Summary
Heat stress (HS) causes substantial damages to worldwide crop production. As a cool season crop, wheat (Triticum aestivum) is sensitive to HS‐induced damages. To support the genetic improvement of wheat HS tolerance (HST), we conducted fine mapping of TaHST1, a locus required for maintaining wheat vegetative and reproductive growth under elevated temperatures. TaHST1 was mapped to the distal terminus of 4AL chromosome arm using genetic populations derived from two BC6F6 breeding lines showing tolerance (E6015‐4T) or sensitivity (E6015‐3S) to HS. The 4AL region carrying TaHST1 locus was approximately 0.949 Mbp and contained the last 19 high confidence genes of 4AL according to wheat reference genome sequence. Resequencing of E6015‐3S and E6015‐4T and haplotype analysis of 3087 worldwide wheat accessions revealed heightened deletion polymorphisms in the distal 0.949 Mbp region of 4AL, which was confirmed by the finding of frequent gene losses in this region in eight genome‐sequenced hexaploid wheat cultivars. The great majority (86.36%) of the 3087 lines displayed different degrees of nucleotide sequence deletions, with only 13.64% of them resembling E6015‐4T in this region. These deletions can impair the presence and/or function of TaHST1 and surrounding genes, thus rendering global wheat germplasm vulnerable to HS or other environmental adversities. Therefore, conscientious and urgent efforts are needed in global wheat breeding programmes to optimize the structure and function of 4AL distal terminus by ensuring the presence of TaHST1 and surrounding genes. The new information reported here will help to accelerate the ongoing global efforts in improving wheat HST.
Keywords: genetic mapping, haplotype analysis, heat stress tolerance, gene deletion, TaHST1, wheat
Introduction
Heat stress (HS) is a major and constant threat to crop production (Lesk et al., 2016). As global warming continues, HS is expected to become more frequent and more severe (Reinman, 2013). Consequently, substantial worldwide efforts are being devoted to study and improve the genetic basis underlying HS tolerance (HST) in crop plants (Driedonks et al., 2016; Singh et al., 2019). As a typical cool season crop, wheat (Triticum aestivum, AABBDD) is vulnerable to HS, particularly at the flowering and grain‐filling stages (Asseng et al., 2015; Cossani and Reynolds, 2012; Kaur et al., 2019; Stratonovitch and Semenov, 2015; Zampieri et al., 2017). Many experiments have shown significant yield losses associated with HS in wheat (Balla et al., 2011; Li et al., 2019; Lizana and Calderini, 2013; Telfer et al., 2018; Wardlaw et al., 2002), and it is estimated that worldwide wheat yield may decrease by 4.1%–6.4% with each increase of 1 °C in global temperature (Liu et al., 2016a). Clearly, urgent efforts are needed to understand the mechanisms underlying wheat HST and to develop HS tolerant cultivars for maintaining global wheat production at present and trying to increase it in the future (Kaur et al., 2019; Posch et al., 2019).
To date, several approaches have been used to characterize the genetic and molecular mechanisms underlying wheat HST. Quantitative trait locus (QTL) mapping and genome‐wide association study (GWAS) suggest that wheat HST is under polygenetic control and affected by environmental conditions (Guan et al., 2018; Hassouni et al ., 2019; Li et al., 2019; Mason et al., 2010; Maulana et al., 2018; Paliwal et al., 2012; Sall et al., 2018; Shirdelmoghanloo et al., 2016; Tadesse et al ., 2019). For example, Li et al. (2019) conducted a comprehensive GWAS analysis and uncovered 27, 65 and 30 chromosomal loci controlling the tolerance to heat, drought and combined heat and drought stresses, respectively. Meanwhile, reverse genetic studies have revealed the involvement of a number of genes in wheat response and tolerance to HS (Kaur et al., 2019; Ni et al., 2018). Many of these genes encode regulatory proteins that participate in heat shock signalling and responsive pathways, for example TaHsfA2d, TaHSF3, TaHsfA6f, TaHsfC2a, Hsp16.9 and sHsp26 (Agarwal and Khurana, 2019; Chauhan et al., 2012, 2013; Comastri et al., 2018; Duan et al., 2019; Garg et al., 2012; Hu et al., 2018; Liu et al., 2020; Xue et al., 2015; Zhang et al., 2013), while others code for various types of proteins with different functional specificities (Agarwal and Khurana, 2018; Fu et al., 2008; Guo et al., 2015; He et al., 2016; Li et al., 2018; Qin et al., 2015; Singh and Khurana, 2016; Tian et al., 2019; Wang et al., 2014; Zang et al., 2017a,b, 2018, 2017a, 2017b; Zhang et al., 2017a). Consistent with the findings made in above research, transcriptomic and proteomic studies disclosed diverse genes and proteins whose expression is altered by heat treatment (Ni et al., 2018; Su et al., 2019; Wang et al., 2019; Zhang et al., 2017b).
Despite the progress outlined above, further efforts are still needed to better understand the genetic and molecular processes controlling wheat HST. To achieve this goal efficiently, it is desirable to isolate the genes that function directly in wheat HST by forward genetic analysis, which has proved to be highly effective in characterizing plant genes controlling important biological processes and agronomic traits (Bettgenhaeuser and Krattinger, 2019; Schneeberger and Weigel, 2011). However, it is challenging to isolate wheat genes controlling HST by positional cloning because heat response phenotypes are difficult to score, varying among genotypes and growth stages and requiring substantial inputs of time and labour (Li et al., 2019). Moreover, the hexaploid genome of wheat is both large (~16 Gb) and complex, with three subgenomes and ~85% repetitive sequences (IWGSC et al., 2018). Additionally, complicated chromosomal alterations, for examples large indels, translocations, inversions and alien introgressions, are frequently found among wheat cultivars (Cheng et al., 2019; He et al., 2019). These difficulties may explain why HST controlling genes have not been isolated by map‐based cloning in wheat despite the fact that heat tolerant wheat mutants were reported nearly 20 years ago (Mullarkey and Jones, 2000).
A major challenge in dissecting the genetic factors controlling HST is phenotypic characterization of a large number of genotypes (or individuals) in a short period of time. One convenient method is to conduct HST test at seedling stage in controlled growth chamber (Maulana et al ., 2018; Mullarkey and Jones, 2000), but the results obtained may not reflect HST at adult plant stage or under field conditions. Consequently, many researchers have evaluated wheat HST at adult stage in the field by covering flowering plants with heat stress shelters (Hassouni et al ., 2019; Li et al ., 2019; Tadesse et al., 2019). In this work, we conducted HST test under both growth chamber and field conditions to identify TaHST1, a chromosomal locus required for wheat HST at both seedling and adult stages. Following the scheme outlined in Figure S1, TaHST1 was fine‐mapped to a genomic region in the distal end of 4AL chromosome arm, which was ~0.949 Mbp according to the reference genome sequence of Chinese Spring (CS) (IWGSC et al., 2018). Further analysis revealed an unexpectedly high level of deletion polymorphisms in the terminal 0.949 Mbp region of 4AL, which was validated using genome sequence information generated by the 10+ Wheat Genomes Project (http://www.10wheatgenomes.com/). Our findings provide new information on the genetic basis of wheat HST, shed light on the structural variation of 4AL distal terminus and suggest the necessity to improve wheat HST by enhancing the structure and function of 4AL distal terminus.
Results
Characterization of the HS phenotypes of E6015‐3S and E6015‐4T
E6015‐3S and E6015‐4T were two BC6F6 spring wheat breeding lines derived from a cross between the Chinese cultivar Longmai 20 and the Canadian cultivar Glenlea (Figure 1a). To systematically characterize their difference in HST, we compared their responses to elevated temperature treatment at both seedling and adult plant stages. For the test at juvenile stage, the seedlings (at three‐leaf stage) were subjected to heat treatment (38°C) for three days followed by recovery at 20°C for three days. At the end of the recovery period, the leaves of E6015‐3S seedlings, but not those of E6015‐4T individuals, generally showed a pronounced wilting phenotype (Figure 1b), and while these seedlings apparently recovered to some extent after transferring to normal growth conditions, seed setting was substantially decreased. Physiological analysis conducted on the second day of recovery showed that E6015‐4T had a higher value of maximum quantum efficiency of photosystem II photochemistry (Fv /Fm ratio, 180.2% higher, P < 0.0001), more chlorophyll pigments (SPAD value, 386.4% higher, P < 0.0001), and better membrane stability (electrolyte leakage, 93.7% lower, P < 0.0001) than E6015‐3S, while in control seedlings (before HS) E6015‐4T and E6015‐3S did not differ significantly for the three measured parameters (Figure 1c).
Figure 1.
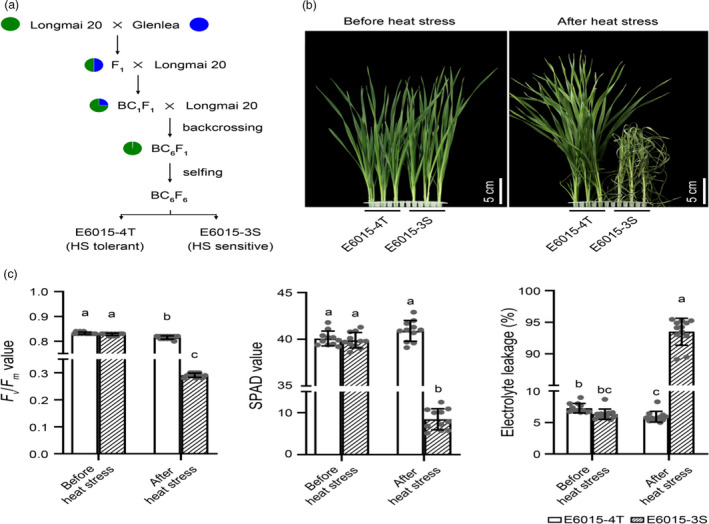
Origin of E6015‐3S and E6015‐4T and assessment of their HS responses at seedling stage. (a) Origin of E6015‐3S and E6015‐4T from the genetic cross between Longmai 20 and Glenlea. Longmai 20 was the recurrent parent and also the donor of heat tolerant allele (see also Figure 6b). (b) Morphology of E6015‐3S and E6015‐4T seedlings before HS or at the end of the recovery after HS treatment. (c) Comparison of maximal photochemistry efficiency (Fv /Fm ), level of chlorophyll (SPAD value), and electrolyte leakage between E6015‐3S and E6015‐4T seedlings before or after HS. The assays were executed either just before HS or at the second day of the recovery after HS treatment. The values, presented as means ± SE (n ≥ 10), were analysed using one‐way ANOVA and LSD multiple comparison method, with those marked by different letters being statistically significant (P < 0.05). The data shown were representative of three independent experiments.
In field test, E6015‐4T and E6015‐3S plants were covered with manually constructed thermal stress tents since the heading stage at early May, with uncovered individuals grown alongside as controls (Figure 2a). In a typical clear day in late May, the temperatures inside the tents became higher than those outside from 8 to 18 h, with the highest temperature differences occurred from 12 to 13 h, which were approximately 10–13 °C (Figure 2b). Compared with controls, the thermo‐stressed plants exhibited earlier leaf and spike senescence, which was considerably more severe in E6015‐3S (Figure 2c). No significant differences were found in plant height (PH), spike length (SL) and spikelet number (SN) between E6015‐3S and E6015‐4T under either thermo‐stressed or control conditions (Figure S2). However, E6015‐3S had significantly lower values of grain number per spike (GNS), grain weight per spike (GWS), thousand grain weight (TGW), grain length (GL) and grain width (GW) than E6015‐4T under either thermo‐stressed or control conditions (Figure 2d). Heat stress decreased GNS, GWS, TGW, GL and GW in both lines, with the percentages of reduction found for E6015‐3S being generally larger than those for E6015‐4T (Table S1). Clearly, E6015‐3S was significantly more susceptible to HS than E6015‐4T in both vegetative and reproductive growth.
Figure 2.
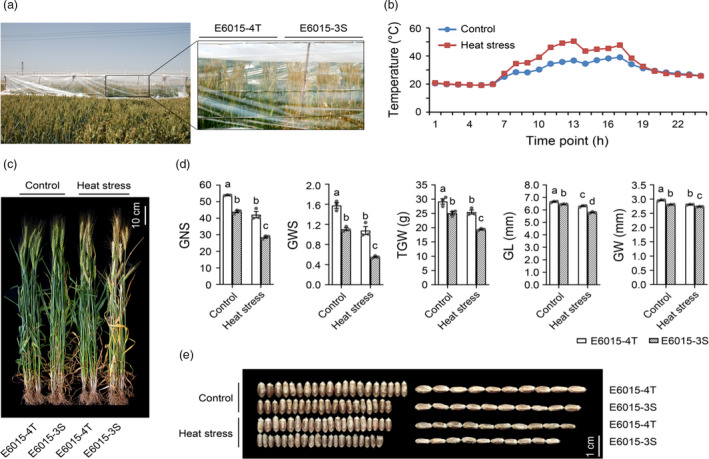
Assessment of the high temperature responses of E6015‐3S and E6015‐4T in the field. (a) Thermal stress tents used for heat treatment of adult E6015‐3S and E6015‐4T plants in the field. (b) Temperature differences inside (HS) and outside (Control) the thermal stress tents in a typical clear day in late May as monitored using Mini T data logger. (c) Morphology of E6015‐3S and E6015‐4T adult plants grown under control or heat stressed field conditions. (d) Comparison of grain number per spike (GNS), grain weight per spike (GWS), thousand grain weight (TGW), grain length (GL) and grain width (GW) of E6015‐3S and E6015‐4T plants grown under control or heat stressed field conditions. The values, presented as means ± SE of three independent measurements (each using 10 plants), were analysed by one‐way ANOVA and LSD multiple comparison method, with those marked by different letters being statistically significant (P < 0.05). (e) Morphological differences of E6015‐3S and E6015‐4T grains developed under control or heat stressed conditions in the field.
Genetic inheritance and fine mapping of TaHST1
The F1 seedlings, produced by E6015‐4T × E6015‐3S, exhibited a medium level of HST (Figure S3a). A subsample of the F2 population (272 seedlings) was phenotyped, which showed three types of HS responses, with 64 being tolerant (as displayed by E6015‐4T), 73 being sensitive (as shown by E6015‐3S), and 135 being intermediately tolerant (as exhibited by F1 seedlings). These values fitted well with a segregation ratio of 1 : 2 : 1 (χ 2 = 0.610; P > 0.05), suggesting that the HST displayed by E6015‐4T was conditioned by a single chromosomal locus. This locus was designated as TaHST1 to facilitate further analysis.
Five steps were taken to achieve fine mapping of TaHST1 locus (Figure 3). In the first step, E6015‐3S and E6015‐4T were comparatively analysed using the Affymetrix 55K SNP chip contained 66 835 SNP markers (Liu et al., 2018). Of the 64 905 SNPs with successful genotype calling, 64 557 (99.46%) were non‐polymorphic, with only 348 being polymorphic between the two lines (Table S2). Genome‐wide mapping of SNP marker positions using the genomic sequence of CS (IWGSC RefSeq v1.0, 14.789 Gb) (IWGSC et al., 2018) showed that the total physical distances covered by the 64 905 and 64 557 SNP markers were approximately 14.055 and 14.003 Gb, respectively. Among the 348 polymorphic SNPs, 250 were located in six contiguous polymorphic chromosomal segments, that is 1A.1 (45 SNPs, covering 10.721 Mbp), 2A.1 (23 SNPs, 6.008 Mbp), 3A.1 (16 SNPs, 0.906 Mbp), 4A.1 (12 SNPs, 4.851 Mbp), 4A.2 (137 SNPs, 27.068 Mbp) and 4B.1 (17 SNPs, 2.685 Mbp) (Figure 3a; Table S2). Thus, E6015‐3S and E6015‐4T were 99.46% similar in the assayed SNP markers, and about 99.63% similar in their genomes.
Figure 3.
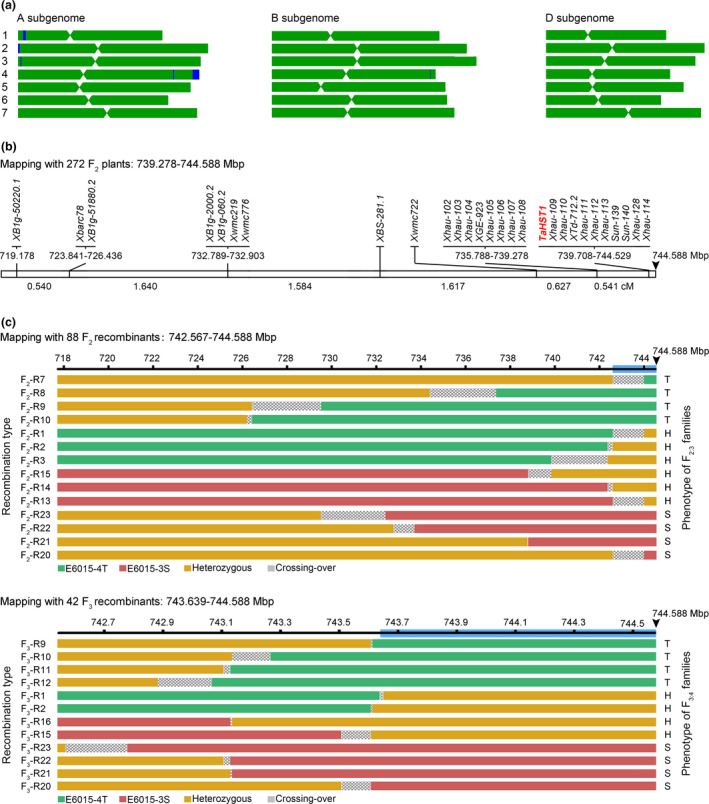
Major steps in TaHST1 mapping. (a) Genomic similarity and difference between E6015‐3S and E6015‐4T analysed using an Affymetrix 55K chip. The six regions shown in blue contained continuous stretches of polymorphic SNPs. (b) Mapping of TaHST1 to an interval of 5.310 Mbp (739.278–744.588 Mbp) using 272 F2 plants. (c) Reduction of TaHST1‐containing interval to 2.021 Mbp (742.567–744.588 Mbp) by analysing 88 F2 recombinants (top panel) and narrowing down TaHST1‐containing interval to 0.949 Mbp (743.639–744.588 Mbp) by analysing 42 F3 recombinants (lower panel). Arrowhead indicates the end position of 4AL (744.588 Mbp) according to CS genome sequence (IWGSC RefSeq assembly v1.0). This reference genome sequence was also used to infer the physical positions of DNA markers used in the mapping.
In the second step, E6015‐3S and E6015‐4T were scanned with 756 genome‐wide simple sequence repeat (SSR) markers; the resultant 26 polymorphic markers were used to analyse 93 F2 plants with known heat response phenotypes. This mapped TaHST1 to a location very close to the distal end of 4AL (Figure S3b), which was 744.588 Mbp according to IWGSC RefSeq v1.0 (IWGSC et al., 2018). Although five SSRs (Xbarc78, Xwmc776, Xwmc219, Xwmc497 and Xwmc722) were anchored upstream of TaHST1, we failed to identify downstream DNA markers (Figure S3b). Of the five upstream SSR markers, reliable physical position information was obtained for only Xbarc78 (723.841 Mbp) and Xwmc219 (732.903 Mbp) by searching CS reference genome sequence (Figure S3b). Thus, TaHST1 locus was assigned to the distal terminal region of 4AL (from 732.903 to 744.588 Mbp) after this step (Figure S3b).
In the third step, 272 F2 plants with known heat response phenotypes were genotyped using 27 SSR markers residing in the distal portion of 4AL (719.178–744.529 Mbp), 14 of which were newly developed and validated in this work (Table S3). TaHST1 co‐segregated with the 10 markers located from 739.708 to 744.529 Mbp, thus placing TaHST1 into a physical interval of 4.880 Mbp (739.708–744.588 Mbp) (Figure 3b). Notably, the recombination frequency was quite low, being 0.26 cM/Mbp for the 25.351 Mbp region bordered by markers XB1g‐50220.1 and Xhau114, which was further decreased to 0.10 cM/Mbp for the very distal end region (5.21 Mbp) delineated by Xhau‐108 and Xhau‐114 (Figure 3b).
In the fourth step, 88 F2 recombinants, selected from 1006 F2 plants using the SSR markers XB1g‐50220.1 (located at 719.178 Mbp), XB1g‐2000.2 (732.789 Mbp) and Sun‐140 (744.411 Mbp) (Figure 3c, top panel), were genotyped using the Affymetrix 55K SNP chip. This analysis validated the 137 polymorphic SNPs initially found for the 4A.2 region (Table S2), and these SNPs allowed classification of the 88 F2 recombinants into 28 types (Table S4), for which F2:3 families were obtained and phenotyped for HS responses. Combined analysis of the genotypic and phenotypic data of the 28 types of F2 recombinants (Table S4) reduced the physical interval of TaHST1 to 2.021 Mbp (742.567–744.588 Mbp) (Figure 3c, top panel). We also selected 466 F2 heterozygotes in this step based on the genotyping information of XB1g‐50220.1, XB1g‐2000.2 and Sun‐140, which were selfed and yielded 21 024 F3 plants for further mapping of TaHST1.
Lastly, 42 F3 recombinants were identified by screening the 21 024 F3 plants with the DNA markers Xhau‐111 (located at 742.548 Mbp) and Xhau‐128 (744.528 Mbp) (Figure 3b). The 42 F3 recombinants were divided into 24 types after genotyping with 40 DNA markers (including 38 newly developed) located in the 4AL region from 742.548 to 744.531 Mbp (Table S3), with their heat response phenotypes determined using derivative F3:4 families (Table S5). By integrating the genotypic and phenotypic data of these F3 recombinant types (Table S5), the physical interval of TaHST1 was narrowed down to 0.949 Mbp (743.639–744.588 Mbp) (Figure 3c, lower panel). According to CS RefSeq v1.0 (IWGSC et al., 2018), this region harbours the last 19 high confidence (HC) protein‐coding genes of 4AL (Table 1).
Table 1.
Functional annotations of the last 19 HC genes in the terminal 0.949 Mbp of Chinese Spring 4AL and comparison of their PCR amplicons in E6015‐4T and E6015‐3S
| Gene ID a | Annotation b | PCR amplicons from gene specific markers c | |
|---|---|---|---|
| E6015‐4T | E6015‐3S | ||
| TraesCS4A02G497900 | Receptor‐like kinase | +, +, +, + | −, −, −, − |
| TraesCS4A02G498000 | Thaumatin‐like protein | +, +, +, +, + | −, −, −, −, − |
| TraesCS4A02G498100 | Late embryogenesis abundant protein | +, + | −, − |
| TraesCS4A02G498200 | Terpene cyclase/mutase family member | +, +, +, + | −, −, −, − |
| TraesCS4A02G498300 | Cytochrome P450 protein | +, +, + | −, −, − |
| TraesCS4A02G498400 | WD repeat‐containing protein | +, +, +, +, + | −, −, −, −, − |
| TraesCS4A02G498600 | PHD zinc finger protein | +, +, + | +, +, − |
| TraesCS4A02G498700 | U‐box domain‐containing protein | + | + |
| TraesCS4A02G498800 | Phosphoenolpyruvate carboxylase | +, +, +, + | −, +, −, − |
| TraesCS4A02G498900 | Terpene cyclase/mutase family member | +, +, +, +, + | A, +, −, +, − |
| TraesCS4A02G499000 | Receptor‐like kinase | +, +, + | −, −, − |
| TraesCS4A02G499100 | Terpene cyclase/mutase family member | +, +, +, +, + | +, −, −, −, + |
| TraesCS4A02G499200 | RING/U‐box superfamily protein | +, +, +, + | +, −, A, − |
| TraesCS4A02G499300 | Methyltransferase‐like protein | +, +, +, + | −, −, −, − |
| TraesCS4A02G499400 | Receptor‐like kinase | +, +, +, +, + | +, +, +, A, A |
| TraesCS4A02G499500 | Plastocyanin | +, +, +, + | +, −, −, − |
| TraesCS4A02G499600 | ARF GAP‐like zinc finger protein | + | + |
| TraesCS4A02G499800 | Oxoglutarate/Fe(II)‐dependent oxygenase | +, +, +, + | −, −, −, − |
| TraesCS4A02G499900 | No apical meristem (NAM) protein | +, +, + | −, −, + |
Gene ID was obtained from the EnsemblPlants website (http://plants.ensembl.org/index.html).
Annotation information was derived from IWGSC RefSeq v1.1 (https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Annotations/v1.1/).
The 19 genes were each analysed with one or more gene specific DNA markers by PCR. ‘+’ and ‘−’ represent positive or negative amplification of the expected product deduced according to genomic DNA sequences of the 19 genes annotated in CS. ‘A’ indicates altered size of the amplicon from E6015‐3S relative to its counterpart from E6015‐4T. A total of 69 markers were used in this analysis, with marker names and locations of amplicons in the 19 target genes given in Table S8.
Comparative analysis of allelic genomic regions carrying TaHST1 locus
To gain a deeper insight into TaHST1 containing genomic region, we conducted two further analyses. First, the 2.043 Mbp terminal region (742.545–744.588 Mbp) of 4AL to which TaHST1 was mapped was analysed using 176 diagnostic SSR markers (Table S3), the majority of which were newly developed according to CS genome sequence. Of the 154 markers polymorphic between E6015‐3S and E6015‐4T, 109 (70.78%) were dominant and yielded PCR amplicons in E6015‐4T but not E6015‐3S; the remaining 45 co‐dominant markers tended to distribute in discrete patches (Figure 4a). This result indicated possible occurrence of nucleotide sequence deletions in the 4AL distal terminus of E6015‐3S as compared to that of E6015‐4T.
Figure 4.
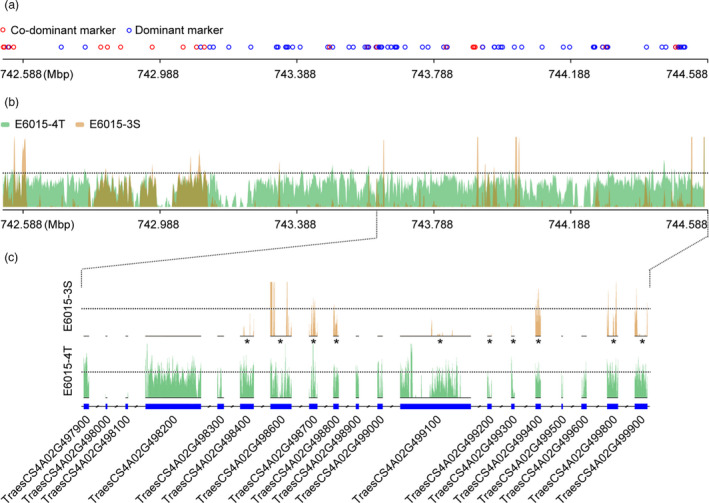
Comparative analyses of 4AL distal terminal regions of E6015‐3S and E6015‐4T using diagnostic DNA markers and through mapping resequencing reads. (a) Schematic representation of differences of marker amplifications in the compared genomic regions of the two lines. The co‐dominant markers amplified products in both lines, whereas the dominant markers amplified positively in only E6015‐4T. (b) Different patterns of resequencing read mapping found for E6015‐3S and E6015‐4T. The reads from E6015‐4T (green bars) covered the target genomic region much more extensively than those from E6015‐3S (brown bars). (c) Mapping the resequencing reads of E6015‐3S and E6015‐4T onto the last 19 HC genes of 4AL terminal region annotated by CS reference genome sequence. E6015‐4T reads (green bars) covered 17 of the 19 annotated genes, but those of E6015‐3S (brown bars) were found on only 10 of them (indicated by asterisks). TraesCS4A02G498000 and TraesCS4A02G498100 were poorly covered by the reads from either E6015‐4T or E6015‐3S.
Second, we conducted genome resequencing of E6015‐3S and E6015‐4T to check probable nucleotide sequence deletions in the 4AL distal terminus of E6015‐3S. The clean reads obtained for the two lines, being 18–20× coverage of common wheat genome (Table S6), were mapped to CS genome sequence. From Figure 4b, it is clear that the reads from E6015‐4T covered 4AL distal terminus extensively, indicating high similarity between E6015‐4T and CS in this region. However, the reads from E6015‐3S covered the examined region poorly, with many locations devoid of coverage (Figure 4b). Regarding the 19 HC genes located in the terminal 0.949 Mbp region of 4AL, the reads from E6015‐4T covered 17 of them (Figure 4c). In contrast, the reads from E6015‐3S covered only ten of the 19 genes (Figure 4c).
TraesCS4A02G498000 and TraesCS4A02G498100 were poorly covered by the reads from either E6015‐3S or E6015‐4T (Figure 4c). Bioinformatic analysis revealed that TraesCS4A02G498000 and TraesCS4A02G498100 were single‐exon genes, and they had three and two highly identical homologs (>97% identity), respectively, on other wheat chromosomes according to CS reference genome sequence (Table S7). Therefore, poor coverage of TraesCS4A02G498000 and TraesCS4A02G498100 by the reads of E6015‐3S or E6015‐4T may be caused by the presence of multiple closely related homologs. To investigate this possibility and the status of the remaining 17 genes in E6015‐3S and E6015‐4T, we performed PCR analysis using 69 DNA markers specific for the 19 genes (Tables S3 and S8), with CS as a control. The 69 markers all yielded anticipated amplicons identical between E6015‐4T and CS (Table 1; Table S8). But in E6015‐3S, only 15 of the 69 markers yielded the expected amplicons, four of them produced amplicons with altered size, and 50 of them did not show positive amplification (Table 1; Table S8). Based on these results, we deduced that the 19 HC genes were all and similarly present in E6015‐4T and CS, but at least 17 of them were affected by sequence deletion, alteration or both in E6015‐3S (Table 1; Table S8).
Since we used CS reference genome sequence to design the PCR markers for investigating nucleotide sequence and gene deletions in 4AL distal terminal region in E6015‐3S, there was a possibility that lack of amplification for certain markers in E6015‐3S may be caused by SNP polymorphisms and small indels in E6015‐3S genomic DNA, which prohibited efficient primer binding and thus PCR. To examine this possibility, we aligned the primers of all 264 PCR markers, designed for 4AL distal terminal region (Table S3), to the genome resequencing reads of E6015‐4T and E6015‐3S using Blastn (Figure S4). In E6015‐4T, perfect matching between PCR primers and resequencing reads was found for 257 markers (~97% of the 264 markers used), with imperfect matching observed for only seven markers (Table S3). Of the seven cases, four were caused by SNPs in E6015‐4T reads and three by the lack of matching resequencing reads (Figure S4, Table S3). This indicated high nucleotide sequence similarity between CS and E6015‐4T in 4AL distal terminus. However, in E6015‐3S, the corresponding figures were 60 (perfect matching), 73 (imperfect matching because of SNPs in E6015‐3S reads) and 131 (imperfect matching due to the lack of corresponding resequencing reads), respectively (Table S3). Thus, compared to CS, abundant nucleotide sequence and gene deletions did occur in the 4AL distal terminus of E6015‐3S. The diagnostic PCR markers we used were effective in revealing these deletions.
Presence/absence of the last 19 HC genes of CS 4AL in other genome‐sequenced wheat cultivars
Apart from CS, the 10+ Wheat Genomes Project also sequenced nine other diverse common wheat cultivars (http://www.10wheatgenomes.com/). To gain more insight into the deletion polymorphisms of 4AL distal terminus, we examined the presence/absence of the last 19 HC genes of CS 4AL in the nine genome‐sequenced wheat cultivars. Of them, ArinaLrFor carried all of the 19 HC genes, while Jagger having only six, making them most similar to or divergent from CS, respectively (Figure 5; Table S9). Gene losses in the remaining cultivars varied from three to five (Figure 5; Table S9). This result, plus the findings depicted in Figure 4 and Figure S4, indicated the occurrence of extensive nucleotide sequence and gene deletions in the distal end of 4AL in many wheat genotypes including E6015‐3S.
Figure 5.
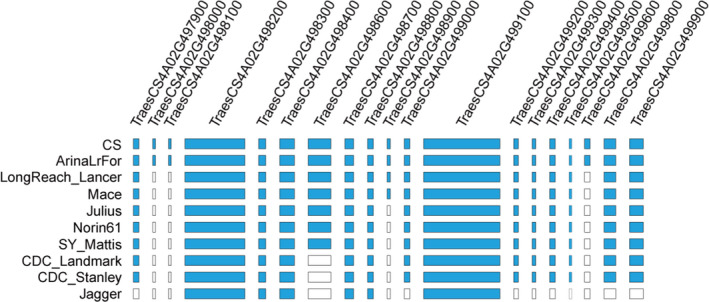
Presence/absence of the last 19 HC genes annotated for the 4AL distal terminus (743.639–744.588 Mbp) of CS in nine other sequenced common wheat cultivars. Gene losses are shown as blank rectangles. The names of the compared cultivars are displayed on the left side. The genome sequence of CS was downloaded from https://www.wheatgenome.org/Tools‐and‐Resources, whereas that of the other nine wheat cultivars was retrieved from http://www.10wheatgenomes.com/.
Haplotype analysis of 4AL distal terminal region in global wheat accessions
A panel of 3087 common wheat accessions, including 1852 spring and facultative lines and 1235 winter entries (Table S10) and representing a subset of the global common wheat germplasm core collection (Bulli et al., 2016; Maccaferri et al., 2015), was analysed with five DNA markers (Xhau‐1, Xhau‐2, Xhau‐3, Xhau‐4 and Xhau‐5, Table S3) that were distributed in the 0.949 Mbp interval harbouring TaHST1 (Figure 6a). Based on marker amplification patterns (Figure 6b), a total of 15 haplotypes were distinguished, with four major ones (Hap1 to Hap4) covering 2703 accessions (Table 2). E6015‐4T, CS and Longmai 20 belonged to Hap1, while E6015‐3S, Glenlea and Cadenza were assigned to Hap2 (Figure 6b; Table 2). Of the five DNA markers, Xhau‐1, Xhau‐2, Xhau‐3 and Xhau‐5 were dominant whereas Xhau‐4 was either co‐dominant or dominant depending on the haplotypes compared (Table 2). All five markers showed positive amplifications in Hap1, but there were one or more markers that failed to amplify in the remaining 14 haplotypes (Table 2). The finding of four unamplified markers in E6015‐3S conformed to the occurrence of multiple deletions in its 4AL terminal region (Figure 4; Table 1).
Figure 6.
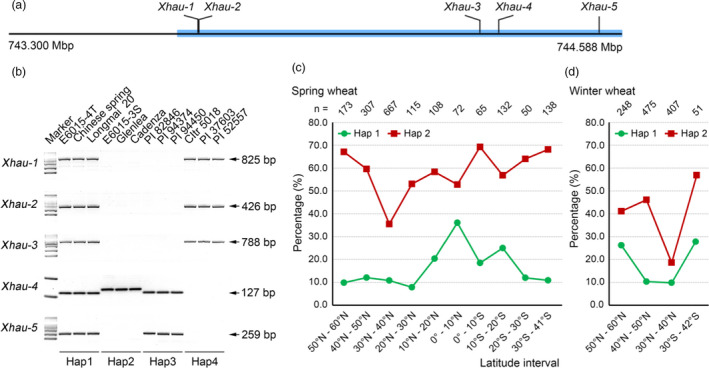
Haplotype analysis of 4AL distal terminal region to which TaHST1 was mapped in global common wheat accessions. The region analysed was 0.949 Mbp according to CS genome sequence (IWGSC RefSeq assembly v1.0). (a) Positions of the five diagnostic DNA markers (Xhau‐1, −2, −3, −4, and −5) used for haplotype analysis. The blue rectangle represents the terminal 0.949 Mbp of 4AL. (b) Amplicon patterns of four major haplotypes (Hap1–Hap4) exemplified using 12 common wheat genotypes. (c) Frequencies of Hap1 and Hap2 in the spring wheat lines originated from ten latitude intervals. (d) Frequencies of Hap1 and Hap2 in the winter wheat materials originated from four latitude intervals. In (c) and (d), n denotes the number of wheat lines analysed for each latitude interval.
Table 2.
Description of 15 haplotypes revealed for the terminal 0.949 Mbp of 4AL to which TaHST1 was mapped a
| Haplotype | Xhau‐1 | Xhau‐2 | Xhau‐3 | Xhau‐4 | Xhau‐5 | Deleted site | Number of lines | Frequency (%) |
|---|---|---|---|---|---|---|---|---|
| Hap1 | + | + | + | + (T) | + | 0 | 421 | 13.64 |
| Hap2 | − | − | − | + (S) | − | 4 | 1409 | 45.64 |
| Hap3 | − | − | − | + (T) | + | 3 | 491 | 15.91 |
| Hap4 | + | + | + | − | − | 2 | 382 | 12.38 |
| Hap5 | + | + | + | + (T) | − | 1 | 131 | 4.24 |
| Hap6 | − | − | + | + (T) | − | 3 | 106 | 3.43 |
| Hap7 | − | − | + | + (T) | + | 2 | 53 | 1.72 |
| Hap8 | − | − | − | + (S) | + | 3 | 36 | 1.17 |
| Hap9 | − | − | − | − | − | 5 | 17 | 0.55 |
| Hap10 | + | + | − | − | + | 2 | 3 | 0.10 |
| Hap11 | − | − | + | − | + | 3 | 10 | 0.32 |
| Hap12 | + | + | + | − | + | 1 | 9 | 0.29 |
| Hap13 | + | + | + | + (S) | − | 1 | 8 | 0.26 |
| Hap14 | − | − | + | − | − | 4 | 5 | 0.16 |
| Hap15 | − | − | − | + (T) | − | 4 | 6 | 0.19 |
| Total | 3087 | 100.00 |
A total of 3087 global common wheat accessions were analysed using five PCR‐based DNA markers, including Xhau‐1 (chr4A: 743680033‐743680857 bp), Xhau‐2 (chr4A: 743680939‐743681364 bp), Xhau‐3 (chr4A: 744277785‐744278572 bp), Xhau‐4 (chr4A: 744311304‐744311430 bp), and Xhau‐5 (chr4A: 744530489‐744530747 bp). Xhau‐1, −2, −3, and −5 were dominant markers, while Xhau‐4 was either co‐dominant or dominant depending the lines analysed. ‘+’ and ‘−’ denote positive and negative amplifications, respectively. ‘T’ and ‘S’ indicate the amplified alleles resembling that of E6015‐4T or E6015‐3S.
Globally, Hap2 was the most frequent haplotype (45.64%), followed by Hap3 (15.91%), Hap1 (13.64%) and Hap4 (12.38%) (Table 2). Because E6015‐3S and E6015‐4T were spring wheat, we thus performed a more detailed analysis of haplotype frequencies in the 1827 spring and facultative lines for which there are latitude information (Table S11), with the emphasis on Hap1 and Hap2. Among the ten latitude intervals, Hap1 frequencies were clearly elevated (18.46%–36.11%; Table S11) in the four latitude intervals (0°–10°N, 0°–10°S, 10°N–20°N and 10°S–20°S) with tropical or subtropical climates, with the highest frequency (36.11%) found for the interval 0°–10°N (Figure 6c). In contrast, Hap1 frequencies in the remaining intervals were comparatively low (Figure 6c), varying from 7.83%–12.05% (Table S11). Hap2 frequencies were substantially higher than those of Hap1 for all ten latitude intervals, and tended to rise as latitude increased towards the north or the south, with very high frequencies detected for the intervals 50°N–60°N (67.05%) and 30°S–41°S (68.12%) (Figure 6c; Table S11). The highest and lowest Hap2 frequencies were found in the latitude intervals 0°–10°S (69.23%) and 30°N–40°N (35.53%), respectively (Figure 6c; Table S11). Further inspection showed that haplotype diversity was lowest in the interval 0°–10°S (with only five haplotypes found) but highest in the interval 30°N–40°N (with all 15 haplotypes detected) (Table S11).
We next evaluated Hap1 and Hap2 frequencies in the winter wheat lines from three latitude intervals in the north globe (30°N–40°N, 40°N–50°N and 50°N–60°N) and one interval (30°S–42°S) in the south globe (Table S12). The frequencies of Hap2 were generally higher than those of Hap1, and both Hap1 and Hap2 frequencies were inclined to rise as latitude climbed towards the north or the south (Figure 6d). Hap1 and Hap2 frequencies were lowest for the interval 30°N–40°N (Figure 6d), which was correlated with considerably increased frequencies of Hap3, Hap5 and Hap7 in this interval (Table S12).
Regarding the haplotypes of TaHST1 containing genomic region in the accessions from ten major wheat producing countries, the frequency of Hap2 was much higher than that of Hap1, except in the winter lines from China and Germany where Hap1 frequency was larger than that of Hap2 (Table S13). Remarkably, Hap2 frequencies in the spring wheat from eight countries were all much higher than the global frequency of Hap2 (i.e. 45.64%), which was particularly evident for Russia (73.33%), Canada (72.97%), India (72.09%), United States (67.74%) and China (60.00%). In winter wheat, Hap1 frequency was low in Russia (0.00%), Ukraine (5.56%) and United States (6.19%), but was relatively high in China (22.22%) and Germany (45.45%); Hap2 frequency was exceptionally high in France (75.86%) and fairly high in United States (61.86%) (Table S13).
Discussion
In this study, we identified a novel chromosomal locus (TaHST1) required for wheat HST and mapped its location to the distal 0.949 Mbp region of 4AL. Furthermore, we discovered heightened deletion polymorphisms in the distal end of 4AL, a phenomenon that has not been reported before. Our findings have important and immediate implications on further study of TaHST1 and ongoing global efforts in the genetic improvement of wheat HST.
TaHST1 confers HST at both vegetative and reproductive growth stages of wheat
In the era of global warming with increasing incidences of heat waves, the type of HST that is active at both vegetative and reproductive stages is highly valuable for maintaining plant growth and final yield level. However, no prior studies have reported the identification of a wheat HST locus that is active at both seedling and adult plants stages and its fine mapping to a sub‐megabase level. From the data generated in this work, it is clear that our HS tests, conducted at both seedling stage and in the field as described previously (Hassouni et al ., 2019; Li et al., 2019; Maulana et al ., 2018; Tadesse et al., 2019), are effective in revealing HST differences between E6015‐3S and E6015‐4T, and that TaHST1 is required for wheat HST at both vegetative and reproductive stages. Lack of TaHST1 not only causes pronounced sensitivity to elevated temperatures at the seedling stage under laboratory conditions, but also leads to significant decreases in multiple yield‐related traits, that is GNS, GWS, TGW, GL and GW, in the field. Furthermore, TaHST1 is also likely required for maintaining efficient grain production in normal growth environments, because E6015‐3S showed lower values of yield‐related traits than E6015‐4T under control field temperatures. This may be due to the multifunctionalities of TaHST1 or the gene(s) tightly linked with TaHST1 locus (see also below).
Novel features in 4AL terminal region carrying TaHST1
First, the terminal 0.949 Mbp region of 4AL shows a very low recombination rate, which was estimated to be approximately 0.10 cM/Mbp using the F2 population derived from E6015‐4T × E6015‐3S (Figure 3b). A recent study employing the SynOpDH92 population suggests that recombination frequency is uneven along wheat chromosomes; although the average recombination frequency of chromosome 4A is 0.21 cM/Mbp, the frequency drops sharply to nearly zero towards the far end of 4AL (Gutierrez‐Gonzalez et al., 2019). This low recombination rate might explain our inability to map polymorphic DNA markers downstream of TaHST1.
Second, the 4AL distal terminus exhibits heightened deletion polymorphisms in wheat. Judging from the haplotype analysis data (Table 2), the great majority of global common wheat lines (86.36%) may contain one to five deletions in 4AL terminal region, with only a small proportion of them (13.64%) lacking the observed deletions. As exemplified by our analysis data on E6015‐3S (Figure 4; Table 1), these deletions can negatively affect gene composition in 4AL terminus, thus disrupting important wheat processes and traits including HST. The high gene deletion polymorphisms in 4AL distal terminus were also seen in the diverse cultivars sequenced by the 10+ Wheat Genomes Project (Figure 5), thus confirming our findings made using E6015‐3S, E6015‐4T and the 3087 common wheat accessions.
Finally, the intact 0.949 Mbp terminal region of 4AL, as annotated in CS and detected in E6015‐4T, is gene‐rich and carries multiple genes likely involved in stress tolerance. For example, the three genes (TraesCS4A02G497900, TraesCS4A02G499000 and TraesCS4A02G499400) encoding predicted RLKs, as well as the two genes (TraesCS4A02G498700 and TraesCS4A02G499200) specifying putative proteins involved in protein ubiquitination (Table 1), may regulate tolerance to various stressors including high temperature. This proposition is inferred from the improvement of thermotolerance in rice and tomato by overexpressing the ERECTA RLK (Shen et al., 2015), regulation of thermotolerance in African rice by OsTT1 that is a component of the ubiquitin/26S proteasome system (Li et al., 2015), and a positive role in rice seedling tolerance to heat stress played by OsHTAS that has a ubiquitin ligase activity (Liu et al., 2016b). Thus, the terminal 0.949 Mbp of 4AL may well play active roles in wheat responses to environmental stresses, which is consistent with the mapping of TaHST1 to this important region.
The distal terminus of present‐day 4AL of hexaploid wheat was originally derived from 7BS through translocation (Devos et al., 1995; Dvorak et al., 2018; Hernandez et al., 2012; Ma et al., 2013; Miftahudin et al., 2004). This chromosomal rearrangement event might confer instability to the translocated 7BS segment, thus leading to frequent deletions that may vary in number, size and location in different genetic backgrounds. Although this kind of phenomenon has not been studied in depth in plants, it is well known in animals that chromosomal rearrangements (including translocation) can accelerate the evolution of the segments involved and the genes carried by them, resulting in enhanced sequence polymorphisms and mutation rates (Bacolla et al., 2016; Marques‐Bonet et al., 2007; Navarro and Barton, 2003).
Environmental and breeding selections of Hap1‐type of TaHST1 locus
Elevation of Hap1 frequencies in the four latitude intervals (0°–10°N, 0°–10°S, 10°N–20°N, and 10°S–20°S) with tropical or subtropical climates (Figure 6c; Table S12) is indicative of possible environmental selection of Hap1‐type of TaHST1 locus in spring wheat. The much higher Hap1 frequencies (22.22%–45.45%, Table S13) in the winter wheat from China and Germany are probably the result of breeding selection. Consistent with this idea, an additional analysis of 270 Chinese modern wheat cultivars and advanced lines developed by intense breeding selection in recent years showed that the mean Hap1 frequency was increased to 35.93% (Table S14).
Despite potential environmental and breeding selections of Hap1‐type of TaHST1 locus, the frequencies of Hap2‐type of TaHST1 locus were alarmingly high, especially in the spring wheat from Russia (73.33%), Canada (72.97%), India (72.09%), United States (67.74%) and China (60.00%) and in the winter wheat from France (75.86%) and United States (61.86%). We speculate that this may be caused by the lack of vigorous selection of Hap1‐type of TaHST1 locus in past wheat breeding programmes. The likely hazardous effect(s) conferred by Hap2‐type of TaHST1 locus and surrounding gene deletions might have been compensated by homoeologous genes located in the terminal regions of 7AS and 7DS. However, this compensation may not be highly effective because the yield traits of E6015‐3S, which lacked TaHST1 and carried extensive deletions in 4AL distal terminus, were decreased under control conditions in the field relative to those of E6015‐4T (Table S1). In line with this proposition, a recent study in tetraploid cotton shows that the functional deficiency of two glycosyltransferase genes, caused by a terminal deletion in the short arm of chromosome 18, could not be compensated by their homoeologs, thus leading to a defective short fibre phenotype (Patel et al., 2020). From this finding and the contrasting performance of E6015‐3S and E6015‐4T under field conditions (Figure 2), we reckon that the deletions in 4AL distal end should be corrected timely so as to enhance wheat tolerance to more severe HS episodes that are likely to occur in the coming years because of global warming (Lesk et al., 2016; Reinman, 2013).
Implications for future research
The findings in this work have several implications for further research. First, because of low recombination rate in the distal terminus of 4AL, it is difficult to isolate TaHST1 by continuously decreasing its map interval. However, there are other ways that can be followed to isolate and characterize TaHST1. For example, potential candidate genes may be selected using publically available wheat gene expression data, which can then be validated by genome editing (GE), particularly multiplex GE (Chen et al., 2019).
Second, E6015‐3S may be used to identify and clone additional new genes controlling HST in common wheat. Briefly, heat tolerant wheat lines identified under field or laboratory conditions are firstly screened using the five diagnostic markers (Figure 6a), which excludes the genotypes with a Hap1‐type of TaHST1 locus from further analysis. A non‐Hap1 heat tolerant genotype is then crossed with E6015‐3S, with the resultant F2 seedlings treated according to the procedure depicted in Figure 1a. If a new HST locus is introduced from the non‐Hap1 heat tolerant genotype, the F2 seedlings would segregate monogenically for heat tolerant or sensitive phenotypes. Then, the new HST locus can be finely mapped, with the acting gene isolated by positional cloning. Moreover, E6015‐4T seeds may be mutagenized, and the resulting M2 (or M3) families are subjected to heat treatment, followed by more detailed analysis of heat sensitive mutants. Hence, appropriate utilization of E6015‐3S and E6015‐4T may enable more systematic dissection of the complex genetic basis of HST in wheat.
Third, wheat lines from the latitude interval 30°N–40°N showed unique haplotype characteristic in the terminal 0.949 Mbp region of 4AL, with all 15 haplotypes detected in the spring wheat accessions (Table S11). This may be due to the fact that the 30°N–40°N latitude interval encompasses many Fertile Crescent (FC) countries (e.g. Turkey, Iran and Iraq). It is well known that common wheat evolved in FC, and wheat germplasm from the FC countries often show enhanced genetic diversity (Balfourier et al., 2019; Venske et al., 2019). These haplotypes may be useful for studying the origin and evolution of TaHST1 locus in further research.
Finally, we suggest that from now on more rigorous efforts should be taken to raise the frequency of Hap1‐type TaHST1 locus in wheat breeding materials because of its associations with heat tolerance and a structurally intact 4AL terminal region. To this end, the wheat lines identified to carry Hap1‐type TaHST1 locus by this work may be used as donor materials. The PCR markers developed in this work may speed up the breeding processes involved.
Experimental procedures
Phenotyping HS response
HS responses of wheat seedlings were tested as described in the preceding section. Physiological assays and HS phenotyping were carried out before HS (as control) as well as at the second day of the recovery period. Chlorophyll fluorescence (Fv /Fm ) and content were evaluated as reported previously (Rosyara et al., 2010). Electrolyte leakage was quantified as detailed by Shan et al. (2015). The adult plants of E6015‐3S and E6015‐4T were examined for high temperature responses in the field using thermal stress tents (Figure 2), which is an effective method for simulating terminal heat stress under field conditions (Hassouni et al., 2019; Li et al., 2019). For both E6015‐3S and E6015‐4T, three different plots were covered by thermal tents from heading stage. The shelters remained in place until seed harvest. Adjacent replicates were uncovered as controls. Temperatures inside and outside the thermal tents were recorded using Mini T data logger (TL100, Zoglab, Hangzhou, China). No rainfall occurred after the application of heat stress tents. But two irrigations were provided at the middle and late grain‐filling stages, respectively. PH, SL, SN, GNS, GWS, TGW, GL and GW were assessed using 10 plants from each plot (Zhai et al., 2016, 2018). The field experiment was conducted in two wheat crop years (2018 and 2019, respectively) with similar results obtained.
SNP chip assay
Genomic DNA was extracted from plant materials using the Trelief™ Plant Genomic DNA Kit (http://www.tsingke.net) and quantified with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington). The resulting DNA samples were genotyped using the 55K SNP Array, which was designed to analyse 66 835 SNPs, by CapitalBio Technology Company (Beijing, China) as described previously (Liu et al., 2018). The physical positions of SNP markers were obtained by blasting their flanking sequences against the IWGSC RefSeq assembly v1.0 with the following parameters: e‐value ≥ 1e‐10, identity ≥ 95%, mismatches ≤ 5.
Mapping of TaHST1
Briefly, the initial mapping used 26 polymorphic SSR markers (https://wheat.pw.usda.gov), which included 93 F2 plants and was executed as described before (Somers et al., 2004; Zhai et al., 2016). Subsequent mapping necessitated the development of new DNA markers (i.e., Xhau markers, Table S3) in the 4AL terminal region (719.178–744.588 Mbp) according to Chinese Spring genome sequence. The various populations used in the mapping were developed as follows. First, 20 F1 hybrids derived from E6015‐4T × E6015‐3S were selfed to produce 1278 F2 individuals, 272 of which were used in the initial mapping. Second, the remaining 1006 F2 individuals were searched for recombinants and heterozygotes occurred within the 719.178–744.411 (Mbp) interval with the markers XB1g‐50220.1 (719.2 Mbp), XB1g‐2000.2 (732.8 Mbp) and Sun‐140 (744.3 Mbp). The resultant 88 F2 recombinants were genotyped with the 55K SNP chip and selfed to produce F2:3 families. Third, 466 F2 heterozygotes obtained in the previous step were selfed to produce 21 024 F3 individuals, which were screened for recombinants by genotyping with the markers Xhau‐111 and Xhau‐128. The resulting 42 F3 recombinants were further genotyped with 40 DNA markers, followed by selfing to produce F3:4 families. The F2:3 and F3:4 families were evaluated for HS phenotypes as described above.
Whole‐genome resequencing of E6015‐3S and E6015‐4T
About 10 μg of genomic DNA, extracted from the young leaves (Murray and Thompson, 1980), was used to construct a paired‐end sequencing library for each genotype following Illumina’s standard pipeline. The insert size was approximately 350 bp, with the read length being 150 bp. The libraries were sequenced on an Illumina® HiSeq X Ten platform. The raw reads were processed with Trimmomatic (version 0.36) (Bolger et al., 2014), with the resultant clean reads (~ 20× genome coverage for each line) aligned to CS genome sequence (IWGSC RefSeq assembly v1.0) using BWA‐MEM (version 0.7.17‐r1188) (Li and Durbin, 2009). Duplicate reads were removed using MarkDuplicates in GATK tools (version 4.0.10.1). Reads with low mapping quality (Q < 40) or multiple hits were removed with Samtools (version 1.9) (Li et al., 2009). The read mapping depth was approximately 20× genome coverage for both lines.
Examination of gene losses in 4AL distal terminus
The last 19 HC genes annotated for CS 4AL were examined for their collinear counterparts in the nine wheat cultivars sequenced by the 10+ Wheat Genomes Project (http://www.10wheatgenomes.com/). The MCscan package [https://github.com/tanghaibao/jcvi/wiki/MCscan‐(Python‐version)] was used with default parameters for this investigation.
Haplotype analysis and PCR detection of HC genes in 4AL distal terminal region
Xhau‐1, Xhau‐2, Xhau‐3, Xhau‐4 and Xhau‐5, located in the terminal 0.949 Mbp region of 4AL (Table S3), were employed for haplotype analysis (Zhai et al., 2016). A total of 69 gene specific markers were designed for the 19 HC genes annotated for the terminal 0.949 Mbp of 4AL of CS (Tables S3 and S8), with their 4A chromosome specificity confirmed using CS and the nulli‐tetrasomic line N4AT4B (Yu et al., 2010). They were then employed for analysing the 19 genes in E6015‐3S, E6015‐4T and CS as described previously (Zhai et al., 2018).
Statistical analysis
Numerical values were presented as means ± SE. Statistical analysis was performed using one‐way ANOVA and LSD comparison installed in the SPSS software package (SPSS version 19.0 for Windows; SPSS Inc., Chicago, IL, USA).
Conflict of interest
The authors declare no conflict of interests.
Author contributions
D.W. and K.Z. perceived the project. H.Z., C.J., Y.Z. and S.Y. performed the mapping. H.Z., K.Y., S.W., B.L. and Y.D. took part in phenotype screening and field experiment. H.Z., G.L., K.Z. and H.J. conducted haplotype, bioinformatic, and gene detection analyses. Y.Z., K.Z. and X.L. selected and maintained E6015‐3S and E6015‐4T lines. F.L. and Y.L. multiplied and managed global wheat accessions. X.O., W.Q., Z.J., T.P., D.G., W.H., J.W., H.G. and G.Y. evaluated Hap1 frequencies in modern Chinese wheat materials. H.Z., D.W. and M.R. wrote the manuscript.
Supporting information
Figure S1 Schematic representation of the actions taken to characterize TaHST1.
Figure S2 Comparison of three agronomic traits between E6015‐3S and E6015‐4T grown in the field.
Figure S3 Heat stress phenotype of F1 seedlings and preliminary mapping of TaHST1.
Figure S4 Mapping PCR primer sequences to E6015‐3S and E6015‐4T genome resequencing reads.
Table S1 Assessment of yield related traits of E6015‐3S and E6015‐4T grown in the field.
Table S2 List of 348 SNPs polymorphic between E6015‐3S and E6015‐4T.
Table S3 PCR markers used in this work and mapping their primer sequences to E6015‐3S and E6015‐4T genome resequencing reads.
Table S4 Description of 28 F2 recombinant types and their corresponding F2:3 families.
Table S5 Description of 24 F3 recombinant types and their corresponding F3:4 families.
Table S6 Statistics of genome resequencing reads of E6015‐3S and E6015‐4T.
Table S7 Highly similar homologs of TraesCS4A02G498000 and TraesCS4A02G498100 in wheat genome.
Table S8 PCR detection of the last 19 high confidence genes of 4AL in E6015‐3S and E6015‐4T.
Table S9 Comparison of the last 19 high confidence genes of 4AL in ten wheat varieties.
Table S10 List of the 3087 global common wheat accessions used for haplotype analysis.
Table S11 Frequencies of 15 haplotypes in the spring and facultative wheat lines.
Table S12 Frequencies of 15 haplotypes in the winter wheat materials.
Table S13 Hap1 and Hap2 frequencies in the wheat lines from 10 major wheat production countries.
Table S14 Haplotypes of TaHST1 containing genomic region in 270 Chinese modern wheat materials.
Acknowledgements
This work was supported by the Ministry of Science and Technology of China (2017YFD0100600 and 2017YFD0101000), China Postdoctoral Science Foundation (2018M630820), and Henan Postdoctoral Science Foundation. We thank USDA‐ARS Small Grains and Potato Germplasm Research Unit (https://www.ars‐grin.gov) for providing the wheat accessions used in the haplotype analysis. The authors are grateful to Dr. Fuhao Lu (Henan University, China) for advice on genome resequencing analysis.
Zhai, H. , Jiang, C. , Zhao, Y. , Yang, S. , Li, Y. , Yan, K. , Wu, S. , Luo, B. , Du, Y. , Jin, H. , Liu, X. , Zhang, Y. , Lu, F. , Reynolds, M. , Ou, X. , Qiao, W. , Jiang, Z. , Peng, T. , Gao, D. , Hu, W. , Wang, J. , Gao, H. , Yin, G. , Zhang, K. , Li, G. and Wang, D. (2021) Wheat heat tolerance is impaired by heightened deletions in the distal end of 4AL chromosomal arm. Plant Biotechnol. J., 10.1111/pbi.13529
Contributor Information
Kunpu Zhang, Email: zkp66@126.com.
Guangwei Li, Email: wheatwill@126.com.
Daowen Wang, Email: dwwang@henau.edu.cn.
References
- Agarwal, P. and Khurana, P. (2018) Characterization of a novel zinc finger transcription factor (TaZnF) from wheat conferring heat stress tolerance in Arabidopsis . Cell Stress Chaperon. 23, 253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal, P. and Khurana, P. (2019) Functional characterization of HSFs from wheat in response to heat and other abiotic stress conditions. Funct. Integr. Genom. 19, 497–513. [DOI] [PubMed] [Google Scholar]
- Asseng, S. , Ewert, F. , Martre, P. , Rötter, R.P. , Lobell, D.B. , Cammarano, D. , Kimball, B.A. et al. (2015) Rising temperatures reduce global wheat production. Nat. Clim. Chang. 5, 143–147. [Google Scholar]
- Bacolla, A. , Tainer, J.A. , Vasquez, K.M. and Cooper, D.N. (2016) Translocation and deletion breakpoints in cancer genomes are associated with potential non‐B DNA‐forming sequences. Nucleic Acids Res. 44, 5673–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balfourier, F. , Bouchet, S. , Robert, S. , De Oliveira, R. , Rimbert, H. , Kitt, J. , Choulet, F. et al. (2019) Worldwide phylogeography and history of wheat genetic diversity. Sci. Adv. 5, eaav0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla, K. , Rakszegi, M. , Li, Z. , Bekes, F. , Bencze, S. and Veisz, O. (2011) Quality of winter wheat in relation to heat and drought shock after anthesis. Czech J. Food Sci. 29, 117–128. [Google Scholar]
- Bettgenhaeuser, J. and Krattinger, S.G. (2019) Rapid gene cloning in cereals. Theor. Appl. Genet. 132, 699–711. [DOI] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulli, P. , Zhang, J. , Chao, S. , Chen, X. and Pumphrey, M. (2016) Genetic architecture of resistance to stripe rust in a global winter wheat germplasm collection. G3: Genes ‐ Genomes ‐ Genet. 6, 2237–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, H. , Khurana, N. , Agarwal, P. , Khurana, J.P. and Khurana, P. (2013) A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment. PLoS One, 8, e79577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, H. , Khurana, N. , Nijhavan, A. , Khurana, J.P. and Khurana, P. (2012) The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 35, 1912–1931. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Wang, Y. , Zhang, R. , Zhang, H. and Gao, C. (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. [DOI] [PubMed] [Google Scholar]
- Cheng, H. , Liu, J. , Wen, J. , Nie, X. , Xu, L. , Chen, N. , Li, Z. et al. (2019) Frequent intra‐ and inter‐species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol. 20, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comastri, A. , Janni, M. , Simmonds, J. , Uauy, C. , Pignone, D. , Nguyen, H.T. and Marmiroli, N. (2018) Heat in wheat: exploit reverse genetic techniques to discover new alleles within the Triticum durum sHsp26 family. Front. Plant Sci. 9, 1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossani, C.M. and Reynolds, H.P. (2012) Physiological traits for improving heat tolerance in wheat. Plant Physiol. 160, 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K. , Dubcovsky, J. , Dvorak, J. , Chinoy, C. and Gale, M. (1995) Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor. Appl. Genet. 91, 282–288. [DOI] [PubMed] [Google Scholar]
- Driedonks, N. , Rieu, I. and Vriezen, W.H. (2016) Breeding for plant heat tolerance at vegetative and reproductive stages. Plant Reprod. 29, 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, S. , Liu, B. , Zhang, Y. , Li, G. and Guo, X. (2019) Genome‐wide identification and abiotic stress‐responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genom. 20, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak, J. , Wang, L. , Zhu, T. , Jorgensen, C.M. , Luo, M.C. , Deal, K.R. , Gu, Y.Q. et al. (2018) Reassessment of the evolution of wheat chromosomes 4A, 5A, and 7B. Theor. Appl. Genet. 131, 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J. , Momčilović, I. , Clemente, T.E. , Nersesian, N. , Trick, H.N. and Ristic, Z. (2008) Heterologous expression of a plastid EF‐Tu reduces protein thermal aggregation and enhances CO2 fixation in wheat (Triticum aestivum) following heat stress. Plant Mol. Biol. 68, 277–288. [DOI] [PubMed] [Google Scholar]
- Garg, D. , Sareen, S. , Dalal, S. , Tiwari, R. and Singh, R. (2012) Heat shock protein based SNP marker for terminal heat stress in wheat (Triticum aestivum L.). Aust. J. Crop Sci. 6, 1516–1521. [Google Scholar]
- Guan, P. , Lu, L. , Jia, L. , Kabir, M.R. , Zhang, J. , Lan, T. , Zhao, Y. et al. (2018) Global QTL analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield‐related traits across different environments in wheat (Triticum aestivum L.). Front. Plant Sci. 9, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W. , Zhang, J. , Zhang, N. , Xin, M. , Peng, H. , Hu, Z. , Ni, Z. and Du, J. (2015) The wheat NAC transcription factor TaNAC2L is regulated at the transcriptional and post‐translational levels and promotes heat stress tolerance in transgenic Arabidopsis . PLoS One, 10, e0135667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez‐Gonzalez, J.J. , Mascher, M. , Poland, J. and Muehlbauer, G.J. (2019) Dense genotyping‐by‐sequencing linkage maps of two Synthetic W7984 × Opata reference populations provide insights into wheat structural diversity. Sci. Rep. 9, 1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassouni, K.E. , Belkadi, B. , Filali‐Maltouf, A. , Tidiane‐Sall, A. , Al‐Abdallat, A. , Nachit, M. and Bassi, F.M. (2019) Loci controlling adaptation to heat stress occurring at the reproductive stage in durum wheat. Agronomy, 9, 414. [Google Scholar]
- He, F. , Pasam, R. , Shi, F. , Kant, S. , Keeble‐Gagnere, G. , Kay, P. , Forrest, K. et al. (2019) Exome sequencing highlights the role of wild‐relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet. 51, 896–904. [DOI] [PubMed] [Google Scholar]
- He, G.H. , Xu, J.Y. , Wang, Y.X. , Liu, J.M. , Li, P.S. , Chen, M. , Ma, Y.Z. et al. (2016) Drought‐responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis . BMC Plant Biol. 16, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, P. , Martis, M. , Dorado, G. , Pfeifer, M. , Gálvez, S. , Schaaf, S. , Jouve, N. et al. (2012) Next generation sequencing and syntenic integration of flow‐sorted arms of wheat chromosome 4A exposes the chromosome structure and gene content. Plant J. 69, 377–386. [DOI] [PubMed] [Google Scholar]
- Hu, X.J. , Chen, D. , Lynne Mclntyre, C. , Fernanda Dreccer, M. , Zhang, Z.B. , Drenth, J. , Kalaipandian, S. et al. (2018) Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA‐mediated regulatory pathway. Plant Cell Environ. 41, 79–98. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium , Appels, R. , Eversole, K. , Feuillet, C. , Keller, B. , Rogers, J. and Stein, N. et al. (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, 361, eaar7191. [DOI] [PubMed] [Google Scholar]
- Kaur, R. , Sinha, K. and Bhunia, R.K. (2019) Can wheat survive in heat? Assembling tools towards successful development of heat stress tolerance in Triticum aestivum L. Mol. Biol. Rep. 46, 2577–2593. [DOI] [PubMed] [Google Scholar]
- Lesk, C. , Rowhani, P. and Ramankutty, N. (2016) Influence of extreme weather disasters on global crop production. Nature, 529, 84–87. [DOI] [PubMed] [Google Scholar]
- Li, X. , Chao, D. , Wu, Y. , Huang, X. , Chen, K. , Cui, L. , Su, L. et al. (2015) Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 47, 827–833. [DOI] [PubMed] [Google Scholar]
- Li, H. and Durbin, R. (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , Marth, G. et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Mao, X. , Wang, J. , Chang, X. , Reynolds, M. and Jing, R. (2019) Genetic dissection of drought and heat‐responsive agronomic traits in wheat. Plant Cell Environ. 42, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Wang, W. , Wang, W. , Zhang, G. , Liu, Y. , Wang, Y. and Wang, W. (2018) Wheat F‐Box protein gene TaFBA1 is involved in plant tolerance to heat stress. Front. Plant Sci. 9, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Asseng, S. , Müller, C. , Ewert, F. , Elliott, J. , Lobell, D.B. , Martre, P. et al. (2016. a) Similar estimates of temperature impacts on global wheat yield by three independent methods. Nat. Clim. Chang. 6, 1130–1136. [Google Scholar]
- Liu, Z. , Li, G. , Zhang, H. , Zhang, Y. , Zhang, Y. , Duan, S. , Sheteiwy, M.S.A. et al. (2020) TaHsfA2‐1, a new gene for thermotolerance in wheat seedlings: Characterization and functional roles. J. Plant Physiol. 246–247, 153135. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Luo, W. , Qin, N. , Ding, P. , Zhang, H. , Yang, C. , Mu, Y. et al. (2018) A 55 K SNP array‐based genetic map and its utilization in QTL mapping for productive tiller number in common wheat. Theor. Appl. Genet. 131, 2439–2450. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Zhang, C. , Wei, C. , Liu, X. , Wang, M. , Yu, F. , Xie, Q. et al. (2016b) The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2‐induced stomatal closure in rice. Plant Physiol. 170, 429–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizana, X. and Calderini, D. (2013) Yield and grain quality of wheat in response to increased temperatures at key periods for grain number and grain weight determination: considerations for the climatic change scenarios of Chile. J. Agric. Sci. 151, 209–221. [Google Scholar]
- Ma, J. , Stiller, J. , Berkman, P.J. , Wei, Y. , Rogers, J. , Feuillet, C. , Dolezel, J. et al. (2013) Sequence‐based analysis of translocations and inversions in bread wheat (Triticum aestivum L.). PLoS One, 8, e79329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri, M. , Zhang, J. , Bulli, P. , Abate, Z. , Chao, S. , Cantu, D. , Bossolini, E. et al. (2015) A genome‐wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3: Genes ‐ Genomes ‐ Genet. 5, 449–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques‐Bonet, T. , Sànchez‐Ruiz, J. , Armengol, L. , Khaja, R. , Bertranpetit, J. , Lopez‐Bigas, N. , Rocchi, M. et al. (2007) On the association between chromosomal rearrangements and genic evolution in humans and chimpanzees. Genome Biol. 8, R230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, R.E. , Mondal, S. , Beecher, F.W. , Pacheco, A. , Jampala, B. , Ibrahim, A.M. and Hays, D.B. (2010) QTL associated with heat susceptibility index in wheat (Triticum aestivum L.) under short‐term reproductive stage heat stress. Euphytica, 174, 423–436. [Google Scholar]
- Maulana, F. , Ayalew, H. , Anderson, J.D. , Kumssa, T.T. , Huang, W. and Ma, X. (2018) Genome‐wide association mapping of seedling heat tolerance in winter wheat. Front. Plant Sci. 9, 1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftahudin , Ross, K. , Ma, X‐f , Mahmoud, A.a , Layton, J. , Milla, M.A.R. , Chikmawati, T. et al. (2004) Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics, 168, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullarkey, M. and Jones, P. (2000) Isolation and analysis of thermotolerant mutants of wheat. J. Exp. Bot. 51, 139–146. [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, A. and Barton, N.H. (2003) Chromosomal speciation and molecular divergence‐accelerated evolution in rearranged chromosomes. Science, 300, 321–324. [DOI] [PubMed] [Google Scholar]
- Ni, Z. , Li, H. , Zhao, Y. , Peng, H. , Hu, Z. , Xin, M. and Sun, Q. (2018) Genetic improvement of heat tolerance in wheat: recent progress in understanding the underlying molecular mechanisms. Crop J. 6, 32–41. [Google Scholar]
- Paliwal, R. , Röder, M.S. , Kumar, U. , Srivastava, J. and Joshi, A.K. (2012) QTL mapping of terminal heat tolerance in hexaploid wheat (T. aestivum L.). Theor. Appl. Genet. 125, 561–575. [DOI] [PubMed] [Google Scholar]
- Patel, J.D. , Huang, X. , Lin, L. , Das, S. , Chandnani, R. , Khanal, S. , Adhikari, J. et al. (2020) The Ligon lintless‐2 short fiber mutation is located within a terminal deletion of chromosome 18 in cotton. Plant Physiol. 183, 277–288. 10.1104/pp.19.01531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch, B.C. , Kariyawasam, B.C. , Bramley, H. , Coast, O. , Richards, R.A. , Reynolds, M.P. , Trethowan, R. et al. (2019) Exploring high temperature responses of photosynthesis and respiration to improve heat tolerance in wheat. J. Exp. Bot. 70, 5051–5069. [DOI] [PubMed] [Google Scholar]
- Qin, D. , Wang, F. , Geng, X. , Zhang, L. , Yao, Y. , Ni, Z. , Peng, H. et al. (2015) Overexpression of heat stress‐responsive TaMBF1c, a wheat (Triticum aestivum L.) multiprotein bridging factor, confers heat tolerance in both yeast and rice. Plant Mol. Biol. 87, 31–45. [DOI] [PubMed] [Google Scholar]
- Reinman, S.L. (2013) Intergovernmental Panel on Climate Change (IPCC). Encyclopaedia Energy Natural Resource Environ. Econom. 26, 48–56. [Google Scholar]
- Rosyara, U.R. , Subedi, S. , Duveiller, E. and Sharma, R.C. (2010) Photochemical efficiency and SPAD value as indirect selection criteria for combined selection of spot blotch and terminal heat stress in wheat. J. Phytopathol. 158, 813–821. [Google Scholar]
- Sall, A.T. , Bassi, F.M. , Cisse, M. , Gueye, H. , Ndoye, I. , Filali‐Maltouf, A. and Ortiz, R. (2018) Durum wheat breeding: In the heat of the Senegal river. Agriculture, 8, 99. [Google Scholar]
- Schneeberger, K. and Weigel, D. (2011) Fast‐forward genetics enabled by new sequencing technologies. Trends Plant Sci. 16, 282–288. [DOI] [PubMed] [Google Scholar]
- Shan, C. , Zhou, Y. and Liu, M. (2015) Nitric oxide participates in the regulation of the ascorbate‐glutathione cycle by exogenous jasmonic acid in the leaves of wheat seedlings under drought stress. Protoplasma, 252, 1397–1405. [DOI] [PubMed] [Google Scholar]
- Shen, H. , Zhong, X.B. , Zhao, F.F. , Wang, Y.M. , Yan, B.X. , Li, Q. et al. (2015) Overexpression of receptor‐like kinase ERECTA improves thermotolerance in rice and tomato. Nat. Biotechnol. 33, 996–1003. [DOI] [PubMed] [Google Scholar]
- Shirdelmoghanloo, H. , Taylor, J.D. , Lohraseb, I. , Rabie, H. , Brien, C. , Timmins, A. , Martin, P. et al. (2016) A QTL on the short arm of wheat (Triticum aestivum L.) chromosome 3B affects the stability of grain weight in plants exposed to a brief heat shock early in grain filling. BMC Plant Biol. 16, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. and Khurana, P. (2016) Molecular and functional characterization of a wheat B2 protein imparting adverse temperature tolerance and influencing plant growth. Front. Plant Sci. 7, 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, B. , Salaria, N. , Thakur, K. , Kukreja, S. , Gautam, S. and Goutam, U. (2019) Functional genomic approaches to improve crop plant heat stress tolerance. F1000Research, 8, 1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.J. , Isaac, P. and Edwards, K. (2004) A high‐density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109, 1105–1114. [DOI] [PubMed] [Google Scholar]
- Stratonovitch, P. and Semenov, M.A. (2015) Heat tolerance around flowering in wheat identified as a key trait for increased yield potential in Europe under climate change. J. Exp. Bot. 66, 3599–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, P. , Jiang, C. , Qin, H. , Hu, R. , Feng, J. , Chang, J. , Yang, G. et al. (2019) Identification of potential genes responsible for thermotolerance in wheat under high temperature stress. Genes (Basel), 10, pii: E174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadesse, W. , Suleiman, S. , Tahir, I. , Sanchez‐Garcia, M. , Jighly, A. , Hagras, A. , Thabet, S. et al. (2019) Heat‐tolerant QTLs associated with grain yield and its components in spring bread wheat under heat‐stressed environments of Sudan and Egypt. Crop Sci. 59, 199–211. [Google Scholar]
- Telfer, P. , Edwards, J. , Bennett, D. , Ganesalingam, D. , Able, J. and Kuchel, H. (2018) A field and controlled environment evaluation of wheat (Triticum aestivum) adaptation to heat stress. Field Crop Res. 229, 55–65. [Google Scholar]
- Tian, X. , Wang, F. , Zhao, Y. , Lan, T. , Yu, K. , Zhang, L. , Qin, Z. et al. (2019) Heat shock transcription factor A1b regulates heat tolerance in wheat and Arabidopsis through OPR3 and jasmonate signalling pathway. Plant Biotechnol. J. 18, 1109–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venske, E. , Dos Santos, R.S. , Busanello, C. , Gustafson, P. and Costa de Oliveira, A. (2019) Bread wheat: a role model for plant domestication and breeding. Hereditas, 156, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Chen, S. , Shi, X. , Liu, D. , Zhao, P. , Lu, Y. , Cheng, Y. et al. (2019) Hybrid sequencing reveals insight into heat sensing and signaling of bread wheat. Plant J. 98, 1015–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Zang, X.S. , Kabir, M.R. , Liu, K.L. , Liu, Z.S. , Ni, Z.F. , Yao, Y.Y. et al. (2014) A wheat lipid transfer protein 3 could enhance the basal thermotolerance and oxidative stress resistance of Arabidopsis . Gene, 550, 18–26. [DOI] [PubMed] [Google Scholar]
- Wardlaw, I.F. , Blumenthal, C. , Larroque, O. and Wrigley, C.W. (2002) Contrasting effects of chronic heat stress and heat shock on kernel weight and flour quality in wheat. Funct. Plant Biol. 29, 25–34. [DOI] [PubMed] [Google Scholar]
- Xue, G.P. , Drenth, J. and McIntyre, C.L. (2015) TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 66, 1025–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C. , Li, Y. , Li, B. , Liu, X. , Hao, L. , Chen, J. , Qian, W. et al. (2010) Molecular analysis of phosphomannomutase (PMM) genes reveals a unique PMM duplication event in diverse Triticeae species and the main PMM isozymes in bread wheat tissues. BMC Plant Biol. 10, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampieri, M. , Ceglar, A. , Dentener, F. and Toreti, A. (2017) Wheat yield loss attributable to heat waves, drought and water excess at the gloabl, national and subnational scales. Environ. Res. Lett. 12, 064008. [Google Scholar]
- Zang, X. , Geng, X. , He, K. , Wang, F. , Tian, X. , Xin, M. , Yao, Y. et al. (2018) Overexpression of the wheat (Triticum aestivum L.) TaPEPKR2 gene enhances heat and dehydration tolerance in both wheat and Arabidopsis . Front. Plant Sci. 9, 1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang, X. , Geng, X. , Liu, K. , Fei, W. , Liu, Z. , Zhang, L. , Yue, Z. et al. (2017a) Ectopic expression of TaOEP16‐2‐5B, a wheat plastid outer envelope protein gene, enhances heat and drought stress tolerance in transgenic Arabidopsis plants. Plant Sci. 258, 1–11. [DOI] [PubMed] [Google Scholar]
- Zang, X. , Geng, X. , Wang, F. , Liu, Z. , Zhang, L. , Zhao, Y. , Tian, X. et al. (2017b) Overexpression of wheat ferritin gene TaFER‐5B enhances tolerance to heat stress and other abiotic stresses associated with the ROS scavenging. BMC Plant Biol. 17, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, H. , Feng, Z. , Du, X. , Song, Y. , Liu, X. , Qi, Z. , Song, L. et al. (2018) A novel allele of TaGW2‐A1 is located in a finely mapped QTL that increases grain weight but decreases grain number in wheat (Triticum aestivum L.). Theor. Appl. Genet. 131, 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, H. , Feng, Z. , Li, J. , Liu, X. , Xiao, S. , Ni, Z. and Sun, Q. (2016) QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high‐density SNP and SSR‐based linkage map. Front. Plant Sci. 7, 1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Geng, X. , Zhang, H. , Zhou, C. , Zhao, A. , Wang, F. , Zhao, Y. et al. (2017a) Isolation and characterization of heat responsive gene TaGASR1 from wheat (Triticum aestivum L.). J. Plant Biol. 60, 57–65. [Google Scholar]
- Zhang, Y. , Pan, J. , Huang, X. , Guo, D. , Lou, H. , Hou, Z. , Su, M. et al. (2017b) Differential effects of a post‐anthesis heat stress on wheat (Triticum aestivum L.) grain proteome determined by iTRAQ. Sci. Rep. 7, 3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Xu, Z.S. , Li, P. , Yang, L. , Wei, Y. , Chen, M. , Li, L. et al. (2013) Overexpression of TaHSF3 in transgenic Arabidopsis enhances tolerance to extreme temperatures. Plant Mol. Biol. Rep. 31, 688–697. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Schematic representation of the actions taken to characterize TaHST1.
Figure S2 Comparison of three agronomic traits between E6015‐3S and E6015‐4T grown in the field.
Figure S3 Heat stress phenotype of F1 seedlings and preliminary mapping of TaHST1.
Figure S4 Mapping PCR primer sequences to E6015‐3S and E6015‐4T genome resequencing reads.
Table S1 Assessment of yield related traits of E6015‐3S and E6015‐4T grown in the field.
Table S2 List of 348 SNPs polymorphic between E6015‐3S and E6015‐4T.
Table S3 PCR markers used in this work and mapping their primer sequences to E6015‐3S and E6015‐4T genome resequencing reads.
Table S4 Description of 28 F2 recombinant types and their corresponding F2:3 families.
Table S5 Description of 24 F3 recombinant types and their corresponding F3:4 families.
Table S6 Statistics of genome resequencing reads of E6015‐3S and E6015‐4T.
Table S7 Highly similar homologs of TraesCS4A02G498000 and TraesCS4A02G498100 in wheat genome.
Table S8 PCR detection of the last 19 high confidence genes of 4AL in E6015‐3S and E6015‐4T.
Table S9 Comparison of the last 19 high confidence genes of 4AL in ten wheat varieties.
Table S10 List of the 3087 global common wheat accessions used for haplotype analysis.
Table S11 Frequencies of 15 haplotypes in the spring and facultative wheat lines.
Table S12 Frequencies of 15 haplotypes in the winter wheat materials.
Table S13 Hap1 and Hap2 frequencies in the wheat lines from 10 major wheat production countries.
Table S14 Haplotypes of TaHST1 containing genomic region in 270 Chinese modern wheat materials.


