Abstract
Resource Description Framework (RDF) is one of the three standardized data formats in the HL7 Fast Healthcare Interoperability Resources (FHIR) specification and is being used by healthcare and research organizations to join FHIR and non-FHIR data. However, RDF previously had not been integrated into popular FHIR tooling packages, hindering the adoption of FHIR RDF in the semantic web and other communities. The objective of the study is to develop and evaluate a Java based FHIR RDF data transformation toolkit to facilitate the use and validation of FHIR RDF data. We extended the popular HAPI FHIR tooling to add RDF support, thus enabling FHIR data in XML or JSON to be transformed to or from RDF. We also developed an RDF Shape Expression (ShEx)-based validation framework to verify conformance of FHIR RDF data to the ShEx schemas provided in the FHIR specification for FHIR versions R4 and R5. The effectiveness of ShEx validation was demonstrated by testing it against 2693 FHIR R4 examples and 2197 FHIR R5 examples that are included in the FHIR specification. A total of 5 types of errors including missing properties, unknown element, missing resourceType, invalid attribute value, and unknown resource name in the R5 examples were revealed, demonstrating the value of the ShEx in the quality assurance of the evolving R5 development. This FHIR RDF data transformation and validation framework, based on HAPI and ShEx, is robust and ready for community use in adopting FHIR RDF, improving FHIR data quality, and evolving the FHIR specification.
Keywords: Fast Healthcare Interoperability Resources (FHIR), Resource Description Framework (RDF), Shape Expression (ShEx), Semantic Web, Data Transformation, Quality Assurance
Introduction
HL7 Fast Healthcare Interoperability Resources (FHIR) is rapidly becoming the standards framework for the exchange of electronic health record (EHR) data.(1) The FHIR specification defines a common language and mechanism for sharing EHR data using building blocks called “Resources”. Every major EHR vendor has produced standard FHIR interfaces to their clinical and patient data. In particular, the HL7 Argonaut Project has published a FHIR-based API and accompanying Data Services specification to enable expanded information sharing for EHRs and other health information technology based on Internet standards and architectural patterns and styles.(2) One of the exciting aspects of FHIR is that, through the RDF specification, FHIR has become one of the first clinical data standards to incorporate the vision of the “Semantic Web” described by Tim Berners-Lee in his seminal 2001 Scientific American article.(3) Berners-Lee envisions the Semantic Web as the next “killer app” and outlines a formula for success, in which RDF and accompanying ontologies are key enablers. RDF, in particular, serves as the lingua franca for exchanging machine-processable information, with ontologies providing the formal definitions that allow both machines and human beings to understand the intent of the information, even when isolated from its original communities and contexts. RDF defines a simple “triples”-based structure that can represent data, metadata, terminology, and ontology as a single unified whole. The atomicity of the RDF graph representation allows information from disparate sources and structures to be joined as a single, federated collection of “linked data”. The Semantic Web vision is intimately associated with the realization of artificial intelligence (AI) in developing smart agents.(3, 4)
The Yosemite Project positions RDF as the “Universal Healthcare Exchange Language” and asserts that “existing standard healthcare vocabularies, data models, and exchange languages should be leveraged by defining standard mappings to RDF, and any new standards should have RDF representations”.(5) The Yosemite Project also asserts that healthcare information should either have a “standard mapping to RDF” or should already be in an RDF format.
Before FHIR included an RDF format -- i.e., prior to the Standard for Trial Use 3 (STU3) release, when it only had JSON and XML formats -- FHIR provided two of the three components of Berners-Lee’s vision: a well-defined structural foundation for the recording of clinical data and a collection of ontological components (eg, SNOMED, LOINC, ICD) that formalize the meaning of much of what can be expressed in the clinical space. The FHIR RDF format fills in that third component, providing a standard machine-processable semantic foundation for clinical data to be linked with other data using ontologies. The potential availability of large stores of FHIR data in standardized RDF has led to a renewed interest in the FHIR RDF data representations.
The RDF representation of FHIR was first specified in STU3. FHIR RDF is being used by healthcare and research organizations to join FHIR and non-FHIR data. For example, in a collaboration with the Global Alliance for Genomics and Health (GA4GH), we explored the harmonization of the Phenopackets data model with the FHIR standard using the FHIR RDF technology.(6, 7) The United Kingdom (UK) National Health Service (NHS) is using FHIR RDF-enabled Social Linked Data (SOLID) technology to provide patient-centric data representation, which has been deployed in live patient in the Greater Manchester General Practices.(8–11) In addition, FHIR RDF has been used to represent cancer data for developing predictive models.(12, 13) However, RDF has not been integrated in many of the popular FHIR tooling packages, hindering the broader adoption of FHIR RDF in the semantic web and other communities. This work addresses this gap by providing an open-source library to convert between FHIR/RDF and other FHIR formats.
In a previous study, we explored the use of JSON for Linked Data, version 1.1 (JSON-LD 1.1) as a means of both documenting and automating the conversion of FHIR data to and from RDF.(6, 14) Although that approach yields one convenient and flexible way to convert FHIR data to and from RDF, it was not based on popular FHIR tooling and lacks some of the error-reporting and performance features expected in production-ready tooling.
The objective of this study was to develop and evaluate a robust, production-ready FHIR RDF data transformation and validation framework, based on a popular FHIR toolkit, to facilitate FHIR data use and quality improvement. We added FHIR RDF parsing and serialization to the widely adopted HAPI FHIR implementation, which enables bidirectional FHIR RDF data transformation. We also implemented an RDF data validation mechanism using the FHIR Shape Expression (ShEx) schemas to validate the RDF data converted from FHIR instance data.
Materials and Methods
Materials
HAPI FHIR Java APIs
The HAPI FHIR library is an open source implementation of the HL7 FHIR specification for Java.(15) It has over 120 contributors and has been used in over 800 FHIR projects.(16) HAPI defines a model class for every resource type and datatype defined by the FHIR specification. It consists of a number of modules: a core library module; a structure module containing model classes for different FHIR versions; a client framework including an HTTP implementation; a validation module which is used to validate resource instances against FHIR Profiles (StructureDefinition, ValueSet, CodeSystem, etc.); and a server module that can be used to develop FHIR compliant servers against your own data storage layer. HAPI FHIR already provided FHIR JSON and XML parsers for parsing the FHIR Resource instances into Java objects, and it also already provided serializers for writing out those Java objects in FHIR JSON and XML formats. In this study, we developed and integrated a FHIR RDF parser and serializer for HAPI, to enable this popular FHIR tool to read RDF, write RDF, or convert FHIR JSON or XML data to or from FHIR RDF.
FHIR Core Library
The FHIR publication process entails the generation and publication of Java libraries which capture the core FHIR StructureDefinition and provide Java objects to capture and manipulate them.(15) These libraries include functions to test equivalence between two Java objects, which we used for our round-trip processing
HAPI parsers are driven from a core parser which uses the FHIR Core Library to navigate these definitions to populate those core objects. During parsing, the core parser generates entities not directly expressed in the serialized representation, such as an array of included resources which is aggregated from multiple places in a serialized document. The order of these objects is not defined but at present (there’s no formula specifying that eg, a contained Patient precedes a contained Specimen), testing equivalence between two Java objects is sensitive to the order and leads to arbitrary rejection.
Jena Java API for RDF
Apache Jena is a free and open source Java framework for building Semantic Web and Linked Data applications.(17) It provides a Java API to create and read RDF graphs, and serialize RDF triples in the XML and turtle formats. In this study, the Jena Java API was integrated into HAPI as a library component, to parse and serialize FHIR RDF.
ShEx and FHIR ShEx Schemas
The RDF Shape Expressions (ShEx) language, developed in a W3C Community Group, is a language for formally describing RDF structures and can serve the same role for RDF as that W3C XML Schema serves for XML.(18) A collection of RDF triples -- an RDF “graph” or “dataset” -- can be validated against a given ShEx definition (schema) to determine whether the collection meets the requirements defined in the schema. A ShEx schema contains one or more data shape definitions. Validating an RDF node against a shape tests the adjacent nodes against the constraints in the shape. The constraints can be atomic triple constraints or boolean combinations thereof. The ShEx validators have been implemented in different programming languages, including JavaScript (shex.js), Java (ShExJava), Scala (shex-s), Python (PyShEx), and Ruby (Ruby ShEx). (19–23)
ShEx proved useful for describing a standard model of FHIR RDF data.(24) The combination of a formal model and a succinct format enabled comprehensive review and automated validation. FHIR has adopted the ShEx schemas to describe the FHIR data models (i.e., FHIR resources) since its STU3 release. Figure 1 illustrates the Patient resource with its ShEx schema and a Patient instance in turtle.
Figure 1.
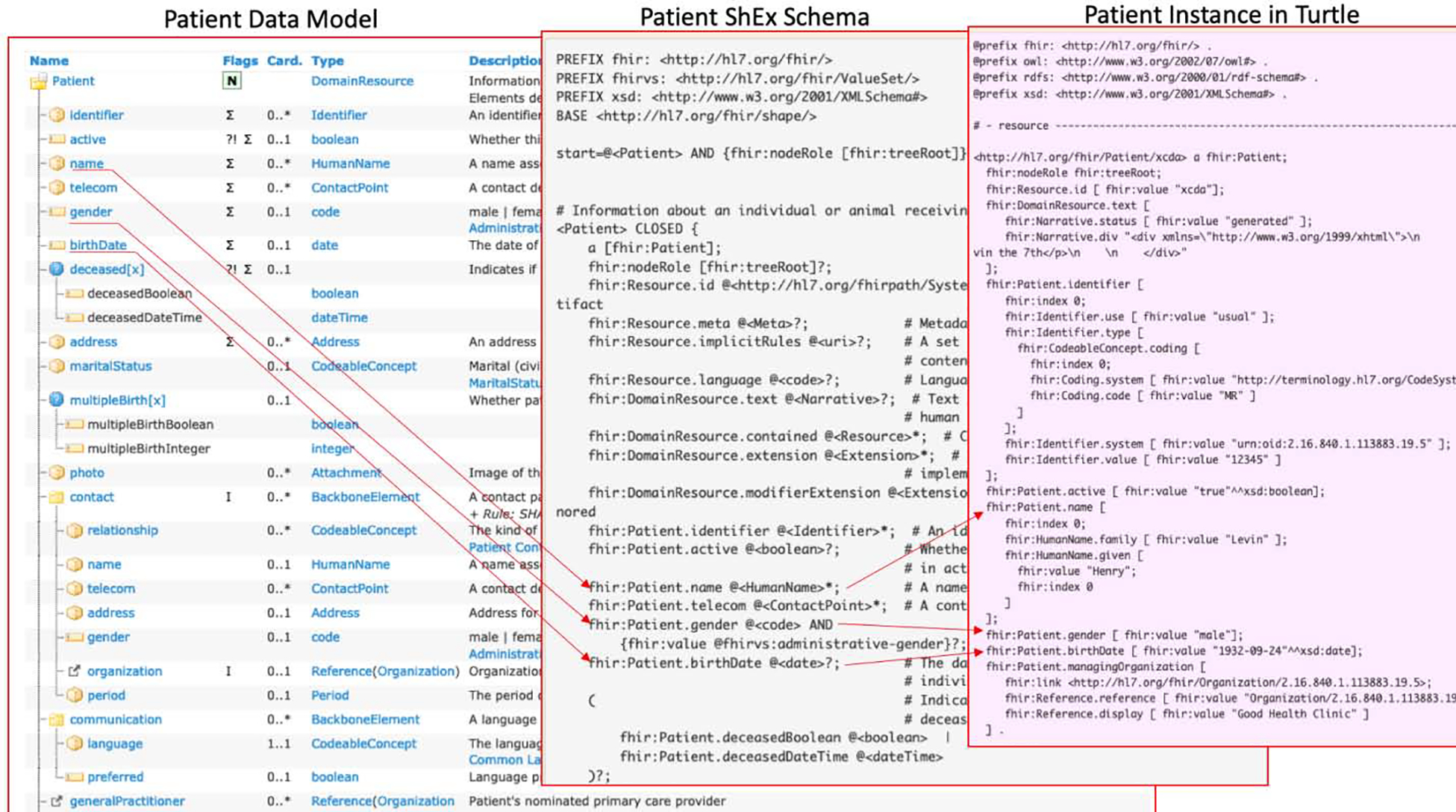
An illustration from the FHIR specification of the Patient resource with its ShEx schema and a Patient instance in the Turtle RDF format.
ShExJava now uses the Commons RDF API that supports RDF4J, Jena, JSON-LD-Java, OWL API and Apache Clerezza.(20, 25) It can parse ShEx schema written in multiple ShEx formats -- ShExJ (a JSON representation serving as the abstract syntax), ShExR (an equivalent RDF representation) and ShExC (a human-facing “compact syntax”) -- and can serialize a schema in ShExJ format. Two different algorithms are available in ShExJava for validating data against a ShEx schema: the refine algorithm, which computes the typing for the whole graph and retains the results; or the recursive algorithm, which computes only the typing required to answer a particular validate(node, ShapeLabel) call, and forgets the results.
Methods
We extended the HAPI FHIR library by adding an RDF (Turtle) parser and serializer, which enables bidirectional conversion of FHIR instance data to and from RDF.
Figure 2 shows the system architecture of the data transformation and validation mechanism. We added two main components to HAPI: (1) an RDF/Turtle Java Parser; and (2) an RDF/Turtle Java Serializer.
Figure 2.
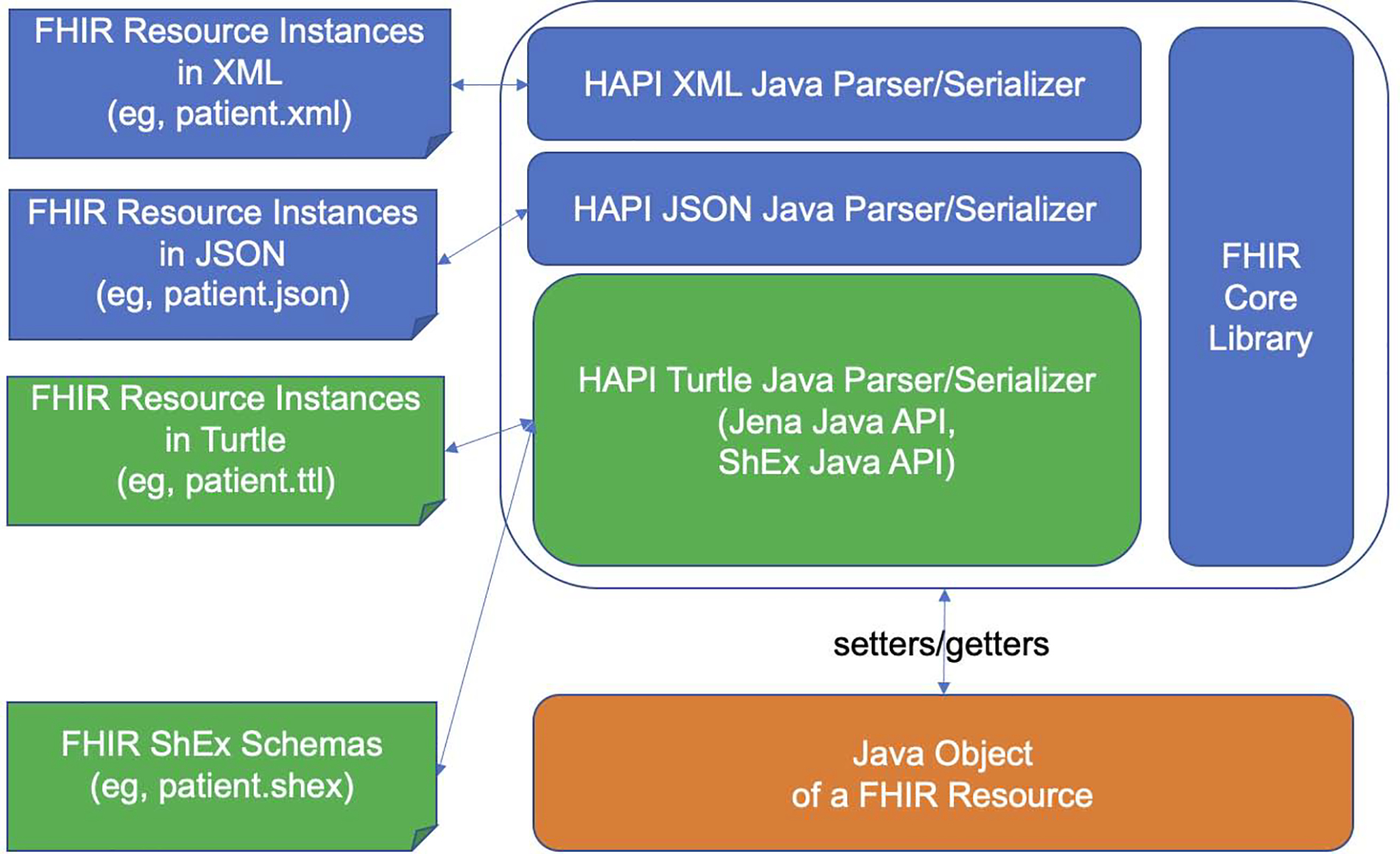
System architecture of the FHIR RDF data transformation and validation framework. Blue and orange represent the state of the art in HAPI FHIR Library; Green represents the contribution of the study.
The HAPI RDF/Turtle Parser takes a FHIR RDF resource instance (in Turtle format), parses that instance using Jena Java API, and sets the result into a Java object representing that FHIR resource instance. This Java object is then available for other processing using the HAPI API -- including serialization using any of HAPI’s JSON, XML or RDF serializers.
Conversely, the HAPI RDF/Turtle Serializer takes a Java object representing a FHIR resource instance, which may have been created by any of the HAPI parsers and uses RDF4J to serialize it into conforming FHIR RDF in Turtle format.
By parsing one FHIR format and serializing to a different FHIR format, this enables HAPI to transform arbitrary FHIR data to and from FHIR RDF.
We also implemented a FHIR RDF data validation mechanism that uses ShExJava to validate FHIR RDF instance data against the ShEx schema defined in the FHIR specification. This validation mechanism can be used either before a FHIR RDF resource instance is consumed by the HAPI RDF parser and/or after the HAPI RDF serializer has produced FHIR RDF.
Evaluation Design
To evaluate the FHIR RDF transformation and validation capabilities that we implemented, we created a test suite based on the examples that are included in the FHIR specification. The examples include RDF, JSON and XML formats, and are generated by templates specific to the FHIR publication process. FHIR R4 included 2912 JSON examples, but to keep the test suite performant we excluded 219 large example files and ran our tests on the remaining 2693 files. To help ensure that the approach will be future-proof against changes to the FHIR specification, we also ran our tests against examples from the R5 release, which contains 2870 JSON examples. We again excluded 673 large examples and ran our tests on the remaining 2197 examples.
Our goal in testing was to ensure the correctness of RDF serializer and parser, and to verify the conformance of serialized FHIR RDF to the FHIR specification. To do this, we compared RDF-sourced Java objects against JSON-sourced Java objects to ensure that format conversions are “round trippable”, meaning that no information is lost, added or changed during bidirectional data transformation between FHIR data formats. To compare the RDF-sourced Java objects against the JSON-sourced Java objects, we used the èqualsDeep` function provided by FHIR in the òrg.hl7.fhir.r5.model.Basè class, which performs a complete attribute-by-attribute equality comparison of two Java FHIR objects. Figure 3 illustrates the two components of the evaluation design, ie, ShEx validation and Round-trippable check.
Figure 3.
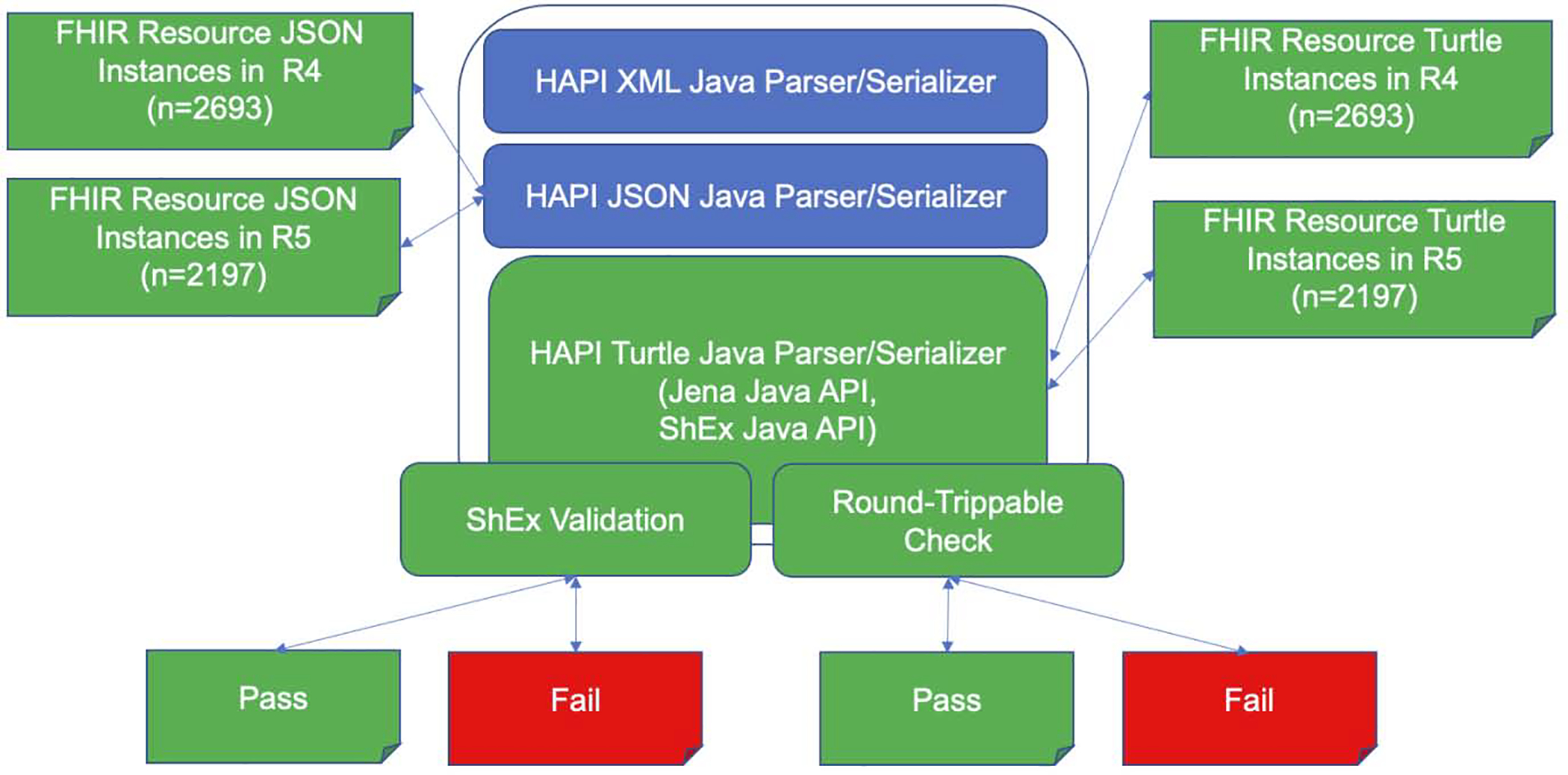
A diagram illustrating the two components of the evaluation design, ie, ShEx validation and Round-Trippable Check. Blue represents the state of the art in HAPI FHIR Library; Green and red represent the contribution of the study.
To verify conformance of FHIR RDF to the FHIR specification, we also performed ShEx validation of each example against the ShEx schemas provided in the FHIR specification.
The following testing steps were performed on each example:
Parse the JSON version of the example into a Java object (the “reference Java instance”).
Serialize that Java object with the new FHIR RDF (Turtle) serializer.
Parse the resulting Turtle file into an RDF graph using Jena Java API.
Validate that RDF graph against the FHIR ShEx schemas, using ShExJava.
Parse the same Turtle file into a new Java object, using the new FHIR RDF parser.
Use the èqualsDeep` comparator to verify that the new Java object is identical to the reference Java instance.
This process verifies the integrity of the serialized Turtle and verifies that the RDF serializer and parser round-trips back to an identical Java object.
Results
HAPI FHIR RDF Implementation Status
The FHIR RDF implementation in HAPI has been completed, and the source code has been merged into the HAPI FHIR master code repository. The FHIR RDF features have been incorporated into the latest release of HAPI FHIR 5.2.0 (Numbat) as of December 28, 2020.(26)
Evaluation Results for the FHIR R4 Dataset
For the R4 dataset, a total of 2693 JSON example files were converted into the RDF Turtle files, all of which (100%) passed the ShEx validation. Those 2693 RDF files were then converted back to Java objects, and 2675 (99.3%) of the RDF-sourced Java objects were the same as the original JSON-sourced data objects. Performing this same sequence with the JSON serializer and parser revealed 18 files which, when serialized back to JSON, had changed the order of some arrays of nested objects. Because RDF, JSON (and XML) use the same functions to order nested objects, we could not test RDF round-tripping on those 18 instances as they too would have the order scrambled.
Evaluation Results for the FHIR R5 Dataset
While passing 100% of the R4 tests demonstrates that the RDF library emits valid FHIR/RDF and parses it back to the same structures, the objective for the R5 tests is to show its ability to catch errors, in this case, those that arise from version skew between the developing FHIR example data and the episodic publication of the FHIR core structure library. For reference, about 90% of the examples (1998 out of 2197) we downloaded during development conformed to the last published FHIR core library by ShEx validation. The ordering problem described above caused another 337 to parse to a semantically equivalent Resource rather than exact copy, whereas 1860 (93.1%) of the passing instances arrived unscathed.
For errors detected in 199 RDF turtle files from the R5 dataset, an error analysis indicated that there were 5 types of errors including missing properties, unknown element, missing resourceType, invalid attribute value, and unknown resource name. Table 1 shows the error type and their cause analysis with examples.
Table 1.
Different types of errors identified by validating FHIR R5 examples (n=2197) against FHIR ShEx schemas. These may be hard to replicate as the JSON examples are subject to continuous updates and fixes.
| Error Type | Num. Of Instances with error | Cause |
|---|---|---|
| Missing an property linkId on nested Questionnaire items | 182 | The example violates the FHIR specification (detected by ShEx validation). Many of the JSON example questionnaires available in the downloadable ZIP include extensions with no linkId, which contradicts the Resource definition. |
| Unknown element | 12 | The example violates the FHIR specification (detected by ShEx validation). These have unknown elements and fail RDF validation. HAPI’s core parser, which is driven by the FHIR model, has a lenient mode which allows the parsers to ignore unknown elements with warnings like: medicinalproductdefinition-acetaminophen-500mg-tablets-box-of-20.json [LenientErrorHandler.java:150] - Unknown element ‘code’ [LenientErrorHandler.java:150] - Unknown element ‘reference’ |
| Missing resourceType | 2 | The example violates the FHIR specification (detected by ShEx validation). These included nested resources with no resourceType. |
| Invalid attribute value | 1 | The example violates the FHIR specification (detected by ShEx validation). The RDF (ShEx) schema includes all permissible values for enumerable value sets. This example had an invalid code in a value set. |
| Unknown resource name | 1 | The example violates the FHIR specification (detected by ShEx validation). This file (evidencereport-example.json) had an unrecognized resource type: “EvidenceReport”. |
Discussion
Since the FHIR STU3 release, RDF (in Turtle format) has officially been adopted by FHIR as one of its exchange formats, and ShEx schemas have been used to describe the FHIR resources and as a validation mechanism for FHIR RDF resource instances. However, tooling to support the transformation of FHIR data to and from the RDF representation has been very limited. Therefore, there has been great interest in the FHIR RDF community, to develop such RDF data transformation tools to facilitate FHIR data use in the context of semantic web applications.
Notably, there are two parallel efforts in this area. The first one is an effort to develop and evaluate a JSON-LD 1.1 approach to translate FHIR data between JSON and RDF formats. The focus of that effort was flexibility rather than production-ready tooling. Its goal was to support the process of developing the R5 version of FHIR RDF by making it easy to experiment with different JSON<->RDF mappings. That effort yielded a web-based FHIR JSON-LD Playground, implemented in JavaScript, and a stand-alone FHIR JSON-LD data transformation command line tool, implemented in Java. This paper describes the second of these parallel efforts. It focused on extending popular production-ready FHIR tooling -- the HAPI FHIR library -- to support FHIR RDF parsing and serialization. The robustness of HAPI’s RDF implementation was demonstrated by testing it on two suites of FHIR examples (R4 and R5), both by round tripping each example between JSON and RDF, and by performing ShEx validation. These tests also identified limitations of the implementation for future improvement.
For the FHIR R4 tests, all examples passed the ShEx validation. For the FHIR R5 tests, the ShEx validator correctly detected 5 types of modeling errors in the examples. Because R5 is still in development, and the FHIR publication process automatically generates core Java classes (org.hl7.fhir.r5.model.Base) and from a master definition of the FHIR resource models, it is to be expected to get validation errors representing temporary skew between the examples and the Java classes. These validation errors could be viewed as a (probably obsolete) “to do” list for the FHIR specification maintainers. With the FHIR R5 becoming more mature, the FHIR resource models and examples are likely to become more synchronized, resulting in fewer validation errors. However, the breadth of these tests demonstrates how the HAPI RDF support can be used to verify developer consensus about FHIR Resources and Profile definitions.
In addition to validation, the majority of the test examples passed the round tripping tests: out of 2693 R4 tests, 2675 passed and 18 failed; out of 2197 R5 tests, 1860 passed and 337 failed. These tests revealed limitations in the HAPI implementation with regard to preservation of order when round-tripping FHIR Resources. In the interest of providing a convenient API, HAPI’s data structures for contained Resources and entries in a Bundle are destroyed and reassembled during parsing. After parsing, the order of contained resources reflects the order of the attributes referencing them, which in turn is dictated by the property iterators provided by the FHIR Java model. These failed tests were skipped because the JSON representation did not round-trip due to re-ordering of repeated resources, demonstrating the value of the ShEx in the quality assurance of the evolving R5 development.
The FHIR specification outlines eight aspects of validation that could be performed against a FHIR resource instance: structure; cardinality; value domains (eg, enumerated codes); Coding/CodeableConcept bindings; invariants (eg, co-occurrence rules), profiles; questionnaires, and business rules.(27) Currently, ShEx can validate the first four of these aspects, but has the potential to validate some other aspects. FHIR Resource specifications include a large number of consistency constraints beyond the enumeration of properties and cardinalities visible in StructureDefinition. For instance, a FHIR Timing object has nine constraints at http://build.fhir.org/datatypes#Timing-inv. 7 of these are co-occurrence constraints, eg tim-1: if there’s a duration, there needs to be duration units, and 2 of these are range constraints, eg duration SHALL be a non-negative value, and one of the co-occurrence constraints includes extra value set constraints: If there’s an offset, there must be a when (and not C, CM, CD, CV). Not all of these constraints are currently expressed in the ShEx schema -- or any other schema language, eg XML Schema -- but are well within ShEx’s expressivity, although it would require a translation effort to quantify how many this may be. In addition, although the ShEx supports the Coding/CodeableConcept binding validation, it needs a specific implementation of the FHIR terminology services.(28) Therefore, we consider this implementation is beyond the scope of this study and treat it as future work.
As demonstrated above, ShEx schemas and either RDF examples or other examples translated to RDF with the HAPI FHIR library offer communities a streamlined way to capture consensus in a machine-readable way and test that consensus with a corpus of examples expected to be accepted or rejected by a validation process. Some additional tooling and release engineering would make this process available to non-RDF experts, including 1) separating the FHIR ShEx generation from the FHIR publication process so it can be used in more light-weight development processes; 2) developing a methodology to express FHIR Profiles using ShEx language features for extending one shape with another; and 3) creating a tool to translate FHIR-specific ShEx schemas back to the FHIR StructureDefintion resource.
Conclusion
We demonstrated that the FHIR RDF data transformation and validation framework developed for HAPI is robust for community use. The framework can be used to facilitate FHIR RDF data use for clinical and research applications, and for quality improvement of the FHIR specification.
Highlights.
RDF is one of the three standard data formats in the HL7 FHIR
A Java based FHIR RDF data transformation and validation toolkit is developed
The FHIR RDF implementation in HAPI FHIR is completed
Conformance of FHIR RDF data to its ShEx schema is evaluated for FHIR R4 and R5
The round-trippable check highlights an ordering issue in HAPI FHIR
Acknowledgments
This work was supported in part by funding from the NIH FHIRCat (R56EB028101) and the NCATS CD2H (U24 TR02306).
Footnotes
Declaration of Interest Statement
Authors declare that there is no interest conflict.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Eric Prud’hommeaux, Janeiro Digital, Boston, MA; W3C/MIT, Cambridge, MA.
Josh Collins, Janeiro Digital, Boston, MA.
David Booth, Yosemite Project, Somerville, MA.
Kevin J. Peterson, Mayo Clinic, Rochester, MN
Harold R. Solbrig, Johns Hopkins University, Baltimore, MD
Guoqian Jiang, Mayo Clinic, Rochester, MN.
References
- 1.HL7 FHIR 2020 [December 28, 2020]. Available from: https://www.hl7.org/fhir.
- 2.HL7 Argonaut Project 2020 [December 28, 2020]. Available from: https://argonautwiki.hl7.org/Main_Page.
- 3.Berners-Lee T, Hendler J, Lassila O. The Semantic Web - A new form of Web content that is meaningful to computers will unleash a revolution of new possibilities. Sci Am. 2001;284(5):34–+. doi: DOI 10.1038/scientificamerican0501-34.11396337 [DOI] [Google Scholar]
- 4.Lawrynowicz A Creative AI: A new avenue for the Semantic Web? Semant Web. 2020;11(1):69–78. doi: 10.3233/Sw-190377. [DOI] [Google Scholar]
- 5.The Yosemite Project 2020 [December 28, 2020]. Available from: https://yosemiteproject.org/. [Google Scholar]
- 6.Solbrig HR, Jiao Z, Prud’hommeaux E, Booth D, Endle CM, Stone DJ, Jiang G, editors. Exploring JSON-LD as an Executable Definition of FHIR RDF to Enable Semantics of FHIR Data. AMIA 2020 Annual Symposium (in press); 2020. [PMC free article] [PubMed] [Google Scholar]
- 7.Phenopackets 2021 [March 4, 2021]. Available from: https://phenopacketsschema.readthedocs.io/en/latest/
- 8.Solid POD Platform 2021 [March 4, 2021]. Available from: https://bidstats.uk/tenders/2019/W43/713556009.
- 9.Social Linked Data 2021 [March 4, 2021]. Available from: https://en.wikipedia.org/wiki/Solid_(web_decentralization_project).
- 10.Greater Manchester Digital Platform 2021 [March 4, 2021]. Available from: https://www.gmdigitalplatform.nhs.uk/projects/personal-online-data-stores-pods/
- 11.Iacobucci G Manchester expands seven day general practice to three million people. BMJ. 2015;350:h3192. Epub 2015/06/13. doi: 10.1136/bmj.h3192. [DOI] [PubMed] [Google Scholar]
- 12.Zong N, Ngo V, Stone DJ, Wen A, Zhao Y, Yu Y, Liu S, Huang M, Wang C, Jiang G. Leveraging Genetic Reports and Electronic Health Records for Predicting Primary Cancers Based on FHIR and RDF. JMIR Medical Informatics (in press). 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Prediction on FHIR 2021 [March 4, 2021]. Available from: https://github.com/fhircat/cancer-prediction-on-fhir-rdf
- 14.JSON-LD 1.1 Specification 2020 [December 28, 2020]. Available from: https://w3c.github.io/json-ld-syntax/.
- 15.The HAPI FHIR Library 2020 [December 28, 2020]. Available from: https://hapifhir.io/hapi-fhir/.
- 16.The HAPI FHIR GitHub Site 2020 [December 28, 2020]. Available from: https://github.com/hapifhir/hapi-fhir.
- 17.Apache Jena 2020 [December 28, 2020]. Available from: https://jena.apache.org/.
- 18.The RDF Shape Expressions (ShEx) Language 2020 [December 28, 2020]. Available from: http://shex.io/.
- 19.ShEx JavaScript Implementation 2020 [December 28, 2020]. Available from: https://github.com/shexSpec/shex.js.
- 20.ShEx Java Implementation 2020 [December 28, 2020]. Available from: http://shexjava.lille.inria.fr/.
- 21.ShEx Scala Implementation 2020 [December 28, 2020]. Available from: https://github.com/weso/shex-s.
- 22.ShEx Python Implementation 2020 [December 28, 2020]. Available from: https://github.com/hsolbrig/PyShEx.
- 23.ShEx Ruby Implementation 2020 [December 28, 2020]. Available from: https://github.com/ruby-rdf/shex.
- 24.Solbrig HR, Prud’hommeaux E, Grieve G, McKenzie L, Mandel JC, Sharma DK, Jiang G. Modeling and validating HL7 FHIR profiles using semantic web Shape Expressions (ShEx). J Biomed Inform. 2017;67:90–100. Epub 2017/02/19. doi: 10.1016/j.jbi.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Commons RDF API 2020 [December 28, 2020]. Available from: https://commons.apache.org/proper/commons-rdf/.
- 26.HAPI FHIR 5.2.0 (Numbat) 2020 [December 28, 2020]. Available from: https://github.com/hapifhir/hapi-fhir/releases/tag/v5.2.0.
- 27.FHIR Validation 2020 [December 28, 2020]. Available from: https://www.hl7.org/fhir/validation.html.
- 28.FHIR Terminology Services 2020 [December 28, 2020]. Available from: https://www.hl7.org/fhir/terminology-service.html.


