Abstract
Our previous work demonstrated that two commonly used fluorescent dyes that were accumulated by wild-type Escherichia coli MG1655 were differentially transported in single-gene knockout strains, and also that they might be used as surrogates in flow cytometric transporter assays. We summarize the desirable properties of such stains, and here survey 143 candidate dyes. We eventually triage them (on the basis of signal, accumulation levels and cost) to a palette of 39 commercially available and affordable fluorophores that are accumulated significantly by wild-type cells of the ‘Keio’ strain BW25113, as measured flow cytometrically. Cheminformatic analyses indicate both their similarities and their (much more considerable) structural differences. We describe the effects of pH and of the efflux pump inhibitor chlorpromazine on the accumulation of the dyes. Even the ‘wild-type’ MG1655 and BW25113 strains can differ significantly in their ability to take up such dyes. We illustrate the highly differential uptake of our dyes into strains with particular lesions in, or overexpressed levels of, three particular transporters or transporter components (yhjV, yihN and tolC). The relatively small collection of dyes described offers a rapid, inexpensive, convenient and informative approach to the assessment of microbial physiology and phenotyping of membrane transporter function.
Keywords: transporters, bioenergetics, E. coli, fluorescence, assays, energization
Introduction
Notwithstanding that the entire genome of strain MG1655 of Escherichia coli K12 was sequenced more than 20 years ago [1], some 35 % of its genes are still of unknown function [2]. These genes are known as y-genes [3], and transporters are pre-eminent among them [2]. An important general problem [4, 5] thus involves the related questions of (i) what are the potential substrates for a given transporter, and (ii) what are the membrane transporters for a molecule that cells take up and/or efflux? Our focus here is on finding methods to help provide answers to these questions, in particular for the (approximately) 124 y-genes of E. coli that are considered from sequence analysis to be transporters.
Fluorescence flow cytometry provides a convenient and high-throughput means for assessing the extent of uptake and accumulation of a given fluorophore (e.g. [6–18]), and in recent work [19, 20] we have found this to be true in E. coli in vivo. In particular, the availability of the Keio collection of single-gene knockouts in ‘non-essential’ genes [21, 22] allowed us to assess the contribution of many of these genes to the uptake of given fluorophores. The chief findings were that the carbocyanine dyes diSC3(5) and SYBR Green I could be taken up and/or effluxed differentially relative to the reference strain by a very great number of strains harbouring single-gene knockouts, consistent with the views that (i) any electron transport-mediated membrane potential was not largely responsible for their steady-state uptake [23–25], and (ii) any non-transporter-mediated transport through the phospholipid bilayer was negligible [26–31].
The fluorophores chosen in this earlier work [20] were two dyes that have been used in the assessment of the numbers and physiological status of intact bacteria, as they seem to permeate wild-type membranes more or less easily (presumably via a variety of transporters). While the strong dependence on transporter activity meant that such dyes did not faithfully reflect either bioenergetic parameters or nucleic acid content, we noted [20] that they opened up the possibility of high-throughput screening of transporter activity, including of competitive or inhibitory (non-fluorescent) substrates of membrane transporters (as in [32] for uptake and [33–38] for efflux). We recognize, of course, that such dyes are unlikely to be the ‘natural’ substrates of these transporters (see also [39]), but that an assessment of differential uptake can provide useful hints as to the kinds of molecular structures with which a given transporter interacts. We recognize too that individual dyes may use multiple transporters, that the number of transporters in E. coli significantly exceeds the number of dyes, that multiple dyes may be substrates for the same transporter, and that as with the uptake and efflux of natural substrates we are initially simply seeking an understanding of the activities of transporters of largely unknown potential. Here high-throughput methods come to the fore.
The desirable properties of dyes (and assays) of this type therefore include the following:
They are taken up intracellularly more or less rapidly by the target species.
They do not interfere significantly with the host’s biochemistry at the concentrations used and on the timescale of interest.
They have a high fluorescence signal (usually involving a high absorbance at the excitation wavelength and a high quantum yield).
They are not interfered with by autofluorescence (often implying a large Stokes shift or an absorbance nearer the red).
They show a fluorescence that is linear with intracellular concentration at the time point(s) of interest (consistent with point 2).
Assays should be performed at constant cell number, or if they are not the number of cells used does not materially affect the external concentration of the dye.
When used as a surrogate for a particular transporter, the bulk of the flux of interest is mediated by that transporter.
We also recognize that in some cases (not least when binding is to DNA or to metal ions) the binding of such fluorophores to intracellular targets can induce changes in both the magnitude and the fluorescence spectrum of a given dye, or that dyes might exhibit concentration-dependent self-quenching; this will in some cases need to be considered when the uptake of these dyes is used as a surrogate measure for transporter activity. Normally, however, it will be the case that the magnitude of a dye’s specific fluorescence is the same both inside and outside the cell, such that its uptake cannot be registered reliably simply by measuring total fluorescence in a cuvette or a plate reader, but requires the use of an instrument where the signals can be triggered by other means (see also [40]). It goes without saying that the expression of particular transporters (or anything else) depends on the growth conditions used, and these are typically controlled in E. coli by transcriptional regulatory networks (e.g. [41–44]). Consequently, in any given condition there is a danger of ‘false negatives’ [45, 46], i.e. rejection of dyes that under other circumstances might be taken up well. Thus, we have sought to be inclusive of potentially useful dyes so far as is possible. We note that neither flow cytometry nor filtration assays can easily discriminate binding from uptake; doing this relies on the use of mutants and other evidence (e.g. where the extent of uptake far exceeds plausible binding sites). We note too that no dye (nor any other substance) is necessarily likely to be a substrate of just a single transporter; again this is established via the use of mutant strains and/or of other conditions varying expression profiles substantially.
Dyes with most or all of the desirable properties given above are not particularly common as applied to unfixed cells, and we recognized that a wider survey of potentially useful bacterial membrane-permeating dyes of different structures might prove of considerable value for many purposes. Further, most dyes applied to biological cells are used on mammalian cells, and so may not be pertinent to bacteria. Nonetheless, we surveyed the catalogues of a wide variety of biological stain suppliers, plus other sources such as food colours, laser dyes, dyes used in the water industry for tracking, and even the core scaffolds used in organic light-emitting diodes. Thus, the present study represents an initial survey to this end, and provides a significant number of dyes that do seem to have most or all of the desired properties in the reference strain. We triaged our initial set down to 39 dyes based on a combination of uptake, chemical structure and cost (Fig. 1, Table 1), and these provide our main palette, although if resources are at a premium we also provide a cheminformatics analysis that permits further triaging to as few as six dyes (Fig. 2). In addition, to illustrate their utility in actual cases, we assess their utility in profiling three membrane transporters using appropriate mutants.
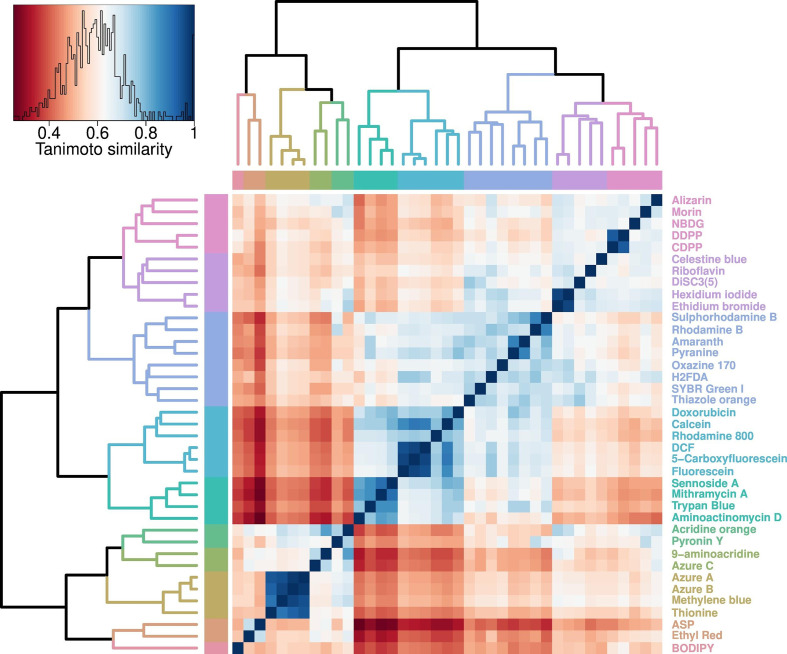
Fig. 1.
Cheminformatic analysis of fluorophores accumulated by E. coli BW25113. Heatmap representing the Tanimoto similarities (derived as described in the Methods section) of the palette of 39 compounds. Scale from zero (least similarity) to 1 (greatest similarity). Stars indicate clusters with a Tanimoto similarity exceeding 0.8 (seen as a cutoff for similar bioactivities).
Table 1.
Fluorophore set specifically accumulated by E. coli BW25113. List of fluorescent (Fluor) molecules that recorded fluorescence signals two-fold or higher (Signal strength) than the median values of autofluorescence. SMILES (Simplified Molecular Input Line Entry System [135]) Channel (as listed in Methods): Intellicyt® flow cytometer channel that registered the highest median for each fluorophore. The excitation and emission profiles of these channels should have matched the excitation and emission wavelengths for those fluorophores (Table S1). The structures of some of these fluorophores are unknown or unavailable (NA). Concentration: concentration in micromolar at which the highest median values were found for each fluorophore in the range tested. SD: standard deviation of the sample, CI95: 95% confidence interval for the data of signals from each fluorophore at the given channel. BODIPY tetra-methyl: Difluoro{2-[(3,5-dimethyl-2H-pyrrol-2-ylidene-N)methyl]-3,5-dimethyl-1H-pyrrolato-N}boron
|
Fluor |
Channel |
Mean |
Concentration |
Median |
SD |
CI95 |
Signal strength |
|---|---|---|---|---|---|---|---|
|
2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)−2-Deoxyglucose |
BL3 |
10 609 |
2.5 |
11 615 |
3311 |
[9256–11 962] |
4.16 |
|
2',7′-Dichlorofluorescein |
BL3 |
16 332 |
3 |
16 550 |
4821 |
[14361–18 302] |
5.93 |
|
3,6-Bis(4-chlorophenyl)−2,5-dihydropyrrolo[3,4 c]pyrrole-1,4-dione |
RL1 |
12 552 |
3 |
12 303 |
1956 |
[11695–13 410] |
2.92 |
|
3,6-Diphenyl-2,5-dihydropyrrolo[3,4 c]pyrrole-1,4-dione |
RL1 |
10 731 |
3.0 |
9415 |
3065 |
[9388–12 074] |
2.24 |
|
4-(4-(Dimethylamino)styryl-N-methylpyridinium (ASP+)- |
RL1 |
26 815 |
0.3 |
9288 |
42 128 |
[17216–36 413] |
2.21 |
|
5-Carboxyfluorescein |
BL3 |
14 787 |
1.0 |
18 608 |
11 353 |
[11787–17 788] |
6.67 |
|
7-Aminoactinomycin D |
VL4 |
11 609 |
2.5 |
12 250 |
2399 |
[10629–12 589] |
2.47 |
|
9-aminoacridine |
VL1 |
1458 |
5.0 |
1597 |
426 |
[1349–1566] |
2.83 |
|
Acridine orange |
BL3 |
7508 |
10 |
7467 |
3265 |
[6675–8341] |
2.68 |
|
Alizarin |
BL3 |
7380 |
3.0 |
6862 |
3767 |
[6419–8341] |
2.46 |
|
Amaranth |
BL3 |
6330 |
5.0 |
6303 |
2604 |
[5665–6994] |
2.26 |
|
ATTO 430LS carboxy |
BL3 |
5203 |
5.0 |
6191 |
1436 |
[4616–5790] |
2.22 |
|
ATTO 488 carboxy |
BL3 |
19 619 |
1.3 |
19 556 |
5804 |
[18138–21 100] |
7.01 |
|
ATTO 490LS carboxy |
BL5 |
10 272 |
3.0 |
8232 |
7447 |
[8046–12 498] |
9.66 |
|
ATTO 633 carboxy |
RL2 |
20 712 |
3.0 |
18 035 |
11 004 |
[17423–24 001] |
11.1 |
|
ATTO 647 carboxy |
RL1 |
46 880 |
10 |
38 512 |
67 804 |
[29579–64 181] |
9.15 |
|
ATTO Rho14 carboxy |
RL1 |
30 855 |
10 |
12 507 |
37 080 |
[19772–41 938] |
2.97 |
|
Azure A |
RL1 |
10 631 |
3.0 |
9994 |
3830 |
[9654–11 608] |
2.37 |
|
Azure B |
RL1 |
11 507 |
10 |
10 951 |
3462 |
[10623–12 390] |
2.60 |
|
Azure C |
RL1 |
16 418 |
2.5 |
13 675 |
14 301 |
[12639–20 197] |
3.25 |
|
Calcein |
BL3 |
16 131 |
10 |
16 616 |
4493 |
[14984–17 277] |
5.96 |
|
Celestine blue |
RL1 |
14 754 |
10 |
12 252 |
7653 |
[12352–17 156] |
2.91 |
|
CruzFluor 405 succinimidyl ester |
BL3 |
15 887 |
10 |
16 691 |
4278 |
[14139–17 636] |
5.98 |
|
BODIPY tetra-methyl |
VL5 |
7969 |
1.0 |
9639 |
5286 |
[6310–9628] |
2.31 |
|
DiSC3(5) |
RL1 |
205 602 |
1.3 |
189 686 |
152 318 |
[166736–244 468] |
45.1 |
|
Doxorubicin |
BL3 |
8589 |
1.0 |
7583 |
4062 |
[7552–9625] |
2.72 |
|
Ethidium bromide |
BL3 |
11 501 |
10 |
11 246 |
3136 |
[10701–12 301] |
4.03 |
|
Ethyl Red |
RL1 |
29 928 |
10 |
18 738 |
25 518 |
[21920–37 937] |
4.45 |
|
Fluorescein |
BL3 |
16 869 |
5.0 |
17 035 |
3560 |
[15961–17 777] |
6.11 |
|
H2FDA |
BL1 |
9137 |
5.0 |
8708 |
4036 |
[8107–10 167] |
3.54 |
|
Hexidium iodide |
BL3 |
12 993 |
10 |
13 090 |
3819 |
[9250–16 735] |
4.69 |
|
Methylene blue |
RL1 |
10 259 |
1.0 |
9259 |
4009 |
[9236–11 282] |
2.20 |
|
Mithramycin A |
VL4 |
11 137 |
5.0 |
11 616 |
3723 |
[10187–12 087] |
2.34 |
|
Morin |
BL3 |
7223 |
3.0 |
7437 |
732 |
[6506–7940] |
2.67 |
|
Oxazine 170 |
RL1 |
9014 |
3.0 |
10 024 |
3831 |
[8002–10 026] |
2.38 |
|
Pyranine |
VL4 |
11 124 |
10 |
11 047 |
6960 |
[9285–12 964] |
2.23 |
|
Pyronin Y |
BL3 |
11 652 |
1.3 |
9030 |
6434 |
[10011–13 294] |
3.24 |
|
Rhodamine 800 |
RL2 |
13 444 |
1.3 |
8436 |
13 241 |
[9288–17 599] |
5.21 |
|
Rhodamine B |
BL3 |
7012 |
1.3 |
6580 |
3552 |
[6106–7918] |
2.36 |
|
Riboflavin |
BL3 |
7020 |
1.0 |
7254 |
2762 |
[6315–7724] |
2.60 |
|
Sennoside A |
VL4 |
9506 |
10 |
10 634 |
2645 |
[8715–10 297] |
2.15 |
|
Sulphorhodamine B |
BL3 |
9440 |
10 |
8117 |
4183 |
[8373–10 508] |
2.91 |
|
SYBR Green I |
BL1 |
16 690 |
2.5 |
8517 |
19 703 |
[11663–21 718] |
3.46 |
|
SYTO13 |
BL1 |
281 386 |
5.0 |
197 317 |
235 885 |
[178007–384 765] |
80.1 |
|
Thiazole orange |
BL1 |
25 740 |
10 |
10 600 |
32 357 |
[17484–33 997] |
4.30 |
|
Thionine |
RL1 |
9878 |
10 |
10 166 |
5068 |
[8539–11 218] |
2.42 |
|
Trypan Blue |
RL1 |
19 833 |
5.0 |
15 151 |
16 464 |
[15482–24 184] |
3.60 |
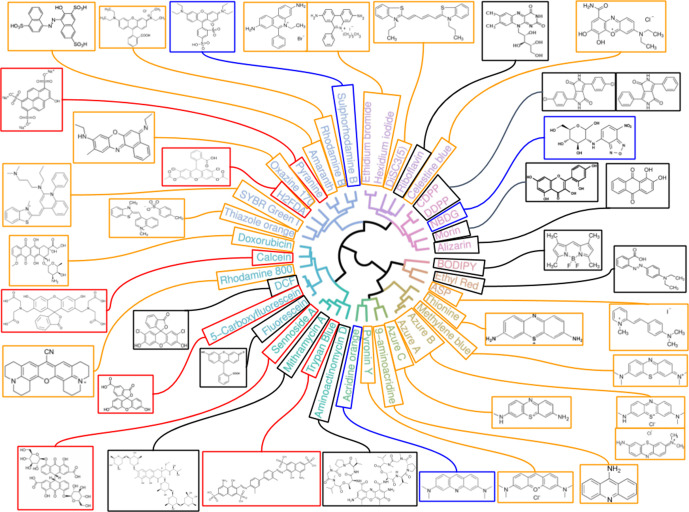
Fig. 2.
Chemical diversity among 39 fluorophores taken up by E. coli BW25113. Circularized topology of the dendrogram in Fig. 1 for the visualization of the structures of this set of fluorophores. The colour of the box indicates the molecular type and/or the expected charge at pH 7.4: black, uncharged; blue, zwitterionic; red, negative; orange, positive.
Methods
E. coli strain BW25113 [Keio collection reference strain: Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ −, rph-1, Δ(rhaD-rhaB)568, hsdR514] and certain other representatives of the Keio collection [21, 22] used here included the following gene knockouts: yhjV [F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, ΔyhjV722::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514); yihN (F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, rph-1, ΔyihN736::kan, Δ(rhaD-rhaB)568, hsdR514]; and tolC [F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, ΔtolC732::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514]. We also used the strain overexpressing yhjV from the ASKA collection: E. coli K-12, strain AG1 [recA1 endA1 gyrA96 thi-1 hsdR17 (r K− m K+) supE44 relA1] carrying recombinant constructs in the IPTG-inducible plasmid pCA24N (CmR, lacIq). The induction with IPTG was optimized to 250 µM for 3 h (37 °C, shaking at 200 r.p.m.) previous to the fluorophore uptake assays with the ASKA strain. Both the Keio and the ASKA collection (pmid16769691) were provided by the National Institute of Genetics, Mishima, Shizuoka, Japan.
For each fluorophore uptake assay the starting point was the spread of a small flake from a frozen culture stock of E. coli onto a fresh plate of complex solid media (Merck LB 110283) containing selective antibiotics when appropriate. Kanamycin was used at 50 µg ml−1 final concentration for the Keio knockout strains and chloramphenicol at 30 µg ml−1 for the ASKA strain. A single colony from a fresh solid media plate was then incubated in liquid complex media (Merck LB 110285) overnight at 37 °C with shaking at 200 r.p.m. in the absence of antibiotics. The overnight cultures were diluted 1 : 5000 in fresh liquid complex media and grown for 2 h at 37 °C with shaking at 200 r.p.m. The cell density was then adjusted to ~2000 cells µl−1, as judged by turbidity (OD600). Cells were then exposed to fluorophores (37 °C, 15 min, or as indicated, with shaking at 1300 r.p.m.) in 384-well plates and final volumes of 50 µl.
The experiments with chlorpromazine (CPZ) were performed using two different protocols. For the first set of experiments, the protocol was replicated from a previously described one [20], where 5 µl CPZ (1 mM in DMSO) was added to the wells of a 96-well plate in triplicate. A vacuum centrifuge was used to dry all the DMSO in the plates. Thereafter, 200 µl of overnight-cultured E. coli (MG1655 or BW25113) was added to each well at 1000 cells µl−1 final concentration in complex media. The plates were sealed and incubated at 30 °C with 900 r.p.m. shaking for 30 min. Then, DiSC3(5) or SYBR Green I were added to a final concentration of 3 µM and 1× (a 10 000-fold dilution of the stock material supplied), respectively [20]. The plate with DiSC3(5) was incubated for 2 min, while the one with SYBR Green I was incubated for 15 min at 37 °C before sampling in the flow cytometer. For the second set of experiments, CPZ was added to overnight-cultured E. coli (BW25113; 2000 cells µl−1) cells at a final concentration of 10 µM in complex media. This culture was then incubated with different fluorophores (3 µM) at different pH values for 15 min. To modify the pH of the media the following buffers were added individually (cf. [47]) to the media at final concentrations of 100 mM: MES (pH 6, 6.5), MOPS (pH 7.0, 7.5, 8.0) and Bicine (pH 8.5). Fluorophores were purchased from Sigma Aldrich, Thermo Fisher, TCI America, or ATTO-TEC GmbH. Other reagents were purchased from Sigma Aldrich unless stated otherwise.
We used a high-throughput flow cytometer, the Intellicyt iQue Screener Plus (Sartorius, Göttingen, Germany) [19, 20], with the following protocol: buffer equilibration (QSol, Sartorius) and plate shaking at 2000 r.p.m. for 50 s, sampling for 2 s with 1 s upload time, 5 s wash in Qsol buffer every three wells, and further probe wash for 10 s every 12 wells. The instrument has three LED lasers (405 nm, 488 nm, 640 nm) and collects data for 2 light scattering and 13 fluorescence channels. Once the light from any of three lasers has reached the samples, these channels collect the fluorescent signals back from the samples in the following spectral ranges (in nm): VL1 (445±45), VL2 (530±30), VL3 (572±28), VL4 (615±24), VL5 (675±30), VL6 (780±60), BL1 (530±30), BL2 (572±28), BL3 (615±24), BL4 (675±30), BL5 (780±60), RL1(675±30) and RL2(780±60). In contrast to some instruments [40] (and as is evident from the data), this instrument is highly resistant to the detection of extracellular fluorescence. As with any other transport assay method, such as those based on filtration or on flow dialysis [48], we do not directly discriminate molecules that are bound from those that are intracellular; other arguments given below serve to do that.
Data were analysed and displayed using a combination of the instrument’s Forecyt software, FlowJo, and routines written by the first author in R (all scripts available upon request to the first author).For the dose–response regression analyses the data were fitted with a non-parametric approach (locally weighted scatterplot smoothing) as implemented in R’s ggplot2. The hierarchical dendrogram and heatmap of the palette of 39 fluorophores were scripted with R’s dendextend and ggplot’s heatmap.2 packages. The hclust function of dendextend produced hierarchical clusters from a Tanimoto similarity matrix derived as follows: fingerprints of 39 fluorophores were derived from their SMILES [49] using the Patterned algorithm within the RDkit (www.rdkit.org/) nodes in KNIME (http://knime.org/) [39, 50]. Fingerprints are binary strings (a vector of 1 and 0 s), from which a Tanimoto distance can be calculated in terms of the fraction of positions in which the pairwise elements match (e.g. [51–54]). A Tanimoto distance (TD) matrix was generated from these fingerprints (KNIME’s Distance Matrix Calculate); and the scores of this matrix were converted to Tanimoto similarity indices (1-TD) using KNIME’s Similarity Search. In one case, the hierarchical dendrogram was circularized with R’s circlize package to allow space for the composite with the structures of these 39 fluorophores.
Results
Baseline analysis
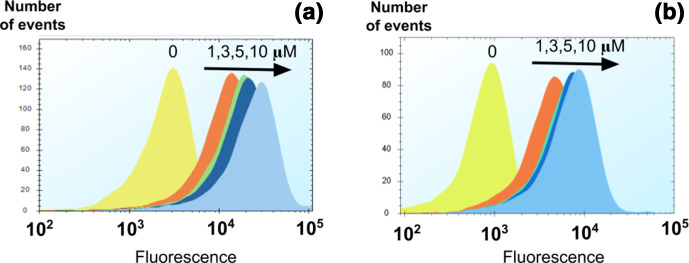
A total of 143 fluorophores were tested in E. coli BW25113 (Table S1, Fig. S1, available in the online version of this article). Fig. 3 shows typical cytograms of stained and unstained cells at four concentrations of two particular dyes, in this case calcein (A) and fluorescein (B). The typical existence of a small population with a very high fluorescence biases the mean and so we normally use the median fluorescence for data analysis.
Fig. 3.
Cytograms of calcein and fluorescein. Cells were grown, harvested and reinoculated at a certain concentration, as described in the Methods section. Both dyes detected in channel BL3 of Intellicyt (ex=488, em=615/24). (a) Calcein fluorescence signals at 1, 3, 5 and 10 µM against the non-dye control (0). (b) Fluorescein signals at 1, 3, 5 and 10 µM against the non-dye control (0).
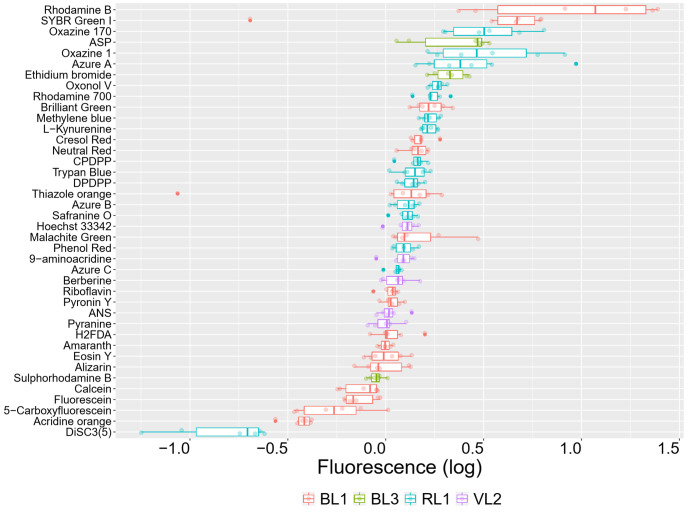
The fluorescence signals across the entire set of fluorophores spans some three orders of magnitude, with a median value approximately 1×103.6 (Fig. S2a). Autofluorescence (the fluorescence from cells to which no dye has been added) provides a ‘baseline’ (orange line for each channel in Fig. S2b), above which any fluorescence resulting from dye uptake must be resolved (though note that some dyes can act to quench autofluorescence). The channel that captured fluorescence emissions in the region of 530±30 nm (VL2) exhibits the highest values of autofluorescence relative to the other channels (Fig. S2b). The same data after correction for autofluorescence showed a median of approximately 1×103.3 (Fig. S2c). (Note that because dyes can potentially quench autofluorescence, the fluorescence changes induced by dyes include such phenomena.). Also, a fractional carry over between wells was observed in one of the red channels. The latter is observed as a separate cluster at the very highest values in channel RL1 (Fig. S2d). To avoid this, future experiments were designed, and the data filtered, accordingly.
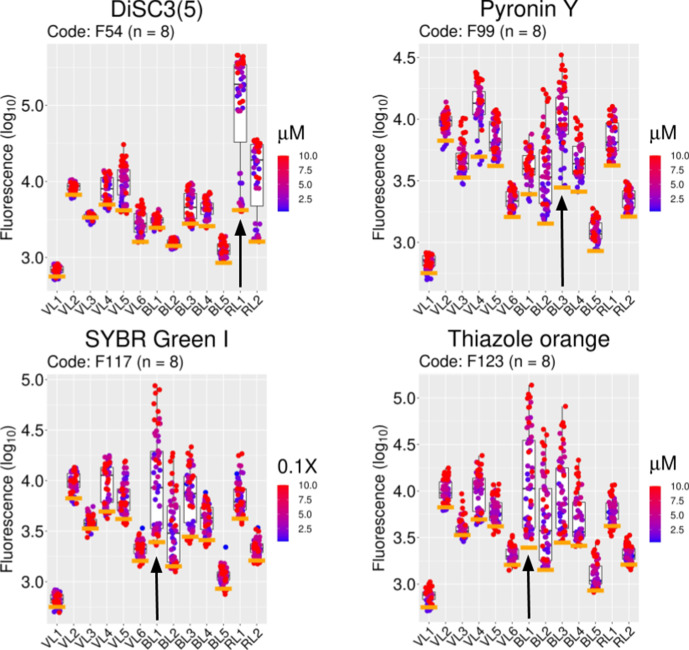
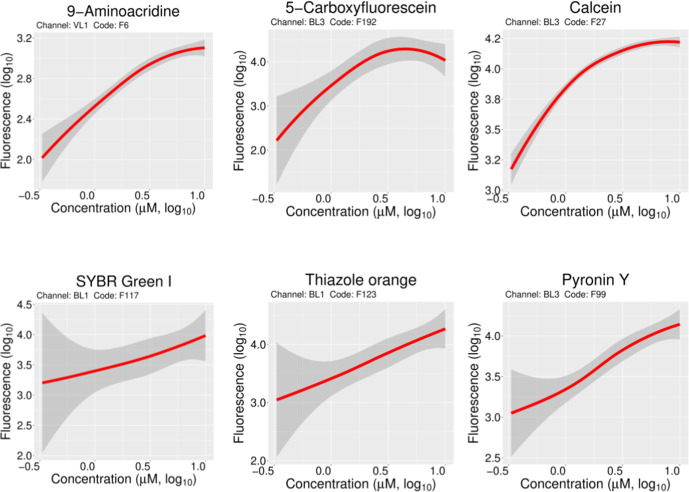
A set of 47 fluorophores that showed median values at least twofold (log10 greater than ~0.3) more than the median of the autofluorescence for a given channel at the concentration used was the core of the set for the next phases of the study (Tables 1 and S1). Inspection of the data showed no particular bias towards either low MW, polarity, or excitation wavelengths. Two of these dyes are the two that we had used previously (DiSC3(5) and SYBR Green I) [19, 20]. The compiled data for four of these molecules (DiSC3(5), pyronin Y, SYBR Green I and thiazole orange) are shown in Fig. 4. They display their strongest signals in channels RL1, BL3, BL1 and BL1, respectively. The set of 47 molecules displayed a reasonable dose response, as judged by accumulated fluorescence (ignoring autofluorescence) in the concentration range 0.1 to 10 µM (Figs 5 and S3).
Fig. 4.
Examples of specific fluorophore uptake in E. coli BW25113. Fluorescence signals from 4 fluorophores selected from the set of 47 molecules (Table 1) with values twofold or higher above autofluorescence. The signals for all fluorophores (log10 values, ordinate) are compiled against autofluorescence (orange bars) for every channel (abscissa). The light signal range for each channel is given in the Methods section. Colour-coded distributions of data by concentration of fluorophore (micromolar) are shown. The median values (log10) in channel RL1 for DiSC3 and BL3 for pyronin Y are 50-fold (at 1.3 µM) and 4-fold (at 1.3 µM) above autofluorescence, respectively. In channel BL1 SYBR Green I (stock diluted 10000×) and thiazole orange (10 µM) showed log10 values above autofluorescence equivalent to 3.2-fold and 4-fold, respectively. The chosen channel for each fluorophore is mapped with a black vertical arrow. Each concentration is represented by eight biological replicates taken over a period of 5 months.
Fig. 5.
Effect of concentration on fluorophore uptake. Locally weighted scatterplot smoothing was applied to the fluorescence signal versus fluorophore concentration (1, 2.5, 3, 5, 7.5 and 10 µM). These data were from 6 of the 47 initially chosen fluorophores (see also Fig. S2). Three fluorophores detected in channels VL1 (9-aminoacridine) and BL3 (5-carboxyfluorescein and calcein) only showed linearity up to 1 µM. The dose–response trend was essentially linear for SYBR Green I (channel BL1), thiazole orange (channel BL1) and pyronin Y (channel BL3) up to 10 µM.
Eight dyes that were quite expensive or of unknown/unavailable structure (i.e. SYTO-13, Cruz Fluor 405 and the six ATTO fluorophores) from the set of 47, were usually not used in follow-up experimentation (except for the gene knockout strain screening shown later). When evaluating the structural similarity of the resultant 39 molecules (Fig. 1), a number were phenothiazine derivatives: acridine orange, pyronin Y, 9-aminoacridine, azure A, azure B, azure C, methylene blue and thionine. Xanthene dyes [55] include rhodamines, calcein, fluoresceins and eosin.
Fluorescein derivatives were the second largest group: fluorescein, 5-carboxyfluorescein and 2′,7′-dichlorofluorescein (DCF), rhodamine 800, calcein and doxorubicin (an antineoplastic antibacterial) (Fig. 2). The predicted molecular charges (seen as a decisive property for membrane transport) for these fluorophores showed that NBDG, acridine orange and sulphorhodamine B are expected to be zwitterionic at neutral pH (Fig. 2). Six other fluorophores are expected to have a net negative charge and 18 to have a positive charge (cationic dyes) (Fig. 2). An anticipated example of the latter included the nucleic acid-binding fluorophores (i.e. SYBR Green I, thiazole orange, ethidium bromide and pyronin Y), some of which increase their fluorescence considerably upon such binding. It is of interest that a number are natural products, some with significant molecular masses (e.g. sennoside A, MW 862.7; 7-aminoactinomycin D, MW 1270.4, Table S1), consistent with the view [39] that many transporters evolved and were selected to transport natural products. Pyranine and amaranth are trisulphonated dyes (and they cluster), and how they might get into cells is of some interest.
Overall, the dyes cover a heterogeneous swathe of chemical space (an overall median Tanimoto similarity below 0.6), and very few form clusters with a Tanimoto similarity greater than 0.75, the lower end of the cutoff region in typical cheminformatic analyses for similar bioactivities [39, 56–59]. The structural heterogeneity is reasonably equated with functional heterogeneity in uptake via different transporters, and this is assessed next.
Membrane transport mediates intracellular fluorophore accumulation
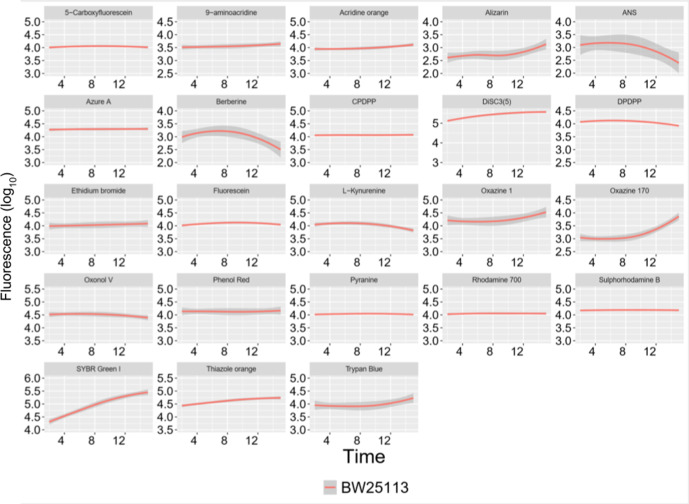
Most of the fluorophores of interest were taken up rapidly by E. coli BW25113, and the intracellular fluorescence as determined flow cytometrically had reached levels within 2 min that did not vary substantially at 15 min (Fig. 6). A few fluorophores such as DiSC3(5), oxazine 170, SYBR Green I, and thiazole orange were accumulated more slowly, but had reached an approximate steady state by the end of 15 min.
Fig. 6.
Representative time courses for fluorophore uptake. Locally weighted scatterplot fitting was applied to the fluorescence signal versus time data to follow the levels of fluorophore uptake in the time frame relevant to the experimental settings. x-axis: time in minutes (up to 15). y-axis: the log10 of fluorescence signals. For the fluorescence uptake experiments with different incubation times. The subset shown consisted of 23 dyes out of the 39 dyes used earlier.
Another important parameter that can affect membrane transport is external pH. The accumulation of all fluorophores tested could vary somewhat with pH in the range tested (pH 6–8.5), with the highest effect (up to two orders of magnitude) seen in the DPP-based dyes CPDPP and DPDPP (Fig. S4). For these two molecules the reduction in their signals is likely due in part to the quenching of their fluorescence because of the deprotonation of the lactam nitrogen in the DPP core [60]. The pH-related behaviour of the other fluorophores is also presented in Fig. S4.
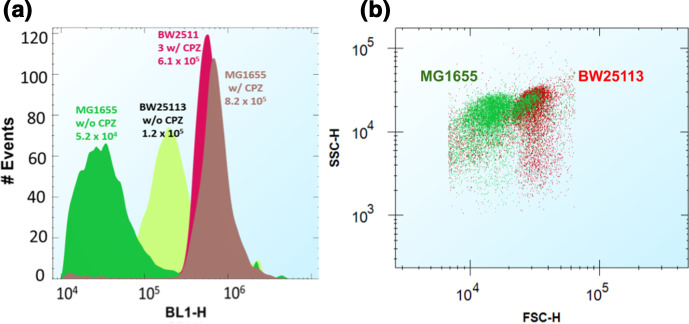
Chlorpromazine is a known efflux inhibitor in E. coli [20, 61–63]. In strain BW25113, CPZ increased the accumulation of rhodamine B by an order of magnitude and that of SYBR Green by four- or fivefold (Fig. 7). The fluorescence signal from oxazine 1, oxazine 170 and ASP [4-(4-(dimethylamino)-styryl-N-methylpyridinium (ASP+)] was increased threefold. Here, however, the opposite effect was shown for DiSC3(5), with CPZ inducing a decreased uptake of some threefold. We previously published significantly greater effects of CPZ on SYBR Green I uptake in E. coli MG1655 [20] (almost a 20-fold increase). We repeated these experiments for strain MG1655, with the same results as previously. The explanation for the difference observed with strain BW25113 is that BW25113 can accumulate far more di-SC3(5) in the absence of CPZ than does strain MG1655 under the same conditions. The uptake in the presence of CPZ is fairly similar for the two strains, implying a much lowered basal expression of efflux pumps such as acrAB/tolC in strain BW32113 (Fig. 8a). The breadth of the distribution of uptake is consistent with this. Such data illustrate the potential for very substantial variation in the uptake of individual dyes between strains of the same organism. Furthermore, we observed significant differences in the light scattering of these two strains of E. coli (Fig. 8b), especially in the forward scattering (which reflects differences in cell size distribution [7, 64]).
Fig. 7.
Effect of CPZ in the uptake of fluorophores in LB at pH 8.5. Fluorescence signals are presented as the log10 of ratios of the fluorescence from treated cells ( E. coli BW25113 and 10 µM CPZ) over the fluorescence signals from untreated cells. This time the fluorophore uptake incubation (37 °C, 15 min) was carried out at pH 8.5, which showed the strongest deviations from a ratio of 1 (log10=0). Boxplots are ordered by median values. Colours encode the IntelliCyt’s fluorescence channels used.
Fig. 8.
Difference in fluorophore accumulation and light scattering between E. coli strains MG1655 and BW25113. (a) Cytograms showing the effects of CPZ on the uptake of SYBR Green into the two strains. The experiments were performed using the same settings published in [20], as also mentioned in the Methods section. (b) Dot plots of light scattering for these two strains. FSC-H, forward scattering; SSC-H, side scattering. Both strains were processed identically and in parallel for these fluorophore uptake assays.
Profiling membrane transporters using fluorophores
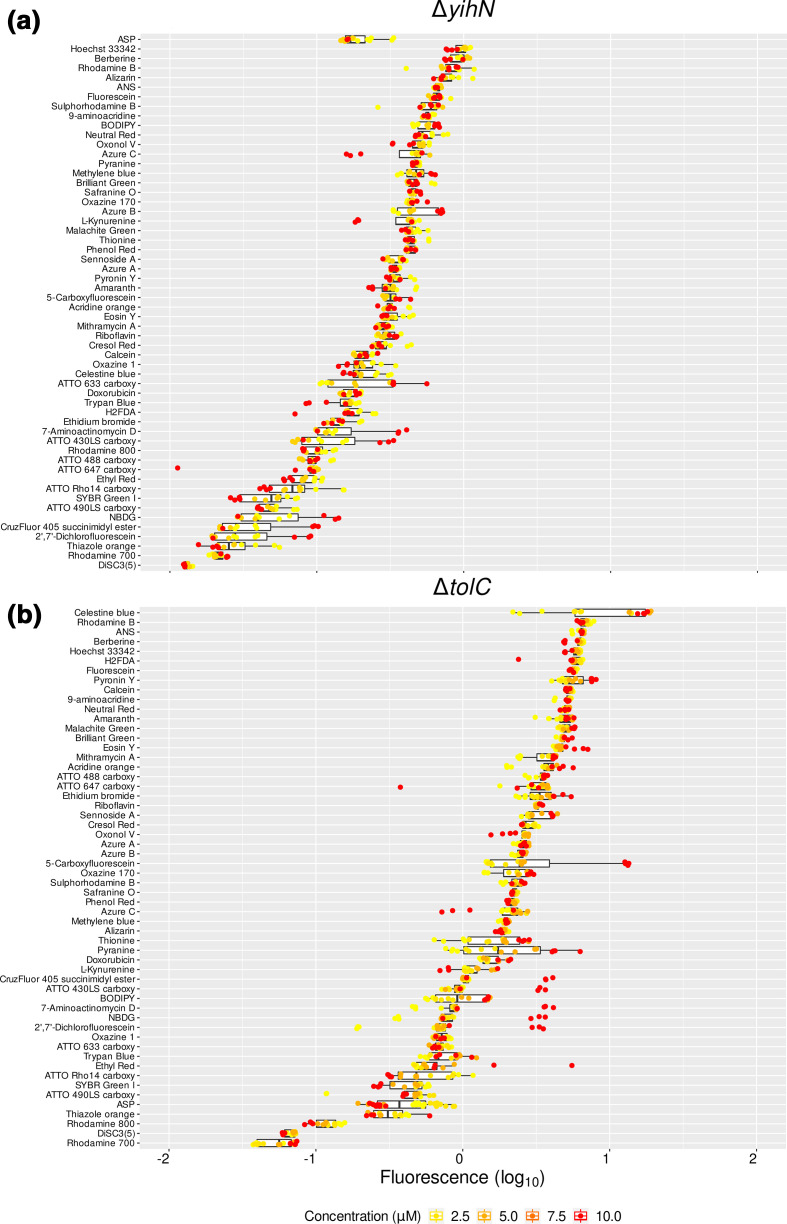
As indicated, one of our interests is in the deorphanization of y-gene transporters. An expanded set of fluorophores was used to interrogate the functional traits of strains lacking one of three genes encoding membranes transporters, viz yhjV, yihN and tolC. The success of the gene knockouts was confirmed by PCR (not shown). Two of these genes, yhjV and yihN, encode transporters with unknown substrates. The third one, tolC, encodes an ancillary protein component that is used by a number of different membrane transporters, mainly effluxers. After normalizing for autofluorescence, the y-gene knockouts ΔyhjV (Fig. S5) and ΔyihN (Fig. 9a) showed a clear ‘influxer’ pattern of fluorescence whereby the median signals of the tested fluorophores were below that of the reference strain (i.e. below a ratio of 1, log10=0). For the majority of dyes the uptake responded to concentrations of up to 10 µM. The fluorescence signal ratios of KO strains versus the reference strains are given in Table 2 for all three strains, together with their standard deviations and the 95 % confidence interval.
Fig. 9.
E. coli membrane influxers and effluxers have discriminatory fluorophore accumulation profiles. (a) Fluorophore uptake in the E. coli gene knockout strain yihN. Fluorescence signals are presented as the log10 of ratios of the fluorescence from the yihN knockout cells (∆yihN) over the fluorescence signals from the reference cells (BW25113). x-axis: log10 of the ratios (log10=0). No difference against the reference strain would have had a ratio of 1 (log10=0). (b) Fluorophore uptake in the E. coli gene knockout strain tolC. Fluorescence signals are presented as the log10 of ratios of the fluorescence from the tolC knockout cells (∆tolC) over the fluorescence signals from the reference cells (BW25113). x-axis: log10 of the ratios (log10=0). No difference against the reference strain would have had a ratio of 1 (log10=0). Boxplots are ordered by median (highest on top of the plot). The legend lists the concentrations used for each fluorophore.
Table 2.
Fluorophore uptake profile for three different membrane transporters of E. coli BW25113. Fluorescence (Signal Ratios) data are presented as the ratios of the fluorescence (median) from the three different E. coli knockout strains (∆yhjV, ∆yihN, and ∆tolC) over the fluorescence (median) from the reference strain ( E. coli BW25113). No difference between a KO strain and the reference would have had a ratio of 1. SD: standard deviation of the population, CI95: 95% confidence interval. The data for each knockout strain are sorted by incremental signal ratios
|
yhjV |
yihN |
tolC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Fluor |
Median |
SD |
CI95 |
Fluor |
Median |
SD |
CI95 |
Fluor |
Median |
SD |
CI95 |
|
Thiazole orange |
0.29 |
0.06 |
[0.26–0.31] |
DiSC3(5) |
0.01 |
0 |
[0.01–0.01] |
Rhodamine 700 |
0.06 |
0.01 |
[0.05–0.06] |
|
ATTO Rho14 carboxy |
0.37 |
0.08 |
[0.34–0.41] |
Rhodamine 700 |
0.02 |
0 |
[0.02–0.02] |
DiSC3(5) |
0.07 |
0.01 |
[0.07–0.07] |
|
SYTO13 |
0.44 |
0.24 |
[0.32–0.55] |
2',7′-Dichlorofluorescein |
0.03 |
0.03 |
[0.01–0.04] |
Rhodamine 800 |
0.12 |
0.02 |
[0.11–0.13] |
|
SYBR Green I |
0.46 |
0.09 |
[0.43–0.49] |
CruzFluor 405 succinimidyl ester |
0.03 |
0.03 |
[0.01–0.05] |
Thiazole orange |
0.31 |
0.1 |
[0.26–0.36] |
|
Rhodamine 800 |
0.55 |
0.12 |
[0.49–0.61] |
Thiazole orange |
0.03 |
0.01 |
[0.02–0.03] |
DMAMP |
0.38 |
0.2 |
[0.31–0.45] |
|
Ethyl Red |
0.58 |
0.09 |
[0.53–0.62] |
NBDG |
0.04 |
0.04 |
[0.02–0.06] |
ATTO 490LS carboxy |
0.47 |
0.14 |
[0.4–0.54] |
|
Sennoside A |
0.6 |
0.07 |
[0.57–0.64] |
ATTO 490LS carboxy |
0.05 |
0.01 |
[0.04–0.05] |
ATTO Rho14 carboxy |
0.48 |
0.28 |
[0.35–0.62] |
|
Rhodamine B |
0.61 |
0.19 |
[0.54–0.68] |
SYBR Green I |
0.05 |
0.02 |
[0.04–0.06] |
SYBR Green I |
0.48 |
0.12 |
[0.42–0.54] |
|
Alizarin |
0.63 |
0.06 |
[0.61–0.65] |
ATTO Rho14 carboxy |
0.07 |
0.04 |
[0.05–0.09] |
Ethyl Red |
0.55 |
1.26 |
[-0.07–1.16] |
|
Amaranth |
0.68 |
0.13 |
[0.63–0.72] |
Ethyl Red |
0.08 |
0.02 |
[0.07–0.09] |
Trypan Blue |
0.68 |
0.22 |
[0.57–0.79] |
|
Rhodamine 700 |
0.69 |
0.17 |
[0.63–0.75] |
ATTO 488 carboxy |
0.09 |
0.02 |
[0.08–0.1] |
ATTO 633 carboxy |
0.69 |
0.07 |
[0.65–0.72] |
|
Cresol Red |
0.74 |
0.12 |
[0.7–0.78] |
ATTO 647 carboxy |
0.09 |
0.02 |
[0.08–0.11] |
2',7′-Dichlorofluorescein |
0.72 |
1.1 |
[0.18–1.26] |
|
DiSC3(5) |
0.75 |
0.2 |
[0.68–0.82] |
Rhodamine 800 |
0.1 |
0.02 |
[0.09–0.11] |
Oxazine 1 |
0.72 |
0.07 |
[0.69–0.75] |
|
Oxonol V |
0.75 |
0.15 |
[0.7–0.8] |
ATTO 430LS carboxy |
0.11 |
0.1 |
[0.05–0.16] |
NBDG |
0.75 |
1.08 |
[0.22–1.27] |
|
Phenol Red |
0.75 |
0.23 |
[0.67–0.83] |
7-Aminoactinomycin D |
0.12 |
0.12 |
[0.05–0.19] |
7-Aminoactinomycin D |
0.88 |
1.23 |
[0.28–1.49] |
|
Malachite Green |
0.76 |
0.17 |
[0.7–0.82] |
Ethidium bromide |
0.14 |
0.03 |
[0.13–0.16] |
BODIPY |
0.92 |
0.39 |
[0.73–1.11] |
|
Eosin Y |
0.77 |
0.05 |
[0.75–0.78] |
H2FDA |
0.16 |
0.05 |
[0.14–0.19] |
ATTO 430LS carboxy |
0.95 |
1.02 |
[0.45–1.45] |
|
Sulphorhodamine B |
0.77 |
0.22 |
[0.7–0.85] |
Doxorubicin |
0.17 |
0.02 |
[0.16–0.18] |
CruzFluor 405 succinimidyl ester |
1.07 |
1.12 |
[0.52–1.62] |
|
Brilliant Green |
0.79 |
0.12 |
[0.75–0.84] |
Trypan Blue |
0.17 |
0.04 |
[0.15–0.19] |
l-Kynurenine |
1.08 |
0.3 |
[0.94–1.22] |
|
Doxorubicin |
0.79 |
0.12 |
[0.75–0.84] |
ATTO 633 carboxy |
0.18 |
0.14 |
[0.1–0.26] |
Doxorubicin |
1.47 |
0.28 |
[1.33–1.61] |
|
Trypan Blue |
0.79 |
0.12 |
[0.74–0.83] |
DMAMP |
0.18 |
0.05 |
[0.16–0.21] |
Pyranine |
1.74 |
1.65 |
[0.94–2.55] |
|
l-Kynurenine |
0.8 |
0.12 |
[0.75–0.84] |
Celestine blue |
0.19 |
0.06 |
[0.16–0.23] |
Thionine |
1.89 |
0.74 |
[1.53–2.25] |
|
Pyronin Y |
0.8 |
0.17 |
[0.74–0.85] |
Oxazine 1 |
0.19 |
0.06 |
[0.16–0.23] |
Alizarin |
1.92 |
0.1 |
[1.87–1.96] |
|
Celestine blue |
0.82 |
0.05 |
[0.8–0.85] |
Calcein |
0.2 |
0.03 |
[0.18–0.22] |
Methylene blue |
1.97 |
0.09 |
[1.93–2.01] |
|
ATTO 488 carboxy |
0.83 |
0.18 |
[0.77–0.9] |
Cresol Red |
0.26 |
0.07 |
[0.23–0.3] |
Celestine blue |
13.82 |
6.56 |
[10.61–17.03] |
|
Thionine |
0.84 |
0.1 |
[0.81–0.88] |
Eosin Y |
0.3 |
0.06 |
[0.27–0.34] |
Azure C |
2.2 |
0.61 |
[1.9–2.5] |
|
Hoechst 33 342 |
0.85 |
0.05 |
[0.83–0.87] |
Mithramycin A |
0.3 |
0.02 |
[0.28–0.31] |
Phenol Red |
2.21 |
0.12 |
[2.15–2.26] |
|
Neutral Red |
0.85 |
0.09 |
[0.81–0.88] |
Riboflavin |
0.3 |
0.04 |
[0.28–0.32] |
Safranine O |
2.27 |
0.08 |
[2.23–2.31] |
|
Safranine O |
0.85 |
0.14 |
[0.79–0.9] |
Acridine orange |
0.31 |
0.05 |
[0.28–0.34] |
Sulphorhodamine B |
2.29 |
0.26 |
[2.17–2.42] |
|
Mithramycin A |
0.86 |
0.18 |
[0.8–0.92] |
5-Carboxyfluorescein |
0.32 |
0.04 |
[0.29–0.34] |
Oxazine 170 |
2.4 |
0.59 |
[2.11–2.69] |
|
9-aminoacridine |
0.88 |
0.06 |
[0.86–0.9] |
Amaranth |
0.32 |
0.08 |
[0.27–0.36] |
5-Carboxyfluorescein |
2.44 |
4.99 |
[-0.01–4.89] |
|
Azure B |
0.88 |
0.11 |
[0.84–0.92] |
Pyronin Y |
0.33 |
0.05 |
[0.3–0.36] |
Azure B |
2.53 |
0.17 |
[2.45–2.62] |
|
DMAMP |
0.88 |
0.14 |
[0.83–0.93] |
Azure A |
0.34 |
0.02 |
[0.33–0.35] |
Azure A |
2.61 |
0.18 |
[2.52–2.7] |
|
ASA |
0.9 |
0.02 |
[0.89–0.91] |
Sennoside A |
0.37 |
0.04 |
[0.35–0.39] |
Oxonol V |
2.63 |
0.35 |
[2.45–2.8] |
|
ATTO 647 carboxy |
0.9 |
0.17 |
[0.84–0.95] |
Phenol Red |
0.41 |
0.03 |
[0.4–0.43] |
Cresol Red |
2.67 |
0.27 |
[2.54–2.81] |
|
Berberine |
0.9 |
0.08 |
[0.87–0.93] |
Thionine |
0.42 |
0.06 |
[0.39–0.46] |
Sennoside A |
2.93 |
0.64 |
[2.62–3.25] |
|
5-Carboxyfluorescein |
0.91 |
0.09 |
[0.88–0.94] |
l-Kynurenine |
0.43 |
0.12 |
[0.37–0.5] |
Riboflavin |
3.21 |
0.12 |
[3.15–3.27] |
|
ATTO 490LS carboxy |
0.91 |
0.13 |
[0.85–0.97] |
Malachite Green |
0.43 |
0.05 |
[0.4–0.46] |
Ethidium bromide |
3.33 |
0.88 |
[2.9–3.76] |
|
H2FDA |
0.91 |
0.12 |
[0.87–0.95] |
Azure B |
0.44 |
0.16 |
[0.35–0.53] |
ATTO 647 carboxy |
3.44 |
0.92 |
[2.99–3.89] |
|
Acridine orange |
0.92 |
0.17 |
[0.86–0.98] |
Oxazine 170 |
0.44 |
0.04 |
[0.42–0.47] |
ATTO 488 carboxy |
3.56 |
0.34 |
[3.39–3.72] |
|
Methylene blue |
0.92 |
0.17 |
[0.86–0.98] |
Safranine O |
0.45 |
0.03 |
[0.43–0.46] |
Acridine orange |
3.8 |
1 |
[3.31–4.29] |
|
Riboflavin |
0.92 |
0.12 |
[0.88–0.96] |
Brilliant Green |
0.46 |
0.07 |
[0.42–0.5] |
Mithramycin A |
4.05 |
0.75 |
[3.68–4.42] |
|
Azure A |
0.93 |
0.15 |
[0.88–0.98] |
Methylene blue |
0.47 |
0.1 |
[0.42–0.53] |
Eosin Y |
4.5 |
0.87 |
[4.07–4.93] |
|
Calcein |
0.95 |
0.13 |
[0.9–0.99] |
Azure C |
0.48 |
0.15 |
[0.4–0.57] |
Brilliant Green |
4.78 |
0.26 |
[4.65–4.91] |
|
Ethidium bromide |
0.95 |
0.05 |
[0.93–0.96] |
Pyranine |
0.48 |
0.02 |
[0.47–0.49] |
Malachite Green |
4.97 |
0.45 |
[4.75–5.19] |
|
Fluorescein |
0.95 |
0.1 |
[0.91–0.98] |
Oxonol V |
0.51 |
0.09 |
[0.47–0.56] |
Amaranth |
5.02 |
0.74 |
[4.66–5.39] |
|
Azure C |
0.97 |
0.04 |
[0.95–0.98] |
Neutral Red |
0.52 |
0.1 |
[0.47–0.58] |
Neutral Red |
5.03 |
0.2 |
[4.93–5.13] |
|
Oxazine 1 |
0.98 |
0.14 |
[0.93–1.03] |
9-aminoacridine |
0.57 |
0.03 |
[0.56–0.59] |
9-aminoacridine |
5.18 |
0.43 |
[4.97–5.4] |
|
Oxazine 170 |
0.99 |
0.15 |
[0.94–1.04] |
BODIPY |
0.57 |
0.08 |
[0.53–0.62] |
Calcein |
5.26 |
0.2 |
[5.16–5.36] |
|
ATTO 633 carboxy |
1 |
0.17 |
[0.92–1.09] |
Sulphorhodamine B |
0.6 |
0.12 |
[0.53–0.67] |
Pyronin Y |
5.27 |
1.27 |
[4.65–5.9] |
|
CPDPP |
1.01 |
0.04 |
[0.99–1.03] |
ASA |
0.66 |
0.03 |
[0.64–0.67] |
Fluorescein |
5.62 |
0.2 |
[5.52–5.72] |
|
Pyranine |
1.01 |
0.15 |
[0.96–1.07] |
Fluorescein |
0.66 |
0.06 |
[0.62–0.69] |
H2FDA |
5.86 |
0.97 |
[5.39–6.34] |
|
DPDPP |
1.04 |
0.2 |
[0.94–1.14] |
Alizarin |
0.73 |
0.14 |
[0.65–0.81] |
Hoechst 33 342 |
6.1 |
0.51 |
[5.85–6.35] |
|
BODIPY |
1.19 |
0.05 |
[1.17–1.21] |
Rhodamine B |
0.79 |
0.18 |
[0.69–0.9] |
Berberine |
6.36 |
0.61 |
[6.06–6.66] |
|
Berberine |
0.96 |
0.12 |
[0.89–1.03] |
ASA |
6.51 |
0.42 |
[6.3–6.71] |
||||
|
Hoechst 33 342 |
1.01 |
0.12 |
[0.94–1.08] |
Rhodamine B |
6.8 |
0.49 |
[6.56–7.04] |
||||
All fluorophores had a reduced uptake in ΔyihN, with most of them accumulating less than threefold the amount in comparison to E. coli BW25113 (Fig. 9a). Both rhodamine 700 and DiSC3(5) showed the lowest uptake for any of the concentrations tested (up to 3 µM). Thiazole orange and SYBR Green I were also poorly accumulated by ΔyihN, with a more than 10-fold difference relative to the reference strain (Fig. 9a, Table 2).
E. coli contains a great many ‘efflux’ pumps, mutations that can cause substantial resistance to multiple drugs and antibiotics (e.g. [65–71]). Many are driven by ATP hydrolysis [72], although others obtain free energy via electron transport-linked membrane energization [73]. A variety of antiporters can also, under some circumstances, appear to act as ‘effluxers’ [32, 74, 75]. TolC is an outer membrane component that interacts with many inner membrane-located transporters (exemplified by acrAB) [67, 76–81]. The involvement of TolC with efflux systems was evident in the pattern of fluorophore uptake observed for 56 dyes in this knockout strain (Fig. 9b, Table 2). Most fluorophores were over-accumulated (ratios over 1, or log10 >0). Five fluorophores over accumulated mainly at the higher concentration of 10 µM: fluorescent glucose (NBDG), 2′,7′-dichlorofluorescein, 7-aminoactinomycin D, ATTO 430LS and CruzFluor 450. On the other hand, nine fluorophores had a decreased accumulation of 2-fold or lower in ΔtolC, with 10-fold or lower differences in the cases of rhodamine 800, DiSC3(5) and rhodamine 700 (Fig. 9b, Table 2). That the lack of tolC seems to impair the transport of the latter dyes points at the interaction or involvement of this protein with outer membrane influx systems (e.g. porins), either directly or via pleiotropic effects [82, 83]. The data for the three knockout strains and the 39 dyes are shown in Fig. S6 (with the six major dyes for the ΔyihN knockout being labelled, and with all the data being given in Table 2).
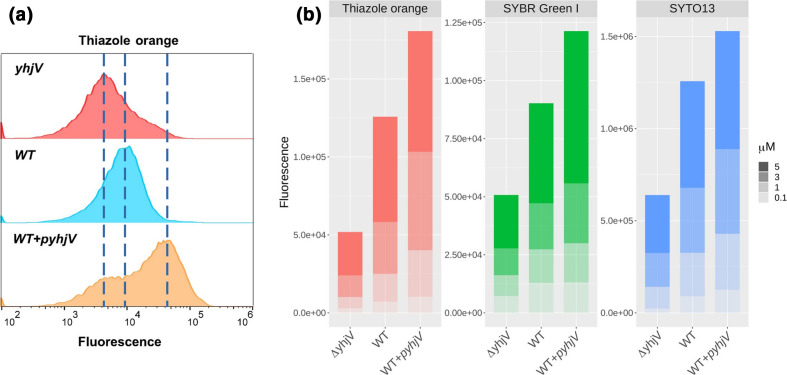
Finally, we also considered the opposite kind of experiment, in which specific transporter genes are carefully overexpressed, using the equivalent ASKA overexpression clones [84, 85]. Fig. 10a shows the cytograms for the uptake of thiazole orange into the deletant, wild-type and overexpression strains for yhjV when the concentration of the dye was 1 µM, while Fig. 10b shows more extensive data for three dyes at four concentrations. Each of these dyes is clearly capable of providing a surrogate transport assay for the YhjV transporter.
Fig. 10.
Fluorophores accumulate differentially in E. coli overexpressing the YhjV transporter. (a) Cytograms of the uptake of thiazole orange by E. coli BW25113 with differing expression levels of yhjV. The cell population distribution for the accumulation of thiazole orange (1 µM, 37 °C, 15 min) is presented for the gene knockout of yhjV (∆yhjV) of the Keio collection. The fluorescence signal of reference strain E. coli BW25113 (WT) is approximately 2.5-fold higher than the signal from the gene knockout strain (∆yhjV). The strain of E. coli expressing the yhjV gene from the ASKA collection (WT +pyhjV) shows a further increase in the accumulation of thiazole orange equivalent to approximately 4.5-fold over the signal from the WT (data taken from the assay where the highest differences were observed for 1 µM). (b) E. coli BW25113 overexpressing yhjV displays increased uptake of thiazole orange, SYBR Green I and SYTO13. The fluorescence signals of cells accumulating these fluorophores (37 °C, 15 min) are presented for the gene knockout for yhjV (∆yhjV), the reference strain E. coli BW25113 (WT) and the strain overexpressing the ASKA construct of yhjV (WT +pyhjV). Cells were exposed to four different concentrations of fluorophores up to 5 µM, as indicated. Fluorescence data are the averages of the median values from four biological replicates. The accumulation of these fluorophores in all three strains increased monotonically with ascending dye concentrations. For each fluorophore WT cells gave higher fluorescence signals than strains deleted in yhjV, and likewise cells expressing episomal yhjV (WT +pyhjV) over the reference strain (WT). The latter relation for thiazole orange at 1 µM is somewhat smaller (approximately 2.62-fold) than that shown in (a) (4.5-fold), since here we have the average of four experimental replicates.
Discussion
Role of fluorescent stains in understanding microbial physiology
Fluorescent molecules continue to be instrumental in the versatile, sensitive and quantitative study of microbial structure and physiology. However, the use of fluorophores in the characterization of membrane transporters continues to be sparse (it is done mainly in mammalian cell lines) and is largely directed to the study of drug resistance mediated by efflux pumps (e.g. [36, 86, 87]). Indeed, many drugs show structural similarities with fluorophores [88]. Nonetheless, early evidence has shown that fluorescent dyes such as rhodamine derivatives used in the study of drug efflux require mediated membrane transport influx in the first place [89], and the same is true for nucleic acid stains such as ethidium bromide [90, 91].
Previously, we only studied diSC3(5) [92, 93], which is subject to quenching at high concentrations [94], and SYBR Green I [9, 95–101], where quenching is intramolecular and is stopped (and fluorescence massively enhanced) by binding mainly to dsDNA [102]. Here we have surveyed a large set of fluorophores to assess their suitability as probes for the study of mediated membrane transport in bacteria, initially using the reference strain BW25113 of the Keio E. coli gene knockout collection [21].
Regarding the desirable properties of such dyes, as listed in the introduction, we note two caveats: (i) the excitations of those dyes surveyed somewhat reflected the wavelengths of the lasers that we used, and (ii) the eventual choice was largely based on the properties of the wild-type strain. Thus, other dyes might have been included if other excitation wavelengths were available, and/or if other strains showing lower expression of (or lacking) particular efflux pumps had been used.
Out of 143 fluorophores surveyed, the median values of 47 provided signals significantly above autofluorescence to consider pursuing. If those with mean values just 2-fold above autofluorescence are included, this set can be larger (60 fluorophores). However, the latter group of fluorophores are mainly accumulated by the population of higher uptake cells in the right-skewed distributions of fluorescence (Fig. S2c). The accumulation of a core of 39 fluorophores was of particular interest, and these were further characterized after selecting against those of higher cost or with unknown structures. These dyes stained the wild-type BW25113 E. coli on short time scales (Fig. 6) in a manner that was suitably dependent on substrate concentration (Figs 5 and S3). Their signals were mostly sensitive to membrane efflux transport inhibitors such as chlorpromazine (Fig. 7), and in some cases to the pH of incubation (Fig. S4).
Cheminformatics of chosen fluorophores
Despite the reasonable number of available fluorophores, many are not particularly cheap, and our survey was largely confined to the more inexpensive ones available commercially. Cheminformatics [52–54] (sometimes called chemoinformatics [52, 103–107]) describes the discipline that helps researchers assess questions such as the degrees of similarity between individual molecules [108–110] or the molecular diversity within a chemical library [111, 112]. We applied standard cheminformatics methods [39, 50, 113, 114] to the analysis of the relative diversity of our palette of 39 dyes. Given that a pairwise Tanimoto similarity below 0.8 (or a Tanimoto difference exceeding 0.2) is usually taken to mean a significant difference in bioactivity [39, 56–59], it is encouraging that while there were some small clusters (Figs 1 and 2), the median Tanimoto similarity was just 0.6, implying strong orthogonality in the behaviour of our palette, as was borne out experimentally.
This said, many of the diverse dyes were still either phenothiazines or xanthene family dyes, and one conclusion is that the need for a much greater variety of fluorescent scaffolds remains, in order to broaden the present collection yet further.
Strain differences
It is well known that strain differences between even non-pathogenic E. coli can have massive effects even on simple traits such as recombinant protein production [115]. In the present work, in some cases, we observed quite striking differences between the uptake of particular dyes into the two wild-type strains BW25113 and MG1655. In particular, the observed differences in uptake of SYBR Green I in E. coli MG1655 and BW25113 might be due to their known genomic differences: E. coli BW25113 is Δ(araD-araB)567 Δ(rhaD-rhaB)568 ΔlacZ4787 (::rrnB-3) hsdR514 rph-1, with the deletion of araBAD and rhaDAB and the replacement of a section of lacZ with four tandem rrnB terminators as well as a frameshift mutation in hsdR resulting in a premature translation stop codon. E. coli BW25113 also contains the lacI+ allele and not lacIq. Other known differences found from genome sequencing are the presence of the rph-1 allele as well as 20 substitutions and 11 indels [116]. Some of those substitutions directly affect membrane transport systems: nagE (N-acetyl glucosamine specific PTS enzyme IIC, IIB and IIA components), gatC (subunit of galactitol PTS permease), btuB (vitamin B12/cobalamin outer membrane transporter). Furthermore, any single or combined genomic difference can have a number of pleiotropic cellular effects that could easily account for broad phenotypic differences between these two strains of E. coli [117, 118]. This is a typical issue of complex systems biology that can cause difficulties yet also has advantages: the difficulties can occur because of an ostensible non-reproducibility of experiments that are in fact different (but in unknown ways), while the advantage is that it shows that our palette of dyes is a particularly strong discriminator of the physiology of different, and even closely related strains. Having previously seen that even single-gene knockouts could induce massive and completely uncorrelated effects in the uptake of particular dyes [20], we sought to assess the utility of this phenomenon in analysing the general dye uptake properties of three membrane transporters, viz YhjV, YihN and TolC.
The profiles observed with gene knockouts discriminate influx from efflux systems
It is reasonable that the genetic knockout of an import transporter (including antiporters [32, 75]) will tend to result in lower uptake of members of the palette than knockouts of predominantly efflux transporters, that may be expected to have the opposite effect. We assessed this expectation using three transporters, with the expected general results.
As an orphan transporter (marked at Uniprot https://www.uniprot.org/uniprot/P37660 as ‘inner membrane transport protein YhjV’, and as a possible ‘amino acid transmembrane transport protein’), we decided to use YhjV as a first test case for exploiting our dyes. It was chosen because in our previous work [20] the KO strain for yhjV accumulated one of the lowest amounts of SYBR Green. There is next to no literature on it, however [119]. Although its main substrates are not known, YhjV is considered to be an uncharacterized member of the hydroxy/aromatic amino acid permease (HAAAP) family within the amino acid/polyamine/organocation (APC) superfamily [120, 121]. To assess other dyes, we compared our palette in terms of uptake between the wild-type and ΔyhjV strains (Fig. S5). With the exception of BODIPY, all dyes were taken up less in the knockout than in the wild-type, although only a few by less than threefold. Thiazole orange was an even better (more discriminatory) substrate than SYBR Green. Despite their rather different names, these two dyes are reasonably similar structurally (Fig. 6 and 11), with both including a benzothiazole moiety linked to dual-ring systems. This similarity in behaviour is consistent with the principle of molecular similarity [108], by which similar molecular structures are expected to possess similar bioactivities.
The product encoded by yihN is another membrane transporter (https://www.uniprot.org/uniprot/P32135) of unknown function [122, 123]. Here again, essentially no dyes were taken up more in the knockout than in the wild-type (Fig. 9a), again implying that YihN is mainly an influx transporter. In this case, about a dozen dyes were accumulated less than 10-fold in the knockout relative to accumulation in the wild-type.
TolC is well known to be involved in the efflux of a great many substances because it is linked to a variety of inner-membrane efflux transporters [76, 79, 124–126]. In this case the uptake of the majority of the palette was, as expected, greater than in the wild-type in the tolC deletion strain. Rhodamines 700 and 800 are hydrophobic (six-ringed) cations, closely related structurally, and are the most and third-most dyes in terms of lowered uptake in the ΔtolC strain (Fig. 9b).
Consequently, the effects of a knockout of a putative transporters on the palette give a clear indication as to whether it is mainly an influx or an efflux transporter (Fig. 9) (although we note that under the conditions of individual assays we cannot discriminate symporters and antiporters; that requires the use of multiple conditions including the putatively antiported substrates [32]).
The above experiments only included gene knockout strains in comparison to the behaviour of the wild-type strain. Overexpression strains provide an arguably more powerful, and at least complementary, approach to understanding the biology of individual proteins. To this end (Fig. 10a), we compared the uptake of thiazole orange in the deletant and overexpression strains of yhjV, finding an approximately ninefold variation in uptake of the dye in the two strains, providing a clear use for it as a surrogate dye in assays for this transporter. The same was true of the other three dyes (Fig. 10b) chosen from the top four in the palette (the ATTO dyes were ignored on grounds of cost).
Pleiotropy
As with any other system based on specific genetic manipulations (e.g. [127, 128]), changes in the expression profile of an individual gene lead to changes in those of many others, a phenomenon referred to as pleiotropy. If X changes Y and Y changes Z, but Y is not measured, one might naively infer that X causes Z directly. Clearly, in our case X is a transporter activity and Z the extent of uptake of a dye of interest. Of course, one might add that the vast majority of studies designed to infer gene function simply perform the equivalent of varying X and only measuring Z. However, we have built confidence in our method for identifying true transporter substrates in a variety of ways. For instance, we can vary the concentration of our target gene both up and down; pleiotropic effects are not monotonic, although gene activities largely mirror gene expression levels. Similarly, we recognize that by using a palette of dyes we can observe the ordering of changes in dye uptake contingent on changing the expression of a gene; if (say) the effects of yihN were mediated solely by its effects on tolC, then their ordering profiles would be identical (and they are not). Ultimately, understanding a transporter’s activity in isolation from others (if that is possible) will rely on methods such as its expression in a system that lacks the activity of interest (e.g. often Xenopus oocytes [129, 130]). However, these are very far from being high-throughput methods. That proposed here provides the wherewithal for choosing the transporter of interest for such more detailed studies.
Concluding remarks
The present results and analysis have, for the first time, provided a reasonably comprehensive set of stains for E. coli , and have illustrated how sensitive their uptake can be to genotype and physiology. Clearly, the same strategy can be applied to the phenotyping of other microbes of interest, where different preferred dyes are certain to be found (note that rhodamine 123 [131] and hexidium iodide [132] readily enter intact cells of Gram-positive but not Gram-negative bacteria). The differential uptake of specific dyes in strains knocked out for or overexpressing particular transporters provides a clear means for high-throughput assay of these transporters and libraries of potential variants [133–135]; this will be the subject of future communications.
Supplementary Data
Funding information
We thank the BBSRC (grants BB/R000093/1 and BB/P009042/1) and the Novo Nordisk Foundation (grant NNF10CC1016517) for financial support. The funding bodies had no role or involvement in the design of the study and collection, analysis and interpretation of data or in writing the manuscript.
Acknowledgements
We thank Drs Kate Baker, Lachlan Munro and Lei Yang for useful discussions. We also thank the Editor for some very helpful comments.
Author contributions
J. E. S-S. performed most of the experiments. S. J. performed some of them. S. O’H. performed some of the cheminformatics. D. B. K. contributed the overall design of the study and obtained funding for it. All authors contributed to the writing of the manuscript and approved it.
Conflicts of interest
D. B. K. and S. J. are named inventors on a vaguely connected patent application relating to the use [19] of flow cytometry in antimicrobial detection. The authors declare that they otherwise have no conflicts of interest beyond the funding of their projects given in the Funding information section.
Footnotes
Abbreviations: ANS, 8-anilinonaphthalene-1-sulfonic acid; ASP, 4-(4-(dimethylamino)styryl-N-methylpyridinium (ASP+) diS-C3(5): 3-propyl-2-{(1E,3E,5E)-5-(3-propyl-1,3-benzothiazol-2(3H)-ylidene)-1,3-pentadien-1-yl}-1,3-benzothiazol-3-ium iodide; BODIPY, difluoro{2-[(3,5-dimethyl-2H-pyrrol-2-ylidene-N)methyl]-3,5-dimethyl-1H-pyrrolato-N}boron; CCCP, carbonyl cyanide m-chlorophenyl hydrazone; CPDPP, 3,6-bis(4-chlorophenyl)-2,5-dihydropyrrolo[3,4-c]pyrrole-1,4-dione; CPZ, chlorpromazine; DMSO, dimethyl sulfoxide; DPDPP, 3,6-diphenyl-2,5-dihydropyrrolo[3,4-c]pyrrole-1,4-dione; H2FDA, dihydrofluorescein diacetate; I, N,N-dimethyl-N′-[(4E)-4-[(3-methyl-1,3-benzothiazol-3-ium-2-yl)methylidene]-1-phenyl-2,3-dihydroquinolin-2-yl]-N′-propylpropane-1,3-diamine; IPTG, isopropyl β- d-1-thiogalactopyranoside; NBDG, 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose SYBR Green.
Six supplementary figures and one supplementary table are available with the online version of this article.
References
- 1.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Ghatak S, King ZA, Sastry A, Palsson BO. The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function. Nucleic Acids Res. 2019;47:2446–2454. doi: 10.1093/nar/gkz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riley M. Systems for categorizing functions of gene products. Curr Opin Struct Biol. 1998;8:388–392. doi: 10.1016/S0959-440X(98)80074-2. [DOI] [PubMed] [Google Scholar]
- 4.César-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, et al. A call for systematic research on solute carriers. Cell. 2015;162:478–487. doi: 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 5.César-Razquin A, Girardi E, Yang M, Brehme M, Sáez-Rodríguez J, et al. In silico prioritization of Transporter–Drug relationships from drug sensitivity screens. Front Pharmacol. 2018;9 doi: 10.3389/fphar.2018.01011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davey HM, Kell DB. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev. 1996;60:641–696. doi: 10.1128/MR.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shapiro HM. Practical Flow Cytometry. 4th ed, 3rd ed. New York: John Wiley; 2003. [Google Scholar]
- 8.Müller S, Davey H. Recent advances in the analysis of individual microbial cells. Cytometry. 2009;75A:83–85. doi: 10.1002/cyto.a.20702. [DOI] [PubMed] [Google Scholar]
- 9.Van Nevel S, Koetzsch S, Weilenmann H-U, Boon N, Hammes F. Routine bacterial analysis with automated flow cytometry. J Microbiol Methods. 2013;94:73–76. doi: 10.1016/j.mimet.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Hutter K-J, Eipel HE. Microbial determinations by flow cytometry. J Gen Microbiol. 1979;113:369–375. doi: 10.1099/00221287-113-2-369. [DOI] [PubMed] [Google Scholar]
- 11.Mason D, Gant VA. The application of flow cytometry to the estimation of bacterial antibiotic susceptibility. J Antibiotic Chemother. 1995;36:441–443. doi: 10.1093/jac/36.2.441. [DOI] [PubMed] [Google Scholar]
- 12.Nebe-von-Caron G, Stephens PJ, Hewitt CJ, Powell JR, Badley RA. Analysis of bacterial function by multi-colour fluorescence flow cytometry and single cell sorting. J Microbiol Methods. 2000;42:97–114. doi: 10.1016/S0167-7012(00)00181-0. [DOI] [PubMed] [Google Scholar]
- 13.Ambriz-Aviña V, Contreras-Garduño JA, Pedraza-Reyes M. Applications of flow cytometry to characterize bacterial physiological responses. Biomed Res Int. 2014;2014:1–14. doi: 10.1155/2014/461941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LN W, Wang S, Song YY, Wang X, Yan XM. Applications and challenges for single-bacteria analysis by flow cytometry. Sci China Chem. 2016;59:30–39. [Google Scholar]
- 15.Props R, Rubbens P, Besmer M, Buysschaert B, Sigrist J, et al. Detection of microbial disturbances in a drinking water microbial community through continuous acquisition and advanced analysis of flow cytometry data. Water Res. 2018;145:73–82. doi: 10.1016/j.watres.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 16.García-Timermans C, Rubbens P, Heyse J, Kerckhof FM, Props R. Discriminating bacterial phenotypes at the population and single-cell level: a comparison of flow cytometry and Raman spectroscopy fingerprinting. Cytometry A. 2019 doi: 10.1002/cyto.a.23952. [DOI] [PubMed] [Google Scholar]
- 17.Nerada Z, Hegyi Z, Szepesi Áron, Tóth S, Hegedüs C, et al. Application of fluorescent dye substrates for functional characterization of ABC multidrug transporters at a single cell level. Cytometry. 2016;89:826–834. doi: 10.1002/cyto.a.22931. [DOI] [PubMed] [Google Scholar]
- 18.Sinclair LV, Neyens D, Ramsay G, Taylor PM, Cantrell DA. Single cell analysis of kynurenine and system L amino acid transport in T cells. Nat Commun. 2018;9:1981. doi: 10.1038/s41467-018-04366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jindal S, Thampy H, Day PJR, Kell DB. Very rapid flow cytometric assessment of antimicrobial susceptibility during the apparent lag phase of microbial (re)growth. Microbiology. 2019;165:439–454. doi: 10.1099/mic.0.000777. [DOI] [PubMed] [Google Scholar]
- 20.Jindal S, Yang L, Day PJ, Kell DB. Involvement of multiple influx and efflux transporters in the accumulation of cationic fluorescent dyes by Escherichia coli . BMC Microbiol. 2019;19:195.:also bioRxiv 603688v603681. doi: 10.1186/s12866-019-1561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, et al. Update on the Keio collection of Escherichia coli single‐gene deletion mutants. Mol Syst Biol. 2009;5:335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kell DB. A protet-based, protonic charge transfer model of energy coupling in oxidative and photosynthetic phosphorylation. OSF preprint. 2020 doi: 10.1016/bs.ampbs.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Kell DB. Protonmotive energy-transducing systems: some physical principles and experimental approaches. In: Anthony CJ, editor. Bacterial Energy Transduction. London: Academic Press; 1988. pp. 429–490. editor. [Google Scholar]
- 25.Kell DB. The protonmotive force as an intermediate in electron transport-linked phosphorylation: problems and prospects. Curr Top Cell Reg. 1992;33:279–289. doi: 10.1016/b978-0-12-152833-1.50021-6. [DOI] [PubMed] [Google Scholar]
- 26.Dobson PD, Kell DB. Carrier-mediated cellular uptake of pharmaceutical drugs: an exception or the rule? Nat Rev Drug Discov. 2008;7:205–220. doi: 10.1038/nrd2438. [DOI] [PubMed] [Google Scholar]
- 27.Kell DB, Dobson PD, Oliver SG. Pharmaceutical drug transport: the issues and the implications that it is essentially carrier-mediated only. Drug Discov Today. 2011;16:704–714. doi: 10.1016/j.drudis.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Kell DB, Dobson PD, Bilsland E, Oliver SG. The promiscuous binding of pharmaceutical drugs and their transporter-mediated uptake into cells: what we (need to) know and how we can do so. Drug Discov Today. 2013;18:218–239. doi: 10.1016/j.drudis.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Kell DB, Oliver SG. How drugs get into cells: tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion. Front Pharmacol. 2014;5:231. doi: 10.3389/fphar.2014.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kell DB. What would be the observable consequences if phospholipid bilayer diffusion of drugs into cells is negligible? Trends Pharmacol Sci. 2015;36:15–21. doi: 10.1016/j.tips.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Kell DB, Kell DB. How drugs pass through biological cell membranes – a paradigm shift in our understanding? Beilstein Magazine. 2016;2 doi: 10.3762/bmag.5. [DOI] [Google Scholar]
- 32.Zhang S, Sakuma M, Deora GS, Levy CW, Klausing A, et al. A brain-permeable inhibitor of the neurodegenerative disease target kynurenine 3-monooxygenase prevents accumulation of neurotoxic metabolites. Commun Biol. 2019;2:271. doi: 10.1038/s42003-019-0520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohnert Jürgen A., Karamian B, Nikaido H. Optimized Nile red efflux assay of AcrAB-TolC multidrug efflux system shows competition between substrates. Antimicrob Agents Chemother. 2010;54:3770–3775. doi: 10.1128/AAC.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards BS, Young SM, Ivnitsky-Steele I, RD Y, Prossnitz ER. High-content screening: flow cytometry analysis. Methods Mol Biol. 2009;486:151–165. doi: 10.1007/978-1-60327-545-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivnitski-Steele I, Larson RS, Lovato DM, Khawaja HM, Winter SS, et al. High-throughput flow cytometry to detect selective inhibitors of ABCB1, ABCC1, and ABCG2 transporters. Assay Drug Dev Technol. 2008;6:263–276. doi: 10.1089/adt.2007.107. [DOI] [PubMed] [Google Scholar]
- 36.Strouse JJ, Ivnitski-Steele I, Waller A, Young SM, Perez D, et al. Fluorescent substrates for flow cytometric evaluation of efflux inhibition in ABCB1, ABCC1, and ABCG2 transporters. Anal Biochem. 2013;437:77–87. doi: 10.1016/j.ab.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer R, Ferrari A, Rijnbrand R, Erwin AL. A fluorescent microplate assay quantifies bacterial efflux and demonstrates two distinct compound binding sites in AcrB. Antimicrob Agents Chemother. 2015;59:2388–2397. doi: 10.1128/AAC.05112-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer R, Erwin AL. Direct measurement of efflux in Pseudomonas aeruginosa using an environment-sensitive fluorescent dye. Res Microbiol. 2015;166:516–524. doi: 10.1016/j.resmic.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 39.O'Hagan S, Kell DB. Consensus rank orderings of molecular fingerprints illustrate the most genuine similarities between marketed drugs and small endogenous human metabolites, but highlight exogenous natural products as the most important ‘natural’ drug transporter substrates. Admet Dmpk. 2017;5:85–125. doi: 10.5599/admet.5.2.376. [DOI] [Google Scholar]
- 40.Wurm M, Ilhan S, Jandt U, Zeng A-P. Direct and highly sensitive measurement of fluorescent molecules in bulk solutions using flow cytometry. Anal Biochem. 2019;570:32–42. doi: 10.1016/j.ab.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli . Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 42.Yan K-K, Fang G, Bhardwaj N, Alexander RP, Gerstein M. Comparing genomes to computer operating systems in terms of the topology and evolution of their regulatory control networks. Proc Natl Acad Sci U S A. 2010;107:9186–9191. doi: 10.1073/pnas.0914771107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sastry AV, Gao Y, Szubin R, Hefner Y, Xu S, et al. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat Commun. 2019;10 doi: 10.1038/s41467-019-13483-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos-Zavaleta A, Salgado H, Gama-Castro S, Sánchez-Pérez M, Gómez-Romero L, et al. RegulonDB v 10.5: tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 2019;47:D212–D220. doi: 10.1093/nar/gky1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broadhurst DI, Kell DB. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics. 2007;2:171–196. doi: 10.1007/s11306-006-0037-z. [DOI] [Google Scholar]
- 46.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data mining, Inference and Prediction. 2nd ed. Berlin: Springer-Verlag; 2009. [Google Scholar]
- 47.Kell DB, Morries JG. Formulation and some biological uses of a buffer mixture whose buffering capacity is relatively independent of pH in the range pH 4–9. J Biochem Biophys Methods. 1980;3:143–150. doi: 10.1016/0165-022X(80)90013-5. [DOI] [PubMed] [Google Scholar]
- 48.Kell DB, Ferguson SJ, John P. Measurement by a flow dialysis technique of the steady-state proton-motive force in chromatophores from Rhodospirillum rubrum. Comparison with phosphorylation potential. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1978;502:111–126. doi: 10.1016/0005-2728(78)90136-6. [DOI] [PubMed] [Google Scholar]
- 49.Weininger D. Smiles, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Model. 1988;28:31–36. doi: 10.1021/ci00057a005. [DOI] [Google Scholar]
- 50.O′Hagan S, Swainston N, Handl J, Kell DB. A ‘rule of 0.5’ for the metabolite-likeness of approved pharmaceutical drugs. Metabolomics. 2015;11:323–339. doi: 10.1007/s11306-014-0733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willett P. Similarity‐based data mining in files of two‐dimensional chemical structures using fingerprint measures of molecular resemblance. WIREs Data Min Knowl. 2011;1:241–251. doi: 10.1002/widm.26. [DOI] [Google Scholar]
- 52.Gasteiger J. Handbook of Chemoinformatics: From Data to Knowledge. Weinheim: Wiley/VCH; 2003. [Google Scholar]
- 53.Guha R, Bender A. Computational Approaches in Cheminformatics and Bioinformatics. Hoboken, NJ: Wiley; 2012. [Google Scholar]
- 54.Todeschini R, Consonni V. Molecular Descriptors for Cheminformatics. Weinheim: WILEY-VCH Verlag GmbH; 2009. [Google Scholar]
- 55.Shabir G, Saeed A, Ali Channar P. A review on the recent trends in synthetic strategies and applications of xanthene dyes. Mini Rev Org Chem. 2018;15:166–197. doi: 10.2174/1570193X14666170518130008. [DOI] [Google Scholar]
- 56.Baldi P, Nasr R. When is chemical similarity significant? the statistical distribution of chemical similarity scores and its extreme values. J Chem Inf Model. 2010;50:1205–1222. doi: 10.1021/ci100010v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hähnke V, Rupp M, Hartmann AK, Schneider G. Pharmacophore Alignment Search Tool (PhAST): significance assessment of chemical similarity. Mol Inform. 2013;32:625–646. doi: 10.1002/minf.201300021. [DOI] [PubMed] [Google Scholar]
- 58.Riniker S, Landrum GA. Open-source platform to benchmark fingerprints for ligand-based virtual screening. J Cheminform. 2013;5:26. doi: 10.1186/1758-2946-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Boyle NM, Sayle RA. Comparing structural fingerprints using a literature-based similarity benchmark. J Cheminform. 2016;8:36. doi: 10.1186/s13321-016-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aigner D, Ungerböck B, Mayr T, Saf R, Klimant I, et al. Fluorescent materials for pH sensing and imaging based on novel 1,4-diketopyrrolo-[3,4-c]pyrrole dyes. J Mater Chem C Mater. 2013;1:5685–5693. doi: 10.1039/c3tc31130a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bailey AM, Paulsen IT, Piddock LJV. RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob Agents Chemother. 2008;52:3604–3611. doi: 10.1128/AAC.00661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lawler AJ, Ricci V, Busby SJW, Piddock LJV. Genetic inactivation of acrAB or inhibition of efflux induces expression of ramA . J Antimicrob Chemother. 2013;68:1551–1557. doi: 10.1093/jac/dkt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baugh S, Phillips CR, Ekanayaka AS, Piddock LJV, Webber MA. Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J Antimicrob Chemother. 2014;69:673–681. doi: 10.1093/jac/dkt420. [DOI] [PubMed] [Google Scholar]
- 64.Davey HM, Davey CL, Kell DB. On the determination of the size of microbial cells using flow cytometry. In: Lloyd D, editor. Flow Cytometry in Microbiology. London: Springer-Verlag; 1993. pp. 49–65. editor. [Google Scholar]
- 65.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 66.X-Z L, Elkins CA, Zgurskaya HI. Efflux-mediated Antimicrobial Resistance in Bacteria: Mechanisms, Regulation and Clinical Implications. Berlin: Springer; 2016. [Google Scholar]
- 67.The PLJV. 2019 Garrod Lecture: MDR efflux in Gram-negative bacteria-how understanding resistance led to a new tool for drug discovery. J Antimicrob Chemother. 2019;74:3128–3134. doi: 10.1093/jac/dkz370. [DOI] [PubMed] [Google Scholar]
- 68.Schuldiner S. The Escherichia coli effluxome. Res Microbiol. 2018;169:357–362. doi: 10.1016/j.resmic.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 69.Law CJ, Alegre KO. Clamping down on drugs: the Escherichia coli multidrug efflux protein MdtM. Res Microbiol. 2018;169:461–467. doi: 10.1016/j.resmic.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 70.Puzari M, Chetia P. RND efflux pump mediated antibiotic resistance in Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa: a major issue worldwide. World J Microbiol Biotechnol. 2017;33:24. doi: 10.1007/s11274-016-2190-5. [DOI] [PubMed] [Google Scholar]
- 71.Anes João, McCusker MP, Fanning Séamus, Martins M, Anes J, et al. The ins and outs of RND efflux pumps in Escherichia coli . Front Microbiol. 2015;6:587. doi: 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Darbani B, Kell DB, Borodina I. Energetic evolution of cellular transportomes. BMC Genomics. 2018;19:418. doi: 10.1186/s12864-018-4816-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Griffith JM, Basting PJ, Bischof KM, Wrona EP, Kunka KS. Experimental evolution of Escherichia coli K-12 in the Presence of Proton Motive Force (PMF) uncoupler carbonyl cyanide m-Chlorophenylhydrazone selects for mutations affecting PMF-driven drug efflux pumps. Appl Environ Microbiol. 2019;85 doi: 10.1128/AEM.02792-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kell DB, Swainston N, Pir P, Oliver SG. Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol. 2015;33:237–246. doi: 10.1016/j.tibtech.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 75.Kell DB. Control of metabolite efflux in microbial cell factories: current advances and future prospects. In: El-Mansi EMT, Nielsen J, Mousdale D, Allman T, Carlson R, editors. Fermentation Microbiology and Biotechnology. 4th ed. Boca Raton: CRC Press; 2019. pp. 117–138. [Google Scholar]
- 76.Koronakis V. TolC - the bacterial exit duct for proteins and drugs. FEBS Lett. 2003;555:66–71. doi: 10.1016/S0014-5793(03)01125-6. [DOI] [PubMed] [Google Scholar]
- 77.Du D, Wang Z, James NR, Voss JE, Klimont E, et al. Structure of the AcrAB–TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krishnamoorthy G, Tikhonova EB, Dhamdhere G, Zgurskaya HI. On the role of TolC in multidrug efflux: the function and assembly of AcrAB-TolC tolerate significant depletion of intracellular TolC protein. Mol Microbiol. 2013;87:982–997. doi: 10.1111/mmi.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zgurskaya HI, Krishnamoorthy G, Ntreh A, Lu S. Mechanism and function of the outer membrane channel TolC in multidrug resistance and physiology of enterobacteria. Front Microbiol. 2011;2:189. doi: 10.3389/fmicb.2011.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zgurskaya HI, Rybenkov VV, Krishnamoorthy G, Leus IV. Trans-envelope multidrug efflux pumps of Gram-negative bacteria and their synergism with the outer membrane barrier. Res Microbiol. 2018 doi: 10.1016/j.resmic.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamamoto K, Tamai R, Yamazaki M, Inaba T, Sowa Y, et al. Substrate-dependent dynamics of the multidrug efflux transporter AcrB of Escherichia coli . Sci Rep. 2016;6:21909. doi: 10.1038/srep21909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Zwaig RN, Luria SE. Genetics and physiology of colicin-tolerant mutants of Escherichia coli . J Bacteriol. 1967;94:1112–1123. doi: 10.1128/JB.94.4.1112-1123.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morona R, Reeves P. The tolC locus of Escherichia coli affects the expression of three major outer membrane proteins. J Bacteriol. 1982;150:1016–1023. doi: 10.1128/JB.150.3.1016-1023.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF Archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 85.Mori H, Baba T, Yokoyama K, Takeuchi R, Nomura W. Identification of essential genes and synthetic lethal gene combinations in Escherichia coli K-12. Methods Mol Biol. 2015;1279:45–65. doi: 10.1007/978-1-4939-2398-4_4. [DOI] [PubMed] [Google Scholar]
- 86.Forster S, Thumser AE, Hood SR, Plant N. Characterization of rhodamine-123 as a tracer dye for use in in vitro drug transport assays. PLoS One. 2012;7:e33253. doi: 10.1371/journal.pone.0033253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haynes MK, Garcia M, Peters R, Waller A, Tedesco P. High-Throughput flow cytometry screening of multidrug efflux systems. Methods Mol Biol. 1700;2018:293–318. doi: 10.1007/978-1-4939-7454-2_16. [DOI] [PubMed] [Google Scholar]
- 88.O'Hagan S, Kell DB. Structural similarities between some common fluorophores used in biology and marketed drugs, endogenous metabolites, and natural products. bioRxiv. 2019;834325 doi: 10.3390/md18110582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Troutman MD, Thakker DR. Rhodamine 123 requires carrier-mediated influx for its activity as a P-glycoprotein substrate in Caco-2 cells. Pharm Res. 2003;20:1192–1199. doi: 10.1023/A:1025096930604. [DOI] [PubMed] [Google Scholar]
- 90.Jernaes MW, Steen HB. Staining ofEscherichia coli for flow cytometry: influx and efflux of ethidium bromide. Cytometry. 1994;17:302–309. doi: 10.1002/cyto.990170405. [DOI] [PubMed] [Google Scholar]
- 91.Walberg M, Gaustad P, Steen HB. Uptake kinetics of nucleic acid targeting dyes in S. aureus, E. faecalis and B. cereus: a flow cytometric study. J Microbiol Methods. 1999;35:167–176. doi: 10.1016/S0167-7012(98)00118-3. [DOI] [PubMed] [Google Scholar]
- 92.Sims PJ, Waggoner AS, Wang C-H, Hoffman JF. Mechanism by which cyanine dyes measure membrane potential in red blood cells and phosphatidylcholine vesicles. Biochemistry. 1974;13:3315–3330. doi: 10.1021/bi00713a022. [DOI] [PubMed] [Google Scholar]
- 93.te Winkel JD, Gray DA, Seistrup KH, Hamoen LW, Strahl H. Analysis of antimicrobial-triggered membrane depolarization using voltage sensitive dyes. Front Cell Dev Biol. 2016;4:29. doi: 10.3389/fcell.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Šíp M, Heřman P, Plášek J, Hrouda V. Transmembrane potential measurement with carbocyanine dye diS-C3-(5): fast fluorescence decay studies. J Photochem Photobiol B. 1990;4:321–328. doi: 10.1016/1011-1344(90)85037-W. [DOI] [PubMed] [Google Scholar]
- 95.Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR green I, its structure determination and methodological implications. Nucleic Acids Res. 2004;32:e103. doi: 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nescerecka A, Hammes F, Juhna T. A pipeline for developing and testing staining protocols for flow cytometry, demonstrated with SYBR green I and propidium iodide viability staining. J Microbiol Methods. 2016;131:172–180. doi: 10.1016/j.mimet.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 97.Van Nevel S, Koetzsch S, Proctor CR, Besmer MD, Prest EI, et al. Flow cytometric bacterial cell counts challenge conventional heterotrophic plate counts for routine microbiological drinking water monitoring. Water Res. 2017;113:191–206. doi: 10.1016/j.watres.2017.01.065. [DOI] [PubMed] [Google Scholar]
- 98.Noble RT, Fuhrman JA. Use of SYBR green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. doi: 10.3354/ame014113. [DOI] [Google Scholar]
- 99.Patel A, Noble RT, Steele JA, Schwalbach MS, Hewson I, et al. Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR green I. Nat Protoc. 2007;2:269–276. doi: 10.1038/nprot.2007.6. [DOI] [PubMed] [Google Scholar]
- 100.Grégori Gérald, Citterio S, Ghiani A, Labra M, Sgorbati S, et al. Resolution of viable and membrane-compromised bacteria in freshwater and marine waters based on analytical flow cytometry and nucleic acid double staining. Appl Environ Microbiol. 2001;67:4662–4670. doi: 10.1128/AEM.67.10.4662-4670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Habtewold T, Duchateau L, Christophides GK. Flow cytometry analysis of the microbiota associated with the midguts of vector mosquitoes. Parasit Vectors. 2016;9:167. doi: 10.1186/s13071-016-1438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dragan AI, Pavlovic R, McGivney JB, Casas-Finet JR, Bishop ES, et al. SYBR green I: fluorescence properties and interaction with DNA. J Fluoresc. 2012;22:1189–1199. doi: 10.1007/s10895-012-1059-8. [DOI] [PubMed] [Google Scholar]
- 103.Bajorath J. Chemoinformatics for Drug Discovery. Hoboken: Wiley; 2014. [Google Scholar]
- 104.Bunin BA, Siesel B, Morales GA, Bajorath J. Chemoinformatics: Theory, Practice and Products. Heidelberg: Springer; 2007. [Google Scholar]
- 105.Faulon J-L, Bender A. Handbook of Chemoinformatics Algorithms. London: CRC Press; 2010. [Google Scholar]
- 106.Leach AR, Gillet VJ. An Introduction to Chemoinformatics. Revised edition. Springer: Dordrecht; 2007. [Google Scholar]
- 107.Oprea TI. Chemoinformatics in Drug Discovery. Weinheim: Wiley/VCH; 2004. [Google Scholar]
- 108.Bender A, Glen RC. Molecular similarity: a key technique in molecular informatics. Org Biomol Chem. 2004;2:3204–3218. doi: 10.1039/b409813g. [DOI] [PubMed] [Google Scholar]
- 109.Monev V. Introduction to similarity searching in chemistry. MATCH-Commun Math Comput Chem. 2004;51:7–38. [Google Scholar]
- 110.Willett P. The calculation of molecular structural similarity: principles and practice. Mol Inform. 2014;33:403–413. doi: 10.1002/minf.201400024. [DOI] [PubMed] [Google Scholar]
- 111.Landon MR, Schaus SE. JEDA: joint entropy diversity analysis. An information-theoretic method for choosing diverse and representative subsets from combinatorial libraries. Mol Divers. 2006;10:333–339. doi: 10.1007/s11030-006-9042-4. [DOI] [PubMed] [Google Scholar]
- 112.Weidlich IE, Filippov IV. Using the Gini coefficient to measure the chemical diversity of small-molecule libraries. J Comput Chem. 2016;37:2091–2097. doi: 10.1002/jcc.24423. [DOI] [PubMed] [Google Scholar]
- 113.O'Hagan S, Kell DB. MetMaxStruct: a Tversky-similarity-based strategy for analysing the (sub)structural similarities of drugs and endogenous metabolites. Front Pharmacol. 2016;7:266. doi: 10.3389/fphar.2016.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O’Hagan S, Kell DB. Analysing and navigating natural products space for generating small, diverse, but representative chemical libraries. Biotechnol J. 2018;13:1700503. doi: 10.1002/biot.201700503. [DOI] [PubMed] [Google Scholar]
- 115.Hockney RC. Recent developments in heterologous protein production in Escherichia coli . Trends Biotechnol. 1994;12:456–463. doi: 10.1016/0167-7799(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 116.Grenier F, Matteau D, Baby V, Rodrigue S. Complete genome sequence of Escherichia coli BW25113. Genome Announc. 2014;2:e01038-14. doi: 10.1128/genomeA.01038-14. 16 10 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cardinale S, Tueros FG, Sommer MOA. Genetic-Metabolic coupling for targeted metabolic engineering. Cell Rep. 2017;20:1029–1037. doi: 10.1016/j.celrep.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 118.Cardinale S, Cambray G. Genome-wide analysis of E. coli cell-gene interactions. BMC Syst Biol. 2017;11:112. doi: 10.1186/s12918-017-0494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sargentini NJ, Gularte NP, Hudman DA. Screen for genes involved in radiation survival of Escherichia coli and construction of a reference database. Mutat Res. 2016;793-794:1–14. doi: 10.1016/j.mrfmmm.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 120.Saier MH, Reddy VS, Tamang DG, Västermark A, Västermark A. The transporter classification database. Nucleic Acids Res. 2014;42:D251–D258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Saier MH, Reddy VS, Tsu BV, Ahmed MS, Li C, et al. The transporter classification database (tcdB): recent advances. Nucleic Acids Res. 2016;44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Herzberg M, Kaye IK, Peti W, Wood TK. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J Bacteriol. 2006;188:587–598. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yamamoto K, Nonaka G, Ozawa T, Takumi K, Ishihama A. Induction of the Escherichia coli yijE gene expression by cystine. Biosci Biotechnol Biochem. 2015;79:218–222. doi: 10.1080/09168451.2014.972328. [DOI] [PubMed] [Google Scholar]
- 124.Masi M, Pagès J-M. Structure, function and regulation of outer membrane proteins involved in drug transport in Enterobactericeae: the OmpF/C - TolC Case. Open Microbiol J. 2013;7:22–33. doi: 10.2174/1874285801307010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Müller RT, Pos KM. The assembly and disassembly of the AcrAB-TolC three-component multidrug efflux pump. Biol Chem. 2015;396:1083–1089. doi: 10.1515/hsz-2015-0150. [DOI] [PubMed] [Google Scholar]
- 126.Weston N, Sharma P, Ricci V, Piddock LJV. Regulation of the AcrAB-TolC efflux pump in Enterobacteriaceae. Res Microbiol. 2018;169:425-431. doi: 10.1016/j.resmic.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 127.Featherstone DE, Broadie K. Wrestling with pleiotropy: genomic and topological analysis of the yeast gene expression network. Bioessays. 2002;24:267–274. doi: 10.1002/bies.10054. [DOI] [PubMed] [Google Scholar]
- 128.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Theodoulou FL, Miller AJ. Xenopus oocytes as a heterologous expression system. Methods Mol Biol. 1995;49:317–340. doi: 10.1385/0-89603-321-X:317. [DOI] [PubMed] [Google Scholar]
- 130.Darbani B, Stovicek V, van der Hoek SA, Borodina I. Engineering energetically efficient transport of dicarboxylic acids in yeast Saccharomyces cerevisiae . Proc Natl Acad Sci U S A. 2019;116:19415–19420. doi: 10.1073/pnas.1900287116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kaprelyants AS, Kell DB. Rapid assessment of bacterial viability and vitality by rhodamine 123 and flow cytometry. J Appl Bacteriol. 1992;72:410–422. doi: 10.1111/j.1365-2672.1992.tb01854.x. [DOI] [Google Scholar]
- 132.Mason DJ, Shanmuganathan S, Mortimer FC, Gant VA. A fluorescent Gram stain for flow cytometry and epifluorescence microscopy. Appl Environ Microbiol. 1998;64:2681–2685. doi: 10.1128/AEM.64.7.2681-2685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Currin A, Swainston N, Day PJ, Kell DB. SpeedyGenes: a novel approach for the efficient production of error-corrected, synthetic gene libraries. Protein Eng Design Sel. 2014;27:273–280. doi: 10.1093/protein/gzu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Currin A, Swainston N, Day PJ, Kell DB. Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently. Chem Soc Rev. 2015;44:1172–1239. doi: 10.1039/C4CS00351A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Currin A, Kwok J, Sadler JC, Bell EL, Swainston N, et al. GeneORator: an effective strategy for navigating protein sequence space more efficiently through Boolean OR-type DNA libraries. ACS Synth Biol. 2019;8:1371–1378. doi: 10.1021/acssynbio.9b00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.