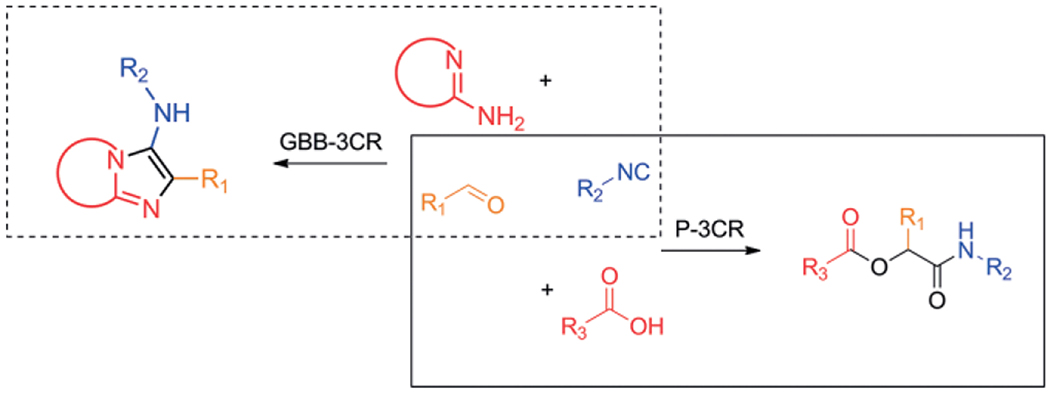
Abstract
Imidazo[1,2-a]pyridine is a well-known scaffold in many marketed drugs, such as Zolpidem, Minodronic acid, Miroprofen and DS-1 and it also serves as a broadly applied pharmacophore in drug discovery. The scaffold revoked a wave of interest when Groebke, Blackburn and Bienaymé reported independently a new three component reaction resulting in compounds with the imidazo[1,2-a]-heterocycles as a core structure. During the course of two decades the Groebke Blackburn Bienaymé (GBB-3CR) reaction has emerged as a very important multicomponent reaction (MCR), resulting in over a hundred patents and a great number of publications in various fields of interest. Now two compounds derived from GBB-3CR chemistry received FDA approval. To celebrate the first 20 years of GBB-chemistry, we present an overview of the chemistry of the GBB-3CR, including an analysis of each of the three starting material classes, solvents and catalysts. Additionally, a list of patents and their applications and a more in-depth summary of the biological targets that were addressed, including structural biology analysis, is given.
Keywords: Imidazo[1,2-a]pyridine; Multicomponent reactions; Isocyanides; Chemical space; Biological activity
Introduction: Medicinal Chemistry and Multicomponent Reactions
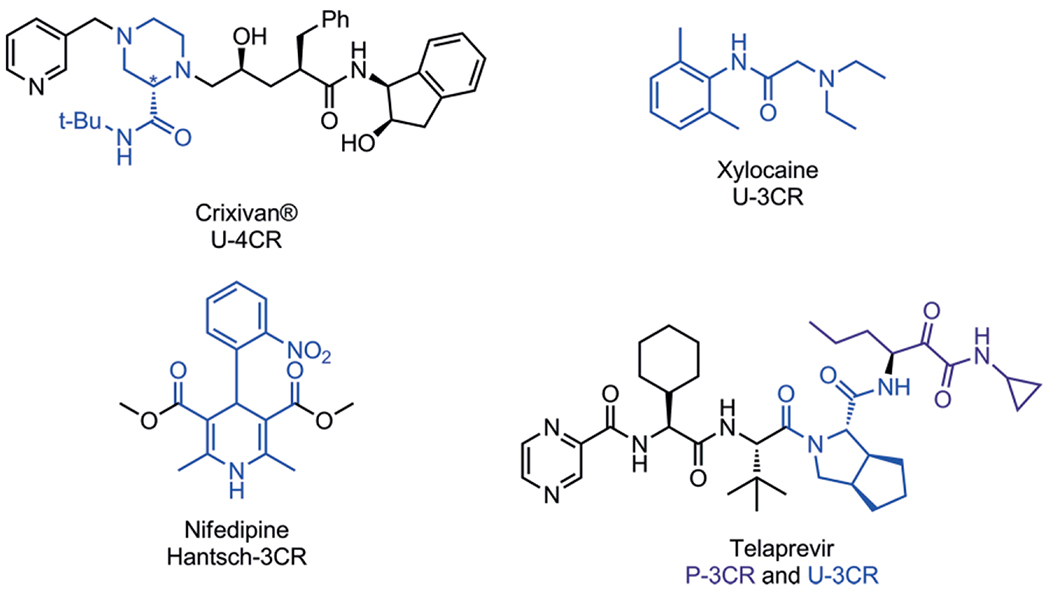
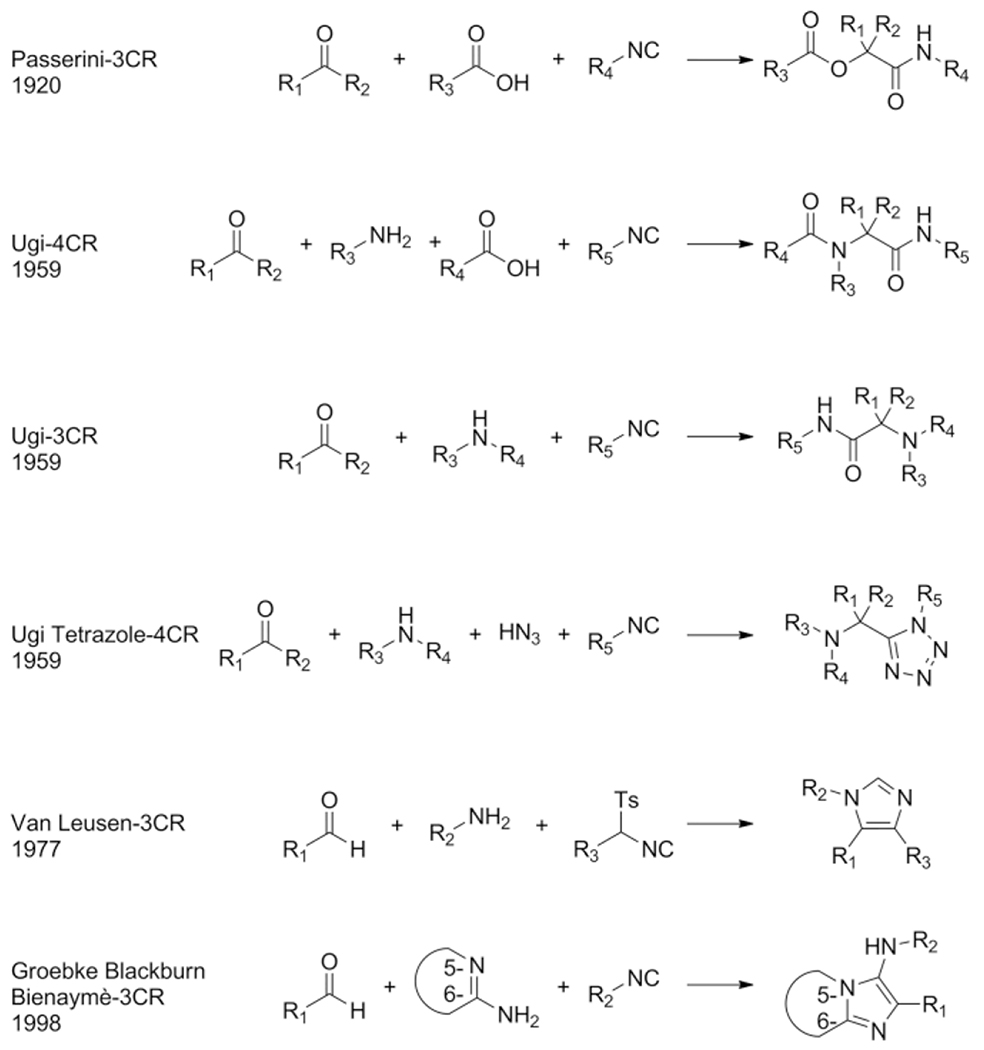
Design and synthesis of biological active compounds are an important field of chemistry as to date there are still many conditions, lacking treatment possibilities. Introduction of novel drugs continuously improve human health despite dramatic increase in world population. The continuous discovery of new biological pathways and the protein targets involved are a great source for medicinal chemist to fill the void in drugs for unmet medical needs. Where the work of structural biologists ends, in elucidating the dynamics and crystallographic structures of large biological structures, medicinal chemists start by the design of agonists and antagonist for those proteins and enzymes. By mimicking the shape and electrostatics of a small natural ligand for example, small molecules can be used to influence those natural processes resulting in inhibition or activation of the associated pathways. Diversity oriented synthesis is often applied to discover small molecules and to optimize their binding affinity in receptor binding sites.[1] Introduction of structural diversity is usually accomplished by the stepwise introduction of the individual moieties in the target molecule and each structural variation requires repetition of a part or even whole synthesis route. This divergent and sequential approach for the synthesis of compound libraries can be quite challenging and material and time consuming. A more convenient way would be to assemble a versatile scaffold and obtaining the variations in a single reaction step. With two component reactions the divergent synthesis of very large chemical space and size based on available building blocks is naturally limited. Multicomponent reactions (MCR), however, allow for the concomitant variation of three or more building blocks at the same time and consequently span a very large chemical space. Since the degree of the reaction enters the exponential, the chemical space is naturally much larger the higher the degree of the reaction. E.g. for 2-component and 5-component reaction based on 1.000 building blocks of each variable class, 1.000.000 and 1.000.000.000.000.000 products can be expected, respectively. This exponential explosion of chemical space is key to the philosophy of MCRs.[2] MCRs allow for the introduction of various scaffold shapes introduced through several MCR variants such as Ugi 3-component reaction (U-3CR),[3] Passerini (P-3CR),[4] Gewald (G-3CR),[5] Groebke-Blackburn-Bienaymé (GBB-3CR),[6–8] Biginelli (B-3CR)[9] and many other MCR name reactions. Decorating and derivatization of the scaffolds is simply done by choosing different starting materials (building blocks). For example, in the Ugi-4CR, with 100 different amines, aldehydes, carboxylic acids and isocyanides as reagents, 100 × 100 × 100 × 100 = 108 different compounds can be synthesized, all connected to the dipeptidic Ugi scaffold. Such information rich chemical space has found recently applications in advanced information technology such as molecular steganography.[10] Clearly, the large chemical MCR space comprise an excellent playground for drug hunters. Of course there are more different starting materials available, and other MCR reactions, where bi-functional orthogonal starting materials could lead to even more complex (poly)cyclic structures in post modifications, such as UDC-procedures (Ugi-Deprotection-Cyclization) and Passerini-reaction-Amine-Deprotection-Acyl-Migration (PADAM) and giving access to a nearly unlimited number of compounds of most diverse shape and electrostatics and composition of 3D pharmacophores.[11,12] The isocyanide-based multicomponent reactions (IMCR), U-3CR, P-3CR, van Leusen (vL-3CR),[13] GBB-3CR and variations, are nowadays the most popular types of MCR because of the versatile behavior of the isocyanides, carbene-type, α-anion and radical reactivity as well as the excellent atom economy. Examples of marketed drugs and lead compounds made by an MCR approach are Xylocaine (Ugi-3CR),[14] Nifedipine (Hantzsch),[15] Telaprevir (Passerini-3CR)[16] and Crixivan (Ugi-4CR)[17] (Scheme 1). It is estimated that approximately 5 % of the currently marketed drugs can be advantageously assembled by an MCR. Thus MCR scaffolds are clearly “drug-like”.
Scheme 1.
Drug synthesized using MCR reactions. The scaffold originating from the MCR is marked in blue or purple. The central piperazine element of the HIV protease inhibitor Crixivan was enantioselectively synthesized by U-4CR, the local anesthetic Xylocaine can be advantageously synthesized in one step using U-3CR, the cardiovascular blockbuster drug Nifedipin by a Hantzsch-3CR and the stereochemical complex HCV protease inhibitor Telaprevir by a combination of P-3CR (purple) and U-3CR (blue) thus reducing the total number of steps by half.
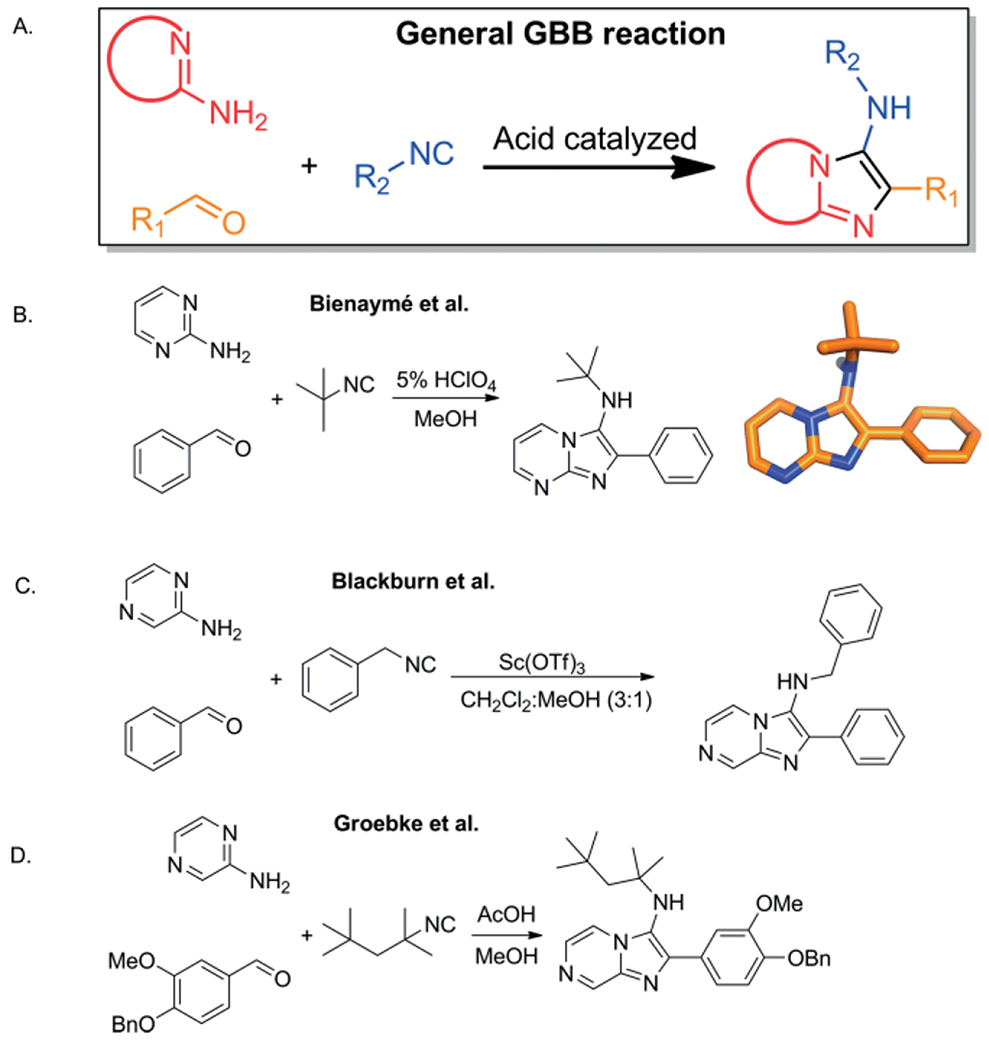
Here we will focus on one of the youngest IMCRs, the GBB-3CR based on its enormous interest in applied chemistry. The development of the reaction started by Groebke et al. (Hoffman-La Roche), initially published as a side reaction of the U-4CR. While studying the effect of various amine components, it was found that amines with a cyclic H2N-C=N substructure (2-aminoazines or amidines), such as 2-aminopyridine, 2-aminopyrazine and 2-aminopyrimidine were yielding the corresponding 3-amino-substituted imidazo[1,2-a]-pyridines, -pyrazines and -pyrimidines respectively.[6] In this report, they made the reference to Sugiura et al. and suggests that the very first report of the GBB-3CR methodology was made almost 30 years earlier by condensing 2-aminopyrazine with formaldehyde, the nitrile compound and sodium or potassium cyanide. This was in contrast to the conventional method to access imidazo[1,2-a] annulated pyridines, pyrazines or pyrimidines whereby a heterocyclization reaction between α-haloketones with the corresponding amidines was performed.[18,19]
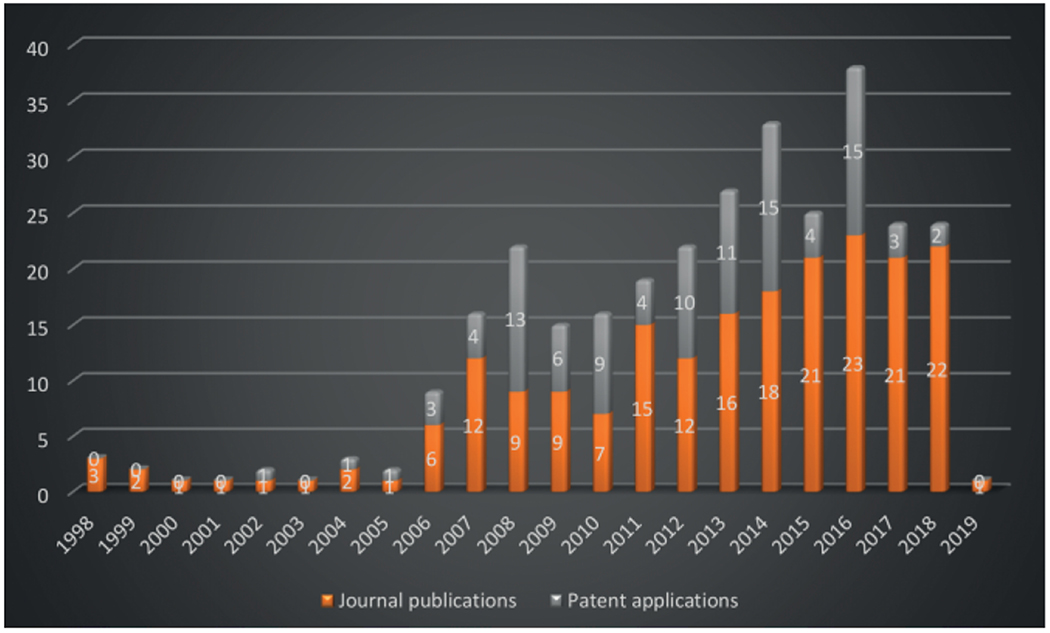
At the very same time Blackburn et al. from Millennium Pharmaceuticals (USA) published their work. The additional value of this work is the use of scandium triflate as Lewis acid catalyst.[20] It follows the finding of Groebke et al. that the presence of an Brønsted acid, such as acetic acid is resulting in higher yields. This pH dependency results from the requirement of proton-assisted activation of the Schiff base to enable attack of the isocyanide component. As a third inventor of the GBB-3CR, Bienaymé et al. (Rhône-Poulenc Technologies (France)) reported in search of new MCR chemistry an approach to apply two covalent bonded or bridged reagents and found a 3-component reaction of 2-aminopyridine or -pyrimidine with aldehydes and isocyanides in the presence of a catalytic amount of perchloric acid in methanol (Scheme 2). This approach was applied to produce 31 examples with variation points in all three components with yields between 33 and 98 %.[8] Due to the nearly simultaneous reports on the discovery of this three component reaction the reaction was called later-on the Groebke-Blackburn-Bienaymé 3-component reaction or in short the GBB-3CR. Interestingly, since its initial description, it took ca. 8 years until an exponential increase in reports was observed using the GBB-3CR methodology broadly in chemistry and also leading to multiple patents (Figure 1).
Scheme 2.
The discovery of the GBB-3CR: A, General description of the GBB-3CR using amidines, aldehydes and isocyanides. B, one of the examples of Bienaymé using 2-aminopyrimidine, benzaldehyde, tert-butyl isocyanide and perchloric acid as catalyst, the formation was verified by X-ray structure refinement (CCDC-101255). C, Blackburn et al. reporting scandium triflate as optional catalyst. D, one of the Groebke examples; with 2-aminopyrazine, catalyzed with acetic acid.
Figure 1.
Development of the GBB-3CR over the last two decades as indicated by increasing number of publications and patents. The numbers were collected through a SciFinder search (April 2019) using GBB-3CR related keywords and structure search of 5- or 6 membered GBB-3CR products including all variations of hetero atoms of the amidine component, corrected for the non GBB-3CR originated 3-amino-substituted imidazo[1,2-a]-pyridines, -pyrazines and -pyrimidines.
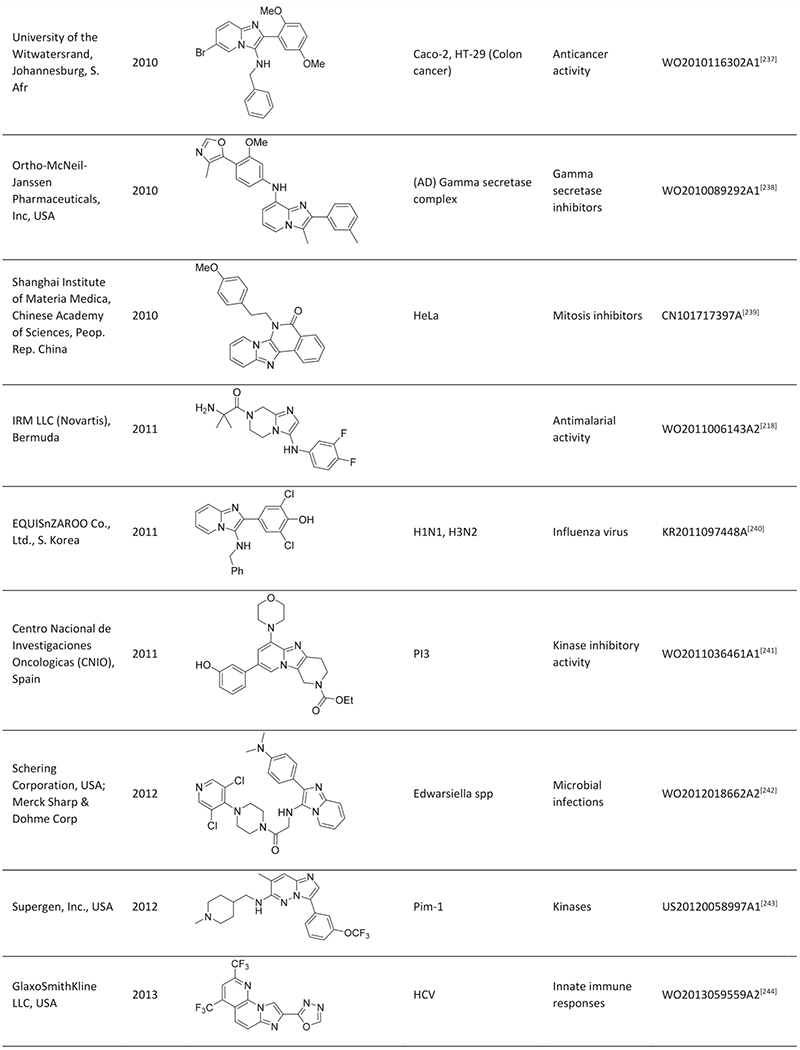
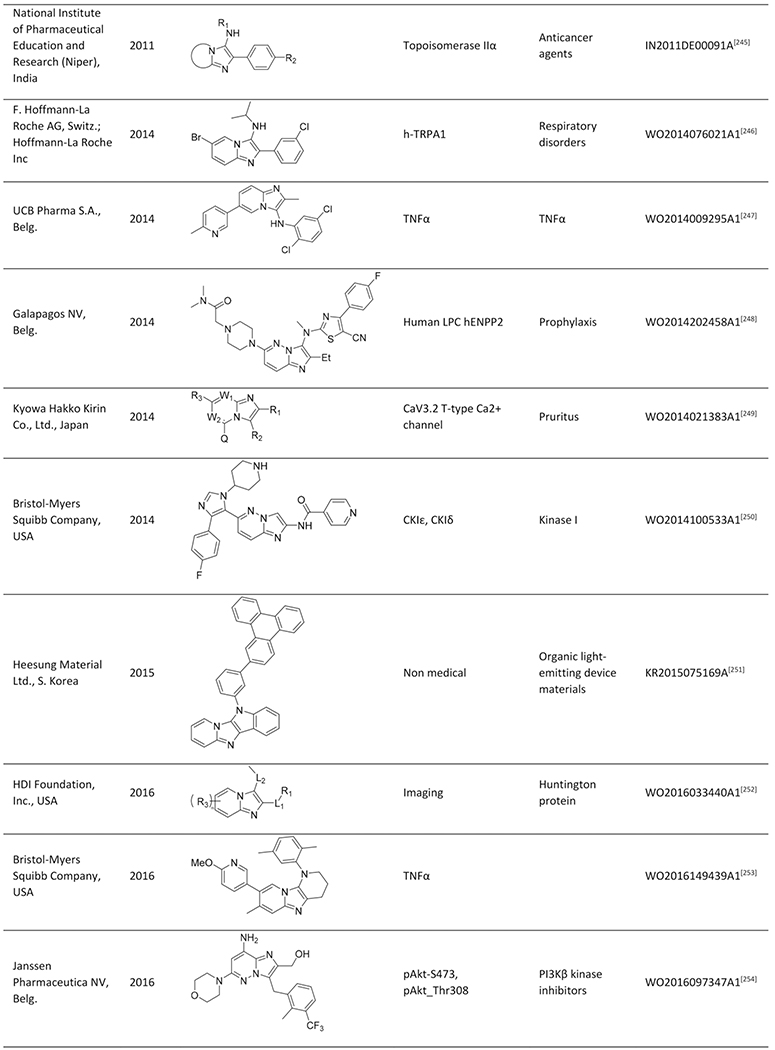
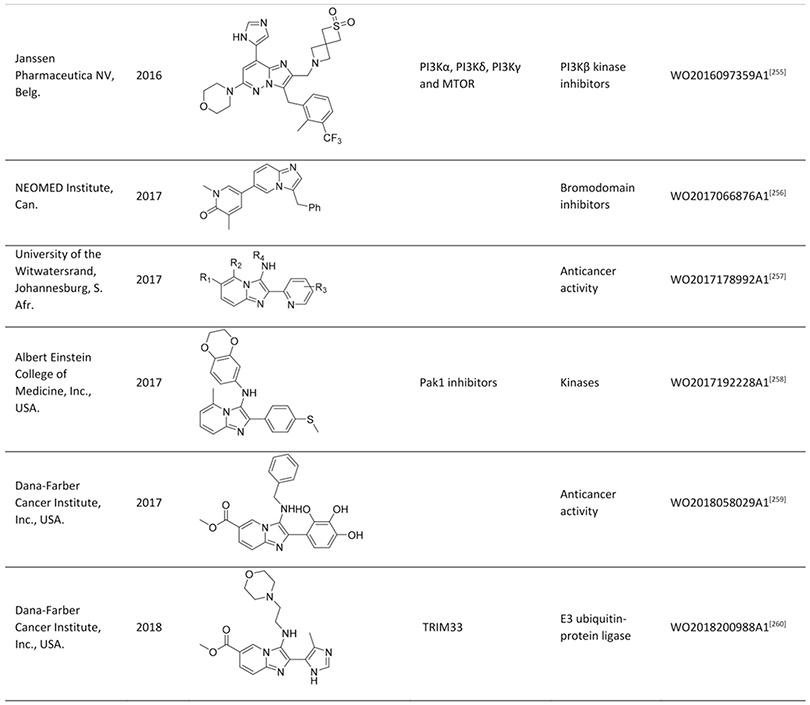
Quite impressively, so far, more than 200 publications and >100 patent applications have been reported, exploiting the GBB-3CR, with a clear trend of further increasing interest. With this in mind a few reviews were published, dedicated to the GBB-3CR as an MCR approach to access 3-amino-substituted imidazo[1,2-a]-heterocycles. Singh et al. were the first to cover the reaction and addressed the used catalysts and the general chemistry behind the GBB-3CR.[21] Abdel/Wahab et al. reported a more comprehensive review, highlighting biological targets the GBB scaffold could be used for, possible post-modifications and to a certain extend the scope and limitations were discussed with some examples of amidines, isocyanides, aldehydes used in the GBB-3CR.[22] In addition Liu published an overview on the Asinger[23] and GBB-3CR, in a historical fashion with emphasis on the possible post modifications and applications of both reactions.[24] Also Liu dedicated a mini review solely on potent applications of the imidazo[1,2-a]-heterocycles.[25] To the best of our knowledge patents were never discussed and our review tries to give a complementary twist by giving insight into all the used reactants, all the catalysts and solvents used in the GBB-3CR, detailed discussion of the structural biology, in order to get more insight in how GBB scaffolds interact with therapeutically relevant targets.
Mechanism
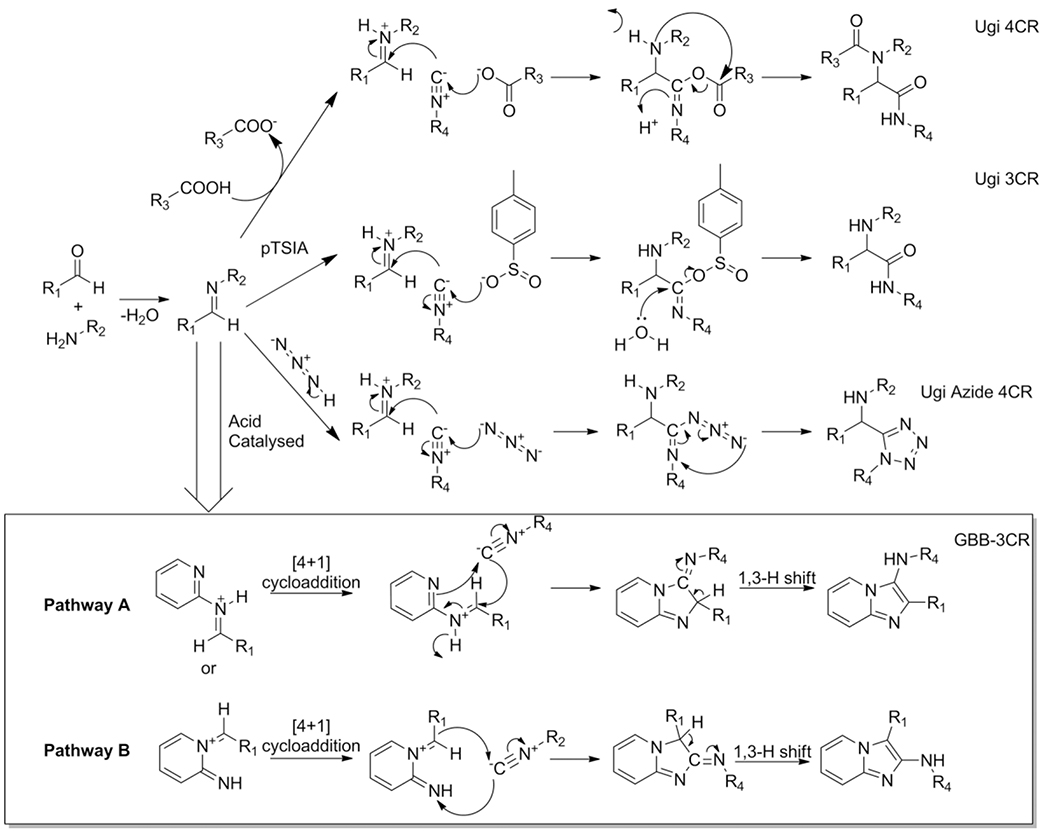
Examples of isocyanide based MCR’s (IMCR’s) are the P-3CR, vL-3CR, U-4CR, Ugi-azide-4CR, U-3CR, and GBB-3CR (Scheme 3). Aside from the P-3CR and the vL-3CR, the latter 3 IMCR’s are mechanistical variations on the U-4CR. The acid component is decisive in how the iminium species will react towards other components, rearrange and ultimately is incorporated into the scaffold.
Scheme 3.
Overview of the most common IMCR’s.
For example, the U-3CR is only taking the proton of specific acidic reagents (e.g. p-toluenesulfinic acid (pTsIA)) and eliminating the Mumm rearrangement that otherwise concludes the U-4CR. The Ugi azide-4CR reaction adheres to a similar mechanism as the U-3CR, hydrazoic acid is introduced by using sodium azide or TMSN3. In the GBB-3CR the additional reactivity of the endocyclic nitrogen in the amidine component allows for the formation of a different scaffold, whereas the acid component is not incorporated in the final product and serves only catalytic purposes instead. In the GBB-3CR, the imine intermediate is activated by a Lewis or Brønsted acid, and follows a formal [4+1] cycloaddition sequence concluding with aromatization via a 1,3-H shift (Scheme 4 Pathway A) to form the imidazo[1,2-a]pyrimidine when 2-aminopyridine is used. The GBB-3CR could follow two possible pathways leading to regioisomers as indicated in Scheme 4. Pathway A is the most common and is referred to as the GBB-3CR and pathway B as the “inverse” GBB-3CR.
Scheme 4.
Variations of the Ugi reaction and the Groebke Bienaymé Blackburn reaction. In the GBB mechanism one of the most commonly used amidines; 2-aminopyridine as R2-group was used in the mechanistic overview to simplify the scheme.
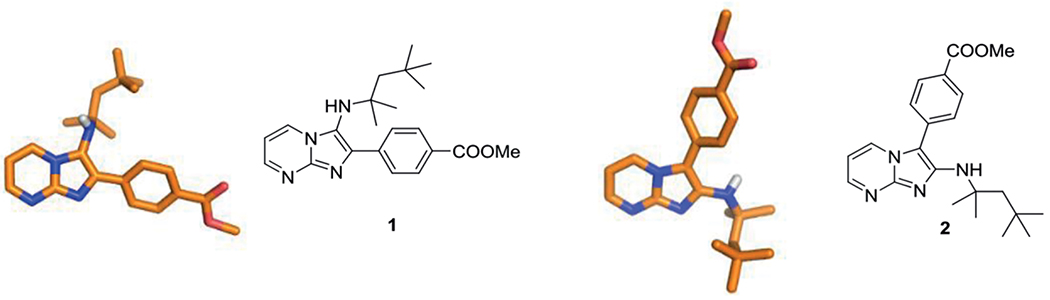
2-Aminopyrimidines tend to form both regioisomers as it was first described by Bradley et al., trying to explain the low yields of some of their reactions while the starting materials were fully consumed.[26] Isolation of a second product having nearly identical TLC-Rf values, mass and 1H NMR spectra gave reason to perform an X-ray structure analysis, uncovering the two regioisomers 1 and 2 depicted in Scheme 5. Luckily, the normal GBB product is the major product for most cyclic amidine building blocks, thus rendering it a quite useful synthetic reaction. In fact, in most cases not even traces of inverse GBB products can be observed.
Scheme 5.
Two possible regioisomers of the GBB-3CR. Left 1 (CCDC-189548) a typical GBB-3CR imidazo[1,2-a]pyrimidine and on the right 2 (CCDC-189549) its inverse GBB regioisomer.
Proceedings in the Development of the GBB-3CR
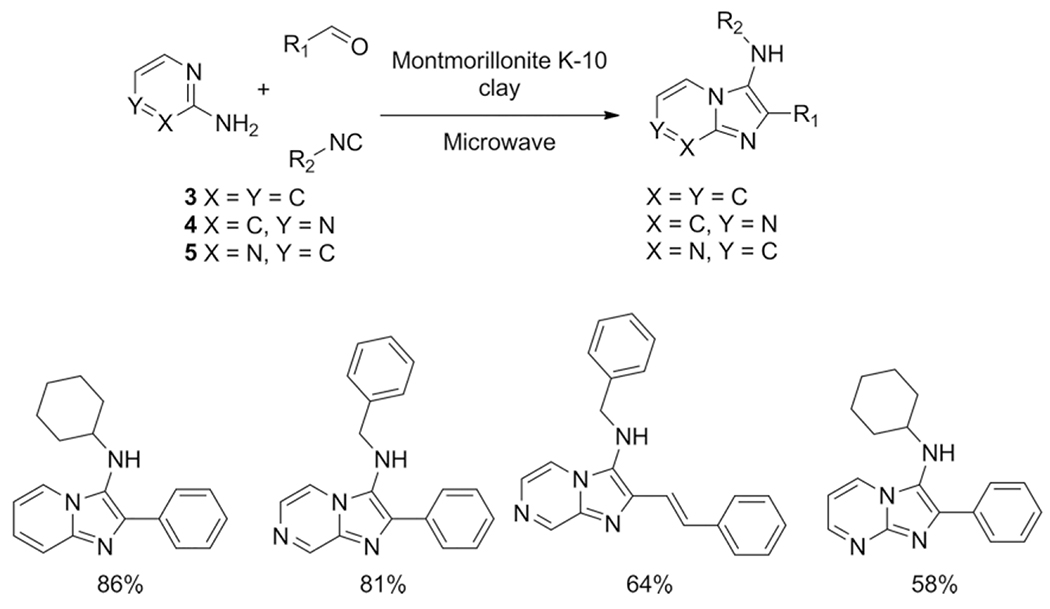
Based on the pharmacological relevance of imidazo[1,2-a]-heterocyclic compounds and their easy access through the GBB-3CR, many scientists explored the scope and limitations of this reaction, trying a large variety of catalysts, solvents and temperature conditions, resulting in a wide variety of publications with a confusing high number of different conditions. Herein, some of the more relevant reports are discussed, showing important developments, noteworthy applications and several reaction conditions used are described to fully comprehend the versatile character of the GBB-3CR. For example, Varma et al. showed that a microwave (MW) assisted solvent-free method could be applied by the aid of montmorillonite K-10 clay.[19] The conditions were quite unusual that time since microwave assisted procedures were not yet broadly applied in 1999. Here a household microwave was used with an unsealed test tube irradiated at 900 W for 3 minutes. Together with the clay catalyst and variations of simple aromatic aldehydes, various isocyanides and 2-amino-pyridine (3), pyrazine (4) or pyrimidine (5) yields of >80 % were obtained (Scheme 6).
Scheme 6.
Montmorillonite K-10 clay-catalyzed and solvent free GBB-3CR.
Whittaker et al. showed a very similar scandium triflate catalyzed reaction via a microwave assisted GBB-3CR in 10 minutes with yields between 33–93 % and using substituted benzaldehydes and mostly aminopyridines, interestingly also a 5-membered aminothiazole substrate was introduced, giving lower yields mostly related to side products (6) formed by the addition of methanol to the intermediate Schiff base (Scheme 7).[8,27] The side product formation could, however be suppressed by the use trifluoroethanol as a less nucleophilic solvent.
Scheme 7.
Application of electron-poor cyclic amidines in the GBB-3CR results in poor conversion and side reactions such as addition of MeOH to the Schiff base intermediate.
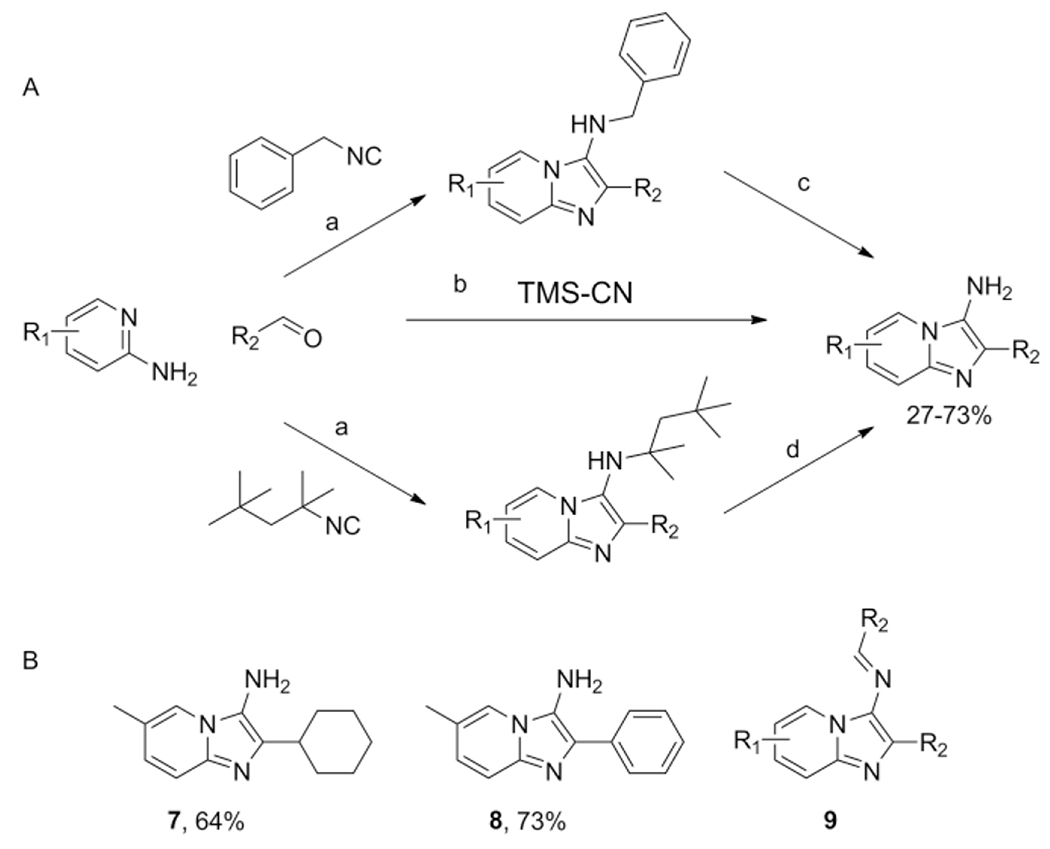
Hulme et al. showed TMSCN as an equivalent to the simplest isocyanide HNC, to directly access 3-aminoimidazo[1,2-a]pyridines 7–8, which otherwise would require an additional deprotection step when a convertible isocyanide such as the Walborski’s reagent (1,1,3,3-tetramethylbutyl isocyanide) is used in the GBB-3CR (Scheme 8).[28]
Scheme 8.
Access to N-exocyclic unsubstituted GBB products. A: reagents and conditions: a) aminopyridine (1 equiv.), R2CHO (1 equiv.), 2 (1 equiv.) or benzyl isocyanide (1 equiv.), Sc(OTf)3 (5 mol-%), 16 h; b) aminopyridine (1.2equiv.), R2CHO (1 equiv.), TMSCN (1 equiv.), Sc(OTf)3 (5 mol-%), MeOH, microwave, 10 min, 140 °C. Followed by Si-trisamine (5 equiv.); c) H2, EtOAc, 24 h; d) 10 % TFA/CH2Cl2, 18 h B: representative examples and their yields and the observed Schiff base as side product.
Some side product was found to be the Schiff base 9, formation could be prevented by using an excess of amidine. After the reaction the residual catalyst Sc(OTf)3 was removed using 5 equivalents of Si-trisamine, which is a powerful scavenger of electrophiles as well as an effective scavenger for transition metals.
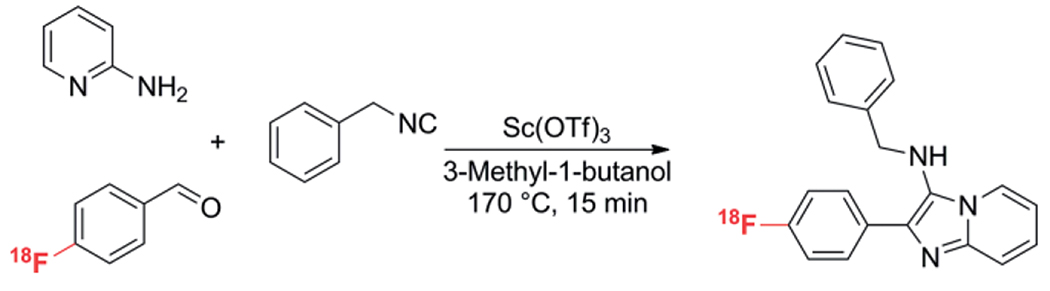
GBB-3CR products are often observed as being strongly fluorescent. Balakirevet al. introduced “Flugis”; fluorescent Ugi products as drug-like probes for the identification and visualization of potential targets.[29] The compounds described as U-3CR compounds arise from the GBB-3CR and were used with the rationale to incorporate drug-like scaffolds into fluorescent molecules. The often fluorescent nature of GBB products should be kept In mind during biophysical receptor-ligand screens based on fluorescence principles! For example, in a recent screen for inhibitors of the NS3/4A serine protease of the hepatitis C virus, some of the compounds with the [1,2-a]pyridine scaffold were found to exhibit auto-fluorescence in UV that interfered in the enzymatic fluorescence detection assay.[29] This led to their approach to incorporate this scaffold that is synthesized using the GBB-3CR in their search of fluorophores in a 1600 compound containing microarray, resulting in fluorescent imidazo[1,2-a]-pyridine compounds, which are known to interact with the peripheral benzodiazepine receptor (known as the translocator protein TPSO) and GABAA benzodiazepine receptors. The compounds were tested for their affinity with, and use as TPSO imaging probes (Scheme 9).
Scheme 9.
18F introduction in GBB-3CR products.
The introduction of a 18F-label to PET radiotracers via 18F-labelled prosthetic groups using MCR chemistry was discussed by Gouverneur et al.[30] Labeling of 18F requires reaction conditions which might not always be compatible with the substrate. By using 18F-benzaldehydes in MCR assisted radiochemistry, mild conditions could easily be performed in the U-4CR, P-3CR, B-3CR and GBB-3CR while maintaining a swift reaction necessary for the rather short half-life time of 110 min for 18F.
The “hot” GBB-3CR was performed in 3-methyl-1-butanol under conventional heating in most cases and some under MW conditions, where 150–170 °C was found to give the highest yields; 64–85 %.
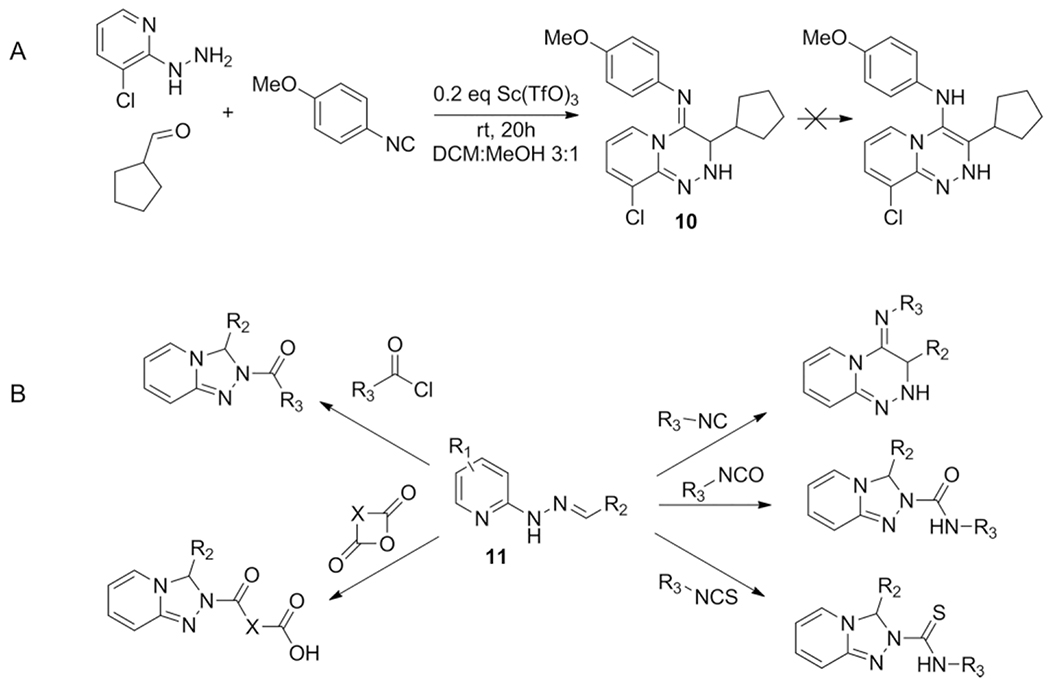
The use of hydrazines in the GBB-3CR has not been explored until 2014. A variation in which 2-hydrazinopyridine is used to obtain bicyclic pyridotriazines was introduced by Hulme et al.[31] The proposed mechanism is quite similar to that of the GBB-3CR reaction, it involves a non-concerted [5+1]-cycloaddition of the Schiff base and isocyanide.
Comparable to the example of ketones (Kumar et al. example in chapter 3.2) in the GBB-3CR discussed below, there is no rearomatization seen with both aldehydes and ketones, which otherwise concludes the GBB-3CR. The appendant moiety originating from the isocyanide component remains in the product as a stable imine (10). The reactive nature of the Schiff base intermediate 11 allows introduction of not only isocyanide, but isocyanates, acyl chlorides and cyclic anhydrides as well (Scheme 10).
Scheme 10.
A: hydrazines undergo a GBB-like [5+1] cycloaddition to furnish bicyclic triazines. This given example was obtained in 85 % yield, aromatic aldehydes will not result in product formation B: The reactivity of the hydrazine derived Schiff base could be exploited for other transformations with various electrophiles.
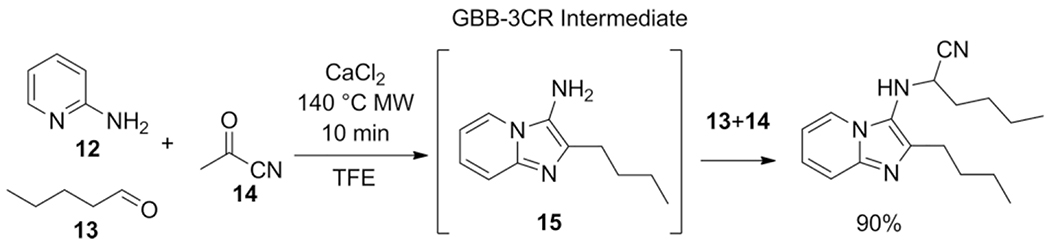
Hulme et al. reported the use of acyl cyanide (14) as isocyanide replacement in the GBB-3CR. The resulting primary amine (15) subjected to the excess of aldehyde (4 eq.) and acyl cyanide (3 eq.) undergoes a domino/tandem acyl-Strecker reaction.[32] A great example of a modified GBB-3CR: on one hand the acetyl cyanide serves as a less toxic replacement for TMSCN, on the other hand a great catalyst, acetic acid is freed during the course of this one-pot 3-step cascade reaction (Scheme 11).
Scheme 11.
Acetyl cyanide assisted one-pot three-step GBB-Strecker cascade.
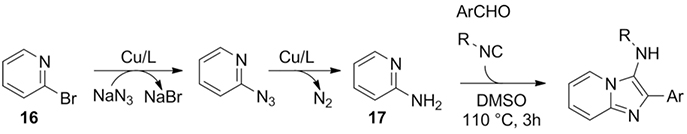
In situ generation of amidines for use in the GBB-3CR was demonstrated by Mahdavi et al., using 2-bromopyridine 16, sodium azide, and aldehyde and isocyanide in a copper-catalyzed 4CR.[33] The activated bromine is converted to amine 17 via a reductive amination as depicted in Scheme 12, prior to take part in the GBB-3CR. The reaction sequence is described a one pot four component reaction, the reaction could as well be considered as a three-component reaction with in situ generated 2-aminopyridine.
Scheme 12.
The scope of the GBB-3CR was expanded by the use of 2-bromopyridine, in situ converted into the essential amidine.
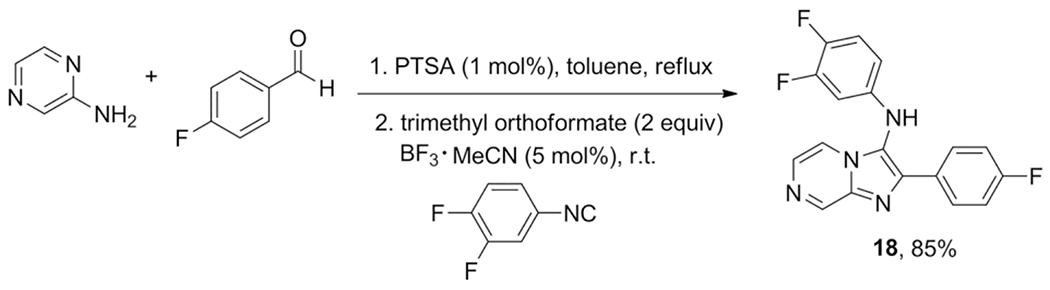
Procedures in process chemistry are necessarily studied in depth for optimal conversion to the desired products. In this respect the report of Mathes et al. is very useful, as it investigates the driving forces of the GBB-3CR, introducing alternative purification methods that circumvent labor-intensive chromatography.[34] Apart from the usual reagents, the Schiff base preformation was promoted by adding a catalytic amount of pTsA. The following cycloaddition was in turn catalyzed by borontrifluoride-acetonitrile complex (BF3·MeCN) and two equivalents of trimethyl orthoformate as dehydrating agent was added to increase the rate of the reaction significantly, giving good yields in less than a day of reaction time (Scheme 13). Purification was done by adding 1,3 equivalents of sulfuric acid to precipitate the GBB-3CR products from i-PrOH as sulfate salts in high purity. From the optimized method, a large scale reaction was performed at 100 mmol scale, yielding the GBB product 18 in 82 % yield, against 85 % on 1 mmol scale, proving the scalability of the reaction.
Scheme 13.
The optimized one-pot two-step process of a BF3·MeCN catalyzed GBB-3CR.
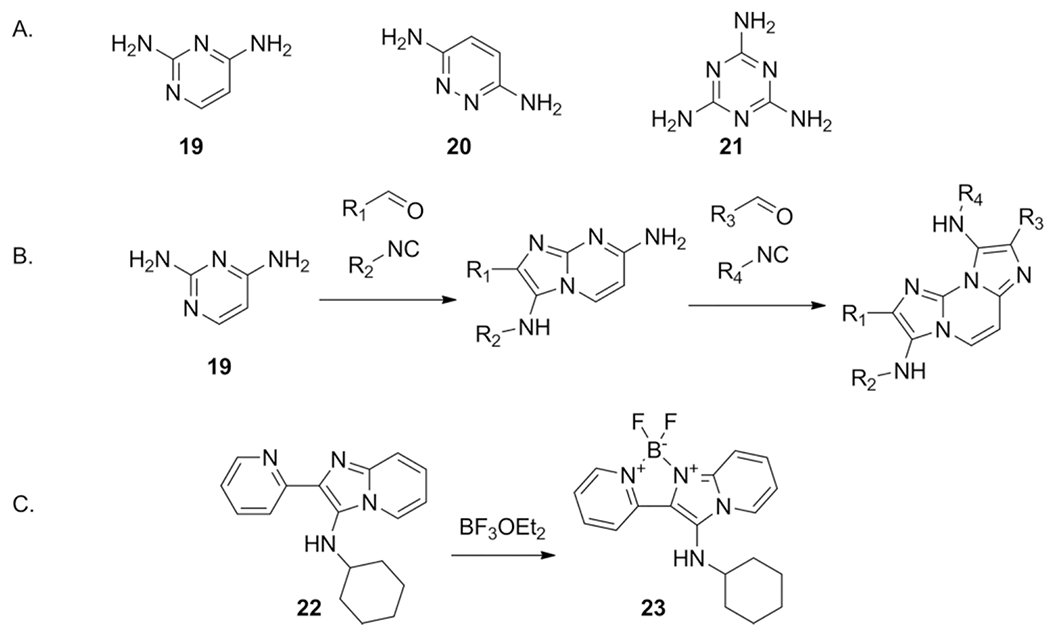
Using bis- (19–20) or tris amidines in the GBB-3CR allows for multiple MCRs in a single transformation as demonstrated by Lavilla et al.[35] The position of each amidine when applying asymmetric bis-amidine 19 determines the reactivity and therefore the selectivity of the first GBB-3CR and enables introduction of different aldehyde and isocyanide components in the second GBB-3CR. This regioselectivity is also seen in symmetric bis-amidines, but require stoichiometric amounts of the aldehyde and isocyanide components. Reactions with the tris-amidine melamine 21 exclusively yielded symmetric products. The fluorescence abilities of the synthesized compounds was highlighted and through introduction of EWG or EDG functionality at specific locations, the fluorescence emission wavelengths could be tuned. Additionally BODIPY (93) like fluorophores were made by reacting α-pyridyl GBB-3CR compounds with BF3OEt2, to create a BF2 bridge between the pyridine and imidazo rings. This bridged compound (23) showed a shift in the emission wavelength by 60 nm, a 40-fold emission increase and pH insensitivity as compared to unbridged (22) (Scheme 14).
Scheme 14.
Bis-amidines in the GBB-3CR: A. The bis- and tris amidines reacted in the GBB-3CR. B. Asymmetric product formation using different aldehydes and isocyanides in two separate MCRs. C. Enhancement of fluorescent properties by introduction of a BF2 bridge.
Scope and Limitations of the GBB-3CR
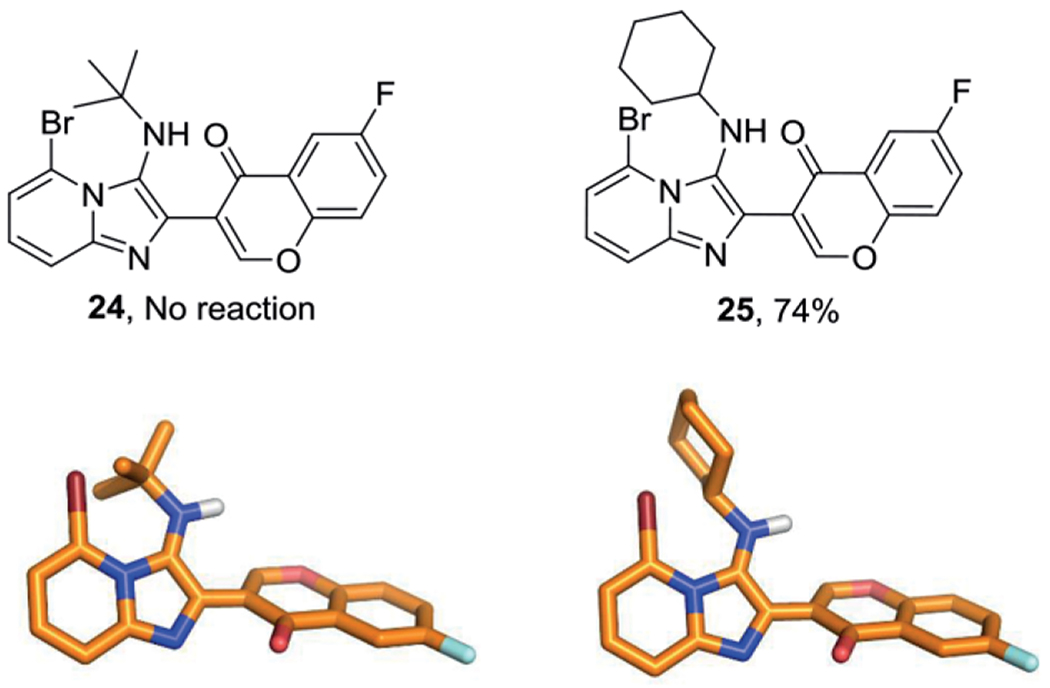
Immediately after the introduction of the GBB-3CR approach to access imidazo[1,2-a] heterocycles, the scope and limitations were elaborated. However, in each of the attempts only a small selection of variables was assessed. This went from solvent screening, varying reaction conditions, catalysts and reagent use to solid phase approaches. A good example is the use of glyoxylic acid to obtain an uncatalyzed formaldehyde-based product. In the general scope and limitation of the GBB-3CR it was found that the reaction is best performed at room temperature in MeOH, with a concentration ranging between 0.3–1.0 m, using arylaldehydes, 2-aminopyridines, and aliphatic isocyanides in stoichiometric amounts, in the presence of a catalytic amount of 10 mol-% Sc(OTf)3. Difficulties in predicting reactivity were found with some combinations of starting materials and cannot always be attributed to electronic factors, as steric effects have a significant effect as well, illustrated in Figure 2.[35]
Figure 2.
Steric effects in the GBB-3CR. The bulk effect of tert-butyl prevents the reaction from happening, the optimized geometry was calculated for compound 24, exchanging the tert-but- for a cyclohexyl moiety, affording compound 25 in a good yield.
Aliphatic aldehydes give good yields when aromatic isocyanides are used, however, when both aldehyde and isocyanide are both electron rich, a reduction in yield was described recently.[34] Although such findings are valuable starting point for better understanding the GBB-3CR, we’ve tried to illustrate the scope and limitations in a broader sense, including the effect of substituents on each of the reported starting materials. Therefore, a tabular summary of amidines, aldehydes, isocyanides and catalysts used in the GBB-3CR is presented in the following. Additionally, the tables act as reference guide to which components are suitable, including every report the component was successfully used in. The benefit here is a complete and simple access to the possible starting materials, grouped according to aromatic, hetero aromatic and aliphatic nature and electron donating or withdrawing substituents (EDG and EWG respectively) and bulkiness. Thus a quick look in the tables can reveal unambiguously which component has been used previously and will likely work in other instances.
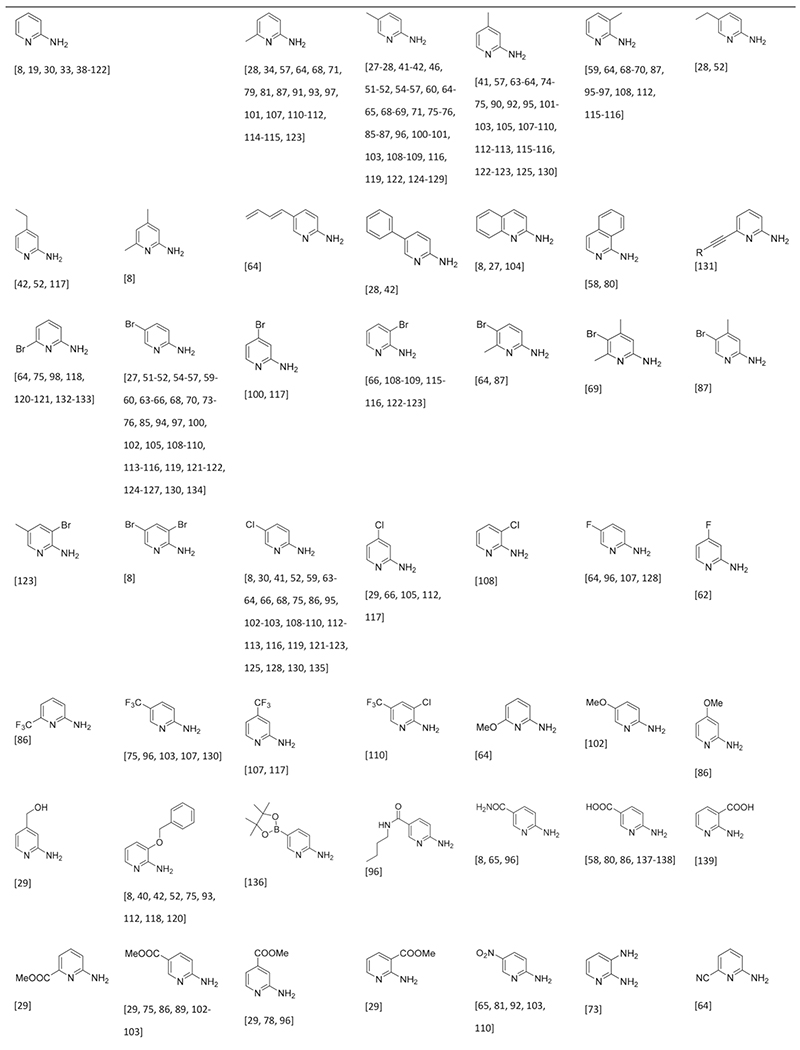
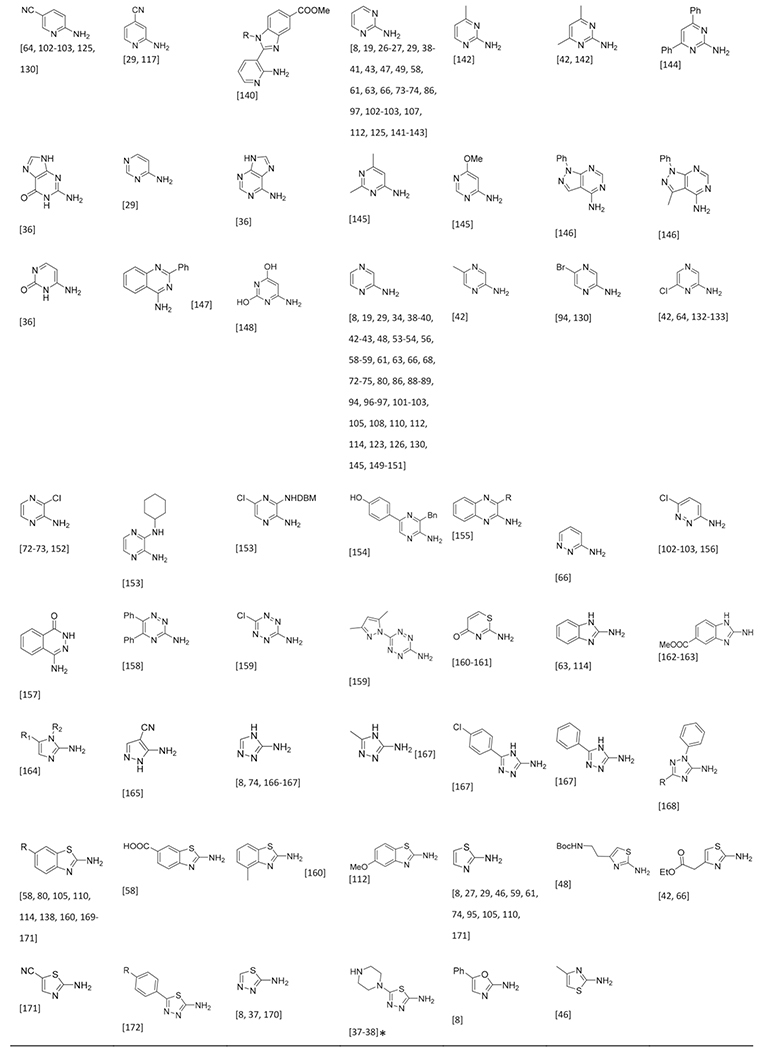
Cyclic Amidines in the GBB-3CR
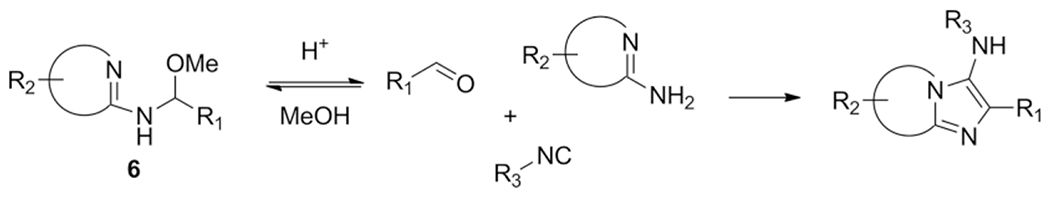
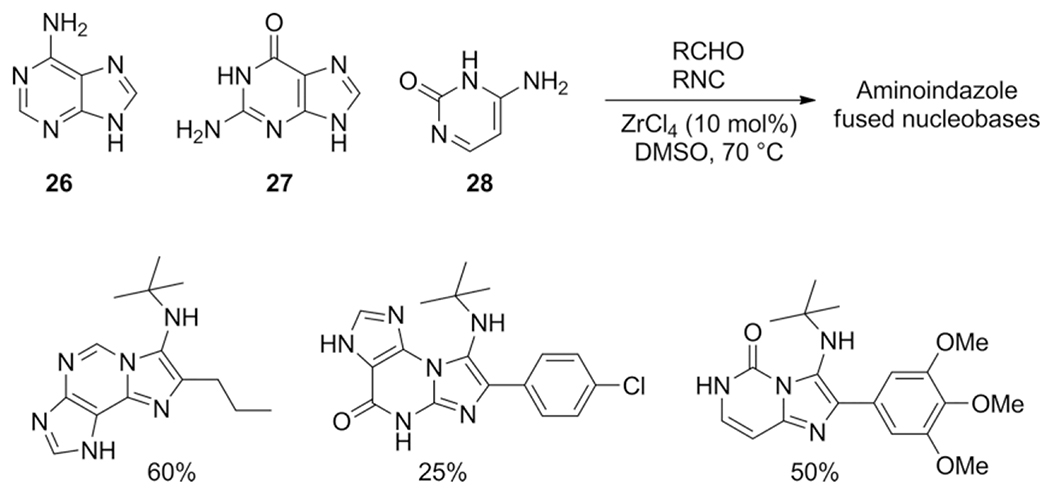
The amidines are key to give the scaffold its typical imidazo-[1,2-a] heterocyclic form, whereas the aldehyde and isocyanide components are more appendant substituents, sometimes called scaffold decoration. Nearly 90 different amidines were used in GBB-3CR, of which 22 are five membered and 66 are six membered amidines. All of them were sorted in Table 1 according to ring size, type and additional substituents. The most abundant amidine in the Table 2-aminopyridine is where the GBB-3CR was discovered with and is not surprisingly used in many model reactions to test other solvents and catalysts. Substituted 2-aminopyridines also show good reactivity, giving products in good yields. Electron withdrawing substituents, mostly halogens seem to increase the yield, with the exception of nitro-groups. Alkyl groups and other more EDG generally give lower yields. More electron deficient pyrimidines give lower yields in the GBB-3CR, both 2- and 4-aminopyrimidine with EWG substituents were reported as low yielding. Aminopyrazines are overall reported as good yielding. A broad range of 5-membered amidines appears often with other hetero atoms, such as oxazoles and thiazoles, and are not very reactive resulting in less good yields. 3-Amino-1,2,4-triazole, on the other hand, is reported as good yielding amidine. Nucleobase derived compounds are popular as they possess good hydrophilicity and solubility, which are therefore potentially therapeutically relevant heterocycles. Adenine 26, guanine 27 and cytosine 28 are nucleobases that bear an amidine moiety which was exploited in the GBB-3CR by Madaan et al. to furnish aminoimidazole-fused nucleobases.[36] The polar nature of these amidines required a more polar solvent than methanol, as this solvent did not yield any product at all. The screened solvents PEG-400, ethylene glycol, DMSO, DMA and DMF all give product, with yields varying between 23 to 68 % with ZrCl4 in DMSO as a catalyst (Scheme 15). Even the otherwise difficult to modify guanine is giving GBB-3CR products albeit in rather low yields.
Table 1.
Table 2.
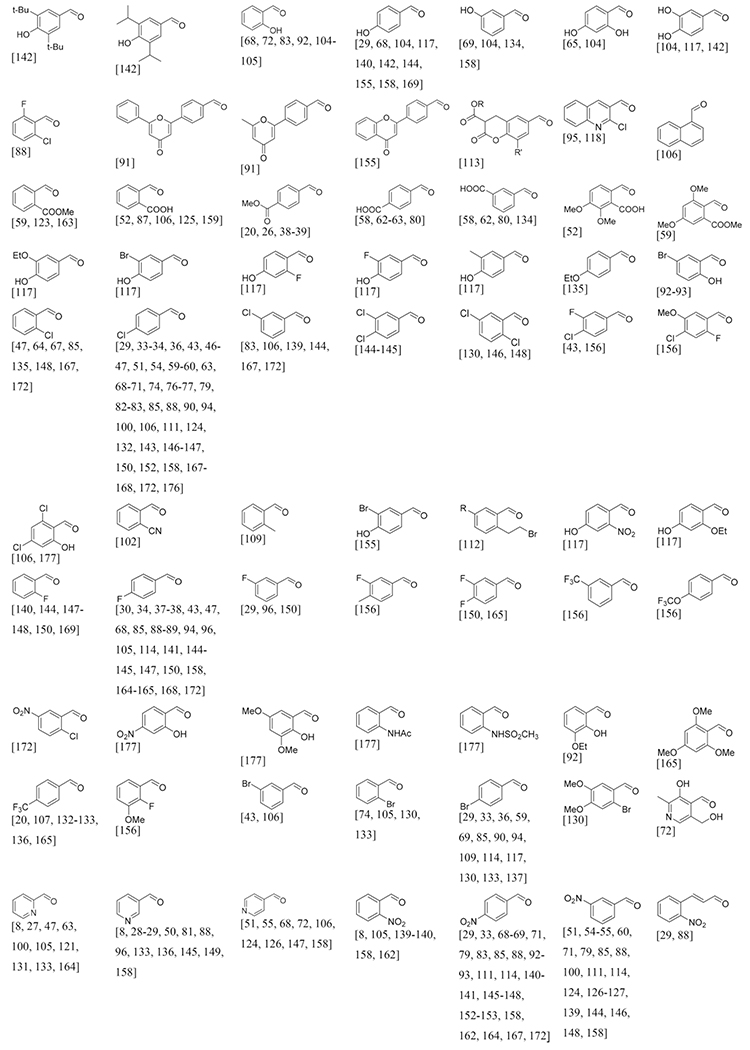
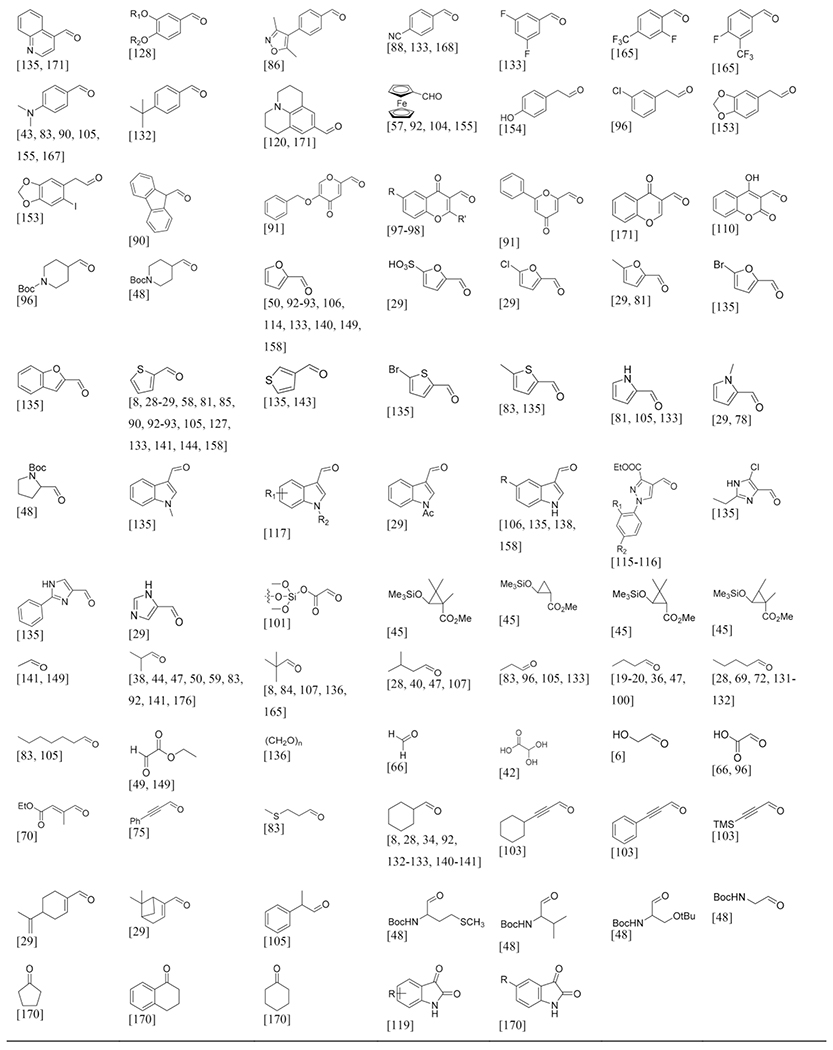
The large variety of aldehydes sorted by origin and substituents.
Scheme 15.
Synthesis of aminoimidazole-condensed nucleobases.
A feature which can be generally observed with many MCRs is their great functional group compatibility. Thus, heteroaromatic amidines can comprise all halogens, and pseudo halogens such as nitrile, nitro, methoxy, free carboxylic acids, esters, unprotected primary and secondary amines, unprotected phenolic and aliphatic hydroxyl, amides, alkynes, alkenes, and boronic acid esters. This great functional group compatibility is important for further reactivity of the initial GBB-3CR products and also for optimal interaction within a receptor pocket.
Aldehydes
Aldehydes are common building blocks which are cheap and have good commercial availability and with 180 different records they are very broadly applicable in the GBB-3CR reaction with great variability (Table 2). The diversity of aldehydes found is broad, from aromatic to aliphatic aldehydes with many different substituents, both electron withdrawing and donating and bulky and small groups and also aldehydes containing reactive to labile protective groups that allow secondary modifications. Aromatic aldehydes generally form stable Schiff bases, where an effective conjugation system is beneficial for its stability. Schiff bases from aliphatic aldehydes are found to be less stable and readily polymerize, which translates to reduced yields of subsequent transformations. Benzaldehyde is the mostly used aldehyde for reasons such as detectability on TLC for reaction monitoring and good reactivity. Substituents on benzaldehydes do affect their reactivity. Using substituted benzaldehydes bearing an electron donating group usually increases the yields, whereas withdrawing substituents reduce the yields.
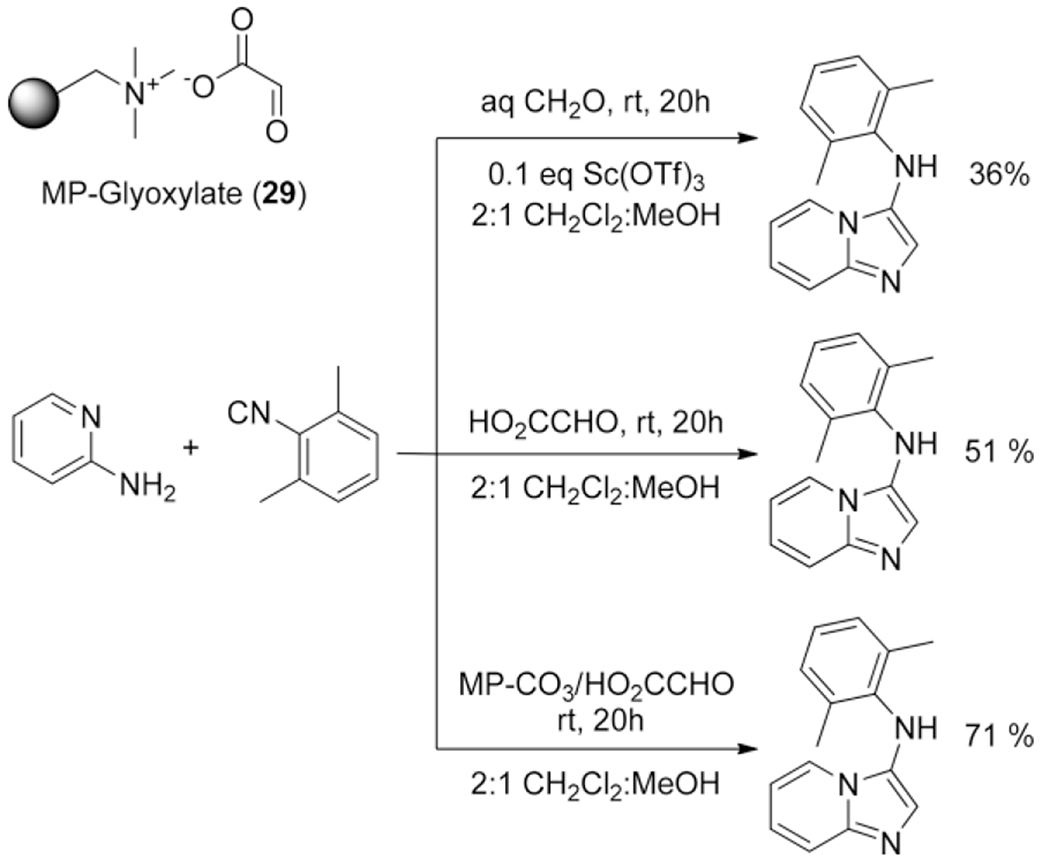
It would be an extensive exercise to discuss every variant in detail, therefore the more interesting examples were highlighted. The use of formaldehyde in order to obtain 2-unsubstituted-3-amino-imidazoheterocycles in the GBB-3CR was not reported until 2004 by Kercher et al.[42] Successful preparations of the unsubstituted imidazole were actually scarce and the few reports that did, showed low yields in non-efficient synthetic routes.[173–175] In this report formaldehyde (aqueous) and paraformaldehyde were used, resulting in poor yields of 36 and 44 % respectively. Other formaldehyde substitutes were screened, showing that glyoxylic acid is giving good yields up to 71 % with glyoxylic acid immobilized on macroporous polystyrene carbonate (MP-CO3) 29 in an uncatalyzed GBB-3CR reaction (Scheme 16).
Scheme 16.
Formaldehyde and formaldehyde surrogates in the GBB-3CR and their associated yields.
Kennedy et al. reported three examples with the successful use of paraformaldehyde with varying yields between 68–78 % in a microwave assisted GBB-3CR in MeOH, catalyzed with 4 mol-% MgCl2 at 160 °C in just 10 minutes.[136]
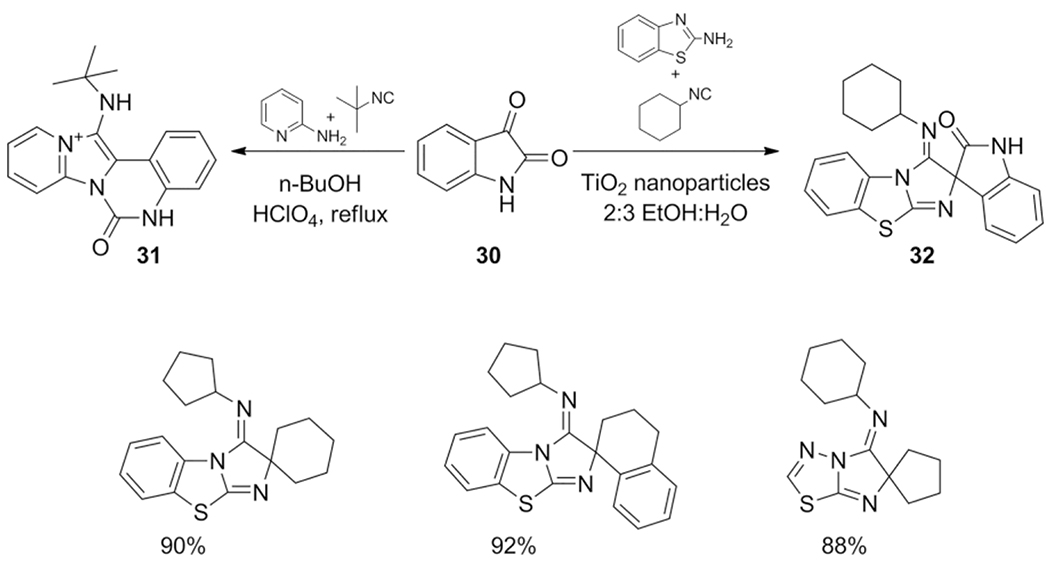
The first ketone example in a tetracyclic fused imidazo-[1,2-a]pyridines from isatin 30 was reported by Che et al.[119] The special multi-reactive nature of isatin is allowing for a formal [4+1] cycloaddition with different isocyanides and subsequent rearomatization via [1,5]-H shift through a retro-aza-ene reaction compound 31 (Scheme 17).
Scheme 17.
Two possible reaction pathways with the α-ketoamide isatin and some examples of the GBB-3CR ran with other ketones.
Using other ketones, Kumar et al. reports the synthesis of spiro-heterocycles catalyzed with TiO2 nanoparticles in excellent yields.[170] The quaternary spiro carbon lacks, however, the proton required for [1,3]-H shift, therefore rearomatization of the unstable [4+1] adduct could not take place. These adducts were reported as the products and also contains an example of an isatin containing product 32, wherein the structure was found to be surprisingly different from the isatin products described by Che et al.[119] The conflicting structures are not likely to be attributed to the amidine and isocyanide components and the right structure is without appropriate structure elucidation such as X-ray diffraction spectroscopy difficult to confirm.
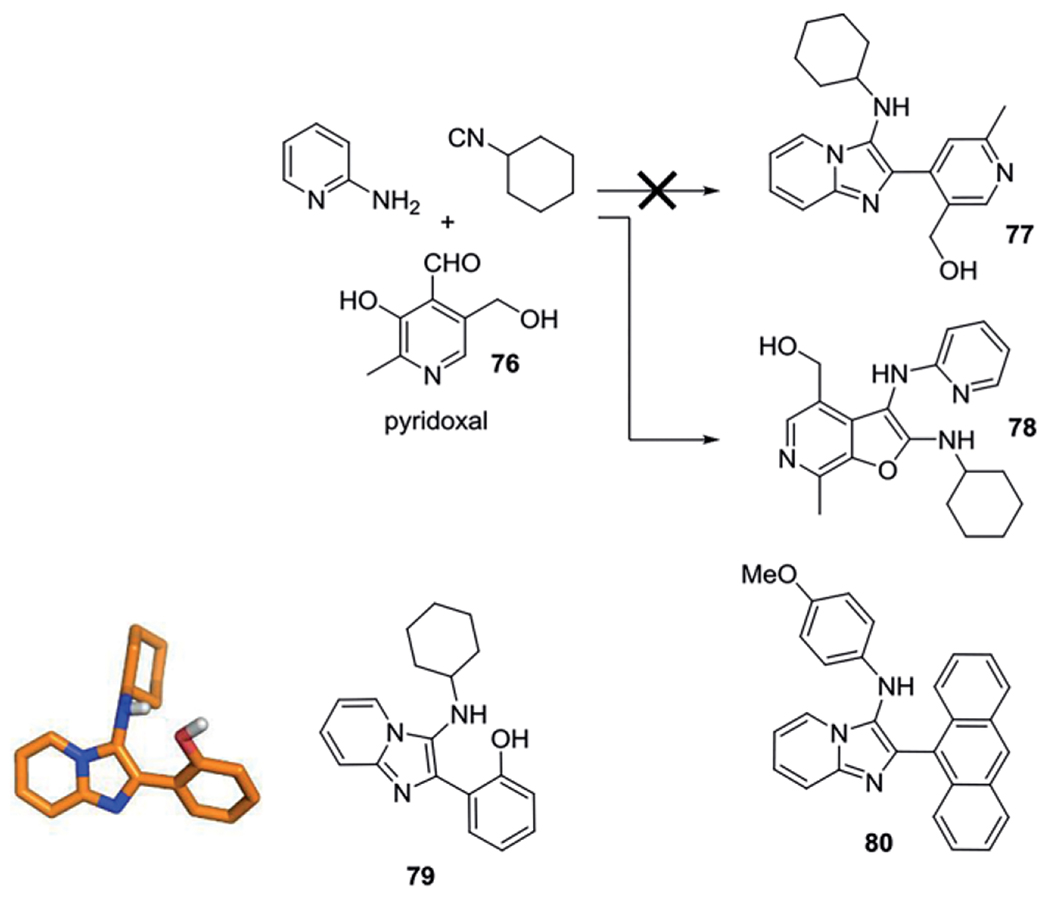
The aldehyde pyridoxal 76 has a different outcome and will not result in the typical GBB-3CR scaffold when reacted with an amidine and isocyanide. The resulting product formed instead, a furo[2,3c]pyridine 78, is shown in Scheme 32 and discussed further in the biologically active compounds section.
Scheme 32.
Discovery of side reactions accompanied with use of pyridoxal as aldehyde component, compound 79 (CCDC-893566) confirmed by X-ray structure analysis.
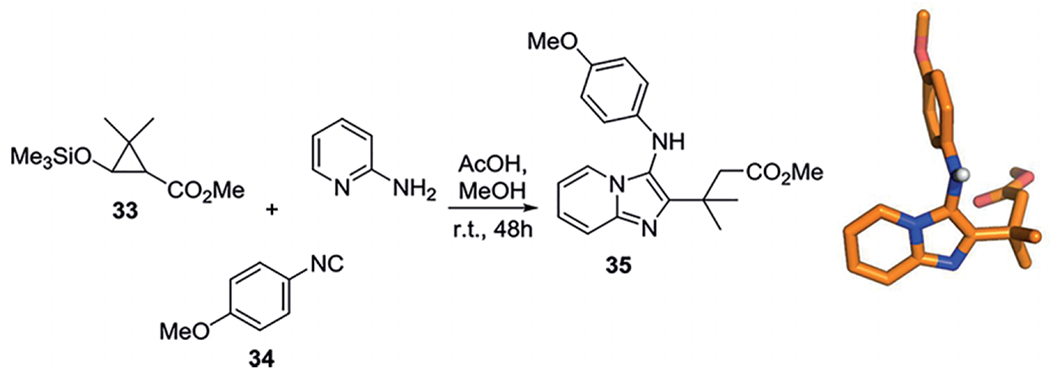
Interestingly, 2-siloxypropanecarboxylates were applied as aldehyde substitutes to obtain GBB-3CR compounds with a δ-amino acid backbone.[45] The siloxanes go through a ring opening pathway and behave as the aldehyde component 33, together with 2-aminopyridine and p-methoxyphenyl isocyanide 34, catalyzed with acetic acid in MeOH to give methyl (3-aminoimidazo[1,2-a]pyridin-2-yl)propanoates in moderate to good yields (Scheme 18).
Scheme 18.
2-Siloxycyclopropanecarboxylates as aldehyde substitutes in the GBB-3CR. One example of a GBB product 35 synthesized from 2-siloxypropanecarboxylates and its resolved X-ray structure (CCDC-601760), in total 8 products were synthesized with variations on the amines, isocyanides with yield varying 40–79 %.
Again the functional group compatibility of the GBB-3CR aldehyde component is amazing and comprises aromatic benzaldehydes include substitutions on all positions with alkyl, aryls, alkoxy groups, alcohols, acids, nitro groups, cyano groups, tertiary amines, polyaromatics and fused heterocycles. Aliphatic aldehydes are also widely represented, including saturated and unsaturated alkyl groups (1°, 2° and 3°), substituted with alcohols, esters, adjacent carbonyls, thiols and cyclic alkyl groups. Thus, heteroaromatic amidines can comprise all halogens, and pseudo halogens such as nitrile, nitro, methoxy, free carboxylic acids, esters, unprotected primary and secondary amines, unprotected phenolic and aliphatic hydroxyl, amides, alkynes, alkenes, and boronic acid esters. This great functional group compatibility is important for further reactivity of the initial GBB-3CR products and also for optimal interaction within a receptor pocket.
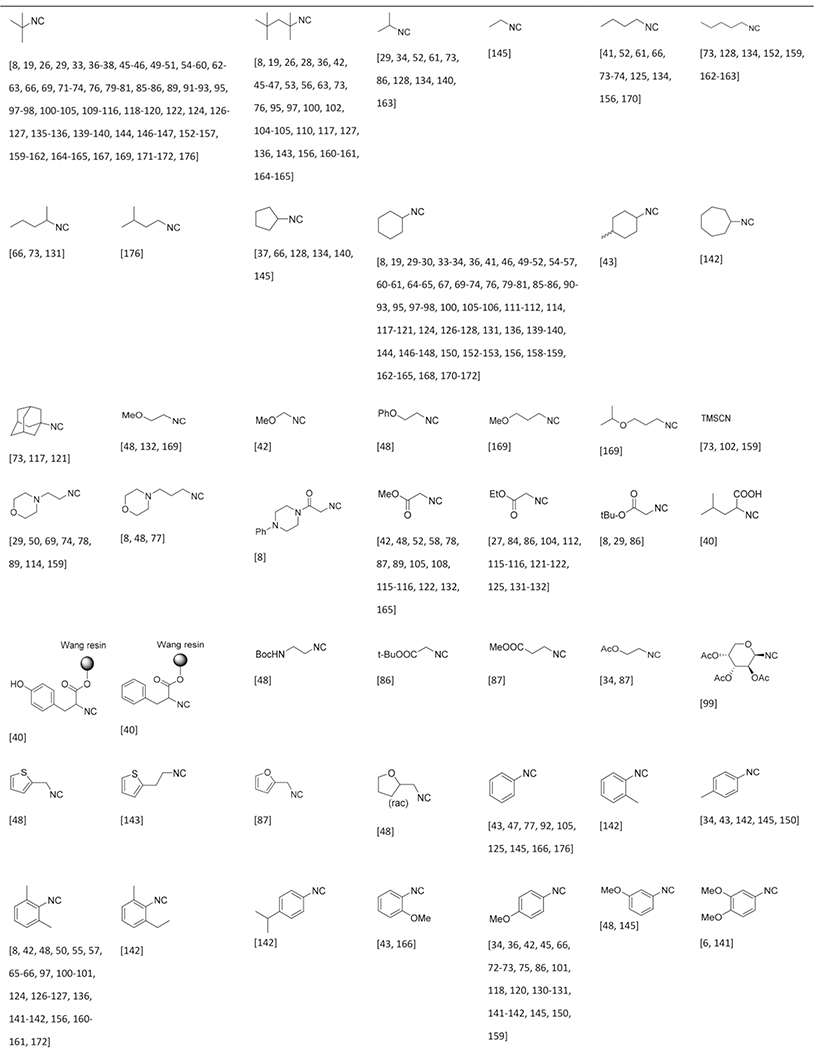
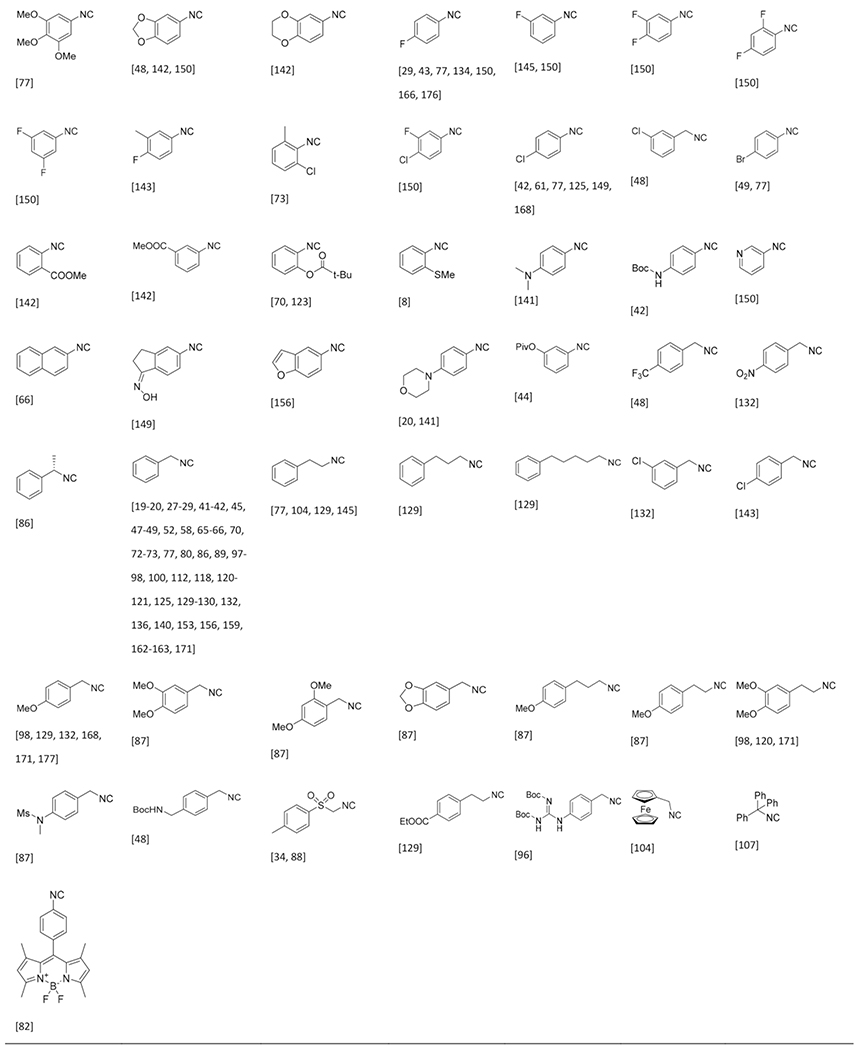
Isocyanides Used in the GBB-3CR
The commercial availability of isocyanides is limited and those available are often expensive, likely because of the stability, applicability and the undesirable smell of most liquid isocyanides. Therefore only a few dozen of isocyanides are commonly used in many isocyanide-based MCRs. Due to the pricing and availability isocyanides are often prepared in house. Preparation of isocyanide from primary amines via the Hoffman- or Ugi route (formylation & dehydration) is most common and recently the substrate scope has been broadened by conversion of cheap and broadly available oxo-compounds (aldehydes and ketone) to isocyanides by the repurposed Leuckart-Wallach reaction as described by Dömling et al.[178] Altogether an astonishing 100 different isocyanides were reported in the GBB-3CR, proving that the isocyanide variation point is broadly exploited and is hardly to be considered as a limitation of variability in this three component reaction (Table 3). Aromatic and aliphatic isocyanides participate with equal ease in the GBB-3CR, although it must be noted that combinations of bulk substituents affect the yield when bulky amidines and aldehydes are used simultaneously. Not surprisingly, there is again a great functional group compatibility. This includes aliphatic isocyanides widely substituted, heterocycles, substituted with esters, alcohols and ethers. Also a broad selection of substituted phenyl isocyanides are compatible, with substitutions on all positions with alkyls, halogens, ethers, esters and thiols.
Table 3.
Nearly 100 reported isocyanides used in the GBB-3CR and sorted by type and substituents.
Catalysts in the GBB-3CR
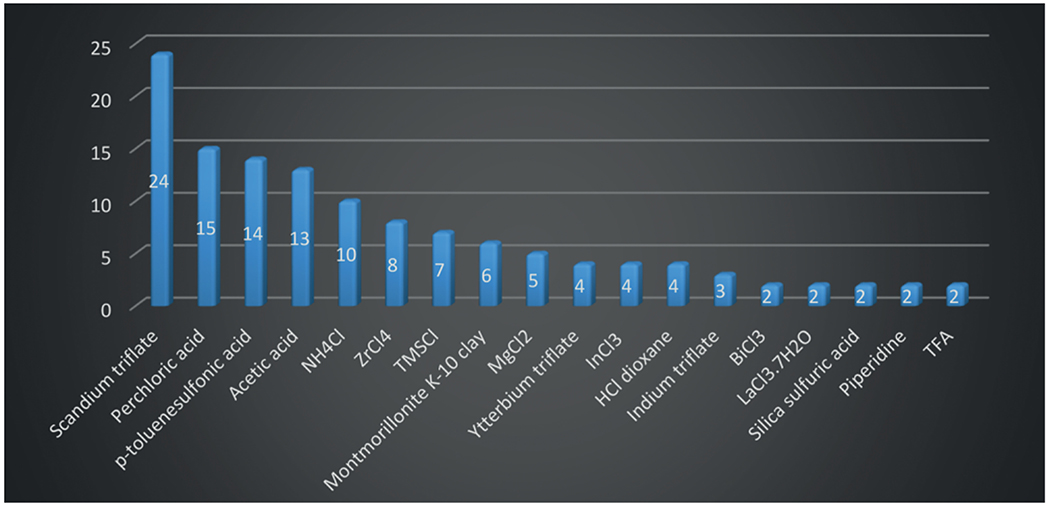
From the moment the GBB-3CR was discovered, Lewis and Brønsted acid catalyst were used for an efficient transformation. Saying this, it is possible to run the GBB-3CR without the aid of a catalyst, it generally depends on the nature of the reagents. Low electrophilic aldehydes such as various benzaldehydes are suspected to be unable to condensate with electron deficient amidines, whereas the addition of Lewis acids would increase the reactivity of the imine formation considerably.[145] In Figure 3 the occurrence of the catalysts used in the GBB-3CR are mapped. Clearly scandium triflate has the highest success rate followed by the Brønsted acids HClO4, pTsA and acetic acid. However, when taking into account that early adaptors of the GBB-3CR were basically reproducing reaction conditions, the frequency of some of the applied catalysts is artificially higher and are possibly not the best catalysts in the given reactions. Examples of such catalysts are acetic acid and Montmorillonite k-10 clay which appear mostly in the first few years after the discovery of the GBB-3CR. Additionally some catalysts were reported multiple times, however from the same research groups, introducing a biased success rate.
Figure 3.
Overview of GBB catalysts that performed best in reported catalyst screenings (single hits were omitted for clarification).
Table 4 summarizes the catalysts applied in the GBB-3CR, which give the highest yield in the described model reaction. Surprisingly not every catalyst screening includes the often excellent performing catalysts (Sc(OTf)3, HClO4, and pTsA). Nearly 50 different catalysts were described as high yielding, indicating a great scope and freedom for selecting the catalyst of choice. There are some reports of catalyst free GBB-3CR’s, that run perfectly fine without any additional reagent. An uncatalyzed GBB-3CR was first reported by Lyon et al. employing immobilized glyoxylic acid on macroporous polystyrene carbonate (MP-glyoxylate) as a formaldehyde equivalent. The decarboxylation leaves a 2-unsubstituted 3-amino-imidazo[1,2-a] heterocycle without use of any catalysts in a yield of 71 % (Scheme 16).[42] Further reports of catalyst free GBB-3CR’s employ bifunctional building blocks that hold a carboxylic acid which probably catalyzes the reaction. Truly catalyst free reactions are only recently reported by Sharma et al., but seem to work exclusively in a reaction using 2-aminothiazole under microwave conditions.[160,161,171,172,196] In some of the catalyst-free reactions, a carboxylic acid functional group was found in one of the three components that facilitated the reaction in good yields, although it must be noted, that the use of carboxylic acids will show Passerini poisoning. This side reaction is driven by the presence of 3 components; aldehyde, isocyanide and carboxylic acid that allow for the P-3CR to happen (Scheme 19).
Table 4.
Every catalyst previously used in the GBB-3CR arranged by occurrence.
| Catalysts (formula) | Occurrence | Reference | Catalysts (formula) | Occurrence | Reference |
|---|---|---|---|---|---|
| Scandium triflate (Sc(OTf)3) | 24 | [20,26–30,39,41,47,48,58,61,62,67,77,86,125,137,138,140,146,149,168,177] | Calcium chloride (CaCl2) | 1 | [32] |
| Perchloric acid (HClO4) | 15 | [8,44,66,75,78,89,96,102,103,119,123,150,167,179,180] | Cellulose sulfuric acid | 1 | [51] |
| p-toluenesulfonic acid, PTSA, pTsOH | 14 | [34,40,55,56,74,85,88,97,107,130,141,164,181,182] | Cellulose@Fe2O3 | 1 | [183] |
| Acetic acid (AcOH) | 13 | [6,45,68,110,128,129,142,154,184–188] | CuFe2O4@SiO2–SO3H | 1 | [115] |
| Ammonium chloride (NH4Cl) | 10 | [43,49,54,95,144,147,156,158,166,189,190] | (MWCNTs) | 1 | [100] |
| ZrCl4 | 8 | [36,55,59,63,94,99,143,191] | TiO2 NPs (nanopowders) | 1 | [170] |
| TMSCl | 7 | [37,38,53,152,155,169,176] | ZnO NPs | 1 | [116] |
| Montmorillonite K-10 clay | 6 | [19,64,65,70,121,192] | Methanesulfonic acid | 1 | [52] |
| Magnesium chloride (MgCl2) | 5 | [134,136,193,194] | Zeolite HY | 1 | [195] |
| Ytterbium triflate (Yb(OTf)3) | 4 | [105,131,132,153] | SnCl2·2H2O | 1 | [127] |
| InCl3 | 4 | [57,98,122,145] | Heteropolyacid H3PMo12O40 | 1 | [60] |
| HCl dioxane | 4 | [72,73,84,117] | Bromodimethylsulfonium bromide (BDMS) | 1 | [69] |
| Indium triflate (In(OTf)3) | 3 | [108,109,122] | γ-Fe2O3@SiO2-OSO3H | 1 | [71] |
| Trifluoroacetic acid (TFA) | 2 | [82,165] | Cationic polyurethane dispersions (CPUDs) | 1 | [76] |
| Bismuth chloride (BiCl3) | 2 | [81,91] | pTsCl | 1 | [77] |
| Silica sulfuric acid | 2 | [126,148] | Iodine | 1 | [135] |
| Lanthanum chloride (LaCl3·7H2O) | 2 | [92,104] | (TBBDA) | 1 | [83] |
| Piperidine | 2 | [162,163] | (PBBS) | 1 | [83] |
| Silver triflate (AgOTf) | 1 | [113] | nano-LaMnO3 | 1 | [90] |
| Ionic liquid [bmim]Br | 1 | [124] | NH2-MIL-53(Al) | 1 | [111] |
| Ionic liquid [bmim]BF4 | 1 | [70] | CuI L-Proline | 1 | [33] |
| Aluminum chloride (AlCl3) | 1 | [136] | BF3·MeCN | 1 | [34] |
| Ruthenium chloride (RuCl3) | 1 | [79] | 2-chloroacetic acid (ClCH2COOH) | 1 | [120] |
Scheme 19.
The P-3CR is competing with the GBB-3CR when carboxylic acids are used as catalyst, consuming the aldehyde and isocyanide components that otherwise would be consumed by the GBB-3CR.
As most Lewis acids rapidly decompose or get deactivated when they come in contact with water, anhydrous conditions are usually required when running a Lewis acid catalyzed GBB-3CR. Sc(OTf)3 is, however, more stable and even applied in aqueous media as an Lewis acid.[197] The GBB-3CR is a condensation reaction, the formation of a water molecule would explain why scandium triflate is such a successful catalyst in this reaction. The only disadvantage noteworthy is that scandium triflate has the ability to polymerize isocyanides, explaining the darkening of some reaction mixtures when this catalyst is applied.[141] Phenyl isocyanides are prone to polymerization and amongst them unsubstituted phenyl isocyanide most.[198–200]
Base Catalyzed GBB-3CR
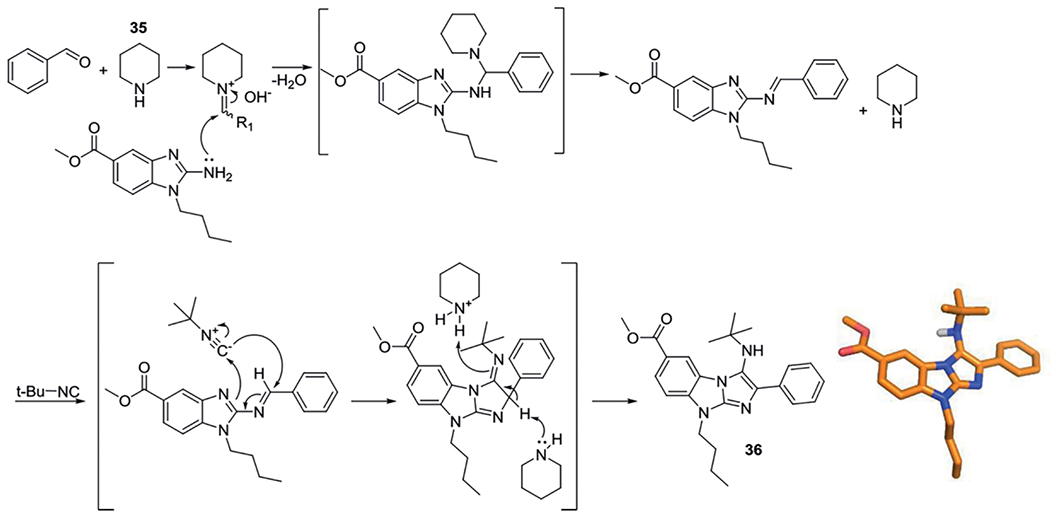
Sun et al. reported in 2013 the first application of piperidine (35) as a Brønsted base catalyst for the GBB-3CR, which was applied to overcome the need to run a deprotection-cyclization-alkylation sequence to obtain their tetracyclic compound. Later in 2013, Sun et al. reported the same strategy without the post modifications and therefore these base catalyzed conditions could be applied for the standard GBB-3CR.[162,163] In the plausible mechanism, piperidine is initially involved in the formation of the Schiff base, then with the introduction of the isocyanide component, the [4+1] cycloaddition takes place, where piperidine facilitates the 1,3-H shift resulting in rearomatization, giving the GBB-3CR product as depicted in Scheme 20.
Scheme 20.
Plausible mechanism for the piperidine catalyzed GBB-3CR projected on methyl 9-butyl-3-(tert-butylamino)-2-phenyl-9H-benzo[d]imidazo[1,2-a]imidazole-6-carboxylate (36) and its corresponding X-ray structure.
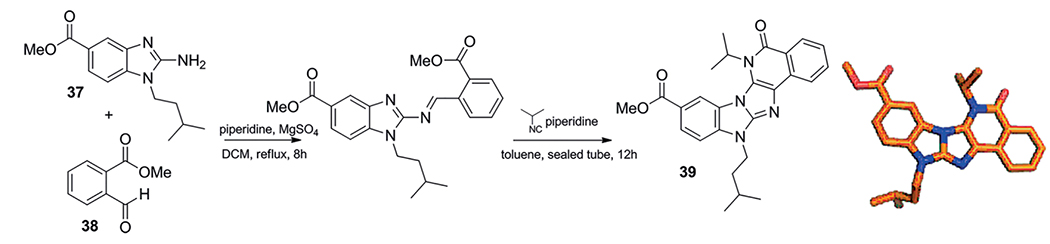
Sun et al. reports the first Brønsted base catalyzed GBB-3CR of 2-aminobenzimidazoles 37, methyl 2-formylbenzoate 38 and isocyanides, piperidine was herein used as catalyst.[163] In their search for a post modification on the MCR product, an intramolecular cyclization of the secondary amine with the methyl ester was performed. The secondary amine was arising from the isocyanide input as mostly cleavable isocyanides were utilized. They envisioned a tandem GBB-3CR and post modification sequence in a single step resulting in 20 compounds with a yield varying between 34–95 % (Scheme 21). The heterocyclic benzimidazole and dihydropyrimidine fragments combined are promising compounds as these are associated with PARP, topoisomerase I, INOS, PI3K and PrCP inhibition.
Scheme 21.
Preparation of polyheterocyclic 39 unambiguously confirmed by X-ray structure analysis (CCDC 850793).
Solvents Used in the GBB-3CR
Around 30 solvents and solvent mixtures used in the GBB-3CR were analyzed and discussed. Looking at the occurrence, methanol is by far the most popular solvent for the GBB-3CR reaction. This is largely attributed to the fact that the highest yields without many side products can be achieved with this solvent, the ease of removal and the ability to dissolve quite some polar catalysts and starting materials as well as more apolar reactants. Discussion of the solvents such as methanol trapping and methanol mediated intermediates rise the question on how broad the solvent range is and whether other solvents than methanol should be applied in the GBB-3CR. Furthermore, the stability of the intermediates from pathway A and B (Scheme 4) are predicted to have different stabilities that vary with the used solvent and therefore would yield either one of the two isomers or the trapped side product.[43] While focusing on solvent screens and suitable solvents for the GBB-3CR, it is remarkable that the 2nd most frequent applied reaction conditions is actually solvent-less. The solvent-less reports show that neat reactions with simple stirring or mechanical mixing such as ball-milling will yield the GBB-3CR compounds as well, but often require more exotic catalysts such as zeolite HY, Montmorillonite K-10 clay and nano-magnetically modified sulfuric acid.[19,71,195] The use of water in a catalyst free GBB-3CR is not only working, but also affords GBB-3CR products in high yields. The aforementioned mechanism should be different from the reaction usually following the concerted [4+1] cycloaddition through iminium ions but rather a non-concerted 5-exo-dig pathway. The protonation of the imine is in general performed with Brønsted acids, in the case of water without catalysts this is unlikely to happen since the pKa of water is higher than pKa’s found for iminium ions.[171] Additionally the Woodward-Hoffmann rules which indicates that the uncatalyzed concerted [4+1] cycloaddition is in fact symmetry-forbidden. The GBB-3CR is, however, mostly performed with a catalyst, which disrupts the symmetry and therefore allowing the [4+1] cycloaddition.[201] Considering the possibility of using water or a percentage of water in EtOH; it might be odd that several reports, indicate that anhydrous conditions (dry methanol, dry MeCN and so on) are required for the GBB-3CR while the individual starting materials and catalysts aren’t sensitive to water, with the exception of some Lewis acids catalysts. Water, however, has the downside to negatively affect the speed of the reaction, anhydrous solvents and the use of a dehydrating agent could greatly increase the reaction rate.[34] The use of non-protic apolar solvents would suppress the intermediate in pathway B, yielding exclusively the product of pathway A; imidazo[1,2-a]-pyrimidines. Toluene is such a solvent and is found quite often to be used together with ammonium chloride as catalyst with heating between 80 °C toward reflux temperatures, ruling out formation of any regioisomers getting exclusively the pathway A product (Scheme 4). Interestingly, while attempts were made to find conditions to control the regioselectivity towards pathway B, Stephenson et al. reported an alternative route, applying a base assisted Dimroth rearrangement giving access to the inverse GBB-3CR regioisomer with full conversion as depicted in Scheme 22.[47] The stability of the regioisomer is the driving force behind this rearrangement, resulting in a preferred single isomer.[202]
Scheme 22.
The Dimroth rearrangement gives access to both regioisomers. A: the rearrangement is performed under basic conditions while heating. B: the proposed mechanism, driven by the stability of the product and properties of the initial starting material, such as aromaticity of the ring, bulkiness of substituents and solvent.
Solvents such as DMSO, DMF and PEG-400 are in general not the solvents of choice because of their high boiling points and difficulties to remove during workup (Table 5). They prove, however, to be quite useful when solubility issues with highly polar starting materials are present such as the aforementioned amidine containing nucleobases.[36,146]
Table 5.
All the solvents used in the GBB-3CR.
| Solvent | References | Occurence | Solvent | References | Occurence |
|---|---|---|---|---|---|
| MeOH | [6,8,26–28,30,39,45,51,54–56,61,66,68,74,75,78,87,96–100,103,108,110,118,120,123,125,126,128,130,136,141,142,150,153,154,156,166–168,177,181,183–185,187,188] | 51 | 1:1 EtOH:H2O | [165,180] | 2 |
| No solvent | [19,60,71,79,81,83,85,90,92,93,101,105,111,112,127,133,140,147,148,161,172,192,195] | 23 | Water | [46,76] | 2 |
| Toluene | [43,49,52,70,95,109,144,145,158,159,163,164,171,186,189,190] | 17 | DMSO | [33,36] | 2 |
| EtOH | [57,84,89,104,113,115,116,121,122,131,132,134,135,139,160,193] | 16 | DMF | [146,180] | 2 |
| MeCN | [53,69,72,73,77,117,152,155,169,176,182] | 9 | MeOH:DCE | [42] | 1 |
| 2:3 MeOH:CH2Cl2 | [58,62,80,137,138] | 5 | TFE/DCE 1:1 | [67] | 1 |
| nBuOH | [59,94,119,194] | 4 | DCE | [162] | 1 |
| 3:1 MeOH:CH2Cl2 | [20,39,41] | 3 | 1:1 MeOH:CH2Cl2 | [129] | 1 |
| 1:3 MeOH:CH2Cl2 | [31,86,143] | 3 | 1:4 MeOH:CH2Cl2 | [47] | 1 |
| Ionic liquid [bmim]Br; DES | [106,114,124] | 3 | 1:2 MeOH:CH2Cl2 | [48] | 1 |
| 1,4-Dioxane | [50,64,65] | 3 | 1:1 EtOH:CH2Cl2 | [107] | 1 |
| MEOH/MeCN | [37,38,203] | 3 | 2:3 ethanol/H2O | [170] | 1 |
| PEG-400 | [63,191,204] | 3 | 1:1:1 CHCl3/TMOF/MeOH | [40] | 1 |
| TFE | [32,180] | 2 | DMSO:H2O 1:1 | [29] | 1 |
| CH2Cl2 | [34,102] | 2 | Et2O | [88] | 1 |
Biologically active Compounds
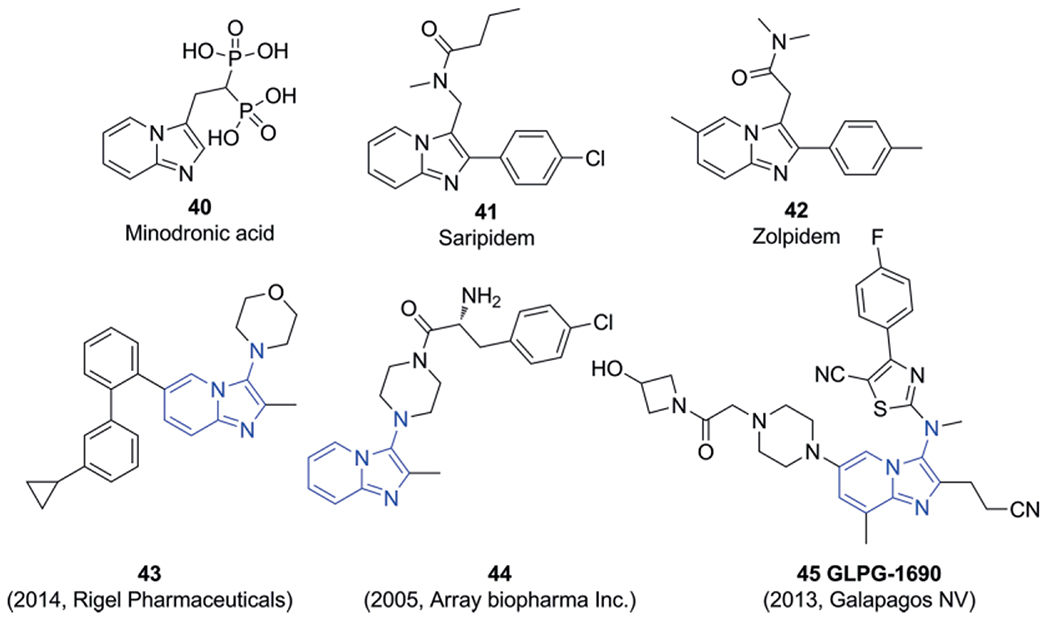
Quite often the imidazo[1,2-a]-heterocycles that arise from the GBB-3CR are linked to known drugs such as Minodronic acid (40), Saripidem (41) Zolpidem (42) and other members of the so-called Z-drug group of tranquilizers. There are however no reports that GBB-3CR compounds act in a similar way as Z-drugs on the GABAA receptor. The difference between Z-drugs and GBB-3CR compounds is found in the exocyclic secondary amine, originating from the isocyanide component. This amine enables ionic or hydrogen bonding, thus having its own influence on the binding affinity with various protein targets. As a whole it might be better to address real examples of the GBB-3CR scaffold and their associated biological activity. Examples of 3-aminoimidazo[1,2-a]pyridines are shown in multiple pharmaceutical drugs or candidates 43–45 (Scheme 23).[205] Most notably, the late stage autotaxin inhibitor GLPG-1690 (45) for the treatment of idiopathic pulmonary fibrosis (IPF) which has been discovered and developed whereby the GBB-3CR chemistry played a key role.[186,194]
Scheme 23.
Examples of imidazo[1,2-a]pyridines 40–42 and 3-aminoimidazo[1,2-a]pyridines, containing the GBB-3CR scaffolds (blue) reported as bioactive leads; 43 anti-inflammatory 44 anticancer, 45 anti-fibrosis.
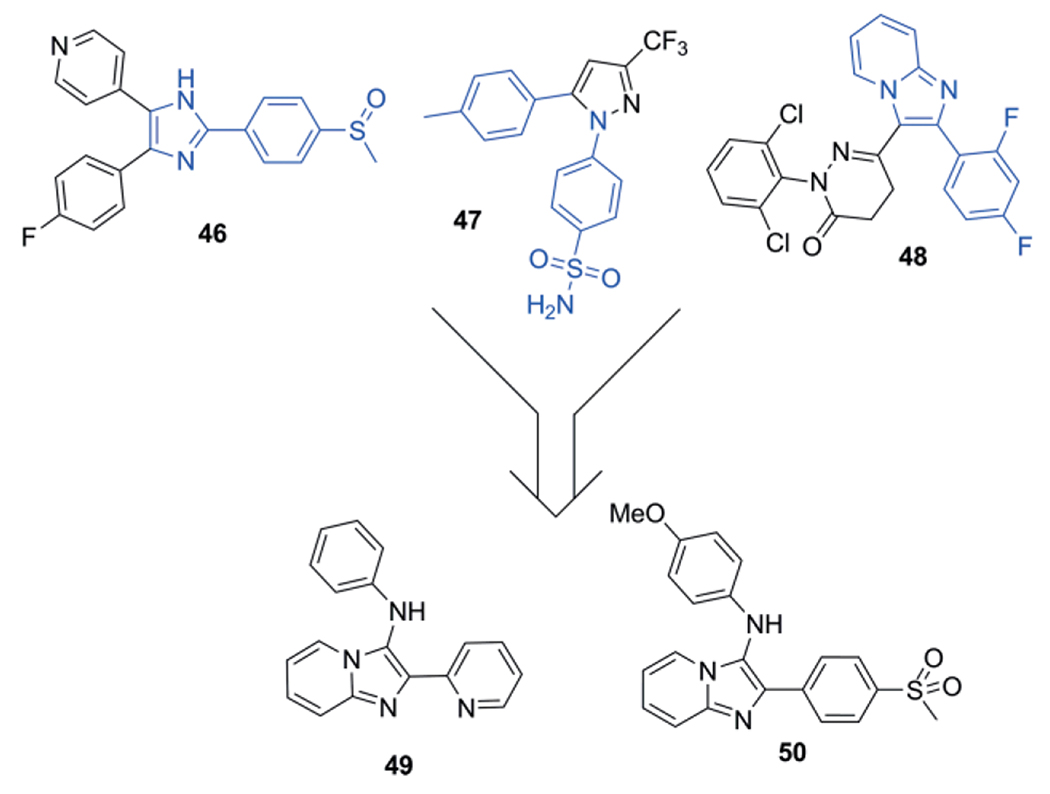
Only in 2009 the first biological target was addressed and published by using a true GBB scaffold as reported by Fraga et al.[184] In their efforts to combine the structural features of (46), a selective p38 MAPK inhibitor, (47) celecoxib a PGHS-2 inhibitor and (48) also a p38 MAPK inhibitor a GBB-3CR scaffold was used. The main target was to obtain polypharmacological 3-arylamine-imidazo[1,2-a]pyridine derivatives with anti-inflammatory and analgesic MOA (Scheme 24).
Scheme 24.
Synthesis of substituted imidazo[1,2-a]pyridine derivatives as anti-inflammatory and analgesic drugs.
This approach resulted in compound 49 which was identified as PGHS-2 inhibitor (IC50 = 18.5 μM) and acts similar to the p38 MAPK inhibitor SB-203580, reverting capsaicin-induced thermal hyperalgesia, a pain model to assess analgesic properties. Compound 50 is a novel PGHS-2 inhibitor, with an ED50 = 22.7 μmol·kg−1, 10-fold more potent than celecoxib.
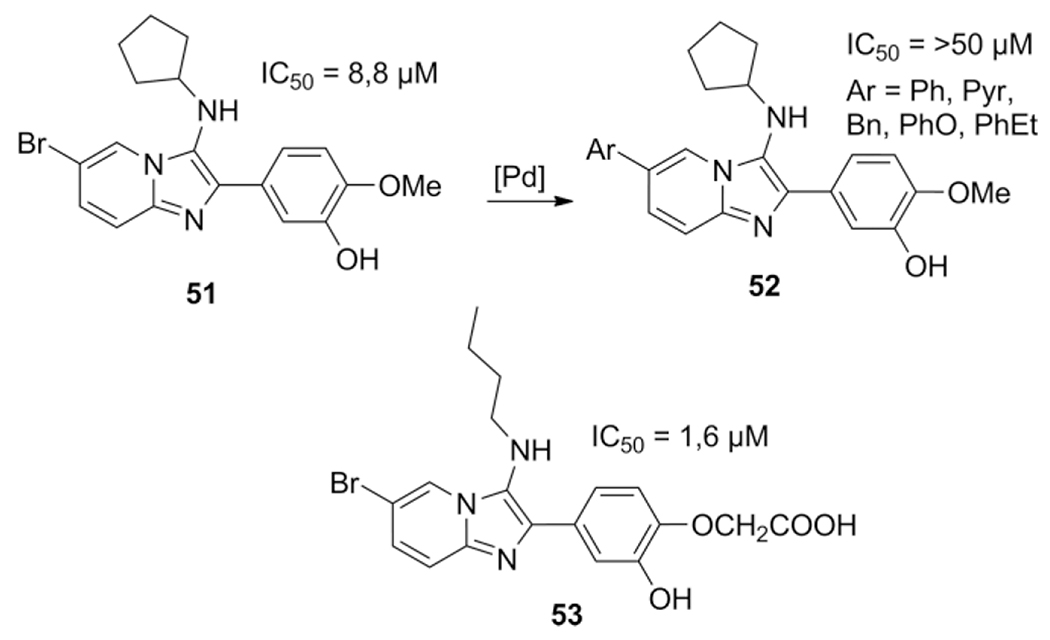
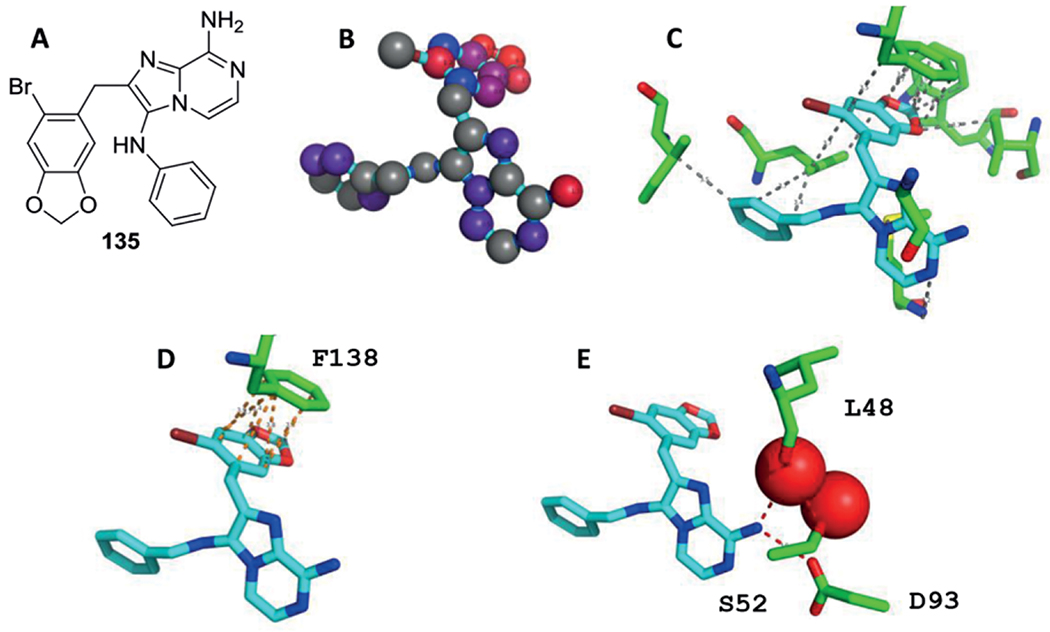
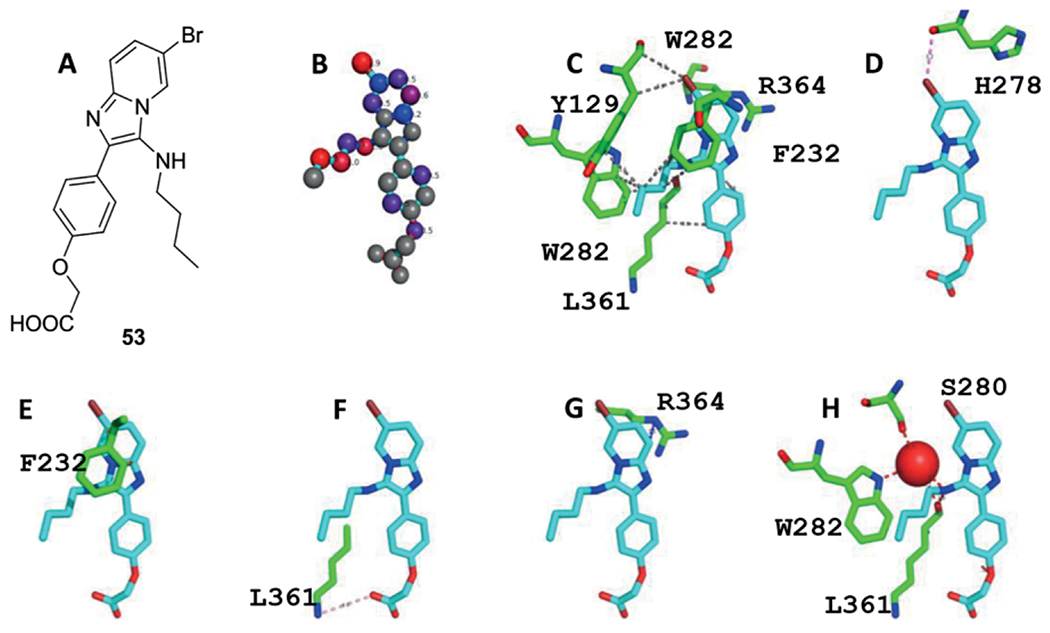
During a study on the TB targets M. tuberculosis glutamine synthetase (MtGS) inhibitory effect, a hit to lead exercise was performed on 3-amino-imidazo[1,2-a]pyridines as these were identified as MtGS inhibitors after a high-throughput screening.[193] Two libraries were synthesized the first via a microwave assisted GBB-3CR sequence for 20–30 min at 160 °C and the second via a post-modification on the pyridine 6-position halogen under Suzuki coupling conditions to study the effect of hydrophobic groups such as aryl moieties 52 (Scheme 25). Unfortunately, the Suzuki coupling did not lead to an improvement in IC50 value, leaving the aryl halides 51 themselves as potent inhibitors for this target. In a subsequent paper, lead optimization did result in a series of potent compounds, 53 was showing a five-fold improvement with an IC50 = 1,6μM.[134] The mode of binding of 53 was discussed according to its co-crystalstructure with MtGS in the structural biology section.
Scheme 25.
Synthesis and optimizing of MtGS inhibitors.
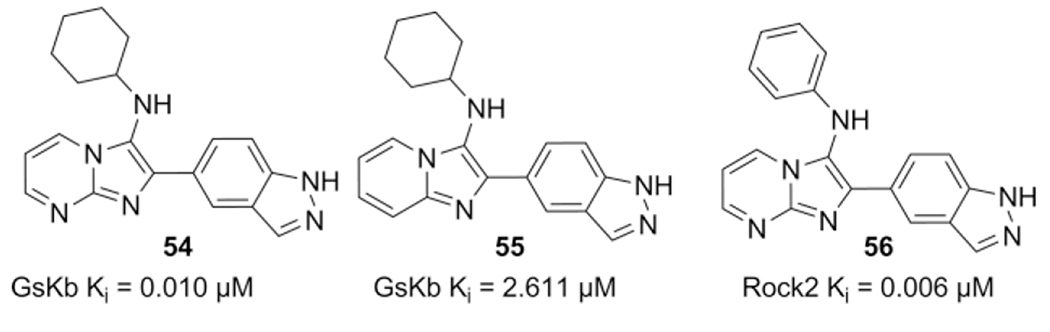
Three years after their patent application, Djuric et al. published the work of 5-substituted indazoles as kinase inhibitors such as Gsk3β, Rock2, and Egfr,[61] by using MCR methodologies such as the GBB-3CR with pyrimidines for imidazopyrimidines, thioamides for thiazolyl-indazoles and van Leusen TosMICs for imidazole substituents.
Introduction of the indazole was achieved through the use of indazole-5-carbaldehyde as reactant in different multicomponent reactions and afforded compounds as 54–56. The imidazole[1,2-a]pyrimidine 54, GBB-3CR product, showed the highest inhibition against Gsk3β, imidazopyridine 55 was however, much less potent. Choosing different substituents on the amino group, coming from the isocyanide input (56), selective inhibition for Rock2 could be fine-tuned as well (Scheme 26).
Scheme 26.
5-substituted indazoles as kinase inhibitors.
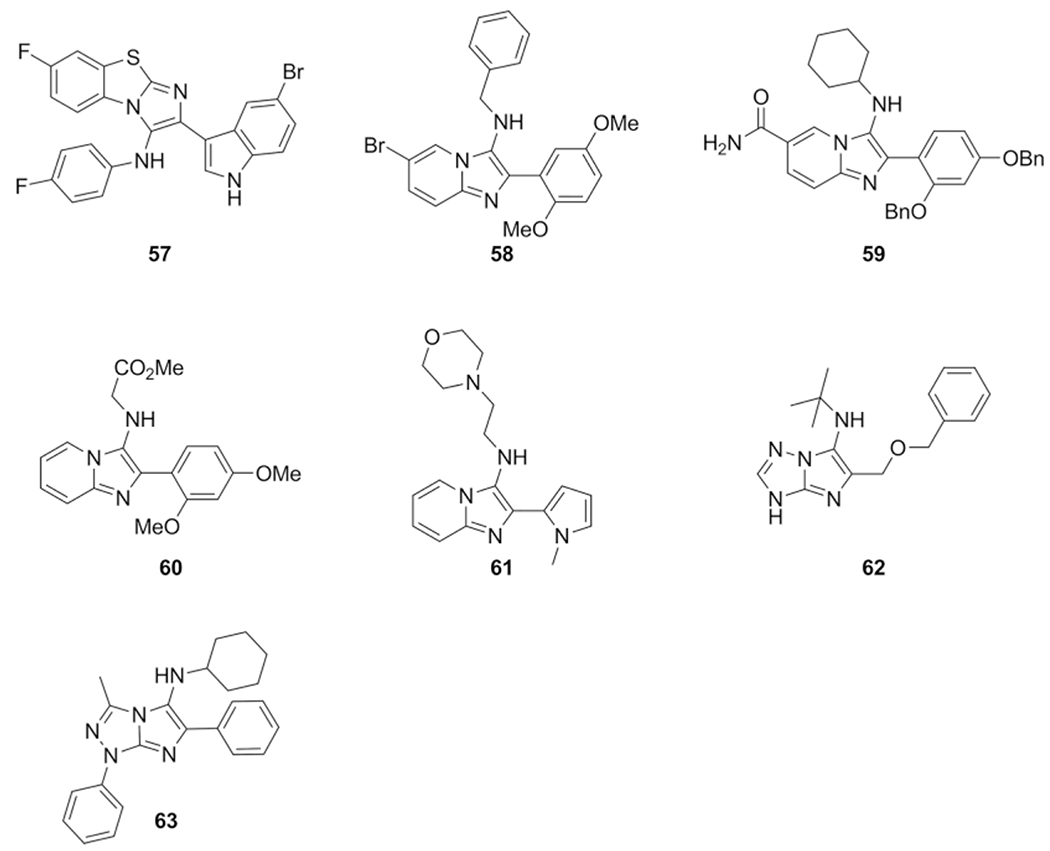
Al-Tel et al. described the synthesis of a series of imidazo-[1,2-a]pyridine and imidazo[2,1-b][1,3]benzothiazole carrying quinolone and indole moieties introduced via the aldehyde component. Many of the synthesized compounds showed both antibacterial and antifungal activities. A very potent broad-spectrum antibiotic was identified as compound 57 depicted in Scheme 27.[138]
Scheme 27.
Various GBB-3CR bioactive compounds.
The effect of imidazo[1,2-a]pyridines was studied on colon cancer cell lines by Koning et al. by showing apoptosis inducing effects in HT-29 and Caco-2 cancer cells.[65] Compounds 58 and 59 showed a cytotoxic effect towards the Huh7 cell line at low μM concentrations and interestingly minimal cytotoxicity against white blood cells.
Schneider et al. performed a chemical advanced template search (CATS) for target profiling. CATS use topological descriptors generated from known binders and to these descriptors it assigns features such as lipophilic, aromatic, hydrogen-bond donors and hydrogen-bond acceptors to find new chemotypes. This virtual target screening yielded a library in which the starting point was the GBB-3CR. From the results, 3840 virtual products, the top 9 compounds were synthesized for PI3Ka inhibition.[78] Four out of nine exhibited activity, with the most active compound 60 showing an IC50 = 131 μM confirming CATS usefulness as a tool to predict targets of virtually generated compounds. In addition, a set of 57 predicted compounds, with highest joint prediction scores for PI3K and DNA topoisomerases, were synthesized using a microwave assisted approach with perchloric acid as catalyst. Although no human DNA topoisomerase II inhibition was observed, four new compounds showed inhibitory effects on bacterial DNA gyrase such a compound 61, a bacterial type II topoisomerase.
In silico screening was initiated by Brenk et al. to produce a library of novel kinase inhibitors derived from commercially available fragments.[151] As a core structure they’ve chosen a ring system with heteroatom containing functional groups. After applying several selection steps, the number of hits was brought down from over two million compounds to 265 that yielded in 186 compounds being successfully synthesized. 17 of those were prepared using the GBB-3CR, starting from 3-amino-1,2,4-triazole, aldehydes and isocyanides, catalyzed with HClO4 in MeOH at room temperature. One of the obtained compounds 62 showed activity against the kinases EPH-B3 (158μM) and FGF-R1 (469 μM).
Similar imidazo[2,1-c][1,2,4]triazoles derivatives were reported to possess antimicrobial and antioxidant activities by El Kaim et al.[168] The reaction conditions were different as compared to the method described by Akbarzadeh.[158] A Sc(OTf)3 catalyzed reaction with substituted benzaldehydes, various isocyanides and substituted 5-amino-1,2,4-triazoles was performed in DMF (80 °C, 30h) to produce a small collection of compounds with yields between 54 and 72 %, the best performing compound 63 possesses both antifungal and microbial activity.
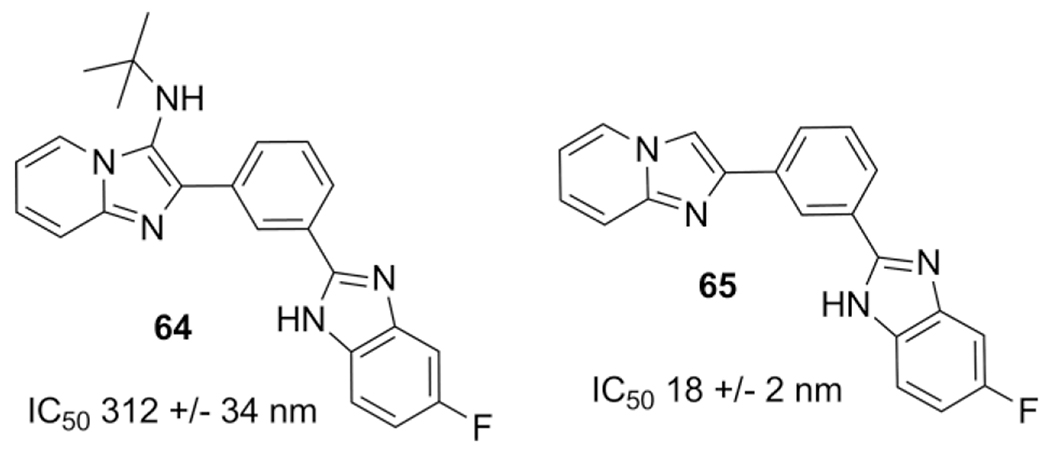
Another report from 2011 by Al-Tel et al. describes the identification of imidazopyridines derivatized with benzimidazole and/or arylimidazole as potent β-secretase inhibitors.[62] In multiple rounds the GBB-3CR compound 64 gave a sub micro molar activity, post modification by removal of the tert-butylamine group, a 40-fold increase in activity was achieved. The best compound 65 showed nanomolar potency for the BACE1 enzyme associated with Alzheimer’s disease (Scheme 28).
Scheme 28.
Highly selective β-Secretase inhibitors of the BACE1 enzyme.
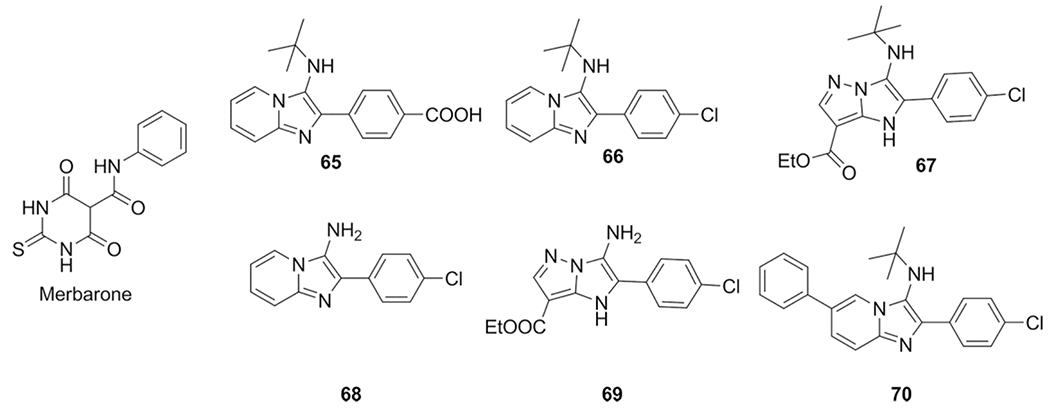
Baviskar et al. published in 2011 their work on novel topoisomerase IIα inhibitors as anticancer agents. With the aid of molecular docking studies they predicted bicyclic N-fused aminoimidazoles as potential human topoisomerase IIα (hTopoIIα).[63,94] Based on the previous reported topo II inhibitors in combination with known drugs containing an imidazo[1,2-a] scaffold, a library was synthesized using the GBB-3CR. The result is a GBB-3CR scaffolds, by heating the corresponding heterocyclic amidine, aldehyde and isocyanide with a catalytic amount of ZrCl4 at 50 °C in PEG-400 or under MW conditions at 140 °C in n-butanol. In some examples the tert-butyl group was cleaved using HBF4 yielding primary amines exhibiting similar activities, multiple compounds 65–70 shown higher potency anticancer activity as compared to 5-fluorouracil and etoposide in kidney cancer cells. Introduction of C2-biaryl and C6-aryl substitutions on the same imidazo[1,2-a]-pyridine/pyrazine scaffold resulted in compound 69 which showed inhibitory effects in the catalytic cycle of hTopoIIα, additional studies suggests that its binds similar as compared to merbarone (a known catalytic inhibitor of hTopoIIα) overlapping the etoposide binding domain (Scheme 29).
Scheme 29.
Synthesis, screening and optimizing of hTopoIIα inhibitor.
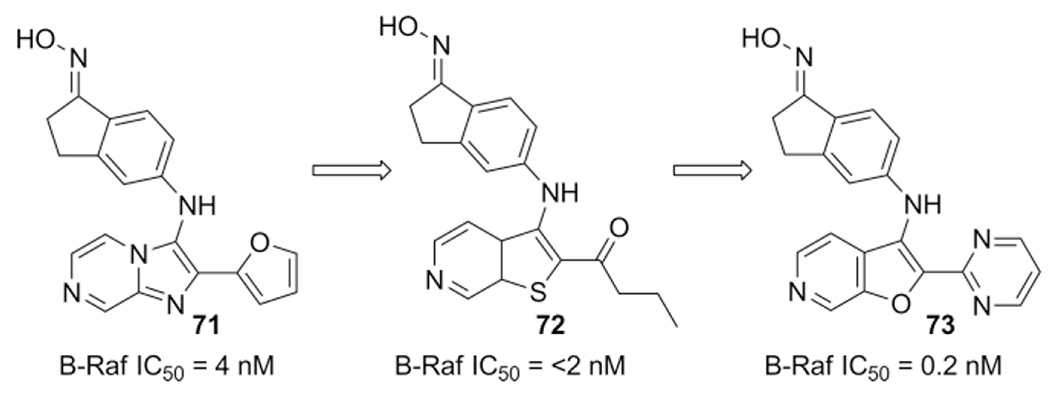
Callejo et al. reported a new cancer therapeutic approach by inhibition of the kinase B-Raf.[149] In a virtual screening they identified imidazo[1,2-a]pyrazines as a binder of B-Raf, in a subsequent synthesis of a small library they found the derivative with an indanone oxime substituent 71 to be highly potent with an IC50 of 4 nm. Exchange of the furfural moiety lead to compounds 72 and 73 showing improvement on B-Raf with an inhibition <2 and 0.2 nm respectively.
Further SAR optimizing led to replacement of the imidazo[1,2-a]pyrazine moiety by thienopyridines 72 with improved inbibition (<2nm) and ultimately with furopyridines 73 bearing superior sub-nanomolar activity (Scheme 30).
Scheme 30.
Imidazo[1,2-a]pyrazine based B-Raf inhibitors and its SAR assisted evolution towards sub-nanomolar inhibitors.
Chatterjee et al. described a screening campaign using the public malaria database for the discovery of novel chemotypes that could inhibit Plasmodium falciparum, a selection was made of compounds that matched criteria such as good potency (IC50 of <1μM), good safety index, (>20-fold in a six-cell line toxicity panel) and easy synthesis. The imidazolopiperazine moiety was found to be an attractive hit, based on the aforementioned criteria.[150]
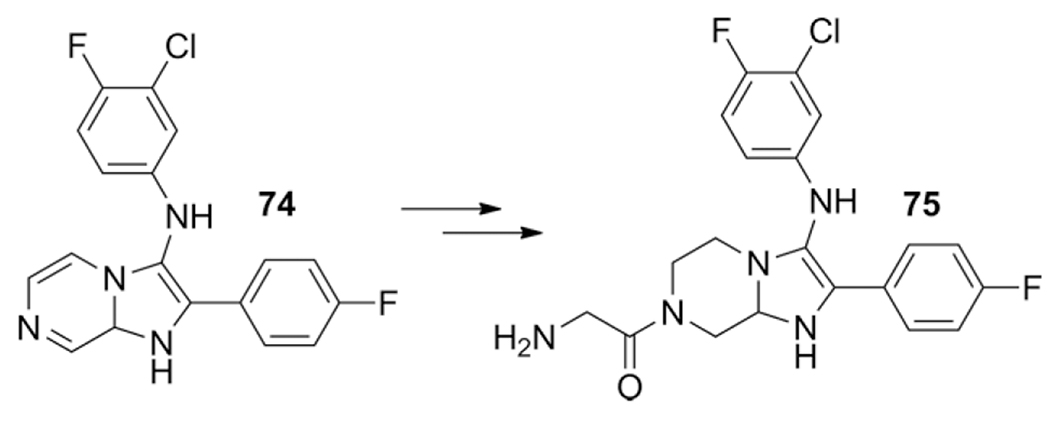
The imidazolopiperazines 74 were accessed by performing a GBB-3CR in MeOH at r.t. catalyzed with HClO4, followed by reduction with PtO2 under hydrogen atmosphere. The most potent compound 75 exhibited an IC50 of 4 and 3 nm on the P. falciparum strains W2 and 3D7 respectively (Scheme 31).
Scheme 31.
Post modifications of GBB-3CR product 74 lead to potent anti-malarial compound 75.
David et al. reported the use of pyridines, isocyanides and benzaldehydes in a microwave assisted GBB-3CR. Some analogues of the imidazo[1,2-a]pyridine products were found to exhibit activation of the TLR8-dependent NF-κB signaling. As the GBB-3CR was discovered by using pyrimidines as a variation of the amine component in the U-4CR, a similar phenomenon that was observed when performing a GBB-3CR with pyridoxal 76 as aldehyde component. Surprisingly the use of pyridoxal 76 as benzaldehyde component did not yield imidazo[1,2-a]-pyrazines/pyridine 77, but the unexpected furo[2,3c]pyridine scaffold 78. Beyond the scope of this paper, they proposed a mechanism and explored scope and limitation of this reaction. The reaction follows the typical GBB-3CR mechanism of the imine formation and attack of the isocyanide, but the otherwise [4+1] cycloaddition concerning the nucleophilic attack of the endocyclic amine with the carbene of the isocyanide does not take place. The proposed mechanism rather proceeds in attack of the phenolic hydroxyl directed by steric hindrance of the apposing benzylic hydroxyl group. The use of anilines together with pyridoxal resulted in formation of the furo[2,3c]pyridine as well. Replacement of pyridoxal with the very similar salicyl aldehyde did however yield the usual GBB-3CR scaffold 79, likely due to the absence of the bulky hydroxymethylene group in this otherwise similar aldehyde (Scheme 32). The obtained imidazo[1,2-a]pyridin-3-amines were TLR7/8 inactive, but showed bacteriostatic activity against Gram-positive bacteria. SAR studies led to the synthesis of a library of 24 compounds with the emphasis on the size of aryl aldehydes, difference between 2-aminopyridines vs. 2-aminopyrazines and aliphatic or aromatic isocyanides.[73] The aim was to assess whether modifications were possible without loss of antibacterial activity, resulting in 80 as best compound.
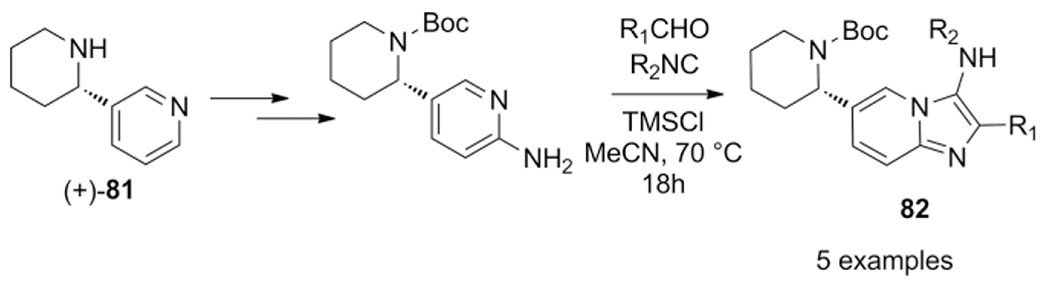
Anabasine 81 a natural product was used by Krasavin et al. as a template to prepare hydrazo-Ugi and GBB-3CR derivatives were evaluated as agonists and antagonists of nAChR. Targeting this family of neurotransmission receptors could lead to drugs for the treatment of associated with PD, AD, schizophrenia and depression.[176]
The five GBB-3CR products were synthesized in low to moderate yields 20–53 %, employing various isocyanides, modified anabasine 2-aminopyridine and substituted benzaldehydes in MeCN, 70 °C using an stoichiometric amount of TMSCl as Lewis acid promoter (Scheme 33).
Scheme 33.
Anabasine derived GBB-3CR products as binders of nAChR.
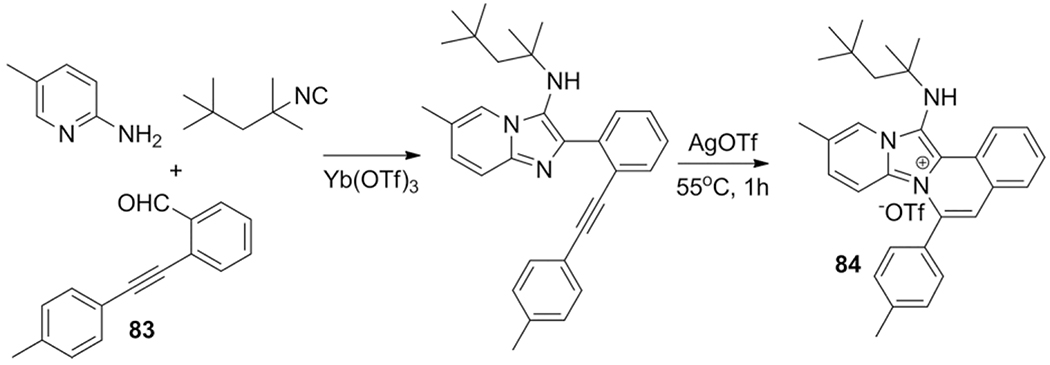
The use of a alkynylbenzaldehydes 83 in the GBB-3CR allows for post modifications such as metal catalyzed intramolecular cyclization yielding a library of compounds, some with strong cytotoxicity against the HeLa cell line, due to binding affinity with the G-quadruplex DNA motif, thus exhibiting anti-proliferative activity as described by Shen et al.[206] The cyclization to the tricyclic compounds, such as 84, was performed in a domino one pot approach of the Yb(OTf)3 catalyzed GBB-3CR between 2-aminopyridines, isocyanides and 2-alkynylbenzaldehyde followed by a AgOTf catalyzed intramolecular cyclization (Scheme 34).
Scheme 34.
In total a library of 23 compounds was synthesized, in which 84 showed the best HeLa cell potency (IC50 = 0.58 μM).
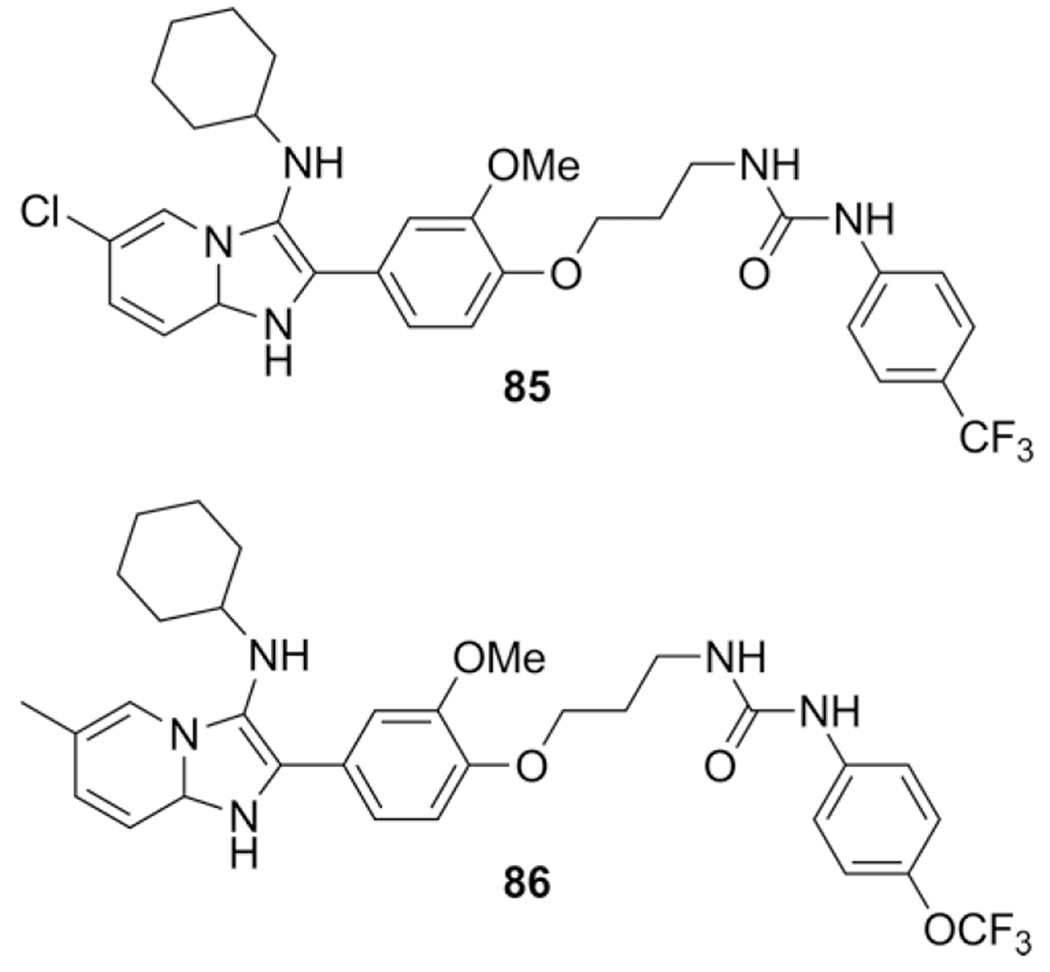
Proschak et al. reports hybrid imidazo[1,2-a]pyridine and an urea moiety as dual soluble epoxide hydrolase (sHE)/ 5-lipoxygenase (5-LOX) inhibitors.[128] A three or four step approach was used to introduce both the urea and imidazo[1,2-a]pyridine elements. In total a library of 22 compounds were synthesized in an AcOH catalyzed GBB-3CR approach in MeOH in yields varying between 7–64 % and showing good inhibition potential on recombinant enzyme on both targets. The best 5-LOX inhibitor 85 was not the best sHE inhibitor (86), and it is to be evaluated whether equipotent inhibition is favored over higher inhibition of one of the two targets (Scheme 35).
Scheme 35.
Combined urea and GBB-3CR products synthesized in an 8-step sequence.
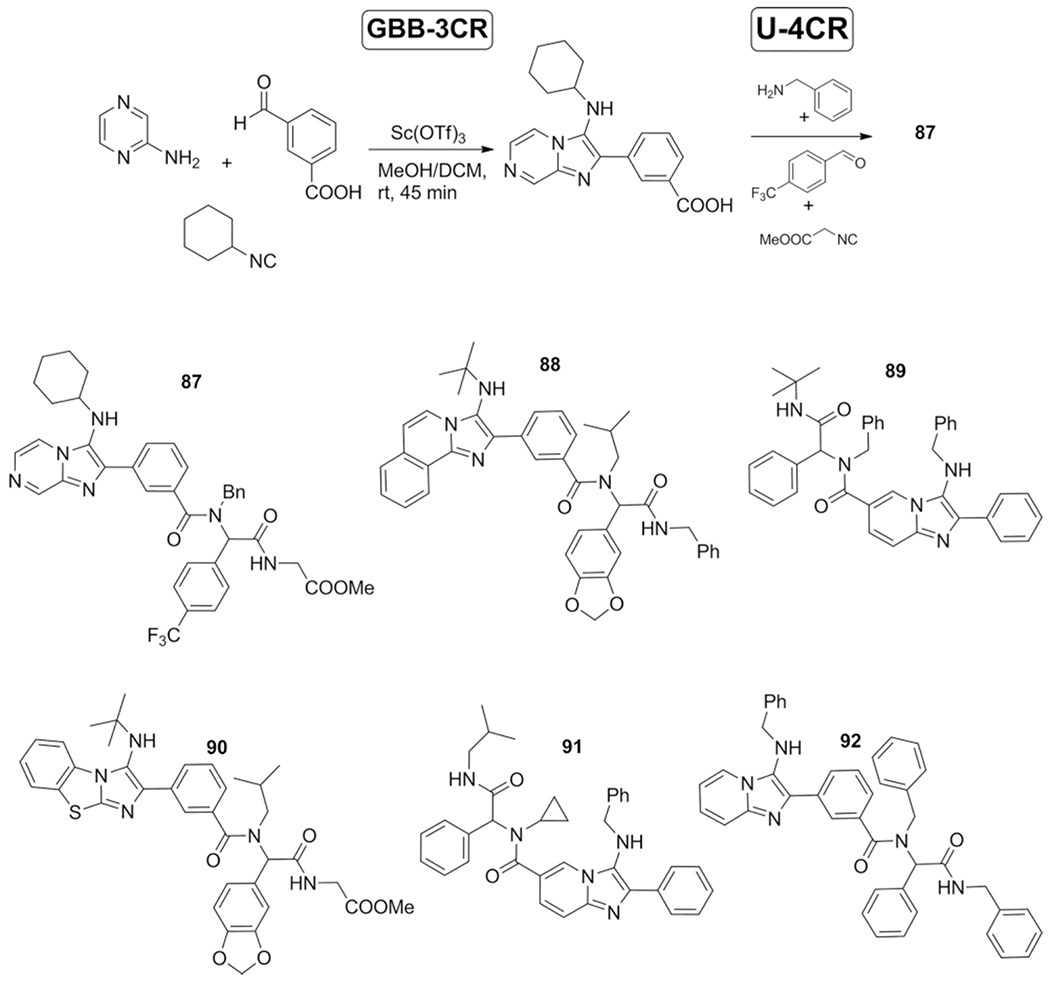
In 2011 Al-Tell et al. described the application of various subsequent MCR’s.[58] The carboxylic acid group attached to one of the three typical GBB-3CR starting materials would not participate in this MCR reaction, but is useful for follow up MCR chemistry via for example the U-4CR to synthesize antibiotics.[80] The six components introduced in two sequential reactions, yielded 40 polyfunctionalized imidazopyridine, pyrimidine and pyrazine derivatives and were screened against multiple bacterial strains such as Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa and screened against two human cancer cell lines; breast carcinoma (MCF7), A549 (lung cancer cell line) and melanoma (M8).
Five of the synthesized compounds 87–91 were found to exhibit lower IC50values compared with the internal standard doxorubicin, against A549, M8 and MCF7 cell lines. Moreover, compound 92 exhibits a high potency against breast cancer (Scheme 36).
Scheme 36.
Application of a subsequent MCR approach to conveniently obtain complex compounds, exhibiting both antibacterial and anticancer properties.
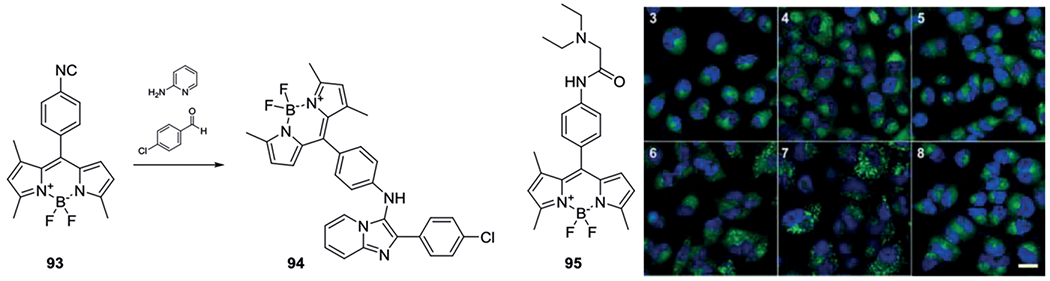
Vendrell et al. reported the synthesis of functionalized BODIPY probes to introduce fluorescence in otherwise difficult accessible structures.[82] While BODIPY dyes were previously derivatized via BODIPY containing an amine or carboxylic acid substituent respectively, this paper focuses on the preparation of isocyanide functionalized BODIPY (93).[207–211] The application of this dye was found through isocyanide-based MCR chemistry for the aid of in vivo imaging of phagocytic macrophages. Different IMCR compounds were synthetized as the GBB-3R variant 94, and incubated with A549 cells and although all the adducts readily entered cells at concentrations in the nanomolar range, the Ugi adduct 95 exhibited the best coloured images (Scheme 37).
Scheme 37.
BODIPY isocyanide 93 used as fluorescent tag. Compound 95 exhibit fluorescence in A549 cells as depicted in image 7 on the right. Reprinted (adapted) with permission from reference[82] Copyright 2013 American Chemical Society.
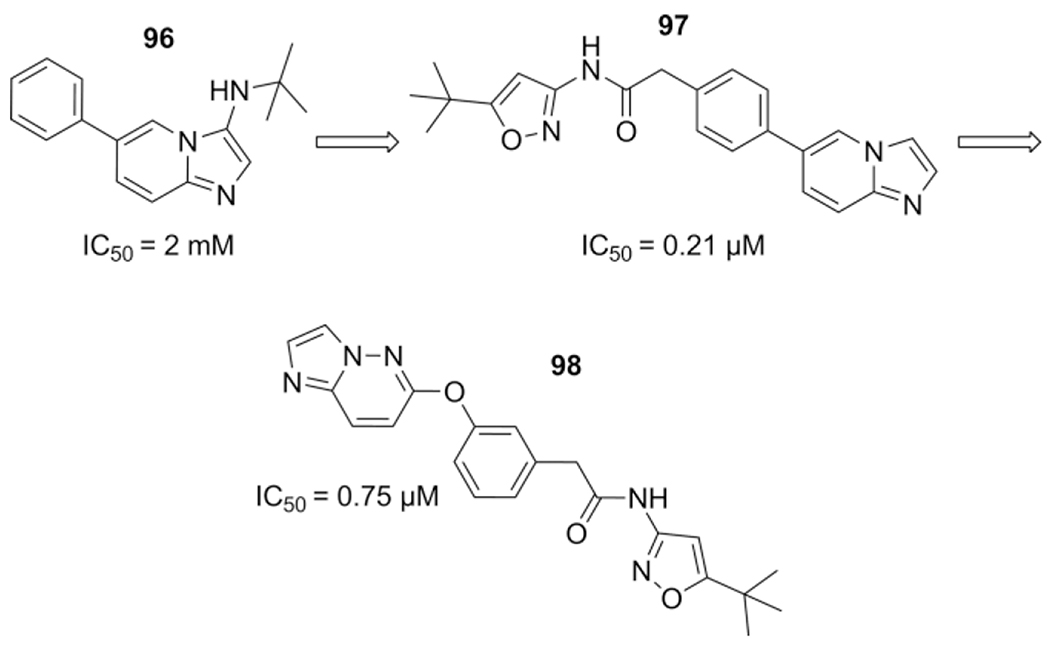
Glyoxylic acid based GBB was employed by Li el al. to obtain a series of mono-substituted imidazopyridines that target the RET kinase class, a known oncogene.[212] In their first series, 100 MCR fragments were synthesized by a acetic acid catalyzed GBB-3CR using substituted 2 aminopyridines, isocyanides and glyoxylic acid. Screening resulted in a hit 96 which was subjected to computational binding analysis in the RET kinase. The phenyl moiety accesses a lyophilic pocket that could be extended from the allosteric pocket. Another non-MCR series of compounds were synthesized to engage this adjacent lipophilic pocket, resulting in an increase of inhibitory potency as shown by two compounds (97 = 0.21 μM and 98 = 0.75 μM) thus identified as novel RET kinase inhibitors (Scheme 38).
Scheme 38.
Initial GBB-3CR hit design towards novel non-MCR RET inhibitors.
Imidazo[1,2-a]pyridine-N-glycinylhydrazone derivatives were applied as inhibitors of tumor necrosis factor alpha (TNF-α) by Fraga et al.[84] The design of the scaffold was performed by combining two earlier described TNF-α inhibitors wherein the N-phenyl-pyrazole nucleus 99 was replaced with the heterocyclic imidazo[1,2-a]pyridine isoster, easily accessible by the GBB-3CR.
Via an acetic acid catalyzed reaction of benzaldehyde or pivaldehyde, 2-aminopyridine and ethyl 2-isocyanoacetate two GBB-3CR products were obtained with yields of 65 and 75 % respectively. Post modification with hydrazine and subsequent hydrazone formation yielded 11 products that were screened in a TNF-α production assay, ultimately leading to the most active 100, equipotent with SB-203580 (101) a known p38 MAPK inhibitor herein applied as anti TNF-α agent (Scheme 39).
Scheme 39.
Imidazo[1,2-a]pyridine-N-glycinylhydrazone derivatives as TNF-α inhibitors.
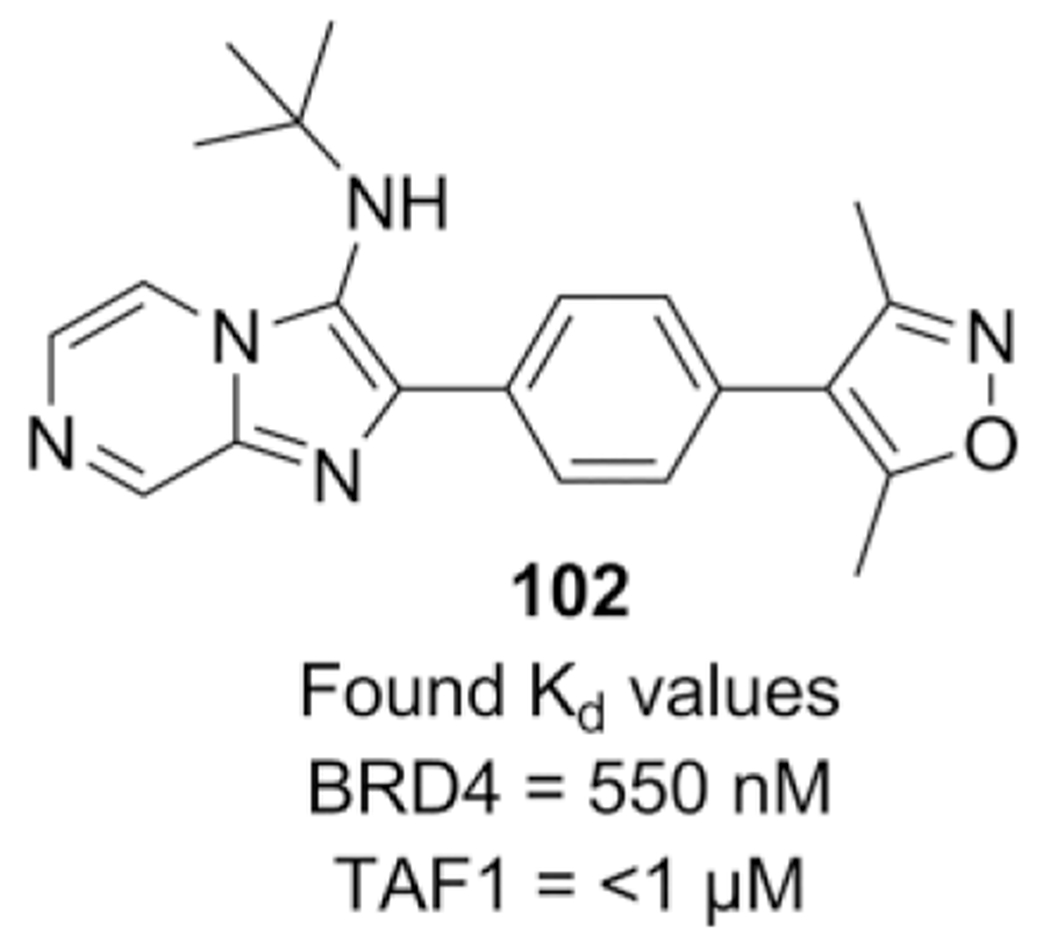
Inhibition of the BET bromodomain is a promising approach as anti-cancer therapy. In a fragment based optimizing strategy, Bradner et al. used the dimethylisoxazole biasing element as a key fragment and screened on BRD4 and BRD4-dependent lines, using different heterocyclic substituents to gather information on preferred regiochemistry.[86] Bromo substituted benzaldehydes were employed in the μW assisted, Sc(OTf3) catalyzed GBB-3CR to perform a follow-up Suzuki coupling in order to introduce the 3,5-dimethylisoxazole warhead. The best compound is built around the para-substituted phenyl-imidazo-[1,2-a]pyrazine scaffold and a small library of 29 compounds was synthesized for SAR optimizing, resulting in lead compound 102 (Scheme 40).
Scheme 40.
GBB-3CR compound 102 as potent BET bromodomain inhibitor.
In continuation of Shen et al.[206] another HeLa cytotoxic series of 6H-pyrido[2′,1′:2,3]imidazo[4,5-c]isoquinolin-5(6H)-ones was reported by the same research group[87] In this report the tetracyclic profile was maintained, however the alkynyl moiety was replaced with a carboxylic acid (phthaldehydic acid) which direct reacts with the secondary amine, formed in the GBB-3CR (Scheme 41).
Scheme 41.
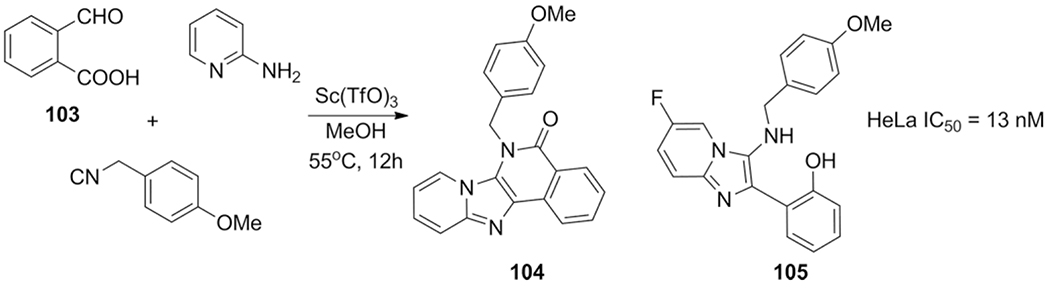
Application of the bi-functional phthaldehydic acid 103 giving access to tetracyclic heterocycles with example 104 exhibiting HeLa toxicity (IC50 = 1.8 μM).
Shen et al. published, more work on cytotoxic compounds while abandoning the tetracyclic scaffold, switching to a bicyclic scaffold exhibiting intramolecular hydrogen bonding to form pseudo-tetracycles.[177] A more straightforward methodology, as it only comprises a single GBB-3CR step. The library was step by step optimized with variations of all three components. The hydrogen bonding ability was most successful when an ortho substituted hydroxyl- or methoxybenzaldehyde component was used. A 100-fold activity increase over compound 104 was realized with compound 105.
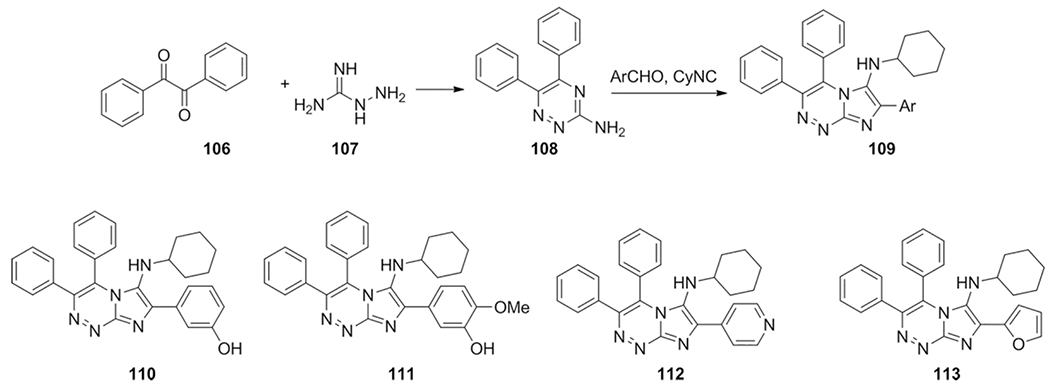
Foroumadi et al. reported novel anticancer agents based on N-fused aminoimidazole derivatives with triazine analogs.[158] The basis of this compound were a combination of potent topoisomerase IIα inhibitors[63] and triazines[213] to yield the N-cyclohexyl-3,4-diphenylimidazo[2,1-c][1,2,4]triazin-6-amine scaffold 109 (Scheme 42). The aminotriazine 108 was prepared from condensation of benzil 106 with aminoguanidine 107. Noteworthy are the conditions that were found whilst optimizing the GBB model reaction. The reaction with toluene under reflux conditions gave the highest yield of 70 % in the presence of 1 equivalent ammonium chloride as compared to 45 % without.
Scheme 42.
A series of anticancer agents, through the clever condensation benzyl 106 and aminoguanidine 107 and subsequent GBB-3CR.
A small library of 20 compounds was prepared and their cytotoxicity was assessed on HL60, MCF-7 and MOL-4 by the MTT reduction assay against cisplatin and doxorubicin. The highest potency of compounds 110 and 111 was observed on MCF-7 and HL60 cells respectively. SAR optimizing showed that replacing the 7-aryl moiety with pyridyl or furan-2-yl (compounds 112 and 113) gave improvement of the potency against MOLT-4.
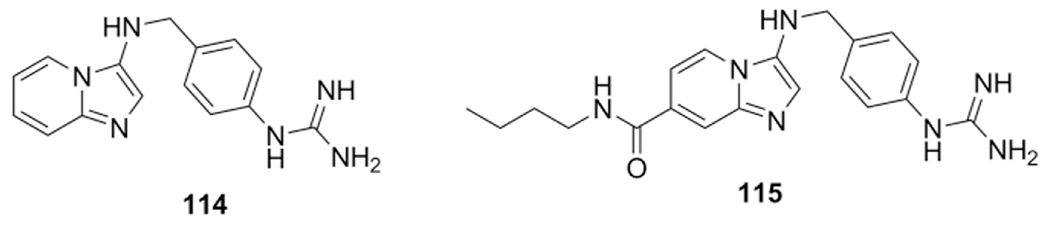
In a substrate activity screening, Veken et al. decorated an imidazo-[1,2-a]pyridine scaffold with a library of fragments that bind to the urokinase plasminogen activator (uPA) S1 pocket.[96] This target is a biomarker and therapeutic target for cancer types such as breast, ovarian and pancreatic cancer. In two rounds the imidazopyridine scaffold was optimized. The first round focused on mono-substitution with fragments on the 3 position of the imidazopyridine scaffold using a GBB-3CR to identify fragments that bind to the S1 pocket. In a second round structural optimization was performed by introducing substituents through the aminopyridine and aldehyde component. This resulted from an IC50 (uPA) value (114) of 9.04 μM towards 0.097 μM (115) improvement between the first and second round respectively (Scheme 43).
Scheme 43.
Binders of the urokinase Plasminogen Activator (uPA) S1 pocket, round 1 and 2 top performing products.
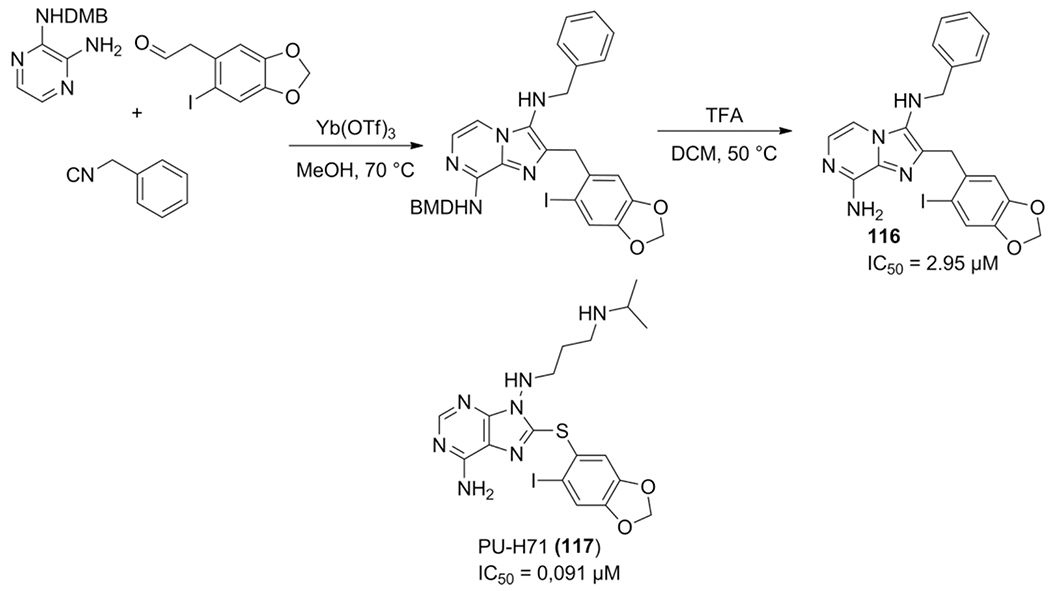
The imidazo[1,2-a]pyrazine scaffold is found in Heat shock protein 90 (Hsp90) inhibitor PU-H71, using the Zolpidem rationale that absence of the amine moiety in the 3 position wouldn’t affect the inhibitory effects, Shen et al. synthesized a library of substituted 3,8-diaminoimidazo[1,2-a]pyrazines to target this protein.[153] The biggest challenge in these compounds was the incorporation of the adenine motif with an amine in the 8 position, which will in unprotected form participate in the GBB-3CR. Introduction of a chlorine at position 8 followed by aminolysis as well as protected amines were tried by the authors. The resulting lead compound 116 exhibits moderate Hsp90 inhibition as compared to PU-H71 (117), however, the co-crystallization of the reported 3,8-diaminoimidazo[1,2-a]pyrazines have shown to bind in the same way as the known inhibitor (Scheme 44).
Scheme 44.
Development of novel imidazo[1,2-a]pyrazine based Hsp90 inhibitors.
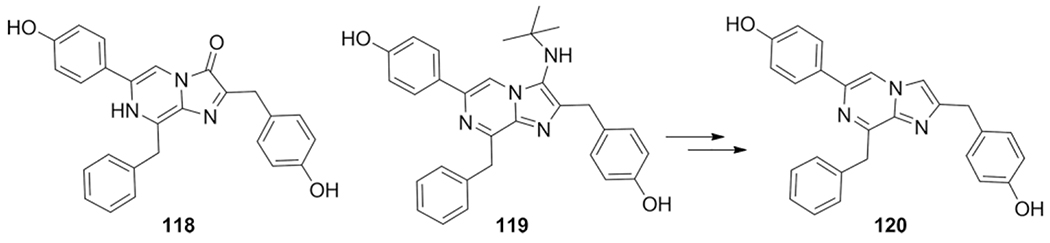
Coelenterazione 118 is a bioluminescent imidazopyrazine and can be found in several marine organisms. The properties such as bioluminescent and antioxidant are useful and utilized as optical signaling agents for cancer behavior studies. Vuocolo et al. reports the synthesis of 3 deoxy- derivatives of coelenterazine to improve some of its properties, such as its low stability.[154] The 3 deoxy approach brings room to apply the GBB-3CR with substituents on the C-3 position.
In this report only tert-butyl isocyanide was used in a acetic acid catalyzed GBB-3CR under dry conditions (3Å molecular sieves) yielding compound 119 in 64 % and in follow up reactions a deprotection and reductive deamination (NaNO2/H2SO4) routine was used to obtain compound 120 (Scheme 45).
Scheme 45.
3 deoxy- derivatives of coelenterazine as bioluminescent and antioxidant.
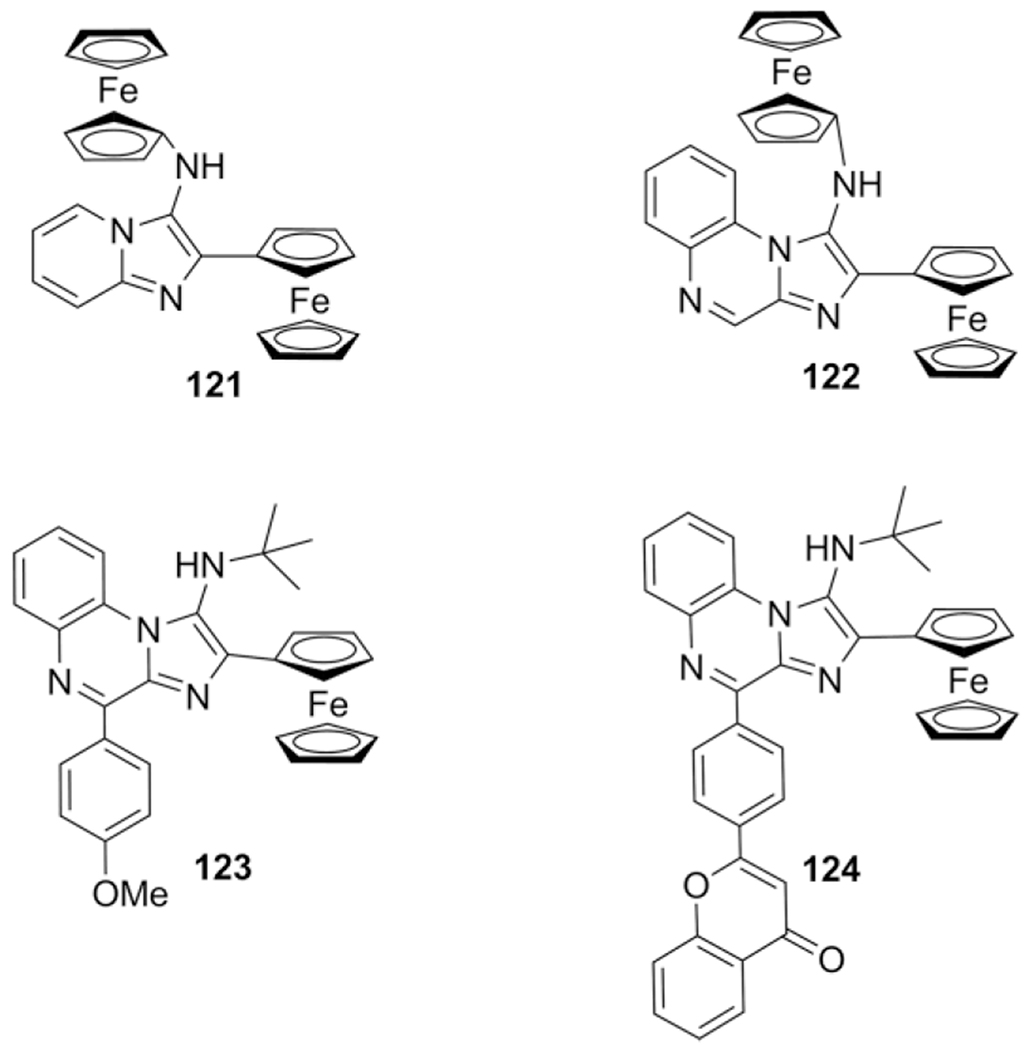
Liu et al. describes a library of imidazopyridine and quinoxaline derived compounds and introduces ferrocene incorporation via the aldehyde or isocyanide component through the GBB-3CR, with the aim of developing new antioxidants to reduce oxidative stress/DNA damage due to reactive oxygen species.[104]
The LaCl3·7H2O has proven to be an excellent catalyst with yields >84 % at a reaction temperature of 60 °C in EtOH for 2h. Compounds 121 and 122, the ferrocene bearing compounds, were the best in inhibiting DNA oxidation. More improvement was made by derivatization of the imidazo[1,2-a]quinoxaline scaffold.[155] Decoration with ferrocenyl and p-methoxyphenyl (123) or flavonyl (124) groups on either the 2 or 4 position boosted the radical suppression/inhibition of DNA oxidation (Scheme 46).
Scheme 46.
Ferrocene based anti-oxidants.
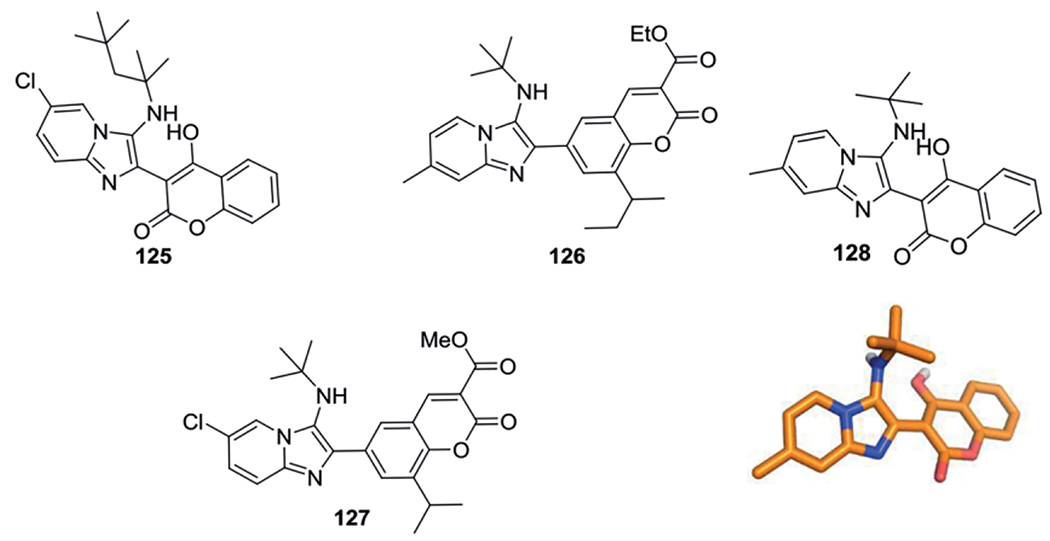
A hybrid structure between imidazo[1,2-a]pyridine and coumarins were synthesized by Shah et al. as potential Hepatitis C virus replicator inhibitor.[110] The 20 compounds were synthesized using a AcOH (20 mol-%) catalyzed GBB-3CR in MeOH at 70 °C for 2h in yields between 67–92 %. Molecular docking studies indicate good binding interactions of compound 125 with nonstructural protein 5B.
Trivedi et al. combined the structural properties of anticancer agents that commonly have a coumarin structure and anti-osteoporotic agents holding pyridine or imidazole heterocycles into coumarin-imidazo[1,2-a]pyridine hybrids for the treatment of cancer induced osteoporosis.[113] 18 GBB-3CR products were obtained using ethanol reflux conditions and 20 mol-% AgOTf with excellent yields (71–88 %), including compound 126. Compound 127 was found to possess anti-breast cancer activity (MDA-MB-231 IC50 = 14.12 + /− 3.69 μM) as well as disrupting the differentiation of osteoblast and osteoclast cells and therefore indicating the compound as promising hybrid for both aforementioned diseases (Scheme 47).
Scheme 47.
Coumarine based GBB-3CR products, compound 128 was crystallized and the structure was unambiguously verified by X-ray analysis (CCDC-1053723).
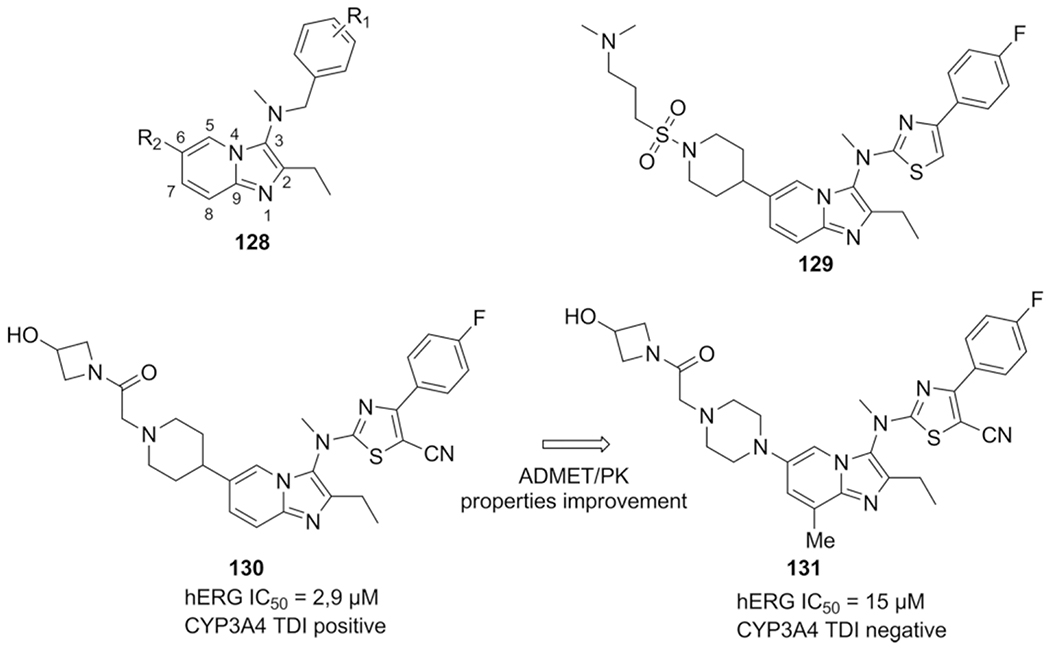
The first-in-class autotaxin (ATX) inhibitor GLPG1690 was described by Heckmann et al. and was discovered and optimized through the synthesis of a 2,3,6 trisubstituted imidazo[1,2-a]-pyridine series, identified by HTS.[186,194] Approximately 100.000 compounds were subjected to screening in which ATX activity is promoting release of the fluorophore FS-3, an HTS suitable substitute of LPC, thus easily identifying inhibitors by a fluorescence detection assay. The N-benzyl-2-ethyl-N-methyl-imidazo[1,2-a]pyridin-3-amine hit series 128 displayed a IC50 around 30 nm in the FS-3 assay (Scheme 48).
Scheme 48.
Autotaxin inhibitors; from hit to lead.
The metabolic stability of this series was however poor and a second series with improved pharmacokinetic properties was developed, resulting in an early lead compound 129 (IC50 = 122 nm (FS-3 assay). The reduced potency gave reason to continue SAR analysis and synthesis. Furthermore the ATX activity could not be reproduced with biochemical assays; using the natural substrate LPC displayed only micromolar activity. Extensive optimizing on all substituents, assisted by analysis of the binding mode using compound 129, resulted in lead compound 130 with a potency of 27 nm in the biochemical assay and 22 nm in a rat plasma assay. The improved potency was achieved through the 6 position by extending the substituent and the addition of a basic amine and an H-bond donor or acceptor. The addition of a nitrile group on the thiazole moiety pushes out a high energy water by full occupation by the ligand, increasing the potency even more. In a second report Heckmann et al. describes improvement of lead compound 130, focusing on ADMET (hERG and CYP3A4) and PK properties (clearance and bioavailability), ultimately resulting in the clinical candidate GLPG1690 (131) (IC50 = 131 nm). The five-fold loss in activity of 131 was outweighed by the absence of CYP3A4 TDI and the compound is overall the best combination of activity, pharmacokinetic, ADMET and PK properties. The ATX inhibitor was further discussed in the structural biology section.
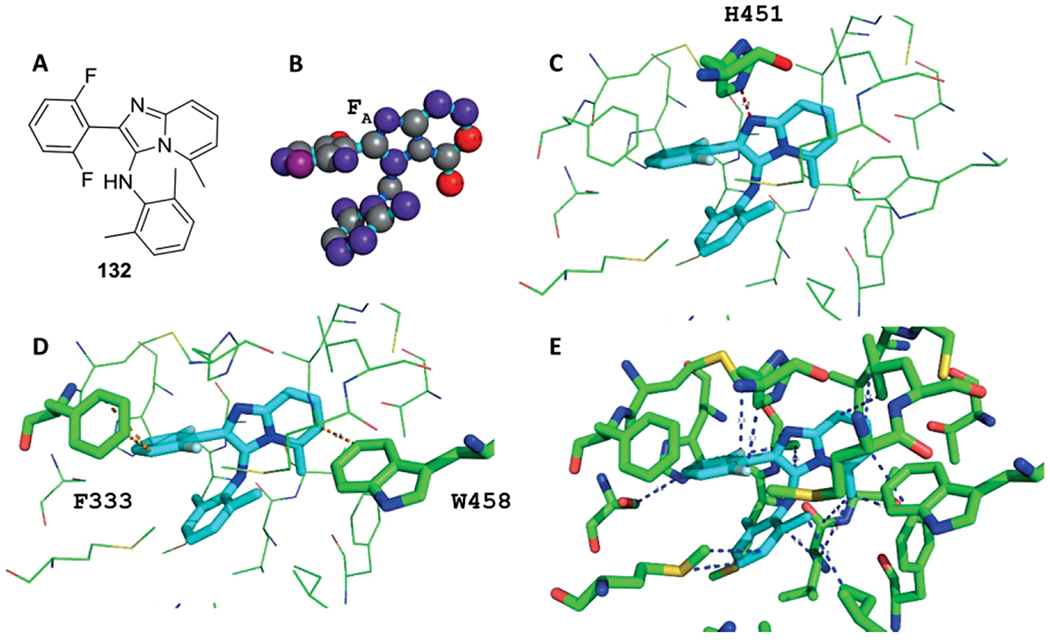
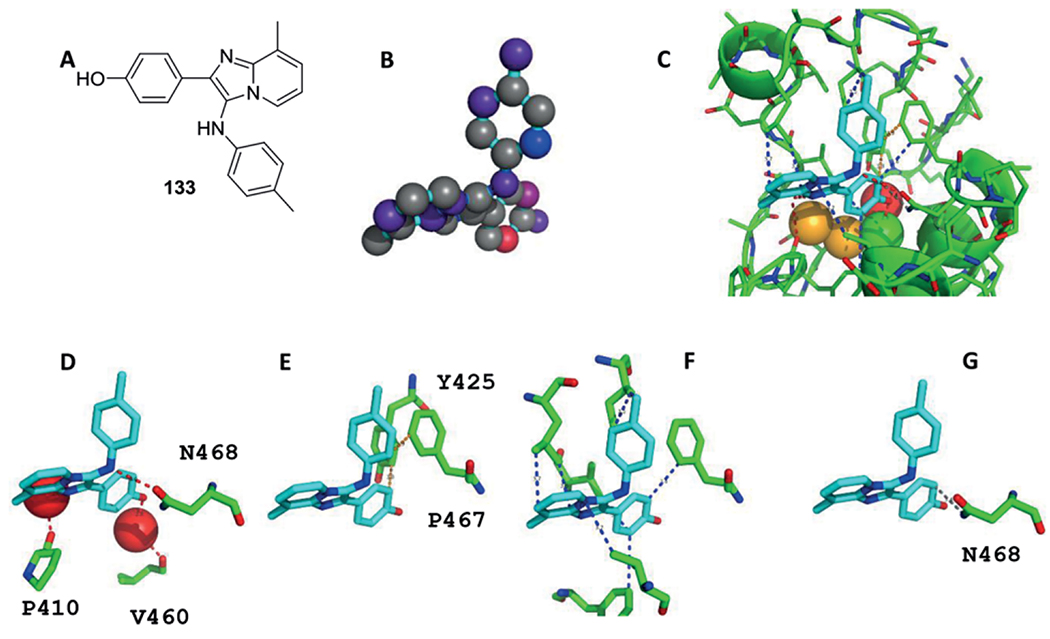
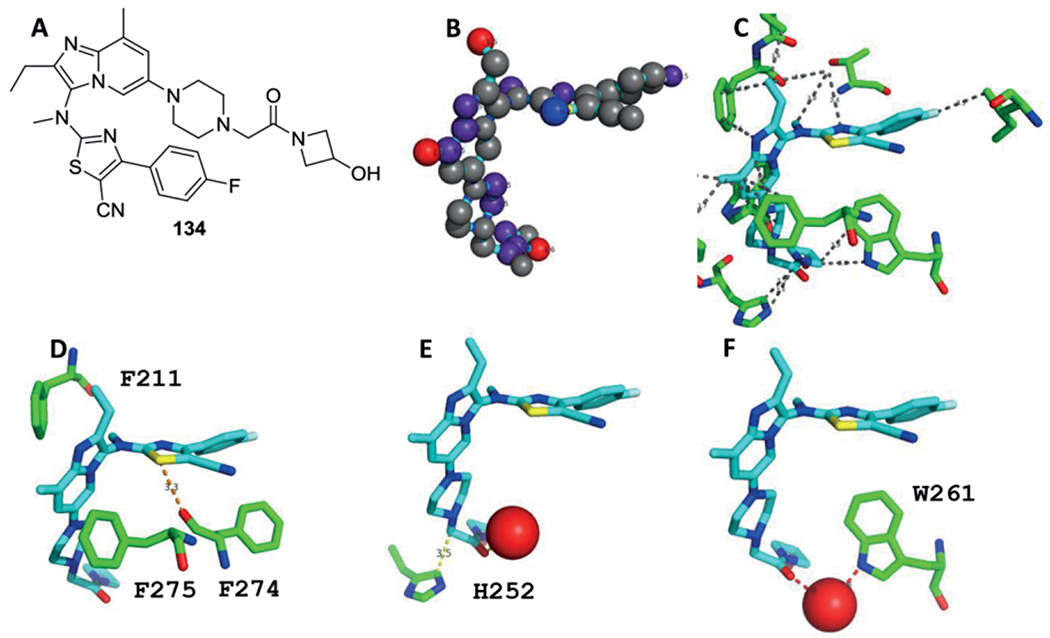
Structural Biology
Farnesoid X receptors (FXRs) are nuclear hormone receptors expressed in high concentration in tissues that participate in bilirubin metabolism including the kidneys, intestines, and liver. In animal models of nonalcoholic fatty liver disease (NAFLD), FXR activation protects against fatty liver injury and nonalcoholic steatohepatitis (NASH), and improved hyperlipidemia, glucose intolerance, and insulin sensitivity. Thus, FXR agonists could potentially be used as drugs to treat nonalcoholic fatty and cholestatic liver diseases. A GBB product 132 was described in a cocrystal structure bound to FXR (Figure 4A).[214] Remarkably, 132 fits tightly in the pocket undergoing a range of hydrogen, pi-stacking and van der Waals interactions (Figure 4. C–E).[214] The 2,6-dimethyl phenyl moiety (derived from the isocyanide) ensures an out-of-plane orientation of the two phenyl substituents with regard to the imidazopyridine bicycle through sterically repulsion which can help to increase binding affinity through preorganisation of the binding conformation (Figure 4B).
Figure 4.
GBB product 132 bound to farnesoid X receptor (FXR, PDB ID 5Q12). A) 2D structure of 132; B) scorpion score (14.0) and its heavy atom distribution on 132 (color code: red strong to purple weak contribution, grey minor contribution); C) the imidazole-N of 132 acts as a hydrogen-bond acceptor towards the His451 (2.8 Å); D) 132 forms two pi-stacking interactions between Phe333 and the 2,6-difluorophenyl moiety and the Trp458 and the 2-methyl pyridine moiety; E) multiple hydrophobic interactions can be found between all parts of 132 and the surrounding hydrophobic receptor amino acids; specifically to mention is FA (red in B) which makes multiple contacts with His298, Met332 and Ala295.
The protein bromodomain (BD) serves the recognition of acetylated lysine as in histones and is composed of approximately 110 amino acids. In the context of signal transduction, the bromodomain function as a “reader” of the lysine acetylation. For a family of bromodomains, the BET (bromodomain and extra terminal domain) family, several compounds are in clinical evaluation. Members of the BET family are targets in both human cancer and multiple sclerosis. The bromodomain has also been investigated as antifungal target. Screening a library of ca. 80.000 chemically diverse compounds by a HTRF inhibition assay revealed GBB 133.[215] Compound 133 inhibits the CaBdf1 BD2 with high selectivity: HTRF and ITC assays yielded IC50 and Kd values of 1.1 and 2.1 μM, respectively, whereas human Brd4 BD2 and CaBdf1 BD1 were only weakly inhibited (IC50 ≥ 40 μM). Moreover, no significant inhibition of any human BDs was observed for compound 133 by BROMO-scan profiling and by an ITC assay on the two most sensitive BDs identified by this screen. Only mild cytotoxicity was observed towards mammalian cells (EC50 ca. 60 μM) for compound 133. A cocrystal structure of 133 in Candida albicans Bdf1 reveals shape complementarity to the receptor and a network of hydrogen bonding, pi-stacking, van der Waals interactions (Figure 5).
Figure 5.
Second Bromodomain (BD2) from Candida albicans Bdf1 binding to an GBB imidazopyridine (PDB 5N18). A) 2D structure of 133; B) scorpion score (5.6) and its heavy atom distribution on 133 (color code: red strong to purple weak contribution, grey minor contribution);; C) overall GBB receptor binding site featuring multiple electrostatic interactions and a string of four signature structural waters (green, yellow and red balls); D) two water mediated hydrogen bondings including Pro410 carbonyl-H2O-N imidazole and Val460 carbonyl-H2O-p-hydroxyphenyl and Asn468 side chain amide-CO and exocyclic NH of isocyanide moiety; E) T-shaped pi-pi interactions between Phe467 and Tyr425 and the phenyl groups of the isocyanide and aldehyde components; F) multiple van der Waals interactions involving Phe467, Val415, Ph411, Val474 and Leu420; G) a H-bond donor-pi interaction involving the side chain amide NH of Asn468 and the aldehyde phenyl.
Autotaxin (ATX) is a secreted lysophophospholipase D that converts lysophosphatidyl choline (LPC) into the bioactive phospholipid derivative lysophosphatidic acid (LPA). LPA exerts its biological activities through activation of the LPA receptors: LPA1–6. The ATX/LPA axis is involved in numerous physiological and pathophysiological processes. Autotaxin inhibitors have been proposed to be useful in various pathologies such as cancer, pain, and cholestatic pruritus, as well as fibrotic, inflammatory, and cardiovascular diseases.
Appropriate structural modifications of the initial GBB hit lead to 134 with improved ADMET (hERG and CYP3A4 TDI) and PK properties (bioavailability and clearance).[186] Compound 134 was able to decrease LPA levels in mouse plasma. Additionally, compound 134 demonstrated significant activity in a mouse fibrosis model at doses of 10 and 30 mg/kg twice a day, with an efficacy comparable or superior to that of the reference compound pirfenidone. Compound 134 is currently being evaluated in an exploratory phase 2a study in idiopathic pulmonary fibrosis patients. Compound 134 was also cocrystallized with its target to better understand the molecular basis of its disease modifying activity (Figure 6).
Figure 6.
GBB Imidazo[1,2-a]pyridine derivatives with potent autotaxin/ENPP2 inhibitor activity (PDB ID 5MHP). A) 2D structure of 134; B) scorpion score (10.6) and its heavy atom distribution on 134 (color code: red strong to purple weak contribution, grey minor contribution); C) overall topology of the interaction of 134 with its receptor featuring multiple hydrophobic, hydrogen bonding and stacking interactions; D) x is embedded in a aromatic cage including Phe’s211, 275, 274, Trp261 and His252; Phe274 carbonyl forms a pi-pi interaction with the thiazole sulfur; E) two H bond donor pi interactions involve His252 and a structural water molecule; F) a water mediated hydrogen bond involves the azetidine carbonyl and the NH of Trp261.
Heat shock protein 90 (HSP-90) is a so-called chaperone protein that assists other proteins to fold properly, stabilizes proteins against heat stress, and aids in protein degradation. It has shown to stabilize proteins during cancer development. The natural product geldanamycin and derivatives and totally synthetic compounds show promising antitumor activity in clinical trials and therefore HSP-90 is a validated cancer target. The GBB compound 135 was synthesized based on the observation of ATP-competitive Hsp90 inhibitors containing the 8,9-disubstituted adenine motif, which might be replaced with the 2,3-disubstituted 8-aminoimidazo[1,2-a]pyrazine scaffold (GBB).[153] Additionally, computational scaffold hopping exercises suggested that a similarly substituted 8-aminoimidazo[1,2-a]pyrazine might possess Hsp90 inhibitory activity due to the remarkable resemblance to PU-H71, an Hsp90 inhibitor in Phase I clinical trial. The hypothesis proved to be trough and multiple GBB compounds with this adenosine mimicking patterns showed competitive binding to HSP-90.135 has been cocrystallized with HSP-90 (Figure 7) and shows multiple contacts to the ATP binding site including hydrogen bonding, pi-stacking of the piperonyl moiety and van der Waals interactions.
Figure 7.
GBB compound 135 binding to HSP-90 (PDB ID 4R3M). A) 2D structure of 135; B) scorpion score (14.8) and its heavy atom distribution on 135 (color code: red strong to purple weak contribution, grey minor contribution); C) overall receptor ligand interaction involving multiple van der Waals, stacking and hydrogen bonding interactions; D) a parallel pi-pi stacking interaction between Phe138 and the moiety 1,2- methylenedioxybenzen e moiety is a strong affinity contributor; E) the adenine-NH2 forms a direct hydrogen bond towards Asp93 carboxylate and a tripartite hydrogen bond Leu48 backbone carbonyl and Ser52-OH mediated by two structural water molecules accounting for the adenine-NH2 importance for binding.
Glutamine synthetase catalyzes the synthesis of glutamine from glutamate and ammonia under hydrolysis of adenosine 5′-triphosphate (ATP). The enzyme is involved in nitrogen metabolism and cell wall biosynthesis in pathogenic mycobacteria and thus comprise a potential anti tuberculosis target. ATP-competitive MtGS inhibitors were identified in a high-throughput screening (HTS). One hit compound was a GBB 3-amino-imidazo[1,2-a]pyridines. This was an ideal candidate class for structure–activity relationship (SAR) studies, because diverse analogs can easily be obtained in one step via GBB-3CR.[134] The best compound showed an IC50 of 0.6 μM. Co-crystallization of one of the best inhibitors with MtGS 136, helped to rationalize the observed SAR (Figure 8).
Figure 8.
Crystal structure of Mycobacterium tuberculosis glutamine synthetase in complex with GBB 53 (PDB ID 4ACF). A) 2D structure of X; B) scorpion score (10.3) and its heavy atom distribution on 53 (color code: red strong to purple weak contribution, grey minor contribution); C) 53 is embedded in a hydrophobic site involving Arg364, Phe232, Lys361, Trp282 and Tyr129; D) a halogen bonding between Hi278 backbone carbonyl and the Br is identified; E) a parallel pi-pi stacking interaction between Phe231 and the pyridine central part of GBB backbone is formed: F) a ionic interaction is spotted between the ligands carboxy group and the Lys361; G) a cation-pi interaction is formed between Arg364 and the pyridine part of the GBB backbone; H) the aminobutyl side chain-NH is involved in a direct hydrogen bonding with Lys361 carbonyl and a tripartite water mediated network with Trp282 indole NH and Ser280-OH.
Patents
The first appearance of the GBB-3CR in patents took roughly 3 years when a nitric oxide synthase inhibitor was reported by Gruenenthal GmbH.[216] About 100 patents were filed in the course of 18 years, in which some had the imidazo[1,2-a]pyridine/pyrazine as main decorated scaffold obtained using the GBB methodology, while others reported the scaffold either by sequential synthesis and as one of many different claimed structures. The GBB-3CR was found in the pharmaceutical industry and shortly after its discovery patents concerning the [1,2-a]-imidazo scaffold appeared in patent applications, either directly or indirectly as seen in the 2004 Patent from Merck & Co. Inc.[217] where the double bonds in the originating pyrazine were reduced to obtain a piperazine. The drug was with various structural modifications brought to the market as Sitagliptin, also known under the tradename Januvia. As many patents were reported in the last decades, not many true GBB-3CR scaffolds were marketed as actual drugs. Recently compound KAF156in development by Novartis AG, completed phase 2 clinical trials ( NCT01753323), being effective against the Plasmodium falciparum and Plasmodium vivax forms of the malaria parasite with clearance of the parasites within 2 days.[218,219] More examples of patents that leverage the GBB scaffold were listed in Table 6.
Table 6.
The structures found in the patent were found as GBB products, whereas the listed structures comprise the in general most active compound, shown as the active component of interest.
AnchorQuery
The scaffold of the GBB-3CR is one of the 27 MCR scaffolds that is included in the AnchorQuery library which was recently introduced as powerful tool for rational structure-based design of protein-protein interaction (PPI) inhibitors.[261] The often deeply buried amino acids comprise a major energetic hotspot which is leveraged in a pharmacophore model search that rapidly screens libraries yielding a possible 31 million synthesizable compounds, each containing an anchor motif that is bioisosteric to amino acid residues, including D, W, Y, F, V and E anchor side chains. Important examples of the power of AnchorQuery are demonstrated through the role it played in the development of novel p53-MDM2 inhibitors and an allosteric inhibitor.[262–264] In the virtual screening the suggested scaffolds are amenable in one to maximal three reaction steps. More specifically, the screening platform contains xyz GBB in xyz different conformations. Using AnchorQuery as a tool is free of charge, accessible via http://anchorquery.csb.pitt.edu and will provide all the tools necessary to interactively perform online virtual screening, rapidly going through millions of compounds with pharmacophore elucidation and enrichment analysis queries. A great benefit of the screening platform is obviously that a binding hypothesis can be immediately tested by resynthesizing the virtual hit (Figure 9).
Figure 9.
Workflow of the online virtual screening platform ANCHOR.QUERY.
Conclusions
The GBB-3CR has been described the first-time 20 years ago and is currently one of the mostly used MCR reactions. According to the definition of the Ugi reaction as an α-addition of a Schiff base and a suitable acid component and a rearrangement reaction it belongs formally to the same class of reactions. Similar to the classical Ugi MCR the GBB-3CR also shows a great scope in terms of diversity of starting materials. For mostly any combination between aldehydes, isocyanides and heterocyclic amidines, with the help of a suitable catalyst, the reaction is giving the expected products. Not surprisingly the GBB scaffold is described in multiple publications and patents. The first derivatives of GBB products have entered clinical trials. This is very well in line with the saying that after 15–20 years since the discovery of a new reaction first products based on new reactions will enter the market. It can be predicted that several more bioactive compounds will enter the market in the future. An interesting freely accessible virtual screening platform will be useful in the discovery of tool compounds against difficult to drug targets. Currently still underdeveloped are materials science application of the GBB-3CRs, such as functional polymers or organic conductors or light to energy transformers. Clearly the GBB scaffold has the potential that many interesting electronic and optical applications will be described in the future.
Acknowledgments
Research in the Dömling laboratory is supported by the National Institutes of Health (NIH) (2R01GM097082–05), the European Lead Factory (IMI), under grant agreement number 115489, the Qatar National Research Foundation (NPRP6–065–3–012), ITN “Accelerated Early stage drug dIScovery” (AEGIS, grant agreement No 675555), and COFUND ALERT (grant agreement No 665250), COFUND PROMINENT (grant agreement No 754425), Hartstichting (ESCAPE-HF, 2018B012), and KWF Kankerbestrijding grant (grant agreement No 10504).
Abbreviations
A list of abbreviations is given in Table 7.
Table 7.
List of abbreviations.
| AcOH | Acetic acid |
|---|---|
| AD | Alzheimers disease |
| ADMET | Absorption, distribution, metabolism, and excretion |
| ATX | Autotaxin |
| 3CR | 3-component reaction |
| BODIPY | Boron-dipyrromethene |
| CB2 | Cannabinoid receptor type 2 |
| CH2Cl2 | Dichloromethane |
| CK2 | Casein Kinase II |
| DMA | Dimethylaceetamide |
| DMF | Dimethylformamide |
| DMSO | Dimethyl sulfoxide |
| ERK | Extracellular signal-regulated kinases |
| FDA | Food and Drug Administration |
| FGFR3 | Fibroblast growth factor receptor 3 |
| GABA | Gamma aminobutyric acid |
| GLP | Glucagon-like peptide-1 |
| h-TRPA1 | Transient receptor potential cation channel, subfamily A, member 1 |
| HClO4 | Perchloric acid |
| HCV | Hepatitis C |
| HDAC | Histone deacetylase |
| hENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 (ATX) |
| HTS | High throughput screening |
| IMCR | Isocyanide-based multicomponent reactions |
| JNK1 | See MAPK8 |
| LPC | Lysophosphatidylcholine |
| MAPK8 | Mitogen-activated protein kinase 8 |
| MCR | Multicomponent reactions |
| MDM2 | Murine double minute 2 |
| MeCN | Acetonitrile |
| MeOH | Methanol |
| MP-CO3 | Macroporous polystyrene carbonate |
| MTOR | Mammalian target of rapamycin |
| MW | Microwave |
| NMR | Nuclear magnetic resonance |
| P-3CR | Passerini 3-component reaction |
| PADAM | Passerini-reaction-Amine-Deprotection-Acyl-Migration |
| PD | Parkinsons Disease |
| PDGFR | Platelet-derived growth factor receptor |
| PEG | Polyethylene glycol |
| PET | Positron emmision tomography |
| PGHS-2 | Prostaglandin-endoperoxide synthase 2 |
| PI3 | Phosphoinositide 3-kinase |
| Pim-1 | Proto-oncogene serine/threonine-protein kinase |
| PK | Pharmacokinetic |
| PTSA | para-Toluenesulfonic acid |
| pTsIA | para-Toluenesulfinic acid |
| RET | Rearranged during transfection |
| SAR | Structure-activity relationship |
| Sc(OTf)3 | Scandium triflate |
| SGLT | Sodium-glucose transport proteins |
| SKM | Leukemic cell line |
| TDI | Time-dependent inhibition |
| Tf | Triflate |
| TFE | Trifluoroethanol |
| TLC | Thin Layer Chromatography |
| TMOF | Trimethyl orthoformate |
| TMSN3 | Trimethylsilyl azide |
| TNFa | Tumor necrosis factor alpha |
| TPSO | Translocator protein |
| TRIM33 | Tripartite motif-containing 33 |
| U-3CR | Ugi 3-component reaction |
| U-4CR | Ugi 4 component reaction |
| UDC | Ugi-deprotection-cyclize |
| vL-3CR | van Leusen 3 component reaction |
Biographies

André Boltjes was born in the Netherlands. He received his Bachelor’s Degree in chemistry in 2007 from Hanze University and started working at the University of Groningen, developing dopamine agonists, HAT inhibitors, doxorubicin prodrugs, and performing various contract synthesis projects. In 2011 he joined Dömling’s group as a technician in University of Groningen. Over the years he was heavily involved in many research projects and is expecting to obtain his Ph.D. at the end of 2019.

Alexander Dömling studied chemistry & biology at the Technical University Munich. After performing his Ph.D. under the supervision of Ivar Ugi he spent his postdoctoral year at the Scripps Research Institute in the group of Barry Sharpless under a prestigious Feodor Lynen research fellowship from the Alexander von Humboldt society. He is founder of several biotech companies, including Morphochem, Telesis and SMIO. In 2004 he performed his habilitation in chemistry at the Technical University of Munich. Since 2006 he was Professor at the University of Pittsburgh in the Department of Pharmacy and Chemistry and in 2011 he became Chair of Drug Design at the University of Groningen. He is author of more than 200 papers, reviews, book contributions and patents. His research interest focuses on novel aspects of multicomponent reaction chemistries and its applications to drug design.
Footnotes
ORCID(s) from the author(s) for this article is/are available on the WWW under https://doi.org/10.1002/ejoc.201901124.
References
- [1].Schreiber SL, Science 2000, 287, 1964–1969. [DOI] [PubMed] [Google Scholar]
- [2].Dömling A, Wang W, Wang K, Chem. Rev 2012, 112, 3083–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ugi I, Meyr R, Fetzer U, Steinbrückner C, Angew. Chem. Int. Ed. Engl 1959, 71, 386. [Google Scholar]
- [4].Passerini M, Simone L, Gazz. Chim. Ital 1921, 51, 126–129. [Google Scholar]
- [5].Gewald K, Schinke E, Bottcher H, Chem. Ber-Recl 1966, 99, 94. [Google Scholar]
- [6].Groebke K, Weber L, Mehlin F, Synlett 1998, 661–663. [Google Scholar]
- [7].Blackburn C, Guan B, Fleming P, Shiosaki K, Tsai S, Tetrahedron Lett. 1998, 39, 3635–3638. [Google Scholar]
- [8].Bienayme H, Bouzid K, Angew. Chem. Int. Ed 1998, 37, 2234–2237; [DOI] [PubMed] [Google Scholar]; Angew. Chem 1998, 110, 2349. [Google Scholar]
- [9].Biginelli P, Ber. Dtsch. Chem. Ges 1891, 24, 2962–2967. [Google Scholar]
- [10].Boukis AC, Reiter K, Frölich M, Hofheinz D, Meier MAR, Nat. Commun 2018, 9, 1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Banfi L, Guanti G, Riva R, Basso A, Calcagno E, Tetrahedron Lett 2002, 43, 4067–4069. [Google Scholar]
- [12].Keating TA, Armstrong RW, J. Am. Chem. Soc 1996, 118, 2574–2583. [Google Scholar]
- [13].Van Leusen AM, Wildeman J, Oldenziel OH, J. Org. Chem 1977, 42, 1153–1159. [Google Scholar]
- [14].Ugi I, Steinbrückner C, DE-B1 1959, 103, 337. [Google Scholar]
- [15].Mulder P, Litwinienko G, Lin S, MacLean PD, Barclay LRC, Ingold KU, Chem. Res. Toxicol 2006, 19, 79–85. [DOI] [PubMed] [Google Scholar]
- [16].Moni L, Banfi L, Basso A, Carcone L, Rasparini M, Riva R, J. Org. Chem 2015, 80, 3411–3428. [DOI] [PubMed] [Google Scholar]
- [17].Rossen K, Sager J, DiMichele LM, Tetrahedron Lett. 1997, 38, 3183–3186. [Google Scholar]
- [18].Goto T, Inoue S, Sugiura S, Tetrahedron Lett. 1968, 9, 3873–3876. [DOI] [PubMed] [Google Scholar]
- [19].Varma RS, J. Heterocycl. Chem 1999, 36, 1565–1571. [Google Scholar]
- [20].See ref.[7].
- [21].Devi N, Rawal RK, Singh V, Tetrahedron 2015, 71, 183–232. [Google Scholar]
- [22].Shaaban S, Abdel-Wahab BF, Mol. Diversity 2016, 20, 233–254. [DOI] [PubMed] [Google Scholar]
- [23].Asinger F, Thie-683l M, Angew. Chem 1958, 70, 667. [Google Scholar]
- [24].Liu Z-Q, Curr. Org. Synth 2015, 12, 20–60. [Google Scholar]
- [25].Zai-Qun L, Mini-Rev. Org. Chem 2016, 13, 166–183. [Google Scholar]
- [26].Mandair GS, Light M, Russell A, Hursthouse M, Bradley M, Tetrahedron Lett. 2002, 43, 4267–4269. [Google Scholar]
- [27].Ireland SM, Tye H, Whittaker M, Tetrahedron Lett. 2003, 44, 4369–4371. [Google Scholar]
- [28].Schwerkoske J, Masquelin T, Perun T, Hulme C, Tetrahedron Lett. 2005, 46, 8355–8357. [Google Scholar]
- [29].Burchak ON, Mugherli L, Ostuni M, Lacapere JJ, Balakirev MY, J. Am. Chem. Soc 2011, 133, 10058–10061. [DOI] [PubMed] [Google Scholar]
- [30].Li L, Hopkinson MN, Yona RL, Bejot R, Gee AD, Gouverneur V, Chem. Sci 2011, 2, 123–131. [Google Scholar]
- [31].Martinez-Ariza G, Ayaz M, Medda F, Hulme C, J. Org. Chem 2014, 79, 5153–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Martinez-Ariza G, Nunez-Rios J, Lee Y-S, Hulme C, Tetrahedron Lett. 2015, 56, 1038–1040. [Google Scholar]
- [33].Allahabadi E, Ebrahimi S, Soheilizad M, Khoshneviszadeh M, Mahdavi M, Tetrahedron Lett. 2017, 58, 121–124. [Google Scholar]
- [34].Baenziger M, Durantie E, Mathes C, Synthesis 2017, 49, Ahead of Print. [Google Scholar]
- [35].Ghashghaei O, Caputo S, Sintes M, Reves M, Kielland N, Estarellas C, Luque FJ, Avino A, Eritja R, Serna-Gallego A, Marrugal-Lorenzo JA, Pachon J, Sanchez-Cespedes J, Treadwell R, de Moliner F, Vendrell M, Lavilla R, Chem. Eur. J 2018, 24, 14513–14521. [DOI] [PubMed] [Google Scholar]
- [36].Guchhait SK, Madaan C, Tetrahedron Lett. 2011, 52, 56–58. [Google Scholar]
- [37].Krasavin M, Tsirulnikov S, Nikulnikov M, Kysil V, Ivachtchenko A, Tetrahedron Lett. 2008, 49, 5241–5243. [Google Scholar]
- [38].Krasavin M, Tsirulnikov S, Nikulnikov M, Sandulenko Y, Bukhryakov K, Tetrahedron Lett. 2008, 49, 7318–7321. [Google Scholar]
- [39].Blackburn C, Guan B, Tetrahedron Lett. 2000, 41, 1495–1500. [Google Scholar]
- [40].Chen JJ, Golebiowski A, Klopfenstein SR, McClenaghan J, Peng SX, Portlock DE, West L, Synlett 2001, 1263–1265. [Google Scholar]
- [41].Lu Y, Zhang W, QSAR Comb. Sci 2004, 23, 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lyon MA, Kercher TS, Org. Lett 2004, 6, 4989–4992. [DOI] [PubMed] [Google Scholar]
- [43].Parchinsky VZ, Shuvalova O, Ushakova O, Kravchenko DV, Krasavin M, Tetrahedron Lett. 2006, 47, 947–951. [Google Scholar]
- [44].Pirrung MC, Ghorai S, J. Am. Chem. Soc 2006, 128, 11772–11773. [DOI] [PubMed] [Google Scholar]
- [45].Veljkovic I, Zimmer R, Reissig H-U, Bruedgam I, Hartl H, Synthesis 2006, 2677–2684. [Google Scholar]
- [46].Adib M, Mahdavi M, Noghani MA, Mirzaei P, Tetrahedron Lett. 2007, 48, 7263–7265. [Google Scholar]
- [47].Carballares S, Cifuentes MM, Stephenson GA, Tetrahedron Lett. 2007, 48, 2041–2045. [Google Scholar]
- [48].Kercher T, Rao C, Bencsik JR, Josey JA, J. Comb. Chem 2007, 9, 1177–1187. [DOI] [PubMed] [Google Scholar]
- [49].Nenajdenko VG, Reznichenko AL, Balenkova ES, Russ. Chem. Bull 2007, 56, 560–562. [Google Scholar]
- [50].Rousseau AL, Matlaba P, Parkinson CJ, Tetrahedron Lett. 2007, 48, 4079–4082. [Google Scholar]
- [51].Shaabani A, Maleki A, Rad JM, Soleimani E, Chem. Pharm. Bull 2007, 55, 957–958. [DOI] [PubMed] [Google Scholar]
- [52].Mert-Balci F, Conrad J, Meindl K, Schulz T, Stalke D, Beifuss U, Synthesis 2008, 3649–3656. [Google Scholar]
- [53].Sandulenko Y, Komarov A, Rufanov K, Krasavin M, Tetrahedron Lett. 2008, 49, 5990–5993. [Google Scholar]
- [54].Shaabani A, Rezazadeh F, Soleimani E, Monatsh. Chem 2008, 139, 931–933. [Google Scholar]
- [55].Shaabani A, Soleimani E, Maleki A, Moghimi-Rad J, Synth. Commun 2008, 38, 1090–1095. [Google Scholar]
- [56].Shaabani A, Soleimani E, Maleki A, Moghimi-Rad J, Mol. Diversity 2009, 13, 269–274. [DOI] [PubMed] [Google Scholar]
- [57].Akbarzadeh R, Shakibaei GI, Bazgir A, Monatsh. Chem 2010, 141, 1077–1081. [Google Scholar]
- [58].Al-Tel TH, Al-Qawasmeh RA, Voelter W, Eur. J. Org. Chem 2010, 2010, 5586–5593, S5586/5581-S5586/5516. [Google Scholar]
- [59].Guchhait SK, Madaan C, Org. Biomol. Chem 2010, 8, 3631–3634. [DOI] [PubMed] [Google Scholar]
- [60].Oskooie HA, Amini M, Heravi MM, Bamoharram FF, Chin. J. Chem 2010, 28, 299–302. [Google Scholar]
- [61].Akritopoulou-Zanze I, Wakefield BD, Gasiecki A, Kalvin D, Johnson EF, Kovar P, Djuric SW, Bioorg. Med. Chem. Lett 2011, 21, 1480–1483. [DOI] [PubMed] [Google Scholar]
- [62].Al-Tel TH, Semreen MH, Al-Qawasmeh RA, Schmidt MF, El-Awadi R, Ardah M, Zaarour R, Rao SN, El-Agnaf O, J. Med. Chem 2011, 54, 8373–8385. [DOI] [PubMed] [Google Scholar]
- [63].Baviskar AT, Madaan C, Preet R, Mohapatra P, Jain V, Agarwal A, Guchhait SK, Kundu CN, Banerjee UC, Bharatam PV, J. Med. Chem 2011, 54, 5013–5030. [DOI] [PubMed] [Google Scholar]
- [64].Bode ML, Gravestock D, Moleele SS, van der Westhuyzen CW, Pelly SC, Steenkamp PA, Hoppe HC, Khan T, Nkabinde LA, Bioorg. Med. Chem 2011, 19, 4227–4237. [DOI] [PubMed] [Google Scholar]
- [65].Dahan-Farkas N, Langley C, Rousseau AL, Yadav DB, Davids H, de Koning CB, Eur. J. Med. Chem 2011, 46, 4573–583. [DOI] [PubMed] [Google Scholar]
- [66].Sharma A, Li H.-y, Synlett 2011, 1407–1412. [Google Scholar]
- [67].Elleder D, Baiga TJ, Russell RL, Naughton JA, Hughes SH, Noel JP, Young JAT, Virol. J 2012, 9, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hieke M, Roedl CB, Wisniewska JM, Buscato E, Stark H, Schubert-Zsilavecz M, Steinhilber D, Hofmann B, Proschak E, Bioorg. Med. Chem. Lett 2012, 22, 1969–1975. [DOI] [PubMed] [Google Scholar]
- [69].Khan AT, Sidick Basha R, Lal M, Tetrahedron Lett. 2012, 53, 2211–2217. [Google Scholar]
- [70].Mert-Balci F, Conrad J, Beifuss U, ARKIVOC 2012, 243–256. [Google Scholar]
- [71].Rostamnia S, Lamei K, Mohammadquli M, Sheykhan M, Heydari A, Tetrahedron Lett. 2012, 53, 5257–5260. [Google Scholar]
- [72].Salunke DB, Yoo E, Shukla NM, Balakrishna R, Malladi SS, Serafin KJ, Day VW, Wang X, David SA, J. Med. Chem 2012, 55, 8137–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Shukla NM, Salunke DB, Yoo E, Mutz CA, Balakrishna R, David SA, Bioorg. Med. Chem 2012, 20, 5850–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tyagi V, Khan S, Bajpai V, Gauniyal HM, Kumar B, Chauhan PMS, J. Org. Chem 2012, 77, 1414–1421. [DOI] [PubMed] [Google Scholar]
- [75].Arnould M, Hiebel M-A, Massip S, Leger JM, Jarry C, Berteina-Raboin S, Guillaumet G, Chem. Eur. J 2013, 19, 12249–12253. [DOI] [PubMed] [Google Scholar]
- [76].Daemi H, Rad RR, Barikani M, Adib M, Appl. Catal. A 2013, 468, 10–17. [Google Scholar]
- [77].Guchhait SK, Priyadarshani G, Chaudhary V, Seladiya DR, Shah TM, Bhogayta NP, RSC Adv. 2013, 3, 10867–10874. [Google Scholar]
- [78].Reutlinger M, Koch CP, Reker D, Todoroff N, Schneider P, Rodrigues T, Schneider G, Mol. Inf 2013, 32, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Rostamnia S, Hassankhani A, RSC Adv. 2013, 3, 18626–18629. [Google Scholar]
- [80].Semreen MH, El-Awady R, Abu-Odeh R, Saber-Ayad M, Al-Qawasmeh RA, Chouaib S, Voelter W, Al-Tel TH, Curr. Med. Chem 2013, 20, 1445–1459. [DOI] [PubMed] [Google Scholar]
- [81].Shahrisa A, Esmati S, Synlett 2013, 24, 595–602. [Google Scholar]
- [82].Vazquez-Romero A, Kielland N, Arevalo MJ, Preciado S, Mellanby RJ, Feng Y, Lavilla R, Vendrell M, J. Am. Chem. Soc 2013, 135, 16018–16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ghorbani-Vaghei R, Amiri M, J. Heterocycl. Chem 2014, 51, E372–E379. [Google Scholar]
- [84].Lacerda RB, Sales NM, da Silva LL, Tesch R, Miranda ALP, Barreiro EJ, Fernandes PD, Fraga CAM, PLoS One 2014, 9, 91610e91660/91661-e91660/91610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Maleki A, Javanshir S, Naimabadi M, RSC Adv. 2014, 4, 30229–30232. [Google Scholar]
- [86].McKeown MR, Shaw DL, Fu H, Liu S, Xu X, Marineau JJ, Huang Y, Zhang X, Buckley DL, Kadam A, Zhang Z, Blacklow SC, Qi J, Zhang W, Bradner JE, J. Med. Chem 2014, 57, 9019–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Meng T, Wang W, Zhang Z, Ma L, Zhang Y, Miao Z, Shen J, Bioorg. Med. Chem 2014, 22, 848–855. [DOI] [PubMed] [Google Scholar]
- [88].Rahmati A, Moazzam A, Khalesi Z, Tetrahedron Lett. 2014, 55, 3840–3843. [Google Scholar]
- [89].Reutlinger M, Rodrigues T, Schneider P, Schneider G, Angew. Chem. Int. Ed 2014, 53, 582–585; [DOI] [PubMed] [Google Scholar]; Angew. Chem 2014, 126, 593. [Google Scholar]
- [90].Sanaeishoar T, Tavakkoli H, Mohave F, Appl. Catal. A 2014, 470, 56–62. [Google Scholar]
- [91].Shahrisa A, Safa KD, Esmati S, Spectrochim. Acta 2014, 117, 614–621. [DOI] [PubMed] [Google Scholar]
- [92].Shinde AH, Srilaxmi M, Satpathi B, Sharada DS, Tetrahedron Lett. 2014, 55, 5915–5920. [Google Scholar]
- [93].Vidyacharan S, Shinde AH, Satpathi B, Sharada DS, Green Chem. 2014, 16, 1168–1175. [Google Scholar]
- [94].Baviskar AT, Amrutkar SM, Trivedi N, Chaudhary V, Nayak A, Guchhait SK, Banerjee UC, Bharatam PV, Kundu CN, ACS Med. Chem. Lett 2015, 6, 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Dianat S, Mahdavi M, Moghimi S, Mouradzadegun A, Shafiee A, Foroumadi A, Mol. Diversity 2015, 19, 797–805. [DOI] [PubMed] [Google Scholar]
- [96].Gladysz R, Adriaenssens Y, De Winter H, Joossens J, Lambeir A-M, Augustyns K, Van der Veken P, J. Med. Chem 2015, 58, 9238–9257. [DOI] [PubMed] [Google Scholar]
- [97].Keshipour S, Shaabani A, Shojaei S, Nosrati H, Ng SW, J. Iran. Chem. Soc 2015, 12, 1655–1663. [Google Scholar]
- [98].Kishore KG, Basavanag UMV, Islas-Jacome A, Gamez-Montano R, Tetrahedron Lett. 2015, 56, 155–158. [Google Scholar]
- [99].Neochoritis CG, Zhang J, Doemling A, Synthesis 2015, 47, 2407–2413. [Google Scholar]
- [100].Shaabani A, Seyyedhamzeh M, Shaabani S, Ganji N, Res. Chem. Intermed 2015, 41, 2377–2383. [Google Scholar]
- [101].Surkar P, Kaur T, Sharma A, Synlett 2015, 26, 1403–1407. [Google Scholar]
- [102].Tber Z, Hiebel MA, Allouchi H, El Hakmaoui A, Akssira M, Guillaumet G, Berteina-Raboin S, RSC Adv. 2015, 5, 35201–35210. [DOI] [PubMed] [Google Scholar]
- [103].Tber Z, Hiebel MA, El Hakmaoui A, Akssira M, Guillaumet G, Berteina-Raboin S, J. Org. Chem 2015, 80, 6564–6573. [DOI] [PubMed] [Google Scholar]
- [104].Xi G-L, Liu Z-Q, Tetrahedron 2015, 71, 9602–9610. [Google Scholar]
- [105].Ansari AJ, Sharma S, Pathare RS, Gopal K, Sawant DM, Pardasani RT, ChemistrySelect 2016, 1, 1016–1021. [Google Scholar]
- [106].Azizi N, Dezfooli S, Environ. Chem. Lett 2016, 14, 201–206. [Google Scholar]
- [107].Cioc RC, Preschel HD, van der Heijden G, Ruijter E, Orru RVA, Chem. Eur. J 2016, 22, 7837–7842. [DOI] [PubMed] [Google Scholar]
- [108].Devi N, Singh D, Honey, Mor S, Chaudhary S, Rawal RK, Kumar V, Chowdhury AK, Singh V, RSC Adv. 2016, 6, 43881–43891. [Google Scholar]
- [109].Devi N, Singh D, Sunkaria RK, Malakar CC, Mehra S, Rawal RK, Singh V, ChemistrySelect 2016, 1, 4696–4703. [Google Scholar]
- [110].Manvar P, Shaikh F, Kakadiya R, Mehariya K, Khunt R, Pandey B, Shah A, Tetrahedron 2016, 72, 1293–1300. [Google Scholar]
- [111].Rostamnia S, Jafari M, Appl. Organomet. Chem 2016, Ahead of Print. [Google Scholar]
- [112].Sagar A, Nagarjuna Babu V, Shinde AH, Sharada DS, Org. Biomol. Chem 2016, 14, 10366–10370. [DOI] [PubMed] [Google Scholar]
- [113].Sashidhara KV, Singh LR, Choudhary D, Arun A, Gupta S, Adhikary S, Palnati GR, Konwar R, Trivedi R, RSC Adv. 2016, Ahead of Print. [Google Scholar]
- [114].Shaabani A, Hooshmand SE, Tetrahedron Lett. 2016, 57, 310–313. [Google Scholar]
- [115].Swami S, Agarwala A, Shrivastava R, New J. Chem 2016, 40, 9788–9794. [Google Scholar]
- [116].Swami S, Devi N, Agarwala A, Singh V, Shrivastava R, Tetrahedron Lett. 2016, 57, 1346–1350. [Google Scholar]
- [117].Tazeem, Han X, Zhou Q, Wei J, Tien P, Yang G, Wu S, Dong C, RSC Adv. 2016, 6, 95177–95188. [Google Scholar]
- [118].Unnamatla MVB, Islas-Jacome A, Quezada-Soto A, Ramirez-Lopez SC, Flores-Alamo M, Gamez-Montano R, J. Org. Chem 2016, 81, 10576–10583. [DOI] [PubMed] [Google Scholar]
- [119].Yang B, Tao C, Shao T, Gong J, Che C, Beilstein J. Org. Chem 2016, 12, 1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Basavanag Unnamatla MV, Islas-Jacome A, Renteria-Gomez A, Conejo AS, Kurva M, Jimenez-Halla JOC, Velusamy J, Ramos-Ortiz G, Gamez-Montano R, New J. Chem 2017, Ahead of Print. [Google Scholar]
- [121].Dam J, Ismail Z, Kurebwa T, Gangat N, Harmse L, Marques HM, Lemmerer A, Bode ML, de Koning CB, Eur. J. Med. Chem 2017, 126, 353–368. [DOI] [PubMed] [Google Scholar]
- [122].Devi N, Singh D, Kaur G, Mor S, Putta VPRK, Polina S, Malakar CC, Singh V, New J. Chem 2017, 41, 1082–1093. [Google Scholar]
- [123].Song G-T, Xu Z-G, Tang D-Y, Li S-Q, Xie Z-G, Zhong H-L, Yang Z-W, Zhu J, Zhang J, Chen Z-Z, Mol. Diversity 2016, 20, 575–580. [DOI] [PubMed] [Google Scholar]
- [124].Shaabani A, Soleimani E, Maleki A, Tetrahedron Lett. 2006, 47, 3031–3034. [Google Scholar]
- [125].Meng T, Zhang Z, Hu D, Lin L, Ding J, Wang X, Shen J, J. Comb. Chem 2007, 9, 739–741. [DOI] [PubMed] [Google Scholar]
- [126].Shaabani A, Soleimani E, Maleki A, Monatsh. Chem 2007, 138, 73–76. [Google Scholar]
- [127].Shaabani A, Soleimani E, Sarvary A, Rezayan AH, Maleki A, Chin. J. Chem 2009, 27, 369–371. [Google Scholar]
- [128].Meirer K, Roedl CB, Wisniewska JM, George S, Haefner A-K, Buscato E. l., Klingler F-M, Hahn S, Berressem D, Wittmann SK, Steinhilber D, Hofmann B, Proschak E, J. Med. Chem 2013, 56, 1777–1781. [DOI] [PubMed] [Google Scholar]
- [129].Flesch D, Schubert-Zsilavecz M, Pharmazie 2015, 70, 507–510. [PubMed] [Google Scholar]
- [130].El Akkaoui A, Hiebel M-A, Mouaddib A, Berteina-Raboin S, Guillaumet G, Tetrahedron 2012, 68, 9131–9138. [Google Scholar]
- [131].Sun H, Zhou H, Khorev O, Jiang R, Yu T, Wang X, Du Y, Ma Y, Meng T, Shen J, J. Org. Chem 2012, 77, 10745–10751. [DOI] [PubMed] [Google Scholar]
- [132].Yang A, Jiang R, Khorev O, Yu T, Zhang Y, Ma L, Chen G, Shen J, Meng T, Adv. Synth. Catal 2013, 355, 1984–1988. [Google Scholar]
- [133].Qian Z, Yang A, An W, Yu T, Wang X, Zhang Y, Shen J, Meng T, RSC Adv. 2014, 4, 50947–50949. [Google Scholar]
- [134].Nordqvist A, Nilsson MT, Lagerlund O, Muthas D, Gising J, Yahiaoui S, Odell LR, Srinivasa BR, Larhed M, Mowbray SL, Karlen A, Med. Chem. Commun 2012, 3, 620–626. [Google Scholar]
- [135].Puttaraju KB, Shivashankar K, RSC Adv. 2013, 3, 20883–20890. [Google Scholar]
- [136].DiMauro EF, Kennedy JM, J. Org. Chem 2007, 72, 1013–1016. [DOI] [PubMed] [Google Scholar]
- [137].Al-Tel TH, Al-Qawasmeh RA, Eur. J. Med. Chem 2010, 45, 5848–5855. [DOI] [PubMed] [Google Scholar]
- [138].Al-Tel TH, Al-Qawasmeh RA, Zaarour R, Eur. J. Med. Chem 2011, 46, 1874–1881. [DOI] [PubMed] [Google Scholar]
- [139].Marandi G, Saghatforoush L, Mendoza-Merono R, Garcia-Granda S, Tetrahedron Lett. 2014, 55, 3052–3054. [Google Scholar]
- [140].Maiti B, Chanda K, Selvaraju M, Tseng C-C, Sun C-M, ACS Comb. Sci 2013, 15, 291–297. [DOI] [PubMed] [Google Scholar]
- [141].Mironov MA, Tokareva MI, Ivantsova MN, Mokrushin VS, Russ. Chem. Bull 2006, 55, 1835–1839. [Google Scholar]
- [142].Polyakov AI, Eryomina VA, Medvedeva LA, Tihonova NI, Voskressensky LG, J. Heterocycl. Chem 2008, 45, 1589–1596. [Google Scholar]
- [143].Butler AJE, Thompson MJ, Maydom PJ, Newby JA, Guo K, Adams H, Chen B, J. Org. Chem 2014, 79, 10196–10202. [DOI] [PubMed] [Google Scholar]
- [144].Mahdavi M, Dianat S, Khavari B, Moghimi S, Abdollahi M, Safavi M, Mouradzadegun A, Kabudanian Ardestani S, Sabourian R, Emami S, Akbarzadeh T, Shafiee A, Foroumadi A, Chem. Biol. Drug Des 2017, Ahead of Print. [DOI] [PubMed] [Google Scholar]
- [145].Umkehrer M, Ross G, Jaeger N, Burdack C, Kolb J, Hu H, Alvim-Gaston M, Hulme C, Tetrahedron Lett. 2007, 48, 2213–2216. [Google Scholar]
- [146].Agrebi A, Allouche F, Chabchoub F, El-Kaim L, Alves S, Baleizao C, Farinha JP, Tetrahedron Lett. 2013, 54, 4781–784. [Google Scholar]
- [147].Akbarzadeh T, Ebrahimi A, Saeedi M, Mahdavi M, Foroumadi A, Shafiee A, Monatsh. Chem 2014, 145, 1483–1487. [Google Scholar]
- [148].Bakherad M, Keivanloo A, Siavashi M, Omidian M, Chin. Chem. Lett 2014, 25, 149–151. [Google Scholar]
- [149].Buckmelter AJ, Ren L, Laird ER, Rast B, Miknis G, Wenglowsky S, Schlachter S, Welch M, Tarlton E, Grina J, Lyssikatos J, Brandhuber BJ, Morales T, Randolph N, Vigers G, Martinson M, Callejo M, Bioorg. Med. Chem. Lett 2011, 21, 1248–1252. [DOI] [PubMed] [Google Scholar]
- [150].Wu T, Nagle A, Kuhen K, Gagaring K, Borboa R, Francek C, Chen Z, Plouffe D, Goh A, Lakshminarayana SB, Wu J, Ang HQ, Zeng P, Kang ML, Tan W, Tan M, Ye N, Lin X, Caldwell C, Ek J, Skolnik S, Liu F, Wang J, Chang J, Li C, Hollenbeck T, Tuntland T, Isbell J, Fischli C, Brun R, Rottmann M, Dartois V, Keller T, Diagana T, Winzeler E, Glynne R, Tully DC, Chatterjee AK, J. Med. Chem 2011, 54, 5116–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Urich R, Wishart G, Kiczun M, Richters A, Tidten-Luksch N, Rauh D, Sherborne B, Wyatt PG, Brenk R, ACS Chem. Biol 2013, 8, 1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Guasconi M, Lu X, Massarotti A, Caldarelli A, Ciraolo E, Tron GC, Hirsch E, Sorba G, Pirali T, Org. Biomol. Chem 2011, 9, 4144–4149. [DOI] [PubMed] [Google Scholar]
- [153].Ren J, Yang M, Liu H, Cao D, Chen D, Li J, Tang L, He J, Chen Y-L, Geng M, Xiong B, Shen J, Org. Biomol. Chem 2015, 13, 1531–1535. [DOI] [PubMed] [Google Scholar]
- [154].Vece V, Vuocolo G, Tetrahedron 2015, 71, 8781–8785. [Google Scholar]
- [155].Chen J-F, Liu Z-Q, Tetrahedron 2016, 72, 1850–1859. [Google Scholar]
- [156].Lamberth C, Synlett 2011, 1740–1744. [Google Scholar]
- [157].Vlaar T, Ruijter E, Znabet A, Janssen E, de Kanter FJJ, Maes BUW, Orru RVA, Org. Lett 2011, 13, 6496–6499. [DOI] [PubMed] [Google Scholar]
- [158].Akbarzadeh T, Noushini S, Taban S, Mahdavi M, Khoshneviszadeh M, Saeedi M, Emami S, Eghtedari M, Sarrafi Y, Khoshneviszadeh M, Safavi M, Divsalar K, Moshafi MH, Asadipour A, Sabourian R, Edraki N, O Firouzi R Miri A Shafiee A Foroumadi, Mol. Diversity 2015, 19, 273–281. [DOI] [PubMed] [Google Scholar]
- [159].Pellegatti L, Vedrenne E, Hiebel M-A, Buron F, Massip S, Leger J-M, Jarry C, Routier S, Tetrahedron Lett. 2011, 52, 5224–5228. [Google Scholar]
- [160].Kaur T, Gautam RN, Sharma A, Chem. Asian J 2016, Ahead of Print. [DOI] [PubMed] [Google Scholar]
- [161].Kaur T, Saha D, Singh N, Singh UP, Sharma A, ChemistrySelect 2016, 1, 434–439. [Google Scholar]
- [162].Hsiao Y-S, Narhe BD, Chang Y-S, Sun C-M, ACS Comb. Sci 2013, 15, 551–555. [DOI] [PubMed] [Google Scholar]
- [163].Lee C-H, Hsu W-S, Chen C-H, Sun C-M, Eur. J. Org. Chem 2013, 2013, 2201–2208. [Google Scholar]
- [164].Pereshivko OP, Peshkov VA, Ermolat’ev DS, Van der Eycken EV, Synlett 2013, 24, 351–354. [Google Scholar]
- [165].Demjen A, Gyuris M, Wolfling J, Puskas LG, Kanizsai I, Beilstein J. Org. Chem 2014, 10, 2338–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Parchinsky VZ, Koleda VV, Shuvalova O, Kravchenko DV, Krasavin M, Tetrahedron Lett. 2006, 47, 6891–6894. [Google Scholar]
- [167].Huang Y, Hu X-Q, Shen D-P, Chen Y-F, Xu P-F, Mol. Diversity 2007, 11, 73–80. [DOI] [PubMed] [Google Scholar]
- [168].Aouali M, Mhalla D, Allouche F, El Kaim L, Tounsi S, Trigui M, Chabchoub F, Med. Chem. Res 2015, 24, 2732–2741. [Google Scholar]
- [169].Tsirulnikov S, Kysil V, Ivachtchenko A, Krasavin M, Synth. Commun 2010, 40, 111–119. [Google Scholar]
- [170].Tailor YK, Khandelwal S, Kumari Y, Awasthi K, Kumar M, RSC Adv. 2015, 5, 46415–46422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Kishore KG, Islas-Jacome A, Renteria-Gomez A, Conejo AS, Basavanag UMV, Wrobel K, Gamez-Montano R, Tetrahedron Lett. 2016, 57, 3556–3560. [Google Scholar]
- [172].Wadhwa P, Kaur T, Sharma A, RSC Adv. 2015, 5, 44353–44360. [Google Scholar]
- [173].Kaminski JJ, Hilbert JM, Pramanik BN, Solomon DM, Conn DJ, Rizvi RK, Elliott AJ, Guzik H, Lovey RG, J. Med. Chem 1987, 30, 2031–2046. [DOI] [PubMed] [Google Scholar]
- [174].Groziak MP, Wilson SR, Clauson GL, Leonard NJ, J. Am. Chem. Soc 1986, 108, 8002–8006. [Google Scholar]
- [175].Katritzky AR, Xu Y-J, Tu H, J. Org. Chem 2003, 68, 4935–4937. [DOI] [PubMed] [Google Scholar]
- [176].Sandulenko Y, Krasavin M, Chem. Heterocycl. Compd 2012, 48, 606–612. [Google Scholar]
- [177].An W, Wang W, Yu T, Zhang Y, Miao Z, Meng T, Shen J, Eur. J. Med. Chem 2016, 112, 367–372. [DOI] [PubMed] [Google Scholar]
- [178].Neochoritis CG, Zarganes-Tzitzikas T, Stotani S, Domling A, Herdtweck E, Khoury K, Domling A, ACS Comb. Sci 2015, 17, 493–499. [DOI] [PubMed] [Google Scholar]
- [179].Pirrung MC, Ghorai S, Ibarra-Rivera TR, J. Org. Chem 2009, 74, 4110–4117. [DOI] [PubMed] [Google Scholar]
- [180].Murlykina MV, Kornet MN, Desenko SM, Shishkina SV, Shishkin OV, Brazhko AA, Musatov VI, Van der Eycken EV, Chebanov VA, Beilstein J Org. Chem 2017, 13, 1050–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Che C, Xiang J, Wang G-X, Fathi R, Quan J-M, Yang Z, J. Comb. Chem 2007, 9, 982–989. [DOI] [PubMed] [Google Scholar]
- [182].Shao T, Gong Z, Su T, Hao W, Che C, Beilstein J Org. Chem 2017, 13, 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Shaabani A, Nosrati H, Seyyedhamzeh M, Res. Chem. Intermed 2015, 41, 3719–3727. [Google Scholar]
- [184].Lacerda RB, de Lima CKF, da Silva LL, Romeiro NC, Miranda ALP, Barreiro EJ, Fraga CAM, Bioorg. Med. Chem 2009, 17, 74–84. [DOI] [PubMed] [Google Scholar]
- [185].Polyakov AI, Eryomina VA, Medvedeva LA, Tihonova NI, Listratova AV, Voskressensky LG, Tetrahedron Lett. 2009, 50, 4389–4393. [Google Scholar]
- [186].Desroy N, Housseman C, Bock X, Joncour A, Bienvenu N, Cherel L, Labeguere V, Rondet E, Peixoto C, Grassot J-M, Picolet O, Annoot D, Triballeau N, Monjardet A, Wakselman E, Roncoroni V, Le Tallec S, Blanque R, Cottereaux C, Vandervoort N, Christophe T, Mollat P, Lamers M, Auberval M, Hrvacic B, Ralic J, Oste L, van der Aar E, Brys R, B Heckmann J Med. Chem 2017, 60, 3580–3590. [DOI] [PubMed] [Google Scholar]
- [187].Flesch D, Cheung S-Y, Schmidt J, Gabler M, Heitel P, Kramer J, Kaiser A, Hartmann M, Lindner M, Lueddens-Daemgen K, Heering J, Lamers C, Lueddens H, Wurglics M, Proschak E, Schubert-Zsilavecz M, D Merk J Med. Chem 2017, 60, 7199–7205. [DOI] [PubMed] [Google Scholar]
- [188].Gladow D, Senf D, Wiecko J, Lentz D, Zimmer R, Reissig H-U, Chem. Heterocycl. Compd 2017, 53, 416–421. [Google Scholar]
- [189].Sun C, Ji S-J, Liu Y, J. Chin. Chem. Soc. (Taipei, Taiwan) 2008, 55, 292–296. [Google Scholar]
- [190].Azimi S, Zonouzi A, Firuzi O, Iraji A, Saeedi M, Mahdavi M, Edraki N, Eur. J. Med. Chem 2017, 138, 729–737. [DOI] [PubMed] [Google Scholar]
- [191].Guchhait SK, Madaan C, Synlett 2009, 628–632. [Google Scholar]
- [192].Varma RS, Kumar D, Tetrahedron Lett. 1999, 40, 7665–7669. [Google Scholar]
- [193].Odell LR, Nilsson MT, Gising J, Lagerlund O, Muthas D, Nordqvist A, Karlen A, Larhed M, Bioorg. Med. Chem. Lett 2009, 19, 4790–4793. [DOI] [PubMed] [Google Scholar]
- [194].Joncour A, Desroy N, Housseman C, Bock X, Bienvenu N, Cherel L, Labeguere V, Peixoto C, Annoot D, Lepissier L, Heiermann J, Hengeveld WJ, Pilzak G, Monjardet A, Wakselman E, Roncoroni V, Le Tallec S, Galien R, David C, Vandervoort N, Christophe T, Conrath K, Jans M, Wohlkonig A, Soror S, Steyaert J, Touitou R, Fleury D, Vercheval L, Mollat P, Triballeau N, van der Aar E, Brys R, Heckmann B, J. Med. Chem 2017, 60, 7371–7392. [DOI] [PubMed] [Google Scholar]
- [195].Thompson MJ, Hurst JM, Chen B, Synlett 2008, 3183–3187. [Google Scholar]
- [196].Kaur T, Wadhwa P, Bagchi S, Sharma A, Chem. Commun 2016, 52, 6958–6976. [DOI] [PubMed] [Google Scholar]
- [197].Kobayashi S, Eur. J. Org. Chem 1999, 15–27. [Google Scholar]
- [198].Millich F, Chem. Rev 1972, 72, 101–113. [Google Scholar]
- [199].Millich F, in Polymerization Reactions, Springer Berlin Heidelberg, Berlin, Heidelberg, 1975, pp. 117–141. [Google Scholar]
- [200].Saegusa T, Kobayashi S, Ito Y, Yasuda N, J. Am. Chem. Soc 1968, 90, 4182–4182.5655885 [Google Scholar]
- [201].Hoffmann R, Woodward RB, J. Am. Chem. Soc 1965, 87, 2046–2048. [Google Scholar]
- [202].El Ashry ESH, Nadeem S, Shah MR, Kilany YE, in Advances in Heterocyclic Chemistry, Vol. 101 (Ed.: Katritzky AR), Academic Press, 2010. pp. 161–228. [Google Scholar]
- [203].Krasavin M, Shkavrov S, Parchinsky V, Bukhryakov K, J. Org. Chem 2009, 74, 2627–2629. [DOI] [PubMed] [Google Scholar]
- [204].Guchhait SK, Madaan C, Thakkar BS, Synthesis 2009, 3293–3300. [Google Scholar]
- [205].Regnier S, Bechara WS, Charette AB, J. Org. Chem 2016, 81, 10348–10356. [DOI] [PubMed] [Google Scholar]
- [206].Zhou H, Wang W, Khorev O, Zhang Y, Miao Z, Meng T, Shen J, Eur. J. Org. Chem 2012, 2012, 5585–5594, S5585/5510-S5585/5530. [Google Scholar]
- [207].Myochin T, Hanaoka K, Komatsu T, Terai T, Nagano T, J. Am. Chem. Soc 2012, 134, 13730–13737. [DOI] [PubMed] [Google Scholar]
- [208].Michel BW, Lippert AR, Chang CJ, J. Am. Chem. Soc 2012, 134, 15668–15671. [DOI] [PubMed] [Google Scholar]
- [209].Komatsu T, Urano Y, Fujikawa Y, Kobayashi T, Kojima H, Terai T, Hanaoka K, Nagano T, Chem. Commun 2009, 7015–7017. [DOI] [PubMed] [Google Scholar]
- [210].Isik M, Ozdemir T, Turan IS, Kolemen S, Akkaya EU, Org. Lett 2013, 15, 216–219. [DOI] [PubMed] [Google Scholar]
- [211].Gavande N, Kim HL, Doddareddy MR, Johnston GAR, Chebib M, Hanrahan JR, ACS Med. Chem. Lett 2013, 4, 402–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Frett B, Moccia M, Carlomagno F, Santoro M, Li H.-y, Eur. J. Med. Chem 2014, 86, 714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Labouta IM, Eshba NH, Salama HM, Monatsh. Chem 1988, 119, 591–596. [Google Scholar]
- [214].Gaieb Z, Liu S, Gathiaka S, Chiu M, Yang H, Shao C, Feher VA, Walters WP, Kuhn B, Rudolph MG, Burley SK, Gilson MK, Amaro RE, J. Comput.-Aided Mol. Des 2018, 32, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Mietton F, Ferri E, Champleboux M, Zala N, Maubon D, Zhou Y, Harbut M, Spittler D, Garnaud C, Courcon M, Chauvel M, d’Enfert C, Kashemirov BA, Hull M, Cornet M, McKenna CE, Govin J, Petosa C, Nat. Commun 2017, 8, 15482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Sundermann B, Maul C, Hennies H-H, Schneider J, Gruenenthal Gmbh, Germany: 2002, p. 114 pp. [Google Scholar]
- [217].Duffy JL, Edmondson SD, Kim D, Kirk BA, Wang L, Weber AE, Merck & Co., Inc., USA, 2004, p. 118 pp. [Google Scholar]
- [218].Chatterjee AK, Nagle A, Wu T, Tully D, Kuhen KL, IRM LLC, Bermuda, 2011. p. 313pp. [Google Scholar]
- [219].White NJ, Duong TT, Uthaisin C, Nosten F, Phyo AP, Hanboonkunupakarn B, Pukrittayakamee S, Jittamala P, Chuthasmit K, Cheung MS, Feng YY, Li RB, Magnusson B, Sultan M, Wieser D, Xun XL, Zhao R, Diagana TT, Pertel P, Leong FJ, New Engl. J. Med 2016, 375, 1152–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Berset C, Audetat S, Tietz J, Gunde T, Barberis A, Schumacher A, Traxler P, Esbatech AG, Switz. 2005, p. 44 pp. [Google Scholar]
- [221].Laird E, Topalov G, Lyssikatos JP, Welch M, Grina J, Hansen J, Newhouse B, Array BioPharma Inc., USA, 2006, p. 103 pp. [Google Scholar]
- [222].Meng W, Hamann LG, Brigance RP, Bristol-Myers Squibb Company, USA, 2006, p. 180 pp. [Google Scholar]
- [223].Mederski W, Beier N, Cezanne B, Gericke R, Klein M, Tsaklakidis C, Merck Patent GmbH, Germany, 2007, p. 113pp. [Google Scholar]
- [224].Berdini V, Besong GE, Callaghan O, Carr MG, Congreve MS, Gill AL, Griffiths-Jones CM, Madin A, Murray CW, Nijjar RK, O’ Brien MA, Pike A, Saxty G, Taylor RD, Vickerstaffe E, Astex Therapeutics Limited, UK, 2008, p. 404pp. [Google Scholar]
- [225].Klein M, Gericke R, Beier N, Cezanne B, Tsaklakidis C, Mederski W, Merck Patent GmbH, Germany, 2008, p. 41pp. [Google Scholar]
- [226].Lee KC, Sun ET, Wang H, S bio Pte. Ltd., Singapore, 2008, pp. 129 pp., Cont.-in-part of Appl. No. PCT/SG2006/000064. [Google Scholar]
- [227].Marmsaeter FP, Munson MC, Rizzi JP, Robinson JE, Schlachter ST, Topalov GT, Zhao Q, Lyssikatos JP, Array BioPharma Inc., USA, 2008, p. 107 pp. [Google Scholar]
- [228].Muci A, Finer JT, Morgan BP, Russell AJ, Morgans DJ Jr., Cytokinetics, Incorporated, USA, 2008, p. 75 pp. [Google Scholar]
- [229].Olsson R, Burstein E, Knapp AE, Eskildsen J, Castillo J, ACADIA Pharmaceuticals Inc., USA, 2008, p. 95 pp. [Google Scholar]
- [230].Reddy PAP, Siddiqui AM, Tadikonda PK, Mansoor UF, Shipps GW Jr., Belanger DB, Zhao L, Schering Corporation, USA, 2008, p. 362pp. [Google Scholar]
- [231].Van Niel MB, Miah A, Merck Sharp & Dohme Ltd., UK, 2008, p. 28pp. [Google Scholar]
- [232].Bando K, Taguchi K, Fuji Film RI Pharma Co., Ltd., Japan; Daiichi Sankyo Co., Ltd., 2009, p. 70 pp. [Google Scholar]
- [233].Barreiro E. J. d. L., Fraga CAM, Miranda ALP, Freitas CK, Louback L, Lacerda RB, Universidade Federal do Rio de Janeiro - UFRJ, Brazil, 2009, p. 95pp. [Google Scholar]
- [234].Fink BE, Chen L, Chen P, Dodd DS, Gavai AV, Kim S-H, Vaccaro W, Zhang LH, Zhao Y, Bristol-Myers Squibb Company, USA, 2009, p. 433pp. [Google Scholar]
- [235].Klein M, Mederski W, Tsaklakidis C, Beier N, Merck Patent GmbH, Germany, 2009, p. 75pp.; Chemical Indexing Equivalent to 150:472736 (DE). [Google Scholar]
- [236].Capraro H-G, Oncalis AG, Switz., 2010, p. 81pp. [Google Scholar]
- [237].Farkas ED, Davids H, Langley C, De Koning CB, University of the Witwatersrand, S. Afr., 2010, p. 36pp. [Google Scholar]
- [238].Gijsen HJM, Velter AI, MacDonald GJ, Bischoff FP, Wu T, Van Brandt SFA, Surkyn M, Zaja M, Pieters SMA, Berthelot DJ-C, De Cleyn MAJ, Oehlrich D, Ortho-Mcneil-Janssen Pharmaceuticals, Inc, USA, 2010, p. 164pp. [Google Scholar]
- [239].Shen J, Meng T, Zhang Z, Miao Z, Ding J, Ma L, Hu D, Zhang Y, Jin Y, Wang X, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Peop. Rep. China, 2010, p. 30pp. [Google Scholar]
- [240].Han CG, Yoon JH, Kim ND, Park GJ, Kim HM, Sung BR, Shin UJ, Lee HM, Noh GT, Nam GY, Kim SH, EQUISnZAROO Co., Ltd., S. Korea, 2011, p. 22pp. [Google Scholar]
- [241].Pastor Fernandez J, Martinez Gonzalez S, Alvarez Escobar RM, Rodriguez Aristegui S, Gonzales Cantalapiedra E, Hernandez Higueras AI, Varela Busto C, Centro Nacional de Investigaciones Oncologicas CNIO, Spain, 2011, p. 150pp. [Google Scholar]
- [242].Chan T-Y, Vaccaro HM, Schering Corporation, USA; Merck Sharp & Dohme Corp., 2012, p. 82 pp. [Google Scholar]
- [243].Xu Y, Brenning BG, Kultgen SG, Liu X, Saunders M, Ho K-K, Supergen, Inc., USA, 2012, pp. 106pp., Cont.-in-part of U. S., Ser. No. 785,322. [Google Scholar]
- [244].Banka AL, Botyanszki J, Burroughs EG, Catalano JG, Chern WH, Dickson HD, Gartland MJ, Hamatake R, Hofland H, Keicher JD, Moore CB, Shotwell JB, Tallant MD, Therrien J-P, You S, GlaxoSmithKline LLC, USA, 2013, p. 224pp. [Google Scholar]
- [245].Guchhait SK, Kundu CN, Banerjee UC, Baviskar A, Madaan C, Agarwal A, Preet R, Mohapatra P, National Institute of Pharmaceutical Education and Research Niper, India, 2013, p. 53pp. [Google Scholar]
- [246].Bachmann S, Erickson SD, Laine DI, Qian Y, Hoffmann-La Roche AG F, Switz.; Hoffmann-La Roche Inc., 2014, p. 34pp. [Google Scholar]
- [247].Bentley JM, Brookings DC, Brown JA, Cain TP, Chovatia PT, Foley AM, Gallimore EO, Gleave LJ, Heifetz A, Horsley HT, Hutchings MC, Jackson VE, Johnson JA, Johnstone C, Kroeplien B, Lecomte FC, Leigh D, Lowe MA, Madden J, Porter JR, Quincey JR, Reed LC, Reuberson JT, Richardson AJ, Richardson SE, Selby MD, Shaw MA, Zhu Z, UCB Pharma S. A., Belg., 2014, p. 458pp. [Google Scholar]
- [248].Desroy N, Heckmann B, Brys RCX, Joncour AM, Peixoto C, Bock XM, Housseman CG, Galapagos NV, Belg., 2014, p. 262pp. [Google Scholar]
- [249].Furuta T, Sawada T, Danjo T, Nakajima T, Uesaka N, Kyowa Hakko Kirin Co., Ltd., Japan, 2014, p. 251pp. [Google Scholar]
- [250].Velaparthi U, Darne CP, Dodd DS, Sampognaro AJ, Wittman MD, Kumaravel S, Mullick D, Reddy R C, Liu P, Bristol-Myers Squibb Company, USA, 2014, p. 379pp. [Google Scholar]
- [251].Jang SH, Noh YS, Kim DJ, Jang HG, Eum SJ, Lee JD, Heesung Material Ltd., S. Korea, 2015, p. 69pp. [Google Scholar]
- [252].Dominguez C, Wityak J, Bard J, Kiselyov A, Brown CJ, Prime ME, Johnson PD, Clark-Frew D, Schaertl S, Herrmann F, Grimm SK, Kahmann JD, Scheich C, CHDI Foundation, Inc., USA, 2016, p. 105pp. [Google Scholar]
- [253].Marcin LR, Wrobleski ST, Dhar TGM, Bristol-Myers Squibb Company, USA, 2016, p. 93pp. [Google Scholar]
- [254].Mevellec LA, Meerpoel L, Coupa S, Poncelet VS, Pilatte INC, Pasquier ETJ, Berthelot DJ-C, Querolle OAG, Meyer C, Angibaud PR, Demestre CGM, Mercey GJM, Janssen Pharmaceutica NV, Belg., 2016, p. 302pp. [Google Scholar]
- [255].Mevellec LA, Meerpoel L, Coupa S, Poncelet VS, Pilatte INC, Pasquier ETJ, Berthelot DJ-C, Querolle OAG, Meyer C, Angibaud PR, Demestre CGM, Mercey GJM, Janssen Pharmaceutica NV, Belg., 2016, p. 203pp. [Google Scholar]
- [256].Pourashraf M, Beaulieu M-A, Claridge S, Bayrakdarian M, Johnstone S, Albert JS, Griffin A, NEOMED Institute, Can: 2017, p. 135pp. [Google Scholar]
- [257].De Koning CB, Harmse L, Dam J, University of the Witwatersrand, Johannesburg, S. Afr., 2017, p. 46pp. [Google Scholar]
- [258].Steidl U, Eckel AM, Stanley RF, Rogovoy B, Okun I, Albert Einstein College of Medicine, Inc., USA, 2017, p. 81pp. [Google Scholar]
- [259].Bradner JE, Qi J, Federation A, Jacobson Z, Varca A, Dana-Farber Cancer Institute, Inc., USA, 2018, p. 125pp. [Google Scholar]
- [260].Qi J, Pei C-K, Dana-Farber Cancer Institute, Inc., USA, 2018, p. 128pp. [Google Scholar]
- [261].Koes DR, Domling A, Camacho CJ, Protein Sci. 2018, 27, 229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Surmiak E, Neochoritis CG, Musielak B, Twarda-Clapa A, Kurpiewska K, Dubin G, Camacho C, Holak TA, Domling A, Eur. J. Med. Chem 2017, 126, 384–407. [DOI] [PubMed] [Google Scholar]
- [263].Shaabani S, Neochoritis CG, Twarda-Clapa A, Musielak B, Holak TA, Domling A, Med. Chem. Commun 2017, 8, 1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Kroon E, Schulze JO, Suss E, Camacho CJ, Biondi RM, Domling A, Angew. Chem. Int. Ed 2015, 54, 13933–13936; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem 2015, 127, 14139. [Google Scholar]