Abstract
Background
Age-related macular degeneration (AMD) is one of the leading causes of vision loss in the elderly population. The application of artificial intelligence (AI) provides convenience for the diagnosis of AMD. This systematic review and meta-analysis aimed to quantify the performance of AI in detecting AMD in fundus photographs.
Methods
We searched PubMed, Embase, Web of Science and the Cochrane Library before December 31st, 2020 for studies reporting the application of AI in detecting AMD in color fundus photographs. Then, we pooled the data for analysis. PROSPERO registration number: CRD42020197532.
Findings
19 studies were finally selected for systematic review and 13 of them were included in the quantitative synthesis. All studies adopted human graders as reference standard. The pooled area under the receiver operating characteristic curve (AUROC) was 0.983 (95% confidence interval (CI):0.979–0.987). The pooled sensitivity, specificity, and diagnostic odds ratio (DOR) were 0.88 (95% CI:0.88–0.88), 0.90 (95% CI:0.90–0.91), and 275.27 (95% CI:158.43–478.27), respectively. Threshold analysis was performed and a potential threshold effect was detected among the studies (Spearman correlation coefficient: -0.600, P = 0.030), which was the main cause for the heterogeneity. For studies applying convolutional neural networks in the Age-Related Eye Disease Study database, the pooled AUROC, sensitivity, specificity, and DOR were 0.983 (95% CI:0.978–0.988), 0.88 (95% CI:0.88–0.88), 0.91 (95% CI:0.91–0.91), and 273.14 (95% CI:130.79–570.43), respectively.
Interpretation
Our data indicated that AI was able to detect AMD in color fundus photographs. The application of AI-based automatic tools is beneficial for the diagnosis of AMD.
Funding
Capital Health Research and Development of Special (2020–1–2052).
Keywords: Artificial intelligence, Deep learning, Convolutional neural networks, Algorithm, Agerelated macular degeneration
Research in context.
Evidence before this study
Artificial intelligence (AI) has shown high prospects in biomedical science, particularly in the diagnosis of ocular diseases. Some AI -based investigations have focused on the detection of age-related macular degeneration (AMD) from color fundus images, while the results have been inconsistent due to various confounding factors, such as databases, methods, and sample sizes. The assessment of AI performance has significant clinical and public health impacts for primary prevention and policy making.
Added value of this study
In this systematic review and meta-analysis, we searched electronic databases for studies reporting the application of AI in detecting AMD from retinal images. 19 studies were selected for systematic review and 13 of them were included in the meta-analysis. Reference standard was labeled by human graders in all included studies. The pooled area under the receiver operating characteristic curve (AUROC), sensitivity, specificity, and diagnostic odds ratio (DOR) with 95% confidence intervals (CIs) were 0.983 (95% CI: 0.979–0.987), 0.88 (95% CI: 0.88–0.88), 0.90 (95% CI: 0.90–0.91), and 275.27 (95% CI: 158.43–478.27), respectively. The main cause for the high heterogeneity among the studies was threshold effects (Spearman correlation coefficient: −0.600, P = 0.030). Age-Related Eye Disease Study (AREDS) database was the most commonly used data set for the development and validation of AI models. For studies applying convolutional neural networks (CNNs) in AREDS database, the pooled AUROC, was 0.983 (95% CI: 0.978–0.988).
Implications of all the available evidence
Our study found that AI is promising in detecting AMD from color fundus photographs. The application of AI-based automatic tools can provide substantial benefits for the screening and diagnosis of AMD. However, since the diagnostic power of the AI-based algorithms decreases in larger data sets, caution is needed when applying these algorithms in a larger population under different settings and conditions.
Alt-text: Unlabelled box
1. Introduction
Age-related macular degeneration (AMD) is an ocular disorder that affects the macular region of the retina. With increasing lifespans, AMD has emerged as one of the leading causes of vision impairment in the elderly population in both developing and developed countries. [1] By 2020, the number of people with AMD is projected to be approximately 196 million globally, and the number will increase to 288 million by 2040, [2] representing a major public health issue with substantial socioeconomic impacts.
Early AMD includes clinical signs such as drusen and abnormalities of the retinal pigment epithelium (RPE), while advanced AMD presents neovascular (also called wet or exudative AMD) or central geographic atrophy (also called dry or nonexudative AMD). Advanced AMD often leads to the loss of central visual acuity, which causes considerable impacts on quality of life. [3,4] The pooled global prevalence of any AMD, early AMD, and advanced AMD in the population aged 45–85 years old is 8.69%, 8.01%, and 0.37%, respectively. [2] It has also been estimated that the 15-year incidence was 22.7% for early AMD and 6.8% for advanced AMD in subjects aged more than 49 years old. [5] Due to the high incidence and risk, it is urgent to improve the efficiency of the screening and diagnosis of AMD.
Artificial intelligence (AI) is a branch of computer science that aims to build machines to mimic brain function, which has attracted considerable global interest. [6] Machine learning is a kind of AI process in which the machine writes its programming and learns to achieve a task on its own. [7] Deep learning (DL) is a subset of machine learning and is based on the framework of an artificial neural network (ANN), which is composed of multiple inputs and a single output. The neuron between the input and output layers (known as hidden layers) receives multiple signals from the dendrites and sends a single stream of action through the axon. [8] Each hidden layer can learn different features for the stimuli, which allows the model to complete complex tasks. Among the various DL architectures, convolutional neural networks (CNNs) show the best performance in analyzing imaging data. [9] CNNs include special layers that apply a mathematical filtering procedure called convolution, which makes each neuron process data only for its receptive field and response to visual stimuli. [9] The development of CNNs plays a critical role in bringing DL into the spotlight.
To date, AI has shown high prospects in biomedical science, particularly in the diagnosis of ocular diseases. AI techniques have been applied for detecting diabetic retinopathy (DR…), AMD, retinopathy of prematurity (ROP), glaucoma, and papilledema from multimodality imaging, including fundus photographs, optical coherence tomography (OCT), and fundus fluorescence angiography (FFA). [10], [11], [12], [13], [14] Although some investigations have tried to assess the performance of AI in detecting AMD, the results have been inconsistent due to various confounding factors, such as databases, methods, and sample sizes. The assessment of AI performance has significant clinical and public health impacts for primary prevention and policy making. Therefore, we performed this systematic review and meta-analysis to quantify the performance of AI for the detection of AMD in color fundus photographs.
2. Methods
2.1. Literature search
The protocol of the meta-analysis was registered in PROSPERO website (University of York, York, UK) with a registration number of CRD42020197532. We searched PubMed, Embase, Web of Science and the Cochrane Library using the following keywords with various combinations: “deep learning”, “DL”, “artificial intelligence”, “AI”, “algorithm”, “neural networks”, “CNN”, “age-related macular degeneration”, “macular degeneration”, “geographic atrophy”, and “AMD”. The searches were from inception to December 31st, 2020, and were limited to human studies.
2.2. Study selection
The inclusion criteria were as follows: (1) studies reporting an outcome of the AI-based algorithm and AMD detection; (2) studies presenting a clear definition of AMD; (3) studies providing clear information about the database and number of images in various data sets; (4) studies including more than 50 fundus photographs for validation; (5) studies providing information on evaluation indices, such as sensitivity (SEN), specificity (SPE), accuracy, and area under the curve (AUC); (6) studies describing the algorithms and procedures used in AMD detection; (7) studies presenting clear information of the reference standard; and (8) English-language literature only.
The exclusion criteria were as follows: (1) ongoing investigations or unpublished studies; (2) studies applying multimodality imaging, such as OCT and FFA; (3) publication forms including reviews, meta-analyses, comments, letters, and editorials; and (4) no access to obtain the original data. The articles were independently screened and selected by two researchers (LD, RHZ), and any disagreements between them were resolved through consensus.
2.3. Quality assessment
The articles that passed the primary screening were then reviewed by the two reviewers (LD, RHZ) individually. They independently assessed the quality of the studies according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement. [15] Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool was applied for the risk of bias assessment of the included studies. [16] The QUADAS-2 scale consists of 4 aspects for risk of bias including patient selection, index test, reference standard, and flow & timing as well as 3 domains for applicability concerns including patient selection, index test, and reference standard. The risk of bias was classified into 3 categories (i.e. low, high, and unclear risk bias). Studies with low quality or with evident defects in design and procedure were excluded from this survey. Any disagreements between the two authors were resolved by discussion or judged by senior researchers (WBW).
2.4. Data extraction
The following data were extracted: (1) the basic characteristics of the included studies and participants, including the methods, algorithms, databases, sample sizes, outcomes, and procedures; and (2) the evaluation indices of the algorithms, including the number of true positive (TP), true negative (TN), false positive (FP), and false negative (FP) outcomes as well as the SEN, SPE, accuracy, and AUC.
2.5. Statistical analysis
The pooled quantitative analysis, threshold analysis, meta-regression, and subgroup analysis were performed using Meta-Disc 1.4 software (U. de Bioestadística, Madrid (España)). The flow diagram for literature selection and quality assessment for the included studies were performed using RevMan 5.3 software (Cochrane Collaboration, Denmark). Some included studies adopted referable AMD as an outcome, which was defined as intermediate and advanced AMD (Table 1). The pooled area under the receiver operating characteristic curve (AUROC), SEN, SPE, positive likelihood ratio (LR+), and negative likelihood ratio (LR-) were calculated with 95% confidence intervals (CIs) and were presented in forest plots. The diagnostic odds ratio (DOR) was calculated to evaluate how much greater the odds of having AMD were for the participants with a positive test result than for those with a negative test result. The statistical heterogeneity among studies was analyzed using the chi-squared test and was presented as the I2 statistic (less than 50%: low heterogeneity; 50%−75%: moderate heterogeneity; and more than 75%: high heterogeneity). Fixed-effects models were used when the heterogeneity was lower than 50%; otherwise, random-effects models were applied. Threshold analysis was applied to test whether the heterogeneity resulted from the threshold effects. [17] Meta-regression with the backward method was used to detect the cause of heterogeneity. Then, subgroup analysis was performed according to the various methods (CNN and support vector machine (SVM)), number of images, definition of AMD, publication year, and regions (Asian countries and the western countries). Two-tailed P<0.05 was considered statistically significant.
Table 1.
Definition of referable AMD and non-referable AMD in this study.
| Category | Stage | Definition | Classification |
|---|---|---|---|
| 1 | No AMD | No drusen or only small drusen ≤63 μm, and no pigment abnormalities | Non-referable AMD |
| 2 | Early AMD | Medium drusen >63 μm and ≤125 μm, and no pigment abnormalities | |
| 3 | Intermediate AMD | Large drusen >125 μm or any pigment abnormalities | Referable AMD |
| 4 | Advanced AMD | Neovascular AMD or geographical atrophy |
AMD: age-related macular degeneration.
2.6. Role of funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, and writing of the manuscript. The corresponding author had full access to all study data and had final responsibility for the decision to submit for publication.
3. Results
3.1. Study selection
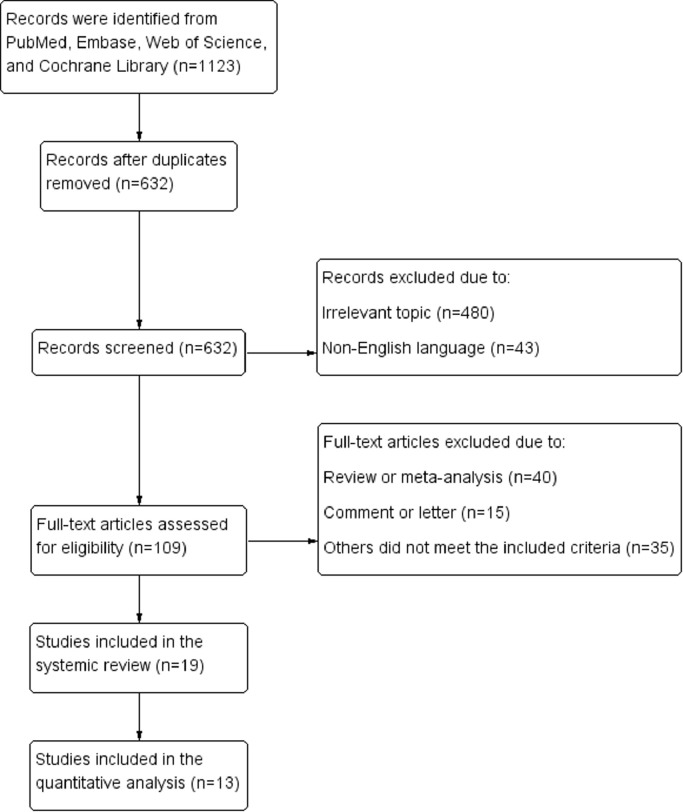
Fig. 1 shows the literature selection process. At the initial searches, a total of 1123 articles were potentially eligible for inclusion (432 from PubMed, 373 from Embase, 317 from Web of Science, and 1 from the Cochrane Library). After primary screening and the removal of duplicates, 109 potentially eligible articles were selected. After full-text reviews, 19 eligible studies with supervised learning approaches were finally selected for inclusion in the systematic review, [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36] and 13 of them were included in the quantitative synthesis. [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30]
Fig. 1.
Flow diagram of literature selection.
3.2. Study characteristics
The basic characteristics of the included studies were presented in Table 2. These studies included more than 1.2 million color fundus images for training, validation, and testing. CNN was applied in 12 studies, SVM was used in 6 studies, ANN was used in 1 study. And 1 study applied both SVM and random forest (RF). The Age-Related Eye Disease Study (AREDS) database was the most commonly used database and was adopted in 9 studies. [37] Referable AMD was regarded as the primary outcome in 8 investigations, and AMD severity with various classes was evaluated in 8 studies. In addition, all studies adopted human graders as the reference standard.
Table 2.
Basic characteristics of the included studies.
| First author | Publication year | Country | Database | Total images | Method | Outcome | Classification | Performance |
|---|---|---|---|---|---|---|---|---|
| Keenan [18] | 2019 | United States | AREDS | 59,812 | CNN | Dry AMD | Disease/no disease | ACC: 0.965; AUC: 0.976 |
| Zapata [19] | 2020 | Spain | Optretina | 306,302 | CNN | AMD | Disease/no disease | ACC: 0.863; AUC: 0.936 |
| Zheng [20] | 2012 | United Kingdom | ARIA, STARE | 258 | SVM | AMD | Disease/no disease | ACC: 0.996 |
| Kunumpol [21] | 2017 | Thailand | STARE | 106 | ANN | AMD | Disease/no disease | ACC: 0.989 |
| Mookiah [22] | 2014a | Singapore | Private dataset | 540 | SVM | Dry AMD | Disease/no disease | ACC: 0.937 |
| Keel [23] | 2019 | Australia | LabelMe | 56,113 | CNN | Wet AMD | Disease/no disease | ACC: 0.965; AUC: 0.995 |
| González-Gonzalo [24] | 2019 | The Netherlands | 1. DR…-AMD 2. AREDS |
134,421 | CNN | Referable AMDa | Disease/no disease | ACC1: 0.880; AUC1: 0.949 ACC2: 0.859; AUC2: 0.927 |
| Burlina [25] | 2017a | United States | AREDS | 133,821 | CNN | Referable AMD | Disease/no disease | ACC: 0.916; AUC: 0.96 |
| Burlina [26] | 2017b | United States | AREDS | 5664 | CNN | Referable AMD | 1. Disease/no disease 2. AMD severity (4 classes) |
ACC1: 0.934 ACC2: 0.794 |
| Ting [27] | 2017 | Singapore | SIDRP | 108,558 | CNN | Referable AMD | Disease/no disease | ACC: 0.888; AUC: 0.932 |
| Kankanahalli [28] | 2013 | United States | AREDS | 2772 | CNN | Referable AMD | 1. Disease/no disease 2. AMD severity (3 classes) |
ACC1: 0.955 ACC2: 0.918 |
| Burlina [29] | 2011 | United States | Private dataset | 66 | SVM | AMD | Disease/no disease | ACC: 0.955 |
| Bhuiyan [30] | 2020 | United States | AREDS | 116,875 | CNN | 1. Referable AMD 2. AMD |
1. Disease/no disease 2. AMD severity (4 classes) |
ACC1: 0.992 ACC2: 0.961 |
| Phan [31] | 2016 | Canada | Private dataset | 279 | SVM, RF | 1. AMD 2. Referable AMD |
Disease/no disease | AUC1: 0.877 AUC2: 0.899 |
| Govindaiah [32] | 2018 | United States | AREDS | 116,875 | CNN | 1. Referable AMD 2. AMD |
1. Disease/no disease 2. AMD severity (4 classes) |
ACC1: 0.953 ACC2: 0.861 |
| Grassmann [33] | 2018 | German | AREDS | 120,656 | CNN | AMD | AMD severity (13 classes) | ACC: 0.633 |
| Mookiah [34] | 2014b | Singapore | 1. Private dataset 2. ARIA 3. STARE |
784 | SMV | AMD | AMD severity (4 classes) | ACC1: 0.902 ACC2: 0.951 ACC3: 0.950 |
| Peng [35] | 2019 | United States | AREDS | 59,302 | CNN | AMD | AMD severity (6 classes) | ACC: 0.671 |
| Mookiah [36] | 2015 | Singapore | 1. Private dataset 2. ARIA 3. STARE |
784 | SMV | AMD | AMD severity (4 classes) | ACC1: 0.935 ACC2: 0.914 ACC3: 0.978 |
AREDS: Age-Related Eye Disease Study, CNN: convolutional neural networks, AMD: age-related macular disease, ACC: Accuracy, AUC: area under curve, ARIA: Automated Retinal Image Analysis, STARE: Structured Analysis of the Retina, SVM: support vector machine, ANN: artificial neural network, DR…: diabetic retinopathy, SIDRP: Singapore National Diabetic Retinopathy Screening Program, RF: random forest.
Referable AMD was defined as intermediate and advanced AMD.
3.3. Quality assessment
In the present study, we also evaluated the risk of bias of the included studies based on the QUADAS-2 tool (Fig. 2). Ten included studies were of high quality with low risk of bias and applicability concerns. The risk of bias for patient selection was unclear for 8 studies, and only 1 study had an unclear risk of bias for the reference standard. High risk of bias or applicability concerns was not detected in any included study
Fig. 2.
Bias assessment of the included studies via Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool.
3.4. Performance of AI in AMD detection
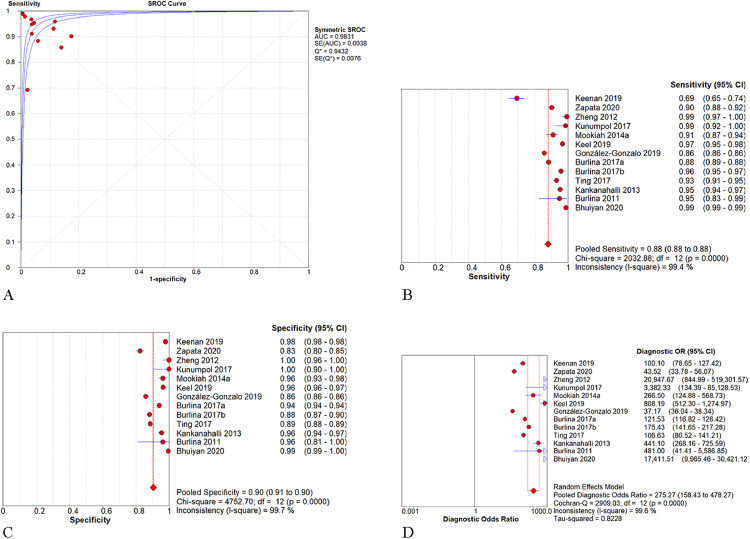
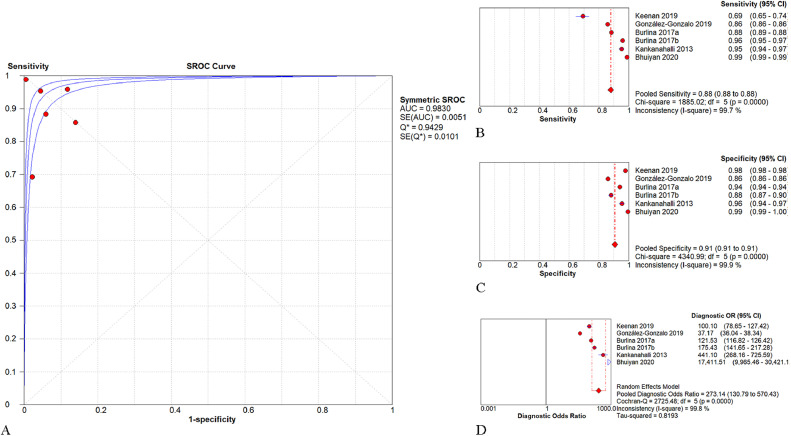
As shown in Fig. 3, the pooled AUROC of AI-based algorithms in detecting AMD or referable AMD was 0.983 (95% CI: 0.979–0.987). The pooled SEN, SPE, and DOR were 0.88 (95% CI: 0.88–0.88; I2=98.7%), 0.90 (95% CI: 0.90–0.91; I2=99.7%), and 275.27 (95% CI: 158.43–478.27; I2=99.6%), respectively. For studies applying CNN in the AREDS database, the pooled AUROC, SEN, SPE, and DOR were 0.983 (95% CI: 0.978–0.988), 0.88 (95% CI: 0.88–0.88; I2=99.0%), 0.91 (95% CI: 0.91–0.91; I2=99.2%), and 273.14 (95% CI: 130.79–570.43; I2=90.0%), respectively (Fig. 4).
Fig. 3.
Performance of artificial intelligence for the detection of AMD. -Fig. 3A. The pooled area under the receiver operating characteristic curve (AUROC) was 0.983 (95% CI: 0.979–0.987). -Fig. 3B. The pooled sensitivity was 0.88 (95% CI: 0.88–0.88). -Fig. 3C. The pooled specificity was 0.90 (95% CI: 0.90–0.91). -Fig. 3D. The pooled diagnostic odds ratio was 275.27 (95% CI: 158.43–478.27).
Fig. 4.
Performance of the convolutional neural network (CNN) models for the detection of AMD in the AREDS database. -Fig. 4A. The pooled area under the receiver operating characteristic curve (AUROC) was 0.983 (95% CI: 0.978–0.988). -Fig. 4B. The pooled sensitivity was 0.88 (95% CI: 0.88–0.88). -Fig. 4C. The pooled specificity was 0.91 (95% CI: 0.91–0.91). -Fig. 4D. The pooled diagnostic odds ratio was 273.14 (95% CI: 130.79–570.43).
3.5. Heterogeneity analysis
Since high heterogeneity was found among the studies, we first applied threshold analysis to test whether there was a threshold effect. The results showed a potential threshold existed among the included studies (Spearman correlation coefficient: −0.600, P = 0.030). Then, meta-regression was performed to analyze the cause of heterogeneity. Potential factors included various methods (classified as CNN, SVM, and others), databases (classified as AREDS and others), number of images for validation (classified as <500, 500–5000, and >5000), outcomes (classified as AMD and referable AMD), publication year (classified as before 2015 and after 2015), and regions (classified as Asian countries and Western countries). The results showed that the DOR was not correlated with any factors (all P values>0.10). However, when excluding Bhuiyan's study, [29] the DOR was significantly lower in studies with larger validation data sets (P = 0.018), which contributed most to the heterogeneity.
3.6. Subgroup analysis
Subgroup analysis was performed according to different methods, number of images for validation, definition of AMD, publication year, and regions (Table 3). The results showed that SVM had a higher DOR (917; 95% CI: 97–8861; I2=71.4%) than CNN (225; 95% CI: 123–409; I2=99.7%). The pooled AUC for detection of AMD and referable AMD was 0.993 (95% CI: 0.984–1.000) and 0.983 (95% CI: 0.978–0.988), respectively. The DOR and AUC were lower in studies with larger validation data sets. Similar AUCs were detected for studies from Asian countries and studies from Western countries (0.979 versus 0.984).
Table 3.
Subgroup analysis showing the performance of the artificial intelligence for the detection of AMD.
| Variables | No. of study | AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | LR+ (95% CI) | LR- (95% CI) | DOR (95% CI) | Heterogeneity for DOR |
|
|---|---|---|---|---|---|---|---|---|---|
| I2,% | P value | ||||||||
| Methods | |||||||||
| CNN | 9 | 0.980 (0.975–0.985) | 0.88 (0.88–0.88) | 0.90 (0.90–0.91) | 16.0 (11.1–23.1) | 0.07 (0.06–0.10) | 225 (123–409) | 99.7 | <0.001 |
| SVM | 3 | 0.994 (0.988–1.000) | 0.94 (0.92–0.96) | 0.97 (0.95–0.99) | 30.9 (11.9–79.8) | 0.04 (0.01–0.16) | 917 (97–8861) | 71.4 | 0.030 |
| Images for validation | |||||||||
| <500 | 3 | 0.997 (0.995–1.000) | 0.99 (0.96–1.00) | 0.99 (0.97–1.00) | 54.6 (13.9–215.0) | 0.02 (0.01–0.07) | 2656 (286–24,635) | 42.0 | 0.178 |
| 500–5000 | 4 | 0.982 (0.970–0.994) | 0.93 (0.92–0.94) | 0.93 (0.93–0.94) | 16.3 (5.7–46.8) | 0.07 (0.03–0.13) | 252 (50–1274) | 98.1 | <0.001 |
| >5000 | 6 | 0.980 (0.974–0.985) | 0.88 (0.88–0.88) | 0.90 (0.90–0.91) | 17.0 (10.9–26.5) | 0.08 (0.06–0.11) | 216 (105–445) | 99.8 | <0.001 |
| Outcomes | |||||||||
| AMDa | 4 | 0.993 (0.984–1.000) | 0.92 (0.90–0.93) | 0.85 (0.83–0.87) | 29.2 (3.4–248.4) | 0.04 (0.01–0.14) | 853 (39–18,403) | 88.2 | <0.001 |
| Referable AMDb | 6 | 0.983 (0.978–0.988) | 0.88 (0.88–0.88) | 0.90 (0.90–0.90) | 15.8 (10.2–24.3) | 0.06 (0.05–0.08) | 276 (132–579) | 99.8 | <0.001 |
| Publication year | |||||||||
| Before 2015 | 4 | 0.989 (0.985–0.993) | 0.95 (0.94–0.96) | 0.96 (0.95–0.97) | 22.7 (16.9–30.3) | 0.05 (0.03–0.10) | 474 (197–1142) | 58.1 | 0.067 |
| After 2015 | 9 | 0.980 (0.975–0.985) | 0.88 (0.88–0.88) | 0.90 (0.90–0.91) | 15.9 (10.8–23.3) | 0.08 (0.06–0.10) | 224 (120–418) | 99.7 | <0.001 |
| Regions | |||||||||
| Asian countries | 3 | 0.979 (0.970–0.988) | 0.93 (0.91–0.94) | 0.89 (0.88–0.89) | 20.6 (2.6–163.5) | 0.08 (0.06–0.11) | 212 (73–613) | 77.9 | 0.011 |
| The western countries | 10 | 0.984 (0.980–0.988) | 0.88 (0.88–0.88) | 0.91 (0.91–0.91) | 19.0 (12.0–30.2) | 0.07 (0.05–0.09) | 288 (155–536) | 99.7 | <0.001 |
AUC: area under curve, CI: confidence interval, LR+: positive likelihood ratio, LR-: negative likelihood ratio, DOR: diagnostic odds ratio, CNN: convolutional neural networks, SVM: support vector machine.
Studies detecting dry-AMD only or wet-AMD only were excluded in this analysis.
Referable AMD was defined as intermediate and advanced AMD.
4. Discussion
Our results demonstrate that AI-based algorithms are able to detect AMD in fundus images with a pooled AUC, SEN, and SPE of 0.983 (95% CI: 0.979–0.987), 0.88 (95% CI: 0.88–0.88), and 0.90 (95% CI: 0.90–0.91), respectively, which is almost comparable to the performance of retinal specialists. [18,26,33,35] Although AMD remains one of the leading causes of irreversible vision impairment worldwide, the incidence of wet AMD with visual loss has decreased due to the introduction of treatment targeting vascular endothelial growth factor (VEGF). [38] With available effective treatment, early diagnosis and treatment are crucial for these patients to retain functional vision. Therefore, the application of AI-based tools for AMD detection may provide substantial benefits in disease management.
In this study, the pooled DOR of AI models for detecting AMD was 275.27 (95% CI: 158.43–478.27). The value of a DOR ranges from 0 to infinity, with higher values indicating better discriminatory test performance. A value of 1 means that a test does not discriminate between patients with the disorder and those without it. And values lower than 1 mean improper test interpretation (more negative tests among the diseased). The DOR offers considerable advantages in diagnostic meta-analysis that pools data from various studies into summary estimates with increased precision. [39]
The AREDS database is the largest publicly available database with more than 130 thousand fundus photographs and has been broadly applied for investigating AMD. [37] In this study, the AREDS database was used in 9 included studies. For studies applying CNN in the AREDS database, the pooled AUROC, SEN, SPE, and DOR were 0.983 (95% CI: 0.978–0.988), 0.88 (95% CI: 0.88–0.88), 0.91 (95% CI: 0.91–0.91), and 273.14 (95% CI: 130.79–570.43), respectively. However, it should be noted that some of the nuances of hard drusen and age-related changes for clinical classification of AMD as enlightened by Ferris et al. [40] did not exist in the 1980s during AREDS, which might make AREDS database inadequate for develop AI. Moreover, these photographs were all film images that were digitized.
The present study shows that the diagnostic power of AI is lower in studies with larger validation data sets, with only Bhuiyan's study being an exception. [30] As a more recent research, Bhuiyan et al. trained and validated the CNN-based algorithm in AREDS database, which finally achieved an accuracy of 99.2% for detecting referable AMD. So far, this is the best screening accuracy among such existing models. However, it should be also noticed that these models are tested in research data sets rather than real-world data. Caution is needed when applying AI-based screening in larger populations under different settings and conditions.
In this study, CNN and SVM were the most commonly used models, both of which showed high SEN and SPE. CNN contains multilayer neurons that can recognize visual patterns and learn the features directly from the raw image pixels. [41] There are various types of CNN architectures, such as AlexNet, Inception v1 (GoogLeNet), and CifarNet. [42] SVM is a machine learning that classifies data in categories with supervised learning. [43] CNN and SVM are both good at data handling, and the optimal choice for use depends on the study aims and data types.
The performance of different AI-based algorithms varies a lot in the included studies, with accuracy from 0.633 to 0.996. Many factors may account for it. First, different architectures of algorithms are basic cause for the performance variation. Second, data size for training and validation of the algorithms, as mentioned above, is another reason. Third, the quality of the included images for algorithm development is also an important factor. Fourth, there still lacks reference standards to define AMD and threshold effects exist among the studies. Therefore, comprehensive evaluation should be placed when we compare the performance of the different AI system.
It is interesting that all included studies were performed in Asia, western Europe, and the United States, while no study from Africa, eastern Europe, and the Middle East was found. This may imply that AMD has become one of the leading causes for vision loss in those countries and the automatic tools for AMD detection are more needed in regions with more populations.
Other than fundus images, it has also been reported that AI can learn to detect AMD from multimodality imaging data. Some researchers have succeeded in developing CNN models to detect advanced AMD based on spectral domain optical coherence tomography (SD-OCT) images. [44,45] Yoo et al. [46] demonstrated that the combination of OCT and fundus images could improve the diagnostic accuracy of their DL models for detection of AMD over fundus images alone. Another study further detected a higher accuracy for CNN-based models trained by multimodality imaging (fundus photographs, OCT, and angio-OCT) than those trained by a single imaging modality. [47] Moreover, a DL algorithm was trained to identify geographic atrophy in fundus autofluorescence (FAF) images. [48] Future interest may focus on the methods to improve diagnostic power or disease progression prediction using multimodality image analysis. However, it should be clarified that, so far at least, none of these techniques is applicable for screening in the primary care setting due to the much higher cost of the devices than non-mydriatic automatic cameras. Additionally, they may not be useful for retinal specialists who can read the images themselves.
Our results have some significant clinical and public health implications. First, a fundus camera with AI-based software may help ophthalmologists reduce the workload as well as the rates of misdiagnosis and missed diagnosis. Second, implementation of the AI system in the community can help to detect AMD at an early stage so that necessary management will be applied to prevent the conversion to advanced AMD. Third, AI significantly improves the efficiency for screening ocular disorders, particularly in remote areas where skilled ophthalmologists are not always available. However, several challenges also exist and should be addressed. First, algorithms are commonly developed to detect only one disease or sign; thus, some other important eye conditions may be missed. Second, most algorithms are trained on limited data sets, and the performance remains doubtful when validated in larger cohorts under different settings and conditions. Third, the diagnostic power of AI algorithms depends on the quality of the data, and image quality software is needed to reject images that are unreadable. Fourth, the feasibility and performance of AI software compared with those of clinical physicians are still unclear, and whether patients will trust the machines is another important question. Furthermore, since AI is a “black box”, [49] it may affect the perception and acceptance of AI in further applications. The main obstacle to deploy AI may be the risk of missing false negative cases and no action would be taken until routine physical examinations. Participants undergoing AI-assisted screening should be informed that referrals are needed if any symptoms occur. So far, therefore, physicians cannot be free from reading thousands of normal tests.
The limitations of the present study should also be noted. First, different definitions of AMD among the included studies might have influenced the pooled analysis, though subgroup analysis was performed. Second, some included studies involved relatively small sample sizes, which may reduce the representativeness of AI performance. Third, the heterogeneity among those investigations was large, which mainly resulted from threshold effects. To reduce the effects of heterogeneity for the analysis, we applied random-effects models for the pooled analysis. We also performed subgroup analysis to dig out the potential factors that resulted in the high heterogeneity. Fourth, we did not compare the performance between AI and human experts since limited data were available. To some extent, the diagnostic performance of AI models cannot be well presented unless comparing to human ophthalmologists. Fifth, we evaluated only the diagnostic power of AI in detecting AMD, while the performance for classifying AMD severity was not assessed. AMD is a spectrum of presentations with various classifications, such as referable/non referable AMD, dry/wet AMD, and early/advanced AMD, etc. Future interest may focus on optimizing AI models in assisting AMD classifications for clinical application. Sixth, the search of this study was only restricted to standard sources, and other sources including conference abstracts, ongoing clinical trials were excluded, which might increase the risk of publication bias. Seventh, a potential bias for pooled analysis might exist since 9 included studies used the same database (AREDS database), while this could be also an advantage of being able to compare performance of different algorithms in the same population. Additionally, we failed to provide data on AI-based prediction of AMD progression. Predicting the AMD progression may help to improve the therapeutic regimens and management of disease.
Our study found that AI is promising in detecting AMD from color fundus photographs. The application of AI-based automatic tools can provide substantial benefits for the diagnosis of AMD. However, AI is likely to have better ability to detect advanced AMD than early AMD, similarly to humans, which may have contributed to the very high AUCs observed. Since the diagnostic power of the AI system decreases in larger data sets and the performance has not been tested in the real world, caution is needed when applying these algorithms in populations under different settings and conditions. And particularly if such algorithms are applied autonomously, additional safeguards must be implemented.
Author contributions
Conception and design of the research: LD and WBW; Acquisition and interpretation of the data: LD, QY, RHZ and WBW; Statistical analysis and writing of the manuscript: LD, QY and RHZ; Critical revision of the manuscript: LD, QY and WBW.
Funding
This study was supported by the Capital Health Research and Development of Special (2020-1-2052).
Data sharing statement
The original data generated in the current study are available from the corresponding author on reasonable request.
Declaration of Competing Interest
All authors declare there is no conflict of interest.
References
- 1.Mitchell P., Liew G., Gopinath B., Wong T.Y. Age-related macular degeneration. Lancet. 2018;392(10153):1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 2.Wong W.L., Su X., Li X., Cheung C.M., Klein R., Cheng C.Y., Wong T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2(2):e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 3.Coleman H.R., Chan C.C., Ferris F.L., 3rd, Chew E.Y. Age-related macular degeneration. Lancet. 2008;372:1835–1845. doi: 10.1016/S0140-6736(08)61759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim L.S., Mitchell P., Seddon J.M., Holz F.G., Wong T.Y. Age-related macular degeneration. Lancet. 2012;379:1728–1738. doi: 10.1016/S0140-6736(12)60282-7. [DOI] [PubMed] [Google Scholar]
- 5.Joachim N., Mitchell P., Burlutsky G., Kifley A., Wang J.J. The incidence and progression of age-related macular degeneration over 15 years: the blue mountains eye study. Ophthalmology. 2015;122(12):2482–2489. doi: 10.1016/j.ophtha.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Rahimy E. Deep learning applications in ophthalmology. Curr Opin Ophthalmol. 2018;29:254–260. doi: 10.1097/ICU.0000000000000470. [DOI] [PubMed] [Google Scholar]
- 7.Samuel A.L. Some studies in machine learning using the game of checkers. IBM J Res Dev. 2000;44:206–226. [Google Scholar]
- 8.Kriegeskorte N., Golan T. Neural network models and deep learning. Curr Biol. 2019;29(7):R231–R236. doi: 10.1016/j.cub.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt-Erfurth U., Sadeghipour A., Gerendas B.S., Waldstein S.M., Bogunović H. Artificial intelligence in retina. Prog Retin Eye Res. 2018;67:1–29. doi: 10.1016/j.preteyeres.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Gulshan V., Peng L., Coram M. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 11.Gargeya R., Leng T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology. 2017;124:962–969. doi: 10.1016/j.ophtha.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Li Z., He Y., Keel S. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology. 2018;125:1199–1206. doi: 10.1016/j.ophtha.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Brown J.M., Campbell J.P., Beers A. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol. 2018;136:803–810. doi: 10.1001/jamaophthalmol.2018.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milea D., Najjar R.P., Zhubo J. Artificial intelligence to detect papilledema from ocular fundus photographs. N Engl J Med. 2020;382(18):1687–1695. doi: 10.1056/NEJMoa1917130. [DOI] [PubMed] [Google Scholar]
- 15.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 16.Whiting P.F., Rutjes A.W., Westwood M.E. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 17.Carpenter C.R., Hussain A.M., Ward M.J. Spontaneous subarachnoid hemorrhage: a systematic review and meta-analysis describing the diagnostic accuracy of history, physical examination, imaging, and lumbar puncture with an exploration of test thresholds. Acad Emerg Med. 2016;23(9):963–1003. doi: 10.1111/acem.12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keenan T.D., Dharssi S., Peng Y. A deep learning approach for automated detection of geographic atrophy from color fundus photographs. Ophthalmology. 2019;126(11):1533–1540. doi: 10.1016/j.ophtha.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zapata M.A., Royo-Fibla D., Font O. Artificial intelligence to identify retinal fundus images, quality validation, laterality evaluation, macular degeneration, and suspected glaucoma. Clin Ophthalmol. 2020;14:419–429. doi: 10.2147/OPTH.S235751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Y., Hijazi M.H., Coenen F. Automated "disease/no disease" grading of age-related macular degeneration by an image mining approach. Invest Ophthalmol Vis Sci. 2012;53(13):8310–8318. doi: 10.1167/iovs.12-9576. [DOI] [PubMed] [Google Scholar]
- 21.Kunumpol P., Umpaipant W., Kanchanaranya N. Automated age-related macular degeneration screening system using fundus images. Conf Proc IEEE Eng Med Biol Soc. 2017;2017:1469–1472. doi: 10.1109/EMBC.2017.8037112. [DOI] [PubMed] [Google Scholar]
- 22.Mookiah M.R., Acharya U.R., Koh J.E. Decision support system for age-related macular degeneration using discrete wavelet transform. Med Biol Eng Comput. 2014;52(9):781–796. doi: 10.1007/s11517-014-1180-8. [DOI] [PubMed] [Google Scholar]
- 23.Keel S., Li Z., Scheetz J. Development and validation of a deep-learning algorithm for the detection of neovascular age-related macular degeneration from colour fundus photographs. Clin Exp Ophthalmol. 2019;47(8):1009–1018. doi: 10.1111/ceo.13575. [DOI] [PubMed] [Google Scholar]
- 24.González-Gonzalo C., Sánchez-Gutiérrez V., Hernández-Martínez P. Evaluation of a deep learning system for the joint automated detection of diabetic retinopathy and age-related macular degeneration. Acta Ophthalmol. 2020;98(4):368–377. doi: 10.1111/aos.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burlina P.M., Joshi N., Pekala M., Pacheco K.D., Freund D.E., Bressler N.M. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017;135(11):1170–1176. doi: 10.1001/jamaophthalmol.2017.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burlina P., Pacheco K.D., Joshi N., Freund D.E., Bressler N.M. Comparing humans and deep learning performance for grading AMD: a study in using universal deep features and transfer learning for automated AMD analysis. Comput Biol Med. 2017;82:80–86. doi: 10.1016/j.compbiomed.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ting D.S.W., Cheung C.Y., Lim G. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318(22):2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kankanahalli S., Burlina P.M., Wolfson Y., Freund D.E., Bressler N.M. Automated classification of severity of age-related macular degeneration from fundus photographs. Invest Ophthalmol Vis Sci. 2013;54(3):1789–1796. doi: 10.1167/iovs.12-10928. [DOI] [PubMed] [Google Scholar]
- 29.Burlina P., Freund D.E., Dupas B., Bressler N. Automatic screening of age-related macular degeneration and retinal abnormalities. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:3962–3966. doi: 10.1109/IEMBS.2011.6090984. [DOI] [PubMed] [Google Scholar]
- 30.Bhuiyan A., Wong T.Y., Ting D.S.W., Govindaiah A., Souied E.H., Smith R.T. Artificial intelligence to stratify severity of age-related macular degeneration (AMD) and predict risk of progression to late AMD. Transl Vis Sci Technol. 2020;9(2):25. doi: 10.1167/tvst.9.2.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan T.V., Seoud L., Chakor H., Cheriet F. Automatic screening and grading of age-related macular degeneration from texture analysis of fundus Images. J Ophthalmol. 2016;2016 doi: 10.1155/2016/5893601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Govindaiah A., Smith R.T., Bhuiyan A. A new and improved method for automated screening of age-related macular degeneration using ensemble deep neural networks. Conf Proc IEEE Eng Med Biol Soc. 2018;2018:702–705. doi: 10.1109/EMBC.2018.8512379. [DOI] [PubMed] [Google Scholar]
- 33.Grassmann F., Mengelkamp J., Brandl C. A DEEP LEARNING ALGORITHM FOR PREDICTION OF AGE-RELATED EYE DISEASE STUDY SEVERITY SCALE FOR AGE-RELATED MACULAR DEGENERATION FROM COLOR FUNDUS PHOTOGRAPHy. Ophthalmology. 2018;125(9):1410–1420. doi: 10.1016/j.ophtha.2018.02.037. [DOI] [PubMed] [Google Scholar]
- 34.Mookiah M.R., Acharya U.R., Koh J.E. Automated diagnosis of age-related macular degeneration using greyscale features from digital fundus images. Comput Biol Med. 2014;53:55–64. doi: 10.1016/j.compbiomed.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 35.Peng Y., Dharssi S., Chen Q. DeepSeeNet: a deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs. Ophthalmology. 2019;126(4):565–575. doi: 10.1016/j.ophtha.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mookiah M.R., Acharya U.R., Fujita H. Local configuration pattern features for age-related macular degeneration characterization and classification. Comput Biol Med. 2015;63:208–218. doi: 10.1016/j.compbiomed.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 37.Liew G., Joachim N., Mitchell P., Burlutsky G., Wang J.J. Validating the AREDS simplified severity scale of age-related macular degeneration with 5- and 10-year incident data in a population-based sample. Ophthalmology. 2016;123(9):1874–1878. doi: 10.1016/j.ophtha.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 38.Al-Zamil W.M., Yassin S.A. Recent developments in age-related macular degeneration: a review. Clin Interv Aging. 2017;12:1313–1330. doi: 10.2147/CIA.S143508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glas A.S., Lijmer J.G., Prins M.H., Bonsel G.J., Bossuyt P.M. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56(11):1129–1135. doi: 10.1016/s0895-4356(03)00177-x. [DOI] [PubMed] [Google Scholar]
- 40.Ferris F.L., 3rd, Wilkinson C.P., Bird A. Beckman initiative for macular research classification committee. clinical classification of age-related macular degeneration. Ophthalmology. 2013;120(4):844–851. doi: 10.1016/j.ophtha.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anwar S.M., Majid M., Qayyum A., Awais M., Alnowami M., Khan M.K. Medical image analysis using convolutional neural networks: a review. J Med Syst. 2018;42(11):226. doi: 10.1007/s10916-018-1088-1. [DOI] [PubMed] [Google Scholar]
- 42.Ragab D.A., Sharkas M., Marshall S., Ren J. Breast cancer detection using deep convolutional neural networks and support vector machines. PeerJ. 2019;7:e6201. doi: 10.7717/peerj.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brereton R.G., Lloyd G.R. Support vector machines for classification and regression. Analyst. 2010;135(2):230–267. doi: 10.1039/b918972f. [DOI] [PubMed] [Google Scholar]
- 44.Treder M., Lauermann J.L., Eter N. Automated detection of exudative age-related macular degeneration in spectral domain optical coherence tomography using deep learning. Graefes Arch Clin Exp Ophthalmol. 2018;256(2):259–265. doi: 10.1007/s00417-017-3850-3. [DOI] [PubMed] [Google Scholar]
- 45.Venhuizen F.G., van Ginneken B., van Asten F. Automated staging of age-related macular degeneration using optical coherence tomography. Invest Ophthalmol Vis Sci. 2017;58(4):2318–2328. doi: 10.1167/iovs.16-20541. [DOI] [PubMed] [Google Scholar]
- 46.Yoo T.K., Choi J.Y., Seo J.G., Ramasubramanian B., Selvaperumal S., Kim D.W. The possibility of the combination of OCT and fundus images for improving the diagnostic accuracy of deep learning for age-related macular degeneration: a preliminary experiment. Med Biol Eng Comput. 2019;57(3):677–687. doi: 10.1007/s11517-018-1915-z. [DOI] [PubMed] [Google Scholar]
- 47.Vaghefi E., Hill S., Kersten H.M., Squirrell D. Multimodal retinal image analysis via deep learning for the diagnosis of intermediate dry age-related macular degeneration: a feasibility study. J Ophthalmol. 2020;2020 doi: 10.1155/2020/7493419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Treder M., Lauermann J.L., Eter N. Deep learning-based detection and classification of geographic atrophy using a deep convolutional neural network classifier. Graefes Arch Clin Exp Ophthalmol. 2018;256(11):2053–2060. doi: 10.1007/s00417-018-4098-2. [DOI] [PubMed] [Google Scholar]
- 49.Castelvecchi D. Can we open the black box of AI? Nature. 2016;538:20–23. doi: 10.1038/538020a. [DOI] [PubMed] [Google Scholar]