Gutiérrez et al. describe a novel function for the molecular motor KIF13A, in complex with centaurin-α1, for the insertion of AMPA receptors at synapses during long-term potentiation. This process is mediated by the remodeling of intracellular endosomal compartments in the dendritic shaft.
Abstract
The regulated trafficking of AMPA-type glutamate receptors (AMPARs) from dendritic compartments to the synaptic membrane in response to neuronal activity is a core mechanism for long-term potentiation (LTP). However, the contribution of the microtubule cytoskeleton to this synaptic transport is still unknown. In this work, using electrophysiological, biochemical, and imaging techniques, we have found that one member of the kinesin-3 family of motor proteins, KIF13A, is specifically required for the delivery of AMPARs to the spine surface during LTP induction. Accordingly, KIF13A depletion from hippocampal slices abolishes LTP expression. We also identify the vesicular protein centaurin-α1 as part of a motor transport machinery that is engaged with KIF13A and AMPARs upon LTP induction. Finally, we determine that KIF13A is responsible for the remodeling of Rab11-FIP2 endosomal structures in the dendritic shaft during LTP. Overall, these results identify specific kinesin molecular motors and endosomal transport machinery that catalyzes the dendrite-to-synapse translocation of AMPA receptors during synaptic plasticity.
Introduction
Activity-dependent changes in synaptic strength, known as synaptic plasticity, are believed to constitute the cellular basis for learning and memory. Important forms of synaptic plasticity, such as long-term potentiation (LTP) and long-term depression in the hippocampus, are critically dependent on the regulated transport of AMPA-type glutamate receptors (AMPARs) to and from synapses (Shepherd and Huganir, 2007; Henley et al., 2011; Bredt and Nicoll, 2003; Greger and Esteban, 2007). The control of this dynamic behavior depends on a variety of proteins that interact with AMPARs and determine their transport, intracellular localization, endosomal mobilization, and anchoring to the synaptic membrane.
It is now well recognized that the regulated transport of glutamate receptors is highly dependent on the endosomal machinery and motor proteins. In particular, recycling endosomes driven by Rab11 have been shown to carry AMPARs for synaptic delivery during LTP (Park et al., 2004; Brown et al., 2007). This process is powered by molecular motors of the myosin V family (Correia et al., 2008; Wang et al., 2008), which are thought to confer directionality and calcium dependence to the transport event. The coupling between recycling endosomes and cytoskeletal-dependent motor elements occurs through a variety of vesicular adaptors (Welz et al., 2014). Among them, the Rab11 family of interacting proteins (FIPs) participate in endosomal compartmentalization and trafficking, as well as in the regulation of specific Rab11 functions (Lindsay and McCaffrey, 2004; Jing and Prekeris, 2009; Baetz and Goldenring, 2013; Schafer et al., 2014). In the context of synaptic plasticity, Rab11-FIP2 (hereon referred to as FIP2 for simplicity) associates with the actin-based motor myosin Vb (Hales et al., 2002; Kneussel and Wagner, 2013; Schafer et al., 2014) and is mobilized and recruited to dendritic spines, together with AMPARs, after LTP induction (Wang et al., 2008). Moreover, FIP2-carrying endosomes undergo rapid rearrangements upon LTP induction (Royo et al., 2019), reminiscent of the redistribution of AMPARs after NMDA receptor (NMDAR) activation during tetanic stimulation (Shi et al., 1999).
Microtubule-based motor complexes are also Rab effector proteins, allowing membrane association and directional movement of various vesicular cargoes along the microtubule cytoskeleton (Goldstein and Yang, 2000; Hirokawa and Takemura, 2005; Kapitein and Hoogenraad, 2011; Granger et al., 2014; Horgan and McCaffrey, 2011). In the case of AMPARs, microtubule-dependent trafficking has been typically associated with long-range transport along the dendrites, supporting the continuous supply and redistribution of AMPARs among synapses (Hoerndli et al., 2013a; Heisler et al., 2014; Hoogenraad et al., 2005; Monteiro et al., 2012; Setou et al., 2002; Shin et al., 2003), which can also be regulated by neuronal activity (Maas et al., 2009; Hoerndli et al., 2013a). In addition to this dendritic transport, there is increasing evidence for transient microtubule entry into spines in an activity-dependent manner (Hu et al., 2008; Jaworski et al., 2009) that is facilitated by F-actin remodeling (Merriam et al., 2013; Schätzle et al., 2018). These local actions of microtubules raise the possibility of microtubule-dependent transport in the regulation of synaptic function. This has been shown to happen through the transport of NMDARs (Setou et al., 2000; Yin et al., 2011) and signaling regulators (Morikawa et al., 2018), as well as through the delivery of cargo important for spine maturation (Muhia et al., 2016; Zhao et al., 2020). However, no kinesin motor protein has yet been linked directly to AMPAR trafficking in LTP.
In this study, we have focused on the kinesin-3 family motor KIF13, as it catalyzes anterograde trafficking of AMPARs in Caenorhabditis elegans (Monteiro et al., 2012). Furthermore, KIF13 associates with early (Bentley et al., 2015) and recycling endosomes (Delevoye et al., 2014) and is required for endosomal rearrangements reminiscent of those described for FIP2 during LTP (Royo et al., 2019). In this work, we have found that KIF13A (but not KIF13B) is required for LTP in hippocampal slices and mediates the activity-dependent delivery of AMPARs to the spine surface. In addition, we identify centaurin-α1 as a vesicular linker, which associates with KIF13A and AMPARs upon LTP induction and is also required for LTP expression. Finally, we propose that the remodeling of recycling endosomes in the dendritic shaft, as monitored by the endosomal recruitment of FIP2, is the key mechanism underlying KIF13A’s role in LTP.
Results
KIF13A, and not KIF13B, is required specifically for LTP
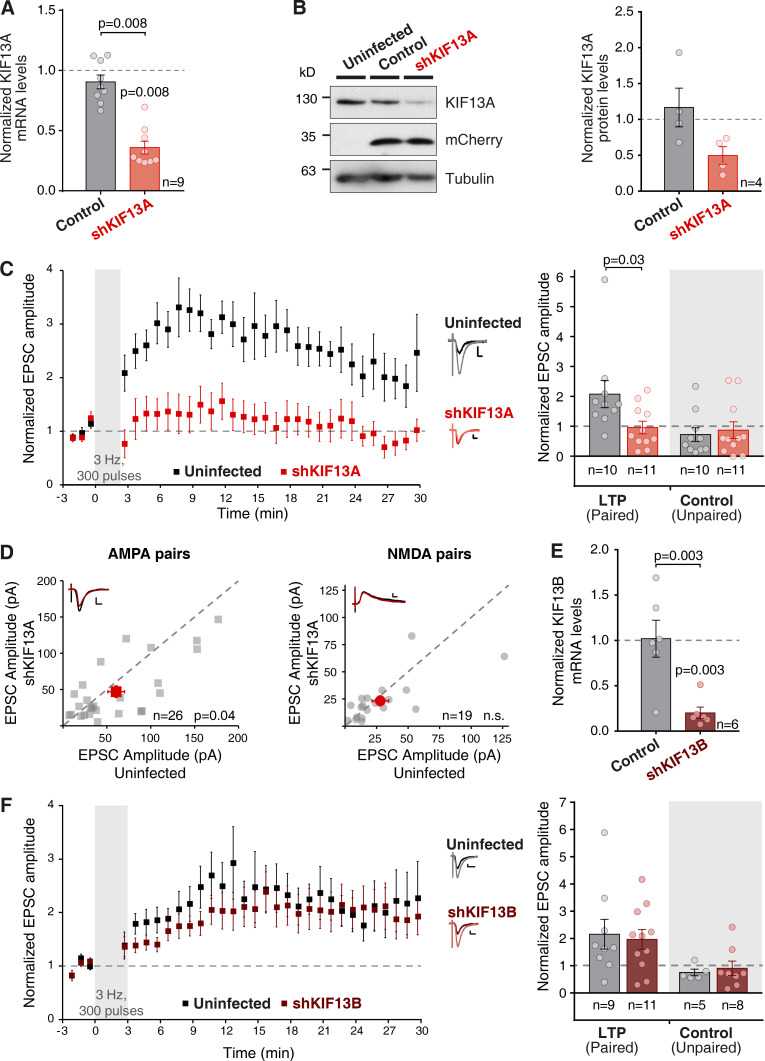
As a first step to evaluate the role of KIF13A in synaptic function, we employed an shRNA knockdown to deplete KIF13A in hippocampal neurons. A specific shRNA (shKIF13A) was designed against the rat mRNA sequence of KIF13A and expressed using a lentivirus system (see Methods for the targeting sequence). Dissociated hippocampal neurons were infected, and the expression levels of endogenous KIF13A were evaluated 7 d after infection. Both mRNA (Fig. 1 A) and protein (Fig. 1 B) levels were strongly reduced, as compared with uninfected neurons (used for normalization) or neurons infected with a control lentivirus (lacking an shRNA sequence).
Figure 1.
KIF13A is required for LTP. (A) Quantification of KIF13A mRNA levels from dissociated hippocampal neurons infected with the shKIF13A lentivirus (red) or control lentivirus (lacking the shRNA sequence; black) relative to the uninfected condition. Cultures were lysed after 15 DIV and 7 d after infection. Bars show mean ± SEM, together with individual values from each experiment. n = 9 independent experiments; P = 0.008, Wilcoxon test. (B) Left: Representative Western blot result from protein lysates from conditions described in A. Apparent protein size was lower than expected, perhaps as a consequence of proteolytic degradation. Right: Quantification of KIF13A protein levels relative to uninfected condition. Bars show mean ± SEM, together with individual values from each experiment; n = 4 independent experiments. (C) Left: Time-course of the averaged AMPAR-mediated evoked synaptic currents (mean ± SEM per condition). LTP was induced (pairing protocol of 300 pulses at 3 Hz) in organotypic hippocampal slices (7‒10 DIV) infected in the CA1 layer with a lentivirus expressing shKIF13A and analyzed at 7‒10 d after infection. Uninfected (black) and infected (red) cells were recorded. Amplitude of the synaptic responses is normalized to a 3-min baseline. Right: Average of AMPAR-mediated responses from the last 5 min of the recording and normalized to the baseline. Bar plots show mean ± SEM, together with individual values for each experiment. Left bars (LTP, paired) correspond to the stimulation pathway in which postsynaptic depolarization (0 mV) was paired to presynaptic stimulation (3 Hz, 300 pulses). Right (shaded) bars (control, unpaired) correspond to the pathway that was not stimulated during depolarization. Inset: Representative traces from uninfected (black lines) and shKIF13A (red lines), averaged from baseline (dark lines) or from the last 5 min of the recording (light lines). Scale bars: vertical, 10 pA; horizontal, 10 ms. n = 10 uninfected cells, and n = 11 shKIF13A-expressing cells. Uninfected neurons are significantly potentiated from baseline (P = 0.01, Wilcoxon test). Uninfected and shKIF13A-expressing neurons were significantly different (P = 0.03, Mann–Whitney test). (D) Basal synaptic responses mediated by AMPARs (left) and NMDARs (right) from adjacent uninfected and shKIF13A-infected CA1 pyramidal neurons at −60 mV and at +40 mV (at 65 ms), respectively. Gray symbols represent each individual pair while red symbols show the mean ± SEM of the measured currents. n = 26 cell pairs for AMPAR currents and n = 19 cell pairs for NMDAR currents; P value according to the Wilcoxon test; n.s., not significantly different. Insets: Representative traces for uninfected (black lines) and shKIF13A (red lines) cells; scale bars: vertical, 50 and 10 pA respectively; horizontal, 10 ms. (E) RT-qPCR quantification of KIF13B mRNA levels from dissociated hippocampal neurons infected with shKIF13B (burgundy) or control (lacking shRNA sequence; black) lentivirus, relative to uninfected condition. Bars show mean ± SEM, together with individual values for each experiment. n = 6 independent experiments; P = 0.03, Wilcoxon test. (F) Similar to C, but with shKIF13B-expressing (burgundy) neurons. Inset: scale bars: vertical, 10 pA; horizontal, 20 ms. n = 9 uninfected cells and n = 11 shKIF13B-expressing cells for the LTP (paired) pathway, and n = 5 uninfected cells and n = 8 shKIF13B-expressing cells for the control (Unpaired) pathway. Uninfected and shKIF13B-expressing neurons are significantly different from baseline (P = 0.02 and P = 0.03, respectively) according to the Wilcoxon test.
Having established the efficiency of the shRNA, the control lentivirus or shKIF13A were expressed in the CA1 region of organotypic hippocampal slice cultures for 7 d. We then performed whole-cell patch-clamp electrophysiological recordings from CA1 pyramidal neurons where endogenous KIF13A was knocked down (shKIF13A) and from neighboring control (uninfected) neurons. LTP was induced using a standard pairing protocol (Hayashi et al., 2000) in which the postsynaptic neuron is depolarized to 0 mV during the delivery of a 3-Hz presynaptic stimulation (300 pulses) of the Schaffer collateral fibers. This protocol induced robust LTP in control uninfected cells (Fig. 1 C, black), but synaptic potentiation was virtually abolished in shKIF13A-expressing neurons (Fig. 1 C, red). Importantly, the synapses that were not stimulated during LTP induction were not affected by shKIF13A expression (Fig. 1 C, unpaired control pathways). These results strongly suggest that KIF13A-mediated microtubule-dependent transport is required for LTP expression.
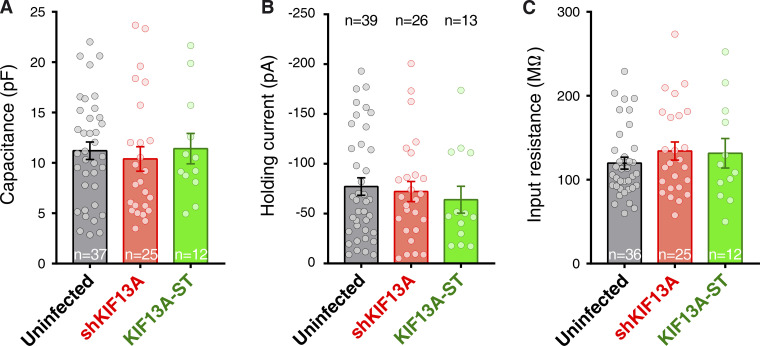
It has been previously described that a different motor protein, KIF17, mediates the microtubule-dependent transport of NMDARs into synapses (Wong et al., 2002; Guillaud et al., 2003; Setou et al., 2000) and in this manner has a direct impact on NMDAR-dependent synaptic plasticity (Yin et al., 2011). Therefore, we evaluated the potential role of KIF13A in NMDAR (and AMPAR) synaptic function under basal conditions. We performed simultaneous electrophysiological recordings of neighboring control (uninfected) and shKIF13A-expressing CA1 neurons from organotypic hippocampal slices. Synaptic transmission was evoked by Schaffer collateral stimulation. AMPAR responses were measured at −60 mV holding membrane potential and NMDAR responses at +40 mV at a latency of 65 ms after stimulation. As shown in Fig. 1 D, knockdown of KIF13A produced a slight but significant decrease in AMPAR basal transmission (left), without any significant effect on NMDAR synaptic responses (right). These results indicated that the impairment of NMDAR-mediated LTP in KIF13A-depleted neurons was not due to an altered function of NMDARs at synapses. On the other hand, the small decrease in AMPAR responses may be due to the impairment of spontaneous LTP-like events occurring in the organotypic culture (see for example, Correia et al., 2008). Importantly, passive membrane properties, such as input resistance and holding current (related to basal ionic conductances), and whole-cell capacitance (related to cell size) were similar in control and in shKIF13A-expressing neurons (Fig. S1).
Figure S1.
Basal physiological properties of neurons expressing shKIF13A and KIF13A-ST. (A–C) Capacitance (A), holding current (B), and input resistance (C) were measured from whole-cell patch-clamp experiments of uninfected (black) or shKIF13A- (red) and KIF13-ST–expressing (green) CA1 pyramidal neurons. Whole-cell capacitance was calculated from the exponential decay of the current in response to a square voltage step. Input resistance is calculated from Ohm's law using the same voltage step. Bars represent the mean ± SEM, together with individual values for each cell. n = 39, 26, or 13 cells for uninfected, shKIF13A, or KIF13-ST neurons, respectively.
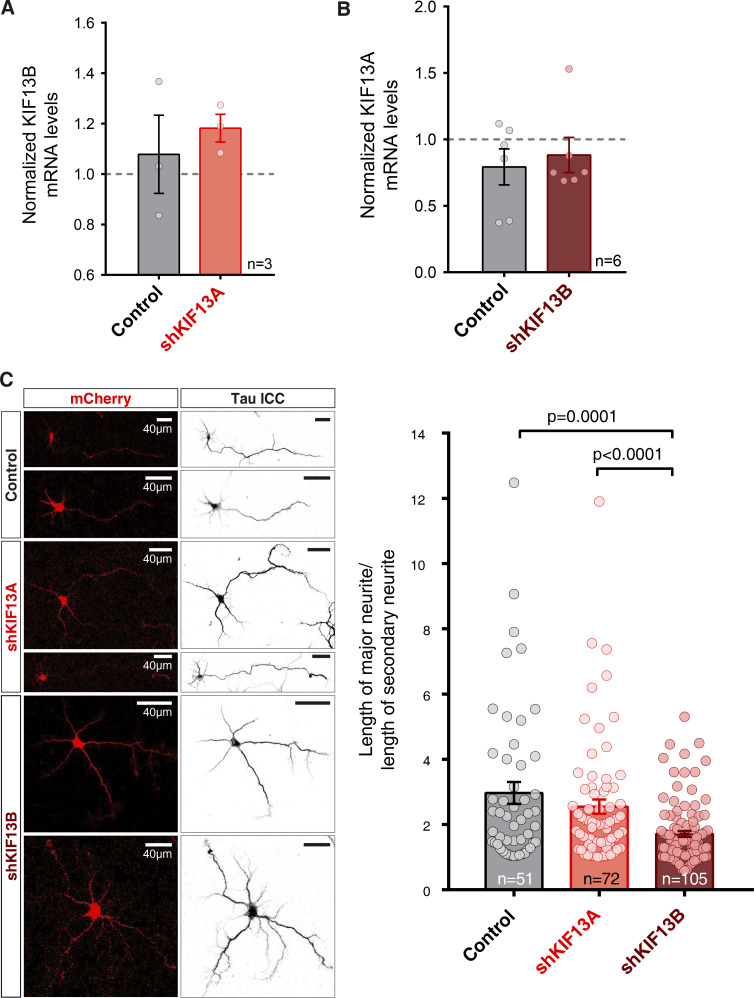
Another kinesin-3 subfamily member, KIF13B, is also expressed in neurons and presents a high degree of homology with KIF13A. Consequently, we also analyzed the potential role of KIF13B in LTP. A previously described shRNA (Yoshimura et al., 2010) was used to knock down endogenous KIF13B in hippocampal neurons using a lentivirus approach (shKIF13B). Knockdown efficiency was tested in dissociated primary hippocampal neuronal cultures and evaluated by quantitative PCR (qPCR). As shown in Fig. 1 E, at 7 d after infection, the levels of KIF13B mRNA were strongly reduced compared with those present in both uninfected (used for normalization) and control lentivirus neurons. To note, commercially available antibodies for KIF13B were not specific enough for Western blot analysis, but protein knockdown had already been reported for this shKIF13B target (Yoshimura et al., 2010). It is also worth noting that the reduction in the expression levels of KIF13B did not alter KIF13A expression and vice versa (Fig. S2, A and B). Additionally, we observed that axon specification and polarization in developing neurons, a process that has been shown to rely on KIF13B function (Horiguchi et al., 2006), was impaired by shKIF13B and not by shKIF13A (Fig. S2 C), supporting the functionality of the KIF13B knockdown.
Figure S2.
Specificity of shKIF13A and shKIF13B knockdown, and effect on axonal polarization. (A) RT-qPCR quantification of KIF13B mRNA levels from dissociated hippocampal neurons infected with the shKIF13A lentivirus (red) or control lentivirus (lacking the shRNA sequence; black) relative to the uninfected condition. Cultures were lysed after 15 DIV and 7 d after infection. Bars show mean ± SEM, together with individual values from each experiment. n, number of independent experiments; not significantly different from uninfected (Wilcoxon test). (B) RT-qPCR quantification of KIF13A mRNA levels from dissociated hippocampal neurons infected with shKIF13B (burgundy) or control (lacking shRNA sequence; black) lentivirus relative to the uninfected condition. Bars show mean ± SEM, together with individual values for each experiment. n = 3 (skKIF13A) and n = 6 (shKIF13B) independent experiments; not significantly different from uninfected (Wilcoxon test). (C) Left: Representative confocal images of 3 DIV dissociated hippocampal neurons infected with a control lentivirus (control; top panels) or lentiviruses expressing shKIF13A (middle panels) and shKIF13B (bottom panels), all expressing mCherry (red, left panels). Tau immunocytochemistry (ICC) was performed to assess neurite length (right panels) in all conditions. Scale bars in right panels are identical to those in left panels. Right: Quantification of axonal polarization was calculated from the ratio between the length of the major neurite compared with the length of the secondary neurite. Bars represent the mean ± SEM, together with individual values for each experiment. shKIF13B neurons (burgundy) show significantly less axonal polarization with respect to control (black) and shKIF13A (red) neurons (P = 0.0001 and P < 0.0001, respectively, Mann–Whitney test). n = 51, 72, or 105 cells for control, shKIF13A- or shKIF13B-expressing neurons, respectively.
Finally, the function of KIF13B in LTP was tested in organotypic hippocampal slices, as described above for KIF13A. As shown in Fig. 1 F, control (black) and shKIF13B-expressing (burgundy) neurons displayed similar synaptic potentiation. These data were in clear contrast with those obtained after KIF13A knockdown (Fig. 1 C), indicating that the KIF13B motor is not involved in LTP. Importantly, this result also serves to verify that lentiviral infection or shRNA expression per se does not affect LTP expression. Additionally, the unpaired pathway remained unaltered in the presence or absence of KIF13B protein (Fig. 1 F, unpaired control pathways).
KIF13A mediates AMPAR transport upon LTP induction
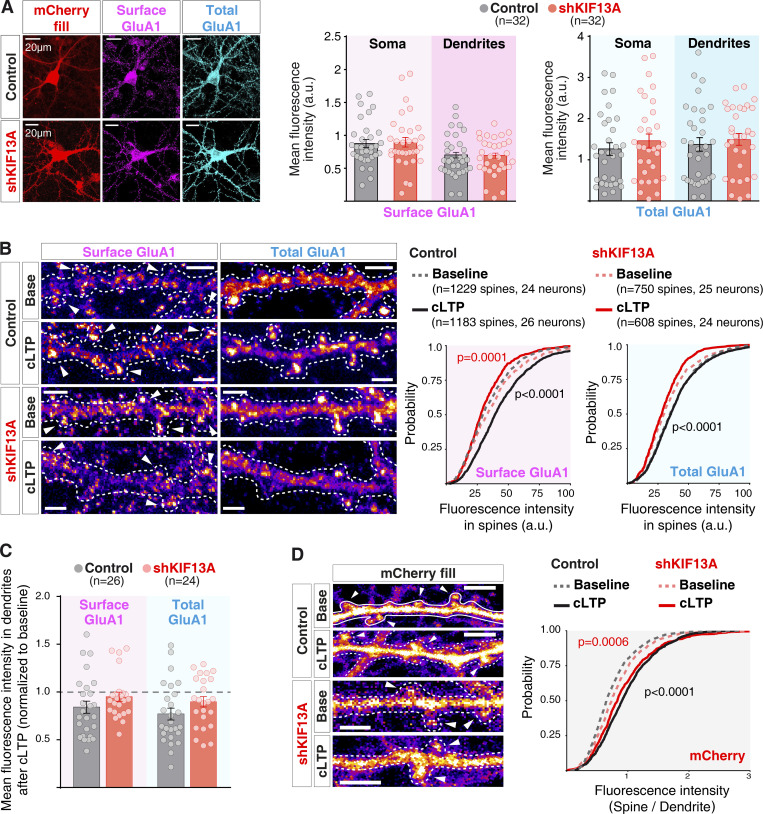
After establishing the importance of KIF13A for LTP, we wished to determine whether this motor protein was mediating a specific transport step involved in synaptic potentiation. Previous studies have shown that kinesin-3 molecular motors contribute to the long-range transport of glutamate receptors from the cell bodies along dendrites (Monteiro et al., 2012). Therefore, we tested the effect of KIF13A depletion on AMPAR basal distribution. To this end, we evaluated the surface expression of endogenous GluA1-containing AMPARs in basal conditions. Dissociated hippocampal neurons (7 d in vitro [DIV]) were infected either with a control lentivirus or with the lentivirus expressing shKIF13A for 7 d. Subsequently, cells were fixed and labeled for endogenous surface GluA1 using an antibody against an N-terminal (extracellular) epitope under nonpermeabilizing conditions. Following a permeabilization step, the whole pool of endogenous GluA1 receptors was also labeled with an antibody against the C-terminal (intracellular) epitope of GluA1 (representative images in Fig. 2 A, left). The quantifications of the fluorescence intensity levels at different cellular compartments (soma and dendrites) revealed no significant difference between control and KIF13A-depleted conditions (Fig. 2 A, right panels). Altogether, these results suggest that KIF13A is not required for the constitutive export of AMPARs from the soma into dendrites or receptor expression at the neuronal surface.
Figure 2.
Activity-dependent transport of AMPARs is mediated by KIF13A. (A) Left: Representative images of hippocampal neuronal cultures after 7 d in vitro, infected either with a control (top row) or the shKIF13A-expressing lentivirus (bottom row; red channel) for 7 d. Cultures were fixed and stained for surface GluA1 (nonpermeabilizing conditions, magenta) and total GluA1 (after permeabilization, cyan). Scale bars, 20 µm. Right: Quantifications of fluorescence intensity, measured separately in the somatic and dendritic compartments for both surface (magenta) and total (cyan) GluA1 channels. Plots show mean fluorescence intensity for the individual experiments and mean ± SEM for each pool of receptors in the different compartments. n = 32 for both control neurons and shKIF13A-expressing neurons from six independent experiments. (B) Left: Representative images for dissociated hippocampal neurons infected with lentivirus, as described for A. Immunocytochemistry was performed after baseline or 15 min of cLTP induction. Scale bars, 2.5 µm. Dashed lines outline neuron morphology (from mask of the mCherry channel). Arrowheads indicate individual spines used for the quantification of both surface (left) and total (right) GluA1 immunostaining. Plots on the right show cumulative probability distributions of fluorescence intensity from individual spines for each condition described above. Gray traces represent spines from neurons infected with a control lentivirus, and red traces show spines from neurons infected with a lentivirus expressing shKIF13A. Dashed lines represent baseline values, while continuous lines represent values after cLTP induction. P values compare cLTP versus baseline condition (black for control and red for shKIF13A), according to the Kolmogorov–Smirnov test (left graph, surface GluA1) or control versus shKIF13A in baseline condition (right graph, total GluA1). n = 1,229 spines (24 neurons) for baseline conditions and n = 1,183 spines (26 neurons) for cLTP conditions, from control neurons. n = 750 spines (25 neurons) for baseline conditions and n = 608 spines (24 neurons) for cLTP conditions from shKIF13A-expressing neurons, analyzed from three independent experiments. (C) Quantifications of fluorescence intensity in the adjacent dendritic shaft from each condition described in B. Plots show mean fluorescence intensity after cLTP normalized to mean baseline intensity for each condition for the individual experiments and mean ± SEM for each pool of receptors (surface GluA1, magenta; total GluA1, cyan). n = 26 control neurons and n = 24 shKIF13A-expressing neurons from three independent experiments. (D) Left: Representative images of spine morphology from mCherry channel of the experiments described in B. Scale bars, 2.5 µm. Right, cumulative probability distribution of fluorescence intensity at spines over the value at the adjacent dendrite from the different conditions described for B. Dashed lines represent baseline values, while continuous lines represent values after cLTP induction. P values compare cLTP versus baseline condition (black for control and red for shKIF13A) according to the Kolmogorov–Smirnov test.
Finally, we tested whether KIF13A may specifically mediate the local synaptic transport of AMPARs generated with LTP. To this end, we employed a pharmacological approach for LTP induction (chemical LTP [cLTP]; Otmakhov et al., 2004), which allows us to maximize the number of synapses undergoing plasticity. cLTP was induced on dissociated hippocampal neurons infected with a control lentivirus or shKIF13A lentivirus. After fixation, immunocytochemistry was performed as described above to label both the surface and total pools of GluA1-containing AMPARs. Images were acquired to specifically quantify surface and total GluA1 levels at dendritic spines (see representative images in Fig. 2 B, left). In control neurons, there was a significant increase in surface GluA1 expression in dendritic spines after 15 min of cLTP treatment (Fig. 2 B; “Surface GluA1,” rightward shift in continuous versus dashed black curves; P < 0.0001, Kolmogorov–Smirnov test). In contrast, this mobilization of AMPARs to the surface of the spine was no longer observable in the absence of KIF13A (Fig. 2 B; “Surface GluA1,” red curves). In fact, AMPAR accumulation in the spine surface was decreased after cLTP induction in the absence of KIF13A (leftward shift in continuous red curve with respect to dashed red curve; P < 0.0001, Kolmogorov–Smirnov test). These data strongly suggest that KIF13A participates in the export of AMPARs onto the spine surface during LTP.
In addition, we observed that KIF13A knockdown caused a decrease in the total pool of GluA1 subunits in dendritic spines under basal conditions (Fig. 2 B; “Total GluA1,” leftward shift in dashed red curve versus dashed gray curve; P < 0.0001, Kolmogorov–Smirnov test). This result suggests an impairment in the basal translocation of GluA1 into dendritic spines in the absence of KIF13A. Nevertheless, this was not reflected in a reduction of surface GluA1 under basal conditions (Fig. 2 B; “Surface GluA1,” compare dashed gray and red curves).
Interestingly, this GluA1 trafficking role of KIF13A during LTP appeared to be restricted to spines. Thus, when we quantified the amount of surface and total GluA1 in the dendritic shaft adjacent to the spines used for the previous quantification, no significant differences were observed between control and shKIF13A neurons in response to cLTP induction (Fig. 2 C, compare black and red bars, normalized to baseline distribution).
Finally, we tested whether KIF13A was involved in spine structural plasticity. To this end, we monitored spine size from the intensity of mCherry fluorescence (expressed in both control and shKIF13A lentiviral vectors) with respect to the fluorescence signal in the adjacent dendrite (representative images in Fig. 2 D, left panels). As shown in Fig. 2 D (right panel), LTP induction was accompanied by spine enlargement (right shift in continuous versus dashed curves), and this structural plasticity was similar in control and shKIF13A neurons (compare black and red curves).
Overall, these results strongly suggest that KIF13A specifically catalyzes the transport of AMPARs into the spine surface during LTP, without a detectable participation in basal dendritic trafficking or in structural plasticity.
The stalk domain of KIF13A is needed for LTP
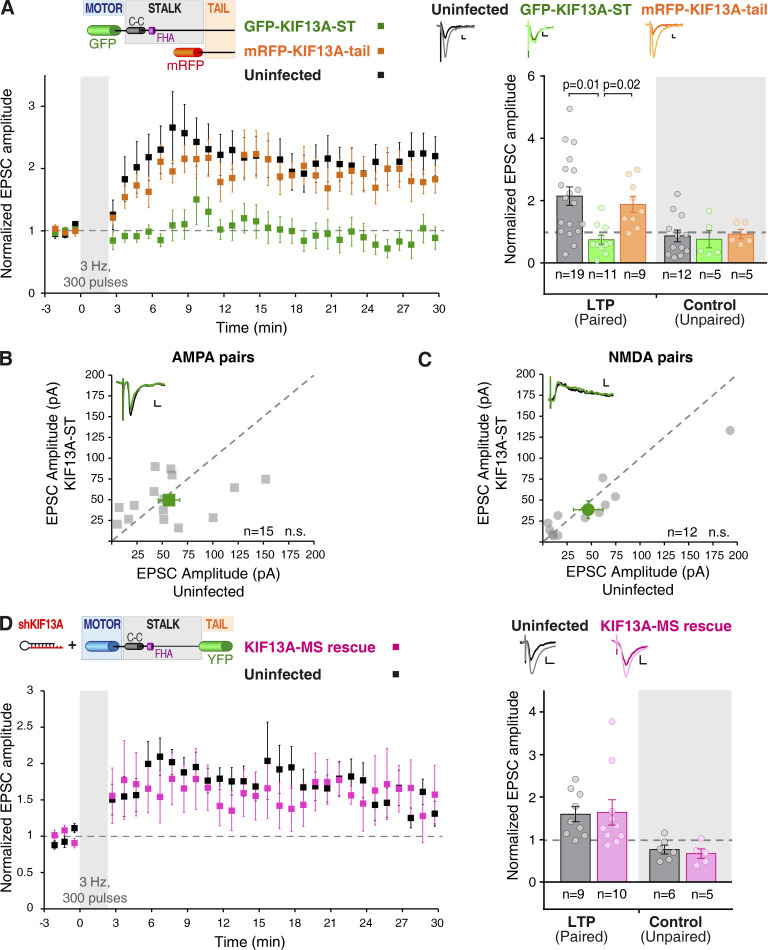
The structure of most kinesin motors (particularly N-kinesins) presents a cargo binding domain in its C-terminal domain (Hirokawa et al., 2009). Specifically for KIF13A, different cargo interactions have been described with the stalk or C-tail regions of the protein (Zhou et al., 2013; Nakagawa et al., 2000). To start dissecting potential molecular interactions of KIF13A for synaptic plasticity, we generated two recombinant proteins (schematic representation in Fig. 3 A, top left inset): GFP-KIF13A-ST (“stalk-tail” lacking the motor domain; see protein expression in Fig. S3, A and B) and mRFP-KIF13A-tail (lacking the motor and stalk domains; see expression in Fig. S3, A and C). We then tested the effect of these recombinant proteins in LTP of CA1 pyramidal neurons from organotypic slice cultures. The rationale of these experiments is that these truncated motor proteins will act as dominant negatives if they contain domains that compete with endogenous protein interactions required for LTP. As control, responses from uninfected neurons from the same slices were also recorded. LTP was induced using the pairing protocol described before.
Figure 3.
KIF13A domain analysis for LTP. (A) Time course (mean of the normalized excitatory postsynaptic currents [EPSCs] ± SEM per condition) of the LTP induction in organotypic hippocampal slices (7‒10 DIV) expressing GFP-KIF13A-ST (green) or mRFP-KIF13A-tail (orange) at 24 h after infection. Top left: Inset shows schematic representation of the GFP-KIF13A-ST and mRFP-KIF13A-tail recombinant proteins with known domains (coiled-coil [C-C] and FHA). Top right: Insets show representative traces for uninfected (black lines), GFP-KIF13A-ST (green lines), and mRFP-KIF13A-tail (orange lines) neurons, averaged from baseline (dark lines) or from the last 5 min of the recording (light lines). Scale bars: vertical, 20 pA; horizontal, 10 ms. Bars show mean ± SEM, together with individual values for each experiment, from the last 5 min of the recording and normalized to the baseline. n = 19, 11, or 9 cells for control, GFP-KIF13A-ST– or mRFP-KIF13A-tail–expressing neurons, respectively, for the LTP (paired) pathway. n = 12, 5, or 5 cells for control, GFP-KIF13A-ST–,or mRFP-KIF13A-tail–expressing neurons, respectively, for the control (unpaired) pathway. Uninfected and KIF13A-tail–expressing neurons are significantly potentiated (P = 0.002 and P = 0.01, respectively) with respect to baseline (Wilcoxon test). P values displayed in the panel represent KIF13A-ST significant difference from uninfected (P = 0.001) and KIF13A-tail (P = 0.002), respectively (Mann–Whitney test). (B and C) Basal synaptic responses mediated by AMPARs (B) and NMDARs (C) from organotypic hippocampal slices (7‒10 DIV) infected with a Sindbis virus expressing GFP-KIF13A-ST. Synaptic responses were recorded from pairs of uninfected and infected neighboring CA1 neurons. Gray circles represent values from each individual pair, while green symbols show the mean ± SEM of the measured currents. n = 15 cell pairs for AMPAR currents and n = 12 cell pairs for NMDAR currents; n.s., not significantly different (Wilcoxon test). Insets: Representative traces for uninfected (black lines) and GFP-KIF13A-ST (green lines) cells; scale bars: vertical, 10 pA; horizontal, 10 ms. (D) Time course (mean of the normalized EPSCs ± SEM per condition) of the LTP induction in organotypic hippocampal slices (7‒9 DIV) transfected with a biolistic approach to allow simultaneous expression of shKIF13A and KIF13A-MS-YFP (pink; KIF13A-MS-rescue) for 7–9 d. Top left: Inset shows schematic representation of the KIF13A-MS-YFP recombinant protein. Top right: Insets show representative traces for uninfected (black lines) and KIF13A-MS-rescue (pink lines) neurons, averaged from baseline (dark lines) or from the last 5 min of the recording (light lines). Scale bars: vertical, 10 pA; horizontal, 10 ms. Bars show mean ± SEM, together with individual values for each experiment, from the last 10 min of the recording and normalized to the baseline. n = 9 uninfected cells and n = 10 KIF13A-MS rescue neurons for the LTP (paired) pathway and n = 6 uninfected cells and n = 5 KIF13A-MS rescue neurons for the control (unpaired) pathway. Uninfected and KIF13A-MS-rescue neurons are significantly potentiated (P = 0.008 and P = 0.02, respectively) with respect to baseline (Wilcoxon test).
Figure S3.
Expression of the recombinant proteins GFP-KIF13A-ST, mRFP-KIF13A-tail, and KIF13A-MS-YFP. (A) Representative confocal images of GFP-KIF13A-ST (green), mRFP-KIF13A-tail (orange), and KIF13A-MS-YFP (pink) expressed in organotypic hippocampal slices. (B) Western blot analysis of GFP-KIF13A-ST expression with antibodies against GFP and KIF13A. Infection with a GFP virus is used as control. Actin is used as loading control. (C) Similar Western blot analysis with slices expressing mRFP-KIF13A-tail or tdTomato, as control. (D) Western blot analysis of dissociated hippocampal neurons showing the reduction on the endogenous KIF13A protein levels (shKIF13A and KIF13A-MS-rescue conditions; KIF13A antibody) together with the expression of the recombinant and shKIF13A-resistant KIF13A-MS-YFP protein (KIF13A-MS-rescue condition; GFP antibody). Recombinant KIF13A-MS-YFP is not recognized by the KIF13A antibody, as the epitope is located in the absent globular tail domain. Both control (lacking the shRNA sequence) and shKIF13A lentiviruses express mCherry. Actin is used as loading control.
Responses obtained after LTP induction both in control uninfected neurons and neurons expressing mRFP-KIF13A-tail were potentiated to a similar extent when compared with the basal currents (Fig. 3 A, black and orange traces). In contrast, synaptic potentiation was abolished in GFP-KIF13A-ST–expressing neurons (Fig. 3 A, green traces). As control, the unpaired pathways remained unaltered in all cases along the experiment (Fig. 3 A, unpaired control pathways). Additionally, neither basal synaptic transmission (AMPAR, Fig. 3 B; NMDAR, Fig. 3 C) nor passive membrane properties (Fig. S1) were affected by expressing GFP-KIF13A-ST. This dominant-negative approach strongly suggests that the stalk domain is responsible for the protein interactions of KIF13A required for LTP.
As a complementary approach to test the role of stalk and tail domains of KIF13A during LTP, we generated a truncated protein containing motor and stalk domains and lacking the tail domain (KIF13A-MS; top left inset in Fig. 3 D). This recombinant protein was fused to YFP and expressed together with shKIF13A to knock down endogenous KIF13A in CA1 pyramidal neurons from organotypic slice cultures. To note, KIF13A-MS is resistant to shRNA knockdown, because it was cloned from human KIF13A, which harbors two point mutations with respect to the endogenous rat sequence in the shRNA target (see protein expression in Fig. S3, A and D). Therefore, this approach allows us to knock down the endogenous KIF13A protein while replacing it with a recombinant motor protein lacking the tail domain. In contrast to what was observed with shKIF13A (Fig. 1 C), responses obtained after LTP induction in neurons coexpressing shKIF13A with KIF13A-MS (Fig. 3 D, pink traces) were potentiated to a similar extent as control, uninfected neurons (Fig. 3 D, black traces). In both cases, no potentiation was observed in the unpaired pathways (Fig. 3 D, unpaired control pathways).
Taken together, these data strongly argue that the stalk domain of KIF13A establishes protein interactions that are required for LTP, and interactions mediated by the KIF13A tail appear to be dispensable. Incidentally, this rescue experiment also serves to verify that the impairment of LTP produced by shKIF13A is not due to shRNA off-target effects.
Activity-dependent association of the motor transport machinery
Since KIF13A is required for the delivery of AMPARs to the spine surface after LTP induction but does not affect AMPAR transport under basal conditions, we reasoned that an activity-induced interaction between these proteins may occur. To test this hypothesis, we performed coimmunoprecipitation experiments using organotypic hippocampal slices. The recombinant GFP-KIF13A-ST protein was initially used as bait for these experiments. This protein preserves the domains required for LTP interactions, as it acted as a dominant negative for the endogenous KIF13A protein, but also presented the advantage of the GFP tag for a more efficient immunoprecipitation.
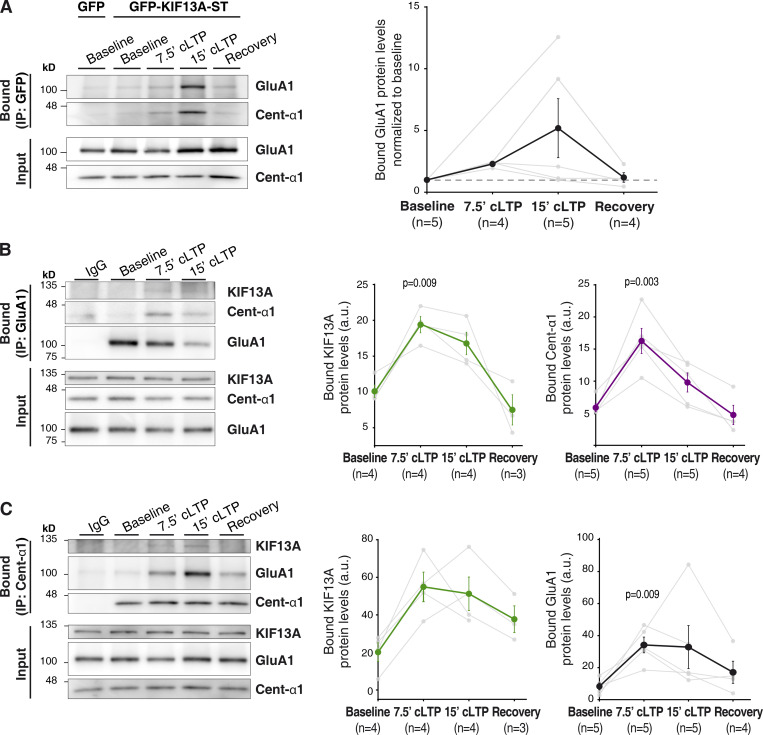
The CA1 region of organotypic hippocampal slices was infected with Sindbis viruses expressing either GFP-KIF13A-ST or GFP (as control for nonspecific interactions). At 24 h after infection, cLTP treatment was applied and slices were lysed under different conditions: at baseline, during (7.5 min) and after (15 min) cLTP induction, and after a recovery period of 15 min after induction. As shown in Fig. 4 A (quantification on the right panel), GFP-KIF13A-ST coimmunoprecipitated weakly GluA1 under basal conditions, but this interaction was strongly enhanced upon cLTP induction. Interestingly, the interaction appears to be transient, as it reverted back to basal levels after a recovery period of 15 min. Immunoprecipitation of the bait protein, GFP-KIF13A-ST, over the course of the cLTP experiment is shown in Fig. S4 A.
Figure 4.
Protein interactions between GluA1, KIF13A and centaurin-α1 upon LTP induction. (A) GFP-immunoprecipitation from protein extracts of 7‒10 DIV organotypic hippocampal slices expressing GFP-KIF13A-ST protein or GFP as control. Extracts were obtained from slices maintained at basal conditions (aCSF) or from slices undergoing cLTP induction for different times (7.5 min, 15 min) or from slices recovered for 15 min following cLTP (Recovery), as indicated. Coimmunoprecipitated proteins (“Bound”) and input fractions were analyzed by Western blot using antibodies for GluA1 and centaurin-α1. IP, immunoprecipitation. Left, representative Western blots. Right, quantification of the amount of coprecipitated GluA1 normalized to baseline (dashed line). Dark symbols represent mean ± SEM; gray symbols represent individual values from each experiment. n = 5, 4, 5, and 4 experiments for baseline, 7.5-min cLTP, 15-min cLTP, and recovery, respectively. (B) Left: Similar to A, with the immunoprecipitation of endogenous GluA1. Nonimmune rabbit IgGs were used as immunoprecipitation control. GluA1, centaurin-α1, and KIF13A were detected by Western blot. Right: Quantification of the coprecipitated amount of KIF13A (green circles) and centaurin-α1 (purple circles). P values (P = 0.009 and P = 0.003) represent significant difference from baseline (Friedman test). n = 4, 4, 4, and 3 experiments for baseline, 7.5-min cLTP, 15-min cLTP, and recovery, respectively, in the case of KIF13A coimmunoprecipitation. n = 5, 5, 5, and 4 experiments for baseline, 7.5-min cLTP, 15-min cLTP, and recovery, respectively, in the case of centaurin-α1 coimmunoprecipitation. (C) Left: Similar to B, with the immunoprecipitation of endogenous centaurin-α1 and Western blot detection of endogenous KIF13A, GluA1, and centaurin-α1. Nonimmune goat IgGs were used as immunoprecipitation control. Right: Quantifications of the coprecipitated KIF13A (green circles) and GluA1 (black circles). P value represents significant difference from baseline (P = 0.009, Friedman test). n = 4, 4, 4, and 3 experiments for baseline, 7.5-min cLTP, 15-min cLTP, and recovery, respectively, in the case of KIF13A coimmunoprecipitation. n = 5, 5, 5, and 4 experiments for baseline, 7.5-min cLTP, 15-min cLTP, and recovery, respectively, in the case of GluA1 coimmunoprecipitation.
Figure S4.
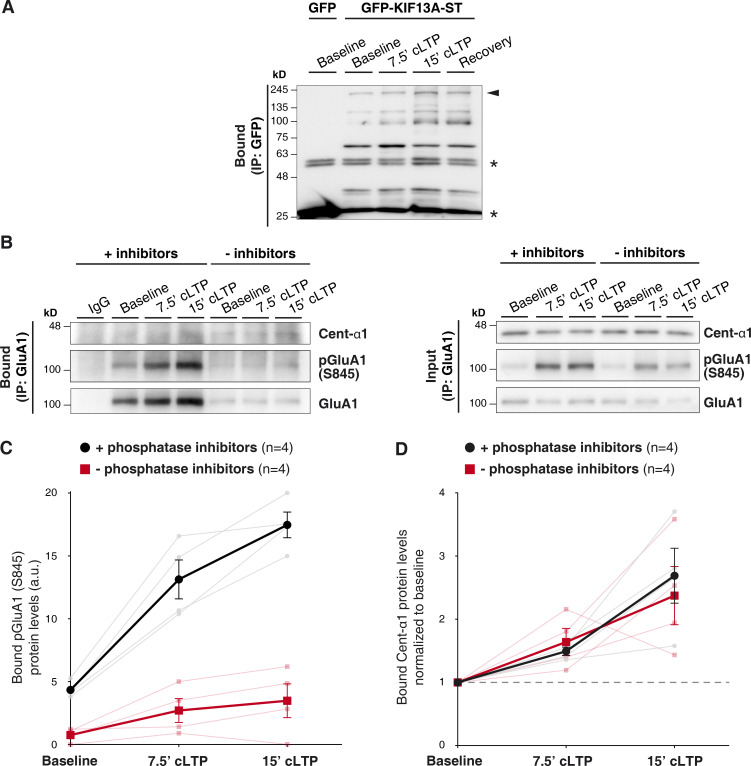
Immunoprecipitation of GFP-KIF13A-ST and effect of phosphatase inhibitors on centaurin-α1 coprecipitation. (A) GFP-immunoprecipitation from protein extracts of 7–10 DIV organotypic hippocampal slices expressing GFP-KIF13A-ST protein, or GFP as control, as described for Fig. 4 A of the main text. The immunoprecipitation of the recombinant protein was detected by Western blot with an anti-GFP antibody. GFP-KIF13A-ST appears as multiple bands, probably due to proteolytic degradation. Arrowhead indicates full-length recombinant protein; asterisks indicate light and heavy chains of the IgG used for immunoprecipitation. (B) Representative Western blots from experiments similar to Fig. 4 B of the main text but in the presence (+inhibitors) or absence (−inhibitors) of phosphatase inhibitors in the homogenization buffer. Nonimmune rabbit IgGs were used as immunoprecipitation control. GluA1, centaurin-α1, and pGluA1 (S845) were detected by Western blot. (C) Quantification of the amount of pGluA1 (S845) immunoprecipitated in both conditions (black circles, with phosphatase inhibitors; red squares, without phosphatase inhibitors). Darker lines represent the mean ± SEM, while lighter lines show individual values for each experiment. n = 4 experiments. (D) Quantification of the amount of centaurin-α1 protein coprecipitated in both conditions (black circles, with phosphatase inhibitors; red squares, without phosphatase inhibitors) and normalized to baseline coprecipitated protein. Darker lines represent the mean ± SEM, while lighter lines show individual values for each experiment. n = 4 experiments.
Additionally, the interaction of endogenous GluA1 and KIF13A was also analyzed (Fig. 4 B; quantification in the middle panel). When endogenous GluA1 was immunoprecipitated, the coprecipitation of endogenous KIF13A was more clearly detected during cLTP. Again, this interaction was transient, since it was no longer detectable after the recovery period following cLTP induction. Even though the coprecipitation of endogenous proteins is weak under these experimental conditions, altogether, these data indicated a transient interaction between KIF13A and AMPARs during LTP. These results further reinforce the notion of KIF13A being part of the molecular machinery required to deliver AMPARs to synapses during LTP.
It is worth noting that these results did not necessarily imply a direct interaction between KIF13A and GluA1. Indeed, the association between motor proteins and their cargo is typically mediated by adaptor molecules. Our data suggested that the stalk region of KIF13A is involved in LTP, as this domain acted as a dominant negative and was sufficient, together with the motor domain, to rescue LTP. Among the described domains present in the stalk, the forkhead-associated (FHA) domain has been previously linked to the transport of serotonin receptors (Zhou et al., 2013) and has been shown to interact with the adaptor protein centaurin-α1 (Tong et al., 2010; also known as ADAP1 and p42IP4). Additionally, using membrane fractionations from hippocampal extracts, we found that centaurin-α1 is enriched in extrasynaptic (Triton-soluble) membranes within synaptosomal compartments, in agreement with previous reports (Moore et al., 2007), and is also present in microsomal membranes, together with KIF13A and GluA1 (Fig. S5). Therefore, we tested a potential activity-dependent binding of centaurin-α1 to KIF13A and/or GluA1 using coimmunoprecipitation assays as described above. Indeed, endogenous centaurin-α1 coprecipitated with recombinant GFP-KIF13A-ST (Fig. 4 A, left panel) and endogenous GluA1 (Fig. 4 B; quantification in the rightmost panel) only after the induction of cLTP. Moreover, immunoprecipitation of endogenous centaurin-α1 coprecipitated endogenous GluA1 and KIF13A, most prominently under cLTP conditions (Fig. 4 C; quantifications on the right panels).
Figure S5.
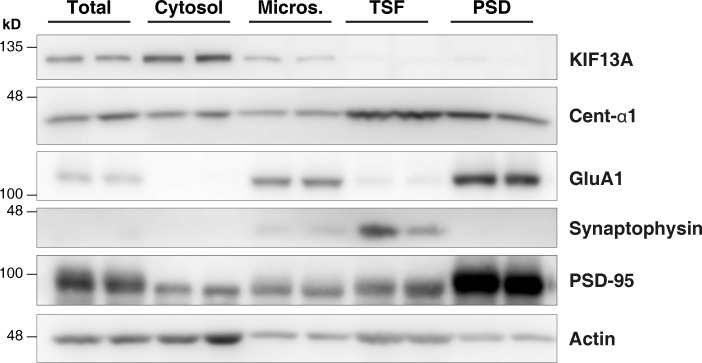
Subcellular fractionation of hippocampal slices. Hippocampal slices were homogenized (total fraction) and subsequently underwent different centrifugation steps to isolate different subcellular fractions: Micros., light membranes plus microsomes; PSD, postsynaptic density; TSF, Triton-soluble fraction and presynaptic plus extrasynaptic fraction. Blots show two different experiments performed in parallel. Synaptophysin and PSD-95 are used as pre and postsynaptic markers, respectively.
In all cases, these interactions were transient, as they returned to basal levels after a recovery period following cLTP treatment. Given this transient association, we tested whether the association between centaurin-α1 and GluA1 may be regulated by phosphorylation mechanisms. To this end, we performed similar coimmunoprecipitation assays in the presence or absence of phosphatase inhibitors in the homogenization buffer (Fig. S4 B). Importantly, phosphatases were very active in the immunoprecipitation assay, as the expected increase in pGluA1 (S845) following LTP induction (Fig. S4 C, black symbols) was strongly attenuated in the absence of phosphatase inhibitors (Fig. S4 C, red symbols). Nevertheless, the extent of protein phosphorylation/dephosphorylation did not alter the increase in centaurin-α1 coprecipitated with GluA1 following cLTP treatment (Fig. S4, B and D). Therefore, this transient association of the membrane transport machinery does not appear to be regulated by phosphorylation/dephosphorylation mechanisms.
The adaptor protein centaurin-α1 is required for LTP
To evaluate the requirement of centaurin-α1 for LTP, we employed a knockdown approach in organotypic hippocampal slices. A specific shRNA (shCentaurin-α1) was designed against the rat centaurin-α1 mRNA sequence, and lentiviral particles were produced for delivery into neurons (see Methods for the specific centaurin-α1 targeting sequence). The efficiency of the knockdown was confirmed in dissociated hippocampal neuronal cultures 7 d after infection (Fig. 5 A), normalized to uninfected neurons, and compared with neurons infected with a control (without shRNA) lentivirus. We also generated an shCentaurin-α1 virus coexpressing an shRNA-resistant recombinant centaurin-α1 protein for rescue experiments (Fig. 5, B and C). Simultaneous expression of shCentaurin-α1 together with GFP-centaurin-α1 allowed the knockdown of the endogenous protein while restoring protein levels with the exogenous centaurin-α1.
Figure 5.
LTP requires centaurin-α1. (A) Left: Representative Western blot of protein extracts from dissociated hippocampal neurons (15 DIV) infected for 7 d with shCentaurin-α1 lentivirus (purple) or with a control lentivirus (lacking the shRNA sequence, black). Right: Quantification of centaurin-α1 protein levels relative to uninfected condition. Bars show mean ± SEM, together with individual values from each experiment; n = 6 independent experiments; shCentaurin-α1 values were significantly different from uninfected and control (P = 0.03, Wilcoxon test). (B) Western blot similar to the one shown in A, including neuronal cultures infected with the rescue GFP-centaurin-α1 lentivirus. (C) GFP-channel (top) and immunohistochemistry (IHC) against centaurin-α1 (bottom) from organotypic hippocampal slices infected with the rescue GFP-centaurin-α1 lentivirus. (D) Left: Time course of the LTP induction in organotypic hippocampal slices (7‒11 DIV) infected for 7‒10 d with lentivirus for the expression of shCentaurin-α1 (purple symbols) or shCentaurin-α1 plus shRNA-resistant GFP-centaurin-α1 (blue symbols), compared with uninfected neurons (gray symbols). The amplitude of the synaptic response is normalized to a 3-min baseline before the LTP induction (300 pulses at 3 Hz, coupled to postsynaptic depolarization at 0 mV). Each point represents the mean of the normalized EPSCs ± SEM per condition. Right: Average response from the last 5 min of the recording and normalized to the baseline. Bars represent the mean ± SEM, together with individual values for each experiment. Left bars (LTP, paired) correspond to the stimulation pathway in which LTP protocol was induced; right (shaded) bars (control, unpaired) correspond to the pathway that was not stimulated during induction. n = 20, 10, or 12 cells for control, shCentaurin-α1, or centaurin-α1 rescue neurons, respectively, for the LTP (paired) pathway. n = 20, 6, or 6 cells for control, shCentaurin-α1, or centaurin-α1 rescue neurons, respectively, for the control (unpaired) pathway. Uninfected and rescue neurons are significantly potentiated with respect to baseline (P = 0.0003 and P = 0.005, respectively, Wilcoxon test). P values displayed in the figure indicate significant difference between shCentaurin-α1 with respect to uninfected (P = 0.001) and rescue (P = 0.009) conditions, respectively (Mann–Whitney test). (E and F) Basal synaptic responses mediated by AMPARs (E) and NMDARs (F) recorded simultaneously from adjacent neurons, uninfected and infected with either shCentaurin-α1 (purple) or GFP-centaurin-α1 rescue (blue). Gray symbols represent absolute values of the responses from each individual pair. Colored symbols show the mean ± SEM of the measured currents. n = 16 cell pairs for shCentaurin-α1 and n = 15 for centaurin-α1 rescue, in the case of AMPAR currents (E). n = 12 cell pairs for shCentaurin-α1 and n = 15 for centaurin-α1 rescue in the case of NMDAR currents (F). n.s., not significantly different (Wilcoxon test). Insets: Representative traces for uninfected (black lines), shCentaurin-α1 (purple lines), and centaurin-α1 rescue (blue lines) neurons; scale bars: vertical, 10 pA; horizontal, 10 ms.
Whole-cell patch-clamp electrophysiology experiments were performed on organotypic hippocampal slices infected with the lentiviruses described before. LTP induction produced robust synaptic potentiation in uninfected neurons (Fig. 5 D, black traces). In contrast, centaurin-α1–depleted neurons failed to potentiate after LTP induction (Fig. 5 D, purple traces). Importantly, LTP expression was rescued when recombinant GFP-centaurin-α1 was reintroduced using the lentiviral vector (Fig. 5 D, blue traces). The pathways not paired during induction (control) showed no alteration as compared with the corresponding baseline responses in all cases. Finally, we also observed that centaurin-α1 depletion (shCentaurin-α1) or rescue with the recombinant GFP-centaurin-α1 did not alter AMPAR- (Fig. 5 E) or NMDAR-mediated (Fig. 5 F) synaptic responses. These results indicate that the impairment in NMDAR-mediated LTP in the absence of centaurin-α1 is not due to altered function of NMDARs or AMPARs.
Altogether, these data indicate that centaurin-α1 is an active player required for the KIF13A-dependent trafficking of AMPARs during LTP.
KIF13A is required for FIP2 endosomal remodeling during cLTP
One of the cell-biological features of LTP is the redistribution of endosomal compartments within the spine and dendrites (Park et al., 2006) for the synaptic delivery of AMPARs (Park et al., 2004). This endosomal mobilization has been traditionally linked to the actin cytoskeleton (Wang et al., 2008). However, KIF13A has been shown to mediate the formation of tubular recycling endosomes from sorting endosomes in cell cultures (Delevoye et al., 2014). Therefore, we decided to test whether this activity of KIF13A is recruited during LTP induction in hippocampal neurons.
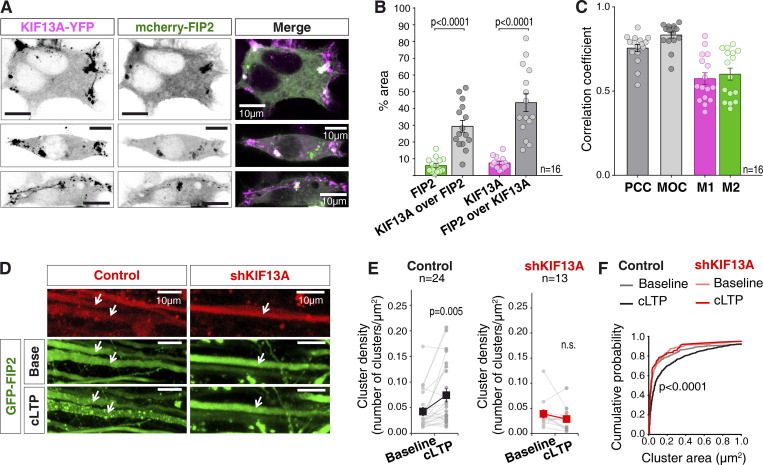
To visualize endosomal rearrangements, we decided to monitor the Rab11 adaptor FIP2, as we have recently shown that it undergoes a fast and reversible redistribution in response to LTP induction (Royo et al., 2019). As a first step, we analyzed the potential association between KIF13A and FIP2. To this end, we cotransfected human embryonic kidney (HEK) 293T cells with mCherry-FIP2 and KIF13A-YFP. After 24 h of expression, the relative intracellular distribution of these two proteins was evaluated by confocal fluorescence microscopy (Fig. 6 A). Both KIF13A and FIP2 presented a diffuse expression pattern with a fraction of the protein localized in clusters. The analysis of the clustered distribution revealed a degree of colocalization between FIP2 and KIF13A significantly higher than that expected from a random distribution throughout the cell (as calculated from the relative areas of the cell covered by clusters, green and magenta bars; Fig. 6 B). Additionally, colocalization within clusters was further assessed through intensity correlation with Pearson correlation coefficient (PCC; dark gray) and Manders overlap coefficient (MOC; light gray), as well as signal cooccurrence calculated as individual Manders coefficients (M1 for KIF13A and M2 for FIP2; Fig. 6 C). All these parameters indicated a partial but significant colocalization between recombinant KIF13A and FIP2 (to note, the colocalization of the endogenous proteins could not be evaluated because of the lack of specificity of the available antibodies for immunocytochemical techniques, as assessed with our shRNA controls; not shown).
Figure 6.
Role of KIF13A in LTP-induced endosomal redistribution. (A) Representative images of KIF13A-YFP (magenta) and mCherry-FIP2 (green) fluorescence in transfected HEK-293T cells 24 h after transfection. Scale bars, 10 µm. (B) Quantification of colocalization between KIF13A-YFP and mCherry-FIP2 in clustered areas compared with the expected colocalization for a random distribution of the proteins (percentage of mCherry-FIP2 and KIF13A-YFP clustered area over whole-cell area, respectively). Plots represent percentage of the area for each individual cell and show mean ± SEM for each analysis. n = 16 cells from two independent experiments; P < 0.0001, Wilcoxon test. (C) Quantification of colocalization between KIF13A-YFP and mCherry-FIP2 clusters from the experiments described in A using PCC an MOC. Individual Manders coefficients (M1 and M2, KIF13A and FIP2, respectively) are also shown. Plots represent correlation coefficient for each individual cell and show mean ± SEM for each analysis. n = 16 cells from two independent experiments. (D) Representative images of a cLTP experiment showing dendritic branches from an organotypic hippocampal slice infected with GFP-FIP2 (green) together with a control lentivirus (control; left panels) or a lentivirus expressing shKIF13A (right panels), both expressing mCherry (red). Arrows indicate coinfected dendrites. Scale bars, 10 µm. (E) Quantifications of cluster density per dendrite for each condition before and after cLTP induction. Analysis of cluster density was performed blind with respect to the lentivirus identity. Gray lines represent each analyzed dendrite, and black and red symbols show the mean ± SEM for each condition. n = 24 (control) and n = 13 (shKIF13A) dendrites from 10 and 6 independent experiments, respectively; P = 0.005, Wilcoxon test. (F) Cumulative probability distribution of cluster area of GFP-FIP2 for each condition described in D and E. Lighter lines represent baseline conditions. Darker lines represent conditions after cLTP induction. Gray lines represent data from slices infected with the control lentivirus (lacking shRNA sequence). Red lines represent data from slices expressing shKIF13A. P value < 0.0001 (Kolmogorov–Smirnov test) compares baseline and 15-min LTP in control conditions. Cluster size did not change in dendrites lacking KIF13A (shKIF13A; P = 0.92, Kolmogorov–Smirnov test).
After determining the colocalization between FIP2 and KIF13A in heterologous cells, we set out to study the potential role of KIF13A in the recruitment of FIP2 to endosomal structures in hippocampal neurons during LTP as a reporter for LTP-induced endosomal remodeling. The control lentivirus or shKIF13A was expressed in the CA1 region of organotypic hippocampal slice cultures for 7 d. Subsequently, GFP-FIP2 was expressed for 24 h in the same CA1 region using Sindbis virus (see representative images from apical dendrites in Fig. 6 D). Organotypic hippocampal slices were maintained in continuously perfused artificial cerebrospinal fluid (aCSF) and monitored under a confocal microscope. Under basal conditions (aCSF perfusion), GFP-FIP2 presented a mostly diffuse expression in dendrites, with a small fraction of clusters (Fig. 6, D and E), as expected (Royo et al., 2019). This basal expression pattern was similar in control and shKIF13A-infected neurons (Fig. 6, D and E; “Baseline”). Perfusion solution was then changed for cLTP induction, and images were acquired after the 15 min of the cLTP protocol (“cLTP”). As shown in Fig. 6 D, bottom left panel (quantification in Fig. 6 E; “Control”), the number of clusters of GFP-FIP2 along the dendritic shaft approximately doubled after the induction of cLTP (see also Video 1). This was also accompanied by a significant increase in cluster size (Fig. 6 F, rightward shift in the dark-gray curve). In contrast, in the absence of KIF13A (shKIF13A), cLTP failed to increase cluster density (Fig. 6 D, bottom right panel; quantification in Fig. 6 E, “shKIF13A”; Video 2) or produce cluster enlargement (Fig. 6 F, red curves). To note, analysis was performed blind regarding the identity of the lentivirus (KIF13A knockdown or control).
Video 1.
Representative time-lapse video of a cLTP experiment showing dendritic branches from an organotypic hippocampal slice infected with GFP-FIP2. Arrows highlight coinfected dendrites with a control lentivirus. Images were acquired every 5 min, and the green circle indicates the onset of cLTP induction. Related to Fig. 6 D.
Video 2.
Representative time-lapse video of a cLTP experiment showing dendritic branches from an organotypic hippocampal slice infected with GFP-FIP2, but lacking KIF13A. In this case, arrows highlight coinfected dendrites with a lentivirus expressing shKIF13A. Images were acquired every 5 min, and the green circle indicates the onset of cLTP induction. Related to Fig. 6 D and similar to Video 1.
These results indicate that KIF13A is required for the redistribution of FIP2 into endosomal structures in response to LTP induction. Altogether, these data suggest that LTP expression relies on the dendritic remodeling of endosomal compartments powered by KIF13A.
Discussion
In this work, we have demonstrated that a specific microtubule-dependent motor protein, namely KIF13A, catalyzes the remodeling of dendritic endosomal compartments for the synaptic delivery of AMPARs during LTP. Specifically, we have shown that genetic depletion of KIF13A (by shRNA) impairs the reorganization of FIP2-associated endosomes that occurs during LTP induction. This results in a failure to deliver AMPARs to the spine surface, and consequently prevents synaptic potentiation.
The microtubule cytoskeleton has been previously shown to respond to neuronal activity (Jaworski et al., 2009; Hu et al., 2008) and organize the signaling machinery that assists synaptic plasticity (Morikawa et al., 2018). However, this is the first time that microtubule-dependent transport in the dendritic shaft was linked to the local mobilization of AMPARs for their synaptic insertion during plasticity. In fact, most previous studies have focused on the actin cytoskeleton for the activity-dependent transport of AMPARs in spines during LTP (Wang et al., 2008; Correia et al., 2008). Therefore, our results are most compatible with a handover transport mechanism, in which AMPARs are recruited together with the rearrangement of an endosomal sorting station at the dendritic shaft (visualized by FIP2 redistribution in this work) in a process powered by KIF13A on the microtubule cytoskeleton. Then, these mobilized endosomes would be transferred to the actin cytoskeleton (via myosin V motor proteins) for their final delivery to the spine surface. KIF13A would be well suited to carry over this relay transport, as it preferentially associates with early endosomes (Bentley et al., 2015) and participates in the biogenesis and transport of recycling endosomes in a Rab11-dependent manner (Delevoye et al., 2014). Importantly, this model does not restrict where the switch between cytoskeletal elements takes place, and therefore, it is also compatible with a transient entry of microtubules into the spine (Schätzle et al., 2018; Merriam et al., 2013) for cargo transfer. Nevertheless, it is important to highlight that the rate of microtubule entry into spines is relatively small, and nocodazole treatment has been shown to have only a mild effect on endosomal dynamics in spines (Esteves da Silva et al., 2015). Therefore, considering the effects we have observed of KIF13A on the rearrangement of dendritic endosomal structures, a spine-specific transport of AMPARs catalyzed by KIF13A would not be necessary to account for its role in LTP.
Admittedly, the specific dynamic interplay between KIF13A and endosomal compartments containing AMPARs and FIP2 still remains to be characterized, particularly when considering that AMPARs and FIP2 undergo separate trafficking during LTP (Royo et al., 2019).
Another interesting conclusion from this study is that the synaptic function of KIF13A is restricted to the activity-dependent transport of AMPARs during LTP. The basal dendritic trafficking and surface expression of AMPARs did not appear to rely on KIF13A, in agreement with our observations of a mainly unaltered basal synaptic function in the absence of this motor protein. There is a large number of kinesin motors in the mammalian genome (Hirokawa et al., 2009), and some of them have been linked with glutamate receptor transport (Hoerndli et al., 2013b; Heisler et al., 2014; Shin et al., 2003; Maas et al., 2009; Setou et al., 2002; Hoogenraad et al., 2005). Therefore, it is conceivable that one (or several) of these proteins supports the continuous supply of AMPARs to maintain synaptic function under basal conditions. This selectivity of the role of KIF13A during LTP is also reflected in the observation that structural plasticity (spine growth) was not affected by KIF13A depletion. This result would suggest that KIF13A is not required for delivering the bulk of endosomal membrane needed for spine expansion (Park et al., 2006). In fact, there is already evidence that AMPARs delivered during synaptic plasticity are contained in a specialized pool of endosomes (Zheng et al., 2015; Jurado et al., 2013). Another point of contention is the source of receptors for synaptic insertion during LTP. Convincing evidence has been provided for activity-dependent insertion from an intracellular reserve pool (Kennedy et al., 2010; Ahmad et al., 2012; Jurado et al., 2013; Park et al., 2004), as well as for lateral diffusion from the extrasynaptic surface (Makino and Malinow, 2009; Kneussel and Hausrat, 2016; Tang et al., 2016; Penn et al., 2017). Some of these conflicting results may be explained by differences in the protocols to induce synaptic plasticity (a faster potentiation induced by high-frequency or theta-burst stimulation versus a slower pairing protocol at 3 Hz). Nevertheless, and irrespective of the delivery pathway, our results would argue that KIF13A-mediated transport is required for the activity-dependent transport of AMPARs that results in their delivery to the synaptic surface.
This study has also allowed us to define some of the molecular machinery that assists AMPAR transport during LTP. Specifically, we have found that KIF13A, AMPARs, and the vesicular protein centaurin-α1 associate during LTP induction and that both KIF13A and centaurin-α1 are required for LTP expression. Nevertheless, we should mention that LTP has been recently reported to be preserved in a centaurin-α1 knockout (Szatmari et al., 2021). We are not sure for the reason of this discrepancy, but it may be related to compensatory mechanisms or secondary changes in the knockout animal, which may be absent with the semiacute manipulation of the shRNA knockdown.
The participation of centaurin-α1 in a KIF13A-AMPAR transport mechanism is particularly fitting. On the one hand, while KIF13A is ubiquitously expressed (Nakagawa et al., 2000), centaurin-α1 is a brain-specific protein (Stricker and Reiser, 2014). Therefore, it provides neuronal specificity for this function of KIF13A. On the other hand, centaurin-α1 contains two pleckstrin homology domains that bind phosphatidylinositol-(3,4,5)-trisphosphate (Stricker and Reiser, 2014; Tong et al., 2010). Phosphatidylinositol-(3,4,5)-trisphosphate is a key signaling molecule transiently up-regulated during LTP induction (Arendt et al., 2014) and required for LTP expression (Arendt et al., 2010). Consequently, it is tempting to speculate that centaurin-α1 may act as a link between phosphoinositide 3-kinase activation and endosomal trafficking during synaptic potentiation. In fact, we hypothesize that centaurin-α1 may provide a mechanism for the recruitment of specific endosomal compartments “tagged” by phosphoinositide 3-kinase activity. In summary, this work provides mechanistic insight to understand how cytoskeletal-dependent transport and intracellular membrane sorting are coupled for the regulation of synaptic strength during plasticity.
Materials and methods
Animals and ethics statement
All biosafety procedures and animal experimental protocols were approved by the Ethical Committee from the Consejo Superior de Investigaciones Científicas in strict accordance with Spanish (RD 53/2013, 32/2007) and European Union guidelines set out in the European Community Council Directives (2010/63/EU, 86/609/EEC). Wistar rats of both genders were used for all experiments. Animals were housed in standard laboratory cages with ad libitum access to food and water under a 12:12-h dark/light cycle in temperature-controlled rooms.
Cell culture
Organotypic hippocampal slices
Cultures were prepared as previously described (Gähwiler et al., 1997; Fuller and Dailey, 2007). In brief, hippocampal slices were dissected from young rats (postnatal days 5–7) in cold dissection medium (10 mM D-glucose, 4 mM KCl, 20 mM NaHCO3, 233.7 mM sucrose, 5 mM MgCl2, and 1 mM CaCl2) gassed with carbogen (5% CO2/95% O2). 400-µm-thick slices were prepared in sterility using a tissue slicer (Stoelting), and individual slices were separated and placed in culture on porous membranes (Merk Millipore) over culture medium (0.8% [wt/vol] MEM powder, 20% [vol/vol] horse serum, 1 mM L-glutamine, 1 mM CaCl2, 2 mM MgSO4, 1 mg/liter insulin, 0.0012% [vol/vol] ascorbic acid, 30 mM Hepes, 13 mM D-glucose, and 5.2 mM NaHCO3). Slices were maintained in vitro at 35.5°C and 5% CO2 for 6‒11 d until use, replacing with fresh medium every 2‒3 d.
Primary dissociated hippocampal neurons
Dissociated cultures were prepared as previously described (Banker and Cowan, 1977; Dotti et al., 1988; Goslin and Banker, 1989). Hippocampi were dissected from rat embryos at embryonic day 19 in Hank’s balanced salt solution (Gibco) supplemented with 10 mM Hepes, pH 7.4 (Gibco), 100 IU/ml penicillin, and 100 µg/ml streptomycin. Cells were disaggregated with 0.25% trypsin (Gibco) and mechanically by repeated passage through a constricted Pasteur pipette. Neurons were then plated on glass coverslips coated with 1 mg/ml poly-L-lysine (Sigma-Aldrich) for imaging experiments or on 0.1 mg/ml poly-L-lysine–coated culture plates for biochemical analysis. Cells were allowed to recover and attach to the substrate in MEM containing 1.5% D-glucose and 10% FBS (Hyclone) for 3‒4 h at 37°C and 5% CO2. Subsequently, the media was replaced with Neurobasal medium (Gibco) supplemented with 1× B27 (Gibco) and 2 mM glutamine. Cultures were maintained at 37°C and 5% CO2 and used after 2 wk in vitro.
DNA constructs for recombinant protein expression and biochemical analysis
KIF13A-YFP plasmid, containing full-length human KIF13A sequence under the Gateway system (Invitrogen), was generated by Dr. Cédric Delevoye, in the laboratory of Dr. Graça Raposo (Institut Curie, Paris, France). pSinRrep5-GluA1-GFP construct was generously provided by Dr. Roberto Malinow (University of California, San Diego, San Diego, CA), and pSinRep5-GFP-FIP2 (containing the rat FIP2 cDNA sequence) was previously generated in our laboratory (Royo et al., 2019). This cDNA sequence was also used to generate the mCherry-FIP2 fusion protein under the control of the cytomegalovirus promoter. The KIF13A-YFP sequence was used to generate the corresponding plasmids for the expression of GFP-KIF13A-ST (residues 351–1,770), mRFP-KIF13A-tail (residues 1,307–1,770), and KIF13A-MS-YFP (residues 1–957) fusion proteins. Subsequently, GFP-KIF13A-ST and mRFP-KIF13A-tail were cloned into the pSinRep5 vector (Invitrogen) for Sindbis virus expression and KIF13A-MS-YFP into the lentiviral vector for shKIF13A coexpression (see below).
Protein expression with Sindbis virus was performed for 24 h in organotypic slice cultures. For the lentiviral expression of specific shRNAs, two complementary oligonucleotides (Sigma-Aldrich) with the specifically designed target sequences, hairpin loops, and restriction sites were annealed and cloned into the KH1-LV-mCherry vector, which was a gift from Dr. María S. Soengas (Centro Nacional de Investigaciones Oncológicas, Madrid, Spain). As negative control, a lentivirus with no hairpin expression was used. In all cases, mCherry is also expressed. The following specific shRNA sequences were used: shKIF13A (designed using GeneLink and sequence NM_001107462 from GenBank), 5′-AAATATGCCGGTCAAGAAG-3′; shKIF13B (Yoshimura et al., 2010), 5′-CCACTGATGGAAGAGTGCATATTGT-3′; shCentaurin-α1 (designed using siRNA Wizard v3.1 and sequence NM_133567 from GenBank), 5′-GCCCAAGCTCTCTAGGAACTA-3′. For the generation of the centaurin-α1 rescue plasmid, the human sequence of centaurin-α1 was amplified by PCR from the Addgene plasmid CENTA1 (3FM8, plasmid #25334). This sequence was cloned into the pEGFP-C1 vector (Clontech) and recloned into the KH1-LV-shCentaurin-α1 vector, substituting the mCherry sequence. Lentiviral infections in organotypic slice cultures were performed for 7‒10 d. For neurite polarization assays, dissociated primary hippocampal neurons were infected 18 h after plating with the corresponding lentiviral vectors, and neurite extension was assessed 3 d later.
KIF13A-MS coexpression with shKIF13A was performed by biolistic transfection (gene-gun) of the corresponding lentiviral vector in organotypic slice cultures. Expression was allowed for 7–9 d before performing electrophysiological recordings on transfected CA1 neurons and adjacent nontransfected ones.
To obtain protein extracts for Western blot analyses, samples from the indicated sources were homogenized in cold lysis buffer containing 10 mM Hepes, pH 7.4, 150 mM NaCl, 10 mM EDTA, 1% Triton X-100, and protease and phosphatase inhibitor cocktails (Complete Mini EDTA-free and PhosSTOP; Roche). The following primary antibodies were used: α-tubulin (mouse; T6199; Sigma-Aldrich), actin (mouse; MAB1501R; Merck Millipore), centaurin-α1 (goat; ab27476; Abcam), GFP (mouse; 11814460001; Roche), GluA1 (rabbit; ab31232; Abcam), KIF13A (goat; sc-16789; Santa Cruz), mCherry (rabbit; GTX59788; GeneTex), pGluA1-S845 (rabbit; OPA1-04118; Thermo Fisher Scientific), PSD-95 (mouse; MA1-046; Thermo Fisher Scientific), and synaptophysin (mouse; Sigma-Aldrich; S5768).
For immunoprecipitation, samples were homogenized in a buffer containing 10 mM Hepes, pH 7.4, 150 mM NaCl, 1% Triton-X100, 1 mM EGTA, 5 mM MgCl2, and protease and phosphatase inhibitor cocktails (Complete Mini EDTA-free and PhosSTOP; Roche) and prepared at 0.5 µg/µl (400‒500 µl). Primary antibody was immobilized using 50 µl Protein G Sepharose beads (GE Healthcare) per condition, equilibrated at 50% in homogenization buffer. Antibodies for immunoprecipitation were anti-GFP (4 µg/ml), anti-GluA1 (10 µg/ml; rabbit; AB1504; Merck Millipore), or anti–centaurin-α1 (5 µg/ml). For infected slices, the specificity control used was slices infected with a Sindbis virus expressing just the fluorescent protein (GFP) to which the recombinant protein was tagged to in the rest of the conditions. Consequently, the same anti-GFP antibody was used also in the control condition. For uninfected slices, the same amount of nonspecific serum was used as a specificity control in the corresponding sample: goat serum (catalog no. G9023; Sigma-Aldrich) or rabbit IgG (catalog no. I5006; Sigma-Aldrich).
Subcellular fractionation
Subcellular fractions were prepared as previously described (Gardoni et al., 2009). All centrifugation steps were performed at 4°C, and buffers were maintained on ice. All buffers contained protease and phosphatase inhibitor cocktails (Complete Mini EDTA-free and PhosSTOP; Roche). Briefly, hippocampal slices were homogenized on a glass-Teflon potter (25 strokes) with buffer containing 0.32 M sucrose, 1 mM Hepes, 1 mM MgCl2, 1 mM NaHCO3, and 1 mM EDTA. Nuclei and myelin were removed by 10-min centrifugation at 10,000 g. Subsequently, the supernatant was centrifuged at 13,000 g for 15 min to yield the crude synaptosomal fraction (pellet) and the cytosol and light membrane/microsome fraction (supernatant). After washes with the homogenization buffer, the pellet was resuspended in a buffer containing 75 mM KCl and 1% Triton X-100. Both the resuspended pellet and supernatant were centrifuged for 1.5 h at 100,000 g. From the centrifuged supernatant, we obtained the cytosolic fraction and a pellet corresponding to the microsomal fraction. The centrifuged crude synaptosomal fraction yielded a supernatant corresponding to the Triton-soluble fraction (presynaptic and extrasynaptic fraction) and a pellet enriched in the postsynaptic density. Both pellets were washed extensively and resuspended in 0.1% NP-40 and 20 mM Hepes for western analysis.
Quantitative RT-PCR
Total RNA from dissociated hippocampal neuronal cultures was extracted with TRIzol reagent (Ambion/Life Technologies) following the manufacturer’s procedures. Quantification of phenol/chloroform–extracted and purified RNA was performed by absorbance at 260 nm using a NanoDrop ND-100 (Thermo Fisher Scientific). Equal amount (500 ng) of total RNA for all samples was retrotranscribed to first-strand cDNA using the RevertAid H Minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific) following the indicated protocol with random hexamer primers. For qPCR analysis, 0.2 µl of the synthesized cDNA was used per condition (10 µl). GoTaq quantitative PCR Master Mix (Promega) was used following the manufacturer’s specifications. The following primers were purchased from Sigma-Aldrich at 0.5 µM final concentration and used for rat transcript detection: KIF13A forward (5′-TGCACGACTCTCTTCACCTG-3′) and reverse (5′-CTGCGTGAAACTCTGCTTGT-3′); KIF13B forward (5′-ATGGCTACAATGCCTGTATCTTT-3′) and reverse (5′-TCGGCTGTTCCCATCATAGT-3′); and two housekeeping genes as endogenous controls: Gapdh forward (5′-ATGACTCTACCCACGGCAAG-3′) and reverse (5′-GATCTCGCTCCTGGAAGATG-3′) and GusB forward (5′-ATACTGGAGGATTGCCAACG-3′) and reverse (5′-ACGGTCTGCTTCCCATACAC-3′). Data were acquired with a 7900HT real-time PCR system (Applied Biosystems) and analyzed with SDS software, version 2.4. The relative quantifications were performed using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Electrophysiology
Whole-cell voltage-clamp recordings were obtained from CA1 pyramidal neurons in organotypic hippocampal slice cultures, evoking synaptic responses via stimulation of Schaffer collateral fibers from CA3 pyramidal neurons. The CA3 region was removed from the slices to prevent seizures. Only CA1 cells were infected with viral vectors, thus ensuring that recombinant proteins or shRNAs were expressed exclusively in the postsynaptic cell. Organotypic hippocampal slices were maintained in an immersion recording chamber that was continuously perfused with aCSF (119 mM NaCl, 2.5 mM KCl, 4 mM CaCl2, 4 mM MgCl2, 26 mM NaHCO3, 1 mM NaH2PO4, 11 mM glucose, pH 7.4 and osmolarity adjusted to 290 ± 5 mOsm), constantly gassed with 5% CO2/95% O2 (carbogen), and maintained at 29°C. For all recordings, aCSF was supplemented with 4 µM 2-chloroadenosine and 0.1 mM picrotoxin. Patch recording pipettes (3–8 MΩ) were filled with a solution containing 115 mM CsMeSO3, 20 mM CsCl, 10 mM Hepes, 2.5 mM MgCl2, 4 mM Na2-ATP, 0.4 mM Na-GTP, 10 mM sodium phosphocreatine, 0.6 mM EGTA, and 10 mM lidocaine N-ethyl bromide, pH 7.25, osmolarity 290 mOsm. Stimulating bipolar electrodes were placed over Schaffer collateral fibers between 300 and 500 µm from the recorded cells with visual guidance using fluorescence and transmitted light illumination under a microscope equipped with a 60× water immersion objective. Synaptic responses were evoked using single-voltage pulses (200 µs, up to 25 V), and electrophysiological recordings and data acquisition were performed with Multiclamp 700A/B amplifiers, a digitizer and pClamp software (Molecular Devices). Data analysis was performed with custom-made Excel (Microsoft) macros (Brachet et al., 2015).
For basal synaptic transmission, simultaneous whole-cell recordings were obtained from nearby pairs of infected and uninfected (control) neurons. Synaptic AMPAR-mediated responses were measured at −60 mV and NMDAR-mediated responses at +40 mV at a latency when AMPAR-mediated responses had fully decayed (65 ms after stimulation). Responses were averaged over 60‒80 trials. NMDAR-dependent LTP was induced after stable baseline synaptic responses with a pairing protocol by coupling a 3-Hz presynaptic stimulation (300 pulses) of the Shaffer collaterals with a 0-mV postsynaptic depolarization (Chen et al., 1999; Hayashi et al., 2000). Acquisition of basal synaptic responses was performed during a relatively short time (1.5‒2 min) to prevent washout of LTP in whole-cell recordings. Slices were discarded after one induction of the LTP protocol.
cLTP treatments
Experiments in hippocampal organotypic slices were performed following the previously described protocol (Otmakhov et al., 2004). Briefly, hippocampal slices were transferred to a submersion-type holding chamber containing aCSF gassed with 5% CO2/95% O2 at 29°C. For biochemical analysis, slices were equilibrated for 5 min before each experiment while for microscopy experiments images were acquired for a longer baseline period (10‒15 min). For cLTP induction, slices were transferred (or solution exchanged) to a modified aCSF lacking Mg2+ and containing 0.1 µM rolipram, 50 µM forskolin, and 100 µM picrotoxin for 15 min (or less as indicated in each experiment). After the induction protocol, for some experiments, slices were returned back to regular aCSF for a recovery period of 15 min. For biochemical analysis, slices were immediately homogenized and processed after each indicated point of the treatment. In the case of dissociated hippocampal cultures, neurons were equilibrated for 30 min at 37°C and 5% CO2 in a modified aCSF containing Hepes buffer (125 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 33 mM glucose, and 25 mM Hepes buffer, pH 7.4‒7.5, with osmolarity adjusted to 320 mOsm). cLTP was induced by placing the neurons in Hepes-based Mg2+-free aCSF containing 0.1 µM rolipram, 50 µM forskolin, and 100 µM picrotoxin for 15 min (Molnár, 2011; Fernández-Monreal et al., 2016). Cells were fixed immediately after cLTP induction or after baseline as indicated in each experiment.
Microscopy experiments
For live imaging experiments, hippocampal slices were placed on a chamber continuously perfused with aCSF gassed with 5% CO2/95% O2 at 29°C. Z-stack confocal fluorescence images were acquired with a confocal microscope (LSM710, Zeiss) using a 63×, NA 1.2 water C-Apochromat objective and 488-nm and 561-nm lasers. For cLTP experiments, after acquiring images every 5 min during a baseline period of 10‒15 min, the perfusion solution was switched to cLTP aCSF, as described above, and images were acquired after the 15 min of induction.
For immunofluorescence experiments, samples were fixed with 4% paraformaldehyde and 4% sucrose in PBS for 10 min (for dissociated cultures and HEK cells) or for 2 h (for slices) at room temperature, followed by extensive washing with PBS. Nonspecific staining was blocked with 0.2% fish-gelatin (Sigma-Aldrich) in PBS for 30 min. Triton was omitted for immunostaining under nonpermeabilized conditions to label only surface GluA1. Neurons were incubated for 1 h with an antibody against the GluA1 extracellular N-terminal domain (MAB2263; Merck Millipore) in the blocking solution. Total GluA1 was then immunodetected after washes and a permeabilization step (30 min incubation in 0.1% Triton X-100 and 0.2% fish gelatin in PBS) with an antibody against the GluA1 C-terminal domain (ab31232; Abcam) in the permeabilizing solution. For neurite polarization assays, following permeabilization and blocking steps after fixation, neurons were incubated with an antibody against Tau5 (577801; Merck Millipore) in blocking solution overnight at 4°C. After repeated washing with PBS, samples were incubated with the corresponding secondary antibodies, coupled to Alexa Fluor 488 or Alexa Fluor 647. All samples were mounted on microscope slides with Mowiol/DABCO (Confocal Microscopy Facility at the Centro de Biología Molecular Severo Ochoa) or Prolong Glass antifade medium (P36982; Thermo Fisher Scientific). Fluorescence images were acquired with a confocal microscope (LSM710 or LSM800; Zeiss) using a 10× NA 0.3, 20× NA 0.8, 40× NA 1.3, or 63× NA 1.4 oil-immersion objectives and 488-nm, 514-nm, 561-nm, and 633/640-nm lasers as Z-stacks. Image acquisition was performed at room temperature for fixed tissue experiments. All images corresponding to the same experiment were acquired using the same microscope settings and conditions using ZEN Blue 3.2 (LSM800) and Zeiss Zen2010B sp1 (LSM710) software.
Image analysis and quantification
Acquired Z-stacks were reconstructed (maximum intensity projection) and analyzed using Fiji v1.52 (Schindelin et al., 2012) and MetaMorph (Molecular Devices). For illustration of representative pictures, image editing was performed using Fiji and was limited to linear contrast enhancement.
For the axonal polarity assay, the length of longest Tau-positive neurite was divided by the length of the second longest Tau-positive neurite.
For surface/total GluA1 expression (Fig. 2, A–C), regions of interest (ROIs) corresponding to neurons were automatically selected from a thresholded mCherry channel image for either control or shKIF13A-expressing neurons. ROIs for individual somas were manually drawn on the mCherry channel and subtracted from the cell-wide ROI to measure separately dendritic and somatic fluorescence in the total and surface GluA1 channels. ROIs for individual spines and adjacent dendritic shafts were also manually drawn on the mCherry channel in the indicated experiments to measure fluorescence in the total and surface GluA1 channels. Therefore, image quantification was essentially blind with respect to both GluA1 channels. All intensities were corrected for unspecific background labeling.
To analyze spine volume (Fig. 2 D), the same ROIs for individual spines determined for Fig. 2 B were used to measure mCherry fluorescence intensity (expressed in both control and shKIF13A lentiviral vectors). Spine intensity was normalized with respect to the fluorescence signal from the adjacent dendrite.
For the analysis of colocalization of KIF13A-YFP and mCherry-FIP2 (Fig. 6, A and B), clusters were defined on the calibrated projection images for each channel (KIF13A-YFP or mCherry-FIP2) using and inclusive threshold defined with the MetaMorph software. The overlapping colocalized cluster area was calculated from the thresholded images using the Measure Colocalization tool from the same program. To calculate the total cell area, a different threshold was applied on the same projection images to include the whole cell analyzed. The Morphometry Analysis tool allowed measuring the area of the object defined with the previous threshold. PCCs, MOCs, and individual M1 and M2 coefficients (Fig. 6 C) were calculated on thresholded projection images using the JACoP plugin for Fiji (Bolte and Cordelières, 2006).
FIP2 activity–dependent clustering (Fig. 6, D–F) was analyzed by manual selection of ROIs for each coinfected dendrite. A threshold for the GFP-FIP2 channel was established from the baseline images for individual dendrites. The same threshold was then applied to the projection of the 15 min cLTP image. Using the analyze particles plugin on these masks, the number of clusters and their size was measured within the ROI of each dendrite. The area of the ROI of each dendrite was also measured to calculate cluster density. To avoid bias, ROI definition and quantifications were performed using the mCherry channel as the reference, either from control or shKIF13A-expressing neurons.
Statistical analysis
Graphs represent mean values ± SEM or cumulative distributions. Statistical differences (P values) were calculated according to nonparametric tests, including two-tailed Mann–Whitney tests (for unpaired data) or Wilcoxon/Friedman tests (for paired data). Comparisons between cumulative distributions were calculated with the Kolmogorov–Smirnov test.
Online supplemental material
Fig. S1 shows the basal physiological properties of neurons expressing shKIF13A and KIF13A-ST. Fig. S2 reports the specificity of shKIF13A and shKIF13B knockdown and the effect on axonal polarization. Fig. S3 displays the expression of the recombinant proteins GFP-KIF13A-ST, mRFP-KIF13A-tail, and KIF13A-MS-YFP. Fig. S4 shows the immunoprecipitation of GFP-KIF13A-ST and the effect of phosphatase inhibitors on centaurin-α1 coprecipitation. Fig. S5 shows the subcellular fractionation of hippocampal slices. Video 1 is a time-lapse video of a cLTP experiment showing dendritic branches from an organotypic hippocampal slice infected with GFP-FIP2. Video 2 is a time-lapse video of a cLTP experiment showing dendritic branches from an organotypic hippocampal slice infected with GFP-FIP2 and expressing shKIF13A.
Supplementary Material
Acknowledgments
We thank Raquel Jiménez for technical assistance, Ernest Palomer for aiding with the first qPCR experiments, Jonathan Draffin and Víctor Briz for critical reading of the manuscript, and the personnel at the facilities (Confocal Microscopy and Genomics) of the Centro de Biología Molecular Severo Ochoa for expert technical assistance.
This work was supported by the Spanish Ministry of Science and Innovation (grants SAF2014-57233-R and SAF2017-86983-R) and the European Union (PCIN-2016-095) to J.A. Esteban, and by the Institut National de la Santé et de la Recherche Médicale, Institut Curie, and the Centre National de la Recherche Scientifique (LabEx Cell(n)Scale, ANR-11-LABX-0038) to C. Delevoye. Y. Gutiérrez and S. López-García are recipients of predoctoral contracts from the Spanish Ministry of Education. A. Lario was the recipient of a predoctoral contract from the Spanish Ministry of Economy and Competitiveness.
The authors declare no competing financial interests.
Author contributions: Y. Gutiérrez carried out most of the experimental work. S. López-García performed the axonal polarization experiments, the electrophysiological recordings with the rescue KIF13A-MS construct, and most of the immunoprecipitation assays. A. Lario designed and cloned the shRNA for KIF13A. S. Gutiérrez-Eisman cloned the rescue construct with KIF13A-MS. C. Delevoye contributed the KIF13A-YFP plasmid and helped edit the manuscript. Y. Gutiérrez, S. López-García, and J.A. Esteban analyzed data. Y. Gutiérrez and J.A. Esteban designed the research and wrote the manuscript.
References
- Ahmad, M., Polepalli J.S., Goswami D., Yang X., Kaeser-Woo Y.J., Südhof T.C., and Malenka R.C.. 2012. Postsynaptic complexin controls AMPA receptor exocytosis during LTP. Neuron. 73:260–267. 10.1016/j.neuron.2011.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt, K.L., Royo M., Fernández-Monreal M., Knafo S., Petrok C.N., Martens J.R., and Esteban J.A.. 2010. PIP3 controls synaptic function by maintaining AMPA receptor clustering at the postsynaptic membrane. Nat. Neurosci. 13:36–44. 10.1038/nn.2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt, K.L., Benoist M., Lario A., Draffin J.E., Muñoz M., and Esteban J.A.. 2014. PTEN counteracts PIP3 upregulation in spines during NMDA-receptor-dependent long-term depression. J. Cell Sci. 127:5253–5260. 10.1242/jcs.156554 [DOI] [PubMed] [Google Scholar]
- Baetz, N.W., and Goldenring J.R.. 2013. Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Mol. Biol. Cell. 24:643–658. 10.1091/mbc.e12-09-0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker, G.A., and Cowan W.M.. 1977. Rat hippocampal neurons in dispersed cell culture. Brain Res. 126:397–42. 10.1016/0006-8993(77)90594-7 [DOI] [PubMed] [Google Scholar]
- Bentley, M., Decker H., Luisi J., and Banker G.. 2015. A novel assay reveals preferential binding between Rabs, kinesins, and specific endosomal subpopulations. J. Cell Biol. 208:273–281. 10.1083/jcb.201408056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte, S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Brachet, A., Norwood S., Brouwers J.F., Palomer E., Helms J.B., Dotti C.G., and Esteban J.A.. 2015. LTP-triggered cholesterol redistribution activates Cdc42 and drives AMPA receptor synaptic delivery. J. Cell Biol. 208:791–806. 10.1083/jcb.201407122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt, D.S., and Nicoll R.A.. 2003. AMPA receptor trafficking at excitatory synapses. Neuron. 40:361–379. 10.1016/S0896-6273(03)00640-8 [DOI] [PubMed] [Google Scholar]
- Brown, T.C., Correia S.S., Petrok C.N., and Esteban J.A.. 2007. Functional compartmentalization of endosomal trafficking for the synaptic delivery of AMPA receptors during long-term potentiation. J. Neurosci. 27:13311–13315. 10.1523/JNEUROSCI.4258-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.X., Otmakhov N., and Lisman J.. 1999. Requirements for LTP induction by pairing in hippocampal CA1 pyramidal cells. J. Neurophysiol. 82:526–532. 10.1152/jn.1999.82.2.526 [DOI] [PubMed] [Google Scholar]
- Correia, S.S., Bassani S., Brown T.C., Lisé M.F., Backos D.S., El-Husseini A., Passafaro M., and Esteban J.A.. 2008. Motor protein-dependent transport of AMPA receptors into spines during long-term potentiation. Nat. Neurosci. 11:457–466. 10.1038/nn2063 [DOI] [PubMed] [Google Scholar]
- Delevoye, C., Miserey-Lenkei S., Montagnac G., Gilles-Marsens F., Paul-Gilloteaux P., Giordano F., Waharte F., Marks M.S., Goud B., and Raposo G.. 2014. Recycling endosome tubule morphogenesis from sorting endosomes requires the kinesin motor KIF13A. Cell Rep. 6:445–454. 10.1016/j.celrep.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti, C.G., Sullivan C.A., and Banker G.A.. 1988. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8:1454–1468. 10.1523/JNEUROSCI.08-04-01454.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves da Silva, M., Adrian M., Schätzle P., Lipka J., Watanabe T., Cho S., Futai K., Wierenga C.J., Kapitein L.C., and Hoogenraad C.C.. 2015. Positioning of AMPA Receptor-Containing Endosomes Regulates Synapse Architecture. Cell Rep. 13:933–943. 10.1016/j.celrep.2015.09.062 [DOI] [PubMed] [Google Scholar]
- Fernández-Monreal, M., Sánchez-Castillo C., and Esteban J.A.. 2016. APPL1 gates long-term potentiation through its plekstrin homology domain. J. Cell Sci. 129:2793–2803. 10.1242/jcs.183475 [DOI] [PubMed] [Google Scholar]
- Fuller, L., and Dailey M.E.. 2007. Preparation of rodent hippocampal slice cultures. CSH Protoc. 2007:pdb.prot4848. doi:. 10.1101/PDB.PROT4848 [DOI] [PubMed]
- Gähwiler, B.H., Capogna M., Debanne D., McKinney R.A., and Thompson S.M.. 1997. Organotypic slice cultures: a technique has come of age. Trends Neurosci. 20:471–477. 10.1016/S0166-2236(97)01122-3 [DOI] [PubMed] [Google Scholar]
- Gardoni, F., Mauceri D., Malinverno M., Polli F., Costa C., Tozzi A., Siliquini S., Picconi B., Cattabeni F., Calabresi P., and Di Luca M.. 2009. Decreased NR2B subunit synaptic levels cause impaired long-term potentiation but not long-term depression. J. Neurosci. 29:669–677. 10.1523/JNEUROSCI.3921-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, L.S.B., and Yang Z.. 2000. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu. Rev. Neurosci. 23:39–71. 10.1146/annurev.neuro.23.1.39 [DOI] [PubMed] [Google Scholar]
- Goslin, K., and Banker G.. 1989. Experimental observations on the development of polarity by hippocampal neurons in culture. J. Cell Biol. 108:1507–1516. 10.1083/jcb.108.4.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger, E., McNee G., Allan V., and Woodman P.. 2014. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin. Cell Dev. Biol. 31:20–29. 10.1016/j.semcdb.2014.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger, I.H., and Esteban J.A.. 2007. AMPA receptor biogenesis and trafficking. Curr. Opin. Neurobiol. 17:289–297. 10.1016/j.conb.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Guillaud, L., Setou M., and Hirokawa N.. 2003. KIF17 dynamics and regulation of NR2B trafficking in hippocampal neurons. J. Neurosci. 23:131–140. 10.1523/JNEUROSCI.23-01-00131.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales, C.M., Vaerman J.P., and Goldenring J.R.. 2002. Rab11 family interacting protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J. Biol. Chem. 277:50415–50421. 10.1074/jbc.M209270200 [DOI] [PubMed] [Google Scholar]
- Hayashi, Y., Shi S.-H., Esteban J.A., Piccini A., Poncer J.-C., and Malinow R.. 2000. Driving AMPA receptors into synapses by LTP and CaMKII: Requirement for GluR1 and PDZ domain interaction. Science. 287:2262–2267. [DOI] [PubMed] [Google Scholar]
- Heisler, F.F., Lee H.K., Gromova K.V., Pechmann Y., Schurek B., Ruschkies L., Schroeder M., Schweizer M., and Kneussel M.. 2014. GRIP1 interlinks N-cadherin and AMPA receptors at vesicles to promote combined cargo transport into dendrites. Proc. Natl. Acad. Sci. USA. 111:5030–5035. 10.1073/pnas.1304301111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley, J.M., Barker E.A., and Glebov O.O.. 2011. Routes, destinations and delays: recent advances in AMPA receptor trafficking. Trends Neurosci. 34:258–268. 10.1016/j.tins.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, N., and Takemura R.. 2005. Molecular motors and mechanisms of directional transport in neurons. Nat. Rev. Neurosci. 6:201–214. 10.1038/nrn1624 [DOI] [PubMed] [Google Scholar]
- Hirokawa, N., Noda Y., Tanaka Y., and Niwa S.. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10:682–696. 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- Hoerndli, F.J., Maxfield D.A., Brockie P.J., Mellem J.E., Jensen E., Wang R., Madsen D.M., and Maricq A.V.. 2013a. Kinesin-1 regulates synaptic strength by mediating the delivery, removal, and redistribution of AMPA receptors. Neuron. 80:1421–1437. 10.1016/j.neuron.2013.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerndli, F.J., Maxfield D.A., Brockie P.J., Mellem J.E., Jensen E., Wang R., Madsen D.M., and Maricq A.V.. 2013b. Kinesin-1 regulates synaptic strength by mediating the delivery, removal, and redistribution of AMPA receptors. Neuron. 80:1421–1437. 10.1016/j.neuron.2013.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad, C.C., Milstein A.D., Ethell I.M., Henkemeyer M., and Sheng M.. 2005. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat. Neurosci. 8:906–915. 10.1038/nn1487 [DOI] [PubMed] [Google Scholar]
- Horgan, C.P., and McCaffrey M.W.. 2011. Rab GTPases and microtubule motors. Biochem. Soc. Trans. 39:1202–1206. 10.1042/BST0391202 [DOI] [PubMed] [Google Scholar]
- Horiguchi, K., Hanada T., Fukui Y., and Chishti A.H.. 2006. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J. Cell Biol. 174:425–436. 10.1083/jcb.200604031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X., Viesselmann C., Nam S., Merriam E., and Dent E.W.. 2008. Activity-dependent dynamic microtubule invasion of dendritic spines. J. Neurosci. 28:13094–13105. 10.1523/JNEUROSCI.3074-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski, J., Kapitein L.C., Gouveia S.M., Dortland B.R., Wulf P.S., Grigoriev I., Camera P., Spangler S.A., Di Stefano P., Demmers J., et al. 2009. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 61:85–100. 10.1016/j.neuron.2008.11.013 [DOI] [PubMed] [Google Scholar]
- Jing, J., and Prekeris R.. 2009. Polarized endocytic transport: the roles of Rab11 and Rab11-FIPs in regulating cell polarity. Histol. Histopathol. 24:1171–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado, S., Goswami D., Zhang Y., Molina A.J.M., Südhof T.C., and Malenka R.C.. 2013. LTP requires a unique postsynaptic SNARE fusion machinery. Neuron. 77:542–558. 10.1016/j.neuron.2012.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein, L.C., and Hoogenraad C.C.. 2011. Which way to go? Cytoskeletal organization and polarized transport in neurons. Mol. Cell. Neurosci. 46:9–20. 10.1016/j.mcn.2010.08.015 [DOI] [PubMed] [Google Scholar]
- Kennedy, M.J., Davison I.G., Robinson C.G., and Ehlers M.D.. 2010. Syntaxin-4 defines a domain for activity-dependent exocytosis in dendritic spines. Cell. 141:524–535. 10.1016/j.cell.2010.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneussel, M., and Hausrat T.J.. 2016. Postsynaptic Neurotransmitter Receptor Reserve Pools for Synaptic Potentiation. Trends Neurosci. 39:170–182. 10.1016/j.tins.2016.01.002 [DOI] [PubMed] [Google Scholar]
- Kneussel, M., and Wagner W.. 2013. Myosin motors at neuronal synapses: drivers of membrane transport and actin dynamics. Nat. Rev. Neurosci. 14:233–247. 10.1038/nrn3445 [DOI] [PubMed] [Google Scholar]
- Lindsay, A.J., and McCaffrey M.W.. 2004. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane. J. Cell Sci. 117:4365–4375. 10.1242/jcs.01280 [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Maas, C., Belgardt D., Lee H.K., Heisler F.F., Lappe-Siefke C., Magiera M.M., van Dijk J., Hausrat T.J., Janke C., and Kneussel M.. 2009. Synaptic activation modifies microtubules underlying transport of postsynaptic cargo. Proc. Natl. Acad. Sci. USA. 106:8731–8736. 10.1073/pnas.0812391106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino, H., and Malinow R.. 2009. AMPA receptor incorporation into synapses during LTP: the role of lateral movement and exocytosis. Neuron. 64:381–390. 10.1016/j.neuron.2009.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriam, E.B., Millette M., Lumbard D.C., Saengsawang W., Fothergill T., Hu X., Ferhat L., and Dent E.W.. 2013. Synaptic regulation of microtubule dynamics in dendritic spines by calcium, F-actin, and drebrin. J. Neurosci. 33:16471–16482. 10.1523/JNEUROSCI.0661-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár, E. 2011. Long-term potentiation in cultured hippocampal neurons. Semin. Cell Dev. Biol. 22:506–513. 10.1016/j.semcdb.2011.07.017 [DOI] [PubMed] [Google Scholar]
- Monteiro, M.I., Ahlawat S., Kowalski J.R., Malkin E., Koushika S.P., and Juo P.. 2012. The kinesin-3 family motor KLP-4 regulates anterograde trafficking of GLR-1 glutamate receptors in the ventral nerve cord of Caenorhabditis elegans. Mol. Biol. Cell. 23:3647–3662. 10.1091/mbc.e12-04-0334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, C.D., Thacker E.E., Larimore J., Gaston D., Underwood A., Kearns B., Patterson S.I., Jackson T., Chapleau C., Pozzo-Miller L., and Theibert A.. 2007. The neuronal Arf GAP centaurin α1 modulates dendritic differentiation. J. Cell Sci. 120:2683–2693. 10.1242/jcs.006346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa, M., Tanaka Y., Cho H.S., Yoshihara M., and Hirokawa N.. 2018. The Molecular Motor KIF21B Mediates Synaptic Plasticity and Fear Extinction by Terminating Rac1 Activation. Cell Rep. 23:3864–3877. 10.1016/j.celrep.2018.05.089 [DOI] [PubMed] [Google Scholar]
- Muhia, M., Thies E., Labonté D., Ghiretti A.E., Gromova K.V., Xompero F., Lappe-Siefke C., Hermans-Borgmeyer I., Kuhl D., Schweizer M., et al. 2016. The Kinesin KIF21B Regulates Microtubule Dynamics and Is Essential for Neuronal Morphology, Synapse Function, and Learning and Memory. Cell Rep. 15:968–977. 10.1016/j.celrep.2016.03.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T., Setou M., Seog D., Ogasawara K., Dohmae N., Takio K., and Hirokawa N.. 2000. A novel motor, KIF13A, transports mannose-6-phosphate receptor to plasma membrane through direct interaction with AP-1 complex. Cell. 103:569–581. 10.1016/S0092-8674(00)00161-6 [DOI] [PubMed] [Google Scholar]
- Otmakhov, N., Khibnik L., Otmakhova N., Carpenter S., Riahi S., Asrican B., and Lisman J.. 2004. Forskolin-induced LTP in the CA1 hippocampal region is NMDA receptor dependent. J. Neurophysiol. 91:1955–1962. 10.1152/jn.00941.2003 [DOI] [PubMed] [Google Scholar]
- Park, M., Penick E.C., Edwards J.G., Kauer J.A., and Ehlers M.D.. 2004. Recycling Endosomes Supply AMPA Receptors for LTP. Science. 305:1972–1975. [DOI] [PubMed] [Google Scholar]
- Park, M., Salgado J.M., Ostroff L., Helton T.D., Robinson C.G., Harris K.M., and Ehlers M.D.. 2006. Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron. 52:817–830. 10.1016/j.neuron.2006.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn, A.C., Zhang C.L., Georges F., Royer L., Breillat C., Hosy E., Petersen J.D., Humeau Y., and Choquet D.. 2017. Hippocampal LTP and contextual learning require surface diffusion of AMPA receptors. Nature. 549:384–388. 10.1038/nature23658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo, M., Gutiérrez Y., Fernández-Monreal M., Gutiérrez-Eisman S., Jiménez R., Jurado S., and Esteban J.A.. 2019. A retention-release mechanism based on RAB11FIP2 for AMPA receptor synaptic delivery during long-term potentiation. J. Cell Sci. 132:jcs234237. 10.1242/jcs.234237 [DOI] [PubMed] [Google Scholar]
- Schafer, J.C., Baetz N.W., Lapierre L.A., McRae R.E., Roland J.T., and Goldenring J.R.. 2014. Rab11-FIP2 interaction with MYO5B regulates movement of Rab11a-containing recycling vesicles. Traffic. 15:292–308. 10.1111/tra.12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schätzle, P., Esteves da Silva M., Tas R.P., Katrukha E.A., Hu H.Y., Wierenga C.J., Kapitein L.C., and Hoogenraad C.C.. 2018. Activity-Dependent Actin Remodeling at the Base of Dendritic Spines Promotes Microtubule Entry. Curr. Biol. 28:2081–2093.e6. 10.1016/j.cub.2018.05.004 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setou, M., Nakagawa T., Seog D.H., and Hirokawa N.. 2000. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 288:1796–1802. 10.1126/science.288.5472.1796 [DOI] [PubMed] [Google Scholar]
- Setou, M., Seog D.-H., Tanaka Y., Kanai Y., Takei Y., Kawagishi M., and Hirokawa N.. 2002. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 417:83–87. 10.1038/nature743 [DOI] [PubMed] [Google Scholar]
- Shepherd, J.D., and Huganir R.L.. 2007. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 23:613–643. 10.1146/annurev.cellbio.23.090506.123516 [DOI] [PubMed] [Google Scholar]
- Shi, S.H., Hayashi Y., Petralia R.S., Zaman S.H., Wenthold R.J., Svoboda K., and Malinow R.. 1999. Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science. 284:1811–1816. [DOI] [PubMed] [Google Scholar]
- Shin, H., Wyszynski M., Huh K.-H., Valtschanoff J.G., Lee J.-R., Ko J., Streuli M., Weinberg R.J., Sheng M., and Kim E.. 2003. Association of the kinesin motor KIF1A with the multimodular protein liprin-α. J. Biol. Chem. 278:11393–11401. 10.1074/jbc.M211874200 [DOI] [PubMed] [Google Scholar]
- Stricker, R., and Reiser G.. 2014. Functions of the neuron-specific protein ADAP1 (centaurin-α1) in neuronal differentiation and neurodegenerative diseases, with an overview of structural and biochemical properties of ADAP1. Biol. Chem. 395:1321–1340. 10.1515/hsz-2014-0107 [DOI] [PubMed] [Google Scholar]
- Szatmari, E.M., Moran C., Cohen S., Jacob A., Parra-Bueno P., Kamasawa N., Guerrero-Given D., Klein M., Stackman R. Jr., and Yasuda R.. 2021. Adap1/centaurin-α1 negatively regulates dendritic spine function and memory formation in the hippocampus. eNeuro. 8:1–14. 10.1523/ENEURO.0111-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, A.H., Chen H., Li T.P., Metzbower S.R., MacGillavry H.D., and Blanpied T.A.. 2016. A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature. 536:210–214. 10.1038/nature19058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, Y., Tempel W., Wang H., Yamada K., Shen L., Senisterra G.A., MacKenzie F., Chishti A.H., and Park H.-W.. 2010. Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1. Proc. Natl. Acad. Sci. USA. 107:20346–20351. 10.1073/pnas.1009008107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Edwards J.G., Riley N., Provance D.W. Jr., Karcher R., Li X.D., Davison I.G., Ikebe M., Mercer J.A., Kauer J.A., and Ehlers M.D.. 2008. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 135:535–548. 10.1016/j.cell.2008.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz, T., Wellbourne-Wood J., and Kerkhoff E.. 2014. Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 24:407–415. 10.1016/j.tcb.2014.02.004 [DOI] [PubMed] [Google Scholar]
- Wong, R.W.-C., Setou M., Teng J., Takei Y., and Hirokawa N.. 2002. Overexpression of motor protein KIF17 enhances spatial and working memory in transgenic mice. Proc. Natl. Acad. Sci. USA. 99:14500–14505. 10.1073/pnas.222371099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, X., Takei Y., Kido M.A., and Hirokawa N.. 2011. Molecular motor KIF17 is fundamental for memory and learning via differential support of synaptic NR2A/2B levels. Neuron. 70:310–325. 10.1016/j.neuron.2011.02.049 [DOI] [PubMed] [Google Scholar]
- Yoshimura, Y., Terabayashi T., and Miki H.. 2010. Par1b/MARK2 phosphorylates kinesin-like motor protein GAKIN/KIF13B to regulate axon formation. Mol. Cell. Biol. 30:2206–2219. 10.1128/MCB.01181-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Fok A.H.K., Fan R., Kwan P.-Y., Chan H.-L., Lo L.H.-Y., Chan Y.-S., Yung W.-H., Huang J., Lai C.S.W., and Lai K.-O.. 2020. Specific depletion of the motor protein KIF5B leads to deficits in dendritic transport, synaptic plasticity and memory. eLife. 9:e53456. 10.7554/eLife.53456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, N., Jeyifous O., Munro C., Montgomery J.M., and Green W.N.. 2015. Synaptic activity regulates AMPA receptor trafficking through different recycling pathways. eLife. 4:e06878. 10.7554/eLife.06878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, R., Niwa S., Guillaud L., Tong Y., and Hirokawa N.. 2013. A molecular motor, KIF13A, controls anxiety by transporting the serotonin type 1A receptor. Cell Rep. 3:509–519. 10.1016/j.celrep.2013.01.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.