Summary
Cancer cells acquire genotypic and phenotypic changes over the course of the disease. A minority of these changes enhance cell fitness, allowing a tumor to evolve and overcome environmental constraints and treatment. Cancer evolution is driven by diverse processes governed by different rules, such as discrete and irreversible genetic variants and continuous and reversible plastic reprogramming. In this perspective, we explore the role of cell plasticity in tumor evolution through specific examples. We discuss epigenetic and transcriptional reprogramming in “disease progression” of solid tumors, through the lens of the epithelial-to-mesenchymal transition, and “treatment resistance”, in the context endocrine therapy in hormone-driven cancers. These examples offer a paradigm of the features and challenges of cell plastic evolution, and we investigate how recent technological advances can address these challenges. Cancer evolution is a multi-faceted process, whose understanding and harnessing will require an equally diverse prism of perspectives and approaches.
Subject areas: Evolutionary Biology, Cancer
Evolutionary Biology; Cancer
Introduction
Human solid tumors can be described as a population of molecularly and/or phenotypically heterogeneous cancer cells. The genetic component of this intrinsic diversity was initially defined as intra-tumor heterogeneity (ITH) (Greaves and Maley, 2012; Swanton, 2012). ITH has been studied through sampling multiple regions (Jamal-Hanjani et al., 2017; Turajlic et al., 2018) or even single cells (Gaiti et al., 2019; Klein et al., 2002; Suva et al., 2013) from the same tumor. These studies demonstrated the existence of multiple cancer cell populations (or clones) sharing common molecular features, proof of a common ancestor, but also harboring distinctive alterations acquired during tumor progression. In the context of species evolution, analysis of common and distinct traits allowed tracing the evolutionary history of different species, i.e., their phylogeny. Similar approaches applied to ITH can be used to reconstruct cancer evolution in individual patients (Schwartz and Schäffer, 2017). Tracing cancer evolution in human tumors has so far mostly focused on DNA mutations (here intended with the broad meaning of alterations of the DNA sequence) treated as “passive”, “discrete”, “irreversible”, and “inheritable” molecular features. From this perspective, models inspired by Darwinian species evolution have provided a suitable framework to study cancer evolution: DNA mutations passively and randomly emerge as a consequence of diverse mutational processes (Degasperi et al., 2020), by chance a minority will provide some sort of advantage to the cells where they occurred (generally thought as a proliferative or survival advantage), which will outgrow the siblings lacking the advantageous mutations (Greaves and Maley, 2012). Based on this model, cancer evolution is thus determined by random emergence and functional selection of somatic DNA mutations. Although powerful, this model has a fundamental limitation: genotype is not necessarily phenotype.
The proper homeostasis of the human body, intended as coordinated interactions of specialized cell communities, is a reflection of pervasive phenotypic heterogeneity. As such, human cells can access a series of mechanisms which allow inheritable phenotypical changes in the absence of somatic DNA changes. Cancer originates from cells which share this multicellularity heritage but proliferate via asexual cell division. In this context, vertical transmission through the germline is bypassed, and epigenetic modifications become the primary constrain on cell identity but also function as an inheritable unit of information which can be actively encoded (as a direct response to microenvironment cues) and transmitted from mother to daughter cells. Therefore, in cancer, ITH is not exclusively a genetic phenomenon, and cancer cell populations within a tumor often exhibit great epigenetic and transcriptional diversity (Filbin et al., 2018; Gaiti et al., 2019; Landau et al., 2014; Patel et al., 2014; Puram et al., 2017; Tirosh et al., 2016). Histone modifications and DNA methylation can indeed sustain inheritable and heterogeneous gene expression among cancer cells that can be in itself a substrate for Darwinian selection. Additionally, epigenetic reprogramming can result in changes of cell state, sometimes identity, and enable “cell plasticity”. Cell plasticity is the ability of a cell to change its identity by altering its differentiation status and/or lineage commitment (Suva et al., 2013). Importantly, cell plasticity is not necessarily driven or associated with specific genetic modifications and is a reversible process. Plastic reprogramming in cancer has been categorized into two broad classes: “de-differentiation”, i.e., the acquisition by a fully differentiated cell of features observed in progenitor or stem cells, and “trans-differentiation”, i.e., the acquisition by a cell of a given lineage of features characteristic of a distinct lineage (Figure 1A) (Magnen et al., 2017). In contrast to DNA mutations, plastic reprogramming can hardly be categorized into discrete units (or phenotypes), and it can be reverted, and its emergence is often not random, as it can be induced by intrinsic or extrinsic factors, such as interactions with the tumor microenvironment or drug treatment (Figure 1B). Nonetheless, similarly to DNA mutations, cell plasticity influences the evolution of the disease through heritable transcriptional and/or epigenetic changes. In this perspective, we will discuss a few representative examples of epigenetic heterogeneity and cell plasticity, which exemplify these features, and we will examine the molecular tools that can be used to deconvolute plasticity and explore its role in cancer evolution and adaptation.
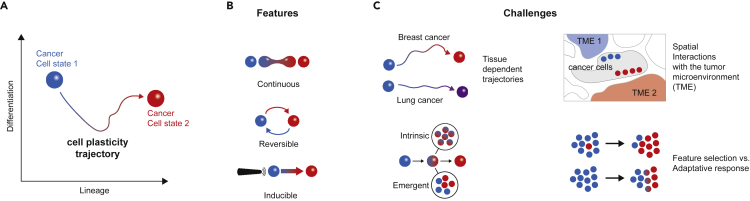
Figure 1.
Cancer cell plasticity and cancer evolution
(A) Schematic representation of the space of cell plasticity trajectories: a cancer cell (colored sphere) moves along two major axes: differentiation status (Y axis) and lineage commitment (X axis).
(B) Cell plasticity features. Change of cell state for a cancer cell is color coded (initial state: blue, new state: red).
(C) Challenges to address in the study of cell plasticity.
The plastic identity of cancer cells
The most evident and possibly most studied example of “cancer cell plasticity” is the epithelial-to-mesenchymal transition (EMT) and its reverse, mesenchymal-to-epithelial transition (MET). The dynamic acquisition (and loss) of epithelial and mesenchymal markers is a key property of cells during embryonic development and wound healing (Park et al., 2018). Therefore, cell plasticity is pre-built in the system as a required physiological process that can be hijacked by cancer cells. Downregulation of the epithelial marker and adherent junction molecule E-cadherin leads to loss of cell adhesion and together with expression of mesenchymal markers (such as vimentin and N-cadherin) is associated with increased migratory potential. This program is frequently rebooted by cancer cells where it increases their invasive and possibly metastatic potential (Thiery, 2002), although the latter has sometimes been debated (Bornes et al., 2019; Fischer et al., 2015; Stanger et al., 2017). In epithelial tumors, EMT features are associated with tumor grade, discriminating between poorly and well-differentiated histology. Interestingly, during cancer evolution, EMT is possible and has been studied at multiple phases of disease progression, from initiation (Rhim et al., 2012) to metastatic invasion. Through these phases, partial EMT states have been documented, with cells concurrently exhibiting epithelial and mesenchymal markers (Brabletz et al., 2018). These intermediate and heterogeneous states highlight the non-discrete nature of EMT.
Importantly, EMT is “reversible”, and the same cell lineage can undergo EMT and MET; it is not associated with the acquisition of specific genetic alterations, but it involves complex epigenetic and transcriptional reprogramming that can be induced by interactions with the tumor microenvironment (Hanahan and Weinberg, 2011; Thiery, 2002; Tsai and Yang, 2013). EMT plastic reprogramming involves the coordinate actions of several transcription factors (TFs) (most notably Snail, Zeb, and Twist), whose activation is coordinated and not necessarily simultaneous. For example, mutually exclusive and subsequent expression of Snail and Prrx1 has been show to first repress epithelial markers (by Snail) and then activate mesenchymal markers (by Prrx1) (Fazilaty et al., 2019). It should be noted that cancer cells from different tissues have been shown to express different EMT-associated TFs and EMT dependencies. For example, Snail and Zeb have been shown to have a different role in breast and pancreatic tumors (Brabletz et al., 2018). Transcriptional changes during EMT are tightly associated with epigenetic ones, especially concerning histone post-translational modifications (McDonald et al., 2017; Peixoto et al., 2019). Here, tissue-specific chromatin accessibility might confer different EMT potentials associated with the cell of origin. For example, squamous cell carcinomas of the skin driven by KRAS oncogenic mutations and p53 loss led to epithelial tumors when originating from the interfollicular epidermis, whereas acquired mesenchymal features when derived from hair follicles (Latil et al., 2017). In these contexts, bivalent chromatin states might be indicative of plastic potential. Indeed, concurrent activating and repressing histone modifications have been reported at promoters of both Snail (Fazilaty et al., 2019) and Zeb (Chaffer et al., 2013). The extent to which bivalent marks characterize other critical genes involved in cancer progression is currently unknown, but it is tempting to speculate that a similar arrangement could offer a great deal of flexibility to cancer cells in time of need. Overall, altered histone modifications and transcriptional programs driven by microenvironmental cues, e.g., as a result of inflammation, or stochastic epigenetic changes lead to heterogeneous cell states and tissue dependencies. This heterogeneity makes it hard to univocally define the EMT transition.
Consistent with a progressive rewiring of epigenetic and transcriptional programs orchestrated by multiple factors, EMT does not occur as a switch from one stable state to another, i.e., it cannot be modeled as a discrete or, even less, binary event. In recent years, this process has been shown to encompass a continuum of cell states between two archetypal epithelial and mesenchymal cell identities (Park et al., 2018; Zhang and Weinberg, 2018). Gene expression signature approaches have been proposed to quantify this continuum (Chakraborty et al., 2020; Li et al., 2017). However, when applied to bulk RNA-seq, these approaches face two main challenges: (1) expression of EMT markers might come from tumor and non-tumor cells (e.g. cancer-associated fibroblasts) and the latter might lead to misinterpret the state of the former (Li et al., 2017), and (2) it is impossible to determine whether a given EMT score reflects the intrinsic state of individual tumor cells or the average state emerging from a heterogeneous cancer cell population. Indeed, single-cell analyses by either RNA-seq or mass cytometry data have confirmed heterogeneous EMT states within individual tumors (Karacosta et al., 2019; McFaline-Figueroa et al., 2019; Puram et al., 2017), and such heterogeneity was also observed spontaneously emerging in vivo (Beerling et al., 2016). Similarly, specific chromatin states, such as bivalent promoters, will need to be assessed in single cells (Cheung et al., 2018) to determine when these are intrinsic or emergent properties of the cancer cell population. Induction of EMT in distinct cancer cell lines has been recently shown to drive heterogeneous transcriptional changes, in a tissue-dependent manner, although converging on EMT-like cell phenotypes (Cook and Vanderhyden, 2020). These results challenged again the notion of a universal, i.e. tissue independent, EMT program. Hence, similarly to heterogeneous genetic variants converging on deregulating the same pathway (Sanchez-Vega et al., 2018), the same plastic phenotype might be implemented through heterogeneous transcriptional and epigenetic modifications.
EMT in cancer has been directly linked with the emergence of cancer stem cells (CSCs) (Shibue and Weinberg, 2017). CSCs have been defined as tumor cells with unlimited self-renewal and tumor initiating/regenerating capacity, although their molecular features may differ across tumor types (Wainwright and Scaffidi, 2017). Initial CSC models posited a unidirectional differentiation trajectory: CSCs give rise to both CSCs and non-CSCs, vice versa the differentiated offspring of CSCs cannot return into a stem-like state. Recent evidence disproved this theory. Indeed, similarly to normal differentiated cells (Takahashi and Yamanaka, 2006), intrinsic and extrinsic cues can lead more differentiated tumor cells to acquire stem features and generate novel CSCs (Chaffer et al., 2013; Guen et al., 2017; Li et al., 2019; Mani et al., 2008). This de-differentiation trajectory has been shown to partly overlap with EMT, as exemplified by Zeb transcriptional program and Wnt signaling (Shibue and Weinberg, 2017). Intriguingly, given the continuum of cell states that defines EMT, the CSC model might actually encompass a similar continuum of differentiation states, which are for the most part yet to be characterized.
Instances of plastic evolution have been observed in multiple tumor types, often associated with morphological changes of cancer cells. In lung adenocarcinomas, the tumor tissue architecture is often heterogeneous, especially in early-stage diseases, and it gives rise to histologic patterns recognized by histopathology analyses (Travis et al., 2015). Recent evidence from histopathology-guided multiregional sampling has shown that this heterogeneity does not correspond to emergence of specific genetic variants, but it rather reflects epigenetic and transcriptional reprogramming associated with cell de-differentiation and increased proliferation (Tavernari et al., 2021). Additional examples include reprogramming to a progenitor state of luminal breast cancer cells (Domenici et al., 2019; Poli et al., 2018), intermixing and transition between differentiated and progenitor states in melanoma (Cheli et al., 2011; Roesch et al., 2010), epithelial plasticity in pancreatic cancer (Genovese et al., 2017; Reichert et al., 2018), de-differentiation trajectories in glioblastoma (Gallo et al., 2015), or lineage reprogramming from adeno to neuroendocrine carcinoma in prostate and lung cancer (Park et al., 2018), which, similarly to EMT, shows evidence of a continuum of states along which individual tumors may transition and evolve (Ireland et al., 2020). Several examples of plastic reprogramming have been associated with cancer progression; however, pre-cancerous lesions can arise through similar processes. Indeed, injury or inflammation can lead to change of cell identity and organization of normal tissues, a condition referred to as “metaplasia”. Metaplasia is accompanied by profound epigenetic changes and increased risk of cancer development, potentially by favoring activation and tolerance of oncogenic signaling (Yuan et al., 2019). A notable example of cancer predisposing metaplasia is Barrett esophagus. Here, inflammation driven by gastroesophageal reflux leads to trans-differentiation of esophageal squamous cells into columnar cells (Que et al., 2019) and increased risk of developing esophageal carcinoma. Although driven by different transcriptional programs, often dependent on the tissue or cell of origin, all these instances of cell plasticity involve de-differentiation and trans-differentiation trajectories (Figure 1A). These trajectories represent therefore a common denominator of cell plasticity and determine phenotypic heterogeneity among cancer cells in a rather dynamic and reversible manner, enabling rapid adaptation during the evolution of the disease.
Under the light of these findings, the epigenetic landscape originally proposed by Conrad Waddington in 1957 (Goldberg et al., 2007) appears flatter than initially imagined for many malignancies. Pushed by intrinsic and extrinsic forces, cancer cells move in a space whose dimensions are broadly defined by differentiation status and lineage commitment (Figure 1A) but whose features vary among tumor types and evolutionary pressures. In this context, EMT can be thought as a subgroup of trajectories along these dimensions. Importantly, plasticity features are likely to depend on the cell of origin, oncogenic variants, interactions with the tumor microenvironment, and therapeutic intervention. In addition, as a cancer cell population evolves toward the achievement of different objectives (Hanahan and Weinberg, 2011), evolutionary trade-offs might be required, e.g., between cell growth and survival (Hausser et al., 2019; Shoval et al., 2012). These objectives are likely to change during tumor progression requiring adaptive reprogramming of gene expression (Aktipis et al., 2013). This is particularly critical for treatment efficacy. Indeed, cell plasticity programs, such as EMT and CSC, have been for long associated with resistance to therapy (Shibue and Weinberg, 2017) (Byers et al., 2013), and treatment strategies are in development and trial to prevent the adaptive transitions (Sheridan, 2013). Overall, to understand the contribution of cell plasticity to cancer evolution, several questions remain to be answered (Figure 1C): How does plastic reprogramming depend on the tumor cell of origin and/or selected genetic variants? Are plastic phenotypes the result of intrinsic cell changes or emergent properties of a heterogeneous cell population? Do they correlate with spatial feature and interactions? Finally, is cell plasticity the result of feature selection or adaptive response? Critically, addressing these questions will not only inform on tumor evolution but also help us understand how cell plasticity influences response to therapy.
Evolutionary trajectories under clinical intervention
Systemic therapies are essential for the management of patients with cancer. Their goal is to prevent or delay tumors progression, often by targeting cancer-specific vulnerabilities (personalized medicine). This creates significant bottlenecks which cancer cells have to evade (Acar et al., 2020). Therefore, we believe it is essential to differentiate between the evolutionary ark of cancer before and after treatment, with a particular consideration on the anticipated effect of the therapy on individual cancer cells within the tumor ecosystem. Most patients with solid tumors undergo surgery with curative intent, but almost all cancer types receive additional layers of treatments. Some treatment modalities (surgery and radiotherapy) aim to physically eradicate cancer cells, thus precluding future adaptation/evolution. On the other hand, the vast majority of neo-adjuvant (before surgery) or adjuvant (after surgery) intervention relies on the systemic delivery of therapeutics. The ambition of precision medicine is to develop chemotherapy strategies that restrict “collateral casualties”, by leveraging cancer-specific functional, or “driver”, alterations. In the last 20 years or so, the field has focused mostly on genetic drivers with the hope of developing an array of silver bullets. Unfortunately, only a minor set of cancer drivers can be targeted (Tannock and Hickman, 2016), and the benefit of targeted therapies is invariably limited in time. Indeed, genetic ITH and evolution foster disease relapse through either pre-existing or emergent mechanisms of resistance (Tannock and Hickman, 2016). Efforts such as TRACERx (Jamal-Hanjani et al., 2017) aim at tracking genetic evolution of cancers treated with targeted therapies, with the hope of predicting resistance and eventually anticipate it altogether (Turajlic et al., 2019). Yet, we believe it is important to highlight that genetic heterogeneity is not the only player in the evolutionary arm race between the oncologist and the disease. Cancers might be thought as populations of single cancer cells sharing a multicellular past: each cancer cell has been developed via a series of plastic developmental stages before the onset of the tumor, and it is unlikely that these memories are completely eradicated by tumor initiation. Multicellularity and cell identity are governed by epigenetic processes and chemical modifications decorating the DNA and associated with inheritable gene expression (Consortium et al., 2015). Of note, epigenetic processes are perfectly suited for quick and stable phenotypic switches during normal development, even if epigenetic modifications are by definition reversible. It is worth mentioning that epigenetic changes have been linked to trans-generational effects in response to sudden and drastic changes in the environment such as famine in humans (Horsthemke, 2018). Therefore, transcriptional plasticity and epigenetic memory represent two instruments cancer cells inherit from their cell of origin and are likely to play a role in adaptation and evolutionary processes in response to rapid disruptive changes in the tumor microenvironment (therapy and metastasis).
Several groups have now started to explore how cancer cells treated with targeted therapies might exploit developmental strategies to “persist and adapt” to treatment in addition to classical mutational processes (Acar et al., 2020; Echeverria et al., 2019; Gaiti et al., 2019; Hinohara et al., 2018; Hong et al., 2019; Kim et al., 2018; Sharma et al., 2010). These mechanisms might explain, for example, the evolutionary trajectories of a vast subset of patients diagnosed with hormone-dependent cancers (breast and prostate cancers) where relapsing tumors can occur decades after surgery (Pan et al., 2017). In these instances, expansion via Darwinian selection of genetically defined clones appears at odds with clinical records.
Nongenetic adaptation in dormant cells
A striking example of this paradigm comes from the study of patients with estrogen receptor-positive (ERα) breast cancer. ERα breast cancer is the most common form of the disease (70%), and it is generally treated with surgery. These patients then receive various forms of adjuvant targeted therapies (endocrine treatment) designed to de-activate ERα, the hormone receptor driving gene proliferation in these cells in disseminated tumors cells, which are thought to leave the primary tumor from the early stages of transformation (Hüsemann et al., 2008; Klein, 2009; Sosa et al., 2014). Of note, the receptor is rarely mutated at the onset of the disease (~1%), providing an interesting example of a targeted treatment designed against a developmental program rather than an actionable mutation. In only about 40% of returning tumors, the field has managed to identify genetic events possibly linked to treatment resistance (Razavi et al., 2018), and even in these cases, there was little evidence for selective processes favoring de novo pre-existent genetic clones, strongly suggesting that resistance in most instances is acquired. These observations lead to a critical question: since endocrine therapies act mostly in a cytostatic fashion (i.e. inducing/extending dormancy), how do they acquire mutations in absence of significant cycling? One possibility could be stress-induced adaptive mutability (Cipponi et al., 2020; Russo et al., 2019), a situation in which cells transiently lower DNA repair and increase DNA replication errors. Nevertheless, it is not known if these processes are directly responsible in originating resistance-associated mutations, and the clinical timeline of relapse does not appear to fit with this mechanism. A second possibility is that cells undergo nongenetic phenotypic transitions while dormant.
Several groups including ours have hypothesized that acquired resistance has significant nongenetic components. For example, in vitro studies have highlighted how epigenetic reprogramming occurs during the acquisition of resistance (Hinohara et al., 2018; Magnani et al., 2013; Nguyen et al., 2015; Perone et al., 2019). These studies provide a theoretical framework for adaptation, where only certain epigenetic clones could efficiently enter in a dormant state and begin an adaptive journey toward epigenetic-mediated awakening. Currently, we do not know if dormancy is one epigenetic state, a set of directional transition similar to normal development, or a continuum of states navigated stochastically by cancer cells. In one particular model, epigenetic awakening is ascribed to metabolic rewiring, with cells acquiring the ability of autonomously producing cholesterol ligands carrying estrogenic activity (Nguyen et al., 2015; Perone et al., 2019; Perone and Magnani, 2016; Simigdala et al., 2016). Epigenetic reprogramming has been also documented in vivo by comparing patients with primary and metastatic ERα breast cancer (Patten et al., 2018). Many in vitro studies have also repeatedly shown that resistance emerges only after months of treatment (Aguilar et al., 2010; Yue et al., 2003), strongly implicating the existence of treatment-induced dormancy (Aguirre-Ghiso, 2007). One of the immediate outcomes of these observations is that non-cycling cells would involve a dramatically reduced ability of cementing new mutations, similar to lung quiescent cells (Yoshida et al., 2020). It is less clear however how dormancy would impact on DNA damage and if adaptive mutability (Russo et al., 2019) could still occur during extended G0-G1 intervals.
These studies however could not determine if the changes in the epigenome leading to transcriptional changes and cell identity switch were truly induced by the treatment (true plasticity) or are more accurately modeled by Darwinian selection of epigenetic subpopulations. This limitation is exacerbated by the nature of these studies which tend to measure the start and the end of the adaptive journey. With the advent of single-cell technologies, this question is finally becoming accessible, by mapping epigenetic and transcriptional features longitudinally, in single lineages within a population under therapeutic stress (see next section for technical details). Using some of these emerging approaches, it has been shown that transcriptional (nongenetic) heterogeneity is common even in ERα breast cancer cell lines cultured in the absence of therapeutic challenges (Chen et al., 2020; Hong et al., 2019). More importantly, these studies have shown that the initial phases of drug resistance invoke the selection of a well-defined set of cells, which rely on a dormant developmental program to avoid cell death. The transcriptome of these “pre-adapted cells” is nevertheless vastly different from the final proliferative stages of resistance and lacks the mutational features that characterize these latter stages (Magnani et al., 2017). Altogether, these reports suggest that ERα breast cancer cells might adapt to treatment, while being dormant at a minimal residual disease stage, initially exploiting nongenetic mechanisms. These data advocate for a refined mutational timeline for hormone-dependent cancers, where resumed proliferation (awakening) would re-instate more conventional mutational processes that could feed classical Darwinian evolution (Figure 2).
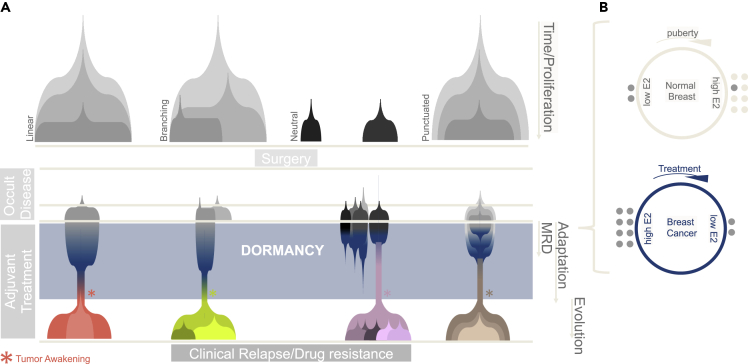
Figure 2.
Dormancy in the context of clonal evolution
(A) Many alternative models have been proposed to explain emergent genetic heterogeneity commonly observed in newly diagnosed cancers (gray subclones). By definition, curative surgery would remove the substrate for future evolution in the primary lesion. Further evolution of minimal residual disease (MRD) might be dependent on “nongenetic mechanisms”, especially under cytostatic treatment (gray→color transition). In this particular example, the color gradient represents phenotypic adaptation (multiple state transitions, i.e., dormancy entrance to tumor awakening) which can occur while mutational processes and generation of additional ITH are disfavored. Once cells resume proliferation, classical clonal evolution resumes on the backbone of a reprogrammed epigenome.
(B) An example of nongenetic adaptation is the selection/induction of dormancy under therapeutic stress. During periods of low circulating hormone (before puberty and after menopause), estrogen-dependent cells appear to survive in a prolonged dormancy status (dark gray dots). We hypothesize that the transcriptional programs underlying endocrine therapies, which effectively mimic hormone deprivation, might possibly be a reboot of tissue-specific developmental pathways (i.e. preservation of hormone-sensing ERα during periods of low hormone circulation via induced/selected dormancy).
Is this paradigm confined to patients with ERα breast cancer and endocrine therapies? And how does adaptation play out? The idea of drug-tolerant persister cells (conceptually compatible with the idea of dormant), i.e., cells characterized by adaptive features that transcend simple genetic mechanisms, has been scrutinized in several other cancer types. For example, Sharma and colleagues described quite some time ago how PC9 lung NSCLC cancer cells contain a persister population that plays a central role in the development of resistance toward a targeted EGFR inhibitor (gefitinib) (Sharma et al., 2010). More recently, a similar phenomenon was documented in a melanoma model system (Shaffer et al., 2017). Shaffer and colleagues show how transient transcriptional programs associated with BRAF inhibitor (vemurafenib) ultimately can be fixed and inherited. In this case, the cell of origin of the resistant epigenetic clone did not show a classical dormant/persister phenotype raising an interesting point: What is the balance between stable/inheritable and transient/reversible transcriptional and epigenetic programs associated with de novo drug resistance? Is it possible that epigenetic inheritance and stochastic fate switching depend on cell or tissue of origin? For example, it is conceivable that endocrine treatments might induce a clinical response down to a minimal residual disease approaching the development stage of adult breast progenitor cells (ERα positive, capable of sensing estradiol but quiescent in hormone-deprived animals: pre-puberty [Dall et al., 2018]). These ERα cells are developmentally programmed to respond to hormonal chances and contribute to continuously remodeling the breast in response to developmental changes (after puberty, during menstrual cycle, pregnancy, and after labor), while they must also be capable of withstanding a long period of physiological hormone deprivation (before puberty and possibly after menopause Figure 2B). Individuals with ERα breast cancer might have variable numbers of cells capable of returning to a dormant, progenitor cell phenotype which could progressively adapt to the new conditions until these cells awaken and resume proliferation and mutational processes. On the other hand, non-inheritable plasticity might also occur in certain contexts, with therapy tilting the balance of coordinated TF levels up until the fixation of a drug-resistant phenotype (Shaffer et al., 2017). Lineage tracing transcriptional profile and single-cell longitudinal assays will surely advance our understanding of these processes. Additionally, introducing these molecular strategies in clinically relevant models, especially in patient-derived organoids or xenografts, will potentially reveal nongenetic vulnerabilities which are currently underappreciated considering the strong focus on actionable mutations.
Single cell in the fourth (time) dimension
Evolutionary studies focusing on genetic events can leverage the waves of expansion and contraction of single nucleotide variants in the cell population to reconstruct the history and possibly predict the future of the disease (Werner et al., 2020). DNA changes can be accurately tracked by next-generation sequencing. High-depth sequencing of (possibly) multiple samples per tumor allowed us to accurately reconstruct the history of a tumor represented by clone phylogenies (Williams et al., 2018). Within this context, a clone can be defined as a population of genetically identical cells or cells exhibiting the same set of functional mutations. Under the (reasonable) assumption that DNA mutations are discrete and irreversible, it is possible to time these events from the most recently acquired back to the last emerging clonal mutation, i.e., observed in all tumor cells.
The non-discrete and reversible nature of transcriptional and epigenetic changes makes similar analyses and definitions considerably more challenging, if not impossible. How can a “clone” be defined from dynamic and reversible features? As discussed in the previous sections, cell populations found in distinct meta-stable states by single-cell experiments could fit the definition. Although, these should be considered as distinct populations (or “clones”) only at the specific time point of the measurement (Patten et al., 2018). Indeed, gene transcription is in itself a continuous process, in which genes can be expressed at variable levels, for certain period of times, and the mRNA degraded at variable rates (Fukaya et al., 2016; Larsson et al., 2019; Rodriguez et al., 2018). Even in post-mitotic, well-differentiated tissues, it is expected to observe oscillating changes in gene expression when sampling many thousands of cells. Hence, apparently distinct clones at a given time point might actually reflect transition states of a dynamically changing population of cells.
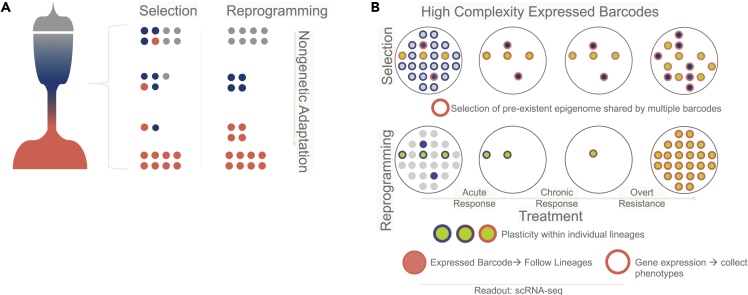
To track dynamic evolution of transcriptionally defined cell populations, an important advance is the recent implementation of genetic (Ba et al., 2019) or lentiviral delivery-based (Biddy et al., 2018) barcode strategies. These approaches can be coupled with transcriptomics profiles and modified fluctuation analysis (e.g. Memory-seq (Shaffer et al., 2018)) to either directly (transcribed barcodes) or indirectly (genetic barcodes) monitor the transcriptomes of individual lineages at different time points. Importantly, these technologies might allow us to address what is in our opinion a central issue in the field: measure and disentangle the contribution of feature selection and plastic reprogramming in cancer evolution. In principle, by coupling lineage tracing with single-cell RNA-seq profiles of longitudinal samples, it could be possible to estimate whether transcriptional reprogramming observed at the population level is driven by selection of a subpopulation of cells (barcodes tracking persistent phenotypes) or plastic reprogramming within a cell population (modified transcriptional program of cells with identical barcodes) (Figure 3). These strategies must be coupled with epigenetic annotation, which might mitigate some of the issue related to transcriptional noise while providing evidence on how epigenetic memory is transferred through the population. One of the first attempts in this direction leveraged the notion that histone occupancy at a given time has a discrete nature (normally, one histone for each DNA copy) (Patten et al., 2018). Since epigenetic modifications are thought to contribute to inheritable transmission of transcriptional statuses, they are considered more stable and potentially trackable. Patten and colleagues have exploited these concepts to deconvolute bulk tumor ChIP-seq signal for the histone 3 lysine 27 acetylation mark into clonal and subclonal signals and showed that epigenetic clonal landscapes of tumors evolve during progression and treatment (Probert and Curtis, 2018). While most epigenetic analyses are not yet available at the single-cell level, chromatin occupancy is (scATAC-seq). Tracking adaptation via scATAC-seq analysis might reveal the transcriptional driver of state transitions and measure epigenetic memory in adapting cells. A recent study on lung cancer progression aptly shows the feasibility of these strategies (LaFave et al., 2020).
Figure 3.
New technologies can disentagle nongenetic selection from trancriptional reprogramming
(A) Nongenetic inheritance can operate as classical genetic Darwinian selection (i.e. a fitter phenotype is randomly generated before a change in the environment and sweeps through the population) or could be interpreted in a more Lamarckian way (i.e. a change in the external environment elicits a series of coordinate transcriptional responses which can then be fixed and inherited via epigenetic changes).
(B) Feature selection or adaptive response (or a combination of thereof) can be tracked and traced by transforming classical genetic barcodes into transcriptional barcodes (inner colors) which can be captured at the same time of single-cell transcriptional profiles (outer colors). By matching lineages and transcriptomes at multiple time points, scientists can potentially deconvolute intermediate states driving the final resistant phenotype.
Implementing these experimental designs and interpreting the data generated through them will not however be straightforward, as the data might be cofounded by a number of factors. For example, when examining changes in barcode representation in a cell pool, neutral drifts of cell populations and sampling biases occurring both in vitro and in vivo need to be taken into account. Furthermore, both plastic reprogramming and feature selection might concurrently affect shifts of cell populations, especially in highly dynamic environments (after cell migration or during therapy switching). Integrating mathematical models and data from these experiments could alleviate these issues. So far, there have only been a few in vivo attempts in limited model organisms with similar technologies owing to a number of technical limitations (Wagner et al., 2018).
Importantly, characterization of cell populations by lineage tracing and single-cell experiments still fails to capture interactions among these populations. These interactions might play a key role in adaptive behaviors, as is the case of EMT driven by interactions between tumor cells and the surrounding microenvironment. In this context, integrating spatial genomics analyses or updates in single-cell technologies such as physically interacting cells with single-cell RNA-sequencing (Giladi et al., 2020) could provide the possibility of mapping cell populations to their physical location in the tumor [refs] and thus observe how distinct cell populations co-localize and change over time.
In the future, we anticipate that cancer researchers will be able to measure dynamic changes in real time via alternative methods. Parallelized reporter assays have the inconvenience of being limited by the emission/excitation spectra and thus will not be applicable to a large set of genes. An alternative might be the development of single-cell biopsies from previously developed nanotechnologies. For example, Edel, Ivanov and colleagues have already implemented miniaturized nanotweezers capable of performing nano-surgery on cells with the added value of leaving them viable for additional intervention (Nadappuram et al., 2019). In the same publication, they showed how these instruments can be used to subsample the mRNA from living cells (Nadappuram et al., 2019). It is conceivable that similar approaches could be scaled up to perform RNA-seq studies on individual cellular biopsies on cancer cells exposed to any number of perturbations.
Conclusions
Cancer evolution can be fueled by nongenetic mechanisms adding complexity, challenges, but also opportunities to our hope of solving the cancer puzzle.
Recurrent genetic mutations driving tumor initiation and progression have been reported in most tumor types. The majority of these “driver” mutations have been proposed to be early events, whereas a minor fraction of subclonal drivers have been identified across tumor types (McGranahan et al., 2015). Although these observations vary among tumor types, possibly in association with clinical management of the disease (Iacobuzio-Donahue et al., 2020), evolution of advanced and treatment-naive tumors could be heavily influenced by nongenetic mechanisms. For example, cancer cell reprogramming to stem-like states or metastatic spread cannot be traced back to specific genetic mutations but is rather sustained by transcriptional and epigenetic rewiring. In this context, interactions between tumor cells and the surrounding microenvironment, as well as among heterogeneous cancer cell populations, are likely to play a key role.
Strong evolutionary bottlenecks, such as treatment administration, have the power to reshape the cell fitness landscape and, therefore, the rules of selection. Here, subclonal genetic variants might become a critical asset for tumor survival conferring resistance to therapy (Schmitt et al., 2015). It should be noted that, even if pre-existing, these events might had not been providing a selective advantage in untreated tumors, assuming the role “subclonal driver” only upon therapeutic intervention. Even in this setting, nongenetic alterations can implement rapid adaptation and survival strategies.
Adaptation might also offer additional insights around our limited capacity to achieve full curative status despite extremely successful targeted therapies in prevalent pathologies such as hormone-dependent cancers. Indeed, it might be that established therapeutic strategies (endocrine therapies) and new ones (CDK4/6i) are simply not suitable for this particular job. Upon treatment, certain cancers might regress to persister stages that strongly mimic resilient development programs, requiring a completely new approach to drug discovery. Understanding the nongenetic vulnerabilities of minimal residual disease might prove successful. It is not clear if similar developmental regression to dormant, persister cells happens in other malignancies, but it is not unlikely that other carrying the relic of the developmental processes of the tissue of origin (i.e. glioblastoma and colon cancer) might also benefit from the study of nongenetic inheritance in treatment adaptation. Is there a common thread between all these cancers? Is minimal residual disease in solid tumor the epicenter on developmentally borrowed adaptation processes? Which processes should we target to steer adaptation? A plethora of epigenetic drugs have been already designed with some in clinical use already, but they all have the limitation of targeting all cells and the entire epigenome (Morel et al., 2019). Ultimately, is plasticity a cell autonomous/constant feature that could be measured (the same way we measure the constant of deformation on a spring) or is this property a function of the cell population that requires active information transfer across the ecosystem?
In recent years, technological advances are providing new ways of interrogating cancer evolution. Single-cell multi-omics, spatial genomics, and lineage tracing approaches are among the most promising to study the balance and interplay of tumor evolutionary mechanisms: genetic and nongenetic, intrinsic and emergent, selection and adaptation. On the one hand, these technologies will need to be coupled with theoretical and data-driven frameworks to convert high-throughput data into information. On the other hand, new perspectives and questions need to be advanced to exploit the potential of these approaches. Overall, to understand, predict, and steer tumor evolution will require integrating the many fragmented views of a rather heterogeneous scientific community into a new coherent and comprehensive picture.
Resource availability
Lead contact
Further information and requests should be directed to the Lead contact, Giovanni Ciriello (giovanni.ciriello@unil.ch).
Materials availability
This study did not generate new unique reagents.
Data and code availability
There are no data and no code associated with this.
Acknowledgments
Luca Magnani is funded by a CRUK Career Development Fellowship (A23110).
Declaration of interests
The authors declare no competing interests.
Contributor Information
Giovanni Ciriello, Email: giovanni.ciriello@unil.ch.
Luca Magnani, Email: l.magnani@imperial.ac.uk.
References
- Acar A., Nichol D., Fernandez-Mateos J., Cresswell G.D., Barozzi I., Hong S.P., Trahearn N., Spiteri I., Stubbs M., Burke R. Exploiting evolutionary steering to induce collateral drug sensitivity in cancer. Nat. Commun. 2020;11:1923. doi: 10.1038/s41467-020-15596-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar H., Solé X., Bonifaci N., Serra-Musach J., Islam A., López-Bigas N., Méndez-Pertuz M., Beijersbergen R.L., Lázaro C., Urruticoechea A., Pujana M.A. Biological reprogramming in acquired resistance to endocrine therapy of breast cancer. Oncogene. 2010;29:6071. doi: 10.1038/onc.2010.333. [DOI] [PubMed] [Google Scholar]
- Aguirre-Ghiso J.A. Models, mechanisms and clinical evidence for cancer dormancy. Nat. Rev. Cancer. 2007;7:nrc2256. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktipis C.A., Boddy A.M., Gatenby R.A., Brown J.S., Maley C.C. Life history trade-offs in cancer evolution. Nat. Rev. Cancer. 2013;13:883–892. doi: 10.1038/nrc3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba A.N.N., Cvijović I., Echenique J.I.R., Lawrence K.R., Rego-Costa A., Liu X., Levy S.F., Desai M.M. High-resolution lineage tracking reveals travelling wave of adaptation in laboratory yeast. Nature. 2019;575:494–499. doi: 10.1038/s41586-019-1749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerling E., Seinstra D., de Wit E., Kester L., van der Velden D., Maynard C., Schäfer R., van Diest P., Voest E., van Oudenaarden A. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cel Rep. 2016;14:2281–2288. doi: 10.1016/j.celrep.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddy B.A., Kong W., Kamimoto K., Guo C., Waye S.E., Sun T., Morris S.A. Single-cell mapping of lineage and identity in direct reprogramming. Nature. 2018;564:219–224. doi: 10.1038/s41586-018-0744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornes L., Scheppingen R.H.van, Beerling E., Schelfhorst T., Ellenbroek S.I.J., Seinstra D., Rheenen J.van. Fsp1-Mediated lineage tracing fails to detect the majority of disseminating cells undergoing EMT. Cell Rep. 2019;29:2565–2569.e3. doi: 10.1016/j.celrep.2019.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T., Kalluri R., Nieto A.M., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–324. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- Byers D., Diao L., Wang J., Saintigny P., Girard L., Peyton M., Shen L., Fan Y., Giri U., Tumula P.K. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer C.L., Marjanovic N.D., Lee T., Bell G., Kleer C.G., Reinhardt F., D’Alessio A.C., Young R.A., Weinberg R.A. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P., George J.T., Tripathi S., Levine H., Jolly M.K. Comparative study of transcriptomics-based scoring metrics for the epithelial-hybrid-mesenchymal spectrum. Biorxiv. 2020 doi: 10.1101/2020.01.02.892604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli Y., Guiliano S., Botton T., Rocchi S., Hofman V., Hofman P., Bahadoran P., Bertolotto C., Ballotti R. Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny. Oncogene. 2011;30:2307–2318. doi: 10.1038/onc.2010.598. [DOI] [PubMed] [Google Scholar]
- Chen F., Ding K., Priedigkeit N., Elangovan A., Levine K.M., Carleton N., Savariau L., Atkinson J.M., Oesterreich S., LEE A.V. Single cell transcriptomic heterogeneity in invasive ductal and lobular breast cancer cells. Biorxiv. 2020 doi: 10.1101/2020.02.21.959023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P., Vallania F., Warsinske H.C., Donato M., Schaffert S., Chang S.E., Dvorak M., Dekker C.L., Davis M.M., Utz P.J. Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell. 2018;173:1385–1397. doi: 10.1016/j.cell.2018.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipponi A., Goode D.L., Bedo J., McCabe M.J., Pajic M., Croucher D.R., Rajal A.G., Junankar S.R., Saunders D.N., Lobachevsky P. MTOR signaling orchestrates stress-induced mutagenesis, facilitating adaptive evolution in cancer. Science. 2020;368:1127–1131. doi: 10.1126/science.aau8768. [DOI] [PubMed] [Google Scholar]
- Consortium R.E., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D.P., Vanderhyden B.C. Context specificity of the EMT transcriptional response. Nat. Commun. 2020;11:2142. doi: 10.1038/s41467-020-16066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall G.V., Hawthorne S., Seyed-Razavi Y., Vieusseux J., Wu W., Gustafsson J.-A., Byrne D., Murphy L., Risbridger G.P., Britt K.L. Estrogen receptor subtypes dictate the proliferative nature of the mammary gland. J. Endocrinol. 2018;237:323–336. doi: 10.1530/joe-17-0582. [DOI] [PubMed] [Google Scholar]
- Degasperi A., Amarante T.D., Czarnecki J., Shooter S., Zou X., Glodzik D., Morganella S., Nanda A.S., Badja C., Koh G. A practical framework and online tool for mutational signature analyses show intertissue variation and driver dependencies. Nat. Cancer. 2020:1–15. doi: 10.1038/s43018-020-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenici G., Aurrekoetxea-Rodríguez I., Simões B.M., Rábano M., Lee S.Y., Millán J.S., Comaills V., Oliemuller E., López-Ruiz J.A., Zabalza I. A Sox2-Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene. 2019;38:3151–3169. doi: 10.1038/s41388-018-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria G.V., Ge Z., Seth S., Zhang X., Jeter-Jones S., Zhou X., Cai S., Tu Y., McCoy A., Peoples M. Resistance to neoadjuvant chemotherapy in triple-negative breast cancer mediated by a reversible drug-tolerant state. Sci. Transl. Med. 2019;11:eaav0936. doi: 10.1126/scitranslmed.aav0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilaty H., Rago L., Youssef K.K., Ocaña O.H., Garcia-Asencio F., Arcas A., Galceran J., Nieto M.A. A gene regulatory network to control EMT programs in development and disease. Nat. Commun. 2019;10:5115. doi: 10.1038/s41467-019-13091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbin M.G., Tirosh I., Hovestadt V., Shaw M.L., Escalante L.E., Mathewson N.D., Neftel C., Frank N., Pelton K., Hebert C.M. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018;360:331–335. doi: 10.1126/science.aao4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T.C., Choi H., Rayes T.E., Ryu S., Troeger J. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T., Lim B., Levine M. Enhancer control of transcriptional bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiti F., Chaligne R., Gu H., Brand R.M., Kothen-Hill S., Schulman R.C., Grigorev K., Risso D., Kim K.-T., Pastore A. Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature. 2019;569:576–580. doi: 10.1038/s41586-019-1198-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo M., Coutinho F.J., Vanner R.J., Gayden T., Mack S.C., Murison A., Remke M., Li R., Takayama N., Desai K. MLL5 orchestrates a cancer self-renewal state by repressing the histone variant H3.3 and globally reorganizing chromatin. Cancer Cell. 2015;28:715–729. doi: 10.1016/j.ccell.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Genovese G., Carugo A., Tepper J., Robinson F.S., Li L., Svelto M., Nezi L., Corti D., Minelli R., Pettazzoni P. Synthetic vulnerabilities of mesenchymal subpopulations in pancreatic cancer. Nature. 2017;542:362–366. doi: 10.1038/nature21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi A., Cohen M., Medaglia C., Baran Y., Li B., Zada M., Bost P., Blecher-Gonen R., Salame T.-M., Mayer J.U. Dissecting cellular crosstalk by sequencing physically interacting cells. Nat. Biotechnol. 2020;38:629–637. doi: 10.1038/s41587-020-0442-2. [DOI] [PubMed] [Google Scholar]
- Goldberg A.D., Allis D.C., Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Greaves M., Maley C.C. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guen V.J., Chavarria T.E., Kröger C., Ye X., Weinberg R.A., Lees J.A. EMT programs promote basal mammary stem cell and tumor-initiating cell stemness by inducing primary ciliogenesis and Hedgehog signaling. Proc. Natl. Acad. Sci. U S A. 2017;114:E10532–E10539. doi: 10.1073/pnas.1711534114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hausser J., Szekely P., Bar N., Zimmer A., Sheftel H., Caldas C., Alon U. Tumor diversity and the trade-off between universal cancer tasks. Nat. Commun. 2019;10:5423. doi: 10.1038/s41467-019-13195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinohara K., Wu H.-J., Vigneau S., McDonald T.O., Igarashi K.J., Yamamoto K.N., Madsen T., Fassl A., Egri S.B., Papanastasiou M. KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic Resistance. Cancer Cell. 2018 doi: 10.1016/j.ccell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.P., Chan T.E., Lombardo Y., Corleone G., Rotmensz N., Bravaccini S., Rocca A., Pruneri G., McEwen K.R., Coombes R.C. Single-cell transcriptomics reveals multi-step adaptations to endocrine therapy. Nat. Commun. 2019;10:3840. doi: 10.1038/s41467-019-11721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke B. A critical view on transgenerational epigenetic inheritance in humans. Nat. Commun. 2018;9:2973. doi: 10.1038/s41467-018-05445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüsemann Y., Geigl J.B., Schubert F., Musiani P., Meyer M., Burghart E., Forni G., Eils R., Fehm T., Riethmüller G., Klein C.A. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue C.A., Litchfield K., Swanton C. Intratumor heterogeneity reflects clinical disease course. Nat. Cancer. 2020;1:3–6. doi: 10.1038/s43018-019-0002-1. [DOI] [PubMed] [Google Scholar]
- Ireland A.S., Micinski A.M., Kastner D.W., Guo B., Wait S.J., Spainhower K.B., Conley C.C., Chen O.S., Guthrie M.R., Soltero D. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell. 2020 doi: 10.1016/j.ccell.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal-Hanjani M., Wilson G.A., McGranahan N., Birkbak N.J., Watkins T.B., Veeriah S., Shafi S., Johnson D.H., Mitter R., Rosenthal R. Tracking the evolution of non–small-cell lung cancer. New Engl. J. Med. 2017;376:2109–2121. doi: 10.1056/nejmoa1616288. [DOI] [PubMed] [Google Scholar]
- Karacosta L.G., Anchang B., Ignatiadis N., Kimmey S.C., Benson J.A., Shrager J.B., Tibshirani R., Bendall S.C., Plevritis S.K. Mapping lung cancer epithelial-mesenchymal transition states and trajectories with single-cell resolution. Nat. Commun. 2019;10:5587. doi: 10.1038/s41467-019-13441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Gao R., Sei E., Brandt R., Hartman J., Hatschek T., Crosetto N., Foukakis T., Navin N.E. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018 doi: 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- Klein C.A., Blankenstein T.J., Schmidt-Kittler O., Petronio M., Polzer B., Stoecklein N.H., Riethmüller G. Genetic heterogeneity of single disseminated tumour cells in minimal residual cancer. Lancet. 2002;360:683–689. doi: 10.1016/s0140-6736(02)09838-0. [DOI] [PubMed] [Google Scholar]
- LaFave L.M., Kartha V.K., Ma S., Meli K., Priore I.D., Lareau C., Naranjo S., Westcott P.M.K., Duarte F.M., Sankar V. Epigenomic state transitions characterize tumor progression in mouse lung adenocarcinoma. Cancer Cell. 2020;38:212–228.e13. doi: 10.1016/j.ccell.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau D.A., Clement K., Ziller M.J., Boyle P., Fan J., Gu H., Stevenson K., Sougnez C., Wang L., Li S. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–825. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A.J., Johnsson P., Hagemann-Jensen M., Hartmanis L., Faridani O.R., Reinius B., Segerstolpe Å., Rivera C.M., Ren B., Sandberg R. Genomic encoding of transcriptional burst kinetics. Nature. 2019;565:251–254. doi: 10.1038/s41586-018-0836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latil M., Nassar D., Beck B., Boumahdi S., Wang L., Brisebarre A., Dubois C., Nkusi E., Lenglez S., Checinska A. Cell-type-specific chromatin states differentially prime squamous cell carcinoma tumor-initiating cells for epithelial to mesenchymal transition. Cell Stem Cell. 2017;20:191–204. doi: 10.1016/j.stem.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Courtois E.T., Sengupta D., Tan Y., Chen K.H., Goh J.J.L., Kong S.L., Chua C., Hon L.K., Tan W.S. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 2017;49:708–718. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- Li N., Babaei-Jadidi R., Lorenzi F., Spencer-Dene B., Clarke P., Domingo E., Tulchinsky E., Vries R.G.J., Kerr D., Pan Y. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis. 2019;8:13. doi: 10.1038/s41389-019-0125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani L., Frigè G., Gadaleta R., Corleone G., Fabris S., Kempe H., Verschure P.J., Barozzi I., Vircillo V., Hong S.-P. Acquired CYP19A1 amplification is an early specific mechanism of aromatase inhibitor resistance in ERα metastatic breast cancer. Nat. Genet. 2017;49:444. doi: 10.1038/ng.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani L., Stoeck A., Zhang X., Lánczky A., Mirabella A.C., Wang T.-L., Gyorffy B., Lupien M. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proc. Natl. Acad. Sci. U S A. 2013;110:E1490–E1499. doi: 10.1073/pnas.1219992110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnen C.L., Shen M.M., Abate-Shen C. Lineage plasticity in cancer progression and treatment. Annu. Rev. Cancer Biol. 2017;2:271–289. doi: 10.1146/annurev-cancerbio-030617-050224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.-J., Eaton E.Ng., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald O.G., Li X., Saunders T., Tryggvadottir R., Mentch S.J., Warmoes M.O., Word A.E., Carrer A., Salz T.H., Natsume S. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 2017;49:367–376. doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFaline-Figueroa J.L., Hill A.J., Qiu X., Jackson D., Shendure J., Trapnell C. A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition. Nat. Genet. 2019;51:1389–1398. doi: 10.1038/s41588-019-0489-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N., Favero F., Bruin E.C.de, Birkbak N.J., Szallasi Z., Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 2015;7:283ra54. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel D., Jeffery D., Aspeslagh S., Almouzni G., Postel-Vinay S. Combining epigenetic drugs with other therapies for solid tumours — past lessons and future promise. Nat. Rev. Clin. Oncol. 2019;17:91–107. doi: 10.1038/s41571-019-0267-4. [DOI] [PubMed] [Google Scholar]
- Nadappuram B., Cadinu P., Barik A., Ainscough A.J., Devine M.J., Kang M., Gonzalez-Garcia J., Kittler J.T., Willison K.R., Vilar R. Nanoscale tweezers for single-cell biopsies. Nat. Nanotechnology. 2019;14:80–88. doi: 10.1038/s41565-018-0315-8. [DOI] [PubMed] [Google Scholar]
- Nguyen V.T.M., Barozzi I., Faronato M., Lombardo Y., Steel J.H., Patel N., Darbre P., Castellano L., Győrffy B., Woodley L. Differential epigenetic reprogramming in response to specific endocrine therapies promotes cholesterol biosynthesis and cellular invasion. Nat. Commun. 2015;6:10044. doi: 10.1038/ncomms10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Gray R., Braybrooke J., Davies C., Taylor C., McGale P., Peto R., Pritchard K.I., Bergh J., Dowsett M. 20-Year risks of breast-cancer recurrence after stopping endocrine therapy at 5 Years. New Engl. J. Med. 2017;377:1836–1846. doi: 10.1056/nejmoa1701830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.W., Lee J.K., Sheu K.M., Wang L., Balanis N.G., Nguyen K., Smith B.A., Cheng C., Tsai B.L., Cheng D. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science. 2018;362:91–95. doi: 10.1126/science.aat5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten D.K., Corleone G., Győrffy B., Perone Y., Slaven N., Barozzi I., Erdős E., Saiakhova A., Goddard K., Vingiani A. Enhancer mapping uncovers phenotypic heterogeneity and evolution in patients with luminal breast cancer. Nat. Med. 2018;24:1469–1480. doi: 10.1038/s41591-018-0091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto P., Etcheverry A., Aubry M., Missey A., Lachat C., Perrard J., Hendrick E., Delage-Mourroux R., Mosser J., Borg C. EMT is associated with an epigenetic signature of ECM remodeling genes. Cell Death Dis. 2019;10:205. doi: 10.1038/s41419-019-1397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perone Y., Farrugia A.J., Rodríguez-Meira A., Győrffy B., Ion C., Uggetti A., Chronopoulos A., Marrazzo P., Faronato M., Shousha S. SREBP1 drives Keratin-80-dependent cytoskeletal changes and invasive behavior in endocrine-resistant ERα breast cancer. Nat. Commun. 2019;10:2115. doi: 10.1038/s41467-019-09676-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perone Y., Magnani L. Going off the grid: ERα breast cancer beyond estradiol. J. Mol. Endocrinol. 2016;57:F1–F5. doi: 10.1530/jme-16-0062. [DOI] [PubMed] [Google Scholar]
- Poli V., Fagnocchi L., Fasciani A., Cherubini A., Mazzoleni S., Ferrillo S., Miluzio A., Gaudioso G., Vaira V., Turdo A. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat. Commun. 2018;9:1024. doi: 10.1038/s41467-018-03264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probert C., Curtis C. A role for chromatin regulatory dynamics in breast cancer evolution. Nat. Med. 2018;24:1309–1311. doi: 10.1038/s41591-018-0182-8. [DOI] [PubMed] [Google Scholar]
- Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J., Garman K.S., Souza R.F., Spechler S.J. Pathogenesis and cells of origin of barrett's esophagus. Gastroenterology. 2019;157:349–364. doi: 10.1053/j.gastro.2019.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi P., Chang M.T., Xu G., Bandlamudi C., Ross D.S., Vasan N., Cai Y., Bielski C.M., Donoghue M., Jonsson P. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34:427–438.e6. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M., Bakir B., Moreira L., Pitarresi J.R., Feldmann K., Simon L., Suzuki K., Maddipati R., Rhim A.D., Schlitter A.M. Regulation of epithelial plasticity determines metastatic organotropism in pancreatic cancer. Dev. Cell. 2018;45:696–711.e8. doi: 10.1016/j.devcel.2018.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim A.D., Mirek E.T., Aiello N.M., Maitra A., Bailey J.M., McAllister F., Reichert M., Beatty G.L., Rustgi A.K., Vonderheide R.H. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J., Ren G., Day C.R., Zhao K., Chow C.C., Larson D.R. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell. 2018 doi: 10.1016/j.cell.2018.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A., Fukunaga-Kalabis M., Schmidt E.C., Zabierowski S.E., Brafford P.A., Vultur A., Basu D., Gimotty P., Vogt T., Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo M., Crisafulli G., Sogari A., Reilly N.M., Arena S., Lamba S., Bartolini A., Amodio V., Magrì A., Novara L. Adaptive mutability of colorectal cancers in response to targeted therapies. Science. 2019;366:eaav4474. doi: 10.1126/science.aav4474. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M.W., Loeb L.A., Salk J.J. The influence of subclonal resistance mutations on targeted cancer therapy. Nat. Rev. Clin. Oncol. 2015;13:335–347. doi: 10.1038/nrclinonc.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R., Schäffer A.A. The evolution of tumour phylogenetics: principles and practice. Nat. Rev. Genet. 2017;18:213–229. doi: 10.1038/nrg.2016.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer S.M., Dunagin M.C., Torborg S.R., Torre E.A., Emert B., Krepler C., Beqiri M., Sproesser K., Brafford P.A., Xiao M. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546:431. doi: 10.1038/nature22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer S.M., Emert B.L., Sizemore A.E., Gupte R., Torre E., Bassett D.S., Raj A. Memory sequencing reveals heritable single cell gene expression programs associated with distinct cellular behaviors. bioRxiv. 2018:379016. doi: 10.1101/379016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M.A. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. First Axl inhibitor enters clinical trials. Nat. Biotechnol. 2013;9:775–776. doi: 10.1038/nbt0913-775a. [DOI] [PubMed] [Google Scholar]
- Shibue T., Weinberg R.A. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoval O., Sheftel H., Shinar G., Hart Y., Ramote O., Mayo A., Dekel E., Kavanagh K., Alon U. Evolutionary trade-offs, pareto optimality, and the geometry of phenotype space. Science. 2012;336:1157–1160. doi: 10.1126/science.1217405. [DOI] [PubMed] [Google Scholar]
- Simigdala N., Gao Q., Pancholi S., Roberg-Larsen H., Zvelebil M., Ribas R., Folkerd E., Thompson A., Bhamra A., Dowsett M., Martin L.-A. Cholesterol biosynthesis pathway as a novel mechanism of resistance to estrogen deprivation in estrogen receptor-positive breast cancer. Breast Cancer Res. 2016;18:58. doi: 10.1186/s13058-016-0713-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa M.S., Bragado P., Aguirre-Ghiso J.A. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger B.Z., Longmore G.D., Yang J., Nieto A.M., Weinberg R.A., Brabletz T., Ye X., Kang Y. Upholding a role for EMT in breast cancer metastasis. Nature. 2017;547:E1. doi: 10.1038/nature22816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva M.L., Riggi N., Bernstein B.E. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72:4875–4882. doi: 10.1158/0008-5472.can-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tannock I.F., Hickman J.A. Limits to personalized cancer medicine. New Engl. J. Med. 2016;375:1289–1294. doi: 10.1056/nejmsb1607705. [DOI] [PubMed] [Google Scholar]
- Tavernari D., Battistello E., Dheilly E., Petruzzella A., Mina M., Sordet-Dessimoz J., Peters S., Krueger T., Gfeller D., Riggi N. Non-genetic evolution drives lung adenocarcinoma spatial heterogeneity and progression. Cancer Discov. 2021 doi: 10.1158/2159-8290.CD-20-1274. [DOI] [PubMed] [Google Scholar]
- Thiery J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis W.D., Brambilla E., Nicholson A.G., Yatabe Y., Austin J.H.M., Beasley M., Chirieac L.R., Dacic S., Duhig E., Flieder D.B. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- Tsai J.H., Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Gene Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turajlic S., Sottoriva A., Graham T., Swanton C. Resolving genetic heterogeneity in cancer. Nat. Rev. Genet. 2019;20:404–416. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- Turajlic S., Xu H., Litchfield K., Rowan A., Horswell S., Chambers T., O’Brien T., Lopez J.I., Watkins T., Nicol D. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell. 2018;173:595–610.e11. doi: 10.1016/j.cell.2018.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D.E., Weinreb C., Collins Z.M., Briggs J.A., Megason S.G., Klein A.M. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science. 2018;360:eaar4362. doi: 10.1126/science.aar4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright E.N., Scaffidi P. Epigenetics and cancer stem cells: unleashing, hijacking, and restricting cellular plasticity. Trends Cancer. 2017;3:372–386. doi: 10.1016/j.trecan.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner B., Case J., Williams M.J., Chkhaidze K., Temko D., Fernández-Mateos J., Cresswell G.D., Nichol D., Cross W., Spiteri I. Measuring single cell divisions in human tissues from multi-region sequencing data. Nat. Commun. 2020;11:1035. doi: 10.1038/s41467-020-14844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.J., Werner B., Heide T., Curtis C., Barnes C.P., Sottoriva A., Graham T.A. Quantification of subclonal selection in cancer from bulk sequencing data. Nat. Genet. 2018;50:895–903. doi: 10.1038/s41588-018-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Gowers K.H.C., Lee-Six H., Chandrasekharan D.P., Coorens T., Maughan E.F., Beal K., Menzies A., Millar F.R., Anderson E. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature. 2020;578:1–7. doi: 10.1038/s41586-020-1961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Norgard R.J., Stanger B.Z. Cellular plasticity in cancer. Cancer Discov. 2019;9:857–951. doi: 10.1158/2159-8290.CD-19-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W., Wang J.-P., Conaway M.R., Li Y., Santen R.J. Adaptive hypersensitivity following long-term estrogen deprivation: involvement of multiple signal pathways. J. Steroid Biochem. Mol. Biol. 2003;86:265–274. doi: 10.1016/s0960-0760(03)00366-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Weinberg R.A. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front. Med-prc. 2018;12:361–373. doi: 10.1007/s11684-018-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no data and no code associated with this.