Abstract
D614G genotype of SARS-CoV-2 virus is highly infectious and responsible for almost all infection for 2nd wave. However, there are currently no reports with D614G as vaccine candidate. Here we report the development of an mRNA-LNP vaccine with D614G variant and characterization in animal model. We have used special mRNA-architecture and formulation that provides suitable response of the product. The surface plasmon resonance (SPR) data with spike protein (S) revealed that immunization generated specific antibody pools against the whole extracellular domain (RBD and S2) of the spike protein. The anti-sera and purified IgGs from immunized mice neutralized SARS-CoV-2-pseudoviruses in ACE2-expressing HEK293 cells in a dose dependent manner. Importantly, single-dose immunization protected mice-lungs from homotypic-pseudovirus entry and cytopathy. The immunologic responses have been implicated by a balanced and stable population of CD4+ cells with a Th1 bias. The data suggested great promise for immediate translation of the technology to the clinic.
Keywords: COVID, Coronavirus, D614G, Lipid nanoparticle, LNP, Vaccination, Immunization
1. Introduction
A new infectious coronavirus (SARS-CoV-2) has been first reported from Wuhan, China in December 2019 that causes COVID-19 [1]. The World Health Organization (WHO) declared the COVID-19 a global public health emergency situation on February 5, 2020 after getting growing evidence of continuous person-to-person transmission [2]. The virus has been spread worldwide quickly, and consequently WHO has declared it pandemic on March 11, 2020. As of December 21, 2020, the pandemic has resulted in 1,701,187 deaths among over 77,243,764 patients in 220 countries, with a case-fatality rate of 2.20%.
There will be a risk of pandemic as long as there is COVID-19 epidemic situation in any area of the world unless people are properly vaccinated. Therefore, effective vaccines against SARS-CoV-2 are immediately required to control morbidity and mortality related with COVID-19. Generally, non-replicating viral vectors, inactivated virus, DNA-based and protein-based vaccines have been the major approaches for the development of stable and effective vaccines; though they have their inherent limitations [3]. Recently, mRNA-based vaccines have become a promising approach because of their opportunity for rapid development, comparative low dose, logical better safety profile in terms of no potential risk of infection or insertional mutagenesis, and low capital expenditure (CAPEX) [4]. Several other leading vaccines under development against SARS-CoV-2 are also mRNA-based [5], [6], [7], [8]. Lipid nanoparticle technology has been developed for effective delivery of single-stranded therapeutics like siRNA, antisense oligo, mRNA etc. The first RNA-LNP therapeutic was approved in 2018 and has set the example for clinical safety of LNP-formulated RNA [9]. Therefore, we have also opted for mRNA-based LNP-mediated vaccine development technology to support the initiative for preventing the ongoing wave of the COVID-19 pandemic. Two of the mRNA vaccines (developed by Pfizer-BioNTech and Moderna Inc.) very recently have got conditional approvals for general use from regulators, which is suggesting that the mRNA-LNP vaccine technology should be promoted to satisfy the need of billions of doses to the world population within the shortest possible time to restore normal life.
The candidate mRNA vaccine ‘GBPD060’ is an LNP–encapsulated, unmodified nucleoside, mRNA–based vaccine that encodes the SARS-CoV-2 spike (S) glycoprotein stabilized in its prefusion conformation. Coronaviruses have genetic proofreading mechanisms, and SARS-CoV-2 sequence diversity is comparatively low [10], [11]; though, natural selection can adopt rare but favorable mutations. Since the outbreak in China, SARS-CoV-2 has gone through numerous mutations. The D614G amino acid change, among these, in the spike protein of Wuhan reference strain is caused by an A-to-G nucleotide substitution at position 23,403 of the relevant nucleotide sequence. Currently, D614G is the most prevalent circulating isotype of SARS-CoV-2 worldwide (more than 95%) [12], [13]. To date, there is no published report about the D614G-relevant vaccine development. Few studies have shown that antibody generated using D614 variant-target did not show significant difference between D614 and G614 variants in terms of cellular entry [14]. These studies did not use G614-specific antibody, and applied artificial systems for characterizing relevant functional experiments. Furthermore, how G614 variant vaccine behave in immunization in human and what would be the impact of relevant antibody on SARS-CoV-2 is not known. Therefore, developing of G614 variant-specific vaccine is a prime importance, and warrant characterization. To address this, we have incorporated D614G variant-targeted nucleic acid sequence, as well as few other immunogen-enhancing aspects in our mRNA design consideration. In this study, we described the design and preclinical characterization of ‘GBPD060’ mRNA-LNP vaccine candidate.
2. Materials and methods
2.1. Target gene and vector cloning
The target DNA was identified using data mining and analysis using bioinformatics. The target DNA was amplified from a patient sample, sequence confirmed, modified to achieve the desired design architecture as such that it harbors a suitable 5′ UTR, ORF to express the S protein with G614 and double proline (2P) mutations (K986P and V987P) with a IgE-secretory signal sequence, a special 3′ UTR constructed with modified alpha and beta globin in tandem, and finally a 130 residue-long poly-A tail. The detail of the design, construction, and confirmation is described in supplementary document.
2.2. mRNA production
The in vitro transcription (IVT) of mRNA was performed in presence with 3́-O-Me-m7G(5′)ppp(5′)G RNA Cap Structure Analog and S-adenosylmethionine (NEB, USA) using MEGAscript™ T7 Transcription Kit (ThermoFisher, USA), and Ribonucleotide Solution Set (NEB, USA). IVT mRNA synthesis reaction was optimized into 4 steps (supplementary document); final concentration of ribonucleotides was as follows: ATP and UTP – 13.13 mM, and GTP and CTP – 9.38 mM. The reaction was run for 2 h at 37 °C followed by a DNase treatment at 37 °C for 15 min and dephosphorylation using Antarctic Phosphatase (NEB, USA) according to supplier’s manual. IVT capped mRNA was purified using phenol:chloroform:isoamyl alcohol, and using MEGAclear™ Transcription Clean-Up Kit (ThermoFisher, USA).
2.3. mRNA identification
Purified mRNA, purified capped mRNA and formulated LNPs were treated with RNase samples, and analyzed by size exclusion chromatography (SEC). SEC was performed in Ultimate 3000 (ThermoFisher, USA) system using 10 mM Disodium hydrogen phosphate (Wako, Japan), 10 mM Sodium dihydrogen phosphate (Wako, Japan), 100 mM Sodium chloride (Merck, Germany), pH 6.6 as mobile phase. Biobasic SEC-300 (300 × 7.8 mm, particle size; 5 µm, ThermoFisher, USA) column was used with 1.0 mL/minute flow rate, 260 nm wavelength, 10 µL sample injection volume for 20 min.
2.4. Formulation of mRNA
Purified capped mRNAs were first diluted with sodium acetate buffer at desired concentration. The lipid molecules were dissolved in ethanol and mixed well. Lipids (MC3: DSPC: Cholesterol: DMG-PEG2000) were combined in the molar ratio of 50:10:38.5:1.5 [15], [16], [17], [18]. Then, sodium acetate buffer containing mRNA and lipid samples were mixed at a ratio of 3:1 and passed through the liposome extruder (Genizer, USA) to encapsulate the mRNA. The size distribution was checked after encapsulation of mRNA into nanoparticles. Then the formulations were dialyzed against 50 mM HEPES/sodium acetate buffer and phosphate-buffered saline for 18 h. The size distribution was again checked after dialysis by Zetasizer Nano ZSP (Malvern, USA). LNP samples were analyzed for size distribution in PBS. The formulation was concentrated using Ultra centrifugal filters (Merck, Germany), passed through 0.22 µm filter, and stored at 5 ± 3 °C [19]. The formulation was confirmed by quality control for the particle size, encapsulation efficiency, endotoxin limit and sterility.
2.5. Animal management and vaccination
The study procedures were performed according to local and international regulation. The detailed process is explained in the supplementary document.
2.6. Local tolerance
Local tolerance was confirmed by clinical signs, macroscopic and histopathology evaluations of injection sites in animals. Euthanasia and evaluation of lesions was performed in one representative mouse from placebo and control group and 3 from the treatment group at 48 h post treatment. The inner thigh muscle of injected site of each mouse was excised and placed in 10% neutral buffered formalin until adequately fixed. After trimming, processing and paraffin embedding, the sections (6 µm) are HE stained and observed for erythema and edema under microscope.
2.7. Immunogenicity
Approximately 200 µL blood was collected from facial vein and centrifuged at 1500 ×g for serum isolation (10 min at 4 °C). All serums were aliquoted, frozen immediately and stored at −80 °C until analysis. The reactivity of the sera from each group of mice immunized with GBPD060 was measured against SARS-CoV-2 S antigen (SinoBiologicals, China). The serum IgG binding endpoint titers (EPTs) were measured in mice immunized with GBPD060. EPTs were observed in the sera of mice at day 7 and day 14 after immunization with a single dose of the vaccine candidate.
2.8. Toxicity
Whole blood (~50 µL) from each mouse was collected in 2% EDTA at 3 days pre-immunization and 14 days post-immunization. Complete blood count (CBC) analysis was measured using auto hematology analyzer BK-6190-Vet (Biobase, China). Samples were used for blood-chemistry analysis viz., alanine transferase (ALT), aspartate transaminase (AST) and blood urea nitrogen (BUN) using semi-automatic chemistry analyzer (Biobase, China).
2.9. Pseudovirus preparation and in vitro neutralization
Adenovirus and retrovirus based pseudovirus were prepared for SARS-CoV-2 in vitro and in vivo neutralization assay. The detailed process is explained in the supplementary document.
2.10. In vivo neutralization
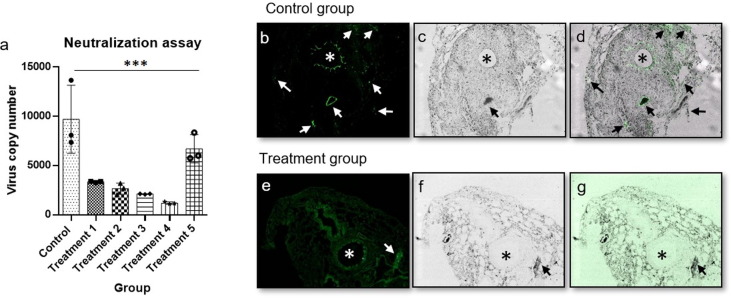
18 albino male mice of 6–8 weeks were selected divided into 6 groups, 1 control and 5 treatment, comprising of 3 male mice in each group. The control group mice were immunized intramuscularly with 50 µL of placebo and treatment group mice were immunized with 1 µg/50 µL of GBPD060 vaccine. GFP pseudotyped homotypic SARS-CoV-2 adenovirus (or treated as indicated in Fig. 6) were sprayed in the nasopharynx on 21-day post immunization. Nasopharynx and lung aspirate samples from mice were collected and analyzed for virus copy number using qPCR at indicated time point. Animals were sacrificed and lung sections were performed and microscopic slides were prepared for fluorescence imaging (GFP) to detect virus load.
Fig. 6.
In vivo neutralization assay, lung section, * indicates trachea and arrow indicate infection. A representative data set from 2 independent experiments are shown, where n = 3 for each experiment. (a) the inhibition and neutralization of the SARS-CoV-2-S pseudo-type virus, data were analyzed using one-way ANNOVA method, ***= p-value < 0.001. (b) fluorescence image of lung section of control group mouse, (c) trans image of lung section of control group mouse, (d) overlay image of lung section of control group mouse, (e) fluorescence image of lung section of treatment group mouse, (f) trans image of lung section of treatment group mouse, (g) overlay image of lung section of treatment group mouse, intentional green color enhancement was done to observe any GFP intensity for panel g.
2.11. Antibody analysis by ELISA
Serum from the mice of different groups were analyzed by standard enzyme‐linked immunosorbent assay (ELISA) to determine antibody titers (supplementary document).
2.12. Antibody binding affinity
The BIAcore T200 equipment (GE Healthcare, USA) and Amine coupling kit (GE Healthcare, USA) were used for immobilization of SARS-CoV-2 Spike S1 + S2 ECD-His recombinant protein (Sino Biological, China) in Series S Sensor Chips CM5 (GE Healthcare, USA). The detailed process is explained in the supplementary document.
2.13. SARS-CoV-2 surface glycoprotein peptide pool generation and mapping
SARS-CoV-2 S-ECD His recombinant protein (Sino Biological, China), S2-ECD-His Recombinant Protein (Sino Biological, China), and RBD(S1)-His recombinant proteins (Sino Biological, China) were digested with Serine Protease (MS grade, Pierce, ThermoFisher, USA). The detailed process is explained in the supplementary document.
2.14. Mouse splenocyte isolation, peptide stimulation and flow cytometric analysis of T cell populations
Mice were sacrificed and splenocytes were harvested, surface antigen and intracellular cytokine staining of cells were performed for day 14 and day 91 samples. The detailed process is explained in the supplementary document.
3. Results
3.1. Bioinformatics analysis to initiate the designing of ‘GBPD060’
We have started with alignment of available sequences of SARS-CoV-2 spike (S) protein. In march 2020, we found a total of 15 D614G sequences out of 170 reference sequences of SARS-CoV-2 (supplementary Fig. 1a). Hydropathy profile showed a minor variation in relevant protein between D614 and G614 genotypes (supplementary Fig. 1b and 1c). Relevant 3D modeling suggested that there might be higher angular strain on G614 than the D614, which could affect the stability and atomic distance with the neighboring atoms (supplementary Fig. 1e and 1f). Our observation has been recently experimentally validated by others [13], [20], [21].
3.2. Construction, antigen expression and formulation of ‘GBPD060’
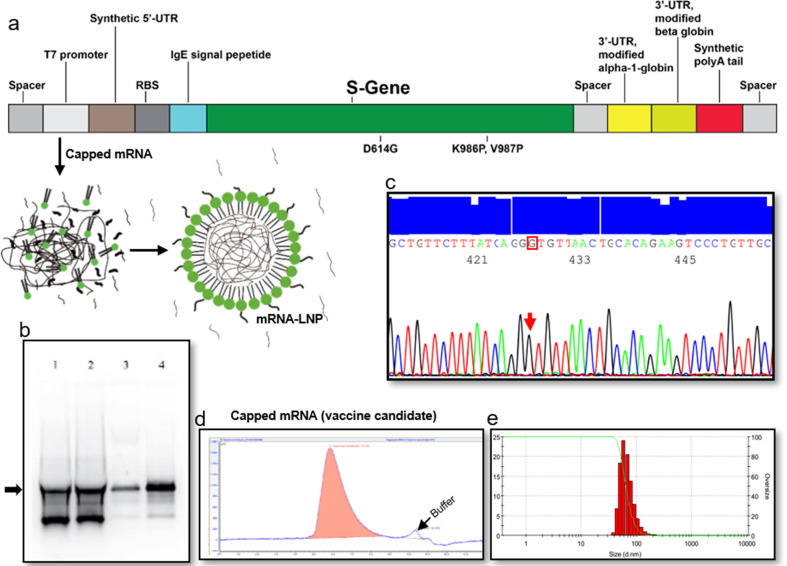
We have obtained the ORF for the SARS-CoV-2 spike with G614-translating codon from a clinically confirmed COVID-19 patient through PCR amplification (Accession No. MT676411.1). Necessary modifications were performed to obtain the desired clone in pET31b vector as described in ‘Materials and Method' section. The schematic diagram of the target gene and construction scheme are shown in Fig. 1 a and supplementary Fig. 2 a and 2b, respectively. The in vitro transcription (IVT) process was modified to obtain high yield and desired quality of mRNA (Fig. 1b). We have obtained the capped-mRNA with a 130-nucleotide residue-long poly A tail. The mRNA sequence with poly A tail was confirmed by DNA sequencing after reverse transcription (Fig. 1c); Accession No. MWO45214. The IVT process was tuned and validated to obtain desired mRNA with high yield and quality (Fig. 1b). The mRNA was encapsuled in lipid nano particle (LNP) ranging from 60 to 140 nm with the final pH of 7.2. We did a pilot study with limited numbers of mice to identify the suitable mRNA-LNP size for our formulation. mRNA-LNP either smaller than 60 nm or larger than 120 nm did not generate considerable immunological response even with a dose of 10 µg/mouse (data not shown). To obtain the best process control for the dose production, we therefore, set our mRNA-LNP size range at 85 ± 20 nm. We used mRNA-LNP of this range throughout the rest of the experiments (Fig. 1e). LNP without SARS-CoV-2 S-mRNA was used as placebo.
Fig. 1.
Target construction, amplification, IVT optimization, purification, and LNP formation. A representative data set from 3 independent experiments are shown. (a) Graphical representation of linear DNA construct for mRNA transcription, (b) IVT optimization where Lane 4 is the optimized condition, (c) DNA sequencing electropherogram data of D614G sequence in the target, (d) Identification of purified capped mRNA by SEC-HPLC, (e) size distribution of mRNA-LNP dose formulation.
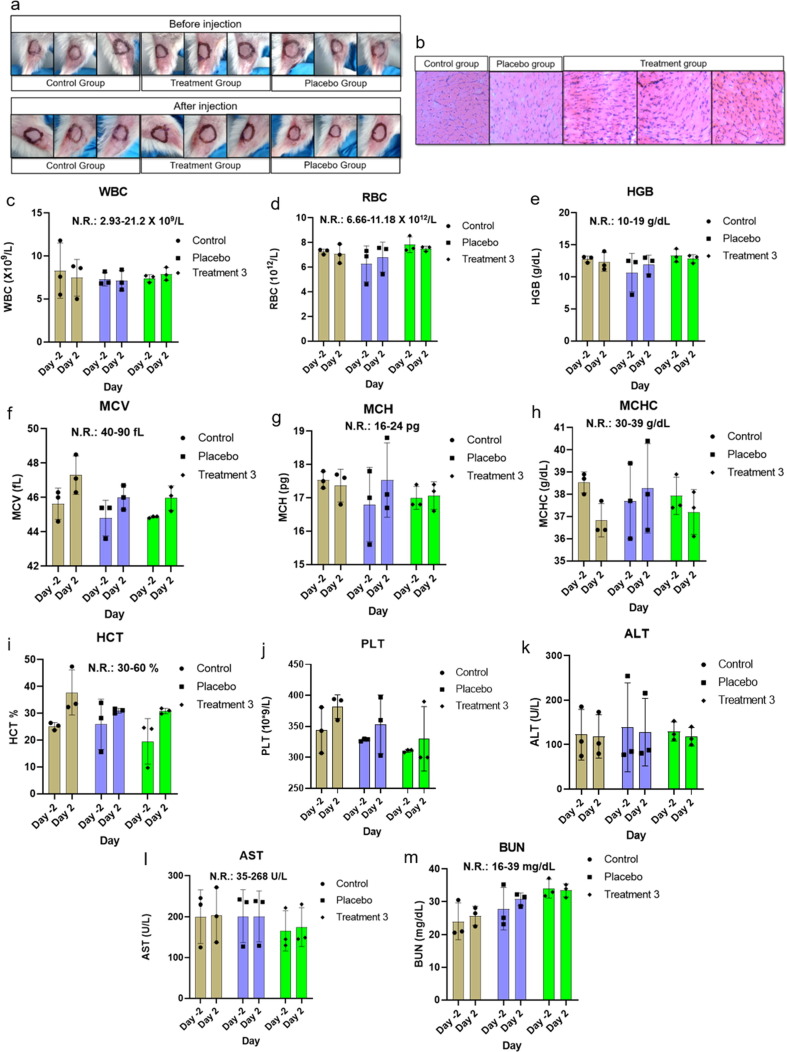
3.3. Local tolerance and toxicity
Control, treatment and placebo group comprising 3 male mice each were used for local tolerance testing. Pictures of the site of injection before and 24 h after injection are shown in Fig. 2a (top and bottom panels, respectively). No detrimental physical consequences of administration were observed such as, local trauma following injection and/or physicochemical actions of the vaccine from local toxicological or pharmacodynamics effects. No sign of erythema or erythredema were observed in muscle tissue section from the site of injection (Fig. 2b). Complete blood count (CBC) from different groups indicated good health status of mice; all parameters were in normal physiological range (Fig. 2c – j). There were no signs for anemia, infection, inflammation, and bleeding disorder. Liver function tests (LFTs) such as alanine transaminase (ALT) and aspartate aminotransferase (AST) were performed to confirm clinical suspicion of potential liver injury or disease and to distinguish between hepatocellular damage and cholestasis (Fig. 2k and l). Blood urea nitrogen (BUN) was tested to evaluate the health of kidneys, such as kidney disease or damage (Fig. 2m). Data for ALT, AST and BUN were in normal range and no significant changes were observed between pre-immunization and after immunization.
Fig. 2.
Local tolerance and CBC analysis. A representative data set from 2 independent experiments are shown, where n = 3 for each experiment. (a) check for sign of visible adverse reaction of administration before and after injection, (b) HE stained tissue from site of injection for erythema and edema, (c) WBC, white blood count, (d) RBC, red blood cell, (e) HGB, hemoglobin, (f) MCV, mean corpuscular volume, (g) MCH, mean corpuscular hemoglobin, (h) MCHC, mean corpuscular hemoglobin concentration, (i) HCT, hematocrit, (j) PLT, platelet, (k) ALT/SGPT, alanine transaminase, (l) AST/SGOT, aspartate aminotransferase, (m) BUN, blood urea nitrogen. Data were analyzed using one-way ANNOVA method, and found statistically non-significant. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. ‘GBPD060’ induces high and Th-1 biased antibodies against full-length SARS-CoV-2 S-protein
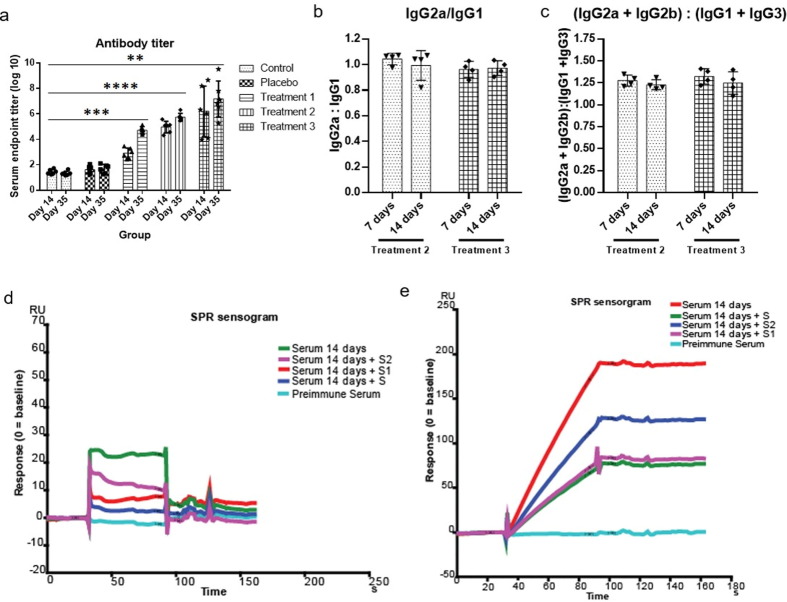
Immunization of mice with mRNA-LNP produced specific titer at a dose dependent manner (Fig. 3 a). Low dose (0.1 µg/mouse) immunization produced moderate level of antibody response (Fig. 3a, Treatment 1). We found the best antibody response with 1 µg/mouse dose (Fig.: 3a, Treatment 2). High dose (10 µg/mouse) immunization produced higher level of titer with wide-spread distribution, and therefore, we have considered this dose unacceptable (Fig. 3a, Treatment 3). The subtyping analysis revealed that the titer contains balanced ratio of IgG2a and IgG1 in 7-day post immunization sera, and it remains stable for 14-day post immunization sera (Fig. 3b, Treatment 2). Similar trend was observed for (IgG2a + IgG2b) and (IgG1 + IgG3) (Fig. 3c, Treatment 2), which has suggested that the antigenic response was CD4 + Th1-biased. High dose (10 µg/mouse) injected mice sera also produced similar response (Fig. 3b and c, Treatment 3). To check whether the immunization have generated antibody pool spanning for the whole antigen or for any specific domain (either S1 or S2), we have chosen surface plasmon resonance (SPR) experiment. The S protein chip recognized high-affinity antibody from anti-sera pool (Fig. 3d). The response was attenuated significantly for S-protein(s) (S, S1 and S2) pretreated sera (Fig. 3d). S and S1 pretreatment showed similar and strong inhibitory response while S2 pretreatment showed comparatively moderate inhibitory response. The purified Ig from the pooled anti-sera produced significantly pronounced response (Fig. 3e). The SPR data clearly showed that the vaccination has produced specific antibody pool against the full-length of S protein.
Fig. 3.
Antibody titer and affinity analysis. A representative data set from 2 independent experiments are shown, where n = 4 for each experiment (unless mentioned otherwise). (a) antibody titer analysis from serum of different groups after 14 and 35 days of immunization (n = 6), data were compared by Mann-Whitney test, ****= p-value < 0.0001, ***= p-value < 0.001, **= p-value < 0.01 (b) ratio of IgG2a and IgG1 in treatment 2 and treatment 3 group, (c) ratio of IgG2a + IgG2b and IgG1 + IgG3 in treatment 2 and treatment 3 group, (d) serum antibody affinity analysis, (e) resin pull-down serum antibody affinity analysis.
3.5. Cellular and cytokine responses to ‘GBPD060’
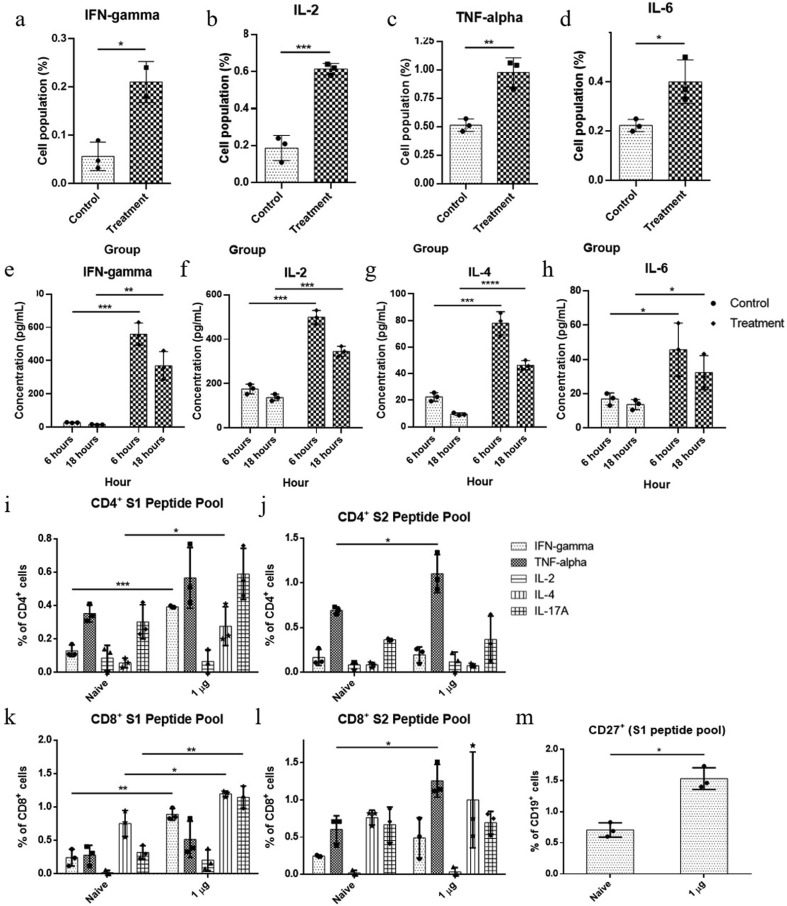
We further characterized the cellular response and induction of specific cytokines in response to vaccination after 14 and 91 days of first immunization. The splenocytes obtained from vaccinated mice were re-stimulated with a library of SARS-CoV-2-S peptide pool. 14 days after first immunization, the stimulated splenocytes generated significantly higher population of CD4 + Th1 cytokine IFN-gamma, IL-2 and TNF-α expressing cells (0.21 ± 0.03, 0.61 ± 0.03 and 0.95 ± 0.11, respectively) compare to the placebo treated group (0.06 ± 0.03, 0.18 ± 0.07 and 0.52 ± 0.06, respectively) (Fig. 4 a-c.). CD4 + Th2 cytokine IL-6-expressing cells were moderately increased in stimulated splenocytes of vaccinated mice compare to the placebo-injected mice (0.42 ± 0.08 and 0.23 ± 0.02, respectively) (Fig. 4d). The secreted cytokines were found modulated over the placebo group as follows: IFN-gamma (treatment group, 559.87 ± 70.76 pg/ml and 303.47 ± 156.53 pg/ml; placebo, 28.29 ± 2.03 pg/ml and 16.04 ± 2.52 pg/ml), IL2 (treatment group, 499.10 ± 30.80 pg/ml and 345.17 ± 22.85 pg/ml; placebo group, 175.71 ± 21.92 pg/ml and 136.87 ± 15.18 pg/ml), IL4 (treatment group, 77.94 ± 7.7 pg/ml and 46.36 ± 3.7 pg/ml; placebo group, 22.5 ± 3.25 pg/ml and 9.5 ± 1.08 pg/ml) and IL6 (treatment group, 45.78 ± 15.52 pg/ml and 32.61 ± 15.52 pg/ml; placebo group, 16.96 ± 3.53 pg/ml and 14.87 ± 3.08 pg/ml), respectively for 6- and 18-h, which have suggested vaccine specific response (Fig. 4e-h).
Fig. 4.
Cellular immune response analysis (cellular and secretory cytokine) in control and treatment group; unpaired T-test were performed between control and treatment groups; ***= p-value < 0.001, **= p-value < 0.01, *= p-value < 0.05. A representative data set from 2 independent experiments are shown, where control n = 3 and treatment n = 3 for each experiment (unless mentioned otherwise). (a) IFN-gamma expressing cell population percentage at Day 14, treatment n = 2 (b) IL-2 expressing cell population percentage at Day 14, (c) TNF-α expressing cell population percentage at Day 14, (d) IL-6 expressing cell population percentage at Day 14, (e) secretory IFN-gamma concentration at 6 and 18 h, (f) secretory IL-2 concentration at 6 and 18 h, (g) secretory IL-4 concentration at 6 and 18 h, (h) secretory IL-6 concentration at 6 and 18 h, (i) CD4 + cell population at Day 91, stimulated with S1 peptide pool, (j) CD4 + cell population at Day 91, stimulated with S2 peptide pool, (k) CD8 + cell population at Day 91, stimulated with S1 peptide pool, (l) CD8 + cell population at Day 91, stimulated with S2 peptide pool, (m) memory B cell population at Day 91, stimulated with S1 peptide pool.
Again, 91 days after first immunization, the stimulated (S1 and S2) splenocytes generated significantly higher population of CD4 + Th1 cytokine IFN-γ, TNF-α and IL-2-expressing cells (0.39 ± 0.01 and 0.19 ± 0.08, 0.6 ± 0.17 and 1.13 ± 0.21, and 0.07 ± 0.07 and 0.11 ± 0.11, respectively) compare to the placebo treated group (0.14 ± 0.03 and 0.18 ± 0.08, 0.36 ± 0.05 and 0.69 ± 0.04, and 0.07 ± 0.07 and 0.06 ± 0.05, respectively) (Fig. 4i and 4j). Similarly, S1 and S2 stimulated splenocytes also generated significantly higher population of CD8 + Th1 cytokine IFN-γ, TNF-α and IL-2-expressing cells (0.91 ± 0.08 and 0.48 ± 0.27, 0.58 ± 0.25 and 1.3 ± 0.21, and 0.3 ± 0.08 and 0.05 ± 0.05, respectively) compare to the placebo treated group (0.24 ± 0.13 and 0.25 ± 0.1, 0.26 ± 0.15 and 0.55 ± 0.16, and 0.03 ± 0.03 and 0.03 ± 0.03, respectively) (Fig. 4k and 4l). S1 and S2 stimulated splenocytes generated moderate CD4 + and CD8 + Th2 cytokine IL-4 and IL-17A-expressing cells compare to the placebo-injected mice (Fig. 4i-l). Higher level of sustained Th1 specific cytokine response over Th2 specific cytokine suggested a stable and balanced Th1-biased immunologic response after administration of ‘GBPD060’ vaccine. S1 stimulated splenocytes generated significantly higher population of CD19 + CD27 + expressing cells (1.57 ± 0.17) compare to the placebo treated group (0.72 ± 0.12) (Fig. 4m).
3.6. ‘GBPD060’ induces neutralization of SARS-CoV-2-S pseudo-type viruses
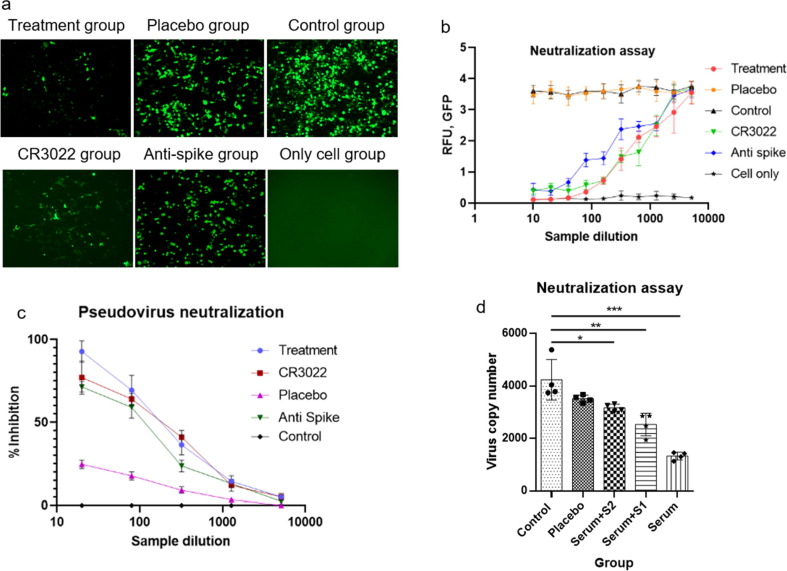
Sera of vaccinated mice inhibited infection of GFP-expressing pseudo-type SARS-CoV-2-S adenovirus in hACE2-expressing HEK293 (ACE2-HEK293) cell in a dose dependent manner (Fig. 5 a and 5b). Neutralization assay demonstrated that there is a correlation between the intensity of GFP and SARS-CoV-2 specific antibody for vaccinated mice. Higher concentration of SARS-CoV-2 antibody efficiently neutralize the entry of the pseudovirus into the ACE2-HEK293 cell. IC50 value for GFP-inhibition were found significantly higher for the anti-sera (~3 µg/mL) compared to the CR3022 and a commercially available polyclonal mouse antibody against S-protein (~7 µg/ml). In parallel with the GFP analysis, to confirm the virion copy number, we have implemented real-time PCR as an orthologous method (supplementary document). The data showed correlation with the GFP and gene copy analysis (Fig. 5d). HIV1-based SARS-CoV-2-S pseudo type virus infection was also significantly inhibited by 1 µg/mouse dose anti-sera compared to the placebo anti-sera (Fig. 5d, Serum). Either S1 or S2 protein pretreatment nullified the inhibition capacity of anti-sera (Fig. 5d, Serum + S1 and Serum + S2) confirmed that the inhibition property for the HIV1-based SARS-CoV-2-S pseudo type virus is related to the vaccination.
Fig. 5.
In vitro neutralization assay. A representative data set from 3 independent experiments are shown, where n = 4 for each experiment. (a) Image of Green fluorescence protein (GFP) expression after adeno-based SARS-CoV-2 pseudovirus neutralization assay from 2 to 4 sample dilution, (b) correlation between SARS-CoV-2 antibody from mice sera and intensity of GFP in different experimental group. For treatment group with the decrease of the antibody concentration, the intensity of GFP expression increased, which indicated the inhibition of SARS-Cov-2 pseudovirus into ACE2 overexpressed HEK293 cell (ACE2-HEK293 cell), (c) adeno-based SARS-CoV-2 pseudovirus neutralization percentage at different sample dilution, analyzed by real-time PCR, (d) HIV-1 based SARS-CoV-2 pseudovirus copy number analysis by real-time PCR; all the samples were compared by one-way ANNOVA method, ***= p-value < 0.001, **= p-value < 0.01, *= p-value < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, we attempted to check whether immunization can protect mice from GFP-expressing homotypic pseudo-type adenovirus [22]. Virus were sprayed through nasopharyngeal space of mice either in buffer or pretreated with immunized sera. The anti-sera-treated SARS-CoV-2-S adenovirus produced lower copy of virus compared to the buffer-treated virus (Fig. 6 a, Treatment 2 and Treatment 1, respectively). The copy number of virus was found reduced further from day 2 to day 3 (Fig. 6a, Treatment 4 and Treatment 3, respectively). These data clearly revealed that though the anti-serum has significant inhibitory capacity against viral infection but systemic immune-protection is essential for better protection. Lower copy number of virus over the time indicated that the cellular immunity is also necessary, along with humoral immunity, for efficient viral clearance. The anti-sera treated with S1 + S2 protein failed to inhibit SARS-CoV-2-S adenovirus infection in the placebo-injected mice (Fig. 6a, Treatment 5), and proved that the inhibition and neutralization of the SARS-CoV-2-S pseudo-type virus is correlated with the immunogenic response generated due to ‘GBPD060’.
We further collected the lungs of control- and GBPD060-immunized mice (single-dose day 14 postimmunization), which were challenged with homotypic SARS-CoV-2 pseudovirus (day-5 post challenge). GFP-labelled viruses were clearly found spread throughout the lungs of control group mice (Fig. 6b – d, arrowhead). Peritracheal viral infection was predominant and suggesting that the virus entry pathway is the airways of the lung (Fig. 6b – d, *). The immunized mice lungs showed few viral-spots on day-5 post challenge tissue (Fig. 6e – g, arrowhead), which were completely disappeared by day-6 of post challenge (data not shown). No peritracheal viral signal was observed in these lungs (Fig. 6e – g, *). The data provided visual and conclusive evidence that GBPD060 immunization can neutralize SARS-CoV-2 virus and protect the animal from infection of the virus into the lungs.
4. Discussion
The G614 variant was first identified in China and Germany in January 2020. It was a rare genotype before March 2020, which then quickly became the major circulating genotype worldwide [13]. Cardozo et al., reported in May 2020 that G614 genotype is associated with increased case fatality rate over D614 [23]. Scientific findings evidently demonstrated that the G614 variant is ~10 times more infectious over the D614 genotype [24], [25]. It has been revealed from in vitro and clinical data that G614 variant has a distinct phenotype, and there likely be a huge impact of this mutation on infection, transmission, disease onset, disease prognosis, as well as on vaccine and therapeutic development [13], [26], [27]. We found the sequence G614 from a PCR-confirmed patient in May, 2020 (Accession No. MT676411.1). Based on the scientific information available then, we have predicted that this variant may become dominant in future, and at that period of time there was no information for any vaccine candidate in development considering G614 genotype. We, therefore, decided to develop the vaccine considering this mutation.
The design consideration for the vaccine was to obtain high-expressing spike protein as antigen in a putative perfusion stabilized condition. Comparative design features of ‘GBPD060’ with the available published information for relevant vaccine candidates in development are shown in Table 1 . ‘GBPD060’ mRNA has few features along with the G614-targeted mutations, which are different than the other relevant known candidates. Ribosome binding site, IgE-signal sequence by replacing the native 13 amino acids from the N-terminal of the SARS-CoV-2 S protein, 3′ UTR constituted with the 3′UTRs of alpha and beta globin gene in tandem are worth to mention. We have used 65 – 105 nm LNP to deliver the mRNA. LNPs out of this range did not elicit considerable antibody response in a separate pilot study (data not shown); the best range was observed with 85 ± 20 nm of LNP. The LNP-sizes determine the delivery efficiency of the cargo to the target cells [28]. The pH (7.2) of our formulation buffer for mRNA-LNP is lower than the other relevant references (7.4–8.0) [5], [6], [7], [8]. Lower pH helps quick release of the cargo from endosomal compartment and protects mRNA from acid hydrolysis and lysosomal digestion in intracellular milieu [29]. Together, numbers of minute changes in the design context likely playing in concert and produced quick, balanced, stable Th1-IgG2-biased antibody response.
Table 1.
Comparative design features of ‘GBPD060’.
| Parameter | GBPD060 | Others |
|---|---|---|
| Construct | T7 promoter | T7 promoter [6] |
| 51 bp 5′-UTR | 5′-UTR [5], [6] | |
| Ribosome binding sequence | Not specified | |
| IgE signal peptide in the ORF | ORF [5], [6], [7], [8] | |
| D614G mutation | Not in consideration | |
| K986P and V987P mutations | K986P and V987P mutations [5], [6] | |
| Modified alpha and beta globin in 3′-UTR | 3′-UTR [5], [6] | |
| 130 bp poly A tail | Poly A tail [5], [7] poly(A) tail (100 nucleotides) interrupted by a linker [6] |
|
| LNP |
LNP composition: MC3, DSPC, Cholesterol and DMG-PEG2000 (50:10:38.5:1.5). Stabilization buffer: 1x PBS, pH 7.2 LNP size range: 85 ± 20 nm |
LNP composition: ionizable lipid, DSPC, Cholesterol and PEG2000-DMG [7] Stabilization buffer: HEPES buffer, pH 8.0 [7] LNP Size: ~75 nm [8] and average size 100 nm [32] |
| IgG2a/IgG1 ratio | ~ 1.0 | ~ 0.8 [5], 1.6 [8] |
‘GBPD060’ immunization did not produce any noticeable effect for local or systemic toxicity as primarily evident by the absence of four cardinal signs of inflammation: redness (Latin rubor), heat (calor), swelling (tumor), and pain (dolor). There was no erythema or erythredema as well in any injection site. The CBC and blood chemistry data did not show significant changes in relevant profiles and has been suggesting that the vaccine behaves safely in animal.
A balanced response between Th1 and Th2 is desired to achieve safe and effective humoral immunity performance.[30] ‘GBPD060’ has produced well-balanced IgG1 and IgG2 response by 7th day postimmunization and remained similar on 14th day postimmunization sera, which is suggesting a stable antibody response during the sampling period. Along with opsonizing characteristics, IgG2 has higher affinity to its receptors and have superior complement system activation potential over IgG1 [30], [31]. Accordingly, ‘GBPD060’-mediated higher ratio of IgG2 than IgG1 has suggested that higher capacity of the antibody pool to clear antigen from the system. The ratio of IgG2a and IgG1, and cytokine-stained CD4 + and CD8 + T cell population showed a Th1-bias response. Since mouse IgG2 is equivalent to human IgG1 [30], [31] therefore, it is plausible that ‘GBPD060’ will elicit effective cellular and humoral response against SARS-CoV-2 in human.
Importantly, ‘GBPD060’ has elicited high level of specific antibody with a single immunization, which is comparable with the level of antibody response observed after administration of 2nd dose of relevant vaccine developed by Moderna. Additionally, we have observed significant level of S1 peptide-pool-specific CD27+ memory B cell after 91 days of first immunization. Therefore, it is highly likely that GBPD060 may provide suitable protection against SARS-CoV-2 in human with a single dose of injection. A single dose for effective immunization against SARS-CoV-2 will be highly rewarding in terms of satisfying mass vaccination to the global community to stop unwanted death and restore normal life; an immediate clinical trial is therefore necessary that can reveal the fact and opportunity.
The early vaccine development initiatives were taken before the G614 variant became predominant. Therefore, there are no specific vaccine tested so far in human with G614 variant-related molecule. It has been shown that immunization might not have any role for selection of D614 or G614 variant [33], [34]. Studies with the sera obtained from COVID-19 patients showed variable results regarding the neutralization propensity of D614 and G614 genotype. With 88 sera from a high-incidence community, Hicks et al., showed that antibody pool did not differentiate between D614 and G614 binding [14], [35]. However, a few data point stayed out of the correlation trend in their study, which might be linked with the functional variations associated with the SARS-CoV-2 variants. Korber et al., with 6 convalescent sera, showed that D614 and G614 both types of VSV-pseudovirus can be inhibited by the sera; though G614 and D614 showed little variation in their responses to the assay [13]. They further showed that the G614 genotype produced higher titers against pseudoviruses from in vitro experiments. The variations observed in both of the studies might not be just a coincident rather suggesting potential differences in modus operandi between the G614 and D614 variants. The proposition is supported by the Huang et al.; they have showed that 7% of the convalescent sera out of their 70 samples failed to neutralize G614 variant of pseudovirus [36]. All of these studies did not identify whether the subjects were infected by either D614 or G614 variant, which could have revealed better insight for the correlation of the observation.
The roles of G614 mutation on constitutive infection have been attributed to its conformational change. It has been proposed that the –COOH group of D614 forms hydrogen bond with the –OH group of T859 across the S1/S2 interface, which cannot form in G614 [13]. On the contrary, structural modeling studies revealed that “the D614G substitution creates a sticky packing defect in subunit S1, promoting its association with subunit S2 as a means to stabilize the structure of S1 within the S1/S2 complex [26]. In other words, the D614G mutation in fact promotes the S1/S2 association and stabilize the spike [26]. The finding is in accordance with the observation that G614 has a greater stability originating from less S1 domain shedding and greater accumulation of the intact S protein into the pseudovirion [25]. It has also been reported that G614 mutation promoted the interaction of two of the three S glycoprotein chains with the RBD, whereas only one chain from D614 interacts with RBD [21]. This interaction creates favorable conformation of the RBD to its partners resulting higher access for effective binding and infection.
Predecessor SARS-CoV virus entry and infection was shown promoted by protease-mediated processing of the spike protein [37]. It has been postulated that SARS-CoV-2 also likely be adopting such properties by acquiring G614 genotype by incorporating protease processing site [38]. Consequently, it has been shown that indeed G614 protein has been cleaved by serine protease elastase-2 more efficiently than the D614 variant [36]. They further showed that G614 pseudovirus infection of 293T-ACE2 was potentiated in the presence of elastase-2, which can be blocked by elastase inhibitor. These findings corroborate the fact that G614-targetted vaccine is necessary.
Two prominent antigenic sites on the S-protein have been proposed those are spanning 614 position: V595-D614 and D614-A647 [39], [26]. The role of V595-D614 domain is not known but the D614-A647 is a dehydron wrapped intramolecularly by residues D614, V615, T645, R646 and A647, and forms a salt bridge with D614. The salt bridge contributes to stabilize D614-A647 in the uncomplexed S1 and inhibits the S1/S2 association. G614 diminishes the salt bridge formation and S1/S2 association resulting interaction with the RBD to facilitate higher infection [26]. Therefore, blocking of G614 with a specific antibody would inhibit such acquired fitness of SARS-CoV-2. ‘GBPD060’ immunization has produced a pool of antibody that covers the whole length of the spike protein suggesting that highly likely relevant antibody-mix against these domains have been developed. Since relevant domains are highly glycosylated therefore, we could not obtain homotypic peptides for definitive characterization of the purified antibody pool against these predicted antigens. Further study will be attempted to reveal the relevant scientific aspects. Nevertheless, the findings clearly demonstrated that ‘GBPD060’ is safe for in vivo administration, and elicits balanced and stable cellular and humoral response that neutralize SARS-CoV-2 spike protein-mediated infection. Currently, we have been preparing for the phase-1 clinical trial. Appropriate clinical trial will reveal further insight regarding the significance of G614-targeted vaccine for the efficient management of COVID-19 pandemic.
The recent metadata analysis on more than 5000 clinical samples revealed that 100% of the second-wave of infection has been associated with G614 variant, which is emphasizing the need for G614-targetted vaccine for managing this uncontrolled infection [12]. Therefore, the rapid transition of the 1st G614-targetted vaccine ‘GBPD060’ to clinical trial would be highly rewarding.
5. Data availability
The data that support the findings of this study are available within the article and its supplementary document file, or are available from the corresponding author upon reasonable request.
Author Contributions
K.N., N.S., A.M., and J.C.B. conceived the idea, designed the experiment, analyzed the data and wrote the manuscript. J.C.B., M.M.R.K., M.J.I., S.M., M.R.A., M.E.H.S., S.K., M.M.H.C., R.R, F.I., U.B., H.K., S.C., M.A.B., M.M.H., S.A., M.M.R.C., P.G., R.I.S., R.A., M.E.H.B., B.K.B., and M.M. performed experiments. M.M.R.K., M.J.I., S.M., M.R.A., M.E.H.S., S.K., M.E.H.B., and M.M. contributed to manuscript writing. M.R.A., and U.B. performed the animal husbandry and animal studies.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The study was funded by Globe Biotech Limited. We thank Md. Harunur Rashid, the chairman of Globe Pharmaceuticals Group of Companies, Ahmed Hossain, Md. Mamunur Rashid and Md. Shahiduddin Alamgir, Abdullah Al Rashid, the directors of Globe Pharmaceuticals Group of Companies for their continuous support and encouragement. We also thank Md. Raihanul Hoque, Dibakor Paul, Biplob Biswas, Zahir Uddin Babor, Mithun Kumar Nag and Imran Hossain for their support for facility and information management system.
Funding
Globe Biotech Limited funded this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2021.05.035.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Feng W., Zong W., Wang F., Ju S. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review. Mol. Can. 2020;19(1):100. doi: 10.1186/s12943-020-01218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.“COVID-19 Public Health Emergency of International Concern (PHEIC) Global research and innovation forum,” Feb. 2020. Accessed: Sep. 09, 2020. [Online]. Available: https://www.who.int/publications/m/item/covid-19-public-health-emergency-of-international-concern-(pheic)-global-research-and-innovation-forum.
- 3.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbett K.S., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020 doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A.B. Vogel et al., A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates, Immunology, preprint, Sep. 2020. doi: 10.1101/2020.09.08.280818.
- 7.R. de Alwis et al., A Single Dose of Self-Transcribing and Replicating RNA Based SARS-CoV-2 Vaccine Produces Protective Adaptive Immunity In Mice, Immunology, preprint, Sep. 2020. doi: 10.1101/2020.09.03.280446. [DOI] [PMC free article] [PubMed]
- 8.P. F. McKay et al., Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice, Nat. Commun., vol. 11, no. 1, p. 3523, 09 2020, doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed]
- 9.Garber K. Alnylam launches era of RNAi drugs. Nat. Biotechnol. Oct. 2018;36(9):777–778. doi: 10.1038/nbt0918-777. [DOI] [PubMed] [Google Scholar]
- 10.Sevajol M., Subissi L., Decroly E., Canard B., Imbert I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith E.C., Blanc H., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9(8) doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S.W. Long et al., Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area, Pathology, preprint, Sep. 2020. doi: 10.1101/2020.09.22.20199125. [DOI] [PMC free article] [PubMed]
- 13.Korber B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klumpp-Thomas C., et al. D614G spike variant does not alter IgG, IgM, or IgA spike seroassay performance. MedRxiv Prepr. Serv. Health Sci. 2020 doi: 10.1101/2020.07.08.20147371. [DOI] [Google Scholar]
- 15.Evers M.J.W., Kulkarni J.A., van der Meel R., Cullis P.R., Vader P., Schiffelers R.M. State-of-the-art design and rapid-mixing production techniques of lipid nanoparticles for nucleic acid delivery. Small Methods. 2018;2(9):1700375. doi: 10.1002/smtd.201700375. [DOI] [Google Scholar]
- 16.Corbett K.S., et al. SARS-CoV-2 mRNA vaccine development enabled by prototype pathogen preparedness. Immunology. 2020 doi: 10.1101/2020.06.11.145920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R.L. Ball, C.M. Knapp, K. A. Whitehead, Lipidoid Nanoparticles for siRNA Delivery to the Intestinal Epithelium: In Vitro Investigations in a Caco-2 Model, PLOS ONE, vol. 10, no. 7, p. e0133154, Jul. 2015, doi: 10.1371/journal.pone.0133154. [DOI] [PMC free article] [PubMed]
- 18.A. A., M. A., P. F., Lipid Nanoparticulate Drug Delivery Systems: A Revolution in Dosage Form Design and Development, in Recent Advances in Novel Drug Carrier Systems, A. D. Sezer, Ed. InTech, 2012.
- 19.Ball R., Bajaj P., Whitehead K. Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int. J. Nanomedicine. 2016;12:305–315. doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilleman M.R. Recombinant vector vaccines in vaccinology. Dev. Biol. Stand. 1994;82:3–20. [PubMed] [Google Scholar]
- 21.I.O. Omotuyi et al., Atomistic simulation reveals structural mechanisms underlying D614G spike glycoprotein-enhanced fitness in SARS-COV-2, J. Comput. Chem., vol. 41, no. 24, pp. 2158–2161, 15 2020, doi: 10.1002/jcc.26383. [DOI] [PMC free article] [PubMed]
- 22.Dinnon K.H., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586(7830):560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.M. Becerra-Flores, T. Cardozo, SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate, Int. J. Clin. Pract., p. e13525, May 2020, doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed]
- 24.Daniloski Z., et al. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. Genetics. 2020 doi: 10.1101/2020.06.14.151357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. Microbiology. 2020 doi: 10.1101/2020.06.12.148726. [DOI] [Google Scholar]
- 26.Fernández A. Structural impact of mutation D614G in SARS-CoV-2 spike protein: enhanced infectivity and therapeutic opportunity. ACS Med. Chem. Lett. 2020;11(9):1667–1670. doi: 10.1021/acsmedchemlett.0c00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi P.-Y., et al. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. Res. Sq. 2020 doi: 10.21203/rs.3.rs-70482/v1. [DOI] [Google Scholar]
- 28.T. Nakamura, M. Kawai, Y. Sato, M. Maeki, M. Tokeshi, H. Harashima, The effect of size and charge of lipid nanoparticles prepared by microfluidic mixing on their lymph node transitivity and distribution, Mol. Pharm., vol. 17, no. 3, pp. 944–953, 02 2020, doi: 10.1021/acs.molpharmaceut.9b01182. [DOI] [PubMed]
- 29.Lin Q., Chen J., Zhang Z., Zheng G. Lipid-based nanoparticles in the systemic delivery of siRNA. Nanomed. 2014;9(1):105–120. doi: 10.2217/nnm.13.192. [DOI] [PubMed] [Google Scholar]
- 30.Spellberg B., Edwards J.E. Type 1/Type 2 immunity in infectious diseases. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2001;32(1):76–102. doi: 10.1086/317537. [DOI] [PubMed] [Google Scholar]
- 31.Collins A.M. IgG subclass co-expression brings harmony to the quartet model of murine IgG function. Immunol. Cell Biol. 2016;94(10):949–954. doi: 10.1038/icb.2016.65. [DOI] [PubMed] [Google Scholar]
- 32.Lu J., et al. A COVID-19 mRNA vaccine encoding SARS-CoV-2 virus-like particles induces a strong antiviral-like immune response in mice. Cell Res. 2020 doi: 10.1038/s41422-020-00392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie X., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021 doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 34.Dearlove B., et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. 2020;117(38):23652–23662. doi: 10.1073/pnas.2008281117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hicks J., et al. Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal Betacoronaviruses. Infectious Diseases (except HIV/AIDS) 2020 doi: 10.1101/2020.06.22.20137695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J., et al. D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity. Microbiology. 2020 doi: 10.1101/2020.06.20.161323. [DOI] [Google Scholar]
- 37.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.C. Bhattacharyya et al., Global Spread of SARS-CoV-2 Subtype with Spike Protein Mutation D614G is Shaped by Human Genomic Variations that Regulate Expression of TMPRSS2 and MX1 Genes, Genomics, preprint, May 2020. doi: 10.1101/2020.05.04.075911.
- 39.Banerjee A., Santra D., Maiti S. Energetics and IC50 based epitope screening in SARS CoV-2 (COVID 19) spike protein by immunoinformatic analysis implicating for a suitable vaccine development. J. Transl. Med. 2020;18(1):281. doi: 10.1186/s12967-020-02435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the article and its supplementary document file, or are available from the corresponding author upon reasonable request.