Abstract
The search for new bioactive molecules remains an open challenge limiting our ability to discover new drugs to treat disease and chemical probes to comprehensively study biological processes. The vastness of chemical space renders its exploration unfeasible by synthesis alone. Historically, chemists have tended to explore chemical space unevenly without committing to systematic frameworks for navigation. This minireview covers a range of approaches that take inspiration from the structure or origin of natural products, and help focus molecular discovery on biologically-relevant regions of chemical space. All these approaches have enabled the discovery of distinctive and novel bioactive small molecules such as useful chemical probes of biological mechanisms. This minireview comments on how such approaches may be developed into more general frameworks for the systematic identification of currently unexplored regions of biologically-relevant chemical space, a challenge that is central to both chemical biology and medicinal chemistry.
Natural products serve as starting points for the systematic exploration of biologically-relevant chemical space to afford bioactive molecules which can be used to study biological processes.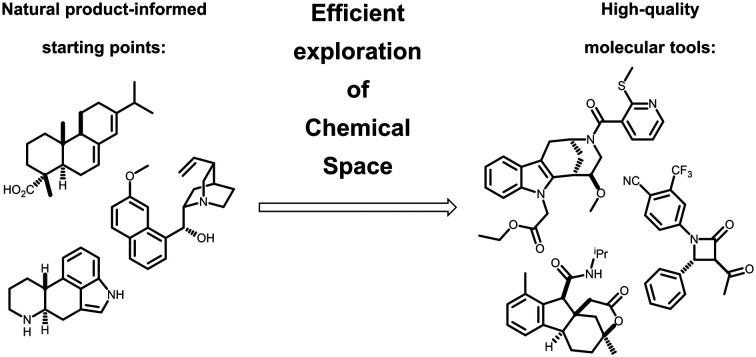
1. Introduction
Small molecules dominate our ability to treat disease1 and can facilitate our understanding of complex biological mechanisms.2 The discovery of bioactive small molecules is facilitated by the ability to navigate biologically-relevant chemical space effectively and efficiently, including previously unexplored regions. However, the vastness of chemical space3 prevents exploration by synthesis alone. Although estimates vary widely,4–6 extrapolation of the exhaustive fragments database GDB17 has suggested that there are ca. 1033 possible drug-like small molecules.7 The historic exploration of chemical space has been uneven and sparse,8 which has hampered the discovery of bioactive molecules based on novel molecular scaffolds.9 This uneven exploration may stem from the over-reliance on a limited palette of established, reliable chemical transformations10 despite the recent development of many novel synthetic methodologies.11 Furthermore, in the context of natural products, there has been an historic focus on the target-oriented synthesis of specific complex molecules.12 To enable diverse chemical space to be explored, approaches such as diversity-oriented synthesis (DOS)13 and lead-oriented synthesis (LOS)14 have been developed. Although DOS and LOS can enable diverse and novel regions of chemical space to be explored efficiently, they tend not to be informed by biology (see below). In contrast, fragment-based ligand discovery, which has been reviewed extensively elsewhere,15,16 starts with fragments (typically with MW < 250) that bind weakly, yet efficiently to a target protein. The exploration of chemical space is rendered more tractable by focusing on fragment-like chemical space; in a few cases, fragment sets based on natural product (NP) substructures have been designed17,18 and productively exploited.18
In this minireview, we focus on approaches in which the exploration of chemical space is informed by the structures or origin of natural products (see section 2). In contrast to approaches that enable the optimisation of structure–function relationships, the featured approaches enable new regions of biologically-relevant chemical space to be identified and explored. In each case, the approaches that can drive the discovery of structurally distinctive functional molecules such as drugs and chemical probes.
2. Natural product-informed approaches for the discovery of functional molecules
Several approaches to help identify novel biologically-relevant chemical space have been developed that are informed by known bioactive compounds or the evolution of biosyntheses of natural products (NPs). Additionally, ingenious platforms have been developed to enable the discovery of molecules that bind/modulate a specific target.19 A number of these platforms harness evolved biological mechanisms to enable molecular discovery. These include, but are not limited to, the split-intein circular ligation of peptides and proteins (SICLOPPS) technology20 for the discovery of bioactive peptidic marcocycles, the flexizyme platform for the expression of peptides consisting of unnatural amino acids,21 and phage-display technology that has enabled the discovery of protein–protein interaction inhibitors.22 In this review, we focus on approaches which are underpinned by synthetic chemistry, and have enabled the discovery of novel and distinctive series of bioactive compounds which lie in new regions of biologically-relevant chemical space. In each case, the approaches are informed by the structures or the origin of natural products. All of these approaches have enabled useful tools to be developed that may facilitate the investigation of complex biological processes.
2.1. Biology-oriented synthesis
Biology-oriented synthesis (BIOS) takes inspiration from the structures of NP scaffolds. NPs are inherently biologically relevant as they have evolved to interact dynamically with multiple proteins during their biosynthesis, and bind to evolved small-molecule binding sites. Using the computational algorithm SCONP to enable systematic simplification of NP scaffolds,23 it is possible to select NP-inspired scaffolds which may retain biological relevance. An interactive computational tool, termed Scaffold Hunter,24 has been developed to facilitate the identification of NP-inspired scaffolds that are sub-structures of NP scaffolds. In this manner, BIOS identifies and exploits the gaps in the coverage of chemical space by NPs, focusing synthetic effort on simplified, yet unexplored, molecular scaffolds. Compound libraries resulting from BIOS scaffolds can be regarded as more biologically relevant than conventional combinatorial compound libraries.25
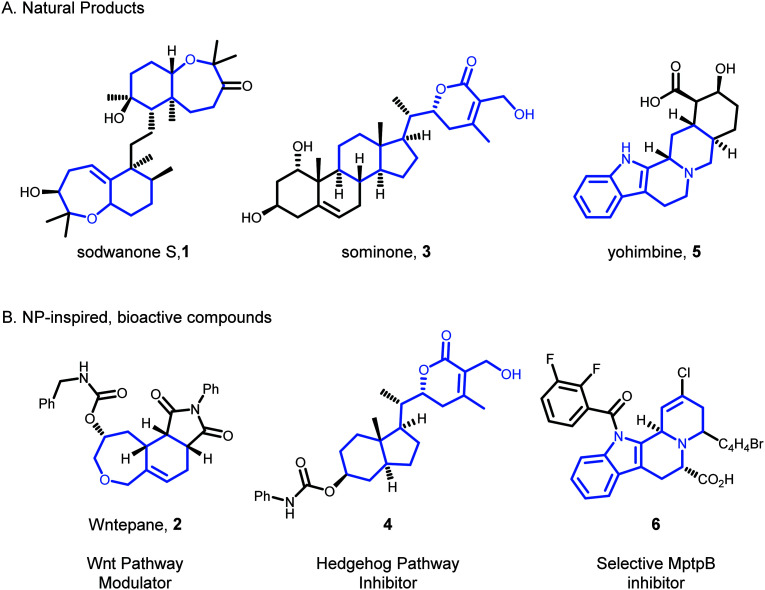
Inspired by NPs such as sodwanone S26 (1, Fig. 1 panel A), a compound library based on a bicyclic oxepane scaffold was designed. The development of a multistep, one-pot synthetic sequence was crucial, and enabled the preparation of 91 derivatives. Utilising a reporter gene assay, 50 of these compounds were found to modulate the Wnt pathway.27 The Wnt signaling pathway is involved in biological processes such as cell migration, renewal, and polarity, and is implicated in the proliferation of cancerous tissue.28 The number of active compounds allowed structure–activity relationships to be established, and ten modulators with low micromolar activity to be discovered. The synthetic route also enabled the preparation of biotinylated analogues which were used to validate the observed bioactivity of the most active analogue, Wntepane (2, Fig. 1 panel B), by means of competitive immunoblotting. Additional, immunoenrichment investigations revealed that Wntepane activates the Wnt pathway by binding reversibly to the protein Vangl1,29 for which no previous small-molecule ligands had been reported.
Fig. 1. NPs inspire the design of BIOS libraries. BIOS exploits structurally simplified molecular scaffolds derived from natural products and has been effective in the identification of biologically relevant small molecules with diverse bioactivities. Panel A: Structures of natural products that have provided inspiration for BIOS. Panel B: Structures of corresponding bioactive compounds which retain conserved portions of the NPs (blue).
The hedgehog (Hh) pathway is another conserved, developmental signaling pathway involved in a number of biological processes such as tissue regeneration and repair, and has been linked with birth defects.30 Inspired by NPs such as sominone31 (3, Fig. 1, panel A), a BIOS approach, involving five linear steps and three parallel derivatisations, resulted in the preparation of 30 compounds. Four analogues were identified as Hh pathway inhibitors of which the most potent compound (4, Fig. 1, panel B) was revealed to act through modulation of the protein Smoothened.32 Novel chemotypes for modulation of the Hh pathway are in high demand as they may potentially have direct clinical applications.33
BIOS has yielded small molecule inhibitors for non-mammalian biological targets as well, demonstrating the general applicability of this approach in exploring biologically-relevant chemical space.34 MptpB is a protein tyrosine phosphatase in M. Tuberculosis, which alters host signaling pathways and is a key biological target for the development of new drugs against this pathogen.35 Brachiation along the branch of the NP yohimbine (5, Fig. 1, panel A), suggested a tetracyclic indoloquinolizidine scaffold. A solid-support synthesis enabled the preparation of a library with 188 compounds. Biological evaluation then revealed that eleven compounds inhibited MptpB, with compound 6 (Fig. 1, panel B) being active in the low micromolar range. Notably, this activity was not shared by the NP yohimbine itself. Moreover, 6 was selective for MptpB over its isoform MptpA, and other mammalian-derived protein tyrosine phosphatases such as Cdc25A and PTP1B.
Overall, it has been demonstrated that BIOS can afford selective compounds which reside in previously unexplored portions of biologically-relevant chemical space. Crucially, bioactive compounds based on BIOS scaffolds can have functions that are wholly distinct from those of the parent NP.
2.2. Complexity to diversity
In contrast to NP synthesis research, “Complexity-to-Diversity” (CtD) exploits NPs themselves as starting materials for the preparation of diverse scaffolds. The resulting scaffolds are related to the guiding NPs, but are distinct from those generated through scaffold simplification in BIOS. Reactions that retain certain parts of a NP structure, while modifying other portions, lead to unprecedented structures which are likely to retain the biological relevance of NPs. Through chemoselective reactions, the core scaffold of a given NP, can be systematically transformed into new scaffolds, for example through ring expansion, fusion or rearrangement (or combinations of these processes). The resulting compounds may also have favourable key properties for bioactive compound discovery, such as high number of stereogenic centres,36 number of sp3 hybridised carbons,37 and solubility.38 Additionally, as the structural complexity is already embedded into the resulting scaffold, synthetic effort can be focused on the preparation of diverse analogues.
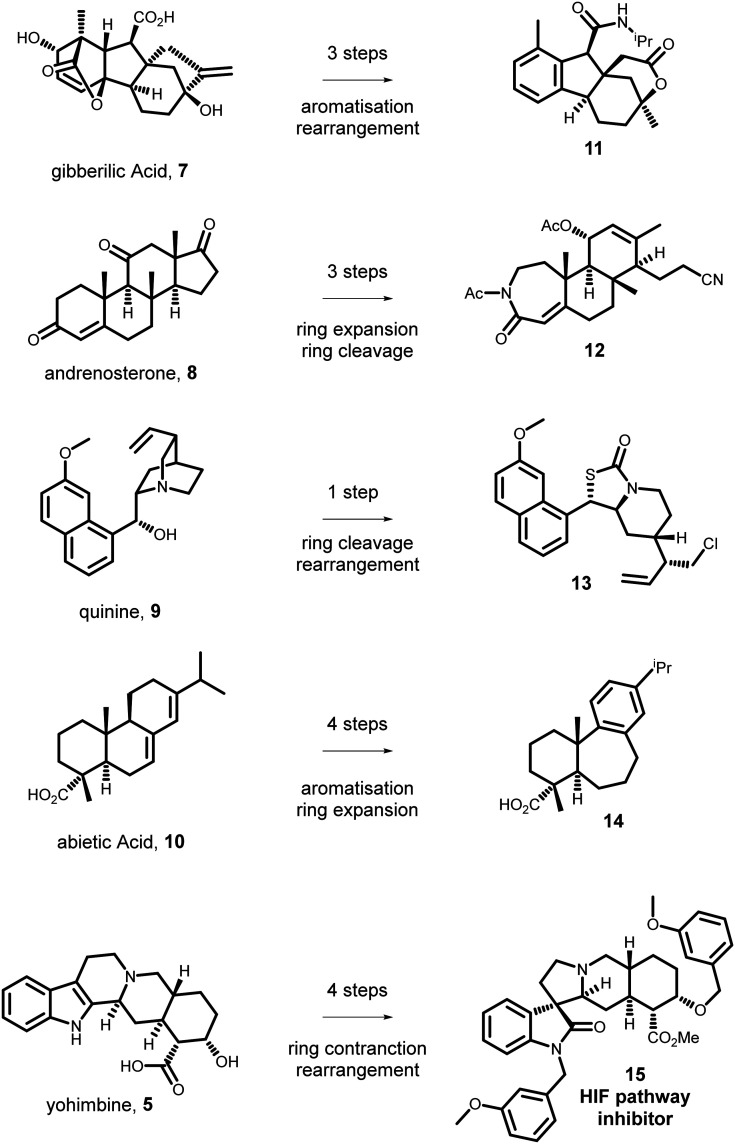
Novel scaffolds were prepared from the readily-available diterpene gibberellic acid (7, Fig. 2, panel A), the steroid andrenosterone (8, Fig. 2, panel A), and the alkaloid quinine (9, Fig. 2, panel A).39 Compounds were prepared in three to five synthetic steps using suitably applicable chemoselective ring cleavage or ring expansion reactions. A compound library based on all three starting NPs 7–9 was subjected to a cheminformatic analysis, looking at desired molecular properties metrics. This analysis showed that CtD compounds had higher three-dimensionality than compounds in the ChemBridge MicroFormat Library. Further pairwise Tanimoto similarity40 analysis provided additional evidence for the high diversity of the CtD library.
Fig. 2. From complex NPs to diverse scaffolds. CtD exploits NPs as starting materials for organic synthesis. Chemoselective transformations, can yield structurally complex molecules which are not accessible by nature. Examples of NPs (left) that have been transformed into novel complex molecular scaffolds (right) are shown. The approach enabled identification of a HIF pathway inhibitor with antiproliferative effects.
Demonstrating the general applicability of CtD approach, the preparation of compound libraries using the diterpene abietic acid (10, Fig. 2)41 or the alkaloid yohimbine (5)42 has been reported. In both cases, compounds where prepared by the application of known chemical transformations following a general pattern. Ring-cleavage (e.g. → 12, 13, Fig. 2) reactions enabled dramatic structural changes in one chemical step and provided new functional groups that can be further diversified. Ring-expansions, (e.g. → 12, 14Fig. 2) for example via the Baeyer–Villiger reaction, led to the formation of novel ring systems, and also preceded ring-cleavage reactions. Ring-fusion reactions provided further diversification by connecting distal groups in the pre-existing scaffold or by merging a new ring to it. Finally, ring-rearrangement (e.g. → 11, 13, 15, Fig. 2) reactions, which drastically change the core scaffold, were used on a case-by-case basis. The molecular scaffolds prepared were demonstrated to be structurally diverse between them and have high fraction of sp3 carbons and number of stereogenic centres. The yohimbine based library was subjected to a range of phenotypic screens for example related to inflammation, proliferation, and anti-bacterial activity. This comprehensive biological evaluation led to the identification of a compound (15, Fig. 2), which demonstrated anti-inflammatory and anticancer activities by modulating the HIF functional pathway.43 Notably, yohimbine does not share this bioactivity profile, further validating CtD as an approach for exploring novel biologically-relevant chemical space in the pursuit of small molecules with diverse biological functions.
2.3. Pseudo-natural products
Fragment-based ligand discovery44 (FBLD) is an established approach for biomolecular discovery, enabling the rapid exploration of chemical space. NPs have been exploited directly as fragments,45 and have inspired sets of NP-derived fragments.18 However, it is also possible to merge NP-derived fragments to yield unprecedented molecular scaffolds. The resulting compounds have been termed pseudo-natural products as they are inspired by NPs and may be inherently biologically relevant. Moreover, these scaffolds lie in portions of chemical space which are not accessible via biosynthesis, and are not simplified versions of NP scaffolds.
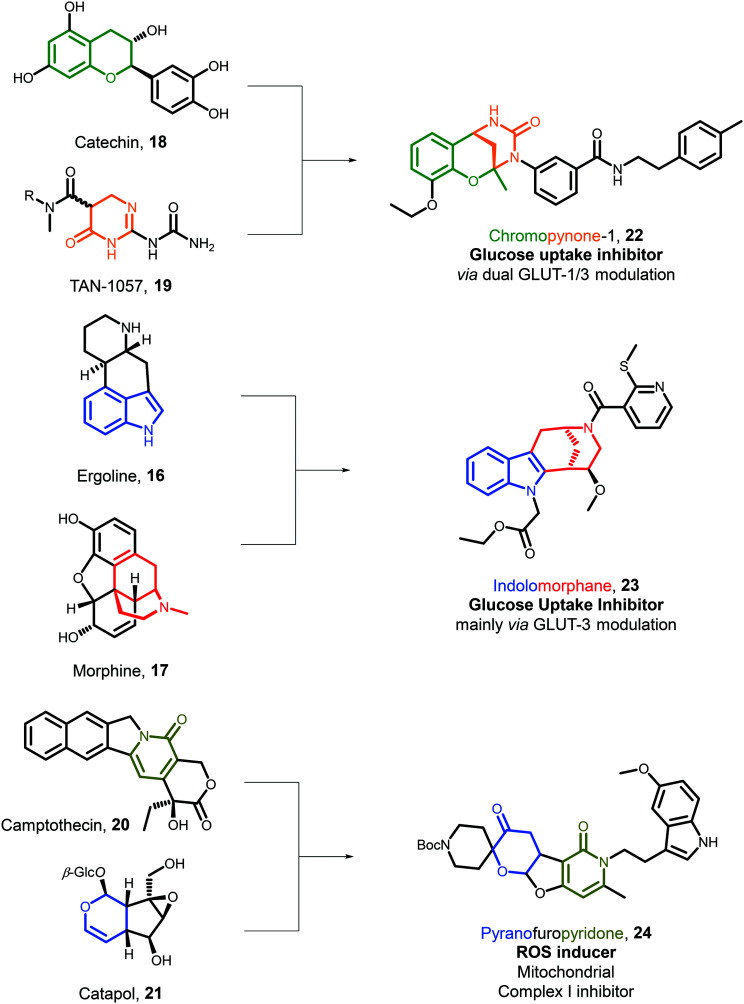
To design new pseudo-NPs, it has been proposed to increase the high fraction of stereogenic centres, combine fragments of diverse biological relevance, and fragments with complementary heteroatoms, e.g. oxygen and nitrogen. Following these basic principles, a new bridged molecular scaffold based on the chromane and tetrahydropyrimidinone (THP) NP-derived fragments was designed.46 An efficient synthesis enabled the preparation of 44 chromopynone derivatives (22, Fig. 3). A cheminformatic analysis showed that these compounds occupied a different portion of chemical space compared to NPs as judged by their NP-score distribution,47 and possessed desired molecular properties for bioactive molecular discovery. Biological evaluation in a broad range of phenotypic and cell-based assays monitoring macroscopic effects in cell signalling and metabolic processes revealed that some chromopynones were active glucose uptake inhibitors. Malignant cells are over-reliant on glucose as an energy source48 and selective modulators of glucose uptake can have clinical applications in cancer therapy.49 A more in-depth biological investigation revealed that the most potent compound (22) was a dual GLUT-1/-3 inhibitor. Additionally, compounds containing either a chromane or a THP fragment alone did not inhibit glucose uptake, indicating that chromopynones display a novel bioactivity profile and are more than just the sum of their constituent parts. This observation demonstrates the potential of this approach to enable the discovery of bioactive small molecules with new functions. Notably, indolomorphane pseudo-NP compounds such as 23 (Fig. 3,), stemming from the combination of the indole and morphan fragments were discovered to display a complementary glucose uptake inhibition profile, engaging primarily with GLUT-3.50
Fig. 3. From NPs to pseudo-NPs. Merging NP-derived fragments yields novel pseudo-NP scaffolds. These compounds retain the biological relevance of NPs, yet reside in chemical space that is not accessible through biosynthesis. Embedded fragments in NPs (left) were fused to yield pseudo-NPs (right). The embedded and merged NP-derived fragments are indicated by colour.
Merging biosynthetically-unrelated NP-derived fragments can also afford pseudo-NP scaffolds and can enable the identification of compounds with novel bioactivity profiles. For example, combining the pyridone and dihydropyran fragments resulted in the design of pyrano-furo-pyridones (24, Fig. 3) which were readily prepared by the application of a Tsuji–Trost oxa-Michael cascade reaction.51 Related cheminformatic analysis demonstrated that pyrano-furo-pyridones reside in a different portion of chemical space compared to NPs, and they may have optimal physiochemical properties as judged by the distribution of molecular weight and estimated lipophilicity (ALogP). The pyrano-furo-pyridones were evaluated for bioactivity in multiple bioassays covering a range of biological processes. This broad evaluation led to the identification of compound 24 as a potent inducer of reactive oxygen species (ROS). ROS have been implicated in several diseases52 and compounds which allow modelling their generation in a controlled manner are of high value. Once more, the observed bioactivity was not shared by compounds containing either the pyridone or pyran fragments, indicating that the biological effect is unique to the pseudo-NP scaffold. Further biological studies showed that 24 induces the production of ROS by inhibiting mitochondrial complex I.
From the above case studies and others,53 more specific guidelines for the design of pseudo-NPs have been developed.54 In brief, these guidelines build on connectivity patterns between fragments that are observed in NPs themselves. Designing pseudo-NP scaffolds following these patterns, allows the resulting scaffold to inherit the biological relevance of its constituent NP-derived fragments. Crucially, bioactive molecules based on pseudo-NP scaffolds have tended to have biological functions that are distinct from compounds based on either of the parent fragments.
2.4. Activity-directed synthesis
The discovery of non-naturally occurring bioactive small molecules is broadly achieved through iterative rounds of design, synthesis and testing. Within this context, medicinal chemists tend to rely on a narrow ensemble of well-established chemical transformations with predictable outcomes such as metal-catalysed biaryl couplings and heteroatom functionalisations.10 In stark contrast, NPs have evolved in tandem with their respective biological targets and their associated biosynthetic pathways, in a function-driven manner i.e. to provide evolutionary advantage to the host organism.55 Activity-directed synthesis (ADS) is a function-driven discovery approach that draws inspiration from the evolution of biosynthetic pathways to NPs (rather than the structures of specific NPs). However, ADS exploits reactions which are not catalyzed by biosynthetic machinery, instead harnessing promiscuous reactions with multiple potential outcomes. Through iterative rounds of reaction arrays, screening and array redesign, ADS focuses attention on active compounds, rather investing resources equally on all compounds within a designed array.
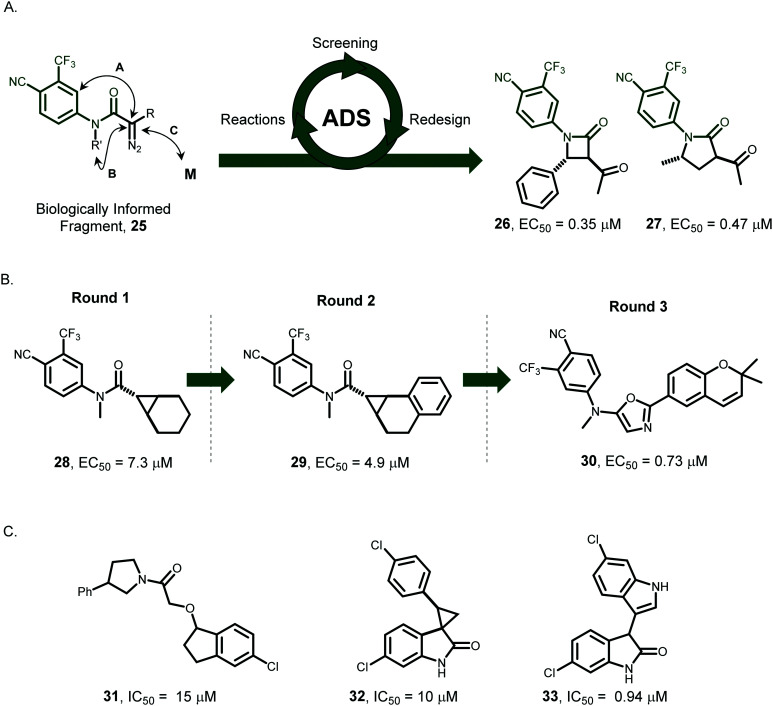
Starting with a structural motif56 (25) found in known ligands, ADS enabled the discovery of androgen receptor modulators (26 and 27) that were based on scaffolds with no previously annotated activity for this target (Fig. 4).57,58 Both intra- and intermolecular reactions were exploited in sequential rounds of carbenoid reactions which had many alternative outcomes. Three iterative rounds of screening crude product mixtures and design of subsequent reaction arrays enabled the rapid discovery of reactions that yielded bioactive products. When harnessing intramolecular reactions, a total of 272 microreactions was performed. Initially, an array of 36 reactions was performed in which 12 diazo substrates 25 were treated with 3 catalysts. The crude reaction mixtures were screened, enabling the identification of four hit reactions. In round 2, the most promising substrates were treated with an expanded range of 8 catalysts in 4 different solvents. Based on ten hit reactions from round 2, 4 additional diazo substrates were prepared. Finally, in round 3 the selection was focused on six catalysts and three solvents, leading to the identification of eight promising reactions that were prioritised for scale-up. This iterative approach enabled the parallel discovery of multiple ligand series. Retrospective analysis showed that ADS enabled parallel optimisation of both the structure of bioactive molecules, and the routes for their syntheses. It was subsequently shown that intermolecular reactions could also be harnessed to drive molecular discovery.57 Here, using non-exhaustive reaction arrays totalling 326 microreactions, additional structurally-diverse sub-micromolar modulators of the androgen receptor (30, Fig. 4, panel B) were also discovered.
Fig. 4. Applications of activity-directed synthesis. ADS is a function-driven approach where bioactive molecules emerge in tandem with their associated synthetic routes, through iterative rounds of activity optimisation. Panel A: ADS was used to discover AR modulators based on scaffolds with no previously annotated AR activity. Panel B: ADS was used to “grow” a known fragment to yield structurally-diverse ligands with increased activity against AR. Panel C: Structures of recently-reported inhibitors of the p53/hMDM2 interaction discovered through ADS. AR: Androgen receptor.
More recently, the applicability of ADS to more challenging targets was demonstrated by the discovery of diverse inhibitors of the p53/hDM2 protein–protein interaction (PPI).59 In this case, the co-substrates and diazo-substrates contained motifs that were intended to mimic p53 hotspot residues.60 In just two rounds of ADS, a total of 346 microscale reactions was performed leading to the identification of four diverse novel p53/hDM2 inhibitors (31–33, Fig. 4, panel C). The structures of these inhibitors were shown to be dissimilar to each other, as judged by Tanimoto similarity index values, as well as to over 1000 reported hDM2 ligands. This observation demonstrates that ADS can enable the parallel discovery of multiple distinct and novel chemotype. It was noted that ADS had enabled scaffold-hopping: that is, the discovery of ligands in which a common pharmacophore is displayed in the context of different scaffolds. In addition, this study showed that ADS was useful approach for lead generation against a target without an evolved small-molecule binding site.
3. Towards systematic frameworks for exploring biologically-relevant chemical space
The chemocentric approaches presented (BIOS, CtD, pseudo-NPs, ADS) can enable the discovery of functional molecules based on novel small-molecule scaffolds. Although these approaches have been described here in the context of specific case studies, they may also enable more systematic identification and exploration of biologically-relevant chemical space. The progress of each approach highlighted in this minireview towards enabling the systematic exploration of biologically-relevant chemical space has been summarised in Table 1.
Progress towards the systematic exploration of biologically-relevant chemical space.
| Approach | How informed by NPs | Scaffold selection | Synthesis |
|---|---|---|---|
| Biology-oriented synthesis | Simplified NP scaffolds | SCONP algorithm and Scaffold Hunter tool help scaffold identification | Tailored synthesis developed for each prioritised scaffold |
| Complexity-to-diversity | NPs are starting materials for synthesis | Tools not currently available to identify NPs that may be suitable substrates | Synthetic methods developed for each NP starting material |
| Pseudo-NPs | Scaffolds are merged NP fragments | Algorithms to generate merged structures not currently available | Tailored synthesis developed for each prioritised scaffold |
| Activity-directed synthesis | Broadly parallels emergence of biosynthetic pathways to NPs | Scaffolds are not designed: functionalised scaffolds emerge on the basis of their biological function | Common protocols used for each reaction class used. Reaction arrays are informed by the activity of products obtained in previous rounds. Array design is currently human-driven |
BIOS and pseudo-NPs focus on molecular scaffolds that are informed by specific NP structures. Crucially, both approaches can identify fertile regions of chemical space for exploration, and can enable discovery of molecules with functions other than those of the parental NPs. For BIOS, the SCONP algorithm can enable systematic navigation of NP-related scaffolds, and can inspire the selection of specific scaffolds for synthesis. This analysis may be facilitated by the computational tool, Scaffold Hunter.18 Unfortunately, a similar tool is not available to facilitate the generation of pseudo-NP structures by fusion of NP-derived fragments, preventing systematic navigation of pseudo-NP scaffolds. Additionally, both approaches require specific scaffolds to be selected for synthesis, which is currently a human-driven process. We note, however, that computational approaches have been developed to identify large numbers of likely synthetically-accessible structures61 from which future BIOS and pseudo-NP scaffolds could be selected systematically. Finally, considerable resource is required to address the synthetic challenges tend to arise (and be solved) on a scaffold-by-scaffold basis. Screening of NP-informed libraries against multiple targets can be achieved through phenotypic, cell-based assays including multi-parametric screening platforms.62 “Cell-painting” assays combine the high information content that is captured in the form of a phenotypic fingerprint. Cell-painting has enabled the systematic assessment of the functional diversity of specific pseudo-NP classes to be captured.51 Whilst BIOS and pseudo-NP approaches can identify fertile chemical space for the discovery of bioactive small molecules, it remains an open question whether the guidance from NP scaffolds is more effective than from other classes of bioactive molecules (for example, human-designed FDA drugs). It has been shown that BIOS and pseudo-NP libraries have relatively high hit rates compared to conventional compound libraries used in high-throughput screening campaigns.63–66 However, rigorous assessment of the productivity of the BIOS and pseudo-NP approaches would require statistical analysis of the performance of many libraries across many assays. It is possible that large-scale screening efforts, such as the European Lead Factory,67 may enable such analyses to be performed.
CtD focuses on the design of transformations that exploit the functionality within complex NPs as starting materials for synthesis. Thus far, the approach has required the selection of specific NP/reaction combinations by individually. Similar to BIOS and pseudo-NPs, a tool for the systematic selection of NP starting materials, together with transformations that may enable the preparation of diverse scaffolds, have not been yet reported. Again, we note the potential value of computational tools that enable systematic assessment of synthetic feasibility.61 As with BIOS and Pseudo-NPs, in each example reported, significant resource was invested in the synthesis of each novel scaffold. In addition, learnings from the optimisation of reactions involving specific substrate/transformation combinations are unlikely to be transferrable to other scaffolds.
In contrast, ADS focuses on the exploitation of promiscuous reactions to generate compounds in situ, rather than on specifically designed target structures. ADS thereby links actual synthetic availability with biological relevance. The design of subsequent reaction arrays is informed by the function of product mixtures obtained in previous rounds. Although this design has been human-driven to date, the approach may lend itself to being algorithmically-driven. For example, one can envisage computational tools that enable reactant combinations to be selected on the basis of the reaction hits from previous rounds. Furthermore, having designed these arrays, their execution could be performed by a liquid handling robot. Crucially, the experimental stages in the workflow are all performed in parallel and may be readily integrated (and potentially automated). As such ADS raises the prospect of realising fully autonomous molecular discovery. We note that autonomous robotic approaches have been developed to address chemical discovery problems such as the discovery of ligands for supramolecular chemistry,68 and the identification of novel chemical reactivity.69
Finally, novel chemistry has always been at the heart of biomolecular discovery. Innovative chemical transformations can prove crucial in our ability to efficiently explore chemical space in general. Recent advances in late-stage functionalisations11,70 can be implemented in the context of either of the highlighted discovery approaches to expedite their coverage of chemical space. For example, methods which allow chemoselective ring formations from functionalised fragments71 or linear precursors,71 could expand the current scope of CtD and pseudo-NPs. Additionally, new multicomponent72 or cascade reactions could also be beneficially as part of the workflow of any of these approaches.73 Modifying NP-derived fragments or larger structural motifs creates opportunities for the application of biocatalytic methods due to the fact that NPs have evolved to interact with multiple protein binding sites. Together these new reactions may facilitate the productive exploration of biologically-relevant chemical space.
4. Summary and outlook
Small molecules are powerful tools for the study of complex biological processes and treating disease, but their discovery requires exploration of relevant chemical space. Approaches which are informed by biology have met great success in navigating the vast chemical space and identifying unexplored, yet biologically-relevant regions. BIOS, CtD, pseudo-NPs, and ADS exploit natural or non-naturally occurring compounds, building on their biological function by trimming, merging, or expanding the original structures. Each approach has led to the successful identification of bioactive small molecules with novel compounds which, in selected cases, have been used as probes to uncover key biological targets.
Although these approaches have been proven to be successful on a case-by-case basis, they have generally not been implemented in a systematic manner: we have commented in section 3 how this might be done in the future. In particular, tools which integrate the design of new scaffolds with the assessment of synthetic accessibility may have great value in opening up broader swathes of unexplored chemical space of direct relevance to molecular discovery. The resulting integrated approaches will thereby address central challenges at the chemistry/biology interface. As such they would have a transformative effect on our collective ability to discover novel small-molecule drugs and chemical tools for elucidating complex biological mechanisms.
Conflicts of interest
The authors declare no conflict of interest. G. K. is an employee of AstraZeneca, UK.
Acknowledgments
We thank EPSRC (EP/N025652/1) for funding.
References
- Fuentes A. V. Pineda M. D. Venkata K. C. N. Pharmacy. 2018;6:43. doi: 10.3390/pharmacy6020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. M. Isserlin R. Bader G. D. Frye S. V. Willson T. M. Yu F. H. Nature. 2011;470:163. doi: 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- Dobson C. M. Nature. 2004;432:824–828. doi: 10.1038/nature03192. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick P. Ellis C. Nature. 2004;432:823. doi: 10.1038/432823a. [DOI] [Google Scholar]
- Reymond J.-L. van Deursen R. Blum L. C. Ruddigkeit L. MedChemComm. 2010;1:30–38. doi: 10.1039/C0MD00020E. [DOI] [Google Scholar]
- Reymond J.-L. Acc. Chem. Res. 2015;48:722–730. doi: 10.1021/ar500432k. [DOI] [PubMed] [Google Scholar]
- Polishchuk P. G. Madzhidov T. I. Varnek A. J. Comput.-Aided Mol. Des. 2013;27:675–679. doi: 10.1007/s10822-013-9672-4. [DOI] [PubMed] [Google Scholar]
- Lipkus A. H. Yuan Q. Lucas K. A. Funk S. A. Bartelt W. F. Schenck R. J. Trippe A. J. J. Org. Chem. 2008;73:4443–4451. doi: 10.1021/jo8001276. [DOI] [PubMed] [Google Scholar]
- Langdon S. R. Brown N. Blagg J. J. Chem. Inf. Model. 2011;51:2174–2185. doi: 10.1021/ci2001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. G. Boström J. J. Med. Chem. 2016;59:4443–4458. doi: 10.1021/acs.jmedchem.5b01409. [DOI] [PubMed] [Google Scholar]
- Boström J. Brown D. G. Young R. J. Keserü G. M. Nat. Rev. Drug Discovery. 2018;17:709–727. doi: 10.1038/nrd.2018.116. [DOI] [PubMed] [Google Scholar]
- Corey E. J. Angew. Chem., Int. Ed. Engl. 1991;30:455–465. doi: 10.1002/anie.199104553. [DOI] [Google Scholar]
- Burke M. D. Schreiber S. L. Angew. Chem., Int. Ed. 2004;43:46–58. doi: 10.1002/anie.200300626. [DOI] [PubMed] [Google Scholar]
- Doveston R. Marsden S. Nelson A. Drug Discovery Today. 2014;19:813–819. doi: 10.1016/j.drudis.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Murray C. W. Rees D. C. Nat. Chem. 2009;1:187–192. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- Murray C. W. Rees D. C. Angew. Chem., Int. Ed. 2016;55:488–492. doi: 10.1002/anie.201506783. [DOI] [PubMed] [Google Scholar]
- Hanby A. R. Troelsen N. S. Osberger T. J. Kidd S. L. Mortensen K. T. Spring D. R. Chem. Commun. 2020;56:2280–2283. doi: 10.1039/C9CC09796A. [DOI] [PubMed] [Google Scholar]
- Over B. Wetzel S. Grütter C. Nakai Y. Renner S. Rauh D. Waldmann H. Nat. Chem. 2013;5:21–28. doi: 10.1038/nchem.1506. [DOI] [PubMed] [Google Scholar]
- Ebo J. S. Saunders J. C. Devine P. W. A. Gordon A. M. Warwick A. S. Schiffrin B. Chin S. E. England E. Button J. D. Lloyd C. Bond N. J. Ashcroft A. E. Radford S. E. Lowe D. C. Brockwell D. J. Nat. Commun. 2020;11:1816. doi: 10.1038/s41467-020-15667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asby D. J. Cuda F. Beyaert M. Houghton F. D. Cagampang F. R. Tavassoli A. Chem. Biol. 2015;22:838–848. doi: 10.1016/j.chembiol.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Hirose H. Tsiamantas C. Katoh T. Suga H. Curr. Opin. Biotechnol. 2019;58:28–36. doi: 10.1016/j.copbio.2018.10.012. [DOI] [PubMed] [Google Scholar]
- Hughes D. J. Tiede C. Penswick N. Tang A. A.-S. Trinh C. H. Mandal U. Zajac K. Z. Gaule T. Howell G. Edwards T. A. Duan J. Feyfant E. McPherson M. J. Tomlinson D. C. Whitehouse A. Sci. Signaling. 2017;10:eaaj2005. doi: 10.1126/scisignal.aaj2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M. A. Schuffenhauer A. Scheck M. Wetzel S. Casaulta M. Odermatt A. Ertl P. Waldmann H. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17272–17277. doi: 10.1073/pnas.0503647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel S. Klein K. Renner S. Rauh D. Oprea T. I. Mutzel P. Waldmann H. Nat. Chem. Biol. 2009;5:581–583. doi: 10.1038/nchembio.187. [DOI] [PubMed] [Google Scholar]
- van Hattum H. Waldmann H. J. Am. Chem. Soc. 2014;136:11853–11859. doi: 10.1021/ja505861d. [DOI] [PubMed] [Google Scholar]
- Dai J. Fishback J. A. Zhou Y.-D. Nagle D. G. J. Nat. Prod. 2006;69:1715–1720. doi: 10.1021/np060278q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S. Ellinger B. Rizzo S. Deraeve C. Schürmann M. Preut H. Arndt H.-D. Waldmann H. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6805–6810. doi: 10.1073/pnas.1015269108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastas J. N. Moon R. T. Nat. Rev. Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- Torban E. Patenaude A.-M. Leclerc S. Rakowiecki S. Gauthier S. Andelfinger G. Epstein D. J. Gros P. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3449–3454. doi: 10.1073/pnas.0712126105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J. Therond P. P. Nat. Rev. Mol. Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Chen L.-X. He H. Qiu F. Nat. Prod. Rep. 2011;28:705–740. doi: 10.1039/C0NP00045K. [DOI] [PubMed] [Google Scholar]
- Lipinski R. J. Gipp J. J. Zhang J. Doles J. D. Bushman W. Exp. Cell Res. 2006;312:1925–1938. doi: 10.1016/j.yexcr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Seifert K. Büttner A. Rigol S. Eilert N. Wandel E. Giannis A. Bioorg. Med. Chem. 2012;20:6465–6481. doi: 10.1016/j.bmc.2012.08.026. [DOI] [PubMed] [Google Scholar]
- Nören-Müller A. Reis-Corrêa I. Prinz H. Rosenbaum C. Saxena K. Schwalbe H. J. Vestweber D. Cagna G. Schunk S. Schwarz O. Schiewe H. Waldmann H. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10606–10611. doi: 10.1073/pnas.0601490103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A. Herget T. Klebl B. Ullrich A. Nat. Rev. Microbiol. 2004;2:189–202. doi: 10.1038/nrmicro840. [DOI] [PubMed] [Google Scholar]
- Lovering F. Bikker J. Humblet C. J. Med. Chem. 2009;52:6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- Veber D. F. Johnson S. R. Cheng H.-Y. Smith B. R. Ward K. W. Kopple K. D. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A. Lombardo F. Dominy B. W. Feeney P. J. Adv. Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Huigens III R. W. Morrison K. C. Hicklin R. W. Flood Jr T. A. Richter M. F. Hergenrother P. J. Nat. Chem. 2013;5:195–202. doi: 10.1038/nchem.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D. J. Tanimoto T. T. Science. 1960;132:1115–1118. doi: 10.1126/science.132.3434.1115. [DOI] [PubMed] [Google Scholar]
- Rafferty R. J. Hicklin R. W. Maloof K. A. Hergenrother P. J. Angew. Chem., Int. Ed. 2014;53:220–224. doi: 10.1002/anie.201308743. [DOI] [PubMed] [Google Scholar]
- Paciaroni N. G. Ratnayake R. Matthews J. H. Norwood IV V. M. Arnold A. C. Dang L. H. Luesch H. Huigens III R. W. Chem. – Eur. J. 2017;23:4327–4335. doi: 10.1002/chem.201604795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkitt K. Chun S. Y. Dang D. T. Dang L. H. Mol. Cancer Ther. 2009;8:1148–1156. doi: 10.1158/1535-7163.MCT-08-0944. [DOI] [PubMed] [Google Scholar]
- Erlanson D. A. Fesik S. W. Hubbard R. E. Jahnke W. Jhoti H. Nat. Rev. Drug Discovery. 2016;15:605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- Vu H. Pedro L. Mak T. McCormick B. Rowley J. Liu M. Di Capua A. Williams-Noonan B. Pham N. B. Pouwer R. Nguyen B. Andrews K. T. Skinner-Adams T. Kim J. Hol W. G. J. Hui R. Crowther G. J. Van Voorhis W. C. Quinn R. J. ACS Infect. Dis. 2018;4:431–444. doi: 10.1021/acsinfecdis.7b00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgis G. Reckzeh E. S. Ceballos J. Schwalfenberg M. Sievers S. Ostermann C. Pahl A. Ziegler S. Waldmann H. Nat. Chem. 2018;10:1103–1111. doi: 10.1038/s41557-018-0132-6. [DOI] [PubMed] [Google Scholar]
- Vanii Jayaseelan K. Moreno P. Truszkowski A. Ertl P. Steinbeck C. BMC Bioinf. 2012;13:106–112. doi: 10.1186/1471-2105-13-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scatena R. Bottoni P. Pontoglio A. Giardina B. Proteomics: Clin. Appl. 2010;4:143–158. doi: 10.1002/prca.200900157. [DOI] [PubMed] [Google Scholar]
- Barron C. C. Bilan P. J. Tsakiridis T. Tsiani E. Metabolism. 2016;65:124–139. doi: 10.1016/j.metabol.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Ceballos J. Schwalfenberg M. Karageorgis G. Reckzeh E. S. Sievers S. Ostermann C. Pahl A. Sellstedt M. Nowacki J. Carnero Corrales M. A. Wilke J. Laraia L. Tschapalda K. Metz M. Sehr D. A. Brand S. Winklhofer K. Janning P. Ziegler S. Waldmann H. Angew. Chem., Int. Ed. 2019;58:17016–17025. doi: 10.1002/anie.201909518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforow A. Wilke J. Binici A. Pahl A. Ostermann C. Sievers S. Waldmann H. Angew. Chem., Int. Ed. 2019;58:14715–14723. doi: 10.1002/anie.201907853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori I. Russo G. Curcio F. Bulli G. Aran L. Della-Morte D. Gargiulo G. Testa G. Cacciatore F. Bonaduce D. Abete P. Clin. Interventions Aging. 2018;13:757–772. doi: 10.2147/CIA.S158513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. Golz C. Strohmann C. Antonchick A. P. Waldmann H. Angew. Chem., Int. Ed. 2016;55:7761–7765. doi: 10.1002/anie.201602084. [DOI] [PubMed] [Google Scholar]
- Karageorgis G. Foley D. J. Laraia L. Waldmann H. Nat. Chem. 2020;12:227–235. doi: 10.1038/s41557-019-0411-x. [DOI] [PubMed] [Google Scholar]
- Firn R. D. Jones C. G. Nat. Prod. Rep. 2003;20:382–391. doi: 10.1039/B208815K. [DOI] [PubMed] [Google Scholar]
- Gao W. Bohl C. E. Dalton J. T. Chem. Rev. 2005;105:3352–3370. doi: 10.1021/cr020456u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgis G. Dow M. Aimon A. Warriner S. Nelson A. Angew. Chem., Int. Ed. 2015;54:13538–113544. doi: 10.1002/anie.201506944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgis G. Warriner S. Nelson A. Nat. Chem. 2014;6:872–876. doi: 10.1038/nchem.2034. [DOI] [PubMed] [Google Scholar]
- Green A. Hobor F. Tinworth C. Warriner S. Wilson A. Nelson A. Chem. – Eur. J. 2020;26:10682–10689. doi: 10.1002/chem.202002153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Aguilar A. Bernard D. Wang S. J. Med. Chem. 2015;58:1038–1052. doi: 10.1021/jm501092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewska E. P. Szymkuć S. Dittwald P. Startek M. Popik O. Mlynarski J. Grzybowski B. A. Chem. 2020;6:280–293. [Google Scholar]
- Caicedo J. C. Cooper S. Heigwer F. Warchal S. Qiu P. Molnar C. Vasilevich A. S. Barry J. D. Bansal H. S. Kraus O. Wawer M. Paavolainen L. Herrmann M. D. Rohban M. Hung J. Hennig H. Concannon J. Smith I. Clemons P. A. Singh S. Rees P. Horvath P. Linington R. G. Carpenter A. E. Nat. Methods. 2017;14:849–863. doi: 10.1038/nmeth.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breinbauer R. Vetter I. R. Waldmann H. Angew. Chem., Int. Ed. 2002;41:2878–2890. doi: 10.1002/1521-3773(20020816)41:16<2878::AID-ANIE2878>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Drewry D. H. Macarron R. Curr. Opin. Chem. Biol. 2010;14:289–298. doi: 10.1016/j.cbpa.2010.03.024. [DOI] [PubMed] [Google Scholar]
- Wetzel S. Bon R. S. Kumar K. Waldmann H. Angew. Chem., Int. Ed. 2011;50:10800–10826. doi: 10.1002/anie.201007004. [DOI] [PubMed] [Google Scholar]
- Sukuru S. C. K. Jenkins J. L. Beckwith R. E. J. Scheiber J. Bender A. Mikhailov D. Davies J. W. Glick M. J. Biomol. Screening. 2009;14:690–699. doi: 10.1177/1087057109335678. [DOI] [PubMed] [Google Scholar]
- Karawajczyk A. Giordanetto F. Benningshof J. Hamza D. Kalliokoski T. Pouwer K. Morgentin R. Nelson A. Müller G. Piechot A. Tzalis D. Drug Discovery Today. 2015;20:1310–1316. doi: 10.1016/j.drudis.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Porwol L. Kowalski D. J. Henson A. Long D.-L. Bell N. L. Cronin L. Angew. Chem. 2020;59:11256–11261. doi: 10.1002/anie.202000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granda J. M. Donina L. Dragone V. Long D.-L. Cronin L. Nature. 2018;559:377–381. doi: 10.1038/s41586-018-0307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernak T. Dykstra K. D. Tyagarajan S. Vachal P. Krska S. W. Chem. Soc. Rev. 2016;45:546–576. doi: 10.1039/C5CS00628G. [DOI] [PubMed] [Google Scholar]
- Hennessy E. T. Betley T. A. Science. 2013;340:591–595. doi: 10.1126/science.1233701. [DOI] [PubMed] [Google Scholar]
- Slobbe P. Ruijter E. Orru R. V. A. MedChemComm. 2012;3:1189–1218. doi: 10.1039/C2MD20089A. [DOI] [Google Scholar]
- Devine P. N. Howard R. M. Kumar R. Thompson M. P. Truppo M. D. Turner N. J. Nat. Rev. Chem. 2018;2:409–421. doi: 10.1038/s41570-018-0055-1. [DOI] [Google Scholar]