Abstract
Microorganisms including bacteria, fungi, viruses, protists and archaea live as communities in complex and contiguous environments. They engage in numerous inter- and intra- kingdom interactions which can be inferred from microbiome profiling data. In particular, network-based approaches have proven helpful in deciphering complex microbial interaction patterns. Here we give an overview of state-of-the-art methods to infer intra-kingdom interactions ranging from simple correlation- to complex conditional dependence-based methods. We highlight common biases encountered in microbial profiles and discuss mitigation strategies employed by different tools and their trade-off with increased computational complexity. Finally, we discuss current limitations that motivate further method development to infer inter-kingdom interactions and to robustly and comprehensively characterize microbial environments in the future.
Keywords: Microbial co-occurrence networks, Microbial interactions, Network analysis, Trans-kingdom interactions
1. Introduction
The human body acts as a host for complex microbial communities consisting of bacteria, protozoa, archaea, viruses and fungi [1]. Next-generation sequencing techniques proved very effective for characterizing microbial communities by sequencing suitable molecular targets such as 16S ribosomal RNA gene amplicons for bacteria, internal transcribed spacer regions of ribosomal RNA genes for fungi and shotgun metagenomics for viruses (Fig. 1A). Since these organisms share the same host, they are in constant competition, where some organisms develop symbiotic relationships in which they cooperate or synergize with each other for gaining a fitness advantage that may or may not benefit the host organism [2], [3]. Thus far, microbiome research has mostly focused on interactions between the host and its microbiome, mostly on the level of bacteria. However, trans-kingdom interactions between bacteria, fungi and viruses, as well as their joint effect on the host, have only recently been studied [4].
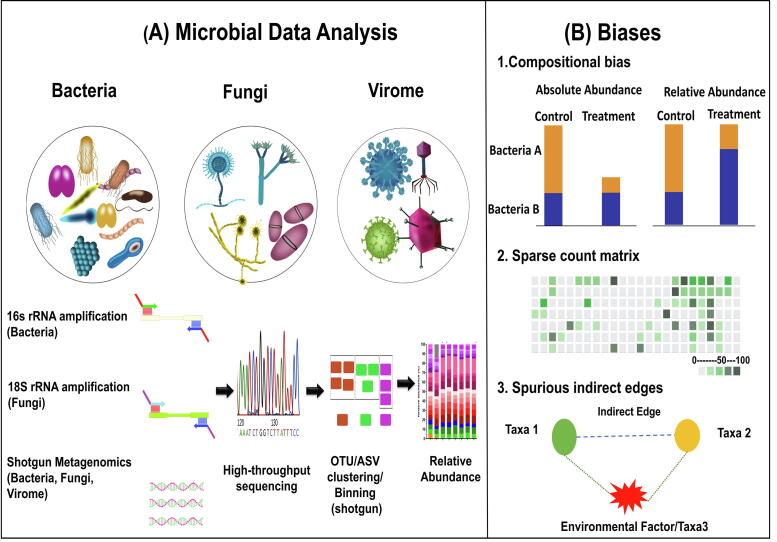
Fig. 1.
(A) Schematic overview on taxonomic profiling of bacteria, fungi and the virome. (B) Illustrates three important biases: compositionality, sparsity and spurious correlations in microbial co-occurrence network analysis.
Network-based analytical approaches have proven useful to study systems with complex interactions and represent a powerful tool in systems biology to infer gene-regulatory and other complex networks [5], [6], [7]. The complex interactions between thousands of individual species across kingdoms as found, for instance, in the human gut microbiome, suggests that such network analysis methods are also useful in the microbiome field. In this review, we first highlight network analysis methods that have already been used successfully for inferring community structures from bacterial abundances. Next, we focus on recently developed and repurposed methods that have been used for trans-kingdom analysis. Finally, we will discuss why more concerted efforts in network method development are necessary to address the unique aspects of microbial data.
2. Network methods for microbial communities
Until now, bacterial co-occurrence patterns were studied extensively while fungal or viral interactions have received less attention [8], [9]. Systems and network biology approaches have been used to decipher microbial co-occurrence patterns and range from correlation methods to complex graph-based models. A recent study investigating the earth microbial co-occurrence network identified connections across fourteen different environments, including plants, animals, water and soil [10]. The earth microbial co-occurrence network thus highlights the importance of studying microbial interactions across microbial niches using suitable tools.
Decoding complex microbial co-occurrence relationships is associated with three main challenges. Firstly, microbiome data are compositional [11]; i.e. microbial counts represent proportions instead of absolute abundances. Secondly, sparsity in the dataset can lead to false associations of microorganisms. A zero indicates either the absence of a microorganism, or an insufficient sequencing depth. Thirdly, it is challenging to differentiate between direct and indirect associations, in particular if these are related to environmental factors (Fig. 1B).
Correlation-based techniques, including Pearson or Spearman correlation, are among the most popular methods for studying microbial interactions in human gut [12], oral [13] and soil [14] microbiomes. Weiss et al. [15] evaluated the strengths and weaknesses of eight different correlation methods and provided recommendations based on the nature of the data and identified sparsity as a key issue not sufficiently addressed by these approaches. Correlation analysis often results in artefactual and spurious associations between low-abundant microbial members in a community as it fails to account for compositionality [11]. As Lovell et al. [16], [17] showed, correlation-based methods are not subcompositionally coherent such that, for instance, depleting rare taxa is expected to change the outcome of correlation analysis. To overcome this issue, compositional data analysis can be employed. Various proportionality measures [16], [17] have been proposed some of which are implemented in the R package propr [18] and can be used for network construction. A frequently used method to account for compositionality is centered log ratio transformation (CLR) [19], [20], where the geometric mean of the sample vector is used as the reference. CLR transformation maps the relative counts from simplex into Euclidean space and hence makes these data compatible with linear analysis methods. Apart from these classical approaches, more complex methods have been proposed based on probabilistic graphical, Gaussian graphical and complex multiple regression models to construct microbial interaction networks [6], [21]. Most methods take compositionality into account either by performing CLR transformation as a pre-processing step or by using a Dirichlet multinomial model to directly account for compositionality. Existing methods differ with respect to sensitivity, specificity and computational complexity and can be grouped into four different categories (Fig. 2). In the following, we describe the underlying concepts of tools (Table 1 and Supplemental Material) that have been successfully applied in the analysis of microbial data in humans [22] as well as other environments [23].
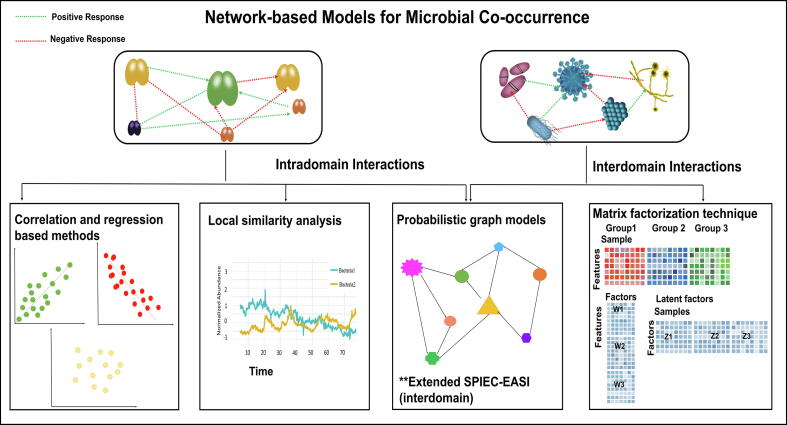
Fig. 2.
Overview of network approaches for microbial intra- and inter-kingdom interactions.
Table 1.
Overview of microbial co-occurrence network methods.
| Tools | Principle/Models | Advantages | Limitation | Applications |
|---|---|---|---|---|
| Correlation based Methods | ||||
| SparCC (2012) python r-sparcc |
|
|
|
|
| CCLasso (2015) R package |
|
|
|
|
| REBACCA (2015) |
|
|
|
|
| CoNet (2016) Cytoscape Command line tool |
|
|
|
|
| Meta-Network (2019) |
|
|
|
|
| Correlation-Centric Network (2020) Command line tool |
|
|
|
|
| MENAP (2012) online tool |
|
|
|
|
| Conditional Dependence/graphical Models | ||||
| gCoda (2017) R package |
|
|
|
|
| MDiNE (2019) R package |
|
|
|
|
| MixMPLN (2019) R package |
|
|
|
|
| NetComi (2020) R package |
|
|
|
|
| Environmentally-Driven Edge Detection (2020) |
|
|
|
|
| Mint (2015) R package |
|
|
|
|
| mLDM (2016) R package |
|
|
|
|
| HARMONIES (2020) R package webtool |
|
|
|
|
| SPIEC-EASI (2015) R package |
|
|
|
|
| Hubs weighted graphical lasso (2020) |
|
|
|
|
| FlashWeave (2019) |
|
|
|
|
| COZINE R package (2020) |
|
|
|
|
| Network-based methods for trans-kingdom analysis | ||||
| SPIEC-EASI Extension (2018) R package |
|
|
|
|
| Multi-Omics Factor Analysis R package (2018) |
|
|
|
|
| DIABLO R package (2019) |
|
|
|
|
3. Correlation based methods
Many correlation-based methods employ variants of Pearson or Spearman correlation to obtain an estimate of microbial interaction between pairs of taxa [24], [25]. However, these measures do not account for compositionality, where, for instance, an increase in absolute abundance of just a single taxon is followed by a decrease in relative abundances of all other taxa even if their absolute abundance does not change (Fig. 1B) [11]. This can be mitigated by ratio transformation of the data. Ratio transformations ensure that the ratios between two features are the same whether the data are absolute counts or proportions. Taking the logarithm of these counts makes the data further symmetric and linearly related [19]. The resulting correlation coefficients are thus compositionally coherent, i.e. the log ratio of two taxa is completely independent of other taxa. Sparse Correlations for Compositional data (SparCC) [26] is a popular method employing this strategy with applications ranging from human gut microbiome studies [27], [28], [29] to environmental studies. SparCC is based on an iterative approximation approach and uses log-ratio transformed data to infer the correlations between the components. Under the assumption that the underlying networks are large-scale and sparse. SparCC was shown to be better suited to avoid spurious correlations compared to direct Pearson correlations [15] at the cost of higher computational complexity [30]. Another strategy that was proposed to improve the robustness of correlation coefficients is bootstrapping as implemented in CoNet [24]. CoNet further employs similarity (Steinhaus, distance correlation) and dissimilarity measures (Euclidean, Jensen-Shannon, Kullback Leibler, Bray Curtis) as alternatives to correlation coefficients. Another challenge in correlation-based networks is the choice of a suitable correlation cut-off which controls the sparsity of the resulting network. While the choice for the cut-off is often left to the user, the Molecular Ecological Network Analysis Pipeline (MENAP) [25] offers an automated selection of the optimal correlation threshold via a random matrix theory-based method [31] to simulate a random background.
4. Regularized linear regression
An alternative to correlation methods is to build linear regression models in which the abundance of each taxon is modelled as a response variable using the abundance of all other taxa as explanatory variables. Here, the coefficient of each taxon serves as a linear measure for the interaction strength of two taxa. However, due to the large number of features, such models are generally prone to overfitting. A common strategy to mitigate this issue is to introduce a penalty term, yielding regularized regression models. Here, the ℓ1-penalty, also known as lasso, is typically used to drive the coefficients of taxa with negligible contribution to zero, thus increasing the sparsity of the solution. For instance, Correlation inference for compositional data through Lasso (CCLasso) [32] and Regularized Estimation of the BAsis Covariance based on Compositional dAta (REBACCA) [33] use this strategy to build a regularized correlation network of microbiome data. CCLasso also adapts CLR transformation to address compositionality, while REBACCA models the log basis covariance structure to directly account for compositionality. While CCLasso and REBACCA perform similar to SparCC in terms of reproducibility and consistency, regularization appears to be beneficial for avoiding the detection of spurious relations [32]. In addition to the existing lasso methods, Bates and Tibshirani [34] proposed a new ℓ1-penalized regression model based on all-pairs log-ratios for sparse estimation. The all-pairs log-ratio model overcomes compositionality, increases accuracy and leads to improved interpretability. Further, Lu et al. [35] introduced ℓ1-penalized generalized linear regression models (GLMs) with linear constraints that achieve sub-compositional coherence.
5. Association rule mining
Instead of regularization, Meta-Network [36] uses advanced association rule mining [37] to detect intricate (i.e. including indirect and non-linear) correlations. To this end, Meta-Network first generates presence-absence indicator matrices for each sample. Subsequently, the co-occurrence frequencies of taxa pairs are computed yielding a co-occurrence probability matrix. This matrix is then used to construct a network with a co-occurrence probability of e.g. 80%. (default threshold in Meta-Network). Following this loose definition, Meta-Network uses the graph-based Functional Similarity Weight (FS-Weight) [38] algorithm to detect indirect relationships and the PCA-PMI [39] method (Path Consistency Algorithm) to infer non-linear associations. These two methods (FS-Weight and PCA-PMI) are able to independently capture many of the same nodes and edges which, according to the authors, indicates that they both can depict the complex nature of the microbial relationships.
6. Conditional dependence and graphical methods
Correlation based methods typically fail to differentiate between direct and indirect associations. To account for this, a plethora of methods have been developed to model conditional dependence which usually have a higher computational complexity and run-time than correlation-based methods. Partial correlation [40] and related approaches are used here to distinguish between direct and indirect interactions, resulting in an undirected weighted graph where the edges imply the conditional dependency between two taxa.
Most of these methods can also account for confounders such as biological covariates and technical biases such as sequencing depth. For instance, Mint (MicrobialInteraction) employs a Poisson-multivariate normal hierarchical model to identify direct microbial interactions while controlling for user-provided confounders at the multivariate normal layer using an ℓ1-penalized precision matrix [41].
Two different strategies are implemented in SPIEC-EASI (SParse InversE Covariance Estimation for Ecological Association Inference) [42] after applying a CLR transformation to the data to address compositionality. The first method generates a graphical network by estimating a sparse inverse covariance matrix (sparse graphical model inference with Glasso) [43] and the second method employs the Meinshausen-Bühlman method, a node wise regression model [44], [45]. SPIEC-EASI infers the appropriate amount of sparsity of a network by using the Stability Approach to Regularization Selection [46].
A number of methods were inspired by SPIEC-EASI and mostly differ in the models they employ to infer conditional independence. For instance, gCoda [47] also performs CLR transformation on the relative abundance but then uses a logistic normal distribution to model the counts and a maximum likelihood model with ℓ1-penalty to deal with sparsity. According to the authors, gCoda surpasses SPIEC-EASI in terms of stability, accuracy and runtime.
Another method, metagenomic Lognormal-Dirichlet-Multinomial (mLDM) [48] affords a more complex hierarchical Bayesian model with three layers. First, mLDM models the count matrix by using a multinomial distribution. Second, a Dirichlet distribution is used to model the multinomial probabilities and, finally, mLDM utilizes a multivariate log-normal distribution to model the absolute microbial abundance [48]. The authors could show that mLDM performed favourably compared to Pearson and Spearman correlation, SparCC, CCLasso, CCREPE, glasso and SPIEC-EASI in terms of finding true taxa-taxa and environmental factors and taxa associations. However, this multi-layer approach leads to high computational complexity and limits both scalability and interpretability [47].
Hybrid Approach foR MicrobiOme Network Inferences via Exploiting Sparsity (HARMONIES) [49] employs the zero-inflated negative binomial distribution (ZINB) and a Dirichlet prior to deal with overdispersion and the large number of zero counts. HARMONIES then uses a graphical lasso approach to infer interactions with favourable results compared to SPIEC-EASI (using both Glasso and the Meinhausen-Bühlmann method), and CClasso on synthetic data, in particular when additional zeros were added.
Most of the methods which try to solve zero-inflation, introduce pseudo counts before log transformation. Ha et al. [50] discussed that introducing pseudo counts may have a huge impact on downstream analysis and also may lead to spurious associations i.e. neglecting the fact that some taxa are completely absent in the data. To overcome this, Ha et al. [50] proposed a new COmpositional Zero-Inflated Network Estimation (COZINE) model where they generate a binary incidence matrix and a compositional abundance matrix in which CLR is applied only to the non-zero count data. A Multivariate Gaussian Hurdle model [51] with group-lasso penalty is then fitted into the combined form of binary and continuous matrix to infer the three types of interactions: binary-binary, binary-continuous and continuous-continuous relations. By doing so, COZINE tries to accommodate both compositionality and zero inflation.
7. Addressing network topology bias
Topological features of the network such as the node degree may introduce a bias in statistical inference, where highly connected taxa (hubs) have disproportionate influence. The vast majority of methods do not consider topological network features since they implicitly assume conditional independence [52]. Recently, McGillivray [53] proposed a weighted graphical lasso approach that incorporates row/column sums as weights to penalize hub edges. The method performed significantly better compared to competing methods including graphical lasso, adaptive graphical lasso or hubs graphical lasso.
8. Methods scaling to large-scale data
A general issue with probabilistic graphical model approaches is their lack of computational scalability. FlashWeave [54] mitigates this issue by using a modified version of the semi-interleaved HITON-PC algorithm [55], a causal inference algorithm which infers for each taxon its Markov blanket. Given its Markov blanket, a taxon is conditionally independent of every other taxon in the graph. The algorithm starts by labelling for each taxon T significantly associated taxa, based either on Pearson correlation or mutual information, as candidate neighbours. Then only taxa which are conditional dependent from T given all combinations of other neighbour taxa are kept. Individual neighbourhoods are subsequently connected. To achieve scalability FlashWeave uses a set of heuristics and optionally incorporates metadata to disentangle direct microbial associations from confounding factors introduced in cross study analyses.
9. Multi-view networks
Most of the network methods assume that the sample-taxa matrix is associated with a single network, i.e. there is only one network topology with a set of edge weights. However, a sample-taxa matrix may be derived from a larger number of biological samples where taxa may be associated with more than one network topology. Especially the human gut microbiome is associated with various factors including diet, age and health and, hence, the associated microbial network may vary according to the influence of these factors. Tavakoli et al. [56] used a mixture model based on the Multivariate Poisson Log-Normal (MPLN) distribution [57] to build K microbial networks from a sample-taxa matrix associated with K underlying distributions. Similar to Mint [41], MixMPLN uses a Poisson-multivariate normal hierarchical model to capture the direct microbial interactions. However, different to Mint, MixMPLN constructs one network for each confounder and infers the parameters of the distributions using a maximum likelihood framework based on the Minorization–Maximization (MM) algorithm [58]. In addition, MixMPLN also uses the ℓ1-penalty to regularize the sparsity of the networks. The authors extended this idea also to other algorithms such as MixGGM and MixMCMC [59]. MixMCMC utilizes Markov Chain Monte Carlo model to evaluate the latent parameters in the MPLN mixture framework. MixGGM utilizes Gaussian distributions on CLR transformed- abundance matrix to overcome compositionality bias.
10. Differential network analysis
While most methods construct a single co-occurrence network irrespective of study conditions such as disease, treatment or control, it is in many cases the differences between such conditions that are of greatest interest. To account for this, a few methods have been developed for differential network analysis.
Microbiome Differential Network Estimation (MDINE) [60] generates differential networks to show how microbial relationships vary between two conditions based on an estimation of the precision matrix. MDiNE addresses compositionality by utilizing a Dirichlet-multinomial logistic-normal distribution model [61], [62]. Apart from handling compositionality, multinomial logistic models are also suited to handle the large number of zeros in microbial abundances without reverting to pseudo counts. In contrast to MDiNE, NetCoMi [63] utilizes permutation tests to evaluate the significantly different taxa between the groups. More specifically, NetCoMi performs differential association analysis using Fisher’s z-test [64], a non-parametric resampling procedure [65] and the discordant method [66] to build differential networks that are limited to differentially associated taxa.
11. Inferring interaction types
In ecological networks, microbial interactions are shown as directed edges from a source to a target species, where different types of interactions can be modelled, e.g. competition, mutualisms or parasitism [67]. In contrast, microbial association networks are typically undirected and not all interactions represent true ecological relationships. EnDED [68] aims to differentiate direct and indirect associations based on environmental factors which may affect the dynamics of the ecosystem, such as temperature, turbidity, salinity and nutrients. It employs four different approaches, such as Sign Pattern [69], Overlap [68], Interaction Information [69], [70], and Data Processing Inequality [71], [72] to identify indirect (environmentally-driven) edges. It classifies an edge as indirect due to environment factor, only if all four methods classify it as indirect.
Alternatively, Lotka–Volterra models are commonly used to predict different types of interactions. While classical Lotka-Volterra models are used to predict predator–prey (competition) interaction between two species, the generalized Lotka–Volterra (gLV) [73], [74] uses a logistic model to simulate the growth of microbes and to infer whether an interaction of two species is competitive, amensalistic or predator–prey [75]. However, since gLV-based models estimate dynamics with respect to absolute abundance, a new nonlinear dynamical system called compositionally aware Lotka-Volterra method (cLV) [76] was developed. cLV predicts microbial dynamics in-terms of ratio of relative abundance between taxa. Joseph TA et al. [76] compared the performance of cLV against gLV using simulated and real datasets and showed that cLV forecasts microbial interactions more accurately compared to gLV.
12. Studying microbiome time-series dynamics
Microbiomes tend to change their compositions in response to perturbations of their environment. Time-series analysis aims to study dynamic interaction changes in microbial compositions to reveal contemporaneous patterns and factors which are responsible for changes in the community behaviour. Faust et al. [77] discussed different network inference techniques to investigate temporal changes in microbiome studies including local similarity analysis (LSA) [78], Time-decay analysis [79], Augmented Dickey Fuller test [80], Cross correlation [80], Time-varying network inference [81], Hurst exponent [82], Bistability analysis [83], as well as Extended LSA (eLSA) [84], which offers support for replicates. Among these techniques, LSA is the most commonly used method to study dynamic changes. It utilizes dynamic programming to detect changes between the time series and to identify associations based on a similarity score. Alternatively, Dynamic Bayesian networks and temporal event networks can be used to study the temporal changes in microbial data. Dynamic Bayesian networks have been successfully used to study the changes in microbial compositions of the infant gut microbiome [85], other longitudinal microbiome data including vaginal and oral cavity microbiome [86].
The majority of the microbial network tools emphasize nodes (representing taxa at different taxonomic levels) but only limited attention is given to the edges capturing their associations [87], [88]. Although these may delineate important dynamic changes of microbial co-occurrence. Correlation-Centric Network (CCN) [89] transforms the node into an edge graph, where nodes represent the co-occurrence of two taxa while edges represent one of the two co-occurring taxa, respectively. This correlation-centric network representation is hence suited to capture dynamic changes in the microbial environment [89].
13. Network-based methods for trans-kingdom analysis
During the last years, the continuously dropping costs for high throughput sequencing technologies allowed scientists to go beyond the mere characterization of the bacterial part of the microbiome and to investigate the role of viruses and fungi within the microbial community. Methods using the information of several data modalities concurrently are thus sought to deliver insights into the relation between taxa from different kingdoms and to gain a more comprehensive understanding of the microbial system. Since such multi-modal data is still relatively scarce in the microbiome community, only few methods have been developed and applied for this purpose. For instance, Tripton et al. [90] adapted the SPIEC-EASI method for trans-kingdom analysis by concatenating two or more data sets which were independently CLR-transformed. The combined data is then used to estimate a sparse inverse covariance matrix which can be interpreted as an intra- and cross-domain interaction network. Applying their method on data from lung and skin bacteria as well as from the fungal microbiome, the authors showed that cross-kingdom networks had a higher overall connectivity and that the modularity was reduced compared to the single-domain networks.
However, the SPIEC-EASI extension does not offer insights into underlying factors driving the variation across samples or different groups of samples. To achieve this, Argelaguet et al. [91] proposed a method called Multi-Omics Factor Analysis (MOFA) which uses group factor analysis [92] to provide an integrative analysis of a set of samples with measurements from different data modalities, making it an attractive tool for trans-kingdom analysis as demonstrated by Haak et al. [93].
Similarly, Data Integration Analysis for Biomarker discovery using Latent cOmponents (DIABLO) [94] is a multi-omics integration tool based on partial least squares (PLS) regression, a technique to reduce the number of predictors by finding a small set of uncorrelated variables which are then used to perform least squares regression and was successfully applied to multi omics data set consisting of microbiome, metabolome, proteome und mRNA measurements, where it revealed discriminatory biomarkers for fibromyalgia patients [95]. However, in contrast to MOFA, DIABLO supports only continuous variables and assumes a linear relationship between the selected variables which may not be given, in particular in such complex scenarios.
14. Discussion
Microorganisms have built complex and robust ecosystems in various environments ranging from soil or sea water to various organs of the human body. Understanding the nature of microbial co-occurrence and correlation patterns within and between kingdoms may thus provide insights into the robustness of ecological systems and offer insights into complex human diseases such as inflammatory bowel disease, which is known to be influenced by the microbiome. However, to study microbial interactions, we need suitable computational tools that can robustly infer the microbial interaction network and subsequently disentangle it to interpret the contribution of microorganisms and their interactions with respect to their environment. This is of particular importance in medical research, where microbial interactions may be associated with the onset of certain diseases. Microbial communities, or key members thereof, are thus attractive drug targets in precision medicine [96]. Network-based approaches are powerful concepts to model and study complex relationships which can be employed in this context. Currently, the majority of network-based tools and models are used to study intra-kingdom interactions, mostly between bacteria. Most of these methods employ linear models based on correlation, regression, and probabilistic graphical models. Only few tools consider confounding factors in spite of their importance in microbiome studies. A frequently addressed bias is compositionality which results in artefactual correlations and is typically countered by pre-processing relative abundances with CLR transformation or by using a Dirichlet multinomial model. Few methods are able to distinguish direct and indirect effects using more complex conditional models and regularization. Nevertheless, simple linear models appear to be used more frequently in the literature, likely because more complex models that account for biases are plagued by a steep increase in computational complexity which leads to intolerable runtime. Moreover, network inference is hindered by a large fraction of zeros in the abundance matrices as well as by an unfavourable ratio of samples to features (curse of dimensionality). To counter these issues, relative abundances are often grouped on a higher taxonomic level, precluding insights into smaller communities. This may be mitigated by recently proposed tools such as FlashWeave, which offer bias-aware inference of microbial networks for hundreds of thousands of samples.
Species- or strain-level network analysis is not only prohibited by the additional computational complexity but also limited by the sequencing method employed. 16S rRNA amplicon sequencing method provides reliable taxonomic resolution up to genus level. In contrast, shotgun metagenomics enables species-level, and potentially strain-level resolution. Species-level network analysis may be subject to additional challenges that are currently not addressed. For instance, strain/species-level associations can be dominated by a single species as one species may comprise more than 100 different strains, yet this may not imply that all members of a particular species should be associated. This is the common phenomena observed in microbial network studies [42], [97] and is known as assortativity [98]. In other words, taxa are more likely to interact with other phylogenetically related taxa. Ha et al. [50] proposed using a standardized assortative coefficient [99] to quantify the extent of assortativity in the constructed networks of various methods but only investigated this issue for genus level and higher.
None of the existing tools successfully addresses all issues of microbial network inference. For instance, very few existing network approaches cannot reliably separate the actual ecological relationships from other pseudo (artifact) relationships. Furthermore, they typically fail to detect the nature of microbial relationships, i.e. they are unable to distinguish between competition and cooperation [21].
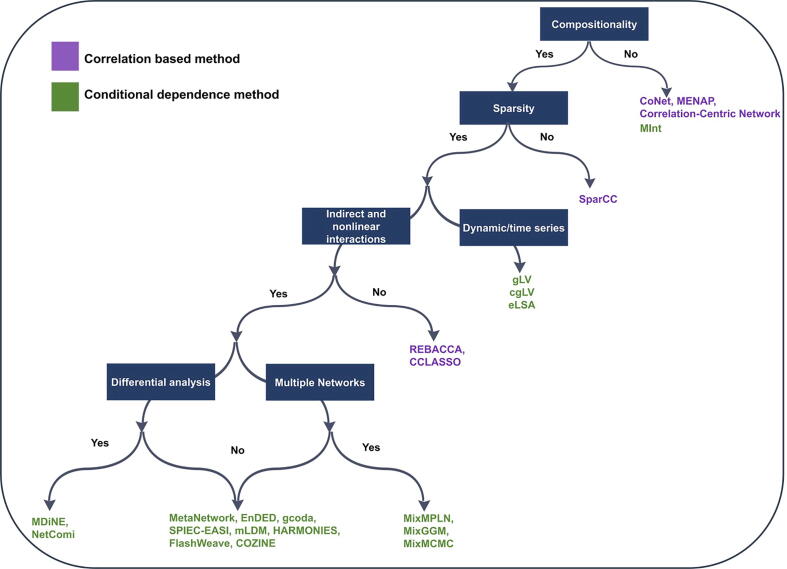
A plethora of methods have already been developed for studying microbial interactions from a network-level perspective. Considering that many of the available tools employ similar strategies, the choice of method is a daunting task for microbiome researchers. Comprehensive guidelines are currently missing due to the lack of a suitable and comprehensive benchmark datasets or commonly accepted simulated datasets which could be used as a gold standard to systematically evaluate the performance of existing network models. Thus, users can currently choose an optimal method for their analysis only based on the trade-off between complex models that address existing biases and reveal the differences of direct and indirect relationships and faster methods which can reliably infer networks even when hundreds of taxa and thousands of samples are given. We have summarized the trade-offs in Table 1 and Fig. 3 illustrates the best options to use depending on various challenges.
Fig. 3.
Workflow indicating the suitable network approaches depending upon different challenges.
Microbial interactions go far beyond within-kingdom interactions of bacteria alone. With the availability of trans-kingdom and multi-modal data sets including the transcriptome, metabolome and proteome, integrative network approaches are urgently needed to study trans-kingdom and functional interactions in the microbiome. However, very few methods have been adapted for trans-kingdom analysis, motivating the use of general methods developed for multi-modal integrative analysis such as MOFA+ [100]. Moreover, several data integration techniques have been used to integrate nodes from different networks, including a bipartite network [101] approach, which was used to build the community fungal-bacterial networks on the root microbiome [102] and a deep learning model allowing the integration of microbiome and metabolome [103]. Many of the existing tools currently used for inferring bacterial interaction networks have yet to be adapted for trans-kingdom interactions, including Gaussian graphical models, graphical lasso and mixed graphical models [104].
15. Conclusion
Network analysis provides valuable insights into microbial interaction networks. However, the currently available methods are not able to overcome all of the challenges associated with microbiome data including compositionality bias, overdispersion, a poor sample to feature ratio and trans-kingdom interactions. Analysis methods should be carefully selected based on the computational complexity that can be afforded with the data set and also with respect to the biological question that defines if it is acceptable to study the microbiome on a higher taxonomic level. In addition, further studies have to be carried out to validate these methods using universal benchmark datasets. While network analysis methods often suggest plausible hypotheses and interpretations of the data, they cannot infer causality. Integrative methods utilizing shotgun sequencing, metatranscriptomics and meta-metabolomics data are thus needed. Finally, efforts in computational method development need to be matched with experimental studies of microbial communities in, e.g., gnotobiology to be able to validate findings and to ultimately unravel the complexity of microbial interactions.
CRediT authorship contribution statement
Monica Steffi Matchado: Writing - original draft, Visualization, Resources. Michael Lauber: Writing - original draft, Resources. Sandra Reitmeier: Writing - review & editing. Tim Kacprowski: Writing - review & editing. Jan Baumbach: Writing - review & editing. Dirk Haller: Writing - review & editing. Markus List: Conceptualization, Supervision, Project administration, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German 582 Research Foundation) – Projektnummer 395357507 – SFB 1371. Michael Lauber was supported by the Hanns-Seidel-Stiftung.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.05.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Huseyin C.E., O’Toole P.W., Cotter P.D., Scanlan P.D. Forgotten fungi-the gut mycobiome in human health and disease. FEMS Microbiol Rev. Jul. 2017;41(4):479–511. doi: 10.1093/femsre/fuw047. [DOI] [PubMed] [Google Scholar]
- 2.Salazar N, de los Reyes-Gavilán CG. Editorial: insights into microbe–microbe interactions in human microbial ecosystems: strategies to be competitive, Front Microbiol 2016;7. doi: 10.3389/fmicb.2016.01508. [DOI] [PMC free article] [PubMed]
- 3.Kamada N., Seo S.-U., Chen G.Y., Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. May 2013;13(5):321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 4.Rowan-Nash AD, Korry BJ, Mylonakis E, Belenky P. Cross-domain and viral interactions in the microbiome. Microbiol Mol Biol Rev Feb. 2019;83(1). doi: 10.1128/MMBR.00044-18. [DOI] [PMC free article] [PubMed]
- 5.Proulx S.R., Promislow D.E.L., Phillips P.C. Network thinking in ecology and evolution. Trends Ecol Evol. Jun. 2005;20(6):345–353. doi: 10.1016/j.tree.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Layeghifard M., Hwang D.M., Guttman D.S. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol. Mar. 2017;25(3):217–228. doi: 10.1016/j.tim.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidal M., Cusick M.E., Barabási A.-L. Interactome networks and human disease. Cell. Mar. 2011;144(6):986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sogin M.L. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA. Aug. 2006;103(32):12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghannoum MA, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog Jan. 2010; 6(1). doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed]
- 10.Ma B. Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome. Jun. 2020;8(1):82. doi: 10.1186/s40168-020-00857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gloor G.B., Macklaim J.M., Pawlowsky-Glahn V., Egozcue J.J. Microbiome datasets are compositional: and this is not optional. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature May 2011;473(7346) Art. no. 7346. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed]
- 13.Gross E.L. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. Nov. 2010;48(11):4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. Feb. 2012;6(2). Art. no. 2. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed]
- 15.Weiss S. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. Jul. 2016;10(7):1669–1681. doi: 10.1038/ismej.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovell D., Pawlowsky-Glahn V., Egozcue J.J., Marguerat S., Bähler J. Proportionality: a valid alternative to correlation for relative data. PLoS Comput Biol. Mar. 2015;11(3):e1004075. doi: 10.1371/journal.pcbi.1004075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erb I., Notredame C. How should we measure proportionality on relative gene expression data? Theory Biosci. 2016;135:21–36. doi: 10.1007/s12064-015-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinn T.P., Richardson M.F., Lovell D., Crowley T.M. propr: an R-package for identifying proportionally abundant features using compositional data analysis. Sci Rep. Dec. 2017;7(1):16252. doi: 10.1038/s41598-017-16520-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aitchison J. The statistical analysis of compositional data. J R Stat Soc Ser B Methodol. 1982;44(2):139–177. [Google Scholar]
- 20.Aitchison J. A concise guide to compositional data analysis. 2do Compos Data Anal Workshop CoDaWork. 2005;5:17–21. [Google Scholar]
- 21.Faust K., Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. Jul. 2012;10(8):538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 22.Flemer B. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. Aug. 2018;67(8):1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toju H., Yamamoto S., Tanabe A.S., Hayakawa T., Ishii H.S. Network modules and hubs in plant-root fungal biomes. J R Soc Interface. Mar. 2016;13(116):20151097. doi: 10.1098/rsif.2015.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faust K., Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Research. 2016;5:Oct. doi: 10.12688/f1000research.9050.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng Y., Jiang Y.-H., Yang Y., He Z., Luo F., Zhou J. Molecular ecological network analyses. BMC Bioinf. May 2012;13(1):113. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman J., Alm E.J. Inferring correlation networks from genomic survey data. PLoS Comput Biol. Sep. 2012;8(9):e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Y.-N., Fang J.-Y. Gut microbiota and colorectal cancer. Gastrointest Tumors. May 2015;2(1):26–32. doi: 10.1159/000380892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorvitovskaia A., Holmes S.P., Huse S.M. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome. Apr. 2016;4(1):15. doi: 10.1186/s40168-016-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McHardy I.H. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome. Jun. 2013;1(1):17. doi: 10.1186/2049-2618-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano H., Takemoto K. Difficulty in inferring microbial community structure based on co-occurrence network approaches. BMC Bioinf. Jun. 2019;20(1):329. doi: 10.1186/s12859-019-2915-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo F. Constructing gene co-expression networks and predicting functions of unknown genes by random matrix theory. BMC Bioinf. Aug. 2007;8(1):299. doi: 10.1186/1471-2105-8-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang H., Huang C., Zhao H., Deng M. CCLasso: correlation inference for compositional data through Lasso. Bioinformatics. Oct. 2015;31(19):3172–3180. doi: 10.1093/bioinformatics/btv349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ban Y., An L., Jiang H. Investigating microbial co-occurrence patterns based on metagenomic compositional data. Bioinforma Oxf Engl. Oct. 2015;31(20):3322–3329. doi: 10.1093/bioinformatics/btv364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bates S., Tibshirani R. Log-ratio lasso: scalable, sparse estimation for log-ratio models. Biometrics. Jun. 2019;75(2):613–624. doi: 10.1111/biom.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J., Shi P., Li H. Generalized linear models with linear constraints for microbiome compositional data. Biometrics. Mar. 2019;75(1):235–244. doi: 10.1111/biom.12956. [DOI] [PubMed] [Google Scholar]
- 36.Yang P., Yu S., Cheng L., Ning K. Meta-network: optimized species-species network analysis for microbial communities. BMC Genomics. Apr. 2019;20(2):187. doi: 10.1186/s12864-019-5471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price M.N. Indirect and suboptimal control of gene expression is widespread in bacteria. Mol Syst Biol. Apr. 2013;9:660. doi: 10.1038/msb.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chua H.N., Sung W.-K., Wong L. Exploiting indirect neighbours and topological weight to predict protein function from protein–protein interactions. Bioinformatics. Jul. 2006;22(13):1623–1630. doi: 10.1093/bioinformatics/btl145. [DOI] [PubMed] [Google Scholar]
- 39.Marino S., Baxter N.T., Huffnagle G.B., Petrosino J.F., Schloss P.D. Mathematical modeling of primary succession of murine intestinal microbiota. Proc Natl Acad Sci. Jan. 2014;111(1):439–444. doi: 10.1073/pnas.1311322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erb I. Partial correlations in compositional data analysis. Appl Comput Geosci. Jun. 2020;6:100026. doi: 10.1016/j.acags.2020.100026. [DOI] [Google Scholar]
- 41.Biswas S, Mcdonald M, Lundberg DS, Dangl JL, Jojic V. Learning microbial interaction networks from metagenomic count data. J Comput Biol J Comput Mol Cell Biol Jun. 2016;23(6) 526–535. doi: 10.1089/cmb.2016.0061. [DOI] [PubMed]
- 42.Kurtz Z.D., Müller C.L., Miraldi E.R., Littman D.R., Blaser M.J., Bonneau R.A. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. May 2015;11(5):e1004226. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman J., Hastie T., Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. Jul. 2008;9(3):432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meinshausen N., Bühlmann P. High-dimensional graphs and variable selection with the Lasso. Ann Stat. Jun. 2006;34(3):1436–1462. doi: 10.1214/009053606000000281. [DOI] [Google Scholar]
- 45.Tibshirani R. Regression Shrinkage and Selection via the Lasso. J R Stat Soc Ser B Methodol. 1996;58(1):267–288. [Google Scholar]
- 46.Liu H., Roeder K., Wasserman L. Stability approach to regularization selection (StARS) for high dimensional graphical models. Adv Neural Inf Process Syst. Dec. 2010;24(2):1432–1440. [PMC free article] [PubMed] [Google Scholar]
- 47.Fang H., Huang C., Zhao H., Deng M. gCoda: conditional dependence network inference for compositional data. J Comput Biol. Jul. 2017;24(7):699–708. doi: 10.1089/cmb.2017.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y., Chen N., Chen T. Inference of environmental factor-microbe and microbe-microbe associations from metagenomic data using a hierarchical Bayesian statistical model. Cell Syst. Jan. 2017;4(1):129–137.e5. doi: 10.1016/j.cels.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 49.Jiang S. HARMONIES: a hybrid approach for microbiome networks inference via exploiting sparsity. Front Genet. Jun. 2020;11 doi: 10.3389/fgene.2020.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ha M.J., Kim J., Galloway-Peña J., Do K.-A., Peterson C.B. Compositional zero-inflated network estimation for microbiome data. BMC Bioinf. Dec. 2020;21(21):581. doi: 10.1186/s12859-020-03911-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDavid A., Gottardo R., Simon N., Drton M. Graphical models for zero-inflated single cell gene expression. Ann Appl Stat. Jun. 2019;13(2):848–873. doi: 10.1214/18-AOAS1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan K.M., London P., Mohan K., Lee S.-I., Fazel M., Witten D. Learning graphical models with hubs. J Mach Learn Res JMLR. Oct. 2014;15:3297–3331. [PMC free article] [PubMed] [Google Scholar]
- 53.McGillivray A., Khalili A., Stephens D.A. Estimating sparse networks with hubs. J Multivar Anal. Sep. 2020;179:104655. doi: 10.1016/j.jmva.2020.104655. [DOI] [Google Scholar]
- 54.Tackmann J, Matias Rodrigues JF, von Mering C. Rapid inference of direct interactions in large-scale ecological networks from heterogeneous microbial sequencing data. Cell Syst Sep. 2019;9(3):286–96.e8. doi: 10.1016/j.cels.2019.08.002. [DOI] [PubMed]
- 55.Aliferis C.F., Statnikov A., Tsamardinos I., Mani S., Koutsoukos X.D. Local causal and markov blanket induction for causal discovery and feature selection for classification part I: algorithms and empirical evaluation. J Mach Learn Res. 2010;11:171–234. [Google Scholar]
- 56.Tavakoli S., Yooseph S. Learning a mixture of microbial networks using minorization–maximization. Bioinformatics. Jul. 2019;35(14):i23–i30. doi: 10.1093/bioinformatics/btz370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aitchison J., Ho C.H. The multivariate Poisson-log normal distribution. Biometrika. Dec. 1989;76(4):643–653. doi: 10.1093/biomet/76.4.643. [DOI] [Google Scholar]
- 58.Zhou H, Lange K. MM algorithms for some discrete multivariate distributions. J Comput Graph Stat Jt Publ Am Stat Assoc Inst Math Stat. Interface Found N Am Sep. 2010;19(3):645–65. doi: 10.1198/jcgs.2010.09014. [DOI] [PMC free article] [PubMed]
- 59.Tavakoli S, Yooseph S. Algorithms for inferring multiple microbial networks. In: 2019 IEEE international conference on bioinformatics and biomedicine (BIBM), San Diego, CA, USA; Nov. 2019, p. 223–7. doi: 10.1109/BIBM47256.2019.8983194.
- 60.McGregor K., Labbe A., Greenwood C.M.T. MDiNE: a model to estimate differential co-occurrence networks in microbiome studies. Bioinformatics. Mar. 2020;36(6):1840–1847. doi: 10.1093/bioinformatics/btz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS ONE Feb. 2012;7(2). doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed]
- 62.Chen J., Li H. Variable selection for sparse Dirichlet-multinomial regression with an application to microbiome data analysis. Ann Appl Stat. Mar. 2013;7(1):418–442. doi: 10.1214/12-AOAS592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peschel S., Müller C.L., von Mutius E., Boulesteix A.-L., Depner M. NetCoMi: network construction and comparison for microbiome data in R. Brief Bioinform. 2020;no. bbaa290:Dec. doi: 10.1093/bib/bbaa290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fisher R.A. Statistical methods for research workers. In: Kotz S., Johnson N.L., editors. Breakthroughs in statistics: methodology and distribution. Springer; New York, NY: 1992. pp. 66–70. [Google Scholar]
- 65.Gill R., Datta S., Datta S. A statistical framework for differential network analysis from microarray data. BMC Bioinf. Feb. 2010;11:95. doi: 10.1186/1471-2105-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siska C., Bowler R., Kechris K. The discordant method: a novel approach for differential correlation. Bioinformatics. Mar. 2016;32(5):690–696. doi: 10.1093/bioinformatics/btv633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao Y, Angulo MT, Friedman J, Waldor MK, Weiss ST, Liu Y-Y. Mapping the ecological networks of microbial communities. Nat Commun 2017;8(1):2042, 11. doi: 10.1038/s41467-017-02090-2. [DOI] [PMC free article] [PubMed]
- 68.Deutschmann I.M. Disentangling environmental effects in microbial association networks. Review, preprint, Aug. 2020 doi: 10.21203/rs.3.rs-57387/v1. [DOI] [Google Scholar]
- 69.Lima-Mendez G. Determinants of community structure in the global plankton interactome. Science. May 2015;348(6237):1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 70.Ghassami A., Kiyavash N. Interaction information for causal inference: the case of directed triangle. IEEE international symposium on information theory (ISIT) 2017;2017:1326–1330. doi: 10.1109/ISIT.2017.8006744. [DOI] [Google Scholar]
- 71.Thomas M. Cover, Joy A. Thomas. Inequalities in Information theory. In: Elements of information theory. John Wiley & Sons, Ltd; 2001, p. 482–509.
- 72.Margolin A.A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinf. Mar. 2006;7(1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mounier J. Microbial interactions within a cheese microbial community. Appl Environ Microbiol. Jan. 2008;74(1):172–181. doi: 10.1128/AEM.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuntal B.K., Gadgil C., Mande S.S. Web-gLV: a web based platform for Lotka-Volterra based modeling and simulation of microbial populations. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li C., Chng K.R., Kwah J.S., Av-Shalom T.V., Tucker-Kellogg L., Nagarajan N. An expectation-maximization algorithm enables accurate ecological modeling using longitudinal microbiome sequencing data. Microbiome. Aug. 2019;7(1):118. doi: 10.1186/s40168-019-0729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joseph T.A., Shenhav L., Xavier J.B., Halperin E., Pe’er I. Compositional Lotka-Volterra describes microbial dynamics in the simplex. PLoS Comput Biol. May 2020;16(5):e1007917. doi: 10.1371/journal.pcbi.1007917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Faust K., Lahti L., Gonze D., de Vos W.M., Raes J. Metagenomics meets time series analysis: unraveling microbial community dynamics. Curr Opin Microbiol. Jun. 2015;25:56–66. doi: 10.1016/j.mib.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 78.Ruan Q., Dutta D., Schwalbach M.S., Steele J.A., Fuhrman J.A., Sun F. Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinf Oxf Engl. Oct. 2006;22(20):2532–2538. doi: 10.1093/bioinformatics/btl417. [DOI] [PubMed] [Google Scholar]
- 79.Shade A, Gregory Caporaso J, Handelsman J, Knight R, Fierer N. A meta-analysis of changes in bacterial and archaeal communities with time. ISME J Aug. 2013;7(8). Art. no. 8. doi: 10.1038/ismej.2013.54. [DOI] [PMC free article] [PubMed]
- 80.David L.A. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15(7):R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trifonova N., Duplisea D., Kenny A., Tucker A. A spatio-temporal bayesian network approach for revealing functional ecological networks in fisheries. In: Blockeel H., van Leeuwen M., Vinciotti V., editors. vol. 8819. Springer International Publishing; Cham: 2014. pp. 298–308. (Advances in intelligent data analysis XIII). [Google Scholar]
- 82.Hekstra D.R., Leibler S. Contingency and statistical laws in replicate microbial closed ecosystems. Cell. May 2012;149(5):1164–1173. doi: 10.1016/j.cell.2012.03.040. [DOI] [PubMed] [Google Scholar]
- 83.Lahti L, Salojärvi J, Salonen A, Scheffer M, de Vos WM. Tipping elements in the human intestinal ecosystem. Nat Commun Jul. 2014;5(1). Art. no. 1. doi: 10.1038/ncomms5344. [DOI] [PMC free article] [PubMed]
- 84.Xia L.C. Extended local similarity analysis (eLSA) of microbial community and other time series data with replicates. BMC Syst Biol. Dec. 2011;5(Suppl 2):S15. doi: 10.1186/1752-0509-5-S2-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McGeachie MJ, et al. Longitudinal prediction of the infant gut microbiome with dynamic Bayesian networks. Sci Rep Feb. 2016;6(1). Art. no. 1. doi: 10.1038/srep20359. [DOI] [PMC free article] [PubMed]
- 86.Lugo-Martinez J., Ruiz-Perez D., Narasimhan G., Bar-Joseph Z. Dynamic interaction network inference from longitudinal microbiome data. Microbiome. Apr. 2019;7(1):54. doi: 10.1186/s40168-019-0660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang J.K. Systematic evaluation of molecular networks for discovery of disease genes. Cell Syst. Apr. 2018;6(4):484–495.e5. doi: 10.1016/j.cels.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Smet R., Marchal K. Advantages and limitations of current network inference methods. Nat Rev Microbiol. Oct. 2010;8(10):717–729. doi: 10.1038/nrmicro2419. [DOI] [PubMed] [Google Scholar]
- 89.Yang P, Tan C, Han M, Cheng L, Cui X, Ning K. Correlation-Centric Network (CCN) representation for microbial co-occurrence patterns: new insights for microbial ecology. NAR Genomics Bioinf Jun. 2020;2(2). doi: 10.1093/nargab/lqaa042. [DOI] [PMC free article] [PubMed]
- 90.Tipton L. Fungi stabilize connectivity in the lung and skin microbial ecosystems. Microbiome. Jan. 2018;6(1):12. doi: 10.1186/s40168-017-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Argelaguet R. Multi-omics factor analysis—a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol. Jun. 2018;14(6):e8124. doi: 10.15252/msb.20178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Virtanen S, Klami A, Khan S, Kaski S. Bayesian group factor analysis. In: Proceedings of the fifteenth international conference on artificial intelligence and statistics, La Palma, Canary Islands, Apr. 2012, vol. 22, p. 1269–77, [Online]. Available: http://proceedings.mlr.press/v22/virtanen12.html.
- 93.Haak BW, et al. Integrative transkingdom analysis of the gut microbiome in antibiotic perturbation and critical illness. mSystems Mar. 2021;6(2). doi: 10.1128/mSystems.01148-20. [DOI] [PMC free article] [PubMed]
- 94.Singh A. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics. Sep. 2019;35(17):3055–3062. doi: 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clos-Garcia M. Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine. Aug. 2019;46:499–511. doi: 10.1016/j.ebiom.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Petrosino J.F. The microbiome in precision medicine: the way forward. Genome Med. Feb. 2018;10(1):12. doi: 10.1186/s13073-018-0525-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Faust K. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. Jul. 2012;8(7):e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hall CV, et al. Co-existence of network architectures supporting the human gut microbiome. iScience Dec. 2019;22:380–91. doi: 10.1016/j.isci.2019.11.032. [DOI] [PMC free article] [PubMed]
- 99.Newman M.E.J. Mixing patterns in networks. Phys Rev E. Feb. 2003;67(2):026126. doi: 10.1103/PhysRevE.67.026126. [DOI] [PubMed] [Google Scholar]
- 100.Argelaguet R. MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. May 2020;21(1):111. doi: 10.1186/s13059-020-02015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dong X, Yambartsev A, Ramsey SA, Thomas LD, Shulzhenko N, Morgun A. Reverse enGENEering of regulatory networks from big data: a roadmap for biologists. Bioinf Biol Insights Jan. 2015;9:BBI.S12467. doi: 10.4137/BBI.S12467. [DOI] [PMC free article] [PubMed]
- 102.Bonito G. Fungal-bacterial networks in the populus rhizobiome are impacted by soil properties and host genotype. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reiman D., Layden B., Dai Y. MiMeNet: exploring microbiome-metabolome relationships using neural networks. bioRxiv. 2020 doi: 10.1371/journal.pcbi.1009021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hawe J.S., Theis F.J., Heinig M. Inferring interaction networks from multi-omics data. Front Genet. 2019;10 doi: 10.3389/fgene.2019.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nash A.K. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. Nov. 2017;5(1):153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Coker O.O. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. Jun. 2018;67(6):1024–1032. doi: 10.1136/gutjnl-2017-314281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ai D., Li X., Pan H., Chen J., Cram J.A., Xia L.C. Explore mediated co-varying dynamics in microbial community using integrated local similarity and liquid association analysis. BMC Genomics. Apr. 2019;20(2):185. doi: 10.1186/s12864-019-5469-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xie W. Localized high abundance of Marine Group II archaea in the subtropical Pearl River Estuary: implications for their niche adaptation: High abundance of MGII in an estuary. Environ Microbiol. Feb. 2018;20(2):734–754. doi: 10.1111/1462-2920.14004. [DOI] [PubMed] [Google Scholar]
- 109.Fettweis JM, et al. The vaginal microbiome and preterm birth. Nat Med Jun. 2019;25(6). Art. no. 6. doi: 10.1038/s41591-019-0450-2. [DOI] [PMC free article] [PubMed]
- 110.Durán P. Microbial interkingdom interactions in roots promote arabidopsis survival. Cell. Nov. 2018;175(4):973–983.e14. doi: 10.1016/j.cell.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang B., Zhang J., Liu Y., Shi P., Wei G. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol Biochem. Mar. 2018;118:178–186. doi: 10.1016/j.soilbio.2017.12.011. [DOI] [Google Scholar]
- 112.Mandakovic D, et al. Structure and co-occurrence patterns in microbial communities under acute environmental stress reveal ecological factors fostering resilience. Sci Rep Apr. 2018;8(1), Art. no. 1. doi: 10.1038/s41598-018-23931-0. [DOI] [PMC free article] [PubMed]
- 113.Coretti L. Gut microbiota features in young children with autism spectrum disorders. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.03146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu X., Wang Y., Ji H., Aihara K., Chen L. Personalized characterization of diseases using sample-specific networks. Nucleic Acids Res. Dec. 2016;44(22):e164. doi: 10.1093/nar/gkw772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao H, et al. Variations in oral microbiota associated with oral cancer. Sci Rep Sep. 2017;7(1). Art. no. 1. doi: 10.1038/s41598-017-11779-9. [DOI] [PMC free article] [PubMed]
- 116.Ling N. Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil Biol Biochem. Aug. 2016;99:137–149. doi: 10.1016/j.soilbio.2016.05.005. [DOI] [Google Scholar]
- 117.Li X. Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ Pollut. Dec. 2017;231:908–917. doi: 10.1016/j.envpol.2017.08.057. [DOI] [PubMed] [Google Scholar]
- 118.Cong J, et al. Analyses of soil microbial community compositions and functional genes reveal potential consequences of natural forest succession. Sci Rep May 2015;5(1). Art. no. 1. doi: 10.1038/srep10007. [DOI] [PMC free article] [PubMed]
- 119.Yuan H., He S., Deng M. Compositional data network analysis via lasso penalized D-trace loss. Bioinformatics. Sep. 2019;35(18):3404–3411. doi: 10.1093/bioinformatics/btz098. [DOI] [PubMed] [Google Scholar]
- 120.Röttjers L., Faust K. From hairballs to hypotheses–biological insights from microbial networks. FEMS Microbiol Rev. Nov. 2018;42(6):761–780. doi: 10.1093/femsre/fuy030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.He S., Deng M. Direct interaction network and differential network inference from compositional data via lasso penalized D-trace loss. PLoS ONE. Jul. 2019;14(7):e0207731. doi: 10.1371/journal.pone.0207731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jiang S. HARMONIES: a hybrid approach for microbiome networks inference via exploiting sparsity. Front Genet. 2020;11 doi: 10.3389/fgene.2020.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ruiz VE, et al. A single early-in-life macrolide course has lasting effects on murine microbial network topology and immunity. Nat Commun Sep. 2017;8(1). Art. no. 1. doi: 10.1038/s41467-017-00531-6. [DOI] [PMC free article] [PubMed]
- 124.Gregory A.C., Zablocki O., Zayed A.A., Howell A., Bolduc B., Sullivan M.B. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe. Nov. 2020;28(5):724–740.e8. doi: 10.1016/j.chom.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Murray AE, et al. Uncovering the core microbiome and distribution of palmerolide in synoicum adareanum across the Anvers Island Archipelago, Antarctica. Mar Drugs Jun. 2020;18(6). Art. no. 6. doi: 10.3390/md18060298. [DOI] [PMC free article] [PubMed]
- 126.Martin O. Haem iron reshapes colonic luminal environment: impact on mucosal homeostasis and microbiome through aldehyde formation. Microbiome. 2019;7(1):1–18. doi: 10.1186/s40168-019-0685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.