Abstract
The Alinity m (Abbott Molecular, Des Plaines, IL) automated molecular analyzer allows continuous loading of samples and sample-to-result molecular detection of several microorganisms. The detection of SARS-CoV-2 by the Alinity m was compared with that of the cobas 6800 (Roche Molecular Systems, Branchburg, NJ; standard comparator) in a manufacturer-independent clinical evaluation on 2157 consecutive nasopharyngeal swab samples. Valid initial results on Alinity m and cobas 6800 were obtained from 2129 (98.7%) and 2157 (100%) samples, respectively. The overall percent agreement (95% CI) was 98.3% (2092/2129 [97.6%–98.7%]); positive percent agreement, 100% (961/961 [99.6%–100%]); negative percent agreement, 96.8% (1131/1168 [95.7%–97.7%]); and high κ value, 0.965 (0.954–0.976). There were 37 discordant results on Alinity m and, based on discordant analyses, including previous and/or follow-up PCR results, 22 could be considered analytically true positive with high probability. Due to a lack of additional information and an inability to perform repeated/further testing, the status of the remaining 15 discordant results remained unresolved. The throughput of the two analyzers was compared using testing on 564 samples in parallel across two 8-hour shifts in clinical practice. The turnaround times were compared using processing of 94 routine samples in parallel on each working day for 5 consecutive days. The two analyzers showed similar performance, with certain differences that have potential importance in some laboratory settings.
The coronavirus disease–2019 (COVID-19) pandemic has affected over 135 million individuals, with over 2.9 million COVID-19–related deaths as of April 10, 2021. Highly reliable laboratory diagnostics for COVID-19 are essential for case identification, patient management, and contact tracing. Detecting severe acute respiratory syndrome–coronavirus 2 (SARS-CoV-2) RNA in nasopharyngeal swabs is still considered the COVID-19 reference laboratory diagnostic standard.1 , 2 Although several commercial SARS-CoV-2 RNA assays have received Emergency-Use Authorization (EUA) from the US Food and Drug Administration (FDA), only a few have been designed for analyzers with high sample throughput to manage the unprecedented demand for SARS-CoV-2 RNA testing and to allow for significant scaling-up due to the fully automated sample-to-result solution.3 The cobas 6800 and 8800 Systems (Roche Molecular Systems, Branchburg, NJ) are fully integrated and automated analyzers that allow for sample-to-result qualitative and quantitative molecular detection of several microorganisms. The FDA recently approved the use of a range of molecular assays designed for use with the cobas 6800/8800 System (https://www.fda.gov/medical-devices/vitro-diagnostics/nucleic-acid-based-tests, last accessed April 10, 2021), including assays for SARS-CoV-2, which were given FDA EUA on March 12, 2020. Several studies have evaluated the performance of this reliable and robust SARS-CoV-2 assay,4, 5, 6, 7, 8, 9, 10, 11 and it has become a primary comparator in many performance evaluations of novel SARS-CoV-2 RNA assays.
Alinity m (Abbott Molecular, Des Plaines, IL) is another, recently launched, fully integrated, automated, sample-to-result molecular analyzer that allows continuous loading of samples and random-access testing. Seven molecular assays have been developed for use with Alinity m,12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23 including an assay for SARS-CoV-2, which was given FDA EUA on May 11, 2020. Unlike the cobas SARS-CoV-2 assay with much-published performance data, peer-reviewed literature on the Alinity m SARS-CoV-2 assay contains only limited data on verification and validation.20
This article presents the results from a large-scale, manufacturer-independent comparison of the clinical performance of the Alinity m SARS-CoV-2 assay versus the cobas 6800 SARS-CoV-2 assay as a standard comparator. This head-to-head study evaluated the clinical performance of these two high-throughput, automated assays in detecting the presence of SARS-CoV-2 RNA in 2157 consecutive nasopharyngeal swab samples in a routine diagnostic setting. In addition, throughput of the two analyzers was compared in clinical practice using testing on 564 samples across two 8-hour shifts. TATs were compared using assessments of the first 94 samples received in the laboratory on each working day for 5 consecutive days.
Materials and Methods
Samples Used for Head-to-Head Clinical Comparison
The clinical performance of the Alinity m and cobas 6800 SARS-CoV-2 assays was compared using testing on a total of 2157 unselected nasopharyngeal swab samples, routinely collected during a 10-day period in mid-January 2021. All samples were obtained from individuals treated at the University Clinical Center Ljubljana [Ljubljana, Slovenia; the largest (2138 beds) tertiary hospital, and the principal COVID-19 hospital, in Slovenia, with the largest COVID-19 intensive care unit]. Samples were collected in a commercially available 3-mL transport medium (VTM; Liofilchem, Roseto degli Abruzzi, Italy) from 2157 individuals referred for COVID-19 testing during the study period. The median transport time of samples from the collection site to the laboratory was 1:51 hours. On arrival at the laboratory, swabs were vortexed for 1 minute at maximum speed, and two VTM aliquots were prepared: 800 μL for Alinity m SARS-CoV-2 testing and 700 μL for cobas 6800 SARS-CoV-2 testing.
Alinity m SARS-CoV-2 Testing
The Alinity m SARS-CoV-2 assay is a real-time reverse transcriptase PCR–based assay for the qualitative detection of SARS-CoV-2 RNA in nasal, nasopharyngeal, and oropharyngeal swab samples.20 By using target-specific, fluorescence-labeled oligonucleotide probes, the assay allows simultaneous detection and amplification of two SARS-CoV-2–specific sequences targeting the RdRp and N genes, reported as a combined signal in one channel, and individually an internal control target sequence for the evaluation of sample extraction and amplification efficiency reported separately in another channel. The assay was performed on Alinity m—a fully integrated, automated molecular analyzer that allows continuous loading of samples and performs sample preparation, reverse transcriptase PCR assembly, amplification, detection, calculation, and reporting of the results. Alinity m provides two reports: i) "result" [each signal is reported either as "not detected" if a specimen cycle number (CN) is not generated, or as "CN value" if CN is <42], and ii) "interpretation" (either "negative" or "positive"). Prior to transfer of the results into the laboratory information system, the results can be reviewed directly on the system screen or as a printed report.
In this study, 800-μL sample aliquots were transferred into Alinity m aliquot tubes, loaded on Alinity m, and tested following the manufacturer's instructions.
Cobas 6800 SARS-CoV-2 Testing
The cobas SARS-CoV-2 assay is a two-target real-time reverse transcriptase PCR test for the qualitative detection of SARS-CoV-2 RNA in nasopharyngeal and oropharyngeal swab samples.4 One target is viral ORF1, a region unique to SARS-CoV-2 (target 1), and the second is a conserved region in the E gene for pan-Sarbecovirus detection (target 2). The assay utilizes RNA internal control as sample preparation and PCR amplification processing controls and the uracil-N-glycosylase system for the prevention of PCR contamination. The assay is performed on either the cobas 6800 or 8800 System, which consists of the sample supply module, transfer module, processing module, and analytic module. Automated data management is performed using the manufacturer's software, which assigns test results. The results can be reviewed directly on the system screen, as a printed report, or transferred to a laboratory information system. According to the manufacturer's instructions, a tested sample was considered SARS-CoV-2 positive if cobas showed positive results either for both the ORF1 (target 1) and E (target 2) genes or for the ORF1 gene only (target 1). In the case of positivity for the E gene only (target 2), the result was reported as SARS-CoV-2 presumptive positive.
In this study, 700-μL sample aliquots were transferred to barcoded secondary tubes, loaded on the cobas 6800 System, and tested following the manufacturer's instructions. Testing was performed in 94-sample batches plus one negative and positive control each.
Data Analysis
A contingency table was constructed for the assessment of percent overall, positive, and negative agreements with 95% CIs. The level of agreement between tests was assessed using the Cohen κ statistic. All statistical analyses were performed using Excel 2016 version 16.0.5134.1000 (Microsoft, Redmond, WA) and R statistical software version 3.2.5 (Free Software Foundation, Boston, MA). The study protocol conformed with the World Medical Association's Declaration of Helsinki and was approved by the Medical Ethics Committee of the Republic of Slovenia (protocol 0120-211/2020/7).
Comparative Throughput Assessment
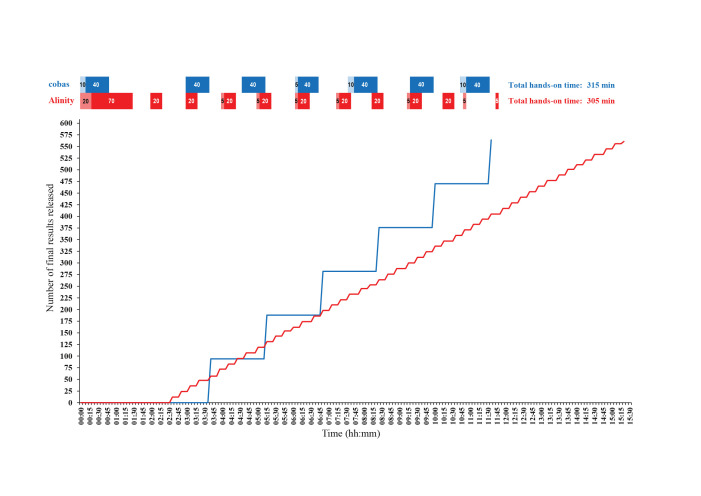
To comparatively assess the throughput of the Alinity m and cobas 6800 systems in clinical practice, 564 samples were processed in parallel on both systems across two 8-hour shifts. Samples were processed by experienced operators starting on the morning of January 19, 2021. Both analyzers were initialized at the same time and run in parallel until all samples were processed and all final results were obtained. The sample-handling time, instrument-handling time, and total hands-on time with each analyzer were meticulously measured by two independent observers (R.K. and A.O.V.). For visualization of the release dynamics of the final results across the two shifts, a time curve from each analyzer was generated, and the time curves obtained were comparatively evaluated.
Comparative Turnaround-Time Assessment
To determine the turnaround times (TATs) of the two SARS-CoV-2 assays in clinical practice, the first 94 routine samples received in the laboratory on each working day for 5 consecutive days were processed in parallel on both analyzers by experienced operators (M.S. and P.K.). The entire testing procedure of each analyzer was meticulously monitored by two independent observers (R.K. and A.O.V.), and the following timepoints were recorded for each of the 470 (94 × 5) samples processed: sample admission time, starting point of sample handling, starting point of instrument processing, and time of result release. From the collected data, total preanalytical time, instrument on-board time, and total TAT were calculated for each of the 470 samples processed.
Results
Head-to-Head Clinical Comparison
Of 2157 nasopharyngeal swab samples tested, valid initial results from the Alinity m and cobas 6800 SARS-CoV-2 assays were obtained from 2129 (98.7%) and 2157 (100%) samples, respectively. A total of 28 samples were excluded from further analysis due to initial invalid results on Alinity m SARS-CoV-2; all invalid results were due to a lack of amplification of the target and internal control failure. Table 1 summarizes the results from the comparative clinical evaluation of the two assays on 2129 samples with initial valid results from both assays. The diagnostic approaches showed an overall percent agreement (95% CI) of 98.3% (2092/2129 [97.6%–98.7%]), a positive percent agreement of 100% (961/961 [99.6%–100.0%]), a negative percent agreement of 96.8% (1131/1168 [95.7%–97.7%]), and a high κ value of 0.965 (0.954–0.976) on 2129 samples with valid results from both assays.
Table 1.
| Alinity m SARS-CoV-2 assay | cobas 6800 SARS-CoV-2 assay |
Overall agreement (95% CI) | κ Value (95% CI) | ||
|---|---|---|---|---|---|
| Positive | Negative | Total | |||
| Positive, n | 961 | 37 | 998 | 98.3% (97.6%–98.7%) | 0.965 (0.954–0.976) |
| Negative, n | 0 | 1131 | 1131 | ||
| Total, n | 961 | 1168 | 2129 | ||
A total of 2129 nasopharyngeal swab samples with valid initial results on both assays were tested; 28 others were excluded from further analysis due to initial invalid results on Alinity m. The interpretation of 37 discordant results, including 4 positive on Alinity m and presumptive-positive on cobas 6800, is provided in detail in the Results and Discussion sections.
Trademark of Abbott Molecular (Des Plaines, IL).
Trademark of Roche Molecular Systems (Branchburg, NJ).
Thirty-seven discordant results were obtained; four samples were positive using Alinity m SARS-CoV-2 and presumptive positive on cobas 6800 SARS-CoV-2, and 33 were positive using Alinity m SARS-CoV-2 and negative on cobas 6800 SARS-CoV-2 (Table 1). With Alinity m SARS-CoV-2, mean and median CN values among 961 concordantly positive samples were 20.6 and 18.8 (range, 8.9 to 39.1), respectively, and among 37 samples with discordant results, 37.8 and 37.9 (range, 31.1 to 41.9), respectively.
An insufficient volume of leftover specimens prevented retesting of samples with discrepant Alinity m/cobas 6800 results. To resolve the status of samples with discrepant results, all previous and follow-up testing results from the 37 individuals with discrepant results recorded in the laboratory information system were reviewed. In 18 of these 37 individuals, previous and/or follow-up SARS-CoV-2 PCR result(s) were identified: 10 of 18 had a recorded previous SARS-CoV-2 PCR–positive result in samples collected on 0, 6, 8, 9, 9, 10, 11, 27, 59, and 90 days (mean, 22.9 days; median, 9.5 days; range, 0 to 90 days) before the sample in this study was collected (for multiple previous samples identified, the first previously positive sample was considered for each individual), and 8 of 18 had a recorded positive result on follow-up SARS-CoV-2 PCR in samples collected on 0, 0, 0, 2, 2, 5, 6, and 7 days (mean, 2.8 days; median, 2 days; range, 0 to 7 days) after the sample in this study was collected (for multiple follow-up samples identified, the first positive follow-up sample was considered for each individual). In 19 individuals with Alinity m/cobas 6800 discrepant results, no previous and/or follow-up SARS-CoV-2 PCR results were identified in the laboratory information system database. Of these 19 individuals, 4 were Alinity m SARS-CoV-2 positive/cobas 6800 SARS-CoV-2 presumptive positive (positive for target 2 only) and 15 were Alinity m SARS-CoV-2 positive/cobas 6800 SARS-CoV-2 negative. Based on the discordant analyses described, of 37 samples with Alinity m SARS-CoV-2–positive/cobas 6800 SARS-CoV-2–negative discrepant results, 22 (18 + 4) samples could be considered Alinity m SARS-CoV-2 analytically true positive with high probability. Due to the lack of additional information and the inability of repeated/further testing, the status of the remaining 15 discordant samples remains unresolved.
Comparative Throughput Assessment
Figure 1 shows the time curves with the Alinity m and cobas 6800 systems, with the release dynamics of the final results from testing 564 samples in parallel across two 8-hour shifts. The first results were produced sooner with Alinity m than with the cobas 6800 System; the first 12 results were released 2:35 hours after initialization. Afterward, Alinity m released results consistently every 16 minutes in batches of 12, releasing 96, 192, 288, 384, 480, and 564 results at 4:25, 6:50, 8:55, 11:05, 13:10, and 15:30 hours after initialization, respectively (Figure 1). On the other hand, the cobas 6800 System required 3:40 hours from initialization to release the first batch of 94 results. Afterward, the cobas 6800 System released results in batches of 94, releasing 188, 282, 376, 470, and 564 results at 5:15, 6:50, 8:25, 10:00, and 11:35 hours after initialization, respectively. The total hands-on times of testing 564 samples were similar between the two instruments: 305 minutes with Alinity m and 315 minutes with the cobas 6800 System. However, the cobas 6800 System required slightly less total instrument-handling time than Alinity m (35 versus 50 minutes), but slightly more total sample-handling time (280 versus 255 minutes). In addition, the cobas 6800 System required laboratory staff presence during seven similar time slots (40 to 50 minutes each), whereas Alinity m required presence during 12 varying time slots (5 to 90 minutes) (Figure 1).
Figure 1.
Time curves visualizing the release dynamics of the final SARS-CoV-2 RNA results from the Alinity m (Abbott Molecular, Des Plaines, IL) and cobas 6800 (Roche Molecular Systems, Branchburg, NJ) analyzers in the testing of 564 nasopharyngeal swab samples in parallel across two 8-hour shifts. The sample-handling times (dark), instrument-handling times (light), and total hands-on times of each analyzer were measured by two independent observers (R.K. and A.O.V).
Comparative TAT Assessment
On parallel routine processing of the first 94 samples received in the laboratory on each working day for 5 consecutive days, the TATs of almost all samples were shorter with Alinity m (143 to 257 minutes) in comparison to those with the cobas 6800 System (243 to 269 minutes). Although the ranges of instrument on-board time were similar between the two analyzers (Alinity m, 130 to 215 minutes; cobas 6800 System, 168 to 174 minutes), the ranges of total preanalytical time of most samples differed (Alinity m, 12 to 86 minutes; cobas 6800 System, 72 to 91 minutes).
Discussion
The demand for SARS-CoV-2 diagnostics has been unprecedented, and has led to the development of a range of molecular assays designed for use with analyzers that provide high sample throughput. This throughput allows for significant scaling-up due to the fully automated sample-to-result solution. Currently, at least six high-throughput SARS-CoV-2 RNA assays are available: cobas SARS-CoV-2,4, 5, 6, 7, 8, 9, 10, 11 Alinity m SARS-CoV-2,20 Panther Fusion SARS-CoV-2 (Hologic, San Diego, CA),5 , 8 , 24, 25, 26, 27 Aptima SARS-CoV-2 (Hologic),5 , 8 , 24, 25, 26, 27 NeuMoDx SARS-CoV-2 (NeuMoDx Molecular, Ann Arbor, MI),28 , 29 and GeneXpert Infinity SARS-CoV-2 (Cepheid, Sunnyvale, CA). Although several evaluations of these assays have been published in addition to the present study, only a single head-to-head comparison of two SARS-CoV-2 high-throughput assays was identified using database searches.8 In that study, the clinical performance of the cobas and Panther Fusion SARS-CoV-2 assays was compared using 389 nasopharyngeal swab samples; the two assays showed a high overall percent agreement of 96.4% and a high κ value of 0.922.8
Among fully automated analyzers used for SARS-CoV-2 RNA testing, Alinity m was most recently launched (2019), and as of April 2021, seven molecular assays had been developed and launched for use with Alinity m. These assays are intended for: i) the quantitative detection of hepatitis B virus DNA13 , 17 , 19 , 21; ii) the quantitative detection of hepatitis C virus RNA12 , 17 , 18 , 21; iii) the quantitative detection of HIV-1 RNA15, 16, 17 , 21; iv) the qualitative detection of SARS-CoV-2 RNA20; v) the qualitative detection of 14 high-risk human papillomaviruses coupled with extended genotyping14 , 22; the vi) qualitative detection and differentiation of Chlamydia trachomatis, Trichomonas vaginalis, Mycoplasma genitalium, and Neisseria gonorrhoeae 23; and vii) the qualitative detection and differentiation of SARS-CoV-2, influenza A and B viruses, and respiratory syncytial virus. Twelve evaluations of Alinity m assays are available from the peer-reviewed literature,12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23 but only a single analytical and clinical evaluation of the Alinity m SARS-CoV-2 assay in a limited number of samples is known to have been published to date.20 In that study, the manufacturer's claim of a lower limit of detection of 100 copies/mL with the Alinity m SARS-CoV-2 assay was verified, with clinical evaluation performed on 203 residual nasopharyngeal swabs from symptomatic and asymptomatic individuals with suspected COVID-19. The testing of samples was compared between the Alinity m and the RealTime (Abbott) SARS-CoV-2 assays, and positive and negative percent agreement between the two assays were 92.2% (95% CI, 85.3%–96.6%) and 92.0% (95% CI, 84.8%–96.5%), respectively. The CN values found in 95 concordantly positive samples showed high correlation (R 2 = 0.95); however, on average, for 14.1 higher CN values were recorded with Alinity m than with RealTime, perhaps due to the first 10 unread cycles with the RealTime assay.20 Some limited performance data are also provided by the manufacturer, which compared Alinity m SARS-CoV-2 with an undeclared reverse transcriptase PCR assay approved for use by FDA EUA; on the testing of 104 clinical samples with positive and negative percent agreements between the two assays of 100% (95% CI, 95.5%–100.0%) and 96.5% (95% CI, 87.9%–99.6%), respectively, with two more samples found positive on Alinity m SARS-CoV-2, both of which had a CN value of >40. The performance of the assays was also assessed using samples from 144 asymptomatic individuals, of which 19 were concordantly positive, yielding percent positive and negative agreements of 100% (95% CIs, 82.4%–100.0% and 97.1%–100.0%, respectively). In addition, no in silico impact on the detection of different SARS-CoV-2 strains/variants or cross-reactivity with similar viruses has been reported. A manufacturer's technical brief dated January 18, 2021, reported that the performance of Alinity m SARS-CoV-2 was unaffected by three emerging variants of concern: i) VOC 202012/01-B.1.1.7, ii) VOC 501Y.V2-B.1.351, and iii) IC-0561-B.1.1248.
In the present study, Alinity m SARS-CoV-2 showed excellent overall, positive, and negative agreement with the comparator, cobas 6800 SARS-CoV-2, in the testing of 2129 samples, with valid results with both assays. Thirty-seven discordant results were identified. Discordance between the assays was completely unidirectional (Table 1), suggesting slightly higher analytical sensitivity with Alinity m SARS-CoV-2 over cobas 6800 SARS-CoV-2. These findings are in agreement with recent data on assay performance from an FDA SARS-CoV-2 reference panel facilitating comparisons of performance among different FDA EUA–approved assays. Those data indicated that the relative sensitivity of Alinity m SARS-CoV-2 was 600 nucleic acid amplification test detectable units (NDUs) per milliliter30 compared to 1800 NDU/mL with cobas 6800 SARS-CoV-2 (https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-reference-panel-comparative-data, last accessed April 10, 2021). Higher sample-volume input with Alinity m SARS-CoV-2 (800 versus 600 μL) might have contributed to the higher analytical sensitivity compared with that of cobas 6800 SARS-CoV-2. Given that the Alinity m and the cobas 6800 SARS-CoV-2 assays target different SARS-CoV-2 genes, there is also a possibility that discordant performance was a consequence of differing assay designs (ORF1 and E genes versus N and RdRp genes, respectively). Namely, it has been reported recently that certain deletions/mutations in the SARS-CoV-2 genome can affect commercial PCRs.31 , 32 However, such a scenario is less likely with the Alinity m and cobas 6800 SARS-CoV-2 assays, given that both were designed as two-target assays, making false-negative results less likely. Nonetheless, there is a need for constant monitoring for the emergence of polymorphisms that might adversely affect PCR systems used in diagnosing COVID-19.
Higher analytical sensitivity with a particular PCR test for SARS-CoV-2 does not necessarily indicate more clinical value: The majority of clinically relevant SARS-CoV-2 RNA–positive samples contain a high viral load (and have relatively low CN/Ct values) and thus are concurrently positive on different PCR-based assays. With PCR-based assays with an analytical sensitivity higher than that of standard PCR assays, the clinical benefit may be evident only in a limited fraction of positive samples having a low SARS-CoV-2 RNA viral load (and having high CN/Ct values), mainly due to sample collection that was suboptimal or that took place in a very early phase of SARS-CoV-2 infection. Discordant analysis of samples weakly positive on one assay and negative on another can be challenging. Due to the high sample-volume requirements of the Alinity m and cobas 6800 SARS-CoV-2 assays, and due to the consequent insufficient volume of leftover specimen, samples showing discrepant results between the Alinity m and cobas 6800 SARS-CoV-2 assays could not be retested. Thus, the true SARS-CoV-2 RNA status of samples with discrepant results could be resolved only using previous and follow-up testing in particular individuals whose data were recorded in the laboratory information system. Based on findings from detailed discordant analyses, of 37 individuals with samples that yielded discordant results, 22 (59.5%) could have been considered true positive on analysis with Alinity m SARS-CoV-2, with high probability. In at least 8 of these 22, the higher analytical sensitivity with Alinity m SARS-CoV-2 compared with that of cobas 6800 SARS-CoV-2 was clinically beneficial because these individuals tested positive on cobas 6800 SARS-CoV-2 in one or more follow-up samples obtained after the initial negative result was obtained with cobas 6800 SARS-CoV-2. This clinical benefit potentially also extends to 4 individuals with Alinity m SARS-CoV-2–positive/cobas 6800 SARS-CoV-2–presumptive positive results in the study sample, but without previous or follow-up results available. The clinical benefit of the higher analytical sensitivity with Alinity m SARS-CoV-2 in the remaining 10 individuals who had a sample that yielded an Alinity m SARS-CoV-2–analytically true positive/cobas 6800 SARS-CoV-2–negative result, but a cobas 6800 SARS-CoV-2–positive result in one or more previous sample(s), is less evident because the clinical value of extremely high–sensitivity molecular tests used for follow-up testing in individuals with clear laboratory-confirmed infection remains controversial. Unfortunately, due to the lack of additional information and the inability of repeated/further testing in 15 of 37 individuals with results discordant between Alinity m SARS-CoV-2/cobas 6800 SARS-CoV-2, the status of the samples that yielded discordant results remains unresolved.
Alinity m SARS-CoV-2 internal control is a noninfectious Armored RNA sequence unrelated to the SARS-CoV-2 sequence and is introduced into each specimen at the beginning of the sample preparation. The internal control is simultaneously amplified by PCR to demonstrate that the entire testing procedure has proceeded correctly in each sample. In the present study, 28 of 2157 samples (1.3%) showed initial invalid results on Alinity m SARS-CoV-2, all due to a lack of amplification of target and internal control failure. Insufficient leftover volume allowed for the retesting of only 10 of 28 study samples with initial invalid results on Alinity m SARS-CoV-2, and after retesting, a valid result on Alinity m SARS-CoV-2 was obtained in 9 of the 10 initially failed samples (90%). The cobas 6800 System was installed at the Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana, Slovenia, 2 years ago, and Alinity m was installed a week before the start of the present study. Although invalid results on SARS-CoV-2 RNA testing were also initially an issue with the cobas 6800 System,4 and were mainly caused by clots, mucus, or physical contamination detected by the instrument during sample aspiration, or by insufficient sample volume identified in sample tubes or processing plates, the instrument soon stabilized, and no significant problems with invalid results on the cobas 6800 System have been recorded during the past 10 months of routine testing of >120,000 samples using the cobas 6800 SARS-CoV-2 assay. After the head-to-head study was closed, monitoring the rate of invalid results on Alinity m was continued for 8 weeks. Interestingly, the rates of initially invalid results on Alinity m SARS-CoV-2 in the first 4 weeks remained similar to those recorded during the head-to-head study [41/1562 (2.6%), 44/2375 (1.9%), 42/2819 (1.5%), and 42/3059 (1.4%) in weeks 1 to 4, respectively]. However, in the subsequent 4 weeks, the rates of initially invalid results were significantly decreased [10/3949 (0.3%), 9/3830 (0.2%), 11/4070 (0.3%), and 6/2761 (0.2%) in weeks 5 to 8, respectively]. The most probable reasons for the improvement might have been a combination of the increased stability of the analyzer and the accumulated experience of the operators. Due to the insufficient leftover volume of most of the routinely processed samples, at the Institute of Microbiology and Immunology all of the samples with initially invalid results on Alinity m SARS-CoV-2 are currently being retested using a protocol that requires a smaller sample volume (200 μL), as described previously.4 However, in the present study, all of the routinely tested samples with initially invalid results that had ≥800 μL of leftover volume available were retested with Alinity m SARS-CoV-2, and a valid result was obtained in 77 of 78 of these initially failed samples (98.7%). Thus, if leftover sample volume is sufficient, simple repeated testing (without further sample manipulation) is recommended in cases of samples with initially invalid results on Alinity m SARS-CoV-2.
When selecting a SARS-CoV-2 RNA assay, virologists must consider not only sensitivity and specificity but also sample throughput, time to result, test complexity, reagent and instrument availability, and cost per reportable result. Assay throughput is a parameter especially crucial for large-scale testing. The Alinity m and the cobas 6800 Systems are both fully integrated and automated analyzers that allow for sample-to-result detection of SARS-CoV-2 RNA, but the present comparative throughput assessment showed potentially important differences (Figure 1). Alinity m produced the first reportable results much sooner than the cobas 6800 System (2:35 versus 3:40 hours), but with the cobas 6800 System, the testing of 564 samples in parallel was finished almost 4 hours faster than with Alinity m (11:35 versus 15:30 hours). Similarly, although the total hands-on time of testing 564 samples was similar between the two instruments, slightly less total instrument-handling time, but slightly more total sample-handling time, was required with the cobas 6800 System than with Alinity m. In addition, during testing of 564 samples in parallel across two 8-hour shifts, the cobas 6800 System required the presence of laboratory staff during seven similar time slots, whereas Alinity m required staff presence during 12 varying time slots (Figure 1). Similar, potentially important, differences between the analyzers were also recorded during in-parallel routine processing of the first 94 samples received at the laboratory on each working day for 5 consecutive days. On this testing of 470 samples, the TATs of most of the samples were somewhat shorter with Alinity m in comparison to the cobas 6800 System. This discrepancy was mainly due to differences in total preanalytical time between the two analyzers, and due to routine (and most economically) processing of 12-sample batches with Alinity m and 94-sample batches with the cobas 6800 System. Thus, in different laboratory settings, the differences between the analyzers observed in the present study might be less evident and less important.
When asked to qualitatively compare their experiences in using the two analyzers for the routine detection of SARS-CoV-2 RNA, laboratory staff agreed that the main comparative advantages of Alinity m were a rapid TAT with smaller batches and flexible STAT prioritization, and that the main disadvantages were the need for prethawing and centrifuge reagents, more frequent instrument interactions and sample loading/unloading, and limited (48-hour) on-board stability of positive and negative controls. The main comparative advantages with the cobas 6800 System were a higher 24-hour throughput, ready-to-use reagents that did not require thawing or mixing, and less frequent instrument interactions and sample loading/unloading; the main disadvantage (in the laboratory setting at the Institute of Microbiology and Immunology) was the requirement of sample centrifugation to avoid problems during sample aspiration due to clots, mucus, or physical contamination.
In conclusion, in the present manufacturer-independent comparison of Alinity m SARS-CoV-2 against cobas 6800 SARS-CoV-2 used in the routine diagnostic testing of 2157 samples, the Alinity m SARS-CoV-2 assay was reliable in the qualitative detection of SARS-CoV-2 from nasopharyngeal swab samples. Assays showed excellent overall, positive, and negative percent agreements, with a high κ value. Slightly higher analytical sensitivity with the Alinity m SARS-CoV-2 assay was clinically beneficial in a limited number of samples. Comparative throughput and TAT assessments in clinical practice showed similar performance of the two assays, with performance differences that could be important in some laboratory settings.
Acknowledgments
We thank Robert Krošelj and Maja Accetto Kos for excellent laboratory assistance with the cobas 6800 System testing and all members of the COVID-19 diagnostic team, who have been working diligently for the past 12 months to ensure timely SARS-CoV-2 results.
Footnotes
Supported by the Faculty of Medicine, Institute of Microbiology and Immunology, University of Ljubljana; Slovenian Research Agency grants P3-0083 and V3-2034; and the European Virus Archive Global project, which has received funding from European Union Horizon 2020 Research and Innovation Program grant 871029.
Disclosures: The Institute of Microbiology and Immunology (Ljubljana, Slovenia) has received research grants from Abbott Molecular (but not for this work); A.O.V. has received travel reimbursement and speaking honoraria from Abbott Molecular. [new line]The manufacturers of the cobas 6800 and Alinity m SARS-CoV-2 assays had no role in the study design, data collection or analysis, interpretation of the results, writing of the manuscript, or the decision to submit the work for publication.
References
- 1.Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loeffelholz M.J., Tang Y.W. Laboratory diagnosis of emerging human coronavirus infections—the state of the art. Emerg Microbes Infect. 2020;20:1–26. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arena F., Pollini S., Rossolini G.M., Margaglione M. Summary of the available molecular methods for detection of SARS-CoV-2 during the ongoing pandemic. Int J Mol Sci. 2021;22:1298. doi: 10.3390/ijms22031298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poljak M., Korva M., Knap Gašper N., Fujs Komloš K., Sagadin M., Uršič T., Avšič Županc T., Petrovec M. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol. 2020;58:e00599-20. doi: 10.1128/JCM.00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieberman J.A., Pepper G., Naccache S.N., Huang M.L., Jerome K.R., Greninger A.L. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol. 2020;58:e00821-20. doi: 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cradic K., Lockhart M., Ozbolt P., Fatica L., Landon L., Lieber M., Yang D., Swickard J., Wongchaowart N., Fuhrman S., Antonara S. Clinical evaluation and utilization of multiple molecular in vitro diagnostic assays for the detection of SARS-CoV-2. Am J Clin Pathol. 2020;154:201–207. doi: 10.1093/ajcp/aqaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pujadas E., Ibeh N., Hernandez M.M., Waluszko A., Sidorenko T., Flores V., Shiffrin B., Chiu N., Young-Francois A., Nowak M.D., Paniz-Mondolfi A.E., Sordillo E.M., Cordon-Cardo C., Houldsworth J., Gitman M.R. Comparison of SARS-CoV-2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS-CoV-2 test and a laboratory-developed real-time RT-PCR test. J Med Virol. 2020;92:1695–1698. doi: 10.1002/jmv.25988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craney A.R., Velu P.D., Satlin M.J., Fauntleroy K.A., Callan K., Robertson A., La Spina M., Lei B., Chen A., Alston T., Rozman A., Loda M., Rennert H., Cushing M., Westblade L.F. Comparison of two high-throughput reverse transcription-PCR systems for the detection of severe acute respiratory syndrome coronavirus 2. J Clin Microbiol. 2020;58:e00890-20. doi: 10.1128/JCM.00890-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smithgall M.C., Scherberkova I., Whittier S., Green D.A. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the rapid detection of SARS-CoV-2. J Clin Virol. 2020;128:104428. doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V., Love N. Detection of SARS-CoV-2 by use of the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 assays. J Clin Microbiol. 2020;58:e00772-20. doi: 10.1128/JCM.00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirden M., Feghoul L., Bertine M., Nere M.L., Le Hingrat Q., Abdi B., Boutolleau D., Ferre V.M., Jary A., Delaugerre C., Marcelin A.G., Descamps D., Legoff J., Visseaux B., Chaix M.L. Multicenter comparison of the cobas 6800 System with the RealStar RT-PCR kit for the detection of SARS-CoV-2. J Clin Virol. 2020;130:104573. doi: 10.1016/j.jcv.2020.104573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevaliez S., Onelia F., Pacenti M., Goldstein E., Galán J.C., Martínez-García L., Vilas A., Glass A., Maree L., Krügel M., Ehret R., Knechten H., Braun P., Naeth G., Bonanzinga S., Jackson K., Abravaya K., Dhein J., Huang S., Joseph A.M., Lucic D., Marlowe N., Palm M.J., Pfeifer K., Toolsie D., Reinhardt B., Obermeier M., Gunson R. Multicenter clinical evaluation of Alinity m HCV assay performance. J Clin Virol. 2020;129:104531. doi: 10.1016/j.jcv.2020.104531. [DOI] [PubMed] [Google Scholar]
- 13.Bonanzinga S., Onelia F., Jackson K., Glass A., Maree L., Krügel M., Pacenti M., Gunson R., Goldstein E., García L.M., Galán J.C., Vilas A., Ehret R., Knechten H., Naeth G., Braun P., Obermeier M., Marlowe N., Palm M.J., Pfeifer K., Joseph A.M., Dhein J., Reinhardt B., Lucic D., Chevaliez S. Multicenter clinical evaluation of Alinity m HBV assay performance. J Clin Virol. 2020;129:104514. doi: 10.1016/j.jcv.2020.104514. [DOI] [PubMed] [Google Scholar]
- 14.Oštrbenk Valenčak A., Šterbenc A., Seme K., Poljak M. Alinity m HR HPV assay fulfills criteria for human papillomavirus test requirements in cervical cancer screening settings. J Clin Microbiol. 2019;58:e01120-19. doi: 10.1128/JCM.01120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun P., Glass A., Maree L., Krügel M., Pacenti M., Onelia F., Gunson R., Goldstein E., Martínez-García L., Galán J.C., Vilas A., D’costa J., Sameer R., Ehret R., Knechten H., Naeth G., Bouvier-Alias M., Marlowe N., Palm M.J., Joseph A.M., Dhein J., Reinhardt B., Pfeifer K., Lucic D., Obermeier M. Multicenter clinical comparative evaluation of Alinity m HIV-1 assay performance. J Clin Virol. 2020;129:104530. doi: 10.1016/j.jcv.2020.104530. [DOI] [PubMed] [Google Scholar]
- 16.Maree L., Krügel M., Reinhardt B., Glass A.J. Evaluation of the Alinity m HIV-1 assay for the quantification of HIV-1 RNA plasma viral load in a high-throughput molecular laboratory in South Africa. J Clin Virol. 2020;132:104644.17. doi: 10.1016/j.jcv.2020.104644. [DOI] [PubMed] [Google Scholar]
- 17.Mouna L., Pallier C., Proust S., Prégermain C., Roque-Afonso A.M. Comparison of the Abbott Alinity m and m2000 assays for the quantification of HIV-1, HCV and HBV in clinical samples. J Clin Virol. 2020;126:104331. doi: 10.1016/j.jcv.2020.104331. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein E.J., Shepherd S.J., Gunson R.N. Investigating utilising the Alinity m platform to detect hepatitis C virus RNA in dried blood spot samples. J Clin Virol. 2020;132:104647. doi: 10.1016/j.jcv.2020.104647. [DOI] [PubMed] [Google Scholar]
- 19.Jackson K., Tekoaua R., Li X., Locarnini S. Real-world application of the Xpert HBV viral load assay on serum and dried blood spots. J Med Virol. 2021;93:3707–3713. doi: 10.1002/jmv.26662. [DOI] [PubMed] [Google Scholar]
- 20.Hirschhorn J.W., Kegl A., Dickerson T., Glen W.B., Jr., Xu G., Alden J., Nolte F.S. Verification and validation of SARS-CoV-2 assay performance on the Abbott m2000 and Alinity m systems. J Clin Microbiol. 2021;59:e03119–e03120. doi: 10.1128/JCM.03119-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galindo L.T., Domingues Hristov A., Girotto Gentil L., Scarpelli L., Santiago J., Levi J.E. Performance evaluation of the fully automated molecular system Alinity m in a high-throughput central laboratory. J Clin Virol. 2021;137:104786. doi: 10.1016/j.jcv.2021.104786. [DOI] [PubMed] [Google Scholar]
- 22.Dhillon S.K., Valenčak A.O., Xu L., Poljak M., Arbyn M. Clinical and analytical evaluation of the Alinity m HR HPV assay within the VALGENT-3 framework. J Clin Microbiol. 2021;59 doi: 10.1128/JCM.00286-21. e00286-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrmann B., Malm K. Comparison between Abbott m2000 RealTime and Alinity m STI systems for detection of Chlamydia trachomatis, Neisseria gonorrhoeae, and Mycoplasma genitalium. Eur J Clin Microbiol Infect Dis. 2021 doi: 10.1007/s10096-020-04135-9. [Epub ahead of print] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhen W., Manji R., Smith E., Berry G.J. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J Clin Microbiol. 2020;58:e00743-20. doi: 10.1128/JCM.00743-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trémeaux P., Lhomme S., Abravanel F., Raymond S., Mengelle C., Mansuy J.M., Izopet J. Evaluation of the Aptima™ transcription-mediated amplification assay (Hologic®) for detecting SARS-CoV-2 in clinical specimens. J Clin Virol. 2020;129:104541. doi: 10.1016/j.jcv.2020.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith E., Zhen W., Manji R., Schron D., Duong S., Berry G.J. Analytical and clinical comparison of three nucleic acid amplification tests for SARS-CoV-2 detection. J Clin Microbiol. 2020;58:e01134-20. doi: 10.1128/JCM.01134-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogan C.A., Sahoo M.K., Huang C., Garamani N., Stevens B., Zehnder J., Pinsky B.A. Comparison of the Panther Fusion and a laboratory-developed test targeting the envelope gene for detection of SARS-CoV-2. J Clin Virol. 2020;127:104383. doi: 10.1016/j.jcv.2020.104383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mostafa H.H., Lamson D.M., Uhteg K., Geahr M., Gluck L., de Cárdenas J.N.B., Morehead E., Forman M., Carroll K.C., Hayden R.T., George K.S. Multicenter evaluation of the NeuMoDx SARS-CoV-2 test. J Clin Virol. 2020;130:104583. doi: 10.1016/j.jcv.2020.104583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lima A., Healer V., Vendrone E., Silbert S. Validation of a modified CDC assay and performance comparison with the NeuMoDx™ and DiaSorin® automated assays for rapid detection of SARS-CoV-2 in respiratory specimens. J Clin Virol. 2020;133:104688. doi: 10.1016/j.jcv.2020.104688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alinity m SARS-CoV-2 amp kit EUA version R7 [package insert] Abbott Molecular; Des Plains, IL: 2020. [Google Scholar]
- 31.Artesi M., Bontems S., Göbbels P., Franckh M., Maes P., Boreux R., Meex C., Melin P., Hayette M.P., Bours V., Durkin K. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol. 2020;58:e01598-20. doi: 10.1128/JCM.01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramírez J.D., Muñoz M., Patiño L.H., Ballesteros N., Paniz-Mondolfi A. Will the emergent SARS-CoV2 B.1.1.7 lineage affect molecular diagnosis of COVID-19? J Med Virol. 2021;93:2566–2568. doi: 10.1002/jmv.26823. [DOI] [PMC free article] [PubMed] [Google Scholar]