Fusarium crown rot (FCR) is one of the most serious diseases in wheat and causes serious yield loss and health problems (Nellist et al., 2019). Lignin in plant tissue exerts a variety of functions such as antimicrobial, antiviral, antioxidant and anti‐cytotoxic (Ralph et al., 2006). Research indicates that dirigent (DIR) proteins are involved in the synthesis of lignin and play important roles in plant defence responses (Hosmani et al., 2013; Ralph et al., 2006). Wang and Fristensky (2001) found that DIR protein enhanced the resistance to the pathogens such as Rhizoctonia solani and Leptosphaeria maculans in Brassica rapa. Reboledo et al. (2015) reported that the DIR gene in Physcomitrella patens was involved in the formation of lignin polymer to harden cell wall and improve the resistance capacity of cells to Colletotrichum gloeosporioides.
In this study, an association panel containing 435 wheat introgression lines (created by crossing of Yanzhan1 and many other elite varieties) was used to investigate FCR resistance in four environments and their disease index (DI) ranged from 14.81 to 72.29 (Figure 1a,b). Genome‐wide association study (GWAS) using the Wheat 660K SNP array identified 65 significant SNPs associated with FCR resistance, and they were mainly distributed on 4B (Figure 1c,d). Of all significant SNPs, 15 were detected in all 4 environments, and 12 on 4B formed a 0.5‐Mb independent block containing five haplotypes (Figure 1e,f). The accessions with haplotype Hap‐1 (TATATATTCAAG, DI = 40.72) and Hap‐3 (CGTACACTCAAG, DI = 38.14) showed an extremely significantly lower DI than those with Hap‐2 (CGGGCGCGTGGC, DI = 44.54) (Figure 1f,g) (P < 0.01). The block ranged from 664 152 928 to 664 679 942 (≈527 kb) with 6 annotation genes in the genome of Chinese Spring (Figure 1i). Sequencing results indicated that two genes (TraesCS4B02G385200 and TraesCS4B02G385500) showed allelic variations with amino acid alteration in the exons between 10 extremely low DI accessions and 10 extremely high DI accessions. The QTL analysis of a double haploid (DH) population (Bainong64/Jingshuang16) containing 181 lines (Chen et al., 2019) genotyped with Wheat 55K SNP array identified a significant QTL flanked by markers TraesCS4B02G385500 and AX‐110467582 with phenotypic variation explained (PVE) of 16.40‐19.97% (Figure 1h). Combined results suggested that TraesCS4B02G385500 [TaDIR‐B1: encoding dirigent (DIR) protein] was the candidate gene (Figure 1i). Sequencing results indicated that the parent Bainong 64 and FCR‐resistant introgression lines (YZ_94, YZ_170, YZ_425, etc.) with low DI showed a G to A mutation at the 15‐bp position of TaDIR‐B1 gene when compared to the Jingshuang 16 with high DI, resulting in a premature stop codon of deduced amino acid of TaDIR‐B1 at the 5th position. The mutant allele with A and the wild type (WT) with G were designated as TaDIR‐B1b and TaDIR‐B1a, respectively (Figure 1j). A dCAPS (Derived Cleaved Amplified Polymorphic Sequences) marker was developed to identify the two TaDIR‐B1 alleles with the help of restriction enzyme BalI (Figure 1k).
Figure 1.
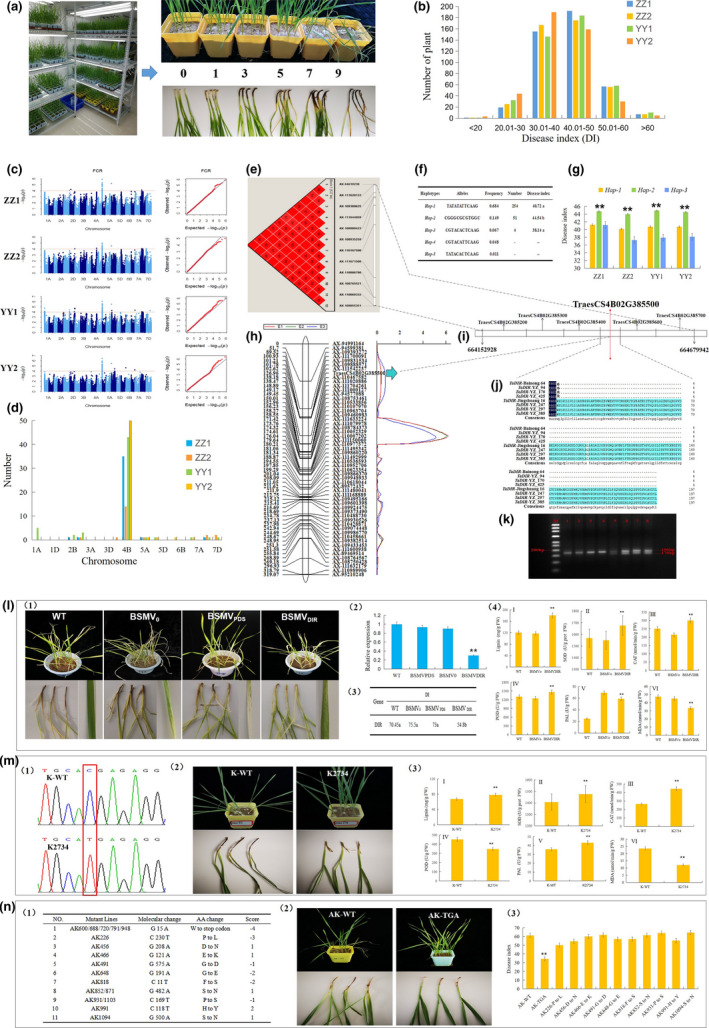
The TaDIR gene was identified by GWAS and QTL mapping of Fusarium crown rot (FCR) resistance and was functionally verified by VIGS and analysis of tetraploid and hexaploid wheat mutants. (a): Classification of FCR disease index in the surveyed cultivars. (b) Number of accessions with different FCR DI in the association panel. (c) Manhattan and Q‐Q plots for FCR DI in different environments. (d) Distribution of significant SNPs revealed by GWAS on various chromosomes. (e) Haplotype analysis of significant SNPs on 4B identified at multiple environments. (f–g) Comparison of FCR DI of wheat accessions with different alleles in the block on 4B. (h) QTL mapping for FCR DI in the Bainong64/Jingshuang16 (BJ) population. (i) The schematic range of 6 candidate genes including TaDIR identified by haplotype analysis and QTL mapping. (j) Full alignment of amino acid of TaDIR‐B1 alleles between low DI and high DI accessions. (k) Development of the dCAPS marker to distinguish TaDIR‐B1 alleles. The 190‐bp fragment from cultivars with TaDIR‐B1a could be digested into 170 and 20 bp by the restriction enzyme BalI, while those from cultivars with TaDIR‐B1b could not be digested. (l) VIGS (virus‐induced gene silencing) experiment verified the function of TaDIR‐B1 gene. (1) The phenotypes of wild type (WT), BSMV0 and BSMV TaDIR‐B1 plants for FCR; (2) the relative expression levels; (3) the averaged DI; and (4) physiological parameters. (m) Function of TaDIR‐B1 gene was verified in Kronos mutants. (1) Mutation point of TaDIR‐B1 gene in K2734; (2) investigation of FCR DI; and (3) physiological parameters. (n) Function of TaDIR‐B1 gene was verified in AK58 mutants. (1) Summary of AK58 mutant lines with amino acid change of the TaDIR‐B1 gene by sequencing; (2) investigation of FCR DI; and (3) comparison of averaged FCR DI.
Furthermore, TaDIR‐B1 gene was silenced in the FCR‐susceptible cultivar Pingyuan 50 using virus‐induced gene silencing (VIGS) by barley stripe mosaic virus (BSMV). qRT‐PCR analysis showed significant down‐regulation of TaDIR‐B1 in VIGS‐silenced plants (Figure 1l‐2). The DI was significantly lower in VIGS‐silenced plants (54.8) compared to the wild type (70.45) and the viral control (BSMV0) (75.50) after inoculation with F. pseudograminearum (Fpg) (Figure 1l‐1, 3). Compared to the WT and BSMV0 plants, the lignin content in the BSMV TaDIR‐B1 plants was remarkedly increased, and the activities of superoxide dismutase (SOD), catalase (CAT) and peroxidase (POD) were also dramatically increased, but the malondialdehyde (MDA) content was significantly decreased in BSMV TaDIR‐B1 plants (Figure 1l‐4). These results indicated that silencing of the TaDIR‐B1 gene resulted in increased FCR resistance and accumulation of secondary metabolites (lignin, PAL) and ROS‐related antioxidant enzymes (SOD, CAT, POD).
The DIR gene was further analysed in the EMS‐mutagenized tetraploid wheat Kronos and hexaploid wheat Aikang 58 (AK58) mutant libraries. The line Kronos2734 showed a C to T mutation at the 220‐bp position of the TaDIR‐B1a gene and caused premature stop codon (Figure 1m‐1). After inoculation with Fpg, Kronos2734 had significantly lower DI (37.56) than the wild type (61.62) (Figure 1m‐2), suggesting that mutation of the TaDIR‐B1 gene significantly enhanced FCR resistance in tetraploid wheat. Additionally, lignin content and activities of phenylalanine ammonia‐lyase (PAL) and antioxidant enzymes (SOD, CAT) were significantly increased in the Kronos2734 plants (Figure 1m‐3). Furthermore, sequencing 862 mutant lines in the AK58 mutant library indicated that 17 of them possessed SNPs at TaDIR‐B1 that caused amino acid change compared with the wild type, and 5 mutant lines showed a G to A change at the 15‐bp position causing premature stop codon at the 5th position and the remaining 12 mutant lines resulted in 10 kinds of amino acid change for the TaDIR‐B1 gene (Figure 1n‐1). Investigation of FCR DI indicated that the 5 mutant lines (AK600, AK688, AK720, AK791 and AK948) with premature stop codon at TaDIR‐B1 gene possessed averaged DI of 34.26 and AK226 with a proline to leucine change possessed DI of 51.34, and their DIs were significantly lower than that of the wild type (60.36) (Figure 1n‐2, 3), suggesting that mutating the TaDIR‐B1 gene significantly increased FCR resistance in hexaploid wheat AK58. In summary, silencing TaDIR‐B1 gene led to enhanced FCR resistance in both tetraploid and hexaploid wheat.
To date, screening FCR‐resistant germplasm and QTL mapping of FCR resistance have been performed in several studies (Liu and Ogbonnaya, 2015; Yang et al., 2019), but no immune or highly resistant cultivars have been identified. In the present study, 4.8%, 82.1% and 13.1% of the evaluated accessions had the average DI of <30, 30‐50 and >50, respectively (Figure 1b), suggesting that vast majority of current wheat cultivars are susceptible to FCR. Some accessions (e.g. YZ‐67, YZ‐106, YZ‐170 and YZ‐425) showed averaged DI < 20, which could be considered as valuable germplasm for improvement of FCR resistance in wheat breeding programme.
In this study, the TaDIR‐B1 gene was identified to be associated with FCR resistance, and its function was verified by VIGS and EMS‐mutagenized wheat lines. As reported previously, DIR is an exocytic protein and first found in the woody plant. Though DIR cannot catalyse lignin synthesis directly, it can enable lignan to produce strict optical rotation and trap free lignin monomer radicals to guide the stereoselective coupling reaction of free radicals with the help of other oxidases (Chen, 2018). Paniagua et al. (2017) found that DIR gene possibly combined with laccase to mediate the polymerization and deposition of lignin in cell wall and lead to the dimerization of monolignols to form lignan resistance to pathogen. In this study, wheat lines with a silenced‐TaDIR‐B1 gene showed a relatively high level of resistance to FCR using VIGS and EMS mutation, suggesting that TaDIR‐B1 gene negatively regulated the FCR resistance in polyploid wheat. Physiological analysis showed that lignin accumulation was dramatically increased in silenced‐TaDIR‐B1 plants. Therefore, improvement of FCR resistance caused by loss of function of the TaDIR‐B1 gene may be attributed to accumulation of lignin in wheat plants. This study provides some important information on genetic loci controlling FCR resistance.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
FC conceived the study. XY, YR and CS collected the data. XY, FC, SZ, QZ, YR and CS analysed the data and interpreted the results. XY, FC and SZ wrote the manuscript with input from all authors.
Acknowledgements
This project was funded by the National Key Research and Development Program (2019YFE0118300), the National Natural Science Foundation (31861143008) Henan Major Science and Technology Projects (181100110200), and Ten Thousand Talents Plan (Z04295) of China.
Yang, X. , Zhong, S. , Zhang, Q. , Ren, Y. , Sun, C. and Chen, F. (2021) A loss‐of‐function of the dirigent gene TaDIR‐B1 improves resistance to Fusarium crown rot in wheat. Plant Biotechnol. J., 10.1111/pbi.13554
References
- Chen, R.B. (2018) Study on the mechanism of the Dirigent proteins to catalyze lignans in Isatis indugotica [D]. PhD dissertation: the Second Military Medical University, Shanghai,13–17. (In Chinese with English abstract).
- Chen, J.H. , Zhang, F.Y. , Zhao, C.J. , Lv, G.G. , Sun, C.W. , Pan, Y.B. , Guo, X.Y. and et al. (2019) Genome‐wide association study of six quality traits reveals the association of the TaRPP13L1 gene with flour color in Chinese bread wheat. Plant Biotechnol. J. 17, 2106–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmani, P.S. , Kamiya, T. , Danku, J. , Naseer, S. , Geldner, N. and Guerinot, M.L. (2013) Dirigent domain ‐containing protein is part of the machinery required for formation of the ligin‐based Casparian strip in the root. Proc. Natl Acad. Sci. USA, 110, 14498–14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. and Ogbonnaya, F.C. (2015) Resistance to Fusarium crown rot in wheat and barley: a review. Plant Breed. 134, 365–372. [Google Scholar]
- Nellist, C.F. , Vickerstaff, R.J. , Sobczyk, M.K. , Marina‐Montes, C. , Wilson, F.M. , Simpson, D.W. and Whitehouse, A.B. et al. (2019) Quantitative trait loci controlling Phytophthora cactorum resistance in the cultivated octoploid strawberry (Fragaria × ananassa). Hortic. Res. 6, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua, C. , Bilkova, A. , Jackson, P. , Dabravolski, S. , Riber, W. , Didi, V. , Houser, J. et al. (2017) Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 68, 3287–3301. [DOI] [PubMed] [Google Scholar]
- Ralph, S.G. , Park, J.Y. , Bohlmann, J. and Mansfield, S.D. (2006) Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound‐ and insect‐induced expression of a family of DIR and DIR‐like genes in spruce (Picea spp.). Plant Mol. Biol. 60, 21–40. [DOI] [PubMed] [Google Scholar]
- Reboledo, G. , Campo, R.D. , Alvarez, A. , Montesano, M. , Mara, H. and Ponce, I.L. (2015) Physcomitrella patens activates defense responses against the pathogen Colletotrichum gloeosporioides . Int. J. Mol. Sci. 16, 22280–22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.P. and Fristensky, B. (2001) Transgenic canola lines expressing pea defense gene DRR206 have resistance to aggressive blackleg isolates and to Rhizoctonia Solani . Mol. Breed. 8, 263–271. [Google Scholar]
- Yang, X. , Pan, Y.B. , Pawan, K.S. , He, X.Y. , Ren, Y. , Zhao, L. , Zhang, N. et al. (2019) Investigation and genome‐wide association study for Fusarium crown rot resistance in Chinese common wheat. BMC Plant Biol. 19, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]


