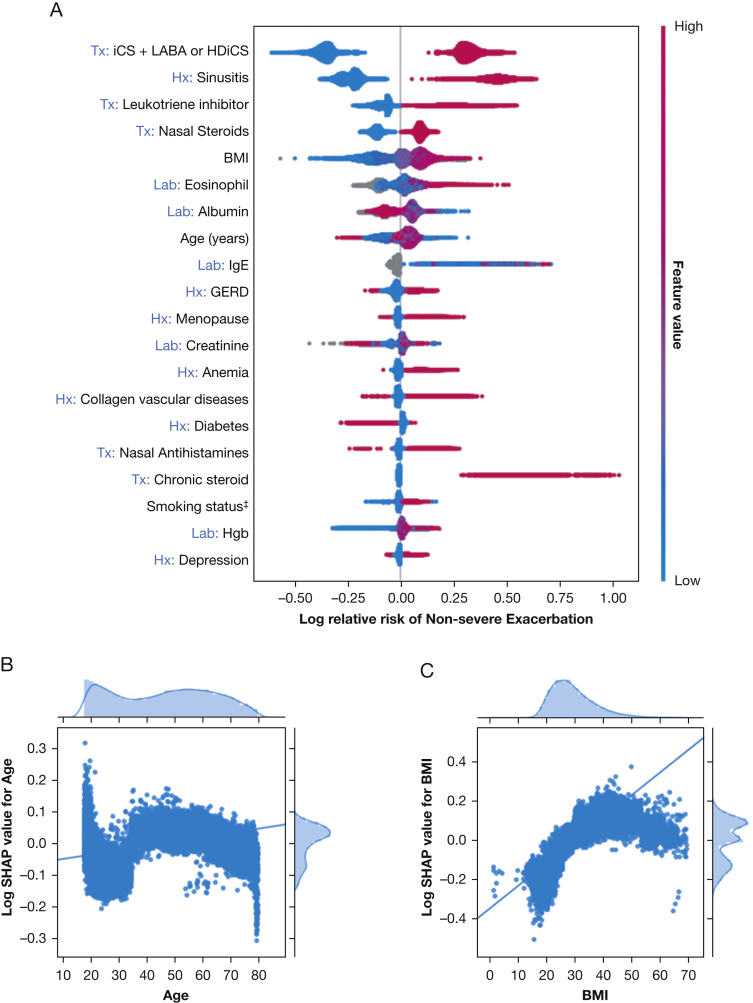
The important features that affect prediction models are shown in Figure 1. The LightGBM prediction model for nonsevere exacerbations showed an AUC of 0.71 (95% CI, 0.70-0.72), a sensitivity of 0.64 (95% CI, 0.62-0.65), and a specificity of 0.67 (95% CI, 0.66-0.68). Given the low prevalence of nonsevere exacerbations, the negative predictive value (0.78 [95% CI, 0.77-0.78]), was higher than the positive predictive value (0.51 [95% CI, 0.49-0.52]). The result is similar to that of logistic regression and better than that of random forests. The most important risk factors for nonsevere exacerbation prediction were a history of sinusitis, treatment with combination inhaled corticosteroids (iCS) and LABA or with HDiCS, and leukotriene inhibitors (Fig 1, e-Fig 2). Patients at higher risk for nonsevere exacerbation showed higher BMI, higher peripheral absolute eosinophil count, and lower albumin level than patients who did not report exacerbations.
Figure 1.
A-C, Prediction model of nonsevere asthma exacerbation. Interpretation of model built with gradient boosting machine algorithm. A, Top 20 risk factors, with iCS + inhaled LABA or HDiCS being the most important one: present (pink) increased the risk of exacerbation, whereas absent (blue) decreased it. B, C, Plots of relative risk (log SHAP value) and age (B) and BMI (C) showed that advanced age and higher BMI associated with higher risk of nonsevere asthma exacerbation. †Race: 0 = other, 1 = White, 2 = Black, and 3 = Asian. ‡Smoking status: 0 = other, 1 = never smoker, 2 = active smoker, and 3 = former smoker. Tx variables were: 1 = present or 0 = absent. Lab variables were the value of the tests. GERD = gastroesophageal reflux disease; HDiCS = high-dose inhaled corticosteroid; Hgb = hemoglobin; iCS = inhaled corticosteroid; Lab = laboratory; LABA = long-acting inhaled β-agonist; SHAP = shapley additive explanation; Tx = treatment.
Predicting ED Visits
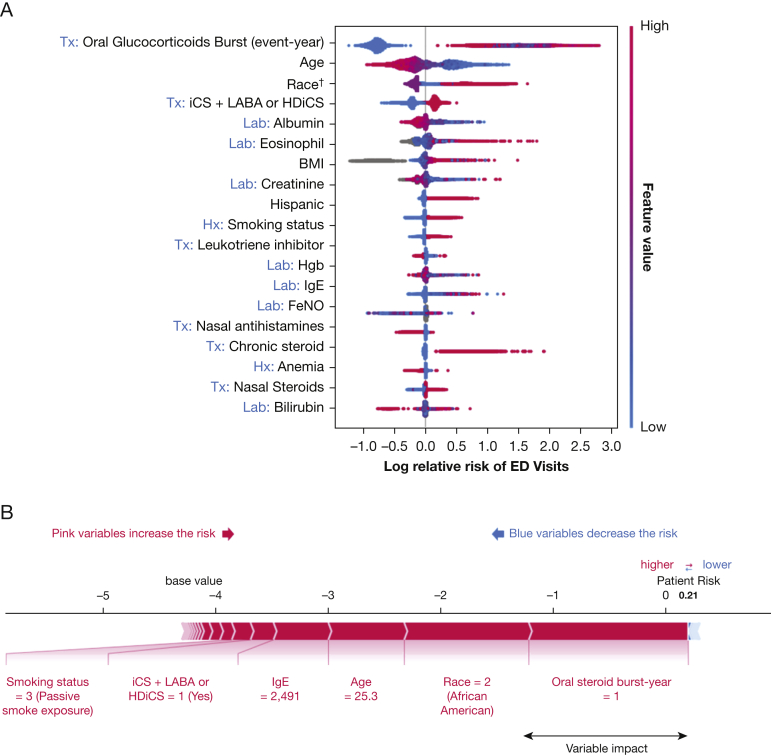
Figure 2A lists all variables important in the prediction of asthma-related ED visits. The LightGBM model showed good predictive value with an AUC of 0.88 (95% CI, 0.86-0.89), a sensitivity of 0.84 (95% CI, 0.81-0.88), and a specificity of 0.76 (95% CI, 0.75-0.77). Similar to the nonsevere exacerbation prediction model, the negative predictive value was high (0.99 [95% CI, 0.99-0.99]), whereas the positive predictive value was low (0.12 [95% CI, 0.11-0.13]). In ED visit prediction, the LightGBM model outperformed logistic regression and random forests algorithms.
Figure 2.
A, B, Prediction model of ED visits. Interpretation of model built with gradient-boosting machine algorithm. A, Top 3 risk factors are the need for oral glucocorticoid bursts, younger age, and Black race: present (pink) increased the risk of ED visit, whereas absent (blue) decreased it. B, Predicted high-risk patient that explained, with additive factors of variables, the width of bar indicated relative impact of one variable. †Race: 0 = other, 1 = White, 2 = Black, and 3 = Asian. ‡Smoking status: 0 = other, 1 = never smoker, 2 = active smoker, and 3 = former smoker. Tx variables were 1 = present or 0 = absent. Lab variables were the value of the tests. FeNO = fractional exhaled nitric oxide; GERD = gastroesophageal reflux disease; HDiCS = high-dose inhaled corticosteroid; Hgb = hemoglobin; iCS = inhaled corticosteroid; Lab = laboratory; LABA = long-acting inhaled β-agonist; SHAP = shapley additive explanation; Tx = treatment.
The most important risk factors in ED visit prediction were: younger age, Black race, a history of nonsevere exacerbations requiring oral glucocorticoid bursts during the study period, and a history of severe asthma treated with combination iCS and LABA therapy or HDiCS (e-Fig 3). Similar to the nonsevere exacerbation prediction model, higher IgE values, higher peripheral blood absolute eosinophil counts, and lower albumin levels were associated with higher risk for ED visits.
To explain the model prediction value better, we used an example to predict the need for an ED visit in one patient, as demonstrated in Figure 2B. We demonstrated that the patient’s risk for ED visits was increased compared with base value because of the cumulative effects of various features, including history of severe asthma, age of 25.3 years, Black race, an IgE level of 2,491 IU/mL, and a history of nonsevere exacerbations requiring oral glucocorticoid bursts.
Predicting Hospitalization
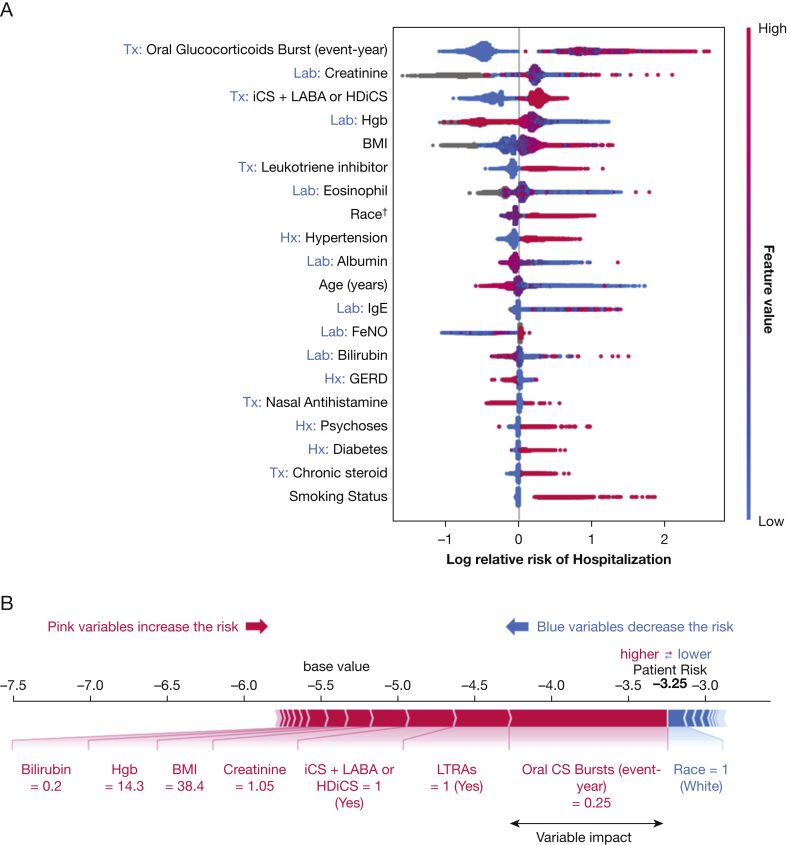
The most important variables for hospitalization prediction are shown in Figure 3A. The LightGBM model showed an AUC of 0.85 (95% CI, 0.82-0.88), a sensitivity of 0.86 (95% CI, 0.81-0.91), and a specificity of 0.73 (95% CI, 0.72-0.73). Similarly, the low prevalence of hospitalizations resulted in a high negative prediction rate (1 [95% CI, 1-1]) and a low positive prediction rate (0.05 95% CI, [0.04-0.05]). Similar to ED visit models, LightGBM outperformed logistic regression and random forest algorithms in predicting hospitalization.
Figure 3.
A, B, Prediction model of hospitalization. Interpretation of model built with gradient boosting machine algorithm. A, SHAP summary plot for hospitalization prediction model ranking from the covariable with highest impact. The plot was colored according to the value of covariables, with pink indicating a higher value and blue indicating a lower value. B, Example of a predicted high-risk patient with interpretable additive effects of various variables. †Race: 0 = other, 1 = White, 2 = Black, and 3 = Asian. ‡Smoking status: 0 = other, 1 = never smoker, 2 = active smoker, and 3 = former smoker. Tx variables were 1 = present or 0 = absent. Lab variables were the value of the tests. CS = corticosteroid; FeNO = fractional exhaled nitric oxide; GERD = gastroesophageal reflux disease; HDiCS = high-dose inhaled corticosteroid; Hgb = hemoglobin; iCS = inhaled corticosteroid; LABA = long-acting inhaled β-agonist; LTRA = leukotriene receptor antagonist.
Similar to our ED visit model, the most important risk factors for hospitalization were a history of nonsevere exacerbations requiring oral glucocorticoid bursts and treatment with combination iCS and LABA or with HDiCS. Lower hemoglobin level and higher BMI also were related to a higher risk of hospitalization. The age-related risk of hospitalization followed a U-shape distribution, in which the risk was highest among young patients (younger than 20 years) and elderly patients (older than 70 years). Other important features are shown in e-Figure 4. For example, a 35-year-old White woman with a high BMI and a history of severe asthma requiring chronic steroid use, iCS plus LABA or HDiCS, a prior history of nonsevere exacerbations requiring oral glucocorticoid bursts, and leukotriene receptor antagonists was found to predict a high risk for hospitalization (Fig 3B). The plot demonstrated the variables and their impact on individual risk prediction.
Subgroup Analysis in Patients Who Underwent Pulmonary Function Tests
In the subgroup analysis of 9,448 patients with spirometry data, low FEV1 and FEV1 to FVC ratio were identified as top risk factors for both ED visit and hospitalization prediction (e-Figs 5, 6). However, adding spirometry data did not significantly improve our model prediction performance (e-Table 3).
Models Performances in a Replicative Cohort
We evaluated the performance of prediction models built with data from individuals seen between 2010 and 2018 (n = 60,302) (e-Fig 7) in a new cohort of 12,093 patients seen for the first time for asthma between January 1, 2019, and November 15, 2020, as an independent test dataset and found that the models’ performances were similar in both cohorts (e-Table 4).
Discussion
In this study, we used ML algorithms and readily available outpatient data from 60,302 asthma patients to build models to predict asthma exacerbation and health-care use. Compared with classic logistic regression, LightGBM demonstrated superior performance in predicting asthma-related emergency visits and hospitalization with improved AUC. The shared risk factors for nonsevere asthma exacerbation, ED visits, and hospitalizations were frequent exacerbation requiring oral glucocorticoid bursts, severe asthma, and age. To our knowledge, we are the first to use outpatient data to build predictive models that enable primary care providers to identify high-risk asthma patients. ML algorithms can be incorporated into EHRs to build predictive models using real-world data that account for local population characteristics. These models subsequently can be used in clinical decision tools at the point of care to guide clinicians to improve patients’ care. In asthma, these clinical decision tools can help to identify at-risk patients who might benefit from escalation of therapy or referral to specialized asthma centers. Theoretically, they also can be used to alert patients about downtrends in their health conditions and encourage them to seek medical attention. This became possible because of the widespread use of EHRs and their easy and instantaneous accessibility to patients.
In a systematic review for prediction models of asthma exacerbation, 24 prediction models were developed that took into account the patient’s demographics, history, PFT results, and asthma risk factors using traditional statistics. These studies were validated externally with two major cohorts in Europe.20 The performance of the prediction models yielded AUCs ranging from 0.50 to 0.77. In contrast, our study used outpatient data from EHRs on more than 60,000 patients and included clinical data such as laboratory measurements and spirometry. More importantly, our models included asthma-specific information such as asthma severity, medication use, and asthma-related laboratory values. Our model’s inclusion of robust clinical parameters available in the EHR could account for the significant improvement in our prediction model compared with others.
Except for fractional exhaled nitric oxide (Feno), prior asthma predictions models did not include biomarkers such as IgE or eosinophil count in risk prediction, despite the strong association of asthma exacerbations with these variables.21,22 Our study included these variables and enhanced the ability of personalized prediction especially in those with a type 2 inflammation phenotype.23 Additionally, our results showed that higher albumin was related to lower risk of asthma exacerbation. The relationship between albumin and exacerbation had not been established in asthma, but severe hypoalbuminemia, commonly seen in chronic illnesses, was reported previously as an independent risk factors for acute respiratory failure in COPD.24 It was unclear in our study whether low albumin was associated directly with severe asthma or was the consequence of other chronic comorbidities.
In an outpatient setting, preventing acute exacerbations and avoiding adverse outcomes have always been the primary goals of physicians taking care of asthma patients. Classical prediction models usually were built using published literature from the ED or extrapolated from clinical trials, which may not be generalizable to local populations.20 In contrast, our approach has the advantage of using locally generated data that takes into consideration demographic distributions and local practice habits that could be extrapolated to a physician‘s individual community. We also graphically demonstrated the interpretability of complicated ML algorithms by plotting the individual risks prediction. These features highlight the population of asthma patients for whom targeted interventions may alter patient-related outcomes significantly.
Compared with traditional statistics, the use of AI provides an alternative approach to enhance the accuracy of prediction models. It is worth noting that ML emphasizes optimization of performance and does not require presumptions, whereas traditional statistical methods usually focus on verifying specific hypothesis (eg, whether high Feno increase the risk of asthma exacerbation).25 To compare ML with traditional statistical methods in an asthma population, Goto et al15 built hospitalization prediction models at EDs and included 3,896 ED visits with variables that included demographics, vitals, chief symptoms, and comorbidities. The author used several ML algorithms (least absolute shrinkage and selection operator [LASSO] regression, random forests, gradient-boosted decision trees, and deep neural networks) to compare with a traditional logistic regression model and concluded that ML algorithms had higher accuracy. Among ML algorithms, Patel et al16 showed gradient-boosting algorithms to have the highest accuracy compared with decision trees, logistic regression, and random forests in predicting hospitalization for pediatric asthma patients in the ED. Our study demonstrated superior results of the LightGBM algorithm in ED visit and hospitalization prediction; however, this was not the case for nonsevere exacerbation prediction. This could be related, at least in part, to prevalence of predicted outcome, which was similar to a previous study.26
Another strength of ML is the capability of handling massive amounts of data, because “velocity” is one of the key components of big data research. With the increasing speed of data accumulation, ML has a significant advantage over traditional statistics with the capability of improving accuracy with new input data. Models like ours can serve as clinician decision support tools in real time27 and have been studied for real-life asthma outpatient monitoring. Finkelstein and Jeong28 used telemonitoring results, including patient’s symptoms and daily asthma diary findings, to build prediction modeling that identified asthma exacerbations in real time with a sensitivity 0.80 to 1 and specificity of 0.77 to 1. In contrast to the study by Finkelstein and Jeong, our study used rich EHR data that was readily available at the point of care to generate general risk profiles. Our data also were independent of patient effort and did not require telemonitoring, which avoided concerns regarding privacy violations.
Additionally, our study has several strengths. First, sample size is key to building an accurate ML model. To our knowledge, this is the largest-scale asthma exacerbation prediction model built with ML algorithms, which have the ability to improve accuracy continually by expanding the data over time.29 Second, our data were collected directly from day-to-day patient care EHRs instead of randomized control trials. Out data reflect the variety of real-world patient care and include all asthma patients with variable severity. Third, we unrevealed the so-called black box myth with interpretable tools. The black box effect refers to the phenomena in which one inputs the data, and the algorithm spits out the results. There are serious concerns because of lack of transparency regarding how these algorithms predicted their decision.30 To solve the problem, we used SHAP to demonstrate graphically the interaction between the risk and variables19 and significantly improved the transparency of ML models.
Our study has several limitations. Our models, which used EHR data from a single health-care system, may be useful for local patients, but may not be generalizable to other health-care institutions. It also is possible that the number of asthma exacerbations, ED visits, and hospitalizations may be underestimated, and therefore subject our results to bias because patients may seek urgent care in other health-care organizations in northeast Ohio. As in many other EHR-based studies, the identification of medical conditions—asthma, in our case—is based on International Classification of Diseases codes, and therefore can be limited by the clinicians’ coding habits. In this study, we used stringent criteria and a multistep method to define asthma properly and to avoid bias introduced by including patients with smoking-related airway obstruction or other concurrent chronic lung diseases. However, disease severity, which commonly is based on well-defined consensus statements, cannot be defined easily in EHR-based research. In this case, we used the intensity of medical therapy as a marker of asthma severity. We defined severe asthma by the need for HDiCS therapy with a second controller medication instead of standardized definitions31 because of limited information on asthma control. EHR-based research is based on real-world data, but commonly is limited by missing clinical information. For example, only 15.7% of patients included in our analysis had spirometry measurements available, and the Asthma Control Test, which is used commonly to define asthma control, also was missing in a large proportion of patients. We believe that the use of the Asthma Control Test as a tool to monitor asthma control and compliance with asthma guidelines is extremely important to the care of patients with asthma and always should be encouraged. We did not include anti-IL5 biologic asthma therapy as a covariate in our analysis because a very limited number of patients received such therapy in our asthma center before 2018. The impact of anti-IL5 therapy on predictive models of asthma exacerbations continues to evolve as more patients are administered this therapy. We also acknowledge that the lack of information on asthma control and guideline compliance negatively impacted the performance of our models and that the analysis of the original and replicative cohorts are limited by their cross-sectional nature. To account for the duration of follow-up, we assessed outcomes as a function of the duration of follow-up and presented those outcomes per patient-year. Our models also were impacted negatively by the quality of clinical information. For example, information regarding medication use in our study was based on medication prescribed through the EHR, rather than dispensed by a pharmacy. As the EHR evolves, it is expected that information on dispensed medications will be available readily in the future to help improve predictive models and to circumvent this important limitation.
ML models are focused on accuracy and cannot define causality. For example, hemoglobin was identified as one of the important factors affecting our models, but it was unknown whether the laboratory value was the cause or the consequence of asthma. In addition, we have PFT and FeNO data for only a small group of patients. However, although our data are limited, including PFT results did not significantly improve the outcome prediction in our subgroup analysis, despite our finding that low FEV1 and FEV1 to FVC ratio were associated with a higher risk of exacerbations. This could be related to the fact that only 16% of patients in our database had PFT results available. However, our study showed that in patients with low FEV1 or FEV1 to FVC ratio, these features significantly affect their individual risk prediction. In real-world EHRs, missing data are inevitable and may vary depending on the patients’ disease severity, health-care providers’ preferences, and variation between local guidance and third-party payers. In traditional statistics, most missing values are handled by imputation or exclusion. The tree-based ML algorithms like LightGBM or extreme gradient boosting (XGBOOST) possess the advantage of identifying missing values as a unique entity and thus increase the overall performance of the prediction model.
In conclusion, we built accurate models from real-world EHR data that can be used to predict asthma exacerbation. Future studies using targeted interventions that apply these risk prediction models to improve patient care are needed.
Acknowledgments
Author contributions: J. G. Z. and C.-P. W. made substantial contributions to the conception or design of the work and acquired, analyzed, and interpreted the data for the work. J. G. Z., C.-P. W., A. H. A., P. Z., and A. N. contributed to drafting the work or revising it critically for important intellectual content. J. G. Z. agrees to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are investigated and resolved appropriately. J. G. Z., C.-P. W., A. H. A., P. Z., and A. N. gave the final approval of the version to be published.
Financial/nonfinancial disclosures: None declared.
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Other contributions: The authors thank Bradly Souder, senior system analyst from business intelligence at the Cleveland Clinic, for his valuable input and help.
Additional information: The e-Appendix, e-Figures, and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
J. G. Zein and C.-P. Wu contributed equally to this manuscript.
FUNDING/SUPPORT: This study was funded by the National Heart, Lung and Blood Institute, National Institutes of Health [Grant K08 HL133381 (J. G. Z.)].
Supplementary Data
References
- 1.Centers for Disease Control and Prevention Most recent national asthma data. 2019. Centers for Disease Control and Prevention website. https://www.cdc.gov/asthma/most_recent_national_asthma_data.htm
- 2.Centers for Disease Control and Prevention Asthma attack prevalence percents among those with current asthma by age, United States: National Health Interview Survey, 2018. Centers for Disease Control and Prevention website. https://www.cdc.gov/asthma/nhis/2018/table6-1.htm Accessed July 19, 2020.
- 3.Nurmagambetov T., Kuwahara R., Garbe P. The economic burden of asthma in the United States, 2008-2013. Ann Am Thorac Soc. 2018;15(3):348–356. doi: 10.1513/AnnalsATS.201703-259OC. [DOI] [PubMed] [Google Scholar]
- 4.Global Initiative for Asthma. (GINA 2019). https://ginasthma.org/wp-content/uploads/2019/06/GINA-2019-main-report-June-2019-wms.pdf. Accessed July 8, 2020.
- 5.State of Ohio Detailed business requirements asthma and COPD acute exacerbation episodes. State of Ohio website. https://medicaid.ohio.gov/Portals/0/Providers/PaymentInnovation/DBR/Asthma-COPD.pdf Published 2017. Accessed June, 21, 2020.
- 6.Loymans R.J.B., Honkoop P.J., Termeer E.H. Identifying patients at risk for severe exacerbations of asthma: development and external validation of a multivariable prediction model. Thorax. 2016;71(9):838–846. doi: 10.1136/thoraxjnl-2015-208138. [DOI] [PubMed] [Google Scholar]
- 7.Howard R., Rattray M., Prosperi M., Custovic A. Distinguishing asthma phenotypes using machine learning approaches. Curr Allergy Asthma Rep. 2015;15(7):38. doi: 10.1007/s11882-015-0542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loymans R.J.B., Honkoop P.J., Termeer E.H. Identifying patients at risk for severe exacerbations of asthma: development and external validation of a multivariable prediction model. Thorax. 2016;71(9):838–846. doi: 10.1136/thoraxjnl-2015-208138. [published correction appears in: Thorax. 2018;73(8):795-796] [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Chen P.H.C., Krause J., Peng L. How to read articles that use machine learning: users’ guides to the medical literature. JAMA. 2019;322(18):1806–1816. doi: 10.1001/jama.2019.16489. [DOI] [PubMed] [Google Scholar]
- 10.Tison G.H., Zhang J., Delling F.N., Deo R.C. Automated and interpretable patient ECG profiles for disease detection, tracking, and discovery. Circ Cardiovasc Qual Outcomes. 2019;12(9):e005289. doi: 10.1161/CIRCOUTCOMES.118.005289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Awan S.E., Sohel F., Sanfilippo F.M., Bennamoun M., Dwivedi G. Machine learning in heart failure: Ready for prime time. Curr Opin Cardiol. 2018;33(2):190–195. doi: 10.1097/HCO.0000000000000491. [DOI] [PubMed] [Google Scholar]
- 12.Xiong X.-L., Zhang R.-X., Bi Y., Zhou W.-H., Yu Y., Zhu D.-L. Machine learning models in type 2 diabetes risk prediction: results from a cross-sectional retrospective study in Chinese adults. Curr Med Sci. 2019;39(4):582–588. doi: 10.1007/s11596-019-2077-4. [DOI] [PubMed] [Google Scholar]
- 13.Spathis D., Vlamos P. Diagnosing asthma and chronic obstructive pulmonary disease with machine learning. Health Informatics J. 2019;25(3):811–827. doi: 10.1177/1460458217723169. [DOI] [PubMed] [Google Scholar]
- 14.Saglani S., Custovic A. Childhood asthma: advances using machine learning and mechanistic studies. Am J Respir Crit Care Med. 2019;199(4):414–422. doi: 10.1164/rccm.201810-1956CI. [DOI] [PubMed] [Google Scholar]
- 15.Goto T., Camargo C.A., Jr., Faridi M.K., Yun B.J., Hasegawa K. Machine learning approaches for predicting disposition of asthma and COPD exacerbations in the ED. Am J Emerg Med. 2018;36(9):1650–1654. doi: 10.1016/j.ajem.2018.06.062. [DOI] [PubMed] [Google Scholar]
- 16.Patel S.J., Chamberlain D.B., Chamberlain J.M. A machine learning approach to predicting need for hospitalization for pediatric asthma exacerbation at the time of emergency department triage. Acad Emerg Med. 2018;25(12):1463–1470. doi: 10.1111/acem.13655. [DOI] [PubMed] [Google Scholar]
- 17.Machado M.R., Karray S., de Sousa I.T. 14th International Conference on Computer Science and Education. ICCSE 2019; 2019. LightGBM: an effective decision tree gradient boosting method to predict customer loyalty in the finance industry. [DOI] [Google Scholar]
- 18.Pedregosa F., Varoquaux G., Gramfort A. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 19.Lundberg S.M., Erion G., Chen H. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2(1):56–57. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loymans R.J.B., Debray T.P.A., Honkoop P.J. Exacerbations in adults with asthma: a systematic review and external validation of prediction models. J Allergy Clin Immunol Pract. 2018;6(6):1942–1952.e15. doi: 10.1016/j.jaip.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 21.ten Brinke A., Sterk P.J., Masclee A.A.M. Risk factors of frequent exacerbations in difficult-to-treat asthma. Eur Respir J. 2005;26(5):812–818. doi: 10.1183/09031936.05.00037905. [DOI] [PubMed] [Google Scholar]
- 22.Sherenian M.G., Wang Y., Fulkerson P.C. Hospital admission associates with higher total IgE level in pediatric patients with asthma. J Allergy Clin Immunol Pract. 2015;3(4):602–603.e1. doi: 10.1016/j.jaip.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuruvilla M.E., Lee F.E.H., Lee G.B. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin Rev Allergy Immunol. 2019;56(2):219–233. doi: 10.1007/s12016-018-8712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C.W., Chen Y.Y., Lu C.L. Severe hypoalbuminemia is a strong independent risk factor for acute respiratory failure in COPD: a nationwide cohort study. Int J COPD. 2015;10:1147–1154. doi: 10.2147/COPD.S85831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bzdok D., Altman N., Krzywinski M. Statistics versus machine learning. Nat Methods. 2018;15(4):233–234. doi: 10.1038/nmeth.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raita Y., Goto T., Faridi M.K., Brown D.F.M., Camargo C.A., Hasegawa K. Emergency department triage prediction of clinical outcomes using machine learning models. Crit Care. 2019;23(1):64. doi: 10.1186/s13054-019-2351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundberg S.M., Nair B., Vavilala M.S. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng. 2018;2(10):749–760. doi: 10.1038/s41551-018-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finkelstein J., Jeong I.C. Machine learning approaches to personalize early prediction of asthma exacerbations. Ann N Y Acad Sci. 2017;1387(1):153–165. doi: 10.1111/nyas.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gianfrancesco M.A., Tamang S., Yazdany J., Schmajuk G. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med. 2018;178(11):1544–1547. doi: 10.1001/jamainternmed.2018.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opening the black box of machine learning. Lancet Respir Med. 2018;6(11):801. doi: 10.1016/S2213-2600(18)30425-9. [DOI] [PubMed] [Google Scholar]
- 31.Chung K.F., Wenzel S.E., Brozek J.L. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43(2):343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.