Abstract
Metabolic engineering is a promising approach for the synthesis of valuable compounds. Transcriptional factor-based biosensors are efficient tools to regulate the metabolic pathway dynamically. Here, we engineered the p-coumaric acid responsive regulator PadR from Bacillus subtilis. We found that yveF and yveG , two previously uncharacterized components in the sensor system, showed positive impacts on the regulation of PadR-PpadC sensor system, mostly on assisting the release of the repression by PadR. By site directed PadR engineering, we obtained two mutants, K64A and H38A, which exhibited increased dynamic range and superior sensitivity. To increase the promoter strength of the sensor system and investigate whether the PadR binding boxes can function in a “plug-and-play” manner, a series of hybrid promoters were constructed. Four of them P1, P2, P7, and P9 showed increased strength compared to PpadC and can be regulated by PadR and p-coumaric acid. The PadR variants and hybrid promoters obtained in this paper would expand the applicability of this sensor system ./in future metabolic engineering research.
Keywords: PadR, p-coumaric acid, hybrid promoter, biosensor
Graphical Abstract
Introduction
Metabolic engineering is a sustainable and promising approach for the synthesis of various compounds, such as natural products1, 2, pharmaceuticals3, biofuels4, and bulk chemicals5, 6. It can be achieved by combining heterologous or non-natural biosynthetic pathways into genetically advantageous microbial cell factories. In these processes, metabolic imbalance and toxic intermediates can affect the cell growth and final productivity. To address this problem, transcriptional factor-based biosensor can be used to dynamically regulate gene expression and release the burden from unbalanced metabolism. To date, transcriptional factor-based biosensors have been widely applied in dynamic regulation for the synthesis of muconic acid7, fatty acids8, and other valuable compounds9–11. In addition to the application in dynamic pathway regulation, transcriptional factor-based biosensors also were applied for high-throughput screening, which were demonstrated efficient for screening high-yield strains of 1-butanol12, ectoine13, benzoic acid, L-phenylalanine, and malonyl-CoA14. While the progress was promising, there are some challenges when utilizing transcriptional factor-based biosensors in metabolic engineering, such as poor substrate specificity, sub-optimal output strength, or narrow dynamic range. Fundamental research focusing on characterization of biosensors and studies on engineering the biosensor systems to increase their usability are thus essential to expand their application in metabolic engineering.
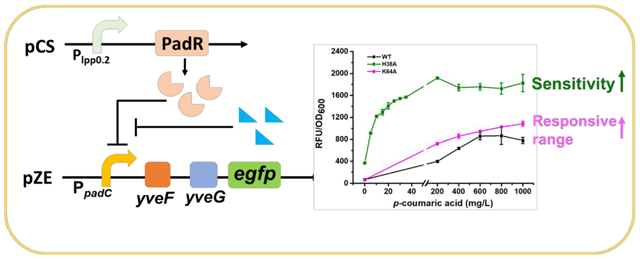
PadR is a transcriptional repressor which can inhibit the expression of phenolic acid decarboxylase (PadC) in Bacillus subtilis by binding to the −10 region in PpadC (the promoter of padC gene) and blocking the access of RNA polymerase. When Bacillus subtilis is exposed to the environment containing p-coumaric acid or ferulic acid, these compounds can bind to PadR and cause conformational change in this repressor, releasing the inhibition. (Fig.1)15, 16. The PadR-PpadC biosensor system can be applied in high-throughput screening for strains with increased p-coumaric acid or ferulic acid titer17. Besides, it shows potential in dynamic regulation to control the synthesis of flavonoids and polyphenols, which are the derivates of p-coumaric acid or ferulic acid. Both flavonoids and polyphenols are valuable natural products, and the market sizes of flavonoids and polyphenols are predicted to be $1.06 billion and $2.08 billion by 2025, respectively. In recent years, researchers paid lots of attention on PadR-PpadC biosensor system. Sung-il Yoon and coworkers obtained the crystal structure of PadR and its complex with specific DNA sequence, which revealed the key residues affecting the binding between PadR and PpadC and provided valuable insights for PadR optimization18. To apply this biosensor system in high-throughput screening, Haakan N. Joensson and colleagues applied RBS engineering to fine-tune the expression level of PadR and increased the dynamic range of the PadR-PpadC biosensor system. The optimized biosensor was then applied to screen yeast variants with high p-coumaric acid titer17. However, some key regulation components of this system have not been fully characterized yet. For example, there are two hypothetic genes, yveF and yveG, located in the intergenic region between the PpadC and padC gene. The specific functions of these two hypothetic genes in the aspect of PadR regulation have not been characterized. Besides, even though the key residues of PadR were reported, how the mutations of these residues affect the dynamic performance of the biosensor system have not been explored and characterized. Moreover, the narrow dynamic range, which caused by strong binding affinity between PadR and corresponding promoter sequences, limited its application in metabolic engineering.
Fig. 1.
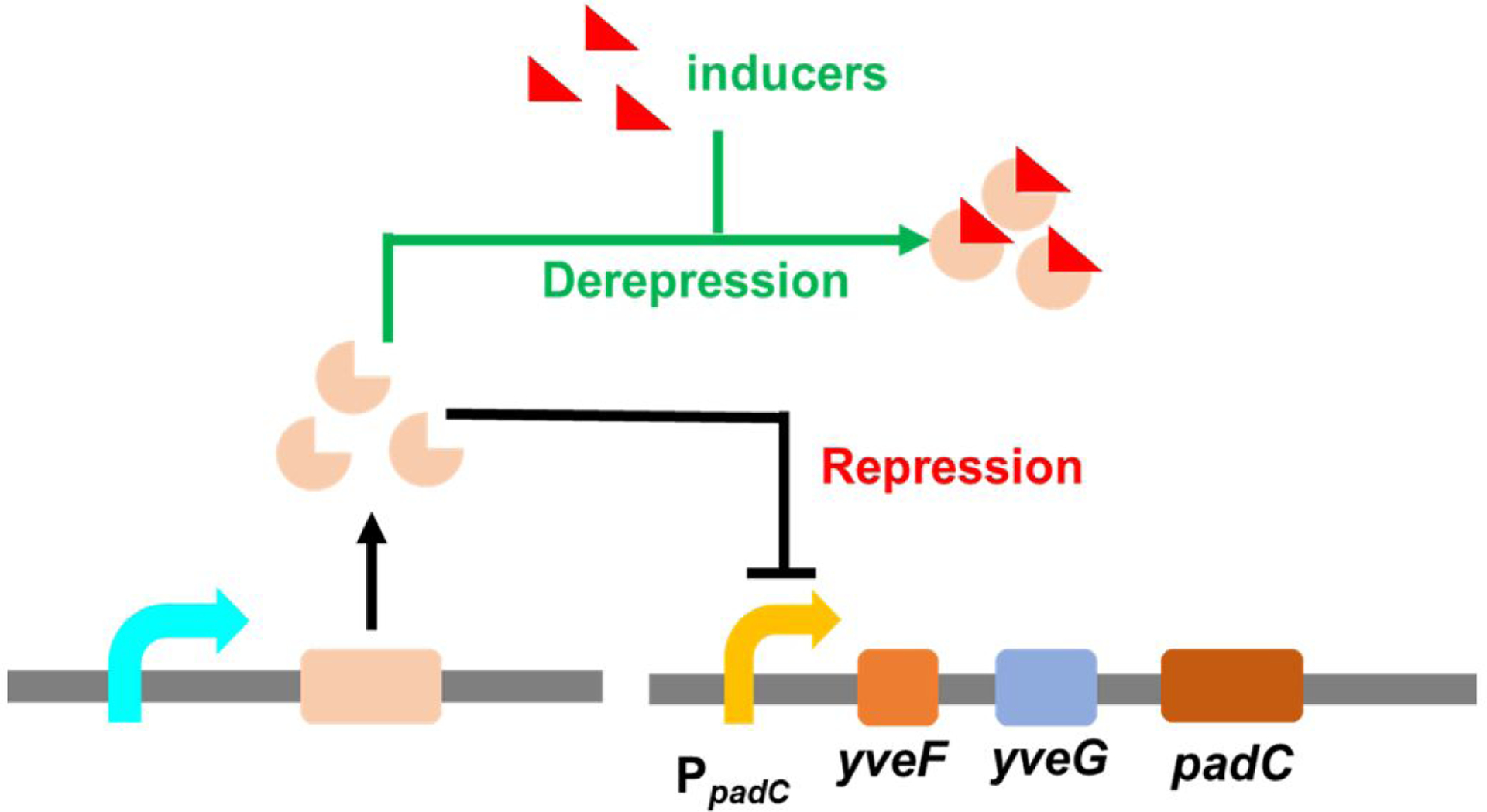
the mechanism of PadR-PpadC biosensor system. Repression: PadR binds in the specific regions in PpadC, which repressed the expression of padC gene. Derepression: In the existence of inducers, they can combine with PadR and released the inhibition of PpadC. padC gene can express.
In this paper, we cloned PadR and PpadC from Bacillus subtilis. Through functional analysis and verification, we characterized that the addition of yveF and yveG strongly enhanced the dynamic range of this biosensor system, which largely expanded its usability and potential application in metabolic engineering. The expression level of PadR on the dynamic range of this biosensor was also systematically analyzed. Then, we investigated the key residues affecting the binding between PadR and PpadC and obtained a PadR single mutant (K64A) which exhibited an increased operational range (0–1000 mg/L) induced by p-coumaric acid compared with the wild type PadR (0–600 mg/L). Besides, another PadR single mutant H38A exhibited the increased sensitivity and strength compared wild type PadR. To increase the promoter strength and investigate whether the PadR binding boxes can function in a “plug-and-play” manner, we designed a series of hybrid promoters by placing the PadR binding boxes into the strong constitutive promoter PL with or without yveF and yveG. By doing this, we demonstrated the feasibility to converting constitutive promoters to PadR-controllable promoters. Moreover, four hybrid promoters without yveF and yveG inserted (P1, P2, P7, and P9) showed increased strength compared to PpadC and can be regulated by PadR and p-coumaric acid. In summary, the engineered PadR-PpadC biosensor system showed versatile dynamic performance or behaviors. These variants can be applied in dynamic regulation or high-throughput screening. Our work also provides valuable insights in how to optimize and engineer natural biosensor systems that are not suitable for immediate metabolic engineering application.
Results
Construction and Verification of PadR-PpadC biosensor system
According to the previously reported mechanism15–17, the expression of PadR can repress PpadC promoter and inhibit the downstream gene padC. Upon induction of p-coumaric acid or ferulic acid, this repression can be released. This is a typical transcriptional factor-based biosensor system. To further understand and engineer the dynamic behavior of this biosensor system, we first constructed the functional biosensor system in E. coli. The padR gene was placed under the control of the constitutive promoter lpp1.0, and the reporter gene egfp was under the control of promoter PpadC cloned from Bacillus subtilis to create a regulator and a reporter plasmid, pCS-lpp1.0-PadR and pZE-PpadC-egfp, respectively (Fig. 2a). To validate the successful construction and regulation of the biosensor system, the previously reported PadR effector, p-coumaric acid, was utilized for induction. The strain containing only the reporter plasmid pZE-PpadC-egfp was used as a positive control (PC1, with no effector added). The fluorescence intensity normalized with OD600 (RFU/OD600) was used to reflect the promoter activity of PpadC. As shown in Fig. 2b, the promoter activity of the uninhibited PpadC in positive control (PC1) was 2619.5 a.u.. The PpadC can work well without PadR inhibition. In the existence of pCS-lpp1.0-PadR (blank 1, with no inducer added), the fluorescence intensity was reduced to 44.5 a.u., indicating that 98.3% of the PpadC activity was repressed by PadR. After the addition of 600 mg/L p-coumaric acid, the fluorescence intensity was increased to 79.4 a.u. (Fig 2b). The results demonstrated that the p-coumaric acid successfully activated the biosensor system, which were consistent with previous studies16, but the activated strength was too low to be applied in dynamic regulation or high-throughput screening compared with the commonly used biosensors. Proceeding further, we hypothesized that a better performance of this biosensor might be achieved by using different inducers. Thus, ferulic acid and the cinnamic acid with a similar structure to p-coumaric acid were used to induce the biosensor system. After the addition of 600 mg/L ferulic acid, the florescence intensity was only 72.8 a.u., which was even lower than the activated strength by p-coumaric acid. The addition of cinnamic acid did not result in any increase expression of egfp , indicating that this compound was unable to activate the biosensor (Fig 2b). To better illustrate the inducibility of different inducers, we tested the promoter performance under different concentrations of each inducer, as shown in Fig. 2e. With the increased concentration of p-coumaric acid and ferulic acid, the activated strength of PpadC increased, which further indicated that these two inducers can release the PadR repression. However, with the increase of cinnamic acid concentration, there was no increase in the egfp expression level (Fig. 2e), which proved that cinnamic acid cannot release the PadR repression. Taken together, there are some issues in the PadR-PpadC biosensor system, especially the limited dynamic range.
Fig. 2.
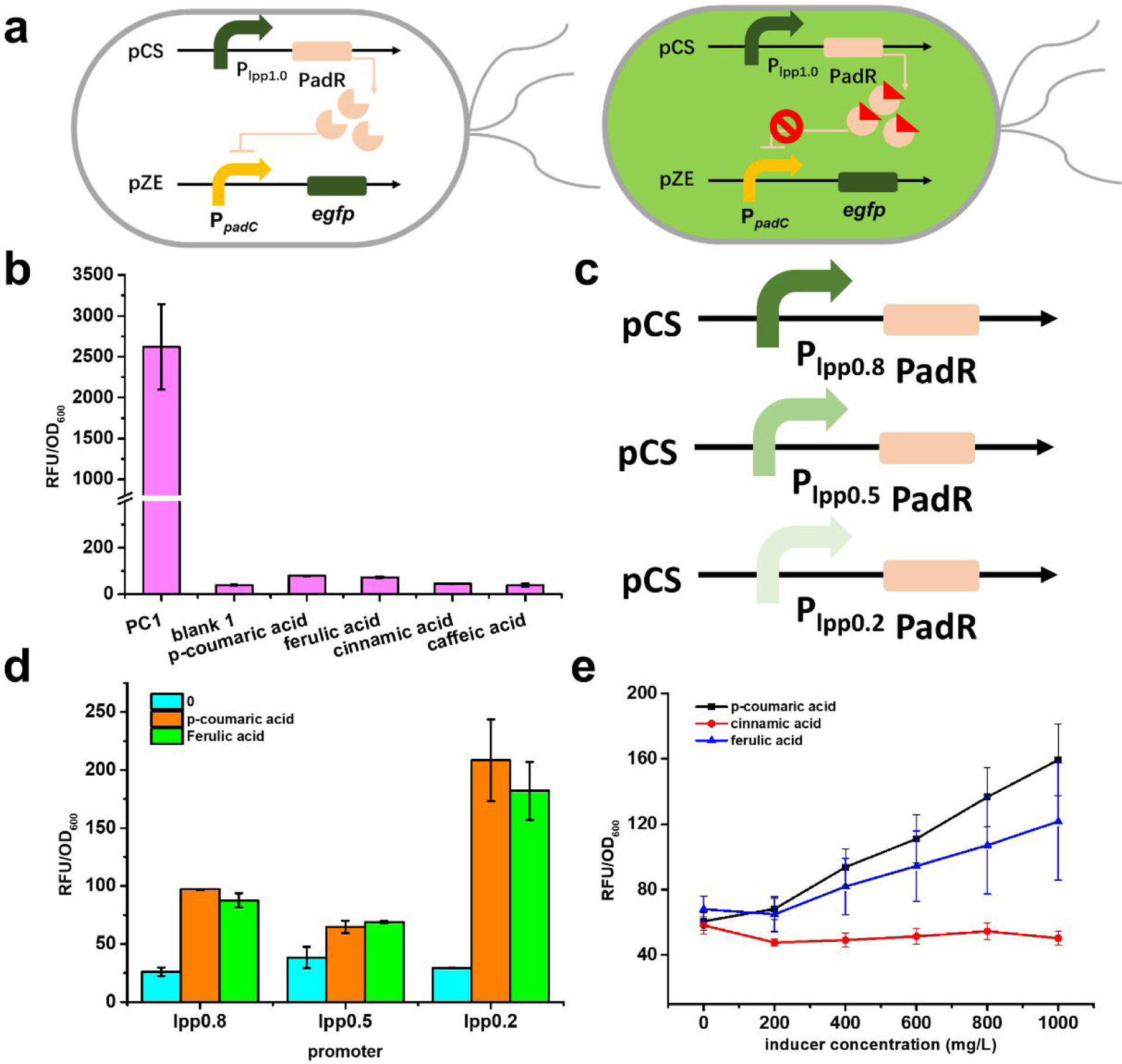
The design of PadR-PpadC biosensor system and its optimization. (a) The genetic circuit of PadR-PpadC biosensor system. There are two modules in this system, reporter module and regulator module. In the absence of p-coumaric acid, PadR can repress the PpadC and egfp cannot express successfully. When p-coumaric acid was added, it can bind to PadR, which released the inhibition of PpadC; the green fluorescence can be detected. (b) The promoter strength with or without PadR inhibition. PC1, without regulator module, was used to test the strength of PpadC; blank, with reporter module and regulator module, which was used to characterize the inhibition efficiency; p-coumaric acid, ferulic acid, and cinnamic acid represent that different inducers were added to release the PadR inhibition. (c) Different inducers in different concentration. P-coumaric acid, ferulic acid, and cinnamic acid were used as inducers to release the inhibition by pCS-lpp1.0-PadR. (d) Differe nt regulator modules with various promoter. (e) The effects of different expression level of PadR in decreasing its inhibition activity. All data are reported as mean±s.d. from three independent experiments (n=3). Error bars are defined as s.d.
The poor performance of the biosensor system was presumably caused by high-level expression of PadR, as demonstrated in a previous study17. To optimize the biosensor system for further application, a series of lpp promoters (lpp0.8, lpp0.5, and lpp0.2) with gradually descending strength were used to fine-tune the PadR expression level19, resulting in three additional regulator plasmids pCS-lpp0.8-PadR, pCS-lpp0.5-PadR, and pCS-lpp0.2-PadR (Fig. 2d). These were each co-transferred with the reporter plasmid pZE-PpadC-egfp to E. coli BW25113 (F’). The p-coumaric acid and ferulic acid were used as inducers. When PadR was expressed under the control of lpp0.8 and lpp0.5 (the promoter strength was 80% and 50% compared with lpp1.0, respectively) and fed with 600 mg/L p-coumaric acid or ferulic acid, the resumed activities were nearly the same as when PadR was controlled by lpp1.0, indicating that the PpadC was still tightly repressed by PadR (Fig. 2d). The PadR expression level was too high under the control of lpp0.8 and lpp0.5. However, when PadR was expressed under the control of lpp0.2 at 20% strength of lpp1.0, the addition of 600 mg/L p-coumaric acid or ferulic acid resulted in increased strengths of PpadC to 208.4 a.u. or 182.0 a.u., respectively, which accounted for 7.96% or 6.95% of the fully induced PpadC activity. The resumed activity was approximately 1.6-fold higher than that when PadR was controlled by lpp1.0. The results demonstrated that the repression by PadR can be partially released by decreasing the PadR expression level to 20%. The decreased expression level of PadR showed a positive effect on releasing the repression, which was consistent with the previously reported results17. However, while the availability of PadR is a related parameter in tuning the dynamic range of this biosensor, the increase of dynamic range obtained by tuning the expression of PadR was limited as the induced PpadC only showed up to 8% of fully induced promoter activity (PC1) when induced by 600 mg/L p-coumaric acid or ferulic acid. The lower activated strength will limit the applicability of this biosensor system in metabolic engineering. To further enhance the dynamic behavior of this biosensor, we sought to investigate whether the genetic components behind PpadC affect the dynamic behavior of the biosensor system.
The downstream yveF and yveG of PpadC affects the dynamic performance of PadR-PpadC
There are two hypothetical genes, yveF and yveG, located in the intergenic region between the promoter PpadC and padC gene in Bacillus subtilis genome. However, the specific functions of these two genes in the regulation of the PadR-PpadC biosensor system have not been characterized thus far. We hypothesized that these two genes may affect the PpadC activity and the dynamic behavior of the system. To test this hypothesis, yveF and/or yveG were inserted between the promoter PpadC and the egfp gene to form three additional reporter plasmids: pZE-PpadC-yveF-egfp, pZE-PpadC-yveG-egfp, and pZE-PpadC-yveF-yveG-egfp (Fig. 3a).
Fig. 3.
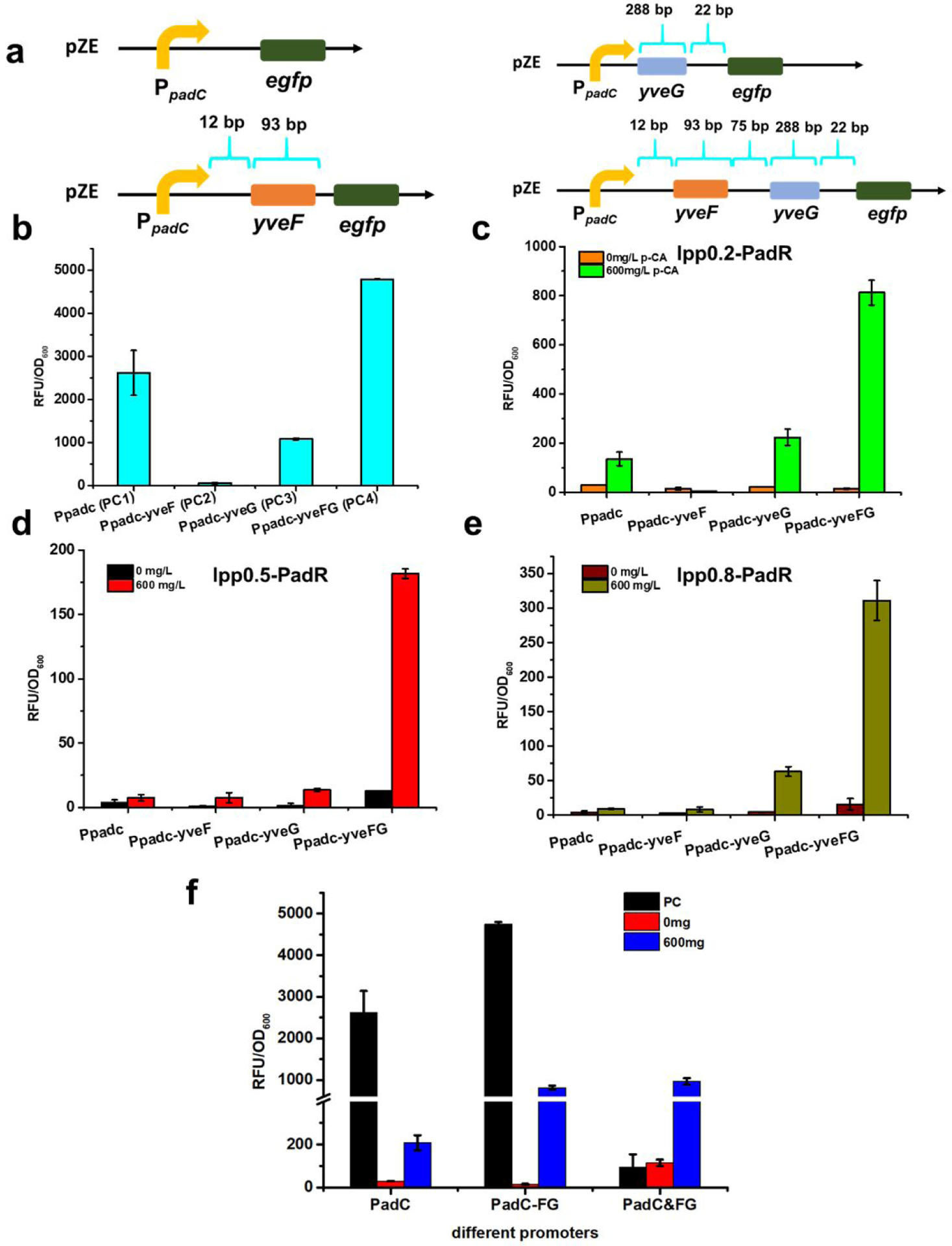
Exploring the function of yveF and yveG in the biosensor system. (a) Different reporter modules with or without yveF and yveG. yveF: 93 bp; yveG: 288 bp. Between yveF and yveG, there is a 75 bp spacer sequence. Between PpadC-yveF, there is a 12 bp spacer sequence. Between yveG and downstream padC gene, there is a 22 bp spacer sequence. To construct pZE-PpadC-yveF-egfp, the sequence of PpadC, yveF, and spacer sequence between PpadC-yveF were inserted before egfp. To construct pZE-PpadC-yveG-egfp, the sequence of PpadC, yveG, and the spacer sequence between yveG and downstream gene padC were inserted before egfp. To construct pZE-PpadC-yveF-yveG-egfp, the sequence of PpadC, spacer sequence between PpadC and yveF, yveF, the spacer sequence between yveF and yveG, yveG, and the spacer sequence between yveG and padC were inserted before egfp. (b) The promoter strength without PadR inhibition. (c) The biosensor performance in the existence of pCS-lpp0.2-PadR and p-coumaric. (d) pCS-lpp0.5-PadR was used as the regulator plasmid. (e) pCS-lpp0.8-PadR was used as the regulator plasmid. (f) The biosensor performance with/without yveF and yveG before egfp, or when yveF and yveG were expressed independently. All data are reported as mean±s.d. from three independent experiments (n=3). Error bars are defined as s.d.
To characterize whether the addition of these two genes will alter the promoter activity and lead to different expression levels of egfp, four reporter plasmids, pZE-PpadC-egfp, pZE-PpadC-yveF-egfp, pZE-PpadC-yveG-egfp, and pZE-PpadC-yveF-yveG-egfp were introduced into E. coli BW25113 (F’) individually to determine the promoter activities. The highest promoter strength was observed when both yveF and yveG were presented (PC4), resulting in a normalized fluorescence intensity of 4855.4 a.u. (Fig. 3b), which is 85.3% higher than the activity of PpadC. As for promoters with either yveF (PC2) or yveG (PC3), the promoter activities only accounted for 2.3% or 40.2% of PC1, respectively (Fig. 3b). YveF and yveG may have a positive effect in increasing the output strength of PpadC. Next, we explored the influence of yveF and yveG on the regulation of this biosensor system. The aforementioned reporter plasmids were each co-transformed with the regulator plasmid pCS-lpp0.2-PadR into E. coli BW255113 (F’). When no inducer was present, all four promoters can be effectively repressed by PadR (Fig. 3c). When induced with 600 mg/L p-coumaric acid, these promoters showed varied activity. The highest fluorescence level was achieved by the promoter PpadC-yveF-yveG (PpadC-FG) with a normalized RFU/OD600 of 812.4 a.u., releasing 16.7% of the PpadC-FG activity (Fig. 3c). However, the promoter activities of the other three promoters induced by p-coumaric acid were relatively low. The promoter strength of PpadC and PpadC-yveG were 136.1 a.u. and 223.8 a.u., respectively. PpadC-yveF could not be activated by p-coumaric acid (Fig. 3c). The results demonstrated that in the existence of both yveF and yveG, the PadR repression can be better released. Similar patterns were observed when the regulator plasmids pCS-lpp0.5-PadR and pCS-lpp0.8-PadR were used to test the function of yveF and yveG (Fig. 3d&e). Therefore, the promoter PpadC-FG was used for our following experiments.
Taken together, the results indicated that only when both yveF and yveG were inserted between the PpadC and the egfp, the output strength will be increased. In the existence of yveF behind PpadC (PC2), it lost almost all the activity. However, when yveG was added behind PpadC (PC3), the promoter activity was a little bit higher than PC2. It is possible that between yveG and downstream gene padC, there is a weaker RBS which was responsible for the expression of egfp. When only adding yveF, the egfp cannot be expressed because of the lack of a functional RBS, and no output strength can be detected. When yveF and yveG were both added, it is possible that there is a weaker RBS which was responsible for the expression of egfp. An alternative possibility is that the successful expression of yveF and yveG may have a synergistic effect on increasing the promoter strength. Some other experiments were needed to verify whether yveF and yveG work as encoding proteins or nucleotide sequence (e.g., 5’-UTR) in the PpadC-PadR sensor system.
To further investigate the function of yveF and yveG, we expressed two genes in a separate operon under the control of lpp1.0 in a high-copy plasmid pSC. We introduced the regulator plasmid pCS-lpp0.2-PadR with the pZE-PpadC-egfp and pSC-lpp1.0-yveF-yveG. The leaking expression of PpadC increased when yveF and yveG were expressed in another plasmid compared to PpadC and PpadC-FG. After adding 600 mg/L p-coumaric acid, the induced activity of PpadC&FG was similarly to PpadC-FG, which was higher than the control PpadC (Fig. 3f). Therefore, even the yveF and yveG were expressed from a separate plasmid, it still enhanced the dynamic range of the PadR-PpadC system, which indicated that yveF and yveG may work as encoding proteins to assist relieving the inhibition of PadR. We noticed that when no PadR was introduced, the strain only exhibited a very low level of fluorescence. This was likely because the high expression of egfp caused significant burden on the cells, as the growth of this strain was notably slower than others.
PadR engineering to decrease the binding affinity with PpadC
Although the addition of yveF and yveG enhanced the activation efficiency of PadR-PpadC biosensor, the activity of PpadC induced by 600 mg/L p-coumaric acid was still less than 20% of the unrepressed promoter. We hypothesized that this was due to the tight binding between PadR and the PpadC. To test this hypothesis, we engineered the key binding residues of PadR to diminish the binding affinity between PadR and PpadC. The crystal structure of PadR was resolved by Yoon et al18. They characterized the key binding residues (K37, H38, S39, Q40, Q61, K64) between PadR and PpadC, and found that the mutation of these residues can decrease the binding affinity between PadR and the synthetic dsDNA18. To explore how the mutation of these residues affect the dynamic performance of PadR-PpadC biosensor system, the key residues were mutated to alanine. Alanine is a chiral amino acid with the shortest side chain, which has the potential to decrease the binding affinity effectively. Six mutants (K37A, H38A, S39A, Q40A, Q61A, K64A) were obtained. These PadR mutants were under the control of lpp0.2 promoter and were each co-transformed with pZE-PpadC-yveF-yveG-egfp to determine the dynamic performance. Although all the PadR mutants can inhibit the activity of PpadC-FG, there were five mutants (K37A, H38A, S39A, Q40A, Q61A) exhibiting the increased leaking expression (Fig. 4a). Especially, when PpadC-FG was repressed by pCS-lpp0.2-H38A, the leaking expression level can account for 14% of the unrepressed promoter activity (Fig. 4a). The potential reason that H38A showed the weakest binding affinity among all the mutants. When PpadC-FG was repressed by pCS-lpp0.2-K64A, only 0.12% leaking activity was observed. This was more stringent than the wild type PadR (Fig. 4a). This result is interesting as K64 was mutated to a smaller residue, but mutation has not caused increased leaking expression compared to wild type.
Fig. 4.
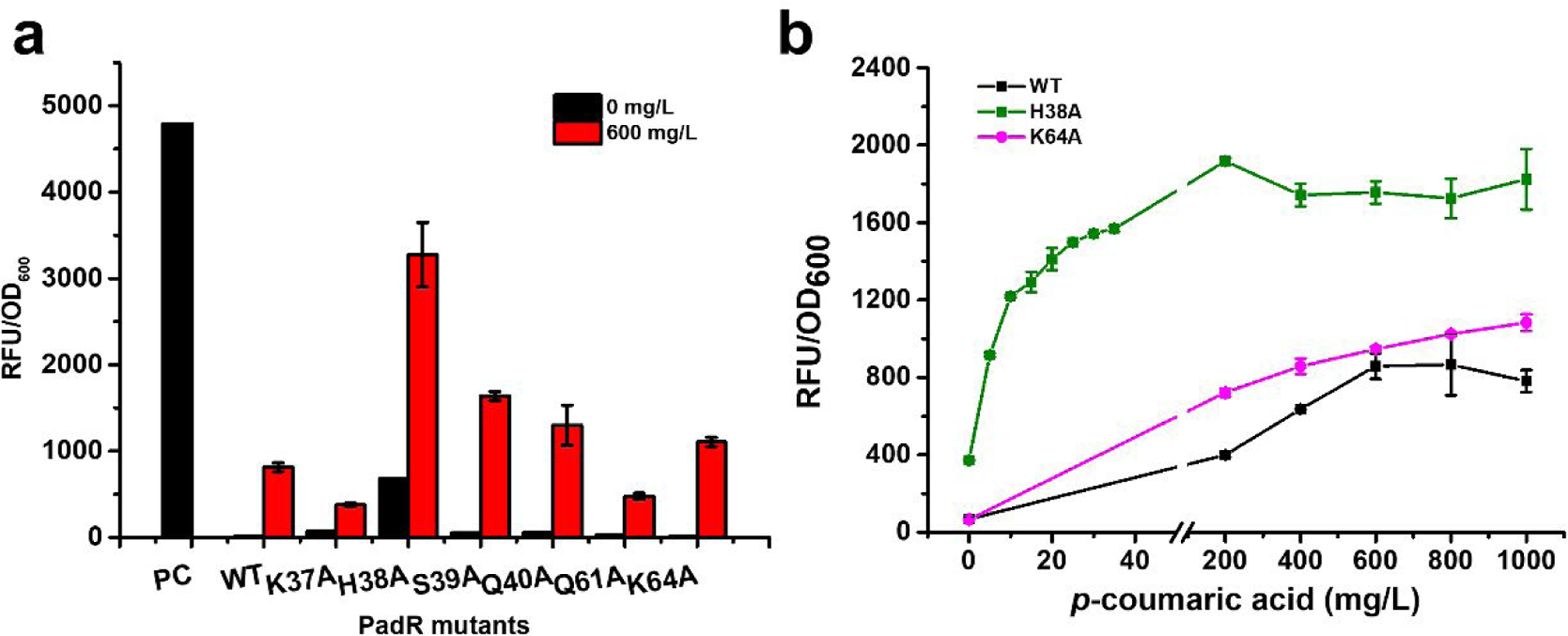
The dynamic behavior of different PadR mutants. (a) The different PadR mutants were tested by 0 and 600 mg/L p-coumaric acid; PC means the strain without regulator module. (b) The dynamic range of WT, H38A, K64A. All data are reported as mean±s.d. from three independent experiments (n=3). Error bars are defined as s.d.
After adding 600 mg/L p-coumaric acid, there were four PadR mutants, H38A, S39A, Q40A, and K64A, showing increased activated strength compared with the wild type, especially the PadR mutants H38A. 68.3% of the PpadC-FG activity can be recovered by 600 mg/L p-coumaric acid (Fig. 4a). Another interesting mutant is the K64A, which was not only more stringent but also showed an elevated activity compared with wild-type PadR after induction. On the other hand, two mutants, K37A and Q61A, exhibited lowered induction activities compared with wild type (Fig. 4a). Taken together, we obtained two PadR mutants, K64A and H38A, which exhibited the increased inhibition efficiency and activated strength, respectively, compared with wild type PadR.
To better characterize the dynamic behavior of these two mutants (H38A and K64A), the dynamic range of wild type PadR, H38A, and K64A was measured with p-coumaric acid. As shown in Fig. 4b, different concentrations (from 0 to 1000 mg/L) of p-coumaric acid were used to induce the biosensor system. In the group of wild type PadR, the activated strength of the promoter increases as the p-coumaric acid concentration increases from 0 to 600 mg/L, with the highest activated strength of 859.0 a. u. in 600 mg/L. There was no obvious increasing in the promoter strength when concentration of p-coumaric acid is higher than 600 mg/L. Thus, the operational range of wild type PadR was from 0 to 600 mg/L and its dynamic range is from 68.4 a.u. to 859.0 a.u.. The variant K64A enabled a broader dynamic range and higher activated strength of the sensor system compared with the wild type PadR. The activated strength of the promoter PpadC-yveF-yveG combined with PadR (K64A) increased from 65.9 to 1083.7 a.u. with p-coumaric acid concentration increasing from 0 to 1000 mg/L. Therefore, this mutant is more suitable for high-throughput screening due to the expanded dynamic range. As for the variant H38A, the highest activated strength can be achieved at 200 mg/L p-coumaric acid. Although the responsive range of PadR (H38A) is narrower (0–200 mg/L) than the wild type PadR (0–600 mg/L), it can be activated with only 5 mg/L p-coumaric acid with the strength reaching to 916.4 a.u.. These results indicated that when equipped with the PadR (H38A), the sensor system became extremely sensitive and exhibited a higher strength. This variant may be suitable for dynamic regulation of the synthesis of p-coumaric acid or its derivatives. In summary, we obtained two PadR mutants H38A and K64A with broader dynamic ranges, increased sensitivity and activated strengths. The applicability of the PadR-PpadC biosensor system in metabolic engineering can be largely expanded by using these variants.
Understanding the molecular mechanism of PadR mutants by modeling structure analysis
To better understand the molecular mechanisms and why these PadR mutants displayed the different performance, the 3D structures of each PadR mutants were necessary. Yoon et al resolved the complex of PadR and the related nucleotide binding sequence in PpadC18. The crystal structure of PadR and related DNA sequence was obtained from PDB database (PDB ID: 5X11) , as shown in Fig.5a. The 3D structures of six PadR mutants were simulated in Pymol. After the structure analysis, we found that, in the wild type PadR, there are two hydrogen bonds between His38 and DNA sequence. However, the mutation in His38 eliminated the interaction between PadR and PpadC based on the simulation result (Fig. 5b&5c). The weaken interaction between H38A and DNA sequence might explain the leaking expression when depressing the PpadC. Besides, p-coumaric acid can better release the inhibition than wild type PadR possibly due to the decreased interaction, which also caused a higher activated strength. As for K64A, there is one hydrogen bond between Lys64 and DNA sequence in wild type PadR. According to the simulation result, though the hydrogen bond still existed after the mutation to Ala, the loop became more flexible than before, which might be favorable to combine with the DNA sequence (Fig. 5d & 5e). The better binding pattern was likely the reason for the increased inhibition efficiency of K64A compared to wild type PadR, and possibly the increased flexibility made the promoter more easily activated by p-coumaric acid. There are four mutants, K37A, S39A, Q40A, and Q61A, which exhibited the increased leaking expression compared to wild type PadR. The simulated structure analysis showed there were one hydrogen bond between residues Ser39, Gln40, Gln61 and their related DNA sequence (Fig. 6). However, after the mutation, this hydrogen bond was eliminated, and this possibly resulted in the decreased binding affinities. Because the Lys37 is positively charged, it can interact with the negatively charged bases, allowing PadR to bind closely to the PpadC (Fig. 6). When Lys37 was mutated to Ala, the electrostatic interaction disappeared and the binding capacity became weaker, which might be the reason for the increased leaking expression of K37A.
Fig. 5.
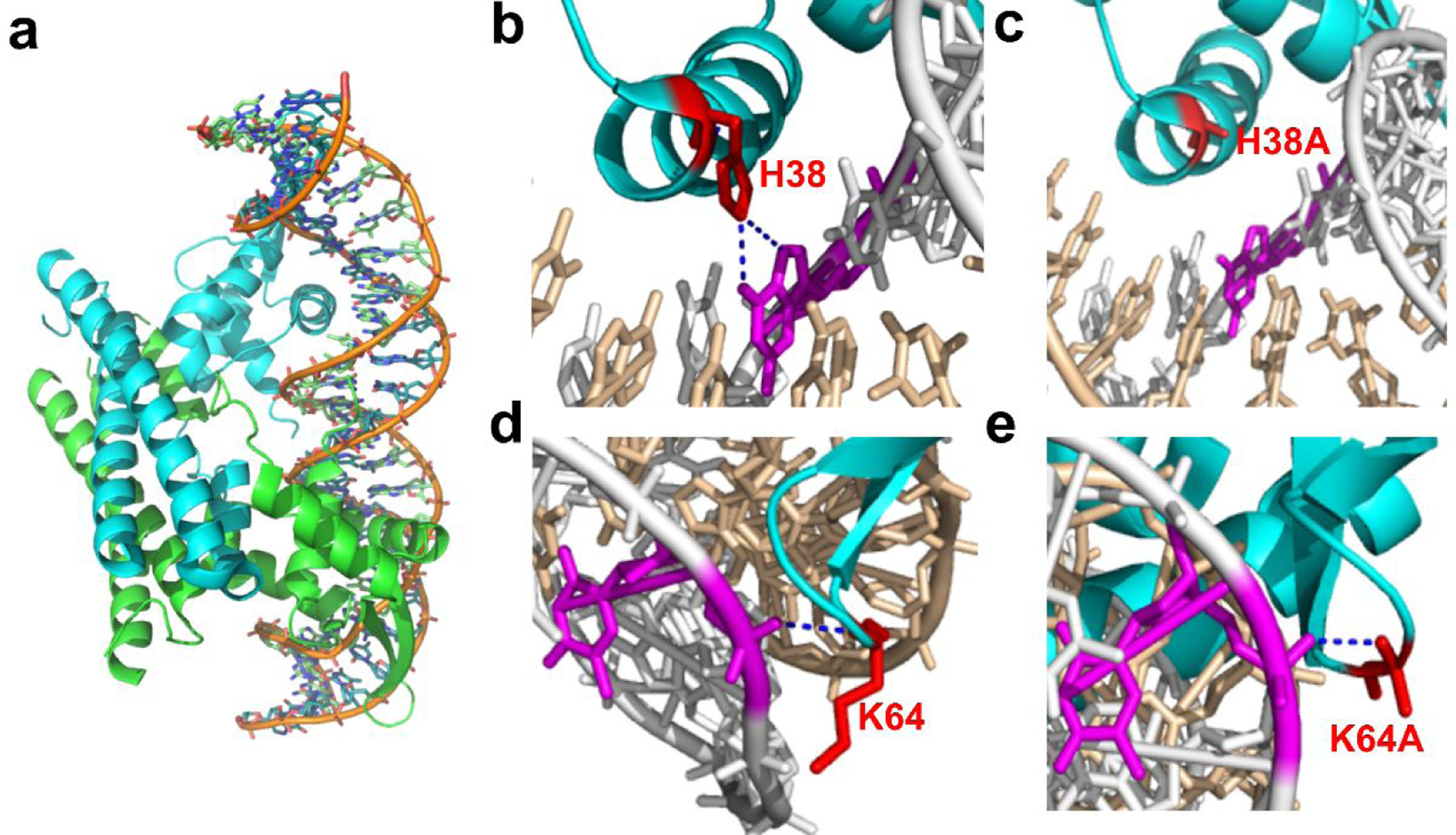
The structure analysis of PadR mutants. (a) The crystal structure of PadR and related DNA sequence. (b) The interaction between H38 and dsDNA. (c) The interaction between H38A and dsDNA. (d) The interaction between K64 and dsDNA. (e) The interaction between K64A and dsDNA.
Fig. 6.
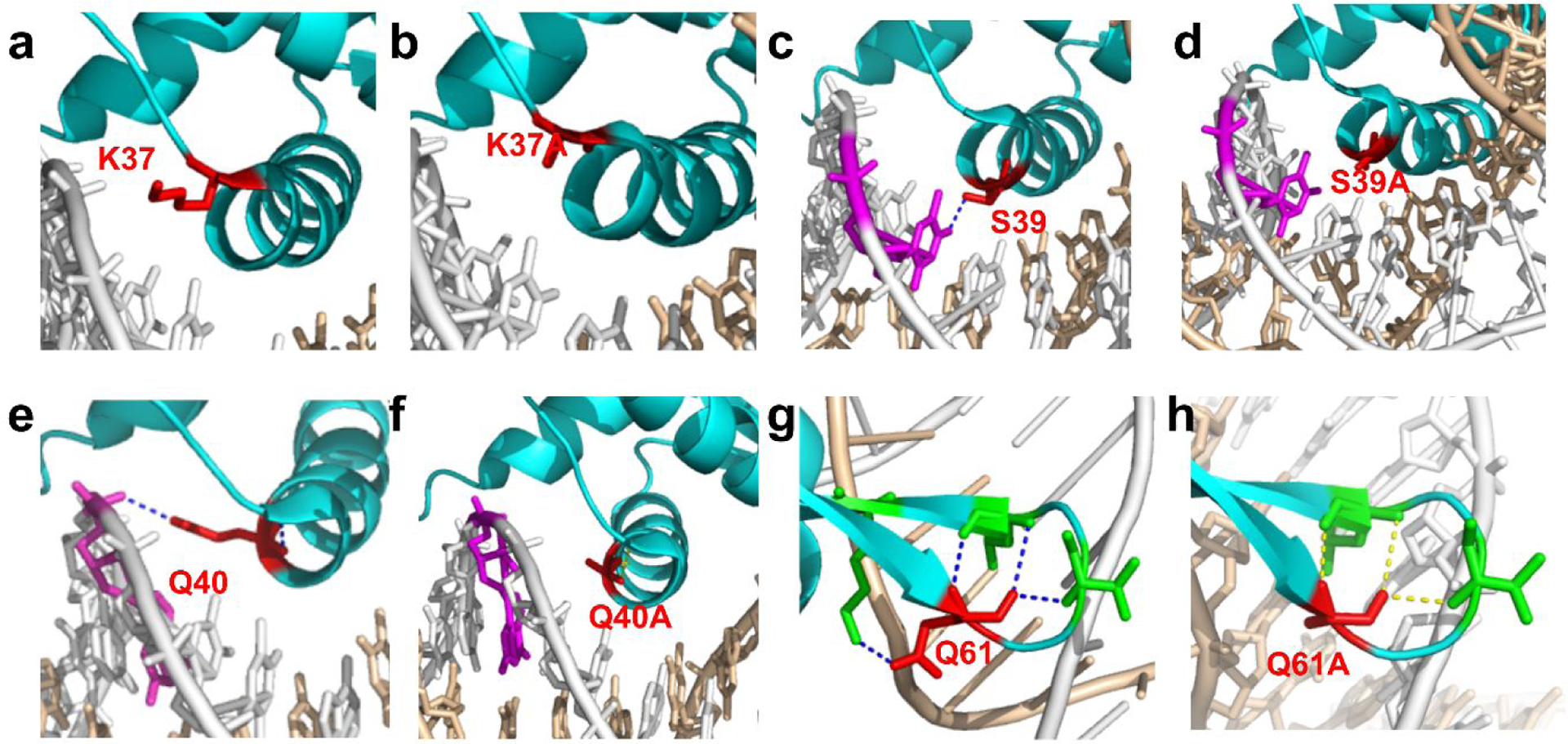
the modeling structure analysis of PadR mutants. (a) The interaction between K37 and dsDNA. (b) The interaction between K37A and dsDNA. (c) The interaction between S38 and dsDNA. (d) The interaction between S39A and dsDNA. (e) The interaction between Q40 and dsDNA. (f) The interaction between Q40A and dsDNA. (g) The interaction between Q61 and dsDNA. (h) The interaction between Q61A and dsDNA.
Constructing hybrid promoters to explore the “plug-and-play” possibility of PadR binding regions and increase the output strength of the biosensor system
There are two binding boxes, PadR-1 and PadR-2 located in the promoter PpadC15 We hypothesized that the PadR binding regions can function in a “plug-and-play” manner, and thus placing these binding boxes in a constitutive promoter would convert it to a PadR-regulatable promoter. Besides, although PpadC-FG exhibited a higher strength compared to PpadC (Fig. 3), the strength was still relatively low compared with other biosensors used in metabolic engineering. Therefore, to investigate our hypothesis and further increase the output strength of the sensor system, we chose the strong constitutive promoter PL from phage lambda as a proof-of-concept demonstration to construct hybrid promoters. We hypothesized that by placing the PadR-1 and PadR-2 in the PL promoter, the newly constructed hybrid promoters can be regulated by p-coumaric acid and PadR, and they may also exhibit increased strength compared to wild type PpadC-FG.
The widely used constitutive promoter PL has been engineered to form different inducible promoters, such as PLlacO1, PLtetO1, and PLara. These inducible promoters were constructed by replacing the upstream 19 bp sequence before −35 region (Position 1) and 17 bp sequence between −35 region and −10 region (Position 2) with corresponding binding sequence (e.g LacO box and tetO box)20. Based on this design principle, we constructed eight hybrid promoters by placing PadR-1 (22 bp) and PadR-2 (22 bp) in Position 1 and Position 2, as shown in Fig. 7a. Because of the previously demonstrated positive effect of yveF and yveG on the performance of PpadC, the two genes were added behind all these eight hybrid promoters as well as the original PL promoter. We first determined the promoter activity of these eight hybrid promoters. Six hybrid promoters (P1-FG, P2-FG, P5-FG, P6-FG, P7-FG, and P8-FG) only accounted for less than 5% activity of the PpadC-FG (Fig. 7b). There were two hybrid promoters, P3 and P4, showed usable activities. We further verified whether the P3 and P4 can be regulated by the PadR and p-coumaric acid. When PadR was present, about 90% of the activities of P3 and P4 can be repressed (Fig. 7b). After adding 600 mg/L p-coumaric acid, about 30% and 68% of the activity can be activated for P3 and P4, respectively (Fig. 7b). On the contrary, the wild-type PL promoter cannot be regulated by PadR and activated by p-coumaric acid. Although there was a slight decrease in fluorescence level of PL promoter when PadR was introduced, this was likely due to the burden caused by the overexpression of PadR. The hybrid promoters P3 or P4 can be regulated by p-coumaric acid and PadR, but the promoter strengths were much lower compared with the PpadC-FG. We suspected that the insertion of yveF and yveG affected the transcription or translation of egfp when it was expressed under the control of hybrid promoters. The promoter structures or transcriptional machineries between PpadC and hybrid promoters may be different. Therefore, the existence of yveF and yveG may pose a detrimental effect on egfp expression because they were co-translated with this reporter gene, even though yveF and yveG have a positive effect on increasing PpadC strength in our previous experiment (Fig 3).
Fig. 7.
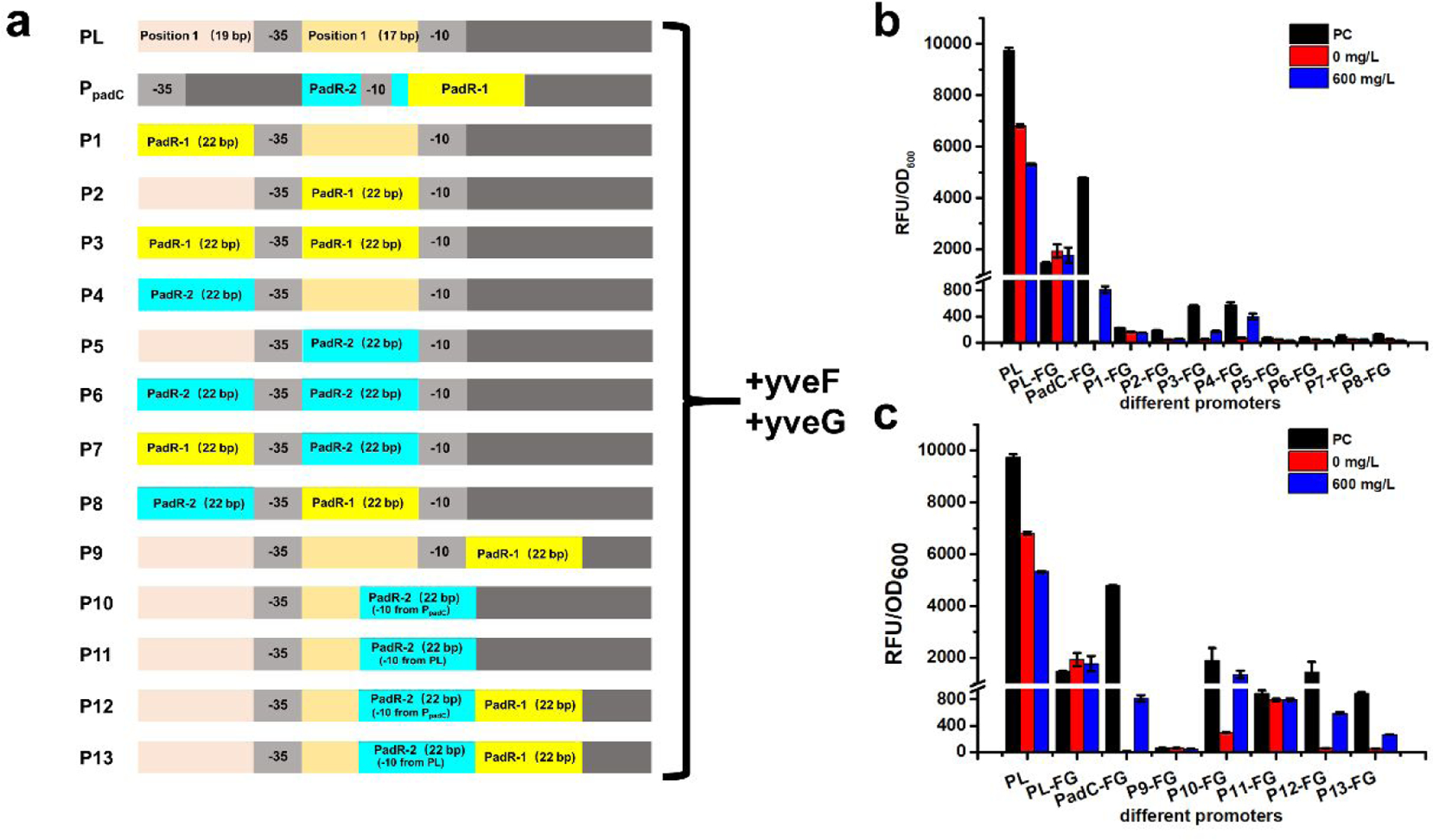
The performance of hybrid promoters with yveF and yveG. (a) The design of hybrid promoters. From P1-FG to P8-FG, PadR-1 and PadR-2 were placed before −35 region or −10 region simultaneously or respectively. The P9-FG and P10-FG contained only PadR-1 and PadR-2, respectively. Because the PadR-2 overlapped with the −10 region, and this may affect the promoter performance, we designed another hybrid promoter P11-FG by keeping the −10 sequence from PL. For hybrid promoters P12-FG and P13-FG, they contained both PadR binding boxes, but in P12-FG the −10 sequence from PpadC was used and in the P13-FG the −10 sequence from PL was used (b) The hybrid promoters designed according to PLlacO1 were rested by 0 and 600 mg/L p-coumaric acid. (c) The hybrid promoters, which were designed based on the relative position in PpadC, were tested by 0 and 600 mg/L p-coumaric acid. All data are reported as mean±s.d. from three independent experiments (n=3). Error bars are defined as s.d.
To test this hypothesis, we removed the yveF and yveG in the eight hybrid promoters. The promoter activities of these new hybrid promoters were determined. There are three hybrid promoters (P3, P5, and P6) which exhibited the decreased strength compared to wild type PpadC, although they can be regulated by PadR and p-coumaric acid. Another five promoters (P1, P2, P4, P7, P8) have increased strength compared to PpadC. In particular, the strength of P1, P2, and P7 increased by 119%, 109%, and 32% compared to PpadC-FG, respectively (Fig. 8b). The results confirmed our hypothesis. The potential reason was that the existence of yveF and yveG affected the expression of egfp because the three genes were co-expressed by one promoter, and the transcriptional machineries between PpadC and hybrid promoters may be very different. When PadR was introduced, more than 92% of the strength of P1, P2, P7, P8 can be repressed except P4, which cannot be repressed when PadR was introduced. After adding 600 mg/L p-coumaric acid, 37%, 26%, and 36% of the activity of P1, P2, and P7 can be obtained. The P8 cannot be induced by p-coumaric acid. This was likely due to stringent bind between PadR and the P8. Taken together, without the insertion of yveF and yveG, we obtained three hybrid promoters which not only can be regulated by PadR and p-coumaric acid but also have increased strength. The results also indicated that these two genes may pose detrimental effects on the egfp expression when they were placed after the hybrid promoters, even their expression can increase the strength of wild type PpadC.
Fig. 8.
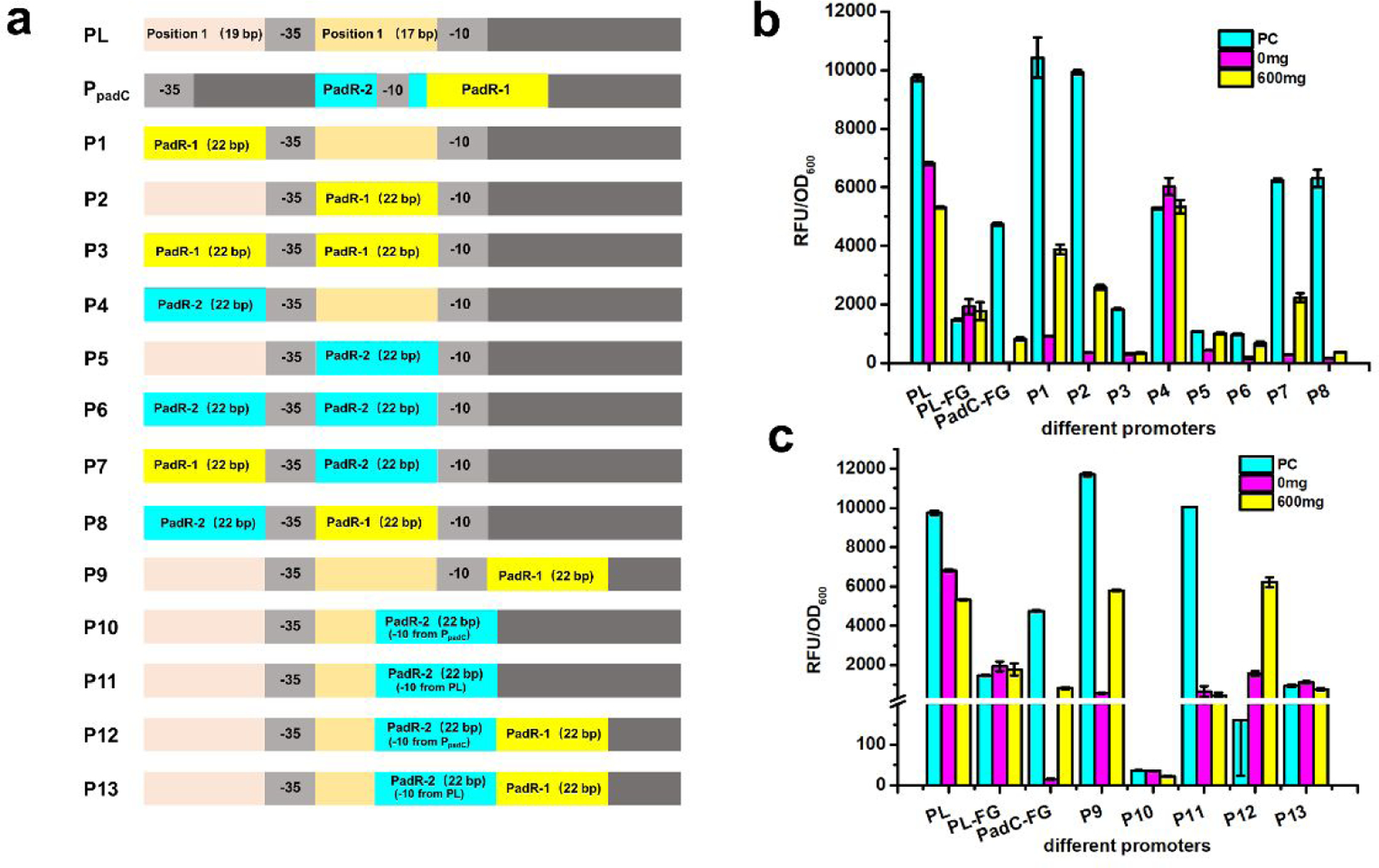
The performance of hybrid promoters without yveF and yveG. (a) The design of hybrid promoters. (b) The hybrid promoters designed according to PLlacO1 were rested by 0 and 600 mg/L p-coumaric acid. (c) The hybrid promoters, which were designed based on the relative position in PpadC, were tested by 0 and 600 mg/L p-coumaric acid. All data are reported as mean±s.d. from three independent experiments (n=3). Error bars are defined as s.d.
We sought to construct a second series of hybrid promoters based on the position of two PadR binding sequences in PpadC. We constructed five hybrid promoters by placing the PadR binding sequences in the same position in PL as where they are in the PpadC promoter. For example, the PadR-1 starts from the +2 position in PpadC and ends in +23, and the sequence starting from the +2 position in PL will be replaced with PadR-1. Similarly, PadR-2, which is located from −13 to +9, can be placed in the corresponding location in PL. All the hybrid promoters using this design have two groups, one group was with yveF an yveG behind the promoters and the other one was without the insertion of these two genes. The design principles for the five hybrid promoters in the first group was shown in Fig. 7a. All the five promoters only accounted for less than 40% activity of the PpadC-FG. The potential reason was likely the same as what in the first round of hybrid promoters that the existence of yveF and yveG behind the hybrid promoters negatively affected the expression of egfp (Fig. 7c). Therefore, the yveF and yveG were removed in the hybrid promoters. After the elimination of yveF and yveG, there were two promoters, P9 and P11, exhibiting 147% and 112% strength increase compared to PpadC-FG. Another three promoters, P10, P12, and P13 have decreased strength compared to PpadC-FG. When co-transferred with pCS-lpp0.2-PadR, 96% of P9’s activity can be repressed and 50% of the activity can be obtained after adding 600 mg/L p-coumaric acid. As for P11, it cannot be activated by 600 mg/L p-coumaric acid though it can be repressed by PadR (Fig. 8c). We also noticed that the positive control of P12 without PadR cannot grow well, which led to very low fluorescence level. These results indicated that the constitutive promoter can be engineered to be regulated by PadR and p-coumaric acid.
So far, four hybrid promoters (P1, P2, P7, and P9) without yveF and yveG insertion exhibited an increased strength compared to PpadC-FG, which showed the best performance among the wild-type promoters. Because of the positive effect of yveF and yveG in PadR-PpadC biosensor system, we further explored the performance of the best four hybrid promoters (P1, P2, P7, and P9) with the existence of pSC-lpp1.0-yveF-yveG. The results indicated that the maximum promoter strength of P1&FG, P2&FG, P7&FG, and P9&FG exhibited 30%, 42%, 24%, and 22% decrease compared to the promoter strength of P1, P2, P7, and P9, respectively. The potential reason was that co-transferring two plasmids caused the burden on cells which might decrease the egfp expression. When PadR was introduced, four hybrid promoters (P1&FG, P2&FG, P7&FG, and P9&FG) can be repressed by PadR with higher leaking activities except P1&FG. When 600 mg/L p-coumaric acid was added, the activity of four hybrid promoters can be obtained with higher strength compared to the promoters without the yveF and yveG overexpression (Fig. 9). These results further confirmed that yveF and yveG can function as proteins and positively assist the inhibition release.
Fig. 9.
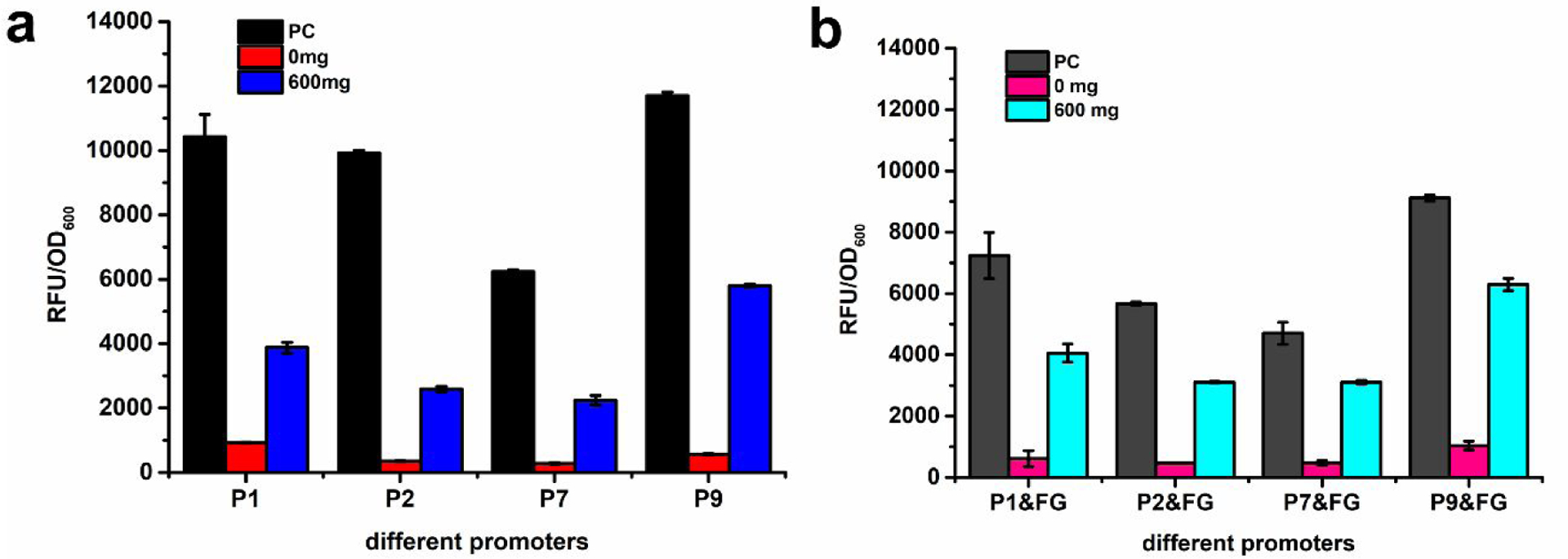
The dynamic performance of hybrid promoters without yveF and yveG or expressing yveF and yveG independently. (a) The dynamic performance of P1, P2, P7, and P9, which without yveF and yveG. (b) The dynamic performance of P1&FG, P2&FG, P7&FG, and P9&FG, which express yveF and yveG independently. All data are reported as mean±s.d. from three independent experiments (n=3). Error bars are defined as s.d.
Conclusion
Transcriptional factor-based biosensors played a significant role in metabolic engineering. They can be utilized to dynamically regulate the pathway for the synthesis of valuable compounds or used for high-throughput screening of high yield strains. In this paper, we cloned and optimized the p-coumaric acid responsive regulator PadR from Bacillus subtilis. After functional analysis and verification, we found that yveF and yveG showed a significant impact on the regulation of PadR-PpadC biosensor system, mostly on increasing the PpadC strength and assisting the repression release. In the existence of yveF and yveG, the promoter strength of PpadC-FG increased 80% compared with PpadC. In addition, when p-coumaric acid added, the activated efficiency increased 116%. To further increase the dynamic range of PadR-PpadC biosensor system, we designed six PadR mutants and obtained two mutants with increased dynamic range and sensitivity. The mutant K64A exhibited a broader operational range (0–1000 mg/L) compared to wild type PadR (0–600 mg/L). The highest activated strength reached 1083.7 a.u., which is 1.26-fold higher than wild type PadR. The other mutant H38A, which achieved the highest activated strength at 200 mg/L p-coumaric acid, was extremely sensitive and it can be activated with even only 5 mg/L p-coumaric acid. We also revealed the molecular mechanism of how these mutants affected the dynamic behavior of this biosensor system based on rational structure analysis of PadR. Finally, a series of hybrid promoters, which were engineered to be regulated by PadR and p-coumaric acid, were constructed. Four of them P1, P2, P7, and P9 showed increased strength compared to PpadC and can be regulated by PadR and p-coumaric acid. The PadR and PpadC variants we obtained in this paper expanded the application of this biosensor system in dynamic regulation and high-throughput screening.
Materials and methods
Strains, plasmids, and hybrid promoter sequence
High-copy number plasmid pZE12-luc and medium-copy number plasmid pCS27 were used for plasmids construction. E. coli strains XL1-Blue and BW25113 (F’) were used for plasmids construction and biosensor characterization, respectively. Strains and plasmids used in this paper were shown in Table 1. Bacillus subtilis was used for PadR, yveF, yveG, and PpadC clone.
Table 1.
List of strains and plasmids used in this study
| Strains | Genotype | Reference |
|---|---|---|
| XL 1-Blue | recA1 endA1gyrA96thi-1hsdR17supE44relA1lac [F’ proAB lacIqZDM15n1O (TetR)] | Stratagene |
| BW25113(F’) | rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 F’ [traD36 proAB lacIqZΔM15 Tn10(Tetr)] | Yale CGSC |
| Plasmids | Description | Reference |
| pZE12-luc | pLlacO-1; luc; ColE1 ori; AmpR | 20 |
| pCS27 | pLlacO-1; p15A ori; KanR | 21 |
| pZE-PpadC-egfp | pZE12-luc carrying PpadC from Bacillus subtilis and egfp | In this study |
| pZE-PpadC-yveF-egfp | pZE12-luc carrying PpadC and yveF from Bacillus subtilis and egfp | In this study |
| pZE-Ppadc-yveG-egfp | pZE12-luc carrying PpadC and yveG from Bacillus subtilis and egfp | In this study |
| pZE-PpadC-yveF-yveG-egfp | pZE12-luc carrying PpadC and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pCS-lpp1.0-PadR | pCS27 carrying promoter lpp1.O and PadR from Bacillus subtilis | In this study |
| pCS-lpp0.8-PadR | pCS27 carrying promoter lpp0.8 and PadR from Bacillus subtilis | In this study |
| pCS-lpp0.5-PadR | pCS27 carrying promoter lpp0.5 and PadR from Bacillus subtilis | In this study |
| pCS-lpp0.2-PadR | pCS27 carrying promoter lpp0.2 and PadR from Bacillus subtilis | In this study |
| pCS-lpp0.2-K37A | pCS27 carrying promoter lpp0.2 and PadR mutant K37A from Bacillus subtilis | In this study |
| pCS-lpp0.2-H38A | pCS27 carrying promoter lpp0.2 and PadR mutant H38A from Bacillus subtilis | In this study |
| pCS-lpp0.2-S39A | pCS27 carrying promoter lpp0.2 and PadR mutant S39A from Bacillus subtilis | In this study |
| pCS-lpp0.2-Q40A | pCS27 carrying promoter lpp0.2 and PadR mutant Q40A from Bacillus subtilis | In this study |
| pCS-lpp0.2-Q61A | pCS27 carrying promoter lpp0.2 and PadR mutant Q61A from Bacillus subtilis | In this study |
| pCS-lpp0.2-K64A | pCS27 carrying promoter lpp0.2 and PadR mutant K64A from Bacillus subtilis | In this study |
| pZE-P1-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P1, and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P2-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P2, and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P3-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P3, and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P4-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P4, and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P5-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P5, and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P6-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P6, and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P7-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P7, and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P8-yveF-yveG-egfp | pZE12-/wc carrying PpadC mutant P8, and yveF andyveG from Bacillus subtilis and egfp | In this study |
| pZE-P9-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P9, and yveF andyveG from Bacillus subtilis and egfp | In this study |
| pZE-P10-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P10, and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P11-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P11, and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P12-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P12, andyveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P13-yveF-yveG-egfp | pZE12-luc carrying PpadC mutant P13, and yveF and yveG from Bacillus subtilis and egfp | In this study |
| pZE-P1-egfp | pZE12-luc carrying PpadC mutant P1 and egfp | In this study |
| pZE-P2-egfp | pZE12-luc carrying PpadC mutant P2 and egfp | In this study |
| pZE-P3-egrp | pZE12-luc carrying PpadC mutant P3 and egfp | In this study |
| pZE-P4-egfp | pZE12-luc carrying PpadC mutant P4 and egfp | In this study |
| pZE-P5-egfp | pZE12-luc carrying PpadC mutant P5 and egfp | In this study |
| pZE-P6-egfp | pZE12-luc carrying PpadC mutant P6 and egfp | In this study |
| pZE-P7-egfp | pZE12-luc carrying PpadC mutant P7 and egfp | In this study |
| pZE-P8-egfp | pZE12-luc carrying PpadC mutant P8 and egfp | In this study |
| pZE-P9-egfp | pZE12-luc carrying PpadC mutant P9 and egfp | In this study |
| pZE-P10-egfp | pZE12-luc carrying PpadC mutant P10 and egfp | In this study |
| pZE-P11-egfp | pZE12-luc carrying PpadC mutant P11 and egfp | In this study |
| pZE-P12-egfp | pZE12-luc carrying PpadC mutant P12 and egfp | In this study |
| pZE-P13-egfp | pZE12-luc carrying PpadC mutant P13 and egfp | In this study |
| pSC-lpp1.0-yveF-yveG | pCS27 carrying lpp1.0, yveF, and yveG | In this study |
Spacer sequence before yveF:
GAGGTGGTTCTC
Spacer sequence between yveF and yveG:
TTCATTCCGATTCGTTCGGGAACAATAACCGGGGAAGTAAAGGGACGTATTTTGC CGGGCGGTGCCGATTCACAA
Spacer sequence between yveG and downstream padC
AAGACTAAGGAGAGTGTGTAAG
Hybrid promoters with yveF and yveG were constructed by adding two genes behind related hybrid promoters directly.
Medium and chemicals
For plasmids construction and biosensor characterization, Luria-Bertani (LB) medium containing 10 g/L NaCl, 5 g/L yeast extract, and 10 g/L tryptone was used in strains inoculation. The antibiotics ampicillin and kanamycin were added in the culture as needed with the final concentration of 100 μg/mL and 50 μg/mL, respectively. For the preparation of inducer solution, 100 mg p-coumaric acid, ferulic acid, cinnamic acid, and caffeic acid were dissolved into 1mL methanol, respectively. p-coumaric acid, ferulic acid, cinnamic acid, and caffeic acid were purchased from MP Biomedicals. Methanol was purchased from Fisher Chemicals. High-Fidelity Phusion DNA polymerase, restriction endonucleases, and Quick Ligation Kit were purchased from New England Biolabs (Beverly, MA, USA). Zyppy™ Plasmid Miniprep Kit, Zymoclean™ Gel DNA Recovery Kit, and DNA Clean & Concentrator™−5 were purchased from Zymo Research (Irvine, CA, USA).
DNA manipulation
PadR, yveF, yveG, and PpadC were amplified from genome DNA of Bacillus subtilis. PpadC digested by XhoI and KpnI was cloned into pCS-lpp1.0-egfp to construct the plasmid pZE-PpadC-egfp. The same methods were used to construct plasmids pZE-PpadC -yveF-egfp, pZE-PpadC -yveF-yveG-egfp. PpadC and yveG were combined by overlap extension PCR to obtain the fragments PpadC-yveG. PpadC-yveG digested by XhoI and KpnI was cloned into pCS-lpp1.0-egfp to construct the plasmid pZE- PpadC -yveG-egfp. PadR digested by KpnI and BamHI was cloned into pCS-lpp1.0-egfp to construct the plasmid pCS-lpp1.0-PadR. Same methods in pCS-lpp1.0-PadR construction were used to construct the plasmids pCS-lpp 0.8-PadR, pCS-lpp 0.5-PadR, and pCS-lpp 0.2-PadR. YveF-yveG were digested by EcoRI and BamHI was cloned into pCS-lpp1.0-egfp to construct the plasmid pCS-lpp1.0-egfp. pCS-lpp1.0-egfp was used as template to get the fragment of lpp1.0-yveF-yveG. lpp1.0-yveF-yveG digested by XhoI and BamHI was cloned to pSC-egfp to get the plasmid pSC-lpp1.0-yveF-yveG.
Mutants construction
Site-directed mutation of PadR was carried out by overlap extension PCR using plasmid pCS-lpp0.2-PadR as template. The primer containing PL was synthesized containing the restriction site of XhoI. The fragment PL-yveF-yveG-egfp digested by XhoI and XbaI was cloned into pZE-PLlacO1 to construct the plasmid pZE-PL-yveF-yveG-egfp, which was used as the template for hybrid promoter’s construction. The hybrid promoter digested by AatII and EcoRI was cloned to pZE-egfp to get the hybrid promoters without yveF and yveG.
Dynamic range characterization of PadR-PpadC biosensor system
Specific plasmids were transformed into E. coli BW25113 (F’), respectively. After 12 hours, single colony was picked and inoculated into 3.5 mL LB medium containing specific antibiotics. After 10 hours of cultivation, the cultures were transferred into 15 mL test tubes containing 3.5 mL LB medium and specific antibiotics, with an inoculation ratio of 5%. When OD600 reached 0.4, different amounts of 100 g/L inducers were added to the specific concentration. After 12 hours, 100 μL cell culture were sampled for fluorescence assay. All experiments were carried out with three groups.
Fluorescence assay
Cell cultures (100 μL) were transferred into a black 96-well plate (BRAND plates) and diluted with equal volume of water. The fluorescence intensity of egfp was detected with Biotek Synergy HT plate reader using excitation filter of 520 nm, and emission filter of 485 nm. The egfp fluorescence intensity of each sample was normalized against its OD600 and background cell fluorescence was subtracted.
Table 2.
the sequence of genes and hybrid promoters
| Name | Sequence |
|---|---|
| PpadC | GGACTGTCTTCAAACAGTCCTTGTTTTTTTATGTTCCTATTGTTTGACAGT TAACTGCAATGGTGTTAAAGTGAACATGTAAATAGTTACATGATTTTTTC TGAAGGT |
| yveF | gtgAAGAAGCCGGTTTTAAAACCATTCGCCTCTTTAGAAATCAAGGTTGAT CCGCCTATCACGATTGGTGAGACAAGCCTGGGACTGAGAtga |
| yveG | atgATTCGCGCTAACGGCAGAACAGATTTATCTGCCAGGTATGTGATTGAA ACAGCAGATCATGAACTGATTTACATTGAAAACAATGGAATACGGCAAG TCAGCAAGCCGTTTCGAAAACAAGCGGCAGCCGGGGAAATTATTGAACC GGAGCATGTTTATTTTCGTACGGTACCGACGTTTGAAACAGGCAGTGAAG TCTATCAATGGCTCCATGACCGCTTGTTTATCGGTTCCGCAGAAAGAACC CCTGATTACGTTCTACTAGACATTTATGAAGTACAGtaa |
| P1 | AACATGTAAATAGTTACATGATttgacaTAAATACCACTGGCGGTgatactGAG CACATCAGCAGGACGCACTGACC |
| P2 | TAAATTATCTCTGGCGGTGttgacaAACATGTAAATAGTTACATGATgatactGA GCACATCAGCAGGACGCACTGACC |
| P3 | AACATGTAAATAGTTACATGATttgacaAACATGTAAATAGTTACATGATgata ctGAGCACATCAGCAGGACGCACTGACC |
| P4 | ATGGTGTTAAAGTGAACATGTAttgacaTAAATACCACTGGCGGTgatactGAG CACATCAGCAGGACGCACTGACC |
| P5 | TAAATTATCTCTGGCGGTGttgacaATGGTGTTAAAGTGAACATGTAgatactGA GCACATCAGCAGGACGCACTGACC |
| P6 | ATGGTGTTAAAGTGAACATGTAttgacaATGGTGTTAAAGTGAACATGTAgata ctGAGCACATCAGCAGGACGCACTGACC |
| P7 | AACATGTAAATAGTTACATGATttgacaATGGTGTTAAAGTGAACATGTAgata ctGAGCACATCAGCAGGACGCACTGACC |
| P8 | ATGGTGTTAAAGTGAACATGTAttgacaAACATGTAAATAGTTACATGATgata ctGAGCACATCAGCAGGACGCACTGACC |
| P9 | TAAATTATCTCTGGCGGTGttgacaTAAATACCACTGGCGGTgatactgAACATGTAAATAGTTACATGAT |
| P10 | TAAATTATCTCTGGCGGTGttgacaTAAATACCACTATGGTGTtaaagtGAACATGTAagcaggacgcactgacc |
| P11 | TAAATTATCTCTGGCGGTGttgacaTAAATACCACTATGGTGTgatactGAACATGTAagcaggacgcactgacc |
| P12 | TAAATTATCTCTGGCGGTGttgacaTAAATACCACATGGTGTtaaagtGAACATGTAAATAGTTACATGAT |
| P13 | TAAATTATCTCTGGCGGTGttgacaTAAATACCACATGGTGTgatactGAACATG TAAATAGTTACATGAT |
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM128620. We also acknowledge the support from the College of Engineering, The University of Georgia, Athens
Footnotes
The authors declare no competing financial interest.
Reference
- [1].Zhou Y, Lin L, Wang H, Zhang Z, Zhou J, and Jiao N (2020) Development of a CRISPR/Cas9n-based tool for metabolic engineering of Pseudomonas putida for ferulic acid-to-polyhydroxyalkanoate bioconversion, Commun Biol 3, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhu M, Wang C, Sun W, Zhou A, Wang Y, Zhang G, Zhou X, Huo Y, and Li C (2018) Boosting 11-oxo-beta-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants, Metab Eng 45, 43–50. [DOI] [PubMed] [Google Scholar]
- [3].Florez AB, Alvarez S, Zabala D, Brana AF, Salas JA, and Mendez C (2015) Transcriptional regulation of mithramycin biosynthesis in Streptomyces argillaceus: dual role as activator and repressor of the PadR-like regulator MtrY, Microbiology 161, 272–284. [DOI] [PubMed] [Google Scholar]
- [4].Humphreys CM, and Minton NP (2018) Advances in metabolic engineering in the microbial production of fuels and chemicals from C1 gas, Curr Opin Biotechnol 50, 174–181. [DOI] [PubMed] [Google Scholar]
- [5].Wang J, Shen X, Yuan Q, and Yan Y (2018) Microbial synthesis of pyrogallol using genetically engineered Escherichia coli, Metab Eng 45, 134–141. [DOI] [PubMed] [Google Scholar]
- [6].Wang J, Zhang R, Zhang Y, Yang Y, Lin Y, and Yan Y (2019) Developing a pyruvate-driven metabolic scenario for growth-coupled microbial production, Metab Eng 55, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang Y, Lin Y, Wang J, Wu Y, Zhang R, Cheng M, Shen X, Wang J, Chen Z, Li C, Yuan Q, and Yan Y (2018) Sensor-regulator and RNAi based bifunctional dynamic control network for engineered microbial synthesis, Nat Commun 9, 3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xu P, Li L, Zhang F, Stephanopoulos G, and Koffas M (2014) Improving fatty acids production by engineering dynamic pathway regulation and metabolic control, Proc Natl Acad Sci U S A 111, 11299–11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gupta A, Reizman IM, Reisch CR, and Prather KL (2017) Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit, Nat Biotechnol 35, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang F, Carothers JM, and Keasling JD (2012) Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids, Nat Biotechnol 30, 354–359. [DOI] [PubMed] [Google Scholar]
- [11].Lan EI, and Liao JC (2012) ATP drives direct photosynthetic production of 1-butanol in cyanobacteria, Proc Natl Acad Sci U S A 109, 6018–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dietrich JA, Shis DL, Alikhani A, and Keasling JD (2013) Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis, ACS Synth Biol 2, 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen W, Zhang S, Jiang P, Yao J, He Y, Chen L, Gui X, Dong Z, and Tang SY (2015) Design of an ectoine-responsive AraC mutant and its application in metabolic engineering of ectoine biosynthesis, Metab Eng 30, 149–155. [DOI] [PubMed] [Google Scholar]
- [14].Xu P, Wang W, Li L, Bhan N, Zhang F, and Koffas MA (2014) Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli, ACS Chem Biol 9, 451–458. [DOI] [PubMed] [Google Scholar]
- [15].Nguyen TK, Tran NP, and Cavin JF (2011) Genetic and biochemical analysis of PadR-padC promoter interactions during the phenolic acid stress response in Bacillus subtilis 168, J Bacteriol 193, 4180–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tran NP, Gury J, Dartois V, Nguyen TK, Seraut H, Barthelmebs L, Gervais P, and Cavin JF (2008) Phenolic acid-mediated regulation of the padC gene, encoding the phenolic acid decarboxylase of Bacillus subtilis, J Bacteriol 190, 3213–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Siedler S, Khatri NK, Zsohar A, Kjaerbolling I, Vogt M, Hammar P, Nielsen CF, Marienhagen J, Sommer MOA, and Joensson HN (2017) Development of a Bacterial Biosensor for Rapid Screening of Yeast p-Coumaric Acid Production, ACS Synth Biol 6, 1860–1869. [DOI] [PubMed] [Google Scholar]
- [18].Park SC, Kwak YM, Song WS, Hong M, and Yoon SI (2017) Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR, Nucleic Acids Res 45, 13080–13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang J, Mahajani M, Jackson SL, Yang Y, Chen M, Ferreira EM, Lin Y, and Yan Y (2017) Engineering a bacterial platform for total biosynthesis of caffeic acid derived phenethyl esters and amides, Metab Eng 44, 89–99. [DOI] [PubMed] [Google Scholar]
- [20].Lutz R, and Bujard H. J. N. a. r. (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements, 25, 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shen CR, and Liao JC (2008) Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways, Metab Eng 10, 312–320. [DOI] [PubMed] [Google Scholar]


