Self‐incompatibility (SI) is a genetic mechanism that prevents inbreeding in hermaphrodite angiosperms via the rejection of self‐pollen. SI lines facilitate the utilization of heterosis (Nettancourt, 2001). In cruciferous plants, SI is determined by the S locus, which harbors genes encoding the stigma papilla cell‐specific S‐receptor kinase (SRK) and the pollen surface‐localized ligand of SRK, S‐locus cysteine‐rich protein/S‐locus protein 11 (SCR/SP11) (Schopfer et al., 1999; Suzuki et al., 1999). Brassica napus is self‐compatible (SC), even though it is generated from two SI species, Brassica rapa and Brassica oleracea. SI B. napus could be generated by introducing the S locus from B. rapa or/and B. oleracea into B. napus by interspecific hybridization or genetic transformation (Fu and Liu, 1975; Gao et al., 2016). However, few B. napus SI lines are currently available due to the instability of offspring produced by interspecific hybridization, and silencing of the introduced BrSCR gene over several generations. One promising method for creating new B. napus SI lines is CRISPR/Cas9 genome editing (Chen et al., 2019).
In B. rapa, the linear dominance hierarchy of class II S haplotype SCR alleles is controlled by SCR‐methylation‐inducing region 2 (Smi2) in the S locus; the Smi2 of the dominant S locus generates small interfering RNAs (siRNAs), which suppresses the expression of the recessive S‐locus SCR by siRNA‐mediated DNA methylation (e.g., BrS44 > BrS60 > BrS40 > BrS29, Yasuda et al., 2016). BoS15 and BrS60 are interspecific pairs between B. oleracea and B. rapa based on their high sequence similarity and incompatibility of B. oleracea BoS15 pollen with BrS60/BoS18 interspecific hybrid stigmas (Sato et al., 2006). Therefore, BrS29 could be deduced to be recessive to BoS15. The B. napus SC line 326 has two S haplotypes, BnS7 (homolog of BrS29) in the A subgenome and BnS6 (homolog of BoS15) in the C subgenome, and low expression of BnSCR7 might confer SC to 326 (Zhai et al., 2014). We speculate that BnS7 should be recessive to BnS6. Then, the Smi2 was identified in BnS6 (Figure 1a), which could target promoter of BnSCR7 (Figure 1b). These results suggest that BnSCR7 might be suppressed by BnS6‐Smi2, and then knockout of BnS6‐Smi2 in 326 might recover BnSCR7 expression and create SI B. napus.
Figure 1.
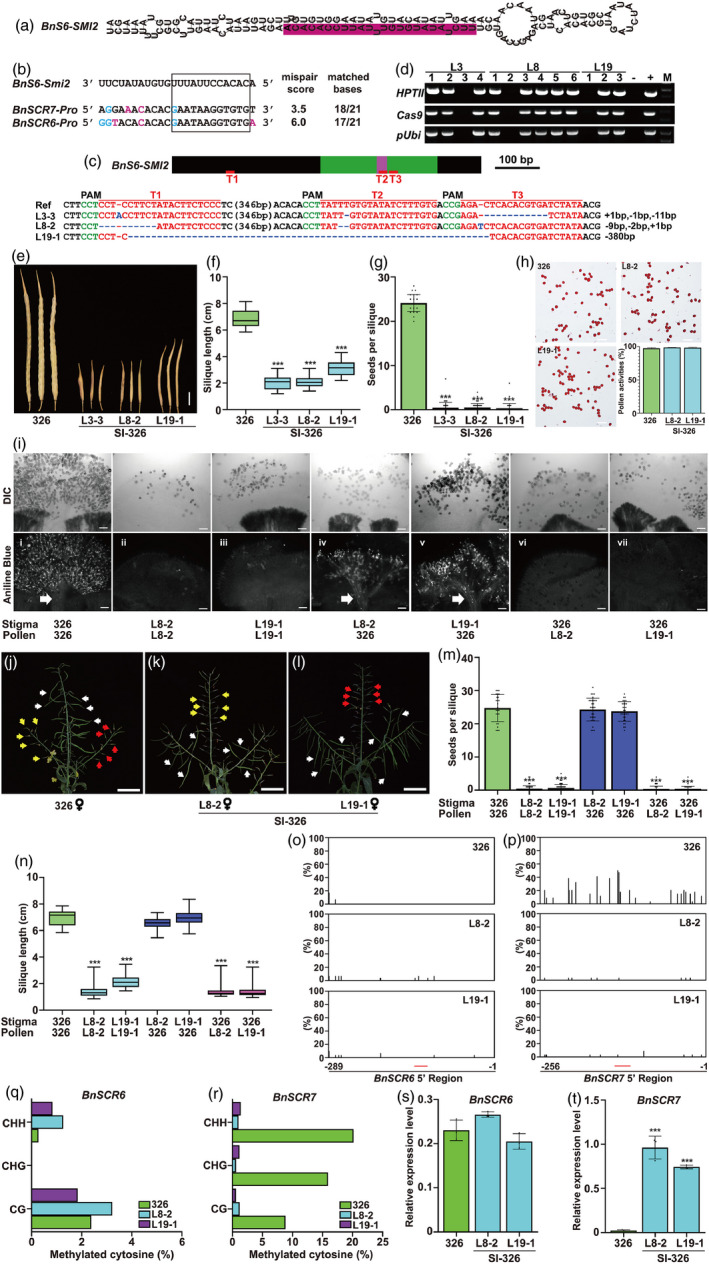
Generation of self‐incompatible B. napus by knocking out BnS6‐Smi2 via CRISPR/Cas9. (a) Stem‐loop structure of BnS6‐Smi2 precursor. The mature BnS6‐Smi2 is shown in prune. (b) Alignment of BnSCR6 and BnSCR7 promoter regions and seed sequence of BnS6‐Smi2. Mismatched bases and G/U pairs are shown in magenta and blue, respectively. (c) BnS6‐Smi2, including the sequences outside the siRNA precursor (black boxes), precursor region (green boxes) and mature siRNA (pink box). The red lines under the gene model indicate the sgRNA target sites. The protospacer adjacent motif (PAM) is shown in green. The sgRNA is shown in red. The mutation sites are indicated in blue. (d) Images showing PCR products of Cas9, HPTII and pUbi in SI‐326 lines in the T1 generation. +: pRGEB32 is used as the positive control. ‐: gDNA of 326 is used as the negative control. (e) The silique phenotypes of 326 and SI‐326 at 8 weeks after self‐pollination. Scale bars = 1 cm. (f) Quantification of silique length of 326 and SI‐326 after self‐pollination. (g) Seeds per silique of 326 and SI‐326 after self‐pollination. (h) Images showing the pollen viability of 326 and SI‐326. Quantification of pollen viability of 326 and SI‐326 is also shown. Scale bars = 100 µm. (i) Typical DIC (upper panel) and aniline blue staining (lower panel) of 326 and SI‐326 pistils after self‐ or cross‐pollination. A bundle of PTs indicates compatible pollination (white arrow). Scale bars = 100 µm. (j)–(l) Images showing the 326 and SI‐326 siliques pollinated by 326 and SI‐326 pollen. Pollens from 326, L8‐2 and L19‐1 are indicated by white, yellow and red arrows, respectively. Scale bars = 5 cm. (m) Seeds per silique of 326 and SI‐326 after self‐ or cross‐pollination. (n) Quantification of silique length of 326 and SI‐326 after self‐ or cross‐pollination. (o) ‐ (p) Percentage of methylation at all cytosine residues in 326 and SI‐326 in the promoter regions of BnSCR6 (nucleotides—289 to −1) (o) and BnSCR7 (nucleotides −256 to −1) (p). The red line indicates the region homologous to BnS6‐Smi2. (q)–(r) Percentage of cytosine methylation of CpG, CpNpG and CpNpN in 326 and SI‐326 in the promoter regions of BnSCR6 (nucleotides—289 to −1) (q) and BnSCR7 (nucleotides—256 to −1) (r). (s)–(t) Expression levels of BnSCR6 (s) and BnSCR7 (t) in 326 and SI‐326. Data are means ± SD (n = 3) obtained from three biological replicates. BnActin is used as the internal control. In (f), (g), (m) and (n), at least three plants per line were sampled for each measurement. Error bars are means ± SD (n ≥ 25), *** indicate significant differences at P < 0.001, based on ANOVA.
To test this hypothesis, three single‐guide RNAs (sgRNAs) were designed to target the regions of BnS6‐SMI2 (Figure 1c). Following transformation of these sgRNAs into 326, ten mutants were isolated in T0 generation, and three independent lines were genotyped in T1 generation. The deletions or insertions at sgRNA target sites were validated by sequencing (Figure 1c). The three transgene‐free homozygous mutants (named SI‐326) based on the absence of amplified transgene fragments were used for further analyses (Figure 1d).
The siliques of the three T1 transgene‐free SI‐326 lines were first observed. Following self‐pollination, their siliques were dramatically shorter than those of 326 (Figure 1e and f), and many siliques were completely empty (33/44 siliques for L3‐3, 30/45 for L8‐2 and 34/41 for L19‐1). The average seed‐set of each line was less than one seed per silique, which was less than 326 (24.15 seeds per silique) (Figure 1g). Like 326, SI‐326 had pollens with normal viability (Figure 1h). These results indicate that the reduced seed number of SI‐326 is not due to reduced pollen viability.
When wild‐type 326 stigmas were pollinated with SI‐326 pollen (L8‐2 or L19‐1) in T2 generation, pollen tube (PT) growth was strongly inhibited (Figure 1i: ii, iii, vi and vii), and the siliques and seed‐set were not significantly different from those of SI‐326 self‐pollination (Figure 1j‐l and m‐n). By contrast, pollination of SI‐326 stigma with 326 pollen resulted in normal PT growth (Figure 1i: i, iv v), and the siliques were normally developed like those of wild‐type 326 self‐pollination (Figure 1j‐l and m‐n). These results indicate that self‐incompatibility rather than fertility defect causes less self‐seed of the SI‐326 lines.
Bisulfite sequencing was performed to investigate relation of the transcriptional regulation of BnSCR7 with DNA methylation in SI‐326 and 326, as reported in B. rapa (Yasuda et al., 2016). The cytosine methylation levels were not significantly different between 326 and SI‐326 in BnSCR6 promoter regions (Figure 1o and q). By contrast, the cytosine methylation levels of CpG, CpNpG and CpNpN sequences in BnSCR7 promoter region in 326 were 8.82%, 15.88% and 20.16%, respectively (Figure 1p and r), which were much higher than those of the BnSCR6 promoter region (2.38%, 0% and 0.28%). However, the levels of methylated cytosines in this region were significantly lower in both SI‐326 lines (Figure 1p and r). Then, there was no difference in BnSCR6 expression between SI‐326 and 326 in unopened floral buds (Figure 1s), whereas the expression of BnSCR7 was 32‐fold higher in SI‐326 than in 326 (Figure 1t). These results suggest that BnS6‐Smi2 specifically suppresses BnSCR7 transcription and that the SI phenotype of SI‐326 is caused by decreased DNA methylation in BnSCR7 promoter region.
To date, very few SI B. napus are available for breeding. Here, we generated new SI B. napus lines by CRISPR/Cas9 mutation of BnS6‐Smi2. We demonstrated that BnS6‐Smi2 was essential for maintaining the SC phenotype of 326. Furthermore, we determined that the levels of methylated cytosines in BnSCR7 promoter were significantly reduced in SI‐326, indicating that BnSCR7 was suppressed by BnS6‐Smi2‐induced DNA methylation. In B. napus, the SC accessions that suffer from the same SC reason as accession 326 could possibly be engineered to be SI. The SI lines also could be generated in other Brassicaceae species including Brassica juncea and Brassica carinata, since the function of siRNA in S‐locus dominant–recessive interactions is highly conserved (Yasuda et al., 2016). Such transgene‐free homozygous mutants containing allele‐specific markers could be applied for the breeding of SI Brassica plants, thereby accelerating the utilization of heterosis.
Conflict of interest
The authors declare no conflicts of interest.
Author contributions
C.D., C.M. and S. D. designed the research. S.D. performed the experiments. J.T., J.S., B.Y., J.W. and T.F. provided laboratory support. C.D., T.Z. and S.D. analyzed the data. C.D., C.M. and S.D. wrote the manuscript. All authors read and approved the manuscript.
Acknowledgements
This research was supported by grants from National Key Research and Development Program of China (2016YFD0101803), and Central Plains Science and Technology Innovation Leading Talents (194200510021).
Dou, S. , Zhang, T. , Tu, J. , Shen, J. , Yi, B. , Wen, J. , Fu, T. , Dai, C. and Ma, C. (2021) Generation of novel self‐incompatible Brassica napus by CRISPR/Cas9. Plant Biotechnol. J, 10.1111/pbi.13577
The authors responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors are: Dr. Cheng Dai (cdai@mail.hzau.edu.cn) and Dr. Chaozhi Ma (yuanbeauty@mail.hzau.edu.cn).
Contributor Information
Cheng Dai, Email: cdai@mail.hzau.edu.cn.
Chaozhi Ma, Email: yuanbeauty@mail.hzau.edu.cn.
References
- Chen, K.L. , Wang, Y.P. , Zhang, R. , Zhang, H.W. and Gao, C.X. (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. [DOI] [PubMed] [Google Scholar]
- Fu, T.D. and Liu, H.L. (1975) Preliminary report on breeding of self‐incompatible lines of Brassica napus . Oil Crop China (in Chinese), 4, 77–85. [Google Scholar]
- Gao, C.B. , Zhou, G.L. , Ma, C.Z. , Zhai, W. , Zhang, T. , Liu, Z.Q. , Yang, Y. et al. (2016) Helitron‐like transposons contributed to the mating system transition from out‐crossing to self‐fertilizing in polyploid Brassica napus L. Sci. Rep. 6, 337–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettancourt, D. (2001) Incompatibility and Incongruity in Wild and Cultivated Plants, 2nd ed. Berlin/Heidelberg, New York: Springer‐Verlag. [Google Scholar]
- Sato, Y. , Sato, K. and Nishio, T. (2006) Interspecific pairs of class II Shaplotypes having different recognition specificities between Brassica oleracea and Brassica rapa . Plant Cell Physiol. 47, 340–345. [DOI] [PubMed] [Google Scholar]
- Schopfer, C.R. , Nasrallah, M.E. and Nasrallah, J.B. (1999) The male determinant of self‐incompatibility in Brassica . Science, 286, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Suzuki, G. , Kai, N. , Hirose, T. , Fukui, K. , Nishio, T. , Takayama, S. , Isogai, A. et al. (1999) Genomic organization of the S locus: identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa) . Genetics, 153, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, S. , Wada, Y. , Kakizaki, T. , Tarutani, Y. , Miura‐Uno, E. , Murase, K. , Fujii, S. et al. (2016) A complex dominance hierarchy is controlled by polymorphism of small RNAs and their targets. Nat. Plants, 3, 16206. [DOI] [PubMed] [Google Scholar]
- Zhai, W. , Zhang, J.F. , Yang, Y. , Ma, C.Z. , Liu, Z.Q. , Gao, C.B. , Zhou, G.L. et al. (2014) Gene expression and genetic analysis reveal diverse causes of recessive self‐compatibility in Brassica napus L. BMC Genom. 15, 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]


