Dendritic spines undergo rapid size expansion upon induction of long-term potentiation. Yang et al. uncover that in response to Ca2+/calmodulin, endophilin A1 quickly associates with the plasma membrane and recruits p140Cap to promote actin polymerization that provides driving force for spine enlargement.
Abstract
Induction of long-term potentiation (LTP) in excitatory neurons triggers a large transient increase in the volume of dendritic spines followed by decays to sustained size expansion, a process termed structural LTP (sLTP) that contributes to the cellular basis of learning and memory. Although mechanisms regulating the early and sustained phases of sLTP have been studied intensively, how the acute spine enlargement immediately after LTP stimulation is achieved remains elusive. Here, we report that endophilin A1 orchestrates membrane dynamics with actin polymerization to initiate spine enlargement in NMDAR-mediated LTP. Upon LTP induction, Ca2+/calmodulin enhances binding of endophilin A1 to both membrane and p140Cap, a cytoskeletal regulator. Consequently, endophilin A1 rapidly localizes to the plasma membrane and recruits p140Cap to promote local actin polymerization, leading to spine head expansion. Moreover, its molecular functions in activity-induced rapid spine growth are required for LTP and long-term memory. Thus, endophilin A1 serves as a calmodulin effector to drive acute structural plasticity necessary for learning and memory.
Introduction
Long-term potentiation (LTP) of synapses contributes to neural mechanisms underlying learning and memory (Nabavi et al., 2014). In the mammalian brain, most glutamatergic synapses are located on dendritic spines, micron-sized membrane protrusions emanating from dendrites of excitatory neurons. In response to input activity, spines undergo changes in both morphology (structural plasticity) and function (functional plasticity), which are tightly correlated during LTP (Harvey and Svoboda, 2007; Matsuzaki et al., 2001; Matsuzaki et al., 2004). Imaging of brain slices has revealed that LTP stimuli triggers a large transient increase in spine volume starting within 1 min of stimulation (acute or initial phase), followed by the decay of enlarged spines (early or transient phase) to a long-lasting size expansion (late or sustained phase; Harvey and Svoboda, 2007; Matsuzaki et al., 2004). This process of spine morphological remodeling is termed structural LTP (sLTP), which would allow physical enlargement of glutamatergic synapses to accommodate more AMPA-type receptors (AMPARs) for synaptic potentiation (Herring and Nicoll, 2016). Indeed, recent studies have not only detected size increase of functionally potentiated spines in the motor cortex of mice trained with motor learning tasks (Hayashi-Takagi et al., 2015; Roth et al., 2020) but also established a direct link between spine expansion and memory trace in vivo by demonstrating erasure of acquired motor skills by optical shrinkage of potentiated spines (Hayashi-Takagi et al., 2015). Nonetheless, whether acute sLTP is necessary for LTP and its physiological relevance to learning and memory remain to be determined. Moreover, although sLTP has been studied intensively, with calcium (Ca2+) signaling–regulated actin remodeling being the central process that governs the stabilization and consolidation of spine enlargement (Nakahata and Yasuda, 2018), the mechanism initiating rapid spine growth remains largely unexplored due to limited spatiotemporal resolution of the molecular events in the acute phase of LTP induction.
Endophilin A1 is a member of the endophilin A protein family characterized by an amino-terminal N-BIN/amphiphysin/Rvs (BAR) domain and a carboxyl-terminal Src homology 3 (SH3) domain. The gene encoding endophilin A1 (EEN1, gene name sh3gl2) is almost exclusively expressed in brain (Ringstad et al., 1997) and has been implicated in epilepsy, schizophrenia, and Alzheimer’s disease (Corponi et al., 2019; Ren et al., 2008; Yu et al., 2018a; Yu et al., 2018b). Originally identified as a component of the endocytic machinery, endophilin As function in synaptic vesicle recycling (Milosevic et al., 2011; Ringstad et al., 1997; Schuske et al., 2003; Verstreken et al., 2003; Watanabe et al., 2018), autophagosome formation, and protein homeostasis at presynaptic terminals (Murdoch et al., 2016; Soukup et al., 2016). Endophilin A2 (gene name sh3gl1) also mediates fast clathrin-independent endocytosis in mammalian epithelial cells (Boucrot et al., 2015; Renard et al., 2015). At the postsynaptic membrane, endophilin A2 and A3 (gene name sh3gl3) interact with Arc/Arg3.1 to accelerate AMPAR endocytosis during late-phase synaptic plasticity (Chowdhury et al., 2006).
During synaptic development, endophilin A1 contributes to dendritic spine morphogenesis and stabilization via interaction with p140Cap (Yang et al., 2015), which recruits cortactin to promote Arp2/3-mediated branched actin polymerization (Uruno et al., 2001; Weaver et al., 2001). EEN1 gene knockout (KO) in the hippocampal CA1 region of mouse brain causes impairment of LTP of the Schaffer collateral (SC)–CA1 pathway and long-term memory (Yang et al., 2018). At the cellular level, endophilin A1, but not A2 or A3, is required for N-methyl-D-aspartate receptor (NMDAR)–mediated synaptic plasticity in mature CA1 pyramidal cells (Yang et al., 2018). Intriguingly, overexpression of p140Cap fails to rescue the spine plasticity phenotypes in EEN1 KO (EEN1−/−) neurons (Yang et al., 2018), suggesting the necessity of spatiotemporal coordination of membrane dynamics and actin polymerization via endophilin A1 during synaptic potentiation. In this study, we investigated the mechanistic roles of endophilin A1 in sLTP. We present evidence that endophilin A1 serves as an immediate effector of Ca2+/calmodulin to promote actin polymerization–dependent membrane expansion in the acute phase of spine structural plasticity.
Results
Ablation of endophilin A1 abolishes acute structural plasticity of dendritic spines
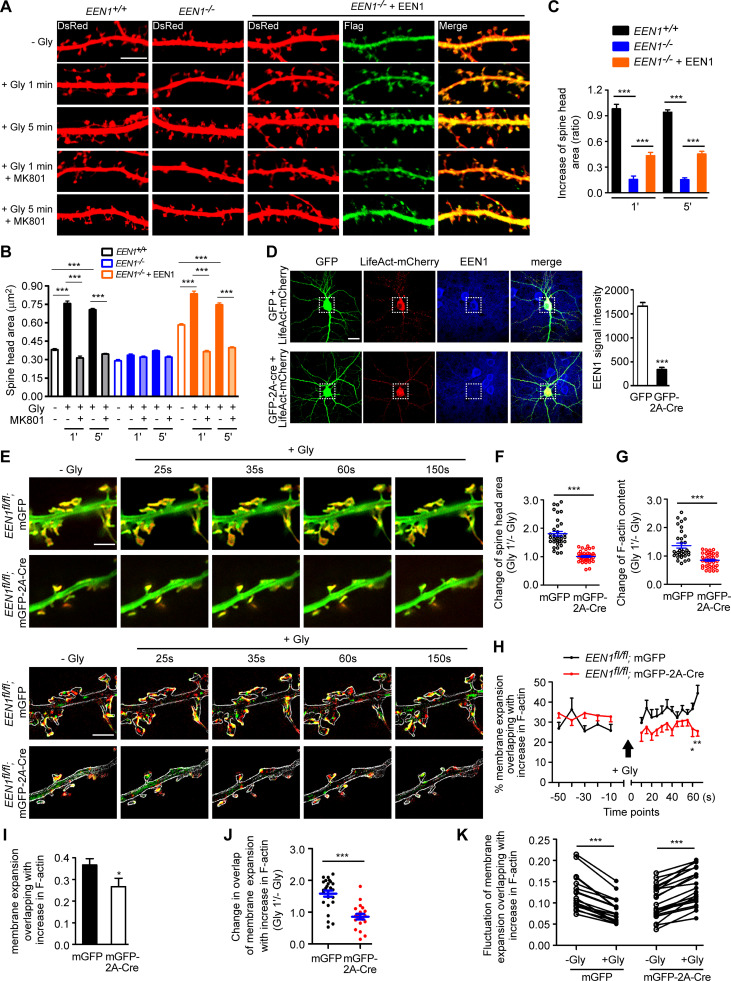
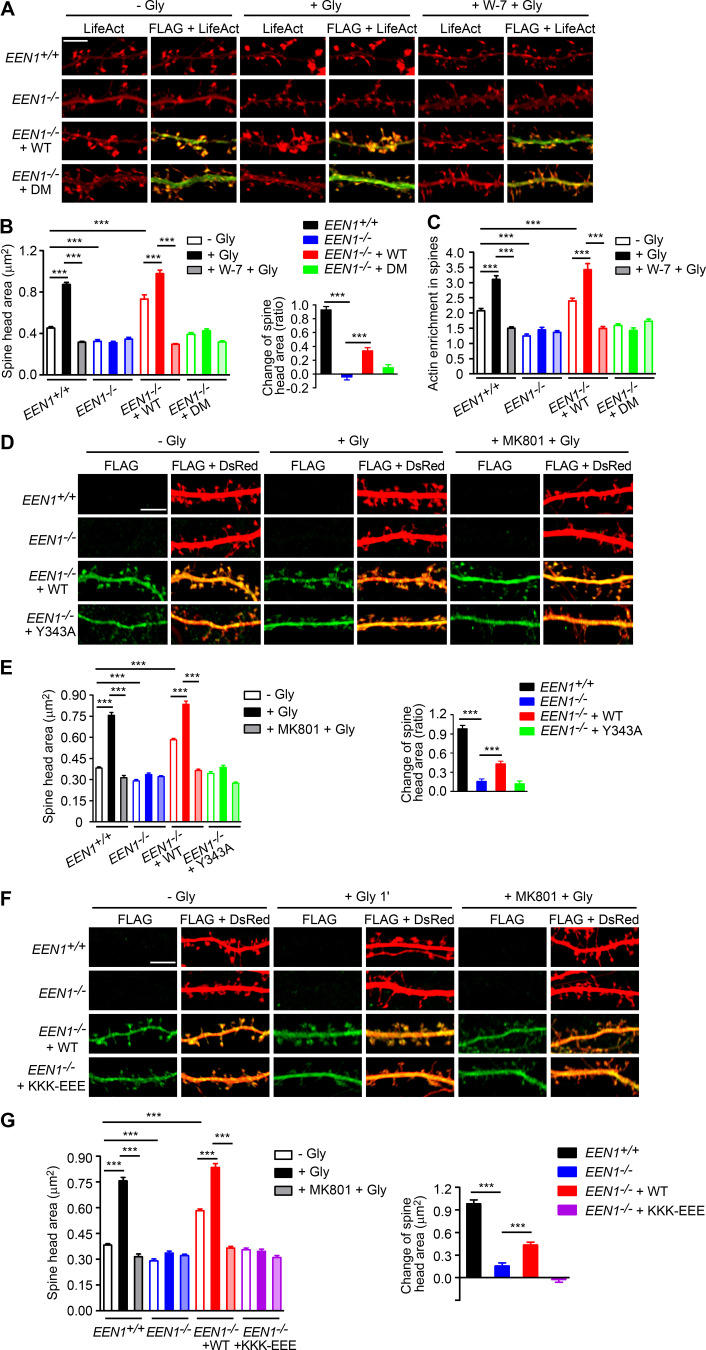
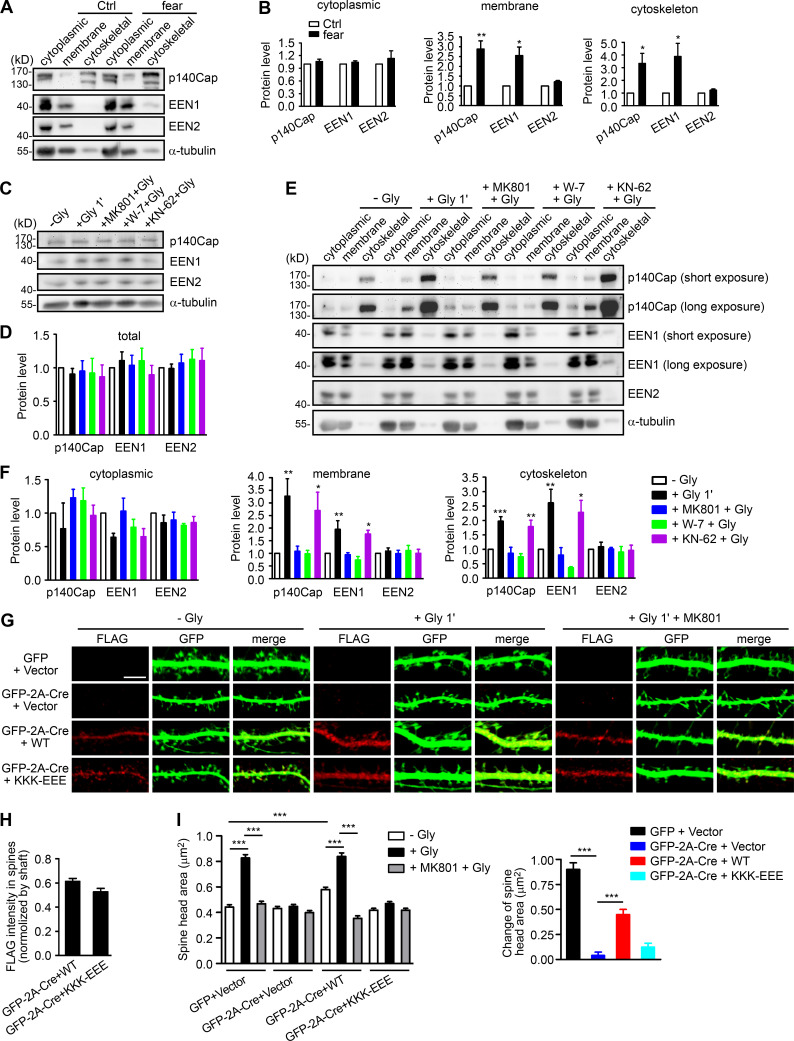
To investigate the mechanistic roles of endophilin A1 in synaptic plasticity, first we determined at which temporal stages of LTP it functions by rescuing the morphological phenotype of EEN1 KO neurons with overexpressed endophilin A1 in the early phase of chemically induced LTP (cLTP; Fig. 1 A). Quantification of spine size indicated that endophilin A1 is required for spine enlargement as early as 1 min after application of the NMDAR coagonist glycine (Fig. 1, B and C).
Figure 1.
Ablation of endophilin A1 abolishes spine expansion and actin polymerization in the acute phase of sLTP. (A) Rescue of sLTP in EEN1−/− neurons. Mouse hippocampal neurons cotransfected with pLL3.7-DsRed (volume marker) and pCMV-Tag2B (FLAG vector) or pCMV-Tag2B-endophilin A1 (FLAG-EEN1) were pretreated with DMSO (vehicle control) or MK801 (NMDAR antagonist), and cLTP was performed using glycine (Gly) on DIV16. Neurons were fixed 1 min or 5 min after glycine application, immunostained for FLAG, and imaged by confocal microscopy. Scale bar, 5 µm. (B and C) Quantification of spine size (B) and changes of spine size (C) in A. Data are expressed as mean ± SEM for each group (EEN1+/+: n [cells] = 20, n [spines] = 851 for − Gly; n = 15, n = 571 for + Gly 1 min; n = 20, n = 871 for + Gly 5 min; n = 14, n = 568 for + Gly 1 min + MK801; n = 17, n = 668 for + Gly 5 min + MK801. EEN1−/−: n = 20, n = 831 for − Gly; n = 13, n = 563 for + Gly 1 min; n = 16, n = 614 for + Gly 5 min; n = 15, n = 573 for + Gly 1 min + MK801; n = 15, n = 569 for + Gly 5 min + MK801. EEN1−/− + EEN1: n = 19, n = 731 for − Gly; n = 14, n = 581 for + Gly 1 min; n = 16, n = 654 for + Gly 5 min; n = 14, n = 572 for + Gly 1 min + MK801; n = 15, n = 604 for + Gly 5 min + MK801). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001. (D) Cre-mediated KO in EEN1fl/fl hippocampal neurons. Neurons from an EEN1fl/fl mouse were transfected with constructs expressing LifeAct-mCherry and GFP or both GFP and the Cre recombinase (GFP-2A-Cre) on DIV12 and immunostained for EEN1 on DIV16. Scale bar, 20 µm. Quantitative data were expressed as mean ± SEM for each group (n = 15 for GFP, n = 13 for GFP-2A-Cre). P values were calculated using two-tailed unpaired t test. ***, P < 0.0001. (E) GI-SIM live imaging of EEN1fl/fl hippocampal neurons expressing LifeAct-mCherry and mGFP or mGFP-2A-Cre. Glycine was applied after imaging for 1 min, and imaging was continued for 5 more minutes. Upper panels are representative still images before and after glycine application. Lower panels are color-coded images showing regions of membrane expansion (increase in mGFP signals, green) and actin polymerization (increase in F-actin signals, red) in dendritic spines (outlined with the Find Edges tool of ImageJ). Scale bar, 2 µm. (F and G) Quantitative analysis of spine size increase (F) or F-actin enrichment in spines (G) of EEN1fl/fl;mGFP (WT) and EEN1fl/fl;mGFP-2A-Cre (KO) neurons imaged by GI-SIM. Data are plotted along with the mean ± SEM for each group (n = 5, n = 36 for EEN1fl/fl;mGFP, n = 5, n = 42 for EEN1fl/fl;mGFP-2A-Cre). P values were calculated using two-tailed unpaired t test. ***, P < 0.0001. (H) Quantification of fractions of expanded membrane overlapping with actin polymerization in individual spines before and after glycine application at 10-s (before) or 5-s (after) intervals. Data for each point are expressed as mean ± SEM (n = 5, n = 18 for EEN1fl/fl;mGFP, n = 5, n = 20 for EEN1fl/fl;mGFP-2A-Cre). P values were calculated using two-tailed unpaired t test. *, P < 0.05; **, P < 0.01. (I) Quantification of fractions of membrane expansion overlapping with actin polymerization at 1 min after glycine application. Data are expressed as mean ± SEM (n = 5, n = 24 for mGFP; n = 5, n = 20 for mGFP-2A-Cre). P values were calculated using two-tailed unpaired t test. *, P < 0.05. (J) Changes in the extent of overlap between membrane expansion and actin polymerization 1 min after glycine application in individual spines. Data are normalized to the time point right before glycine application. The data are plotted along with the mean ± SEM for each group (n = 5, n = 24 for mGFP; n = 5, n = 20 for mGFP-2A-Cre). P values were calculated using two-tailed unpaired t test. ***, P < 0.0001. (K) Mean fluctuation of the overlap between membrane expansion and actin polymerization in individual spines within 1 min before and after glycine application. n = 5, n = 18 for mGFP; n = 5, n = 20 for mGFP-2A-Cre. P values were calculated using two-tailed paired t test. ***, P < 0.0001.
Actin dynamics in dendritic spines is crucial for structural plasticity (Nakahata and Yasuda, 2018). Imaging studies have shown that actin polymerization in spines starts as early as 20 s after LTP induction (Okamoto et al., 2004). As endophilin A1 can recruit p140Cap to spines via its SH3 domain (Yang et al., 2015), we reasoned that it might function to promote actin polymerization in the acute phase of sLTP. To monitor morphological changes and actin dynamics of spines simultaneously, we performed super-resolution live imaging of EEN1fl/fl (WT) and Cre-mediated KO neurons (Fig. 1 D) expressing membrane-anchored GFP (mGFP) and the F-actin probe LifeAct-mCherry by grazing incidence structured illumination microscopy (GI-SIM; Guo et al., 2018). In WT neurons, consistent with our previous study (Guo et al., 2018), we observed rapid increase in both spine size and F-actin signals in dendritic spines within 1 min upon glycine application (Video 1 and Fig. 1, E–G). In contrast, no significant changes in spine size were detected in KO neurons, even though the shape of spines changed constantly (Video 2 and Fig. 1, E and F), indicating that endophilin A1 is required for spine enlargement during the acute phase of sLTP. Moreover, although the spine heads of KO neurons were as motile as those of WT cells (Video 2), the glycine-induced net increase in F-actin content was abolished (Fig. 1, E and G), indicating that endophilin A1 is also required for actin polymerization in the acute phase of sLTP.
Video 1.
Morphological changes and actin dynamics in EEN1 WT dendrite before and during glycine-induced sLTP. Dendrites of DIV16 EEN1fl/fl mouse hippocampal neurons coexpressing mGFP (green) and the F-actin probe LifeAct-mCherry (red) were imaged every 10 s by GI-SIM for 6 frames before and every 5 s for 44 frames after glycine treatment. Arrowheads indicate expanding spine heads. Video plays at 7 frames/s. Scale bar, 2 µm.
Video 2.
Morphological changes and actin dynamics in EEN1 KO dendrite before and during glycine-induced sLTP. Dendrites of DIV16 EEN1fl/fl mouse hippocampal neurons coexpressing mGFP-2A-Cre (green) and the F-actin probe LifeAct-mCherry (red) were imaged every 10 s by GI-SIM for 6 frames before and every 5 s for 47 frames after glycine treatment. Video plays at 7 frames/s. Scale bar, 2 µm.
In line with previous findings (Guo et al., 2018; Honkura et al., 2008), we observed membrane expansion, but not retraction, of the spine head, accompanied by a local increase in F-actin content in the acute phase of sLTP (lower panels, Fig. 1 E; and Videos 3, 4, 5, and 6). Quantitative image analysis clearly revealed that plasma membrane (PM) protrusion of spine head and actin polymerization are tightly coupled spatially and temporally (Fig. 1, H–K; and Video 3). In contrast, membrane protrusion and the increase in local F-actin content were much less coupled in spines of EEN1 KO neurons (Fig. 1, E and H–K; and Video 7), implicating endophilin A1 in the interplay between spine PM and actin cytoskeleton in the acute phase of sLTP.
Video 3.
Spatiotemporal relationship between spine membrane expansion and increase in F-actin in EEN1fl/fl spines before and during glycine-induced sLTP. Increases in fluorescent signals of spine membrane (labeled by mGFP) and F-actin (labeled by LifeAct-mCherry) in sequential differential images are color coded green and red, respectively. Video plays at 7 frames/s. Scale bar, 2 µm.
Video 4.
Spatiotemporal relationship between spine membrane retraction and increase in F-actin in EEN1fl/fl spines before and during glycine-induced sLTP. Decrease in fluorescent signals of spine membrane (labeled by mGFP) and increase in F-actin (labeled by LifeAct-mCherry) in sequential differential images are color coded green and red, respectively. Video plays at 7 frames/s. Scale bar, 2 µm.
Video 5.
Spatiotemporal relationship between spine membrane expansion and decrease in F-actin in EEN1fl/fl spines before and during glycine-induced sLTP. Increase in fluorescent signals of spine membrane (labeled by mGFP) and decrease in F-actin (labeled by LifeAct-mCherry) in sequential differential images are color coded green and red, respectively. Video plays at 7 frames/s. Scale bar, 2 µm.
Video 6.
Spatiotemporal relationship between spine membrane retraction and decrease in F-actin in EEN1fl/fl spines before and during glycine-induced sLTP. Decreases in fluorescent signals of spine membrane (labeled by mGFP) and F-actin (labeled by LifeAct-mCherry) in sequential differential images are color coded green and red, respectively. Video plays at 7 frames/s. Scale bar, 2 µm.
Video 7.
Spatiotemporal relationship between spine membrane expansion and increase in F-actin in EEN1 KO spines before and during glycine-induced sLTP. Increases in fluorescent signals of spine membrane (labeled by mGFP) and F-actin (labeled by LifeAct-mCherry) in sequential differential images of EEN1fl/fl mouse hippocampal neuron coexpressing mGFP-2A-Cre and LifeAct-mCherry are color coded green and red, respectively. Video plays at 7 frames/s. Scale bar, 2 µm.
Ca2+-dependent interaction between calmodulin and endophilin A1 is required for initiation of sLTP
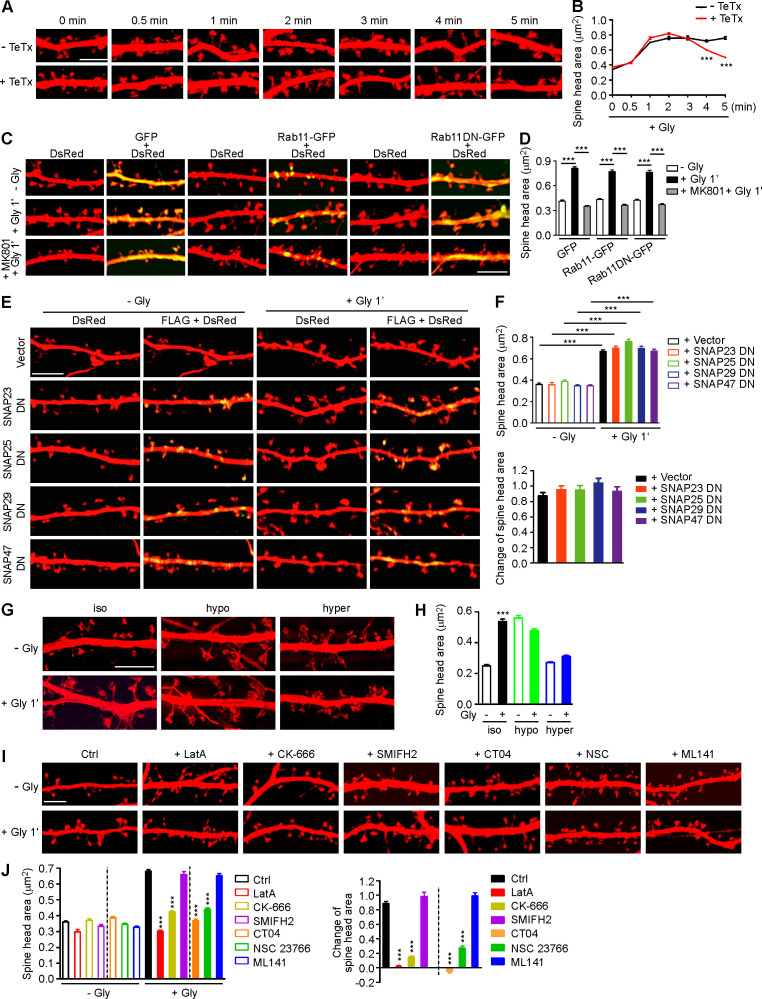
The tight coupling of PM expansion and actin polymerization in spines undergoing synaptic plasticity prompted us to investigate mechanisms underlying initiation of sLTP. Although it was postulated that the membrane source for LTP-induced spine enlargement comes from transport of Rab11-labeled recycling endosomes to the PM (Park et al., 2004), imaging studies revealed that spine head expansion precedes most of the AMPAR exocytic events in the early phase of cLTP (Kopec et al., 2006) and that the light chain of botulinum toxin type B, which inhibits SNARE complex–mediated membrane fusion/exocytosis, had no effect on the initial spine expansion after the theta burst paring protocol of LTP induction (Yang et al., 2008). Similarly, treatment with tetanus toxin, another SNARE inhibitor, did not affect the initial spine growth of glycine-stimulated hippocampal neurons (Hiester et al., 2018; Fig. S1, A and B). Overexpression of the dominant negative (DN) form of Rab11 or the SNARE component SNAP23, 25, 39, or 47 (Ibata et al., 2019) in neurons had no inhibitory effect either (Fig. S1, C–F). Together, these data indicate that vesicle fusion is not the direct source of membrane supply for rapid spine expansion in the acute phase of sLTP.
Figure S1.
Membrane unfolding and branched actin polymerization are required for initiation of sLTP. (A) Mouse hippocampal neurons transfected with pLL3.7-DsRed on DIV12 were pretreated with DMSO or tetanus toxin (TeTx) for 10 min before cLTP induction with glycine on DIV16. Neurons were fixed at different time points after glycine application and imaged by confocal microscopy. Scale bar, 5 µm. (B) Quantification of spine size in A. Data are expressed as mean ± SEM at each point (− TeTx: n = 12, n = 531 for 0 min; n = 12, n = 536 for 0.5 min; n = 15, n = 685 for 1 min; n = 13, n = 558 for 2 min; n = 12, n = 538 for 3 min; n = 15, n = 675 for 4 min; n = 14, n = 580 for 5 min + TeTx: n = 14, n = 561 for 0 min; n = 14, n = 546 for 0.5 min; n = 14, n = 549 for 1 min; n = 15, n = 642 for 2 min; n = 15, n = 687 for 3 min; n = 15, n = 698 for 4 min; n = 15, n = 588 for 5 min). P values were calculated using two-tailed unpaired t test. ***, P < 0.001. (C) Mouse hippocampal neurons were transfected with pLL3.7-DsRed and expression construct for GFP fusion of Rab11 WT or DN (S25N) mutant on DIV12 and pretreated with DMSO or MK801 before cLTP induction on DIV16. Neurons were fixed 1 min after glycine application and imaged by confocal microscopy. Scale bar, 5 µm. (D) Quantification of spine size in C. Data are expressed as mean ± SEM for each group (GFP: n = 10, n = 222 for − Gly; n = 11, n = 274 for + Gly; n = 10, n = 228 for + MK801 + Gly; Rab11-GFP: n = 11, n = 273 for − Gly; n = 10, n = 220 for + Gly; n = 11, n = 236 for + MK801 + Gly; Rab11DN-GFP: n = 10, n = 228 for − Gly; n = 11, n = 234 for + Gly; n = 11, n = 237 for + MK801 + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001. (E) Mouse hippocampal neurons transfected with pLL3.7-DsRed and expression construct for FLAG-tagged SNAP DN mutant on DIV12 and induced cLTP on DIV16. Neurons were fixed 1 min after glycine application, immunostained with antibodies to FLAG, and imaged by confocal microscopy. Scale bar, 5 µm. (F) Quantification of spine size in E. Data are expressed as mean ± SEM for each group (vector: n = 10, n = 378 for − Gly; n = 11, n = 391 for + Gly; SNAP23 DN: n = 11, n = 374 for − Gly; n = 11, n = 386 for + Gly; SNAP25 DN: n = 10, n = 364 for − Gly; n = 11, n = 394 for + Gly; SNAP29 DN: n = 11, n = 393 for − Gly; n = 11, n = 385 for + Gly; SNAP47 DN: n = 11, n = 379 for − Gly; n = 11, n = 382 for + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001. (G) Effect of osmotic shock on spine enlargement 1 min after glycine application. Neurons were pretreated with iso-osmotic, hypo-osmotic, or hyperosmotic solution, respectively, for 10 min, and cLTP was performed in the same solution on DIV16. Neurons were fixed 1 min after glycine application and imaged by 3D-SIM microscopy. Scale bar, 4 µm. (H) Quantification of spine size in G. Data are expressed as mean ± SEM for each group (iso: n = 14, n = 388 for − Gly; n = 12, n = 382 for + Gly; hypo: n = 14, n = 398 for − Gly; n = 15, n = 488 for + Gly; hyper: n = 14, n = 464 for − Gly; n = 13, n = 434 for + Gly). P values were calculated using two-tailed unpaired t test. ***, P < 0.001. (I) Effects of inhibitors for actin remodeling regulators on spine enlargement 1 min after glycine application. Shown are representative confocal images. Scale bar, 5 µm. CK-666, Arp2/3 inhibitor; SMIFH2, formin inhibitor; CT04, RhoA inhibitor; NSC 23766, Rac1 inhibitor; ML141, Cdc42 inhibitor. (J) Quantification of spine size (left) and changes in spine size (right) in I. Data are expressed as mean ± SEM for each group (Ctrl: n = 13, n = 523 for − Gly; n = 13, n = 537 for + Gly. LatA: n = 12, n = 516 for − Gly; n = 12, n = 519 for + Gly; CK-666: n = 13, n = 520 for − Gly; n = 13, n = 533 for + Gly; SMIFH2: n = 13, n = 552 for − Gly; n = 13, n = 561 for + Gly; CT04: n = 13, n = 546 for − Gly; n = 13, n = 522 for + Gly; NSC 23766: n = 13, n = 524 for − Gly; n = 14, n = 562 for + Gly; ML141: n = 13, n = 532 for − Gly; n = 13, n = 530 for + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001 when compared with Ctrl.
Electron microscopy and super-resolution imaging reveal that the surface of mature spines is convoluted (Arellano et al., 2007; Harris and Stevens, 1989; Smith et al., 2014). To determine whether the membrane folds/invaginations in spine head contribute to acute expansion, we increased membrane tension by exposing neurons to hypo-osmotic buffer and found that glycine-induced spine enlargement was abolished (Fig. S1, G and H). As membrane tension increases fusion efficiency (Kliesch et al., 2017), these findings corroborate that membrane fusion does not contribute to rapid spine expansion. Conversely, incubation of neurons with hyperosmotic buffer, which shrunk the spines and generated membrane folds, also antagonized cLTP-induced spine enlargement (Fig. S1, G and H). These data together suggest that the membrane supply for rapid spine enlargement in the acute phase of sLTP comes from local unfolding of spine surface convolutions.
The Ca2+/calmodulin‒CaMKII pathway triggers several signaling cascades to promote actin polymerization during synaptic potentiation (Murakoshi and Yasuda, 2012). In line with previous studies (Honkura et al., 2008; Matsuzaki et al., 2004), inhibition of actin polymerization with latrunculin A (LatA) abolished spine enlargement in the acute phase of sLTP (Fig. S1, I and J). Inhibition of Arp2/3, but not Formin, had a similar effect to that of LatA (Fig. S1, I and J), indicating that branched rather than linear actin polymerization is required for initial spine expansion. As endophilin A1 is required for both spine enlargement and actin polymerization in the acute phase of sLTP, we hypothesized that it promotes branched actin polymerization in spines to provide propulsive force for membrane expansion.
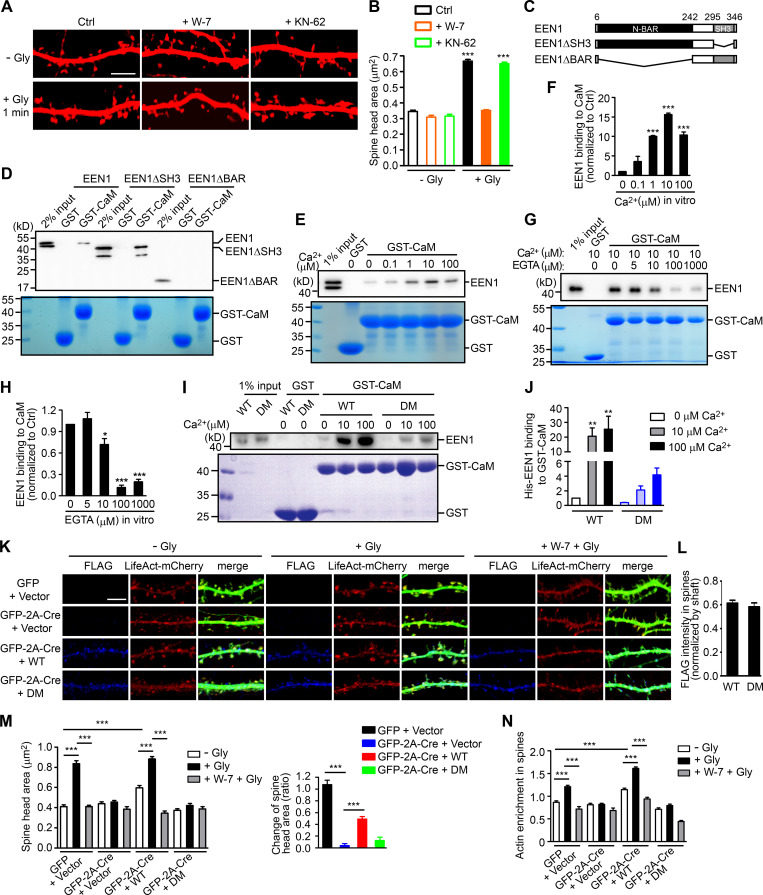
Glutamate uncaging-induced rapid spine enlargement requires NMDAR, calmodulin, and actin polymerization, whereas long-lasting size expansion also requires CaMKII (Matsuzaki et al., 2004). Similarly, inhibition of calmodulin, but not CaMKII, abolished the initial spine enlargement of hippocampal neurons undergoing cLTP (Fig. 2, A and B). We thus reasoned that endophilin A1 functions earlier than CaMKII in molecular events triggered by NMDAR-mediated Ca2+ influx upon LTP induction. Surprisingly, although Ca2+ binding of endophilin A2 inhibits its interaction with dynamin and voltage-gated Ca2+ channels (Chen et al., 2003), no direct binding between endophilin A1 and Ca2+ was detected by isothermal titration calorimetry (ITC) and fluorescence spectrometry (Fig. S2).
Figure 2.
Ca2+-dependent interaction between calmodulin and endophilin A1 is required for acute structural plasticity. (A) Effects of W-7 (calmodulin inhibitor) and KN-62 (CaMKII inhibitor) on spine enlargement 1 min after glycine application. Scale bar, 5 µm. (B) Quantification of spine size in A. Data are expressed as mean ± SEM for each group (control [Ctrl]: n = 12, n = 504 for − Gly; n = 14, n = 563 for + Gly. W-7: n = 13, n = 517 for − Gly; n = 18, n = 749 for + Gly; KN-62: n = 14, n = 526 for − Gly; n = 16, n = 661 for + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.0001. (C) Diagram showing the domain structure and fragments of endophilin A1 used in this study. (D) Binding of His-tagged endophilin A1 (EEN1) full-length, ΔSH3, and ΔBAR fragments to GST and calmodulin (CaM) in the GST pull-down assay. (E) Effect of Ca2+ on EEN1-CaM binding in the GST pull-down assay. (F) Quantification of EEN1 binding to calmodulin in E. n = 3 independent experiments. P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001 when compared with 0 µM Ca2+. (G) Effect of EGTA on EEN1-CaM binding in GST pull-down assay. (H) Quantification of EEN1-CaM binding in G. n = 5 independent experiments. P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. *, P < 0.05; ***, P < 0.001 when compared with 0 µM EGTA. (I) Effect of Ca2+ on binding of EEN1 DM mutant to calmodulin in GST pull-down assay. (J) Quantification of EEN1 DM binding to calmodulin compared with WT in I. The y axis shows two segments. n = 4 independent experiments. P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. **, P < 0.01 when compared with 0 µM Ca2+. (K) Cultured EEN1fl/fl hippocampal neurons cotransfected with LifeAct-mCherry, GFP or GFP-2A-Cre and FLAG vector, or LifeAct-mCherry, GFP-2A-Cre and FLAG-EEN1 WT or DM expression constructs on DIV12 were pretreated with DMSO or W-7 and induced cLTP on DIV16. Neurons were fixed 1 min after glycine application, immunostained for FLAG and imaged by confocal microscopy. Scale bar, 5 µm. (L) Spine/shaft distribution of EEN1 DM compared with WT. Data are expressed as mean ± SEM for each group (n = 11, n = 210 for WT; n = 10, n = 180 for DM). P values were calculated using two-tailed unpaired t test. (M) Quantification of spine size and changes in spine size in K. (N) Quantification of F-actin enrichment in spines in K. Data are expressed as mean ± SEM for each group in M and N (GFP + vector: n = 12, n = 230 for − Gly; n = 12, n = 223 for + Gly; n = 12, n = 246 for + W-7 + Gly. Cre + vector: n = 10, n = 187 for − Gly; n = 10, n = 183 for + Gly; n = 10, n = 181 for + W-7 + Gly. Cre + WT: n = 11, n = 210 for − Gly; n = 11, n = 209 for + Gly; n = 10, n = 189 for + W-7 + Gly. Cre + DM: n = 10, n = 180 for − Gly; n = 10, n = 187 for + Gly; n = 10, n = 192 for + W-7 + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001.
Figure S2.
Endophilin A1 does not bind Ca2+. (A) Purification of recombinant full-length EEN1 expressed in E. coli. Affinity-purified His-tagged EEN1 was supplied with 5 mM EDTA and loaded into Superdex 200 16/300 size exclusion chromatography with buffer (20 mM Tris-Cl, pH 7.0, and 150 mM NaCl). The peak fractions for EEN1 were collected and analyzed by SDS-PAGE. EEN1 displays as monomer and presents negligible degradation without affecting its function. (B) ITC analysis of 1 mM CaCl2 and 30 µM calmodulin. The raw data were fitted to sequential binding site model. The molar ratio of Ca2+ ions (II) to calmodulin is 4, as previously indicated. Ca2+ ions (II) appear to bind to calmodulin with dissociation constants (Kd) of 2.26 ± 0.31 µM, 9.08 ± 1.25 µM, 55.1 ± 11.4 µM, and 2.60 ± 2.05 mM, respectively. Each binding site reveals enthalpy changes (ΔH) of 0.22 ± 0.06 kcal/mol 3.31 ± 0.76 kcal/mol, −1.62 ± 0.14 kcal/mol, and 48.1 ± 4.3 kcal/mol, respectively. DP, power differential. (C) ITC analysis of 1 mM CaCl2, 40 µM EEN1 (left panels) and 3 mM CaCl2, 100 µM EEN1 (right panels). No significant binding was detected. (D and E) Tryptophan fluorescence of Synaptotagmin-1 C2B (positive control) and EEN1 in the presence of CaCl2. Shown are tryptophan fluorescence spectra (310–400 nm) of Synaptagmin-1 C2B (D) and EEN1 (E) respectively with an excitation wavelength of 280 nm. Right panels are enlarged views of the emission peaks in the left panels. The tryptophan fluorescence of Synaptagmin-1 C2B increases with the addition of CaCl2 and saturates at 6 mM final concentration. In contrast, the tryptophan fluorescence of EEN1 fluctuates irregularly with the addition of CaCl2.
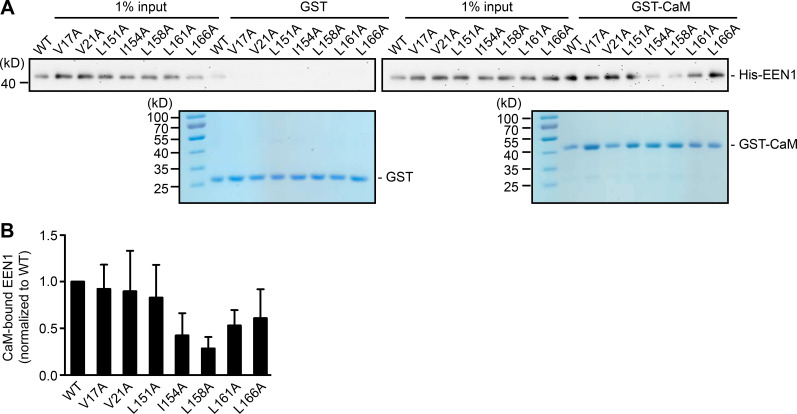
Recent studies found that calmodulin binds to N-BAR proteins, including endophilin A1 and A2 (Myers et al., 2016). Indeed, a GST pull-down assay showed that endophilin A1 binds to calmodulin via its N-BAR domain, and the interaction is strengthened by Ca2+ (Fig. 2, C–H). To determine whether Ca2+/calmodulin regulates its function in plasticity, we generated a calmodulin binding–deficient mutant of endophilin A1 (I154AL158A, hereafter DM) that has much lower affinity for calmodulin and is less responsive to Ca2+ than WT (Fig. 2, I and J; Fig. S3). Indeed, in EEN1 KO neurons, WT but not the DM mutant restored the rapid increase in spine size and F-actin content in spines upon LTP induction (Fig. 2, K–N; and Fig. S4, A–C). Together, these data indicate that Ca2+-regulated calmodulin–endophilin A1 interaction is required for acute structural plasticity.
Figure S3.
I154 and L158 are required for endophilin A1 binding to calmodulin. (A) Binding of EEN1 WT and mutants to GST-calmodulin in pull-down assay. Bound proteins were analyzed by SDS-PAGE and immunoblotting with anti-His antibodies. (B) Quantification of mutants binding to GST-calmodulin in A. Data are represented as mean ± SEM, n = 5 independent experiments. P values were calculated using one-way ANOVA by Dunnett post hoc test.
Figure S4.
Rescue of acute sLTP in EEN1 KO neurons by EEN1 mutants. (A) Cultured EEN1+/+ and EEN1−/− hippocampal neurons cotransfected with LifeAct-mCherry and FLAG vector and EEN1−/− hippocampal neurons cotransfected with LifeAct-mCherry and FLAG-EEN1 WT or DM on DIV12 were pretreated with DMSO or W-7 and induced cLTP on DIV16. Neurons were fixed 1 min after glycine application and imaged by confocal microscopy. Scale bar, 5 µm. (B) Quantification of spine size and changes in spine size in A. (C) Quantification of F-actin enrichment in spines in A. Data are expressed as mean ± SEM for each group in B and C (EEN1+/+: n = 12, n = 472 for − Gly; n = 12, n = 483 for + Gly; n = 12, n = 450 for + W-7 + Gly; EEN1−/−: n = 12, n = 458 for − Gly; n = 12, n = 464 for + Gly; n = 12, n = 461 for + W-7 + Gly; EEN1−/− + WT: n = 12, n = 475 for − Gly; n = 12, n = 466 for + Gly; n = 12, n = 453 for + W-7 + Gly; EEN1−/− + DM: n = 12, n = 467 for − Gly; n = 13, n = 487 for + Gly; n = 12, n = 456 for + W-7 + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001. (D) Cultured EEN1+/+ and EEN1−/− hippocampal neurons cotransfected with pLL3.7-DsRed and FLAG vector and EEN1−/− hippocampal neurons cotransfected with pLL3.7-DsRed and FLAG-EEN1 WT or Y343A on DIV12 were pretreated with DMSO or MK801 and induced cLTP on DIV16. Neurons were fixed 1 min after glycine application and imaged by confocal microscopy. Scale bar, 5 µm. (E) Quantification of spine size and changes in spine size in D. Data are expressed as mean ± SEM for each group (EEN1+/+: n = 15, n = 528 for − Gly; n = 15, n = 574 for + Gly; n = 14, n = 542 for + MK801 + Gly; EEN1−/−: n = 13, n = 522 for − Gly; n = 13, n = 564 for + Gly; n = 12, n = 502 for + MK801 + Gly; EEN1−/− + WT: n = 13, n = 515 for − Gly; n = 13, n = 526 for + Gly; n = 13, n = 523 for + MK801 + Gly; EEN1−/− + Y343A: n = 13, n = 527 for − Gly; n = 13, n = 512 for + Gly; n = 13, n = 504 for + MK801 + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001. (F) Same as D, except that the mutant was KKK-EEE. Scale bar, 5 µm. (G) Quantification of spine size or changes in spine size in F. Data are expressed as mean ± SEM for each group (EEN1+/+: n = 15, n = 528 for − Gly; n = 15, n = 574 for + Gly; n = 14, n = 542 for + MK801 + Gly; EEN1−/−: n = 13, n = 522 for − Gly; n = 13, n = 564 for + Gly; n = 12, n = 502 for + MK801 + Gly; EEN1−/− + WT: n = 13, n = 515 for − Gly; n = 13, n = 526 for + Gly; n = 13, n = 523 for + MK801 + Gly; EEN1−/− + KKK-EEE: n = 12, n = 482 for − Gly; n = 12, n = 491 for + Gly; n = 12, n = 489 for + MK801 + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001.
Ca2+/calmodulin-enhanced endophilin A1-p140Cap interaction is required for initiation of sLTP
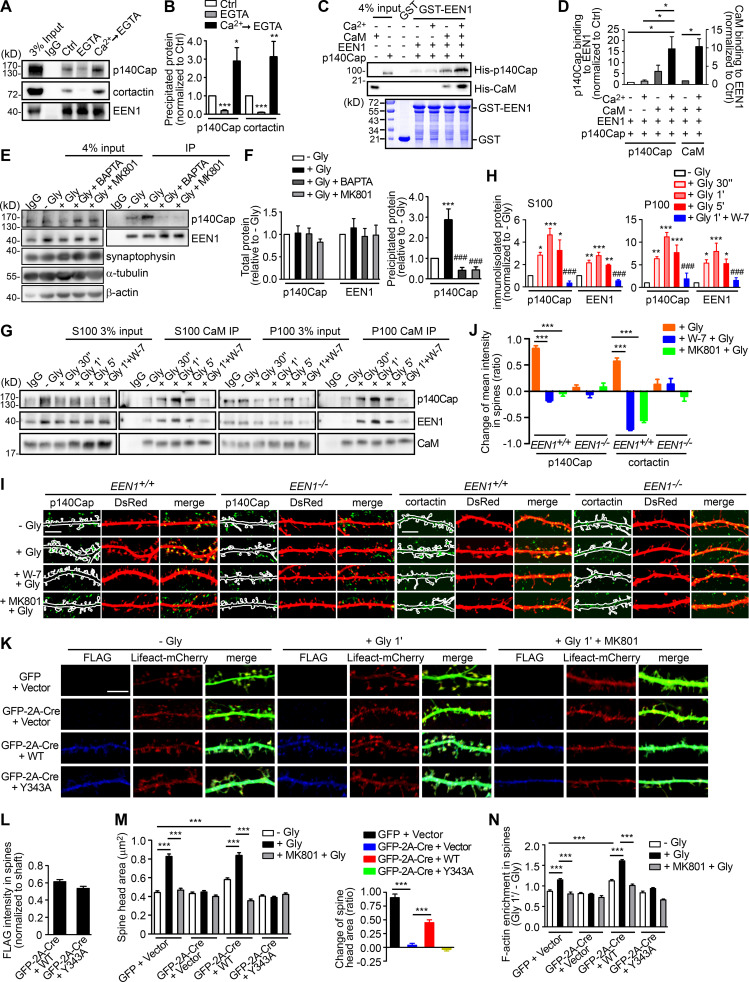
Next, we investigated regulatory mechanisms underlying actin polymerization in spines in the acute phase of sLTP. The Rho GTPases are known effectors of Ca2+/calmodulin and regulators of actin reorganization and structural plasticity (Hedrick and Yasuda, 2017; Spiering and Hodgson, 2011). In agreement with previous findings (Hedrick et al., 2016; Murakoshi et al., 2011), inhibition of RhoA, which stabilizes polymerized actin via inhibition of the actin severing factor ADF/cofilin, abolished glycine-induced rapid spine enlargement (Fig. S1, I and J). Notably, inhibition of Rac1, which promotes actin polymerization via the WAVE-Arp2/3 pathway, only partially inhibited acute spine expansion (Fig. S1, I and J), suggesting the presence of other positive regulators for branched actin polymerization upon sLTP induction. Given that ablation of endophilin A1 inhibits increase in F-actin content in spines, we reasoned that Ca2+/calmodulin might regulate the interaction between endophilin A1 and p140Cap, which recruits cortactin to drive Arp2/3-mediated branched actin polymerization (Jaworski et al., 2009; Schnoor et al., 2018; Yang et al., 2015). Indeed, coimmunoprecipitation (coIP) from mouse brain lysates showed that the interaction between endophilin A1 and p140Cap is Ca2+ dependent (Fig. 3, A and B). Moreover, calmodulin enhanced their interaction in a Ca2+- and NMDAR-dependent manner (Fig. 3, C–F). Further, in both cytosolic and membrane fractions of cultured neurons, the association of endophilin A1 with not only calmodulin but also p140Cap was enhanced acutely upon LTP induction, which was abolished by the calmodulin inhibitor W-7 (Fig. 3, G and H). Together, these data indicate that via either binding-induced conformational change or posttranslational modification, Ca2+/calmodulin enhances the interaction of endophilin A1 with p140Cap in the acute phase of LTP.
Figure 3.
Ca2+/calmodulin promotes rapid spine enlargement via the endophilin A1-p140Cap pathway. (A) Effect of EGTA or Ca2+ on binding of EEN1 to p140Cap and cortactin. Endogenous immunoprecipitation assay was performed from mouse brain lysates with antibodies to EEN1. To mimic a transient increase in intracellular Ca2+, CaCl2 (1 mM) was added to lysates for 10 min on ice, followed by incubation with EGTA (1 mM) to chelate Ca2+ (Ca2+→EGTA). Lysates only (Ctrl) and lysates with EGTA serve as negative control. (B) Quantification of EEN1 binding to p140Cap and cortactin in A. n = 5 independent experiments. P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 when compared with Ctrl. (C) Effect of Ca2+/calmodulin on EEN1-p140Cap binding in GST pull-down assay. (D) Quantification of p140Cap and calmodulin binding to EEN1 in C. n = 3 independent experiments. P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. *, P < 0.05. (E) Effect of BAPTA or MK801 on EEN1-p140Cap binding in neurons upon cLTP induction. (F) Total protein levels of EEN1 and p140Cap and quantification of EEN1-p140Cap binding in E. n = 5 independent experiments. P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001 when compared with − Gly; ###, P < 0.001 when compared with + Gly 1 min. (G) Effect of W-7 on interactions between calmodulin and EEN1/p140Cap upon cLTP induction. DIV16 neurons were collected, and the cytosolic (S100) and membrane (P100) fractions were used for immunoisolation with antibodies to calmodulin. (H) Quantification of EEN1 and p140Cap immunoisolated by anti-calmodulin in G. n = 5 independent experiments. P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 when compared with − Gly; ###, P < 0.001 when compared with + Gly 1 min. (I) Neurons expressing DsRed were pretreated with DMSO, W-7, or MK801 and induced cLTP on DIV16 with glycine for 1 min, and immunostained for p140Cap or cortactin (green) and imaged by confocal microscopy. Scale bars, 5 µm. (J) Quantification of changes in p140Cap and cortactin signal intensities in spines compared with the control (− Gly) group in I. Data are expressed as mean ± SEM for each group. For p140Cap, EEN1+/+: n = 11, n = 431 for − Gly; n = 15, n = 482 for + Gly; n = 14, n = 437 for + W-7 + Gly; n = 14, n = 429 for + MK801 + Gly; EEN−/−: n = 12, n = 447 for − Gly; n = 14, n = 473 for + Gly; n = 14, n = 448 for + W-7 + Gly; n = 14, n = 445 for + MK801 + Gly. For cortactin, EEN1+/+: n = 12, n = 446 for − Gly; n = 15, n = 474 for + Gly; n = 14, n = 451 for + W-7 + Gly; n = 14, n = 443 for + MK801 + Gly; EEN−/−: n = 14, n = 456 for − Gly; n = 15, n = 473 for + Gly; n = 14, n = 464 for + W-7 + Gly; n = 13, n = 435 for + MK801 + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001. (K) Cultured EEN1fl/fl hippocampal neurons cotransfected with LifeAct-mCherry, GFP or GFP-2A-Cre and FLAG vector, or LifeAct-mCherry, GFP-2A-Cre, and FLAG-EEN1 WT or Y343A expression constructs on DIV12 were pretreated with DMSO or MK801 and induced cLTP on DIV16. Neurons were fixed 1 min after glycine application, immunostained for FLAG, and imaged by confocal microscopy. Scale bar, 5 µm. (L) Spine/shaft distribution of EEN1 Y343A compared with WT. Data are expressed as mean ± SEM for each group (n = 10, n = 202 for Cre + WT; n = 10, n = 190 for Cre + Y343A). P values were calculated using two-tailed unpaired t test. (M) Quantification of spine size and changes in spine size in K. (N) Quantification of F-actin enrichment in spines in K. Data are expressed as mean ± SEM for each group in M and N (GFP + vector: n = 11, n = 205 for − Gly; n = 11, n = 213 for + Gly; n = 11, n = 227 for + MK801 + Gly. Cre + Vector: n = 11, n = 197 for − Gly; n = 11, n = 194 for + Gly; n = 11, n = 195 for + MK801 + Gly; Cre + WT: n = 10, n = 202 for − Gly; n = 11, n = 226 for + Gly; n = 10, n = 193 for + MK801 + Gly; Cre + Y343A: n = 10, n = 190 for − Gly; n = 11, n = 207 for + Gly; n = 10, n = 196 for + MK801 + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001.
Notably, coIP of not only p140Cap but also cortactin by anti-endophilin A1 antibody was enhanced by Ca2+ (Fig. 3, A and B). To test the idea that endophilin A1 functions via p140Cap and cortactin in acute sLTP, we first determined whether they are recruited to dendritic spines in a neural activity– and endophilin A1–dependent manner. Quantitative analysis of immunofluorescence confocal images indicated that enrichment of p140Cap and cortactin in spines upon LTP induction requires not only endophilin A1 but also activities of calmodulin and NMDAR (Fig. 3, I and J). Further, the p140Cap-binding deficient Y343A mutant of endophilin A1 (Yang et al., 2015) failed to rescue the sLTP phenotypes of EEN1 KO neurons (Fig. 3, K–N; and Fig. S4, D and E). Thus, Ca2+/calmodulin-enhanced endophilin A1–p140Cap interaction is required for acute structural plasticity.
Ca2+/calmodulin enhances association of endophilin A1 with spine PM in the acute phase of sLTP
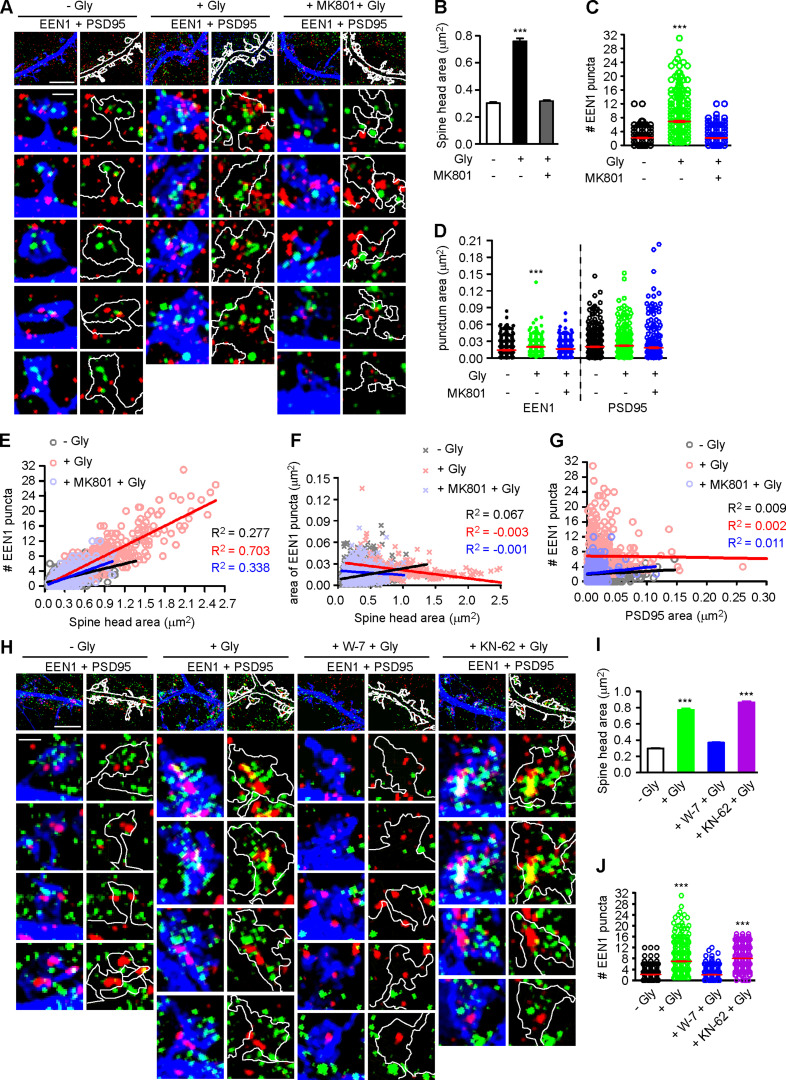
The nanoscale organization of branched F-actin regulators change dynamically in spines during synaptic plasticity (Chazeau et al., 2014). Intriguingly, although the interaction between endophilin A1 and p140Cap is required for sLTP, overexpression of p140Cap could not rescue plasticity phenotypes of EEN1 KO neurons (Yang et al., 2018), suggesting spatiotemporal regulation of their interaction during LTP induction. To this end, we analyzed the subspine localization of endophilin A1 by immunostaining and 3D structured illumination microscopy (3D-SIM). Interestingly, in spines endophilin A1 was organized into nanoscale objects (mean area 0.014 µm2) which did not overlap with PSD95, a marker for postsynaptic density structure (Fig. S5 A). Quantitative analysis revealed an NMDAR-dependent increase in the number of endophilin A1 puncta in spines undergoing sLTP (Fig. S5, A–C). Moreover, the size of spine head correlated with the number but not the area of individual endophilin A1 puncta (Fig. S5, E and F). In contrast, the number of endophilin A1 puncta did not correlate with the size of postsynaptic density structures (Fig. S5, D and G). Further, inhibition of calmodulin but not CaMKII abolished the increase in not only spine size but also the number of endophilin A1 puncta in spines (Fig. S5, H–J). These data together indicate that calmodulin-regulated subsynaptic localization of endophilin A1 correlates with spine enlargement.
Figure S5.
Calmodulin-dependent increase in the number of endophilin A1 puncta correlates with spine size during the acute phase of sLTP. (A) Cultured hippocampal neurons transfected with pLL3.7-DsRed on DIV12 were pretreated with DMSO or MK801 for 30 min before cLTP induction with glycine for 5 min on DIV16, immunostained for EEN1 (pseudocolored green) and PSD95 (pseudocolored red), and imaged by 3D-SIM. Shown are representative images with DsRed pseudocolored blue. Spines were outlined manually. Scale bars represent 4 µm in upper panels and 500 nm in magnified images. (B) Quantification of spine size in A. (C) Quantification of the number of EEN1 puncta in spines in A. (D) Quantification of the area of EEN1 or PSD95 puncta in spines in A. Data are expressed as mean ± SEM or plotted along with mean ± SEM for each group in C and D (n = 13, n = 570 for − Gly; n = 13, n = 502 for + Gly; n = 14, n = 543 for + MK801 + Gly). P values were calculated using one-way ANOVA by Dunnett post hoc test. ***, P < 0.001 when compared with − Gly. (E and F) Scatterplot of the number or area of EEN1 puncta versus the size of spine head with linear fits using linear regression analysis. (G) Scatterplot of the number of EEN1 puncta versus the area of PSD95 puncta in spines with linear fits using linear regression analysis. In E–G, n = 13, n = 570 for − Gly; n = 13, n = 502 for + Gly; n = 14, n = 543 for + MK801 + Gly. (H) Effect of W-7 or KN-62 on EEN1 puncta in spines. Neurons were pretreated with DMSO, W-7 or KN-62, before cLTP induction with glycine for 5 min on DIV16, immunostained and imaged by 3D-SIM. Shown are representative images. Scale bars represent 4 µm in upper panels and 500 nm in magnified images. (I) Quantification of spine size in H. (J) Quantification of the number of EEN1 puncta in spines in H. Data are expressed as mean ± SEM or plotted along with mean ± SEM for each group in I and J (n = 14, n = 577 for − Gly; n = 14, n = 596 for + Gly; n = 14, n = 588 for + W-7 + Gly; n = 14, n = 540 for + KN-62 + Gly). P values were calculated using one-way ANOVA by Dunnett post hoc test. ***, P < 0.001 when compared with − Gly.
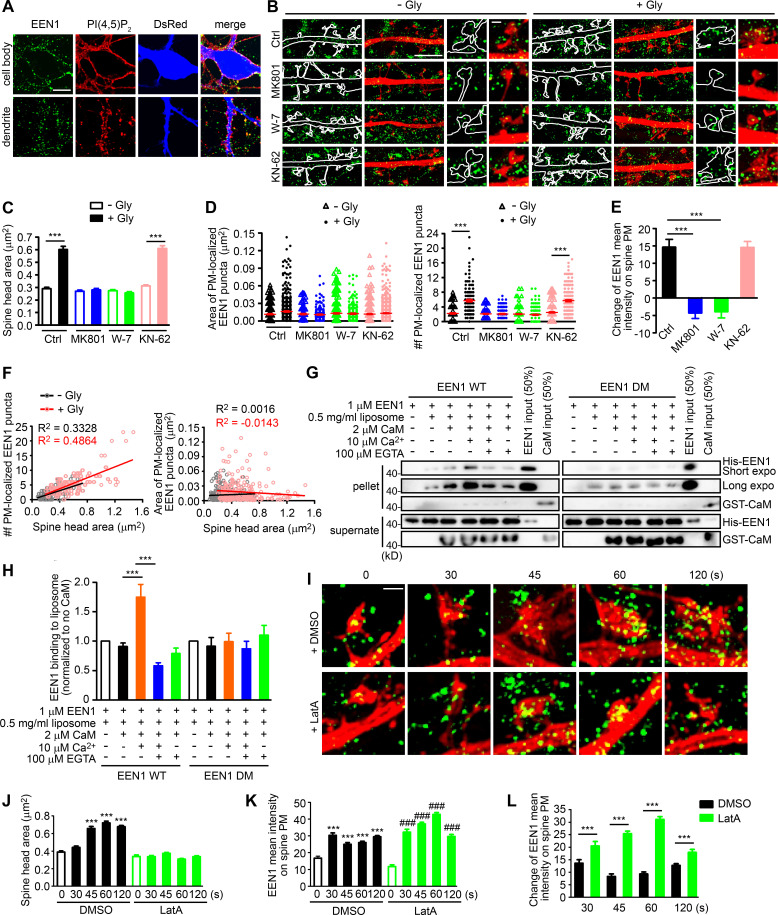
As endophilin A1 contains the positive membrane curvature–sensing and binding N-BAR domain (Gallop et al., 2006) that enables its association with PM invaginations, next we investigated whether its association with spine PM is also regulated in the acute phase of sLTP. To distinguish PM-localized endophilin A1 from those localized to endomembrane structures, we permeabilized cell membrane gently with the mild detergent saponin to limit access of antibodies to the cytosolic leaflet of the PM (Hammond et al., 2009; Fig. 4 A). Indeed, SIM imaging detected an increase in the number of spine PM-localized endophilin A1 puncta as early as 1 min after LTP induction in a NMDAR- and calmodulin-dependent manner (Fig. 4, B and D). Further, the number but not area of the puncta correlated with the size of spine head (Fig. 4, C and F). As endophilin A1 can recruit p140Cap and its effector cortactin (Yang et al., 2015), these data suggest that upon LTP induction, endophilin A1 rapidly localizes to the inner surface of spine head, most likely by association with folds or invaginations of the PM via its N-BAR domain, and promotes local Arp2/3-mediated actin polymerization underneath.
Figure 4.
Ca2+/calmodulin-enhanced PM association of endophilin A1 correlates with spine expansion. (A) Detection of PM-localized endophilin A1. DIV16 hippocampal neurons were immunostained for PM-localized EEN1 and PI(4,5)P2 (+ control). The volume marker DsRed is pseudocolored blue. Scale bar, 10 µm. (B) DIV16 neurons expressing DsRed were pretreated with DMSO, MK801, W-7, or KN-62 and induced cLTP with glycine for 1 min, immunostained for PM-localized EEN1 (green), and imaged by 3D-SIM. Scale bars represent 4 µm in the left and center panels and 500 nm in magnified images in the right panels. (C) Quantification of spine size in B. (D) Quantification of the area and number of individual spine PM-localized EEN1 puncta in B. Data are expressed as mean ± SEM or plotted along with mean ± SEM for each group in C and D (Ctrl: n = 12, n = 278 for − Gly; n = 11, n = 254 for + Gly; MK801: n = 12, n = 256 for − Gly; n = 12, n = 263 for + Gly; W-7: n = 13, n = 295 for − Gly; n = 10, n = 278 for + Gly; KN-62: n = 13, n = 317 for − Gly; n = 12, n = 268 for + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001. (E) Quantification of changes in the amount of spine PM-localized EEN1 in B. Data are expressed as mean ± SEM for each group (n = 11, n = 254 for Ctrl + Gly; n = 12, n = 263 for MK801 + Gly; n = 10, n = 278 for W-7 + Gly; n = 12, n = 268 for KN-62 + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001. (F) Scatterplot of the number or area of PM-localized EEN1 puncta versus the size of spine head for control (− Gly, n = 278 spines) and cLTP (+ Gly, n = 254 spines) groups with linear fits using linear regression analysis. (G) Effect of Ca2+/calmodulin on binding of EEN1 WT or DM protein to curved membrane in liposome (50 nm in diameter) cosedimentation assay. (H) Quantification of EEN1 binding to liposome in F. n = 7 independent experiments. P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001. (I) DIV16 neurons were pretreated with DMSO or LatA, and cLTP was performed. Cells were fixed at 30, 45, 60, or 120 s after glycine application. PM-localized EEN1 was stained and imaged by 3D-SIM. Scale bar, 500 nm. (J) Quantification of spine size in I. (K) Quantification of spine PM-localized EEN1 in I. (L) Quantification of changes in the amount of spine PM-localized EEN1 in I. Data are expressed as mean ± SEM for each group in J–L (DMSO: n = 10, n = 241 for 0 s; n = 11, n = 246 for 30 s; n = 11, n = 252 for 45 s; n = 14, n = 360 for 60 s; n = 14, n = 358 for 120 s; LatA: n = 10, n = 254 for 0 s; n = 11, n = 269 for 30 s; n = 14, n = 368 for 45 s; n = 15, n = 407 for 60 s; n = 14, n = 306 for 120 s). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001 when compared with DMSO 0 s; ###, P < 0.001 when compared with LatA 0 s in J and K. ***, P < 0.001 in L.
Given that subspine and PM accumulation of endophilin A1 requires activation of NMDAR and calmodulin (Fig. 4, B, D, and E; and Fig. S5), we reasoned that binding of Ca2+/calmodulin to endophilin A1 enhances not only its interaction with p140Cap but also its association with the PM. Indeed, in vitro liposome cosedimentation assays revealed that while calmodulin alone did not change the membrane-binding capacity of WT endophilin A1, Ca2+/calmodulin enhanced it significantly (Fig. 4, G and H). In contrast, Ca2+/calmodulin had no effect on the membrane-association ability of the calmodulin binding–deficient DM mutant (Fig. 4, G and H). Notably, although LatA treatment inhibited spine enlargement, it had no effect on LTP-induced increase in PM-localized endophilin A1 (Fig. 4, I–L), indicating that Ca2+/calmodulin-enhanced association of endophilin A1 with the PM precedes actin polymerization during sLTP initiation. Collectively, these data indicate that endophilin A1 associates with the spine PM in response to Ca2+/calmodulin in the acute phase of sLTP.
The membrane- and p140Cap-binding capacities of endophilin A1 coordinate to recruit Arp2/3 to the PM in response to Ca2+/calmodulin
Based on the findings that endophilin A1 associates with spine PM and recruits p140Cap and cortactin in the acute phase of sLTP, we further reasoned that it might transduce the Ca2+ signals instantaneously to enable spine enlargement by promoting branched actin polymerization underneath the PM. If it is true, we should be able to detect more endophilin A1 associated with both membranes and the actin cytoskeleton upon LTP induction. To test this possibility, first we analyzed subcellular fractions of mouse hippocampi from animals subjected to fear conditioning, a physiological learning paradigm associated with synaptic plasticity. Compared with naive mice, there was indeed significant increase in the amount of endophilin A1 and p140Cap in both membrane and cytoskeletal fractions from trained animals (Fig. 5, A and B). Consistently, although the total levels of either protein remained unchanged (Fig. 5, C and D), their association with membrane and cytoskeleton also increased in cultured hippocampal neurons in the acute phase of NMDAR-mediated cLTP (Fig. 5, E and F). Further, inhibition of calmodulin, not CaMKII, abolished the enhanced association of endophilin A1 and p140Cap with both membrane and cytoskeleton (Fig. 5, E and F). Notably, subcellular distribution of endophilin A2 was not affected by either neural activity or Ca2+/calmodulin (Fig. 5, A–F).
Figure 5.
Ca2+/calmodulin enhances the association of endophilin A1 and p140Cap with both membrane and cytoskeleton upon LTP induction. (A) Immunoblotting of subcellular fractionations of mouse hippocampi from naive or fear conditioned animals. (B) Quantification of protein levels of p140Cap, EEN1, or A2 in cytoplasmic, membrane, and cytoskeleton fractions in A. n = 6 animals for each group. P values were calculated using two-tailed paired t test. *, P < 0.05; **, P < 0.01 when compared with Ctrl. (C) DIV16 neurons were pretreated with DMSO, MK801, W-7, or KN-62 and induced cLTP with glycine for 1 min, lysed, and subjected to SDS-PAGE and immunoblotting. (D) Quantification of total protein levels in C. n = 5 independent experiments. (E) Effects of MK801, W-7, and KN-62 on subcellular distribution of proteins. DIV16 neurons were pretreated with DMSO, MK801, W-7, or KN-62 and induced cLTP with glycine for 1 min, collected, and subjected to subcellular fractionation. (F) Quantification of protein levels in subcellular fractions in E. n = 5 independent experiments. P values were calculated using one-way ANOVA by Dunnett post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 when compared with − Gly. (G) EEN1fl/fl hippocampal neurons cotransfected with GFP or GFP-2A-Cre and FLAG vector or GFP-2A-Cre and FLAG-EEN1 WT or KKK-EEE expression constructs on DIV12 were pretreated with DMSO or MK801 and induced cLTP on DIV16 with glycine for 1 min, immunostained for FLAG, and imaged by confocal microscopy. Scale bar, 5 µm. (H) Spine/shaft distribution of EEN1 KKK-EEE compared with WT. Data are expressed as mean ± SEM for each group (n = 10, n = 202 for Cre + WT; n = 11, n = 216 for Cre + KKK-EEE). P values were calculated using two-tailed unpaired t test. (I) Quantification of spine size and changes in spine size in G. Data are expressed as mean ± SEM for each group (GFP + vector: n = 11, n = 205 for − Gly; n = 11, n = 213 for + Gly; n = 11, n = 227 for + MK801 + Gly; Cre + vector: n = 11, n = 197 for − Gly; n = 11, n = 194 for + Gly; n = 11, n = 195 for + MK801 + Gly; Cre + WT: n = 10, n = 202 for − Gly; n = 11, n = 226 for + Gly; n = 10, n = 193 for + MK801 + Gly; Cre + KKK-EEE: n = 11, n = 216 for − Gly; n = 11, n = 211 for + Gly; n = 10, n = 190 for + MK801 + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001.
To test the hypothesis that endophilin A1 associates with the PM and promotes local actin polymerization to drive membrane expansion, we then determined whether the membrane-binding capacity of endophilin A1 is required for the rapid spine enlargement of neurons undergoing sLTP. Compared with WT, the membrane binding–deficient KKK-EEE mutant of endophilin A1 (Gallop et al., 2006) failed to rescue the sLTP phenotype of EEN1 KO neurons (Fig. 5, G–I; and Fig. S4, F and G). Altogether, these data indicate that acute structural plasticity requires Ca2+/calmodulin-dependent enhancement of not only endophilin A1–p140Cap interaction but also the association of endophilin A1 with the membrane.
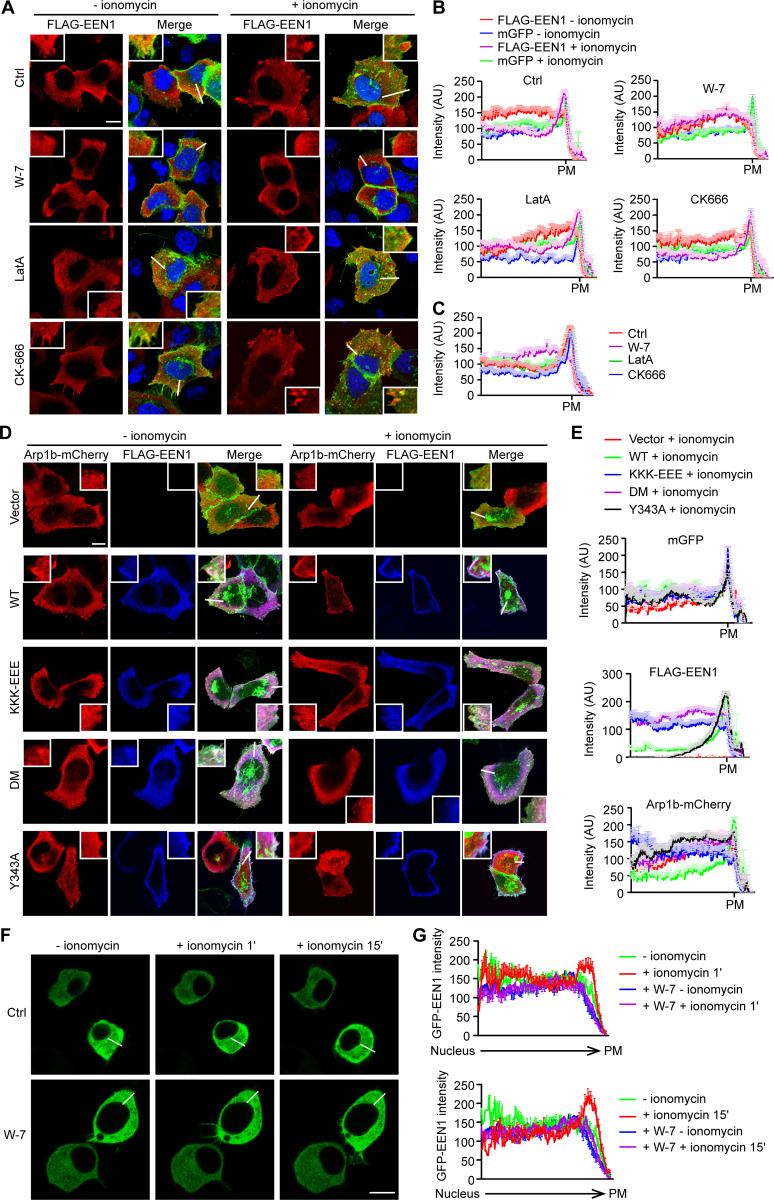
As the small size of dendritic spines and the wide distribution of F-actin and actin polymerization regulators in spines prevent us from better visualizing the spatiotemporal relationship between PM association of endophilin A1 and its effectors, we tested whether Ca2+/calmodulin regulates the subcellular distribution of endophilin A1 and Arp2/3 using HeLa cells as a heterologous model system. In HeLa cells ectopically coexpressing endophilin A1 and p140Cap, upon Ca2+ influx induced by the ionophore ionomycin, we detected enrichment of endophilin A1 signals underneath the PM by confocal microscopy (Fig. 6, A–C). Moreover, consistent with findings that Ca2+/calmodulin directly regulates accumulation of endophilin A1 at the PM (Fig. 4), inhibition of calmodulin but not actin polymerization abolished ionomycin-induced endophilin A1 recruitment to the cell periphery (Fig. 6, A–C). Further, although the strong intrinsic signals for cortical actin did not allow us to quantify changes in F-actin content underneath the PM, the Arp2/3 complex (labeled with fluorescently tagged Arp1b) was also enriched at the cell periphery upon ionomycin application (Fig. 6, D and E). In contrast, ionomycin treatment failed to cause recruitment of either the membrane binding– or calmodulin binding–deficient mutant of endophilin A1 to the cell periphery (Fig. 6, D and E). Intriguingly, although ionomycin application increased PM accumulation of the p140Cap-binding mutant Y343A, no enrichment of Arp1b signals at the cell periphery was detected (Fig. 6, D and E), indicating that PM recruitment of Arp2/3 requires endophilin A1–p140Cap interaction.
Figure 6.
In response to Ca2+/calmodulin, endophilin A1 associates with the PM and recruits Arp2/3 via p140Cap. (A) HeLa cells were cotransfected with constructs expressing mGFP, myc-p140Cap, and FLAG-EEN1 for 24 h; preincubated with DMSO, W-7, LatA, or CK-666 for 30 min; treated with ionomycin for 20 min; and immunostained for FLAG for confocal microscopy. Shown are representative images. Magnified images are shown in white boxes. Scale bar, 10 µm. (B) Quantification of subcellular distribution of FLAG-EEN1 and mGFP in cells by linescan plots of the fluorescence intensity along the white lines indicated in the images in A. AU, arbitrary units. Ctrl: n = 34 for − ionomycin, n = 39 for + ionomycin; W-7: n = 35 for − ionomycin, n = 38 for + ionomycin; LatA: n = 30 for − ionomycin, n = 34 for + ionomycin; CK666: n = 34 for − ionomycin, n = 39 for + ionomycin. (C) Quantification of FLAG-EEN1 distribution in ionomycin-treated cells in B. n = 39 for Ctrl, n = 38 for W-7, n = 34 for LatA, n = 39 for CK666. (D) HeLa cells were cotransfected with constructs expressing mGFP, myc-p140Cap, Arp1b-mCherry and FLAG-EEN1 WT, Y343A, KKK-EEE, or DM for 24 h, treated with ionomycin for 20 min, and immunostained for FLAG for confocal microscopy. Scale bar, 10 µm. (E) Quantification of subcellular distribution of mGFP, FLAG-EEN1 and Arp1b-mCherry in cells in D. n = 32 for vector, n = 31 for WT, n = 31 for KKK-EEE, n = 32 for DM, and n = 31 for Y343A. (F) Neuro-2a cells expressing EGFP-EEN1 were imaged live by confocal microscopy before and after ionomycin application. Shown are still images of Neuro-2a cells at 0, 1, and 15 min of ionomycin treatment. (G) Quantification of EGFP-EEN1 distribution by linescan plots of fluorescence intensity along the white lines indicated in the images in F. Ctrl: 9 cells; W-7: 11 cells. Scale bar, 10 µm.
To corroborate that endophilin A1 associates with the PM in response to Ca2+/calmodulin, we performed live-cell imaging of EGFP-labeled endophilin A1 in Neuro-2a, a mouse neuroblastoma-derived cell line that has been extensively used for neuroscience research. Indeed, ionomycin treatment induced rapid accumulation of endophilin A1 to the cell periphery (Video 8 and Fig. 6, F and G), which was abolished by W-7 (Video 9 and Fig. 6, F and G). Together, these data support the idea that in response to Ca2+/calmodulin, endophilin A1 associates with the PM and promotes branched actin polymerization to initiate spine membrane expansion.
Video 8.
Accumulation of endophilin A1 at the cell periphery in ionomycin-treated Neuro-2a cells. Neuro-2a cells transiently transfected with construct expressing EGFP-EEN1 were imaged by confocal microscopy every minute for 2 or 3 frames before, and imaged every 10 s to 1 min for 25 frames after, ionomycin (1 µM) application. Video plays at 7 frames/s. Scale bar, 10 µm.
Video 9.
W-7 inhibits ionomycin-induced accumulation of endophilin A1 at the cell periphery in Neuro-2a cells. Neuro-2a cells transiently transfected with construct expressing EGFP-EEN1 were treated with W-7 (20 µM) for 20 min and imaged by confocal microscopy every minute for 2 frames, followed by ionomycin (1 µM) addition and continued imaging every 10 s to 1 min for 25 frames. Video plays at 7 frames/s. Scale bar, 10 µm.
Ca2+/calmodulin-regulated functions of endophilin A1 are required for LTP and long-term memory
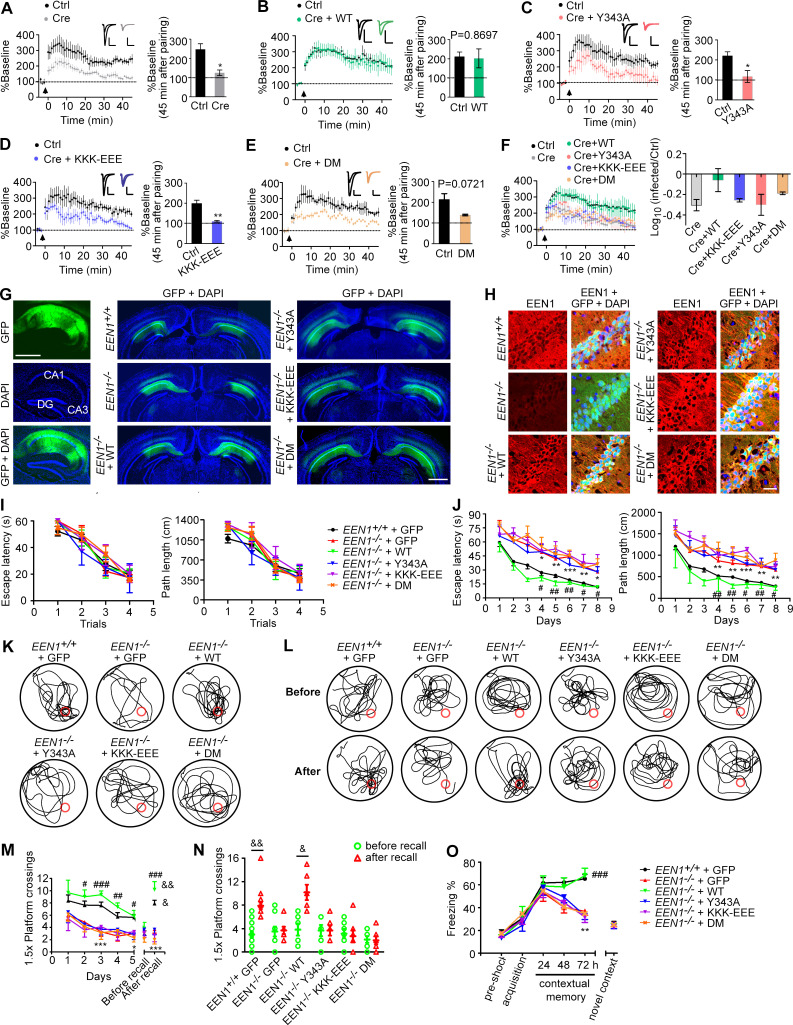
It was proposed that spine enlargement enables formation of a stable F-actin–cofilin complex that serves as a synaptic tag to capture postsynaptic constituent proteins for maintenance and consolidation of the potentiated state (Bosch et al., 2014). To determine the functional significance of endophilin A1–mediated structural plasticity, we next tested whether its molecular functions in activity-induced rapid spine growth are also required for LTP by electrophysiological analyses. To this end, we performed molecular replacement in a small subset of neurons by injection of the hippocampal CA1 region of EEN1fl/fl mice with adeno-associated virus (AAV) vectors encoding the Cre recombinase together with those encoding either WT or mutant endophilin A1 (Y343A, KKK-EEE, or DM) at postnatal day 0 (P0) and induced LTP in SC synapses by double patch whole-cell recording of noninfected and virus-infected CA1 pyramidal cells in acute slices from virus-injected animals at P14–P21. Consistently, Cre-mediated KO of EEN1 caused significant impairment of LTP (Yang et al., 2018), which was fully rescued by reexpression of WT endophilin A1 (Fig. 7, A, B, and F). In contrast, neither the Y343A nor the KKK-EEE mutant could restore the magnitude or the maintenance of LTP in KO neurons, and KO neurons expressing the DM mutant did not exhibit normal LTP with a P value of 0.0721, probably reflecting variations among individual cells recorded (Fig. 7, C–F). Together, the electrophysiological data indicate that molecular functions of endophilin A1 to initiate sLTP are required for expression and stabilization of synaptic potentiation.
Figure 7.
The calmodulin-, membrane-, and p140Cap-binding capacities of endophilin A1 are required for LTP and long-term memory. (A–F) Rescue of the LTP impairment phenotype in EEN1 KO neurons by EEN1 WT and mutants. AAVs expressing Cre (AAV-mCherry-2A-Cre) and GFP (AAV-EGFP) or Cre, GFP, and EEN1 WT or mutant (AAV-EGFP-2A-EEN1) were stereotaxically injected into the CA1 regions of EEN1fl/fl mouse brain at P0. Acute hippocampal slices were prepared on P14–P21 for dual recording analysis of LTP. Shown are pairwise comparisons of LTP in noninfected (Ctrl) and infected neurons of the same slice. For Cre-expressing neurons (Cre) versus Ctrl (A), six recording pairs from three mice, marked as n = 6/3, were analyzed. n = 10/5 (B), n = 5/4 (C), n = 5/3 (D), and n = 4/3 (E). Bar graphs show percentage of baseline at 45 min after pairing. A summary of rescue effects of EEN1 WT and mutants is shown in F. Data are expressed as mean ± SEM for each group. P values were calculated using two-tailed paired t test. *, P < 0.05; **, P < 0.01. (G) AAV was stereotaxically injected into the brain CA1 regions of EEN1+/+ to express GFP alone, EEN1−/− to express GFP alone, or GFP and EEN1 WT, KKK-EEE, Y343A, or DM. Left panels are images of hippocampal sagittal section with GFP signals and DAPI labeling of nuclei. Center and right panels are coronal brain sections showing bilateral injection of AAV. Scale bar, 1 mm. (H) Immunofluorescence staining of EEN1 in CA1 neurons of brain slices from mice in G. Scale bar, 50 µm. (I–N) Morris water maze test of AAV-injected mice. Shown are escape latency or distance traveled before escaping to the platform in the visible platform (I) and invisible platform (J) training, the swim trace in probe trial 3 and recall following training once again 1 mo after training (K and L), number of crossings within the 1.5× platform area in 5-d probe trial and recall test (M), and number of crossings within the 1.5× platform area before and after recall (N). (O) Freezing behavior in AAV-injected mice subjected to contextual fear conditioning. All error bars represent the SEM in I, J, and M–O. Number of animals: 15 EEN1+/+ + GFP, 10 EEN1−/− + GFP, 6 EEN1−/− + EEN1, 6 EEN1−/− + Y343A, 7 EEN1−/− + KKK-EEE, 7 EEN1−/− + DM. P values were calculated using one-way ANOVA by Newman–Keuls post hoc test or two-tailed paired t test. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 when compared with EEN1+/+ + GFP; #, P < 0.05; ##, P < 0.01; and ###, P < 0.001 when compared with EEN1−/− + GFP; and &, P < 0.05 and &&, P < 0.01 when compared with before recall (two-tailed paired t test).
Finally, we determined the physiological significance of the mechanistic roles of endophilin A1 in sLTP by testing whether endophilin A1 mutants can rescue the learning and memory deficits in EEN1 KO mice. While AAV-mediated expression of WT endophilin A1 in the CA1 region restored the long-term memory in KO mice in both Morris water maze and fear conditioning tests, neither of the mutants ameliorated the phenotypes (Fig. 7, G–O). Collectively these data indicate that in CA1 pyramidal cells, Ca2+/calmodulin-regulated functions of endophilin A1 in acute structural plasticity are required for LTP and long-term memory.
Discussion
The temporal phases of sLTP include initiation of dendritic spine expansion (≤1 min after LTP stimulation, acute phase), transient (early phase), and sustained spine enlargement (late phase; Harvey and Svoboda, 2007; Matsuzaki et al., 2004). Although actin polymerization is essential for sLTP (Matsuzaki et al., 2004; Obashi et al., 2019), little is known about the relationship between actin remodeling and membrane dynamics during the acute phase. Here, we uncover a novel mechanism for initiation of sLTP. We show that in direct response to Ca2+/calmodulin, endophilin A1 drives acute spine enlargement in NMDAR-mediated sLTP by localizing to spine PM and recruiting p140Cap to promote actin polymerization (Fig. 8). We further show that the molecular functions of endophilin A1 in acute structural plasticity are also required for LTP and memory, providing evidence that activity-triggered transient spine enlargement is necessary for not only sustained structural and functional plasticity but also long-term memory formation in vivo.
Figure 8.
Model for endophilin A1-mediated initial spine head expansion upon LTP induction. NMDAR-mediated Ca2+ influx into dendritic spines activates calmodulin. Ca2+/calmodulin interacts directly with endophilin A1 and enhances its binding to both the PM and p140Cap, probably by releasing it from the autoinhibitory state caused by the intramolecular interaction between the N-BAR and SH3 domain. As a result, the PM-associated endophilin A1 recruits p140Cap, which in turn recruits cortactin to promote branched actin polymerization underneath the PM, generating propulsive force for rapid spine enlargement during the acute phase of sLTP.
Endophilin A2 and A3 interact with the immediate early protein Arc/Arg3.1 to accelerate AMPAR endocytosis in late-phase synaptic plasticity (Chowdhury et al., 2006). Notably, the plasticity phenotypes of dendritic spines in EEN1 KO neurons cannot be rescued by overexpression of endophilin A2 or A3 (Yang et al., 2018). Although both endophilin A1 and A2 interact with calmodulin (Myers et al., 2016; Fig. 2; this study), endophilin A1, but not endophilin A2 or A3, binds and recruits p140Cap to dendritic spines to promote actin polymerization (Yang et al., 2015). Moreover, in vivo and in vitro LTP stimuli induce increased association of endophilin A1, but not endophilin A2, with both membrane and cytoskeleton. Together these findings indicate that different interaction partners for the endophilin A family members confer them distinct mechanistic roles in the induction and expression of synaptic plasticity.
Several studies found that exocytosis of Rab11 recycling endosomes and SNARE-mediated membrane fusion are not required for acute sLTP (Hiester et al., 2018; Yang et al., 2008; Fig. S1). Instead, imaging analyses have revealed calmodulin-dependent formation of an F-actin pool that associates with spine enlargement (hence referred to as “enlargement pool”; Honkura et al., 2008). Although the observation that the membrane ruffling of the spine head synchronizes with the enlargement pool of F-actin prompted the authors to conclude that spine enlargement is induced by the propulsive force generated by calmodulin-regulated actin polymerization (Honkura et al., 2008), the mechanistic link between Ca2+/calmodulin and actin polymerization was still missing. Here, we demonstrate that endophilin A1 is the direct molecular target of Ca2+/calmodulin, which enhances both its membrane association and its interaction with p140Cap. Our data further indicate that coordination of the membrane-association and p140Cap-binding capacities of endophilin A1 provides the protrusive force for rapid structural remodeling of spines by promoting actin polymerization underneath the relaxed PM, which is also in good agreement with recent findings that the interplay between membrane tension and branched actin polymerization could produce membrane deformations (Simon et al., 2019).
A recent study revealed that although Rac1 activity is not required for the initial spine expansion induced by glutamate uncaging, formation of a reciprocally activating kinase–effector complex between CaMKII and Tiam1, a guanine exchange factor for Rac, converts the transient Ca2+ signal triggered by LTP induction into a persistent kinase signal required for the maintenance of sLTP (Saneyoshi et al., 2019). Given the spinous dynamic reorganization of nanoscale distribution of various F-actin regulators downstream of Ca2+/calmodulin and CaMKII (Chazeau and Giannone, 2016; Chazeau et al., 2014; Noguchi et al., 2016), there might be crosstalk between Ca2+/calmodulin-dependent, endophilin A1–mediated actin polymerization and other Ca2+/calmodulin and/or CaMKII effector–regulated pathways (e.g., Rac1 and ADF/cofilin), which enables spatiotemporally controlled actin reorganization during the acute phase and/or the transition to the early phase of sLTP.
In response to NMDAR-mediated Ca2+ influx, Copine-6 relocalizes from the cytosol to postsynaptic PM and contributes to structural plasticity and LTP, probably stabilizing the actin cytoskeleton by inhibiting ADF/cofilin via the Rac1–PAK–LIMK1 pathway (Reinhard et al., 2016). In addition, by keeping cortactin active to antagonize ADF/cofilin, the fast Ca2+ sensor caldendrin stabilizes an F-actin pool at the spine base required for structural remodeling in the transition from early- to late-phase LTP (Mikhaylova et al., 2018). In our study, rather than stabilizing F-actin, endophilin A1 responds to Ca2+/calmodulin and promotes actin polymerization beneath the PM to achieve rapid spine enlargement. While they all function via CaMKII-independent mechanisms to regulate actin dynamics, how Colpine-6, caldendrin, and endophilin A1 coordinate with each other to ensure spine potentiation and stabilization awaits further investigation.
Materials and methods
Ethics statement
All animal experiments were approved by and performed in accordance with the guidelines of the Animal Care and Use Committee of Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (approval codes AP2013003 and AP2015002) and the Animal Care and Use Committee of the Model Animal Research Center, the host for the National Resource Center for Mutant Mice in China, Nanjing University (approval code AP#SY06). All animals were housed in standard mouse cages at 22–24°C on a 12-h light/dark cycle with free access to food and water.
Animals
Generation of EEN1fl/fl and EEN1−/− (KO first) mice on the C57BL/6J background was as previously described (Yang et al., 2018). Genotyping of mouse lines was performed by genomic PCR of tail prep DNA from offspring with the following primer pairs: loxPF/loxPR, 5′-CAAGGACTCCCAGAGACCTAGCATC-3′ and 5′-GAGATGGCGCAACGCAATTAAT-3′ (a PCR product of 375 bp in EEN1 KO first mice, but not WT mice); zptF/zptR, 5′-GTAAGCGGCTCTAGCGCATGTTCT-3′ and 5′-GCAGGGGCATGTAGGTGGCTCAAC-3′ (a PCR product of 466 bp in WT mice, but not EEN1−/− mice, and 627 bp in EEN1fl/fl mice).
Constructs
DN SNAP23 (deleting the C-terminal 8 residues) and other DN SNAPs (SNAP25, SNAP29, and SNAP47, deleting the C-terminal 20 residues) were generated by PCR amplification of cDNA from mouse brain by RT-PCR and insertion into pCMV-Tag2B. The putative amino acid residues of EEN1 involved in calmodulin binding were predicated using the Binding Site Search and Analysis tool provided at the Calmodulin Target Database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/home.html). pCMV-Tag2B-EEN1 DM (I154AL158A) and pET28a(+)-EEN1 single and DM mutants were created by site-directed mutagenesis using pCMV-Tag2B-EEN1 and pET28a(+)-EEN1 as template, respectively. pET28a(+)-EEN1 ΔBAR (Δaa 6–242) and pET28a(+)-EEN1 ΔSH3 (Δaa 295–346) were subcloned from pGEX4T-1-EEN1 ΔBAR and pGEX4T-1-EEN1 ΔSH3. The pAAV-CaMKIIα-EGFP-2A-MCS-3FLAG-EEN1 WT or mutant viral constructs (KKK-EEE, Y343A, and DM) were generated by cloning EEN1 cDNA amplified from pCMV-Tag2B-EEN1 or mutant constructs into pAOV-CaMKIIα-EGFP-2A-MCS-3FLAG (OBiO Technology). pAOV-CaMKIIα-mGFP-2A-Cre was generated by replacing EGFP in pAOV-CaMKIIα-EGFP-2A-Cre (OBiO Technology) with mGFP. The EGFP-EEN1 construct was generated by cloning EEN1 cDNA with the 3xGGS linker into pEGFP-C2. The bacterial expression construct for His-tagged p140Cap fragment (aa 351–1051) was generated by PCR amplification of the cDNA encoding p140Cap (aa 351–1051) and insertion into pET-28a(+). Bacterial expression constructs for calmodulin were generated by PCR amplification of cDNA from mouse brain by RT-PCR and insertion into pGEX4T-1 and pET28a(+). The Arp1b-mCherry construct was a generous gift from Drs. Na Mi and Li Yu (Tsinghua University, Beijing, China). The Rab11-GFP and Rab11 DN (S225)-GFP constructs were generous gifts from Dr. Jose A. Esteban (Centro de Biología Molecular Severo Ochoa, Madrid, Spain). All other constructs used in this study (pCMV-Tag2B-EEN1 WT, pCMV-Tag2B-EEN1 Y343A, pCMV-Tag2B-EEN1 KKK-EEE, mGFP, and LifeAct-mCherry) were described previously (Guo et al., 2018; Yang et al., 2018). Viral particles of AAV carrying pAOV-CAMKII-mCherry-2A-Cre, pAOV-CaMKIIα-EGFP-2A-Cre, pAAV-CaMKIIα-EGFP-2A-MCS-3FLAG-EEN1 WT, or mutants and the control construct pAAV-CaMKIIα-EGFP-2A-MCS-3FLAG were purchased from OBiO Technology.
Antibodies
The following antibodies were obtained from commercial sources: endophilin A2 (sc-10876), mouse anti-SYP (sc-17750), and mouse anti-cortactin (sc-55588; Santa Cruz Biotechnology) for Western blotting and anti-cortactin (05–180; Sigma-Aldrich) for staining; rabbit anti-endophilin A1 (159002; Synaptic Systems); mouse anti-GST (PM013), rabbit and mouse anti-RFP which recognizes DsRed and mCherry (PM005 and M165-3; Medical & Biological Laboratories); mouse anti-M5 DYKDDDDK-Tag (MF085-02; Mei5 Biotechnology), mouse anti-α-tubulin (T9026), and mouse anti-β-actin (A5441; Sigma-Aldrich); mouse anti-His (CW0285M; CoWin Biosciences), rabbit anti-calmodulin (M06609; Boster Biological Technology) for Western blotting and mouse anti-calmodulin (05–173; Sigma-Aldrich) for immunoprecipitation; mouse anti-phosphatidylinositol 4,5-bisphosphate (anti-PI(4,5)P2; Z-A045; Echelon Biosciences); rabbit anti-p140Cap was described previously (Yang et al., 2015). Alexa Fluor dye–conjugated secondary antibodies for immunofluorescence staining were from Molecular Probes (Invitrogen).
Cell culture, transfection, and drug treatment
Primary hippocampal neurons were cultured as previously described (Yang et al., 2018). Briefly, mouse hippocampi were dissected from P0 C57BL/6J mice, dissociated with 0.125% trypsin in Hank’s balanced salt solution without Ca2+ and Mg2+ at 37°C for 15 min, and triturated in DMEM, 10% F12, and 10% FBS (GIBCO). Hippocampal neurons were plated on poly-D-lysine–coated coverslips in 24-well plates or 30-mm dishes at a density of 2.5–3.0 × 104 cells/well in a 24-well plate or 1.0–1.2 × 105 cells/35-mm dish. The medium was replaced with the serum-free Neurobasal A (NB-A) media supplemented with 2% B27 supplement, GlutaMAX (GIBCO), and 0.3% glucose 4 h after plating. Half of the media were replaced every 3 d until use.
For neuronal morphology and immunofluorescence staining, neuronal transfections were performed using Lipofectamine LTX according to the manufacturer’s instructions (Invitrogen) on 12–14 d in vitro (DIV) after plating. Briefly, DNA (0.5 µg/well) was mixed with 0.5 µl PLUS reagent in 50 µl NB-A medium and then mixed with 1.0 µl Lipofectamine LTX in 50 µl NB-A medium, incubated for 20 min, and added to the neurons in NB-A at 37°C in 5% CO2 for 1 h. Neurons were then rinsed with NB-A and incubated in the original medium at 37°C in 5% CO2 for 4–5 d. For cotransfection, neurons were transfected with 1.0 µg of DNA consisting of two plasmids (0.5 µg each).
DMEM supplemented with 10% FBS and DMEM/F12 supplemented with 10% FBS were used for HeLa (CCL-2; ATCC) and Neuro-2a (CCL-131; ATCC) cell culture, respectively. Cell transfections were performed using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen) after plating.
We referred to literature for working concentrations of actin reorganization inhibitors for cultured neurons (Chen and Firestein, 2007; Jiang et al., 2015; Matsuzaki et al., 2004; Mauceri et al., 2011; Rex et al., 2009; Soykan et al., 2017; van Bommel et al., 2019; Zhou et al., 2011). For drug treatment, HeLa cells or primary neurons cultured on coverslips were preincubated with MK801 (10 µM; Sigma-Aldrich), LatA (100 nM; Sigma-Aldrich), NSC 23766 trihydrochloride (100 µM; Abcam), CK-666 (100 µM; Sigma-Aldrich), W-7 (20 µM; TOCRIS), KN-62 (4 µM; TOCRIS), and BAPTA-AM (10 µM; Sigma-Aldrich) for 30 min. Neurons were preincubated with ML141 (15 µM; Sigma-Aldrich) or SMIFH2 (30 µM; Sigma-Aldrich; van Bommel et al., 2019) for 2 h or with CT04 (2 µg/ml; Cytoskeleton) for 3 h. For tetanus toxin treatment (10 nM; Sigma-Aldrich), neurons were preincubated for 10 min. These drugs were maintained during glycine or ionomycin application.
For ionomycin treatment of HeLa cells, cells were preincubated with modified Krebs-Ringer Hepes (KRH) buffer (containing 120 mM NaCl, 4.8 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 1.3 mM CaCl2, 5.5 mM glucose, and 25 mM Hepes, pH 7.4, at 37°C) for 30 min then treated with ionomycin (2 µM; Beyotime Biotechnology) in KRH buffer for 20 min. For ionomycin treatment of Neuro-2a cells, cells were preincubated with the KRH buffer and treated with ionomycin (1 µM) in KRH buffer for 30 min
cLTP
Chemical induction of LTP was performed as previously described (Fortin et al., 2010; Park et al., 2006). Briefly, neurons were treated with glycine (200 µM) in Mg2+-free extracellular iso-osmotic solution (in mM: 125 NaCl, 2.5 KCl, 2 CaCl2, 5 Hepes, 33 glucose, 0.2 glycine, 0.02 bicuculline, and 0.003 strychnine, pH 7.4; Yang et al., 2018). For experiments performed in the absence of extracellular Ca2+, 10 µM BAPTA-AM was substituted for 2 mM CaCl2 in the Mg2+-free extracellular solution.
For experiment to determine the role of membrane tension in spine expansion, neurons were pretreated with the hypo-osmotic solution (in mM: 80 NaCl, 2.5 KCl, 2 CaCl2, 5 Hepes, and 33 glucose, pH 7.4, mOsm 210) or hyperosmotic solution (in mM: 125 NaCl, 2.5 KCl, 2 CaCl2, 5 Hepes, 33 glucose, and 250 sucrose, pH 7.4, mOsm 600) for 10 min and cLTP in the same solution.
Immunostaining, image acquisition, and analysis
Neurons were fixed with 4% PFA/4% sucrose in PBS at RT for 15 min. After blocking with 1% BSA in PBS containing 0.4% Triton-X 100 for 40 min at RT, neurons were incubated with primary antibodies for 1 h at RT or overnight at 4°C, and appropriate secondary antibodies conjugated with Alexa Fluor 488, Alexa Fluor 555, or Alexa Fluor 647 were applied for detection. Immunostaining of PM-localized EEN1 and PI(4,5)P2 was performed as previously described (Hammond et al., 2009). Briefly, neurons were fixed with 4% formaldehyde and 0.2% glutaraldehyde for 15 min at RT, followed by rinsing with PBS containing 50 mM NH4Cl three times. Coverslips were chilled on ice and blocked with buffer A (20 mM Pipes, 137 mM NaCl, and 2.7 mM KCl, pH 6.8) containing 5% (vol/vol) normal goat serum, 50 mM NH4Cl, and 0.5% (wt/vol) saponin for 45 min on ice. Cell were then incubated with primary antibodies in buffer A containing 5% normal goat serum and 0.1% saponin for 1–2 h and secondary antibodies for 45 min on ice. Cells were postfixed for 10 min on ice and 5 min at RT with 2% formaldehyde in PBS followed by three rinses with PBS containing 50 mM NH4Cl.
Confocal images of DAPI, Alexa Fluor 488, Alexa Fluor 555, or Alexa Fluor 647 channels were collected with a 100× Plan Apochromat VC (NA 1.40) oil objective using the Spectral Imaging Confocal Microscope Digital Eclipse C1Si (Nikon). Images were z projections of images taken at 0.2-µm step intervals. The number of planes (typically four to six) was chosen to encompass the entire dendrite from top to bottom.
The procedure for morphometric analysis of dendritic spines was described previously (Yang et al., 2015; Yang et al., 2018). DsRed or EGFP was used as a cell fill. The final reconstructed spines were obtained using a maximum-intensity projection strategy provided by NIS-Elements AR software (Nikon). DsRed-labeled spines were outlined manually. Dendritic segments 40–120 µm from the neuronal cell body were selected for analysis. All morphological experiments were repeated at least three times with an n ≥ 10 for individual experiments. Quantification of F-actin enrichment in spines was described previously (Yang et al., 2018). To analyze distribution of EEN1 mutants in dendrites, we measured the mean intensity of fluorescent signals in spines and normalized each measurement with the fluorescent signals along the adjacent dendritic shaft to obtain the spine/shaft ratio. For analysis of subcellular distribution of proteins in HeLa and Neuro-2a cells, the fluorescence intensity along the white line in cells was measured with the Color Profiler tool provided by ImageJ (National Institutes of Health; NIH) to generate the linescan plots.
Super-resolution live-cell imaging and data analysis
The GI-SIM live imaging experiments were performed as described by Guo et al. (2018). Briefly, mouse hippocampal neurons cultured on 25-mm coverslips were transfected with constructs expressing mGFP and LifeAct-mCherry on DIV12 and imaged on DIV16 in Mg2+-free extracellular solution (in mM: 125 NaCl, 2.5 KCl, 2 CaCl2, 5 Hepes, 33 glucose, 0.2 glycine, 0.02 bicuculline, and 0.003 strychnine, pH 7.4; Yang et al., 2018). Time-lapse images of the mCherry and GFP channels were obtained with acquisition time of 110 ms for each channel at 5-s intervals. To quantify the area of each spine head and enrichment of F-actin in spines, we measured the fluorescent mean intensity of LifeAct-mCherry within the spines and normalized each measurement by the fluorescence signal along the adjacent dendritic shaft with the NIH ImageJ software. The mGFP-labeled dendrites or spines were outlined manually.
For analysis of the spatiotemporal relationship between actin polymerization and membrane expansion in dendritic spines, spines were segmented from raw GI-SIM images using Otsu’s method (Otsu, 1979). To facilitate the visualization of instantaneous spine growth and localized actin polymerization, differential images of both membrane (mGFP) and F-actin (LifeAct-mCherry) channels were calculated by subtracting the image at time point t from that at t + 1 through the entire time-lapse movie (1-min duration before and 5-min duration after glycine application with 5-s intervals) using MATLAB (R2018a; MathWorks). To highlight the regions with spine growth/membrane expansion or increased F-actin signals, only pixels with a positive difference were displayed in the final differential images shown in Fig. 1 E. To analyze the spatiotemporal coupling of membrane expansion and actin polymerization, we chose spines with increase in F-actin signals, measured the overlap between differential images in the membrane channel and those in the actin channel at individual time points, and determined the extent of overlap by calculating the ratio of overlapped to total changes in the membrane channel using the JACoP plug-in of ImageJ software. Then, we obtained fluctuations in the extent of overlap at different time points before or after glycine treatment to evaluate the randomness of overlap between membrane expansion and actin polymerization.
Immunohistochemical analyses
Mice were anesthetized with 1% sodium pentobarbital and transcardially perfused with normal saline followed by 4% PFA in 0.01 M PBS. Mouse brain was dissected out and postfixed with 4% PFA/PBS for 4 h at 4°C. Fixed brain was incubated with 20% sucrose overnight and then 30% sucrose overnight. The brain was embedded in optimal cutting temperature compound and stored at −80°C until usage. 30-μm cryosections were made using cryostat and collected.
For immunostaining of brain sections, floating 30-µm-thick slices were rinsed with PBS and permeabilized in 0.4% Triton-X 100 in 0.01 M PBS for 30 min. Cryosections were blocked with 1% BSA in PBS containing 0.4% Triton-X 100 for 1 h at RT and then incubated with primary antibodies overnight at 4°C. Secondary antibodies conjugated with Alexa Fluor 555 were used for detection. Sections were then incubated with DAPI (Roche) for nuclear staining for 5 min at RT. Following rinsing, cryosections were mounted on gelatin-coated slides and covered with coverslip with mounting medium. Confocal images were collected using the Spectral Imaging Confocal Microscope Digital Eclipse C1Si (Nikon) with a 10× Plan Apochromat differential interference contrast N1 0.45 objective or 40× Plan Fluo (NA 1.30) oil objective (Yang et al., 2018).
Protein expression and purification
His-EEN1, His-calmodulin, His-p140Cap fragment (aa 351–1051), or GST-calmodulin was expressed in Escherichia coli BL21 (DE3). Cells were grown at 37°C in LB (in g/liters: 10 tryptone, 5 yeast extract, and 10 NaCl) supplemented with ampicillin or kanamycin. Cells were induced at OD600 of ∼0.6 with 0.4 mM IPTG for 4 h at 30°C or 16 h at 16°C. Cells were harvested and stored at −80°C until purification.
For His-tagged proteins, cells were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 15 mM imidazole, pH 8.0) supplemented with 1% Triton-X 100 and 0.1 mM PMSF. Protein were purified with Ni-NTA resin according to the manufacturer’s instructions (R90115; Invitrogen). For GST-tagged proteins, cells were resuspended in PBS supplemented with 0.2% Triton-X 100 and 0.1 mM PMSF. Proteins were purified with glutathione Sepharose 4B (GE17-0756-01; Sigma-Aldrich) according to the manufacturer’s instructions.
For the liposome cosedimentation assay, purified His-calmodulin was dialyzed in Spectra/Por 4 RC Dialysis Membrane Tubing (08-667D; Thermo Fisher Scientific) against 1,000 vol PBS supplemented with 0.5 mM EGTA at 4°C for 4 h and replaced with fresh 1,000 vol PBS twice.
ITC
ITC was performed on a MicroCal PEAQ-ITC (Malvern Panalytical) calorimeter. Calmodulin and EEN1 were purified on a Superdex-200 16/600 column (GE Healthcare) in solution buffer containing 20 mM Tris-Cl at pH 7.0 and 150 mM NaCl. Solution buffer containing 1 mM CaCl2 was injected into a calorimeter cell filled with protein solution, with the cell temperature set to 25°C. Each analysis involved 20 injections of 4-s duration (2 µl per one injection), 120-s spacing, stir with 750 rpm, 5 µcal/s reference power, and high gain feedback mode. Data were processed by Origin software to obtain thermodynamic profiles.
GST pull-down, coIP, and immunoisolation
For GST pull-down assays, 5 µg GST-tagged protein conjugated with glutathione Sepharose beads was incubated with 1 µg His-tagged protein in 0.01 M PBS supplemented with 1% NP-40 at 4°C for 1 h. Beads were washed five times with PBS supplemented with 0.3% Triton-X 100 and boiled in SDS sample buffer.
For coIP experiments, cultured neurons or mouse brain were lysed with lysis buffer 1 (0.05% [vol/vol] NP-40, 15 mM Tris-HCl, pH 7.4, and 50 mM NaCl) supplemented with protease inhibitors. Lysates were then centrifuged at 16,000 ×g for 15 min at 4°C. Antibodies (1 µg) were added to the cell lysates and incubated at 4°C for 2 h on a roller mixer, followed by incubation with Protein A/G agarose (Santa Cruz Biotechnology) preequilibrated in lysis buffer overnight at 4°C. Immunoprecipitates were washed four times in lysis buffer, boiled in SDS sample buffer, and subjected to SDS-PAGE and immunoblotting. To mimic transient increase in intracellular Ca2+, CaCl2 (1 mM) was added to hippocampal neuron cell lysates and incubated for 10 min on ice followed by addition of EGTA (1 mM) to chelate Ca2+ before coIP.
For immunoisolation of membrane proteins, cultured neurons were homogenized with lysis buffer (in mM: 20 Tris-HCl, 10 Hepes, pH 7.4, 150 NaCl, and 250 sucrose) supplemented with protease inhibitors and centrifuged at 800 ×g for 15 min. The supernatants were collected and subjected to high-speed centrifugation at 100,000 ×g for 1 h (TLS-55 rotor, OptimaTMMAX Ultracentrifuge; Beckman Coulter). The supernatants (S100) and pellets (p100, the membrane fraction) resuspended in lysis buffer were subjected to immunoisolation with Dynabeads Protein G (Invitrogen) coupled with 2 µg mouse anti-calmodulin antibody. Bound proteins were eluted by boiling in 2× SDS gel loading buffer and subjected to SDS-PAGE and immunoblotting.
Subcellular fractionation
For subcellular fractionation of mouse hippocampi, mice were sacrificed and hippocampi were removed 30 min after fear conditioning training. Mouse hippocampi or cultured neurons were homogenized to isolate the membrane and cytoskeleton fractions with Subcellular Protein Fractionation Kit for tissues (87790; Thermo Fisher Scientific) or cultured cells (77840; Thermo Fisher Scientific) according to the manufacturer’s instructions. Proteins in different fractions were subjected to SDS-PAGE and immunoblotting.
Liposome cosedimentation assay
Bovine brain extracts (B1502; Sigma-Aldrich) were dissolved in chloroform and dried under vacuum for 30 min. The solvent-free lipid films were rehydrated with liposome buffer (150 mM NaCl, 20 mM Hepes, pH 7.4, and 1 mM DTT) and subjected to seven cycles of flash freezing in liquid nitrogen and thawing in a 37°C bath. Liposomes were then extruded 21 times through a polycarbonate membrane with a 50-nm pore size (Mini-Extruder; Avanti Polar Lipids). Extruded liposomes were centrifuged at 16,000 ×g for 5 min to remove insoluble material. Liposomes (0.5 mg/ml) were then incubated with freshly purified recombinant proteins (1 µM His-EEN1 or 1 µM His-EEN1 and 2 µM GST-calmodulin) in 100 µl liposome buffer for 10 min at 30°C before sedimentation at 140,000 ×g for 30 min at 4°C. The supernatant (unbound) and pellet (bound) were subjected to SDS-PAGE. Ratios of binding to liposomes were determined using NIH ImageJ software.
3D-SIM imaging and image analysis
3D-SIM images of Alexa Fluor 488, Alexa Fluor 555, and Alexa Fluor 647 channels were acquired as previously described (Niu et al., 2013) on the DeltaVision OMX V4 imaging system (Applied Precision) with a 100× 1.4 oil objective (Olympus UPlanSApo), solid-state multimode lasers (488, 593, and 642 nm), and electron-multiplying charge-coupled device cameras (Evolve 512×512; Photometrics). Serial Z-stack sectioning was done at 125-nm intervals. The microscope is routinely calibrated with 100-nm fluorescent spheres to calculate both the lateral and axial limits of image resolution. SIM image stacks were reconstructed using softWoRx 5.0 (Applied Precision) with the following settings: pixel size, 39.5 nm; channel-specific optical transfer functions; Wiener filter, 0.001000; discard negative intensities background; drift correction with respect to first angle; and custom K0 guess angles for camera positions. Pixel registration was corrected to be <1 pixel for all channels using 100-nm Tetraspeck beads. For clarity of display, small linear changes to brightness and contrast were performed on 3D reconstructions.
Live imaging of Neuro-2a cells
Neuro-2a cells expressing EGFP-EEN1 were incubated with modified KRH buffer and imaged live at 37°C using a 100× Plan Apochromat VC (NA 1.40) oil objective with the Spectral Imaging Confocal Microscope DIGITAL ECLIPSE C1Si (Nikon). Cells untreated or treated with W-7 (20 µM) for 20 min were imaged every minute for two frames, supplemented with ionomycin (1 µM), and imaged every 10 s to 1 min for 30 min.
Electrophysiology
Within 24 h after birth, EEN1fl/fl mice were coinjected with high-titer AAV stock carrying pAOV-CAMKIIα-mCherry-2A-Cre (AAV-mCherry-2A-Cre) and pAAV-CaMKIIα-EGFP-2A-MCS-3FLAG-EEN1 or mutants of EEN1 (∼1–5 × 1013 IU/ml). Newborns were anesthetized on ice for 5 min and then mounted in a custom ceramic mold to make the head level in the x and y axes. Lambda was set as (x, y) = (0, 0). Zero point of z axis was the position at which the injecting needle penetrated the skin. Approximately 10 nl viral solution was injected at each of the seven sites ([x, y, z] = [1.2, 1.2, 1.4/1.0/0.6] and [1.5, 1.0, 1.7/1.3/0.9/0.5]) targeting the hippocampus at each cerebral hemisphere with a microsyringe (Sutter Instrument) and beveled glass injection pipette. Injected pups were returned to home cage and used for recording 2–3 wk afterward. Transverse 350-µm hippocampal slices were cut from viral injected EEN1fl/fl mice on a Leica vibratome (VT1000 S) in high-sucrose cutting solution containing (in mM) 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 0.50 CaCl2, 7 MgSO4, 210 sucrose, 10 glucose, and 1.3 Na-ascorbic acid. Freshly cut slices were placed in an incubating chamber containing artificial cerebrospinal fluid, and recovered at 32°C for ∼20 min followed by 60 min at room temperature before recording. The slices were perfused with artificial cerebrospinal fluid containing GABAA receptor antagonists PTX (100 µM)/Bic (10 µM) and saturated with 95% O2/5% CO2 in whole-cell LTP experiments. CA1 pyramidal cells were voltage clamped at −70 mV, and AMPAR excitatory postsynaptic current were evoked by stimulation at SC with concentric electrode (FHC CBBRC75). LTP was induced by stimulating SC axons at 2 Hz for 90 s while clamping the cell at 0 mV, after recording a stable 3- to 5-min baseline, but no more than 6 min after breaking into the cell (Díaz-Alonso et al., 2017; Granger et al., 2013). To minimize run-up of baseline responses during LTP, cells were held cell-attached for ∼1–2 min before breaking into the cell.
Stereotaxic injection and behavioral tests
9-wk-old sexually naive male and female mice were anesthetized and stereotactically injected with viral particles in the hippocampal CA1 region as described previously (Yang et al., 2018). Briefly, mice were anesthetized with isoflurane (1–2% mixed with oxygen) then were sterilized with iodophors and alcohol. Viral particles carrying pAAV-CaMKIIα-EGFP-2A-MCS-3FLAG, pAAV-CaMKIIα-EGFP-2A-MCS-3FLAG-EEN1 WT or mutants (1 µl, 2.0 × 1012 viral genomes/ml) were injected bilaterally into the hippocampal CA1 regions using the following coordinates relative to bregma: 2.0 mm posterior, 1.8 mm lateral, and 1.4 mm ventral at a rate of 0.125 µl/min using a microinjection system (World Precision Instruments). The needle was kept in place for 5 min before withdrawal. The virus-injected mice were tested for behavior 2 wk later. Morris water maze and fear conditioning tests were performed as previously described (Yang et al., 2018). Briefly, mice were exposed to the visible trial when a flag could be visualized on the platform, and then the flag was removed and the platform, submerged 1 cm below the water surface, was kept in a fixed position. Mice were trained for 8 d, and then the number of times that each mouse crossed the 1.5× platform circle area within 60 s was analyzed. The recall ability was tested 1 mo after training. The mouse trajectory in the pool was monitored and analyzed with an automated system (Smart 3.0, Panlab SMART video tracking system). For fear conditioning tests, mice were trained in a standard fear conditioning apparatus (Harvard Apparatus). They were allowed to explore freely for 2 min, and then a 2-s, 0.9-mA footshock (unconditioned stimulus) was delivered, and mice stayed in the chamber for 30 s. Mice were reexposed to the same chamber and a novel chamber for 2 min on the following 3 d. Freezing was scored and analyzed automatically using FREEZING software (Harvard Apparatus). We observed no sex-related differences in behavior, and the results were pooled together.
Statistical analysis
All data are presented as the mean ± SEM. GraphPad Prism 5 (GraphPad Software) was used for statistical analysis. For two-sample comparisons versus controls, a paired or unpaired Student’s t test was used. One-way analysis of variance with a Dunnett’s multiple-comparison or Newman–Keuls multiple-comparison hoc test was used to evaluate statistical significance of three or more groups of samples. A P value of <0.05 was considered statistically significant. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows effects of inhibitors for membrane fusion, recycling endosomal trafficking, and actin dynamics, as well as osmotic shocks on acute sLTP of hippocampal neurons. Fig. S2 shows assays testing binding of endophilin A1 to Ca2+. Fig. S3 shows binding of endophilin A1 point mutants to calmodulin. Fig. S4 shows rescue of acute sLTP in EEN1−/− neurons by overexpression of EEN1 WT and mutants. Fig. S5 shows LTP-induced, calmodulin-dependent change in the subspine distribution of endophilin A1. Videos 1 and 2 show morphological changes and actin dynamics of spines of EEN1 WT and KO neurons during LTP induction, respectively. Videos 3 and 4 show the spatiotemporal relationship between PM expansion or retraction, respectively, and increases in F-actin in EEN1fl/fl spines during LTP induction. Videos 5 and 6 illustrate the spatiotemporal relationship between PM expansion or retraction, respectively, and decreases in F-actin in spines during LTP induction. Video 7 exhibits the spatiotemporal relationship between spine membrane expansion and increases in F-actin in EEN1 KO spines before and during glycine-induced sLTP.Videos 8 and 9 show the effect of ionomycin on subcellular distribution of EGFP-EEN1 in untreated and W-7–treated Neuro-2a cells, respectively.
Acknowledgments
We thank Dr. Anbing Shi (Huazhong University of Science and Technology, Wuhan, China) for critical comments on the manuscript.
This work was supported by funding to J-J. Liu (Ministry of Science and Technology grant 2016YFA0500100; National Natural Science Foundation of China grants 31530039, 91954126, and 31921002; and Chinese Academy of Sciences grant XDB32020100), Y. Yang (National Natural Science Foundation of China grants 32070785 and 31571056), Y.S. Shi (National Natural Science Foundation of China grant 91849112 and Ministry of Science and Technology grant 2019YFA0801603), J. Chen (National Natural Science Foundation of China grant 81901161), Dong Li (National Natural Science Foundation of China grant 91754202), and C. Ma (National Natural Science Foundation of China grant 31670846).
The authors declare no competing financial interests.
Author contributions: conceptualization, Y. Yang and J-J. Liu; investigation, Y. Yang, J. Chen, X. Chen, Di Li, S. Wang, S. Zhao, X. Yang, S. Deng, C. Tong, D. Wang, and Z. Guo; formal analysis, Y. Yang, J. Chen, X. Chen, Di Li, J. He, S. Wang, S. Zhao, and S. Deng; visualization, Y. Yang, J. Chen, X. Chen, J. He, S. Wang, S. Zhao, X. Yang, S. Deng, X. Liang, and J-J. Liu; writing, Y. Yang, J. Chen, S. Wang, S. Zhao, X. Yang, C. Ma, X. Liang, Y.S. Shi, and J-J. Liu; funding acquisition, Y. Yang, J. Chen, Dong Li, C. Ma, Y.S. Shi, and J-J. Liu; supervision, Dong Li, C. Ma, X. Liang, Y.S. Shi, and J-J. Liu; project administration, J-J. Liu.
References
- Arellano, J.I., Benavides-Piccione R., Defelipe J., and Yuste R.. 2007. Ultrastructure of dendritic spines: correlation between synaptic and spine morphologies. Front. Neurosci. 1:131–143. 10.3389/neuro.01.1.1.010.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, M., Castro J., Saneyoshi T., Matsuno H., Sur M., and Hayashi Y.. 2014. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 82:444–459. 10.1016/j.neuron.2014.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot, E., Ferreira A.P., Almeida-Souza L., Debard S., Vallis Y., Howard G., Bertot L., Sauvonnet N., and McMahon H.T.. 2015. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 517:460–465. 10.1038/nature14067 [DOI] [PubMed] [Google Scholar]
- Chazeau, A., and Giannone G.. 2016. Organization and dynamics of the actin cytoskeleton during dendritic spine morphological remodeling. Cell. Mol. Life Sci. 73:3053–3073. 10.1007/s00018-016-2214-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazeau, A., Mehidi A., Nair D., Gautier J.J., Leduc C., Chamma I., Kage F., Kechkar A., Thoumine O., Rottner K., et al. 2014. Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J. 33:2745–2764. 10.15252/embj.201488837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., and Firestein B.L.. 2007. RhoA regulates dendrite branching in hippocampal neurons by decreasing cypin protein levels. J. Neurosci. 27:8378–8386. 10.1523/JNEUROSCI.0872-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Deng L., Maeno-Hikichi Y., Lai M., Chang S., Chen G., and Zhang J.F.. 2003. Formation of an endophilin-Ca2+ channel complex is critical for clathrin-mediated synaptic vesicle endocytosis. Cell. 115:37–48. 10.1016/S0092-8674(03)00726-8 [DOI] [PubMed] [Google Scholar]
- Chowdhury, S., Shepherd J.D., Okuno H., Lyford G., Petralia R.S., Plath N., Kuhl D., Huganir R.L., and Worley P.F.. 2006. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 52:445–459. 10.1016/j.neuron.2006.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corponi, F., Bonassi S., Vieta E., Albani D., Frustaci A., Ducci G., Landi S., Boccia S., Serretti A., and Fabbri C.. 2019. Genetic basis of psychopathological dimensions shared between schizophrenia and bipolar disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry. 89:23–29. 10.1016/j.pnpbp.2018.08.023 [DOI] [PubMed] [Google Scholar]
- Díaz-Alonso, J., Sun Y.J., Granger A.J., Levy J.M., Blankenship S.M., and Nicoll R.A.. 2017. Subunit-specific role for the amino-terminal domain of AMPA receptors in synaptic targeting. Proc. Natl. Acad. Sci. USA. 114:7136–7141. 10.1073/pnas.1707472114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin, D.A., Davare M.A., Srivastava T., Brady J.D., Nygaard S., Derkach V.A., and Soderling T.R.. 2010. Long-term potentiation-dependent spine enlargement requires synaptic Ca2+-permeable AMPA receptors recruited by CaM-kinase I. J. Neurosci. 30:11565–11575. 10.1523/JNEUROSCI.1746-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop, J.L., Jao C.C., Kent H.M., Butler P.J., Evans P.R., Langen R., and McMahon H.T.. 2006. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 25:2898–2910. 10.1038/sj.emboj.7601174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger, A.J., Shi Y., Lu W., Cerpas M., and Nicoll R.A.. 2013. LTP requires a reserve pool of glutamate receptors independent of subunit type. Nature. 493:495–500. 10.1038/nature11775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Li D., Zhang S., Yang Y., Liu J.J., Wang X., Liu C., Milkie D.E., Moore R.P., Tulu U.S., et al. 2018. Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell. 175:1430–1442.e17. 10.1016/j.cell.2018.09.057 [DOI] [PubMed] [Google Scholar]
- Hammond, G.R., Schiavo G., and Irvine R.F.. 2009. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2). Biochem. J. 422:23–35. 10.1042/BJ20090428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, K.M., and Stevens J.K.. 1989. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J. Neurosci. 9:2982–2997. 10.1523/JNEUROSCI.09-08-02982.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, C.D., and Svoboda K.. 2007. Locally dynamic synaptic learning rules in pyramidal neuron dendrites. Nature. 450:1195–1200. 10.1038/nature06416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takagi, A., Yagishita S., Nakamura M., Shirai F., Wu Y.I., Loshbaugh A.L., Kuhlman B., Hahn K.M., and Kasai H.. 2015. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature. 525:333–338. 10.1038/nature15257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, N.G., and Yasuda R.. 2017. Regulation of Rho GTPase proteins during spine structural plasticity for the control of local dendritic plasticity. Curr. Opin. Neurobiol. 45:193–201. 10.1016/j.conb.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Hedrick, N.G., Harward S.C., Hall C.E., Murakoshi H., McNamara J.O., and Yasuda R.. 2016. Rho GTPase complementation underlies BDNF-dependent homo- and heterosynaptic plasticity. Nature. 538:104–108. 10.1038/nature19784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring, B.E., and Nicoll R.A.. 2016. Long-Term Potentiation: From CaMKII to AMPA Receptor Trafficking. Annu. Rev. Physiol. 78:351–365. 10.1146/annurev-physiol-021014-071753 [DOI] [PubMed] [Google Scholar]
- Hiester, B.G., Becker M.I., Bowen A.B., Schwartz S.L., and Kennedy M.J.. 2018. Mechanisms and Role of Dendritic Membrane Trafficking for Long-Term Potentiation. Front. Cell. Neurosci. 12:391. 10.3389/fncel.2018.00391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkura, N., Matsuzaki M., Noguchi J., Ellis-Davies G.C., and Kasai H.. 2008. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 57:719–729. 10.1016/j.neuron.2008.01.013 [DOI] [PubMed] [Google Scholar]
- Ibata, K., Kono M., Narumi S., Motohashi J., Kakegawa W., Kohda K., and Yuzaki M.. 2019. Activity-Dependent Secretion of Synaptic Organizer Cbln1 from Lysosomes in Granule Cell Axons. Neuron. 102:1184–1198.e10. 10.1016/j.neuron.2019.03.044 [DOI] [PubMed] [Google Scholar]
- Jaworski, J., Kapitein L.C., Gouveia S.M., Dortland B.R., Wulf P.S., Grigoriev I., Camera P., Spangler S.A., Di Stefano P., Demmers J., et al. 2009. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 61:85–100. 10.1016/j.neuron.2008.11.013 [DOI] [PubMed] [Google Scholar]
- Jiang, J., Zhang Z.H., Yuan X.B., and Poo M.M.. 2015. Spatiotemporal dynamics of traction forces show three contraction centers in migratory neurons. J. Cell Biol. 209:759–774. 10.1083/jcb.201410068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliesch, T.T., Dietz J., Turco L., Halder P., Polo E., Tarantola M., Jahn R., and Janshoff A.. 2017. Membrane tension increases fusion efficiency of model membranes in the presence of SNAREs. Sci. Rep. 7:12070. 10.1038/s41598-017-12348-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec, C.D., Li B., Wei W., Boehm J., and Malinow R.. 2006. Glutamate receptor exocytosis and spine enlargement during chemically induced long-term potentiation. J. Neurosci. 26:2000–2009. 10.1523/JNEUROSCI.3918-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki, M., Ellis-Davies G.C., Nemoto T., Miyashita Y., Iino M., and Kasai H.. 2001. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat. Neurosci. 4:1086–1092. 10.1038/nn736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki, M., Honkura N., Ellis-Davies G.C., and Kasai H.. 2004. Structural basis of long-term potentiation in single dendritic spines. Nature. 429:761–766. 10.1038/nature02617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauceri, D., Freitag H.E., Oliveira A.M., Bengtson C.P., and Bading H.. 2011. Nuclear calcium-VEGFD signaling controls maintenance of dendrite arborization necessary for memory formation. Neuron. 71:117–130. 10.1016/j.neuron.2011.04.022 [DOI] [PubMed] [Google Scholar]
- Mikhaylova, M., Bär J., van Bommel B., Schätzle P., YuanXiang P., Raman R., Hradsky J., Konietzny A., Loktionov E.Y., Reddy P.P., et al. 2018. Caldendrin Directly Couples Postsynaptic Calcium Signals to Actin Remodeling in Dendritic Spines. Neuron. 97:1110–1125.e14. 10.1016/j.neuron.2018.01.046 [DOI] [PubMed] [Google Scholar]
- Milosevic, I., Giovedi S., Lou X., Raimondi A., Collesi C., Shen H., Paradise S., O’Toole E., Ferguson S., Cremona O., and De Camilli P.. 2011. Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron. 72:587–601. 10.1016/j.neuron.2011.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi, H., and Yasuda R.. 2012. Postsynaptic signaling during plasticity of dendritic spines. Trends Neurosci. 35:135–143. 10.1016/j.tins.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi, H., Wang H., and Yasuda R.. 2011. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 472:100–104. 10.1038/nature09823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch, J.D., Rostosky C.M., Gowrisankaran S., Arora A.S., Soukup S.F., Vidal R., Capece V., Freytag S., Fischer A., Verstreken P., et al. 2016. Endophilin-A Deficiency Induces the Foxo3a-Fbxo32 Network in the Brain and Causes Dysregulation of Autophagy and the Ubiquitin-Proteasome System. Cell Rep. 17:1071–1086. 10.1016/j.celrep.2016.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, M.D., Ryazantsev S., Hicke L., and Payne G.S.. 2016. Calmodulin Promotes N-BAR Domain-Mediated Membrane Constriction and Endocytosis. Dev. Cell. 37:162–173. 10.1016/j.devcel.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi, S., Fox R., Proulx C.D., Lin J.Y., Tsien R.Y., and Malinow R.. 2014. Engineering a memory with LTD and LTP. Nature. 511:348–352. 10.1038/nature13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata, Y., and Yasuda R.. 2018. Plasticity of Spine Structure: Local Signaling, Translation and Cytoskeletal Reorganization. Front. Synaptic Neurosci. 10:29. 10.3389/fnsyn.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, Y., Zhang C., Sun Z., Hong Z., Li K., Sun D., Yang Y., Tian C., Gong W., and Liu J.J.. 2013. PtdIns(4)P regulates retromer-motor interaction to facilitate dynein-cargo dissociation at the trans-Golgi network. Nat. Cell Biol. 15:417–429. 10.1038/ncb2710 [DOI] [PubMed] [Google Scholar]
- Noguchi, J., Hayama T., Watanabe S., Ucar H., Yagishita S., Takahashi N., and Kasai H.. 2016. State-dependent diffusion of actin-depolymerizing factor/cofilin underlies the enlargement and shrinkage of dendritic spines. Sci. Rep. 6:32897. 10.1038/srep32897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obashi, K., Matsuda A., Inoue Y., and Okabe S.. 2019. Precise Temporal Regulation of Molecular Diffusion within Dendritic Spines by Actin Polymers during Structural Plasticity. Cell Rep. 27:1503–1515.e8. 10.1016/j.celrep.2019.04.006 [DOI] [PubMed] [Google Scholar]
- Okamoto, K., Nagai T., Miyawaki A., and Hayashi Y.. 2004. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 7:1104–1112. 10.1038/nn1311 [DOI] [PubMed] [Google Scholar]
- Otsu, N. 1979. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 9:62–66. 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Park, M., Penick E.C., Edwards J.G., Kauer J.A., and Ehlers M.D.. 2004. Recycling endosomes supply AMPA receptors for LTP. Science. 305:1972–1975. 10.1126/science.1102026 [DOI] [PubMed] [Google Scholar]
- Park, M., Salgado J.M., Ostroff L., Helton T.D., Robinson C.G., Harris K.M., and Ehlers M.D.. 2006. Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron. 52:817–830. 10.1016/j.neuron.2006.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard, J.R., Kriz A., Galic M., Angliker N., Rajalu M., Vogt K.E., and Ruegg M.A.. 2016. The calcium sensor Copine-6 regulates spine structural plasticity and learning and memory. Nat. Commun. 7:11613. 10.1038/ncomms11613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y., Xu H.W., Davey F., Taylor M., Aiton J., Coote P., Fang F., Yao J., Chen D., Chen J.X., et al. 2008. Endophilin I expression is increased in the brains of Alzheimer disease patients. J. Biol. Chem. 283:5685–5691. 10.1074/jbc.M707932200 [DOI] [PubMed] [Google Scholar]
- Renard, H.F., Simunovic M., Lemière J., Boucrot E., Garcia-Castillo M.D., Arumugam S., Chambon V., Lamaze C., Wunder C., Kenworthy A.K., et al. 2015. Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature. 517:493–496. 10.1038/nature14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex, C.S., Chen L.Y., Sharma A., Liu J., Babayan A.H., Gall C.M., and Lynch G.. 2009. Different Rho GTPase-dependent signaling pathways initiate sequential steps in the consolidation of long-term potentiation. J. Cell Biol. 186:85–97. 10.1083/jcb.200901084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringstad, N., Nemoto Y., and De Camilli P.. 1997. The SH3p4/Sh3p8/SH3p13 protein family: binding partners for synaptojanin and dynamin via a Grb2-like Src homology 3 domain. Proc. Natl. Acad. Sci. USA. 94:8569–8574. 10.1073/pnas.94.16.8569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, R.H., Cudmore R.H., Tan H.L., Hong I., Zhang Y., and Huganir R.L.. 2020. Cortical Synaptic AMPA Receptor Plasticity during Motor Learning. Neuron. 105:895–908.e5. 10.1016/j.neuron.2019.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saneyoshi, T., Matsuno H., Suzuki A., Murakoshi H., Hedrick N.G., Agnello E., O’Connell R., Stratton M.M., Yasuda R., and Hayashi Y.. 2019. Reciprocal Activation within a Kinase-Effector Complex Underlying Persistence of Structural LTP. Neuron. 102:1199–1210.e6. 10.1016/j.neuron.2019.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoor, M., Stradal T.E., and Rottner K.. 2018. Cortactin: Cell Functions of A Multifaceted Actin-Binding Protein. Trends Cell Biol. 28:79–98. 10.1016/j.tcb.2017.10.009 [DOI] [PubMed] [Google Scholar]
- Schuske, K.R., Richmond J.E., Matthies D.S., Davis W.S., Runz S., Rube D.A., van der Bliek A.M., and Jorgensen E.M.. 2003. Endophilin is required for synaptic vesicle endocytosis by localizing synaptojanin. Neuron. 40:749–762. 10.1016/S0896-6273(03)00667-6 [DOI] [PubMed] [Google Scholar]
- Simon, C., Kusters R., Caorsi V., Allard A., Abou-Ghali M., Manzi J., Di Cicco A., Levy D., Lenz M., Joanny J.F., et al. 2019. Actin dynamics drive cell-like membrane deformation. Nat. Phys. 15:602–609. 10.1038/s41567-019-0464-1 [DOI] [Google Scholar]
- Smith, K.R., Kopeikina K.J., Fawcett-Patel J.M., Leaderbrand K., Gao R., Schürmann B., Myczek K., Radulovic J., Swanson G.T., and Penzes P.. 2014. Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron. 84:399–415. 10.1016/j.neuron.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup, S.F., Kuenen S., Vanhauwaert R., Manetsberger J., Hernández-Díaz S., Swerts J., Schoovaerts N., Vilain S., Gounko N.V., Vints K., et al. 2016. A LRRK2-Dependent EndophilinA Phosphoswitch Is Critical for Macroautophagy at Presynaptic Terminals. Neuron. 92:829–844. 10.1016/j.neuron.2016.09.037 [DOI] [PubMed] [Google Scholar]
- Soykan, T., Kaempf N., Sakaba T., Vollweiter D., Goerdeler F., Puchkov D., Kononenko N.L., and Haucke V.. 2017. Synaptic Vesicle Endocytosis Occurs on Multiple Timescales and Is Mediated by Formin-Dependent Actin Assembly. Neuron. 93:854–866.e4. 10.1016/j.neuron.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Spiering, D., and Hodgson L.. 2011. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adhes. Migr. 5:170–180. 10.4161/cam.5.2.14403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruno, T., Liu J., Zhang P., Fan Yx C., Egile C., Li R., Mueller S.C., and Zhan X.. 2001. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat. Cell Biol. 3:259–266. 10.1038/35060051 [DOI] [PubMed] [Google Scholar]
- van Bommel, B., Konietzny A., Kobler O., Bär J., and Mikhaylova M.. 2019. F-actin patches associated with glutamatergic synapses control positioning of dendritic lysosomes. EMBO J. 38:e101183. 10.15252/embj.2018101183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken, P., Koh T.W., Schulze K.L., Zhai R.G., Hiesinger P.R., Zhou Y., Mehta S.Q., Cao Y., Roos J., and Bellen H.J.. 2003. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 40:733–748. 10.1016/S0896-6273(03)00644-5 [DOI] [PubMed] [Google Scholar]
- Watanabe, S., Mamer L.E., Raychaudhuri S., Luvsanjav D., Eisen J., Trimbuch T., Söhl-Kielczynski B., Fenske P., Milosevic I., Rosenmund C., and Jorgensen E.M.. 2018. Synaptojanin and Endophilin Mediate Neck Formation during Ultrafast Endocytosis. Neuron. 98:1184–1197.e6. 10.1016/j.neuron.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, A.M., Karginov A.V., Kinley A.W., Weed S.A., Li Y., Parsons J.T., and Cooper J.A.. 2001. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11:370–374. 10.1016/S0960-9822(01)00098-7 [DOI] [PubMed] [Google Scholar]
- Yang, Y., Wang X.B., Frerking M., and Zhou Q.. 2008. Spine expansion and stabilization associated with long-term potentiation. J. Neurosci. 28:5740–5751. 10.1523/JNEUROSCI.3998-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Wei M., Xiong Y., Du X., Zhu S., Yang L., Zhang C., and Liu J.J.. 2015. Endophilin A1 regulates dendritic spine morphogenesis and stability through interaction with p140Cap. Cell Res. 25:496–516. 10.1038/cr.2015.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Chen J., Guo Z., Deng S., Du X., Zhu S., Ye C., Shi Y.S., and Liu J.J.. 2018. Endophilin A1 Promotes Actin Polymerization in Dendritic Spines Required for Synaptic Potentiation. Front. Mol. Neurosci. 11:177. 10.3389/fnmol.2018.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Q., Wang Y., Du F., Yan S., Hu G., Origlia N., Rutigliano G., Sun Q., Yu H., Ainge J., et al. 2018a. Overexpression of endophilin A1 exacerbates synaptic alterations in a mouse model of Alzheimer’s disease. Nat. Commun. 9:2968. 10.1038/s41467-018-04389-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Xu T., Ou S., Yuan J., Deng J., Li R., Yang J., Liu X., Li Q., and Chen Y.. 2018b. Endophilin A1 mediates seizure activity via regulation of AMPARs in a PTZ-kindled epileptic mouse model. Exp. Neurol. 304:41–57. 10.1016/j.expneurol.2018.02.014 [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Kim J., Insolera R., Peng X., Fink D.J., and Mata M.. 2011. Rho GTPase regulation of α-synuclein and VMAT2: implications for pathogenesis of Parkinson’s disease. Mol. Cell. Neurosci. 48:29–37. 10.1016/j.mcn.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]