Abstract
Pancreatic ductal adenocarcinoma (PDAC) is the most common form of pancreatic cancer with an abysmal prognosis rate over the last few decades. Early diagnosis and prevention could effectively combat this malignancy. Therefore, it is crucial to discover potential biomarkers to identify asymptomatic premalignant or early malignant tumors of PDAC. Gene expression analysis is a powerful technique to identify candidate biomarkers involved in disease progression. In the present study, five independent gene expression datasets, including 321 PDAC tissues and 208 adjacent non-cancerous tissue samples, were subjected to statistical and bioinformatics analysis. A total of 20 differentially expressed genes (DEGs) were identified in PDAC tissues compared to non-cancerous tissue samples. Gene ontology and pathway enrichment analysis showed that DEGs were mainly enriched in extracellular matrix (ECM), cell adhesion, ECM–receptor interaction, and focal adhesion signaling. The protein–protein interaction network was constructed, and the hub genes were evaluated. Collagen type XII alpha 1 chain (COL12A1), fibronectin 1 (FN1), integrin subunit alpha 2 (ITGA2), laminin subunit beta 3 (LAMB3), laminin subunit gamma 2 (LAMC2), thrombospondin 2 (THBS2), and versican (VCAN) were identified as hub genes. The correlation analysis revealed that identified hub genes were significantly interconnected. Wherein COL12A1, FN1, ITGA2, LAMB3, LAMC2, and THBS2 were significantly associated with PDAC pathological stages. The Kaplan–Meier survival plots revealed that ITGA2, LAMB3, and LAMC2 expression were inversely correlated with a prolonged patient survival period. Furthermore, the Human Protein Atlas database was used to validate the expression and cellular origins of hub genes encoded proteins. The protein expression of hub genes was higher in pancreatic cancer tissue than in normal pancreatic tissue samples, wherein ITGA2, LAMB3, and LAMC2 were exclusively expressed in pancreatic cancer cells. Pancreatic cancer cell-specific expression of these three proteins may play pleiotropic roles in cancer progression. Our results collectively suggest that ITGA2, LAMB3, and LAMC2 could provide deep insights into pancreatic carcinogenesis molecular mechanisms and provide attractive therapeutic targets.
Subject terms: Cancer, Bioinformatics, Gene expression analysis
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most aggressive and common form of pancreatic cancer, accounting for 95% of all pancreatic malignant neoplasms1. The 5-year overall survival rate for patients with PDAC is less than 8% despite advances in medical oncology2. The poor prognosis of PDAC may be due to the lack of precise molecular biomarkers for early diagnosis and prognosis3. Therefore, there is an urgent need for more effective targeted therapies to improve the survival rate of patients with PDAC4.
Gene expression microarrays and gene chips are extensively applied to reveal genetic aspects of diseases. These techniques are routinely used to monitor genome-wide expression levels of genes and are particularly suitable for screening differentially expressed genes (DEGs) between two samples5. The identification of DEGs may elucidate cancer pathogenesis, provide early diagnosis, and improve treatment. Hence, gene expression microarray analysis could be a promising approach to identify candidate biomarkers involved in disease progression.
The gene expression profiles from diverse microarray platforms are submitted to several public databases, including Gene Expression Omnibus (GEO: https://www.ncbi.nlm.nih.gov/gds/). Several previous studies used gene expression microarray technology to underpinning the DEGs of PDAC in recent years6–8. However, the results were inconsistent, and various aspects remain unclear due to sample heterogeneity. Moreover, those studies have not considered ethnic differences, and many studies have proven that ethnic differences may have relevance for disease gene expression profiles9,10. The present study aimed to improve DEGs accuracy and reliability in PDAC compared to adjacent non-cancerous tissue samples using several datasets from different ethnicities.
In the current study, gene expression datasets from PDAC were analyzed to identify DEGs. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were performed using an online toolset. Then, the protein interaction networks were constructed and the hub genes were identified and further verified. The identified hub genes may serve as potential diagnostic and prognostic biomarkers and could be a promising approach for the treatment of PDAC. To the best of our knowledge, this analysis is the first to examine the gene expression microarray database in PDAC tissues and adjacent non-cancerous tissue samples, considering different ethnic groups.
Materials and methods
Microarray datasets information
PDAC datasets were obtained from the Gene Expression Omnibus, a public functional genomic database containing high-throughput gene expression data, chips, and microarrays. The GEO database was searched using the following criteria: “human-derived pancreatic ductal adenocarcinoma tissues and adjacent non-cancerous tissue samples” (study keyword), “Homo sapiens” (organism), “expression profiling by array” (study type), “tissue” (attribute name), and “sample count” > 50. After a systematic review, five independent PDAC microarray datasets were selected, including GSE6245211, GSE2873512, GSE1547113, GSE6216514, GSE10223815, with 321 primary tumor samples and 208 adjacent non-cancerous samples. The dataset GSE62452 was based on the GPL6244 platform (HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array) and included 69 tumor and 61 adjacent non-cancerous tissue samples. The dataset GSE28735 was based on the GPL6244 platform (HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array) and had 45 matched tumor and adjacent non-cancerous samples.
The GSE15471 dataset was produced using the GPL570 Platform [(HG‐U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array], including 39 matched tumors and adjacent non-cancerous samples. The GSE62165 dataset was based on the GPL13667 Platform [(HG‐U219) Affymetrix Human Genome U219 Array], which contained 118 tumors and 13 adjacent non-cancerous samples. The GSE102238 dataset was based on the GPL19072 Platform [Agilent-052909 CBC_lncRNAmRNA_V3], which included 50 matched tumor and adjacent non-cancerous samples. These five gene expression profiles were respectively from different regions, including North America, Europe, and Asia, thus averting the differences caused by sample heterogeneity of single profiles and revealing universal DEGs that apply to different ethnic groups, as it has been reported that ethnic difference may affect disease-associated gene expression profiles9,10. The clinical datasets included 321 tumors and 208 adjacent non-cancerous tissues diagnosed as PDAC (Table 1). Of note, pancreatic tissue samples in microarray datasets were obtained from the patients who underwent surgical resection for PDAC. Subsequently, tissue samples were stored in liquid nitrogen and/or at − 80 °C until further use. Total RNA was extracted from the snap-frozen tissue samples, and further analysis was carried out. The clinicopathological characteristics of the microarray datasets are briefly shown in Supplementary Table 1.
Table 1.
Characteristics of datasets used in meta-analysis of PDAC tissues vs. adjacent non-cancerous tissues.
| Author, year | GEO accession | Region | Platform | Tissue types and sample numbers | ||
|---|---|---|---|---|---|---|
| PDAC | Adjacent | Count | ||||
| Yang et al. (2016)11 | GSE62452 | USA | GPL6244 | 69 | 61 | 130 |
| Zhang et al. (2012)12 | GSE28735 | USA | GPL6244 | 45 | 45 | 90 |
| Badea et al. (2008)13 | GSE15471 | Romania | GPL570 | 39 | 39 | 78 |
| Janky et al. (2016)14 | GSE62165 | Belgium | GPL13667 | 118 | 13 | 131 |
| Yang et al. (2020)8 | GSE102238 | China | GPL19072 | 50 | 50 | 100 |
PDAC pancreatic ductal adenocarcinoma.
Identification of DEGs
DEGs between PDAC and adjacent non-cancerous tissue samples were screened by GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r)16, an online tool that can be used to compare two or more datasets in a GEO series to identify DEGs according to the experimental conditions. Adjusted p values (adj. p) and Benjamini and Hochberg false discovery rates were employed as criteria for statistically significant genes and to limit false positives. The data normalization was applied for the five datasets (Supplementary Fig. 1). Probe sets with no corresponding gene symbols were removed, while genes with multiple gene probe sets were averaged. Log2 FC (fold change) ≥ 1.5 or ≥ − 1.5 and adj. p < 0.01 was considered statistically significant. An online tool (http://www.interactivenn.net) was applied to draw Venn diagrams of the DEGs17. Further, heatmap analysis was visualized with the Heatmapper web application18. A total of 20 DEGs were identified, which consisted of 19 upregulated genes and 1 downregulated gene.
External validation of the identified DEGs mRNA expression level
The external validation was done using the Gene Expression Profiling Interactive Analysis tool19 (http://gepia2.cancer-pku.cn/#index; last access: 14th February 2021) by comparing transcriptomic data from The Cancer Genome Atlas (TCGA) (pancreatic adenocarcinoma), the TCGA normal and the Genotype-Tissue Expression (GTEx) database. p < 0.05 was considered a statistically significant difference.
GO and KEGG pathway analysis of DEGs
To uncover the functional roles of DEGs, the GO was used to perform enrichment analysis, which covers the cellular component (CC), biological process (BP), and molecular function (MF) of the selected genes20. The KEGG is a database that illustrates the selected gene functions and pathways21. The Database for Annotation, Visualization, and Integrated Discovery (DAVID: https://david.ncifcrf.gov; last access: 14th February 2021) is a public online bioinformatics database that contains information on functional biological annotations for genes and proteins20. The cut-off criteria were selected based on p < 0.01. Enrichment of the GO terms and KEGG pathways were performed for the candidate DEGs using DAVID.
Establishment of the PPI network and hub gene identification
To further explore the potential interplay among those DEGs, these were mapped to the STRING (https://string-db.org; version 11.0) database22 and only interactions that enjoyed a minimum required combined score > 0.4 were set as significant. Subsequently, the protein–protein interaction (PPI) networks were visualized using Cytoscape 3.8.2 (https://cytoscape.org/), an open-source bioinformatics software platform23. A combined score of 0.5 and a tissue-specific (pancreas) filter score of 1 was considered for the construction of the PPI network. Subsequently, the MCODE (Molecular Complex Detection) plugin was used to identify hub genes in the constructed network. The standard for selection was set as follows: MCODE scores ≥ 10, degree cut-off = 2, node score cut-off = 0.2, max depth = 100 and k-score = 224.
Oncomine analysis of hub genes in pancreatic cancer
An independent database, namely Oncomine (https://www.oncomine.org/resource/login.html; last access: 14th February 2021), was used to validate hub gene expression. In the Oncomine database, the gene name “COL12A1”, “FN1”, “ITGA2”, “LAMB3”, “LAMC2”, “THBS2” or “VCAN” was entered. The differential gene analysis module (cancer vs. normal analysis) was selected to retrieve the results. This analysis presented a series of pancreatic cancer studies and related COL12A1, FN1, ITGA2, LAMB3, LAMC2, THBS2, and VCAN mRNA expression in cancer and normal tissues. The filters were set as follows: (1) Gene: COL12A1 or FN1 or ITGA2 or LAMB3 or LAMC2 or THBS2 or VCAN. (2) Analysis type: cancer vs. normal analysis. (3) Cancer type: pancreatic carcinoma. (4) Sample type: clinical specimen. (5) Data type: mRNA. (6) Threshold settings: p < 0.01; FC > 2; gene rank, top 10%.
Finding prognostic genes for PDAC
To explore the expression correlation of hub genes in PDAC, the Spearman coefficient correlation was analyzed using the GEPIA2 tool19. The interaction efficiency was represented as an R score. An R score of > 0.8 was considered a significant correlation. Next, the expression levels of hub genes and pathological stages in PDAC tissues were assessed using the GEPIA2 platform. The GEPIA2 was also utilized for overall survival and disease-free survival analyses of the hub genes using the TCGA and GTEx databases. The plots were considered significant when showed in both overall and disease-free survival states. Beta-actin was used to normalize the expression of genes, and the median was selected for group cut-off criteria. p < 0.05 was considered to indicate a statistically significant difference. Further, the expression of proteins encoded by hub genes in pancreatic cancer was validated using the Human Protein Atlas (HPA: https://www.proteinatlas.org) website based on spatial proteomics data and quantitative transcriptomics data (RNA-Seq) obtained from the immunohistochemical analysis of tissue microarrays25.
Literature review of bioinformatics studies associated with pancreatic cancer
PubMed and Scopus databases were searched to explore existing bioinformatics studies in pancreatic cancer (last access: 15th April 2021). The following criteria were set for PubMed: (pancreatic ductal carcinoma [MeSH Terms]) OR (pancreatic cancer [MeSH Terms]) OR (pancreatic neoplasm [MeSH Terms]) AND (bioinformatics [MeSH Terms]) AND (microarray analysis [MeSH Terms]). For Scopus the following criteria were used: TITLE-ABS-KEY (pancreatic AND ductal AND adenocarcinoma OR pancreatic AND cancer OR pancreatic AND neoplasm AND bioinformatics AND microarray AND analysis). Peer-reviewed studies were considered for the last 10-years, and after a comprehensive analysis, nine studies were selected6–8,26–31.
Results
Identification of DEGs in PDAC
The five gene expression microarray datasets for PDAC, GSE62452, GSE28735, GSE15471, GSE62165, and GSE102238, were obtained from GEO. By screening the data with the GEO2R using p < 0.01 and log2FC ≥ 1.5 or ≥ − 1.5 as cut-off criteria, 2636 upregulated and 1103 downregulated genes were obtained. In brief, 90 DEGs, including 45 upregulated and 45 downregulated genes, were obtained in the GSE62452 expression profile data (Fig. 1a). GSE28735, 127 DEGs, including 66 upregulated and 61 downregulated genes, were identified (Fig. 1b). In GSE15471, 706 DEGs, including 622 upregulated and 84 downregulated genes, were identified (Fig. 1c). 1984 DEGs, including 1380 upregulated and 604 downregulated genes, were identified from GSE62165 (Fig. 1d). In addition, 832 DEGs, including 523 upregulated and 309 downregulated genes, were identified from GSE102238 (Fig. 1e). The overview of the DEGs results was briefly presented in Fig. 1f. After a comprehensive analysis of the five datasets, 20 DEGs were identified that were differentially expressed in all of them, with 19 genes up-regulated and 1 down-regulated in PDAC tissues compared to adjacent non-cancerous tissues (Fig. 2a). Figure 2b,c provides a heatmap of the 20 DEGs based on Log2FC. The functions and the involvement of identified DEGs on PDAC tissues are shown in Table 2.
Figure 1.
Differential expression of genes between PDAC tissue and adjacent non-cancerous tissue samples in the datasets. (a) GSE62452; (b) GSE28735; (c) GSE15471; (d) GSE62165; (e) GSE102238. The x-axis indicates the fold-change (log-scaled); the y-axis indicates the p-values (log-scaled). The red data-points represent upregulated genes, while blue data-points represent downregulated genes. The black data-points represent genes with no significant difference in expression. (f) The differential genes screened based on |Log2FC|≥ 1.5/− 1.5 and a corrected p value of < 0.01. FC fold change.
Figure 2.
Identification of DEGs. (a) Venn diagram of the DEGs in the five datasets. A total of 20 DEGs were included in all five datasets. (b) LogFC heatmap image of the 20 commonly changed DEGs of the five datasets. The 5 GEO datasets are denoted on the abscissa, and the gene names are displayed on the ordinate. (c) The average LogFC expression values of 20 DEGs. DEG differentially expressed gene, FC fold change.
Table 2.
Description of differentially expressed genes and functions in pancreatic cancer.
| Gene | Full name | Function |
|---|---|---|
| A. Upregulated differentially expressed genes | ||
| AHNAK2 | AHNAK nucleoprotein 2 | Promotes malignant progression by inducing EMT |
| ANTXR1 | Anthrax toxin receptor 1 | Enhanced CSC renewal and molecular properties |
| CEACAM6 | Carcinoembryonic antigen-related cell adhesion molecule 6 | Promotes malignant progression by inducing EMT; Induction of immune suppression |
| COL12A1 | Collagen type XII alpha 1 chain | Promotes cell migration by remodeling of the ECM |
| FN1 | Fibronectin | Enhanced invasion and metastasis by degrading ECM |
| ITGA2 | Integrin subunit alpha 2 | Promotes progression and immune suppression |
| KRT19 | Keratin 19 | Promotes malignant progression and associated with poor prognosis |
| LAMB3 | Laminin subunit beta 3 | Promotes cell proliferation, invasion, and migration by activating oncogenic pathways |
| LAMC2 | Laminin subunit gamma 2 | Degrades ECM and promotes invasion |
| PLAC8 | Placenta associated 8 | Promotes cell growth and progression |
| POSTN | Periostin | Promotes invasion and metastasis |
| SERPINB5 | Serpin family B member 5 | Promotes invasion and metastasis |
| SLC6A14 | Solute carrier family 6 member 14 | Promotes growth, proliferation, and chemoresistance |
| SLPI | Secretory leukocyte peptidase inhibitor | Promotes growth, proliferation, and inhibition of apoptosis |
| SULF1 | Sulfatase 1 | Promotes invasion and metastasis of tumor |
| THBS2 | Thrombospondin 2 | Enhances glycolytic enzymes activity in CSC |
| TMC5 | Transmembrane channel-like 5 | Contributes to tumor growth, invasion, and metastasis |
| TSPAN1 | Transmembrane serine protease 4 | Promotes invasion and metastasis of tumor |
| VCAN | Versican | Promotes immune suppression and stromal deposition |
| B. Downregulated differentially expressed gene | ||
| PDK4 | Pyruvate Dehydrogenase Kinase 4 | Inhibition of EMT |
EMT epithelial–mesenchymal transition, CSC cancer stem-cell, ECM extracellular matrix.
The mRNA expression level of DEGs in PDAC
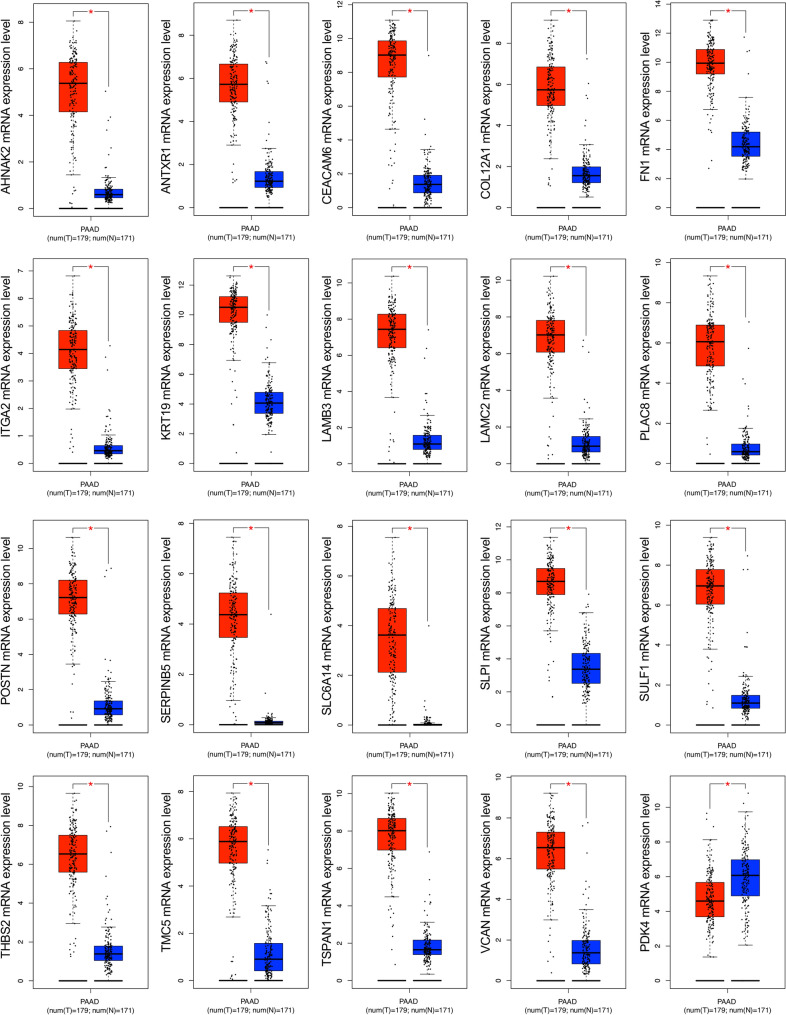
To confirm the mRNA expression levels of identified DEGs in PDAC tissues, TCGA datasets were analyzed using the GEPIA2 platform. Boxplots of the DEGs associated with PDAC were downloaded from the GEPIA2. The results demonstrated that upregulated DEGs were significantly overexpressed in PDAC tissues in comparison to normal pancreatic tissues, while the downregulated DEG, PDK4 was significantly reduced in PDAC tissues in comparison to normal pancreatic tissues (p < 0.05) (Fig. 3).
Figure 3.
The mRNA expression level analysis of 20 DEGs in PDAC tissues. The boxplots were downloaded from the GEPIA2. The red boxes represent the expression levels in PDAC tissues. In contrast, the blue boxes represent the expression levels in normal tissues. p < 0.05 was regarded as statistically significant. DEGs differentially expressed genes, PDAC pancreatic ductal adenocarcinoma.
GO analysis and signaling pathway enrichment of DEGs in PDAC
To elucidate the functions of common DEGs, GO and KEGG pathway enrichment analysis was employed. In the CC category, the upregulated DEGs were mainly enriched in the ECM and extracellular space. In the BP category, the upregulated DEGs were mainly enriched in ECM organization and cell adhesion. While in MF category, upregulated DEGs were enriched with heparin and collagen binding functions. There was no enrichment showed for downregulated DEGs. The ECM–receptor interaction, focal-adhesion, and phosphoinositide-3-kinase-protein kinase B/Akt (PI3K-Akt) signaling were the most enriched pathways for upregulated DEGs. The results of the functional enrichment and KEGG pathway analyses for DEGs are exhibited in Table 3.
Table 3.
Gene ontology and KEGG pathway analysis of differentially expressed genes.
| A. Gene ontology analysis | |||
|---|---|---|---|
| Category | GO ID ~ function | Gene count (%) | p-value |
| GOTERM_BP | GO:0030198 ~ extracellular matrix organization | 7 (36.8) | 3.87E − 08 |
| GOTERM_BP | GO:0007155 ~ cell adhesion | 8 (42.1) | 2.67E − 07 |
| GOTERM_BP | GO:0035987 ~ endodermal cell differentiation | 3 (15.8) | 3.75E − 04 |
| GOTERM_BP | GO:0022617 ~ extracellular matrix disassembly | 3 (15.8) | 0.002951142 |
| GOTERM_BP | GO:0001501 ~ skeletal system development | 3 (15.8) | 0.009281157 |
| GOTERM_CC | GO:0031012 ~ extracellular matrix | 6 (31.6) | 1.16E − 05 |
| GOTERM_CC | GO:0005615 ~ extracellular space | 9 (47.4) | 0.00207988 |
| GOTERM_CC | GO:0070062 ~ extracellular exosome | 9 (47.4) | 0.00207988 |
| GOTERM_MF | GO:0008201 ~ heparin binding | 4 (21.1) | 5.15E − 04 |
| GOTERM_MF | GO:0005518 ~ collagen binding | 3 (15.8) | 0.001632513 |
| B. KEGG pathway analysis | |||
|---|---|---|---|
| Pathway ID | Pathway | Gene count (%) | p-value |
| hsa04512 | ECM–receptor interaction | 5 (26.3) | 8.1175E − 07 |
| hsa04510 | Focal adhesion | 5 (26.3) | 2.5476E − 05 |
| hsa05222 | Small cell lung cancer | 4 (21.1) | 6.1498E − 5 |
| hsa04151 | PI3K-Akt signaling pathway | 4 (21.1) | 0.00019292 |
| hsa05146 | Amoebiasis | 3 (15.8) | 0.00469651 |
| hsa05200 | Pathways in cancer | 4 (21.1) | 0.00544945 |
ECM extracellular matrix, PI3K-Akt phosphatidylinositol 3-kinase/protein kinase B.
PPI network construction and identification of hub nodes
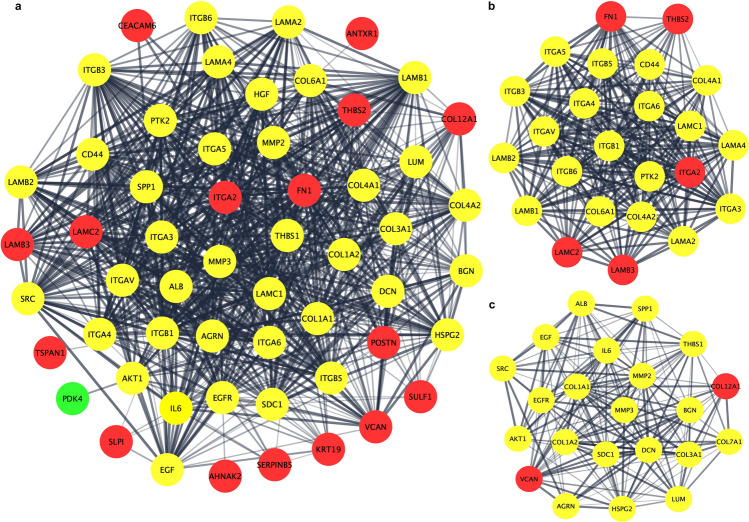
The PPI network of the DEGs was constructed using Cytoscape software and the STRING database. The PPI network of DEGs consisted of 58 nodes and 811 edges (Fig. 4a). The Cytoscape tool MCODE was used to screen hub genes in the network, with a cluster score of ≥ 10 as the inclusion criterion. The MCODE modules included 46 nodes and 432 edges with two clusters. Cluster-1 included 24 nodes and 260 edges with a combined score of 22.6. Wherein cluster-2 included 22 nodes and 172 edges with a cluster score of 16.4. After a comprehensive analysis, hub genes were identified from two clusters highlighted in red color (Fig. 4b,c). COL12A1, FN1, ITGA2, LAMB3, LAMC2, THBS2, and VCAN were finally selected as hub genes. The MCODE plugin scores are briefly shown in Table 4.
Figure 4.
PPI network construction of DEGs and identification of hub genes. (a) PPI network was constructed using Cytoscape. Red nodes represent upregulated genes, whereas green nodes represent downregulated genes. The line represents the interaction relationship between nodes. (b) Significant modules of cluster-1 were identified from the PPI network via the MCODE plug-in. This module consisted of 5 upregulated genes, which are represented by red color. (c) Significant modules of cluster-2 were identified from the PPI network via the MCODE plug-in. This module consisted of 2 upregulated genes, and red nodes represent key genes. PPI protein–protein interaction, DEGs differentially expressed genes.
Table 4.
MCODE cluster scores on PPI network of differentially expressed genes.
| Query term | MCODE score |
|---|---|
| A. Cluster-1, combined score: 22.6 | |
| LAMC2 | 18.3 |
| LAMB3 | 18.0 |
| ITGA2 | 17.4 |
| FN1 | 17.1 |
| THBS2 | 16.9 |
| B. Cluster-2, combined score: 16.4 | |
| VCAN | 14.1 |
| COL12A1 | 13.8 |
MCODE molecular complex detection, PPI protein–protein interaction.
Oncomine analysis of hub genes in pancreatic cancer databases
As COL12A1, FN1, ITGA2, LAMB3, LAMC2, THBS2, and VCAN were selected from the other DEGs, further confirmation of the altered expressions was necessary. Oncomine analysis of cancer vs. normal tissue confirmed that COL12A1, FN1, ITGA2, LAMB3, LAMC2, THBS2, and VCAN were significantly overexpressed in pancreatic cancer from different datasets. A brief overview of those key genes expression in pancreatic cancer was shown by using a heatmap. The color intensity reflects the fold changes between different datasets. Moreover, in the Pei pancreas dataset, COL12A1, FN1, ITGA2, LAMB3, LAMC2, THBS2, and VCAN mRNA expression levels were higher in pancreatic cancer tissue than in normal pancreatic tissue samples (Fig. 5).
Figure 5.
Oncomine analysis of key candidate genes in pancreatic cancer vs. normal tissue. Heat maps of key candidate gene expression in clinical pancreatic cancer samples vs. normal pancreatic tissue samples. [1. Pancreatic ductal adenocarcinoma epithelia vs. normal Badea pancreas; 2. Pancreatic ductal adenocarcinoma epithelia vs. normal Lacobuzio-Donahue pancreas; 3. Pancreatic carcinoma vs. normal Pei pancreas). The color depth represents the intensity of fold changes. Box plots represent the mRNA expression level in pancreatic cancer and normal pancreatic tissues in the Pei pancreas dataset. p < 0.01 was considered statistically significant.
Expression correlation of hub genes in PDAC
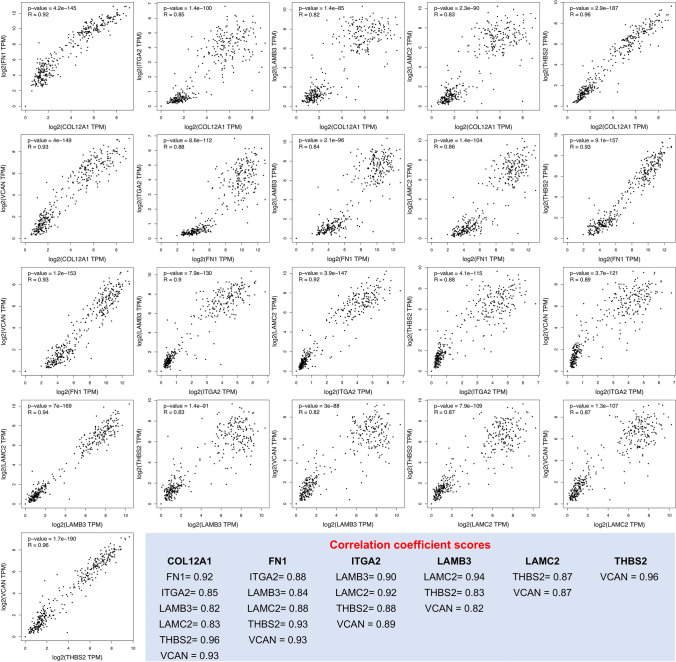
To explore the correlation among the hub genes in PDAC, TCGA datasets were analyzed using the GEPIA2 platform. COL12A1, FN1, ITGA2, LAMB3, LAMC2, THBS2, and VCAN were observed to be significantly correlated (Fig. 6).
Figure 6.
Expression correlation analysis of hub genes in PDAC tissues. The GEPIA2 platform analyzed the expression correlation levels. The Spearman correlation coefficient was used, and an R score of > 0.8 was considered statistically significant. The light blue box represents the correlation coefficient based on R scores.
Association of hub genes in PDAC pathological stages
Further analysis of the TCGA PDAC data in GEPIA2 showed that the hub genes were significantly correlated with the pathological disease stages, underlying their prognostic value for PDAC. COL12A1, FN1, ITGA2, LAMB3, LAMC2, and THBS2 were observed to be significantly associated with PDAC stages (Fig. 7), wherein no significant association on PDAC tumor stages and VCAN was observed (data not shown).
Figure 7.
Pathological stages of hub genes in PDAC tissues. Association of mRNA expression and pathological tumor stages in patients with PDAC. Violin plots were created using the GEPIA2 platform based on the TCGA PDAC dataset. F-value indicates the statistical value of the F test; Pr (> F) indicates p value. A p value of < 0.05 was considered statistically significant.
Survival analysis of hub genes in PDAC
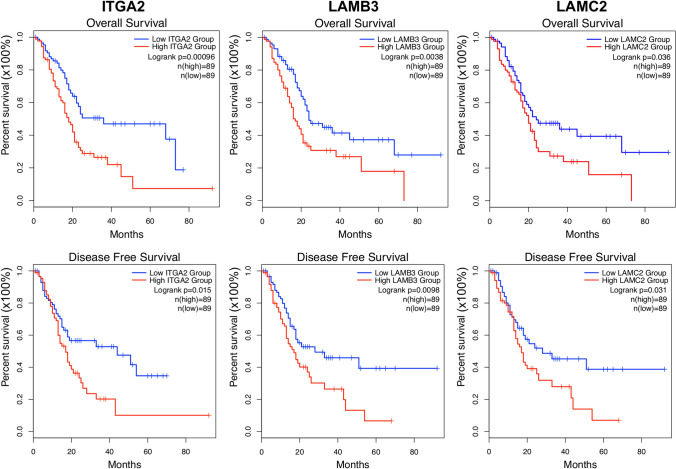
The Kaplan–Meier survival plots were used to observe the overall survival and disease free-survival status of the hub genes in PDAC. Elevated expression levels of ITGA2, LAMB3, and LAMC2 were found to be inversely correlated with prolonged patient survival (Fig. 8), whereas no significant relationship was observed for other genes (data not shown).
Figure 8.
Kaplan–Meier survival plots of hub genes in PDAC tissues. The Kaplan–Meier plots were generated by using the GEPIA2 platform. The overall survival and disease-free survival plots compared a high-risk group (in red) and a low-risk group (in blue) in PDAC tissues. p < 0.05 were regarded as statistically significant.
Validation of expression of hub genes-encoded proteins
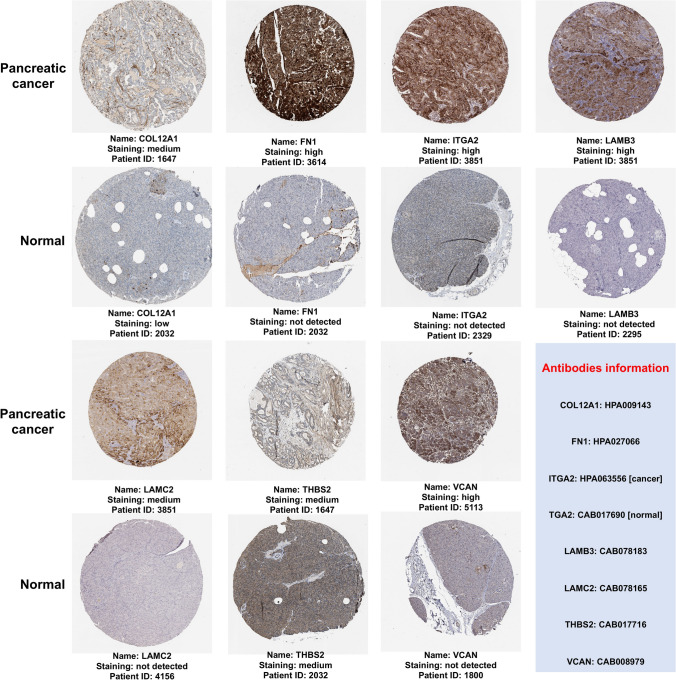
The expression levels of proteins encoded by the COL12A1, FN1, ITGA2, LAMB3, LAMC2, THBS2, and VCAN were obtained. The protein expression profiles in pancreatic cancer clinical specimens are shown in Fig. 9. The antibody intensity for FN1, ITGA2, LAMB3, LAMC2, and VCAN was higher in PDAC tissues, while no staining was observed in corresponding normal tissues. COL12A1 had medium staining intensity with low intensity observed in normal pancreatic tissues. THBS2 had medium staining intensity in both pancreatic cancer and normal pancreatic tissues. Further observations revealed that COL12A1 and FN1 were predominantly expressed by stromal cells. THBS2 and VCAN were expressed in both stromal and pancreatic cancer cells, whereas ITGA2, LAMB3, and LAMC2 were solely expressed by pancreatic cancer cells.
Figure 9.
Immunohistochemical expression of hub genes in human pancreatic cancer specimens. The immunohistochemical data were obtained from the Human Protein Atlas. Staining demonstrated that the protein expression of hub genes was higher in pancreatic cancer tissue than in normal pancreatic tissue samples. The light blue box represents antibodies information. Image courtesy: Human Protein Atlas (http://www.proteinatlas.org).
Identification of hub genes in previous bioinformatics studies associated with pancreatic cancer
The literature review was done to investigate hub genes from previous bioinformatics studies in pancreatic cancer. Nine bioinformatics studies were chosen after a comprehensive analysis based on the criteria which we set. The hub genes, their associated pathways, and potential clinical relevance were explored, which is shown in Table 5. In brief, collagens (COL1A1, COL1A2, COL3A1, COL3A2, and COL5A2), integrins (ITGA2 and ITGB2), laminins (LAMA3, LAMB3, and LAMC2), and fibronectin were the most common hub genes found in those studies. Further, the cell cycle regulation, tissue remodeling, ribosomal protein, and nuclear pore complex-related genes were found to be altered in those studies. The pathways analysis has shown that ECM–receptor interaction, focal adhesion, pathways in cancer, and altered metabolic pathways have been the most commonly involved with those hub genes.
Table 5.
Literature review of the existing bioinformatics studies associated with pancreatic cancer.
| Author, year | Hub genes | Pathways | Observations |
|---|---|---|---|
| Yang et al. (2020)8 | AHNAK2, CDH3, IFI27, ITGA2, LAMB3, SFN, SLC6A14, and TMPRSS4 | Pancreatic secretion, pathways in cancer, p53 signaling pathway, MAPK signaling pathway, Insulin signaling pathway, and pancreatic cancer | AHNAK2, CDH3, IFI27, ITGA2, LAMB3, SLC6A14, and TMPRSS4 may have great diagnostic and prognostic value for pancreatic cancer |
| Jin et al. (2020)7 | TOP2A, CDK1, PRM2, PRC1, NEK2, ZWINNT1, DTL, MELK, CENPF, CEP55, ANLN, ASPM, and ECT2 | ECM–receptor interaction, focal adhesion, pathways in cancer, proteoglycans in cancer, p53 signaling pathway, and PI3-Akt signaling pathway | CDK1, TOP2A, and CEP55 may play pleiotropic roles in the progression of pancreatic cancer |
| Li et al. (2019)26 | ALB, COL1A2, EGF, COL3A1, FN1, CEL, ITGA2, COL5A2, MMP1, and CELA3B | Protein digestion and absorption, ECM–receptor interaction, pancreatic secretion, and fat digestion and absorption | COL1A2, COL3A1, and COL5A2 may promote pancreatic fibrosis and EMT via the ECM–receptor interaction pathway in the early stages of pancreatic cancer |
| Liu et al. (2019)27 | HN1, ITGA2, S100A6, KIF1A, DYM, and BACE1 | Ubiquitin-mediated proteolysis and pathways in cancer | HN1, ITGA2, and S100A6 may be promising potential targets for diagnosing and treating pancreatic cancer |
| Shen et al. (2018)31 | RPL13, RPL17, RPL21, RPL22, RPL23, RPL26, RPL31, RPL35A, RPL36A,RPL37, RPL39, RPL7, RPS17, RPS23, RPS3A,RPS6, RPS7, NUP107, NUP160, and HNRNPU | Ribosome pathway and the spliceosome pathway | Nup170, Nup160, and HNRNPU may be used as possible molecular markers for early diagnosis of pancreatic cancer |
| Pan et al. (2018)28 | CXCL8, ADCY7, ITGAM, ITGB2, ITGB1, IL1A, ICAM1, ITGA2, THBS2, SDC1, COL3A1, COL1A2, COL1A1, MYL9, LAMA3, LAMB3, LAMC2, COL4A1 and FN1 | ECM–receptor interaction, focal adhesion, pathways in cancer, and small cell lung cancer | LAMA3, LAMB3, LAMC2, COL4A1, and FN1 may involve the malignant progression of pancreatic cancer |
| Tang et al. (2018)6 | DKK1 and HMGA2 | Glycine, serine, and threonine metabolism | DKK1 and HMGA2 may be important in the progression of pancreatic cancer |
| Li et al. (2018)29 | ALB, EGF, FN1, ITGA2, COL1A2, SPARC, COL3A1, TIMP1, COL5A1, COL11A1, and MMP7 | ECM–receptor interaction, cell adhesion, and transforming growth factor-beta receptor signaling pathway | ITGA2 and MMP7 may act as potential diagnostic and therapeutic biomarkers for pancreatic cancer |
| Wang et al. (2015)30 | VCAN, SULF1, COL8A1, FAP, COL1A1, THBS2, CTHRC1, COL1A2, COL6A3, FN1, COL10A1, COL3A1, TIMP, AEBP1, and COL5A1 | ECM–receptor interaction, focal adhesion, and complement and coagulation cascades | The collagen family genes and FN1 may play an essential role in the progression of pancreatic cancer |
Discussion
In the present study, 20 DEGs were identified (19 upregulated and 1 downregulated), which were differentially expressed in PDAC tissue compared to the adjacent non-cancerous pancreatic tissue samples. By using an online tool, the mRNA expression levels of DEGs in PDAC tissue samples were validated. The GO and KEGG pathway analysis revealed that DEGs were primarily enriched with ECM-organization, cell adhesion, ECM–receptor interaction, and focal adhesion, especially for the upregulated genes. The PPI network was constructed, and hub genes were selected. COL12A1, FN1, ITGA2, LAMB3, LAMC2, THBS2, and VCAN were identified as hub genes. To verify the expression level of hub genes, an independent database was then used. This confirmed that, compared to normal pancreatic tissues, identified hub genes were highly expressed in pancreatic cancer samples. The correlation analysis revealed that the hub genes in PDAC tissue samples are significantly interconnected. The interaction of hub genes with pathological stages in patients with PDAC showed that the expression of COL12A1, FN1, ITGA2, LAMB3, LAMC2, and THBS2 is negatively associated with disease progression. The survival plots of Kaplan–Meier showed that ITGA2, LAMB3, and LAMC2 expression are inversely correlated with prolonged patient survival. Using histopathological images from the Human Protein Atlas platform, the protein expression profiles of hub genes were validated. It was found that proteins encoded by hub genes are highly expressed in pancreatic cancer tissue compared to normal pancreatic tissue samples. It was also observed that ITGA2, LAMB3, and LAMC2 were the only proteins expressed in pancreatic cancer cells but not in stromal cells. The cancer cells specific expression of these three proteins might be crucial for PDAC pathogenesis and progression. Together, this data suggested that ITGA2, LAMB3, and LAMC2 individually might have high prognostic and diagnostic values, as well as the potential to be therapeutic targets for PDAC.
ITGA2 is a collagen receptor expressed on cell membranes and forms a heterodimer α2β1 with a β subunit, which mediates cell-to-ECM attachment32. The increased ITGA2 level was reported in pancreatic cancer and others, including gastric, liver, prostate, and breast cancer33. The increased ITGA2 expression promotes pancreatic cancer cell migration, invasion, metastasis, and chemoresistance34,35. In contrast, inhibition of ITGA2 abrogated these functions33. Although the exact mechanism by which ITGA2 is involved in pancreatic carcinogenesis remains unclear, it has been suggested that ITGA2 promotes pancreatic cancer progression through ECM remodeling36,37. The reconstituted ECM triggers pancreatic cancer progression by directly promoting cellular transformation and enhancing tumorigenic microenvironment formation by affecting stromal-cell behavior38. In this process, ITGA2 activates fibroblasts to cancer-associated fibroblasts (CAFs), resulting in extensive desmoplasia with ECM deposition39, wherein desmoplasia is a characteristic feature of PDAC and constitutes up to 90% of the tumor volume. Mainly ECM and CAF, immune cells, and vascular components form the desmoplastic microenvironment40,41. ECM is a three-dimensional structural complex consisting of structural and non-structural proteins42,43. ECM-proteins can affect PDAC progression and patient survival by promoting cancer cell proliferation and metastatic spread44. Even though stromal cells produce over 90% of the ECM mass in PDAC, cancer cells produce elevated ECM-proteins, and cancer cell-derived ECM-proteins play important roles in PDAC carcinogenesis45,46. A previous report suggested that ECM proteins originating from cancer cells were the most strongly connected to poor patient survival. In contrast, ECM-proteins derived from stromal cells, include both proteins linked to good and poor patient outcomes47. Hence, using the Human Protein Atlas database, the protein expression profiles and cellular origins of hub genes encoded proteins in pancreatic cancer tissues were observed. ITGA2 is the transmembrane receptor for collagens and related proteins, as mentioned above32, while COL12A1, FN1, LAMB3, LAMC2, THBS2, and VCAN are ECM-related proteins47.
Our histopathological evidence has shown that COL12A1 and FN1 are expressed from stromal cells, THBS2, and VCAN from stromal and cancer cells, while ITGA2, LAMB3, and LAMC2 are expressed solely from the cancer cells. The Kaplan–Meier survival plots showed that ITGA2, among the ECM-proteins LAMB3 and LAMC2 expression, is inversely correlated with the overall and disease-free survival status in PDAC. Interestingly, a previous report confirmed that LAMB3 and LAMC2 were exclusively derived from pancreatic cancer cells47. This study reached a similar conclusion that increased levels of ECM-proteins originated from cancer cells, rather than being solely produced by stromal cells, correlate with poor patient outcomes. However, further studies are needed to clarify this phenomenon. Meanwhile, these results may explain why previous non-selective ECM depletion strategies led to poor patient outcomes and suggest more accurate ECM manipulations as PDAC treatments48. Together, the present data and the previous report suggested that cancer-cell-derived ECM-proteins may be potential therapeutic targets47. Therefore, sorting out the composition and changes of the ECM during PDAC progression would guide the development and application of more effective PDAC therapies.
It is worth noting that DEGs in PDAC have already been demonstrated in several studies6–8,26–31. However, the results were not consistent, which could be due to the differences in the selection of datasets and statistical procedures. Then, using effective search engines, we performed a literature review of existing pancreatic cancer bioinformatics studies and explored hub genes. In brief, the hub genes were mainly involved with ECM remodeling and organization. The predominant expression of collagen, integrin, and laminin family genes was observed in those studies, clarifying their role in ECM remodeling. The reconstituted ECM was reported to promote pancreatic fibrosis and epithelial-mesenchymal transition (EMT) in early stages of PDAC pathogenesis38. Thus, ECM manipulation is an appealing therapeutic strategy for PDAC patients.
While the occurrence of PDAC has been observed to differ between racial/ethnic subpopulations, this disparity may be partially explained by the prevalence of risk factors (smoking and drinking alcohol, obesity, diabetes, and family history) among ethnic groups49,50. These racial/ethnic variations might result in tumor biology differences in PDAC50. Biomarkers that could be useful regardless of racial differences are thus urgently needed. In this study, we selected the datasets from different regions, thus averting the differences caused by the samples heterogeneity and revealing universal DEGs that apply to different ethnic groups. The identified DEGs in this analysis might be applicable irrespective of the ethnicities and may allow the development of more targeted prevention strategies. However, a lack of adequate validation in vitro or in vivo is a limitation of this study. Moreover, due to GEO limitations, the clinicopathological data and demographic variables within this study datasets were not detailed enough. Thus, we failed to consider factors such as the presence of different ethnicities within datasets. Our future research will include experimental verification of this meta-analysis results using different laboratory approaches.
In conclusion, the present meta-analysis identified 20 DEGs. The hub genes are COL12A1, FN1, ITGA2, LAMB3, LAMC2, THBS2, and VCAN. The Kaplan–Meier survival plots indicate that ITGA2, LAMB3, and LAMC2 are inversely correlated with prolonged patient survival. Histopathological evidence shows that ITGA2, LAMB3, and LAMC2 are expressed exclusively from pancreatic cancer cells. The specific expression of these three proteins by cancer cells could make them promising potential targets for diagnosing and treating pancreatic cancer.
Supplementary Information
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by S.I., T.K., Y.A., I.C., and Y.K. The first draft of the manuscript was written by S.I., T.K., B.B., Y.K, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
The R scripts generated during the analysis of microarray datasets were submitted as a PDF file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-90077-x.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel, R., Miller, K. & Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin.68, 7–30 (2018). [DOI] [PubMed]
- 3.Ying H, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30:355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, B. The unsuccessful hunt for pancreatic cancer biomarkers—Time to search deeper in the proteome. Asian J. Sci. Tech.5, 883–891 (2014).
- 5.Parkinson H, et al. ArrayExpress update—An archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2010;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y, Zhang Z, Tang Y, Chen X, Zhou J. Identification of potential target genes in pancreatic ductal adenocarcinoma by bioinformatics analysis. Oncol. Lett. 2018;16:2453–2461. doi: 10.3892/ol.2018.8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin D, et al. Identification of prognostic risk factors for pancreatic cancer using bioinformatics analysis. PeerJ. 2020;8:e9301. doi: 10.7717/peerj.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z-Q, Liu Y-J, Zhou X-L. An integrated microarray analysis reveals significant diagnostic and prognostic biomarkers in pancreatic cancer. Med. Sci. Monitor. 2020;26:e921769-1. doi: 10.12659/MSM.921769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell K, Zingone A, Toulabi L, Boeckelman J, Ryan B. Comparative transcriptome profiling reveals coding and noncoding RNA differences in NSCLC from African Americans and European Americans. Clin. Cancer Res. 2017;23:7412–7425. doi: 10.1158/1078-0432.CCR-17-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardiman G, et al. Systems analysis of the prostate transcriptome in African–American men compared with European–American men. Pharmacogenomics. 2016;17:1129–1143. doi: 10.2217/pgs-2016-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S, et al. A novel MIF signaling pathway drives the malignant character of pancreatic cancer by targeting NR3C2. Cancer Res. 2016;76:3838–3850. doi: 10.1158/0008-5472.CAN-15-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang G, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS ONE. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badea L, Herlea V, Dima S, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 14.Janky R, et al. Prognostic relevance of molecular subtypes and master regulators in pancreatic ductal adenocarcinoma. BMC Cancer. 2016;16:1–15. doi: 10.1186/s12885-016-2540-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang M, et al. Perineural invasion reprograms the immune microenvironment through cholinergic signaling in pancreatic ductal adenocarcinoma. Cancer Res. 2020;80:canres.2689.2019. doi: 10.1158/0008-5472.CAN-19-2689. [DOI] [PubMed] [Google Scholar]
- 16.Shao N, et al. Identification of key candidate genes, pathways and related prognostic values in ER-negative/HER2-negative breast cancer by bioinformatics analysis. J. BUON Off. J. Balkan Union Oncol. 2018;23:891–901. [PubMed] [Google Scholar]
- 17.Heberle H, Meirelles G, da Silva F, Telles G, Minghim R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015;16:1–7. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babicki S, et al. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:147–153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res.49, D545–D551 (2020). [DOI] [PMC free article] [PubMed]
- 22.Szklarczyk, D. et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res.43, D447–D452 (2014). [DOI] [PMC free article] [PubMed]
- 23.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Lei Q, Zhang S, Kong L, Qin B. Screening and identification of key biomarkers in hepatocellular carcinoma: Evidence from bioinformatic analysis. Oncol. Rep. 2017;38:2607–2618. doi: 10.3892/or.2017.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlen, M. et al. A pathology atlas of the human cancer transcriptome. Science357, eaan2507 (2017). [DOI] [PubMed]
- 26.Li, N., Zhao, X. & You, S. Identification of key regulators of pancreatic ductal adenocarcinoma using bioinformatics analysis of microarray data. Medicine98, e14074 (2019). [DOI] [PMC free article] [PubMed]
- 27.Liu L, et al. Identification of differentially expressed genes in pancreatic ductal adenocarcinoma and normal pancreatic tissues based on microarray datasets. Mol. Med. Rep. 2019;20:1901–1914. doi: 10.3892/mmr.2019.10414. [DOI] [PubMed] [Google Scholar]
- 28.Pan Z, et al. Analysis of dynamic molecular networks for pancreatic ductal adenocarcinoma progression. Cancer Cell Int. 2018;18:214. doi: 10.1186/s12935-018-0718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C, Zeng X, Haopeng Y, Gu Y, Zheng W. Identification of hub genes with diagnostic values in pancreatic cancer by bioinformatics analyses and supervised learning methods. World J. Surg. Oncol. 2018;16:1–12. doi: 10.1186/s12957-017-1299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Li Y. Analysis of molecular pathways in pancreatic ductal adenocarcinomas with a bioinformatics approach. Asian Pacif. J. Cancer Prevent. APJCP. 2015;16:2561–2567. doi: 10.7314/APJCP.2015.16.6.2561. [DOI] [PubMed] [Google Scholar]
- 31.Shen Q, et al. Possible molecular markers for the diagnosis of pancreatic ductal adenocarcinoma. Med. Sci. Monitor Int. Med. J. Exp. Clin. Res. 2018;24:2368–2376. doi: 10.12659/MSM.906313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuckwell DS, Calderwood DA, Green LJ, Humphries M. *Integrin α2 I-domain is a binding site for collagens. J. Cell Sci. 1995;108(Pt 4):1629–1637. doi: 10.1242/jcs.108.4.1629. [DOI] [PubMed] [Google Scholar]
- 33.Chuang Y-C, et al. Blockade of ITGA2 Induces Apoptosis and Inhibits Cell Migration in Gastric Cancer. Biol. Procedures Online. 2018;20:1–12. doi: 10.1186/s12575-018-0073-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adorno, V. & Liu, H. Regulation and functions of integrin α2 in cell adhesion and disease. Genes Diseases6, 16–24 (2018). [DOI] [PMC free article] [PubMed]
- 35.Carnevale I, et al. Abstract 4040: α 2 β 1 integrins are potential regulators of chemoresistance through modulation of biomechanical cues in pancreatic cancer. Can. Res. 2018;78:4040. [Google Scholar]
- 36.Ren D, et al. Overexpressed ITGA2 promotes malignant tumor aggression by up-regulating PD-L1 expression through the activation of the STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:1–8. doi: 10.1186/s13046-018-1018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feig C, et al. The pancreas cancer microenvironment. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012;18:4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venning, F., Wullkopf, L. & Erler, J. Targeting ECM disrupts cancer progression. Front. Oncol.5, 224 (2015). [DOI] [PMC free article] [PubMed]
- 39.Zeltz, C. et al. Cancer-associated fibroblasts in desmoplastic tumors: Emerging role of integrins. Semin. Cancer Biol.62, 166–181 (2019). [DOI] [PubMed]
- 40.Whatcott, C., Posner, R., Von Hoff, D. & Han, H. Desmoplasia and chemoresistance in pancreatic cancer. Pancreatic cancer and Tumour microenvironment. Chapter 8, (2021).
- 41.Öhlund D, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017;214:jem.20162024. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hynes R, Naba A. Overview of the matrisome—An inventory of extracellular matrix constituents and functions. Cold Spring Harbor Perspect. Biol. 2011;4:a004903. doi: 10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Humphrey J, Dufresne E, Schwartz M. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 2014;15:802–812. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weniger M, Honselmann K, Liss A. The extracellular matrix and pancreatic cancer: A complex relationship. Cancers. 2018;10:316. doi: 10.3390/cancers10090316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong G-F, Xu R. Function of cancer cell-derived extracellular matrix in tumor progression. J. Cancer Metastasis Treat. 2016;2:357. doi: 10.20517/2394-4722.2016.08. [DOI] [Google Scholar]
- 46.Tian C, et al. Cancer cell-derived matrisome proteins promote metastasis in pancreatic ductal adenocarcinoma. Cancer Res. 2020;80:canres.2578.2019. doi: 10.1158/0008-5472.CAN-19-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian C, et al. Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc. Natl. Acad. Sci. 2019;116:201908626. doi: 10.1073/pnas.1908626116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat. Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 49.Huang B, et al. Interethnic differences in pancreatic cancer incidence and risk factors: The Multiethnic Cohort. Cancer Med. 2019;8:3592–3603. doi: 10.1002/cam4.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nipp R, et al. Disparities in cancer outcomes across age, sex, and race/ethnicity among patients with pancreatic cancer. Cancer Med. 2018;7:525–535. doi: 10.1002/cam4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
The R scripts generated during the analysis of microarray datasets were submitted as a PDF file.