Abstract
The 26S proteasome is responsible for regulated proteolysis in eukaryotic cells. Its substrates are diverse in structure, function, sequence length, and amino acid composition, and are targeted to the proteasome by post-translational modification with ubiquitin. Ubiquitination occurs through a complex enzymatic cascade and can also signal for other cellular events, unrelated to proteasome-catalyzed degradation. Like other post-translational protein modifications, ubiquitination is reversible, with ubiquitin chain hydrolysis catalyzed by the action of deubiquitinating enzymes (DUBs), ~90 of which exist in humans and allow for temporal events as well as dynamic ubiquitin-chain remodeling. DUBs have been known for decades to be an integral part of the proteasome, as deubiquitination is coupled to substrate unfolding and translocation into the internal degradation chamber. Moreover, the proteasome also binds several ubiquitinating enzymes as well as shuttle factors that recruit ubiquitinated substrates. The role of this intricate machinery and how ubiquitinated substrates interact with proteasomes remains an area of active investigation. Here, we review what has been learned about the mechanisms used by the proteasome to bind ubiquitinated substrates, substrate shuttle factors, ubiquitination machinery, and DUBs. We also discuss many open questions that require further study or the development of innovative approaches to be answered. Finally, we address the promise of expanded therapeutic targeting that could benefit from such new discoveries.
Keywords: proteasome, ubiquitin, deubiquitination, ubiquitination, PROTAC
Introduction
The timely and regulated removal of damaged and obsolete proteins is essential to the health of eukaryotic cells. There is an immense cellular investment in this process, which is mediated by the Ubiquitin-Proteasome System (UPS). Approximately 1000 enzymes interact with each other and additional components to form multi-step enzymatic cascades that convert hundreds of proteins (either obsolete, damaged, or misfolded) into substrates for the 26S proteasome, a 2.5 MDa proteolytic machine. At the center of the 26S proteasome is the cylindrical 20S Core Particle (CP) that contains the proteolytic active sites and is capped on either end by a highly dynamic 19S Regulatory Particle (RP), consisting of base and lid sub-assemblies (Fig. 1A). The base sub-assembly includes three intrinsic ubiquitin receptors (Rpn1, Rpn10, and Rpn13), the scaffolding protein Rpn2, and six AAA+ ATPase subunits (Rpt1-Rpt6) that form a heterohexameric ring-shaped motor. The lid sub-assembly contains the essential DUB Rpn11/poh1 and several structural Rpn subunits. To be efficiently recognized and degraded by the 26S proteasome, substrates require a polyubiquitin tag for binding to the proteasomal ubiquitin receptors and an unstructured initiation region for subsequent engagement by the ATPase motor [1–3], which mechanically unfolds and translocates the substrate polypeptides into the CP for degradation (Fig. 1B). Prior to this translocation, ubiquitin modifications must be removed from substrates by the deubiquitinating activity of Rpn11. The Rpt subunits of the base terminate in a 3-amino acid HbYX (hydrophobic-tyrosine-any amino acid) motif [4] and landmark cryo-electron microscopy (cryo-EM) studies from 2012 [5–7] showed that these C-termini dock into hydrophobic pockets on the apical face of the CP, trigger the opening of the CP gate, and thus allow substrate translocation into the degradation chamber (Fig. 1C). These and subsequent studies [8–19] came to reveal the proteasome in distinct states that differ in the conformation of the RP, the alignment of the central processing channel, the position of Rpn11 near the substrate entrance of the proteasome, and the extent of CP gate opening.
Fig. 1.

Structure of the 26S proteasome. (A) A cryo-EM reconstruction of 26S proteasome (bottom, EMDB: 1992) showing the base (upper left) and lid (upper right) sub-complexes and their integration into the RP atop of the CP (bottom image). The DUB Rpn11 (green), together with the components displayed in pink (Rpn3, Rpn5, Rpn6, Rpn7, Rpn8, Rpn9, Rpn12, and Sem1), form the lid sub-complex (top right), whereas the remaining RP components Rpn1 (beige), Rpn2 (beige), Rpn10 (indigo), Rpn13 (light blue), and Rpt1-Rpt6 (yellow) from the base sub-complex (top left). (B) Exterior (left) and CP cross-section (lower right) from cryo-EM reconstruction of the substrate-engaged proteasome (EMDB: 9045; PDB: 6EF3). The hollow degradation chamber of the CP is apparent in the cross section with the α-ring and β-rings in dark and light grey respectively. At the RP, a substrate (red) extends through the central channel of the ATPase ring (yellow) with an attached ubiquitin (orange) bound to the DUB Rpn11 (green). Density maps for Rpt5 and two α-subunits (α6 and α7) is omitted to show the substrate within the ATPase ring and CP entry. (C) Ribbon diagram of the CP:RP interface depicting the α-ring (grey), the C-terminal small AAA+ subdomains of Rpt1–6 (spanning red to green coloring), and a substrate (red) at the center of the substrate processing channel. The C-terminal HbYX motifs of Rpts docked into inter-subunit cavities of the CP α-ring are circled (blue). This figure was generated by using UCSF Chimera [182], UCSF ChimeraX [183], Adobe Illustrator (Adobe), and Adobe Photoshop (Adobe).
Central to the UPS is the small 76 amino acid protein ubiquitin, whose C-terminal glycine can be attached through an isopeptide bond to primary amines, namely the N-terminus (M1) or lysine (K) side chains, in protein substrates or other ubiquitin moieties to form chains. These ubiquitin chains show different topology, depending on whether the subsequent ubiquitin is linked to the N-terminus (M1) or any of the seven surface-dispersed lysines, K6, K11, K27, K29, K33, K48, and K63, of the previous moiety (Fig. 2A). There is an intrinsic polarity to ubiquitin chains, defined by the proximal end where the C-terminal G76 is either conjugated to a protein substrate or unanchored, and the distal end at the other side. Sequential ubiquitin addition during chain formation can occur by using only one of the eight linkage sites in each ubiquitin, leading to homotypic chains, or involve multiple linkage sites to yield heterotypic or branched chains (Fig. 2B). The nature of ubiquitin chains attached to a substrate, referred to as the ubiquitin code [20], is interpreted by the ubiquitin receptors, some of which are linkage-specific, empowering this post-translational modification with the cellular fate of the ubiquitinated protein [21–23].
Fig. 2.
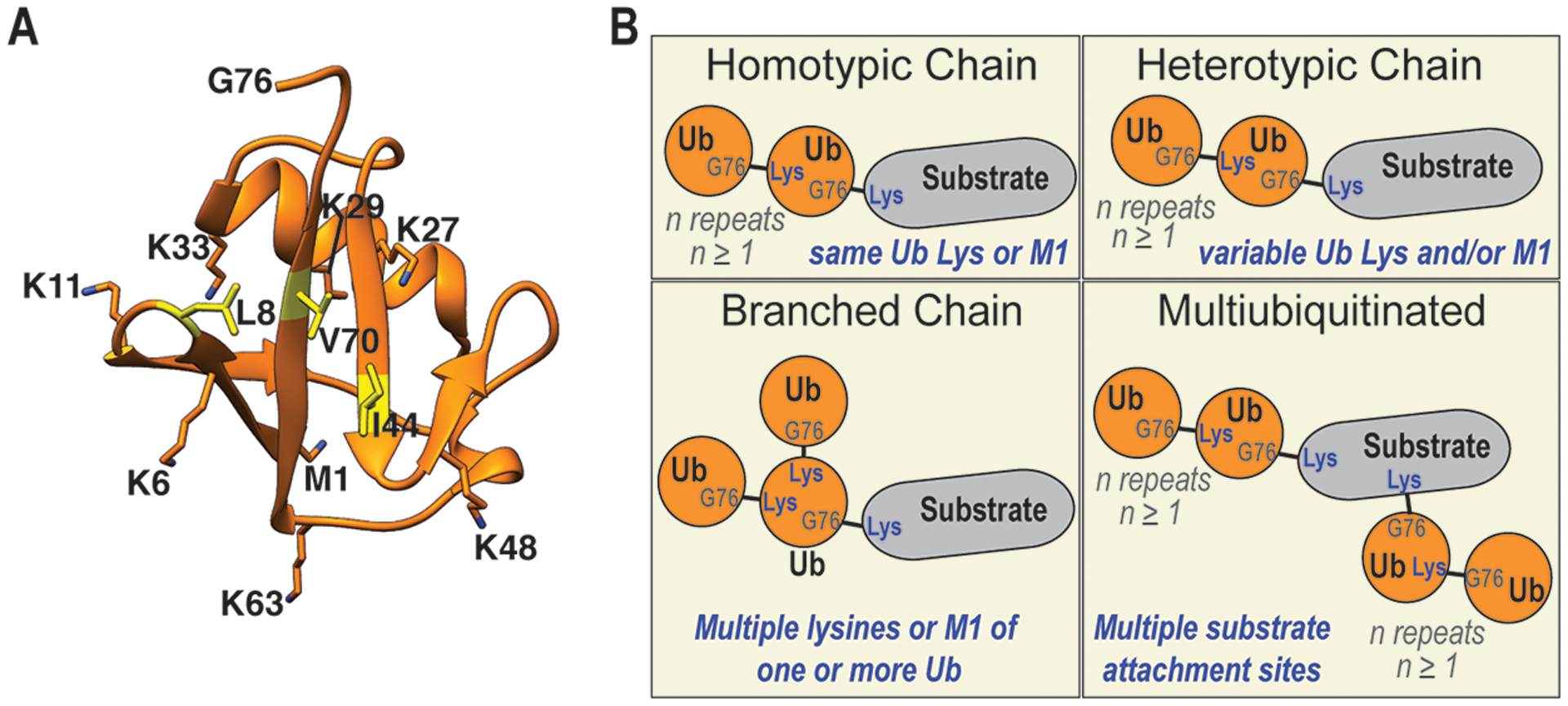
The many forms of ubiquitin modifications. (A) Ribbon diagram of ubiquitin (PDB: 1D3Z) highlighting functional sites, including the C-terminal glycine (G76) that is conjugated to substrate proteins or other ubiquitin molecules; lysine residues (K6, K11, K27, K29, K33, K48, and K63) and the N-terminus (M1), which are used to form ubiquitin chains; and the hydrophobic amino acids L8, I44, and V70 (yellow) that are typically used to bind receptors. Nitrogen is displayed in blue. (B) Depiction of ubiquitin chain types, with homotypic chains containing only one linkage type, heterotypic chains with mixed linkages, and branched or forked chains, in which one or more ubiquitin moieties have multiple ubiquitin molecules conjugated. In multiubiquitinated states more than one ubiquitin or ubiquitin chain is attached to a substrate. This figure was generated by using PyMOL (PyMOL Molecular Graphics System, http://www.pymol.org), Adobe Illustrator (Adobe), and Adobe Photoshop (Adobe).
The configuration of ubiquitin chains is defined by the enzymatic cascade involved in ubiquitination. For example, the anaphase-promoting complex/cyclosome (APC/C) catalyzes formation of heterotypic K11/K48 branched ubiquitin chains, which bind to the proteasome and stimulate substrate degradation [24, 25]. Ubiquitination begins with an E1 ubiquitin-activating enzyme charging ubiquitin in an ATP-dependent manner for thioester transfer to an E2 ubiquitin-conjugating enzyme; in humans, there are two E1 and ~40 E2 enzymes. Aided by scaffolding E3 ubiquitin ligase enzymes, E2 enzymes can modify substrates directly or, alternatively, attach ubiquitin to a catalytic cysteine of the E3 for relayed transfer to a substrate. There are ~600 E3 enzymes in humans as well as ~90 deubiquitinating enzymes (DUBs) that reverse or modulate the ligase effects by ubiquitin chain hydrolysis. Greater than 25% of DUBs bind proteins involved in ubiquitination [26], suggesting that DUBs may rescue ubiquitination machinery from auto-ubiquitination and/or promote dynamic chain remodeling. The proteasome relies on its Rpn11 DUB activity to hydrolyze ubiquitin chains from substrates as they are translocated into the RP.
With its well-recognized β-grasp fold (Fig. 2A), ubiquitin defines a superfamily of ubiquitin-like (UBL) proteins, a subset of which is also covalently attached to protein substrates by analogous enzymatic cascades. There is significant crosstalk between various UBLs and their modifiers, as well as other signaling pathways, including phosphorylation [27, 28]. Thus, in addition to the ubiquitin code, the allosteric amplitude of weak interactions involving protein substrates and other modifications appear to play important roles in determining the signaling outcome of ubiquitination [29, 30]. Several multi-domain proteins bind to different sites of the proteasome RP by using embedded UBL domains, including the DUB Usp14/Ubp6 [31, 32], the E3 ligase parkin [33], the shuttle factors that deliver ubiquitinated substrates to the proteasome [32, 34–39], and co-chaperone proteins [40–42]. The ubiquitin fold is pervasive in the UPS and adapted for multiple purposes. Moreover, interactions with UBL domains can modulate proteasome activity [43–45] through mechanisms that are still not entirely clear.
Here, we review the current understanding of how the 26S proteasome interacts with ubiquitinated substrates via its ubiquitin receptors, the proteasomal AAA+ motor, and the essential deubiquitinase. We also discuss proteasomal interactions with ubiquitination machineries, DUBs, and substrate shuttle factors as well as the many challenges that remain to be addressed for developing a complete mechanistic understanding of substrate processing at the proteasome.
Ubiquitin binding at the proteasome
The proteasome has three established ubiquitin receptors that function in the recruitment of ubiquitinated proteins, namely, Rpn1 [32], Rpn10 [46], and Rpn13 [35, 47]. These three receptors are structurally diverse and use different binding modes to recognize a hydrophobic patch on ubiquitin that contains L8, I44, and V70, and is bound by most ubiquitin receptor proteins (Fig. 2A, [48]. A study of scRpn1 (S. cerevisiae Rpn1) demonstrated this receptor to use an interface within its toroid structure (toroid 1, T1) comprised of three outer helices to bind two ubiquitin moieties [32] (Fig. 3A). The Rpn1 toroid contains an additional recognition site (toroid 2, T2) for the ubiquitin fold at the N-terminal end of deubiquitinating enzyme Ubp6/Usp14 [32] (Fig. 3A), but whether the T1 and T2 sites act independently or are allosterically coupled is not yet known. hRpn10 (human Rpn10) can also bind to two ubiquitin moieties simultaneously and does so at two distinct ubiquitin interaction motifs (UIMs), each of α-helical structure and embedded within an intrinsically disordered region [49, 50] (Fig. 3B). hRpn13 binds diubiquitin at a single ubiquitin-binding site, formed by the loops of a pleckstrin-like receptor for ubiquitin (Pru) domain [35, 47, 51] (Fig. 3C). Like Rpn1, Rpn13 also binds to a deubiquitinating enzyme, namely Uch37/UCHL5, although in this case, through a separate structural domain [52–54] (Fig. 3C). Proteasomes with these three ubiquitin receptors deleted or impaired failed to bind a ubiquitinated model substrate, and the yeast strain containing these mutated proteasomes was highly sensitive to canavanine, an arginine derivative that promotes protein misfolding [32]. The viability of this strain, however, suggests the presence of other ubiquitin receptor sites at the proteasome. Such cryptic binding sites have been proposed to be within the base ATPase subunit Rpt5 [55], the lid subunit Sem1/Dss1 [56], and Rpn10’s globular von Willebrand factor type A (VWA) domain [57]; yet, solid evidence is lacking to substantiate the importance of these regions as ubiquitin receptor sites in the proteasome context. Rpt5 was suggested based on ubiquitin cross-linking, which, however, may have also originated from the crosslinker reacting with ubiquitins bound to other RP subunits nearby, in particular Rpn10’s UIMs and Rpn11. Cryo-EM structures have revealed ubiquitin chains interacting with Rpn11 to extend along the Rpt4/Rpt5 coil coiled domain [58, 59]. Sem1’s ubiquitin-binding region overlaps with its proteasome interface, and no defect in ubiquitin binding was observed upon Sem1 deletion in yeast [32], indicating this subunit’s limited efficacy as a ubiquitin receptor at the proteasome. Rpn10 binds to the proteasome through its N-terminal VWA domain, which is proposed to be ubiquitinated for expulsion from the RP and to also bind ubiquitin non-covalently [57]. Similar to Sem1, future studies are needed to evaluate whether the VWA domain functions as a proteasomal ubiquitin receptor in vivo.
Fig. 3.
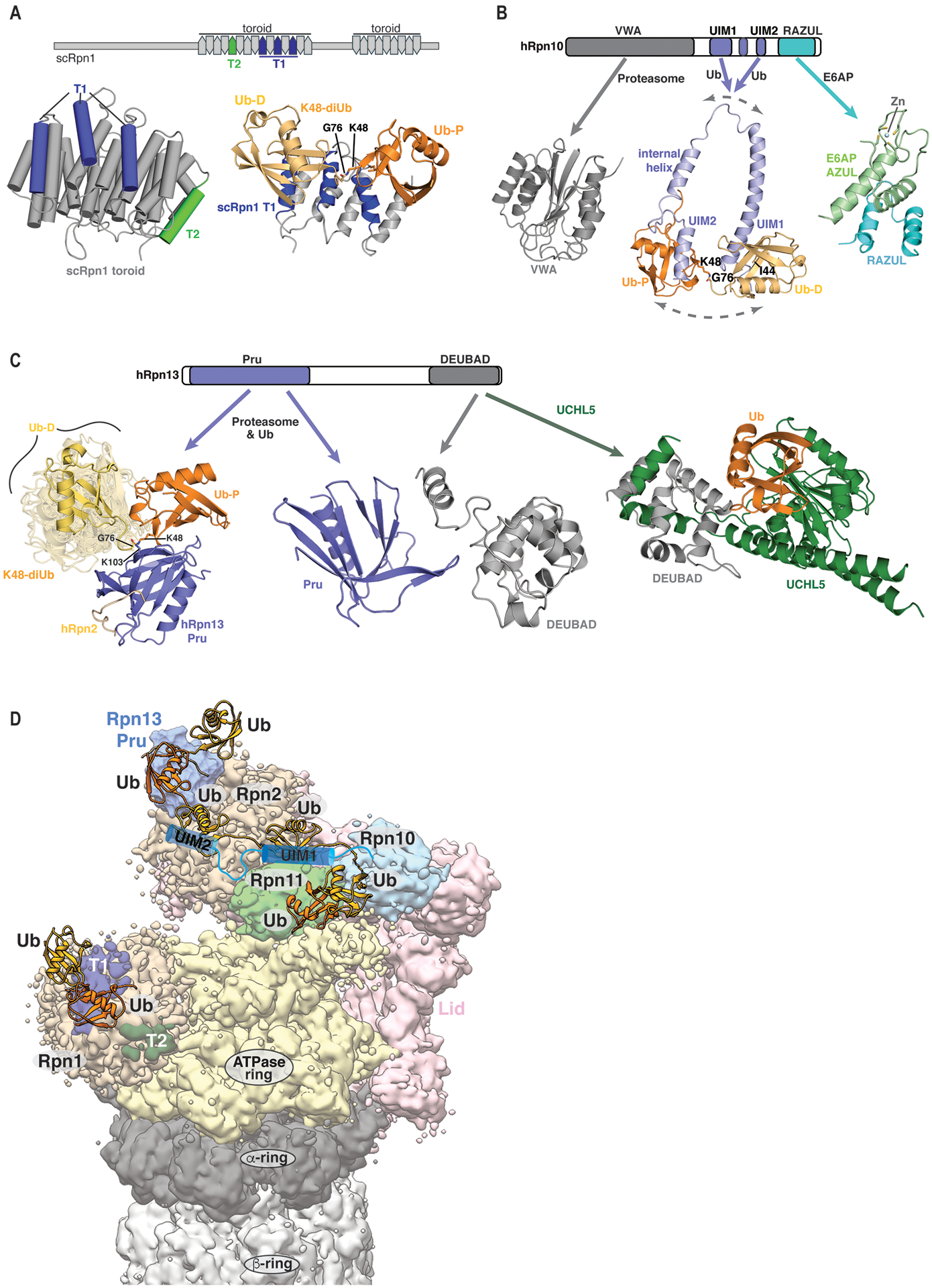
Structure and functional domains of proteasome substrate receptors. (A) Domain layout of scRpn1 (top) illustrating the ubiquitin-binding T1 site (navy) and the T2 site (green), which binds the UBL domain of the DUB Ubp6. The scRpn1 toroid structure is displayed on the lower left, highlighting the T1 and T2 sites (PDB: 4CR2). An expanded ribbon diagram of K48-linked diubiquitin bound to the T1 site is shown on the lower right (PDB: 2N3V). In (A) and (B) the proximal (dark orange) and distal (light orange) ubiquitin moieties are displayed with the K48 linkage site in stick representation (oxygen, red; nitrogen, blue). (B) Domain layout for hRpn10 (top) illustrating the proteasome-binding VWA domain (grey), ubiquitin-binding UIMs and interhelical region (blue), and UBE3A-binding RAZUL domain (cyan). Ribbon diagrams of the VWA domain (lower left), a snapshot of the dynamic UIM region bound to K48-linked diubiquitin with dashed arrows symbolizing flexibility (middle), and the RAZUL:UBE3A AZUL complex (lower right) are displayed. In the AZUL domain, Zn is shown as a blue sphere with coordinating cysteine sulfur atoms in yellow. PDB 2X5N, 2KDE, and 6U19 were used to generate this figure. (C) Domain layout of hRpn13 (top) highlighting the ubiquitin and proteasome binding Pru domain (blue) and UCHL5-binding DEUBAD domain (grey). When free of a binding partner, the two domains interact, as demonstrated in the central ribbon diagram. This structural image has omitted the interdomain region, which is intrinsically disordered. The structure of the Pru domain bound to an extended, dynamic form of K48-linked diubiquitin and 14-amino acid C-terminal region of Rpn2 (gold) is displayed on the lower left. Proximal ubiquitin (orange) is shown bound to the Pru domain loops. The dynamic distal ubiquitin (yellow, eight conformers displayed) exhibits limited order that is defined by interactions at the inter-ubiquitin linker region. A hydrogen bond between the G76 of the distal ubiquitin and K103 of the Pru domain contributes to hRpn13’s preference for K48-linked ubiquitins. The structure of the hRpn13 DEUBAD domain complexed with UCHL5 (green) and a suicide ubiquitin variant (orange) is displayed on the lower right. The DEUBAD domain splits to wrap around UCHL5. PDB 6UYI, 2KR0, and 4WLR were used to generate this figure. (D) Hypothetical model of the ubiquitin-bound 26S proteasome, highlighting the ubiquitin receptors Rpn1 (beige) with T1 (navy) and T2 (green) sites, Rpn10 with the VWA domain (cyan) and UIMs (blue cartoon), and Rpn13’s Pru domain (blue). The ATPase ring, lid (except for Rpn11), Rpn2, Rpn11, and CP are colored yellow, pink, beige, grey, and light green, respectively. Ubiquitin moieties are displayed as yellow or orange ribbon diagrams with the linkage site rendered in stick representation (oxygen, red; nitrogen, blue). This model illustrates how a ubiquitin chain could extend from Rpn10 to Rpn13 and along Rpn1. In addition to the two distinct ubiquitin-chain pathways displayed here, Rpn10’s UIMs or Rpn13’s Pru could bind to different ubiquitin chains of a substrate or alternatively, the ubiquitin chain could bind to just one receptor. The distribution of ubiquitins to the various ubiquitin-binding sites is expected to depend on the number of attached ubiquitin chains, as well as their length and linkage type. PDB 6WJD, 6UYI, 2N3V, 1D3Z, and EMDB 21691 were used to generate this figure. This figure was generated by using UCSF Chimera [182], PyMOL (PyMOL Molecular Graphics System, http://www.pymol.org), Adobe Illustrator (Adobe), and Adobe Photoshop (Adobe).
The receptor sites contributed by Rpn1 T1, Rpn10 UIMs, and Rpn13 Pru share a preference for K48-linked chains and collectively can interact with all ubiquitin chain types in vitro [38]. This combined ability to interact with all chain types is consistent with a quantitative proteomic study in S. cerevisiae that found all non-K63 ubiquitin chains to mediate proteasomal degradation in vivo [60]. More recently, a role even for K63-linked ubiquitin chains in proteasomal degradation was reported; namely, that they can serve as an initiator signal for the production of K63/K48 branched ubiquitin chains to subsequently trigger degradation [61]. Further evidence for branched chains promoting degradation by the proteasome was provided by a study focused on UCHL5, which has poor activity towards homotypic ubiquitin chains [52, 62], but readily hydrolyzes K48 branched chains [63]. In vitro, the proteasome is able to efficiently degrade model substrates modified with K63-linked ubiquitin chains [64], illustrating its promiscuity and suggesting that alternative factors make this chain type a poor signal for proteolysis in vivo [65].
A striking feature of proteasomal substrate binding is the inherently dynamic nature of interaction between receptors and ubiquitin chains, as revealed by the nuclear magnetic resonance (NMR) spectroscopy analyses of the isolated ubiquitin-binding regions in complex with K48-linked diubiquitin [32, 49, 51]. Differential ubiquitin labeling revealed that all three ubiquitin receptors switch in their interactions between the two moieties of K48-linked diubiquitin. K48-linked ubiquitin chains adopt open conformations [51, 66] in dynamic exchange with a ‘closed’ state that is dominant in solution [51] and has the binding sites for proteasomal receptors occluded [67]. NMR studies showed that hRpn13 induces and maintains the extended, open conformational state for K48-diubiquitin, most likely by dynamically switching its interactions between the two ubiquitin moieties [51]. This effect has the potential to also increase the accessibility of K48-linked ubiquitin moieties to the other proteasome receptor binding sites in Rpn1 and Rpn10.
Individually, none of the known ubiquitin receptor sites at the proteasome has high affinity for a single ubiquitin. Rpn13 exhibits increased, yet still moderate, affinity for K48-linked diubiquitin in part due to interactions with the ubiquitin linker region [35, 39, 51, 68]. NMR experiments indicate that scRpn13 binds similarly to ubiquitin compared to hRpn13, but with weaker affinity and a more loosely folded domain structure [35, 47]. However, it is possible that these differences are attenuated when the proteins are assembled into the proteasome. Each of the two ubiquitin-binding sites in the scRpn1 T1 binds to an individual ubiquitin molecule weakly; however, the two sites act in concert to achieve a moderate affinity of 11 μM for K48-diubiquitin [32]. The two Rpn10 UIMs act similarly, binding at the same time to ubiquitin moieties of K48-diubiquitin to yield an affinity of 8.9 μM [32, 49, 69, 70]. Schizosaccharomyces pombe Rpn10 (SpRpn10) and scRpn10 each have only one UIM, but bind to K48-diubiquitin with equivalent affinity compared to hRpn10 [71, 72], perhaps due to interactions with the K48 ubiquitin linker region [71].
The combined actions of Rpn1, Rpn10, and Rpn13 in the context of the proteasome is expected to enhance the affinity for ubiquitinated substrates, while preserving the dynamics observed in the isolated systems. These dynamics are likely critical for enabling the proteasome to bind substrates with varying ubiquitin-chain architectures, allowing multivalent ubiquitin interactions with the three receptors and associated DUBs, and orienting the unstructured initiation regions of substrates in a manner appropriate for engagement by the ATPase motor.
Substrate shuttle factors for the proteasome
In addition to the direct binding of ubiquitinated substrates to the ubiquitin receptors, ubiquitin-binding shuttle factors, namely Rad23, Dsk2, and Ddi1 family members deliver substrates to the proteasome [32, 34–39, 73, 74]. They interact through their UBL domains with receptor sites on Rpn1, Rpn10, and Rpn13 in a manner similar to ubiquitin, but contain amino acid substitutions that result in higher affinity for certain sites [38, 39, 75, 76]. For example, the UBL of Dsk2’s human orthologs preferentially bind hRpn10’s N-terminal UIM [38, 77], whereas hRad23s’ UBLs prefer the C-terminal UIM [36, 75, 76]. scRad23 favors one of the two scRpn1 T1 ubiquitin-binding sites where it forms additional hydrophobic interactions compared to ubiquitin due to substitution of ubiquitin L8 for phenylalanine [78]. The identity of the shuttle factor delivering a substrate thus likely influences how the attached ubiquitin chain(s) are presented to the proteasome receptor sites and how a substrate’s flexible initiation region accesses the central pore. The hRad23 ortholog hHR23b and Dsk2 ortholog hPLIC1 are consistently observed by mass spectrometry to be associated with proteasomes isolated from cells [79, 80]. Similarly, the UBL of Rad23 from S. cerevisiae binds scRpn1 with an affinity of 64 ± 25 nM [32], and how these more strongly binding shuttle factors are released from the proteasome has not yet been revealed.
Binding of shuttle factor UBL domains stimulates the ATP-hydrolysis and proteolytic activities of the proteasome [45], thus further linking binding at substrate receptor sites with induction of proteasome activity. It is therefore possible that proteasomes found to have hPLIC1 or hHR23b present are in activated states. It is not yet clear whether multiple shuttle factors are typically bound simultaneously to a particular proteasome; however, their ubiquitin-associated domains (UBAs) can bind a common substrate [81] and their UBL preferences for different receptor sites [32, 34–39, 73–76] make delivery of a substrate through multiple receptors a possibility.
hPLIC and hHR23 not only bind ubiquitin and ubiquitinated proteins, but also each other through UBL/UBA interactions [82], which for hHR23a occur intramolecularly as well [83]. These UBL/UBA proteins can therefore induce liquid-liquid phase separation [84, 85]. Under hyperosmotic stress, phase separated nuclear foci are formed that contain proteasomes and p97 [84]. Formation of these proteolytic compartments is triggered by multivalent interactions of hHR23b’s two UBA domains [84], which simultaneously bind ubiquitin moieties [69]. These studies highlight the multi-faceted functional roles of proteasome shuttle factors, but how these and their many other activities are integrated is not yet known.
Challenges to studying ubiquitin-chain binding to proteasome receptors
The intrinsic flexibility of proteasomal receptors and the dynamics of their ubiquitin interactions are likely paramount to substrate processing. A technical consequence of these inherent and functionally critical dynamics is the intricacy of studying them with any of the modern structural biology tools. Ubiquitin receptors and ubiquitin chains bound to the proteasome have been at best poorly resolved by cryoEM. Only hRpn1 has been observed with a bound ubiquitin [58], whereas Rpn13 is missing entirely from cryo-EM density maps of the human proteasome and has been visualized only at low resolution in the context of the S. cerevisiae proteasome [5, 7, 10, 15, 17, 86]. hRpn13 binds an intrinsically disordered 14 amino acid C-terminal extension of hRpn2 (Fig. 3C), which enables motion relative to the rest of hRpn2 and the proteasome [62, 68, 87], and likely aids in substrate capture, but challenges observation by cryoEM.
The C-terminal ubiquitin-binding region of Rpn10 is highly dynamic and missing from all cryo-EM density maps, which is unsurprising given that it is preceded by a flexible linker (Fig. 3B). Currently, solving structures by single particle cryo-EM relies on the detection of thousands of particles that are trapped in different orientations, yet a common state – a requirement unachievable for highly dynamic molecules. The applicability of NMR to the intact proteasome, on the other hand, is limited by signal decay due to the slow tumbling of such a large complex [88]. Moreover, the RP has refracted crystallization, presumably due to its dynamic structural heterogeneity, whereas the more rigid CP has been crystallized many times [89]. An interesting thought experiment is to consider the relevance of a structure for the proteasome RP in a substrate-engaged, trapped state, with its receptors rigidly bound to a ubiquitin chain. Such a sample may be more amenable to crystallization or cryoEM study, but how well it would represent functionally relevant states of the proteasome as a highly dynamic proteolytic machine remains unknown.
In vitro biochemical and single molecule experiments with model substrates have provided mechanistic insights that are complementary to the results obtained from structural studies. The proteasome architecture based on structural analyses suggests that two distinct ubiquitin-chain pathways exist, one that extends along an hRpn10/hRpn13 axis and another that is directed across hRpn1 (Fig. 3D). This model is supported by single molecule studies demonstrating more efficient degradation at the proteasome for a model substrate with equivalent number of ubiquitin moieties, but distributed as two separate diubiquitins rather than a single tetraubiquitin chain [90]. A more recent study demonstrated that supernumerary ubiquitin chains can promote degradation of otherwise poor substrates [91], suggesting an advantage in proteasomal degradation for substrates with multiple ubiquitin chains. This study also identified substrate unfolding as the rate-limiting step of degradation occurring on the timescale of seconds [91], which provides insights into the time that the receptors may need to hold on to ubiquitinated substrates. The time of receptor interactions required for successful degradation depends on how stably a substrate’s unstructured initiation region engages with the ATPase motor to prevent substrate release during repeated unfolding attempts, and when, relative to the rate-limiting unfolding, an affinity-conferring ubiquitin chain is removed from the substrate by the Rpn11 DUB.
Substrate engagement and translocation by the ATPase motor
Ubiquitin-chain binding to proteasomal receptors increases the residency time of a substrate at the RP, such that an internal unstructured region or flexible tail of the substrate can passively enter the central processing pore and engage with the ring-shaped AAA+ ATPase motor of the base sub-assembly for ATP-hydrolysis driven translocation and unfolding (Fig. 1B). The AAA+ motor of the proteasome consists of six distinct ATPase subunits in the clockwise order Rpt1, Rpt2, Rpt6, Rpt3, Rpt4, and Rpt5 (Figs. 1C, 4A) [92]. All Rpts have the same domain architecture, which features an N-terminal coil, followed by an oligosaccharide-binding fold (OB-fold) domain and C-terminal AAA+ ATPase domain that in the hexamer assemble into an N-terminal domain ring (N-ring) and an ATPase motor ring, respectively (Fig. 4A). The nucleotide-binding site in the ATPase domain of each Rpt is located at the interface with the clockwise neighboring subunit, allowing coordinated ATP hydrolysis and conformational changes in the hexamer. Motor interactions with the substrate polypeptide are primarily mediated by the so-called pore-1 loops that project from each AAA+ domain into the central channel of the ATPase ring and contain an Aromatic-Nonpolar-Gly motif [93–97]. The highly conserved aromatic residues in this motif, Tyr in scRpts and Tyr or Phe in hRpts, together with the preceding Lys or Met, form pockets to accommodate substrate side chains and directly contact the polypeptide backbone for mechanical pulling during translocation [58, 98].
Fig. 4.

ATPase ring gymnastics. (A) Structural motifs of the proteasome ATPase ring. Left: A top view of the EM density for the Rpt1-Rpt6 hexamer in the s4 state (EMDB: 9045), with the central channel of ATPase ring labeled. Middle: ATPase ring displayed as a ribbon diagram (PDB: 6EF3) rotated 90° relative to the view on the left and highlighting the N-terminal coiled coils, N-ring, ATPase motor ring, and HbYX motifs. Right: ribbon diagram structure of Rpt5 and Rpt6 (PDB: 6EF3) to illustrate the OB fold, Pore-1 loop (red), and nucleotide (oxygen, red; nitrogen, blue; phosphorus, orange). (B) Cutaway representations of the 26S proteasome in s1 (apo, EMDB: 3534) and s4 (engaged, EMDB: 3537) conformations, showing the different position of Rpn11 and change in alignment of the N-ring, ATPase motor ring, and CP. The central channel through the N-ring and ATPase motor ring is indicated by a dashed orange line. (C) Conformational switching of the human 26S proteasome between a ground state (SA, PDB: 5VFS) and substrate processing state (SD, PDB: 5VFP), with the CP aligned. During the transition from SA to SD, hRpn10’s VWA domain rotates by ~30° towards the hRpt4/hRpt5 coiled coil. The color scheme in panels (B) and (C) follows that in Fig. 1B and Fig. 3D. This figure was generated by using UCSF Chimera [182], UCSF ChimeraX [183], Adobe Illustrator (Adobe), and Adobe Photoshop (Adobe).
Biochemical studies have found that proteasomal substrate degradation depends on a flexible initiation region or terminal tail of at least 25 amino acids [3, 91], a length required to reach through the N-ring and into the ATPase ring for engagement by the pore-1 loops [98]. The initiation region also needs some degree of sequence complexity, as stable engagement relies on sufficient grip of the pore-1 loops to prevent substrate escape out of the pore. Substrates with tails that are too short or low in complexity have been shown to be poorly engaged or degraded more slowly [91, 99, 100]. The redundancy of the ubiquitin receptors and the dynamic nature of their ubiquitin binding likely play important roles in properly positioning the substrate for tail insertion and engagement with the pore-1 loops.
Not all proteasome substrates in the cell contain unstructured regions that are long enough for engagement [101], and while most of these proteins may get prepared for proteasomal degradation through ubiquitin-dependent unfolding by Cdc48/p97 [102, 103], a recent study found that ubiquitin attachment itself can destabilize folded domains and induce transient unfolding sufficient for proteasome engagement [104]. Disordered initiation regions can also be obstructed by attached ubiquitin chains that prevent substrate engagement with the translocation machinery prior to deubiquitination [91]. If the obstructing chain is the only ubiquitin modification on a substrate and thus providing the affinity for proteasomal receptors, its removal leads to substrate dissociation and escape from degradation. These findings highlight that individual processing steps at the proteasome need to be strictly coordinated, and substrate engagement has to precede ubiquitin chain removal for efficient degradation, which is accomplished through the direct coupling of mechanical translocation by the ATPase motor and deubiquitination by Rpn11 (see below).
FRET (Förster Resonance Energy Transfer)-based biochemical analyses combined with cryo-EM structures indicated that substrate engagement by the pore-1 loops triggers a large conformational change of the entire RP, leading to the coaxial alignment of the N-ring, the ATPase ring, and CP, and thus facilitating substrate transfer into the degradation chamber [8, 91] (Fig. 4B). In this engaged state, the AAA+ motor domains and their pore-1 loops form a spiral staircase arrangement around the substrate, a feature that was first shown for the 26S proteasome [8] and subsequently described for numerous other translocating AAA+ protein remodelers [105]. Two recent studies have revealed high-resolution cryo-EM structures of the substrate-bound S. cerevisiae and human proteasomes in apparently consecutive states of ATP hydrolysis and substrate translocation [58, 98]. These snapshots suggest that hydrolysis events progress counterclockwise around the ATPase ring and drive substrate translocation by a hand-over-hand mechanism. In this mechanism, 4 or 5 ATP-bound subunits are in contact with the substrate polypeptide, as the post-hydrolysis subunit disengages from the substrate at the bottom of the spiral staircase to move up and, after ATP binding, re-engage at the top; a process that leads to a downward translocation step of two amino acids [58, 98, 105].
Substrate interactions and conformational states of the proteasome
Deep classification of large-scale single-particle cryo-EM data revealed multiple distinct conformations of the proteasome that can be broadly separated into a substrate-free “apo” state (termed s1 for the S. cerevisiae proteasome) and several engaged states (termed s2, s3, s4, s5 and s6), representing the AAA+ motor at various stages of the ATPase cycle and with different registers of the Rpt staircase [5–15, 17–19]. In the apo state, the N-ring, ATPase ring, and CP are offset from a coaxial alignment, leading to a narrow, discontinuous substrate-processing channel (Fig. 4B). Rpn11 is located above the N-ring but shifted laterally relative to the central pore, opening up the accessibility to the flexible initiation regions of substrates that are tethered to ubiquitin receptors (Fig. 4C). Substrate insertion and engagement by the pore-1 loops triggers a major conformational change of the entire RP that aligns the N-ring, ATPase ring, and CP, opens the CP gate, and induces distinct spiral-staircase arrangements of Rpt subunits around the substrate. This conformational change also rotates the lid relative to the base sub-assembly, such that Rpn11 moves to a position directly above the entrance to the processing channel, leaving only a small gap to the N-ring for the already engaged substrate to be pulled through [8, 58, 91, 98]. In this engaged state, Rpn11 is thus ideally positioned to capture and remove ubiquitin modifications from translocating polypeptides, but sterically blocks the pore entrance for newly arriving substrates. Efficient substrate insertion into the central channel and engagement by the ATPase motor therefore seems to depend on the apo state of the proteasome, whereas processive translocation, unfolding, co-translocational deubiquitination, and substrate transfer into the CP are facilitated by the engaged conformational states. This model is supported by mutational studies, showing that the disruption of apo-state specific lid-base interactions shifts the conformational equilibrium towards the engaged states despite the absence of substrate and results in degradation defects that originate from inhibited substrate engagement [19]. Furthermore, FRET-based biochemical analyses indicate that the substrate-induced conformational switch represents a crucial kinetic gateway to select for substrates that contain appropriate ubiquitin modifications and unstructured initiation regions, mechanistically coupling these requirements to the commitment to degradation [91].
Binding of ubiquitin or UBLs to proteasomal receptors or the DUB Usp14/Ubp6 also appears to influence the conformational equilibrium of the proteasome, as various functional studies revealed a consequent increase in CP gate opening, proteolysis, and ATP-hydrolysis activities [45, 106–109]. However, cryo-EM analyses of the yeast proteasome in the presence of K48-linked tetraubiquitin chains showed only very minor differences compared to the conformational states in the absence of ubiquitin [110]. Thus, additional structural and mechanistic studies are needed to investigate how ubiquitin-chain binding affects the conformational dynamics of the proteasome.
Important in vivo evidence regarding the distribution of proteasome states and the conformational switch upon substrate engagement is provided by in situ cryo-electron tomography studies, which revealed that the majority of proteasomes adopt the apo state in intact neurons [111] as well as in the cytosol and nucleus of Chlamydomonas reinhardtii [112, 113]. Introduction of poly-GA containing protein aggregates into neurons results in the recruitment of proteasomes and their enrichment in the engaged states, suggesting that these proteasomes attempt to process and stall on the poly-GA protein aggregates or switch their conformation in response to binding the ubiquitinated aggregates [114].
DUBs at the proteasome
Two independent cryoEM studies of yeast [98] and human [58] proteasome in trapped states bound to model substrates with conjugated K63-linked ubiquitin chains revealed density for the substrate threaded through the center of the ATPase ring and for ubiquitin bound to Rpn11. Rpn11 removes ubiquitin chains en bloc from substrates [115] in a co-translocational manner, with its cleavage activity enhanced by substrate translocation of the ATPase ring [16]. Interaction between ubiquitin and Rpn11 was first observed in a crystal structure of the isolated Rpn11-Rpn8 heterodimer from yeast complexed with monoubiquitin, which revealed that the Rpn11 Insert-1 region transitions from an inhibitory loop across the catalytic groove in the absence of ubiquitin to a β-hairpin that forms a three-stranded β-sheet with the C-terminus of ubiquitin, thereby stabilizing it for isopeptide cleavage [16]. This loop-to-hairpin transition of the Insert-1 region was found to be rate-limiting for ubiquitin cleavage, which explains how mechanical substrate translocation and pulling attached ubiquitin moieties into the Rpn11 catalytic groove can significantly accelerate deubiquitination. Interaction between ubiquitin and hRpn11 is similarly observed by cryoEM for human proteasome complexed with M1-linked hexaubiquitin that is not conjugated to a substrate [59]. Thus, hRpn11 is fully accessible and binds ubiquitin at this location independent of a protein substrate interacting with the ATPase ring.
Not known, however, is the role of hRpn10 in the recruitment of ubiquitin chains to this location. The VWA domain that docks Rpn10 to the proteasome is well-resolved by cryoEM [5–7]. In the apo state, it binds the lid with no observable contacts to the base, yet in the engaged state of the proteasome, a rotation of ~30° causes the VWA domain to interact with the N-terminal coiled coil of Rpt4/Rpt5 (Fig. 4C). This interaction is apparently important for proteasome integrity, as loss of the Rpn10 VWA domain destabilizes lid-base association in both human [116] and yeast [117] proteasomes. The location of the VWA domain is nearby to where ubiquitin binds on hRpn11, however, the hRpn10 portion beyond the VWA, including the UIMs, is not visible in cryoEM density maps of the proteasome [11, 12, 18, 58, 118, 119]. CryoEM proteasome structures show the unanchored M1-linked chains [59] as well as substrate-conjugated K63-linked chains [58] extending beyond Rpn11 and along the Rpt4/Rpt5 coiled. It is therefore likely that hRpn10 contributes to ubiquitin-binding at this location, either through its UIM or VWA domain. This contribution, however, may be less important for substrate-attached ubiquitin chains, as the mechanical translocation of a substrate into the central pore is expected to pull the attached ubiquitin into the Rpn11 active site.
In addition to the essential Rpn11, the DUBs hUsp14 (scUbp6) and hUCHL5/hUCH37 transiently bind to the proteasome. scUbp6 binds via its UBL domain to the Rpn1 T2 site in the base (Fig. 3A and 3D) [12, 32, 107, 120]. Ubp6’s DUB activity is sensitive to the conformational state of the proteasome and stimulated when its catalytic USP domain contacts the coaxially aligned N-ring and ATPase ring in the engaged states of the proteasome [31, 107, 120]. Consistently, ubiquitin-bound Ubp6 stabilizes the engaged state of the proteasome and can therefore prevent subsequent substrate engagement by blocking the apo conformational state required for the insertion of a substrate’s flexible region into the AAA+ motor ring [107, 120]. Although the detailed mechanism is unknown, Ubp6 has been proposed to remove supernumerary ubiquitin chains from proteasomal substrates [121]. hUCHL5 binds to Rpn13 [52–54] and, similar to Usp14, its DUB activity is stimulated through this interaction with the proteasome [52–54, 122–124]. hUCHL5 has been proposed to edit ubiquitin chains on substrates or ubiquitinated proteasomal subunits, and to cleave ubiquitin chains bound to proteasome receptors [125–127]. Both Usp14 and UCHL5 stimulate the ATPase and peptidase activity of the proteasome [128–130], yet the detailed underlying mechanisms and how these effects contribute to substrate degradation remain unclear.
Ubiquitination machinery at the proteasome
As new functional roles continue to be discovered for the shuttle factors, multiple E3 ligases have been reported to associate with the proteasome as well [31, 79, 116, 131, 132]. It is possible that these ligases are recruited to ubiquitinate proteasome components [126, 132, 133]. For UBE3C, the alternative role of remodeling substrate-attached ubiquitin chains at the proteasome has been proposed [134]. UBE3C was the first E3 ubiquitin ligase reported to physically interact with the proteasome [134, 135], but its binding location has yet to be elucidated, and its recruitment to the proteasome appears to be assisted by structurally impaired substrates [133].
Recently, the UBE3A/E6AP AZUL (amino-terminal zinc-binding domain of ubiquitin E3a ligase) was found to bind with 12 nM affinity to a site in the C-terminal flexible region of hRpn10, named RAZUL (Rpn10 AZUL-binding domain) [116]. Although intrinsically disordered on its own, RAZUL adopts a helical structure to form a 4-helix bundle with the E6AP AZUL domain (Fig. 3B) [116]. Despite the presence of ~600 cellular E3 ligases, knockdown of UBE3A by RNAi leads to a reduction of ubiquitinated proteins at the proteasome [116]. Whether this observable reduction is caused by a global effect on all substrates or a specific subset of substrates particularly reliant on UBE3A remains to be elucidated. This finding suggests that UBE3A may belong to a subset of E3 ligases that are prioritized for the proteasome, such as by physically co-localizing substrate ubiquitination and proteasomal degradation.
UBE3A isoform 3 depends on binding to hRpn10 for its nuclear localization [136], although it is not clear whether this interaction occurs with isolated hRpn10 outside of the 26S proteasome context. In the nucleus, UBE3A was found to co-localize with proteasomes within phase separated liquid droplets induced by hyperosmotic stress [84]. UBE3A also appears to have a regulatory role in the formation of these proteolytic compartments, which decrease in abundance by ~30% upon UBE3A knockout [84]. This effect may be caused by reduced substrate ubiquitination, as hHR23b and ubiquitinated proteins are required for proteasome foci formation [84].
UBA3A is at the forefront of multiple diseases. It is hijacked by human papilloma virus (HPV) oncoprotein E6 to trigger p53 degradation, contributing to cervical cancer [137–139]. Furthermore, it appears to be a driver of metastatic prostate cancer [140, 141], loss-of-function mutations are linked to Angelman syndrome [142–144], and elevated gene dosage is correlated with autism spectrum disorders [145]. The recent findings of UBE3A having a dedicated, high affinity binding site at the proteasome, and its involvement in proteasome foci formation spur new research directions towards understanding the role of UBE3A in these diseases.
Huwe1, which also ubiquitinates p53 [146–148], has been found at the proteasome [132], although its mode of interaction remains unclear. Similarly, the N-end rule E3 Ubr4 [149] binds to the proteasome through an unknown mechanism [132]. The E3 ligase parkin is recruited to the proteasome by its UBL domain binding to the Rpn13 Pru domain [33]. Mutations in parkin and its activating protein kinase PINK1 cause defects in mitophagy, the process by which damaged mitochondria are removed from cells, and lead to autosomal-recessive juvenile parkinsonism [150, 151]. The clearance of parkin substrates upon mitochondrial membrane depolarization is reported to be delayed following Rpn13 knockdown [33], suggesting that interaction with the proteasome through Rpn13 plays a role in parkin-mediated protein degradation.
Proteasome as a therapeutic target
Capping the aforementioned advancements in the current understanding of proteasome biology is the breakthrough application of PROTAC (proteolysis targeting chimera) strategies to combat various diseases [152]. Whereas conventional pharmaceutical approaches target and inactivate proteins of interest, PROTACs use a bifunctional architecture to bridge proteins of interest with an E3 ubiquitin ligase for ubiquitination and in turn degradation by the proteasome. This mechanism mimics the activity described for HPV E6-induced ubiquitination of p53 by UBE3A. An outstanding question is whether HPV E6 targets UBE3A among the ~600 E3s because it can directly bind the proteasome and, by extension, whether this strategy of targeting a proteasome-binding E3 would be advantageous as a PROTAC approach. Critical to assess is also why PROTAC approaches lead to rapid degradation of some target proteins, but fail for many others. One aspect to consider is whether ubiquitination of a particular substrate will be sufficient for proteolysis by the proteasome. Some targets may lack appropriate intrinsically unstructured initiation regions, and it is not clear how pervasively the ubiquitin-dependent protein unfoldase Cdc48/p97/VCP can assist by preparing such ubiquitinated proteins of interest for proteasomal engagement and degradation.
Currently, less than ten E3s have been exploited as PROTACs. High affinity binders for the Cullin RING E3 ligase substrate recognition subunits cereblon (CRBN) and von Hippel-Lindau (VHL) tumor suppressor protein have allowed these two E3 complexes to be exploited effectively for the PROTAC technology [153–156]. The success of these PROTACs will likely drive expansion to other E3 ligase complexes, adding new modes of control over protein lifetimes. Alternative targeting strategies are also emerging, including autophagy-targeting chimeras (AUTACs) that use a guanine derivative as a degradation tag [157]. Endogenous 8-nitroguanosine 3’,5’-cyclic monophosphase (8-nitro-cGMP) modification (S-guanylation) of bacteria causes subsequent K63-linked ubiquitination and triggers clearance by ubiquitin-mediated selective autophagic [158, 159]. By exploiting this mechanism, AUTACs have been demonstrated to promote targeted mitochondrial turnover via mitophagy and improve mitochondrial activity in Down syndrome-derived fibroblast cells through preferential degradation of impaired mitochondria [157].
The first application of proteasome biology to medicine was the inhibition of the CP, which is an established treatment for hematological cancers, reviewed in [160]. In an effort to develop alternative proteasome inhibitors, a class of chalcone derivatives, including RA190, were identified to bind to hRpn13, induce apoptosis in cancer cell lines, and restrict growth of tumor xenografts [161, 162]. An independent study further identified an hRpn13-binding peptoid ligand that similarly induces death of multiple myeloma cells [163]. Each of these hRpn13-targeting approaches were found to be synergistic with inhibition of the proteasome CP [163, 164]. RA190-induced accumulation of ubiquitinated proteins at the proteasome is lost upon deletion of hRpn13 [62] or its Pru domain [165]. Loss of cell viability however by either of these two hRpn13-targeting compounds does not seem to require hRpn13 according to hRpn13-knockdown experiments [166, 167] and a cell line with a defective hRpn13 Pru domain [165], although conflicting data on this matter exists [164]. These compounds are able to interact with other cellular components, which likely contribute to the induction of apoptosis [166, 167]. Nonetheless, these findings collectively suggest that hRpn13 is a promising therapeutic target. RA190 was fused to the CRBN binding ligand thalidomide to induce its degradation, but the benefit in this case was found to be modest [168]. Of higher interest may be the development of a PROTAC that targets proteins to hRpn13 at the proteasome and thus substitutes for ubiquitin in the delivery of substrates to the degradation machine.
Since substrate deubiquitination is an essential step for proteasomal degradation, additional proteasome inhibitors have been developed by targeting the zinc-dependent metalloprotease Rpn11. Two small-molecule inhibitors of Rpn11 were discovered by screening a library of metal binding pharmacophores; 8-thioquinoline (8TQ, IC50 ~2.5 μM) and capzimin (a 8TQ derivative, IC50 ~400 nM) [169], which also block proliferation of cancer cells in culture. Additional Rpn11 inhibitors have been described to prevent substrate deubiquitination by acting as zinc chelators, like SOP11 (IC50 ~1.3 μM) and thiolutin. SOP11 was identified via high throughput screening by using a novel proteasome degradation assay, and was found to stabilize some proteasomal substrates, induce the unfolded protein response, and trigger apoptosis [170]. Thiolutin is a disulfide-containing antibiotic and antiangiogenic compound produced by Streptomyces that, in its reduced form, inhibits Rpn11 [171]. SOP11 and reduced thiolutin were found to also inhibit other JAMM metalloproteases, suggesting that additional optimization may be required for improved Rpn11 specificity. A more extensive review of these inhibitors can be found in [172, 173].
Perspectives
Over the past decade, an arsenal of modern scientific technologies has been applied to the UPS, unveiling the proteasome as a sophisticated machine that tightly couples structure and molecular motion to functional activity. It is clear that the UPS relies on many additional layers of regulation and mechanistic action beyond the current understanding, which most likely represents a bare-bones and incomplete comprehension. For example, the role of the many known post-translational modifications to proteasome subunits remain undefined. It has been postulated that the presence of shuttle factors and even intrinsic subunits at the proteasome is influenced by ubiquitination [57, 132], sumoylation [174], and phosphorylation [175, 176]. Nonetheless, what has emerged is a strong appreciation of the dependency of functional activity on dynamic motions, including redistribution of the relative population of conformational states upon binding of substrate or effectors. How this redistribution is implemented remains to be discovered, but single molecule experiments are likely to provide important insights by revealing whether the relative population of distinct proteasome states is defined by longer residency time of the more populated states, shorter times in lesser populated states, and/or changes to activation barriers that affect switching between such states. Additional factors will likely influence how these dynamic relationships play out in a crowded cellular context. Over the next decade, it will be critical to define these aspects of biological function, which will also serve as a catalyst of new emergent technology, just as the need for structural information drove forward technological advancements in cryoelectron microscopy, x-ray crystallography, and NMR spectroscopy. Such interplay between dynamics and function is sure to apply to many other biological systems.
We focused in this article on events that occur after proteasome assembly, beginning with the dynamic binding of ubiquitinated substrates, which occurs at multiple, weak affinity receptor sites that can collectively impart the affinity needed to hold substrates at the proteasome without rigidly locking them into a fixed orientation. The ubiquitin-binding UBA domains of shuttle factors expand this activity, but how all of these binding sites are organized and whether multiple shuttle factors bind to the RP at the same time is not yet known, and further structural studies will be required to answer these questions. Based on current knowledge, it is reasonable to propose that dynamic ubiquitin interactions with various binding sites enables the stochastic motion of the substrate required for its unstructured initiation region to engage with the ATPase motor. Engagement of the substrate’s initiation region with the ATPase motor then drives the conformational switch that facilitates the mechanical translocation and co-translocational deubiquitination needed for substrate entry into the degradation chamber. Given the presence of additional DUBs and E3 ligases at the proteasome, it is likely that ubiquitin chains are remodeled, but future experiments are needed to test this model and assess to what extent proteasome subunits themselves are ubiquitinated (or modified in other ways) as part of the protein degradation cycle.
We have been limited in what we can cover in this article, omitting discussion of proteasome assembly as an important area of research that requires further investigation. We have also focused on the RP-capped 26S proteasome, although there are other ATP-independent regulators capping the proteasome CP. The function of these regulators and whether they act in concert with the RP remains to be elucidated. For example, it is still unclear what fraction of proteasomes exist in hybrid states with different regulators capping each end of the CP. Long-range allosteric interactions have been uncovered between regulators and the CP [177–180], as well as evidence that this long-range allostery does not extend to opposite ends of the CP, suggesting that the CP is capable of being simultaneously sensitive to allosteric signaling at both ends [181]. A greater understanding of these long-range interactions and in particular how they occur between the RP and CP will likely emerge in the next decade and provide a more complete understanding of the functional roles performed by the many CP-binding factors.
The proteasome is an impressive biomachine, the scrutiny of which provides foundational information on how cells apply structural and dynamic properties to execute mechano-chemical events. Yet its detailed study is also motivated by the already existing and growing therapeutic impact of the UPS. PROTAC-mediated turnover of target proteins provides revolutionary opportunity in precision medicine. That this approach mimics viral strategies in interacting with eukaryotic hosts, an evolutionary relationship that spans ~2.5 billion years, is perhaps the most compelling reason to invest heavily in this approach. Finding ways to expand PROTAC targeting to include a larger plethora of E3 ligases will open avenues for precise substrate targeting in specific tissues. The UPS is thus likely to spur myriad new medicines over the next decade directed towards the improvement of human health.
Acknowledgements
The research of X.C. and K.J.W. is supported by the Intramural Research Program through the Center for Cancer Research, National Cancer Institute, National Institutes of Health (1 ZIA BC011490 to K.J.W.). A.M. is an investigator of the Howard Hughes Medical Institute. Z.M.H, E.L.A., and A.M. were funded by HHMI and the National Institutes of Health (R01-GM094497 to A.M.). E.L.A. was also funded by the Ford Foundation Predoctoral Fellowship.
Abbreviations
- 8-nitro-cGMP
8-nitroguanosine 3’,5’-cyclic monophosphase
- 8TQ
8-thioquinoline
- AAA+
ATPases Associated with various cellular Activities
- APC/C
anaphase-promoting complex/cyclosome
- AUTACs
autophagy-targeting chimeras
- AZUL
amino-terminal zinc-binding domain of ubiquitin E3a ligase
- CP
core particle
- CRBN
cereblon
- cryo-EM
cryo-electron microscopy
- DUB
deubiquitinating enzyme
- HbYX
hydrophobic-tyrosine-any amino acid
- HPV
human papilloma virus
- JAMM
Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+)
- NMR
nuclear magnetic resonance
- PROTAC
proteolysis targeting chimera
- Pru
pleckstrin-like receptor for ubiquitin
- RAZUL
Rpn10 AZUL-binding domain
- RP
regulatory particle
- Rpn
Regulatory particle non-ATPase
- Rpt
Regulatory particle triphosphatase
- UBA
ubiquitin-associated
- UBL
ubiquitin-like
- UIM
ubiquitin interaction motif
- UPS
ubiquitin proteasome system
- VHL
von Hippel-Lindau
- VWA
von Willebrand factor type A
Footnotes
Conflicts of Interest
The authors have no conflicts of interest or disclosures to declare.
References
- 1.Prakash S, Tian L, Ratliff KS, Lehotzky RE & Matouschek A (2004) An unstructured initiation site is required for efficient proteasome-mediated degradation, Nat Struct Mol Biol. 11, 830–7. [DOI] [PubMed] [Google Scholar]
- 2.Fishbain S, Prakash S, Herrig A, Elsasser S & Matouschek A (2011) Rad23 escapes degradation because it lacks a proteasome initiation region, Nat Commun. 2, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inobe T, Fishbain S, Prakash S & Matouschek A (2011) Defining the geometry of the two-component proteasome degron, Nat Chem Biol. 7, 161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DM, Chang SC, Park S, Finley D, Cheng Y & Goldberg AL (2007) Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry, Mol Cell. 27, 731–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E & Martin A (2012) Complete subunit architecture of the proteasome regulatory particle, Nature. 482, 186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A & Baumeister W (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach, Proc Natl Acad Sci U S A. 109, 1380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W & Forster F (2012) Near-atomic resolution structural model of the yeast 26S proteasome, Proc Natl Acad Sci U S A. 109, 14870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matyskiela ME, Lander GC & Martin A (2013) Conformational switching of the 26S proteasome enables substrate degradation, Nat Struct Mol Biol. 20, 781–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Śledź P, Unverdorben P, Beck F, Pfeifer G, Schweitzer A, Förster F & Baumeister W (2013) Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation, Proc Natl Acad Sci U S A. 110, 7264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unverdorben P, Beck F, Sledz P, Schweitzer A, Pfeifer G, Plitzko JM, Baumeister W & Forster F (2014) Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome, Proc Natl Acad Sci U S A. 111, 5544–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Wu J, Lu Y, Ma YB, Lee BH, Yu Z, Ouyang Q, Finley DJ, Kirschner MW & Mao Y (2016) Structural basis for dynamic regulation of the human 26S proteasome, Proc Natl Acad Sci U S A. 113, 12991–12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, Luan B, Wu J & Shi Y (2016) An atomic structure of the human 26S proteasome, Nat Struct Mol Biol. 23, 778–85. [DOI] [PubMed] [Google Scholar]
- 13.Luan B, Huang X, Wu J, Mei Z, Wang Y, Xue X, Yan C, Wang J, Finley DJ, Shi Y & Wang F (2016) Structure of an endogenous yeast 26S proteasome reveals two major conformational states, Proc Natl Acad Sci U S A. 113, 2642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding Z, Fu Z, Xu C, Wang Y, Wang Y, Li J, Kong L, Chen J, Li N, Zhang R & Cong Y (2017) High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx, Cell Res. 27, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wehmer M, Rudack T, Beck F, Aufderheide A, Pfeifer G, Plitzko JM, Forster F, Schulten K, Baumeister W & Sakata E (2017) Structural insights into the functional cycle of the ATPase module of the 26S proteasome, Proc Natl Acad Sci U S A. 114, 1305–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Worden EJ, Dong KC & Martin A (2017) An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome, Mol Cell. 67, 799–811 e8. [DOI] [PubMed] [Google Scholar]
- 17.Eisele MR, Reed RG, Rudack T, Schweitzer A, Beck F, Nagy I, Pfeifer G, Plitzko JM, Baumeister W, Tomko RJ Jr. & Sakata E (2018) Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating, Cell Rep. 24, 1301–1315 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y, Wang WL, Yu D, Ouyang Q, Lu Y & Mao Y (2018) Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome, Nat Commun. 9, 1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene ER, Goodall EA, de la Pena AH, Matyskiela ME, Lander GC & Martin A (2019) Specific lid-base contacts in the 26s proteasome control the conformational switching required for substrate degradation, Elife. 8, e49806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komander D & Rape M (2012) The ubiquitin code, Annu Rev Biochem. 81, 203–29. [DOI] [PubMed] [Google Scholar]
- 21.Wertz IE, Newton K, Seshasayee D, Kusam S, Lam C, Zhang J, Popovych N, Helgason E, Schoeffler A, Jeet S, Ramamoorthi N, Kategaya L, Newman RJ, Horikawa K, Dugger D, Sandoval W, Mukund S, Zindal A, Martin F, Quan C, Tom J, Fairbrother WJ, Townsend M, Warming S, DeVoss J, Liu J, Dueber E, Caplazi P, Lee WP, Goodnow CC, Balazs M, Yu K, Kolumam G & Dixit VM (2015) Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation, Nature. 528, 370–5. [DOI] [PubMed] [Google Scholar]
- 22.Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D & Cohen P (2013) Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains, Proc Natl Acad Sci U S A. 110, 15247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Smits AH, van Tilburg GB, Jansen PW, Makowski MM, Ovaa H & Vermeulen M (2017) An Interaction Landscape of Ubiquitin Signaling, Mol Cell. 65, 941–955 e8. [DOI] [PubMed] [Google Scholar]
- 24.Grice GL, Lobb IT, Weekes MP, Gygi SP, Antrobus R & Nathan JA (2015) The Proteasome Distinguishes between Heterotypic and Homotypic Lysine-11-Linked Polyubiquitin Chains, Cell Rep. 12, 545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer HJ & Rape M (2014) Enhanced protein degradation by branched ubiquitin chains, Cell. 157, 910–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sowa ME, Bennett EJ, Gygi SP & Harper JW (2009) Defining the human deubiquitinating enzyme interaction landscape, Cell. 138, 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cappadocia L & Lima CD (2018) Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism, Chem Rev. 118, 889–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swatek KN & Komander D (2016) Ubiquitin modifications, Cell Res. 26, 399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F & Walters KJ (2010) Multitasking with ubiquitin through multivalent interactions, Trends Biochem Sci. 35, 352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh E, Akopian D & Rape M (2018) Principles of Ubiquitin-Dependent Signaling, Annu Rev Cell Dev Biol. 34, 137–162. [DOI] [PubMed] [Google Scholar]
- 31.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H & Finley D (2002) Multiple associated proteins regulate proteasome structure and function., Mol Cell. 10, 495–507. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, Shi Y, Zhang N, de Poot SA, Tuebing F, Sun S, Vannoy J, Tarasov SG, Engen JR, Finley D & Walters KJ (2016) Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome, Science. 351, aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguileta MA, Korac J, Durcan TM, Trempe JF, Haber M, Gehring K, Elsasser S, Waidmann O, Fon EA & Husnjak K (2015) The E3 ubiquitin ligase parkin is recruited to the 26 S proteasome via the proteasomal ubiquitin receptor Rpn13, J Biol Chem. 290, 7492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA & Finley D (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains, Nat Cell Biol. 4, 725–30. [DOI] [PubMed] [Google Scholar]
- 35.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D & Dikic I (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor, Nature. 453, 481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH & Hanaoka F (1999) Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome, J Biol Chem. 274, 28019–25. [DOI] [PubMed] [Google Scholar]
- 37.Walters KJ, Kleijnen MF, Goh AM, Wagner G & Howley PM (2002) Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a, Biochemistry. 41, 1767–77. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Ebelle DL, Wright BJ, Sridharan V, Hooper E & Walters KJ (2019) Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome, J Biol Chem. 431, 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Randles L, Shi K, Tarasov SG, Aihara H & Walters KJ (2016) Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome, Structure. 24, 1257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poulsen EG, Kampmeyer C, Kriegenburg F, Johansen JV, Hofmann K, Holmberg C & Hartmann-Petersen R (2017) UBL/BAG-domain co-chaperones cause cellular stress upon overexpression through constitutive activation of Hsf1, Cell Stress Chaperones. 22, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demand J, Alberti S, Patterson C & Höhfeld J (2001) Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling, Curr Biol. 11, 1569–77. [DOI] [PubMed] [Google Scholar]
- 42.Lüders J, Demand J & Höhfeld J (2000) The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome, J Biol Chem. 275, 4613–7. [DOI] [PubMed] [Google Scholar]
- 43.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW & Finley D (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation, Cell. 127, 99–111. [DOI] [PubMed] [Google Scholar]
- 44.Hanna J, Meides A, Zhang DP & Finley D (2007) A ubiquitin stress response induces altered proteasome composition, Cell. 129, 747–59. [DOI] [PubMed] [Google Scholar]
- 45.Collins GA & Goldberg AL (2020) Proteins containing ubiquitin-like (Ubl) domains not only bind to 26S proteasomes but also induce their activation, Proc Natl Acad Sci U S A. 117, 4664–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deveraux Q, Ustrell V, Pickart C & Rechsteiner M (1994) A 26 S protease subunit that binds ubiquitin conjugates, J Biol Chem. 269, 7059–61. [PubMed] [Google Scholar]
- 47.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ & Groll M (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction, Nature. 453, 548–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Randles L & Walters KJ (2012) Ubiquitin and its binding domains, Front Biosci (Landmark Ed). 17, 2140–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, Kang Y, Haririnia A, Storaska AJ, Cole JL, Fushman D & Walters KJ (2009) Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13, Mol Cell. 35, 280–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Young P & Walters KJ (2005) Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition, J Mol Biol. 348, 727–39. [DOI] [PubMed] [Google Scholar]
- 51.Lu X, Ebelle DL, Matsuo H & Walters KJ (2020) An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure, Structure. 28, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW & Cohen RE (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1, Nat Cell Biol. 8, 994–1002. [DOI] [PubMed] [Google Scholar]
- 53.Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K & Murata S (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes, EMBO J. 25, 4524–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L & Goldberg AL (2006) hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37, EMBO J. 25, 5742–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lam YA, Lawson TG, Velayutham M, Zweier JL & Pickart CM (2002) A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal, Nature. 416, 763–7. [DOI] [PubMed] [Google Scholar]
- 56.Paraskevopoulos K, Kriegenburg F, Tatham MH, Rösner HI, Medina B, Larsen IB, Brandstrup R, Hardwick KG, Hay RT, Kragelund BB, Hartmann-Petersen R & Gordon C (2014) Dss1 is a 26S proteasome ubiquitin receptor, Mol Cell. 56, 453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keren-Kaplan T, Zeev Peters L, Levin-Kravets O, Attali I, Kleifeld O, Shohat N, Artzi S, Zucker O, Pilzer I, Reis N, Glickman MH, Ben-Aroya S & Prag G (2016) Structure of ubiquitylated-Rpn10 provides insight into its autoregulation mechanism, Nat Commun. 7, 12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong Y, Zhang S, Wu Z, Li X, Wang WL, Zhu Y, Stoilova-McPhie S, Lu Y, Finley D & Mao Y (2019) Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome, Nature. 565, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X, Dorris Z, Shi D, Huang RK, Khant H, Fox T, de Val N, Williams D, Zhang P & Walters KJ (2020) Cryo-EM Reveals Unanchored M1-Ubiquitin Chain Binding at hRpn11 of the 26S Proteasome, Structure. 28, 1206–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D & Peng J (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation, Cell. 137, 133–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohtake F, Tsuchiya H, Saeki Y & Tanaka K (2018) K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains, Proc Natl Acad Sci U S A. 115, E1401–E1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu X, Nowicka U, Sridharan V, Liu F, Randles L, Hymel D, Dyba M, Tarasov SG, Tarasova NI, Zhao XZ, Hamazaki J, Murata S, Burke TR & Walters KJ (2017) Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets, Nat Commun. 8, 15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deol KK, Crowe SO, Du J, Bisbee H, Guenette RG & Strieter ER (2020) Proteasome-Bound UCH37/UCHL5 Debranches Ubiquitin Chains to Promote Degradation, Mol Cell. 80, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martinez-Fonts K, Davis C, Tomita T, Elsasser S, Nager AR, Shi Y, Finley D & Matouschek A (2020) The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates, Nat Commun. 11, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jacobson AD, Zhang NY, Xu P, Han KJ, Noone S, Peng J & Liu CW (2009) The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome, J Biol Chem. 284, 35485–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lai MY, Zhang D, Laronde-Leblanc N & Fushman D (2012) Structural and biochemical studies of the open state of Lys48-linked diubiquitin, Biochim Biophys Acta. 1823, 2046–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cook WJ, Jeffrey LC, Carson M, Chen Z & Pickart CM (1992) Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2), J Biol Chem. 267, 16467–71. [DOI] [PubMed] [Google Scholar]
- 68.VanderLinden RT, Hemmis CW, Yao T, Robinson H & Hill CP (2017) Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism, J Biol Chem. 292, 9493–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q, Goh AM, Howley PM & Walters KJ (2003) Ubiquitin recognition by the DNA repair protein hHR23a, Biochemistry. 42, 13529–35. [DOI] [PubMed] [Google Scholar]
- 70.Ryu KS, Lee KJ, Bae SH, Kim BK, Kim KA & Choi BS (2003) Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a, J Biol Chem. 278, 36621–7. [DOI] [PubMed] [Google Scholar]
- 71.Riedinger C, Boehringer J, Trempe JF, Lowe ED, Brown NR, Gehring K, Noble ME, Gordon C & Endicott JA (2010) Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12, J Biol Chem. 285, 33992–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, Glickman MH & Fushman D (2009) Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor, Mol Cell. 36, 1018–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosenzweig R, Bronner V, Zhang D, Fushman D & Glickman MH (2012) Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome, J Biol Chem. 287, 14659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gomez TA, Kolawa N, Gee M, Sweredoski MJ & Deshaies RJ (2011) Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1., BMC Biol. 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mueller TD & Feigon J (2003) Structural determinants for the binding of ubiquitin-like domains to the proteasome, EMBO J. 22, 4634–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujiwara K, Tenno T, Sugasawa K, Jee JG, Ohki I, Kojima C, Tochio H, Hiroaki H, Hanaoka F & Shirakawa M (2004) Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B, J Biol Chem. 279, 4760–7. [DOI] [PubMed] [Google Scholar]
- 77.Ko HS, Uehara T, Tsuruma K & Nomura Y (2004) Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains, FEBS Lett. 566, 110–4. [DOI] [PubMed] [Google Scholar]
- 78.Chen X, Randles L, Shi K, Tarasov SG, Aihara H & Walters KJ (2016) Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome, Structure. 24, 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X, Chen CF, Baker PR, Chen PL, Kaiser P & Huang L (2007) Mass spectrometric characterization of the affinity-purified human 26S proteasome complex, Biochemistry. 46, 3553–65. [DOI] [PubMed] [Google Scholar]
- 80.Yu C, Yang Y, Wang X, Guan S, Fang L, Liu F, Walters KJ, Kaiser P & Huang L (2016) Characterization of Dynamic UbR-Proteasome Subcomplexes by In vivo Cross-linking (X) Assisted Bimolecular Tandem Affinity Purification (XBAP) and Label-free Quantitation, Mol Cell Proteomics. 15, 2279–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ & Walters KJ (2006) UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain, Journal of molecular biology. 356, 1027–35. [DOI] [PubMed] [Google Scholar]
- 82.Kang Y, Zhang N, Koepp DM & Walters KJ (2007) Ubiquitin receptor proteins hHR23a and hPLIC2 interact, J Mol Biol. 365, 1093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walters KJ, Lech PJ, Goh AM, Wang Q & Howley PM (2003) DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a, Proc Natl Acad Sci U S A. 100, 12694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yasuda S, Tsuchiya H, Kaiho A, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T, Baumeister W, Fernández-Busnadiego R, Tanaka K & Saeki Y (2020) Stress- and ubiquitylation-dependent phase separation of the proteasome, Nature. 578, 296–300. [DOI] [PubMed] [Google Scholar]
- 85.Dao TP, Kolaitis RM, Kim HJ, O’Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP & Castaneda CA (2018) Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions, Mol Cell. 69, 965–978 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakata E, Bohn S, Mihalache O, Kiss P, Beck F, Nagy I, Nickell S, Tanaka K, Saeki Y, Forster F & Baumeister W (2012) Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy, Proc Natl Acad Sci U S A. 109, 1479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu X, Liu F, Durham SE, Tarasov SG & Walters KJ (2015) A High Affinity hRpn2-Derived Peptide That Displaces Human Rpn13 from Proteasome in 293T Cells, PLoS One. 10, e0140518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen X & Walters KJ (2012) Identifying and studying ubiquitin receptors by NMR, Methods Mol Biol. 832, 279–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD & Huber R (1997) Structure of 20S proteasome from yeast at 2.4 A resolution, Nature. 386, 463–71. [DOI] [PubMed] [Google Scholar]
- 90.Lu Y, Lee BH, King RW, Finley D & Kirschner MW (2015) Substrate degradation by the proteasome: a single-molecule kinetic analysis, Science. 348, 1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bard JAM, Bashore C, Dong KC & Martin A (2019) The 26S Proteasome Utilizes a Kinetic Gateway to Prioritize Substrate Degradation, Cell. 177, 286–298 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tomko RJ, Funakoshi M, Schneider K, Wang J & Hochstrasser M (2010) Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly, Mol Cell. 38, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martin A, Baker TA & Sauer RT (2008) Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding, Nat Struct Mol Biol. 15, 1147–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamada-Inagawa T, Okuno T, Karata K, Yamanaka K & Ogura T (2003) Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis, J Biol Chem. 278, 50182–7. [DOI] [PubMed] [Google Scholar]
- 95.Erales J, Hoyt MA, Troll F & Coffino P (2012) Functional asymmetries of proteasome translocase pore, J Biol Chem. 287, 18535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW & Horwich AL (2005) Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation, Cell. 121, 1029–41. [DOI] [PubMed] [Google Scholar]
- 97.Beckwith R, Estrin E, Worden EJ & Martin A (2013) Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase, Nat Struct Mol Biol. 20, 1164–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de la Peña AH, Goodall EA, Gates SN, Lander GC & Martin A (2018) Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation, Science. 362, eaav0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fishbain S, Inobe T, Israeli E, Chavali S, Yu H, Kago G, Babu MM & Matouschek A (2015) Sequence composition of disordered regions fine-tunes protein half-life, Nat Struct Mol Biol. 22, 214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu H, Singh Gautam AK, Wilmington SR, Wylie D, Martinez-Fonts K, Kago G, Warburton M, Chavali S, Inobe T, Finkelstein IJ, Babu MM & Matouschek A (2016) Conserved Sequence Preferences Contribute to Substrate Recognition by the Proteasome, J Biol Chem. 291, 14526–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van der Lee R, Lang B, Kruse K, Gsponer J, Sánchez de Groot N, Huynen MA, Matouschek A, Fuxreiter M & Babu MM (2014) Intrinsically disordered segments affect protein half-life in the cell and during evolution, Cell Rep. 8, 1832–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Olszewski MM, Williams C, Dong KC & Martin A (2019) The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome, Commun Biol. 2, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Twomey EC, Ji Z, Wales TE, Bodnar NO, Ficarro SB, Marto JA, Engen JR & Rapoport TA (2019) Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding, Science. 365, eaax1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Carroll EC, Greene ER, Martin A & Marqusee S (2020) Site-specific ubiquitination affects protein energetics and proteasomal degradation, Nat Chem Biol. 16, 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gates SN & Martin A (2020) Stairway to translocation: AAA+ motor structures reveal the mechanisms of ATP-dependent substrate translocation, Protein Sci. 29, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peth A, Besche HC & Goldberg AL (2009) Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening, Mol Cell. 36, 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC & Martin A (2015) Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome, Nat Struct Mol Biol. 22, 712–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li X & Demartino GN (2009) Variably modulated gating of the 26S proteasome by ATP and polyubiquitin, Biochem J. 421, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bech-Otschir D, Helfrich A, Enenkel C, Consiglieri G, Seeger M, Holzhütter HG, Dahlmann B & Kloetzel PM (2009) Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome, Nat Struct Mol Biol. 16, 219–25. [DOI] [PubMed] [Google Scholar]
- 110.Ding Z, Xu C, Sahu I, Wang Y, Fu Z, Huang M, Wong CCL, Glickman MH & Cong Y (2019) Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations, Mol Cell. 73, 1150–1161.e6. [DOI] [PubMed] [Google Scholar]
- 111.Asano S, Fukuda Y, Beck F, Aufderheide A, Förster F, Danev R & Baumeister W (2015) Proteasomes. A molecular census of 26S proteasomes in intact neurons, Science. 347, 439–42. [DOI] [PubMed] [Google Scholar]
- 112.Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko JM, Beck M, Baumeister W & Engel BD (2017) Proteasomes tether to two distinct sites at the nuclear pore complex, Proc Natl Acad Sci U S A. 114, 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Albert S, Wietrzynski W, Lee CW, Schaffer M, Beck F, Schuller JM, Salomé PA, Plitzko JM, Baumeister W & Engel BD (2020) Direct visualization of degradation microcompartments at the ER membrane, Proc Natl Acad Sci U S A. 117, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guo Q, Lehmer C, Martínez-Sánchez A, Rudack T, Beck F, Hartmann H, Pérez-Berlanga M, Frottin F, Hipp MS, Hartl FU, Edbauer D, Baumeister W & Fernández-Busnadiego R (2018) In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment, Cell. 172, 696–705.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yao T & Cohen RE (2002) A cryptic protease couples deubiquitination and degradation by the proteasome, Nature. 419, 403–7. [DOI] [PubMed] [Google Scholar]
- 116.Buel GR, Chen X, Chari R, O’Neill MJ, Ebelle DL, Jenkins C, Sridharan V, Tarasov SG, Tarasova NI, Andresson T & Walters KJ (2020) Structure of E3 ligase E6AP with a proteasome-binding site provided by substrate receptor hRpn10, Nat Commun. 11, 1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA & Finley D (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3, Cell. 94, 615–23. [DOI] [PubMed] [Google Scholar]
- 118.da Fonseca PC, He J & Morris EP (2012) Molecular model of the human 26S proteasome, Mol Cell. 46, 54–66. [DOI] [PubMed] [Google Scholar]
- 119.Schweitzer A, Aufderheide A, Rudack T, Beck F, Pfeifer G, Plitzko JM, Sakata E, Schulten K, Forster F & Baumeister W (2016) Structure of the human 26S proteasome at a resolution of 3.9 A, Proc Natl Acad Sci U S A. 113, 7816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, Goldberg AL, Sakata E, Baumeister W & Förster F (2015) Structural characterization of the interaction of Ubp6 with the 26S proteasome, Proc Natl Acad Sci U S A. 112, 8626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee BH, Lu Y, Prado MA, Shi Y, Tian G, Sun S, Elsasser S, Gygi SP, King RW & Finley D (2016) USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites, Nature. 532, 398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jiao L, Ouyang S, Shaw N, Song G, Feng Y, Niu F, Qiu W, Zhu H, Hung LW, Zuo X, Eleonora Shtykova V, Zhu P, Dong YH, Xu R & Liu ZJ (2014) Mechanism of the Rpn13-induced activation of Uch37, Protein Cell. 5, 616–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vander Linden RT, Hemmis CW, Schmitt B, Ndoja A, Whitby FG, Robinson H, Cohen RE, Yao T & Hill CP (2015) Structural basis for the activation and inhibition of the UCH37 deubiquitylase, Mol Cell. 57, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H & Sixma TK (2015) Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G, Mol Cell. 57, 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lam YA, Xu W, DeMartino GN & Cohen RE (1997) Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome, Nature. 385, 737–40. [DOI] [PubMed] [Google Scholar]
- 126.Jacobson AD, MacFadden A, Wu Z, Peng J & Liu CW (2014) Autoregulation of the 26S proteasome by in situ ubiquitination, Mol Biol Cell. 25, 1824–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang NY, Jacobson AD, Macfadden A & Liu CW (2011) Ubiquitin chain trimming recycles the substrate binding sites of the 26 S proteasome and promotes degradation of lysine 48-linked polyubiquitin conjugates, J Biol Chem. 286, 25540–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peth A, Kukushkin N, Bossé M & Goldberg AL (2013) Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs, J Biol Chem. 288, 7781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim HT & Goldberg AL (2017) The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis, J Biol Chem. 292, 9830–9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim HT & Goldberg AL (2018) UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells, Proc Natl Acad Sci U S A. 115, E11642–E11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Martinez-Noel G, Galligan JT, Sowa ME, Arndt V, Overton TM, Harper JW & Howley PM (2012) Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes, Mol Cell Biol. 32, 3095–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Besche HC, Sha Z, Kukushkin NV, Peth A, Hock EM, Kim W, Gygi S, Gutierrez JA, Liao H, Dick L & Goldberg AL (2014) Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates, EMBO J. 33, 1159–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gottlieb CD, Thompson ACS, Ordureau A, Harper JW & Kopito RR (2019) Acute unfolding of a single protein immediately stimulates recruitment of ubiquitin protein ligase E3C (UBE3C) to 26S proteasomes, J Biol Chem. 294, 16511–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP & Finley D (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities, Cell. 127, 1401–13. [DOI] [PubMed] [Google Scholar]
- 135.You J & Pickart CM (2001) A HECT domain E3 enzyme assembles novel polyubiquitin chains, J Biol Chem. 276, 19871–8. [DOI] [PubMed] [Google Scholar]
- 136.Avagliano Trezza R, Sonzogni M, Bossuyt SNV, Zampeta FI, Punt AM, van den Berg M, Rotaru DC, Koene LMC, Munshi ST, Stedehouder J, Kros JM, Williams M, Heussler H, de Vrij FMS, Mientjes EJ, van Woerden GM, Kushner SA, Distel B & Elgersma Y (2019) Loss of nuclear UBE3A causes electrophysiological and behavioral deficits in mice and is associated with Angelman syndrome, Nat Neurosci. 22, 1235–1247. [DOI] [PubMed] [Google Scholar]
- 137.Huibregtse JM, Scheffner M & Howley PM (1993) Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53, Mol Cell Biol. 13, 775–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huibregtse JM, Scheffner M & Howley PM (1993) Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins, Mol Cell Biol. 13, 4918–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Scheffner M, Huibregtse JM, Vierstra RD & Howley PM (1993) The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53, Cell. 75, 495–505. [DOI] [PubMed] [Google Scholar]
- 140.Gamell C, Bandilovska I, Gulati T, Kogan A, Lim SC, Kovacevic Z, Takano EA, Timpone C, Agupitan AD, Litchfield C, Blandino G, Horvath LG, Fox SB, Williams SG, Russo A, Gallo E, Paul PJ, Mitchell C, Sandhu S, Keam SP, Haupt S, Richardson DR & Haupt Y (2019) E6AP Promotes a Metastatic Phenotype in Prostate Cancer, iScience. 22, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Paul PJ, Raghu D, Chan AL, Gulati T, Lambeth L, Takano E, Herold MJ, Hagekyriakou J, Vessella RL, Fedele C, Shackleton M, Williams ED, Fox S, Williams S, Haupt S, Gamell C & Haupt Y (2016) Restoration of tumor suppression in prostate cancer by targeting the E3 ligase E6AP, Oncogene. 35, 6235–6245. [DOI] [PubMed] [Google Scholar]
- 142.Kishino T, Lalande M & Wagstaff J (1997) UBE3A/E6-AP mutations cause Angelman syndrome, Nat Genet. 15, 70–3. [DOI] [PubMed] [Google Scholar]
- 143.Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM & Beaudet AL (1997) De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome, Nat Genet. 15, 74–7. [DOI] [PubMed] [Google Scholar]
- 144.Cooper EM, Hudson AW, Amos J, Wagstaff J & Howley PM (2004) Biochemical analysis of Angelman syndrome-associated mutations in the E3 ubiquitin ligase E6-associated protein, J Biol Chem. 279, 41208–17. [DOI] [PubMed] [Google Scholar]
- 145.Samaco RC, Hogart A & LaSalle JM (2005) Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3, Hum Mol Genet. 14, 483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kurokawa M, Kim J, Geradts J, Matsuura K, Liu L, Ran X, Xia W, Ribar TJ, Henao R, Dewhirst MW, Kim WJ, Lucas JE, Wang S, Spector NL & Kornbluth S (2013) A network of substrates of the E3 ubiquitin ligases MDM2 and HUWE1 control apoptosis independently of p53, Sci Signal. 6, ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Khoronenkova SV & Dianov GL (2011) The emerging role of Mule and ARF in the regulation of base excision repair, FEBS Lett. 585, 2831–5. [DOI] [PubMed] [Google Scholar]
- 148.Chen D, Kon N, Li M, Zhang W, Qin J & Gu W (2005) ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor, Cell. 121, 1071–83. [DOI] [PubMed] [Google Scholar]
- 149.Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M & Kwon YT (2005) A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons, Mol Cell Biol. 25, 7120–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Harper JW, Ordureau A & Heo JM (2018) Building and decoding ubiquitin chains for mitophagy, Nat Rev Mol Cell Biol. 19, 93–108. [DOI] [PubMed] [Google Scholar]
- 151.Pickles S, Vigié P & Youle RJ (2018) Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance, Curr Biol. 28, R170–R185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Paiva SL & Crews CM (2019) Targeted protein degradation: elements of PROTAC design, Curr Opin Chem Biol. 50, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, Hines J, Winkler JD, Crew AP, Coleman K & Crews CM (2015) Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4, Chem Biol. 22, 755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ottis P, Toure M, Cromm PM, Ko E, Gustafson JL & Crews CM (2017) Assessing Different E3 Ligases for Small Molecule Induced Protein Ubiquitination and Degradation, ACS Chem Biol. 12, 2570–2578. [DOI] [PubMed] [Google Scholar]
- 155.Zengerle M, Chan KH & Ciulli A (2015) Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4, ACS Chem Biol. 10, 1770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S & Bradner JE (2015) DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation, Science. 348, 1376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Takahashi D, Moriyama J, Nakamura T, Miki E, Takahashi E, Sato A, Akaike T, Itto-Nakama K & Arimoto H (2019) AUTACs: Cargo-Specific Degraders Using Selective Autophagy, Mol Cell. 76, 797–810.e10. [DOI] [PubMed] [Google Scholar]
- 158.Sawa T, Zaki MH, Okamoto T, Akuta T, Tokutomi Y, Kim-Mitsuyama S, Ihara H, Kobayashi A, Yamamoto M, Fujii S, Arimoto H & Akaike T (2007) Protein S-guanylation by the biological signal 8-nitroguanosine 3’,5’-cyclic monophosphate, Nat Chem Biol. 3, 727–35. [DOI] [PubMed] [Google Scholar]
- 159.Ito C, Saito Y, Nozawa T, Fujii S, Sawa T, Inoue H, Matsunaga T, Khan S, Akashi S, Hashimoto R, Aikawa C, Takahashi E, Sagara H, Komatsu M, Tanaka K, Akaike T, Nakagawa I & Arimoto H (2013) Endogenous nitrated nucleotide is a key mediator of autophagy and innate defense against bacteria, Mol Cell. 52, 794–804. [DOI] [PubMed] [Google Scholar]
- 160.Cromm PM & Crews CM (2017) The Proteasome in Modern Drug Discovery: Second Life of a Highly Valuable Drug Target, ACS Cent Sci. 3, 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Anchoori RK, Karanam B, Peng S, Wang JW, Jiang R, Tanno T, Orlowski RZ, Matsui W, Zhao M, Rudek MA, Hung CF, Chen X, Walters KJ & Roden RB (2013) A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer, Cancer cell. 24, 791–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Anchoori RK, Jiang R, Peng S, Soong RS, Algethami A, Rudek MA, Anders N, Hung CF, Chen X, Lu X, Kayode O, Dyba M, Walters KJ & Roden RBS (2018) Covalent Rpn13-Binding Inhibitors for the Treatment of Ovarian Cancer, ACS Omega. 3, 11917–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Trader DJ, Simanski S & Kodadek T (2015) A reversible and highly selective inhibitor of the proteasomal ubiquitin receptor rpn13 is toxic to multiple myeloma cells, J Am Chem Soc. 137, 6312–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Song Y, Ray A, Li S, Das DS, Tai YT, Carrasco RD, Chauhan D & Anderson KC (2016) Targeting proteasome ubiquitin receptor Rpn13 in multiple myeloma, Leukemia. 30, 1877–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Osei-Amponsa V, Sridharan V, Tandon M, Evans CN, Klarmann K, Cheng KT, Lack J, Chari R & Walters KJ (2020) Impact of losing hRpn13 Pru or UCHL5 on proteasome clearance of ubiquitinated proteins and RA190 cytotoxicity, Mol Cell Biol. 40, e00122–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dickson P, Abegg D, Vinogradova E, Takaya J, An H, Simanski S, Cravatt BF, Adibekian A & Kodadek T (2020) Physical and Functional Analysis of the Putative Rpn13 Inhibitor RA190, Cell Chem Biol. S2451–9456(20)30304–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dickson P, Simanski S, Ngundu JM & Kodadek T (2020) Mechanistic Studies of the Multiple Myeloma and Melanoma Cell-Selective Toxicity of the Rpn13-Binding Peptoid KDT-11, Cell Chem Biol. S2451–9456(20)30305–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Song Y, Park PMC, Wu L, Ray A, Picaud S, Li D, Wimalasena VK, Du T, Filippakopoulos P, Anderson KC, Qi J & Chauhan D (2019) Development and preclinical validation of a novel covalent ubiquitin receptor Rpn13 degrader in multiple myeloma, Leukemia. 33, 2685–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Perez C, Li J, Parlati F, Rouffet M, Ma Y, Mackinnon AL, Chou TF, Deshaies RJ & Cohen SM (2017) Discovery of an Inhibitor of the Proteasome Subunit Rpn11, J Med Chem. 60, 1343–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li J, Zhang Y, Da Silva Sil Dos Santos B, Wang F, Ma Y, Perez C, Yang Y, Peng J, Cohen SM, Chou TF, Hilton ST & Deshaies RJ (2018) Epidithiodiketopiperazines Inhibit Protein Degradation by Targeting Proteasome Deubiquitinase Rpn11, Cell Chem Biol. 25, 1350–1358.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lauinger L, Li J, Shostak A, Cemel IA, Ha N, Zhang Y, Merkl PE, Obermeyer S, Stankovic-Valentin N, Schafmeier T, Wever WJ, Bowers AA, Carter KP, Palmer AE, Tschochner H, Melchior F, Deshaies RJ, Brunner M & Diernfellner A (2017) Thiolutin is a zinc chelator that inhibits the Rpn11 and other JAMM metalloproteases, Nat Chem Biol. 13, 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wu HQ, Baker D & Ovaa H (2020) Small molecules that target the ubiquitin system, Biochem Soc Trans. 48, 479–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sherman DJ & Li J (2020) Proteasome Inhibitors: Harnessing Proteostasis to Combat Disease, Molecules. 25, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ryu H, Gygi SP, Azuma Y, Arnaoutov A & Dasso M (2014) SUMOylation of Psmd1 controls Adrm1 interaction with the proteasome, Cell Rep. 7, 1842–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hemmis CW, Heard SC & Hill CP (2019) Phosphorylation of Tyr-950 in the proteasome scaffolding protein RPN2 modulates its interaction with the ubiquitin receptor RPN13, J Biol Chem. 294, 9659–9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Liang RY, Chen L, Ko BT, Shen YH, Li YT, Chen BR, Lin KT, Madura K & Chuang SM (2014) Rad23 interaction with the proteasome is regulated by phosphorylation of its ubiquitin-like (UbL) domain, J Mol Biol. 426, 4049–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ruschak AM & Kay LE (2012) Proteasome allostery as a population shift between interchanging conformers, Proc Natl Acad Sci U S A. 109, E3454–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sprangers R, Li X, Mao X, Rubinstein JL, Schimmer AD & Kay LE (2008) TROSY-based NMR evidence for a novel class of 20S proteasome inhibitors, Biochemistry. 47, 6727–34. [DOI] [PubMed] [Google Scholar]
- 179.Li J, Gao X, Ortega J, Nazif T, Joss L, Bogyo M, Steven AC & Rechsteiner M (2001) Lysine 188 substitutions convert the pattern of proteasome activation by REGgamma to that of REGs alpha and beta, EMBO J. 20, 3359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kleijnen MF, Roelofs J, Park S, Hathaway NA, Glickman M, King RW & Finley D (2007) Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites, Nat Struct Mol Biol. 14, 1180–8. [DOI] [PubMed] [Google Scholar]
- 181.Rennella E, Huang R, Yu Z & Kay LE (2020) Exploring long-range cooperativity in the 20S proteasome core particle from, Proc Natl Acad Sci U S A. 117, 5298–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC & Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis, J Comput Chem. 25, 1605–12. [DOI] [PubMed] [Google Scholar]
- 183.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH & Ferrin TE (2020) UCSF ChimeraX: Structure visualization for researchers, educators, and developers, Protein Sci. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]


