Abstract
How far do marine larvae disperse in the ocean? Decades of population genetic studies have revealed generally low levels of genetic structure at large spatial scales (hundreds of kilometres). Yet this result, typically based on discrete sampling designs, does not necessarily imply extensive dispersal. Here, we adopt a continuous sampling strategy along 950 km of coast in the northwestern Mediterranean Sea to address this question in four species. In line with expectations, we observe weak genetic structure at a large spatial scale. Nevertheless, our continuous sampling strategy uncovers a pattern of isolation by distance at small spatial scales (few tens of kilometres) in two species. Individual-based simulations indicate that this signal is an expected signature of restricted dispersal. At the other extreme of the connectivity spectrum, two pairs of individuals that are closely related genetically were found more than 290 km apart, indicating long-distance dispersal. Such a combination of restricted dispersal with rare long-distance dispersal events is supported by a high-resolution biophysical model of larval dispersal in the study area, and we posit that it may be common in marine species. Our results bridge population genetic studies with direct dispersal studies and have implications for the design of marine reserve networks.
Keywords: dispersal, gene flow, isolation by distance, relatedness, marine reserves
1. Introduction
Marine species tend to have high fecundities, pelagic larval stages, large population sizes, and extensive geographic ranges [1,2]. The marine environment is also more homogenous than terrestrial and freshwater environments [3] and presents fewer geographic barriers to the movement of organisms [2]. Altogether, these factors provide the potential for high demographic and genetic connectivity among populations [4], which are key drivers of marine ecosystem resilience and therefore fundamental to establish sound and effective conservation and management practices [5]. Decades of population genetic studies have established that the genetic structure is generally weak across the seascape [6,7], and the advent of high-throughput sequencing has not fundamentally changed this view [8]. Yet, the weak genetic structure does not necessarily imply high gene flow and can even persist for thousands of generations in the absence of gene flow when effective population sizes are large, which is typical of marine populations [9,10]. Furthermore, levels of gene flow that are sufficient to prevent the establishment of a strong genetic structure may be insignificant from a demographic perspective (e.g. just one migrant per generation in the infinite island model, [11]). Thus, while a strong genetic structure can be generally interpreted as a sign of low demographic exchange [12], a weak genetic structure can be compatible with either extensive or restricted dispersal [13–15].
Direct approaches to investigate larval dispersal such as mass mark-recapture or parentage analyses support the predominance of restricted dispersal in a variety of species and indicate that dispersal kernels are often a rapidly decreasing function of distance, with little dispersal beyond a few tens of kilometres [16–23]. This suggests that long-distance dispersal, defined here as dispersal beyond 40 km [24], is rare. Nevertheless, parentage analyses are challenging to scale up because the number of samples that need to be genotyped rises disproportionately with the increasing spatial scale (but see e.g. refs. [16,23]). They also provide a snapshot of dispersal over just one generation and therefore have to be repeated for every new generation analyzed (e.g. refs. [18,20]).
Coupling individual-based sampling designs with robust estimates of relatedness between any pair of individuals provides the potential to estimate dispersal at different spatial and temporal scales. This approach can be applied at any spatial scale and allows one to go deeper into the recent past by identifying relationships that are more distant than parent-offspring. It also offers the possibility to work at the level of individuals in continuous space, as opposed to the population-level analyses that are typically conducted at large spatial scales. Although the norm for the study of marine organisms, the population-level approach can generate biases and artefacts when the sampling design artificially discretizes species distributions that are much more continuous in reality [25–27].
Coupling genetic approaches with individual sampling and high-resolution biophysical models allows us to further refine our understanding of larval dispersal [23,28]. This is the challenge we propose to tackle by combining a continuous sampling design, seascape genomic analyses, and biophysical modelling of larval dispersal. We hypothesize that (i) the genetic structure should be weak at a large spatial scale (hundreds of kilometres), as is often the case in marine species [8]. We also theorize that (ii) if most dispersal is restricted as suggested by direct approaches, a pattern of genetic isolation by distance (IBD) should emerge at small spatial scales (few tens of kilometres). IBD refers to the tendency of individuals or populations to be more similar genetically the closer they are to each other geographically, which is a direct consequence of restricted dispersal [29]. As predicted by the theory [30], IBD is expected to weaken or disappear at larger spatial scales. Finally, we hypothesize that (iii) if long-distance dispersal events do occur, we should be able to capture such events through the identification of pairs of individuals that are geographically distant (hundreds of kilometres) but closely related genetically.
To test our hypotheses, we focused on four marine species along 950 km of coast in the north-western Mediterranean Sea, from Cabo de Gata (Spain) in the south to Cerbères-Banyuls (France) in the north (figure 1). The northern region is mainly influenced by the deep saline Mediterranean current, while the southern region is rich in nutrients from the Atlantic-Mediterranean surface current. The four species are the white seabream (Diplodus sargus, Linnaeus 1758), the striped red mullet (Mullus surmuletus, Linnaeus 1758), the comber (Serranus cabrilla, Linnaeus 1758), and the European spiny lobster (Palinurus elephas, Fabricius 1787, figure 1). The four species are reported over the entire study region [31] and include a variety of pelagic larval durations (PLD) and life-history traits (electronic supplementary material, table S1): P. elephas has the longest PLD (4–6 months), while D. sargus and M. surmuletus have a similar PLD (25–35 days) but may differ in terms of seasonal migratory pattern (present in D. sargus [32], unknown in M. surmuletus). Serranus cabrilla is sedentary [33], solitary, and has the shortest PLD (21–28 days), so it is presumably the species with less dispersal potential among our study species, although we note that PLD is only weakly correlated with dispersal [15]. This comparative approach is adopted to provide a deeper understanding of the determinants of connectivity in the marine realm [34].
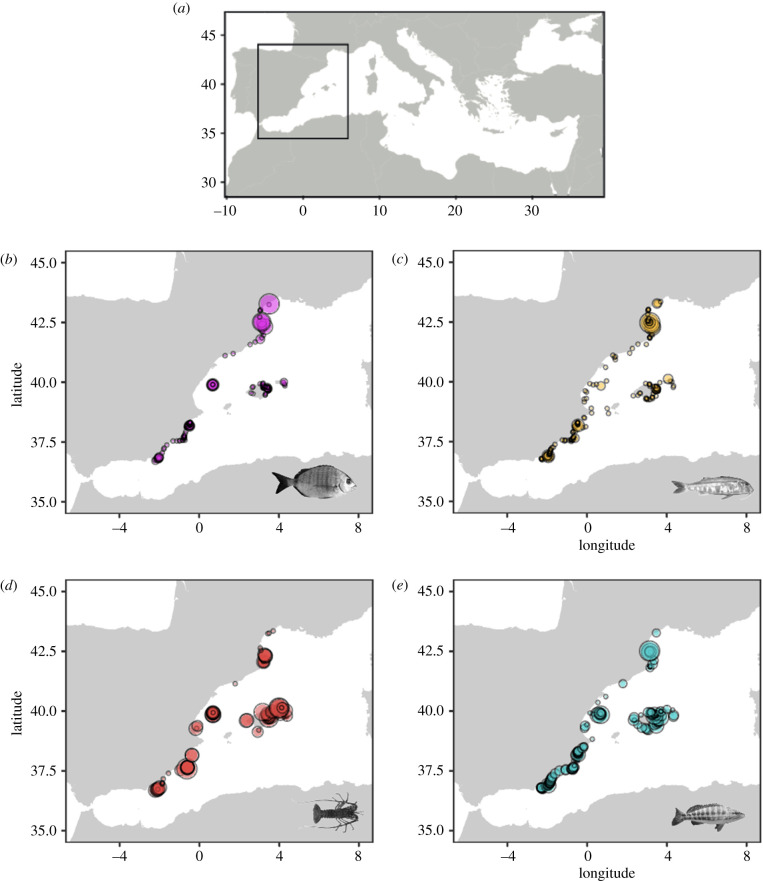
Figure 1.
Sampling design. Four species were sampled as continuously as possible along 950 km of coast in the north-western Mediterranean Sea for a total of 1299 individuals. (a) White seabream (Diplodus sargus) (n = 276), (b) red mullet (Mullus surmuletus) (n = 312), (c) European spiny lobster (Palinurus elephas) (n = 243), and (d) comber (Serranus cabrilla) (n = 456). The size of circles reflects the number of individuals per site (mean = 2, range 1–31 samples). A total of 615 sites were sampled. (Online version in colour.)
2. Material and methods
(a) . Sampling design and study species
We sampled as continuously as possible along 950 km of coast in the north-western Mediterranean Sea (figure 1; electronic supplementary material, figure S1). This strategy provides the opportunity to explore IBD across a continuum of spatial scales, from just a few kilometres to close to 950 km. Mature specimens were mostly obtained from artisanal fishery landings at shallow depths (mean 46 m, range 1.8–693.0 m) between June and November 2017. The fact that we sampled adults implies that our genetic data integrate the effect of dispersal at both the larval and adult stages. This is particularly relevant in our case since the habitats are more or less continuous, which implies that adults can potentially move throughout these habitats [28,32,35,36]. The exact position of each sample was provided by the fishermen. We obtained a total of 2043 samples that consisted of clips from pectoral fins (fishes) or pleopods (lobster). The samples were preserved in 96% ethanol before storage at 4°C. Among these, 1305 samples were selected for genotyping (280 for D. sargus, 312 for M. surmuletus, 245 for P. elephas, and 468 for S. cabrilla). This selection was made to maximize the ratio of continuous spatial coverage over sequencing effort, while also considering several samples per position when possible to increase our power to detect the genetic structure.
(b) . Molecular techniques
We extracted genomic DNA from fin clips (fishes) or pleopods (lobster) using the ReliaPrepTM gDNA Tissue Miniprep System (Promega GmbH, Mannheim, Germany) following the manufacturer's instructions for animal tissue. DNA concentrations were measured with a Qubit™ 2.0 fluorometer (Thermo Fisher Scientific Inc., Waltham, USA). Two different approaches were used to generate single-nucleotide polymorphism (SNP) markers: double-digest restriction-site-associated DNA (ddRAD-seq) sequencing for M. surmuletus and D. sargus and diversity array technology sequencing (DArT-seq, a variant of ddRAD-seq) for S. cabrilla and P. elephas. ddRAD-seq libraries were prepared in-house following a protocol detailed in the study by Fietz et al. [37]. Each library was sequenced on one lane of a HiSeq 4000 Illumina Sequencer (paired-end, 2 × 150 bp) at the Institute of Clinical Molecular Biology, Kiel University, Germany. For S. cabrilla and P. elephas, genomic DNA extracts were standardized to 50 ng µl−1 and sent to Diversity Array Technology (Canberra, Australia) for genotyping with the DArT-seq technology following [38].
(c) . Raw sequence filtering, SNP calling, and filtering
For ddRAD-seq datasets, we removed adapters from raw sequences using Trimmomatic v. 8.25 [39]. Raw sequences were then demultiplexed and filtered using the process_radtags pipeline in STACKS v. 2.2 [40] (see electronic supplementary material, text S1). We then aligned paired-end read sequences with Burrows–Wheeler Aligner (BWA) [41] onto the reference genomes for M. surmuletus and D. sargus, which were assembled for this study specifically and are described in the study by Fietz et al. [37], thereby improving the reliability of genotyping. Aligned reads were sorted using SAMTOOLS 1.9 [42], and loci were built with gstacks. For the DArT-seq data, SNPs were processed using the S. cabrilla reference genome, which was also assembled for this study specifically [37]. Both ddRAD-seq and DArT sequencing data were further filtered with the population pipeline and with vcftools v. 0.1.16 [43]. Each filtering step was performed on each species separately as detailed in the electronic supplementary material, table S2.
(d) . Population structure and isolation by distance—empirical data
We assessed the population structure using ADMIXTURE [1], a maximum-likelihood model to detect population stratification in genome-wide SNP datasets, with 2000 bootstrap replicates. We computed genetic relatedness between pairs of samples, namely, the kinship coefficient of Loiselle et al. [44] using GENODIVE [45]. It was chosen because it uses a correction for a small sample size and tends to show less bias than other coefficients in this case [46]. We note that the Loiselle coefficient correlates well with five other coefficients that we tested on a subset of the data (electronic supplementary material, table S3). This approach was complemented by a sibship inference analysis using COLONY [47], a maximum-likelihood approach to test for the occurrence of siblings in our datasets.
Genetic IBD was tested in the four species. To this end, GEBCO-gridded bathymetry data (https://www.gebco.net) were used to derive the shortest across-water distance (km) between all pairs of samples with a minimum depth of 0.1 m using the marmap package available in R [48]. When samples were collected very close to the coastline, the GEBCO grid sometimes erroneously considered them on land. In these cases, the position of the sample was minimally adjusted to ensure that it is considered in the water and kept in the analysis. The relationships between pairwise individual genetic distances and in-water geographic distances were calculated using the linear regression function lm available in R, and IBD slopes were then calculated at decreasing spatial scales of observation, from the entire sampling area (0–950 km) down to 0–5 km (i.e. considering only pairs of samples separated by ≤5 km) with 5 km decrements. This provided 182–186 spatial intervals per species. To test for significance, we examined whether the observed IBD slope for each spatial interval (i.e. 0–5 km, 0–10 km, etc.) was larger or smaller than expected under a stochastic model. We tested this by running a null model with individual-based genetic distances allocated randomly to each spatial interval. The null modelling procedure was performed for each species and repeated 1000 times (see electronic supplementary material, Text S1). Standardized effect sizes (SES) were calculated for each spatial interval using the observed slope values, the mean and standard deviation of the null distributions: SES = (observed – mean(null))/SD(null).
(e) . Isolation by distance—simulated data
To assess the cause and robustness of the spatial IBD patterns in our empirical data and to explore what kind of dispersal distributions may yield such patterns, we conducted simulations using the coalescent approach implemented in the software IBDSim v. 2.0 [49]. The simulations followed a classic lattice model, with one individual on each node of the lattice. Briefly, each individual produces a large number of gametes and dies. Mutation occurs in the gametes, which then disperse and fuse into diploid individuals. Competition brings the number of individuals in each node back to one and individuals are sampled after the last dispersal step. We considered various dispersal kernels following Puebla et al. [50]. In the extreme case where dispersal occurs only between neighbouring individuals, the lattice model reduces to the stepping-stone model. Different dispersal functions—stepping stone, geometric, and pareto—with varying degrees of short versus long-distance dispersal and emigration rates (from 0.1 to 0.9) were implemented. Simulation settings were set to reflect our empirical sampling design. To this end, we considered a two-dimensional lattice of size 300 × 3000 with absorbing boundaries. We implemented an SNP mutation model and considered only polymorphic loci. Three hundred samples were taken following a line in the centre of the lattice, with single spacing between adjacent samples. We tested the effect of varying the number of loci, the shape of the dispersal function, and the total emigration rate, i.e. the proportion of individuals dispersing to other nodes of the lattice (electronic supplementary material, table S4). The input file of one exemplary simulation is provided in the electronic supplementary material, Text S2. The data generated were analysed and visualized following the same procedure that was used for the empirical SNP data, including the subsampling and SES procedures.
(f) . Lagrangian biophysical model
We built a Lagrangian biophysical model for the three fish species. The model builds on the Lagrangian tracking tool Parcels (oceanparcels.org, [51]) applied to the horizontal surface velocity fields consisting of ocean currents and wave-induced Stokes drift at 1/24° horizontal resolution (approximately 4 km) for the year 2017 from the E.U. Copernicus Marine Service Mediterranean Forecasting System (http://marine.copernicus.eu). Currents were retrieved as daily means from the ocean component of the Mediterranean Forecasting System (Med-Currents, [52]). Stokes drift was obtained as hourly means from the wave component of the Mediterranean Forecasting System (Med-Waves, [53]). Parcels allow one to calculate Lagrangian trajectories by advecting virtual particles approximated as dimensionless and immotile within a given discretized flow field. To ensure no particle flux through boundaries (seafloor, coastlines), we chose a no-slip boundary condition by setting velocities to zero on land so that velocities slow down towards the coast, which is expected to reduce dispersal overall. We then used this Lagrangian model to simulate surface larval dispersal accounting for spawning season, pelagic larval duration, and settlement period of each species, as well as temperature tolerance (electronic supplementary material, table S1). Simulated larvae were seeded in eight marine protected areas from the study region that are known spawning grounds for the selected species and represent a total area of approximately 1072 km2 (electronic supplementary material, figure S2b). Since these areas are shallow, particles were released at the surface, with a uniform random spatial distribution within each marine protected area for a total of 17 160 particles (∼100 per model grid box of 16 km2) per day. As a result, a total of 2 093 520 larvae were simulated for D. sargus, which was seeded over 122 days (March to June), and 2 625 480 for M. surmuletus and S. cabrilla, which were seeded over 153 days (March to July). The biophysical model was not run for the lobster because the drifting parameters (e.g. active vertical migration, resulting in drifting depth) were not known to the degree of certainty needed to be reasonably confident about the simulation results. Subsequently, each virtual larva was passively advected with the horizontal surface velocities (currents and Stokes drift) for the species-specific maximum larval duration period, with an integration time-step of 5 min. The virtual larvae's position and distance travelled in water as well as the ambient temperature were stored hourly. When the ambient temperature fell outside the temperature tolerance, the larva was assumed to die. Further details are available in the electronic supplementary material, Text S3.
3. Results
(a) . Population genetic patterns
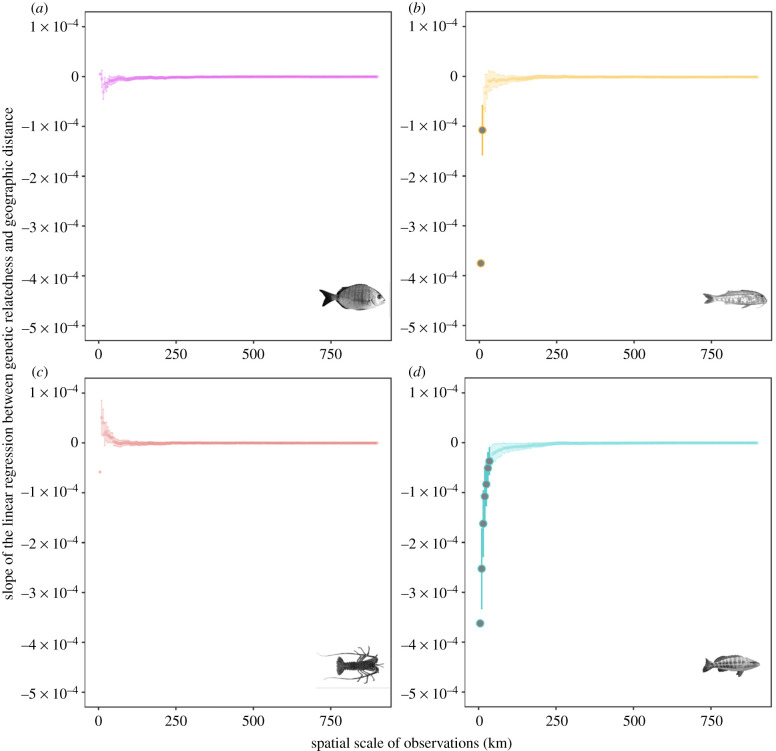
The 1305 selected samples were genotyped at more than 10 000 stringently filtered SNP markers: 18 512 markers for D. sargus (n = 276 samples), 14 318 for M. surmuletus (n = 312), 13 101 for S. cabrilla (n = 468), and 25 230 for P. elephas (n = 243). Six samples were discarded due to a high proportion (more than 30%) of missing data, resulting in a final dataset of 1299 individuals. The distribution of the geographic distance between pairs of samples indicates that our sampling, although not uniform, covers a continuous range of distances from 1 to 950 km (electronic supplementary material, figure S1). As expected, large-scale population genetic structure was absent in three species (D. sargus, M. surmuletus, and P. elephas) and weak in one species (S. cabrilla) (electronic supplementary material, figure S3). Two genetic clusters were identified in S. cabrilla, corresponding to a northern and a southern group separated by the Cabo de la Nao (electronic supplementary material figure S4, fixation index (FST) between the two groups = 0.021). No genetic isolation by distance was observed at the scale of the entire study area in any of the four species (electronic supplementary material, table S5).
In line with our second hypothesis, a significant IBD pattern emerged at small spatial scales (few tens of kilometres) in two species (M. surmuletus and S. cabrilla), with a negative correlation between genetic relatedness (kinship coefficient) and geographic distance among pairs of individuals (figure 2). This pattern was not driven by a small number of highly related and geographically close individuals, which may result from sweepstake recruitment events. Such cases were rare (only seven pairs of individuals across the four datasets), and IBD patterns remained unchanged when these pairs were removed (electronic supplementary material, figure S5). Nevertheless, IBD was weaker—arguably absent—in D. sargus and P. elephas and only statistically significant in M. surmuletus at spatial scales of 0–10 km (SES between −2.91 and −2.31) and in S. cabrilla at spatial scales of 0–35 km (SES between −3.25 and −2.02, considering only the larger northern cluster in the latter species to eliminate the confounding effect of genetic structure; electronic supplementary material, figure S6).
Figure 2.
Empirical spatial genetic patterns. The slope of the linear regression between genetic relatedness and geographic distance among pairs of individuals at various spatial scales of observation. Slope averages and error bars from a resampling procedure (see Methods). Note the logarithmic scale on the slope. The slopes that are statistically significant following the standardized effect size (SES) procedure (see Methods) are highlighted in bold. (a) White seabream (Diplodus sargus), (b) red mullet (Mullus surmuletus), (c) European spiny lobster (Palinurus elephas), and (d) comber (Serranus cabrilla). (Online version in colour.)
To explore the occurrence of this IBD pattern more broadly, we searched the literature for datasets similar to ours (i.e. from marine species sampled continuously at a large spatial scale with more than 100 samples and greater than 1000 SNP 'markers). A single dataset matching these criteria was found [54], confirming the novelty of our approach. This study focused on the summer flounder (Paralichthys dentatus) along 1900 km of coastline in the northwest Atlantic (n = 232 samples, 1137 SNP markers). In line with our results, the authors reported weak genetic structure (FST = 0.0014) and no IBD at the scale of the entire study area (electronic supplementary material, table S5). Nevertheless, a reanalysis of this dataset revealed the emergence of a statistically significant IBD pattern at smaller spatial scales (electronic supplementary material, figure S7).
(b) . Individual-based simulations
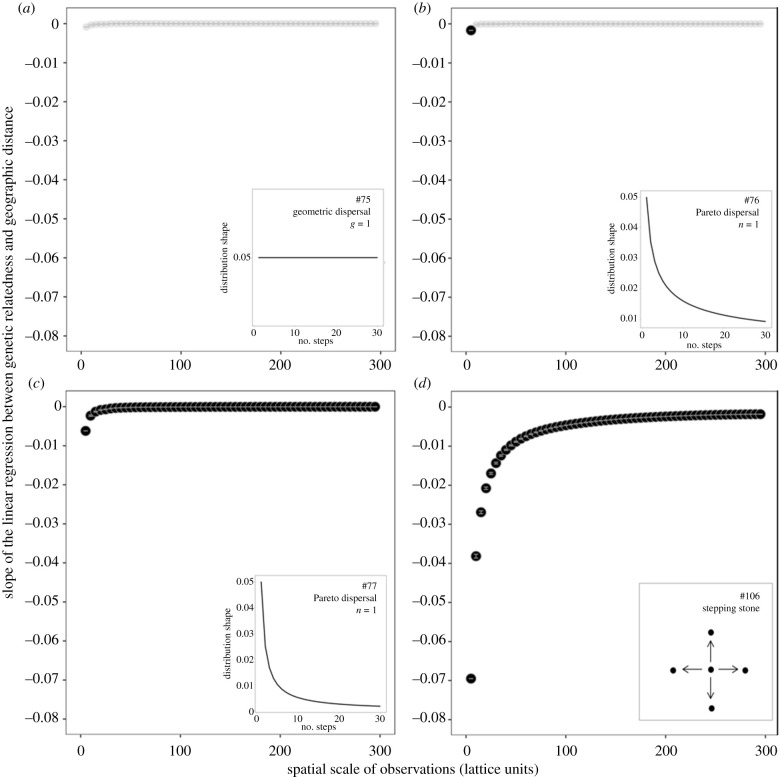
A series of individual-based simulations were run to test whether the emergence of IBD at small spatial scales observed in the empirical data could be generated by simple population genetic models. IBD emerged at small spatial scales for all parameters and dispersal functions that we tested (electronic supplementary material, table S4), indicating that this is a robust pattern (figure 3). Our random subsampling procedure also demonstrated that the emergence of IBD at small spatial scales was not driven by the reduction in sample size (number of pairwise comparisons) at smaller spatial scales. With a flat dispersal function (extensive dispersal, figure 3a), IBD was non-significant at all spatial scales of observation, as observed in D. sargus and P. elephas (figure 2). The small increase of IBD at small spatial scales still observed in this case may be because all simulations presented in figure 3 were standardized to a total emigration rate of 0.1, implying that just 10% of individuals disperse. Dispersal functions with ‘fat tails’ (i.e. that integrate both restricted and long-distance dispersal, figure 3b,c) generated patterns that were similar to what was observed in M. surmuletus, S. cabrilla, and P. dentatus (figure 2, electronic supplementary material, figure S6). The IBD slope also decreased with the increasing spatial scale of observation in the stepping-stone case (figure 3d), where dispersal was restricted to neighbouring nodes only. This implies that the decrease of IBD at large spatial scales does not necessarily imply long-distance dispersal. Nevertheless, IBD decreased more gradually with the increasing spatial scale of observation in the stepping-stone case, plateaued at non-zero values, and presented much stronger levels of significance than the empirical data. The weaker IBD signal that we observed with long-distance dispersal is consistent with analytical IBD models that include long-distance dispersal [55].
Figure 3.
Results from four representative genetic simulations. The slope of the linear regression between genetic relatedness and geographic distance among pairs of samples at various spatial scales of observation. Slope averages and error bars from a resampling procedure (see Methods). The slopes that are statistically significant following the standardized effect size (SES) procedure (see Methods) are highlighted in bold. The four panels show four scenarios with different dispersal kernels that vary in the proportion of long-distance dispersal. Total emigration rate = 0.1 for all simulations with the following dispersal function: (a) geometric, g = 1, (b) Pareto, n = 0.5, (c) Pareto, n = 1, and (d) stepping stone (dispersal between neighbouring nodes only). The dispersal kernel used for each panel is illustrated in the inset (the numbers 75, 76, 77, and 106 refer to the simulation ID in electronic supplementary material table S4, where the details of the simulation parameters are presented). The four simulations illustrate the range of the results that were observed in the simulations, from a slight increase in IBD at small spatial scales and a pattern that is never significant (a) to a marked increase in IBD at small spatial scales and a pattern that is significant at all spatial scales (d).
(c) . Long-distance dispersal
Empirical evidence of long-distance dispersal was provided by two pairs of S. cabrilla individuals that were closely related (i.e. close kin, kinship = 0.106 and 0.108), notwithstanding the fact that they were sampled 292 and 294 km apart (electronic supplementary material, figure S8). This result was confirmed by the sibship inference analysis, which classified these two pairs of individuals as half-sibs (probability = 1.00). This category refers to individuals who share alleles from one parent, so not just half-siblings but also e.g. uncle-niece or grandmother-grandson relationships. In this perspective, the distances of 292 and 294 km would have been covered in one or two generations. It should be noted, however, that the origin of these samples is unknown (it could be between where they were collected or even further away). Another seven pairs of closely related individuals were identified by both the Loiselle coefficient and sibship inference analysis (two in M. surmuletus and five in S. cabrilla), yet these were geographically close (1–6 km).
(d) . Biophysical model
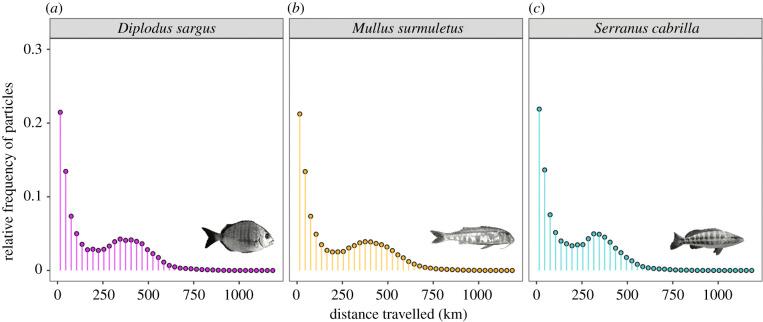
To test whether a combination of restricted and long-distance larval dispersal is consistent with the oceanography of the study area, we ran high-resolution larval dispersal simulations with a biophysical model of the north-western Mediterranean tailored to the biology of our three fish species (electronic supplementary material, table S1). The biophysical model indicates that dispersal is expected to follow a decreasing function of distance for the three species, with dispersal occurring most frequently at distances around 15 km and decreasing thereafter. Dispersal at small distances is greatly enhanced in our simulations including Stokes drift compared to complementary simulations without Stokes drift (not shown), potentially due to wave propagation towards the coast. We also observe a secondary peak centred around 315, 345, and 375 km for S. cabrilla, D. sargus, and M. surmuletus, respectively (figure 4). These are related to advection of larvae with regional currents at speeds in the order of 0.1 m s−1: the longer the PLD, the larger the distances associated with advective dispersal (figure 4). Longer PLD also leads to an increased number of potential dispersal pathways subject to larger current variability, resulting in a broadening of the secondary peak (which is further enhanced through the inclusion of Stokes drift). These results are consistent with the combination of restricted and long-distance dispersal suggested by the empirical and simulated genetic data. Regional oceanographic conditions can either contribute to restricted dispersal at specific locations (e.g. Cap de Creus or Cabo de Gata, see electronic supplementary material, figure S2b in the supplementary electronic material) or facilitate long-distance dispersal. The biophysical model also captured the reduced connectivity observed between the northern and southern groups that we observed in S. cabrilla (electronic supplementary material, figure S2b).
Figure 4.
Dispersal distance distributions as simulated by the high-resolution biophysical model of the north-western Mediterranean. Relative frequencies of binned (30 km bin-width) larval dispersal distances for (a) white seabream (Diplodus sargus, magenta), (b) red mullet (Mullus surmuletus, orange), and (c) comber (Serranus cabrilla, turquoise) reveal that dispersal is occurring most frequently at short distances around 15 km, but long-distance dispersal events with secondary peaks between 300 and 400 km related to the advection of larvae with ocean currents in the order of 0.1 m s−1 are also captured. These results are consistent with the combination of restricted and long-distance dispersal suggested by the empirical and simulated genetic data. (Online version in colour.)
4. Discussion
Generations of population genetic studies have revealed generally low levels of genetic structure in the sea [7], and our results are consistent with this broad picture. Nevertheless, our continuous sampling design uncovers a pattern of genetic IBD at a small spatial scale in two species. Such a pattern is indicative of restricted dispersal and emerges at a surprisingly small spatial scale (few tens of kilometres) considering the weak genetic structure observed at a large spatial scale (close to 1000 km). This result is consistent with the evidence of restricted dispersal provided by direct and local approaches such as mark-recapture or parentage analysis over the last two decades [19,56,57]. In this sense, our approach empirically bridges the results provided by large-scale population genetic [58] and small-scale dispersal [59,60] studies. A step in this direction was taken by Pinsky et al. [57], who showed that dispersal estimates based on parentage analysis are in agreement with those based on IBD in the clownfish Amphiprion percula. Nevertheless, this study considered IBD at the population level and at a fixed spatial scale of 200 km [57]. Our continuous approach shows that IBD can develop at substantially smaller spatial scales. The decrease of IBD at a large spatial scale has been documented both theoretically and empirically [30], but here again at the population level and at large spatial scales (hundreds to thousands of kilometres). Our results show that this pattern holds at a much smaller spatial scale at the individual level. This decrease of IBD at large spatial scales may result from the fact that it takes more time to reach the mutation-drift equilibrium at large spatial scales [61,62] and that factors other than dispersal (e.g. demographic history or selection) may shape genetic variation.
Small-scale IBD was not significant in all species, indicating that this pattern is not universal. The absence of statistically significant IBD in P. elephas conforms with the life history of this species [28]. With a PLD of five to six months, the larvae reside in the water column much longer than the other species, allowing for extensive dispersal and gene flow. Isolation by distance might emerge at a larger spatial scale in such extreme cases (e.g. ref. [63]). The absence of statistically significant IBD in D. sargus was not expected in this perspective considering that this species has a larval duration of 26–30 days [64], which is similar to M. surmuletus and S. cabrilla. We suggest that this non-significance may be due to the known migratory behaviour of D. sargus, which spends the majority of the year in genetically heterogeneous populations [36] before returning to specific and distinct areas for reproduction [32]. Sampling was mostly conducted between June and November, almost exclusively outside of the D. sargus spawning period (March–June [35,65]), when migration may have contributed to obliterate the IBD signal, and the same is true for P. elephas [28,35]. The overlap between sampling and spawning season was greater for M. surmuletus and S. cabrilla (June–July), the two species for which we did detect a significant IBD signal. The combination of restricted larval dispersal, captured with the biophysical model, and limited adult movement during the spawning season is likely to explain our findings. Furthermore, the strongest IBD pattern in terms of both slope and statistical significance was observed in the P. dentatus data from the northwest Atlantic [54] that we reanalysed. This species is characterized by a strong homing behaviour [66] and high residency on spawning grounds [67], where the samples were collected. This illustrates the fact that our approach, which is based on the sampling of adults, integrates both larval and adult movement. It is, therefore, preferable to sample during the spawning season when it comes to detect IBD. All in all, the emergence and statistical significance of IBD at small spatial scales in two species out of four as well as in P. dentatus, the only species for which we could find a previously published dataset similar to ours, suggest that this pattern may be relatively common in marine species. This outcome complements the results of Crandall et al. [14], who provide evidence of stepping-stone dispersal in a variety of marine species.
We detected small-scale IBD with both ddRAD-seq (for M. surmuletus) and DArT-seq (for S. cabrilla) despite the fact that DArT-seq tar- gets gene-rich regions of the genome compared to ddRAD-seq [68]. This suggests that the library type was not a major factor, but we would nonetheless recommend harmonizing library types when possible.
Individual-based simulations indicate that under restricted dispersal, the emergence of IBD at a small spatial scale is a robust pattern that is observed under a broad range of parameter values (figure 3). Nevertheless, this signal is easily missed empirically when considering discrete population samples at the regional scale, which is the standard sampling design in marine population genetic studies [6]. Empirical patterns were more similar to simulated ones when dispersal functions included long-distance dispersal (figure 3c,d). This suggests that long-distance dispersal events also occur, and the observation of two pairs of S. cabrilla individuals that were geographically distant (greater than 290 km), but close kin indicates that this is indeed the case (electronic supplementary material, figure S8). Our empirical and simulated genetic data draw a picture in which the bulk of dispersal is local but where long-distance dispersal events also occur. Such a scenario is also simulated by our high-resolution biophysical model of the western Mediterranean Sea. Our results imply that even though marine populations can be genetically connected over large spatial scales, dispersal is expected to occur mostly at a much smaller spatial scale. The combination of continuous sampling, multispecies genomic data, genetic and biophysical simulations presented here allows us to better understand the relationship between dispersal and gene flow in marine systems. The integrative approach taken here may be extended to a diversity of species whose habitat is more or less continuous.
Supplementary Material
Acknowledgements
Thanks to all the fishermen involved, Eva Maire and Laure Velez (sampling), Virginie Marquez (oceanography) and Veronique Merten (molecular work support).
Data accessibility
All code used for the analysis is available from the Montpellier Bioinformatics Biodiversity platform gitlab at https://gitlab.mbb.univ-montp2.fr/reservebenefit/snakemake_stacks2.
Scripts for the download of all environmental data are available on gitlab at https://gitlab.mbb.univ-montp2.fr/reservebenefit/environmental_assign_data. Scripts for analyses are available on https://github.com/laurabenestan/finescale_ibd at Laura Benestan github page. All data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.8w9ghx3h4 [69] and scripts are available on Reservebenefit Gitlab page (https://gitlab.mbb.univ-montp2.fr/reservebenefit). The metadata from our biological samples were also submitted to geOme under the Reservebenefit name [70].
The data are provided in electronic supplementary material [71].
Authors' contributions
LB: formal analysis, investigation, methodology, visualization, writing—original draft, writing—review and editing; KF: investigation, writing—review and editing; NL: investigation, methodology, visualization; PG: data curation, resources, software; SR: investigation, resources; AB: investigation, resources; WR: investigation, resources; CS: investigation, resources; PL: investigation, resources; AP: resources; PB: resources; AF: resources; SM: resources; RG: resources; LV: resources; MH: resources; SK: resources; DM: conceptualization, writing—review and editing; SM: conceptualization, funding acquisition, methodology, project administration, supervision, writing—review and editing; OP: investigation, methodology, supervision, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This research (RESERVEBENEFIT) was funded through the 2015–2016 BiodivERsA COFUND call for research proposals, with the national funders ANR (France), Formas (Sweden), DLR (Germany), and AEI (Spain). RESERVEBENFIT benefitted from the Montpellier Bioinformatics Biodiversity platform supported by the LabEx CeMEB, an ANR ‘Investissements d'avenir’ program (ANR-10-LABX-04-01).
References
- 1.Kelley JL, Brown AP, Therkildsen NO, Foote AD. 2016. The life aquatic: advances in marine vertebrate genomics. Nat. Rev. Genet. 17, 523-534. ( 10.1038/nrg.2016.66) [DOI] [PubMed] [Google Scholar]
- 2.Grummer JA, Beheregaray LB, Bernatchez L, Hand BK, Luikart G, Narum SR, Taylor EB. 2019. Aquatic landscape genomics and environmental effects on genetic variation. Trends Ecol. Evol. 34, 641-654. ( 10.1016/j.tree.2019.02.013) [DOI] [PubMed] [Google Scholar]
- 3.McCauley DJ, Pinsky ML, Palumbi SR, Estes JA, Joyce FH, Warner RR. 2015. Marine defaunation: animal loss in the global ocean. Science 347, 1255641. ( 10.1126/science.1255641) [DOI] [PubMed] [Google Scholar]
- 4.Lowe WH, Allendorf FW. 2010. What can genetics tell us about population connectivity? Mol. Ecol. 19, 3038-3051. ( 10.1111/j.1365-294X.2010.04688.x) [DOI] [PubMed] [Google Scholar]
- 5.Magris RA, Andrello M, Pressey RL, Mouillot D, Dalongeville A, Jacobi MN, Manel S. 2018. Biologically representative and well-connected marine reserves enhance biodiversity persistence in conservation planning. Conserv. Lett. 11, e12439. ( 10.1111/conl.12439) [DOI] [Google Scholar]
- 6.Riginos C, Crandall ED, Liggins L, Bongaerts P, Treml EA. 2016. Navigating the currents of seascape genomics: how spatial analyses can augment population genomic studies. Curr. Zool. 62, 581-601. ( 10.1093/cz/zow067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selkoe KA, D'Aloia CC, Crandall ED, Iacchei M, Liggins L, Puritz JB, Von Der Heyden S, Toonen RJ. 2016. A decade of seascape genetics: contributions to basic and applied marine connectivity. Mar. Ecol. Prog. Ser. 554, 1-19. ( 10.3354/meps11792) [DOI] [Google Scholar]
- 8.Liggins L, Treml EA, Riginos C. 2019. Seascape genomics: contextualizing adaptive and neutral genomic variation in the ocean environment. In Population genomics: Marine organisms, pp. 171–218. Cham: Springer. [Google Scholar]
- 9.Gagnaire P, Broquet T, Aurelle D, Viard F, Souissi A, Bonhomme F, Arnaud-Haond S, Bierne N. 2015. Using neutral, selected, and hitchhiker loci to assess connectivity of marine populations in the genomic era. Evol. Appl. 8, 769-786. ( 10.1111/eva.12288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauser L, Carvalho GR. 2008. Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. Fish Fish. 9, 333-362. ( 10.1111/j.1467-2979.2008.00299.x) [DOI] [Google Scholar]
- 11.Wright S. 1943. Isolation by distance. Genetics 28, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baguette M, Blanchet S, Legrand D, Stevens VM, Turlure C. 2013. Individual dispersal, landscape connectivity and ecological networks. Biol. Rev. 88, 310-326. ( 10.1111/brv.12000) [DOI] [PubMed] [Google Scholar]
- 13.Purcell JFH, Cowen RK, Hughes CR, Williams DA. 2006. Weak genetic structure indicates strong dispersal limits: a tale of two coral reef fish. Proc. R. Soc. B 273, 1483-1490. ( 10.1098/rspb.2006.3470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crandall ED, Toonen RJ, Selkoe KA. 2019. A coalescent sampler successfully detects biologically meaningful population structure overlooked by F-statistics. Evol. Appl. 12, 255-265. ( 10.1111/eva.12712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selkoe KA, Toonen RJ. 2011. Marine connectivity: a new look at pelagic larval duration and genetic metrics of dispersal. Mar. Ecol. Prog. Ser. 436, 291-305. ( 10.3354/meps09238) [DOI] [Google Scholar]
- 16.Almany GR, et al. 2017. Larval fish dispersal in a coral-reef seascape. Nat. Ecol. Evol. 1, 0148. ( 10.1038/s41559-017-0148) [DOI] [PubMed] [Google Scholar]
- 17.Planes S, Jones GP, Thorrold SR. 2009. Larval dispersal connects fish populations in a network of marine protected areas. Proc. Natl. Acad Sci. USA 106, 5693-5697. ( 10.1073/pnas.0808007106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Aloia CC, Bogdanowicz SM, Francis RK, Majoris JE, Harrison RG, Buston PM. 2015. Patterns, causes, and consequences of marine larval dispersal. Proc. Natl. Acad Sci. USA 112, 13 940-13 945. ( 10.1073/pnas.1513754112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bode M, Leis JM, Mason LB, Williamson DH, Harrison HB, Choukroun S, Jones GP. 2019. Successful validation of a larval dispersal model using genetic parentage data. PLoS Biol. 17, e3000380. ( 10.1371/journal.pbio.3000380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Catalano KA, Dedrick AG, Stuart MR, Puritz JB, Montes HR, Pinsky ML. 2020. Quantifying dispersal variability among nearshore marine populations. Mol. Ecol. ( 10.1111/mec.15732) [DOI] [PubMed] [Google Scholar]
- 21.Mokhtar-Jamaï K, Coma R, Wang J, Zuberer F, Féral JP, Aurelle D. 2013. Role of evolutionary and ecological factors in the reproductive success and the spatial genetic structure of the temperate gorgonian Paramuricea clavata. Ecol. Evol. 3, 1765-1779. ( 10.1002/ece3.588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker D, Porter BA, Avise JC. 2002. Genetic parentage assessment in the crayfish Orconectes placidus, a high-fecundity invertebrate with extended maternal brood care. Mol. Ecol. 11, 2115-2122. ( 10.1046/j.1365-294X.2002.01609.x) [DOI] [PubMed] [Google Scholar]
- 23.Coates JH, Hovel KA, Butler JL, Bohonak AJ. 2014. Recruitment and recovery of pink abalone (Haliotis corrugata) in a historically overexploited kelp forest: are local populations self-sustaining? J. Exp. Mar. Bio. Ecol. 460, 184-192. ( 10.1016/j.jembe.2014.07.004) [DOI] [Google Scholar]
- 24.Manel S, et al. 2019. Long-distance benefits of marine reserves: myth or reality? Trends Ecol. Evol. 34, 342-354. ( 10.1016/j.tree.2019.01.002) [DOI] [PubMed] [Google Scholar]
- 25.Luximon N, Petit EJ, Broquet T. 2014. Performance of individual vs. group sampling for inferring dispersal under isolation-by-distance. Mol. Ecol. Resour. 14, 745-752. ( 10.1111/1755-0998.12224) [DOI] [PubMed] [Google Scholar]
- 26.Prunier JG, Kaufmann B, Fenet S, Picard D, Pompanon F, Joly P, Lena JP. 2013. Optimizing the trade-off between spatial and genetic sampling efforts in patchy populations: towards a better assessment of functional connectivity using an individual-based sampling scheme. Mol. Ecol. 22, 5516-5530. ( 10.1111/mec.12499) [DOI] [PubMed] [Google Scholar]
- 27.Battey CJ, Ralph PL, Kern AD. 2020. Space is the place: effects of continuous spatial structure on analysis of population genetic data. Genetics 215, 193-214. ( 10.1534/genetics.120.303143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goñi R, Latrouite D. 2005. Review of the biology, ecology and fisheries of Palinurus spp. species of European waters: Palinurus elephas (Fabricius, 1787) and Palinurus mauritanicus (Gruvel, 1911). Cah. Biol. Mar. 46, 127-142. [Google Scholar]
- 29.Rousset F. 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145, 1219-1228. ( 10.1093/genetics/145.4.1219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradbury IR, Bentzen P. 2007. Non-linear genetic isolation by distance: implications for dispersal estimation in anadromous and marine fish populations. Mar. Ecol. Prog. Ser. 340, 245-257. ( 10.3354/meps340245) [DOI] [Google Scholar]
- 31.Albouy C, et al. 2015. FishMed: traits, phylogeny, current and projected species distribution of Mediterranean fishes, and environmental data. Ecology 96, 2312-2313. ( 10.1890/14-2279.1) [DOI] [Google Scholar]
- 32.Planes S, Lenfant P. 2002. Temporal change in the genetic structure between and within cohorts of a marine fish, Diplodus sargus, induced by a large variance in individual reproductive success. Mol. Ecol. 11, 1515-1524. ( 10.1046/j.1365-294X.2002.01521.x) [DOI] [PubMed] [Google Scholar]
- 33.Claudet J, Pelletier D, Jouvenel JY, Bachet F, Galzin R. 2006. Assessing the effects of marine protected area (MPA) on a reef fish assemblage in a northwestern Mediterranean marine reserve: identifying community-based indicators. Biol. Conserv. 130, 349-369. ( 10.1016/j.biocon.2005.12.030) [DOI] [Google Scholar]
- 34.Gagnaire PA. 2020. Comparative genomics approach to evolutionary process connectivity. Evol. Appl. 13, 1320-1334. ( 10.1111/eva.12978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abecasis D, Bentes L, Lino PG, Santos MN, Erzini K. 2013. Residency, movements and habitat use of adult white seabream (Diplodus sargus) between natural and artificial reefs. Estuar. Coast. Shelf Sci. 118, 80-85. ( 10.1016/j.ecss.2012.12.014) [DOI] [Google Scholar]
- 36.Hernández-García R, Muñoz I, López-Capel A, Marcos C, Pérez-Ruzafa Á. 2015. The influence of environmental variability of a coastal lagoon ecosystem on genetic diversity and structure of white seabream [Diplodus sargus (Linnaeus 1758)] populations. Mar. Ecol. 36, 1144-1154. ( 10.1111/maec.12210) [DOI] [Google Scholar]
- 37.Fietz K, Trofimenko E, Guerin PE, Arnal V, Torres-Oliva M, Lobréaux S, Pérez-Ruzafa A, Manel S, Puebla O. 2020. New genomic resources for three exploited Mediterranean fishes. Genomics 112, 4297-4303. ( 10.1016/j.ygeno.2020.06.041) [DOI] [PubMed] [Google Scholar]
- 38.Kilian A, et al. 2012. Diversity arrays technology: a generic genome profiling technology on open platforms. Methods Mol. Biol. 888, 67-89. ( 10.1007/978-1-61779-870-2_5) [DOI] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22, 3124-3140. ( 10.1111/mec.12354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Prepr. arXiv (doi:arXiv:1303.3997[q-bio.GN]).
- 42.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078-2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27, 2156-2158. ( 10.1093/bioinformatics/btr330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loiselle BA, Sork VL, Nason J, Graham C. 1995. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 82, 1420-1425. ( 10.2307/2445869) [DOI] [Google Scholar]
- 45.Meirmans PG, Van Tienderen PH.. 2004. Genotype and genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 4, 792-794. ( 10.1111/j.1471-8286.2004.00770.x) [DOI] [Google Scholar]
- 46.Wang J. 2017. Estimating pairwise relatedness in a small sample of individuals. Heredity 119, 302-313. ( 10.1038/hdy.2017.52) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones OR, Wang J. 2010. COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 10, 551-555. ( 10.1111/j.1755-0998.2009.02787.x) [DOI] [PubMed] [Google Scholar]
- 48.Pante E, Simon-Bouhet B. 2013. marmap: a package for importing, plotting and analyzing bathymetric and topographic data in R. PLoS ONE 8, e73051. ( 10.1371/journal.pone.0073051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leblois R, Estoup A, Rousset F. 2009. IBDSim: a computer program to simulate genotypic data under isolation by distance. Mol. Ecol. Resour. 9, 107-109. ( 10.1111/j.1755-0998.2008.02417.x) [DOI] [PubMed] [Google Scholar]
- 50.Puebla O, Bermingham E, McMillan WO. 2012. On the spatial scale of dispersal in coral reef fishes. Mol. Ecol. 21, 5675-5688. ( 10.1111/j.1365-294X.2012.05734.x) [DOI] [PubMed] [Google Scholar]
- 51.Delandmeter P, van Sebille E.. 2019. The Parcels v. 2.0 Lagrangian framework: new field interpolation schemes. Geosci. Model Dev.12, 3571–3584. ( 10.5194/gmd-12-3571-2019) [DOI]
- 52.Clementi E., et al. 2019. Mediterranean Sea Analysis and Forecast (CMEMS MED-Currents 2016-2019) (Version 1) [Data set]. Copernicus Monitoring Environment Marine Service (CMEMS). See 10.25423/CMCC/MEDSEA_ANALYSIS_FORECAST_PHY_006_013_EAS4. [DOI]
- 53.Korres G, Ravdas M, Zacharioudaki A. 2019. Mediterranean Sea Waves Analysis and Forecast (CMEMS MED-Waves 2017-2019) (Version 1) [Data set], Copernicus Monitoring Environment Marine Service (CMEMS), [DOI]
- 54.Hoey JA, Pinsky ML. 2018. Genomic signatures of environmental selection despite near-panmixia in summer flounder. Evol. Appl. 11, 1732-1747. ( 10.1111/eva.12676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith T, Weissman D. 2020. Isolation by distance in populations with long-range dispersal. bioRxiv, 1–36. ( 10.1101/2020.06.24.168211) [DOI]
- 56.Buston PM, D'Aloia CC. 2013. Marine ecology: reaping the benefits of local dispersal. Curr. Biol. 23, 56. ( 10.1016/j.cub.2013.03.056) [DOI] [PubMed] [Google Scholar]
- 57.Pinsky ML, et al. 2017. Marine dispersal scales are congruent over evolutionary and ecological time. Curr. Biol. 27, 149-154. ( 10.1016/j.cub.2016.10.053) [DOI] [PubMed] [Google Scholar]
- 58.Stanley RRE, et al. 2018. A climate-associated multispecies cryptic cline in the northwest Atlantic. Sci. Adv. 4, aaq0929. ( 10.1126/sciadv.aaq0929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schunter C, Pascual M, Raventos N, Garriga J, Garza JC, Bartumeus F, Macpherson E. 2019. A novel integrative approach elucidates fine-scale dispersal patchiness in marine populations. Sci. Rep. 9, 10796. ( 10.1038/s41598-019-47200-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson RE, et al. 2019. Micro-geographic population genetic structure within Arctic cod (Boreogadus saida) in Beaufort Sea of Alaska. ICES J. Mar. Sci. 76, 1713-1721. ( 10.1093/icesjms/fsz041) [DOI] [Google Scholar]
- 61.Slatkin M. 1993. Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47, 264. ( 10.2307/2410134) [DOI] [PubMed] [Google Scholar]
- 62.Hardy OJ, Vekemans X. 1999. Isolation by distance in a continuous population: reconciliation between spatial autocorrelation analysis and population genetics models. Heredity 83, 145-154. ( 10.1046/j.1365-2540.1999.00558.x) [DOI] [PubMed] [Google Scholar]
- 63.Crandall ED, Treml EA, Barber PH. 2012. Coalescent and biophysical models of stepping-stone gene flow in neritid snails. Mol. Ecol. 21, 5579-5598. ( 10.1111/mec.12031) [DOI] [PubMed] [Google Scholar]
- 64.Di Franco A, Guidetti P.. 2011. Patterns of variability in early-life traits of fishes depend on spatial scale of analysis. Biol. Lett. 7, 454-456. ( 10.1098/rsbl.2010.1149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Follesa MC, Cuccu D, Cannas R, Sabatini A, Deiana AM, Cau A. 2009. Movement patterns of the spiny lobster Palinurus elephas (Fabricius, 1787) from a central western Mediterranean protected area. Sci. Mar. 73, 499-506. ( 10.3989/scimar.2009.73n3499) [DOI] [Google Scholar]
- 66.Sackett DK, Able KW, Grothues TM. 2007. Dynamics of summer flounder, Paralichthys dentatus, seasonal migrations based on ultrasonic telemetry. Estuar. Coast. Shelf Sci. 74, 119-130. ( 10.1016/j.ecss.2007.03.027) [DOI] [Google Scholar]
- 67.Desfosse JC. 1995. Movements and ecology of summer flounder, Paralichthys dentatus, tagged in the southern mid-Atlantic bight, p. 187. Virginia Institute of Marine Science.
- 68.Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A. 2004. Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc. Natl. Acad Sci. USA 101, 9915-9920. ( 10.1073/pnas.0401076101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benestan L, et al. 2021. Restricted dispersal in a sea of gene flow. Dryad Digital Repository. ( 10.5061/dryad.8w9ghx3h4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deck J, Gaither MR, Ewing R, Bird CE, Davies N, Meyer C, Riginos C, Toonen RJ, Crandall ED. 2017. The Genomic Observatories Metadatabase (GeOMe): a new repository for field and sampling event metadata associated with genetic samples. PLoS Biol. 15, e2002925. ( 10.1371/journal.pbio.2002925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benestan L, et al. 2021. Restricted dispersal in a sea of gene flow. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Benestan L, et al. 2021. Restricted dispersal in a sea of gene flow. Figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All code used for the analysis is available from the Montpellier Bioinformatics Biodiversity platform gitlab at https://gitlab.mbb.univ-montp2.fr/reservebenefit/snakemake_stacks2.
Scripts for the download of all environmental data are available on gitlab at https://gitlab.mbb.univ-montp2.fr/reservebenefit/environmental_assign_data. Scripts for analyses are available on https://github.com/laurabenestan/finescale_ibd at Laura Benestan github page. All data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.8w9ghx3h4 [69] and scripts are available on Reservebenefit Gitlab page (https://gitlab.mbb.univ-montp2.fr/reservebenefit). The metadata from our biological samples were also submitted to geOme under the Reservebenefit name [70].
The data are provided in electronic supplementary material [71].