Abstract
To advance our understanding of speech motor control, it is essential to image and assess dynamic functional patterns of internal structures caused by the complex muscle anatomy inside the human tongue. Speech pathologists are investigating into new tools that help assessment of internal tongue muscle’s cooperative mechanics on top of their anatomical differences. Previous studies using dynamic magnetic resonance imaging (MRI) of the tongue revealed that tongue muscles tend to function in different groups during speech, especially the floor-of-the-mouth (FOM) muscles. In this work, we developed a method that analyzed the unique functional pattern of the FOM muscles in speech. First, four-dimensional motion fields of the whole tongue were computed using tagged MRI. Meanwhile, a statistical atlas of the tongue was constructed to form a common space for subject comparison, while a manually delineated mask of internal tongue muscles was used to separate individual muscle’s motion. Then we computed four-dimensional motion correlation between each muscle and the FOM muscle group. Finally, dynamic correlation of different muscle groups was compared and evaluated. We used data from a study group of nineteen subjects including both healthy controls and oral cancer patients. Results revealed that most internal tongue muscles coordinated in a similar pattern in speech while the FOM muscles followed a unique pattern that helped supporting the tongue body and pivoting its rotation. The proposed method can help provide further interpretation of clinical observations and speech motor control from an imaging point of view.
Keywords: Tongue function, speech, floor-of-the-mouth muscles, atlas, motion, MRI, tagged, correlation
1. INTRODUCTION
The complex cooperative pattern between various internal tongue muscles during speech is a challenging property to track, identify, and analyze[1]. Quantifying such muscle interactions has been a major topic as a part of speech motor control study[2]. The tongue is a unique organ to analyze because of its meticulously constructed internal muscular structures yielding a wide range of motion capabilities, where interdigitated muscles are able to perform fast and accurate deformations which form specific vocal tract shapes required in speech[3]. We aim to analyze such complex patterns of muscle coordination using both healthy controls and post-glossectomy patients in a common space to reveal regular and group-specific muscle motion patterns required for unique speech sound production processes. We hope such analyses could help understand the tongue muscle anatomy, each muscle’s unique function, and their functional relationships that are responsible for inducing different human oromotor behaviors (e.g., speech, respiration, swallowing, etc.).
With the rapid development of modern imaging techniques, visual analytic methods have been playing an increasingly important role in speech-related research[4,5]. Tagged magnetic resonance imaging (MRI) is a rapidly emerging tool to capture the tongue’s internal motion[6]. MRI slices from different directions are interpolated and combined in post-processing using motion extraction algorithms to produce a spatiotemporal four-dimensional (4D) motion field reflecting the deformation of tissue points in the tongue over time[7]. Motion quantities such as strain tensor and muscle deformation reflecting global or local tissue activities can be calculated from these motion fields.
However, the two-dimensional (2D) MRI slices from each scanning session only cover information of an individual subject under a specifically tuned parameter setting. Three-dimensional (3D) motion can only be initially estimated in each individual subject space without geometric alignment between all subjects in the study group. And all subjects vary in their tongue shapes and scanning positions. As a result, a major difficulty for statistical analysis of a population is to achieve multisubject spatial alignment, which serves as a basis for any following quantitative evaluation. Therefore, a statistical vocal tract atlas derived from the high-resolution (high-res) MRIs of the same study group was previously proposed[8]. An image atlas is a statistical collection of image data that represents the common anatomy and unique subject properties within a study group. This existing atlas has provided a normalized space where subjects in various locations and geometries can be deformed into[9,10]. Moreover, a pre-defined atlas muscle mask can be obtained in this normalized space from manual delineation of speech-language experts. After all internal muscle locations are specified, muscle-specific motion quantities can be calculated from this point. Previous research pointed out that the so-called floor-of-the-mouth (FOM) muscles (mylohyoid, geniohyoid, and digastric muscle) tend to function as a separate group from the other internal muscles[11]. However, the study was carried out to find corporative patterns between pairs of individual muscles without a common pattern to serve as reference, thus any individual muscle group’s function remains unclear.
In this work, we report a pipeline of methods that is capable of revealing and comparing different functional groups of all internal tongue muscles including the FOM muscle group. The genioglossus muscle was used as a major representation of the tongue’s motion pattern, and correlation of each muscle’s motion was computed against genioglossus over time. Henceforth, the FOM muscle functional group was determined and separated from other tongue muscles using the value of dynamic correlation series. Using the dynamic tongue atlas that aligned different subjects in the same space, we performed a correlation analysis among various internal tongue muscles under a specific motion setting. The transformation between each subject space and the atlas space was established on top of the subject-atlas deformation fields computed during atlas construction. With manually labeled high-res MRI muscle masks, muscle motion correlations were computed between each pair of muscles. Results of nineteen subjects including sixteen control subjects and three patients were computed in order to highlight the FOM muscle group’s specific function from the rest of the internal muscles. The proposed pipeline of methods demonstrates a quantitative muscle function analysis by simply using the tools of dynamic and traditional high-res speech MRI.
2. METHODS
2.1. Data acquisition and motion estimation
Sixteen healthy control subjects and three post-glossectomy oral cancer patients participated in the study. During data acquisition, all participants performed a speech task of pronouncing “a souk” (a designed utterance that contains desired motion patterns of this particular study’s interest) in multiple speech repetitions while MRI slices were collected. The acquired frame rate was 26 frames per second. The utterance was designed in such a way that the tongue should start from a neutral position /ә/, go for a forward motion at /s/, and get to an upward position at /k/ after going through an intermediate position of /ʊ/. We process all acquired tagged MRI with the so-called phase vector incompressible registration algorithm[7]. The algorithm estimates an incompressible dense 4D motion field at each of the 26 time frames, which we denote as us,t(X) for subject label s at time frame t (Fig. 1). The corresponding deformation that each motion field represents is denoted as ϕ, where ϕs,t(X) = X + us,t(X). Here we use capitalized X to represent all grid points in the first time frame because the motion is described in a Lagrangian scenario, meaning that all vectors in the motion field root at the grid positions of the static time frame where there is no deformation.
Figure 1.
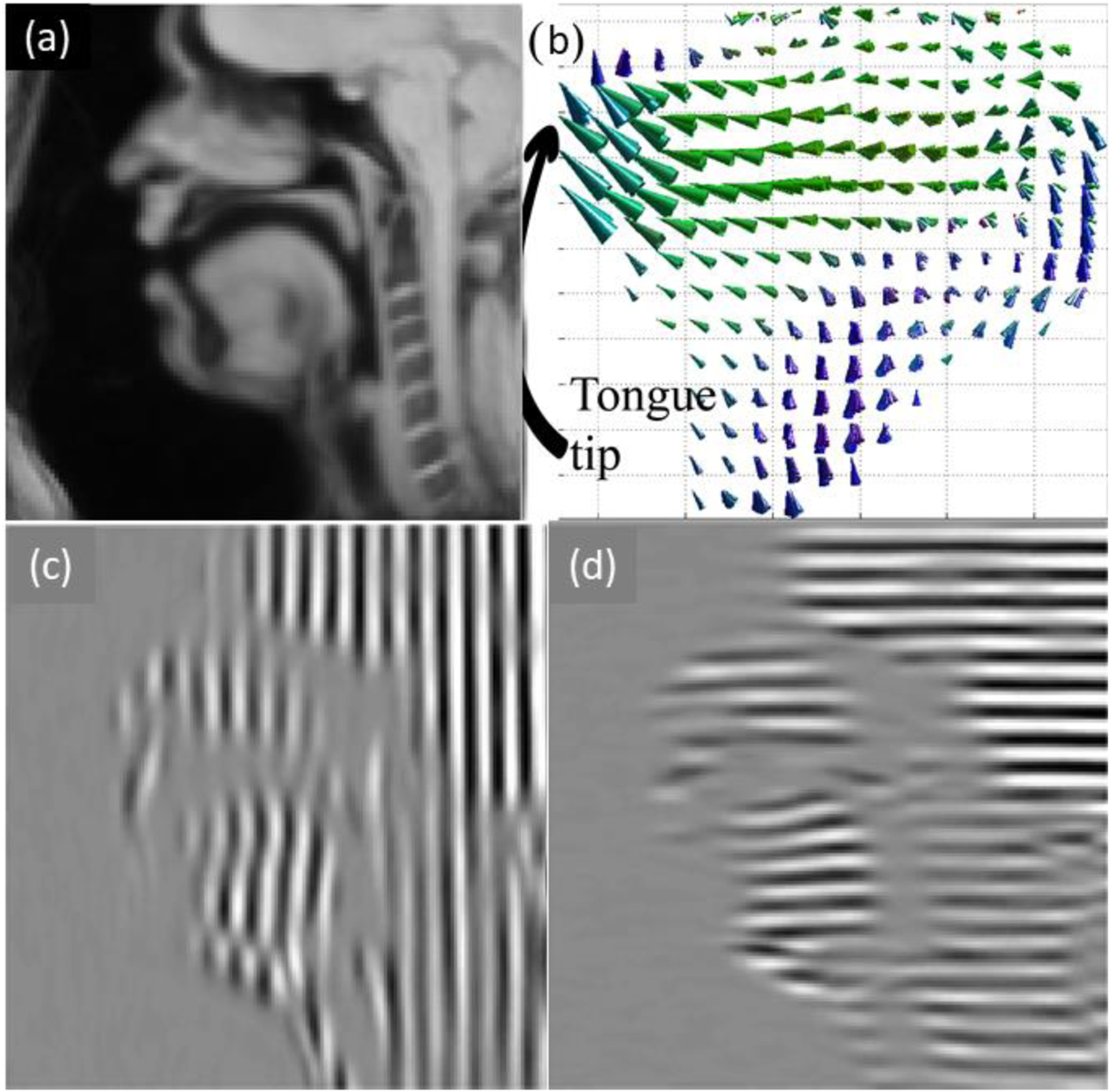
(a) High-resolution MRI of sagittal tongue. (b) Estimated 4D tongue motion in sagittal view. (c)(d) Tagged MRI in two directions.
2.2. Atlas construction and space transfer
For each subject, we utilize its high-res MRI data collected simultaneously with the tagged data for anatomy analysis. Thus, a statistical vocal tract atlas of the same study group was previously constructed using the method pipeline described in [8]. Note that only normal control subjects were used for atlas construction. We adopt this atlas architecture as the common space for all following processes. All 4D motions are then re-allocated in this common space for the following numerical computations and comparisons[8]. When creating the atlas, each subject was eventually deformed into the common space by optimally finding an appropriated spatial alignment. The process was embedded upon an image registration scheme that was symmetric and diffeomorphic[12]. We denote the deformation fields warping each subject to the atlas space by ψs (1 ≤ s ≤ N), where N is the total number of control subjects. Note that these exact deformation fields establish a relationship between each subject and the atlas so that they can be used to deform each subject’s 4D motion field to the atlas space as well. The said motion field deformation is achieved by a mathematical composition of a series of related fields, i.e.,
| (1) |
Similar to our previous notation, we write the new deformation as ϕ′s,t(X) = X + u′s,t(X), and the warped motion fields computed as if they were in the atlas space u′s,t(X) are averaged to represent the entire control group’s 4D motion statistics:
| (2) |
2.3. Mask delineation and muscle motion extraction
A speech expert carried out the delineation task of internal tongue muscles in the atlas space, where each muscle’s label was created on every 2D slice. A 3D rendering can be found after manual labeling of all such 2D slices. This yields a 3D mask for each internal tongue muscle and also physically identifies their locations in space. If we denote muscle label as L, each 3D internal muscle mask can be denoted as ML(X) which is a binary map that separates voxels inside and outside of the mask (values of 1 and 0, respectively). Finally, for muscle L, its unique muscle motion pattern at time frame t is
| (3) |
Note that each muscle’s such motion now exists in the atlas space so that all following numerical analysis is enabled. For example, for each muscle label L and each time frame t, correlation can be computed between these resulting vector fields , where a value towards 1 indicates close muscle cooperation and a value towards 0 indicates little cooperation between muscles.
Since the tongue has a rather complex muscular structure and each muscle behaves a bit differently, we select the genioglossus muscle as the common reference for all other muscles to compare against. Genioglossus is a fan-shaped muscle that occupied a large part of the tongue’s volume. It exists in the tongue center and expands vertically to cover most tongue region from the tongue bottom to the tongue tip. The motion of genioglossus always represents a summarized trend of the whole tongue since its moving direction is the same as the global tongue. If we label the genioglossus as L = 1, the correlation of all other muscles is compared against it with the value of corr.
2.4. Patient data processing
For the three patients, the same pipeline is applied for each individual, expect that it is not appropriate to average all patients’ motion due to everyone’s unique motion pattern. If we label the patients with p (1 ≤ p ≤ M) where M = 3 is the total number of patients, after using Eqn. (1), we have u′p,t(X) for each patient in the atlas space. Eqn. (2) is directly skipped and the muscle motion pattern at time frame t for patient p is
| (4) |
Each patient’s correlation pattern between muscles is also computed independently using the extracted fields u′L,p,t(X).
3. RESULTS
The final 4D atlas was constructed after combining individual spaces of sixteen healthy controls. For this process the high-res MRI was used. Fig. 2 shows the manually delineated muscle masks in this combined atlas space. Note that although all muscle masks are shown in a sagittal view, this is in fact a 3D mesh with depth. And although the muscle masks seem stacked together, they minimally intersect each other and exist in their unique locations.
Figure 2.
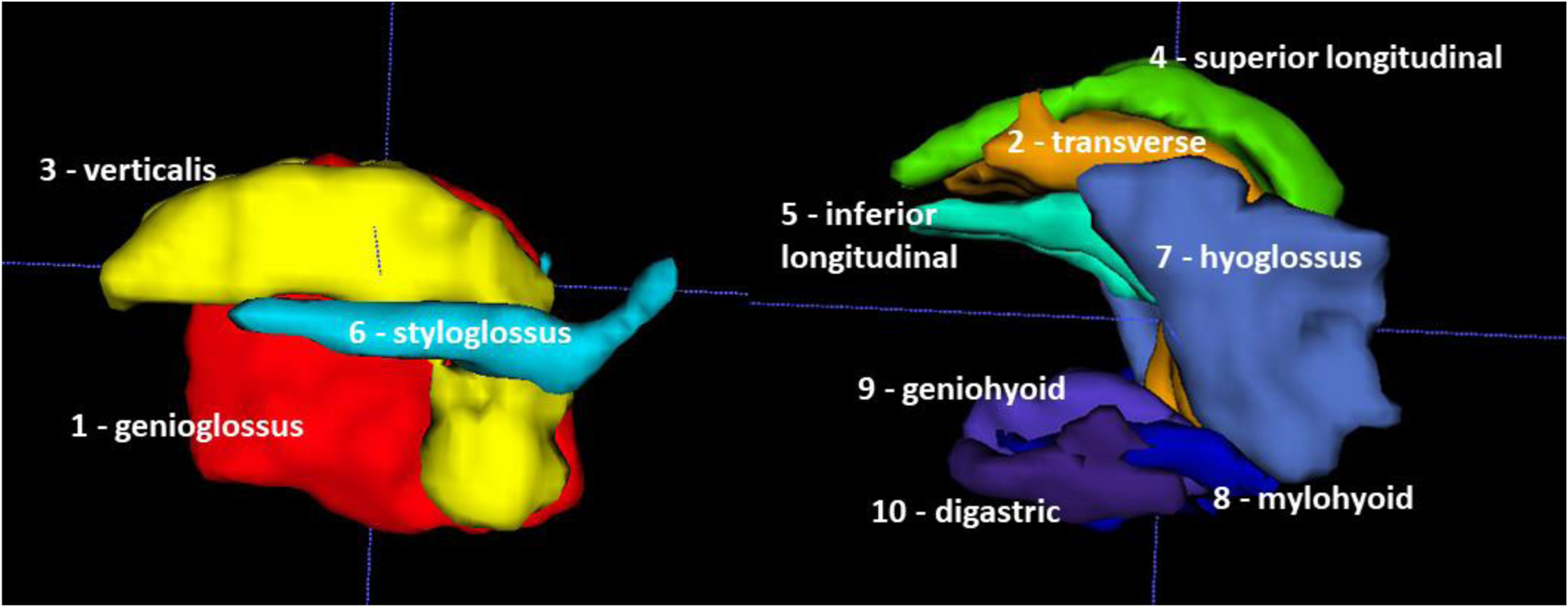
Internal tongue muscle masks in the sagittal view of ten muscles manually labeled from 3D high-resolution MRI.
The atlas space was lined up with the motion fields through image registration. Eventually, motion in individual muscle locations was obtained from Eqn. (3). Note that the curve colors in these plots match the color of the muscles in Fig. 2 for easier visual identification. Figs. 3(a)–3(d) show the correlation score of all internal tongue muscles from the sixteen healthy controls, patient 1, patient 2, and patient 3 at all 26 time frames pronouncing “a souk”. Since we used genioglossus as the anchor point for the whole tongue’s motion field, in the controls shown in Fig. 3(a), the FOM (mylohyoid, geniohyoid, and digastric) muscles functioned as a separate group most of the time, especially when pronouncing /ә/. Quantitatively, the median correlation of the FOM muscles to the genioglossus and the whole tongue was 0.71, while the other internal tongue muscles yielded a median correlation of 0.91 to the genioglossus. Note that we used median instead of mean because the patient data has a few outlier time frames.
Figure 3.
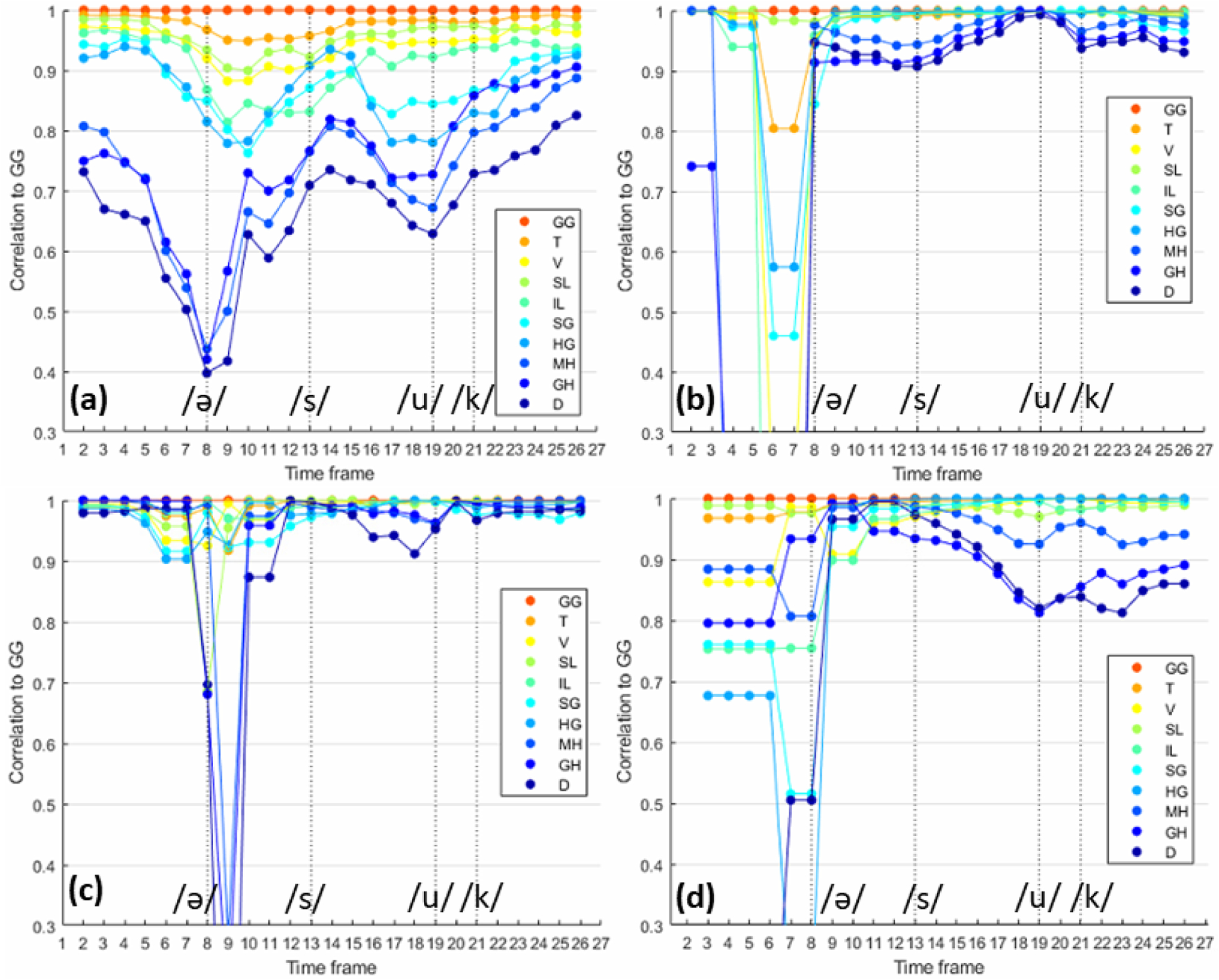
Correlation pattern of ten tongue muscles pronouncing “a souk” over time. (a) Correlation of all control subjects. (b)(c)(d) Correlation of three post-glossectomy patients.
Figs. 3(b)–3(d) show the corresponding results of the three patients. For patient 1, the median correlation of the FOM muscles to the genioglossus was 0.94, while the other internal tongue muscles yielded a median correlation of 0.99 to the genioglossus. For patient 2, the median correlation of the FOM muscles to the genioglossus was 0.96, and the other internal tongue muscles to genioglossus was 0.99. For patient 3, the median correlation of the FOM muscles to the genioglossus was 0.90, and the other internal tongue muscles to genioglossus was 0.98.
4. DISCUSSION
Comparing to the results of normal controls, we observed that the controls’ behavior was similar among each individual while the patients’ behavior varied greatly. 1) The patient’s FOM muscles also seemed to function separately from the rest of the muscles at the beginning of pronouncing “a souk”, but they quickly turned back and co-worked with the other muscles for the rest of the utterance. 2) In the patients, most of their internal muscles functioned as only one unique group. Their correlation score was much higher than that of the control. 3) When the FOM of the patients did cooperate with the remaining muscles, their correlation was usually higher than the controls, indicating a very small functional difference. All these observations are likely due to the compensation strategies used by the patients to account for their post-glossectomy tongue function loss: when a certain pronunciation requires less amount of tongue effort, the FOM muscles tend to stay static letting other parts of the tongue do the deformation; for a more difficult pronunciation requiring more muscle effort, the FOM muscles tend to work together with the genioglossus and almost become a same group with the remaining tongue muscles to strengthen the tongue’s deformation and compensate what was lost after glossectomy surgery.
From a speech pathology point of view, the FOM muscles are usually associated with swallowing as they will pull the hyoid bone up and forward, protecting the airway during the swallow when the jaw is held steady. They also lower the jaw when the hyoid is held steady during speech such as the schwa and also non-speech jaw opening. The FOM are bundled muscles unlike the other muscles in the tongue which are interdigitated, making them easier to see on MRI. They represent two fiber directions: mylohyoid is left-right and the other two are anterior-posterior. Mylohyoid can elevate the tongue as a unit without substantial deformation, which helps ALS and other disorders with reduced motor control. Most tongue muscles are innervated from the XII cranial nerve, but the FOM muscles are innervated by other nerves, which may be less affected in muscle or nerve degenerative diseases.
The proposed pipeline of method has a few strengths. It combines both information from high-res MRI that excels at a detailed anatomical analysis and the information from dynamic MRI that is good at functional analysis. The two parts of information is bridged together by the means of a statistically atlas, making all following quantitative computations possible. Moreover, the selection of the genioglossus muscle as a general representation of the whole tongue’s motion is important to bypass the lack of common standard for comparison. And this operation is only made possible by a detailed manual delineation of the internal tongue muscle masks.
On the other hand, the entire pipeline may lack robustness due to its complex procedure. The computation of deformation fields and motion fields, atlas quality, and manual labeling quality could all affect observations of the final result. Additionally, the conclusion may vary due to the relatively small sample size. Future work should be expanded upon further collection of data in a larger statistical pool.
5. CONCLUSION
In this work, we reported a method pipeline and its analysis result that revealed various functional groups of all internal tongue muscles and especially on the floor-of-the-mouth muscles during speech. The floor-of-the-mouth muscles tended to function as a separate group while most other internal muscles followed genioglossus. Most of patients’ internal muscles functioned as one unique group as a compensation strategy. Assessment of muscle cooperative patterns and general mechanics helps interpretation of clinical observations, providing information for further understanding of speech motor control.
Acknowledgements
This work was supported by NIH R01DC014717, R01DC018511, R21DC016047, R00DC012575, R01CA133015.
REFERENCES
- [1].Sanders I, & Mu L, “A three-dimensional atlas of human tongue muscles,” The Anatomical Record, 296(7), 1102–1114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Takemoto H, “Morphological analyses of the human tongue musculature for three-dimensional modeling,” Journal of Speech, Language, and Hearing Research (2001). [DOI] [PubMed] [Google Scholar]
- [3].Stone M, Woo J, Lee J, Poole T, Seagraves A, Chung M, … & Blemker SS, “Structure and variability in human tongue muscle anatomy,” Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, 6(5), 499–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dawson KM, Tiede MK, & Whalen DH, “Methods for quantifying tongue shape and complexity using ultrasound imaging,” Clinical linguistics & phonetics, 30(3–5), 328–344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Stone M, Davis EP, Douglas AS, NessAiver M, Gullapalli R, Levine WS, & Lundberg A, “Modeling the motion of the internal tongue from tagged cine-MRI images,” The Journal of the Acoustical Society of America, 109(6), 2974–2982 (2001). [DOI] [PubMed] [Google Scholar]
- [6].Parthasarathy V, Prince JL, Stone M, Murano EZ, & NessAiver M, “Measuring tongue motion from tagged cine-MRI using harmonic phase (HARP) processing,” The Journal of the Acoustical Society of America, 121(1), 491–504 (2007). [DOI] [PubMed] [Google Scholar]
- [7].Xing F, Woo J, Gomez AD, Pham DL, Bayly PV, Stone M, & Prince JL, “Phase vector incompressible registration algorithm (PVIRA) for motion estimation from tagged magnetic resonance images,” IEEE Transactions on Medical Imaging (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Woo J, Lee J, Murano EZ, Xing F, Al-Talib M, Stone M, & Prince JL, “A high-resolution atlas and statistical model of the vocal tract from structural MRI,” Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, 3(1), 47–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Perperidis D, Mohiaddin R, & Rueckert D, “Construction of a 4D statistical atlas of the cardiac anatomy and its use in classification,” In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 402–410). Springer, Berlin, Heidelberg: (2005). [DOI] [PubMed] [Google Scholar]
- [10].Martins SB, Spina TV, Yasuda C, & Falcão AX, “A multi-object statistical atlas adaptive for deformable registration errors in anomalous medical image segmentation,” In Medical Imaging 2017: Image Processing (Vol. 10133, p. 101332G). International Society for Optics and Photonics; (2017). [Google Scholar]
- [11].Xing F, Stone M, Goldsmith T, Prince JL, El Fakhri G, & Woo J, “Atlas-Based Tongue Muscle Correlation Analysis From Tagged and High-Resolution Magnetic Resonance Imaging,” Journal of Speech, Language, and Hearing Research, 62(7), 2258–2269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Avants BB, Tustison NJ, Song G, Cook PA, Klein A and Gee JC, “A reproducible evaluation of ANTs similarity metric performance in brain image registration,” Neuroimage, 54(3), pp.2033–2044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]


