Abstract
The Janus kinase (JAK) and epidermal growth factor receptor (EGFR) have been considered as potential targets for cancer therapy due to their role in regulating proliferation and survival of cancer cells. In the present study, the aromatic alkyl-amino analogs of thiazole-based chalcone were selected to experimentally and theoretically investigate their inhibitory activity against JAK2 and EGFR proteins as well as their anti-cancer effects on human cancer cell lines expressing JAK2 (TF1 and HEL) and EGFR (A549 and A431). In vitro cytotoxicity screening results demonstrated that the HEL erythroleukemia cell line was susceptible to compounds 11 and 12, whereas the A431 lung cancer cell line was vulnerable to compound 25. However, TF1 and A549 cells were not sensitive to our thiazole derivatives. From kinase inhibition assay results, compound 25 was found to be a dual inhibitor against JAK2 and EGFR, whereas compounds 11 and 12 selectively inhibited the JAK2 protein. According to the molecular docking analysis, compounds 11, 12 and 25 formed hydrogen bonds with the hinge region residues Lys857, Leu932 and Glu930 and hydrophobically came into contact with Leu983 at the catalytic site of JAK2, while compound 25 formed a hydrogen bond with Met769 at the hinge region, Lys721 near a glycine loop, and Asp831 at the activation loop of EGFR. Altogether, these potent thiazole derivatives, following Lipinski's rule of five, could likely be developed as a promising JAK2/EGFR targeted drug(s) for cancer therapy.
The Janus kinase (JAK) and epidermal growth factor receptor (EGFR) have been considered as potential targets for cancer therapy due to their role in regulating proliferation and survival of cancer cells.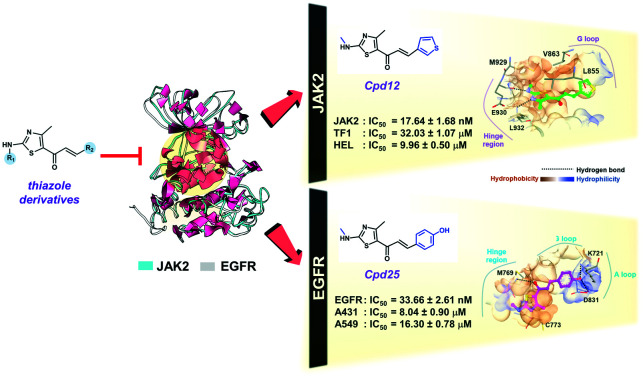
Introduction
Cancer, a group of diseases characterized by uncontrolled growth and spread of abnormal cells,1 is the second leading cause of mortality worldwide.2 Among several types of cancer, myeloproliferative neoplasms and lung cancer are the leading cause of cancer-related death globally.3 The known molecular targets for treating these cancers are the Janus kinases (JAKs) and the epidermal growth factor receptor (EGFR), since they play a major role in regulating proliferation and survival of cancer cells.4,5
JAK, an intracellular tyrosine kinase, is activated by the cytokine(s) binding to its receptor, resulting in the activation of the downstream signal transducer and activator of transcription (STAT), which leads to its dimerization and translocation to the nucleus, promoting cell proliferation, apoptosis and differentiation.6 Among the four JAK family members (JAK1, JAK2, JAK3 and TYK2),7 JAK2 is a critical moderator for hormone-like cytokines such as growth hormone (GH), erythropoietin (EPO), thrombopoietin (TPO) and cytokine receptor ligands involved in hematopoietic cell development such as interleukin-3 (IL-3).8 JAK2 consists of seven homology regions (JH1 to JH7) and the catalytically active domain in JH1 is located at the carboxy-terminus, close to the pseudo-kinase domain in JH2. The overexpression of JAK2, especially the V617F variance in the JH2 domain, has been shown to be related to myeloproliferative disorders (by approximately 50% or more) such as polycythemia vera, essential thrombocythemia and myelofibrosis,9,10 indicating that JAK2 is an important target for the treatment of myeloproliferative diseases.
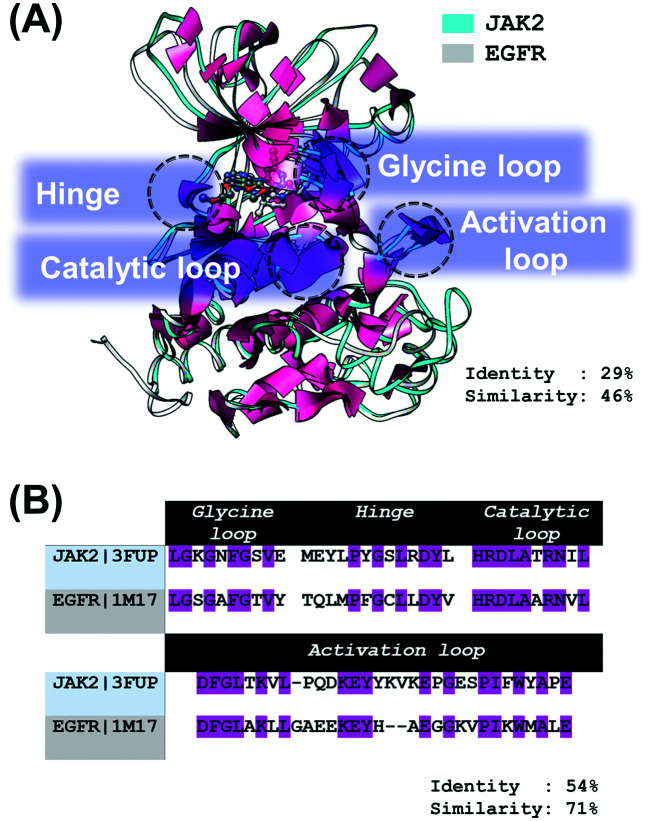
EGFR, a member of the ErbB family of receptor tyrosine kinases,11 is composed of an extracellular domain, a single hydrophobic transmembrane region, an intracellular tyrosine kinase (TK) domain,12 and a C-terminal tyrosine-rich region.13 The EGFR signaling pathways are triggered by the specific growth factors (such as EGF and TGFα) to the extracellular domain, activating autophosphorylation and subsequently initiating downstream signaling pathways, responsible for cell proliferation, survival, differentiation and apoptosis evasion.10,11,14,15 Importantly, more than 60% of non-small cell lung cancer have been involved in the overexpression of EGFR;16 therefore, targeting this protein is an important strategy for lung cancer treatment. As shown in Fig. 1A and B, the sequence identity and similarity of (i) the overall structure and (ii) the conserved regions (catalytic loop, hinge region, glycine-rich loop (G loop) and activation loop (A loop)) between JAK2 and EGFR are 29% and 46% as well as 54% and 71%, respectively.
Fig. 1. (A) Superimposition between JAK2 (PDB ID: 3FUP) and EGFR (PDB ID: 1M17) crystal structures. Tofacitinib (white) and erlotinib (dark gray) are shown in a ball and stick model. The sequence alignment of the four conserved regions: catalytic loop, hinge region, glycine loop and activation loop, between JAK2 and EGFR is given in (B), in which the sequence identity is shaded in magenta.
To date, several JAK2 and EGFR inhibitors have been reported such as lestaurtinib,17 momelotinib,18 and gandotinib19 for JAK2 as well as erlotinib20 and gefitinib21 for EGFR. Among them, ruxolitinib is widely used in clinical treatment for bone marrow cancer (IC50 against JAK2 is 4.5 nM),22,23 while erlotinib has been the first-line drug for lung cancer patients harboring wild-type EGFR (IC50 against EGFR is 2.6 nM).24,25 However, these drugs have several side effects, e.g., anemia, balance impairment and dizziness.26,27 Moreover, acquired drug resistances of ruxolitinib (V617F/L983F) and erlotinib (T790M) have been detected after 1.5 year treatment.28–30 Accordingly, discovery of novel JAK2 and EGFR inhibitors is critically needed.
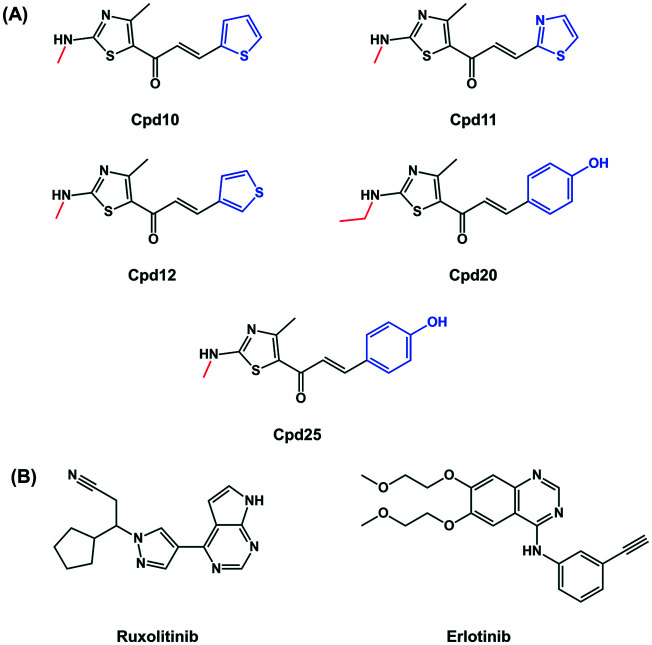
Thiazoles are a class of heterocyclic compounds containing sulfur and nitrogen atoms at positions 1 and 3, respectively. Thiazole derivatives were reported to exert anti-cancer activity against several types of cancer and were identified as JAK2 and EGFR inhibitors.31 Trifluoromethylphenyl thiazole amine inhibited the proliferation of HMC-1.1 cells, which are hematopoietic progenitors in the bone marrow, with an IC50 value of 138 nM.32 In addition, the 4,5-dimethyl thiazole analog potentially inhibited JAK2 activity (IC50 = 2.5 nM) and the JAK2 dependent cell line (SET-2; IC50 = 65 nM).33 From virtual screening against EGFR, the screened thiazole analog, namely 2-(benzo[4,5]imidazo[2,1-b]thiazol-3-yl)-N-(2hydroxyphenyl) acetamide, was found to be a potent EGFR inhibitor (IC50 = 55 mM) by forming a hydrogen bond with Met769 at the hinge region of EGFR-TK.31 Even though several thiazole derivatives have been reported to inhibit JAK2 and EGFR, the aromatic alkyl-amino analogs of thiazole (Fig. 2), possessing antibacterial and anti-cancer activities,34 have not yet been studied in these two proteins.
Fig. 2. Chemical structures of (A) the aromatic alkyl-amino analogs of thiazole obtained from a previous study34 and (B) drugs.
Another interesting class is chalcones. Chalcones are open-chain flavonoids, biosynthesized in a variety of plant species.35 From a chemical point of view they are 1,3-diphenyl-2-propen-1-ones, which consist of two aromatic rings linked by a three-carbon α,β-unsaturated carbonyl system.35 Chalcone is a special chemical template with wide range of biological activities, among which are anti-cancer,36–38 anti-inflammatory,39–41 antioxidant,41,42 antimicrobial,43–45 anti-tubercular,46 anti-HIV,47,48 antimalarial.49 Moreover, it was mentioned in the literatures that chalcones can inhibit kinases essential for tumor cell survival and proliferation such as EGFR,50,51 vascular endothelial growth factor receptor-2 (VEGFR-2) and B-Raf (BRAF) kinase.52,53
In this study, taking all mentioned above into account, in vitro cytotoxicity screening towards erythroleukemia (TF1 and HEL cells expressing JAK2) and lung carcinoma (A549 and A431 cells expressing EGFR) of thiazole-based chalcones (Fig. 2) was performed. Subsequently, the kinase inhibitions of the screened compounds towards both proteins were characterized. Finally, the binding interactions at the atomic level of potent compounds were analyzed using molecular docking.
Results
Cytotoxicity
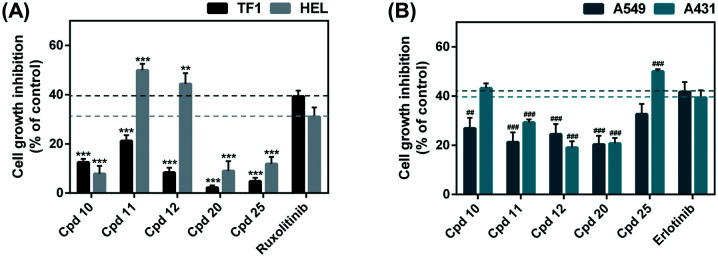
Initially, thiazole derivatives and known drugs (ruxolitinib and erlotinib) at 10 μM were subjected to in vitro cytotoxicity screening against TF1 and HEL cell lines expressing JAK2 as well as against A549 and A431 cell lines expressing EGFR (Fig. 3). The results revealed that the HEL erythroleukemia cell line was susceptible to compounds 11 and 12, whereas the A431 lung cancer cell line was vulnerable to compound 25. However, TF1 and A549 cells were not sensitive to our thiazole-based chalcone derivatives. Therefore, only these three thiazoles-based chalcones (compounds 11, 12 and 25) were selected to evaluate their half maximal inhibitory concentration (IC50) values.
Fig. 3. Cell viability of (A) TF1 and HEL and (B) A549 and A431 cell lines treated with thiazole-based chalcones at 10 μM for 72 h. Data are represented as means ± SEM (n = 2). ** p ≤ 0.01, *** p ≤ 0.001 vs. ruxolitinib and ##p ≤ 0.01, ###p ≤ 0.001 vs. erlotinib.
The IC50 values of thiazole-based chalcones and the known drugs towards the four cancer cell lines are summarized in Table 1, whereas the IC50 curves of these compounds are shown in Fig. S2.† The results revealed that the cytotoxic effect on TF1 cells of compounds 11 (IC50 of 30.23 ± 0.53 μM) and 12 (IC50 of 32.03 ± 1.07 μM) was lower than that of ruxolitinib (IC50 of 14.35 ± 2.03 μM). On the other hand, compounds 11 and 12 (IC50 of ∼7–10 μM) were more susceptible to HEL cells than ruxolitinib (IC50 of 18.62 ± 0.27 μM) by ∼2–3 times. In the case of EGFR-expressing cells, the anti-lung cancer potential of compound 25 (IC50 of 16.30 ± 0.78 μM and 8.04 ± 0.90 μM for A549 and A431, respectively) was higher (∼2–6 fold) than erlotinib used as a reference drug (IC50 of 47.74 ± 6.96 μM and 28.70 ± 6.47 μM for A549 and A431, respectively) in both cell lines. In addition, we found that the IC50 value of the A431 cell line treated with compound 10 (IC50 of 28.49 ± 0.46 μM) is similar to erlotinib (28.70 ± 6.47 μM), while the IC50 value of the A549 cell line treated with compound 10 (IC50 of 62.09 ± 3.30 μM) is higher than erlotinib (47.74 ± 6.96 μM). Altogether, these focused thiazole-based chalcones were then subjected to in vitro kinase inhibitory activity assay against the JAK2 and EGFR-TK proteins.
The IC50 value of thiazole-based chalcones 11, 12 and 25 towards TF1, HEL, A549 and A431 cancer cell lines.
| A549 | A431 | ||
|---|---|---|---|
| 10 | 62.09 ± 3.30 | 28.49 ± 0.46 | >100 |
| 25 | 16.30 ± 0.78 | 8.04 ± 0.90c | >100 |
| Erlotinib | 47.74 ± 6.96 | 28.70 ± 6.47 | >6 |
Data are shown as means ± SEM (n = 3) and ND; not detected.
p ≤ 0.001 vs. ruxolitinib.
p ≤ 0.05 vs. erlotinib.
In addition, the cytotoxicity to Vero cells, which are normal kidney cells, of compounds 10, 11 and 25 was investigated in comparison with the known drugs, ruxolitinib and erlotinib (Table 1 and Fig. S3†). It was found that potent compounds 10, 11 and 25 showed an IC50 value of >100 μM, which is higher than the drugs (>50 μM for ruxolitinib and >6 μM for erlotinib), suggesting that compounds 10, 11 and 25 have a low toxicity against Vero cells. Note that due to the limited amount of compound 12, its cytotoxicity towards Vero cells was not performed.
Kinase inhibition
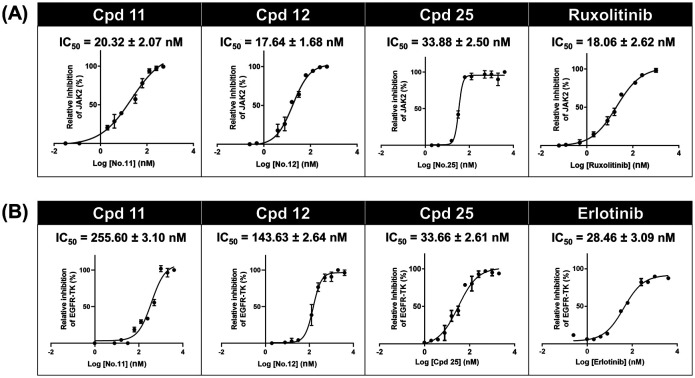
To further support that compounds 11, 12 and 25 can inhibit the JAK2 and EGFR-TK proteins, the kinase inhibitory activity assays were conducted in comparison to the known inhibitors (ruxolitinib and erlotinib) at 1 μM (Fig. S4†). As shown in Fig. 4A, the kinase inhibitory activity against JAK2 of compound 11 (IC50 of 20.32 ± 2.07 nM), compound 12 (17.64 ± 1.68 nM) and compound 25 (IC50 of 33.88 ± 2.50 nM) was in the range of that of ruxolitinib (IC50 of 18.06 ± 2.62 nM). In the case of EGFR-TK (Fig. 4B), only compound 25 (IC50 of 33.66 ± 2.61 nM) inhibited EGFR-TK activity in a manner similar to erlotinib (IC50 of 28.46 ± 3.09 nM), whereas compounds 11 (IC50 of 255.60 ± 3.10 nM) and 12 (IC50 of 143.63 ± 2.64 nM) showed significantly lower inhibitory activity than erlotinib. These results suggested that compound 25 is a dual inhibitor against JAK2 and EGFR, whereas compounds 11 and 12 act as a JAK2 inhibitor (Fig. S6†).
Fig. 4. Kinase inhibitory activity of thiazole-based chalcones towards (A) JAK2 and (B) EGFR-TK. Data are represented as means ± SEM of three independent experiments.
Molecular docking
To investigate the binding mechanism of the focused thiazole derivatives (compounds 11, 12 and 25) towards JAK2 and EGFR-TK in comparison to the known drugs (ruxolitinib and erlotinib), molecular docking was conducted. We found that the fitness score of ruxolitinib in complex with JAK2 (62.13) was higher than those of compounds 11 (56.44), 12 (51.76) and compound 25 (53.57) (Fig. S5†). Similarly, the compound 25/EGFR-TK complex (30.68) gave a fitness score lower than the erlotinib/EGFR complex (36.35).
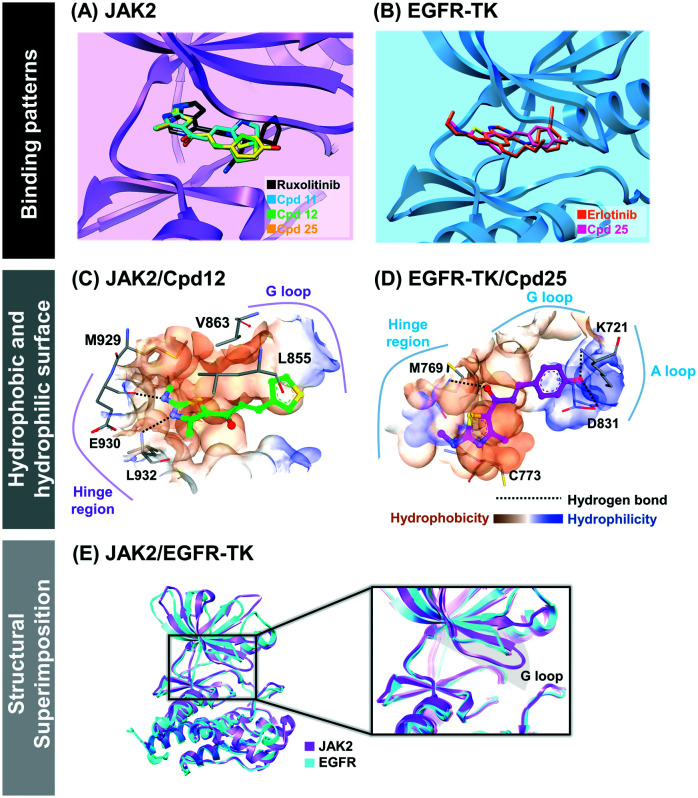
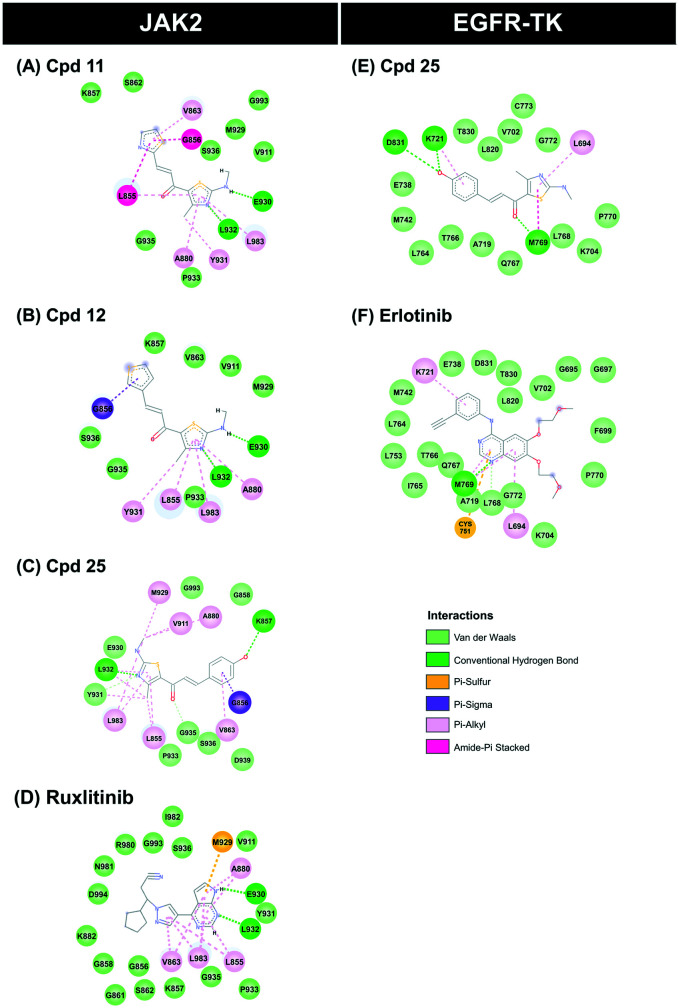
The binding pattern and the underlying interactions of all focused ligands are shown in Table S1,† and Fig. 5 and 6. According to the JAK2 systems, the thiazole core of compounds 11 and 12 aligned well with the pyrrolopyrimidine ring of ruxolitinib at the ATP-binding pocket (Fig. 5A). Moreover, the nitrogen and hydrogen atoms on the thiazole ring of both compounds formed two hydrogen bonds (H-bonds) with Glu930 and Leu932 at the hinge region of JAK2 (Fig. 6A–C), while compound 25 formed H-bonds with Lys858 and Leu932, in a manner similar to ruxolitinib recognition.28 The thiazole ring of compound 11 interacted with Leu855 and Gly856 at the G loop via amide–pi stacking, while the thiophene group of compound 12 and compound 25 bound to Gly856 through pi–sigma interaction. Apart from H-bonds and pi interactions, van der Waals (vdW) forces are also important for ligand binding (Table S1†), and the overlapped vdW contacts between the three thiazole-based chalcones and ruxolitinib were as follows: (i) hinge region: Tyr 931, Pro933, Gly935 and Ser936, (ii) G loop: Leu855, Lys857, Gly858, Ser862 and Val863 (pi–alkyl), (iii) catalytic loop: Leu983 (pi–alkyl), Gly993 and (iv) other regions: Ala880 near the G-loop (pi–alkyl) and Val911 near the hinge region.
Fig. 5. (A and B) binding patterns of thiazole-based chalcones and known drugs within JAK2 and EGFR-TK. (C and D) Hydrophobic and hydrophilic surfaces of compound 12/JAK2 and compound 25/EGFR-TK complexes. (E) Structural superimposition between JAK2 and EGFR-TK, especially at the ATP-binding pocket.
Fig. 6. 2D interactions of JAK2 and EGFR-TK complexed with the thiazole-based chalcones and the known drugs. (A–D) JAK2 complexed with compounds 11, 12, 25 and ruxolitinib (E and F) EGFR-TK complexed with compounds 25 and erlotinib.
In the case of compound 25 complexed with EGFR-TK, its thiazole core occupied the hinge region similar to compounds 11, 12 and 25 complexed with JAK2 (Fig. 5A), and overlapped with the quinazoline ring of erlotinib (Fig. 5B). Interestingly, this thiazole analog shared H-bond formation with erlotinib at the hinge region residue Met769 (Fig. 6D and E).54 In addition, the hydroxyl group of the phenol ring of compound 25 could form a H-bond with other residues, including Lys721 near the glycine loop. The matched vdW contacts between compound 25 and EGFR-TK were as follows: (i) hinge region: Thr766, Gln767, Leu768, Pro770 and G772, (ii) G loop: Leu694 (pi–alkyl), Val702 and Lys704, (iii) catalytic loop: L820 and (iv) other regions: near the G loop (Ala719, Glu738, Met742 and Leu764) and near the activation loop (Thr830).
We further investigated the size of the ligand-binding pocket between JAK2 and EGFR by generating the protein hydrophobic and hydrophilic surfaces around compounds 12 and 25 (Fig. 5C and D). The results showed that the ATP-binding pocket of JAK2 was smaller than that of EGFR, since the glycine loop of JAK2 was positioned closer to the hinge region than EGFR (Fig. 5E).
Drug-likeness prediction
The potent thiazole-based chalcones against JAK2 and EGFR-TK were further analyzed in terms of drug-likeness by assessing their physicochemical properties (i.e., molecular weight (MW), the numbers of hydrogen bond donors (HBD) and acceptors (HBA), rotatable bond (RB), polar surface area (PSA) and Log P) derived from Lipinski's rule of five (Table 2). The obtained result revealed that compounds 11, 12 and 25 showed the acceptable value within the criteria of the rules as follows: (i) molecular weight ≤500 Da, (ii) hydrogen bond donors ≤5 and hydrogen acceptors ≤10, (iii) rotatable bond ≤10, (iv) polar surface area ≤140 Å and (v) lipophilicity (expressed as Log P) ≤5.55 Therefore, these compounds could likely be developed as promising novel JAK2 and EGFR-TK inhibitors.
Predicted Lipinski's rule of five for the thiazole-based chalcones and the known drugs. HBD, hydrogen bond donor; HBA, hydrogen bond accepter; PSA, polar surface area.
| Compound | Lipinski's rule of five | Drug-likeness | |||||
|---|---|---|---|---|---|---|---|
| MW (≤500 Da) | HBD (≤5) | HBA (≤10) | Rotatable bond (≤10) | PSA (≤140 Å) | Log P (≤5) | ||
| 11 | 265.35 | 1 | 3 | 4 | 111.36 | 2.47 | Yes |
| 12 | 264.37 | 1 | 2 | 4 | 98.47 | 3.08 | Yes |
| 25 | 274.34 | 2 | 3 | 4 | 90.46 | 2.65 | Yes |
| Ruxolitinib | 306.37 | 1 | 4 | 4 | 83.18 | 2.40 | Yes |
| Erlotinib | 393.44 | 1 | 6 | 10 | 74.73 | 3.20 | Yes |
Discussion
The thiazole-based chalcones have been previously reported to exhibit antimicrobial and anti-tumor activities,35 and other thiazole derivatives have shown anti-cancer activity against several types of cancer and have been identified as JAK2 and EGFR inhibitors. Therefore, we expected that thiazole-based chalcones can inhibit JAK2 and EGFR-TK activity. Considering the data from the in vitro cytotoxic activity of the five thiazole-based chalcone derivatives against erythroleukemia and lung cancer cell lines, we found that compounds 11 and 12 were more susceptible to mutant JAK2-expressing HEL cells rather than the wild-type JAK2-expressing TF1, since the V617F point mutation in JAK2 maintains its open conformation at the activation loop,56,57 resulting in higher ligand accommodation.58,59 In the case of EGFR-expressing cell lines, only compound 25 significantly inhibited the A431 lung cancer cell line, whereas A549 cells were not sensitive to all thiazole derivatives. In addition, compound 10 showed cytotoxicity to A431 higher than A549. This is because (i) the EGFR expression level found in A431 cells is dramatically higher than that found in A54960 and (ii) A549 cells exhibits KRAS mutation, which constitutively activates downstream MAPK signalling pathways, causing a compensatory mechanism.61 The cytotoxic effect on cancer cell lines of these thiazole derivatives was due to their inhibitory activity against JAK2 and EGFR-TK, in a manner similar to the known inhibitors (Fig. 4). In addition, both compounds 11 and 25 showed higher IC50 than the known drugs in normal kidney Vero cells, suggesting that these thiazole derivatives could be safe for these normal cells, which is consistent with a previous report showing that the cytotoxic effect of thiazole-indenoquinoxaline on normal human cells (WI-38, fibroblast) gave high IC50 (>107 μM).62 Furthermore, compounds 11 and 12 showed kinase inhibitory activity against JAK2 higher than EGFR, whereas compound 25 inhibited both proteins at a similar level (Fig. 4), indicating that compound 25 acts as a dual inhibitor, and compounds 11 and 12 are JAK2 inhibitors. However, the results from the cell-based assay were inconsistent with kinase inhibition in which compounds 11 and 12 were potent against JAK2-expressing cells, whereas compounds 25 showed good inhibitory activity towards EGFR-expressing cells. This is because cells have several factors involved in cell growth inhibition such as cell permeability and compound degradation within the cell.63 Therefore, compounds with good inhibitory activity from both kinase inhibition and cell-based inhibition were selected to study the binding patterns at the atomic level.
Although the fitness score of compounds 11 and 12 in complex with JAK2 as well as compound 25 in complex with EGFR was lower than that of the known inhibitors, their ligand–protein interactions are similar. It has been reported that the pyrrolopyrimidine moiety of ruxolitinib forms H-bonds with Glu930 and Leu932 of JAK2.64–66 In agreement with this evidence, the thiazole core of compounds 11 and 12 interacted with Glu930 and Leu932 residues at the hinge region of JAK2 via hydrogen bonding. The noncovalent pi–alkyl interaction is one of the most important contributions to protein–ligand complexation,67 and the glycine loop residues Ala880, Val863 and Leu855 as well as the catalytic residues Leu983 of JAK2 were found to stabilize both thiazoles through pi–alkyl interaction, in correspondence with the reported ATP binding to JAK2, showing that its adenine and ribose moiety are embedded in pockets stabilized by hydrophobic interactions.64 The thiazole core of compound 25 points into the hinge region, which is a hydrophobic pocket similar to compounds 11 and 12 within JAK2, while its polar phenol group embedded between the glycine loop and catalytic loop, which are hydrophilic regions (e.g. Asp831 and Lys721).68 It has been reported that the p-OH group on the phenyl ring of thiazole derivatives can increase the anti-cancer potential.69 In accordance with this evidence, the p-OH group of compound 25 forms H-bonds with Lys721 near the glycine loop and Asp831 at the activation loop, resulting in higher anti-lung cancer activity than the other analogs (Fig. 3B).
The hinge, deep in the ATP pocket, is an important region called the ‘gatekeeper’, which controls the access to the ‘back-pocket’ of the kinase.70 The ATP-binding pocket of EGFR was quite larger than that of JAK2; therefore, compound 25 is suitable for EGFR because its phenol moiety at the R2 position is bulkier than the thiazole/thiophene ring of compounds 11 and 12. This explains why the same series of thiazole derivatives could bind to the different target proteins. Numerous FDA approved drugs consist of a nitrogen-based heterocyclic moiety (e.g., pyrrolopyrimidine for JAK2 inhibitors and quinazolinamine rings for EGFR-TK inhibitors) similar to the adenosine ring of the ATP substrate, which occupies the hydrophobic pocket and forms H-bonds with the hinge region.65,71 Therefore, the aromatic alkyl-amino analogs of thiazole-based chalcones in this study could be used as an ATP-competitive inhibitors against JAK2 and EGFR, since they contain the nitrogen-based heterocyclic ring similar to that of ATP and the known kinase inhibitors. Furthermore, all of the potent compounds showed good drug-like physicochemical properties, suggesting that these three thiazole-based chalcones could likely be developed as novel anti-cancer drugs.
Conclusions
In this study, we combined experimental and computational studies to identify novel JAK2 and EGFR inhibitors. In vitro cytotoxicity screening results showed that the HEL erythroleukemia cell line was susceptible to compounds 11 and 12, whereas the A431 lung cancer cell line was vulnerable to compound 25. The cytotoxic effect on cancer cell lines of these thiazole-based chalcone derivatives was due to their inhibitory activity against JAK2 and EGFR-TK. From binding interaction analysis, it was found that compounds 11 and 12 formed H-bonds with Leu932 and Glu930 and hydrophobically came into contact with Leu983 at the catalytic site of JAK2, whereas compound 25 formed hydrogen bonds within the ATP-binding pocket of EGFR at Met769 (hinge region), Lys721 (near glycine loop) and Asp831 (activation loop). The bulkiness of the phenol moiety at the R2 position of compound 25 led to the higher selectivity toward EGFR-TK than compounds 11 and 12. All of the potent thiazole derivatives followed Lipinski's rule of five. Altogether, our study successfully identified novel JAK2 and EGFR inhibitors from the aromatic alkyl-amino analogs of thiazole, which can be used as promising starting points for subsequent drug discovery programs against erythroleukemia and lung cancer.
Materials and methods
Cell lines and chemical reagents
The human erythroleukemia HEL 92.1.7 (ATCC TIB-180) and TF1 (ATCC CRL-2003), and lung carcinoma A549 (ATCC CCL-185) and A431 (ATCC CRL-1555) cell lines, and monkey (Cercopithecus aethiops) kidney Vero cell line (ATCC CCL-81) were purchased from the American Type Cell Culture Collection (ATCC, Manassas, VA, USA). Roswell Park Memorial Institute (RPMI), Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), penicillin–streptomycin (Pen–Strep), granulocyte-macrophage colony-stimulating factor (GM-CSF) and trypsin were purchased from Life Technologies (California, USA). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), dimethyl sulfoxide (DMSO) and JAK2 (SRP0171) were purchased from Sigma-Aldrich (Darmstadt, Germany). PrestoBlue™ cell viability reagent was purchased from Thermo Fisher Scientific (Waltham, MA, USA). The ADP-Glo™ kinase assay kit was purchased from Promega (Wisconsin, USA). Poly (glu-tyr) peptide (P61-58) was purchased from SignalChem Biotech (Canada). EGFR-TK was obtained from a previous report.72 A series of thiazole compounds were kindly provided by Dr. Athina Geronikaki from the Aristotle University of Thessaloniki.34 Note that due to limited amounts of thiazole-based chalcones obtained from a previous study,34 we performed JAK2 and EGFR kinase and cytotoxicity assays as well as a binding pattern study at the molecular level on only five thiazole derivatives (Fig. 2 and ESI† section 7).
Cell cultures
The TF1 cells were grown in complete RPMI-1640 medium supplemented with 10% (v/v) FBS, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin and 2 ng ml−1 GM-CSF. The HEL cells were grown in complete RPMI-1640 medium supplemented with 10% (v/v) FBS, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin. The A549, A431 and Vero cells were grown in complete DMEM medium supplemented with 10% (v/v) FBS, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin. All cells were maintained at 37 °C in a 5% (v/v) CO2, 95% (v/v) air humidified incubator.
Cytotoxicity in cancer cell lines
The in vitro cytotoxicity activity of thiazole derivatives against the TF1 and HEL cells, which are suspension cells, was assessed using the Presto Blue assay, while A549 and A431 cell lines, which are adherent cells, were evaluated using the MTT assay. For preliminary screening, 100 μL of TF1 (50 000 cells per well), HEL (25 000 cells per well), A549 (5000 cells per well) and A431 (5000 cells per well) cell suspension was seeded per well in a 96-well microplate and incubated at 37 °C overnight, and the cells were treated with compounds and known drugs (ruxolitinib and erlotinib) at 10 μM. Then, incubated for 72 h. Subsequently, 10 μL of Presto Blue reagent was added in TF1 and HEL cells and incubated at 37 °C for 1 h. The absorbance of the resorufin product was measured. The MTT solution (5 mg mL−1) was added into A549 and A431 cells and incubated at 37 °C for 3 h. The medium was removed, and 50 μL of DMSO was added to each well to lyse the cells and solubilized the formazan crystals. Finally, the absorbance was measured at 570 nm using a microplate reader (Infinite M200 microplate reader, Tecan, Männedorf, Switzerland). Each experiment was performed in duplicate. After screening, the compounds displaying a percentage of cell viability at 10 μM <50 were selected to determine their IC50 value.
In addition, the cytotoxicity of the thiazole derivatives against normal Vero cells (2000 cells per well) was also investigated using the MTT assay.
Kinase inhibition of JAK2 and EGFR-TK
The tyrosine kinase inhibitory activity of JAK2 and EGFR-TK was performed using the ADP-Glo™ kinase assay as previously reported.51,72 The first 8 μL of buffer (40 mM Tris–HCl pH 7.5, 20 mM MgCl2 and 0.1 mg mL−1 bovine serum albumin) was added to a 384-well plate. Then, 5 μL of enzymes (2.5 ng μL−1 for JAK2 and 1.25 ng μL−1 for EGFR) and 2 μL of inhibitors were added, followed by 10 μL of a mixture of 5 μM ATP and 2.5 μM poly(glu-tyr), and incubated for 1 h at room temperature. Next, 5 μL of the ADP-Glo reagent was added and incubated for 40 min. After that, 10 μL of kinase detection reagent was added and incubated at room temperature for 30 min to convert ADP to ATP. ATP was then detected by measuring the luminescence using a microplate reader (Infinite M200 microplate reader, Tecan, Männedorf, Switzerland). All assays were performed in triplicate. The relative inhibition (%) of inhibitors was then calculated and compared to the control with no inhibitor as shown in eqn (1);
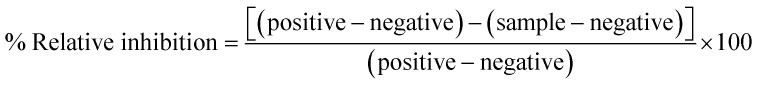 |
1 |
Statistical analysis
The data are represented as mean ± standard error of the mean (SEM). Differences between groups were compared using one-way ANOVA, followed by Tukey's test for multiple comparisons. An independent t-test was used for comparison differences between pairing values. The differences in means were determined at the confidence level P ≤ 0.05.
Molecular docking
The crystal structures of JAK2 complexed with tofacitinib (PDB ID: 3FUP)73 and EGFR complexed with erlotinib (PDB ID: 1M17)71 were downloaded from the Protein Data Bank. These structures were chosen because both of them were crystalized with the known drugs. The missing residues of JAK2 (residues 920-923) were built using the SWISS-MODEL server.74 The 3D structure of the drugs (ruxolitinib and erlotinib) were obtained from the ZINC database, whilst the 3D structure of compounds 11, 12 and 25 were generated using the Gaussian 09 program. All the ligands were optimized using the Gaussian 09 program (HF/6–31d) as per the standard protocol.75–77 The protonation state of all studied ligands was characterized using the ChemAxon.78
For system validation, the crystalized ligands were defined as a center in the active site for redocking using GOLD programs which are based on genetic algorithm (GA),79 and the results are shown in ESI Fig. S1.† The docking protocols were as follows: (i) the JAK2 system was set as 12 Å for sphere docking and GOLD score and ChemScore (rescore) for the scoring function; (ii) the EGFR system was set as 15 Å for sphere docking and ChemScore for the scoring function. All systems were used as 100 docking poses. The binding between proteins and compounds/drugs was visualized using the UCSF Chimera package80 and Accelrys Discovery Studio 2.5 (Accelrys Inc.).
Physicochemical predictions
Physicochemical features such as hydrogen bond donors, hydrogen bond acceptors and drug-likeness play an important role in drug discovery and development.81 Herein, such properties of the potent compounds 11, 12 and 25 were calculated in comparison with drugs (ruxolitinib and erlotinib) using web-based application SwissADME (www.swissadme.ch/).82
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was financially supported by the Ratchadaphiseksomphot Endowment Fund (Grant No. CU_GR_62_96_23_35 for T. R.) and the Thailand Research Fund (DBG6080007 for K.C.). K. S. thanks the Science Achievement Scholarship of Thailand. T. A. acknowledges the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund). P. Mai. thanks Chulalongkorn University for a short-term visit grant.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0md00436g
Notes and references
- DeBerardinis R. J. Genet. Med. 2008;10:267–277. doi: 10.1097/GIM.0b013e31818b0d9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L. Miller K. D. Jemal A. Ca-Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Cancer. Available online: http://www.who.int/en/news-room/factsheets/detail/cancer (accessed on 12 September 2018), 2018
- Dowlati A. Nethery D. Kern J. A. Mol. Cancer Ther. 2004;3:459–463. [PubMed] [Google Scholar]
- Xu P. Shen P. Yu B. Xu X. Ge R. Cheng X. Chen Q. Bian J. Li Z. Wang J. Eur. J. Med. Chem. 2020;192:112155. doi: 10.1016/j.ejmech.2020.112155. [DOI] [PubMed] [Google Scholar]
- O'Shea J. J. Gadina M. Schreiber R. D. Cell. 2002;109:S121–S131. doi: 10.1016/S0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- Schindler C. Levy D. E. Decker T. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Parganas E. Wang D. Stravopodis D. Topham D. J. Marine J. C. Teglund S. Vanin E. F. Bodner S. Colamonici O. R. van Deursen J. M. Grosveld G. Ihle J. N. Cell. 1998;93:385–395. doi: 10.1016/S0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Morgani K. J. Gilliland D. G. Annu. Rev. Med. 2008;59:213–222. doi: 10.1146/annurev.med.59.061506.154159. [DOI] [PubMed] [Google Scholar]
- Levine R. L. Wadleigh M. Cools J. Ebert B. L. Wernig G. Huntly B. J. P. Boggon T. J. Wlodarska L. Clark J. J. Moore S. Adelsperger J. Koo S. Lee J. C. Gabriel S. Mercher T. D'Andrea A. Frohling S. Dohner K. Marynen P. Vandenberghe P. Mesa R. A. Tefferi A. Griffin J. D. Eck M. J. Sellers W. R. Meyerson M. Golub T. R. Lee S. J. Gilliland D. G. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Holbro T. Hynes N. E. Annu. Rev. Pharmacol. Toxicol. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- Choowongkomon K. Carlin C. R. Sonnichsen F. D. J. Biol. Chem. 2005;280:24043–24052. doi: 10.1074/jbc.M502698200. [DOI] [PubMed] [Google Scholar]
- Oliveira S. Bergen en Henegouwen P. M. P. Storm G. Schiffelers R. Expert Opin. Biol. Ther. 2006;6:605–617. doi: 10.1517/14712598.6.6.605. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell. 2002;110:669–672. doi: 10.1016/S0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- Mahalapbutr P. Wonganan P. Chavasiri W. Rungrotmongkol T. Cancers. 2019;11:437. doi: 10.3390/cancers11040437. doi: 10.3390/cancers11040437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha Santos G. Shepherd F. A. Tsao M. S. Annu. Rev. Pathol.: Mech. Dis. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- Hexner E. O. Serdikoff C. Jan M. Swider C. R. Robinson C. Yang S. Angeles T. Emerson S. G. Carroll M. Ruggeri B. Dobrzanski P. Blood. 2008;111:5663–5671. doi: 10.1182/blood-2007-04-083402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardanani A. Lasho T. Smith G. Burns C. J. Fantino E. Tefferi A. Leukemia. 2009;23:1441–1445. doi: 10.1038/leu.2009.50. [DOI] [PubMed] [Google Scholar]
- Verstovsek S. Mesa R. A. Salama M. E. Li L. Pitou C. Nunes F. P. Price G. L. Giles J. L. D'Souza D. N. Walgren R. A. Prchal J. T. Leuk. Res. 2017;61:89–95. doi: 10.1016/j.leukres.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. H. Johnson J. R. Chen Y. F. Sridhara R. Pazdur R. Oncologist. 2005;10:461–466. doi: 10.1634/theoncologist.10-7-461. [DOI] [PubMed] [Google Scholar]
- Mok T. S. Wu Y. L. Thongprasert S. Yang C. H. Chu D. T. Saijo N. Sunpaweravong P. Han B. Margono B. Ichinose Y. Nishiwaki Y. Ohe Y. Yang J. J. Chewaskulyong B. Jiang H. Duffield E. L. Watkins C. L. Armour A. A. Fukuoka M. N. Engl. J. Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- Quintas-Cardama A. Vaddi K. Liu P. Manshouri T. Li J. Scherle P. A. Caulder E. Wen X. Li Y. Waeltz P. Rupar M. Burn T. Lo Y. Kelley J. Covington M. Shepard S. Rodgers J. D. Haley P. Kantarjian H. Fridman J. S. Verstovsek S. Blood. 2010;115:3109–3117. doi: 10.1182/blood-2009-04-214957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H. H. Schroeder G. M. Hart A. C. Inghrim J. Grebinski J. Tokarski J. S. Lorenz M. V. You D. Mcdevie T. Penhallow B. Vuppugalla R. Zhang Y. P. Gu X. M. Iyer R. Lombardo L. J. Trainor G. L. Ruepp S. Lippy J. Blat Y. Sack J. S. Khan J. A. Stefanski K. Sleczka B. Mathur A. Sun J. H. Wong M. K. Wu D. R. Li P. Gupta A. Arunachalam P. N. Pragalathan B. Narayanan S. Nanjundaswamy K. C. Kuppusamy P. Purandare A. V. ACS Med. Chem. Lett. 2015;6:850–855. doi: 10.1021/acsmedchemlett.5b00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. Ning J. F. Meng Q. W. Hu J. Zhao Y. B. Liu C. Cai L. Drug Des., Dev. Ther. 2015;9:3837–3851. doi: 10.2147/DDDT.S85357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguart N. Cardona A. F. Rosell R. Cancer Manage. Res. 2010;2:143–156. doi: 10.2147/CMAR.S5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa R. A. Gotlib J. Gupta V. Catalano J. V. Deininger M. W. Shields A. L. Miller C. B. Silver R. T. Talpaz M. Winton E. F. Harvey J. H. Hare T. Erickson-Viitanen S. Sun W. Sandor V. Levy R. S. Kantarjian H. M. Verstovsek S. J. Clin. Oncol. 2013;31:1285–1292. doi: 10.1200/JCO.2012.44.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methvin A. B. Gausas R. E. Ophthalmic Plast. Reconstr. Surg. 2007;23:63–65. doi: 10.1097/IOP.0b013e31802d97f0. [DOI] [PubMed] [Google Scholar]
- Kesarwani M. Huber E. Kincaid Z. Evelyn C. R. Biesiada J. Rance M. Thapa M. B. Shah N. P. Meller J. Zheng Y. Azam M. Sci. Rep. 2015;5:14538. doi: 10.1038/srep14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C. H. Mengwasser K. E. Toms A. V. Woo M. S. Greulich H. Wong K. K. Meyerson M. Eck M. J. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. L. Lin C. C. Yang S. C. Chen W. L. Chen J. R. Hou Y. H. Lu C. C. Chow N. H. Su W. C. Ho C. L. Front. Oncol. 2019;9:1–8. doi: 10.3389/fonc.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. Tan X. An T. Ma Q. Jin Z. Wang C. Meng Q. Hu C. Molecules. 2019;24:1–13. doi: 10.3390/molecules24040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. Ding K. Wang D. Shen M. Pan J. Cancer Lett. 2014;353:115–123. doi: 10.1016/j.canlet.2014.07.017. [DOI] [PubMed] [Google Scholar]
- Hart A. C. Schroeder G. M. Wan H. Grebinski J. Inghrim J. Kempson J. Guo J. Pitts W. J. Tokarski J. S. Sack J. S. Khan J. A. Lippy J. Lorenzi M. V. You D. McDevitt T. Vuppugalla R. Zhang Y. Lombardo L. J. Trainor G. L. Purandare A. V. ACS Med. Chem. Lett. 2015;6:845–849. doi: 10.1021/acsmedchemlett.5b00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tratrat C. Haroun M. Xenikakis I. Liaras K. Tsolaki E. Eleftheriou P. Petrou A. Aldhubiab B. Attimarad M. Venugopala K. N. Harsha S. Elsewedy H. S. Geronikaki A. Sokovic M. Curr. Top. Med. Chem. 2019;19:356–375. doi: 10.2174/1568026619666190129121933. [DOI] [PubMed] [Google Scholar]
- Orlikova B. Tasdemir D. Golais F. Dicato M. Diederich M. Genes Nutr. 2011;6:125–147. doi: 10.1007/s12263-011-0210-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan C. Moorthy N. S. H. Ramasamy S. Vanam U. Elangovan M. Karunagaran D. Trivedi P. Recent Pat. Anti-Cancer Drug Discovery. 2014;10:97–115. doi: 10.2174/1574892809666140819153902. [DOI] [PubMed] [Google Scholar]
- Wang G. Liu W. Gong Z. Huang Y. Li Y. Peng Z. J. Enzyme Inhib. Med. Chem. 2020;35:139–144. doi: 10.1080/14756366.2019.1690479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavi S. Sreenivasulu R. Yazala J. P. Raju R. R. Saudi Pharm. J. 2017;25:275–279. doi: 10.1016/j.jsps.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ur Rashid H. Xu Y. Ahmad N. Muhammad Y. Wang L. Bioorg. Chem. 2019;87:335–365. doi: 10.1016/j.bioorg.2019.03.033. [DOI] [PubMed] [Google Scholar]
- Wu J. Li J. Cai Y. Pan Y. Ye F. Zhang Y. Zhao Y. Yang S. Li X. Liang G. J. Med. Chem. 2011;54:8110–8123. doi: 10.1021/jm200946h. [DOI] [PubMed] [Google Scholar]
- Abdellatif K. R. Elshemy H. A. Salama S. A. Omar H. A. J. Enzyme Inhib. Med. Chem. 2015;30:484–491. doi: 10.3109/14756366.2014.949255. [DOI] [PubMed] [Google Scholar]
- Wang J. Huang L. Cheng C. Li G. Xie J. Shen M. Chen Q. Li W. He W. Qiu P. Wu J. Acta Pharm. Sin. B. 2019;9:335–350. doi: 10.1016/j.apsb.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan W. Dai J. Eur. J. Med. Chem. 2020;187:111980. doi: 10.1016/j.ejmech.2019.111980. [DOI] [PubMed] [Google Scholar]
- Henry E. J. Bird S. J. Gowland P. Collins M. Cassella J. P. J. Antibiot. 2020;73:299–308. doi: 10.1038/s41429-020-0280-y. [DOI] [PubMed] [Google Scholar]
- Burmaoglu S. Algul O. Gobek A. Aktas Anil D. Ulger M. Erturk B. G. Kaplan E. Dogen A. Aslan G. J. Enzyme Inhib. Med. Chem. 2017;32:490–495. doi: 10.1080/14756366.2016.1265517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandam R. Jadav S. S. Ala V. B. Ahsan M. J. Bollikolla H. B. Med. Chem. Res. 2018;27:1690–1704. doi: 10.1007/s00044-018-2183-z. [DOI] [Google Scholar]
- Al-Hazam H. A. Al-Shamkani Z. A. Al-Masoudi N. A. Saeed B. A. Pannecouque C. Z. Naturforsch., B: J. Chem. Sci. 2017;72:249–256. [Google Scholar]
- Cole A. L. Hossain S. Cole A. M. Phanstiel O. T. Bioorg. Med. Chem. 2016;24:2768–2776. doi: 10.1016/j.bmc.2016.04.045. [DOI] [PubMed] [Google Scholar]
- Syahri J. Yuanita E. Nurohmah B. A. Armunanto R. Purwono B. Asian Pac. J. Trop. Biomed. 2017;7:675–679. doi: 10.1016/j.apjtb.2017.07.004. [DOI] [Google Scholar]
- Al-Anazi M. Al-Najjar B. O. Khairuddean M. Molecules. 2018;23:1–14. doi: 10.3390/molecules23123203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangpheak K. Tabtimmai L. Seetaha S. Rungnim C. Chavasiri W. Wolschann P. Choowongkomon K. Rungrotmongkol T. Molecules. 2019;24:1092. doi: 10.3390/molecules24061092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S. U. Siddiqui H. L. Nisar M. Khan N. Khan I. Bioorg. Med. Chem. Lett. 2012;22:942–944. doi: 10.1016/j.bmcl.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Li Q. S. Li C. Y. Lu X. Zhang H. Zhu H. L. Eur. J. Med. Chem. 2012;50:288–295. doi: 10.1016/j.ejmech.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Park J. H. Liu Y. T. Lemmon M. A. Radhakrishnan R. Biochem. J. 2012;448:417–423. doi: 10.1042/BJ20121513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C. A. Drug Discovery Today: Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Lee T.-S. Ma W. Zhang X. Kantarjian H. Albitar M. BMC Struct. Biol. 2009;9:58. doi: 10.1186/1472-6807-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. S. Ma W. Zhang X. Giles F. Kantarjian H. Albitar M. Cancer. 2009;115:1692–1700. doi: 10.1002/cncr.24183. [DOI] [PubMed] [Google Scholar]
- Quentmeier H. MacLeod R. A. Zaborski M. Drexler H. G. Leukemia. 2006;20:471–476. doi: 10.1038/sj.leu.2404081. [DOI] [PubMed] [Google Scholar]
- Senkevitch E. Durum S. Cytokine+ 2017;98:33–41. doi: 10.1016/j.cyto.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. J. Liu X. J. Xu J. Li L. Li Y. Zhang S. H. Wang J. L. Miao Q. F. Zhen Y. S. Acta Pharmacol. Sin. 2018;39:1777–1786. doi: 10.1038/s41401-018-0069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demiray A. Yaren A. Karagenc N. Bir F. Demiray A. G. Karagur E. R. Tokgun O. Elmas L. Akca H. Balk. J. Med. Genet. 2018;21:21–26. doi: 10.2478/bjmg-2018-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayed E. A. Ammar Y. A. Ragab A. Gohar N. A. Mehany A. B. M. Farrag A. M. Bioorg. Chem. 2020;100:103951. doi: 10.1016/j.bioorg.2020.103951. [DOI] [PubMed] [Google Scholar]
- Housman G. Byler S. Heerboth S. Lapinska K. Longacre M. Snyder N. Sarkar S. Cancers. 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. E. HuangFu W. C. Chao M. W. Sung T. Y. Chang C. D. Chen Y. Y. Hsieh J. H. Tu H. J. Huang H. L. Pan S. L. Hsu K. C. Front. Pharmacol. 2018;9:1–14. doi: 10.3389/fphar.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani M. Huber E. Kincaid Z. Evelyn C. R. Biesiada J. Rance M. Thapa M. B. Shah N. P. Meller J. Zheng Y. Azam M. Sci. Rep. 2015;5:1–19. doi: 10.1038/srep14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T. Georgeon S. Moser R. Moore D. J. Caflisch A. Hantschel O. Leukemia. 2014;28:471–472. doi: 10.1038/leu.2013.299. [DOI] [PubMed] [Google Scholar]
- Toupkanloo H. A. Rahmani Z. Appl. Biol. Chem. 2018;61:209–226. doi: 10.1007/s13765-018-0348-6. [DOI] [Google Scholar]
- Biro J. C. Theor. Biol. Med. Modell. 2006;3:15. doi: 10.1186/1742-4682-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P. Narang R. Nayak S. K. Singh S. K. Judge V. Med. Chem. Res. 2016;25:1717–1743. doi: 10.1007/s00044-016-1610-2. [DOI] [Google Scholar]
- Fabbro D. Cowan-Jacob S. W. Moebitz H. Br. J. Pharmacol. 2015;172:2675–2700. doi: 10.1111/bph.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamos J. Sliwkowski M. X. Eigenbrot C. J. Biol. Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- Seetaha S. Ratanabanyong S. Choowongkomon K. Appl. Microbiol. Biotechnol. 2019;103:8427–8438. doi: 10.1007/s00253-019-10116-6. [DOI] [PubMed] [Google Scholar]
- Williams N. K. Bamert R. S. Patel O. Wang C. Walden P. M. Wilks A. F. Fantino E. Rossjohn J. Lucet I. S. J. Mol. Biol. 2009;387:219–232. doi: 10.1016/j.jmb.2009.01.041. [DOI] [PubMed] [Google Scholar]
- Waterhouse A. Bertoni M. Bienert S. Studer G. Tauriello G. Gumienny R. Heer F. T. de Beer T. A. P. Rempfer C. Bordoli L. Lepore R. Schwede T. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalapbutr P. Thitinanthavet K. Kedkham T. Nguyen H. Theu L. t. h. Dokmaisrijan S. Huynh L. Kungwan N. Rungrotmongkol T. J. Mol. Struct. 2019;1180:480–490. doi: 10.1016/j.molstruc.2018.12.025. [DOI] [Google Scholar]
- Mahalapbutr P. Darai N. Panman W. Opasmahakul A. Kungwan N. Hannongbua S. Rungrotmongkol T. Sci. Rep. 2019;9:10205. doi: 10.1038/s41598-019-46668-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanachai K. Mahalapbutr P. Choowongkomon K. Poo-Arporn R. P. Wolschann P. Rungrotmongkol T. ACS Omega. 2020;5:369–377. doi: 10.1021/acsomega.9b02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin was used for drawing, displaying and characterizing chemical structures, substructures and reactions, Marvin 17.21.0, ChemAxon (https://www.chemaxon.com)
- Jones G. Willett P. Glen R. C. Leach A. R. Taylor R. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Pettersen E. F. Goddard T. D. Huang C. C. Couch G. S. Greenblatt D. M. Meng E. C. Ferrin T. E. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Cheng F. Li W. Zhou Y. Shen J. Wu Z. Liu G. Lee P. W. Tang Y. J. Chem. Inf. Model. 2012;52:3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- Daina A. Michielin O. Zoete V. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.