Abstract
Complete tumor extirpation with clear surgical margins remains a central tenet of oncologic head and neck surgery. Rates of locoregional recurrence and survival are both significantly worse when clear margins are unable to be obtained. Current clinical practice relies on the use of frozen sections intra-operatively, followed by traditional histopathologic analysis post-operatively to assess the surgical margin. However, with improved understanding of tumor biology and advances in technology, new techniques have emerged to analyze margins at a molecular level. Such molecular margin analysis interrogates tissue for genetic, epigenetic, or proteomic changes that may belie tumor presence or aggressive features not captured by standard histopathologic techniques. Intra-operatively, this information may be used to guide resection, while post-operatively, it may help to stratify patients for adjuvant treatment. In this review, we summarize the current state of molecular margin analysis and describe directions for future research.
Keywords: Head and neck cancer, margin analysis, molecular margin, next-generation sequencing, fluorescence, spectroscopy
Successful surgical management of head and neck squamous cell carcinoma (HNSCC) remains contingent upon achieving complete tumor extirpation. Indeed, margin status remains one of the few histopathological features potentially in the surgeon’s control and represents the most significant prognostic factor associated with locoregional control: Positive margins increase locoregional recurrence (LRR) rates from roughly 20% to up to 90%, with grave detriments in overall survival.1–6 However, obtaining clear margins must be carefully balanced against preserving a patient’s function and quality of life, particularly in the context of intricate head and neck anatomy. Current clinical practice relies upon intra-operative frozen sections to guide the extent of resection, whereas the final post-operative histopathological assessment determines appropriate staging and directs decision-making regarding adjuvant therapy.7
Despite the significance of margin status as it relates to prognosis and intra-operative decision-making, a number of controversies persist. There remains heterogeneity in the definition of what constitutes an “adequate” or “negative” margin, and substantial variations exist in how both surgeons and pathologists obtain, sample, and analyze margins. Furthermore, the rate of positive margins in head and neck cancer remains relatively high at between 15–30%.4,8,9 In light of these observations, there has been interest in developing methods for characterizing margins on a molecular level, both intra-operatively and post-operatively. This includes analyzing margins for genetic and epigenetic changes of select molecular markers, as well as utilizing cancer-specific molecules as targets for antibodies coupled to near-infrared (NIR) fluorescent agents. In this review, we aim to systematically characterize the current state of molecular margin analysis, as well as the advancement and potential future applicability of such technologies.
It is clear that additive, progressive genetic mutations ultimately contribute to tumor progression.10 However, during pre-malignant stages of disease, these genetic alterations may not manifest morphologically, and therefore may be undetectable using standard histopathologic margin evaluation. This limitation, in conjunction with the inherent shortcomings of frozen-section analysis, suggest that additional molecular analysis may offer meaningful clinical benefit. Molecular margin analysis combines our expanding understanding of tumor biology with well-established methods of histological evaluation to improve disease identification and accordingly, an individual’s risk of recurrent disease. Through increasing the sensitivity of detecting residual cancer cells or the presence of high-risk pre-malignant changes, molecular margin analysis may also facilitate the identification of patients who would benefit from adjuvant therapy. While the field remains nascent, encouraging findings suggest significant value added from molecular margin analysis as it relates to real-time surgical decision-making and informing patient prognosis and post-operative treatment.
Numerous techniques have been studied, with varying degrees of success and distinct strengths and weaknesses. Broadly, the status of molecular margins can be evaluated through interrogation of genetic mutations, epigenetic changes, and protein markers. Below we outline various molecular methods used to analyze margins, highlighting the most promising techniques under investigation for HNSCC.
Genetic mutations and protein expression
P53
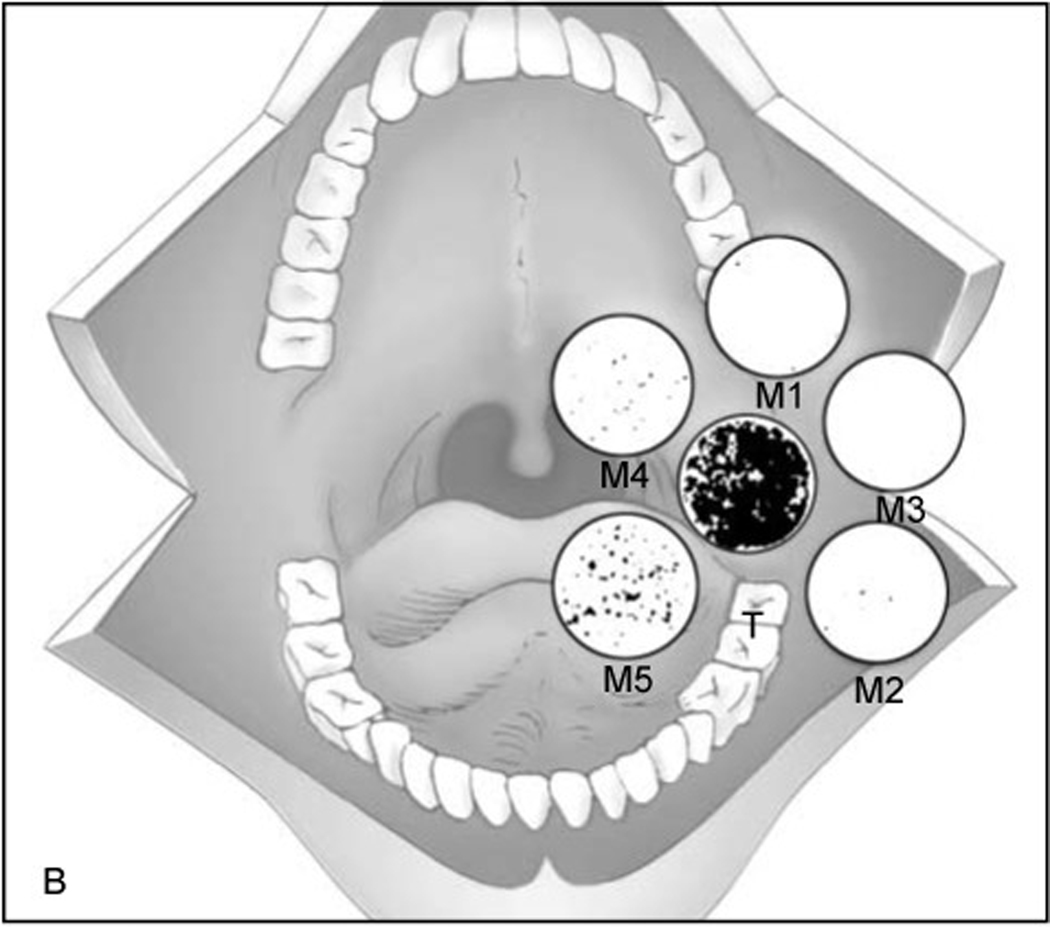
TP53 is a tumor suppressor gene frequently implicated in carcinogenesis, with mutations found in up to 60% of HNSCC tumors.11 Furthermore, such mutations have been established as predictive clonal markers for HNSCC.12 In their seminal study evaluating molecular surgical margins, Brennan and colleagues assessed histologically negative margins for TP53 mutations in 25 TP53-mutant HNSCC tumors by polymerase chain reaction (PCR) (Figure 1). Interestingly, over half of patients harbored TP53 mutations by molecular margin analyses. The authors saw a significantly higher rate of recurrence among those with molecularly positive margins (5 of 13) versus those without (0 of 12).13 These findings led van Houten et al. to evaluate these changes in a larger prospective cohort of 76 patients.14 They found molecularly positive margins to be a strong independent predictor of LRR (Relative Risk (RR) = 7.1; p = 0.021; 95% confidence interval (CI), 0.9 – 56).14 A number of follow-up studies validated these results, demonstrating the presence of TP53 mutations within conventionally clear margins to be associated with increased LRR.15–17 Murillo and colleagues found the presence of TP53 mutations to be a better predictor of recurrence than other established clinicopathological parameters in oral cavity squamous cell carcinoma (OCSCC). Interestingly, this association was significant only if resection margins were ≤ 5mm, suggesting TP53 analysis may be less useful in the context of widely cleared margins.15 Huang et al. found that molecularly positive deep margins were more predictive of LRR than positive mucosal margins.16 In contrast to these DNA-based studies, multiple reports analyzing p53 overexpression and nuclear immunostaining using immunohistochemistry (IHC) have not shown a significant correlation between molecularly positive margins and LRR, perhaps reflecting a greater degree of sensitivity with PCR-based methods than IHC.18–20
Figure 1.
Figure adapted from Brennan et al. T represents the primary tumor and M1-M5 represent margins. Mutant specific oligomers were identified for each patient and oligonucleotide probes were hybridized with phosphorous-32 combined with DNA extracted from margin tissue. Radiographs were obtained with hybridizing plaques being radio-opaque and representing presence of p53 mutation. 38% of patients with positive p53 margins recurred locally compared to 0% of those with negative p53 margins. RightsLink license number 4870271313736.
Several important factors limit the applicability of this method. Only 40–60% of HNSCCs have been shown to harbor TP53 mutations, restricting analysis to this subpopulation.11,21 Furthermore, the myriad of potential TP53 mutations makes it difficult to design a comprehensive assay capable of detecting all variants, and this heterogeneity also limits the ability to prognosticate as distinct mutations may carry variable clinical significance.21–24 TP53 mutations have also been shown to have a strong association with dysplastic lesions, potentially confounding interpretation in the presence of field cancerization.25,26 Utility is also constrained by the capacity to sample and analyze the entire surgical margin for such mutations. However, next-generation high-throughput sequencing has the potential to provide more accelerated, extensive, and quantitative genetic analysis,27–31 offering solutions to some of these impediments and ultimately facilitating the more sizeable prospective studies needed to validate such approaches.
eIF4E
In an effort to overcome some of the limitations posed by TP53 analysis, authors have looked at using eukaryotic translocation initiation factor 4E (eIF4E) as a molecular marker. eIF4E is involved in the initiation of protein synthesis and is overexpressed in most HNSCCs, but not benign processes or normal mucosa.32,33 Nathan and colleagues prospectively assessed 65 HNSCC patients for the presence of eIF4E in histopathologically negative margins using IHC. All primary tumors overexpressed eIF4E, and 36 patients had positive molecular margins. Positive eIF4E margins portended a 7-fold increased risk of LRR (56% recurrence in eIF4E margin-positive patients vs. only 6.9% of margin-negative patients).34 Numerous other studies similarly showed eIF4E overexpression in surgical margins to be associated with LRR.19,35–37 While eIF4E offers a more sensitive and logistically practical method of molecular margin analysis, larger studies are needed to standardize methods and validate these findings.
Other Molecular Markers
In addition to TP53 and eIF4E, Cyclin D1, a G1 checkpoint regulating oncogene, may be another potential biomarker used in molecular margin analysis. Sakashita et al. found that 65.6% of 116 HNSCC patients had cyclin D1-positive tumors, which was overexpressed in 46.6% of histologically negative margins. Cyclin D1-positive margin status was an independent prognostic indicator for local recurrence (hazard ratio (HR) = 4.58) and associated with significantly lower local control rates (77.2% vs 91.5% respectively).38
Similarly, Zhao and colleagues used IHC to retrospectively appraise 112 laryngeal tumors with histologically negative margins for the expression of survivin, a gene involved in cell division and apoptosis regulation, and CD44v6, an adhesion molecule associated with advanced stage malignancies. Survivin and CD44v6 were expressed in 67.9% and 72.3% of the index tumors, respectively. Survivin (39.3%) and/or CD44v6 (31.3%) margin-positivity was independently predictive of recurrence (survivin(+) OR = 8.4; CD44v6(+) OR = 6.9) and disease free survival.39
Schaaij-Visser and colleagues conducted a three-phased retrospective case-control study by performing an initial proteomics-based discovery screen and evaluation of candidate biomarkers in neoplastic and normal tissue, followed by clinical validation of two selected biomarkers.40 They established 40 differentially expressed proteins, identifying keratin 4, a protein involved in differentiation, and cornulin, a protein forming the protective cornified envelope in mucosa and skin, to be the most promising. Among 46 HNSCC cases with histologically negative margins, the loss of expression of keratin 4, cornulin, or both on IHC analysis was associated with the development of local recurrence (HR = 8.8; p = 0.0005).40 Though their results must be validated in larger prospective cohorts, they highlight the potential value of novel biomarker discovery using proteomic analysis.
De Carvalho and colleagues evaluated 55 HNSCC cases for differential gene expression using quantitative reverse transcription (qRT)-PCR analyses in histologically negative margins, focusing on the genes PTHLH (parathyroid hormone-like hormone), EPCAM (epithelial cellular adhesion molecule), MMP9 (metallopreoteinase-9), LGALS1 (galectin 1) and MET (proto-oncogene). MMP9 overexpression within surgical margins was associated with decreased local control (p = 0.022; HR = 4.186; p = 0.035), whereas overexpression of PTHLH, a mediator of EGFR (epidermal growth factor receptor), was associated with the development of second primaries (p = 0.002). They demonstrate that qRT-PCR may be a useful adjunct approach for margin evaluation, though additional validation is necessary.
Microsatellite alterations
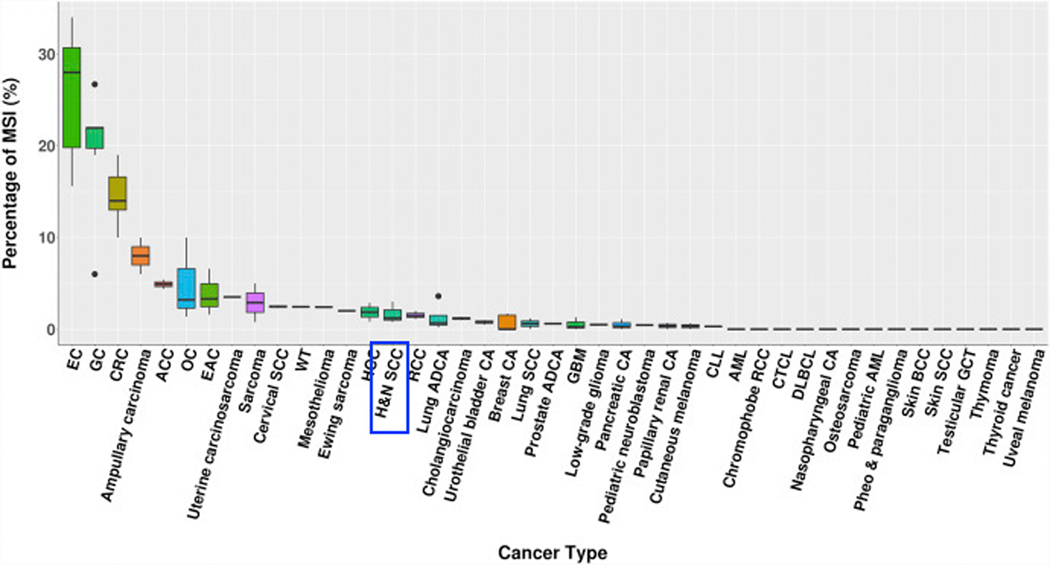
Microsatellites are repetitive segments of DNA found throughout the genome that are inherently vulnerable to mutation. Subsequent mutation accumulation in crucial genes can result in carcinogenesis. Microsatellite instability (MSI) and loss of heterozygosity (LOH) are two forms of microsatellite alterations commonly implicated in tumor formation. Analysis of The Cancer Genome Atlas (TCGA) has demonstrated MSI to play a key role in carcinogenesis in nearly all anatomic sites, most significantly in abdominal malignancies (Figure 2).41,108 MSI occurs secondary to defects in DNA mismatch repair proteins, ultimately resulting in an abnormal length of microsatellite repeats. MSI can be readily detected amongst clonally expanded cells and may permit identification of cells with malignant potential, particularly in the setting of surrounding normal cells.42 LOH signifies the loss of one paired allele or genetic sequence, which is thought to be associated with tumor suppressor gene deactivation.
Figure 2.
Figure adapted from Shirazi and Sepulveda [108]. Rates of tumors with microsatellite instability (MSI) by anatomic site as reported in the literature via analyses of The Cancer Genome Atlas. Head and neck squamous cell carcinoma (HNSCC) is identified; 0.78–3.0% of HNSCC tumors have been shown to demonstrate MSI. Rates of MSI are higher in endometrial (EC), gastric (GC), and colorectal (CRC) carcinomas. RightsLink license number 4870280021085.
Multiple authors have shown the presence of MSI in surgical margins predicts local recurrence.42–45 Temam and colleagues found two tetranucleotide microsatellite markers commonly altered in HNSCC to have a strong independent association with local recurrence when prospectively analyzing histologically negative margins in 54 HNSCCs (p = 0.01; HR = 6.5).42 However, only 48% of tumors harbored MSI, though 5 of 7 recurrences occurred in patients with MSI-positive margins.42 Similarly, Liu et al. found the presence of MSI in histologically negative margins to increase the risk of local recurrence in 145 patients with OCSCC using six dinucleotide microsatellite markers (OR = 7.2, 95% CI, 3.5 – 14.7).45 Fifty-five patients had MSI-positive primaries, of which 41 had MSI-positive margins. Fifty-four percent of those with MSI-positive margins developed recurrence, versus only 29% in the MSI-negative cohort.
A number of authors have also explored the role of LOH in molecular margin analysis.43,45–47 Graveland and colleagues retrospectively evaluated the histologically tumor-free surgical margins of 35 HNSCC patients for LOH at chromosomes 3p, 9p and 17p, as well as for p53 and Ki-67 immunostaining. Seventeen of 35 patients had LOH, while only 13 showed p53-positivity in the surgical margin. LOH at 9p had the strongest positive predict value for local recurrence (HR = 3.2, p = 0.027). However, when combined with the presence of significant p53 staining, it offered even greater prognostic significance (HR = 7.0, p = 0.01). These findings are not surprising, as CDKN2A is located on chromosome 9p21, which encodes cell cycle regulatory proteins p14 and p16, the loss of which has been associated with recurrence in OCSCC.48 Similarly, Sardi and colleagues retrospectively evaluated 41 HNSCC patients using 10 microsatellite markers with various chromosome localizations for the presence of LOH. They found LOH-margin positivity to be independently predictive of recurrence (log rank test, p = 0.0049).43
Microsatellite marker margin analysis offers a relatively simple and sensitive method compared to detecting specific mutations in individual genes. However, while screening for numerous markers theoretically improves feasibility and scalability, substantial limitations persist, as only up to 60% of analyzed specimens tested positive in any of the above studies for selected microsatellite alterations. Furthermore, such biomarkers lack specificity and may exist in pre-malignant lesions.49 They may also lack sensitivity for detecting a few altered cells among a large population of normal cells.43
Copy number alteration
Copy number alteration (CNA), or the gain or loss of segmental or entire chromosomes, is an accepted marker for malignant transformation. More recently, it has been evaluated for use in the molecular detection of neoplastic potential among histologically negative surgical margins. CNA in chromosomes 1 and 7 have been shown to correlate to dysplastic phenotypes in HNSCC. In 20 cases of OCSCC evaluated by interphase fluorescence in situ hybridization (I-FISH), van der Toorn and colleagues found CNA in 88% of samples with carcinoma in situ or SCC, versus only 16% in hyperplastic areas.50 In a follow-up study evaluating 19 of these patients, CNA among chromosomes 1 and 7 in histologically negative margins was associated with LRR (p = 0.018). Seven of 8 molecularly margin-positive and none of the 11 margin-negative patients developed recurrence.20 Handschel and colleagues evaluated histologically negative margins of 40 OCSCC patients for chromosome aneuploidy using DNA-image cytometry. Twenty patients developed LRR, 14 of whom had positive molecular margins. Their margin analysis technique had a sensitivity and specificity of 70% and 90%, respectively.51 While quantitative DNA analysis offers a relatively inexpensive and expedient addition to standard histological evaluation, these findings need prospective validation. Future study is needed to identify the optimal chromosomes for focused I-FISH evaluation to ensure maximally sensitive and specific targets for HNSCC margin analysis.
Epigenetic markers
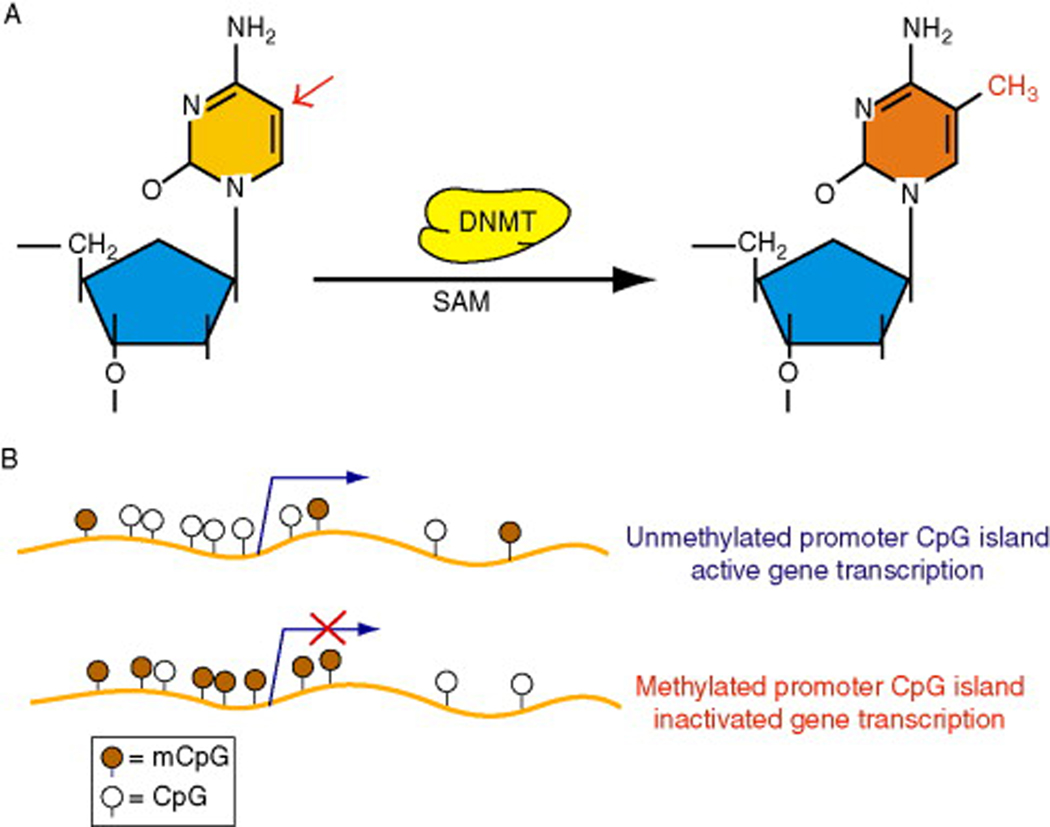
Epigenetic modifications to the genome, such as promoter methylation, can contribute to genetic instability and aberrant gene expression, ultimately leading to carcinogenesis (Figure 3). Epigenetics as it relates to tumor progression has been an area of increasing interest in HNSCC, in part, because mechanisms of resistance often develop through epigenetics. Thus, a deep understanding of epigenetic pathways may enable the development of novel therapeutics that directly prevent escape mechanisms from evolving. Promoter hypermethylation represents one of the most well-characterized epigenetic modifications and has been widely investigated in the context of molecular margin analysis.52–56
Figure 3.
Figure adapted from Kulis and Esteller. DNA methylation is represented. Panel A represents methylation of a cytosine base. In panel B the light brown circles represent an unmethylated promoter region which allows for gene transcription. The dark brown circles represent a methylated promoter region, silencing gene transcription. Methylation of tumor suppressor genes is a known factor in tumor progression. RightsLink license number 4870270425963.
In a recent study, Hayashi and colleagues conducted a prospective evaluation of deep margins in 65 HNSCC patients using quantitative methylation-specific PCR (QMSP). They evaluated tumors for the promoter methylation of six established tumor specific genes (deleted in colorectal cancer [DCC], endothelin receptor type B [EDNRB], homeobox protein A9 [HOXA9], kinesin family member 1A [KIF1A], nidogen-2 [NID2], and N-methyl D-aspartate receptor subtype 2B [NR2B]). Notably, only 66% of tumors had EDRNA and HOXA9 promoter methylation. Fifty-eight percent of EDNRB(+) tumors had EDNRB(+) margins, and 67% of HOXA9(+) tumors had HOXA9(+) margins. Only promoter methylation of the EDRNA and HOXA9 gene combination served as a significant predictor for LRR-free survival (HR = 3.31, p = 0.012).57 Previous studies by the same group established both genes as having high sensitivity and specificity for the diagnosis of HNSCC.58,59 Interestingly, among patients with histologically negative margins not receiving adjuvant radiation, those with positive molecular margins had significantly higher rates of local recurrence than those without. Conversely, no association between outcomes and margin-status was present among those that received adjuvant therapy. Such analyses suggest molecular margins may aid in the selection of appropriate radiation candidates.
Subsequently, this group prospectively evaluated the prognostic associations of PAX5 (paired box 5) promoter methylation in 75 patients; 68% of tumors showed PAX5 promoter methylation. Interestingly, PAX5 promoter methylation only had prognostic implications for patients not undergoing adjuvant radiation. Molecular margin positivity was significantly associated with LRR-free survival (HR = 3.89, p = 0.023) in this group of patients. Furthermore, they found that QMSP by droplet digital PCR (ddQMSP) offered increased sensitivity for methylation marker detection.60 Both QMSP and ddQMSP take roughly three hours to perform for a single patient.61
In the evaluation of 47 patients with histologically negative margins, Supic and colleagues found that DAPK (death associated protein kinase) promoter hypermethylation to be an independent predictor of overall survival (HR = 4.1, p = 0.007). While those with DAPK promoter hypermethylation also showed a tendency towards increased local recurrence, hypermethylation was only seen in 30% of tumors.62
Sinha and colleagues found promoter hypermethylation of the p16 gene (CDKN2A) to be present in 87% of 38 oral tongue SCCs prospectively evaluated in an Indian cohort. Detectable hypermethylation in surgical margins conferred a 6.3-fold increase in local recurrence.63 However, this degree of p16 promoter hypermethylation may be less sensitive in western cohorts, where rates of 21–50% have been reported in HNSCC.53,64 Further demonstration of potential population-based epigenetic variations can be seen in a French study that performed QMSP of CDKN2A, CCNA1, and DCC in 42 HNSCC patients, with hypermethylation of at least one gene present in 64% of tumors. Molecularly positive margins among “hypermethylation informative” tumors were associated with shorter time to local recurrence and disease specific death (p = 0.03 and 0.01, respectively).
While the above studies suggest these types of margin analysis may help distinguish patients that would benefit from the addition of adjuvant therapy, a number of limitations remain. Though potentially financially feasible, time constraints associated with tissue processing and analysis pose possible barriers to the initiation of timely adjuvant treatment. Additionally, the specificity of promoter methylation analysis for any given tumor remains a substantial impediment, as individual HNSCC tumors do not universally undergo consistent epigenetic alterations. Methylation of certain promoters may also be present among preneoplastic cells. Alterations in various promoters play diverse roles in carcinogenesis, and epigenetic changes may evolve differently across populations. Comprehensive studies with heterogenous populations are needed to identify the most predictive target combinations for margin analysis and validate prognostic implications.
Fluorescence molecular imaging
The use of near-infrared (NIR) fluorescence imaging holds much promise in its use for intra-operative assessment of tumor margins. Near-infrared light (650 – 900 nm) is used in conjunction with injected fluorescent agents coupled to cancer-specific molecules and visualized with an optical imaging system that projects emitted fluorescent light as a high definition image, offering a method for intra-operative oncological surgical navigation. Multiple fluorophores, the most common being Irdye800-CW, indocyanine green (ICG), Cy7, and Cy5.5, and both closed- and open-field optical imaging systems are available for use.65 Penetration of NIR light ranges from 5 to 10 mm, allowing immediate evaluation of either the main specimen or separate supplemental margins for areas of concern. Whereas previously discussed non-imaging based molecular margin analyses are typically limited to the post-operative setting due to time constraints, NIR fluorescence imaging facilitates real time margin assessment and guides surgical resection.66,67 As in situ imaging can be associated with substantial challenges related to complex anatomy, heterogenous tissue, and limited exposure, recent efforts have primarily focused on the setting of ex vivo margin assessment.
Though NIR fluorescence imaging in surgical navigation provides encouraging possibilities for future use in head and neck oncology, it is still in the early stages of development. The most promising work to date relates to the use of anti- EGFR antibodies by Rosenthal et al. EGFR is a compelling biomarker, as it is overexpressed in >90% of HNSCCs68 and anti-EGFR targeted therapies showed early success in treatment of HNSCC. Cetuximab and its derivative panitumumab, both anti-EGFR monoclonal antibodies, are FDA approved for the treatment of advanced recurrent and metastatic HNSCC. 69,70 Although enthusiasm for cetuximab as a therapeutic has been somewhat dampened by a high degree of resistance, its therapeutic limitations do not necessarily limit its use as a biomarker.71–76 EGFR has been validated as a promising marker in both preclinical animal and human models, as has the use of anti-EGFR antibodies coupled to fluorescent agents for margin assessment in HNSCC.77–81 The presence of FDA approved targeted antibodies facilitates investigation of broader applications of such technology.
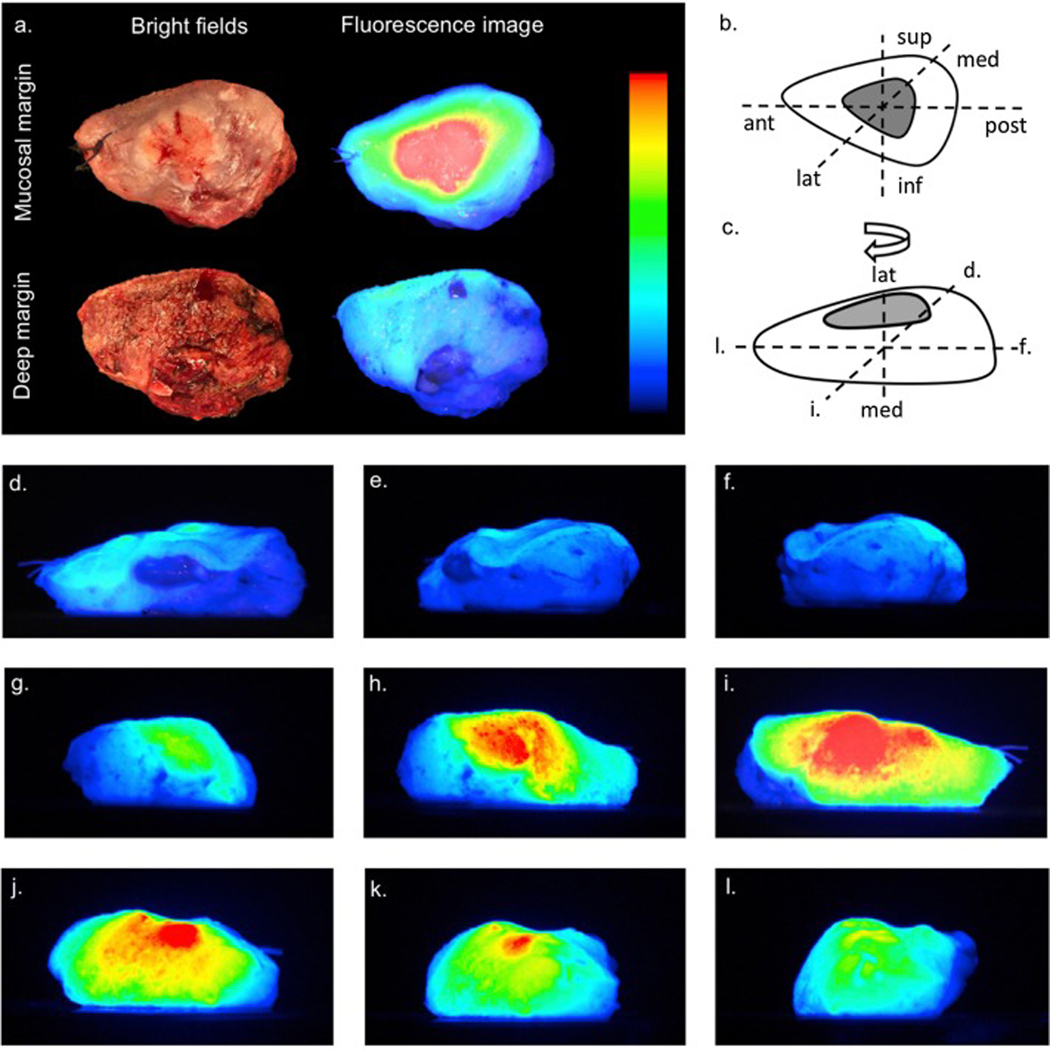
Gao and colleagues conducted a phase I clinical trial in which they administered panitumumab-IRDye800CW intravenously to evaluate margins ex vivo in 21 patients with HNSCC.82 EGFR was heavily expressed among tumor cells and appropriately correlated to fluorescence. Sensitivity and specificity for intra-operative positive margin detection was 100% and 90%, respectively, when compared to standard pathologic analysis. These data demonstrate success in both assessing intra-operative margin samples, as well as identifying close or positive margins on the primary specimen. Remarkably, intra-operative imaging on the back table was completed in only 7 minutes. Given the extraordinary sensitivity and negative predictive value, the authors proposed this method be used as a “rule out” test, suggesting only further assessment of florescent margins with traditional histopathologic methods. By identifying high risk areas, antibody targeted margin analysis theoretically reduces sampling error while avoiding wasted time and resources with frozen section analyses. In a pilot study of 8 HNSCC patients, Van Keulen and colleagues revealed similar findings using closed-field optical specimen mapping (Figure 4), demonstrating the maximal observed penetration depth of panitumumab-IRDye800 to be 6.3mm. Importantly, they were able to map the entire surface area of the tumor specimen, including the deep margin, surmounting a previously established weakness of intra-operative NIR fluorescence imaging. 83 Fakurnejad and colleagues confirmed the ability to identify the “sentinel” mucosal margin using panitumumab-IRDye800 in five OCSCC patients.84 Together, these studies suggest NIR-fluorescence has the potential to quickly identify areas of concern, resulting in more accurate and targeted margin sampling for frozen section and potentially reducing pathology burden and operative times.
Figure 4.
Figure adapted from van Keulen et al. Patients undergoing head and neck oncologic resection were infused with panitumumab-IRDye800 dye 1–4 days prior to surgery and imaged in an Optical Specimen Mapping device with representative images demonstrated here. Panel A demonstrates bright field and fluorescence imaging of the mucosal and deep margins of the resected specimen. Panels D-I demonstrate images acquired from different spatial orientations of the specimen. RightsLink license number 4870280530085.
A number of other biomarkers have been investigated as potential targets for NIR imaging. Preclinical models have recently validated the use of urokinase-like plasminogen activator receptor (uPAR), overexpressed in a variety of carcinomas, as a potential biomarker for the detection of tumor cells in vivo in oral cavity cancer.85,86 Kossatz and colleagues are currently investigating the use of a topically applied fluorescently labeled small-molecule inhibitor, PARPi-FL, which binds to the DNA repair enzyme poly(ADP-ribose)polymerase 1 (PARP1). Based on promising preclinical work, a clinical phase I/II trial is underway to evaluate its use in oral cancer delineation.87 The glucose transporter system (CW800 2-DG), transferrin receptor (TfR), vascular endothelial growth factor (VEGF), and avb3 integrin have also been investigated as potential targets for tumor detection with NIR-fluorescence imaging.77,88–92
While antibody-guided fluorescent imaging techniques hold promise, these approaches do have limitations. Tumor phenotype heterogeneity and variations in expression of EGFR and other potential biomarkers limits the sensitivity across HNSCCs. Additionally, De Boer and others showed that fluorescence was discordantly lower than relative EGFR expression in areas of well-differentiated keratinizing SCC;79 loss of autofluorescence may also occur in areas of bacteria overgrowth or hyperkeratosis.93 Conversely, this group also showed that EGFR expression and subsequent localization of cetuximab-IRDye800CW occurred in normal tissue to some degree.79 In addition, there is a theoretical risk of auto-fluorescence within normal tissues resulting in false-positive results and over-resection, although this was shown to be fairly limited in practice. Scattering and absorption can further negatively affect the signal to noise ratio used to identify malignant cells, but a number of strategies are available to limit these shortcomings.93 Furthermore, current NIR optical imaging systems have limited depth of penetration and are most effective in the context of close margins (< 2 mm). There has been some interest in using adjunct imaging modalities, such as ultrasound or MRI, which could serve to improve penetration depth and will likely be the subject of future studies.65 Further substantiation of optimal imaging systems, fluorescent probes, and biomarkers, as well as phase II and III clinical trials with larger sample sizes are needed to validate and further contextualize the use of this technology in surgical management of HNSCC.
Future directions
Clearly, the future of molecular margin technique is promising with many potential approaches to improve intra-operative and post-operative margin assessment and prognostication. Moving forward, a panoply of technologies have emerged, offering the capacity for novel approaches to molecular margin analysis. While these techniques are maturing, they provide a level of resolution that may not be available with currently utilized technologies and are worthy of further investigation. These technologies include spectroscopy, single cell sequencing, and spatial transcriptomics or multi-spectral imaging.
In general, spectroscopy relies on evaluating the physical properties of a given molecule based on the scattering of incident light. Fluorescence (FS), infrared (IS), and Raman (RS) spectroscopy evaluate the fluorescent, vibrational, and rotational properties of molecules, respectively.94–96 Theoretically, the emission spectra of tumors differ from those of normal tissue, and several groups have demonstrated high sensitivity and specificity when using spectroscopy for diagnostic purposes.97 While there have been numerous studies investigating RS in breast cancer, less work has been completed for margin analysis in the head and neck. Using RS, Barosso et al. demonstrated a higher concentration of water in tumor specimen compared to normal tissue, evaluated over a distance of 4 to 6 mm at the margin.98 In a similar study using FS, Francisco et al. found that in 2 patients with recurrent OCSCC, spectra at the site of recurrence were more similar to that of tumor than healthy tissue.95 Intra-operatively, spectra can be obtained in less than 10 seconds, allowing for rapid and repeated interrogation of margins. However, spectroscopic techniques are limited by several factors including tissue auto-fluorescence, poor depth of penetration and a small sampling area, thereby potentially raising the rate of false negatives. Combining three spectroscopic modalities may mitigate these limitations. In a study of 15 primary and metastatic brain tumors, a combination of FS, RS, and diffuse reflectance spectroscopy provided a tumor detection sensitivity of 100%.99 Fascinatingly, analyzed spectra correlated with biological processes within tumor cells, including increased nuclear volume, changes in the immune response, and adaptation of the extracellular matrix designed to promote cell migration and proliferation.99 These changes in the extracellular matrix are likely part of the epithelial to mesenchymal transition (EMT) which drive tumor progression, drug resistance, and metastasis.100 Thus, while spectroscopy approaches may not offer an individual gene or biomarker based approach, they may provide insights into particular molecular pathways that can be regulated and identified in margins.
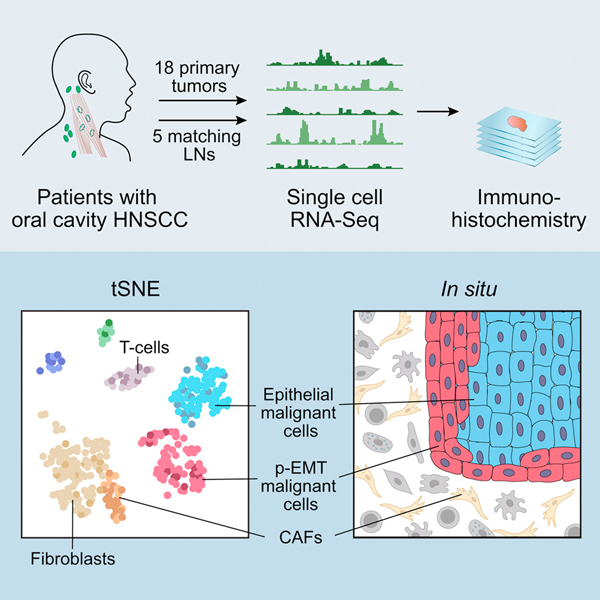
A more nuanced approach to margin assessment would be to comprehensively characterize the present cells and their expression states. Recently, single cell RNA-sequencing (scRNA-seq) has emerged, directly addressing this gap in prior techniques. A specimen is dissociated into individual cells, barcoded, and processed using next-generation sequencing techniques. With this method, even small pieces of tissue can be comprehensively evaluated, accounting for the challenges of tumor heterogeneity common to HNSCC.101–103 However, such approaches are limited by the need for fresh tissue samples, significant manual labor, and need for talented biostatisticians to complete analysis. We recently applied this technique to oral cavity tumors, ultimately identifying a subset of malignant cells in a partial epithelial-to-mesenchymal (p-EMT) state (Figure 5).104 This state has some features of traditional EMT, such as the expression of mesenchymal markers (e.g. vimentin), but retains expression of epithelial genes (e.g. KRTs and EPCAM).105 Strikingly, these cells were localized at the leading edge of tumors and associated with adverse features such as nodal metastasis, extranodal extension, lymphovascular invasion, and higher tumor grade.106 Many of these features currently stand as indications for adjuvant treatment in HNSCC; however, the p-EMT program may suggest the need for adjuvant treatment in the absence of these features, as it may portend an aggressive biology that cannot be captured with existing histopathologic metrics. In the future, it will be important to determine if p-EMT cells can be detected in molecular margins, presumably in close apposition to the leading edge. Detection of p-EMT cells can be done initially with scRNA-seq, but should quickly transition to PCR or protein-based approaches as outlined above for ease of use.
Figure 5.
Figure adapted from Puram et al demonstrating the study workflow. Of note, a group of partial epithelial to mesenchymal (p-EMT) cells were identified and localized to the leading edge of tumor. p-EMT was found to be a predictor of nodal metastasis, grade, and adverse pathologic features. Further work investigating the use of single cell sequencing at tumor margins may also hold promise. RightsLink license number 4870270827236.
One advantage of the scRNA-seq approach is its independence from known gene targets or biomarkers. If tumor specimens and margins could be analyzed with this approach, then particular genes upregulated in the tumor or subpopulations of interest could be specifically probed in margin samples. Thus, an intriguing possibility is the use of scRNA-seq for tumors with de novo identification of relevant biomarkers in a patient-specific manner. Unfortunately, at the time of this writing, such a model is limited by the cost of scRNA-seq and its technical preparation and informatics analysis. As these techniques achieve economies of scale, opportunities for routine implementation may be more readily available and should be fully embraced.
Although scRNA-seq has garnered enthusiasm, with its use for oncologic molecular analyses, the dissociation of samples into individual cells purges spatial information. In the context of margin analysis, this spatial information is critical. Under the current standard-of-care, margins are measured from the main specimen to provide a specific radial margin distance measurement. These metrics have significant implications for adjuvant therapy depending on whether they are positive (< 1 mm), close (2–5 mm), or clear (> 5 mm). Implementation of newer techniques such as scRNA-seq necessarily restricts the evaluation of such metrics. Newer techniques including spatial transcriptomics leverage the pipeline of scRNA-seq but retain an added barcode that captures spatial localization.107 Thus, after library construction and next-generation sequencing, cells (and their expression programs) can be “pinned” to their original spatial localization. In a similar vein, multiplexed, multi-spectral imaging allows the visualization of up to 15 marker genes using standard IHC plus a tyramide amplification step, all while retaining normal tissue architecture. These approaches have the potential to offer a high degree of insight into specific programs and expression states present in tumors and their margins, while retaining the necessary spatial information for downstream oncological decision-making. Certainly, wide scale application of such techniques will be of great interest, but the major hurdle will be the associated accessibility, cost of validation, and bioinformatic demands. Nevertheless, we believe such techniques are bound to make their way into the lexicon of molecular margin analysis over the next several decades.
Conclusion
Obtaining clear margins may be considered the key tenant of successful surgical management of HNSCC as margin status is closely related to LRR and survival. Traditional margin analysis guides adjuvant treatment and predicts survival, however, currently used histopathologic techniques may not capture underlying biologic changes associated with poor outcomes.
Molecular margin analysis allows us to combine our increasing understanding of tumor biology and genetics with standard practices of histological evaluation, ultimately offering the potential to improve the sensitivity, specificity and prognostic value of margin analysis. While promising, HNSCC is genetically heterogeneous and each tumor represents a unique combination of genetic alterations, thereby complicating molecular margin analysis. Routine adoption of molecular margin analysis will require further study and validation of optimal molecular markers, as well as the development of affordable and technically accessible methods. While still maturing as a field, the incorporation of emerging technologies such as spectroscopy, single cell sequencing, and spatial transcriptomics or multi-spectral imaging, have exciting potential to further advance the field in the future.
References
- 1.Jesse RH, Sugarbaker E V. Squamous cell carcinoma of the oropharynx: Why we fail. Am J Surg. 1976. doi: 10.1016/0002-9610(76)90314-7 [DOI] [PubMed] [Google Scholar]
- 2.Eldeeb H, Macmillan C, Elwell C, Hammod A. The effect of the surgical margins on the outcome of patients with head and neck squamous cell carcinoma: single institution experience. Cancer Biol Med. 2012. doi: 10.3969/j.issn.2095-3941.2012.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchakjian MR, Ginader T, Tasche KK, Pagedar NA, Smith BJ, Sperry SM. Independent Predictors of Prognosis Based on Oral Cavity Squamous Cell Carcinoma Surgical Margins. Otolaryngol - Head Neck Surg (United States). 2018. doi: 10.1177/0194599818773070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMahon J, O’Brien CJ, Pathak I, et al. Influence of condition of surgical margins on local recurrence and disease-specific survival in oral and oropharyngeal cancer. Br J Oral Maxillofac Surg. 2003. doi: 10.1016/S0266-4356(03)00119-0 [DOI] [PubMed] [Google Scholar]
- 5.Slootweg PJ, Hordijk GJ, Schade Y, Van Es RJJ, Koole R. Treatment failure and margin status in head and neck cancer. A critical view on the potential value of molecular pathology. Oral Oncol. 2002. doi: 10.1016/S1368-8375(01)00092-6 [DOI] [PubMed] [Google Scholar]
- 6.Eckardt A, Barth EL, Kokemueller H, Wegener G. Recurrent carcinoma of the head and neck: Treatment strategies and survival analysis in a 20-year period. Oral Oncol. 2004. doi: 10.1016/j.oraloncology.2003.09.019 [DOI] [PubMed] [Google Scholar]
- 7.Meier JD, Oliver DA, Varvares MA. Surgical margin determination in head and neck oncology: Current clinical practice. The results of an International American Head and Neck Society member survey. Head Neck. 2005. doi: 10.1002/hed.20269 [DOI] [PubMed] [Google Scholar]
- 8.Woolgar JA, Triantafyllou A. A histopathological appraisal of surgical margins in oral and oropharyngeal cancer resection specimens. Oral Oncol. 2005. doi: 10.1016/j.oraloncology.2005.06.008 [DOI] [PubMed] [Google Scholar]
- 9.Ravasz LA, Slootweg PJ, Hordijk GJ, Smit F, van der Tweel I. The status of the resection margin as a prognostic factor in the treatment of head and neck carcinoma. J Cranio-Maxillofacial Surg. 1991. doi: 10.1016/S1010-5182(05)80339-7 [DOI] [PubMed] [Google Scholar]
- 10.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001. doi: 10.1056/NEJMra001375 [DOI] [PubMed] [Google Scholar]
- 11.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science (80- ). 1991. doi: 10.1126/science.1905840 [DOI] [PubMed] [Google Scholar]
- 12.Tabor MP, Van Houten VMM, Kummer JA, et al. Discordance of genetic alterations between primary head and neck tumors and corresponding metastases associated with mutational status of the TP53 gene. Genes Chromosom Cancer. 2002. doi: 10.1002/gcc.10019 [DOI] [PubMed] [Google Scholar]
- 13.Brennan JA, Mao L, Boyle JO, et al. Molecular assessment of histopathological staging in squamous-cell carcinoma of the head and neck. N Engl J Med. 1995. doi: 10.1056/NEJM199502163320704 [DOI] [PubMed] [Google Scholar]
- 14.Van Houten VMM, Leemans CR, Kummer JA, et al. Molecular diagnosis of surgical margins and local recurrence in head and neck cancer patients: A prospective study. Clin Cancer Res. 2004. doi: 10.1158/1078-0432.CCR-03-0631 [DOI] [PubMed] [Google Scholar]
- 15.Pena Murillo C, Huang X, Hills A, et al. The utility of molecular diagnostics to predict recurrence of head and neck carcinoma. Br J Cancer. 2012. doi: 10.1038/bjc.2012.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X, Pateromichelakis S, Hills A, et al. p53 mutations in deep tissues are more strongly associated with recurrence than mutation-positive mucosal margins. Clin Cancer Res. 2007. doi: 10.1158/1078-0432.CCR-07-1369 [DOI] [PubMed] [Google Scholar]
- 17.Partridge M, Li SR, Pateromichelakis S, et al. Detection of minimal residual cancer to investigate why oral tumors recur despite seemingly adequate treatment. Clin Cancer Res. 2000. [PubMed] [Google Scholar]
- 18.Pierssens DDCG, Borgemeester MC, van der Heijden SJH, et al. Chromosome instability in tumor resection margins of primary OSCC is a predictor of local recurrence. Oral Oncol. 2017. doi: 10.1016/j.oraloncology.2016.12.029 [DOI] [PubMed] [Google Scholar]
- 19.Nathan CAO, Sanders K, Abreo FW, Nassar R, Glass J. Correlation of p53 and the proto-oncogene eIF4E in larynx cancers: Prognostic implications. Cancer Res. 2000. [PubMed] [Google Scholar]
- 20.Bergshoeff VE, Hopman AHN, Zwijnenberg IR, et al. Chromosome instability in resection margins predicts recurrence of oral squamous cell carcinoma. J Pathol. 2008. doi: 10.1002/path.2349 [DOI] [PubMed] [Google Scholar]
- 21.Poeta ML, Manola J, Goldwasser MA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007. doi: 10.1056/NEJMoa073770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science (80- ). 1994. doi: 10.1126/science.8023157 [DOI] [PubMed] [Google Scholar]
- 23.Erber R, Conradt C, Homann N, et al. TP53 DNA contact mutations are selectively associated with allelic loss and have a strong clinical impact in head and neck cancer. Oncogene. 1998. doi: 10.1038/sj.onc.1201690 [DOI] [PubMed] [Google Scholar]
- 24.Mao L, Clark D. Molecular margin of surgical resections - Where do we go from here? Cancer. 2015. doi: 10.1002/cncr.29299 [DOI] [PubMed] [Google Scholar]
- 25.Li MM, Puram SV, Silverman DA, Old MO, Rocco JW, Kang SY. Margin Analysis in Head and Neck Cancer: State of the Art and Future Directions. Ann Surg Oncol. 2019. doi: 10.1245/s10434-019-07645-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang XH, Ding L, Fu Y, et al. P53-positive expression in dysplastic surgical margins is a predictor of tumor recurrence in patients with early oral squamous cell carcinoma. Cancer Manag Res. 2019. doi: 10.2147/CMAR.S192500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science (80- ). 2011. doi: 10.1126/science.1206923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science (80- ). 2011. doi: 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chau NG, Li YY, Jo VY, et al. Incorporation of next-generation sequencing into routine clinical care to direct treatment of head and neck squamous cell carcinoma. Clin Cancer Res. 2016. doi: 10.1158/1078-0432.CCR-15-2314 [DOI] [PubMed] [Google Scholar]
- 30.van Ginkel JH, de Leng WWJ, de Bree R, van Es RJJ, Willems SM. Targeted sequencing reveals TP53 as a potential diagnostic biomarker in the post-treatment surveillance of head and neck cancer. Oncotarget. 2016. doi: 10.18632/oncotarget.11196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pekin D, Skhiri Y, Baret JC, et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011. doi: 10.1039/c1lc20128j [DOI] [PubMed] [Google Scholar]
- 32.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004. doi: 10.1038/sj.onc.1207545 [DOI] [PubMed] [Google Scholar]
- 33.Nathan CAO, Liu L, Li BD, Abreo FW, Nandy I, De Benedetti A. Detection of the proto-oncogene eIF4E in surgical margins may predict recurrence in head and neck cancer. Oncogene. 1997. doi: 10.1038/sj.onc.1201216 [DOI] [PubMed] [Google Scholar]
- 34.Nathan CAO, Franklin S, Abreo FW, Nassar R, De Benedetti A, Glass J. Analysis of surgical margins with the molecular marker eIF4E: A prognostic factor in patients with head and neck cancer. J Clin Oncol. 1999. doi: 10.1200/jco.1999.17.9.2909 [DOI] [PubMed] [Google Scholar]
- 35.Xia L, Zeng J, Guo Z, Rao H, Zeng J, Zeng Z. The prognostic value of pathological and molecular margins marked by p53 and el4E in laryngeal carcinoma. Chinese-German J Clin Oncol. 2005. doi: 10.1007/s10330-004-0241-0 [DOI] [Google Scholar]
- 36.Nathan CAO, Amirghahri N, Rice C, Abreo FW, Shi R, Stucker FJ. Molecular analysis of surgical margins in head and neck squamous cell carcinoma patients. Laryngoscope. 2002. doi: 10.1097/00005537-200212000-00003 [DOI] [PubMed] [Google Scholar]
- 37.SINGH J, JAYARAJ R, BAXI S, et al. Immunohistochemical expression levels of p53 and eIF4E markers in histologically negative surgical margins, and their association with the clinical outcome of patients with head and neck squamous cell carcinoma. Mol Clin Oncol. 2016. doi: 10.3892/mco.2015.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakashita T, Homma A, Suzuki S, et al. Prognostic value of cyclin D1 expression in tumor-free surgical margins in head and neck squamous cell carcinomas. Acta Otolaryngol. 2013. doi: 10.3109/00016489.2013.795287 [DOI] [PubMed] [Google Scholar]
- 39.Zhao H, Ren J, Zhuo X, Ye H, Zou J, Liu S. Prognostic significance of Survivin and CD44v6 in laryngeal cancer surgical margins. J Cancer Res Clin Oncol. 2008. doi: 10.1007/s00432-008-0391-5 [DOI] [PubMed] [Google Scholar]
- 40.Schaaij-Visser TBM, Graveland AP, Gauci S, et al. Differential proteomics identifies protein biomarkers that predict local relapse of head and neck squamous cell carcinomas. Clin Cancer Res. 2009. doi: 10.1158/1078-0432.CCR-09-2134 [DOI] [PubMed] [Google Scholar]
- 41.Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22(11):1342–1350. doi: 10.1038/nm.4191 [DOI] [PubMed] [Google Scholar]
- 42.Temam S, Casiraghi O, Lahaye JB, et al. Tetranucleotide microsatellite instability in surgical margins for prediction of local recurrence of head and neck squamous cell carcinoma. Clin Cancer Res. 2004. doi: 10.1158/1078-0432.CCR-04-0199 [DOI] [PubMed] [Google Scholar]
- 43.Sardi I, Franchi A, Ferriero G, et al. Prediction of recurrence by microsatellite analysis in head and neck cancer. Genes Chromosom Cancer. 2000. doi: [DOI] [PubMed] [Google Scholar]
- 44.Lin JC, Wang CC, Jiang RS, Wang WY, Liu SA. Impact of microsatellite alteration in surgical margins on local recurrence in oral cavity cancer patients. Eur Arch Oto-Rhino-Laryngology. 2017. doi: 10.1007/s00405-016-4215-y [DOI] [PubMed] [Google Scholar]
- 45.Liu SA, Wang CC, Jiang RS, Wang WY, Lin JC. Genetic analysis of surgical margins in oral cavity cancer. Br J Surg. 2018. doi: 10.1002/bjs.10693 [DOI] [PubMed] [Google Scholar]
- 46.Szukała K, Brieger J, Bruch K, et al. Loss of heterozygosity on chromosome arm 13q in larynx cancer patients: Analysis of tumor, margin and clinically unchanged mucosa. Med Sci Monit. 2004. [PubMed] [Google Scholar]
- 47.Graveland AP, Golusinski PJ, Buijze M, et al. Loss of heterozygosity at 9p and p53 immunopositivity in surgical margins predict local relapse in head and neck squamous cell carcinoma. Int J Cancer. 2011. doi: 10.1002/ijc.25523 [DOI] [PubMed] [Google Scholar]
- 48.Shah NG, Trivedi TI, Tankshali RA, et al. Prognostic significance of molecular markers in oral squamous cell carcinoma: A multivariate analysis. Head Neck. 2009. doi: 10.1002/hed.21126 [DOI] [PubMed] [Google Scholar]
- 49.Mao L, Lee JS, Fan YH, et al. Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat Med. 1996. doi: 10.1038/nm0696-682 [DOI] [PubMed] [Google Scholar]
- 50.Van Der Toorn PPG, Veltman JA, Bot FJ, et al. Mapping of resection margins of oral cancer for p53 overexpression and chromosome instability to detect residual (pre)malignant cells. J Pathol. 2001. doi: [DOI] [PubMed] [Google Scholar]
- 51.Handschel J, Öz D, Pomjanski N, et al. Additional use of DNA-image cytometry improves the assessment of resection margins. J Oral Pathol Med. 2007. doi: 10.1111/j.1600-0714.2007.00564.x [DOI] [PubMed] [Google Scholar]
- 52.Martone T, Gillio-Tos A, De Marco L, et al. Association between hypermethylated tumor and paired surgical margins in head and neck squamous cell carcinomas. Clin Cancer Res. 2007. doi: 10.1158/1078-0432.CCR-07-0119 [DOI] [PubMed] [Google Scholar]
- 53.Goldenberg D, Harden S, Masayesva BG, et al. Intraoperative Molecular Margin Analysis in Head and Neck Cancer. Arch Otolaryngol - Head Neck Surg. 2004. doi: 10.1001/archotol.130.1.39 [DOI] [PubMed] [Google Scholar]
- 54.Kato K, Hara A, Kuno T, et al. Aberrant promoter hypermethylation of p16 and MGMT genes in oral squamous cell carcinomas and the surrounding normal mucosa. J Cancer Res Clin Oncol. 2006. doi: 10.1007/s00432-006-0122-8 [DOI] [PubMed] [Google Scholar]
- 55.Laytragoon-Lewin N, Rutqvist LE, Lewin F. DNA methylation in tumour and normal mucosal tissue of head and neck squamous cell carcinoma (HNSCC) patients: New diagnostic approaches and treatment. Med Oncol. 2013. doi: 10.1007/s12032-013-0654-0 [DOI] [PubMed] [Google Scholar]
- 56.Roh JL, Westra WH, Califano JA, Sidransky D, Koch WM. Tissue imprint for molecular mapping of deep surgical margins in patients with head and neck squamous cell carcinoma. Head Neck. 2012. doi: 10.1002/hed.21982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayashi M, Wu G, Roh JL, et al. Correlation of gene methylation in surgical margin imprints with locoregional recurrence in head and neck squamous cell carcinoma. Cancer. 2015. doi: 10.1002/cncr.29303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Demokan S, Chang X, Chuang A, et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int J Cancer. 2010. doi: 10.1002/ijc.25248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guerrero-Preston R, Soudry E, Acero J, et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev Res. 2011. doi: 10.1158/1940-6207.CAPR-11-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi M, Guerrero-Preston R, Sidransky D, Kochz WM. Paired box 5 methylation detection by droplet digital PCR for ultra-sensitive deep surgical margins analysis of head and neck squamous cell carcinoma. Cancer Prev Res. 2015. doi: 10.1158/1940-6207.CAPR-15-0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayashi M, Guerrero-Preston R, Okamura J, et al. Innovative rapid gene methylation analysis of surgical margin tissues in head and neck cancer. Ann Surg Oncol. 2014. doi: 10.1245/s10434-014-3661-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Supic G, Kozomara R, Jovic N, Zeljic K, Magic Z. Prognostic significance of tumor-related genes hypermethylation detected in cancer-free surgical margins of oral squamous cell carcinomas. Oral Oncol. 2011. doi: 10.1016/j.oraloncology.2011.05.014 [DOI] [PubMed] [Google Scholar]
- 63.Sinha P, Bahadur S, Thakar A, et al. Significance of promoter hypermethylation of p16 gene for margin assessment in carcinoma tongue. Head Neck. 2009. doi: 10.1002/hed.21122 [DOI] [PubMed] [Google Scholar]
- 64.Sanchez-Cespedes M, Esteller M, Wu L, et al. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000. [PubMed] [Google Scholar]
- 65.Iqbal H, Pan Q. Image guided surgery in the management of head and neck cancer. Oral Oncol. 2016. doi: 10.1016/j.oraloncology.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 66.Keereweer S, Van Driel PBAA, Snoeks TJA, et al. Optical image-guided cancer surgery: Challenges and limitations. Clin Cancer Res. 2013. doi: 10.1158/1078-0432.CCR-12-3598 [DOI] [PubMed] [Google Scholar]
- 67.Rosenthal EL, Warram JM, Bland KI, Zinn KR. The status of contemporary image-guided modalities in oncologic surgery. Ann Surg. 2015. doi: 10.1097/SLA.0000000000000622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tweardy DJ. Elevated Levels of Transforming Growth Factor a and Epidermal Growth Factor Receptor Messenger RNA Are Early Markers of Carcinogenesis in Head and Neck Cancer. Cancer Res. 1993. [PubMed] [Google Scholar]
- 69.FDA approval for cetuximab. Head and neck cancer. https://www.cancer.gov/about-cancer/treatment/drugs/fda-cetuximab#Anchor-Hea-5647. Accessed April 12, 2020.
- 70.FDA approval for cetuximab. Approved for recurrent or metastatic head and neck cancer. https://www.cancer.gov/about-cancer/treatment/drugs/fda-cetuximab#Anchor-Recur-11711. Accessed April 12, 2020.
- 71.Seiwert TY, Fayette J, Cupissol D, et al. A randomized, phase II study of afatinib versus cetuximab in metastatic or recurrent squamous cell carcinoma of the head and neck. Ann Oncol. 2014. doi: 10.1093/annonc/mdu216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008. doi: 10.1056/NEJMra0707975 [DOI] [PubMed] [Google Scholar]
- 73.Wheeler DL, Huang S, Kruser TJ, et al. Mechanisms of acquired resistance to cetuximab: Role of HER (ErbB) family members. Oncogene. 2008. doi: 10.1038/onc.2008.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008. doi: 10.1056/NEJMoa0802656 [DOI] [PubMed] [Google Scholar]
- 75.Seiwert TY, Kochanny S, Wood K, et al. A randomized phase 2 study of temsirolimus and cetuximab versus temsirolimus alone in recurrent/metastatic, cetuximab-resistant head and neck cancer: The MAESTRO study. Cancer. May 2020:cncr.32929. doi: 10.1002/cncr.32929 [DOI] [PubMed] [Google Scholar]
- 76.Ang KK, Zhang Q, Rosenthal DI, et al. Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J Clin Oncol. 2014. doi: 10.1200/JCO.2013.53.5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keereweer S, Kerrebijn JDF, Mol IM, et al. Optical imaging of oral squamous cell carcinoma and cervical lymph node metastasis. Head Neck. 2012. doi: 10.1002/hed.21861 [DOI] [PubMed] [Google Scholar]
- 78.Warram JM, de Boer E, van Dam GM, et al. Fluorescence imaging to localize head and neck squamous cell carcinoma for enhanced pathological assessment. J Pathol Clin Res. 2016. doi: 10.1002/cjp2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.De Boer E, Warram JM, Tucker MD, et al. In Vivo Fluorescence Immunohistochemistry: Localization of Fluorescently Labeled Cetuximab in Squamous Cell Carcinomas. Sci Rep. 2015. doi: 10.1038/srep10169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Driel PBAA, van de Giessen M, Boonstra MC, et al. Characterization and Evaluation of the Artemis Camera for Fluorescence-Guided Cancer Surgery. Mol Imaging Biol. 2015. doi: 10.1007/s11307-014-0799-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heath CH, Deep NL, Sweeny L, Zinn KR, Rosenthal EL. Use of panitumumab-IRDye800 to image microscopic head and neck cancer in an orthotopic surgical model. Ann Surg Oncol. 2012. doi: 10.1245/s10434-012-2435-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao RW, Teraphongphom NT, van den Berg NS, et al. Determination of tumor margins with surgical specimen mapping using near-infrared fluorescence. Cancer Res. 2018. doi: 10.1158/0008-5472.CAN-18-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Keulen S, van den Berg NS, Nishio N, et al. Rapid, non-invasive fluorescence margin assessment: Optical specimen mapping in oral squamous cell carcinoma. Oral Oncol. 2019. doi: 10.1016/j.oraloncology.2018.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fakurnejad S, Krishnan G, van Keulen S, et al. Intraoperative Molecular Imaging for ex vivo Assessment of Peripheral Margins in Oral Squamous Cell Carcinoma. Front Oncol. 2020. doi: 10.3389/fonc.2019.01476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boonstra MC, Van Driel PBAA, Keereweer S, et al. Preclinical uPAR-targeted multimodal imaging of locoregional oral cancer. Oral Oncol. 2017. doi: 10.1016/j.oraloncology.2016.12.026 [DOI] [PubMed] [Google Scholar]
- 86.Christensen A, Juhl K, Persson M, et al. uPAR-targeted optical near-infrared (NIR) fluorescence imaging and PET for image-guided surgery in head and neck cancer: Proof-of-concept in orthotopic xenograft model. Oncotarget. 2017. doi: 10.18632/oncotarget.14282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kossatz S, Brand C, Gutiontov S, et al. Detection and delineation of oral cancer with a PARP1 targeted optical imaging agent. Sci Rep. 2016. doi: 10.1038/srep21371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Antaris AL, Chen H, Cheng K, et al. A small-molecule dye for NIR-II imaging. Nat Mater. 2016. doi: 10.1038/nmat4476 [DOI] [PubMed] [Google Scholar]
- 89.Jin ZH, Josserand V, Foillard S, et al. In vivo optical imaging of integrin αV- β3 in mice using multivalent or monovalent cRGD targeting vectors. Mol Cancer. 2007. doi: 10.1186/1476-4598-6-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garanger E, Boturyn D, Jin Z, Dumy P, Favrot MC, Coll JL. New multifunctional molecular conjugate vector for targeting, imaging, and therapy of tumors. Mol Ther. 2005. doi: 10.1016/j.ymthe.2005.06.095 [DOI] [PubMed] [Google Scholar]
- 91.Shan L, Hao Y, Wang S, et al. Visualizing head and neck tumors in vivo using near-infrared fluorescent transferrin conjugate. Mol Imaging. 2008. doi: 10.2310/7290.2008.0006 [DOI] [PubMed] [Google Scholar]
- 92.Withrow KP, Newman JR, Skipper JB, et al. Assessment of bevacizumab conjugated to Cy5.5 for detection of head and neck cancer xenografts. Technol Cancer Res Treat. 2008. doi: 10.1177/153303460800700108 [DOI] [PubMed] [Google Scholar]
- 93.Wu C, Gleysteen J, Teraphongphom NT, Li Y, Rosenthal E. In-vivo optical imaging in head and neck oncology: Basic principles, clinical applications and future directions review-Article. Int J Oral Sci. 2018. doi: 10.1038/s41368-018-0011-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramanujam N. Fluorescence spectroscopy of neoplastic and non-neoplastic tissues. Neoplasia. 2000. doi: 10.1038/sj.neo.7900077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Francisco ALN, Correr WR, Pinto CAL, et al. Analysis of surgical margins in oral cancer using in situ fluorescence spectroscopy. Oral Oncol. 2014. doi: 10.1016/j.oraloncology.2014.02.008 [DOI] [PubMed] [Google Scholar]
- 96.Auner GW, Koya SK, Huang C, et al. Applications of Raman spectroscopy in cancer diagnosis. Cancer Metastasis Rev. 2018. doi: 10.1007/s10555-018-9770-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harris AT, Rennie A, Waqar-Uddin H, et al. Raman spectroscopy in head and neck cancer. Head Neck Oncol. 2010. doi: 10.1186/1758-3284-2-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barroso EM, Smits RWH, Van Lanschot CGF, et al. Water concentration analysis by Raman spectroscopy to determine the location of the tumor border in oral cancer surgery. Cancer Res. 2016. doi: 10.1158/0008-5472.CAN-16-1227 [DOI] [PubMed] [Google Scholar]
- 99.Jermyn M, Mercier J, Aubertin K, et al. Highly accurate detection of cancer in situ with intraoperative, label-free, multimodal optical spectroscopy. Cancer Res. 2017. doi: 10.1158/0008-5472.CAN-17-0668 [DOI] [PubMed] [Google Scholar]
- 100.Nieto MA, Huang RYYJ, Jackson RAA, Thiery JPP. EMT: 2016. Cell. 2016. doi: 10.1016/j.cell.2016.06.028 [DOI] [PubMed] [Google Scholar]
- 101.Mroz EA, Patel KB, Rocco JW. Intratumor heterogeneity could inform the use and type of postoperative adjuvant therapy in patients with head and neck squamous cell carcinoma. Cancer. 2020. doi: 10.1002/cncr.32742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013. doi: 10.1016/j.oraloncology.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mroz EA, Tward AD, Pickering CR, Myers JN, Ferris RL, Rocco JW. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer. 2013. doi: 10.1002/cncr.28150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Puram S V, Tirosh I, Parikh AS, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017. doi: 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Puram S V, Parikh AS, Tirosh I. Single cell RNA-seq highlights a role for a partial EMT in head and neck cancer. Mol Cell Oncol. 2018. doi: 10.1080/23723556.2018.1448244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Parikh AS, Puram SV, Faquin WC, et al. Immunohistochemical quantification of partial-EMT in oral cavity squamous cell carcinoma primary tumors is associated with nodal metastasis. Oral Oncol. 2019;99. doi: 10.1016/j.oraloncology.2019.104458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qi Z, Barrett T, Parikh AS, Tirosh I, Puram S V. Single-cell sequencing and its applications in head and neck cancer. Oral Oncol. 2019;99:104441. doi: 10.1016/j.oraloncology.2019.104441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maryam Shirazi, Antonia Sepulveda. Therapy implications of DNA mismatch re-pair deficiency, microsatellite instability, and tumor mutation burden. Adv Mol Pathol 2018;1(1):193–208. [Google Scholar]