Abstract
Here, drug repurposing and molecular docking were employed to screen approved MPP inhibitors and their derivatives to suggest a specific therapeutic agent for the treatment of COVID-19. The approved MPP inhibitors against HIV and HCV were prioritized, while RNA dependent RNA Polymerase (RdRp) inhibitor remdesivir including Favipiravir, alpha-ketoamide were studied as control groups. The target drug surface hotspot was also investigated through the molecular docking technique. Molecular dynamics was performed to determine the binding stability of docked complexes. Absorption, distribution, metabolism, and excretion analysis was conducted to understand the pharmacokinetics and drug-likeness of the screened MPP inhibitors. The results of the study revealed that Paritaprevir (−10.9 kcal/mol) and its analog (CID 131982844) (−16.3 kcal/mol) showed better binding affinity than the approved MPP inhibitors compared in this study, including remdesivir, Favipiravir, and alpha-ketoamide. A comparative study among the screened putative MPP inhibitors revealed that the amino acids T25, T26, H41, M49, L141, N142, G143, C145, H164, M165, E166, D187, R188, and Q189 are at potentially critical positions for being surface hotspots in the MPP of SARS-CoV-2. The top 5 predicted drugs (Paritaprevir, Glecaprevir, Nelfinavir, and Lopinavir) and the topmost analog showed conformational stability in the active site of the SARS-CoV-2 MP protein. The study also suggested that Paritaprevir and its analog (CID 131982844) might be effective against SARS-CoV-2. The current findings are limited to in silico analysis and lack in vivo efficacy testing; thus, we strongly recommend a quick assessment of Paritaprevir and its analog (CID 131982844) in a clinical trial.
Keywords: Main protease protein (MPP) inhibitors, SARS-CoV-2, COVID-19, Drug repurposing, Molecular docking
1. Introduction
COVID-19 is a rapidly spreading viral infectious disease caused by a beta coronavirus – the novel coronavirus SARS-CoV-2. The infection was first described in China in late December 2019, and within three months, 202 countries, territories, and conveyances had reported COVID-19 cases within their boundaries [1], [2]. This emerging human pathogen is causing acute respiratory tract infection with significant morbidity, need intensive care facilities and case fatality is most common. Even the organized health systems of many countries are also facing greatest difficulties to tackle COVID-19. Without delay, COVID-19 has become a global public health emergency and been declared a pandemic by the World Health Organization & this outbreak poses a huge threat to humans [3].
SARS-CoV-2 (also called 2019-nCoV) is an enveloped, single-stranded, positive-sense RNA virus with genome sizes ranging from 26 to 32 kb and virion sizes from 50 to 200 nanometers in diameter [4]. In addition, zoonotic virus SARS-CoV-2 is distantly related to MERS-CoV and SARS-CoV (80.26%), but exhibits dissimilarities that may influence the process of pathogenesis [5], [6], [7]. SARS-CoV-1 and MERS-CoV mainly affect people via nosocomial spread, while this variant of human coronavirus frequently spreads through community transmission [8]. In addition, it infects through the same entry point, the angiotensin-converting enzyme 2 (ACE2) receptor, studies have confirmed a higher affinity of SARS-CoV-2 to human ACE2 than that of previous CoV strains and does not use other receptors such as dipeptidyl peptidase 4, used by MERS-CoV [9]. However, four essential structural proteins, i.e., Spike protein (S), Envelope protein (E), Membrane protein (M), and Nucleocapsid protein (N) are found in SARS-CoV-2 and to generate the major components of the virus particle, polyprotein processing is an essential mechanism [10]. The polyproteins are cleaved and transformed into mature nonstructural proteins by the Main Proteases, 3CL protease and responsible for playing a role in replication/transcription process that’s why main protease could be an effective drug targets [11], [12].
Many efforts are ongoing around the world, but preventive vaccine development usually takes a long time and there are no specific, clear treatment strategies as of now. In the present situation, finding a therapy appropriate for COVID-19 is of paramount importance to reduce the catastrophic effect of the ongoing pandemic. Depending on the target, therapies against SARS-CoV-2 can be divided into two categories: the first acts on the human immune system or human cells, and the second acts on the coronavirus itself [13], [14]. Therapies based on the immune system include blocking the signaling pathways of human cells that include ACE2 receptor protein on the surface of cells required for virus replication. In the case of therapies based on the virus, the retroviral RNA genome encodes for most of the three enzymes essential for virus replication: (i) viral protease (PR), (ii) reverse transcriptase (RT), (iii) integrase (IN) and as of now these three are mainly used as drug targets [15], [16], [17], [18]. In contrast, some of the best characterized, conserved drug targets of coronaviruses are the main protease proteins (MPPs) [19], [20]. Moreover, Inhibition of the activity of MPP would block viral replication, and it would also be nontoxic, as human proteases with similar cleavage specificity have already been reported [21], [22].
In addition, various viral protease inhibitors can inhibit proteases and reduce HIV and HCV to undetectable levels which employ aspartyl and serine proteases, respectively. Some of these drugs are now being repurposed for the treatment of COVID-19, which is also possesses a MPP [23], [24], [25], [26], [27], [28]. A computational drug repurposing study has previously shown that many more drugs including Lopinavir and Ritonavir (also HIV-1 protease inhibitors) are capable of inhibiting SARS-CoV MPP and, therefore, could serve as a homologous target, as the previous SARS-CoV main protease has 96.1% similarity to SARS-CoV-2 MPP [29], [30]. In the United States, four HCV protease inhibitors have also been recognized for use in combination with other specified anti-HCV agents, which also suggests the utilization of similar drugs for the treatment of SARS-CoV-2 infection [31]. On the other hand, eleven compounds such as macrolides antibiotics, proton pump inhibitors, antiarrhythmic agents showing antiviral potency by inhibiting SARS-CoV-2 replication in vitro [32].
The observed impact of COVID-19 is differing among countries. However, the number of global diagnosed cases has exceeded 131.72 million, with a death toll above 2.8 million as of April 4, 2021 [33]. The current trend provides a warning regarding entry into a phase beyond containment of the disease. These circumstances are pushing the global scientific community to determine a suitable drug that can be used for the treatment of COVID-19 patients. The discovery of a new drug is a lengthy process. Thus, there is an urgency for repurposing or reprofiling of drugs. Identifying existing molecules that are suitable for the treatment of COVID-19 would lead to immediate clinical trials and subsequent treatment for patients.
The contribution of computational biology cannot be ignored in the fields of drug discovery and development [34]. Drug repositioning, a promising approach in this field, identifies new therapeutic opportunities for existing drugs and reduces the discovery and development timeline. The approach intends to find alternative uses for a pioneering drug that is made by another innovator. Virtual screening is another computational technique that is widely used in drug discovery to search libraries of small compounds to identify molecules that are most likely to bind to a drug target (e.g., protein receptor or enzyme) [35], [36]. Among different types of virtual screening methods, pharmacophore-based modeling is most often applied to virtual screening, resulting in an abstract description of the molecular features necessary for recognition of a ligand by a biological macromolecule [37]. Despite the abundance of associated successful cases, several demerits are encountered, such as the absence of good scoring metrics [37]. Pharmacophore modeling is also impossible if the target structure or ligands are unknown. Conversely, structure-based virtual screening involves the docking of candidate ligands into a protein target followed by the application of a scoring function to estimate the binding affinity between the target molecules [38], [39]. The pharmaceutical industry now routinely practices examining the effects of a potential drug candidate against particular diseases/pathogens due to its reliability and efficacy [40], [41], [42]. Additionally, computational biology could assist in the field of structural biology, lead molecule optimization, potential drug candidate screening, studying drug surface binding patterns, providing a hypothesis for X-ray crystallography to study the substrates and inhibitors, and establishing a combinatorial library [43], [44]. Thus, computational biology could definitely be utilized for repurposing molecules for the treatment of COVID-19. Repurposing approved drugs against the COVID-19 epidemic also has several advantages, which can help to rapidly identify treatment options [45]. Approved or candidate drugs with repurposing potential as antiviral agents exist for a number of emerging viruses for which urgent, cost-effective therapeutic solutions are required, including influenza virus [46], Ebola [47], HCV [48], Zika virus [49], Japanese encephalitis virus (JEV) [50], human cytomegalovirus [51], dengue virus (DENV) [52], MERS-CoV [53], and SARS-CoV [54]. Recently, a few in silico strategies were also employed against 2019-nCoV, which focused on spike glycoprotein (S‐protein) inhibitors and mainly SARS-CoV-2 MPP inhibitors [55], [56], [57], [58].
However, after one year of the COVID-19 pandemic, no specific antiviral drugs have yet demonstrated 100% effectiveness combating SARS-CoV-2 infections. Scientists are racing to identify effective drugs by continuing many studies worldwide. Developing a novel drug within a short time against an emerging infectious disease is a time-bound challenge. Thus, it would be better to rely on the method of repurposing existing drugs. The present study focuses on screening existing main protease inhibitors and their derivatives, including drug surface hotspots of SARS-CoV-2, using drug repurposing and computational approaches.
2. Materials and methods
2.1. Retrieval of MPPs and MPP inhibitors
The MPP structures of SARS-CoV-2 (PDB ID: 6LU7 and PDB ID: 6Y2E) and HCV (PDB ID: 2P59) were retrieved from the NCBI and RCSB Protein Data Bank (PDB) [59], [60]. The approved MPP inhibitors against HIV and HCV were prioritized for the study. The PDB format for the approved MPP inhibitors was retrieved from the DrugBank database of NCBI (Table S1). For the selection of FDA-approved and widely suggested MPP inhibitors, we used the ‘Drug.com’ database (https://www.drugs.com/drug-class/protease-inhibitors.html). Furthermore, the derivative molecules of the topmost screened MPP inhibitors were collected from the PubChem database (Table S2). Moreover, Favipiravir (DB12466), and alpha-ketoamide (CID 6482451) were also retrieved from the DrugBank database. Remdesivir (DB14761), an approved RNA dependent RNA polymerase inhibitor, was also retrieved from PubChem database, and employed in our study as control group of other MPP.
2.2. Screening of MPP inhibitors against the MPP of SARS-CoV-2
AutoDockVina software was used for molecular docking experiments, which is now being widely used for screening effective therapeutics against the specific drug target of deadly pathogens [60], [61], [62]. Prior to molecular docking, the crystal structure of MPP was cleaned up using PyMOL [63], as it was in a complex structure with an inhibitor. After the removal of unwanted molecules, such as water, ions, and MPP inhibitors, it was docked to 16 approved MPP inhibitors to analyze the lowest binding energy and interactive amino acids. Moreover, 100 derivatives of the top MPP inhibitor candidates found from the initial docking experiment were also docked to the MPP of SARS-CoV-2 to determine the top candidates for the treatment of COVID-19. Paritaprevir and its top derivatives were also employed to molecular docking with the MPP of HCV (ID: 2P59) and another structure of the MPP of SARS-CoV-2 (PDB ID: 6Y2E) suggested by Linlin Zhang et al. [27]. The grid box parameters were set to a size of 60 Å × 70 Å × 62 Å (x × y × z) and center of − 26.289 Å × 13.666 Å × 58.965 Å (x × y × z). Moreover, another newly released crystal structure of SARS-CoV-2 MPP (PDB ID: 6Y2E) was also used to double check the docking results with the experimental data. The grid box parameters were set to a size of 45 Å × 66 Å × 62 Å (x × y × z) and center of − 16.520 Å × −26.112 Å × 17.524 Å (x × y × z) for ‘PDB ID: 6Y2E’. LigPlot+ was used to generate 2D ligand-protein interaction diagrams and identify the involved amino acids with their interactive position in the docked molecules [64]. Discovery Studio and PyMOL were used to visualize and analyze the ligand molecule interactions with the viral proteins [63], [65].
2.3. Structural insights regarding drug surface hotspots in the MPP of SARS-CoV-2
To determine the drug surface hotspot of SARS-CoV-2 MPP, the MPP and MPP inhibitors structure were analyzed by LigPlot+, Discovery Studio, and PyMOL. The alpha-ketoamide inhibitor, recently suggested by Linlin Zhang for the treatment of COVID-19, served as the positive control for the study [66], [67]. The molecular docking approach was employed to study the binding pattern of alpha-ketoamide, Favipiravir, and RdRp inhibitor remdesivir with the MPP of SARS-CoV-2, and the results enabled a comparative structural analysis of screened MPP inhibitors and their derivatives. The conservancy pattern of the predicted drug-binding hotspot was analyzed through multiple sequence alignment of homologous MPPs of SARS-CoV-2. All the sequences of the MPP structure deposited in the PDB were retrieved, and multiple sequence alignment was performed using Clustal Omega [68].
2.4. Absorption, distribution, metabolism, and excretion (ADME) analysis of top drug candidates
To assess the ADME properties of the topmost candidates for the MPP inhibitors, the SwissADME portal was used [69]. The SwissADME portal is an online platform that is being used successfully to evaluate the pharmacokinetics, drug-likeness, and medicinal chemistry friendliness of possible drug candidates [70]. This study examined the physico-chemical parameters (formula molecular weight, molar refractivity, total polar surface area (TPSA)), lipophilicity (Log Po/w (iLOGP), Log Po/w (XLOGP3), Log Po/w (WLOGP), Log Po/w (MLOGP), Log Po/w, (SILICOS-IT), consensus Log Po/w), and water solubility (Log S: SILICOS-IT, solubility) of the screened topmost MPP inhibitors and their derivatives. Drug interaction with cytochromes P450 (CYPs) is crucial in drug discovery. Therefore, the screened MPP inhibitors were employed to study the inhibitory effect of different CYP isoforms (CYP1A2 inhibitor, CYP2C19 inhibitor, CYP2C9 inhibitor, CYP2D6 inhibitor, CYP3A4 inhibitor). However, other relevant pharmacokinetic parameters, such as gastrointestinal (GI) absorption, blood-brain barrier (BBB) permeability, and P-gp substrates, were also investigated for putative MPP drug candidates.
2.5. Molecular dynamics (MD) simulation
MD simulation was employed to analyze the protein-inhibitor complexes. In the MD simulation, successive iterations were generated by the integration of Newton’s low, providing insights into the positions and velocities of each molecule variation over time in the system [71]. In this study, MD simulation was performed by LARMD tools to analyze the MPP in complex with the top 5 predicted drugs and the topmost analog, and the time interval was set to 4 ns, with the influence of water [72]. AMBER16 was the force field, and the minimizations were performed by the Sander module in the AMBER program [73]. In the 4-step minimization, the 2000-step steepest descent method along with the 3000-step conjugated gradient method were adopted, and the system was heated from 10 K to 300 K in 30 ps. Finally, all the atoms were relaxed at 300 K by applying the periodic boundary condition [72]. Moreover, the ligand-receptor root mean square deviation (RMSD), number of H-bonds, and calculation of MM/PB(GB)SA binding free energies are descriptors that were adopted to assess the stability of protein inhibitor complexes during the MD simulation.
3. Results
3.1. Screening of MPP inhibitors against the MPP of SARS-CoV-2
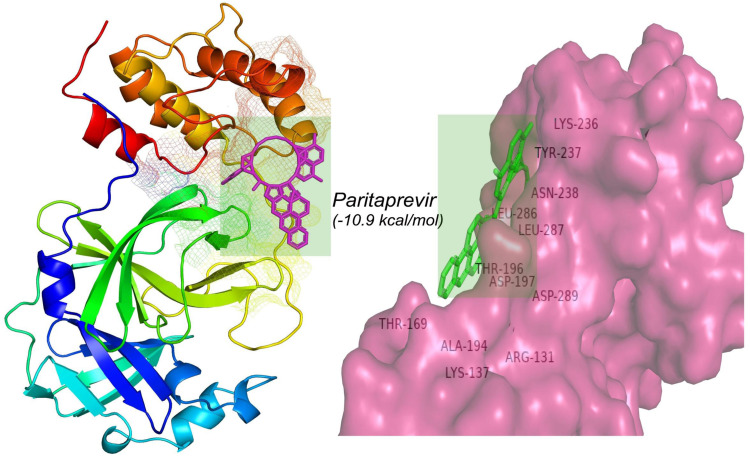
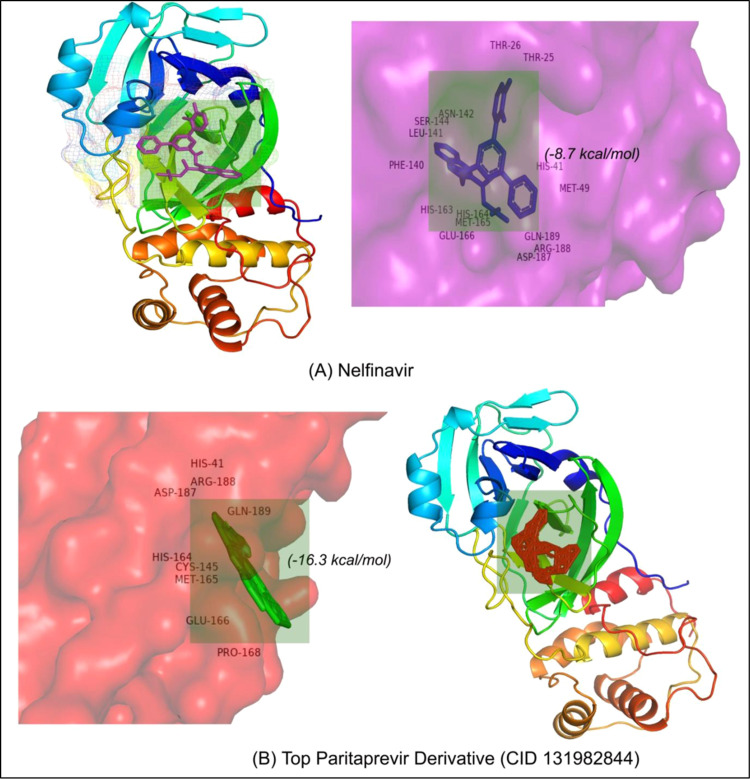
All of the retrieved PDB structures of approved MPP inhibitors were prepared and optimized to allow the molecular docking experiment (Table S1). Widely used common MPP inhibitors of HCV and HIV (https://www.drugs.com/drug-class/protease-inhibitors.html) were prioritized in the study [74]. There were seven HIV MPP inhibitors (Amprenavir, Ritonavir, Nelfinavir, Indinavir, Darunavir, Fosamprenavir, and Saquinavir) and six HCV inhibitors (Paritaprevir, Grazoprevir, Glecaprevir, Lopinavir, Telaprevir, and Boceprevir), as well as Sunaprevir, Atazanavir, and Lopinavir. All of these MPP inhibitors were employed for molecular docking, and the scoring function of AutoDockVina was utilized to predict the interaction between the above-mentioned ligands and the MPP of SARS-CoV-2 (PDB ID: 6LU7). Paritaprevir, an HCV MPP inhibitor, was found to have the highest negative binding energy (−10.9 kcal/mol) when interacting with the MPP of SARS-CoV-2 ( Table 1a and Fig. 1). Moreover, Glecaprevir (−9.3 kcal/mol), Nelfinavir (−8.7 kcal/mol), Lopinavir (−8.6 kcal/mol), and Lopinavir (−8.4 kcal/mol) were also found to be the topmost MPP inhibitors with high binding affinities (Table 1a). The molecular docking results for all retrieved MPP inhibitors against the MPP of SARS-CoV-2 are listed in Table S3. Furthermore, the derivatives of Paritaprevir were used in similar molecular docking studies. The top derivative of Paritaprevir “(3S,9Z,12R,15S,17R)-N-cyclopropylsulfonyl-12-methyl-3-[(5-methylpyrazine-2-carbonyl)amino]-2,14-dioxo-17 phenanthridin-6-yloxy-1,13-diazabicyclo[13.3.0]octadec-9-ene-12-carboxamide” (CID 131982844) demonstrated the highest binding interaction (−16.3 kcal/mol) with the MPP of SARS-CoV-2, which was superior to that of Paritaprevir ( Table 1b, Fig. 2, and Fig. 3).
Table 1b.
Molecular docking results of top ten Paritaprevir derivatives including interactive amino acids from SARS-CoV-2 MPP.
| PubChem ID of Paritaprevir derivatives | IUPAC Name | Binding Energy (kcal/mol) | Involved Amino Acids with Positions |
|---|---|---|---|
| CID 131982844 | (3S,9Z,12R,15S,17R)-N-cyclopropylsulfonyl-12-methyl-3-[(5-methylpyrazine-2-carbonyl)amino]-2,14-dioxo-17-phenanthridin-6-yloxy-1,13-diazabicyclo[13.3.0]octadec-9-ene-12-carboxamide | -16.3 | H41, C145, H164, M165, E166, P168, D187, R188, Q189 |
| CID 117860584 | (1S,4R,6S,7Z,14S,18R)-N-cyclopropylsulfonyl-18-(3,9-difluorophenanthridin-6-yl)oxy-14-[(1-methylpyrazole-4-carbonyl)amino]-2,15-dioxo-3,16-diazatricyclo[14.3.0.04,6]nonadec-7-ene-4-carboxamide | -16.1 | T25, T26, L27, H41, M49, F140, L141, N142, G143, C145, H164, M165, E166, D187, Q189 |
| CID 144881776 | (4S,6R,10S,16Z)-N-cyclopropylsulfonyl-10-[(5-methylpyrazine-2-carbonyl)amino]-3,9-dioxo-6-phenanthridin-6-yloxy-2,8-diazatricyclo[15.2.1.04,8]icos-16-ene-1-carboxamide | -16.1 | T26, H41, M49, F140, L141, N142, C145,H163, H164, M165, E166, R188, Q189 |
| CID 144881777 | (4S,6R,10S,16Z)-N-cyclopropylsulfonyl-10-[(5-methylpyrazine-2-carbonyl)amino]-3,9-dioxo-6-phenanthridin-6-yloxy-2,8-diazatricyclo[15.2.1.04,8]icos-16-ene-1-carboxamide | -16 | H41, M49, F140, L141, N142, C145,H163, H164, M165, E166, R188, Q189 |
| CID 89997958 | (1R,4S,6R,10S,16Z)-N-cyclopropylsulfonyl-10-[(5-methylpyrazine-2-carbonyl)amino]-3,9-dioxo-6-phenanthridin-6-yloxy-2,8-diazatricyclo[15.2.1.04,8]icos-16-ene-1-carboxamide | -15.5 | T26, L27, H41, M49, Y54, F140, L141, N142, G143, C145, H164, M165, E166, D187, Q189 |
| CID 90479564 | (1S,4R,6S,7Z,14S,18R)-N-cyclopropylsulfonyl-14-[(5-methylpyrazine-2-carbonyl)amino]-2,15-dioxo-18-phenanthridin-6-yloxy-3,16-diazatricyclo[14.3.0.04,6]nonadec-7-ene-4-carboxamide;dihydrate | -15.2 | T25, H41, M49, F140, N142, C145, H163, H165, E166, P168, R188, Q189, T190 |
| CID 117930281 | (1S,4R,6S,7Z,14S)-N-cyclopropylsulfonyl-14-[(5-methylpyrazine-2-carbonyl)amino]-2,15-dioxo-18-phenanthridin-6-yloxy-3,16-diazatricyclo[14.3.0.04,6]nonadec-7-ene-4-carboxamide | -15.2 | T25, L27, H41, M49, F140, N142, C145, M165, E166, P168, R188, Q189, T190 |
| CID 126760198 | (1S,4R,7Z,14S,18R)-N-cyclopropylsulfonyl-14-[(5-methylpyrazine-2-carbonyl)amino]-2,15-dioxo-18-phenanthridin-6-yloxy-3,16-diazatricyclo[14.3.0.04,6]nonadec-7-ene-4-carboxamide | -15.2 | T25, L27, H41, M49, F140, N142, C145, M165, E166, P168, R188, Q189, T190 |
| CID 130368695 | (1S,6S,7Z,14S,18R)-N-cyclopropylsulfonyl-14-[(5-methylpyrazine-2-carbonyl)amino]-2,15-dioxo-18-phenanthridin-6-yloxy-3,16-diazatricyclo[14.3.0.04,6]nonadec-7-ene-4-carboxamide | -15.2 | T25, H41, M49, F140, N142, C145, H163, M165, E166, P168, R188, Q189, T190 |
| CID 129606829 | (1R,4S,6R,7Z,14R,18S)-N-cyclopropylsulfonyl-14-[(5-methylpyrazine-2-carbonyl)amino]-2,15-dioxo-18-phenanthridin-6-yloxy-3,16-diazatricyclo[14.3.0.04,6]nonadec-7-ene-4-carboxamide | -15.2 | T25, H41, M49, F140, N142, C145, H163, M165, E166, P168, R188, Q189, T190 |
Table 1a.
Molecular docking results of top five MPP inhibitors including interactive amino acids from SARS-CoV-2 MPP.
| Drug Name | Binding Energy (kcal/mol) | Involved Amino Acid with Position |
|---|---|---|
| Paritaprevir | -10.9 | R131, K137, T169, A194, T196, D197, K236, Y237, N238, L286, L287, D289 |
| Glecaprevir | -9.3 | T199, K236, Y237, L271, G275, M276, N277, G278, L286, L287 |
| Nelfinavir | -8.7 | T25, T26, H41, M49, F140, L141, N142, S144, H163, H164, M165, E166, D187, R188, Q189 |
| Simeprevir | -8.6 | K5, Q12, K137, T199, N238, L272, L286, L287, D289 |
| Lopinavir | -8.4 | Q107, Q110, V202, E240, P241, T243, H246, I249, P293, F294 |
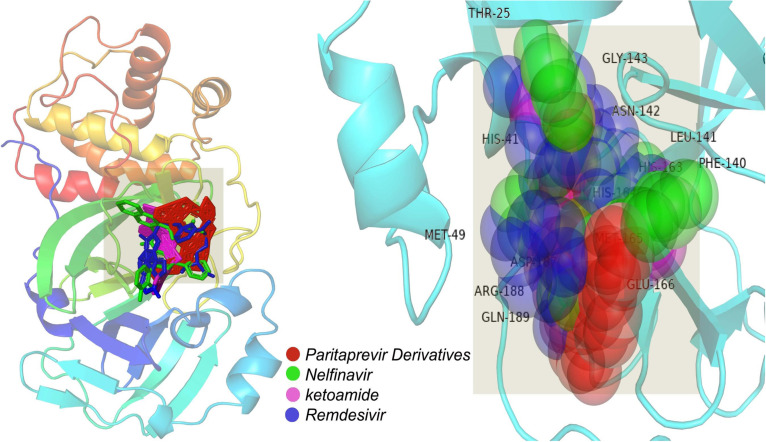
Fig. 1.
Molecular interaction between Paritaprevir and the MPP of SARS-CoV-2.
Fig. 2.
Molecular insights into MPP interactions with (A) Nelfinavir and (B) the top Paritaprevir derivative.
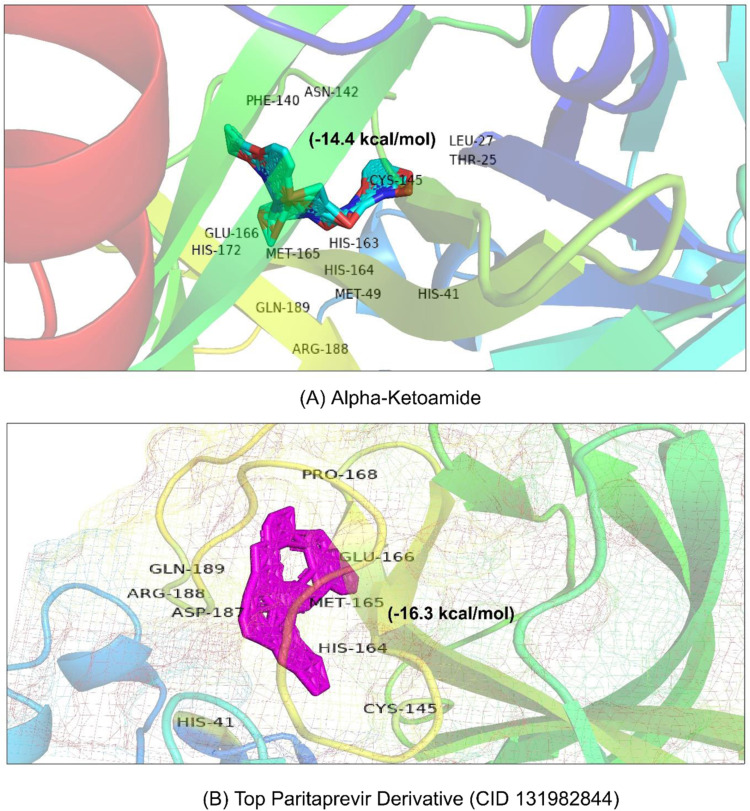
Fig. 3.
Structural overview of molecular interaction of (A) alpha-ketoamide and (B) the top Paritaprevir derivative with the MPP of SARS-CoV-2.
Approximately 98% of the structural analogs of Paritaprevir interacting with the MPP of SARS-CoV-2 showed a higher negative binding energy (>−10 kcal/mol) than Paritaprevir interacting with the MPP of SARS-CoV-2 (Table S4). Moreover, Paritaprevir and its top derivatives (CID 131982844) were also docked to molecular docking with the MPP of HCV (ID: 2P59) and another structure of the MPP of SARS-CoV-2 (PDB ID: 6Y2E) ( Table 2). Paritaprevir showed a similar binding pattern to the MPPs of HCV (−8.4 kcal/mol) and SARS-CoV-2 (PDB ID: 6Y2E, −8.4 kcal/mol), as indicated by the previous molecular binding results found for the Paritaprevir interaction with the MPP of SARS-CoV-2 (PDB ID: 6LU7) [75]. Similar results were also observed in the case of CID 131982844 interactions with the MPPs of HCV (−8.4 kcal/mol) and SARS-CoV-2 (PDB ID: 6Y2E and −8.4 kcal/mol). In addition, alpha-ketoamide (CID 6482451), which has been suggested as an MPP inhibitor based on laboratory experiments, was also docked to the MPP of SARS-CoV-2 (PDB ID: 6Y2E) as a positive control. The results showed that the binding energy and interaction pattern of alpha-ketoamide (−8.4 kcal/mol) with the MPP of SARS-CoV-2 (Fig. 3) were similar to those of Paritaprevir and its top derivatives (CID 131982844). In addition, Favipiravir (DB12466) and remdesivir (ID: DB14761) were employed for docking analysis with the MPP of SARS-CoV-2. Our control RdRp inhibitor Remdesivir showed a higher negative binding energy (−7.4 kcal/mol) than Favipiravir (−4.7 kcal/mol), but these values were lower than those of the MMP inhibitor Paritaprevir (−10.9 kcal/mol) and its derivative (−16.3 kcal/mol) in the present study (Table 2 and Fig. 4).
Table 2.
Molecular docking study of Hydroxychloroquine, Favipiravir, Remdesivir, alpha-ketoamide and MPP Inhibitors with MPPs of SARS-CoV-2 and HCV.
| Ligand Candidates | Main Protease Protein | Molecular Binding Energy (kcal/mol) | Involved Amino Acids with Positions |
|---|---|---|---|
| alpha-ketoamide | SARS-CoV-2 (PDB ID: 6LU7) | -14.4 | T25, L27, H41, M49, F140, N142, C145, H163, H164,pl M165, E166, H172,R188, Q189 |
| HydroxyChloroquine | SARS-CoV-2 (PDB ID: 6LU7) | -5.0 | T198, T199, Y236, Y239, L271, L272, L286, L287 |
| Favipiravir | SARS-CoV-2 (PDB ID: 6LU7) | -4.7 | L141, N142, G143, S144, C145, M165, E166 |
| Remdesivir | SARS-CoV-2 (PDB ID: 6LU7) | -7.8 | T25, T26, H41, M49, L141, N142, G143, C145, H164, M165, E166, D187, R188, Q189 |
| Paritaprevir | SARS-CoV-2 (PDB ID: 6Y2E) | -10.9 | K102, V104, Q107, Q110, N151, I152, D153, S158, T292, F294 |
| Paritaprevir | HCV(PDB ID: 2P59) | -10.6 | V26, K34, A1033, Q1035, R1037, L1039, C1042, S1063, R1135, H1136, S1159 |
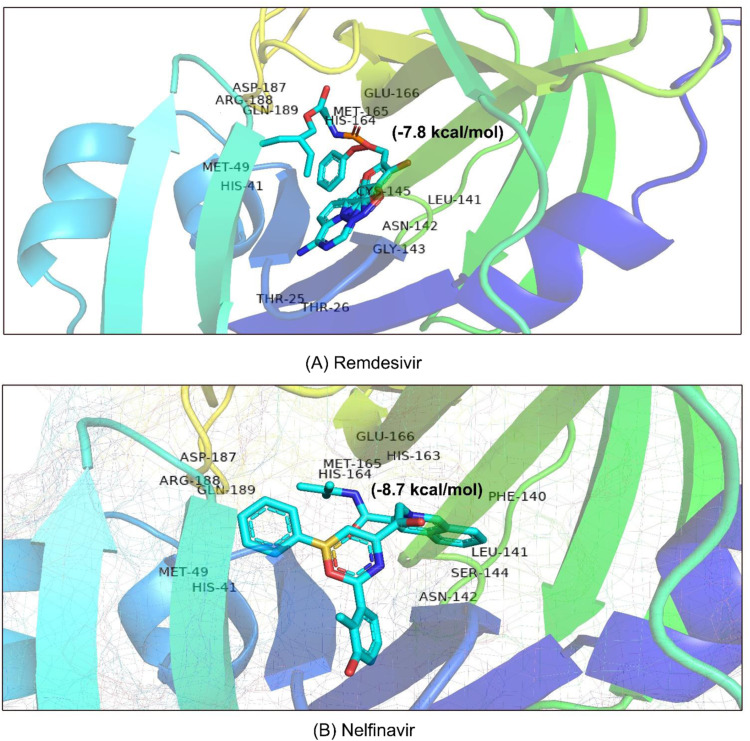
Fig. 4.
Drug surface overview of the interaction of (A) Remdesivir and (B) Nelfinavir with the MPP of SARS-CoV-2.
3.2. Structural insights into drug surface hotspots in the MPP of SARS-CoV-2
To determine common drug surface hotspots of the MPP in SARS-CoV-2, molecular docking interaction of MPP with its inhibitors including control groups were analyzed. Here, basically involved amino acid residues with their respective position were prioritized to target the hotspot of maximum MPP inhibitors against the MPP of SARS-CoV-2. Five approved MPP inhibitors, such as, Paritaprevir, Glecaprevir, Nelfinavir, Lopinavir, and Lopinavir were isolated for the best ligand molecules bound to the MPP of SARS-CoV-2 from docking study, and these interactions were employed for hotspots analysis (Table S1). Moreover, interaction of MPP with Paritaprevir derivatives with highest binding energy were also analyzed to understand the common drug surface hotspots on the MPP of SARS-CoV-2 (Table 1b).
Paritaprevir was found to interact with the amino acids R131, K137, T169, A194, T196, D197, K236, Y237, N238, L286, L287, and D289 in the MPP (PDB ID 6LU7) of SARS-CoV-2 (Fig. 1). The positions N238, L286, and L287 were also crucial for binding of the MPP inhibitors ‘Lopinavir’ (K5, Q12, K137, T199, N238, L272, L286, L287, and D289) and ‘Glecaprevir’ (T199, K236, Y237, L271, G275, M276, N277, G278, L286, and L287) (Table 1a). The most relevant results for the binding with MPP were found in the case of ‘Nelfinavir’. There were 13 amino acid positions (T25, T26, H41, M49, F140, L141, N142, H163, H164, E166, D187, R188, and Q189) in the docking site of Nelfinavir, the positions of which were also abundantly found with most Paritaprevir derivatives (Fig. 2). Moreover, alpha-ketoamide inhibitors, which have been reported to inhibit MPP in laboratory experiments, were also employed to study the interaction with the MPP (PDB ID: 6LU7) of SARS-CoV-2 (Fig. 3). Most surprisingly, amino acids such as T25, L27, H41, M49, F140, N142, C145, H163, H164, M165, E166, H172, R188, and Q189 were found to have critical positions in the alpha-ketoamide inhibitor interactions with MPP, and these amino acids were also found with Nelfinavir and derivatives of Paritaprevir. Moreover, another MPP structure (PDB ID: 6Y2E), reported for alpha-ketoamide inhibitors, was also docked with the topmost MPP inhibitors found from docking study, and Lopinavir showed a similar binding pattern to Paritaprevir for both MPP structures of SARS-CoV-2 (Table 1b and Table 2). Additionally, the binding pattern of remdesivir (T25, T26, H41, M49, L141, N142, G143, C145, H164, M165, E166, D187, R188, and Q189) with the MPP of SARS-CoV-2 (Fig. 4) was highly similar to the binding interaction of Nelfinavir (T25, T26, H41, M49, F140, L141, N142, H163, H164, E166, D187, R188, and Q189), alpha-ketoamide (T25, L27, H41, M49, F140, N142, C145, H163, H164, M165, E166, H172, R188, and Q189), and all the topmost derivatives of Paritaprevir (T25, T26, L27, H41, M49, F140, L141, N142, G143, C145, H164, M165, E166, D187, and Q189). The binding patterns observed for the MPP inhibitors, derivatives, and experimentally suggested molecules indicated that T25, T26, H41, M49, L141, N142, G143, C145, H164, M165, E166, D187, R188, and Q189 could be the critical amino acid positions for drug surface hotspots in the MPP of SARS-CoV-2 ( Fig. 5).
Fig. 5.
Structural insights into the common surface of the drug hotspot in the MPP of SARS-CoV-2.
To check the conservancy level of amino acids in the revealed hotspots of MPP, a total number of 153 MPP sequences were retrieved from the PDB. Multiple sequence alignment (MSA) of these sequences revealed that all the residues predicted as the hotspot of drug binding were conserved (File S1). Multiple sequence alignment revealed no reported mutations or amino acid changes at the drug hotspot positions which revealed the higher conservancy level in the hotspot region. However, three structures (6XB0, 6XB1, and 6XG2) were found to contain X instead of C at position 145. X represents an unknown residue in the protein sequence, which may be due to the poor quality of the protein sequence.
3.3. ADME analysis of top drug candidates
The physico-chemical parameters, lipophilicity, pharmacokinetics properties, and water solubility were studied for the topmost putative MPP inhibitors (Paritaprevir, Glecaprevir, Nelfinavir, Lopinavir, and Lopinavir) and the derivatives of Paritaprevir ( Table 3 and Table S5). The formula, molecular weight, molar refractivity, and TPSA were determined. The lipophilicity and partition coefficient between n-octanol and water (log Po/w) were also calculated by using five common freely available predictive models (XLOGP3, WLOGP, MLOGP, SILICOS-IT, and iLOGP) [76]. The results of the drug interaction with CYP indicated that only Paritaprevir had an inhibitory effect on CYP3A4, while Glecaprevir showed no interaction with the CYP isoforms. Moreover, the cytochromes CYP1A2, CYP2C9, and CYP2D6 had no interaction with the top putative MPP inhibitors. Additionally, there was no interaction possibility of the top Paritaprevir derivatives with the cytochromes CYP1A2, CYP2C19, CYP2C9, or CYP2D6 (Table S5). Additionally, GI absorption was low in the case of Paritaprevir, Glecaprevir, Nelfinavir, and Lopinavir and the top Paritaprevir derivatives. BBB permeability was also calculated by BOILED-Egg models [77], and no BBB permeability was detected among putative MPP inhibitors and Paritaprevir derivatives. This study revealed the water solubility levels of Paritaprevir (1.38e-07 mg/ml; 1.81e-10 mol/l), Glecaprevir (9.22e-06 mg/ml; 1.10e-08 mol/l), Nelfinavir (3.11e-05 mg/ml; 5.48e-08 mol/l), Lopinavir (1.21e-06 mg/ml; 1.61e-09 mol/l), and Lopinavir (5.57e-08 mg/ml; 8.85e-11 mol/l). In addition, the top ten derivatives of Paritaprevir were also subjected to ADME analysis, and the details of the analysis are included in Table S5.
Table 3.
ADME analysis of top five MPP inhibitors by using SwissADME.
| Parameter |
Topmost Main Protease Protein Inhibitors of SARS-CoV-2 |
|||||
|---|---|---|---|---|---|---|
| Paritaprevir | Glecaprevir | Nelfinavir | Simeprevir | Lopinavir | ||
| Physico-chemical parameters | Formula | C40H43N7O7S | C38H46F4N6O9S | C32H45N3O4S | C38H47N5O7S2 | C37H48N4O5 |
| Molecular weight | 765.88 g/mol | 838.87 g/mol | 567.78 g/mol | 749.94 g/mol | 628.80 g/mol | |
| Molar Refractivity | 211.96 | 205.91 | 166.17 | 208.52 | 187.92 | |
| TPSA | 198.03 Å2 | 203.60 Å2 | 127.20 Å2 | 193.51 Å2 | 120.00 Å2 | |
| Lipophilicity | Log Po/w (iLOGP) | 3.34 | 3.71 | 4.24 | 4.3 | 4.22 |
| Log Po/w (XLOGP3) | 4.65 | 4.55 | 5.67 | 4.81 | 5.92 | |
| Log Po/w (WLOGP) | 3.89 | 5.69 | 4.37 | 5.51 | 3.57 | |
| Log Po/w (MLOGP) | 0.88 | 1.32 | 3.2 | 1.48 | 2.93 | |
| Log Po/w (SILICOS-IT) | 2.28 | 2.42 | 4.56 | 4.89 | 6.02 | |
| Consensus Log Po/w | 3.01 | 3.54 | 4.41 | 4.2 | 4.53 | |
| Pharmaco-kinetics | GI absorption | Low | Low | Low | Low | High |
| BBB permeant | No | No | No | No | No | |
| P-gp substrate | Yes | Yes | Yes | Yes | Yes | |
| CYP1A2 inhibitor | No | No | No | No | No | |
| CYP2C19 inhibitor | No | No | Yes | No | Yes | |
| CYP2C9 inhibitor | No | No | No | No | No | |
| CYP2D6 inhibitor | No | No | No | No | No | |
| CYP3A4 inhibitor | Yes | No | Yes | Yes | Yes | |
| Log Kp (skin permeation) | -7.67 cm/s | -8.19 cm/s | -5.74 cm/s | -7.46 cm/s | -5.93 cm/s | |
| Log S (SILICOS-IT) | -9.74 | -7.96 | -7.26 | -8.79 | -10.05 | |
| Water Solubility | Solubility | 1.38e-07 mg/ml; 1.81e-10 mol/l | 9.22e-06 mg/ml; 1.10e-08 mol/l | 3.11e-05 mg/ml; 5.48e-08 mol/l | 1.21e-06 mg/ml; 1.61e-09 mol/l | 5.57e-08 mg/ml; 8.85e-11 mol/l |
3.4. Molecular dynamics (MD) simulation
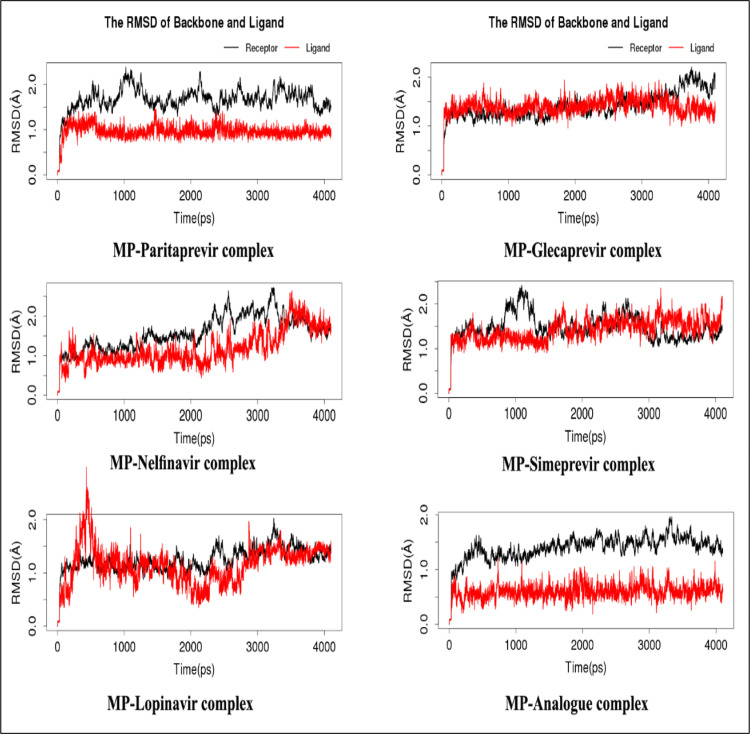
MPP of SARS-CoV-2 complexed with Paritaprevir, Glecaprevir, Nelfinavir, Lopinavir, Lopinavir, and the topmost predicted analog were employed to molecular dynamic simulation study for checking the structural stability at nanosecond scaled cellular environment (Result shown in Table 4). The RMSD value of the topmost predicted analog showed constant binding pattern during the time frame. All of the complex structure of MPP molecules with inhibitors were below 2 Å ( Fig. 6a). Three drugs molecules, such as, Nelfinavir, Lopinavir, and Lopinavir, caused a mostly similar deviation in the MPP of SARS-CoV-2. However, mostly unstable conditions with dramatic increases and decreases had been observed in the RMSD of MPP in complex with Paritaprevir. However, MPP in complex with Glecaprevir showed a constant RMSD that suddenly increased at approximately 4 ns. However, other different features of molecular simulation study, such as, H-bonds, free energies consisting of electrostatic energy (ELE), Van der Waals contributions (VDW), total gas phase energy (GAS), and the final estimated binding energy (deltaPB/deltaGB) were also analyzed for the better understanding of the molecular stability. Paritaprevir (6H-bonds) and its analog (7H-bonds) had a much greater number of H-bonds than the other inhibitors, while Lopinavir and Lopinavir were very poor H-bonding inhibitors (Fig. 6b). Moreover, Nelfinavir-MP complex had a maximum number of 5H-bonds in the interaction while both Lopinavir and Lopinavir had only 2H-bonds with relatively poor interaction pattern. MM/PB(GB)SA calculation of MPP-Paritaprevir showed that the acquired free energy value was deltaPB ≈ −21.48 kcal/mol, while that of its analog was deltaPB ≈ −12.48 kcal/mol (Table 4). Glecaprevir and Nelfinavir had a deltaPB ≈ −6.20 kcal/mol and deltaPB ≈ −4.17 kcal/mol, which were consistent with the docking results showing the lowest values for Lopinavir and Lopinavir.
Table 4.
The results derived from molecular dynamics simulation main protease protein of SARS-CoV-2 in complex with Paritaprevir, Glecaprevir, Nelfinavir, Simeprevir, Lopinavir and the top most predicted analogue.
| Name | ELE | VDW | GAS | deltaPB | deltaGB |
|---|---|---|---|---|---|
| Paritaprevir | -21.79 | -57.56 | -79.35 | -21.48 | -23.49 |
| Glecaprevir | -37.37 | -36.96 | -74.33 | -6.20 | -1.03 |
| Nelfinavir | -2.80 | -49.05 | -51.85 | -4.17 | -17.57 |
| Simeprevir | -32.72 | -27.86 | -60.68 | -3.71 | -2.23 |
| Lopinavir | -0.89 | -48.17 | -49.06 | -2.48 | -18.45 |
| Analogue | -3.59 | -47.92 | -51.51 | -12.68 | -9.53 |
Fig. 6.
(a). RMSD analysis derived from MD simulations of the SARS-CoV-2 main protease and top MPP inhibitors. (b). The statistics for hydrogen bonds derived from MD simulations of the SARS-CoV-2 main protease and top MPP inhibitors.
4. Discussion
COVID-19 has become a great challenge for international policymakers and scientific communities [78], [79]. The newly emerged coronavirus has created a global health crisis. Nevertheless, the virus is spreading worldwide, and a pandemic attitude has been established. The pandemic has already broken previous records set by other types of coronaviruses, such as SARS-CoV and MERS-CoV. The morbidity is increasing, and hospital capacity to handle critical cases is being exhausted. However, no drug to kill the virus or vaccine to protect against it has been identified. The present study aimed to screen and suggest potential drug candidates against SARS-CoV-2 to combat the global pandemic of COVID-19. The study suggested that Paritaprevir, Glecaprevir, Nelfinavir, Lopinavir, and Lopinavir along with their top derivatives might be effective against SARS-CoV-2, and these molecules also had common drug surface hotspots on SARS-CoV-2 MPP.
Extensive research and ongoing discussions have been focused on the efficacy of different drug candidates – namely, RdRp inhibitor remdesivir, Favipiravir, and alpha-ketoamide [80], [81]. Virtual screening for drug repurposing is becoming vital for identifying drugs that could be used for the treatment of COVID-19. Thus, we have endeavored to screen drugs that have been approved and indicated for the treatment of other viral diseases. In the present study, the MPPs of HIV and HCV were prioritized to assess the efficacy of inhibitors of SARS-CoV-2 MPP, which have previously been shown to be effective against different viral pathogens [82], [83]. Here, drug repurposing with the molecular docking approach was employed for comprehensive screening and analysis of the putative drug candidates against SARS-CoV-2. Moreover, to determine the common drug surface hotspots in the MPP of SARS-CoV-2, different recently studied MPP inhibitors of SARS-CoV-2, such as alpha-ketoamide, and Favipiravir [84], [85], [86], were also investigated with the approved MPP inhibitors and their derivatives.
The SARS-CoV-2 main protease is an attractive drug target for pharmacists because of its essential role in processing viral RNA-encoded polyproteins to yield functional viral proteins [87]. Due to the dissimilarity with human proteases, targeting this enzyme may prevent maturation of the viral particle before exiting the host cell [88]. Shamsi and his colleagues [89] also utilized a structure-based drug design approach to screen the existing pool of FDA-approved drugs against SARS-CoV-2 MPP. The study revealed Glecaprevir and maraviroc as potential inhibitors of SARS-CoV-2 main protease. In another study, Sk et al. [90] suggested that alpha-ketoamide could be used as a lead compound in the development of drugs targeting SARS-CoV-2 due to its better binding affinity than other retroviral drugs, including darunavir and Lopinavir. Moreover, membrane fusion of coronavirus with the host cell is triggered by proteolysis of the spike protein [91], [92]. Therefore, the prevention of spike protein trimming in the same treatment may open the possibility for the immune system to present the virus and generate a response in the host [93]. Thus, the development of an antiviral molecule targeting SARS-CoV-2 MPP in combination with other potential strategies might be a promising approach to develop an effective treatment against COVID-19.
From the molecular docking studies, it was found that Paritaprevir, Glecaprevir, Nelfinavir, Lopinavir, and Lopinavir could be potential inhibitors of SARS-CoV-2 MPP, and Lopinavir has been previously reported by different studies for the treatment of COVID-19 [94]. Paritaprevir [95], Glecaprevir [96], Nelfinavir [97], and Lopinavir [98] have already been approved for the treatment of HIV or HCV [99], and the results of this study indicated that they could be potential drug candidates for SARS-CoV-2 rather than Lopinavir. Again, the present study also investigated the binding pattern of the screened MPP inhibitors along with a few experimentally tested or suggested drug molecules for the MPP of SARS-CoV-2. Surprisingly, the screened MPP inhibitors had similar and, in some cases, higher binding affinity to the MPP of SARS-CoV-2. Approximately 100 derivatives of Paritaprevir were also investigated through its molecular docking with SARS-CoV-2, and one of the Paritaprevir derivatives “(3S,9Z,12R,15S,17R)-N-cyclopropylsulfonyl-12-methyl-3-[(5-methylpyrazine-2-carbonyl)amino]-2,14-dioxo-17phenanthridin-6-yloxy-1,13diazabicyclo[13.3.0] octadec-9-ene-12-carboxamide (CID 131982844)’’ was found to have the highest binding affinity in terms of global energy among already approved MPP inhibitors for the treatment of COVID-19. This Paritaprevir derivative (CID 131982844) could be the best drug candidate for inhibition of the MPP of SARS-CoV-2. The docking algorithm search for the potential energy algorithm and rank binding affinity of molecules was based on the global energy minimum [100].
The study of drug surface hotspots is a prerequisite for understanding the molecular interaction between drug candidates and target molecules [101], [102], [103]. The molecular binding site of Nelfinavir, alpha-ketoamide, and all the Paritaprevir derivatives showed a similar pattern that might indicate a drug surface hotspot in the MPP of SARS-CoV-2. Some of these positions were also found to be common to Paritaprevir, Glecaprevir, Lopinavir, and Lopinavir binding. The investigation suggested that the amino acids T25, T26, H41, M49, L141, N142, G143, C145, H164, M165, E166, D187, R188, and Q189 in the MPP could effectively interact with the drug molecules. The residues involved in the hotspot were also checked based on the MSA of homologous protein candidates of SARS-CoV-2 MPPs, which showed that the hotspot positions were conserved in all sequences, thus providing insight into the effectiveness of the drugs targeting MPP to combat SARS-CoV-2.
The top screened drug candidates from MPP inhibitors and their derivatives were also employed for ADME analysis. Physico-chemical parameters, lipophilicity, pharmacokinetics properties, and water solubility were studied, which contribute to the analysis of ADME properties. Menon and his coworkers [104] conducted two parallel double-blind, placebo-controlled phase 1 group studies in healthy volunteers. Single-dose study participants (n = 87) were subjected to one-time administration of Paritaprevir, while multiple-dose study participants (n = 38) received Paritaprevir once or twice daily for 14 days. The study revealed that Paritaprevir exhibited nonlinear pharmacokinetics with greater than dose proportional increases in exposure after single or multiple dosing. Moreover, coadministration of Paritaprevir with Ritonavir increased Paritaprevir exposure and the biological half-life without influencing its tolerability. Regardless of the dose, plasma concentrations reached peak levels within 1.8–2.3 h, and the mean t1/2 was approximately 3 h. In a separate study, Mensing et al. [105] successfully constructed a population pharmacokinetic model for Paritaprevir using phase 2 and phase 3 data from subjects with HCV genotype 1 infection. Safety and efficacy were well characterized in the subjects at higher exposures throughout the treatment periods. Paritaprevir also did not show abnormalities in any liver function tests. In the present study, we also analyzed the ADME properties of Paritaprevir, which did not show any undesirable consequences that could reduce its drug-likeness properties. The study of CYP isoform inhibition revealed that the suggested MPP inhibitors and Paritaprevir derivatives had few possibilities to interact with the CYP isoforms. However, Paritaprevir is predominantly metabolized by CYP3A4 and, to a lesser extent, CYP3A5 [106]. CYP3A4 metabolizes more than 50% of clinically used drugs, and it is most abundant in human liver [107]. Inactivation of CYP3A4 may be responsible for drug toxicity through enhanced exposure to other coadministered drugs [108]. However, proper clinical management may enable professionals to significantly minimize the negative consequences of CYP3A4 inhibition [109]. Thus, proper clinical approaches, such as the rational use of drugs, use of a safe drug combination regimen, dose adjustment, and discontinuation of therapy when toxic drug interactions occur are necessary, are required for using Paritaprevir in clinical phase trials. Furthermore, therapeutic drug monitoring and predicting the risks for potential drug–drug interactions (both qualitatively and quantitatively) should also be prioritized [109]. In the present study, BBB permeability and water solubility of putative MPP inhibitors were also calculated. There was no BBB permeability for the screened MPP inhibitors and Paritaprevir derivatives.
At present, computational approaches have been widely adopted for the prediction of potential drugs that could play a crucial role in further drug development. Thus, it is important to predict accurate protein-inhibitor complexes, which can be achieved by MD simulation. By this MD simulation, the docked pose predicted by molecular docking approaches could be analyzed to predict whether it is stable or not in an aqueous environment [110]. In this study, MD simulation was also employed to assess the stability of MPP in complex with Paritaprevir and its analog, Glecaprevir, Nelfinavir, Lopinavir, and Lopinavir through the analysis of RMSD, H-bonds, and the binding free energies. All the complexes were stable, as the RMSD was below 2 Å. Paritaprevir was found to cause higher conformational changes in MPPs than the others, while its analog was found to form a stable complex with MP. The stability was achieved by the much greater number of H-bonds. Both Paritaprevir and its analog would be more effective for the inhibition of the MPP of SARS-CoV-2, followed by Glecaprevir and Nelfinavir (Table 4). However, Lopinavir and Lopinavir would be less effective for the inhibition of MP.
Paritaprevir, also known as ABT-450, was approved in 2014 for the treatment of HCV genotype 1 and in 2015 for genotype 4 [105]. It showed promising results in combination with Ritonavir and ribavirin and caused a 95% sustained virologic response against HCV genotype 1 [111]. Side effects of Paritaprevir are relatively uncommon, though it is sometimes associated with fatigue, asthenia nausea, insomnia, pruritus, and other skin reactions [106]. Approximately 88% of the drug is eliminated through feces, while the remainder is eliminated via the urine. In a phase 3 trial performed by Poordad et al. [112], less than 1% of the subjects discontinued treatment as a result of side effects in a combination therapy. Paritaprevir does not require dose adjustment for patients with renal impairment [106]. Nevertheless, caution should be taken for the repurposing of Paritaprevir to treat COVID-19 patients with severe hepatic impairment, diabetes, or cardiac diseases to minimize the risk of potential toxicity.
The results concluded that Paritaprevir and its analog (CID 131982844) could be a more promising option for treating SARS-CoV-2 than other previously approved MPP inhibitors, such as Favipiravir. However, the results of the current study are limited to in silico analysis and lack in vivo efficacy testing. At present, multiple MPP inhibitors, such as Favipiravir and other drugs are currently under evaluation through randomized control trials. Thus, we strongly suggest a quick assessment of Paritaprevir and its analog through a clinical trial as a potential candidate for the treatment of COVID-19 patients.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
No funding was received for this work.
CRediT authorship contribution statement
Mahmudul Hasan: Conceptualization, Formal analysis, Investigation, Methodology, Software, Writing - original draft, Writing - review & editing. Md. Sorwer Alam Parvez: Conceptualization, Formal analysis, Investigation, Methodology, Software, Writing - original draft, Writing - review & editing. Kazi Faizul Azim: Data curation, Formal analysis, Investigation, Methodology, Software, Writing - original draft, Writing - review & editing. Md. Abdus Shukur Imran: Data curation, Formal analysis, Investigation, Software, Validation, Writing - review & editing. Topu Raihan: Formal analysis, Methodology, Visualization, Writing - review & editing. Airin Gulshan: Formal analysis, Methodology, Software, Writing - review & editing. Samuel Muhit: Formal analysis, Visualization, Writing - review & editing. Rubaiat Nazneen Akhand: Methodology, Visualization, Writing - review & editing. Syed Sayeem Uddin Ahmed: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Writing - original draft, Writing - review & editing. Md Bashir Uddin: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Writing - original draft, Writing - review & editing. All authors have read and approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank American Journal Experts (www.aje.com) for their voluntary editing services.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2021.111742.
Appendix A. Supplementary material
Supplementary material
.
Data availability
All data generated and analyzed during this study is included in the main manuscript or supplementary materials/files.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren L.-L., Wang Y.-M., Wu Z.-Q., Xiang Z.-C., Guo L., Xu T., Jiang Y.Z., Xiong Y., Li Y.J., Li X.W., Li H., Fan G.H., Gu X.Y., Xiao Y., Gao H., Xu J.Y., Yang F., Wang X.M., Wu C., Chen L., Liu Y.W., Liu B., Yang J., Wang X.R., Dong J., Li L., Huang C.L., Zhao J.P., Hu Y., Cheng Z.S., Liu L.L., Qian Z.H., Qin C., Jin Q., Cao B., Wang J.W. Identification of a novel coronavirus causing severe pneumonia in human. Chin. Med. J. 2020;133:1015–1024. doi: 10.1097/cm9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. (World Health Organization): Coronavirus disease (COVID-2019) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. Retrieved on 4 April 2020, 2020.
- 4.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kannan S., Shaik Syed Ali P., Sheeza A., Hemalatha K. COVID-19 (Novel Coronavirus 2019) - recent trends. Eur. Rev. Med. Pharmacol. Sci. 2020;24:2006–2011. doi: 10.26355/eurrev_202002_20378. [DOI] [PubMed] [Google Scholar]
- 7.Mousavizadeh L.G.S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020;20:30082–30087. doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China - key questions for impact assessment. N. Engl. J. Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 9.Shyr Z.A., Gorshkov K., Chen C.Z., Zheng W. Drug discovery strategies for SARS-CoV-2. J. Pharmacol. Exp. Ther. 2020;375(1):127–138. doi: 10.1124/jpet.120.000123. Oct 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:1–9. doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gildenhuys S. Expanding our understanding of the role polyprotein conformation plays in the coronavirus life cycle. Biochem. J. 2020;477(8):1479–1482. doi: 10.1042/BCJ20200223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer-Almes F.J. Repurposing approved drugs as potential inhibitors of 3CL-protease of SARS-CoV-2: virtual screening and structure based drug design. Comput. Biol. Chem. 2020;88 doi: 10.1016/j.compbiolchem.2020.107351. Oct 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omrani A.S., Saad M.M., Baig K., Bahloul A., Abdul-Matin M., Alaidaroos A.Y., Almakhlafi G.A., Albarrak M.M., Memish Z.A., Albarrak A.M. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect. Dis. 2014;14:1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tözsér J. Comparative studies on retroviral proteases: substrate specificity. Viruses. 2010;2:147–165. doi: 10.3390/v2010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., Gale MJ Jr, Baric R.S., Enjuanes L., Gallagher T., McCray PB Jr, Perlman S. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. USA. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agrawal A.S., Garron T., Tao X., Peng B.-H., Wakamiya M., Chan T.-S., Couch R.B., Tseng C.T. Generation of a transgenic mouse model of middle east respiratory syndrome coronavirus infection and disease. J. Virol. 2015;89:3659–3670. doi: 10.1128/jvi.03427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macchiagodena M., Pagliai M., Procacci P. Inhibition of the Main Protease 3CL-pro of the Coronavirus Disease 19 via Structure-Based Ligand Design and Molecular Modeling, https://arxiv.org/pdf/200209937.pdf, 2020, pp. 1–28. [DOI] [PMC free article] [PubMed]
- 19.García-Fernández R., Ziegelmüller P., González L., Mansur M., Machado Y., Redecke L., Hahn U., Betzel C., Chávez Mde L. Two variants of the major serine protease inhibitor from the sea anemone Stichodactyla helianthus, expressed in Pichia pastoris. Protein Expr. Purif. 2016;123:42–50. doi: 10.1016/j.pep.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;3405:1–9. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 23.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020;30 doi: 10.1016/j.bmcl.2020.127377. Jul 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agbowuro A.A., Huston W.M., Gamble A.B., Tyndall J.D. Proteases and protease inhibitors in infectious diseases. Med. Res. Rev. 2018;38(4):1295–1331. doi: 10.1002/med.21475. [DOI] [PubMed] [Google Scholar]
- 25.Battisti V., Wieder O., Garon A., Seidel T., Urban E., Langer T. A computational approach to identify potential novel inhibitors against the coronavirus SARS CoV 2. Mol. Inform. 2020;39(10) doi: 10.1002/minf.202000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleischmann R., Kremer J., Cush J., Schulze-Koops H., Connell C.A., Bradley J.D., Gruben D., Wallenstein G.V., Zwillich S.H., Kanik K.S., ORAL Solo I. Class-sparing regimens for initial treatment of HIV-1 infection. N. Engl. J. Med. 2008;358:2095–2106. doi: 10.1056/NEJMoa1109071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dayer MR. Old Drugs for Newly Emerging Viral Disease, COVID-19: Bioinformatic Prospective, https://arxiv.org/ftp/arxiv/papers/2003/200304524.pdf, 2020.
- 28.Salazar J., Courter J., Girotto J. Role of tipranavir in treatment of patients with multidrug-resistant HIV. Ther. Clin. Risk Manag. 2010;6:431–441. doi: 10.2147/tcrm.s4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nukoolkarn V., Lee V.S., Malaisree M., Aruksakulwong O., Hannongbua S. Molecular dynamic simulations analysis of ritronavir and Lopinavir as SARS-CoV 3CLpro inhibitors. J. Theor. Biol. 2008;254(4):861–867. doi: 10.1016/j.jtbi.2008.07.030. Oct 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferraz W.R., Gomes R.A., S., Novaes A.L., Goulart Trossini G.H. Ligand and structure-based virtual screening applied to the SARS-CoV-2 main protease: an in silico repurposing study. Future Med. Chem. 2020;12(20):1815–1828. doi: 10.4155/fmc-2020-0165. (Oct) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ann D. Kwong RBPSG. Development and marketing of INCIVEK (Telaprevir; VX-950): a first-generation HCV protease inhibitor, in combination with PEGylated interferon and ribavirin. Top. Med. Chem. 2014;9:1–68. doi: 10.1007/7355. [DOI] [Google Scholar]
- 32.Touret F., Gilles M., Barral K., Nougairède A., van Helden J., Decroly E., de Lamballerie X., Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020;10(1):1–8. doi: 10.1038/s41598-020-70143-6. Aug 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Worldometers. COVID-19 Coronavirus outbreak, 2021, https://www.worldometers.info/coronavirus/. Retrieved 4 April 2021, 2021.
- 34.Hirono S. An introduction to the computer-aided structure-based drug design--applications of bioinformatics to drug discovery. Rinsho Byori. 2002;50:45–51. doi: 10.1155/2013/704806. [DOI] [PubMed] [Google Scholar]
- 35.Rester U. From virtuality to reality - virtual screening in lead discovery and lead optimization: a medicinal chemistry perspective. Curr. Opin. Drug Discov. Dev. 2008;11:559–568. doi: 10.1016/j.cell.2009.01.043. [DOI] [PubMed] [Google Scholar]
- 36.Rollinger J.M., Stuppner H., Langer T. Virtual screening for the discovery of bioactive natural products. Prog. Drug Res. 2008;65:212–249. doi: 10.1007/978-3-7643-8117-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voet A., Qing X., Lee X.Y., De Raeymaecker J., Tame J., Zhang K., De Maeyer M. Pharmacophore modeling: advances, limitations, and current utility in drug discovery. J. Recept. Ligand Channel Res. 2014;7:81–92. doi: 10.2147/JRLCR.S46843. [DOI] [Google Scholar]
- 38.Kooistra A.J., Vischer H.F., McNaught-Flores D., Leurs R., De Esch I.J.P., De Graaf C. Function-specific virtual screening for GPCR ligands using a combined scoring method. Sci. Rep. 2016;6:1–21. doi: 10.1038/srep28288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irwin J.J., Shoichet B.K., Mysinger M.M., Huang N., Colizzi F., Wassam P., Cao Y. Automated docking screens: a feasibility study. J. Med. Chem. 2009;52:5712–5720. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anh Vu L., Thi Cam Quyen P., Thuy Huong N. In silico drug design: prospective for drug lead discovery. Int. J. Eng. Sci. Invent. 2015;4:60–70. [Google Scholar]
- 41.Ivanov A.S., Veselovsky A.V., Dubanov A.V., Skvortsov V.S. Bioinformatics platform development. Bioinform. Drug Discov. 2006;316:389–431. doi: 10.1385/1-59259-964-8:389. [DOI] [PubMed] [Google Scholar]
- 42.Joseph J., Bhaskaran R., Kaliraj M., Muthuswamy M. Molecular docking of phytoligands to the viral protein receptor. Bioinformation. 2017;13:116–121. doi: 10.6026/97320630013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng X.Y., Zhang H.X., Mezei M.C.M. Molecular docking: a powerful approach for structure-baseddrug discovery. Bone. 2008;23:1–7. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brogi S. Computational approaches for drug discovery. Molecules. 2019;24:1–6. doi: 10.3390/molecules24173061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., Yu J., Zhang Z., Ren J., Peluffo AE, Zhang W., et al. Network Bioinformatics Analysis Provides Insight into Drug Repurposing for COVID-2019, https://www.preprints.org/manuscript/2020030286/v1, 2020, doi:10.20944/PREPRINTS202003.0286.V1. [DOI] [PMC free article] [PubMed]
- 46.Pizzorno A., Padey B., Terrier O., Rosa-Calatrava M. Drug repurposing approaches for the treatment of influenza viral infection: reviving old drugs to fight against a long-lived enemy. Front. Immunol. 2019;10:531. doi: 10.3389/fimmu.2019.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A., Davey R., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. Evaluation of ebola virus inhibitors for drug repurposing. ACS Infect. Dis. 2016;1:317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- 48.Gastaminza P., Whitten-Bauer C., Chisari F.V. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc. Natl. Acad. Sci. USA. 2010;107:291–296. doi: 10.1073/pnas.0912966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barrows N.J., Campos R.K., Powell S.T., Prasanth K.R., Schott-lerner G., Soto-acosta R., Galarza-Muñoz G., McGrath E.L., Urrabaz-Garza R., Gao J., Wu P., Menon R., Saade G., Fernandez-Salas I., Rossi S.L., Vasilakis N., Routh A., Bradrick S.S., Garcia-Blanco M.A. A screen of FDA-approved drugs forinhibitors of Zika virus infection. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S., Liu Y., Guo J., Wang P., Zhang L., Xiao G., Wang W. Screening of FDA-approved drugs forinhibitors of Japanese encephalitis virus infection. J. Virol. 2017;91:1–14. doi: 10.1128/JVI.01055-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mercorelli B., Luganini A., Nannetti G., Tabarrini O., Gribaudo G., Gribaudo G. Drug repurposing approach identifies inhibitors of the prototypic viral transcription factor IE2 that article drug repurposing approach identifies inhibitors of the prototypic viral transcription factor IE2 that block human cytomegalovirus replication. Cell Chem. Biol. 2016;23:340–351. doi: 10.1016/j.chembiol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 52.Lai J., Lin Y., Hsieh S. Pharmacological intervention for dengue virus infection. Biochem. Pharmacol. 2017;129:14–25. doi: 10.1016/j.bcp.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Wilde A.H. De, Jochmans D., Posthuma C.C., Zevenhoven-dobbe J.C., Nieuwkoop S. Van. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of middle east respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei D.Q., Zhang R., Du Q.S., Gao W.N., Li Y., Gao H., Wang S.Q., Zhang X., Li A.X., Sirois S., Chou K.C. Anti-SARS drug screening by molecular docking. Amino Acids. 2006;31:73–80. doi: 10.1007/s00726-006-0361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavasotto C.N., Di Filippo J.I. In Silico drug repurposing for COVID 19: targeting SARS CoV 2 proteins through docking and consensus ranking. Mol. Inform. 2021;40(1) doi: 10.1002/minf.202000115. (Jan) [DOI] [PubMed] [Google Scholar]
- 56.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of Lopinavir, oseltamivir and Ritonavir binding with SARS-CoV-2 protease against. J. Biomol. Struct. Dyn. 2020;0:1–6. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 57.Jabeer R., Kumar R., Muluneh G., Jain M., Singh E. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2 0 -O-ribose methyltransferase. J. Biomol. Struct. Dyn. 2020;0:1–14. doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen YW, Yiu CB, Wong K. Prediction of the SARS-CoV-2 ( 2019-nCoV) 3C-like protease ( 3CL pro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates [version 2; peer review: 3 approved]. F1000Res. 2020, 9: 129. [DOI] [PMC free article] [PubMed]
- 59.NCBI Resource C. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2016;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rose P.W., Prlić A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z., Green R.K., Goodsell D.S., Hudson B., Kalro T., Lowe R., Peisach E., Randle C., Rose A.S., Shao C., Tao Y.P., Valasatava Y., Voigt M., Westbrook J.D., Woo J., Yang H., Young J.Y., Zardecki C., Berman H.M., Burley S.K. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017;45:D271–D281. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anil K.T.J.W. Autodock vina: improving the speed and accuracy of docking. J. Comput. Chem. 2019;31:455–461. doi: 10.1002/jcc.21334.AutoDock. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.L DeLano W. Pymol: an open-source molecular graphics tool. Newsl. Protein Crystallogr. 2002:40. [Google Scholar]
- 64.RA L., MB S. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 65.Wang Q., He J., Wu D., Wang J., Yan J., Li H. Interaction of α-cyperone with human serum albumin: determination of the binding site by using Discovery Studio and via spectroscopic methods. J. Lumin. 2015;164:81–85. doi: 10.1016/j.jlumin.2015.03.025. [DOI] [Google Scholar]
- 66.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. α-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 67.Zhang L., Lin D., Sun X., Rox K., Hilgenfeld R. X-ray Structure of Main Protease of the Novel Coronavirus SARS-CoV-2 Enables Design of α-Ketoamide Inhibitors. bioRxiv. 2020, 2020.02.17.952879. doi:10.1101/2020.02.17.952879.
- 68.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:636–641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tripathi P., Ghosh S., Talapatra SN. Bioavailability prediction of phytochemicals present in Calotropis procera (Aiton) R. Br. by using Swiss-ADME tool, 2019, 131,147–163.
- 71.Leach A.R. Molecular Modelling: Principles and Applications. second ed. Pearson Education; 2001. [Google Scholar]
- 72.Yang J., Wang F., Chen Y., Hao G., Yang G. LARMD: integration of bioinformatic resources to profile ligand-driven protein dynamics with a case on the activation of estrogen receptor. Brief. Bioinform. 2020;21:2206–2218. doi: 10.1093/bib/bbz141. [DOI] [PubMed] [Google Scholar]
- 73.Case D.A., Cheatham T.E., Darden T., Gohlke H., Jr., Luo R., Merz K.M., J.r, Merz K.M. Jr, Onufriev A., Simmerling C., Wang B., Woods R.J. The amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agbowuro A.A., Huston W.M., Gamble A.B., Tyndall J.D.A. Proteases and protease inhibitors in infectious diseases. Med. Res. Rev. 2018;38:1295–1331. doi: 10.1002/med.21475. [DOI] [PubMed] [Google Scholar]
- 75.Kadioglu O., Saeed M., Greten H.J., Efferth T. Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Bull. World Health Organ. 2020 doi: 10.2471/BLT.20.255943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arnott J.A., Planey S.L. The influence of lipophilicity in drug discovery and design. Expert Opin. Drug Discov. 2012;7:863–875. doi: 10.1517/17460441.2012.714363. [DOI] [PubMed] [Google Scholar]
- 77.Daina A., Zoete V. A BOILED-Egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11:1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilder-Smith A., Chiew C.J., Lee V.J. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect. Dis. 2020;20:102. doi: 10.1016/S1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuen K.-S., Ye Z.-W., Fung S.-Y., Chan C.-P., Jin D.-Y. SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci. 2020;10:40. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Disco Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 81.Zhou D., Dai S.M.T.Q. COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J. Antimicrob. Chemother. 2020;75:1667–1670. doi: 10.1093/jac/dkaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Asselah T., Kowdley K.V., Zadeikis N., Wang S., Hassanein T., Horsmans Y., Colombo M., Calinas F., Aguilar H., de Ledinghen V., Mantry P.S., Hezode C., Marinho R.T., Agarwal K., Nevens F., Elkhashab M., Kort J., Liu R., Ng T.I., Krishnan P., Lin C.W., Mensa F.J. Efficacy of glecaprevir/pibrentasvir for 8 or 12 weeks in patients with hepatitis C virus genotype 2, 4, 5, or 6 infection without cirrhosis. Clin. Gastroenterol. Hepatol. 2018;16:417–426. doi: 10.1016/j.cgh.2017.09.027. [DOI] [PubMed] [Google Scholar]
- 83.Reau N., Kwo P.Y., Rhee S., Brown R.S., J.r, Agarwal K., Angus P., Gane E., Kao J.H., Mantry P.S., Mutimer D., Reddy K.R., Tran T.T., Hu Y.B., Gulati A., Krishnan P., Dumas E.O., Porcalla A., Shulman N.S., Liu W., Samanta S., Trinh R., Forns X. Glecaprevir/pibrentasvir treatment in liver or kidney transplant patients with hepatitis C virus infection. Hepatology. 2018;68:1298–1307. doi: 10.1002/hep.30046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Al-Tawfiq J.A., Al-Homoud A.H., Memish Z.A. Remdesivir as a possible therapeutic option for the COVID-19. Travel Med. Infect. Dis. 2020;34 doi: 10.1016/j.tmaid.2020.101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Colson P., Rolain J.-M., Lagier J.-C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen C., Huang J., Cheng Z., Wu J., Chen S., Zhang Y., et al. Favipiravir versus Arbidol for COVID-19: A Randomized Clinical Trial. medRxiv. 2020, 2020.03.17.20037432. doi:10.1101/2020.03.17.20037432.
- 87.Krichel B., Falke S., Hilgenfeld R., Redecke L. Processing of the SARS-CoV pp1a / ab nsp7 – 10 region. Biochem. J. 2020;477:1009–1019. doi: 10.1042/BCJ20200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α -ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shamsi A., Mohammad T., Anwar S., Alajmi M.F., Hussain A., Rehman T., et al. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci. Rep. 2020:40. doi: 10.1042/BSR20201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sk F., Roy R., Jonniya N.A., Poddar S., Roy R. Elucidating biophysical basis of binding of inhibitors to SARS-CoV-2 main protease by using molecular dynamics simulations and free energy calculations. J. Biomol. Struct. Dyn. 2020;0:1–13. doi: 10.1080/07391102.2020.1768149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Belouzard S., Millet J.K., Licitra B.N.W.G. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heald-sargent T., Gallagher T. Ready, set, fuse! the coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sulkowski M.S., Eron J.J., Wyles D., Trinh R., Lalezari J., Wang C., Slim J., Bhatti L., Gathe J., Ruane P.J., Elion R., Bredeek F., Brennan R., Blick G., Khatri A., Gibbons K., Hu Y.B., Fredrick L., Schnell G., Pilot-Matias T., Tripathi R., Da Silva-Tillmann B., McGovern B., Campbell A.L., Podsadecki T. Ombitasvir, paritaprevir co-dosed with ritonavir, dasabuvir, and ribavirin for hepatitis C in patients co-infected with HIV-1 a randomized trial. J. Am. Med. Assoc. 2015;313:1223–1231. doi: 10.1001/jama.2015.1328. [DOI] [PubMed] [Google Scholar]
- 96.Gane E., Lawitz E., Pugatch D., Papatheodoridis G., Bräu N., Brown A., Pol S., Leroy V., Persico M., Moreno C., Colombo M., Yoshida E.M., Nelson D.R., Collins C., Lei Y., Kosloski M., Mensa F.J. Glecaprevir and pibrentasvir in patients with HCV and severe renal impairment. N. Engl. J. Med. 2017;377:1448–1455. doi: 10.1056/NEJMoa1704053. [DOI] [PubMed] [Google Scholar]
- 97.Regazzi M., Maserati R., Villani P., Cusato M., Zucchi P., Briganti E., Roda R., Sacchelli L., Gatti F., Delle Foglie P., Nardini G., Fabris P., Mori F., Castelli P., Testa L. Clinical pharmacokinetics of Nelfinavir and its metabolite M8 in human immunodeficiency virus (HIV)-positive and HIV-hepatitis C virus-coinfected subjects. Antimicrob. Agents Chemother. 2005;49:643–649. doi: 10.1128/AAC.49.2.643-649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dieterich D., Rockstroh J.K., Orkin C., Gutiérrez F., Klein M.B., Reynes J., Shukla U., Jenkins A., Lenz O., Ouwerkerk-Mahadevan S., Peeters M., De La Rosa G., Tambuyzer L., Jessner W. Lopinavir (TMC435) with pegylated interferon/ribavirin in patients coinfected with HCV Genotype 1 and HIV-1: a phase 3 study. Clin. Infect. Dis. 2014;59:1579–1587. doi: 10.1093/cid/ciu675. [DOI] [PubMed] [Google Scholar]
- 99.Kumar R.N., Balba G.P. Managing the HIV/HCV-Co-infected patient in the direct-acting antiviral era: a review of pertinent drug interactions. Curr. Treat. Options Infect. Dis. 2017;9:411–424. doi: 10.1007/s40506-017-0138-4. [DOI] [Google Scholar]
- 100.Brooijmans N., Kuntz I.D. Molecular recognition and docking algorithms. Annu Rev. Biophys. Biomol. Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 101.Rosell M., Fernández-Recio J. Hot-spot analysis for drug discovery targeting protein-protein interactions. Expert Opin. Drug Disco. 2018;13:327–338. doi: 10.1080/17460441.2018.1430763. [DOI] [PubMed] [Google Scholar]
- 102.Lionta E., Spyrou G., Vassilatis D., Cournia Z. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr. Top. Med. Chem. 2014;14:1923–1938. doi: 10.2174/1568026614666140929124445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cavasotto C.N., Lamas M.S., Maggini J. Functional and druggability analysis of the SARS-CoV-2 proteome. Eur. J. Pharmacol. 2021;890 doi: 10.1016/j.ejphar.2020.173705. Jan 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Menon R.M., Klein C.E., Podsadecki T.J., Chiu Y.L., Dutta S.A.W. Pharmacokinetics and tolerability of Paritaprevir, a direct acting antiviral agent for hepatitis C virus treatment, with and without Ritonavir in healthy volunteers. Br. J. Clin. Pharmacol. 2016;81:929–940. doi: 10.1111/bcp.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mensing S., Eckert D., Sharma S., Polepally A.R., Khatri A., Podsadecki T.J., Awni W.M., Menon R.M., Dutta S. Population pharmacokinetics of Paritaprevir, ombitasvir, dasabuvir, Ritonavir and ribavirin in hepatitis C virus genotype 1 infection: analysis of six phase III trials. Br. J. Clin. Pharmacol. 2017;83:527–539. doi: 10.1111/bcp.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Burke L.A.M.K. Infectious Diseases. Elsevier; 2017. Drugs to treat viral hepatitis; pp. 1327–1332. fourth ed. 4th ed. [Google Scholar]
- 107.Rendic S.D.C.F. Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab. Rev. 1997;29:413–580. doi: 10.3109/03602539709037591. [DOI] [PubMed] [Google Scholar]
- 108.Dresser G.K., Spence J.D., Bailey D.G. Consequences and clinical relevance of cytochrome P450 3A4 inhibition. Clin. Pharmacokinet. 2000;38:41–57. doi: 10.2165/00003088-200038010-00003. [DOI] [PubMed] [Google Scholar]
- 109.Zhou S., Chan E., Li X.H.M. Clinical outcomes and management of mechanism-based inhibition of cytochrome. Ther. Clin. Risk Manag. 2005;1:3–13. doi: 10.2147/tcrm.1.1.3.53600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sakano T., Mahamood M.I., Yamashita T.F.H. Molecular dynamics analysis to evaluate docking pose prediction. Biophys. Phys. 2016;13:181–194. doi: 10.2142/biophysico.13.0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kowdley K.V., Lawitz E., Poordad F., Cohen D.E., Nelson D.R., Zeuzem S., Everson G.T., Kwo P., Foster G.R., Sulkowski M.S., Xie W., Pilot-Matias T., Liossis G., Larsen L., Khatri A., Podsadecki T., Bernstein B. Phase 2b trial of interferon-free therapy for hepatitis C virus genotype 1. N. Engl. J. Med. 2014;370:222–232. doi: 10.1056/NEJMoa1306227. [DOI] [PubMed] [Google Scholar]
- 112.Poordad F., Hezode C., Trinh R., Kowdley K.V., Zeuzem S., Agarwal K., Shiffman M.L., Wedemeyer H., Berg T., Yoshida E.M., Forns X., Lovell S.S., Da Silva-Tillmann B., Collins C.A., Campbell A.L., Podsadecki T., Bernstein B. ABT-450/r–ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. N. Engl. J. Med. 2014;370:1973–1982. doi: 10.1056/NEJMoa1402869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All data generated and analyzed during this study is included in the main manuscript or supplementary materials/files.