Abstract
Accurate translation termination in bacteria requires correct recognition of the stop codons by the class-I release factors (RFs) RF1 and RF2, which release the nascent peptide from the peptidyl tRNA after undergoing a “compact to open” conformational transition. These RFs possess a conserved Gly-Gly-Gln (GGQ) peptide release motif, of which the Q residue is posttranslationally methylated. GGQ-methylated RFs have been shown to be faster in peptide release than the unmethylated ones, but it was unknown whether this modification had additional roles. Using a fluorescence-based real-time in vitro translation termination assay in a stopped-flow instrument, we demonstrate that methylated RF1 and RF2 are two- to four-fold more accurate in the cognate stop codon recognition than their unmethylated variants. Using pH titration, we show that the lack of GGQ methylation facilitates the “compact to open” transition, which results in compromised accuracy of the unmethylated RFs. Furthermore, thermal melting studies using circular dichroism and SYPRO-orange fluorescence demonstrate that GGQ methylation increases overall stability of the RF proteins. This increased stability, we suspect, is the basis for the more controlled conformational change of the methylated RFs upon codon recognition, which enhances both their speed and accuracy. This GGQ methylation-based modulation of the accuracy of RFs can be a tool for regulating translational termination in vivo.
Keywords: ribosome, translation, release factor, translation termination, methylation, GGQ motif, accuracy
Abbreviations: a.u., arbitrary units; ATP, adenosine triphosphate; BOP, BODIPYTM 576/589; CD, circular dichroism; cryo-EM, cryo-electron microscopy; EF, elongation factor; FRET, fluorescence resonance energy transfer; IF, initiation factor; kcat, maximal rate of catalysis; kcat/KM, catalytic efficiency; KM, Michaelis–Menten constant; M-M, Michaelis–Menten; MD, molecular dynamics; mRF, methylated release factor; RC, release complex; RF, release factor; rNTP, ribonucleoside triphosphate; rRNA, ribosomal ribonucleic acid; RT-PCR, real-time polymerase chain reaction; Tm, melting temperature
Termination is an important step of translation during which the nascent peptides are released from the ribosome. It should be efficient for allowing fast turnover of the translation machinery, and at the same time, accurate to avoid accumulation of the potentially inactive and toxic, truncated, or overlong proteins. Protein synthesis terminates when a translating ribosome encounters one of the three stop codons (UAA, UAG, and UGA) on the mRNA at the ribosomal A site. These codons are recognized in a semispecific manner by the class-I release factors (referred hereafter as RFs) in bacteria, namely release factor 1 (RF1) and release factor 2 (RF2). UAA codon is read by both RF1 and RF2. But, UAG is read specifically by RF1 and UGA by RF2 (1). In eukaryotes, all three stop codons are read by a common class-I RF named eRF1 (2). The class-I RFs in bacteria depend on the class-II RF RF3 for their fast dissociation from the ribosome after peptide release (3).
RF1 and RF2 possess a common universally conserved Glycine–Glycine–Glutamine (GGQ) motif (4) in the tip of domain III, which is involved in release of the nascent peptide by ester bond hydrolysis from the peptidyl tRNA at the peptidyl transferase center (PTC) of the ribosome (5, 6, 7, 8, 9, 10, 11). Mutations of the GGQ motif render RF1 and RF2 inactive in peptide release (8, 12, 13). In both prokaryotes and eukaryotes, the amide group of the Gln (Q) residue in the GGQ motif is posttranslationally methylated at the N5 position by a methyltransferase enzyme encoded by prmC (also known as HemK) (for review see (14, 15, 16, 17)). Knocking out prmC gene is not lethal but results in slow phenotypic growth (18). Earlier in vitro (10, 14, 19, 20) and in vivo studies (18, 21) demonstrated that GGQ methylation significantly enhances the catalytic activity of the RFs. The enhancement is more pronounced for some particular amino acids (20). Using X-ray crystallography and cryo-electron microscopy (cryo-EM), it has been recently shown that the GGQ methylation helps this motif in acquiring a stable conformation in the PTC, which in turn facilitates efficient peptide release by the RFs (10).
Accuracy of translation termination depends on correct stop codon recognition by the class-I RFs. RF1 and RF2 possess different sequence motifs for recognition of the specific stop codons at the decoding center (DC) of the ribosome. Earlier studies based on mutational analysis demonstrated that RF1 uses a Proline-X-Threonine/Alanine/Valine (PXT) motif and RF2 uses a Serine–Proline–Phenylalanine (SPF) motif located in the domain-II for specific recognition of their cognate stop codons (22, 23). Later, molecular dynamics (MD) simulations-based studies (24) identified additional residues beyond the PXT and SPF motifs as crucial for correct stop codon recognition by RF1 and RF2. Among these the role of Arg213 of RF2 has been studied extensively by mutagenesis and fast kinetics (25).
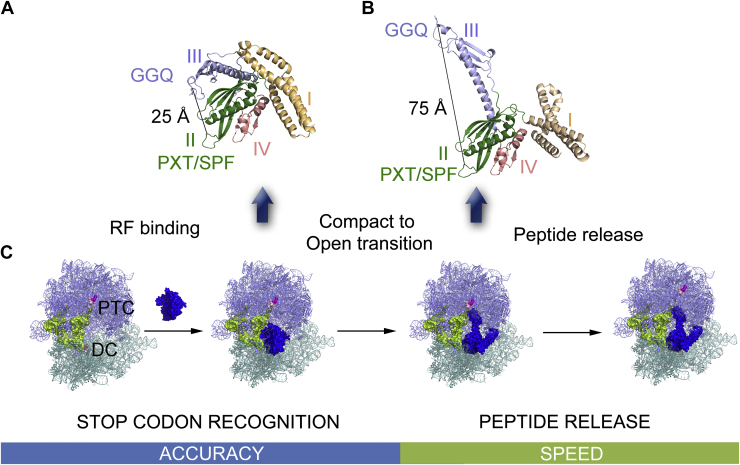
Structural studies of the RFs with X-ray crystallography and cryo-EM have revealed that RF1 and RF2 exist in two different conformations in free state and when bound to the ribosome (5, 6, 7). In the crystal structures, corresponding to the free state, RF1 and RF2 are seen in a compact form, with PXT/SPF motif and GGQ motif separated by a distance of 25 Å (Fig. 1A) (26, 27). In contrast, the class-I RFs mimic aminoacyl-tRNAs on the ribosome and adopt an open conformation spanning ∼75 Å from the DC on the small subunit to the PTC on the large subunit (Fig. 1B) (5, 7, 11, 28, 29, 30, 31). It was proposed earlier that RF1 and RF2 confer the catalytically active open conformation upon correct codon recognition after binding to the ribosome (Fig. 1C) (5). The visual evidence for this hypothesis, however, appeared only very recently through time-resolved cryo-EM, which demonstrated a “compact to open” transition of RF1 and RF2 on the ribosome after proper recognition of the stop codon (32). Contemporary cryo-EM studies have also reported different intermediate conformations (between fully compact and open) of RF1 and RF2 on the ribosome (33, 34, 35). Moreover, kinetics of the “compact to open” transition has been followed by state-of-the-art fluorescence resonance energy transfer (FRET) (36) and in silico simulations (9). Altogether these studies decipher a near complete mechanistic picture of translation termination by the bacterial class-I RFs, where the stop codon recognition and conformational change relate to the accuracy in termination, whereas the peptide release governs the speed of the process (Fig. 1C).
Figure 1.
Class-I release factor-mediated steps of translation termination, which govern the speed and accuracy of the process.A and B, represent compact and open conformations of RFs, respectively, with the domains I, II, III, and IV colored in yellow, green, purple, and pink. In the closed state, the distance between the GGQ and the PXT/SPF motifs is 25 Å, which increases to 75 Å in the open state on the ribosome. C, the scheme describes the main events of translation termination where the RFs bind to the ribosome with an empty A-site harboring a stop codon in the compact state. The RFs undergo “compact to open” transition upon recognition of the stop codon and accommodation in the A-site. Lastly, the peptide is released from the peptidyl tRNA by participation of the GGQ motif of the RFs in the open state. The first two steps contribute to accuracy of the process, whereas the catalytic peptide release state governs speed of translation termination.
The first quantitative analysis of accuracy of stop codon recognition by RF1 and RF2 was performed by Ehrenberg and colleagues. With precise biochemical experiments, they demonstrated the degree of preference of RF1 and RF2 for their cognate stop codons over the near-cognate codons for efficient peptide release (37). Their conclusion was that accuracy in termination depends entirely on the selective recognition of the stop codons by the RFs. However, the new finding that RF1 and RF2 open up on the ribosome upon correct stop codon recognition and thereby become catalytically active (Fig. 1) (32), adds an additional regulatory step in the accuracy process. In addition, it raises the question whether GGQ methylation has any role in modulating this conformational transition and thus, affecting the accuracy of stop codon recognition by RF1 and RF2. Recently, Zeng et al. (10) reported kinetic parameters for peptide release by the methylated and unmethylated RFs on the stop codons, but the accuracy impact has not been explored.
Here, using a fluorescence-based in vitro peptide release assay in stopped-flow, we show that GGQ methylation in RF1 and RF2 enhances both catalytic speed and accuracy of stop codon recognition. Furthermore, by pH titration, we demonstrate that the lack of GGQ methylation expedites the “compact to open” transition of the RFs, thereby making the unmethylated RFs less accurate in peptide release. Finally, by biophysical characterization of the unmethylated and methylated RFs, we find that the GGQ methylation aids in overall stability of the proteins. This increased stability, we suspect, is the basis for more controlled conformational change in the methylated RF1 and RF2 upon codon recognition, which enhances their speed and accuracy.
Results
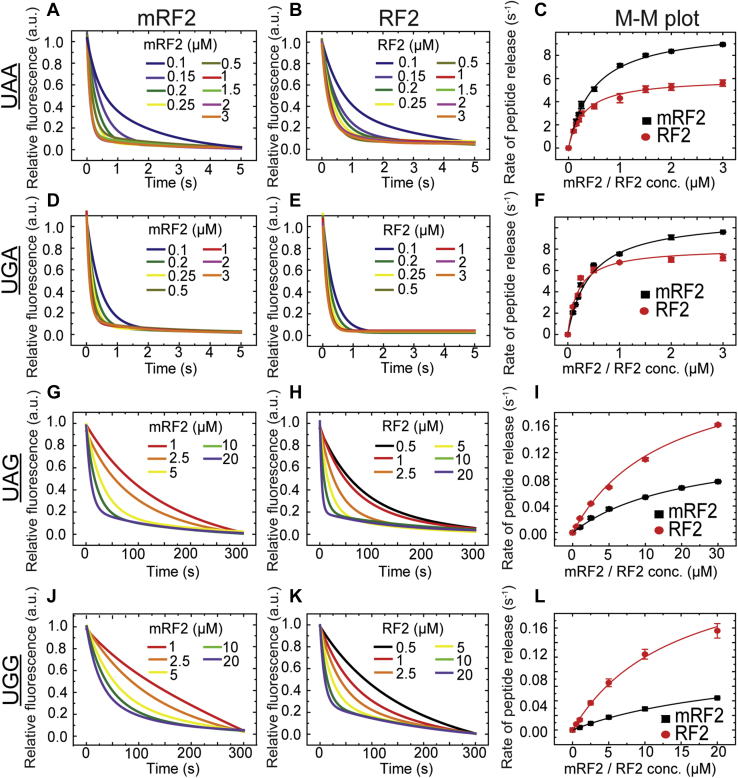
Kinetics of peptide release by mRF2 and RF2 on cognate (UAA, UGA) and near-cognate (UAG, UGG) codons
For peptide release assay, we used a ribosomal release complex (RC), which harbored in the P-site, a peptidyl tRNA carrying fluorescent BODIPY 576/589 (BOP) labeled Met-Leu-Leu tripeptide, and in the A-site, one of the three stop codons or the UGG codon specific for tryptophan (Trp), on the mRNA. The RF2 variants, methylated (mRF2) and unmethylated (RF2), were added in increasing concentration to the RC with either cognate UAA and UGA codons or near-cognate UAG and UGG codons, and the time course of peptide release was monitored by following decrease in BOP fluorescence in a stopped-flow instrument (Fig. 2, left and middle panel). The rates of peptide release were determined by fitting the fluorescence curves with exponential functions (see Experimental procedures for details). The observed rates plotted against concentration of the RFs were fitted with hyperbolic function following Michaelis–Menten equation (Fig. 2, right panel), from which the kinetic parameters kcat, KM, and kcat/KM were derived.
Figure 2.
Stopped-flow-based fluorescent BOP-Met-Leu-Leu tripeptide release kinetics by mRF2 and RF2 on RCs containing cognate/near-cognate codons. The left (A, D, G, and J) and the middle panels (B, E, H, and K) present the time courses of BOP fluorescence change in arbitrary units (a.u.) due to BOP-Met-Leu-Leu tripeptide release from cognate RCUAA and RCUGA and near-cognate RCUAG and RCUGG (0.1 μM), with increasing concentrations of mRF2 (left) and RF2 (middle). The right panels (C, F, I, and L) represent the Michaelis–Menten plots (M-M plot) demonstrating dependence of the observed peptide release rates (derived from the titration curves in the left and middle panels), on the concentration of mRF2 and RF2 on corresponding codons (labeled in the left for each row). The kinetic parameters (kcat, KM, and kcat/KM) are derived by fitting the curves with hyperbolic function. The error bars indicate SEM from minimum three sets of independent experiments.
Our kinetics data (Table 1), consistent with earlier reports (19, 20, 37, 38), show that mRF2 releases peptide from cognate RCUAA and RCUGA with the maximal catalytic rate (kcat) of about 10 s−1, which is about 1.7-fold higher than that of RF2 (Fig. 2, C and F). However, the increase in kcat is accompanied by an equivalent increase in KM by mRF2, thereby resulting in similar catalytic efficiencies (kcat/KM) of mRF2 and RF2 on the cognate UAA and UGA codons (Table 1). This result, in complete agreement with the previous report (38), confirms that the methylation of GGQ does not have any impact on catalytic efficiency of peptide release by RF2 on the cognate codons. However, when the same assay was performed with near-cognate RCUAG, RF2 showed about twofold higher kcat than mRF2 with almost no change in KM (Fig. 2I). Thus, catalytic efficiency of RF2 on UAG codon is about 2.5-fold higher than mRF2 (Table 1). In line with this result, RF2 shows about fourfold higher catalytic efficiency of peptide release than mRF2 on Trp coding UGG codon (Table 1). On RCUGG, RF2 releases peptide with 2.5-fold higher kcat than mRF2 (Fig. 2L). Interestingly, mRF2 shows 1.5-fold higher KM than RF2 on RCUGG. Thus, together these two parameters cause a bigger difference in the catalytic efficiency of RF2 versus mRF2 on UGG than on UAG codon (Table 1).
Table 1.
The Michaelis–Menten parameters for peptide release by the methylated and unmethylated RFs on cognate and near-cognate codons
| Codon | A site codon | Release factor | kcat (s−1) | KM (μM−1) | kcat/KM (s−1 μM−1) |
|---|---|---|---|---|---|
| Cognate | UAA | mRF2 | 10.4 ± 0.04 | 0.49 ± 0.007 | 21.2 ± 0.40 |
| RF2 | 6.0 ± 0.17 | 0.30 ± 0.01 | 20.0 ± 0.008 | ||
| UGA | mRF2 | 10.7 ± 0.03 | 0.38 ± 0.009 | 28.2 ± 0.36 | |
| RF2 | 6.8 ± 0.19 | 0.28 ± 0.04 | 24.3 ± 0.007 | ||
| Near-cognate | UAG | mRF2 | 0.13 ± 0.01 | 15.5 ± 1.76 | 0.008 ± 0.0006 |
| RF2 | 0.28 ± 0.004 | 14.7 ± 0.10 | 0.019 ± 0.0004 | ||
| UGG | mRF2 | 0.10 ± 0.04 | 22 ± 1.4 | 0.0045 ± 0.0001 | |
| RF2 | 0.25 ± 0.03 | 14.4 ± 0.11 | 0.017 ± 0.0006 | ||
| Cognate | UAA | mRF1 | 8.3 ± 0.06 | 0.06 ± 0.005 | 138.3 ± 0.003 |
| RF1 | 2.0 ± 0.04 | 0.03 ± 0.03 | 66.7 ± 0.005 | ||
| Near-cognate | UGA | mRF1 | 0.16 ± 0.004 | 2.7 ± 0.03 | 0.059 ± 0.004 |
| RF1 | 0.12 ± 0.003 | 2.6 ± 0.26 | 0.046 ± 0.004 |
As expected, mRF2 and RF2 show similar kcat and KM values on the two cognate codons UAA and UGA (Table 1). However, both mRF2 and RF2 show slightly higher catalytic efficiency (kcat/KM) for UGA (28.2 ± 0.36 and 24.3 ± 0.007) than UAA (21.2 ± 0.4 and 20 ± 0.008). Our results clearly demonstrate that mRF2 has higher catalytic activity, but similar catalytic efficiency of peptide release on cognate codons. However, on near-cognate codons, RF2 shows higher catalytic activity as well as catalytic efficiency than mRF2.
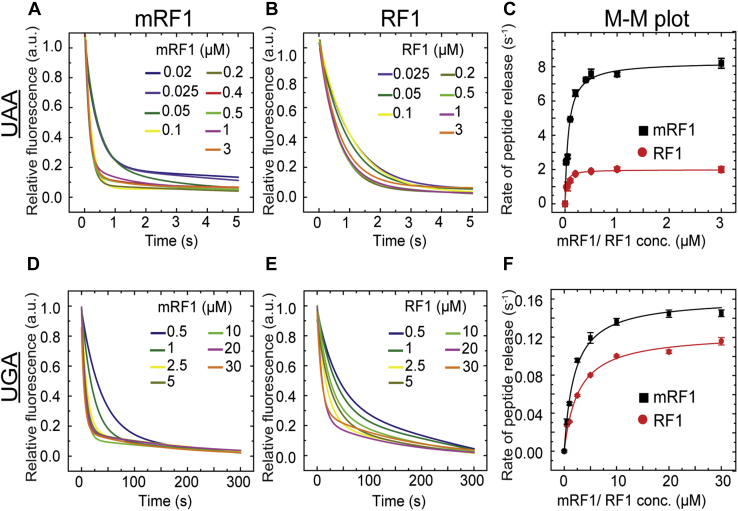
Kinetics of peptide release by mRF1 and RF1 on cognate (UAA) and near-cognate (UGA) codons
We compared mRF1 and RF1 for their catalytic efficiency in peptide release from the RCs containing cognate (UAA) and near-cognate (UGA) codon using the stopped-flow-based Bop-Met-Leu-Leu tripeptide release assay (Fig. 3, left and middle panel). The observed rates estimated from the time course of the peptide release are plotted against mRF1 and RF1 concentration and fitted with hyperbolic function to obtain the Michaelis–Menten parameters (kcat, KM, and kcat/KM) (Fig. 3, right panel).
Figure 3.
Stopped-flow-based fluorescent BOP-Met-Leu-Leu tripeptide release kinetics by mRF1 and RF1 on RC containing cognate/near-cognate codons. The first two panels present the time courses of BOP fluorescence change due to BOP-Met-Leu-Leu tripeptide release from cognate RCUAA and near-cognate RCUGA (0.1 μM), with increasing concentrations of mRF1 (leftpanels: A and D) and RF1 (middlepanels: B and E) in arbitrary units (a.u.). The rightpanels (C and F) are the Michaelis–Menten plots (M-M plot) representing dependence of the observed peptide release rates (derived from the titration curves in the left and middle panels), on the concentration of mRF1 and RF1 on corresponding codons (labeled in left). The kinetic parameters of peptide release (kcat, KM, and kcat/KM) by mRF1 and RF1 are derived by fitting the curves with hyperbolic equation. The error bars indicate SEM from minimum three sets of independent experiments.
Our kinetic data show that mRF1 possesses 4.2-fold higher catalytic activity than RF1 for peptide release from the cognate RCUAA, with kcat 8.3 s−1 (Fig. 3C, Table 1). Along with the increase in kcat, mRF1 also shows a twofold increase in KM, thereby resulting in twofold higher catalytic efficiency (kcat/KM) than RF1 for peptide release on the cognate UAA codon (Table 1). This result confirms that mRF1 has higher catalytic efficiency than RF1 for peptide release on the cognate UAA codon. However, on the near-cognate RCUGA, mRF1 and RF1 show similar catalytic activity with kcat 0.16 s−1 and 0.12 s−1 respectively, with no change in KM (Fig. 3F). Thus, mRF1 and RF1 show similar catalytic efficiency of peptide release on UGA codon (Table 1).
Thus, our results demonstrate that methylation of RF1 results in significantly higher catalytic activity and catalytic efficiency of peptide release on the cognate codon (Table 1).
Accuracy of the class-I RFs
To determine the accuracy of the RFs, we introduce a parameter called “A value,” which is the ratio of their catalytic efficiencies of peptide release (kcat/KM) on cognate versus near-cognate codons (37). In other words, the A value describes the relative preference of the RFs for the cognate codon (reference codon) over the near-cognate codon (or essentially any other codon). Here, we have used the major stop codon UAA as the reference codon.
With regard to the two cognate codons UAA and UGA, both mRF2 and RF2 show slightly higher preference for UGA with the A values 0.74 ± 0.017 and 0.83 ± 0.0004, respectively. However, significantly larger A values are obtained when catalytic efficiencies are compared between UAA and near-cognate UAG and UGG codons. The A values summarized in Table 2 show that mRF2 favors peptide release on UAA over UAG by a factor of 2625 ± 203 and over UGG by a factor of 4200 ± 116. These values are in close agreement with the values previously reported by Freistroffer et al. (37). The unmethylated RF2, however, shows less discrimination for the near-cognate codons, about 1000 ± 25 for both UAG and UGG. Thus, comparison of the respective A value shows that mRF2 is 2.6 and 4.2 times more discriminative than RF2 for near-cognate UAG and UGG codons, respectively (Table 2). These results thus demonstrate that mRF2 is not only more efficient than RF2 for peptide release, but also more accurate for correct recognition of the cognate stop codons.
Table 2.
Accuracy of stop codon recognition by methylated and unmethylated release factors
| Release factor 2 | Cognate versus Cognate |
←Cognate versus near-cognate→ |
|||
|---|---|---|---|---|---|
| A(UAA/UGA) | A(UAA/UAG) | A(UAA/UGG) | Release factor 1 | A(UAA/UGA) | |
| mRF2 | 0.74 ± 0.017 | 2625 ± 203.13 | 4200 ± 116.00 | mRF1 | 2300 ± 153.33 |
| RF2 | 0.83 ± 0.0004 | 1000 ± 20.00 | 1000 ± 30.00 | RF1 | 1340 ± 107.20 |
| mRF2/RF2 | 0.9 | 2.6 | 4.2 | mRF1/RF1 | 1.72 |
The data are presented as “A values.” A = (kcat/KM) cognate/(kcat/KM) near-cognate with SEM estimated from at least three independent sets of experiments.
We have also estimated the accuracy of mRF1 and RF1 for UAA (cognate) versus UGA (near-cognate) codons. The A values summarized in Table 2 show that mRF1 favors UAA codon over UGA, 2300 ± 153 times. In comparison, RF1 shows lesser preference for UAA than UGA, 1340 ± 107 times. Thus, mRF1 is 1.7 times more discriminative than RF1 for the near-cognate UGA codon (Table 2).
It is interesting to compare the basis of the A values for the RF1 and RF2 variants. The mRF2 and RF2 show similar kcat/KM values on the cognate codons, but mRF2 shows lower kcat/KM values than RF2 on the near-cognate codons (Table 1). In the contrary, mRF1 shows higher kcat/KM on the cognate codon than RF1, but similar kcat/KM on the near-cognate codon (Table 1). Thus, higher accuracy of mRF2 originates from its relatively lower efficiency of near-codon recognition than RF2. In contrast, higher accuracy of mRF1 is due to its relatively higher efficiency of cognate codon recognition than RF1. However, in both cases the methylated RFs show higher accuracy than the unmethylated forms.
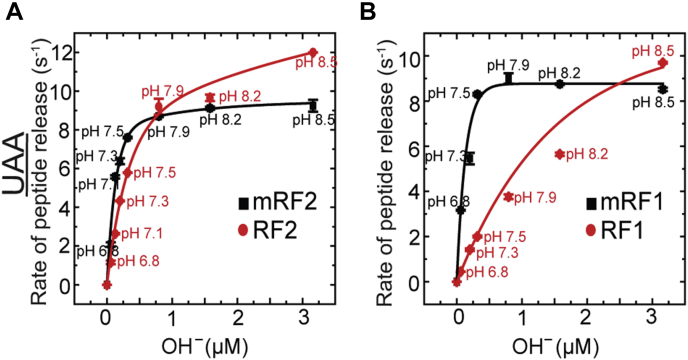
Determination of the rate-limiting step for peptide release by RFs and mRFs using pH titration
As suggested by current literature (19, 32), the “compact to open” conformational transition is the critical step, which makes the RFs catalytically active for peptide release (Fig. 1C). Based on our accuracy measurements, we hypothesized that the RFs lacking GGQ methylation might undergo this transition more easily than mRFs, which compromise their accuracy of peptide release on cognate codons. To test that hypothesis, we followed pH dependence of the maximal rate of peptide release (kcat) by mRFs and RFs on RCUAA (Fig. 4). It has been shown that peptide release by the class-I RFs is rate limited by the chemistry of ester bond hydrolysis at low pH and by a pH-independent conformational change step at high pH (19). Our aim was to find out whether the methylated and unmethylated RFs undergo transition from the “pH-dependent” to the “pH-independent” state in similar pH or not.
Figure 4.
pH dependence of kcatof BOP-Met-Leu-Leu tripeptide release by methylated and unmethylated RFs. The maximal rates of peptide release by mRF2 and RF2 (A) and mRF1 and RF1 (B) on RCUAA are plotted as a function of OH− ion concentration.
The stopped-flow-based BOP-Met-Leu-Leu tripeptide release assay was conducted with RCUAA in HEPES-polymix buffer with pH adjusted to 6.8 to 8.5. To ensure maximal rates, saturating concentration of RFs (4 μM) was used. In good agreement with the published data (19), our results show that kcat for mRF2 increased linearly from 2.2 s−1 at pH 6.8 to 9 s−1 at pH 7.9, after which it did not increase further (Fig. 4A). Interestingly, kcat for RF2 did not saturate in this pH range. Instead, it showed a biphasic linear increase, with slightly different slopes before and after pH 7.5. At pH 8.5 it released peptide with a rate 12 s−1, which is even higher than mRF2 at that pH (Fig. 4A).
For mRF1 and RF1 the difference was more drastic, but the overall trend was same as mRF2 and RF2. For mRF1, the kcat increased linearly from 3.2 s−1 at pH 6.8 to 8.7 s−1 at pH 7.9 after which it saturated. In contrast, RF1 showed pH-dependent biphasic linear increase in the entire pH range tested here and produced higher kcat (10 s−1) than mRF1 (8.5 s−1) at pH 8.5 (Fig. 4B).
Thus, both mRF2 and mRF1 showed “pH-dependent” to “pH-independent” transition of kcat at ∼pH 7.9, while for RF2 and RF1 the kcat increased in a pH-dependent manner even at higher pH. Thus, for RF2 and RF1, the pH-dependent catalytic step remained “rate limiting” and thus “slower” than the pH-independent conformational change step for the entire pH range tested here. Conversely, for mRF2 and mRF1, the catalytic step was rate limiting only at pH less than 7.9. These results suggest that in contrast to the methylated RFs, unmethylated RFs presumably undergo “compact to open” transition faster than the catalytic step even in high pH. It means that RF2 and RF1 are inherently more prone to “compact to open” transition than mRF2 and mRF1.
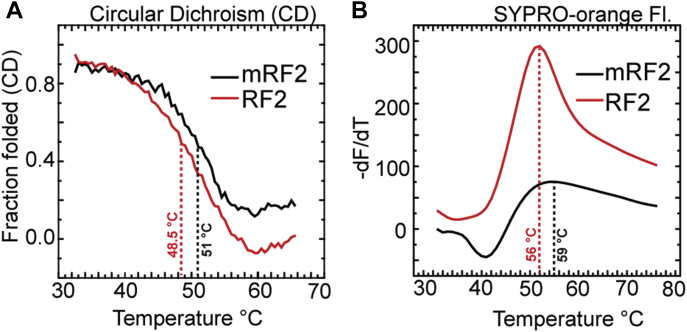
Comparison of the conformational stability of mRF2 and RF2 by thermal melting
To check whether the lack of methylation affects the overall stability of RFs in free state, thermal melting profiles of mRF2 and RF2 were recorded using (i) Circular Dichroism (CD) and (ii) SYPRO-orange thermal shift assay.
CD provides information about secondary structure of a protein. When mRF2 and RF2 were subjected to thermal melting in a JASCO J-1500 CD spectrometer, both showed gradual loss of the intrinsic helicity at 220 nm with increase in temperature. The fraction of the folded protein was estimated from the ratio of the CD value at a given temperature with the total change in CD. As shown in Figure 5A, the fraction of the folded mRF2 and RF2 decreased with increase in temperature and the main transition for both was observed after 40 °C. The melting temperature (Tm) estimated from the midpoint of the transition was 51 °C for mRF2 and 48.5 °C for RF2 (Fig. 5A). Although the observed difference was small, the results were highly reproducible. Also, big difference in the CD-based melting temperature was not expected from the difference of a single methyl group between the two RF2 variants.
Figure 5.
Thermal melting profiles of mRF2 (black traces) and RF2 (red traces) for comparison of their structural stability.A, thermal melting of secondary structure of mRF2 and RF2 followed by CD at 220 nm. The plots represent the fraction of folded proteins, as estimated from the ratio of relative change in ellipticity for a given temperature change over the total change in ellipticity (see Experimental procedures for details). The transition midpoints are recorded as Tm. B, SYPRO-orange thermal shift assay to follow tertiary structure melting of mRF2 and RF2. The plots represent the first derivative of the fluorescence emission at 569 nm as a function of temperature. The peak of the derivative plots is considered as Tm.
Further, SYPRO-orange thermal shift assay was conducted using an RT-PCR machine to compare the thermal stability of the tertiary structures of mRF2 and RF2. The SYPRO-orange fluorescence changes upon binding to the hydrophobic patches of the protein, thereby reflecting on the changes in the tertiary structure of the proteins. The first derivative of the change in the SYPRO-orange fluorescence was plotted for mRF2 and RF2, as a function of temperature (Fig. 5B). The Tm, derived from the peak value of the derivative curves for mRF2 was 59 °C and for RF2 was 56 °C (Fig. 5B). However, RF2 showed noticeably greater fluorescence change than mRF2, suggesting that the dye had larger access to its hydrophobic patches than to mRF2. These results indirectly argue in favor of a relatively flexible structure of RF2 than mRF2.
Discussion
The role of GGQ methylation in the bacterial RFs has been studied extensively with in vivo genetics and in vitro biochemistry (10, 14, 18, 19, 20, 21). These studies unanimously reported increased efficiency of peptide release by virtue of GGQ methylation of the RFs. This is understandable as the GGQ motif places itself in PTC, directly in the site of hydrolysis of the peptidyl tRNA. It is nicely demonstrated by a recent cryo-EM structure (10), which showed that the methylated GGQ motif adopts a highly ordered conformation, which facilitates its interaction with A76 of the peptidyl tRNA as well as with U2506, A2451, and A2452 nucleotides of 23S RNA. However, there was no knowledge so far whether GGQ methylation can also influence other steps of translation termination.
Our results clearly demonstrate the effect of GGQ methylation in the class-1 RFs on both catalytic activity and accuracy of translation termination. On the basis of the A value calculation (Table 2), we infer that accuracy originates from the lower efficiency of recognition of the near-cognate codons than the cognate codons by the RFs. When the catalytic efficiencies were compared for peptide release on cognate versus near-cognate codons, the unmethylated RFs were two- to four-fold less accurate than the methylated RFs by performing less efficient discrimination between a cognate and a near-cognate codon. This applies not only to the near-cognate stop codons, but also to the Trp codon encoded by UGG. However, question remains that how lack of GGQ methylation may affect accuracy of translation termination, when the methylation site is located far from the stop codon recognition site on the ribosome.
We revisited the conformational cycle of the RFs during translation termination to seek an answer to the question above. The RFs are seen in the compact state (Fig. 1A) in complex with the methyltransferase HemK encoded by the gene prmC, which methylates the GGQ motif (17). The RFs also bind to the ribosome in the compact state (33, 34, 35). In this state, the GGQ motif on domain III positions itself closer to the domain II, thereby forming the II–III–IV super domain (26). Upon codon recognition, rearrangement of the switch loop between the domains III and IV directs the opening of domain III carrying the GGQ motif to reach the PTC (30). It has been suggested that stop codon recognition in the DC brings 23S rRNA closer to the RFs, thereby stabilizing their open and active state (39). This “compact to open” transition of the RFs acts as a “check-point” for the RFs before peptide release. Since the GGQ motif shows close interaction with domain II in the compact state, we asked whether GGQ methylation can influence the conformational transition of the RFs from the compact to the open conformation.
It has been shown the rate of the catalytic peptide release step by RFs escalates with an increase in pH until it becomes faster than the pH-independent compact to open transition step, which then becomes rate limiting (19). We conducted peptide release assays at different pH to find out at which pH the transition from pH-dependent to pH-independent kinetics happens in RFs in comparison to mRFs. Our results show that for both mRF1 and mRF2, the pH-independent “compact to open” transition step becomes rate limiting at pH higher than 7.9. In contrast, for unmethylated RF1 and RF2, the rate of peptide release keeps increasing in a pH-dependent manner even at pH 8.5. This means that for the unmethylated RFs, the catalytic step remains rate limiting. Thus, the lack of GGQ methylation likely induces error in RF1/2 by promoting easier transition to the open state, which allows them to bypass the “check-point,” irrespective of the cognate or near-cognate codon.
We further asked whether the lack of GGQ methylation perturbs the overall stability of the RFs. For that we performed thermal melting experiments with mRF2 and RF2 and followed the transition with CD and SYPRO-orange fluorescence. The Tms estimated from the transition midpoints for mRF2 was slightly higher than RF2 in both assays suggesting higher stability of the mRFs. However, much higher binding of SYPRO-orange to RF2 than the same concentration of mRF2 indicated that RF2 allows higher access to the dye to its hydrophobic patches. Thus, unmethylated RFs likely have lesser structural rigidity than mRFs, which may explain why the RFs undergo easier transition from compact to open conformation than the mRFs.
Recent report suggests that ribosomal dynamics during RF1 and RF2 interaction with RF3 play a major role in translation termination (40). It is known that following peptide release, RF3 mediates dissociation of RF1 and RF2 from the ribosome (41). Although most of the RF3 interactions are confined at domain I of RF1 and RF2 (42), it is an interesting question whether structural dynamics of RF1/RF2 can influence the process. Since our results indicate that GGQ methylation modulates structural flexibility of the RFs, whether or not it also has impact on the RF3-mediated steps subsequent to peptide release remains to be seen in future.
In summary, we suspect that a relatively less-rigid structural architecture of the RFs in the absence of GGQ methylation allows them to undergo easier transition from the compact to the open state, thereby compromising their accuracy for codon-specific peptide release. However, the accuracy-loss effects are mild compared with the loss of catalytic activity associated with the lack of GGQ methylation in RFs. Since prmC is not an essential housekeeping gene and its expression varies in different growth conditions, we suspect that GGQ methylation may work like a tool for regulation of speed and accuracy of translation termination, especially in the stress conditions. In a cell when survival becomes a big question, the unmethylated RFs probably manage to terminate translation even on the near-cognate codons with their compromised accuracy, which would rescue the ribosomes from sequestration. We propose that this can be one of the physiological implications of GGQ methylation in the bacterial RFs.
Experimental procedures
Buffer and the components of the translation system
70S ribosome and tRNAfMet were purified from bacteria Escherichia coli MRE600 using a standard protocol (43). The E. coli initiation factors (IF1, IF2, and IF3) and the elongation factors (EF-Tu, EF-Ts, and EF-G) were prepared from the respective C-terminal His-tagged constructs, cloned in pET21a or pET24b, overexpressed in E. coli BL21(DE3), and further purified using Nickel-affinity chromatography HisTrap HP(Cytiva, former GE Healthcare) as described earlier (43). The RFs and mRNA preparations are described below. All experiments were conducted at 37 °C in HEPES-polymix buffer (pH 7.5) containing 5 mM HEPES (pH 7.5), 5 mM Mg(OAc)2, 1 mM dithioerythritol (DTE), 5 mM NH4Cl, 0.5 mM CaCl2, 100 mM KCl, 1 mM spermidine, and 8 mM putrescine (44). The reaction mixes also contained energy pump components 1 mM ATP, 10 mM phosphoenol pyruvate (PEP), 0.05 mg/mL pyruvate kinase (PK), 0.02 mg/mL myokinase (MK) unless mentioned otherwise.
Preparation of methylated and unmethylated RFs
The prfA gene encoding RF1 and the prfB,246T>A gene encoding RF2Ala246 (referred in the manuscript only as RF2) were cloned into pET24a vectors, with C-terminal (His)6 tag. It has been shown that the C-terminal (His)6 tag does not affect the activity of the RFs in peptide release (13). These plasmids were transformed into E. coli BL21-Gold(DE3) strain for overexpression of the unmethylated RFs. For the methylated RFs, the plasmids were cotransformed with the plasmid pACYCDuet-1 prmC expressing HemK methyltransferase (16). The unmethylated and methylated RF1 and RF2 proteins were overexpressed by addition of 1 mM IPTG at OD600 = 0.6 at 37 °C for 3 h. The proteins were purified by affinity chromatography by using a HisTrap High-Performance column (Cytiva, former GE Healthcare) and stored in HEPES-polymix buffer (pH 7.5).
To confirm the state of methylation, the purified RF variants (RF1, RF2, mRF1, and mRF2) were treated with Trypsin/Lys-C Mix (Promega), subjected to mass spectrometry using LC-orbitrap MS/MS, and were analyzed using MaxQuant 1.5.1.2 software as well as Proteome Discoverer 1.4 (Thermo Fisher Scientific) at mass spectrometry–based proteomics facility at Uppsala University. The results summarized in Figs. S1 and S2 confirmed 100% methylation of mRF1 G235 and mRF2 G252 (Figs. S1A and S2A) and also complete absence of the methylation in RF1 and RF2 (Figs. S1B and S2B).
Preparation of the mRNAs
The reverse complementary oligo sequences were ordered from MERCK with sequences 5′- AAGCTTGAAATTAATACGACTCACTATAGGGAATTCGGGCCCTTGTTAACAATTAAGGAGGTATTAAATGCTGCTGTAAGAATTC-3′ and 5′-GAATTCTTACAGCAGCATTTAATACCTCCTTAATTGTTAACAAGGGCCCGAATTCCCTATAGTGAGTCGTATTAATTTCAAGCTT-3′.
The underlined part constitutes T7 promoter. The bold and underlined part of the mRNA sequence encodes Met-Leu-Leu-stop (UAA/UAG/UGA) or Trp (UGG) codon. The part in italics represents a strong Shine-Dalgarno sequence. The oligonucleotides (1 μg/μL) were mixed and heated at 94 °C for proper annealing. Later, the reaction was cooled to room temperature resulting in a double-stranded template for transcription. The mRNA was transcribed at 37 °C using T7 RNA polymerase and transcription buffer. The transcription buffer contains 200 mM HEPES (pH 7.5), 30 mM MgCl2, 30 mM DTE, 2 mM Spermidine, 2.5 mM rNTPs, and 5 μg DNA template. The transcribed mRNA is then extracted by phenol and chloroform (1:1) treatment. The extracted mRNA was precipitated with 75% ethanol for 2 h and then subjected to purification on HiLoad 26/60 Superdex 75 prep grade gel filtration column (45). The concentration of the purified mRNA was determined, and further, the activity of the mRNAs was determined by titrating the mRNAs in dipeptide formation assay as described in (46).
Preparation of BOP-Met-tRNAfMet
The fluorescence dye used in the assay, BODIPY 576/589 (BOP) was purchased from Thermo Scientific. Charging of Met-tRNAfMet was performed in the reaction with 80 μM tRNAfMet, 150 μM Met, 2 U/μL Met-tRNAMet synthetase in a buffer containing 30 mM HEPES (pH 7.5), 1X polymix, 1 mM DTE, 2 mM ATP, 20 mM PEP, 2 mM Mg(OAc)2, 0.05 mg/mL PK, 0.01 mg/mL MK. After incubation of the charging mix for 30 min at 37 °C, the charged tRNA was extracted by phenol and chloroform (1:1) treatment and later precipitated with ethanol. The precipitated tRNA was dissolved in water.
Labeling of the Met-tRNAfMet was performed in 100 mM NaHCO3 (pH 8.0) by mixing Met-tRNAfmet and BOP in excess, for overnight in dark at 4 °C. The labeled tRNA was extracted by phenol (pH 4.3) and chloroform (1:1) treatment, then precipitated with ethanol, and dissolved in ddH20.
The BOP-Met-tRNAfMet was purified using HPLC system (Waters) equipped with LiChospher WP 300 RP-18 column in line UV and fluorescence detectors. A linear gradient of 12% to 54% methanol in a buffer containing 20 mM NH4Ac (pH 5.0), 5 mM MgCl2, and 400 mM NaCl was used to elute BOP-Met-tRNAfMet. Fractions corresponding to the peak of UV (260 nm) and fluorescence (Ex. 565 nm, Em. 610 nm) were pooled together, concentrated in Amicon Ultra-15 centrifugal filters (10 kDa cutoff), and stored as aliquots at −80 °C.
Preparation of release complex (RC)
The RC, a stalled ribosome carrying a peptidyl tRNA with Met-Leu-Leu (MLL) tripeptide in the P site and a stop codon (UAA/UAG/UGA) in the A site, was prepared with the initiation mix (IM) and elongation mix (EM) in HEPES-polymix buffer (pH 7.5) containing energy pump components. IM contained 2 μM 70S ribosomes, 3 μM mRNA, 2 μM BOP-Met-tRNAfMet, 2 μM IF1, 4 μM IF2, and 2 μM IF3. EM contained 20 μM EF-Tu, 20 μM EF-Ts, 10 μM EF-G, 40 μM tRNALeu, 200 uM Leu, and 1 U/μL Leucyl tRNA synthetase. IM and EM were formed by incubating them at 37 °C for 15 min separately. The RC was formed by mixing IM and EM at 37 °C for 5 min and then quenched by putting in ice. Additional 4 mM Mg(OAc)2 was added to stabilize the RC, which was then purified by ultracentrifugation on a 37% sucrose cushion in a swing-out rotor (S55-S; Sorvall) at 258,000g for 2 h at 4 °C. The pellet containing RC was later dissolved in HEPES-polymix buffer (pH 7.5).
Stopped-flow-based fluorescent peptide release assay
The RCs with Met-Leu-Leu-XXX mRNA, where XXX refers to either stop codons (UAA, UGA, UAG) or a Trp codon (UGG), were prepared as described above. Equal volumes of RC and RF1/RF2 were mixed rapidly in a BioLogic (SFM-4000) micro stopped-flow instrument and the time course of BOP-Met-Leu-Leu tripeptide release was measured by following the change of BOP fluorescence (through a long-pass filter of 590 nm) as a function of time. No fluorescence change was observed when RF1/RF2 was not added confirming that the fluorescence change solely indicated release of the BOP-labeled tripeptide with the RFs (Fig. S3). All release experiments were conducted at least in triplicates and standard error of mean (SEM) was estimated.
Estimation of the kinetic and accuracy parameters
The rate of peptide release was estimated by fitting the fluorescence curves with double exponential decay function (y = A1∗exp(-x/t1) + A2∗exp(-x/t2) + y0) where y is the relative fluorescence; x is the time, A is the amplitude of the individual phases; t is the time constant; y0 is the offset, and 1/t is the rate constant (k). The fast phase accounts 80 to 90% of the total peptide release, whereas 10 to 20% of the peptide release happens in the slow phase, possibly due to unknown heterogeneity in the RC. The apparent rates of the fast phase for varying concentrations of RFs were plotted against RF concentration and fitted to the hyperbolic Michaelis–Menten equation from which the Michaelis–Menten parameters (kcat, KM, and kcat/KM) were estimated. The “A value” reflecting accuracy in peptide release by the RFs was derived from the equation A = (kcat/KM)cognate/(kcat/KM)near-cognate. Higher A value indicates higher accuracy. Alternatively, nondiscriminating codons produce A value close to 1.
Thermal melting of mRF2 and RF2 followed by CD and SYPRO-orange fluorescence
CD was used to monitor the change in the secondary structure of the RF2 variants on thermal denaturation. For that, 5 μM mRF2 or RF2 in Phosphate buffered saline (PBS) (pH 7.3) was added in a JASCO J-1500 CD spectrometer. The mean residue ellipticity at 220 nm (θ) was recorded with increase in temperature ranging between 14 °C and 95 °C ramping 0.5 °C at every 5 s. The fraction of the folded proteins was determined by the following equation:
where (θ)obs Temp is the ellipticity at a given temperature, (θ)unfolded is the ellipticity at highest temperature, and (θ)folded is the ellipticity at lowest temperature. The experiments were repeated twice and the melting temperature (Tm) was determined from the transition midpoint.
SYPRO-orange thermal shift assay is an extrinsic fluorescence-based assay to follow the tertiary structure of the proteins. The dye SYPRO-orange binds to the exposed hydrophobic patches of the proteins. Thus, the change in SYPRO-orange fluorescence with increase in temperature reflects access to the otherwise hidden hydrophobic patches during thermal melting. The assay was conducted in a BIO-RAD CFX connect Real-Time PCR system using 10 μM of the RF2s in PBS varying temperature from 15 to 95 °C ramping 1 °C at every 10 s. The first derivatives of the fluorescence scans at 569 nm were plotted against temperature. The Tms was determined from the peak values.
Data availability
All data generated in this study are presented in this article and/or available on request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Prof. Valerie Heurgué-Hamard, Institut de Biologie Physico-Chimique—Sorbonne Université, for the pACYCDuet-1 plasmid containing prmC gene expressing HemK methyltransferse; mass spectrometry–based proteomics facility of Uppsala University for the analysis of RF1 and RF2 methylation; Raymond Fowler for purification of E. coli translation components; Chandra Sekhar Mandava for standardizing the BOP labeling of tRNAfMet; and Soneya Majumder for help with the thermal melting experiments.
Author contributions
S. S. conceived the study, planned the experiments, acquired funding, and coordinated the project. S. P. purified the components and performed all the biochemical experiments. X. G. cloned the RF proteins and developed the fluorescence-based termination assay. S. S. and S. P. wrote the article. All the authors analyzed the results and contributed in the preparation of the article.
Funding and additional information
This work is supported by the grants from Swedish Research Council (grant numbers 2016-06264, 2018-05946 and 2018-05498), Knut and Alice Wallenberg Foundation (grant number KAW 2017.0055), Carl Trygger Foundation (grant numbers CTS 18:338 and CTS 19:806), and Wenner-Gren Stiftelserna (UPD2017:0238) to S. S.
Edited by Ruma Banerjee
Supporting information
References
- 1.Scolnick E., Tompkins R., Caskey T., Nirenberg M. Release factors differing in specificity for terminator codons. Proc. Natl. Acad. Sci. U. S. A. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konecki D.S., Aune K.C., Tate W., Caskey C.T. Characterization of reticulocyte release factor. J. Biol. Chem. 1977;252:4514–4520. [PubMed] [Google Scholar]
- 3.Freistroffer D.V., Pavlov M.Y., MacDougall J., Buckingham R.H., Ehrenberg M. Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frolova L.Y., Tsivkovskii R.Y., Sivolobova G.F., Oparina N.Y., Serpinsky O.I., Blinov V.M., Tatkov S.I., Kisselev L.L. Mutations in the highly conserved GGQ motif of class I polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawat U.B.S., Zavialov A.V., Sengupta J., Valle M., Grassucci R.A., Linde J., Vestergaard B., Ehrenberg M., Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 6.Klaholz B.P., Pape T., Zavialov A.V., Myasnikov A.G., Orlova E.V., Vestergaard B., Ehrenberg M., Van Heel M. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 7.Petry S., Brodersen D.E., Murphy F.V., IV, Dunham C.M., Selmer M., Tarry M.J., Kelley A.C., Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 8.Shaw J.J., Green R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol. Cell. 2007;28:458–467. doi: 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trobro S., Åqvist J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol. Cell. 2007;27:758–766. doi: 10.1016/j.molcel.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 10.Zeng F., Jin H. Conformation of methylated GGQ in the peptidyl transferase center during translation termination. Sci. Rep. 2018;8:2349. doi: 10.1038/s41598-018-20107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korostelev A., Asahara H., Lancaster L., Laurberg M., Hirschi A., Zhu J., Trakhanov S., Scott W.G., Noller H.F. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zavialov A.V., Mora L., Buckingham R.H., Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- 13.Mora L., Heurgué-Hamard V., Champ S., Ehrenberg M., Kisselev L.L., Buckingham R.H. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol. Microbiol. 2003;47:267–275. doi: 10.1046/j.1365-2958.2003.03301.x. [DOI] [PubMed] [Google Scholar]
- 14.Dinçbas-Renqvist V., Engström Å., Mora L., Heurgué-Hamard V., Buckingham R., Ehrenberg M. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graille M., Figaro S., Kervestin S., Buckingham R.H., Liger D., Heurgué-Hamard V. Methylation of class I translation termination factors: Structural and functional aspects. Biochimie. 2012;94:1533–1543. doi: 10.1016/j.biochi.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Heurgu V., Champ S. The hemK gene in Escherichia coli encodes the N5-glutamine methyltransferase that modifies peptide release factors. EMBO J. 2002;21:769–778. doi: 10.1093/emboj/21.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graille M., Heurgué-Hamard V., Champ S., Mora L., Scrima N., Ulryck N., Van Tilbeurgh H., Buckingham R.H. Molecular basis for bacterial class I release factor methylation by PrmC. Mol. Cell. 2005;20:917–927. doi: 10.1016/j.molcel.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 18.Nakahigashi K., Kubo N., Narita S.-I., Shimaoka T., Goto S., Oshima T., Mori H., Maeda M., Wada C., Inokuchi H. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl. Acad. Sci. U. S. A. 2002;5:1473–1478. doi: 10.1073/pnas.032488499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Indrisiunaite G., Pavlov M.Y., Heurgué-Hamard V., Ehrenberg M. On the pH dependence of class-1 RF-dependent termination of mRNA translation. J. Mol. Biol. 2015;427:1848–1860. doi: 10.1016/j.jmb.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Pierson W.E., Hoffer E.D., Keedy H.E., Simms C.L., Dunham C.M., Zaher H.S. Uniformity of peptide release is maintained by methylation of release factors. Cell Rep. 2016;17:11–18. doi: 10.1016/j.celrep.2016.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mora L., Heurgué-Hamard V., De Zamaroczy M., Kervestin S., Buckingham R.H. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J. Biol. Chem. 2007;282:35638–35645. doi: 10.1074/jbc.M706076200. [DOI] [PubMed] [Google Scholar]
- 22.Uno M., Ito K., Nakamura Y. Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie. 1996;78:935–943. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura Y., Ito K., Uno M. A tripeptide “anticodon” deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 24.Sund J., Andér M., Åqvist J. Principles of stop-codon reading on the ribosome. Nature. 2010;465:947–950. doi: 10.1038/nature09082. [DOI] [PubMed] [Google Scholar]
- 25.Korkmaz G., Sanyal S. R213I mutation in release factor 2 (RF2) is one step forward for engineering an omnipotent release factor in bacteria Escherichia coli. J. Biol. Chem. 2017;292:15134–15142. doi: 10.1074/jbc.M117.785238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vestergaard B., Van L.B., Andersen G.R., Nyborg J., Buckingham R.H., Kjeldgaard M. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- 27.Shin D.H., Brandsen J., Jancarik J., Yokota H., Kim R., Kim S.H. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J. Mol. Biol. 2004;341:227–239. doi: 10.1016/j.jmb.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 28.Weixlbaumer A., Jin H., Neubauer C., Voorhees R.M., Petry S., Kelley A.C., Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korostelev A., Zhu J., Asahara H., Noller H.F. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–2585. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laurberg M., Asahara H., Korostelev A., Zhu J., Trakhanov S., Noller H.F. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 31.Jin H., Kelley A.C., Loakes D., Ramakrishnan V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8593–8598. doi: 10.1073/pnas.1003995107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Z., Indrisiunaite G., Kaledhonkar S., Shah B., Sun M., Chen B., Grassucci R.A., Ehrenberg M., Frank J. The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy. Nat. Commun. 2019;10:2579. doi: 10.1038/s41467-019-10608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svidritskiy E., Korostelev A.A. Conformational control of translation termination on the 70S ribosome. Structure. 2018;26:821–828. doi: 10.1016/j.str.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svidritskiy E., Korostelev A.A. Mechanism of inhibition of translation termination by Blasticidin S. J. Mol. Biol. 2018;430:591–593. doi: 10.1016/j.jmb.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svidritskiy E., Demo G., Loveland A.B., Xu C., Korostelev A.A. Extensive ribosome and RF2 rearrangements during translation termination. Elife. 2019;8 doi: 10.7554/eLife.46850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trappl K., Joseph S. Ribosome induces a closed to open conformational change in release factor 1. J. Mol. Biol. 2016;428:1333–1344. doi: 10.1016/j.jmb.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 37.Freistroffer D.V., Kwiatkowski M., Buckingham R.H., Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavlov M.Y., Freistroffer D.V., Dincbas V., MacDougall J., Buckingham R.H., Ehrenberg M. A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol. 1998;284:579–590. doi: 10.1006/jmbi.1998.2220. [DOI] [PubMed] [Google Scholar]
- 39.He S.L., Green R. Visualization of codon-dependent conformational rearrangements during translation termination. Nat. Struct. Mol. Biol. 2010;17:465–470. doi: 10.1038/nsmb.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adio S., Sharma H., Senyushkina T., Karki P., Maracci C., Wohlgemuth I., Holtkamp W., Peske F., Rodnina M.V. Dynamics of ribosomes and release factors during translation termination in E. coli. Elife. 2018;7 doi: 10.7554/eLife.34252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zavialov A.V., Buckingham R.H., Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 42.Pallesen J., Hashem Y., Korkmaz G., Koripella R.K., Huang C., Ehrenberg M., Sanyal S., Frank J. Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1. Elife. 2013;2 doi: 10.7554/eLife.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang C., Mandava C.S., Sanyal S. The ribosomal stalk plays a key role in IF2-mediated association of the ribosomal subunits. J. Mol. Biol. 2010;399:145–153. doi: 10.1016/j.jmb.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Koripella R.K., Holm M., Dourado D., Mandava C.S., Flores S., Sanyal S. A conserved histidine in switch-II of EF-G moderates release of inorganic phosphate. Sci. Rep. 2015;5:12970. doi: 10.1038/srep12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge X., Mandava C.S., Lind C., Åqvist J., Sanyal S. Complementary charge-based interaction between the ribosomal-stalk protein L7/12 and IF2 is the key to rapid subunit association. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4649–4654. doi: 10.1073/pnas.1802001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holm M., Borg A., Ehrenberg M., Sanyal S. Molecular mechanism of viomycin inhibition of peptide elongation in bacteria. Proc. Natl. Acad. Sci. U. S. A. 2016;113:978–983. doi: 10.1073/pnas.1517541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are presented in this article and/or available on request.