Abstract
Copper is a necessary nutrient but quickly becomes toxic at elevated levels. To properly handle environmental copper influxes and maintain metal homeostasis, organisms utilize various methods to chelate, excrete, and metabolize heavy metals. These mechanisms are believed to involve complex signaling pathways mediated by neuropeptides. This study incorporates custom N,N-dimethyl leucine isobaric tags to characterize the neuropeptidomic changes after different time points (1, 2, and 4 hours) of copper exposure in a model organism, blue crab, Callinectes sapidus. Using a modified simplex optimization strategy, the number of identifiable and quantifiable neuropeptides was increased three-fold to facilitate a deeper understanding of the signaling pathways involved in responding to heavy metal exposure. The time course exposure showed many interesting findings, including upregulation of inhibitory allatostatin peptides in the pericardial organs. Additionally, there was evidence of transport of a pigment dispersing hormone from the sinus glands to the brain. Overall, this study improves the multiplexing capabilities of neuropeptidomic studies to understand the temporal changes associated with copper toxicity.
Keywords: Callinectes sapidus, Mass Spectrometry, Copper Toxicity, Neuropeptide, Quantitation, Isobaric Tags
Graphical Abstract
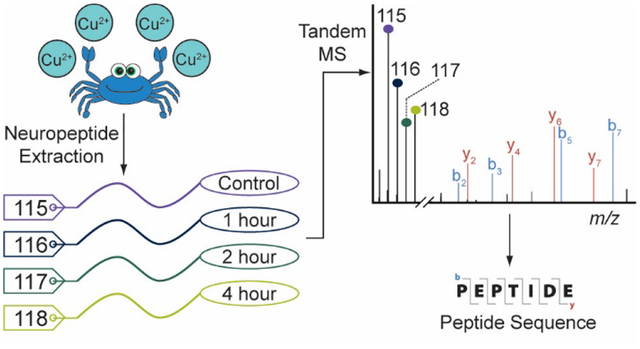
Introduction:
Neuropeptides represent a large, highly diverse class of signaling molecules within the nervous system. These signaling peptides have been shown to play physiological roles in many biochemical processes and can potentially serve as biomarkers for a variety of disease states, such as Alzheimer’s disease, cancers,1 and environmental stress.2–5 The exact function of a particular neuropeptide is difficult to study due to their function being dependent on location, concentration, and the presence of other co-modulating neurotransmitters or neuropeptides.6 As a result, thorough approaches to profile all neuropeptides in a sample is critical to characterize neuropeptidomic changes in a meaningful way. Global profiling of neuropeptides is challenging, however, due to their diverse structures and low in vivo concentrations. Mass spectrometry (MS) offers fast, highly sensitive analyses for probing many biomolecules at once, without needing to know the exact make-up of the sample.
Quantitative analysis of neuropeptides has been achieved using several different quantitative strategies. Label-free methods have been reported but suffer from low throughput and instrumental variability (i.e., signal drift).7 Reductive dimethylation and other isotopic labeling strategies, resulting in a differential mass shift at the MS1 level, have been used to provide relative expression changes in neuropeptidomic studies.3 These methods enable multiple samples to be analyzed simultaneously, but they can also result in complex spectra that make data interpretation difficult. This limits the multiplexing potential of MS1-based labeling strategies. Alternatively, isobaric tags facilitate relative quantitation at the MS2 level by incorporating isotopically encoded tags with the same nominal mass addition, but different reporter ions formed upon fragmentation. These tags do not increase spectral complexity, offering higher multiplexing capabilities. Additionally, they require less sample from a single channel. Commercially available isobaric tags, such as iTRAQ8 and TMT,9 can be expensive for academic lab settings. Instead, the Li Lab has developed N,N-dimethyl leucine (DiLeu) isobaric tags that label primary amines.10,11 These tags are readily synthesized and offer up to a 21-plex experiment,12 enabling a cost-effective experiment with reduced sample needs, required instrument time, and run-to-run variability.
Despite these advantages, the application of isobaric tags in neuropeptidomic analyses has been scarce.13 The low in vivo abundance of neuropeptides often results in a lack of selection for fragmentation through tandem MS analysis using typical data-dependent acquisition settings. While this is a challenge for neuropeptidomics in general, it is especially problematic when using isobaric tags as quantitation is performed exclusively at the MS2 level. Optimization of various data acquisition parameters (e.g., resolution, number of MS2 scans events, and dynamic exclusion window) have been shown to facilitate deeper profiling in -omic studies by reducing redundant analyses and increasing the number of spectra corresponding to low-abundance analytes, as well as reducing scan time to increase the total number of spectra acquired in a single LC-MS run.14,15 As there are numerous parameters affecting the data-dependent acquisition of spectra, it can be laborious to optimize all of them. This is exacerbated by the fact that many of the parameters are interconnected and need to be optimized simultaneously. Design of experiments (DoE) have been used to discern and evaluate the influence of specific parameters on peptide identifications.16–18 A multitude of designs exist, the simplest of which is the factorial DoE.19 In a factorial DoE a number of factors, x, are evaluated at a number of levels, y, leading to xy combinations to be tested. As the number of factors and levels increases, the number of tests required for optimization rapidly increases. As a result, factorial DoE are less useful when trying to determine a precise optimum. Advanced modeling and statistical analyses can be used to ascertain optimum conditions, but still require many tests for confident interpretation.
In this work, the MS1 resolution, MS2 resolution, dynamic exclusion window, and number of MS2 scan events were selected for optimization due to their direct effect on instrument scan time, redundant analyses, and analysis of lower abundance analytes. These factors were evaluated at two levels with a simple factorial DoE to provide a rough evaluation of their effect on neuropeptide identifications. The dynamic exclusion window and MS2 scan events were then systematically optimized using a modified simplex algorithm.20 Using this approach, a set of conditions (i.e., a simplex) is evaluated. The conditions are then ranked, and the next set of conditions are in the geometrically opposed region from the point of worst performance. This simplex algorithm has been modified to include not only the geometric reflection, but also a contracted reflection, elongated reflection, and a reflection contracted in the direction of the low performance points. This modification allows the optimum (determined when additional simplexes provide no improvement) to be reached faster and with fewer points tested overall. Because the resolving power of the orbitrap instrument used in this experiment is fixed at preset values, optimization is constrained and incompatible with the geometric reflections used in the simplex method. Using this modified simplex optimization strategy for the dynamic exclusion window and number of MS2 scan events resulted in a three-fold increase in the number of neuropeptides identified from the crustacean model organism, Callinectes sapidus. This performance improvement could further benefit from optimization of additional parameters (e.g., automatic gain control target, maximum injection time, and isolation window width) and is the subject of ongoing research.
These optimized parameters can be used to study neuropeptidomic changes under various experimental conditions. Herein, we use DiLeu isobaric tags to examine neuropeptide expression after exposure to environmental copper. Concerns over copper in the environment are growing due to increased prevalence of copper in agricultural runoff, industrial pollution, and poorly managed wastewater.21 Moreover, as atmospheric carbon dioxide increases, causing ocean acidification, copper becomes more bioavailable and therefore more toxic.22 Interestingly, although toxic at high levels, copper is also a necessary nutrient and plays roles in neuromodulation, protein structure, and enzymatic activity.23 This dichotomy of being a potent toxin and required nutrient suggests that interesting pathways are involved in handling large effluxes of copper in the environment. In this study, the blue crab is used as a model organism due to its relatively simple nervous system and relevance as it frequently inhabits coastal estuaries plagued by large copper effluxes. To better understand how copper affects neuropeptide expression, a 4-plex experiment was performed using DiLeu tags to characterize expression changes after 1, 2, and 4 hours of exposure to 10 μM copper (Cu2+) relative to a control. Statistically significant changes were observed at all time points in four neuroendocrine tissues (brain, sinus glands, pericardial organs, and thoracic ganglia), demonstrating the feasibility and usefulness of DiLeu multiplex tags for quantifying endogenous expression of neuropeptides.
Methods:
Materials:
Methanol (MeOH), acetonitrile (ACN), glacial acetic acid, crab saline components (described below) and LC-MS solvents were purchased from Fisher Scientific (Pittsburgh, PA). Isotopic leucines and formaldehyde, triethylammonium bicarbonate (TEAB), N,N-dimethylformamide (DMF), 4-(4,6-dimethoxy- 1,3,5-triazin-2-yl)-4-methylmorpholinium tetrafluoroborate (DMTMM), ammonium formate, and copper (II) chloride were purchased from Sigma Aldrich (St. Louis, MO). N-methylmorpholine (NMM) was purchased from TCI America, (Tokyo, Japan). C18 ZipTips were purchased from Millipore (Burlington, MA). Strong cation exchange (SCX) spin tips were purchased from PolyLC (Columbia, MD).
Animals and Copper Exposure Experiment:
Female blue crabs, Callinectes sapidus, were purchased from a local grocery store (Midway Asian Markets, Madison, WI) and maintained in tanks with recirculating artificial seawater (30 ppt salinity) and a 12 h:12 h light:dark cycle. Crabs exposed to copper were placed in a tank of 10 μM Cu2+ (from CuCl2) for 0 (control), 1, 2, or 4 h prior to sacrifice. All crabs, regardless of experimental conditions, were anesthetized on ice for 20 min before sacrificing for its organs as previously described.24 Neuropeptide-rich tissues–brains, paired sinus glands (SG), paired pericardial organs (PO), and thoracic ganglia (TG)–were dissected in chilled saline (440 mM NaCl, 11 mM KCl, 13 mM CaCl2, 26 mM MgCl2, 11 mM trizma base, 5 mM maleic acid, adjusted to pH 7.45 with NaOH) and stored in acidified methanol (90:9:1 MeOH:H2O:glacial acetic acid). Tissues were stored at −80 °C until future processing.
Neuropeptide Extraction:
Neuropeptides were extracted via ultrasonication using a Fisherbrand 120 probe sonicator/sonic dismembrator. Tissues submerged in acidified methanol were sonicated at 50% amplitude for 8 s three times with 15 s rest between pulses. Samples were then centrifuged at 20,000 × g for 20 min at 4°C before collecting the supernatant. The supernatant was then dried down using a Savant SCV100 SpeedVac. Crude extracts were then purified using C18 ZipTips per the manufacturer’s protocol.
Labeling:
For the optimization experiments, neuropeptide extracts from several tissues were pooled together to create enough sample for multiple LC-MS injections. Four equal aliquots of this sample were then differentially labeled using DiLeu tags. For the copper toxicity experiments, extracts were differentially labeled by exposure duration (control, 1 h, 2 h, and 4 h). Each bioreplicate for the copper toxicity experiments had one crab at each time point with four crabs total for each multiplexed sample. A total of four biological replicates (n=4) were analyzed with three technical replicates (repeated LC-MS injections). The labeling procedure has been described previously along with the synthesis of the tags.11 Briefly, purified neuropeptide extracts were resuspended in a 50:50 mixture of ACN:0.5 M triethylammonium bicarbonate. The isotopic dimethyl leucine tags were activated to a triazine ester form by the addition of 50 μL activation solution (15.5 mg DMTMM, 495 μL dry DMF, 5 μL NMM) per 1 mg dimethyl leucine. After being vortexed at room temperature for 30 min, the activated tag was added to the neuropeptide extracts at a 20:1 label-to-peptide ratio. The labeling reaction was quenched after 30 min by the addition of NH2OH for a final concentration of 0.25% NH2OH. The labeled neuropeptide extracts were then pooled together before drying down. Excess labeling reagents were removed using SCX spin tips. Multiplexed samples were loaded to the columns in 0.1% formic acid, 20% ACN, 80% water and excess label was washed away using the same buffer. The sample was eluted using 0.4 M ammonium formate, 20% ACN, 80% water. Multiplexed samples were then desalted once again using C18 ZipTips before being analyzed by LC-MS.
MS Data Collection:
Samples were analyzed in triplicate on a Thermo Q Exactive HF mass spectrometer coupled to a Dionex UltiMate3000 nanoLC system using a homemade C18 column (15 cm). Neuropeptides were eluted over a 90 min gradient from 10% B to 35% B (solvent A = 0.1% formic acid in water; solvent B = 0.1% formic acid in ACN). Spectra were collected at a m/z range of 200–2000 using HCD (high-energy collision dissociation) for fragmentation. The MS1 resolution, MS2 resolution, dynamic exclusion window, and number of MS2 scan events varied from run-to-run and are detailed below. More detailed information about the instrument parameters can be found in Table S1.
Data Analysis:
Raw files were processed using Proteome Discoverer 2.1 to identify neuropeptides from an in-house crustacean neuropeptide database. Static modifications included DiLeu labeling at the N-terminus and lysine residues, along with C-terminal amidation of known amidated neuropeptides (e.g., RFamide and RYamide peptides). Dynamic modifications included oxidation of Met, deamidation of Asn and Gln, methylation of Asp, Glu, His, Lys, Arg, Ser, and Thr, and dehydration of Ala, Asp, Glu, Ser, Thr, Tyr residues. Quantitation and normalization of reporter ions was also performed in Proteome Discoverer 2.1 and exported to Excel. Additional information for the data analysis is provided in Table S2. Neuropeptides described below as “identified” were present in at least two of three technical replicates, identified and quantified across all four channels, and for the copper toxicity experiments, present in at least three bioreplicates. The quantitative ratios were then tested using Dunnett’s test25,26 to determine statistical significance between experimental conditions and the control.
Results and Discussion:
Factorial Design of Experiments
An initial factorial DoE was performed to roughly evaluate the influence of the MS1 and MS2 resolution, dynamic exclusion window, and number of MS2 scan events. These factors were selected as they have a direct influence on the scan time, the total number of acquired spectra, and the ability to reduce redundant analyses. This DoE assessed combinations of high and low values for each of the four parameters for a total of 16 unique sets of conditions. The combinatorial testing parameters and results are given in Figure 1. Overall, more identifications were achieved with reduced MS1 and MS2 resolution and an increased number of MS2 scans events. As the resolution (both MS1 and MS2) is decreased, the scan time with an orbitrap mass analyzer also decreases, allowing more spectra to be acquired. This in turn enables more MS2 scans per precursor scan to be collected to identify and quantify more unique neuropeptides. Analysis of the isolation interference between the experiments in which the resolving power was high compared to low showed the lower resolution not only resulted in more total peptide spectral matches (PSMs), but also that more of those PSMs were high enough quality for identification and quantitation of neuropeptides. More details can be found in the Supplemental Information (Figure S1). This observance could be attributed to the fact that there are simply more spectra to have peptides matched to, and that the spectral purity is more so affected by isolation window width, not resolution. The influence of the dynamic exclusion window was more variable, highlighting the need for a more systematic approach to optimization. As the dynamic exclusion window widens, neuropeptides with high signal are less likely to dominate the acquired MS2 spectra. If the window is too wide, however, there is risk of losing identifications due to collecting MS2 spectra too early in the elution profile (where signal is often too low for meaningful results) and prohibiting future analyses across the peak width. It is also important to note that peptides have variable peak widths, so a reliable dynamic exclusion window cannot be easily predicted.
Figure 1:
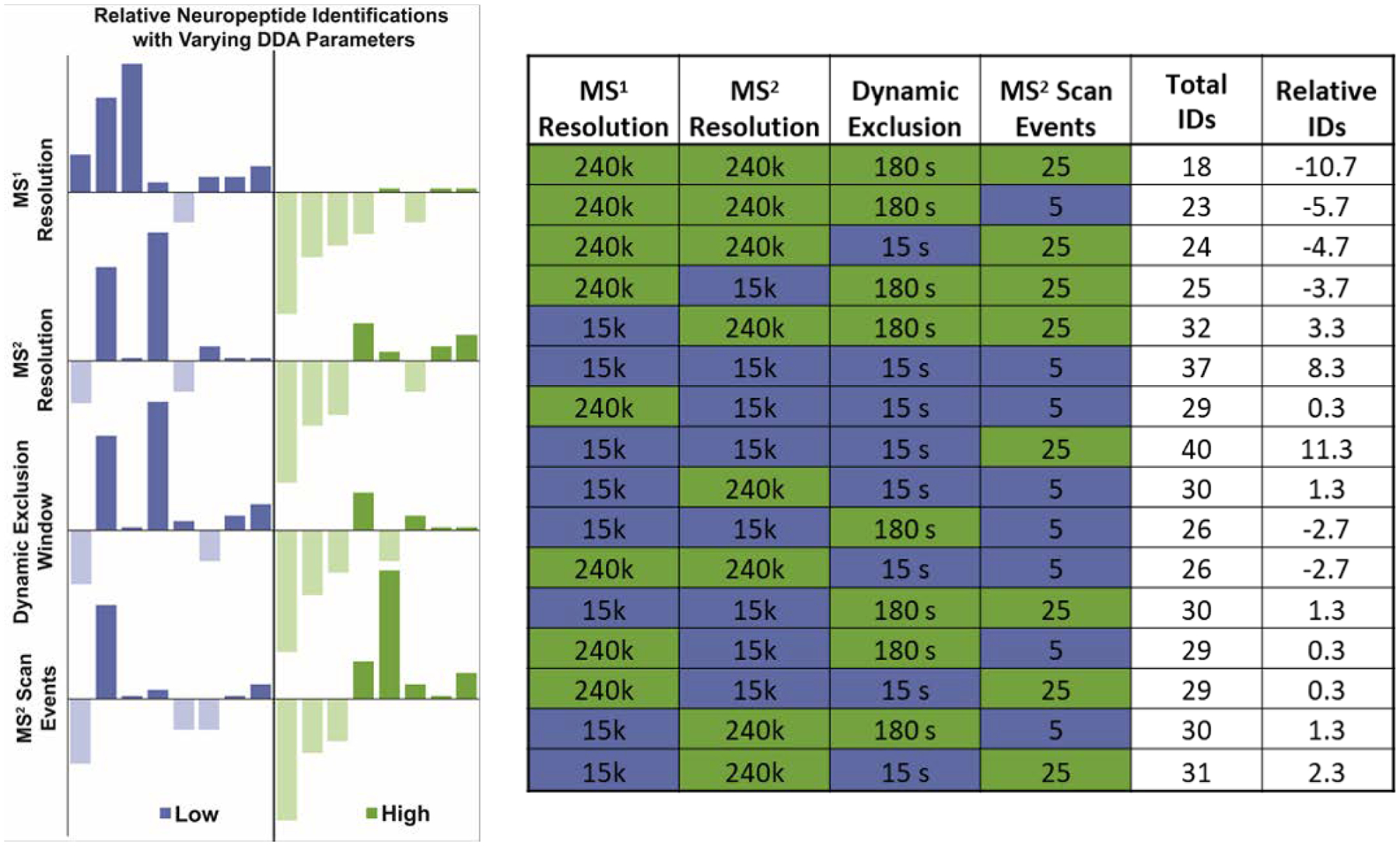
Results from factorial DoE. The data is shown as bar graphs on the left with each of the y-axes representing the deviation from the average number of neuropeptides identified across all 16 conditions. The 16 conditions are represented in each plot; the different plots are merely arranged by low/high (blue/green) values for the selected parameter to more quickly visualize how the number of neuropeptides identified changes when a selected parameter is low/high, regardless of the other parameters. The numeric values are provided in the table shown at right. The Total IDs column represents the total neuropeptides identified and the Relative IDs are the deviations from the average across the 16 conditions.
Modified Simplex Optimization
Many parameters show some degree of interplay and optimization needs to consider this. For example, the number of MS2 scan events affects the optimum dynamic exclusion window (and vice versa), compounding the challenges with efficiently optimizing these parameters. An approach that optimizes these parameters simultaneously is therefore required and can be accomplished via a modified simplex strategy. A comprehensive overview of the simplex optimization strategy has been published by Bezerra, M.A., et al.20 Initially, three sets of conditions (vertices of the graphical representation in Figure 2) were examined (V1, V2, and V3). The second simplex was geometrically reflected away from V2 (the worst performing condition) and additional points (e.g., R1, CR1, etc.) were evaluated. After four iterations, optimum parameters were reached, demonstrated by the fifth simplex showing no improvement over previous simplexes. In total 17 conditions were evaluated. Some conditions were omitted from later simplexes if there was already a set of conditions at or very close to that point. Figure 2 shows the results from these 17 conditions with the relative number of neuropeptides identified as a heat map overlaid on the simplex optimization plot. The table in Figure 2 shows the neuropeptides identified for each vertex of the simplex optimization. A more sequential view of the data in Figure 2 is shown in Figure S2. The conditions near the optimum region (shown in red) offer over a three-fold increase in the number of neuropeptides able to be identified, compared to the initial, unoptimized conditions (Simplex 1 from Figure 2). Although this performance increase is great, it could be further improved in the future by more systematically optimizing the resolving power and other MS parameters.
Figure 2:
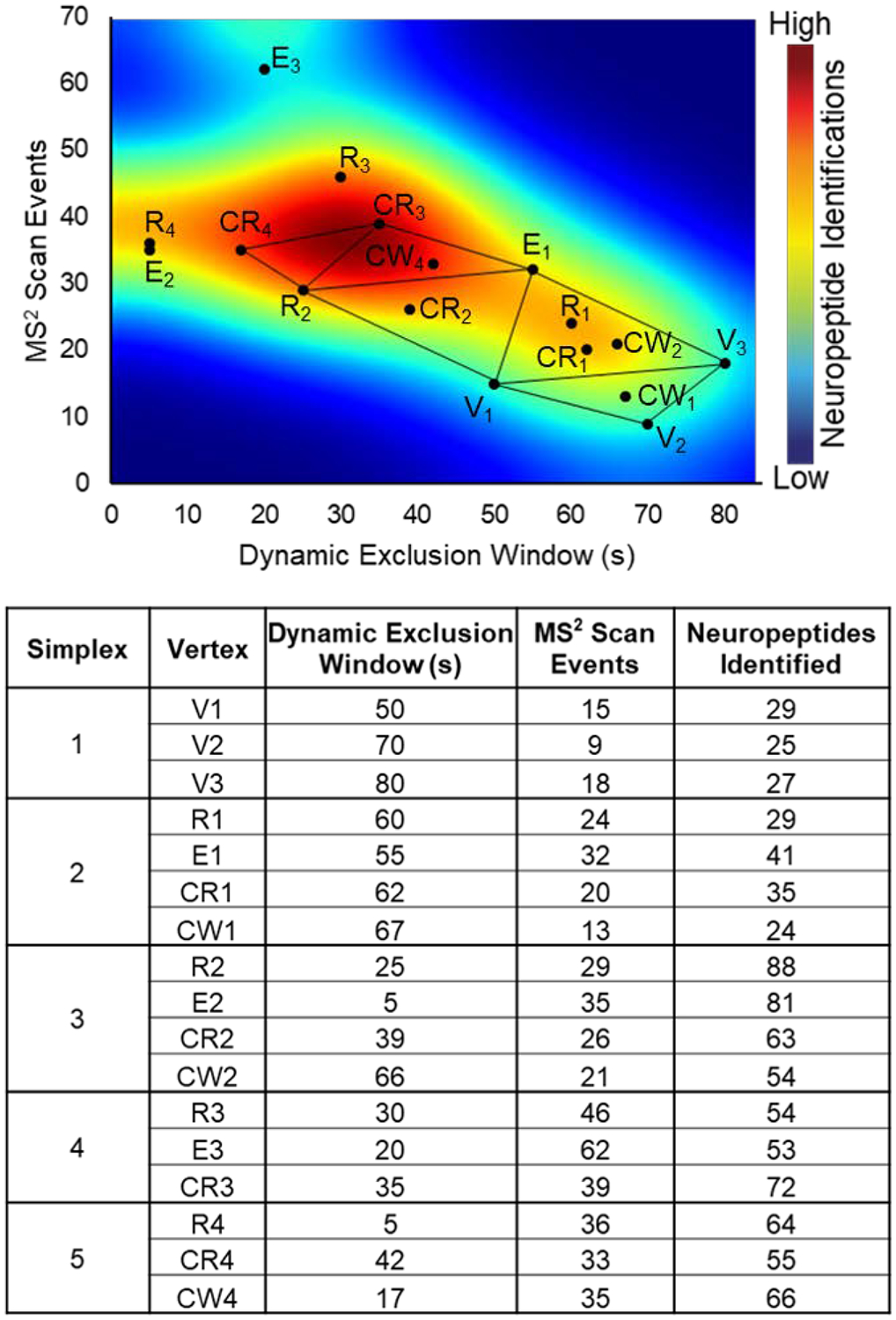
Modified simplex optimization. The optimization of the dynamic exclusion window and number of MS2 scan events is represented graphically with an overlaid heatmap to depict the relative number of identified neuropeptides under each condition. The initial vertices are denoted by V1, V2, and V3; the subsequent simplexes contain points reflected away from the point of worst performance (R), an elongated reflection (E), a contracted reflection (CR), and the contracted reflection closer to the direction of the worst point (CW).
As data-dependent acquisition is known to have limited reproducibility, three conditions (R2, CR3, and CW4) were tested again to determine the ideal parameters to use for later analyses. Vertices R2, CR3, and CW4 were selected based on neuropeptides identified and proximity to the optimum region shown in Figure 2. A separate neuropeptide-rich sample was then analyzed by the three conditions with five technical replicates each. There were 59, 60, and 51 neuropeptides identified across all 4 channels in at least 2 technical replicates for R2, CR3, and CW4, respectively. Condition CR3 was selected for future analyses due to its greater number of neuropeptide identifications and higher reproducibility among technical replicates. It should be noted that R2 would likely be sufficient for most analyses, but CR3 had a slight advantage. The optimum dynamic exclusion window of 35 s appears to match with the chromatographic peak widths for neuropeptides. For the C18 RPLC separations used in this experiment, it was observed that most neuropeptides elute over 30–60 s, but some elute in as little as 15 s and some over 90 s. The dynamic exclusion window seems skewed towards the low range of peak widths, but this could be due to some neuropeptides having similar masses and retention times. A lower dynamic exclusion might enable more of these similar neuropeptides to be differentiated at the MS2 level. The simplex data also showed that higher MS2 scan events facilitated deeper profiling. This is rather intuitive as it allows more low abundance ions to be selected for tandem MS, but 39 MS2 scan events is surprisingly higher than typical LC-MS experiments (often using a Top 5, Top 10, or Top 15 method).
The optimized methods also showed high quantitative accuracy of reporter ion ratios. The differentially labeled samples used for optimization were pooled at a 1:1:1:1 ratio (DiLeu reporter ions 115:116:117:118) and the observed ratio (averaged for all identified neuropeptides) was 1:0.94:0.93:0.89, demonstrating suitable quantitative accuracy. The distribution of neuropeptides by family for the neuropeptides identified under the optimum conditions are shown in Figure 3. The diversity of neuropeptide families demonstrate that the methods are not biased towards a particular sequence motif. The quantitative accuracy and lack of bias will enable the methods to be used to collect meaningful neuropeptidomic data to study crustaceans under environmental stress.
Figure 3:
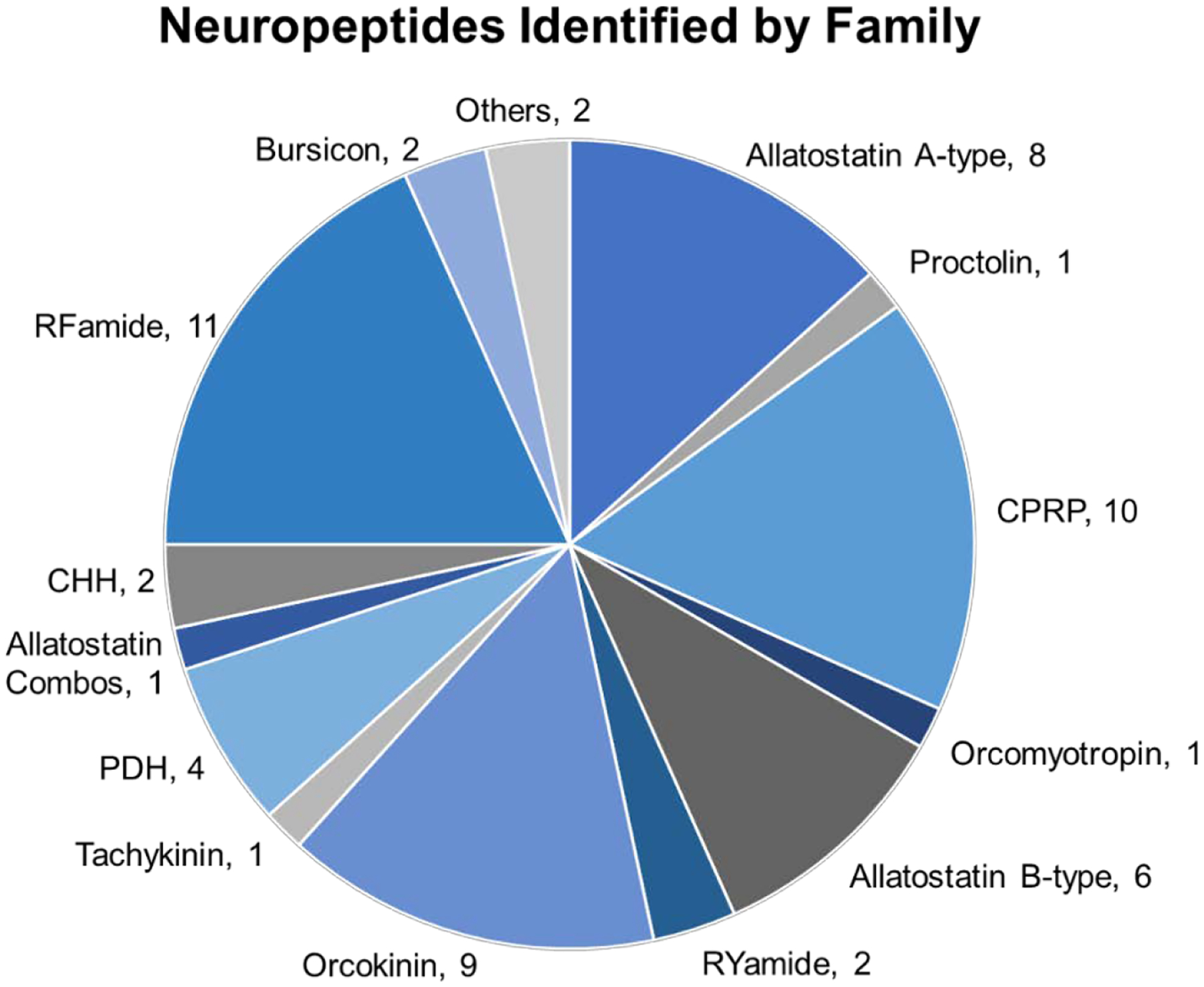
Neuropeptide family distribution. The pie chart shows the distribution of neuropeptides belonging to select families. CHH = crustacean hyperglycemic hormone, CPRP = CHH precursor related peptide, PDH = pigment dispersing hormone.
Copper Toxicity
Using a DiLeu labeling strategy and the optimized acquisition settings, CR3 (low MS1 and MS2 resolving power, 39 MS2 scan events, and a 35 s dynamic exclusion window), this study demonstrates the feasibility of using isobaric tagging to study neuropeptidomic changes in response to acute copper toxicity. Neuropeptides have been shown to be involved in the stress response to many environmental stressors, such as salinity,4 pH,5 and hypoxia.3 Heavy metals can also act as potent environmental stressors by causing oxidative stress and dysregulating metal homeostasis within an organism. Copper is of particular interest as it is an important component of many proteins, such as hemocyanin (analogous to hemoglobin in humans),27,28 and dysregulation of copper homeostasis has been implicated in several diseases, including Alzheimer’s disease, anemia, and liver disease.23,29
In this experiment, crabs were exposed to 10 μM copper (CuCl2) for 0 (control), 1, 2, and 4 hr. The extracted neuropeptides were differentially labeled with 4-plex DiLeu tags to provide relative quantitation of neuropeptides compared to a control sample. Figures 4 and 5 depict the significant neuropeptide expression changes in the TG and POs (Figure 4) and SGs and brain (Figure 5) as a ratio to the control after each exposure duration. These ratios are expressed as a log2 ratio for easier viewing (positive results show upregulation, negative show downregulation). Statistical significance was determined using Dunnett’s test (n = 4) and is denoted by a star in the bar graphs. More precise relative quantitative information for the neuropeptides shown in Figures 4 and 5 are given in Tables S3 and S4 along with the neuropeptides that did not show significant changes.
Figure 4:
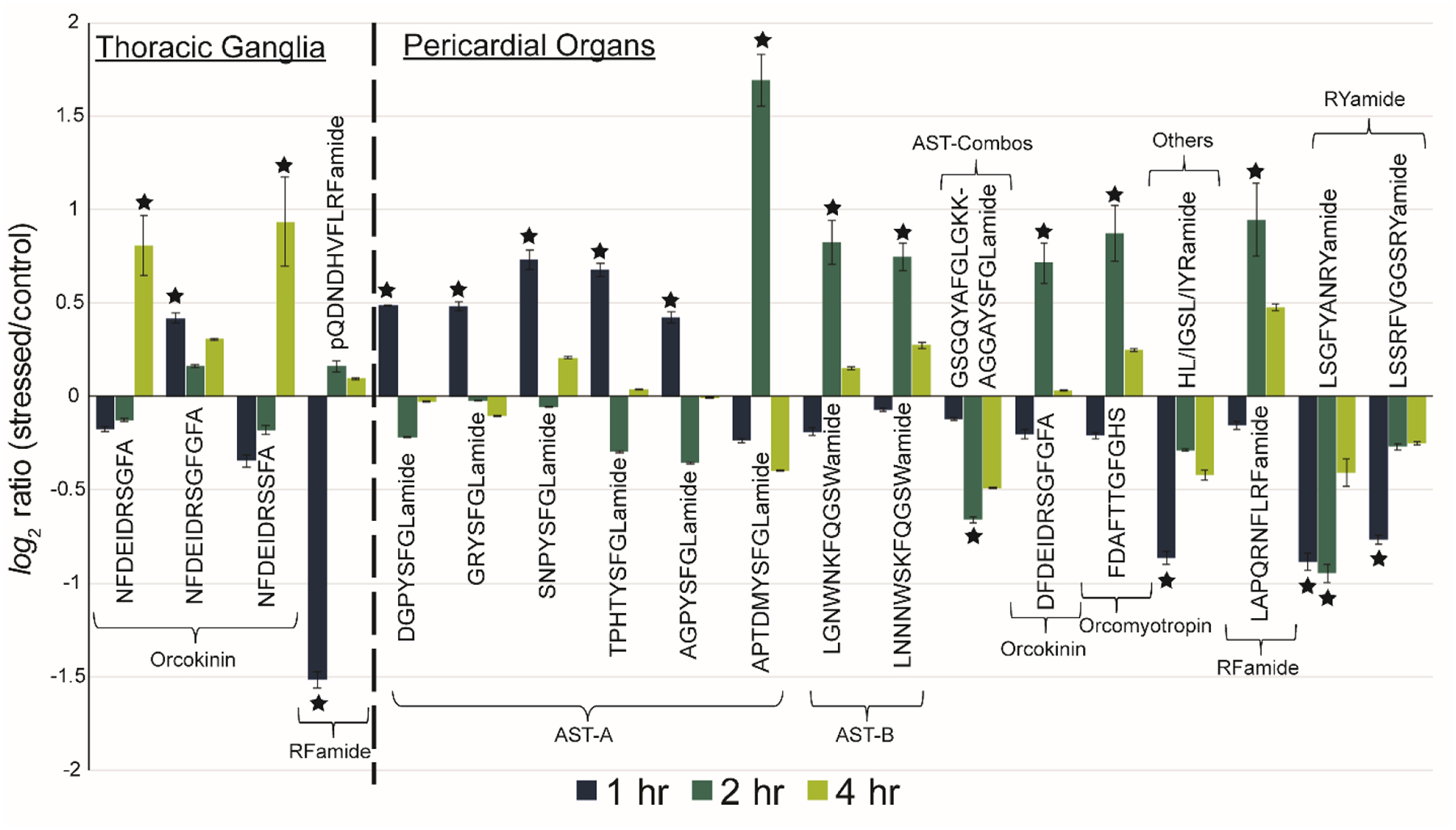
Neuropeptidomic changes in the thoracic ganglia and pericardial organs. Neuropeptide sequences with at least one significant (denoted by star) change are listed across the x-axis and grouped by family. AST-A = allatostatin A-type, AST-B = allatostatin B-type, and AST-Combos = allatostatin combos. The ratio for the intensity of reporter ions for a stress condition (1, 2, or 4 hr copper exposure) relative to the control are depicted on the y-axis as a log2 ratio to clearly see up/downregulation of neuropeptides relative to the x-axis. The error bars denote the standard error of the mean (n=4).
Figure 5:

Neuropeptidomic changes in the sinus glands and brain. Neuropeptide sequences with at least one significant (denoted by star) change are listed across the x-axis and grouped by family. PDH = pigment dispersing hormone, RPCH = red pigment concentrating hormone. The PDH highlighted in blue is identical in sequence in both the sinus glands and brain. The ratio for the intensity of reporter ions for a stress condition (1, 2, or 4 hr copper exposure) relative to the control are depicted on the y-axis as a log2 ratio to clearly see up/downregulation of neuropeptides relative to the x-axis. The error bars denote the standard error of the mean (n=4).
There are interesting dysregulation patterns in neuropeptides with significant changes observed resulting from copper toxicity. Within the POs, which act upon the cardiac system, upregulation of many allatostatin A-type and B-type (AST-A and AST-B, respectively) was observed. AST-A have been well-documented as being inhibitory neuromodulators that slow down a number of processes, including feeding/digestion and cardiac contractions.30,31 The initial burst in AST-A output seen after 1 hour in the POs suggests a hyperarousal signal to decrease cardiac output. Similarly, B-type allatostatin neuropeptides have been shown to act as myoinhibitors.31,32 There is a significant upregulation of AST-B neuropeptides after 2 hr exposure, suggesting the activation of signaling pathways to decrease muscle contractions in the cardiac system. These neuropeptides could also be secreted into the hemolymph (circulating fluid in crustaceans) to act as hormones (rather than locally in the cardiac system). Future experiments to analyze allatostatin neuropeptides in the hemolymph could demonstrate interesting hormonal signaling pathways but are not included in this study due to the challenges of analyzing trace-level neuropeptides in the complex matrix of hemolymph. Although similar in modulation behaviors, the AST-A and AST-B type families have unique structures and signaling pathways.30 The temporal shift in upregulation between these families further supports that distinct pathways are involved in their regulation.
In the brain and SGs, there is an interesting pigment dispersing hormone (PDH) with sequence QELKYQEREMVAELAQQIYRVAQAPWAAAVGPHKRNSELINSILGLPKVMNDAamide that is significantly downregulated in the SGs and upregulated in the brain. This sequence is highlighted in blue shade in Figure 5. As PDH is synthesized in the SGs,31 this pattern is likely indicative of hormonal signaling from the SGs to the brain. Historically, pigment dispersing hormones were shown to primarily regulate the light-adapting pigments in the eyestalks and were named accordingly.33,34 Though typically seen as a result of light-receptor activity, the PDH activity reported here is not believed to be the result of circadian rhythm. Exposures and dissections were not carried out at set times during the day and as a result, are independent of biological changes due to circadian rhythm. Moreover, the exact same neuropeptide has previously been shown by our lab to display the same temporal dysregulation in response to hypoxia (low levels of dissolved oxygen).3 The expression changes of this PDH could therefore be indicative of a general stress response, or perhaps more specifically related to respiratory distress as copper can also impair gill function by disrupting ion channels.27 The insect ortholog for PDH, known as pigment dispersing factor (PDF),35 has been shown to play roles in many processes outside of circadian rhythm, including locomotor activity, courtship, and hormone biosynthesis.31 The findings here support the growing evidence for PDH also acting as a neurotransmitter and neuromodulator in the nervous system and having diverse functional behavior.
There were also interesting general trends observed across the four tissue types. In the TG, there is significant upregulation of two orcokinin neuropeptides (NFDEIDRSGFA and NFDEIDRSSFA), but only after 4 hours of exposure. Conversely, a third orcokinin, NFDEIDRSGFGFA, shows upregulation after only 1 hour of exposure. The differences in dysregulation of neuropeptides within the same family is seen in other tissue types as well (e.g., RFamide neuropeptides in the brain) and highlight the fact that even though they are structurally very similar, neuropeptides of the same family can have different functions, target receptors, degradation pathways, etc. Additionally, while some neuropeptides, like the RYamides in the pericardial organs, show a consistent pattern of up/downregulation over 4 hours, many others show a hyperarousal pattern. In these cases, the neuropeptide is increased or decreased at one time point and returns to baseline. These findings demonstrate the efficacy of multiplexed analyses as the increased throughput and reduced sample needs enables time course studies. Transient shifts in neuropeptide expression that would have gone unnoticed previously are now able to be observed with minimal added effort and cost.
Conclusions and Future Directions:
Quantitative neuropeptidomics can provide many insights into the complex signaling pathways involved in the response to environmental stress like heavy metal toxicity. The deployment of a custom isobaric tag, DiLeu, and systematic optimization of data-dependent acquisition settings has enabled thorough, multiplexed analyses of neuropeptides. In this study, four parameters were examined with two selected for extensive optimization. Omission of MS1 and MS2 resolving power from further optimization reduced experimentation needs but is nonetheless a limitation of this study. Similarly, other optimizable parameters related to MS data acquisition have been shown to improve analyses and are the subject of future work within our lab. Despite these limitations in optimization, a three-fold increase in quantifiable neuropeptides was still observed. The optimized parameters were used to study neuropeptides in response to acute copper toxicity. Several statistically significant trends were observed in key neuroendocrine tissues across different exposure durations. These findings represent the critical first steps in understanding the physiological effects of copper toxicity and the signaling pathways that may be involved in an organism surviving polluted ecosystems. Future work to characterize the neuropeptides in the hemolymph would offer more robust interpretation of the hormonal/paracrine signaling involved. Additionally, by incorporating isobaric tags with mDa mass differences resulting from mass defects, the Li Lab has expanded the DiLeu tagging protocol up to a 21-plex.12 Using these tags in the future will allow more experimental conditions (e.g., different exposure durations or severities of exposure) to be evaluated, facilitating a more in-depth understanding of copper toxicity in aquatic environments.
Supplementary Material
Table S1: LC-MS instrument settings
Table S2: Proteome Discoverer data analysis settings
Table S3: All neuropeptides identified in the thoracic ganglia (TG) and pericardial organs (PO)
Table S4: All neuropeptides identified in the sinus glands (SG) and brain
Figure S1: Comparison of isolation interference between high and low resolving powers
Figure S2: Sequential results from modified simplex algorithm
Acknowledgements:
This work was supported by a National Science Foundation grant (CHE-1710140) and National Institutes of Health grants (R01DK071801, S10RR029531). The instruments used in this project were generously provided by Thermo Fisher Scientific. C.S.S. acknowledges the help and support of the Biotechnology Training Program (T32GM008349) under the National Institutes of General Medical Sciences as well as a National Institute of Environmental Health Sciences fellowship as part of the National Ruth L. Kirschstein Research Service Award fellowship program (F31ES031859)). L.L. acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin−Madison School of Pharmacy.
Footnotes
Supporting Information:
The Supporting Information is available free of charge at https://pubs.acs.org/
Supporting Tables S1-S4 and Supporting Figures S1 and S2 (.docx)
References:
- (1).Sweedler Jonathan V.,Xie Fang, Bora A Protein and Peptide Mass Spectrometry in Drug Discovery. Chapter 1.4. 2012, 221–227. [Google Scholar]
- (2).Chen R; Xiao M; Buchberger A; Li L Quantitative Neuropeptidomics Study of the Effects of Temperature Change in the Crab Cancer borealis. J. Proteome Res 2014, 13 (12), 5767–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Buchberger AR; Sauer CS; Vu NQ; DeLaney K; Li L A Temporal Study of the Perturbation of Crustacean Neuropeptides Due to Severe Hypoxia Using 4-Plex Reductive Dimethylation. J. Proteome Res 2020, 19, 4, 1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zhang Y; Buchberger A; Muthuvel G; Li L Expression and Distribution of Neuropeptides in the Nervous System of the Crab Carcinus maenas and Their Roles in Environmental Stress. Proteomics 2015, 15 (23–24), 3969–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Liu Y; Buchberger AR; Delaney K; Li Z; Li L Multifaceted Mass Spectrometric Investigation of Neuropeptide Changes in Atlantic Blue Crab, Callinectes sapidus, in Response to Low PH Stress. J. Proteome Res 2019, 18, 2759–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Li L; Sweedler JV Peptides in the Brain: Mass Spectrometry–Based Measurement Approaches and Challenges. Annu. Rev. Anal. Chem 2008, 1 (1), 451–483. [DOI] [PubMed] [Google Scholar]
- (7).Romanova EV; Dowd SE; Sweedler JV Quantitation of Endogenous Peptides Using Mass Spectrometry Based Methods. Curr Opin Chem Biol 2013, 17 (5), 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wiese S; Reidegeld KA; Meyer HE; Warscheid B Protein Labeling by ITRAQ: A New Tool for Quantitative Mass Spectrometry in Proteome Research. Proteomics 2007, 7 (3), 340–350. [DOI] [PubMed] [Google Scholar]
- (9).Werner T; Becher I; Sweetman G; Doce C; Savitski MM; Bantscheff M High-Resolution Enabled TMT 8-Plexing. Anal. Chem 2012, 84 (16), 7188–7194. [DOI] [PubMed] [Google Scholar]
- (10).Xiang F; Ye H; Chen R; Fu Q; Li NN,N-Dimethyl Leucines as Novel Isobaric Tandem Mass Tags for Quantitative Proteomics and Peptidomics. Anal. Chem 2010, 82 (7), 2817–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Frost DC; Greer T; Li L High-Resolution Enabled 12-Plex DiLeu Isobaric Tags for Quantitative Proteomics. Anal. Chem 2015, 87 (3), 1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Frost DC; Feng Y; Li L 21-Plex DiLeu Isobaric Tags for High-Throughput Quantitative Proteomics. Anal. Chem 2020, 92 (12), 8228–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jiang X; Xiang F; Jia C; Buchberger AR; Li L Relative Quantitation of Neuropeptides at Multiple Developmental Stages of the American Lobster Using N, N -Dimethyl Leucine Isobaric Tandem Mass Tags. ACS Chem. Neurosci 2018, acschemneuro.7b00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zhang Y; Wen Z; Washburn MP; Florens L Effect of Dynamic Exclusion Duration on Spectral Count Based Quantitative Proteomics. Anal. Chem 2009, 81 (15), 6317–6326. [DOI] [PubMed] [Google Scholar]
- (15).Kalli A; Smith GT; Sweredoski MJ; Hess S Evaluation and Optimization of Mass Spectrometric Settings during Data-Dependent Acquisition Mode: Focus on LTQ- Orbitrap Mass Analyzers. J Proteome Res 2014, 12 (7), 3071–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Randall SM; Cardasis HL; Muddiman DC Factorial Experimental Designs Elucidate Significant Variables Affecting Data Acquisition on a Quadrupole Orbitrap Mass Spectrometer. J. Am. Soc. Mass Spectrom 2013, 24 (10), 1501–1512. [DOI] [PubMed] [Google Scholar]
- (17).Andrews GL; Dean RA; Hawkridge AM; Muddiman DC Improving Proteome Coverage on a LTQ-Orbitrap Using Design of Experiments. J. Am. Soc. Mass Spectrom 2011, 22 (4), 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hecht ES; McCord JP; Muddiman DC Definitive Screening Design Optimization of Mass Spectrometry Parameters for Sensitive Comparison of Filter and Solid Phase Extraction Purified, INLIGHT Plasma N-Glycans. Anal. Chem 2015, 87 (14), 7305–7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lundstedt T; Seifert E; Abramo L; Thelin B; Nyström Å; Pettersen J; Bergman R Experimental Design and Optimization. Chemom. Intell. Lab. Syst 1998, 42 (1–2), 3–40. [Google Scholar]
- (20).Bezerra MA; dos Santos QO; Santos AG; Novaes CG; Ferreira SLC; de Souza VS Simplex Optimization: A Tutorial Approach and Recent Applications in Analytical Chemistry. Microchem. J 2016, 124, 45–54. [Google Scholar]
- (21).Keller AA; Adeleye AS; Conway JR; Garner KL; Zhao L; Cherr GN; Hong J; Gardea-Torresdey JL; Godwin HA; Hanna S; et al. Comparative Environmental Fate and Toxicity of Copper Nanomaterials. NanoImpact 2017, 7 (May), 28–40. [Google Scholar]
- (22).Dietrich AM; Postlethwait N; Gallagher DL Quantifying Bioavailability and Toxicity of Copper to Americamysis Bahia - Mysid Shrimp. Int. J. Environ. Sci. Dev 2013, 4 (1), 37–43. [Google Scholar]
- (23).Kardos J; Héja L; Simon Á; Jablonkai I; Kovács R; Jemnitz K Copper Signalling : Causes and Consequences. 2018, 3, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gutierrez GJ; Grashow RG Cancer Borealis Stomatogastric Nervous System Dissection. J. Vis. Exp 2009, No. 25, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).McHugh ML Multiple Comparison Analysis Testing in ANOVA. Biochem. Medica 2011, 21 (2), 203–209. [DOI] [PubMed] [Google Scholar]
- (26).Jaki T; Hothorn LA Statistical Evaluation of Toxicological Assays: Dunnett or Williams Test - Take Both. Arch. Toxicol 2013, 87 (11), 1901–1910. [DOI] [PubMed] [Google Scholar]
- (27).Martins CDMG; Barcarolli IF; de Menezes EJ; Giacomin MM; Wood CM; Bianchini A Acute Toxicity, Accumulation and Tissue Distribution of Copper in the Blue Crab Callinectes Sapidus Acclimated to Different Salinities: In Vivo and in Vitro Studies. Aquat. Toxicol 2011, 101 (1), 88–99. [DOI] [PubMed] [Google Scholar]
- (28).Paganini CL; Bianchini A Copper Accumulation and Toxicity in Isolated Cells from Gills and Hepatopancreas of the Blue Crab (Callinectes sapidus). Environ. Toxicol. Chem 2009, 28 (6), 1200–1205. [DOI] [PubMed] [Google Scholar]
- (29).Rossbach LM; Shaw BJ; Piegza D; Vevers WF; Atfield AJ; Handy RD Sub-Lethal Effects of Waterborne Exposure to Copper Nanoparticles Compared to Copper Sulphate on the Shore Crab (Carcinus maenas). Aquat. Toxicol 2017, 191 (April), 245–255. [DOI] [PubMed] [Google Scholar]
- (30).Christie AE; Stemmler EA; Dickinson PS Crustacean Neuropeptides. Cell. Mol. Life Sci 2010, 67 (24), 4135–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Takei Y; Ando H; Tsutsui K Handbook of Hormones; 2015.
- (32).Szabo TM; Chen R; Goeritz ML; Maloney RT; Tang LS; Li L; Marder E Distribution and Physiological Effects of B-Type Allatostatins (Myoinhibitory Peptides, MIPs) in the Stomatogastric Nervous System of the Crab, Cancer borealis. J. Comp. Neurol 2011, 519 (13), 2658–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Klein JM; Mohrherr CJ; Sleutels F; Riehm JP; Ranga Rao K Molecular Cloning of Two Pigment-Dispersing Hormone (PDH) Precursors in the Blue Crab Callinectes sapidus Reveals a Novel Member of the PDH Neuropeptide Family. Biochem. Biophys. Res. Commun 1994, 205 (1), 410–416. [DOI] [PubMed] [Google Scholar]
- (34).Sousa GL; Lenz PH; Hartline DK; Christie AE General and Comparative Endocrinology Distribution of Pigment Dispersing Hormone- and Tachykinin-Related Peptides in the Central Nervous System of the Copepod Crustacean Calanus finmarchicus. 2008, 156, 454–459. [DOI] [PubMed] [Google Scholar]
- (35).Nässel DR; Winther ÅME Drosophila Neuropeptides in Regulation of Physiology and Behavior. Prog. Neurobiol 2010, 92 (1), 42–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: LC-MS instrument settings
Table S2: Proteome Discoverer data analysis settings
Table S3: All neuropeptides identified in the thoracic ganglia (TG) and pericardial organs (PO)
Table S4: All neuropeptides identified in the sinus glands (SG) and brain
Figure S1: Comparison of isolation interference between high and low resolving powers
Figure S2: Sequential results from modified simplex algorithm


