Abstract
Background
The increase in global mortality rates from SARS-COV2 (COVID-19) infection has been alarming thereby necessitating the continual search for viable therapeutic interventions. Due to minimal microbial components, subunit (peptide-based) vaccines have demonstrated improved efficacies in stimulating immunogenic responses by host B- and T-cells.
Methods
Integrative immunoinformatics algorithms were used to determine linear and discontinuous B-cell epitopes from the S-glycoprotein sequence. End-point selection of the most potential B-cell epitope was based on highly essential physicochemical attributes. NetCTL-I and NetMHC-II algorithms were used to predict probable MHC-I and II T-cell epitopes for globally frequent HLA-A∗O2:01, HLA-B∗35:01, HLA-B∗51:01 and HLA-DRB1∗15:02 molecules. Highly probable T-cell epitopes were selected based on their high propensities for C-terminal cleavage, transport protein (TAP) processing and MHC-I/II binding.
Results
Preferential epitope binding sites were further identified on the HLA molecules using a blind peptide-docking method. Phylogenetic analysis revealed close relativity between SARS-CoV-2 and SARS-CoV S-protein. LALHRSYLTPGDSSSGWTAGAA242→263 was the most probable B-cell epitope with optimal physicochemical attributes. MHC-I antigenic presentation pathway was highly favourable for YLQPRTFLL269-277 (HLA-A∗02:01), LPPAYTNSF24-32 (HLA-B∗35:01) and IPTNFTISV714-721 (HLA-B∗51:01). Also, LTDEMIAQYTSALLA865-881 exhibited the highest binding affinity to HLA-DR B1∗15:01 with core interactions mediated by IAQYTSALL870-878. COVID-19 YLQPRTFLL269-277 was preferentially bound to a previously undefined site on HLA-A∗02:01 suggestive of a novel site for MHC-I-mediated T-cell stimulation.
Conclusion
This study implemented combinatorial immunoinformatics methods to model B- and T-cell epitopes with high potentials to trigger immunogenic responses to the S protein of SARS-CoV-2.
Keywords: Immunoinformatics, SARS-CoV-2, B-cell epitopes, T-cell epitopes, Vaccine design, High-affinity binding
At a glance commentary
Scientific background on the subject
Immunoinformatics investigations are essential in identifying prospective vaccine candidates for disease treatment. Due to COVID-19's ferocity and fatality, there is need for urgent intervention. Therefore, identifying highly potential B and T-cell epitopes from SARS-CoV-2 Spike protein could be crucial for vaccine development.
What this study adds to the field
The exhaustive and combinatorial approach implemented in this study could align with the high potentials of our predicted epitopes for future anti-COVID-19 vaccine development.
Coronavirus is a pneumonia-related outbreak that intensifies from a milder to a more severe situation. This deadly virus belongs to the family Coronaviridae known to possess a positive-sense, single-stranded polyadenylated RNA virus, more likely to affect humans and animals. Coronaviruses have been identified in several avian hosts as well as in various mammals, including camels; bats, masked palm civets, mice, dogs, and cats. Novel mammalian coronaviruses are now regularly identified. Human coronavirus strains such as HCoV-229E, HCoV-NL63, HCoV-HKU1, HCoV-EMC, and HCoV-OC43 have been epidemic in various continental regions in the past causing multiple respiratory diseases of varying severity, including common cold, pneumonia and bronchiolitis.
Among several coronaviruses that are pathogenic to humans, most are associated with mild clinical symptoms. However, Severe Acute Respiratory Syndrome (SARS) coronavirus (SARS-CoV) and Middle East Respiratory Syndrome (MERS-CoV) emerged to be more lethal compared to the other strains of human corona virus. SARS-CoV is a beta coronavirus that emerged in Southern China in 2002 which led to more than 8000 human infections and 774 deaths in 37 countries during the years 2002–2003 [1,2].
At present, the novel Coronavirus SARS-CoV-2 (2019-nCoV) which has engendered a global panic reportedly originated from the food market in central China metropolis. This, has, in turn, accounted for severe epidemic outbreaks in other provinces of Mainland China, which has further spread to 27 other countries. There are currently rapid increases in the death rates caused by the irate novel coronavirus strain [3,4].
According to research investigations in the Laboratory of Biosafety, National Institute for Viral Disease Control and Prevention, 2019-nCoV is structurally different from SARS-CoV which underlies its identification as a novel host-infecting beta coronavirus with a genome size that ranges from 26 to 32 kilobase in length [5,6]. The phylogenetic studies of the coronavirus indicate that bats might be the original host of this virus [7].
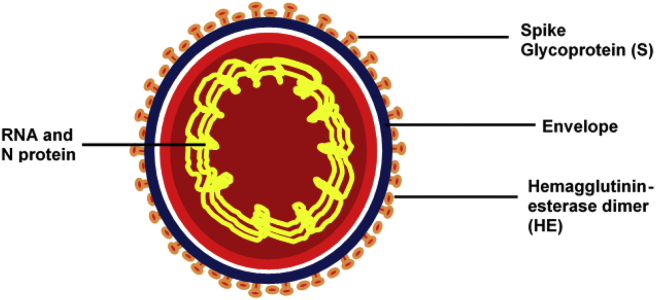
The Coronavirus consists of the following proteins; S-Spike Proteins, Membrane proteins and the Nucleocapsid (N) Proteins [Fig. 1]. These proteins play several crucial roles in the pathogenesis, infection and transmission of the virus in humans. The spike glycoprotein (S) of coronavirus is cleaved into two subunits (S1 and S2). The S1 subunit helps in receptor binding and the S2 subunit facilitates membrane fusion. The spike glycoproteins of coronaviruses are important determinants of tissue tropism and host range. In addition, the spike glycoproteins are critical targets for vaccine development. N is the only protein that functions primarily to bind to the CoV RNA genome, making up the nucleocapsid [[8], [9], [10]].
Fig. 1.
Structural representation of the COVID-19 virion showing antigenic components and their cellular locations.
Although N is largely involved in processes relating to the viral genome, it is also involved in other aspects of the CoV replication cycle and the host cellular response to viral infection. The M protein is the most abundant structural protein and defines the shape of the viral envelope. It is also regarded as the central organizer of viral assembly, interacting with all other major SARS-CoV-2 structural proteins [11].
The SARS-CoV-2 transmits from human-to-human through close contact especially through viral droplets from sneezing and coughing. The symptoms of the viral disease include high fever, dry cough, and breathing difficulties. Virus replication and reproduction occur, as estimated, in a proportion of 3–5 i.e. the virus infects 3 to 5 people per established infection even during the incubation period. Other research groups have estimated the basic reproduction number between 1.4 and 3.8. More so, it has been established that the virus can transmit along a chain of at least four people [12].
Researchers are making assiduous attempts to identify effective treatments for the disease, and currently, Remdesivir and Chloroquine, have been reportedly used in clinical trials to treat patients against COVID-19. Regardless, there is still a need for continued efforts to design strong vaccines to curtail viral spread. Peptide-based vaccines have been promising for treatments against the pathogenic-virulence since it contains minimal components of infectious microbe. This makes it sufficient to effectively trigger immunogenic responses mediated by B-cells and T-cells [13] Peptide-based vaccines are safer amongst other vaccine types due to highly minimal allergic and toxic properties. This explains the implementation of numerous studies aimed at peptide-vaccine design [14,15]. Relatively, immunoinformatics approaches have majorly contributed to the identification of potential vaccine candidates against microbial diseases, by enabling the prediction of highly probable B-cell and T-cell epitopes [16,17].
Immunoinformatics methods incorporate multiple algorithms that assist the predictions of highly potential B-cell and T-cell epitopes that are essential for peptide-vaccine construction. High promising B-cell epitopes are selected based on inherent physicochemical properties such as flexibility, surface-exposure/accessibility, hydrophilicity, and antigenicity. More so, predictions of peptide MHC-I/II binding affinities, proteasome C-terminal cleavage and TAP transport efficiency are essential for identifying the most potential T-cell epitopes for MHC-I and II molecules [18,19].
Therefore, our aim in this study is centered on the use of immunoinformatics methodologies to identify highly potential antiviral peptides (B-cell epitopes and T-cell epitopes) to impede the pathogenic process of SARS-CoV-2. We believe findings from this study will contribute vitally to the vaccine development researches relative to COVID-19 treatment.
Methodology
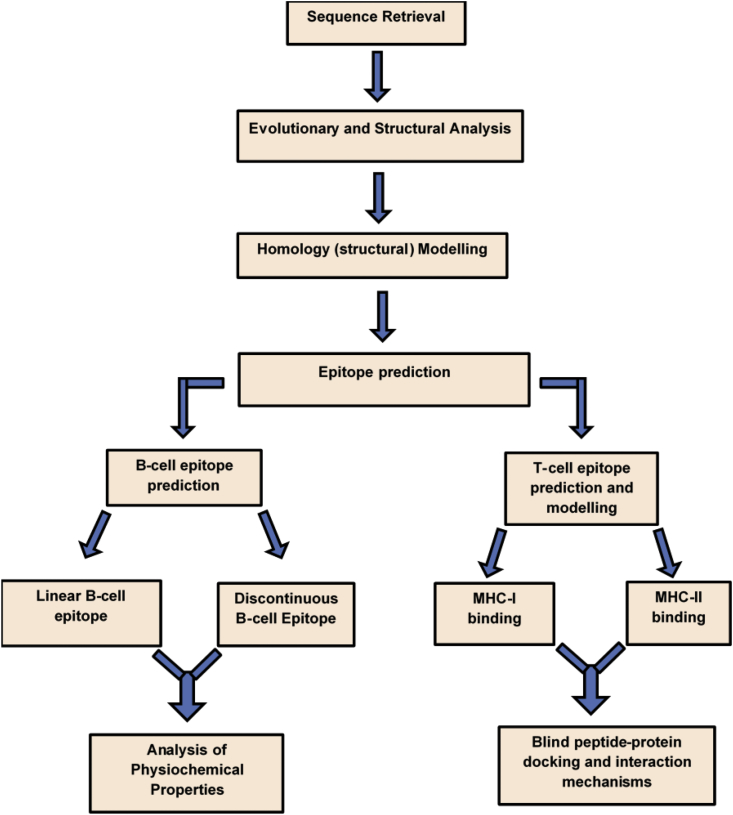
Flowchart presented in Fig. 2 summarizes the paradigmatic approaches employed in this study to identify highly potential B-cell and T-cell epitopes as vaccine candidates for curtailing the pathogenicity of SARS-CoV-2. Incorporated methodologies are subsequently elaborated.
Fig. 2.
Overall workflow of the combinatorial immunoinformatics methodologies implemented in this study.
Protein sequence retrieval
Viral Zone, a database of ExPASy Bioinformatics Resource Portal was utilized to retrieve information such as the host, transmission, ailment, genus, family, genome, and proteome of the virus [20]. The reviewed S-protein sequences of human coronavirus strains (HCOV-229E, HCOV-NL63, HCOV-HKU1, HCOV-EMC, and HCOV-OC43, and the SARS-CoV-2) were obtained as presented in Table 1. The primary sequence of SARS-CoV-2 (QHD43416.1) was obtained from NCBI (National Center for Biotechnology Information) database while the protein sequences of other strains were retrieved from the UniProtKB database in FASTA format for further analysis.
Table 1.
Primary linear sequences of human coronavirus strains used for preliminary studies as obtained from UniProtkb.
| Entry | Entry name | Gene Name | Organism | Length |
|---|---|---|---|---|
| P15423 | SPIKE_CVH22 | S2 | Human coronavirus 229E (HCoV-229E) | 1173 |
| Q14EB0 | SPIKE_CVHN2 | S3 | Human coronavirus HKU1 (isolate N2) (HCoV-HKU1) | 1351 |
| Q0ZME7 | SPIKE_CVHN5 | S3 | Human coronavirus HKU1 (isolate N5) (HCoV-HKU1) | 1351 |
| Q6Q1S2 | SPIKE_CVHNL | S2 | Human coronavirus NL63 (HCoV-NL63) | 1356 |
| Q5MQD0 | SPIKE_CVHN1 | S3 | Human coronavirus HKU1 (isolate N1) (HCoV-HKU1) | 1356 |
| K9N5Q8 | SPIKE_CVEMC | S3 | Middle East respiratory syndrome-related coronavirus (Human coronavirus EMC) | 1353 |
| P36334 | SPIKE_CVHOC | S3 | Human coronavirus OC43 (HCoV-OC43) | 1353 |
| P59594 | SPIKE_CVHSA | S3 | Human SARS coronavirus | 1255 |
| QHD43416.1 | Surface Glycoprotein | S3 | COVID-19 | 1273 |
Evolutionary analysis and structure analysis of protein
The analysis of evolutionary divergence was performed using the Mega7.0 software which was further represented as a Phylogenetic tree. The phylogenetic tree was schemed using a distance of 0.10 with default parameters [21,22]. Furthermore, the selected protein sequence (QHD43416.1) was subjected to secondary and tertiary structural analyses. The secondary structure of the protein was studied using the SOPMA (Self optimized prediction method) algorithm which helped identify the Alpha Helix, Beta Sheet, and coils of the structure [23].
Homology (structural) modelling and validation
Furthermore, the structure of the SARS-CoV-2 spike glycoprotein (QHD43416.1) was modeled using the I-Tasser (Iterative Threading Assembly Refinement) algorithm, which entailed replica-exchange Monte Carlo simulations. This enabled the prediction and modeling of protein structures via an exhaustive search method appropriate for identifying the most matching protein template. Herein, the PDB protein 5 × 58A was identified and used as a template [24] with a z-score of 15.24 indicative of a considerable degree of accuracy. The structural model was validated using the Rampage tool [25].
B-cell epitope prediction
B-cell epitope predictions were performed for constituent linear and discontinuous (conformational) epitopes. The protein sequence (QHD43416.1) was initially subjected to linear B-cell epitope prediction using Ellipro algorithm (combines Thornton's method with a residue clustering algorithm, the MODELLER program, and the JMOL viewer) to identify both the linear and the conformational epitopes where the predictive threshold was set to a minimum of 0.7. In this study, a predictive threshold of 1.000 was set to predict the physicochemical properties of the linear B-cell epitopes. These attributes were defined using the IEDB-integrated Karplus and Schulz flexibility [26], Kolaskar & Tongaonkar Antigenicity [27], Parker Hydrophilicity [28] and Emini Surface accessibility methods [29]. These properties cumulatively account for the immunogenic tendencies of B-cell epitopes [30].
Moreover, the discontinuous epitopes were also predicted from the secondary structures of the antigenic protein based on their protrusion indices (PI), which indicate conformational protrusion. In other words, PI provides a simplistic way of detecting those regions of the protein that bulge from the protein's surface with B-cell recognition potentials. Residues with high protrusion index values are often associated with antigenic sites [31].
Allergenicity of the linear B-cell epitope was evaluated using the Algpred method which integrates support machine vector, motif-based and BLAST-search algorithms, to predict whether or not a particular epitope is an allergen or non-allergen with a reported accuracy of 85%. This makes Algpred tool an exceptionally valuable tool for cross-reactivity prediction of allergens [32].
T-cell epitope prediction and modelling
CD8+ cell epitope prediction
The NetCTL 1.2 method was used for the identification of the CD8+ T cell epitopes from the antigenic S-protein. This tool functions using multiple combinatorial methods such as SNN (Simulated Neural Network), weight matrix and an Artificial Neural Network (ANN). NetCTL prediction method integrates the peptide major histocompatibility complex class I (MHC-I) binding, proteosomal C-terminal cleavage, and TAP transport efficiency. The respective parameters employed for this analysis were set at threshold 0.9 to enhance sensitivity and specificity [19]. This allowed us to identify more potential epitopes for further analysis. A combined algorithm of MHC–I binding, TAP transport efficiency, and proteasomal cleavage efficiency was selected to predict overall scores [17,33]. These describe the crucial stages of the antigenic presentation pathway.
Overall, we performed HLA-T-cell epitope binding prediction for MHC-1 molecules (Human Leukocyte Antigens; HLA-A∗02:01, HLA-B∗35:01 and HLA-B∗51:01) which were selected based on their high global frequency [34]. The most probable potential ligands for these MHC-I molecules were identified (<-E) and presented accordingly.
3-dimensional (3D) structures of the respective HLAs were retrieved from RCSB PDB with the following entries: HLA-A∗02:01 (PDB ID: 3UTQ), HLA-B∗35:01 (PDB ID: 4LNR), HLA-B∗51:01 (PDB ID: 1E28) and HLA-DRB1∗15:01 (1BX2) [[35], [36], [37], [38]].
CD4+ T-cell epitopes identification
CD4+ T-cell epitope prediction was carried out using NetMHC II 2.3; a method that incorporates Artificial Neural Network (ANN) algorithm for binding core and affinity predictions. The parameters employed for NetMHC-II.2.3 were set at a threshold value of 0.7 to maintain high sensitivity and specificity value. T-cell epitope binding prediction was performed for MHC-II molecule; HLA DRB∗15:01 [39]. The crystal structure for HLA-DRB∗15:01 was obtained from PDB with ID 1BX2, for peptide-protein docking studies [40].
Conformational modeling of predicted T-Cell epitopes
Furthermore, the most probable T-cell epitopes (9-mer) were identified and their corresponding 3D structures were modeled using the PEPFOLD3 algorithm. The prediction method utilized a simulation run of 200ns in addition to a sOPEP energy function, which enabled the sampling of multiple conformations predicted [41].
Blind peptide-protein docking and interaction analysis
The pep-ATTRACT method was further utilized to model interactions between the predicted peptides (T-cell epitopes) and HLA molecules using a blind docking approach. This method performs a rigid body global search on the surface of the target protein and also identifies the most appropriate sites for binding [42]. This was more suitable to determine the most preferential binding regions for the epitopes on HLA-A∗02:01, HLA-B∗35:01, HLA-B∗51:01 and HLA-DRB∗15:01. The best protein-peptide complexes were ranked based on global energy scores [43] and the docking results are presented accordingly.
Results
Sequence retrieval and phylogenetic analysis
Amino acid sequences for the SARS-CoV-2 S-protein was retrieved from the NCBI database with entry QHD43416.1, in addition to the primary sequences of other coronavirus strains [Table 1].
Furthermore, the phylogenetic analysis revealed disparities between SARS-CoV-2 and other coronavirus strains throughout evolution. Sequences of the respective spike proteins were mapped out across the selected coronavirus strains and depicted as a phylogenetic tree (Supplementary Figure S1). As shown, results highlighted the close relativity between SARS coronavirus (SARS-CoV) and SARS-CoV-2.
In addition, the secondary structure of the SARS-CoV-2 consists of 1273 amino acids and as estimated, 364 amino acids (28.59%) of the protein were helical, the extended β strand comprises 296 amino acids (23.25%) while 570 amino acids (44.78%) constituted the random coil region of the protein (Supplementary Figure S2).
Structural modelling and validation
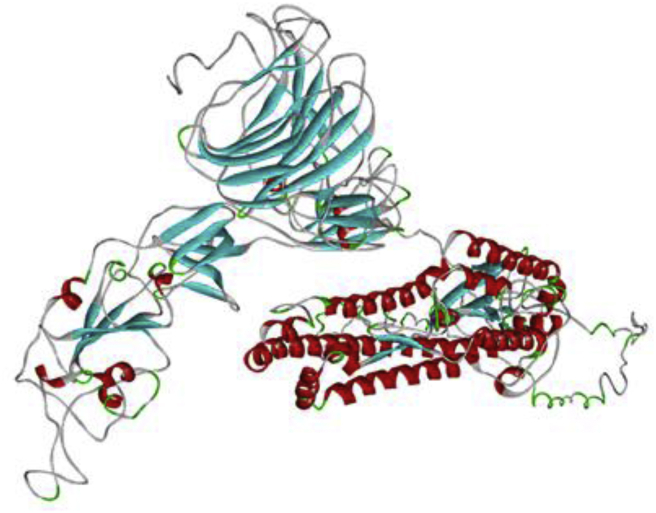
The selection of the 3D structure was based on the obtained C-Scores (confidence score), which is in the normal range of (−5 → +2) [44]. Accordingly, the model with the highest C-score (−1.52) was selected [Fig. 3].
Fig. 3.
Homology 3D model of SARS-CoV-2 S-glycoprotein.
Also, about 1043 residues were located in the favoured region (82.1%) while those in the allowed numbered up to 195 (15.3%) with about 2.6% (33 residues) constituting the outliers. Taken together, a considerable degree of correctness can be presumed for our model since about 97.7% of residues of the predicted model lie within the favoured and allowed regions.
B-cell epitope predictions
Linear B-cell epitope
Five most probable linear epitopes were selected based on their scores relative to the set predictive threshold of 1 as presented in Table 2. Results revealed a 22mer (LALHRSYLTPGDSSSGWTAGAA242-263) peptide as the most potential B-cell epitope with a score of 0.865. Predicted epitopes were further examined based on their physicochemical attributes which underlie their immunogenicity.
Table 2.
Predicted linear B-cell epitopes of COVID-19 S-protein and classification based on physicochemical attributes.
| Physiochemical Properties | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S/No | Sequences | Start → End | Hydrophilicity |
Surface Flexibility |
Surface Accessibility |
Antigenicity |
Allergenicity | ||||
| Epitope | Score (max/min) | Epitope | Score (max/min) | Epitope | Score (max/min) | Epitope | Score (max/min) | ||||
| 1 | LALHRSYLTPGDSSSGWTAGAA | 242 → 263 | PGDSSSG | 6.143 | PGDSSSG | 1.125 | HRSYLTP | 1.956 | LALHRSY | 1.102 | Non allergen |
| ALHRSYL | −0.771 | LALHRSY | 0.964 | GWTAGAA | 0.291 | GWTAGAA | 0.963 | ||||
| 2 | HAIHVSGTNGTKRFD | 66 → 80 | SGTNGTK | 5.857 | SGTNGTK | 1.105 | NGTKRFD | 2.052 | HAIHVSG | 1.099 | Allergen |
| HAIHVSG | 0.971 | HAIHVSG | 0.936 | HAIHVSG | 0.204 | GTNGTKR | 0.878 | ||||
| 3 | VSQPFLMDLEGKQGNFKN | 171 → 188 | DLEGKQG | 4.529 | EGKQGNF | 1.102 | KQGNFKN | 2.496 | VSQPFLM | 1.092 | Allergen |
| QPFLMDL | −1.957 | PFLMDLE | 0.93 | FLMDLEG | 0.271 | KQGNFKN | 0.913 | ||||
| 4 | MFVFLVLLPLVSSQCVNLTTRTQLPP | 1 → 26 | NLTTRTQ | 3.371 | TTRTQLP | 1.044 | TRTQLPP | 3.954 | LVLLPLV | 1.261 | Non allergen |
| FVFLVLL | −7.629 | MFVFLVL | 0.917 | FVFLVLL | 0.066 | NLTTRTQ | 0.949 | ||||
| 5 | TTAPAICHDGKAHFP | 1076 → 1090 | CHDGKAH | 4.157 | CHDGKAH | 1.043 | DGKAHFP | 2.357 | TAPAICH | 1.11 | Allergen |
| TAPAICH | 1.0 | APAICHD | 0.937 | TAPAICH | 0.451 | DGKAHFP | 0.999 | ||||
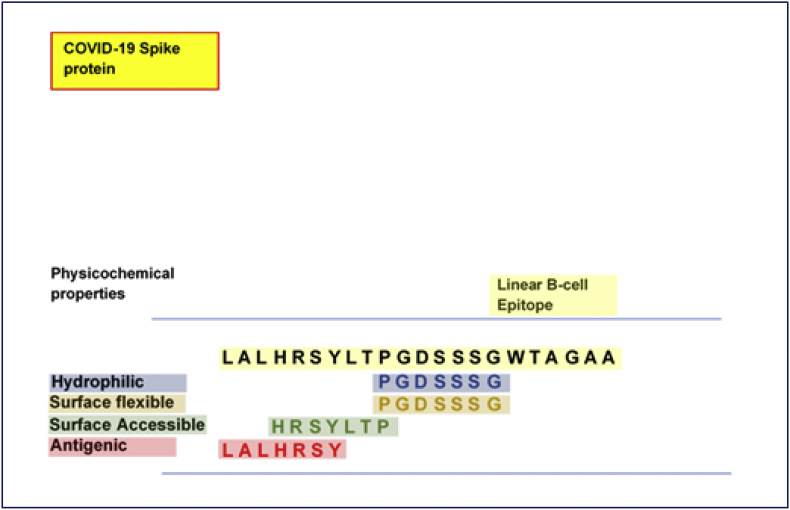
Physicochemical analyses revealed the core PGDSSSG251-257 region to be highly hydrophilic (max score = 6.143) with considerable surface flexibility (max score = 1.125). Moreover, HRSYLTP245-251 was identified for its surface accessibility (max score = 1.956) while regions LALHRSY242-248 were antigenic (max score = 1.102). Prediction of allergenicity also revealed that two among the five probable B-cell epitopes were non-allergenic. We further identified sequence overlaps based on their inherent attributes and how their cumulatively present LALHRSYLTPGDSSSGWTAGAA242-263 as the most potential linear B-cell epitope. This is diagrammatically presented in Fig. 4.
Fig. 4.
Sequential overlapping and analyses of the most probable linear B-cell epitope with characteristic physicochemical properties.
Discontinuous epitope
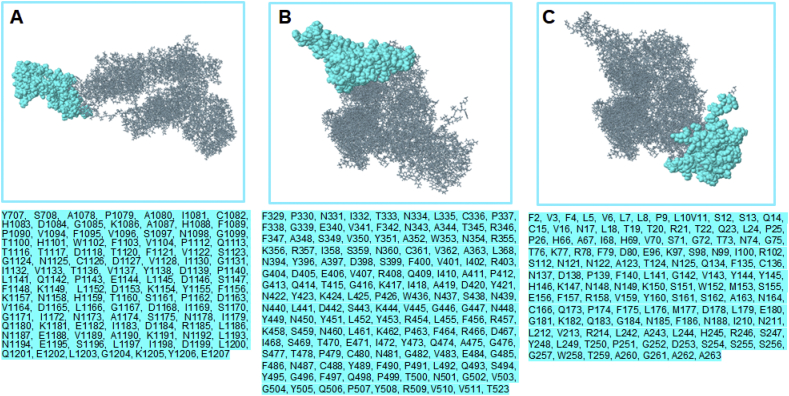
As earlier stated, the 3-D structure of the antigenic spike protein was employed to predict conformational or discontinuous (non-linear) epitopes. Based on PI, predicted non-linear epitopes are represented in Table 3 while their respective positions on the 3-D structure are shown in Fig. 5.
Table 3.
Predicted discontinuous epitopes from COVID-19 S-protein. Overlapping sequence of the most probable linear B-cell epitope are highlighted in red.
| No. | Residues | Number of residues | Scores |
|---|---|---|---|
| 1 | Y707, S708, A1078, P1079, A1080, I1081, C1082, H1083, D1084, G1085, K1086, A1087, H1088, F1089, P1090, V1094, F1095, V1096, S1097, N1098, G1099, T1100, H1101, W1102, F1103, V1104, P1112, Q1113, T1116, T1117, D1118, T1120, F1121, V1122, S1123, G1124, N1125, C1126, D1127, V1128, I1130, G1131, I1132, V1133, T1136, V1137, Y1138, D1139, P1140, L1141, Q1142, P1143, E1144, L1145, D1146, S1147, F1148, K1149, L1152, D1153, K1154, Y1155, F1156, K1157, N1158, H1159, T1160, S1161, P1162, D1163, V1164, D1165, L1166, G1167, D1168, I1169, S1170, G1171, I1172, N1173, A1174, S1175, N1178, I1179, Q1180, K1181, E1182, I1183, D1184, R1185, L1186, N1187, E1188, V1189, A1190, K1191, N1192, L1193, N1194, E1195, S1196, L1197, I1198, D1199, L1200, Q1201, E1202, L1203, G1204, K1205, Y1206, E1207 | 111 | 0.861 |
| 2 | F329, P330, N331, I332, T333, N334, L335, C336, P337, F338, G339, E340, V341, F342, N343, A344, T345, R346, F347, A348, S349, V350, Y351, A352, W353, N354, R355, K356, R357, I358, S359, N360, C361, V362, A363, L368, N394, Y396, A397, D398, S399, F400, V401, I402, R403, G404, D405, E406, V407, R408, Q409, I410, A411, P412, G413, Q414, T415, G416, K417, I418, A419, D420, Y421, N422, Y423, K424, L425, P426, W436, N437, S438, N439, N440, L441, D442, S443, K444, V445, G446, G447, N448, Y449, N450, Y451, L452, Y453, R454, L455, F456, R457, K458, S459, N460, L461, K462, P463, F464, R466, D467, I468, S469, T470, E471, I472, Y473, Q474, A475, G476, S477, T478, P479, C480, N481, G482, V483, E484, G485, F486, N487, C488, Y489, F490, P491, L492, Q493, S494, Y495, G496, F497, Q498, P499, T500, N501, G502, V503, G504, Y505, Q506, P507, Y508, R509, V510, V511, T523 | 144 | 0.842 |
| 3 | F2, V3, F4, L5, V6, L7, L8, P9, L10, V11, S12, S13, Q14, C15, V16, N17, L18, T19, T20, R21, T22, Q23, L24, P25, P26, H66, A67, I68, H69, V70, S71, G72, T73, N74, G75, T76, K77, R78, F79, D80, E96, K97, S98, N99, I100, R102, S112, N121, N122, A123, T124, N125, Q134, F135, C136, N137, D138, P139, F140, L141, G142, V143, Y144, Y145, H146, K147, N148, N149, K150, S151, W152, M153, S155, E156, F157, R158, V159, Y160, S161, S162, A163, N164, C166, Q173, P174, F175, L176, M177, D178, L179, E180, G181, K182, Q183, G184, N185, F186, N188, I210, N211, L212, V213, R214, L242, A243, L244, H245, R246, S247, Y248, L249, T250, P251, G252, D253, S254, S255, S256, G257, W258, T259, A260, G261, A262, A263 | 125 | 0.806 |
Fig. 5.
Structural analysis of conformational or discontinuous B-cell epitopes. The locations of the respective epitopes (surface representation) are shown on the 3D structure of COVID-19 S-protein. Corresponding amino acid sequences, as predicted, are also shown (cyan highlights).
Predictions of high-affinity T-cell epitopes and de-novo structural modeling
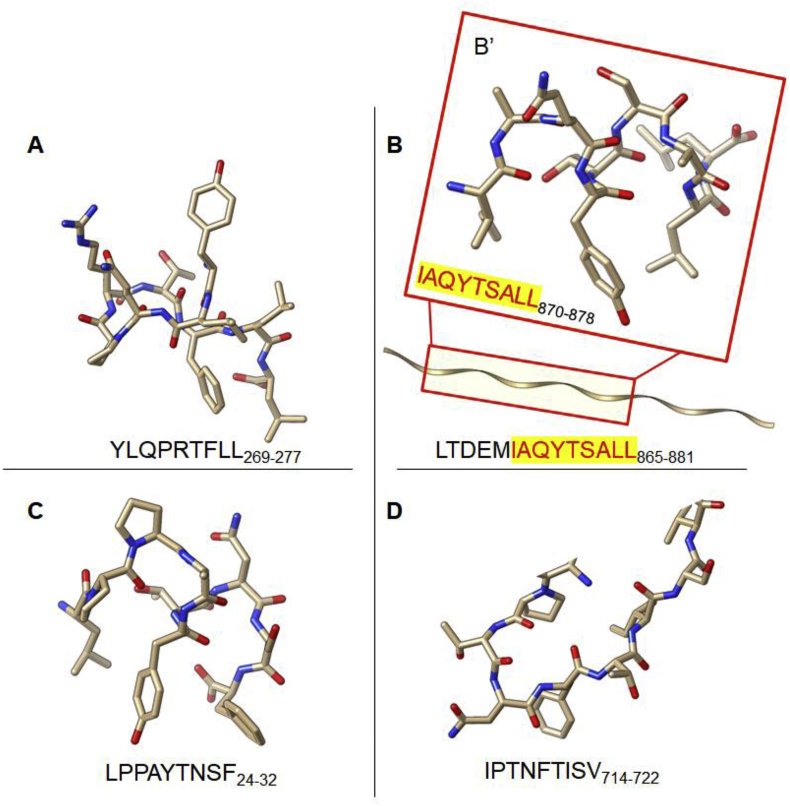
The T-cell epitopes were predicted using the NetCTL-I and NetMHC-II to identify potential T-cell epitopes that interact with HLAs of MHC classes I and II respectively. For our study, we have randomly selected the most frequent HLAs from MHC-I (HLA-A∗02:01, HLA-B∗35:01, HLA-B∗51:01) and MHC-II (HLA-DRB∗15:01). Three most probable T-cell epitopes were selected (<-E) for each of the HLA molecules as presented in Table 5. As shown, YLQPRTFLL269-277 exhibited the highest binding affinity with a score of 0.8882, coupled with a relatively high score for proteasomal C-terminal cleavage and transport affinity. Likewise, for HLA-B∗35:01, binding affinity was highest for LPPAYTNSF 24-32 with a score of 0.6566 while IPTNFTISV714-722 had the highest binding affinity for HLA-B∗51:01 as predicted [Table 4]. For the class II HLA DRB∗15:01, LTDEMIAQYTSALLA demonstrated the highest binding affinity with a score of 8.3 nM. However, it is important to note that the three predicted T-cell epitopes had a uniform core peptide (9mer); IAQYTSALL870-878 that interacted at the binding site of the target HLA DRB∗15:01.
Table 5.
Binding energy estimations for the MHC-epitope complexes. Regions involved in high-affinity interactions are highlighted in yellow.
| Antigenic Proteins | MHC class | Supertypes | Allele | PDB ID | Potential T-cell Epitope | pep-ATTRACT Global energy score (kcal mol −1) |
|---|---|---|---|---|---|---|
| Covid-19 Spike Protein | MHC-I | A2 | HLA-A∗02:01 | 3UTQ | YLQPRTFLL | −16.78 |
| B7 | HLA-B∗35:01 | 4LNR | LPPAYTNSF | −19.09 | ||
| HLA-B∗51:01 | 1E28 | IPTNFTISV | −20.28 | |||
| MHC-II | DR-B1 | HLA-DRB1∗15:01 | 1BX2 | LTDEMIAQYTSALLA | −17.21 |
Table 4.
Prediction of antigenic processing and presentation for potential T-cell epitopes of COVID-19 S-protein.
| Antigenic Protein | MHC Type | Supertypes | Allele | Peptide | Binding Affinity | Rescale Binding Affinity | Proteasomal C-terminal Cleavage | Transport Affinity | Prediction Score | MHC-1 binding |
|---|---|---|---|---|---|---|---|---|---|---|
| Covid-19 Spike Protein | MHC-I | A2 | HLA-A∗02:01 | YLQPRTFLL | 0.8882 | 1.3240 | 0.9774 | 0.8920 | 1.5152 | <-E |
| RLQSLQTYV | 0.7611 | 1.1346 | 0.7484 | 0.5160 | 1.2727 | <-E | ||||
| FIAGLIAIV | 0.7841 | 1.1688 | 0.1792 | 0.3350 | 1.2124 | <-E | ||||
| B7 |
HLA-B∗35:01 | WPWYIWLGF | 0.5309 | 1.0241 | 0.8493 | 2.4760 | 1.2753 | <-E | ||
| LPPAYTNSF | 0.6566 | 1.2666 | 0.9581 | 2.1700 | 1.5189 | <-E | ||||
| MIAQYTSAL | 0.5608 | 1.0820 | 0.9295 | 0.9800 | 1.2704 | <-E | ||||
| HLA-B∗51:01 |
IPTNFTISV | 0.7198 | 1.3887 | 0.9763 | 0.1510 | 1.5427 | <-E | |||
| GPKKSTNLV | 0.5662 | 1.0924 | 0.9405 | 0.1780 | 1.2245 | <-E | ||||
| LPFNDGVYF |
0.4026 |
0.7767 |
0.9761 |
2.3930 |
1.0427 |
<-E | ||||
| MHC-II | Supertype |
Allele |
Peptide |
Core |
1-log 50k (aff) |
Affinity (nM) |
% Rank |
Bind Level |
||
| DR-B1 | HLA-DR B1∗15:01 | DEMIAQYTSALLAGT | IAQYTSALL | 0.8125 | 7.6 | 0.15 | SB | |||
| TDEMIAQYTSALLAG | IAQYTSALL | 0.8104 | 7.8 | 0.15 | SB | |||||
| LTDEMIAQYTSALLA | IAQYTSALL | 0.8049 | 8.3 | 0.20 | SB |
Abbreviation: SB: Strong binding.
3D structures of the selected T-cell epitopes as modeled by the PEP-FOLD3 server are presented in Fig. 6. The interacting core region (9mer) of the 15mer T cell epitope of MHC-II DRB∗15:01 was also modeled.
Fig. 6.
3D structural model of the predicted T-cell epitopes for (A) HLA-A∗02:01 (B) HLA-DRB1∗15:01 (C) HLA-B∗35:01 (D) HLA-B∗51:01.
HLA-docking analysis of potential T cell epitopes and interaction mechanisms
A blind docking approach was employed to investigate the mechanisms of interactions between the predicted T-cell epitopes and selected HLAs of MHC classes I and II. This was an important method since it was suitable to identify regions on the HLA molecules where the epitopes would preferentially bind based on affinity and complementarity.
Hence, the pepATTRACT method was sufficient to identify the most appropriate binding regions on HLA-A∗02:01, HLA-B∗35:01, HLA-B∗51:01 and HLA-DRB1∗15:01 for the respective epitopes. For each peptide-protein complex, 51 clusters were obtained while global energy scoring was used to select the best-docked complexes [Table 5], with structures shown in Fig. 6.
A global energy score of −16.78 kcal mol−1 was estimated for T-cell epitope YLQPRTFLL269-277 when bound to HLA-A∗02:01 while LPPAYTNSF24-32 and IPTNFTISV714-721 complexes with HLA-B∗35:01 and HLA-B∗51:01 had energy values of −21.94 kcal mol−1 and -20.28 kcal mol−1 respectively. For MHC-II, HLA-DRB1∗15:01, while the 15mer T-cell epitope (LTDEMIAQYTSALLA865-881) had a binding energy value of −16.8035 kcal mol−1.
We also enumerated energies associated with interactions between the 9mer core region (IAQYTSALL870-878) for the predicted HLA-DRB1∗15:01 epitope. Findings revealed that the 9mer (IAQYTSALL870-878) had an energy value of −17.21 kcal mol−1, further pinpointing it as the region that majorly mediated complementary interactions at the binding region of HLA-DRB1∗15:01.
Structural analysis of the respective epitope-HLA complexes revealed the most preferential binding regions relative to the crystallized structures.
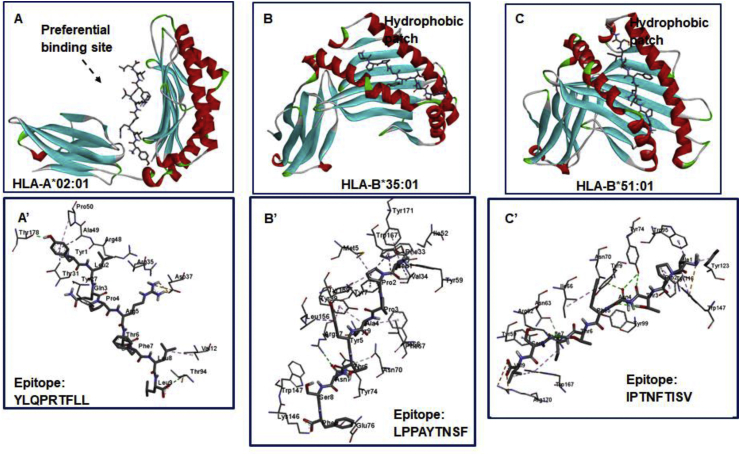
As observed, high-affinity interactions mediated by YLQPRTFLL269-277 occurred at a site adjacent to the primary pocket of HLA-A∗02:01 defined by x-ray crystallization in a previous study by Bulek et al. [35] This could depict a previously undefined high-affinity pocket on HLA-A2 supertypes for peptide binding. At this identified site, high-affinity hydrogen and attractive charge (salt bridge) interactions were observed. H-bonds occurred between A∗02:01Thr94 (→ Leu277), A∗02:01Thr178 (→ Tyr269) while attractive charge interactions were mediated by A∗02:01Asp37 (→ Arg273) [Fig. 7]. Other important interactions that could contribute to the stability of this epitope in this region are ring–ring (π-aromatic interactions) which occurred at A∗02:01Tyr27 (→ Pro272) and A∗02:01Pro50 (→ Tyr269).
Fig. 7.
Predicted MHC binding regions and interactions mechanisms with bound T-cell epitopes.
For HLA-B∗35:01 and HLA-B∗51:01, T cell epitopes; LPPAYTNSF24-32 and IPTNFTISV 714-722 were bound preferentially to the hydrophobic patches similar to the ones experimentally identified by X-ray crystallography in previous studies by Yanaka et al., [37] and Pieper et al. [38] This further validates the predictive ability of the blind-docking approach employed for modeling peptide-protein interactions.
In the LPPAYTNSF24-32 – HLA-B∗35:01 complex, interactions at the hydrophobic pocket was strengthened and stabilized by hydrogen interactions observed among B∗35:01Lys146 (→ Phe32), B∗35:01Arg97 (→ Asn30) and B∗35:01Asn70 (→ Thr29). Important aromatic interactions were also observed among B∗35:01Tyr99 (→ Tyr28), B∗35:01Tyr159 (→ Pro25) and B∗35:01Trp167 (→ Pro25).
Moreover, important hydrogen interactions that contributed to the high-affinity binding of IPTNFTISV714-722 to HLA-B∗51:01 were mediated by B∗51:01Tyr74 and B∗51:01Arg62 with Thr716/Asn717 and Ile720 respectively. Strong attractive charges were also mediated by B∗51:01Arg170 with Val722.
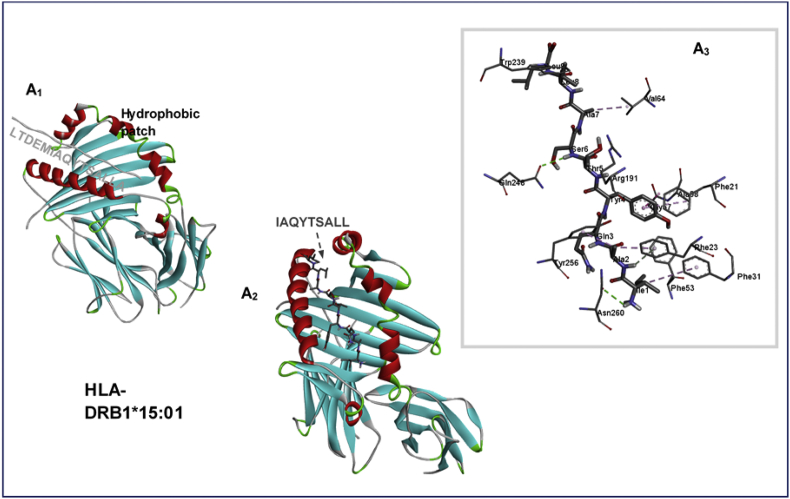
A closer look at the interaction mechanisms of LTDEMIAQYTSALLA865-881 at the hydrophobic patch of MHC-II HLA-DRB1∗15:01 revealed that while the starting LTDEM865-869 region of the 15mer epitope was more extended into the surrounding surface, the remainder core region IAQYTSALLA870-879 was buried in the hydrophobic pocket.
This was further characterized by the occurrence of high-affinity interactions between the pocket residues and the core 9mer IAQYTSALL870-878 epitope. Consequentially strong H-bond interactions were observed among DRB1∗15:01Gln248 (→ Ser876) and DRB1∗15:01Asn260 (→ Ile870) as showed in Fig. 8.
Fig. 8.
Binding site and interaction analysis of HLA-DRB1∗15:01 and LTDEMIAQYTSALLA865-881 epitope. Complementary interaction mediated by the core IAQYTSALLA870-879 epitope region is also shown.
Discussion
The need for novel and highly effective treatments to evade SARS-CoV-2 virulence is highly urgent to help curtail the global pandemic [45]. Although the information on its treatment and management are still elusive, remdesivir and chloroquine are currently being tested for their efficacies since they are most likely to interfere with viral entry and replication in host cells [46].
Vaccines are important treatment modalities that can stimulate immunogenic responses against foreign antigens of the virus in the course of its pathogenesis. Since information on the cellular components of the novel coronavirus is available, the design of highly effective peptide or subunit vaccines is achievable, hence the importance of implementing immunoinformatics methods for predicting highly potential viral T-cell and B-cell epitopes [47]. This approach has been previously used to identify potential T-cell and B-cell epitopes for peptide design against the Zika virus [48], Dengue [49], Chikungunya [50], EBV [51], Ebola Virus [52] and HIV-1 [53,54].
Viral molecules are antigenic in nature, hence during host infection, they are able to drive protective responses, which could in turn lead to the death of infected cells. These immunogenic responses are mediated by B- and T-cells, which via their receptors, identify components of the virus (such as viral proteins) and activate the corresponding cascade of defense to curtail the viral spread. For instance, T-cell receptors (TCRs) require the antigenic presentation pathway which involves antigen-presenting cells (APC) and major histocompatibility complex (MHC) molecules I (MHC-I) and II (MHC-II). While the former is recognized by the cytotoxic CD8+ cells, the helper T cells (CD4+) recognizes the MHC-II molecules [18,35,[55], [56], [57], [58]]. On the other hand, surface-exposed protein antigens are recognized and bound by B cell receptors (BCRs) which in turn activates the humoral adaptive responses.
Proteomic studies on the components of SARS-CoV-2 have revealed various antigenic proteins that perform diverse roles that are crucial to the infectious viral cycle; from viral entry to replication. These components include the spike glycoprotein (S), nucleocapsid, envelope protein, membrane protein, and hemagglutinin-esterase dimer protein (HE). Crucial to viral pathogenesis is the spike glycoprotein which serves as the first point of call for viral entry and attachment to host cells [8,59]. This underlies our rationale and implementation of vaccinomics techniques as performed in this study, complementary to other available data in this regard.
Characteristic epitopic attributes such as antigenicity, hydrophilicity, surface-exposure, surface accessibility among others are essential for B-cell receptor binding and recognition which is essential for provoking B-cell mediated immune responses. These factors were therefore considered for predicting potential linear B-cells epitopes for SARS-CoV-2 S-glycoprotein.
From our findings, B-cell epitopes; LALHRSYLTPGDSSSGWTAGAA242-263, HAIHVSGTNGTKRFD66-80, VSQPFLMDLEGKQGNFKN171-188, MFVFLVLLPLVSSQCVNLTTRTQLPP1-26, and TTAPAICHDGKAHFP1076-1090 were identified for their potentials in eliciting B-cell responses. Amongst all, LALHRSYLTPGDSSSGWTAGAA242-263 demonstrated the highest propensity based on multiple inherent attributes (hydrophilicity, surface-exposure/accessibility, antigenicity, and flexibility) predicted, which are peculiar to B-cell epitopes. Surface-exposure was also an important attribute common to predicted discontinuous/conformational epitopes, which interestingly overlapped with the linear epitope further validating its potentials as a B-cell epitope [Fig. 4/Table 3]. Noteworthy, the majority of residues that constitute the predicted epitopes are hydrophilic with large and aromatic side chains corroborative of the predicted surface-accessibility and immunogenicity [60].
The prediction of T-cell epitopes was further employed to identify 9-mer peptides that are antigenic with innate ability to initiate the activation of CD8 T-cells. Herein, we investigated epitope binding to human MHC-I and MHC-II molecules that are globally frequent; HLA-A∗02:01, HLA-B∗35:01, HLA-B∗51:01, and HLA-DR B1∗15:01 [61]. More so, we enumerated other crucial steps of the antigenic presentation pathway which include TAP processing and C-terminal cleavage for our T-cell epitope prediction. From our findings, YLQPRTFLL269-277 was predicted as the most probable epitope for HLA-A∗02:01 while LPPAYTNSF24-32 was predicted as a high-affinity binder for HLA-B∗35:01, and IPTNFTISV714-721 for HLA-B∗51:01. These were selected based on their high predictive scores for TAP processing and C-terminal cleavage, which are important factors for antigenic presentation.
For MHC-II HLA-DRB1∗15:01, the most potential CD4 T-cell epitope was LTDEMIAQYTSALLA865-881, which presumably mediated its strong affinity binding with its core region; IAQYTSALL. As estimated, their predictive scores were considerably high relative to the 0.9 threshold employed.
It was further important to investigate the mechanisms by which these T-cell epitopes bind to the respective MHC-I and MHC-II molecules, providing primary details into the mechanistic stimulation of CD4 and CD8 T cells by SARS-CoV-2 S protein. Moreover, while binding sites have been previously identified on the target MHC molecules (HLA-A∗02:01, HLA-B∗35:01, HLA-B∗51:01 and HLA-DR B1∗15:01) by X-ray crystallography, it was pertinent to define novel high-affinity sites for epitope binding. This could provide structural details not only for peptide-vaccine design, but also for therapeutic peptides and drug molecules which can to stimulate T cell immunogenic responses.
To this effect, we implemented a blind peptide-docking approach wherein existing binding site information was not considered in the course of complex preparation. Rather, the most probable epitopes predicted were allowed to attach preferentially, without restraints, to sites on the MHC molecules.
Our findings revealed that the S protein T cell epitope YLQPRTFLL269-277 was preferentially bound to a novel site on HLA-A∗02:01, which is adjacent to a previously characterized site. This could represent a novel site for the design of therapeutic T-cell stimulants for HLA-A supertypes relative to impeding SARS-CoV-2 virulence. Further analyses of interaction mechanisms revealed the roles Leu277, Tyr269, Arg273, and Pro272 in stabilizing the epitope at the high-affinity site.
However, epitope binding to the HLA-B molecules occurred at the same binding cleft that has been previously defined in studies by Yanaka et al., [37] and Pieper et al., [38] which further validate the correctness of the blind-peptide docking approach employed.
In HLA-B∗35:01, the binding and stability of the predicted epitope LPPAYTNSF24-32 was enhanced by Phe32, Asn30, Thr29, Tyr28, and Pro25. Also, Thr716, Asn717, Ile720, and Val722 played crucial roles in the affinity binding of IPTNFTISV714-722 to HLA-B∗51:01.
Moreover, while the 15mer MHC-II epitope, LTDEMIAQYTSALLA865-881 was bound to a previously defined pocket in HLA-DRB1∗15:01, important binding interactions were mediated by its core region consisting of IAQYTSALLA870-879, which was buried in the hydrophobic pocket. Taken together, the innate attributes associated with B- and T-cells predicted in this study present them as strong candidates for the design of COVID-19 peptide-vaccines.
Conclusion
Viral Infections by SARS-CoV-2 have translated into a global pandemic since it emerged from the Wuhan region of China in 2019. Ever since, numerous efforts have been put in place to discover effective treatment regimen for curtailing its spread across our national borders. While drug molecules have been integral to several disease treatment, the efficacies of peptide vaccines in pathogenic infections cannot be over-emphasized since it elicits its functionalities by interacting with receptors of B- and T-cells whilst triggering immunogenic responses. More so, peptide vaccines are designed from cellular components of the infectious organisms, hence the ability to trigger immune responses when detected.
Although, no effective treatment option has been discovered, we propose the viability of a peptide vaccine designed from B- and T-cell epitopes derived from the viral spike (S) protein.
In this study, we implemented multiple algorithms to identify highly probable B- and T-cell epitopes for antigenic SARS-CoV-2 S-protein which is crucial for attachment and entry into host cells. Linear and discontinuous (non-linear) epitopes were ranked and predicted using multiple algorithms from the IEDB, which carried out its selection based on the inherent physicochemical attributes of the epitopes. Accordingly, flexibility, surface accessibility/exposure, hydrophilicity and antigenicity were considered for B-cell epitope prediction.
Most probable CD4 and CD8 T-cell epitopes were also predicted, particularly their binding propensities to MHC-I and MHC-II molecules of the HLA-A, HLA-B and HLA-DRB1 supertypes. These predictions were as well performed by taking into consideration the antigenic presentation pathways.
Using a blind peptide docking approach, a novel site was identified for the selective binding of S-protein T-cell epitope on HLA-A∗02:01 while binding sites identified for high-affinity interactions on HLA-B∗35:01, HLA-B∗51:01 and HLA-DR B1∗15:01 have been previously resolved. Binding analyses revealed that complementary interactions were favourable and could account for strong stimulatory interactions.
Findings from this study indicate that B- and T-cells predicted in this study are highly probable which presents them as viable candidates for developing peptide-vaccines relative to COVID-19 treatment.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgement
The authors thank the School of Health Sciences, University of KwaZulu-Natal for infrastructural support.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2021.05.001.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Chafekar A., Fielding B.C. MERS-CoV: Understanding the latest human coronavirus threat. Viruses. 2018;10:93. doi: 10.3390/v10020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China — key questions for impact assessment. N Engl J Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 5.Owusu M., Annan A., Corman V.M., Larbi R., Anti P., Drexler J.F. Human coronaviruses associated with upper respiratory tract infections in three rural areas of Ghana. PloS One. 2014;9:e99782. doi: 10.1371/journal.pone.0099782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim Y., Ng Y., Tam J., Liu D. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microb Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang H.W., Egberink H.F., Halpin R., Spiro D.J., Rottie P.J. Spike protein fusion peptide and feline coronavirus virulence. Emerg Infect Dis. 2012;18:1089–1995. doi: 10.3201/eid1807.120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J Alsaadi E.A., Jones I.M. Membrane binding proteins of coronaviruses. Future Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riou J., Althaus C.L. Pattern of early human-to-human transmission of Wuhan 2019-nCoV. BioRxiv. 2020 doi: 10.2807/1560-7917.ES.2020.25.4.2000058. https://www.biorxiv.org/content/10.1101/2020.01.23.917351v1 2020.01.23.917351 [Preprint] [cited 2020 Jan 23]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malonis R.J., Lai J.R., Vergnolle O. Peptide-based vaccines: current progress and future challenges. Chem Rev. 2020;120:3210–3229. doi: 10.1021/acs.chemrev.9b00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patronov A., Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3:120139. doi: 10.1098/rsob.120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poland G.A., Ovsyannikova I.G., Kennedy R.B., Haralambieva I.H., Jacobson R.M. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. OMICS. 2011;15:625–636. doi: 10.1089/omi.2011.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Backert L., Kohlbacher O. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med. 2015;7:119. doi: 10.1186/s13073-015-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Trincado J.L., Gomez-Perosanz M., Reche P.A. Fundamentals and methods for T- and B-cell epitope prediction. J Immunol Res. 2017;2017:2680160. doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hulo C., de Castro E., Masson P., Bougueleret L., Bairoch A., Xenarios I. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res. 2011;39:D576–D582. doi: 10.1093/nar/gkq901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horiike T. An introduction to molecular phylogenetic analysis. Rob Auton Syst. 2016;4:36–45. [Google Scholar]
- 22.Dubey A.K., Yadav S., Kumar M., Singh V.K., Sarangi B.K., Yadav D. In silico characterization of pectate lyase protein sequences from different source organisms. Enzym Res. 2010;2010:950230. doi: 10.4061/2010/950230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geourjon C., Deléage G. Sopma: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 24.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karplus P.A., Schulz G.E. Prediction of chain flexibility in proteins - a tool for the selection of peptide antigens. Naturwissenschaften. 1985;72:212–213. [Google Scholar]
- 27.Kolaskar A.S., Tongaonkar P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 28.Parker J.M., Guo D., Hodges R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25:5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 29.Emini E.A., Hughes J.V., Perlow D.S., Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Wu W., Negre N.N., White K.P., Li C., Shah P.K. Determinants of antigenicity and specificity in immune response for protein sequences. BMC Bioinform. 2011;12:251. doi: 10.1186/1471-2105-12-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponomarenko J., Bui H.H., Li W., Fusseder N., Bourne P.E., Sette A. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha S., Raghava G.P.S. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34:W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi J., Zhang J., Li S., Sun J., Teng Y., Wu M. Epitope-based vaccine target screening against highly pathogenic MERS-CoV: an in Silico approach applied to emerging infectious diseases. PloS One. 2015;10:e0144475. doi: 10.1371/journal.pone.0144475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardi M.S., Jarduli L.R., Jorge A.J., Camargo R.B., Carneiro F.P., Gelinski J.R. HLA-A, B and DRB1 allele and haplotype frequencies in volunteer bone marrow donors from the north of Parana State. 2012;34:25–30. doi: 10.5581/1516-8484.20120010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bulek A.M., Cole D.K., Skowera A., Dolton G., Gras S., Madura F. Structural basis for the killing of human beta cells by CD8 + T cells in type 1 diabetes. Nat Immunol. 2012;13:283–289. doi: 10.1038/ni.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maenaka K., Maenaka T., Tomiyama H., Takiguchi M., Stuart D.I., Jones E.Y. Nonstandard peptide binding revealed by crystal structures of HLA-B∗5101 complexed with HIV immunodominant epitopes. J Immunol. 2000;165:3260–3267. doi: 10.4049/jimmunol.165.6.3260. [DOI] [PubMed] [Google Scholar]
- 37.Yanaka S., Ueno T., Shi Y., Qi J., Gao G.F., Tsumoto K. Peptide-dependent conformational fluctuation determines the stability of the human leukocyte antigen class I complex. J Biol Chem. 2014;289:24680–24690. doi: 10.1074/jbc.M114.566174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pieper J., Dubnovitsky A., Gerstner C., James E.A., Rieck M., Kozhukh G. Memory T cells specific to citrullinated α-enolase are enriched in the rheumatic joint. J Autoimmun. 2018;92:47–56. doi: 10.1016/j.jaut.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi L., Huang X.Q., Shi L., Tao Y.F., Yao Y.F., Yu L. HLA polymorphism of the Zhuang population reflects the common HLA characteristics among Zhuang-Dong language-speaking populations. J Zhejiang Univ Sci B. 2011;12:428–435. doi: 10.1631/jzus.B1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith K.J., Pyrdol J., Gauthier L., Wiley D.C., Wucherpfennig K.W. Crystal structure of HLA-DR2 (DRA∗0101, DRB1∗1501) complexed with a peptide from human myelin basic protein. J Exp Med. 1998;188:1511–1520. doi: 10.1084/jem.188.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thévenet P., Shen Y., Maupetit J., Guyon F., Derreumaux P., Tufféry P. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012;40:W288–W293. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Vries S.J., Rey J., Schindler C.E.M., Zacharias M., Tuffery P. The pepATTRACT web server for blind, large-scale peptide – protein docking. Nucleic Acids Res. 2017;45:W361–W364. doi: 10.1093/nar/gkx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindler C.E.M., de Vries S.J., Zacharias M. Fully blind peptide-protein docking with pepATTRACT. Structure. 2015;23:1507–1515. doi: 10.1016/j.str.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 44.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joseph A. Springer Nature; New York: 2020. Disease caused by the novel coronavirus officially has a name: Covid-19.https://www.scientificamerican.com/article/disease-caused-by-the-novel-coronavirus-officially-has-a-name-covid-19/ [cited 2013 March 19]. Available from: [Google Scholar]
- 46.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oany A.R., Emran A.A., Jyoti T.P. Design of an epitope-based peptide vaccine against spike protein of human coronavirus: an in silico approach. Drug Des Dev Ther. 2014;8:1139–1149. doi: 10.2147/DDDT.S67861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alam A., Ali S., Ahamad S., Malik M.Z., Ishrat R. From ZikV genome to vaccine: in silico approach for the epitope-based peptide vaccine against Zika virus envelope glycoprotein. Immunology. 2016;149:386–399. doi: 10.1111/imm.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan A.M., Hu Y., Miotto O., Thevasagayam N.M., Sukumaran R., Abd Raman H.S. Analysis of viral diversity for vaccine target discovery. BMC Med Genom. 2017;10:78. doi: 10.1186/s12920-017-0301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.V V.R., Nair A.S., Dhar P.K., Nayarisseri A. Epitope characterization and docking studies on Chikungunya viral Envelope 2 protein. Int J Sci Res Publ. 2015;5:2250–3153. [Google Scholar]
- 51.Olotu F.A., Soliman M.E.S. Immunoinformatics prediction of potential B-cell and T-celll epitopes as effective vaccine candidates for eliciting immunogenic responses against Epstein-Barr virus. Biomed J. 2021;44:317–337. doi: 10.1016/j.bj.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dash R., Das R., Junaid M., Akash M.F., Islam A., Hosen S.Z. In silico-based vaccine design against Ebola virus glycoprotein. Adv Appl Bioinform Chem. 2017;10:11–28. doi: 10.2147/AABC.S115859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sahay B., Nguyen C.Q., Yamamoto J.K. Conserved HIV epitopes for an effective HIV vaccine. J Clin Cell Immunol. 2017;8:518. doi: 10.4172/2155-9899.1000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor-Robinson A.W., Masoud M. Prediction of epitopes of viral antigens recognized by cytotoxic T lymphocytes as an immunoinformatics approach to anti-HIV/AIDS vaccine design. Int J Vaccines Vaccin. 2015;1:00014. [Google Scholar]
- 55.Panahi H.A., Bolhassani A., Javadi G., Noormohammadi Z. A comprehensive in silico analysis for identification of therapeutic epitopes in HPV16, 18, 31 and 45 oncoproteins. PloS One. 2018;13:e0205933. doi: 10.1371/journal.pone.0205933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jensen P.E. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- 57.Vartak A., Sucheck S.J. Recent advances in subunit vaccine carriers. Vaccines. 2016;4:12. doi: 10.3390/vaccines4020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kloetzel P.M. The proteasome and MHC class I antigen processing. Biochim Biophys Acta. 2004;1695:225–233. doi: 10.1016/j.bbamcr.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 59.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rivino L., Tan A.T., Chia A., Kumaran E.A., Grotenbreg G.M., MacAry P.A. Defining CD8 + T cell determinants during human viral infection in populations of Asian ethnicity. J Immunol. 2013;191:4010–4019. doi: 10.4049/jimmunol.1301507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.