Abstract
Introduction
Laboratories worldwide are facing high demand for molecular testing during the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, which might be further aggravated by the upcoming influenza season in the northern hemisphere.
Gap Statement
Given that the symptoms of influenza are largely indistinguishable from those of coronavirus disease 2019 (COVID-19), both SARS-CoV-2 and the influenza viruses require concurrent testing by RT-PCR in patients presenting with symptoms of respiratory tract infection.
Aim
We adapted and evaluated a laboratory-developed multiplex RT-PCR assay for simultaneous detection of SARS-CoV-2 (dual target), influenza A and influenza B (SC2/InflA/InflB-UCT) on a fully automated high-throughput system (cobas6800).
Methodology
Analytical performance was assessed by serial dilution of quantified reference material and cell culture stocks in transport medium, including pretreatment for chemical inactivation. For clinical evaluation, residual portions of 164 predetermined patient samples containing SARS-CoV-2 (n=52), influenza A (n=43) or influenza B (n=19), as well as a set of negative samples, were subjected to the novel multiplex assay.
Results
The assay demonstrated comparable analytical performance to currently available commercial tests, with limits of detection of 94.9 cp ml−1 for SARS-CoV-2, 14.6 cp ml−1 for influenza A and 422.3 cp ml−1 for influenza B. Clinical evaluation showed excellent agreement with the comparator assays (sensitivity of 98.1, 97.7 and 100 % for Sars-CoV-2 and influenza A and B, respectively).
Conclusion
The SC2/InflA/InflB-UCT allows for efficient high-throughput testing for all three pathogens and thus provides streamlined diagnostics while conserving resources during the influenza season.
Keywords: cobas6800, influenza, molecular diagnostics, multiplex, PCR, SARS-CoV-2
Introduction
The upcoming influenza season of 2020/21 will further aggravate the strain on diagnostic laboratories that are already facing unprecedented demand for molecular diagnostics due to the ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. Just like coronavirus disease 2019 (COVID-19), influenza is a major concern for infection control within healthcare facilities, while their symptoms are largely indistinguishable, particularly in the early phase of disease [1, 2]. Consequently, SARS-CoV-2 and the influenza viruses need to be concurrently tested for by RT-PCR before contact precaution measures can be lifted for symptomatic patients. In light of the continuing worldwide shortage of supplies for nucleic acid extraction and PCR diagnostics, it appears desirable to be able to screen for all three viruses (SARS-CoV-2, influenza A and influenza B) within the same reaction.
The cobas6800 system is a fully automated sample-to-result high-throughput platform, requiring minimal hands-on-time, and is able to perform up to 384-tests in an 8 h shift. The instrument was previously evaluated for the detection of influenza viruses in respiratory swabs [3] and is currently seeing increasing use for automated SARS-CoV-2 diagnostics [4, 5].
The aim of this study was to establish and evaluate a multiplex assay for the detection of SARS-CoV-2, influenza A and influenza B on the open mode of the cobas6800 system (cobas omni utility channel).
Methods
SC2/InflA/InflB-UCT setup and preparation
A set of published RT-PCR assays for SARS-CoV-2, influenza A and influenza B virus was selected and adapted for use on the cobas6800 system (Table 1) [6–10]. Primers were modified with 2′O-methylated RNA bases at their penultimate positions to reduce the formation of primer dimers. Double-quenched probes were used to lower background fluorescence. All primers and probes were tested for contamination prior to use, in particular concerning material reactive for SARS-CoV-2, in custom-made commercial primers.
Table 1.
Assays used for the SC2/InflA/InflB-UCT. Primers and probes were custom-made and procured from Integrated DNA Technologies (IL, USA), Biomers.net GmbH (Ulm, Germany) and Ella Biotech GmbH (Martinsried, Germany). OMe (2’O-methyl RNA), NFQ, (non-fluorescent quencher), YakYel (Yakima yellow primers) and probes were diluted in MMX-R2 reagent to final concentrations as indicated (Table 1) to form the MMR2 Master Mix
|
Target |
Primer/probe |
Sequence (5′ - 3′) |
Conc. [nM] |
Inclusivity |
Ref. |
|---|---|---|---|---|---|
|
SARS-CoV-2 E-gene |
Fwd: Rev: Probe: |
ACAGGTACGTTAATAGTTAATAGC(OMe-G)T ATATTGCAGCAGTACGCACA(OMe-C)A Fam- ACACTAGCC(ZEN-NFQ)ATCCTTACTGCGCTTCG -IowaBlack |
400 400 50 |
Sarbecovirus (SARS-CoV, SARS-CoV-2) |
[6] |
|
SARS-CoV-2 RdRp/Hel |
Fwd: Rev: Probe: |
CGCATACAGTCTTACAGG(OMe-C)T TGTGATGTTGATATGATATGG(OMe-U)C Fam- TTAAGATGT(ZEN-NFQ)GGTGCTTGCATACGTAGAC -IowaBlack |
300 300 50 |
SARS-CoV-2 |
[7] |
|
Influenza A M |
Fwd: Rev: Probe: |
CTTCTAACCGAGGTCGAAACG(OMe-U)A GGTGACAGGATTGGTCTTGTCTT(OMe-U)A YakYel- TCAGGCCCC(ZEN-NFQ)CTCAAAGCCGAG -IowaBlack |
300 300 50 |
Pan-influenza A (incl. avian H5 and H7) |
[8, 9] |
|
Influenza B NS2 |
Fwd: Rev: Probe: |
TCCTCAAYTCACTCTTCGAG(OMe-C)G CGGTGCTCTTGACCAAATT(OMe-G)G Atto620- CCAATTCGA(BMN-Q620)GCAGCTGAAACTGCGGTG -BMN-Q620 |
300 300 50 |
Pan-influenza B |
[10] |
The cobas 6800/8800 internal control (IC) is a spike-in (packaged) RNA target, which is automatically added during extraction by the system. MMRX-R2 reagent already contains the internal control assay by default; the respective sequences are not disclosed by the manufacturer. The IC acts as a full process control in the same way as in commercial cobas 6800/8800 IVD tests manufactured by Roche.
Six millilitres of MMR2 Master Mix was loaded into cobas omni utility channel cassettes according to instructions by the manufacturer. The run profile for the SC2/InflA/InflB-UCT multiplex assay was configured using the cobas omni utility channel software, as indicated in Table 2.
Table 2.
Run-profile for the SC2/InflA/InflB, set up using the cobas omni utility channel tool
|
Software settings | |||||
|---|---|---|---|---|---|
|
Sample type |
Alcohol-based sample (400 µl) |
||||
|
Channels |
1: (Not assigned) |
2: SC2 |
3: InflA |
4: InflB |
5: IC |
|
RFI |
|
1.25 |
1.25 |
1.25 |
1.15 |
|
PCR cycling conditions | |||||
|
|
UNG incubation |
Pre-PCR step |
First measurement |
Second measurement |
Cooling |
|
No. of cycles |
Predefined |
1 |
5 |
45 |
Predefined |
|
No. of steps |
3 |
2 |
2 |
||
|
Temperature |
55; 60; 65 °C |
95; 55 °C |
91; 58 °C |
||
|
Hold time |
120; 360; 240 s |
5; 30 s |
5; 25 s |
||
|
Data acquisition |
None |
End of each cycle |
End of each cycle |
||
Limit of detection (LoD), linearity and cross-reactivity
Analytical limit of detection (LoD) was determined for all three targets simultaneously by serial dilution of reference material in Amies medium including cobas PCR media (1 : 1) as matrix. For SARS-CoV-2, a stock of cell culture supernatant containing SARS-CoV-2 [11] was quantified using the SARS-CoV-2 IVD Test for the cobas6800 [12] with the Qnostics ‘SARS-CoV-2 Q Control 01’ as reference for quantification. For influenza A and influenza B, reference material was acquired from Qnostics [‘INFA Medium Q Control 01’ (H1N1) and ‘INFB Medium Q Control 01’ (Victoria lineage)] and used directly for LoD experiments. A total of eight different concentrations was tested with eight repeats each. [SC2 (cp ml−1)-InflA (cp/ml−1)-InflB (cp ml−1), 1000-1000-2000, 333-333-666, 100-100-200, 50-50-100, 25-25-50, 10-10-20, 3-3-6, 1-1-2].
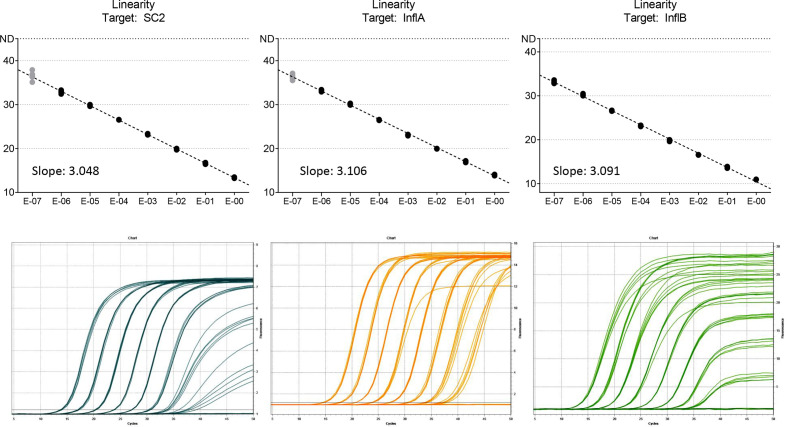
Linearity was determined for all targets simultaneously by 10-fold serial dilution, 5 repeats each step, using cell culture supernatant and Vaxigrip tetravalent influenza vaccine (Sanofi Pasteur, France) to spike Amies medium including cobas PCR media (1 : 1) (Fig. 1).
Fig. 1.
Linearity was determined for all targets simultaneously by serial dilution of SARS-CoV-2 cell culture stocks and Vaxigrip tetravalent influenza vaccine in 1:1 cobas PCR media in eSwab medium. x-axis, dilution factor. Black dots, measurements within linear range, considered for trendline. Grey dots, measurements outside linear range, not considered for trendline.
Inclusivity and cross-reactivity were verified using external quality control panels by INSTAND eV (Düsseldorf, Germany) and clinical samples containing a variety of respiratory pathogens, including endemic human coronaviruses (see Table 3).
Table 3.
Inclusivity and cross-reactivity. A panel of clinical samples containing respiratory pathogens, as well as relevant external quality control panel samples (INSTAND eV) were tested with the SC2/InflA/InflB-UCT. No false positives occurred
|
External quality control panel |
|
|
|
|
|---|---|---|---|---|
|
Species |
No. tested |
Target: SC2 |
Target: InflA |
Target: InflB |
|
Influenza A H1N1 pdm09 (A/Michigan/45/2015) |
1 |
Negative |
Positive |
Negative |
|
Influenza A H7N9 (A/Anhui/1/2013) |
1 |
Negative |
Positive |
Negative |
|
Influenza A H5N8 (A/DE-SH/Reiherente/AR8444/2016) |
1 |
Negative |
Positive |
Negative |
|
Influenza B Yamagata (B/Phuket/3073/2013) |
1 |
Negative |
Negative |
Positive |
|
Influenza B Victoria (B/Colorado/06/2017) |
1 |
Negative |
Negative |
Positive |
|
Human coronavirus 229E |
1 |
Negative |
Negative |
Negative |
|
Human coronavirus OC43 |
1 |
Negative |
Negative |
Negative |
|
MERS coronavirus |
1 |
Negative |
Negative |
Negative |
|
Parainfluenzavirus 2 |
1 |
Negative |
Negative |
Negative |
|
Clinical samples |
|
|
|
|
|
Species |
Target: SC2 |
Target: InflA |
Target: InflB |
|
|
Human coronavirus HKU1 |
2 |
Negative |
Negative |
Negative |
|
Human coronavirus NL63 |
1 |
Negative |
Negative |
Negative |
|
Human coronavirus OC43 |
1 |
Negative |
Negative |
Negative |
|
Bocavirus |
1 |
Negative |
Negative |
Negative |
|
Parainfluenzavirus 3 |
1 |
Negative |
Negative |
Negative |
|
Human metapneumovirus |
2 |
Negative |
Negative |
Negative |
|
Rhino-/enterovirus |
3 |
Negative |
Negative |
Negative |
|
Respiratory syncytial virus |
2 |
Negative |
Negative |
Negative |
|
Mycoplasma pneumoniae |
1 |
Negative |
Negative |
Negative |
|
Chlamydia pneumoniae |
1 |
Negative |
Negative |
Negative |
|
Pneumocystis jirovecii |
1 |
Negative |
Negative |
Negative |
Clinical samples
For clinical evaluation, a total of 164 archived predetermined respiratory swab samples were subjected to the SC2/InflA/InflB-UCT. Clinical samples were oropharyngeal or nasopharyngeal swabs, performed using eSwab sample collection kits (Copan, Italy) containing 1 ml Amies edium. Then 1–2 ml of cobas PCR Media (≤40 % guanidine hydrochloride in Tris/HCL buffer) was added to samples prior to analysis in routine diagnostics. Samples were stored for up to 3 months at −20 °C for SARS-CoV-2 and up to 3 years for influenza A/B.
SARS-CoV-2 samples were included if positive for target 1 and target 2 of the SARS-CoV-2 IVD assay. The Cepheid Xpert Xpress Flu/RSV assay was used to resolve discrepant results for influenza A/B virus. Samples predetermined as positive (by in-house methods) for influenza A or influenza B that tested negative in the SC2/InflA/InflB-UCT were only included in the study if they tested positive for both influenza A targets or positive for influenza B in the Xpert Xpress Flu/RSV assay. (For more details about the in-house influenza A/B assays, see Supplementary File S1, available in the online version of this article)
Results
Analytical performance
Analytical LoD was determined by Probit-Analysis as 94.9 cp ml−1 (95 % CI: 40.5–222.0 cp ml) for SARS-CoV-2, 14.57 cp ml (95 % CI: 6.7–31.6 cp ml) for influenza A and 422.3 cp ml−1 (95 % CI: 213.8–834.4 cp ml−1) for influenza B (Table 4). This implies that analytical sensitivity is nominally slightly lower for influenza B than for influenza A and SARS-CoV-2.
Table 4.
Quantified cell culture stocks and quantified reference material (by Qnostics Ltd) was spiked into 1 : 1 cobas PCR media in eSwab medium. LoD was determined for all targets simultaneously, meaning that every sample contained the indicated concentrations of each pathogen for each dilution step
|
SC2/InflA/InflB-UCT limit of detection (LoD) | |||||
|---|---|---|---|---|---|
|
SARS-CoV-2 |
Influenza A |
Influenza B |
|||
|
Conc. (cp ml−1) |
Pos./rep.* |
Conc.(cp ml−1) |
Pos./rep.* |
Conc. (cp ml−1) |
Pos./rep.* |
|
1000 |
8/8 |
1000 |
8/8 |
2000 |
8/8 |
|
333 |
8/8 |
333 |
8/8 |
666 |
8/8 |
|
100 |
8/8 |
100 |
8/8 |
200 |
7/8 |
|
50 |
8/8 |
50 |
8/8 |
100 |
8/8 |
|
25 |
8/8 |
25 |
8/8 |
50 |
6/8 |
|
10 |
6/8 |
10 |
8/8 |
20 |
5/8 |
|
3 |
0/8 |
3 |
6/8 |
6 |
2/8 |
|
1 |
3/8 |
1 |
4/8 |
2 |
0/8 |
|
0 |
0/8 |
0 |
0/8 |
0 |
0/8 |
*Number of positives/total number of repeats.
The assay showed good linearity for all targets up to a cycle threshold (C T) value of 33 (Fig. 1).
In cross-reactivity experiments, no false positives occurred. Avian influenza A strains H7N9 and H5N8 were correctly detected by the multiplex assay, demonstrating broad coverage (Table 3). Nonetheless, it is recommended to verify inclusivity of primer/probe sequences as new influenza A strains continuously emerge.
Evaluation of clinical performance
The sensitivity in clinical samples containing the respective targets was 98.1 % for SARS-CoV-2 (52 samples, median C T: 31.99, IQR: 27.21–33.96), 97.67 % for influenza A (43 samples of which 15 were subtyped as H1N1, median C T: 27.80, IQR: 24.85–31.1) and 100 % for influenza B (19 samples, median C T: 29, IQR: 28–30) (Table 5).
Table 5.
One hundred and sixty-four clinical samples were tested in total, predefined as positive for SARS-CoV-2 via the SARS-CoV-2 IVD test for the cobas6800 system, or predetermined as positive for influenza A or B by established in-house methods or Xpert Xpress influenza/RSV. The invalid rate was 6.4 % (invalid samples not included in the table). Samples were stored at −20 °C for between 1 and 36 months
|
Predetermined clinical specimen |
|||||
|---|---|---|---|---|---|
|
SARS-CoV2 |
Influenza A |
Influenza B |
Negative* |
||
|
SC2/InflA/InflB-UCT |
SC2-positive |
51/52 |
0/43 |
0/19 |
0/109 |
|
InflA-positive |
0/52 |
42/43 |
0/19 |
0/118 |
|
|
InflB-positive |
0/52 |
0/43 |
19/19 |
0/142 |
|
|
Negative |
1/52 |
1/43 |
0/19 |
47/47 |
|
|
Total: |
52 |
43 |
19 |
369† |
|
*Total number of samples negative for respective targets.
†Sum of all predetermined negative samples for each individual pathogen. This includes 47 samples negative for all three pathogens.
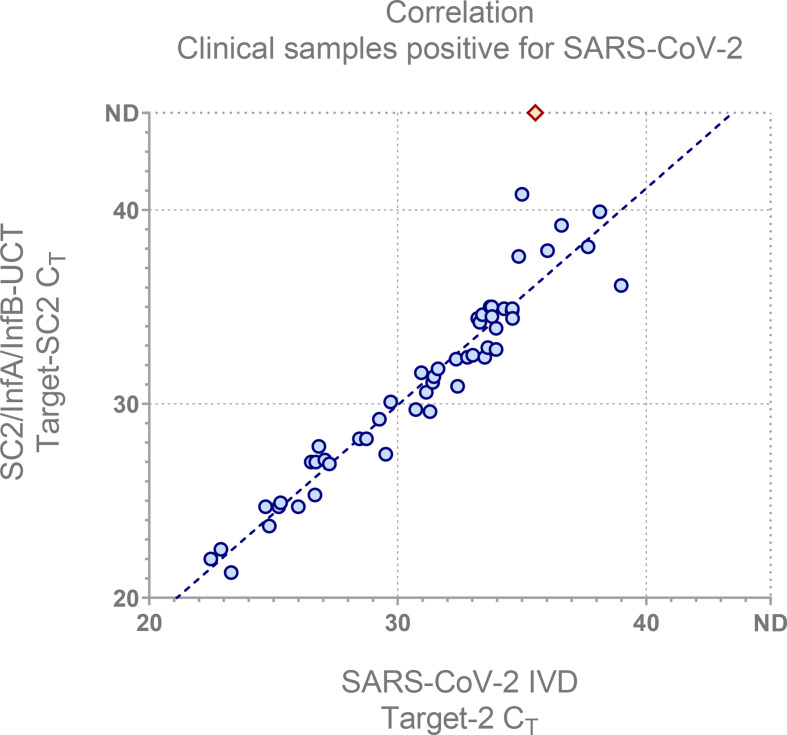
The C T values showed good correlation with the SARS-CoV-2 IVD (Fig. 2). For SARS-CoV-2 a single false negative occurred for a sample containing very low amounts of viral RNA, approximately 184 cp ml−1. Another false negative occurred for influenza A in a sample with low viral load (Xpert Flu/RSV, A1 C T: 33, A2 C T: 35).
Fig. 2.
The The SC2/InflA/InflB-UCT SC2 C T was compared to the the SARS-CoV-2 IVD target 2 C T (used for quantification and best with regard to linear range) to illustrate correlation with a well-established assay. Red diamond indicates false negative for the multiplex assay. nd, not detected.
A total of 50 samples predetermined to be negative for SARS-CoV-2 (by SARS-CoV-2 IVD) and influenza A/B (by LDT assay) were subjected to the SC2/InflA/InflB-UCT. No false positives occurred; the inhibition rate in eSwab samples was 6.4 %.
As a proof of concept, a dilution series of a SARS-CoV-2-positive patient sample was prepared and subjected to testing in the presence and absence of high levels of influenza A and influenza B RNA (Fig. S1). No relevant impact on SARS-CoV-2 detection was observed.
Discussion
In this study we present a functional SARS-CoV-2/influenza A/influenza B multiplex assay for the cobas6800 high-throughput platform, featuring comparable analytical and clinical performance to currently used methods for the detection of these pathogens in clinical specimens. The ability to screen for all three viruses in a single reaction allows for streamlined workflows and conservation of resources during the coming influenza season.
By this time, multiple commercial providers have announced, or are in the process of rolling out, multiplex assays for the detection of SARS-CoV-2 and influenza A/B. Notably, the Centers for Disease Control and Prevention (CDC) has itself recently published primer sets for a SARS-CoV-2/influenza A/influenza B multiplex assay, which relies on manual PCR workflows [10]. Some commercial panels, particularly those designed as point-of-care rapid molecular tests, will also include respiratory syncytial virus (RSV) as a target. As the SC2/InflA/InflB-UCT was mainly designed as a high-throughput screening tool for hospital admissions, RSV was not included, as it is not as important for infection control measures and is rarely requested by clinicians in this context.
Most currently available commercial SARS-CoV-2 tests with FDA emergency authorization are designed as multi-target assays to account for emerging mutations. While the original Sarbeco-E primer set by Corman et al. [6] resides in a particularly stable region of the SARS-CoV-2 genome, single mutations have been reported within the ever-growing catalogue of available whole-genome sequences [13]. The RdRp/Hel assay by Chan et al. [7] was modified and adapted as a second target to provide additional security for inclusivity in SARS-CoV-2 detection. Both assays were allocated to the same channel, as positivity for any single one would constitute a positive result. There was no indication that the presence of influenza A and/or influenza B RNA within the reaction substantially impairs sensitivity for SARS-CoV-2. If necessary, each SARS-CoV-2 assay can be analysed separately by moving RdRp/Hel detection to channel 1, using the following probe (or a comparable one): 5′ Atto425-TTAAGATGT(BMN-Q535)GGTGCTTGCATACGTAGAC-BMN-Q535 3′ (Fig. S2).
It has to be acknowledged that the SARS-CoV-2 assays used for this multiplex setup are technically not specific for SARS-CoV-2, but for the Sarbeco subgenus of betacoronaviruses, including SARS-CoV (from 2003) and SARS-like bat viruses. As of 2020, SARS-CoV-2 is the only Sarbeco subgenus member currently circulating in humans; however, the viability of a pan-Sarbecovirus target for diagnostics would have to be re-evaluated if other sarbecoviruses enter the human population in the future.
Lastly, a relatively high inhibition rate (6.4 %) was observed in clinical samples, likely in part due to the use of Amies medium-based transport medium (eSwab), which is not optimal for use on the cobas6800. This can be mitigated by either adding more guanidine hydrochloride solution (cobas PCR media, 2 : 1 or 3 : 1) or using UTM-based samples. Still, a slightly higher invalid rate can be expected when comparing laboratory-developed assays to commercial IVD tests, as low-level interference between IC and LDT assays cannot be ruled out.
In conclusion, we provide analytical and clinical evaluation of a SARS-CoV-2/influenza A/influenza B multiplex assay for the cobas6800 high-throughput platform. Performance for each target was comparable to that for existing solutions currently in use in diagnostic practice. Our novel assay may prove useful for streamlining diagnostics during the upcoming influenza season.
Supplementary Data
Funding information
The authors received no specific grant from any funding agency.
Author contributions
Conceptualization: D. N., M. A, S. P. and M. L. Methodology and investigation: D. N. Original draft preparation: D. N. and S. P. Review and editing: D. N., A. H., M. A., S. P. and M. L., Supervision: S. P. and M. L. All authors agreed to the publication of the final manuscript.
Conflicts of interest
M. L. received speaker honoraria and related travel expenses from Roche Diagnostics. All other authors declare no conflicts of interest.
Ethical statement
This work was conducted in accordance with §12 of the Hamburg hospital law (§12 HmbKHG). The use of anonymized samples was approved by the Ethics Committee, Freie und Hansestadt Hamburg, PV5626.
Footnotes
Abbreviations: CI, confidence interval; CT, cycle threshold; FDA, United States Food and Drug Administration; IVD, in-vitro diagnostic device; LDT, laboratory developed test; RT-PCR, real-time polymerase chain reaction; UCT, utility channel test; UTM, universal transport medium.
Two supplementary figures and one supplementary file are available with the online version of this article.
References
- 1.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in china: summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA. 2020;323:1239-1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Stokes EK, Zambrano LD, Anderson KN, Marder EP, Raz KM, et al. Coronavirus disease 2019 case surveillance - United States, January 22-May 30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:759–765. doi: 10.15585/mmwr.mm6924e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eigner U, Reucher S, Hefner N, Staffa-Peichl S, Kolb M, et al. Clinical evaluation of multiplex RT-PCR assays for the detection of influenza A/B and respiratory syncytial virus using a high throughput system. J Virol Methods. 2019;269:49–54. doi: 10.1016/j.jviromet.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Pfefferle S, Reucher S, Nörz D, Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25:2000152. doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poljak M, Korva M, Knap Gašper N, Fujs Komloš K, Sagadin M, et al. Clinical evaluation of the COBAS SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan JF-W, Yip CC-Y, To KK-W, Tang TH-C, Wong SC-Y, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel Real-Time reverse transcription-PCR assay validated In Vitro and with clinical specimens. J Clin Microbiol. 2020;58:e00310–00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO Who information for the molecular detection of influenza viruses. WHO Website. 2017 [Google Scholar]
- 9.Terrier O, Josset L, Textoris J, Marcel V, Cartet G, et al. Cellular transcriptional profiling in human lung epithelial cells infected by different subtypes of influenza A viruses reveals an overall down-regulation of the host p53 pathway. Virol J. 2011;8:285-285. doi: 10.1186/1743-422X-8-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CDC Research use only CDC influenza SARS-CoV-2 (flu SC2) multiplex assay real-time RT-PCR primers and probes. CDC Website. 2020 [Google Scholar]
- 11.Pfefferle S, Huang J, Nörz D, Indenbirken D, Lütgehetmann M, et al. Complete genome sequence of a SARS-CoV-2 strain isolated in northern Germany. Microbiol Resour Announc. 2020;9:e00520–20. doi: 10.1128/MRA.00520-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nörz D, Frontzek A, Eigner U, Oestereich L, Wichmann D, Fischer N, et al. Pushing beyond specifications: evaluation of linearity and clinical performance of the COBAS 6800/8800 SARS-CoV-2 RT-PCR assay for reliable quantification in blood and other materials outside recommendations. J Clin Virol. 2020;132:104650. doi: 10.1016/j.jcv.2020.104650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.