Abstract
Todays, COVID-19 has caused much death and its spreading speed is increasing, regarding virus mutation. This outbreak warns diagnosing infected people is an important issue. So, in this research, a computer-aided diagnosis (CAD) system called COV-CAD is proposed for diagnosing COVID-19 disease. This COV-CAD system is created by a feature extractor, a classification method, and a content-based imaged retrieval (CBIR) system. The proposed feature extractor is created by using the modified AlexNet CNN. The first modification changes ReLU activation functions to LeakyReLU for increasing efficiency. The second change is converting a fully connected (FC) layer of AlexNet CNN with a new FC, which results in reducing learnable parameters and training time. Another FC layer with dimensions 1 × 64 is added at the end of the feature extractor as the feature vector. In the classification section, a new classification method is defined in which the majority voting technique is applied on outputs of CBIR, SVM, KNN, and Random Forest for final diagnosing. Furthermore, in retrieval section, the proposed method uses CBIR because of its ability to retrieve the most similar images to the image of a patient. Since this feature helps physicians to find the most similar cases, they could conduct further statistical evaluations on profiles of similar patients. The system has been evaluated by accuracy, sensitivity, specificity, F1-score, and mean average precision and its accuracy for CT and X-ray datasets is 93.20% and 99.38%, respectively. The results demonstrate that the proposed method is more efficient than other similar studies.
Keywords: COVID-19 disease, Computer aided diagnosis (CAD) system, AlexNet CNN, Image classification, Content-based image retrieval (CBIR), X-ray and CT scan
1. Introduction
COVID-19 as a contagious disease, is spreading directly from person to person through coughs and talks with an infected person, or indirectly by touching a surface which is infected by virus and then touching the mouth, nose, and eyes. Some people have symptoms like fever, cough and trouble breathing, whereas others who do not have any symptoms are able to spread the virus too [6]. Therefore, diagnosing and finding infected people play a vital role in curing and also preventing spreading the disease.
For diagnosing many diseases and helping physicians, computer-aided diagnosis (CAD) systems have been considered as an effective approach up to now [19], [26], [28]. Hence, in this study, a computer-aided diagnosis system called COV-CAD is proposed.
The proposed COV-CAD includes three major parts; 1- a feature extractor, 2- a classification method, and 3- content-based image retrieval (CBIR) system. The proposed feature extractor is created by inspiring of AlexNet Convolutional Neural Network (CNN) and it is using the LeakyReLU activation function for increasing efficiency instead of ReLU [9]. In addition, in order to reduce parameters, a fully connected (FC) layer with dimensions 1 × 512 is used instead of the 20th layer of AlexNet CNN that is an FC layer with dimensions 1 × 4096 [28]. Another FC layer with 1 × 64 dimensions is added at the end of the proposed feature extractor as a description of each input image for creating the feature vector.
After feature extraction, the CBIR technique is done and the labels of retrieved first 2-tops images are considered as 2 votes. In addition, SVM, KNN, and Random Forest classifiers are utilized for classifying input images. Then there are 5 votes; 2 votes belong to CBIR, and 3 votes of classifiers (see more detail in Section 3). Finally, the majority voting technique counts the votes in order to make the final decision and diagnosing [33], [7].
Because of the ability of CBIR systems in showing the most similar images to a query image (the image of a patient), the proposed method uses this approach to find the most similar cases [27], [30]. The ability of the proposed COV-CAD system in finding and retrieving the most similar subjects could help physicians to conduct further statistical evaluations on groups of similar patients’ profiles.
The major contributions of the proposed COV-CAD system are divided into the following items.
-
•
Proposing an effective feature extractor by modifying the architecture of AlexNet CNN;
-
•
Proposing a new classification method using applying majority voting on results of CBIR, SVM, KNN, and Random Forest;
-
•
Finding and retrieving the most similar cases by CBIR approach;
-
•
Saving required external memory using optimizing dimensions of feature dataset for CBIR section;
-
•
Reducing training time and ability in running and training the model even with a mediocre computer system.
2. Related works and literature review
Today, many researchers are trying to develop methods for diagnosing COVID-19. One of these effective ways is proposing CAD systems based on convolutional neural networks (CNNs) [28], [7]. In the following, some recent methods which have been proposed for achieving this target are described.
In research [18] CNNs such as ResNet, SqueezeNet, and DenseNet have been used for diagnosing COVID-19 from chest X-ray images. The result of this research shows a high sensitivity for most experiments. Despite the fact that the dataset has been imbalanced, in many experiments, specificity is noticeably lower than sensitivity. This is a weakness in detecting Non-COVID and the confusion matrix clearly indicates it. Also, in research [4] ten well-known CNNs including AlexNet have been utilized for diagnosing COVID-19 from CT scans. This research could achieve significant accuracies for CT images but the results have been reported based on just one running per network and it has not used any effective evaluating method such as cross-validation as well as [18].
For addressing these problems, in this study two types of data (X-ray and CT) are taken into account for a comprehensive evaluation. In addition, cross-validation is considered a reliable evaluation method. More importantly, the proposed feature extractor extracts features in both groups (COVID-19 and Non-COVID) effectively which helps to get high specificity and sensitivity for both groups.
Although state-of-the-arts networks such as EfficientNet, MobileNet, and ResNet have been very successful in different computer vision applications [26], the results of researches [27], [5] demonstrate that AlexNet CNN still has its capability. In research [5], a modified AlexNet CNN and also a hybrid AlexNet CNN have been proposed for diagnosing COVID-19 using chest X-ray images. These modified and hybrid approaches use some extra fully connected and bidirectional long short-term memories (BiLSTM) layers. However, the extra layers not only cannot reduce the connections but also they increase number of learnable parameters and train time.
For addressing such problems, in this study, the architecture of AlexNet CNN has been expertly manipulated that more than 25% reduction in learnable parameters and also 15% saving in training time have achieved over the original AlexNet CNN.
3. Material and methods
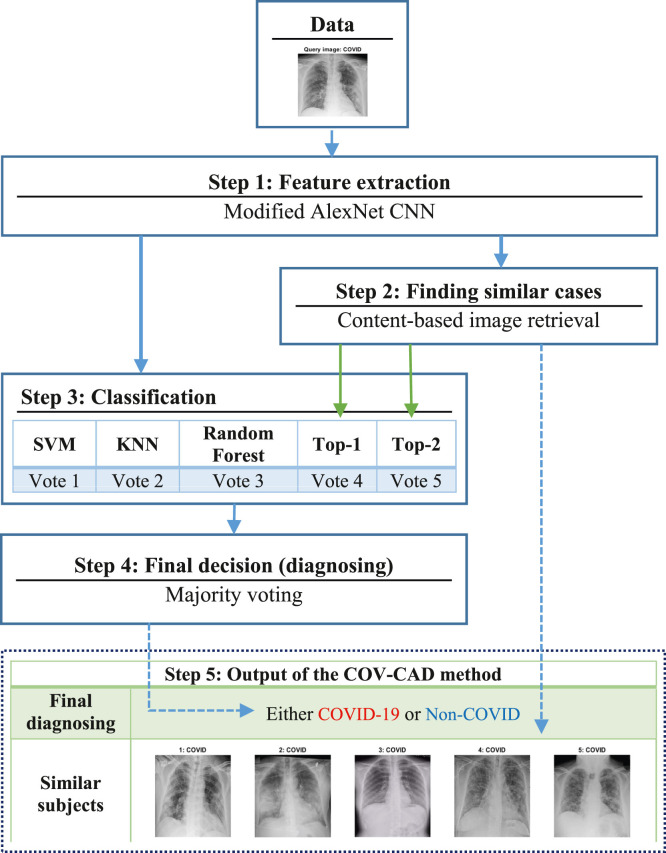
Fig. 1 shows the schematic of the proposed COV-CAD method. By looking at this figure in more detail, it is understood that there are five major steps. At step 1, feature extraction is conducted using a proposed feature extractor created by inspiring of AlexNet CNN [13], [28]. How to build this feature extractor is described in sub-Sections 3.1 in detail.
Fig. 1.
Scheme of the proposed COV-CAD method.
In the conventional classification methods, the classifiers are usually used to classify the data. However, in this research the CBIR not only is used for image retrieval and finding the most similar cases to an input image (query image), but also it is taken into account as an effective solution for classifying images in combination with the classifiers. In this research, when a content-based image retrieval task is carried out, their labels are accessible since the task uses a supervised learning method because of AlexNet CNN. As shown in the step 2 of the Fig. 1, the CBIR section sends labels of the first and second images of training data to the classification section (step 3), which have most similarity to the query image. In addition to that, top-5 retrieved images is sent to step 5 which is defined for showing the output of proposed COV-CAD system. In sub-Sections 3.3, the CBIR approach is explained in detail.
In the third step, images is classified by SVM, KNN, and Random Forests classifiers. After that, the vector of votes is created using the outputs of the classifiers and CBIR technique. Then in the next step, the majority voting technique is applied in order to diagnose patients’ state (see sub-Sections 3.2). Finally, as shown in step 5, similar retrieved images are shown as output of the COV-CAD as well as final diagnosing result.
3.1. Proposed feature extractor
3.1.1. Architecture
In Table 1, the architecture of the proposed feature extractor has been shown and compared with AlexNet CNN [13]. According to this table, in rows that their layer indexes have been mentioned in a bracket show a difference between the proposed feature extractor and AlexNet CNN. By looking at this table in more detail, they resize the inputs (data) to 227 × 227 × 3 and then apply some methods such as convolutional layers, activation functions, normalizations, pooling, and fully connected layers. All changes have happened in activation functions and also fully connected (FC) layers.
Table 1.
Comparison architecture of the proposed feature extractor with AlexNet CNN.
| Layer index | AlexNet CNN |
The proposed feature extractor |
||
|---|---|---|---|---|
| Layer name | Learnable parameters | Layer name | Learnable parameters | |
| 1 | Input: 227 × 227 × 3 | 0 | Input: 227 × 227 × 3 | 0 |
| 2 | Convolution | 34,944 | Convolution | 34,944 |
| [3] | ReLU | 0 | LeakyReLU | 0 |
| 4 | Normalization | 0 | Normalization | 0 |
| 5 | Pooling | 0 | Pooling | 0 |
| 6 | Convolution | 307,456 | Convolution | 307,456 |
| [7] | ReLU | 0 | LeakyReLU | 0 |
| 8 | Normalization | 0 | Normalization | 0 |
| 9 | Pooling | 0 | Pooling | 0 |
| 10 | Convolution | 885,120 | Convolution | 885,120 |
| [11] | ReLU | 0 | LeakyReLU | 0 |
| 12 | Convolution | 663,936 | Convolution | 663,936 |
| [13] | ReLU | 0 | LeakyReLU | 0 |
| 14 | Convolution | 442,624 | Convolution | 442,624 |
| [15] | ReLU | 0 | LeakyReLU | 0 |
| 16 | Pooling | 0 | Pooling | 0 |
| 17 | Fully connected | 37,752,832 | Fully connected | 37,752,832 |
| [18] | ReLU | 0 | LeakyReLU | 0 |
| 19 | Dropout | 0 | Dropout | 0 |
| [20] | Fully connected: 1 × 4096 | 16,781,312 | Fully connected: 1 × 512 | 2,097,664 |
| [21] | – | – | LeakyReLU | 0 |
| [22] | – | – | Dropout | 0 |
| [23] | – | – | Fully connected: 1 × 64 | 32,832 |
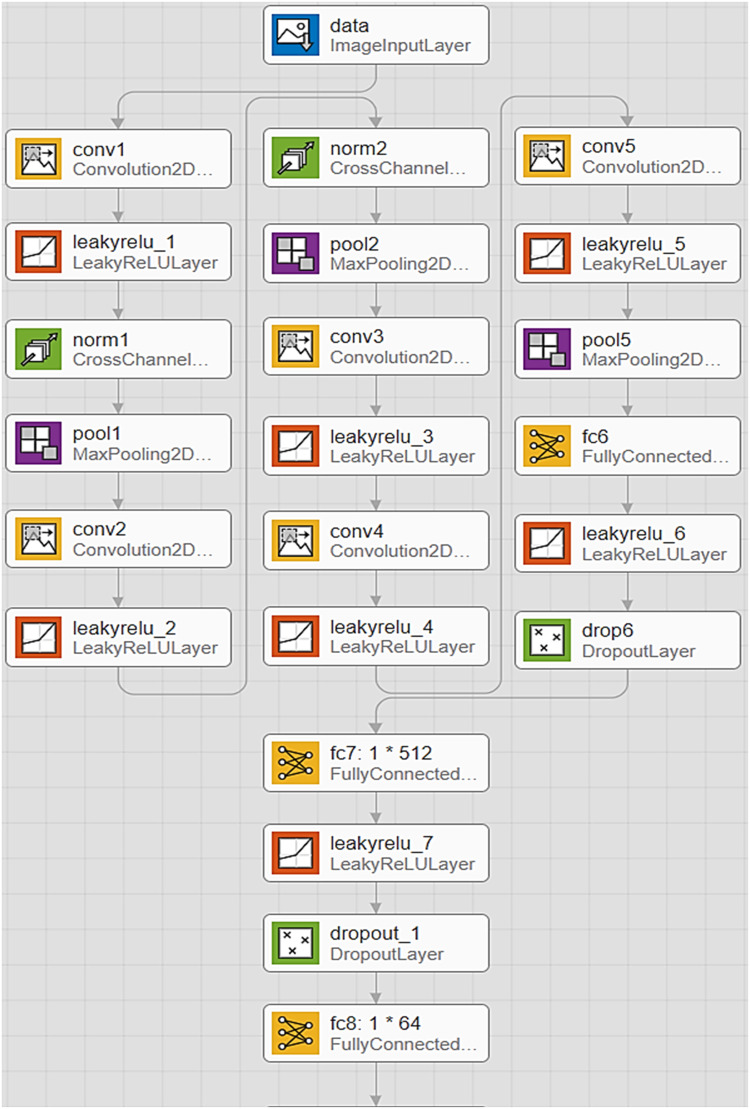
The proposed feature extractor uses the LeakyReLU activation function instead of ReLU (advantages of LeakyReLU are mentioned in sub-Sections 3.1.3) [9]. Besides, the fully connected layer of row [20] in Table 1, which its dimensions are equal to 1 × 512, is applied instead of the fully connected layer of AlexNet CNN with dimensions 1 × 4096. This change not only leads to a significant parameters reduction, but also it reduces the training time. These cases are described in sub-Sections 3.1.2 and also 4.3.1 in detail. The last differences between the proposed feature descriptor and AlexNet CNN have been shown in rows [21], [22] and [23] of Table 1. After adding a LeakyReLU and a Dropout (in layers 21 and 22) then a fully connected layer with dimensions 1 × 64 is added to the end of the extractor as a feature vector of the description of each input data [28]. This feature vector is the output of the proposed feature extractor which is utilized for CBIR as well as image classification [27].
The pictorial form of the proposed feature descriptor has been shown in Fig. 2. As is shown, LeakyReLU is used as an effective activation function. In addition, fully connected layers named “fc7: 1 × 512″ and “fc8: 1 × 64″ are added at the end of this network.
Fig. 2.
Pictorial form of the proposed feature extractor.
3.1.2. Learnable parameters
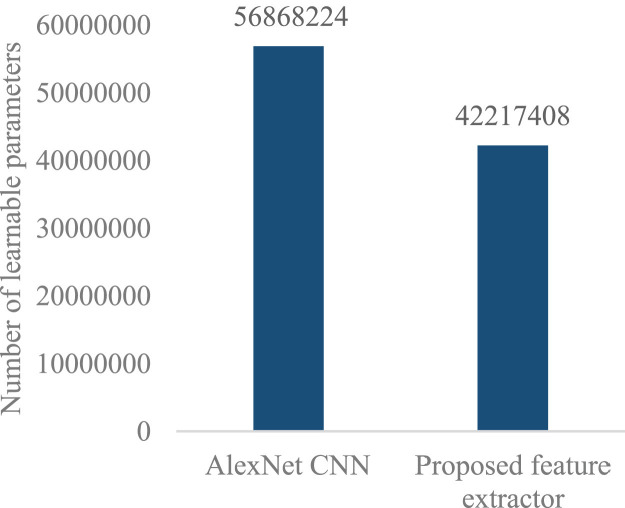
In the proposed feature extractor after applying a fully connected layer with dimensions 1 × 512 instead of fc7 of AlexNet CNN with dimension 1 × 4096 (see row 20 of Table 1 and also the fc7 of Fig. 2), the number of learnable parameters has reduced [13], [28]. According to Fig. 3, which shows the reduction, the number of learnable parameters AlexNet CNN is equal to 56,868,224 while this value is 42,217,408 for the proposed feature extractor. Therefore, the learnable parameters of the proposed feature extractor have 14,650,816 lower than AlexNet CNN. In other words, it could reduce learnable parameters by about 25% than AlexNet CNN. Such reduction could bring benefits such as reducing of training time which is described in sub-Sections 4.3.1 in detail.
Fig. 3.
Comparison learnable parameters’ numbers of the proposed feature extractor with AlexNet CNN.
3.1.3. Activation functions
Dying ReLU is a problem of ReLU in which all negative values are set with zero. In such a situation a negative neuron could not recover. So, all neurons that are stuck on the negative side are literally dead. As a result, they miss their role for discriminating the input and it could say that they will be essentially useless [9].
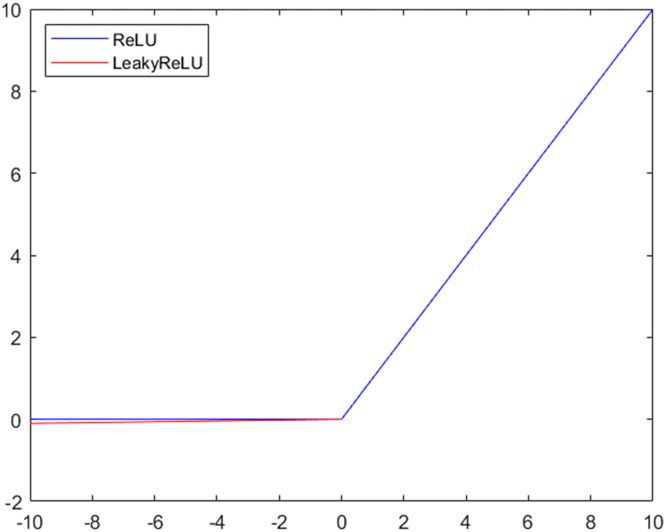
In this research, the LeakyReLU is used for addressing the dying problem of ReLU. The plots of LeakyReLU and ReLU have been shown in Fig. 4. Unlike the ReLU activation function, Leaky ReLU utilizes non-zero mechanism for a negative part. In other words, instead of altogether zero, it uses a small gradient for negative values. For example, when x < 0, LeakyReLU may be defined as y = 0.01x instead of y = x. Accordingly, LeakyReLU could bring two below advantages [17], [9].
-
•
Preventing zero-slope parts in the LeakyReLU can fix the “dying ReLU” problem.
-
•
The LeakyReLU, unlike the ReLU, may learn faster because it is more balanced. Moreover, mean activation to be close to zero makes training faster. So, LeakyReLU speeds up training despite keeping off-diagonal entries of the Fisher information matrix small.
Fig. 4.
Comparisons LeakyReLU with ReLU.
3.2. Classification and diagnosing
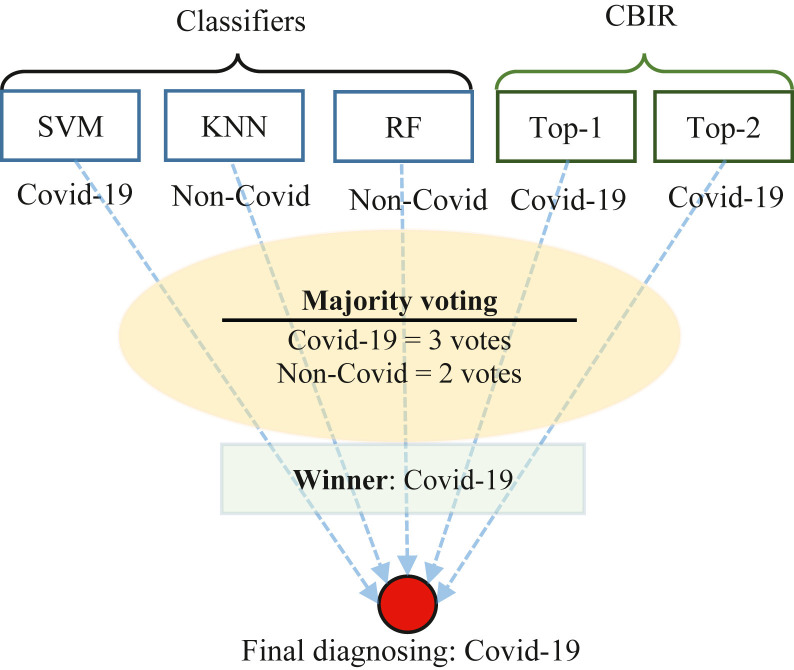
In this study, the proposed classification method is shown in Fig. 5. According to this figure, for a better classification and final diagnosing, the majority voting technique is applied on outputs of SVM, KNN, Random Forest (RF), and CBIR system. CBIR outputs are labels of retrieved first 2-top images [33], [7].
Fig. 5.
The schematic of the proposed classification method.
The majority technique counts the number of votes and then COVID-19 or Non-COVID is declared as a final decision. For instance, if COVID-19 and Non-COVID respectively get three and two votes, the majority voting declares COVID-19 as the winner, as shown in Fig. 5. As a result, the proposed COV-CAD declares that the patient has been diagnosed with COVID-19 like “Final diagnosing” in Fig. 5. Step 5 of Fig. 1 also shows the output of the proposed COV-CAD in more detail.
3.3. CBIR approach
In this research, the proposed COV-CAD system not only uses the CBIR approach as an effective retrieving system, but also it uses in classification section, as mentioned in the previous sub-section [27], [30].
In addition to the advantage of CBIR for increasing the accuracy of classification, its retrieval ability could help physicians to find the most similar cases to a query image (image of a patient). Therefore, they could conduct further statistical evaluations on groups of similar patients.
Creating and saving a feature dataset is an essential step for designing a CBIR system. This feature dataset is used for similarity measurement and must be saved on external memory permanently. In other words, if it is supposed to develop a CAD system as a product, one essential part of the product is saving the feature dataset.
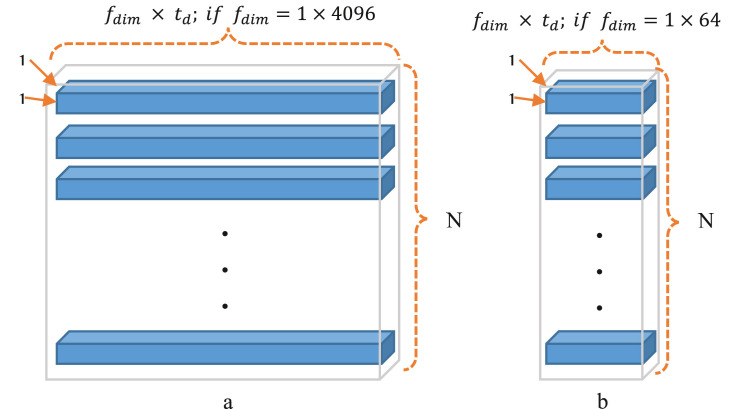
In order to evaluate minimum memory requirement for saving feature dataset, Eq. 1 must be calculated in which denotes the memory required, is the number of images, shows the dimensions of the feature vector for each image, and is the size of the data type. For example, if the data of the feature vector are float, must be set with number 8 as each float number needs 8 bytes (the unit of measurement for this equation is bytes).
| (1) |
As a visual description of Eq. (1) and also showing saving memory by the proposed feature extractor, Fig. 5 has been provided. Looking at this figure, it can be easily understood that the proposed feature extractor ( Fig. 6.b) can save memory significantly rather than AlexNet CNN (Fig. 6.a).
Fig. 6.
Memory required for creating feature dataset by a) AlexNet CNN, b) the proposed feature extractor.
According to Eq. (1) and also Fig. 6, it could be concluded that creating a feature dataset by the proposed feature extractor leads to a significant reduction in terms of required memory because it is 64 times less than AlexNet CNN as .
Another advantage of a shorter feature vector is saving the time that must be spent for similarity measurement between features of the query image and the dataset’s images. In this research Euclidean distance is used for similarity measurement [16], [32]. It is clear that the time of calculation on vectors with dimensions 1 × 64 is noticeably less than 1 × 4096.
4. Results
In this section, sub-Sections 4.1, 4.2, and 4.3 respectively describe implementing details, datasets, and experiments.
4.1. Implementation details
In this study, as it is seen in Table 2, Matlab 2018b has been used for performing experiments. All experiments have been run on a computer system with an NVIDIA GeForce 920 M GPU, an Intel® Core™ i5–7200 U @ 2.50 GHz CPU, and 6 GB RAM. Besides, in order to evaluate the proposed method, criteria such as accuracy, sensitivity, specificity, F1-Score, and mean average precision (mAP) have been measured [24], [8]. Furthermore, the Adam algorithm has been used in order to train the model with 20 epochs and learning rate of 0.0001 [12], [28].
Table 2.
Implementing tools and setting.
| Parameter | Type |
|---|---|
| Environment | Matlab 2018b |
| GPU | NVIDIA GeForce 920M |
| CPU | Intel® Core™ i5-7200U@2.50 GHz |
| RAM | 6 Gigabyte |
| Training optimizer | Adam |
| Evaluation metric | Accuracy, Sensitivity, Specificity, F1-Score, and mAP |
| Data division | 5-fold cross-validation |
| Data augmentation | Used |
As it is shown in Table 2, for setting train and test sets and also evaluating the proposed method, 5-fold cross-validation has been used [25]. Therefore, in this research, all results have been reported an average of 5 folds. Plus, horizontal flip image, converting the image to grayscale, vertical and horizontal translations are applied due to data augmentation [23].
4.2. Datasets
4.2.1. X-ray dataset
The X-ray dataset1 includes 800 chest X-ray images in the pixel domain for two groups: COVID-19 (400 images) and Non-COVID (400 images) [3]. The images of this dataset are in the pixel domain, while if they were in the voxel domain, they would be converted to pixel easily by the slice converting method, which has been proposed in research of [28].
4.2.2. CT dataset
In this research, the CT dataset2 includes 746 chest images in two groups: 349 COVID-19 CT images and 397 Non-COVID, which has two formats.png and.jpg [31]. In this research, the images with.jpg format have been ignored because they do not have a similar resolution to images with.png format, so that if the network uses them for training it might happen a misunderstanding that could reduce accuracy, since they have too low quality. Therefore, 544 images has been considered: 349 COVID-19 and 195 Non-COVID. The images of this imbalanced dataset are also in the pixel domain, but if they were in the voxel domain, they would be transformed into pixel domain easily by the slice converting method which is mentioned in research [28].
4.3. Experiments
4.3.1. Training time
As the proposed feature extractor has lower learnable parameters than the original AlexNet CNN (see Sections 3.1.2), the lower time it needs for training. Table 3 has mentioned these times that are respectively 20 and 17 s per epoch for AlexNet CNN and the proposed feature extractor (on the computer system described in Table 2). This 3 s reduction means that the proposed method in this research could reduce training time to 15% than the original AlexNet CNN.
Table 3.
Training time of feature extractors per epoch.
| Feature extractor | Training time (s) |
|---|---|
| AlexNet CNN | 20 |
| The proposed feature extractor | 17 |
4.3.2. Classification accuracy
In Table 4, the accuracy of the proposed method compared with KNN (1-NN), SVM and Random Forest [27], [28]. As it is seen, Random Forest has achieved a better accuracy on CT images than SVM and KNN, while its accuracy for X-ray images was lower than KNN. But the proposed method has achieved higher accuracy than each classifier for both datasets thanks to the proposed classification method.
Table 4.
Comprising accuracy of the proposed classification method with other classifiers.
| Classifier | Accuracy ± Standard division (%) |
|
|---|---|---|
| CT | X-ray | |
| SVM | 92.83 ± 1.58 | 99.12 ± 0.15 |
| KNN | 92.28 ± 0.75 | 99.38 ± 0.15 |
| Random forest | 93.01 ± 1.41 | 99.25 ± 0.26 |
| The proposed method | 93.20 ± 1.40 | 99.38 ± 0.77 |
4.3.3. CBIR evaluation
Table 5 shows the mean average precisions (mAP) of retrieval 5-top, 10-top, and all relevant images for the CBIR section. For CT images, the mAP for 5-top is 91.93% with standard division 2.72% and these values for 10-top are 91.44% and 2.86%, respectively. The mAP for retrieval of all relevant images is 88.34% as well. The mAP numbers 5-top, 10-top, and all relevant for the X-ray dataset are 99.28%, 99.19%, and 98.62%, respectively. These values demonstrate that the proposed method has been able to retrieve similar images with high precision. Therefore, physicians could trust the system because these mAP guaranty that the proposed COV-CAD could always retrieve a high rate of relevant images with the most similarity.
Table 5.
Mean average precision of the proposed COV-CAD.
| Retrieval radius | mAP ±Standard division (%) |
|||
|---|---|---|---|---|
| 5-top | 10-top | All relevant images | ||
| Image | CT | 91.93 ± 2.72 | 91.44 ± 2.86 | 88.34 ± 2.40 |
| X-ray | 99.28 ± 0.50 | 99.19 ± 0.51 | 98.62 ± 0.47 | |
In Table 5, the meaning of the 5-top is mAP of five top retrieved images which are relevant (have same class labels) to the query image with the most similarity. The 10-top has the same concept for the first to 10th relevant image, and the column “All relevant images” has also the same concept but for all relevant images of the training dataset.
4.3.4. Final diagnosing results
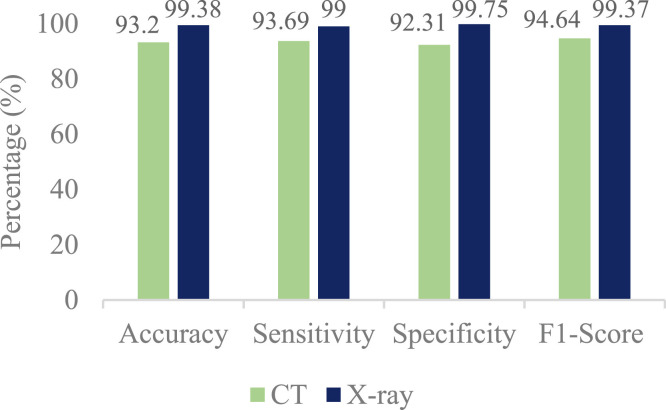
Fig. 7 shows the accuracy, sensitivity, specificity, and F1-Score of the proposed method. According to this figure, the proposed COV-CAD has a high sensitivity besides accuracy on both datasets. So the COV-CAD could diagnose the COVID-19 patients very well. For instance, among 100 subjects infected with COVID-19, which have done CT scans, more than 93 individuals are correctly diagnosed with COVID-19. This number for X-ray is 99. In addition, the high rate of F1-Score demonstrates that the proposed COV-CAD has been successful in terms of harmonic mean of the precision and recall for the CBIR. The high rate of specificities also shows that the proposed method has been completely successful for diagnosing Non-COVID people who actually do not have the conditions of COVID-19.
Fig. 7.
Final diagnosis results of the proposed COV-CAD.
4.3.5. Visual results
Table 6 shows visual results of the CBIR section of the proposed COV-CAD for 4 query images. As it is clear, the COV-CAD could retrieve the top-5 images that have the most similarity to each query.
Table 6.
Outputs of the proposed COV-CAD for subjects’ status.
 |
4.3.6. Comparison results
Table 7 compares the results of the proposed COV-CAD system to another similar method Aslan et al. [5] for X-ray images. According to this table, 5-fold cross validation is used for selecting training and test data. In research Aslan et al. [5] accuracy, specificity and F1-Score have been used for evaluating their method. By comparing these values to the results of the proposed COV-CAD system it is understood that the COV-CAD have a higher efficiency for all criteria.
Table 7.
Comparison results of the proposed COV-CAD with similar researches for X-ray images.
| Method | The feature extractor | Data division | Accuracy (%) | Sensitivity (%) | Specificity (%) | F1-Score (%) |
|---|---|---|---|---|---|---|
| Aslan et al. [5] | AlexNet CNN + BiLSTM | 5-fold CV | 98.70 | Not used | 99.33 | 98.76 |
| The proposed COV-CAD | Modified AlexNet CNN | 5-fold CVa | 99.38 | 99 | 99.75 | 99.37 |
Cross-validation.
Abbas et al. [1] are other researchers that have also used the AlexNet CNN. In their study, cross-validation has not used, instead, 70% of data has been selected for training, and remained 30% has been considered as the test set. Despite this, its accuracy, sensitivity, and specificity had become 92.63%, 89.43%, and 90.18% that are lower than the values of the proposed COV-CAD system. In research [11] a shallow CNN has been used, but the accuracy, sensitivity, and specificity respectively have been equal to 93.20%, 96.10%, and 99.71% that they are lower than the efficiency of the proposed COV-CAD system, which has been mentioned in Table 7. Goel et al. [10] are other researchers that have utilized CNNs for working on COVID-19. In their work, grey wolf optimizer is also used for optimization, but in spite of using 10-fold cross-validation, which in each fold 90% of data is chosen for training and just 10% is considered as test data, the accuracy, sensitivity, specificity, and F1-Score have been 97.78%, 97.75%, 96.25%, and 95.25%, respectively. While, these values for the proposed COV-CAD system are higher and they equal to 99.38%, 99%, 99.75%, and 99.37%, respectively.
5. Discussion
When a CAD system is developed using computer vision methods, in the real clinical situation, small changes on images are an important challenge for medical diagnostics. As, these changes might occur for many reasons, consequently, CAD systems may need to re-train [15], [26].
In this study, the proposed COV-CAD system has utilized convolutional neural networks (modified AlexNet CNN) as a feature extractor. Such networks mostly extract high-level features and are not so sensitive to small changes. Hence, small changes could not reduce the accuracy and efficiency of the method noticeably [14], [29].
Moreover, by transfer learning technique, the proposed COV-CAD system actually does not need a time-consuming and re-training, because most layers of the original AlexNet CNN have been reused in modified AlexNet CNN of the COV-CAD system. Therefore, if the proposed COV-CAD system needs further training with new images after the first training, weight transferring and conducting a light re-training with a small number of epochs could be taken into account as an effective procedure [21], [22].
Besides, the neural networks could solve many new problems very well because of their high function-approximation property and significant generalizability [2], [20]. This is one of the reasons that the proposed COV-CAD system, in most cases, does not need re-training, thanks to the modified AlexNet CNN as the feature extractor.
Conclusively, one can hope that the systems such as the proposed COV-CAD could help physicians widely in finding infected people for preventing more outbreaks.
6. Conclusion
By considering the speed of the COVID-19 virus’ spread and its mutation, diagnosing this infection plays a vital role in preventing more death. Such necessity leads to design a computer-aided diagnosis (CAD) system which is proposed in this study. This system which is called COV-CAD uses a modified AlexNet CNN as an effective feature extractor. The modifications have done in activation functions and fully connected layers. These modifications could bring some advantages such as parameter reduction, and saving training time. As a result, the proposed COV-CAD system is trainable with mediocre computer systems. Besides, a classification method has been proposed in which the majority voting technique is applied on outputs of CBIR approach and also SVM, KNN, and Random Forest for final diagnosis. The results of experiments show that the classification method has proposed a useful proposal due to high accuracy.
Another advantage of this research is that the proposed COV-CAD system could help physicians significantly for seeing similar cases and conducting a further statistical evaluation on similar patients’ profiles, since the CBIR approach is used for finding and retrieving similar images to images of a patient.
The results of experiments demonstrate that the proposed COV-CAD system has a remarkable ability for diagnosing COVID-19. Nevertheless, if networks such as EfficientNet and MobileNet have been more optimized, particularly in terms of the feature vector dimension and reducing learnable parameters, a perfectly optimal COV-CAD system would be developed that could be used in embedded systems and portable equipment. It can be defined as an appropriate future study. Moreover, the proposed classification method can be a basis for further studies about performing classification just using CBIR systems. More importantly, if a high accuracy unsupervised learning method is used for feature extraction, the proposed COV-CAD system will have the ability to develop as a self-learning system and it could be considered as another future research.
Declaration of Competing Interest
The authors report no declarations of interest.
Footnotes
https://github.com/AlaaSulaiman/COVID19-vs-Normal-dataset.
https://github.com/UCSD-AI4H/COVID-CT.
References
- 1.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2021;51(2):854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.B. Adcock, N. Dexter, The Gap between Theory and Practice in Function Approximation with Deep Neural Networks. arXiv preprint arXiv:2001.07523. 2020.
- 3.Al-Waisy A.S., Al-Fahdawi S., Mohammed M.A., Abdulkareem K.H., Mostafa S.A., Maashi M.S., Garcia-Zapirain B. COVID-CheXNet: hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images. Soft Comput. 2020:1–16. doi: 10.1007/s00500-020-05424-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aslan M.F., Unlersen M.F., Sabanci K., Durdu A. CNN-based transfer learning–BiLSTM network: a novel approach for COVID-19 infection detection. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20(5):269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Coronavirus disease (COVID-19) detection in chest X-ray images using majority voting based classifier ensemble. Expert Syst. Appl. 2021;165 doi: 10.1016/j.eswa.2020.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J. Davis, M. Goadrich, The relationship between precision-recall and ROC curves, in: Proceedings of the 23rd International Conference on Machine Learning. 2006, June, pp. 233–240.
- 9.Dubey A.K., Jain V. Applications of Computing, Automation and Wireless Systems in Electrical Engineering. Springer; Singapore: 2019. Comparative study of convolution neural network’s relu and leaky-relu activation functions; pp. 873–880. [Google Scholar]
- 10.Goel T., Murugan R., Mirjalili S., Chakrabartty D.K. Automatic screening of COVID-19 using an optimized generative adversarial network. Cogn. Comput. 2021:1–16. doi: 10.1007/s12559-020-09785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassantabar S., Ahmadi M., Sharifi A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung X-ray image using convolutional neural network approaches. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D.P. Kingma, J. Ba, Adam: A Method for Stochastic Optimization. arXiv preprint arXiv:1412.6980. 2014.
- 13.Krizhevsky A., Sutskever I., Hinton G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012;25:1097–1105. [Google Scholar]
- 14.Lai Z., Deng H. Medical image classification based on deep features extracted by deep model and statistic feature fusion with multilayer perceptron. Comput. Intell. Neurosci. 2018;2018:1–13. doi: 10.1155/2018/2061516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch C.J., Liston C. New machine-learning technologies for computer-aided diagnosis. Nat. Med. 2018;24(9):1304–1305. doi: 10.1038/s41591-018-0178-4. [DOI] [PubMed] [Google Scholar]
- 16.Majhi M., Pal A.K., Islam S.H., Khurram Khan M. Secure content‐based image retrieval using modified Euclidean distance for encrypted features. Trans. Emerg. Telecommun. Technol. 2020;32 [Google Scholar]
- 17.S. Mastromichalakis, ALReLU: A Different Approach on Leaky ReLU Activation Function To Improve Neural Networks Performance. arXiv preprint arXiv:2012.07564. 2020.
- 18.Minaee S., Kafieh R., Sonka M., Yazdani S., Soufi G.J. Deep-covid: predicting covid-19 from chest x-ray images using deep transfer learning. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed. Signal Process. Control. 2021;64 doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohn I., Kim Y. Smooth function approximation by deep neural networks with general activation functions. Entropy. 2019;21(7):627. doi: 10.3390/e21070627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009;22(10):1345–1359. [Google Scholar]
- 22.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S. Deep transfer learning based classification model for COVID-19 disease. Ing. Rech. Biomed. IRBM Biomed. Eng. Res. 2020 doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L. Perez, J. Wang, The Effectiveness Of Data Augmentation In Image Classification Using Deep Learning. arXiv preprint arXiv:1712.04621. 2017.
- 24.D.M. Powers, Evaluation: From Precision, Recall and F-measure to ROC, Informedness, Markedness and Correlation. 2011.
- 25.Z. Reitermanova, Data splitting, In: WDS. vol. 10, 2010. pp. 31–36.
- 26.Shakarami A., Menhaj M.B., Mahdavi-Hormat A., Tarrah H. A fast and yet efficient YOLOv3 for blood cell detection. Biomed. Signal Process. Control. 2021;66 [Google Scholar]
- 27.Shakarami A., Tarrah H. An efficient image descriptor for image classification and CBIR. Optik. 2020;214 doi: 10.1016/j.ijleo.2020.164833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shakarami A., Tarrah H., Mahdavi-Hormat A. An efficient image descriptor for image classification and CBIR. Optik. 2020;214 doi: 10.1016/j.ijleo.2020.164833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M. Tan, Q.V. Le, Efficientnet: Rethinking Model Scaling For Convolutional Neural Networks. arXiv preprint arXiv:1905.11946. 2019.
- 30.Tzelepi M., Tefas A. Deep convolutional learning for content based image retrieval. Neurocomputing. 2018;275:2467–2478. [Google Scholar]
- 31.Yang X., He X., Zhao J., Zhang Y., Zhang S., Xie P. Measurements of the cross section for e(+)e(-) --> hadrons at center-of-mass energies from 2 to 5 GeV. Phys. Rev. Lett. 2002;88 doi: 10.1103/PhysRevLett.88.101802. [DOI] [PubMed] [Google Scholar]
- 32.D. Zhang, G. Lu, Evaluation of similarity measurement for image retrieval, In: International Conference on Neural Networks and Signal Processing, 2003. Proceedings of the 2003 (Vol. 2, pp. 928–931). IEEE. 2003, December.
- 33.Żabiński G., Gramacki J., Gramacki A., Miśta-Jakubowska E., Birch T., Disser A. Multi-classifier majority voting analyses in provenance studies on iron artefacts. J. Archaeol. Sci. 2020;113 [Google Scholar]