Abstract
An engineered cyanovirin-N homolog that exhibits specificity for high mannose N-glycans has been constructed to aid type I α-1,2-mannosidase inhibitor discovery and development. Engineering the lectins C-terminus permitted facile functionalization with fluorophores via a sortase and click strategy. The resulting lectin constructs exhibit specificity for cells presenting high mannose N-glycans. Importantly, these lectin constructs can also be applied to specifically assess changes in cell surface glycosylation induced by type I mannosidase inhibitors. Testing the utility of these lectin constructs led to the discovery of type I mannosidase inhibitors with nanomolar potency. Cumulatively, these findings reveal the specificity and utility of the functionalized cyanovirin-N homolog constructs, and highlight their potential to fill the need for high mannose-specific lectins that can be applied for analytical purposes.
Keywords: Glycoscience, lectin, mannosidase inhibitors, bioconjugation, protein modifications, natural products
Graphical Abstract
Lectins that can be applied to specifically detect high mannose N-glycans on cell surfaces have been prepared by engineering the C-terminus of a naturally occur cyanovirin-N homolog. The functionalized lectins can be utilized to aid type I α-1,2-mannosidase inhibitor discovery and development. Here we utilize the functionalized lectin to prepare kifunensine analogues that are >75-fold more potent than kifunensine.
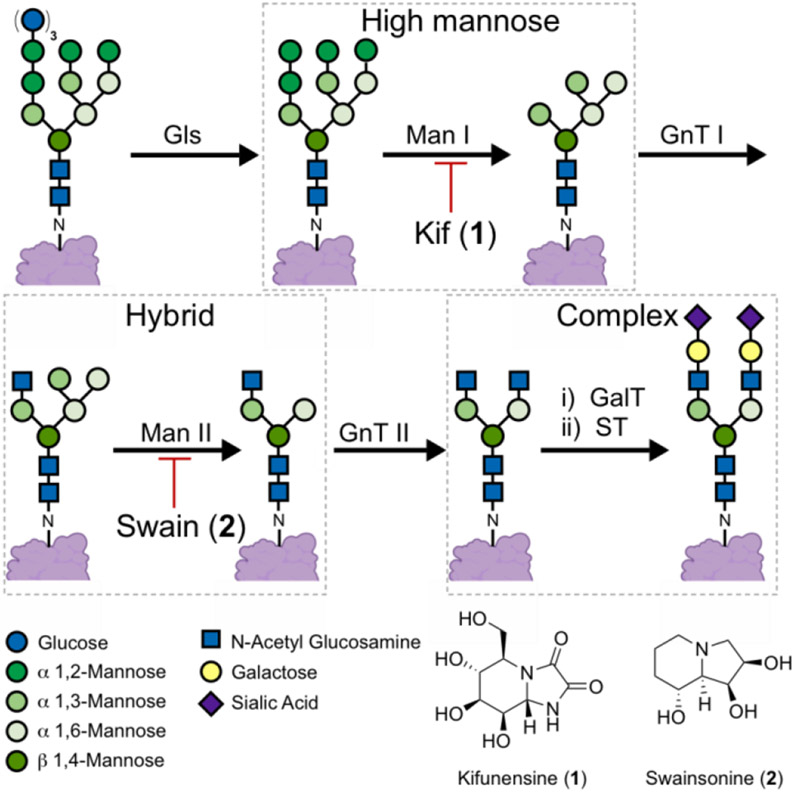
N-linked glycans are present in the glycocalyx of human cells, and play important roles in various processes.[1] N-glycans are covalently attached to proteins via asparagine residues, and are modified by successive enzymatic transformations, culminating in the formation of complex N-glycans (Figure 1). Modulating N-glycosylation has shed light on the physiological and pathophysiological importance of N-glycans.[2] These studies have also uncovered the therapeutic benefits of modulating the enzymes that facilitate N-glycan attachment and maturation.[3]
Figure 1.
N-glycan maturation pathway. N-glycans attached to proteins are trimmed by α-glycosidase (Gls), type I α-1,2-mannosidase (Man I) and type II α-mannosidase (Man II) enzymes. The latter steps can be inhibited by kifunensine (1) and swainsonine (2) respectively. Elaboration of the N-glycans is facilitated by N-acetyl glucosamine transferase I (GnT I), galactosyl transferase (GalT) and sialyl transferase (ST) enzymes.
Among these enzymes, the type I mannosidase (MAN I) enzymes have been described as promising targets for cancer,[4] viral infections,[5] sarcoglycanopathies,[6] lysosomal storage disorders,[7] and for the preparation of therapeutic high mannose bearing glycoproteins.[8] Despite this potential, kifunensine (1, Kif), an alkaloid isolated in the 1980’s, remains the predominantly applied MAN I inhibitor.[9] Efforts to identify Kif analogues or novel scaffolds have yet to provide an inhibitor with superior activity in cellular assays.[10] The development of cell-based assays to aid MAN I inhibitor development is hampered by the scarcity of tools available to specifically detect the high mannose N-glycans induced by MAN I inhibitors. In the absence of said tools, the discovery and development of MAN I inhibitors will remain limited.
Lectins, labelled with fluorophores, are prevalently applied to detect specific carbohydrate motifs on cell surfaces;[11] however, the lectins commonly applied to detect high mannose N-glycans (i.e., concanavalin A, Galanthus nivalis agglutinin and Hippeastrum lectin) bind to motifs that are not unique to the high mannose N-glycans that predominate upon MAN I inhibition.[12] These lectins lack the specificity required to differentiate between the glycans induced by MAN I (e.g., Kif) and MAN II (e.g., swainsonine, 2) inhibitors.[13] For example, concanavalin A (Con A) binds to α 1,3- and α 1,6-mannose motifs that are present in complex, hybrid, and high mannose N-glycans.[12c]
Lectins that bind to motifs uniquely present in high mannose N-glycans have been investigated for their anti-viral properties (e.g., cyanovirin-N and griffithsin);[14] but surprisingly, there are limited examples of these lectins being utilized for analytical purposes. The development and application of cyanovirin-N from Nostoc ellipsosporum has been hampered by this lectin’s tendency to aggregate and precipitate from solution;[15] however, a cyanovirin-N homolog that exhibits favorable solution phase properties has been isolated from Cyanothece7424 (i.e., Cyt-CVNH).[16] We reasoned that labelled Cyt-CVNH derivatives could be applied to detect high mannose N-glycans on cell surfaces.
Here, we describe the design, synthesis and characterization of a series of novel fluorophore labelled Cyt-CVNH derivatives. We demonstrate that these lectins preferentially bind to cells expressing elevated levels of high mannose N-glycans. Importantly, these lectins specifically detect the high mannose N-glycans that predominate on the surface of cells post treatment with MAN I inhibitors. We have applied these Cyt-CVNH derivatives in our efforts to develop Kif analogues, which has led us to identify MAN I inhibitors that exhibit nanomolar activity in cellular assays.
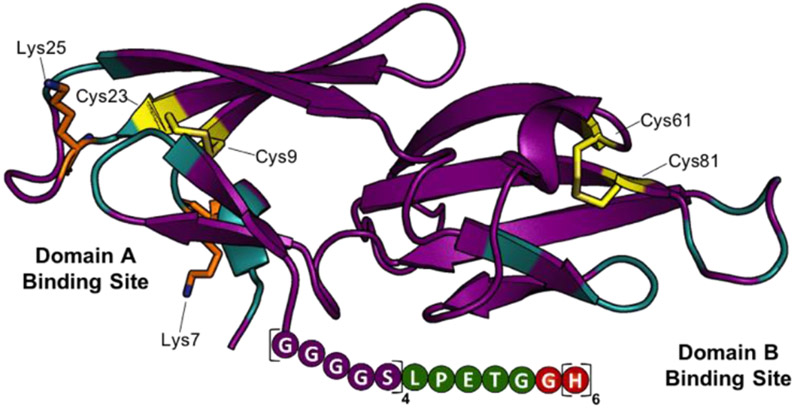
Our initial analysis of Cyt-CVNH, lead us to realize that the residues amenable to facile functionalization are present in or are proximal to the lectin’s carbohydrate binding domains (Figure 2 and SI Figure S1). Unfortunately, conjugation of a fluorophore to these nucleophilic residues hampered binding to high mannose N-glycan expressing cells (SI Figure S1). In search of an alternative strategy, we noted that the C-terminus of cyanovirin-N can be modified without impacting carbohydrate binding.[17] Given the structural similarity between these two lectins (SI Figure S2), we set out to design a C-terminus modified Cyt-CVNH derivative that would permit site selective C-terminus conjugation via a sortase mediated ligation (SML).[18] The C-terminus of Cyt-CVNH was thus modified to incorporate a sortase motif (i.e., LPETG) flanked by a glycine-serine Linker, and a His tag, referred to as Cyt-CVNH-LSH (3, Figure 2). The expression of 3 was performed in an analogous manner to that previously described for the expression of the parental lectin Cyt-CVNH,[16] and appreciable quantities were readily prepared.
Figure 2.
Structure guided design of Cyt-CVNH-LSH (3). The structure of Cyt-CVNH (PDB: 5K79) annotated to highlight binding domain residues (cyan), nucleophilic residues (Lys = orange; Cys = yellow), and the modified C-terminus.
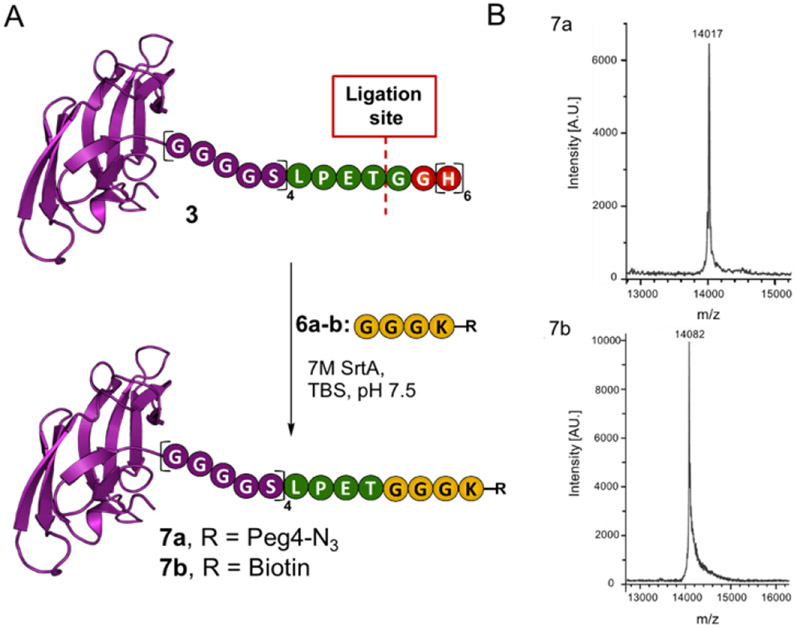
Attention turned to the preparation of the other components required for the SML. The sortase A heptamutant (7M SrtA) was chosen to catalyze these reactions as it works independently of Ca2+ and has enhanced catalytic activity.[19] Additionally, we pre-pared acyl acceptors 6a and 6b that would be utilized to optimize the SML with Cyt-CVNH-LSH (Scheme 1). To gain access to 6a and 6b, previously reported intermediate 4 was functionalized with NHS esters 5a and 5b.[20] The resulting intermediates were subjected to acidic conditions to remove the Boc groups, thus delivering GGGK-PEG4-N3 (6a) and GGGK-PEG4-Biotin (6b).
Scheme 1.
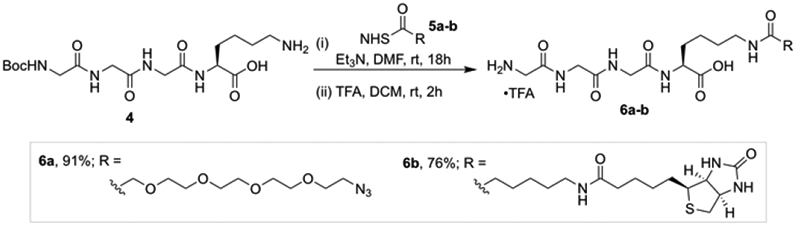
The synthesis of azido and biotin functionalized acyl acceptors.
Next, we subjected the acyl donor CVNH-LSH 1 and acyl acceptor 6a to SML conditions. Initial attempts provided the desired product 7a, however, a threonine terminated lectin by-product was also produced (SI Figure S3). Unfortunately, this by-product cannot be readily separated from the desired product due to modest differences in molecular weight and the absence of an affinity tag on both the desired and undesired products. Upon further experimentation, we found that applying 0.02 equivalents of 7M SrtA and an excess of 6a for 24 hours led to the formation of Cyt-CVNH-N3 (7a) in the absence of the hydrolyzed byproduct (Figure 3 and SI Figures S4-5). The general applicability of these conditions was validated using acyl acceptor 6b.
Figure 3.
Sortase mediated ligation. A) A schematic depiction of the SML involving Cyt-CVNH-LSH (3) and 6a-b to provide Cyt-CVNH-N3 (7a) and Cyt-CVNH-Biotin (7b); B) MALDI spectra of the r products. A.U. = arbitrary units.
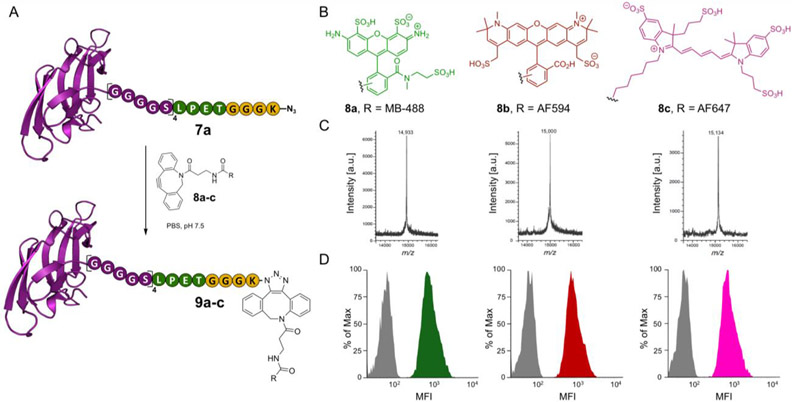
With Cyt-CVNH-N3 (7a) in hand, we reasoned that this is an ideal intermediate to facilitate the preparation of fluorophore labelled Cyt-CVNH constructs. The azide permits the use of strain-promoted azide–alkyne cycloadditions (SPAAC), which proceed in aqueous solutions at room temperature in the absence of additives.[21] Furthermore, numerous dibenzocyclooctyne (DBCO) functionalized fluorophores are commercially available which provides great flexibility. As such, CVNH-N3 (7a) was functionalized with DBCO-fluorophores 8a-c in PBS at room temperature to provide Cyt-CVNH MB 488 (9a), Cyt-CVNH-AFDye 647 (9b) and Cyt-CVNH-AFDye 594 (9c) in high purity after a simple dialysis (Figure 4C and SI Figure S7).
Figure 4.
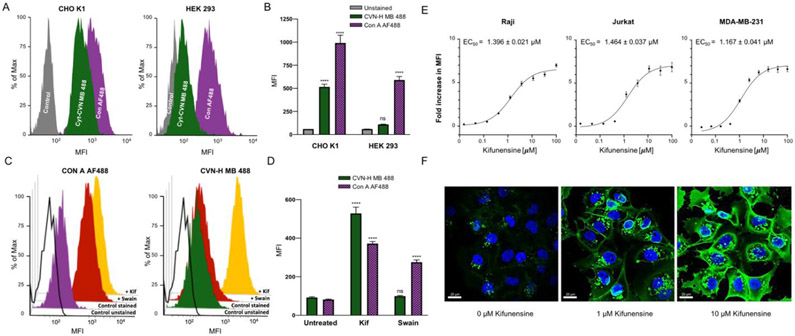
Functionalization of Cyt-CVNH-N3 (7a) with fluorophores using SPACC. A) A schematic depiction of the click reaction between Cyt-CVNH-N3 (7a) and DBCO-fluorophores 8a-c; B) The structure of the respective fluorophores. C) MALDI spectra of the reaction products 9a (left), 9b (middle), and 9c (right). D) Histograms depicting the binding of 9a-c to CHO-K1 cells (Grey = unstained controls; Green = 9a; Red = 9b; and Pink = 9c). MFI = Median Fluorescence Intensity.
The ability of these functionalized lectins to detect high mannose N-glycans on the surface of cells was subsequently investigated (Figure 4D). Previous studies demonstrate that high mannose N-glycans are prevalent in the glycocalyx of CHO K1 cells,[22] thus making them an ideal cell line for our initial experiments. Gratifyingly, staining CHO K1 cells with the Cyt-CVNH fluorophore conjugates 9a-c and subsequent flow cytometry analysis revealed an increase median fluorescence intensity (MFI) relative to unstained controls.
Next, we compared the specificities of Cyt-CVNH MB 488 (9a) and Con A AF488 using cell lines with differential N-glycan profiles (Figure 5a-b). For this, we chose to use CHO K1 cells, in addition to HEK 293 cells, which have a relatively low abundance of glycocalyx localized high mannose N-glycans, but alternatively, have a relatively high abundance of complex N-glycans.[22] Both cell lines were stained with equal concentrations of the respective lectins, and lectin staining was assessed relative to unstained controls. As expected, the high mannose N-glycan expressing CHO K1 cells were stained significantly with both lectins relative to unstained controls; however, staining of the complex N-glycan expressing HEK 293 cells with Con A AF488 is significant, while Cyt-CVNH MB 488 (9a) staining is not significant relative to unstained controls (Figure 5b).
Figure 5.
Cyt-CVNH MB 488 specificity. The binding of Cyt-CVNH MB 488 (green) and Con A AF488 (purple) to CHO K1 (left) and HEK 293 (right) cells depicted with (A) histograms and (B) a bar chart; C) Histograms depicting the binding of Con A AF488 (left) and Cyt-CVNH MB 488 (right) to Raji cells that have been treated with DMSO (purple/green), 25 μM of Swain (red) or Kif (yellow); D) Bar chart depicting the binding of Con A AF488 (purple) and Cyt-CVNH MB 488 (green) to Raji cells that have been treated with DMSO, 25 μM of Kif or Swain; E) Fold change in MFI of Cyt-CVNH MB 488 staining induced by Kif relative to DMSO treated controls. Experiments were carried out using Raji (left), Jurkat (middle), and MDA-MB-231 (right) cells; F) Assessing Kif induced changes in the prevalence of high mannose N-glycans by staining with Cyt-CVNH MB 488 and analyzing microscopy. Statistical significance was determined as described in the SI.
The observed specificity prompted us to investigate if Cyt-CVNH MB 888 (9a) could differentiate between the N-glycans that prevail upon the treatment of cells with MAN I and MAN II inhibitors (Figure 5c-d). As such, Raji cells were treated with 25 μM Kif (1) and swainsonine (2, Swain) for 48 hours prior to staining with Cyt-CVNH MB 488 (9a) and Con A AF488. Treating cells with either Kif (1) or Swain (2) significantly increased Con A AF488 staining. Conversely, staining of Swain (2) treated cells using Cyt-CVNH MB 488 (9a) was not significantly different to DMSO treated controls, but this lectin readily stained Kif (1) treated cells (Figure 5d). As such, the functionalized Cyt-CVNH lectins have the ability to differentiate between the glycans induced by MAN I and MAN II inhibitors.
Next, we investigated if Cyt-CVNH MB 488 (9a) staining correlates to the concentration of Kif (1) that is applied to cells (Figure 5e). To evaluate this, Raji cells were treated with different concentrations of Kif (1) for 48 hours before staining and analysis by flow cytometry. The fold change in staining was assessed relative to DMSO treated controls, and we were pleased to observe that the intensity of cell staining with Cyt-CVNH MB 488 (9a) increased proportionally to the concentration of Kif. Importantly, we could use this assay to determine the EC50 of Kif in Raji, Jurkat, and MDA-MB-231 cells. In addition to flow cytometry, we assessed if changes in the prevalence of high mannose N-glycans on cell surfaces could be visualized by microscopy (Figure 5f). MDA-MB-231 cells were treated with different concentrations of Kif (0, 1, and 10 μM) and were subsequently stained with Cyt-CVNH MB 488 (9a). Using confocal microscopy, an increase in staining, correlating to the concentration of Kif (1), is evidently visualized.
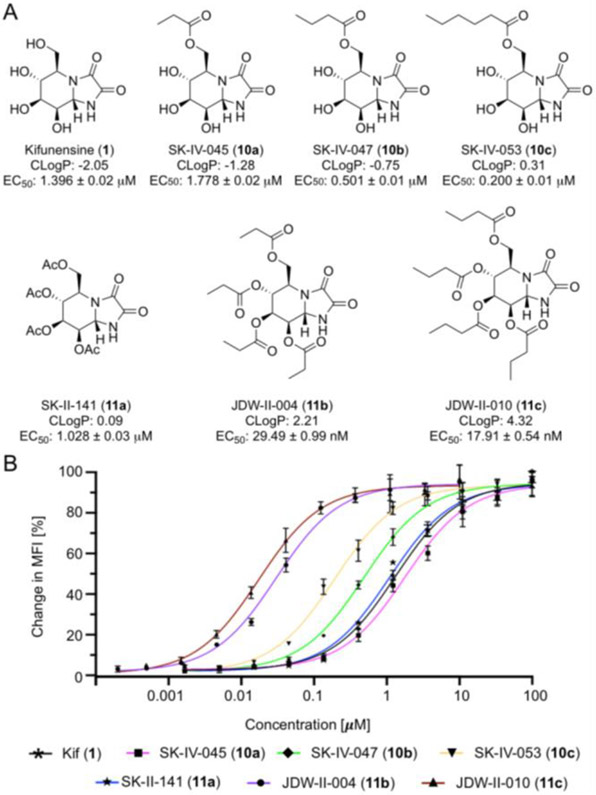
Finally, using a small panel of Kif derivatives, we examined if the functionalized lectins could differentiate between the activity of different MAN I inhibitors. Kif has been reported as a nanomolar inhibitor in biochemical assays;[9c] however, Kif exhibits micromolar activity in the cellular assays. We hypothesized that the lipophilicity (i.e., cLogP = −2.05) of this molecule was likely preventing it from efficiently entering cells. As such, we prepared Kif analogues 10a-c functionalized at the primary hydroxyl with esters (Figure 6a). We envisioned that intracellular esterases would cleave the esters appended to Kif once the molecules enter cells, thus providing Kif. The ability of these compounds to increase the prevalence of high mannose N-glycans in the glycocalyx of Raji cells after treatment for 48 hours was assessed by staining with Cyt-CVNH MB 488 (9a) and analysis by flow cytometry. The addition of a propionate ester (10a) negatively impacted activity, but the more lipophilic butyrate (10b) and hexanoate (10c) esters considerably increased activity (Figure 6b). Spurred on by these results, we prepared three additional analogues (11a-c) by modifying all of the hydroxyl groups with esters. The per-O-acetylated derivative (11a) exhibited a marginal increase in activity relative to Kif (1), however, the per-O-propionate (11b) and butyrate (11c) derivatives exhibit nanomolar activity. The activity of 11b and 11c was further assessed in Jurkat cells with similar results being observed (SI Figure S6).
Figure 6.
Kifunensine analogues. A) The structure, CLogP, and activity of the Kif analogues 10a-c and 11a-c; B) Percentage change in MFI relative to DMSO control (0%) and 200 μM Kif (100%). Experiments were carried out using Raji cells.
In summary, engineering the C-terminus of Cyt-CVNH to include a sortase motif has facilitated the preparation of tools to analyze the prevalence of high mannose N-glycans on cell surfaces. These lectin-fluorophore conjugates can differentiate between the glycans induced by Man I and Man II inhibitors, and can be used to assess the activity of Man I inhibitors; thus, their utility as tools for the discovery and development of Man I inhibitors in evident. Said utility, facilitated our efforts to develop Kif analogues that are >75-fold more potent than Kif (1), and to the best of our knowledge, these molecules are the most potent MAN I inhibitors in cellular assays described to date.
Supplementary Material
Acknowledgements
We thank Prof. A. M. Gronrenborn and Dr. E. Matei for useful discussions. This work was supported by the National Institute of General Medical Sciences (NIGMS) under award numbers P20GM113117 and P30 GM110761. We thank Dr. E. Go and the Synthetic Chemical Biology Core Facility at the University of Kansas for providing the MALDI-TOF service. This facility is supported by NIGMS grants P20GM113117 and P20GM103638. We thank Dr. J. Douglas for providing NMR support. Support for the NMR instrumentation was provided by the NIH award number P50 GM069663.
References
- [1].a) Helenius A, Aebi M, Science 2001, 291, 2364–2369; [DOI] [PubMed] [Google Scholar]; b) Bieberich E, Adv Neurobiol 2014, 9, 47–70; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Lee HS, Qi Y, Im W, Sci Rep 2015, 5, 8926; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Ferreira IG, Pucci M, Venturi G, Malagolini N, Chiricolo M, Dall'Olio F, Int J Mol Sci 2018, 19; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) van Kooyk Y, Rabinovich GA, Nature immunology 2008, 9, 593–601; [DOI] [PubMed] [Google Scholar]; f) Zhou JY, Oswald DM, Oliva KD, Kreisman LSC, Cobb BA, Trends in immunology 2018, 39, 523–535; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Marth JD, Grewal PK, Nat Rev Immunol 2008, 8, 874–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Inoue N, Ikawa M, Okabe M, Biochemical and biophysical research communications 2008, 377, 910–914; [DOI] [PubMed] [Google Scholar]; b) Gibbs GM, Lo JC, Nixon B, Jamsai D, O'Connor AE, Rijal S, Sanchez-Partida LG, Hearn MT, Bianco DM, O'Bryan MK, Endocrinology 2010, 151, 2331–2342; [DOI] [PubMed] [Google Scholar]; c) Diekman AB, Norton EJ, Klotz KL, Westbrook VA, Herr JC, Immunol Rev 1999, 171, 203–211; [DOI] [PubMed] [Google Scholar]; d) Scott H, Panin VM, Adv Neurobiol 2014, 9, 367–394; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Negreiros E, Herszterg S, Kang KH, Câmara A, Dias WB, Carneiro K, Bier E, Todeschini AR, Araujo H, Development 2018, 145; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Parkinson W, Dear ML, Rushton E, Broadie K, Development 2013, 140, 4970–4981; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Vigerust DJ, Cent Eur J Biol 2011, 6, 802; [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Watanabe Y, Bowden TA, Wilson IA, Crispin M, Biochim Biophys Acta Gen Subj 2019, 1863, 1480–1497; [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Lujan AL, Croci DO, Gambarte Tudela JA, Losinno AD, Cagnoni AJ, Mariño KV, Damiani MT, Rabinovich GA, Proc Natl Acad Sci U S A 2018, 115, E6000–e6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Kurosu M, J Mol Pharm Org Process Res 2018, 6; [PMC free article] [PubMed] [Google Scholar]; b) Dawood AA, Altobje MA, Microb Pathog 2020, 149, 104586; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang L, Liu Y, Wu L, Sun XL, Biochim Biophys Acta 2016, 1864, 143–153; [DOI] [PubMed] [Google Scholar]; d) Montefiori DC, Robinson WE Jr., Mitchell WM, Proc Natl Acad Sci U S A 1988, 85, 9248–9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Hamester F, Legler K, Wichert B, Kelle N, Eylmann K, Rossberg M, Ding Y, Kürti S, Schmalfeldt B, Milde-Langosch K, Oliveira-Ferrer L, Br J Cancer 2019, 121, 944–953; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang HF, Wu JH, Gai JW, Yang SQ, Ma QT, Ma HS, Feng Q, Gene 2018, 679, 314–319; [DOI] [PubMed] [Google Scholar]; c) Shi S, Gu S, Han T, Zhang W, Huang L, Li Z, Pan D, Fu J, Ge J, Brown M, Zhang P, Jiang P, Wucherpfennig KW, Liu XS, Clin Cancer Res 2020, 26, 5990–6002; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Silva MC, Fernandes Â, Oliveira M, Resende C, Correia A, de-Freitas-Junior JC, Lavelle A, Andrade-da-Costa J, Leander M, Xavier-Ferreira H, Bessa J, Pereira C, Henrique RM, Carneiro F, Dinis-Ribeiro M, Marcos-Pinto R, Lima M, Lepenies B, Sokol H, Machado JC, Vilanova M, Pinho SS, Cancer Immunol Res 2020, 8, 1407–1425. [DOI] [PubMed] [Google Scholar]
- [5].a) Yang Q, Hughes TA, Kelkar A, Yu X, Cheng K, Park S, Huang WC, Lovell JF, Neelamegham S, Elife 2020, 9; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lavine CL, Lao S, Montefiori DC, Haynes BF, Sodroski JG, Yang X, J Virol 2012, 86, 2153–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Bartoli M, Gicquel E, Barrault L, Soheili T, Malissen M, Malissen B, Vincent-Lacaze N, Perez N, Udd B, Danos O, Richard I, Hum Mol Genet 2008, 17, 1214–1221; [DOI] [PubMed] [Google Scholar]; b) Soheili T, Gicquel E, Poupiot J, N'Guyen L, Le Roy F, Bartoli M, Richard I, Hum Mutat 2012, 33, 429–439. [DOI] [PubMed] [Google Scholar]
- [7].Wang F, Song W, Brancati G, Segatori L, J Biol Chem 2011, 286, 43454–43464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Roychowdhury S, Oh YJ, Kajiura H, Hamorsky KT, Fujiyama K, Matoba N, Front Plant Sci 2018, 9, 62; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Macharoen K, Li Q, Márquez-Escobar VA, Corbin JM, Lebrilla CB, Nandi S, McDonald KA, Int J Mol Sci 2020, 21; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tian W, Ye Z, Wang S, Schulz MA, Van Coillie J, Sun L, Chen YH, Narimatsu Y, Hansen L, Kristensen C, Mandel U, Bennett EP, Jabbarzadeh-Tabrizi S, Schiffmann R, Shen JS, Vakhrushev SY, Clausen H, Yang Z, Nat Commun 2019, 10, 1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Iwami M, Nakayama O, Terano H, Kohsaka M, Aoki H, Imanaka H, J Antibiot (Tokyo) 1987, 40, 612–622; [DOI] [PubMed] [Google Scholar]; b) Kayakiri H, Takase S, Shibata T, Okamoto M, Terano H, Hashimoto M, Tada T, Koda S, The Journal of Organic Chemistry 1989, 54, 4015–4016; [Google Scholar]; c) Elbein AD, Tropea JE, Mitchell M, Kaushal GP, J Biol Chem 1990, 265, 15599–15605. [PubMed] [Google Scholar]
- [10].a) Chen H, Li R, Liu Z, Wei S, Zhang H, Li X, Carbohydr Res 2013, 365, 1–8; [DOI] [PubMed] [Google Scholar]; b) Hering KW, Karaveg K, Moremen KW, Pearson WH, J Org Chem 2005, 70, 9892–9904; [DOI] [PubMed] [Google Scholar]; c) Koyama R, Kano Y, Kikushima K, Mizutani A, Soeda Y, Miura K, Hirano T, Nishio T, Hakamata W, Bioorganic & Medicinal Chemistry 2020, 28, 115492; [DOI] [PubMed] [Google Scholar]; d) Koyama R, Hakamata W, Hirano T, Nishio T, Chem Pharm Bull (Tokyo) 2018, 66, 678–681. [DOI] [PubMed] [Google Scholar]
- [11].Cummings RD, Etzler ME, in Essentials of Glycobiology (Eds.: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME), Cold Spring Harbor Laboratory Press; Copyright © 2009, The Consortium of Glycobiology Editors, La Jolla, California., Cold Spring Harbor (NY), 2009. [Google Scholar]
- [12].a) Fouquaert E, Smith DF, Peumans WJ, Proost P, Balzarini J, Savvides SN, Damme EJ, Biochemical and biophysical research communications 2009, 380, 260–265; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kaku H, Van Damme EJ, Peumans WJ, Goldstein IJ, Arch Biochem Biophys 1990, 279, 298–304; [DOI] [PubMed] [Google Scholar]; c) Moothoo DN, Canan B, Field RA, Naismith JH, Glycobiology 1999, 9, 539–545. [DOI] [PubMed] [Google Scholar]
- [13].Dorling PR, Huxtable CR, Colegate SM, Biochem J 1980, 191, 649–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mitchell CA, Ramessar K, O'Keefe BR, Antiviral Res 2017, 142, 37–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].a) Barrientos LG, Matei E, Lasala F, Delgado R, Gronenborn AM, Protein Engineering, Design and Selection 2006, 19, 525–535; [DOI] [PubMed] [Google Scholar]; b) Gao X, Chen W, Guo C, Qian C, Liu G, Ge F, Huang Y, Kitazato K, Wang Y, Xiong S, Applied Microbiology and Biotechnology 2010, 85, 1051–1060; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shenoy SR, Barrientos LG, Ratner DM, O'Keefe BR, Seeberger PH, Gronenborn AM, Boyd MR, Chemistry & Biology 2002, 9, 1109–1118. [DOI] [PubMed] [Google Scholar]; d) Our efforts to functionalize cyanovirin-N from Nostoc ellupsosperum led so significant protein perciptiation, while our efforts to utilize these constructs to detect high mannose N-glycans on cell surfaces led to highly inconsistant results. These observations led us to conclude that said constructs are not suitable for our endeavours.
- [16].Matei E, Basu R, Furey W, Shi J, Calnan C, Aiken C, Gronenborn AM, J Biol Chem 2016, 291, 18967–18976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Contarino M, Bastian AR, Kalyana Sundaram RV, McFadden K, Duffy C, Gangupomu V, Baker M, Abrams C, Chaiken I, Antimicrob Agents Chemother 2013, 57, 4743–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].a) Mao H, Hart SA, Schink A, Pollok BA, J Am Chem Soc 2004, 126, 2670–2671; [DOI] [PubMed] [Google Scholar]; b) Dai X, Böker A, Glebe U, RSC Advances 2019, 9, 4700–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].a) Swee LK, Lourido S, Bell GW, Ingram JR, Ploegh HL, ACS Chem Biol 2015, 10, 460–465; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wu Q, Ploegh HL, Truttmann MC, ACS Chem Biol 2017, 12, 664–673; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hirakawa H, Ishikawa S, Nagamune T, Biotechnol J 2015, 10, 1487–1492. [DOI] [PubMed] [Google Scholar]
- [20].Brumby T, Friebe M, Lehmann L, Platzek J, Suelzle D , Vol. US20040208828A1, BAYER SCHERING PA, US, 2004.
- [21].a) Agard NJ, Prescher JA, Bertozzi CR, J Am Chem Soc 2004, 126, 15046–15047; [DOI] [PubMed] [Google Scholar]; b) Adronov A, Li K, Fong D, Meichsner E, Chemistry 2020. [DOI] [PubMed] [Google Scholar]
- [22].Hamouda H, Kaup M, Ullah M, Berger M, Sandig V, Tauber R, Blanchard V, J Proteome Res 2014, 13, 6144–6151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.