Abstract
Botrytis cinerea is a world-wide occurring plant pathogen, causing pre- and post-harvest gray mold rot on a large number of fruit, vegetable, and flower crops. B. cinerea is closely related to Botrytis pseudocinerea, another broad host range species which often occurs in sympatry with B. cinerea, and to several host-specific species including Botrytis fabae and Botrytis calthae. B. cinerea populations have been shown to be genetically heterogeneous, and attempts have been made to correlate genetic markers to virulence and host adaptation. Here, we present the development of a multilocus sequence typing (MLST) scheme, with 10 genes selected for high variability and phylogenetic congruence, to evaluate the genetic diversity of B. cinerea, B. fabae, and B. pseudocinerea. Using PacBio-assisted simultaneous mass sequencing of PCR products, MLST analysis of about 100 strains from diverse geographical origins and years of isolation was performed, which resulted in high-resolution strain differentiation and robust species separation. Several B. cinerea strains formed an as yet unknown population, referred to as group B, which was well separated from all other B. cinerea strains. Furthermore, the gene cluster for biosynthesis of the phytotoxin botcinic acid was missing in B. cinerea B strains. B. cinerea strains from the monocot Iris pseudacorus were found to form a genetically distinct population, and contained an intact gene cluster for production of the red pigment bikaverin, which is usually degenerated in B. cinerea. Remarkably, these strains were much more aggressive on Iris than other B. cinerea strains, which is the first unequivocal example for host specialization in B. cinerea. Our data reveal new insights into the genetic diversity of B. cinerea and provide evidence for intraspecific differentiation and different degrees of host adaptation of this polyphagous necrotrophic pathogen.
Keywords: multilocus sequence typing (MLST), indel, taxonomy, host adaptation, PacBio sequencing, bikaverin, botcinic acid
Introduction
The genus name Botrytis, coined by Micheli in 1729, is one of the first described genera of fungi. The major species of this genus is Botrytis cinerea Pers.:Fr., a ubiquitous plant pathogen causing gray mold on more than 1000 plant species, including many economically important fruit, vegetable, and ornamental flower crops (Dean et al., 2012; Elad et al., 2016). Up to now, more than 30 species of Botrytis have been described, which are all necrotrophic pathogens except for Botrytis pyriformis which appears to be a saprophytic species (Hyde et al., 2014; Zhou et al., 2014; Zhang et al., 2016; Richards et al., 2020). A first comprehensive molecular classification of Botrytis spp. was performed by Staats et al. (2005), using sequence comparisons of three conserved genes (RPB2, G3PDH, and HSP60). In later studies, nep1 and nep2 encoding necrosis and ethylene inducing proteins 1 and 2 have been added to provide higher resolution (Staats et al., 2007; Hyde et al., 2014). Based on the molecular classification, Botrytis species can be divided into two phylogenetic clades. Botrytis clade 1 includes species that infect mainly or exclusively dicotyledonous plants, namely, the broad host range species B. cinerea and Botrytis pseudocinerea, and the host-specific species Botrytis fabae, Botrytis calthae, Botrytis sinoviticola, and Botrytis eucalyptii (Staats et al., 2005, 2007; Hyde et al., 2014; Liu et al., 2016). The previously described species Botrytis pelargonii has been found to be genetically very close or indistinguishable from B. cinerea (Staats et al., 2005) and is therefore not included as separate species in this study. Botrytis clade 2 is phylogenetically more diverse and contains predominantly monocot-specific, but also several dicot-specific Botrytis species, usually with restricted host range (Staats et al., 2007; Hyde et al., 2014).
Botrytis cinerea strains show considerable morphological variability, including differences in mycelial growth, conidiation, and sclerotium formation (Kerssies, 1990; Martinez et al., 2003). Numerous studies have also documented a high degree of genetic variability in B. cinerea populations (Giraud et al., 1999; Fournier et al., 2003, 2005; Rowe and Kliebenstein, 2007). Early studies performed with B. cinerea strains from French vineyards and other host plants based on the analysis of PCR-RFLP patterns, fungicide resistance, and the detection of transposable elements indicated the existence of genetically distinct groups within B. cinerea (Giraud et al., 1997, 1999). In particular, the presence or absence of the long terminal repeat retrotransposon Boty (Diolez et al., 1995) and the DNA transposon Flipper (Levis et al., 1997) was used to divide strains into four transposon types called transposa (strains having both elements), vacuma (strains having neither element), boty, and flipper. Evidence was obtained that the presence of transposable elements was correlated with higher virulence (Martinez et al., 2005). This was further supported by a seminal study which revealed that B. cinerea releases small RNAs derived from retrotransposons that are translocated into host cells and suppress expression of plant defense genes (Weiberg et al., 2013). However, while B. cinerea strains differ in their content of transposon types, later studies revealed that this feature was not very useful for genetic classification. Nevertheless, a subgroup of vacuma strains, originally called group I, was found to be clearly distinct from all other B. cinerea strains (group II) on the basis of genetic markers and DNA sequence data, and later defined as a new species called B. pseudocinerea (Fournier et al., 2003, 2005; Walker et al., 2011). B. pseudocinerea is often found as a minor species in vineyards and strawberry fields in sympatry with B. cinerea, but infects also a variety of other plant species, and has been found occasionally as the dominating gray mold pathogen on oilseed rape, apple, cherry, broad bean, and Caltha palustris (Plesken et al., 2015a, b).
The enormous host range of B. cinerea and its phenotypic variability would be consistent with a corresponding genetic variability, and at least some degree of host adaptation of the species. Population genetic studies have provided evidence for the existence of clusters within B. cinerea populations from different host plants with reduced but still existing gene exchange. Investigations of B. cinerea populations in France revealed that they were structured by their host plants tomato and grapevine (Walker et al., 2015). Further studies including pathogenicity tests indicated different degrees of host specialization of the populations, depending on their host origin (Mercier et al., 2019). In German strawberry fields, which are frequently treated with fungicides, B. cinerea strains resistant to multiple fungicides have been identified. Several of these strains carried mutations in a transcription factor gene (mrr1) leading to increased expression of a drug efflux transporter gene (atrB) and partial resistance to two commonly used Botrytis fungicides, fludioxonil and cyprodinil (Kretschmer et al., 2009; Leroch et al., 2013). Sequencing of mrr1 and several other genes indicated that these strains belonged to a new subgroup of B. cinerea called Botrytis group S (subsequently referred to as B. cinerea S), which was separated from the sequenced strains B05.10 and T4 (Leroch et al., 2013). In Germany, B. cinerea S strains were found to be predominating in strawberry fields, but almost absent in vineyards. Analysis of B. cinerea populations in vineyards of New Zealand revealed the occurrence of strains similar to B. cinerea S; however, these studies did not support a clear separation from other B. cinerea strains (Johnston et al., 2014).
Multilocus sequence typing (MLST) and multilocus sequence analysis, in the following referred to as MLST, has been established in pathogenic bacteria for studying the epidemiology and phylogenetic relationships of populations (Maiden, 2006). MLST is performed by partial sequencing of coding regions of several genes encoding conserved proteins, for genotyping or deducing phylogenetic relationships. MLST has also been applied for human pathogenic fungi, such as Candida albicans and Cryptococcus spp. (Odds and Jacobsen, 2008; Alanio et al., 2017). The use of next generation sequencing has been shown to improve the performance and speed of MLST of Cryptococcus neoformans (Chen et al., 2015).
The conserved genes hsp60, g3pdh, and rpb2 have been used for differentiation between Botrytis species (Staats et al., 2005; Hyde et al., 2014). However, they provide poor resolution between B. cinerea strains. Higher inter-species variability was observed for nep1 and nep2, but these genes encode host necrosis-inducing proteins and encode some amino acids that appear to be under positive selection (Staats et al., 2007). In this study, we have tested and confirmed the suitability of genes to provide high resolution, phylogenetically reliable differentiation between strains of B. cinerea and closely related Botrytis species. For this, an MLST scheme with 10 genes was developed and applied for analysis of a large number of Botrytis field strains from diverse geographic and host plant origins. The resulting data confirm the genetic diversity of B. cinerea and revealed the existence of separated group, called B. cinerea B, which has significantly diverged from the majority of B. cinerea strains. We also demonstrate that a genetically distinct B. cinerea population isolated from the monocot plant Iris pseudacorus shows a high degree of host adaptation.
Materials and Methods
Cultivation and DNA Isolation of Botrytis Strains
Botrytis strains were collected in different years from a variety of infected host tissues in different locations and countries. For purification, cultures were transferred several times via hyphal tips or single spores to new malt extract (ME) agar plates. As confirmed by sequencing, all isolates in this study were genetically pure, and can therefore be referred to as strains, consistent with previous reports (Giraud et al., 1999). The strains are listed in Table 1. They were cultivated on ME agar plates under black light bulb illumination to induce rapid sporulation. To induce production of the red pigment bikaverin, B. cinerea strains were cultivated in the dark on CD-B agar medium (Schumacher et al., 2013). Growth tests were performed on Gamborg’s B5 agar medium supplemented with 10 mM (NH4)H2PO4 and one of the following carbon sources: 50 mM glucose, 0.5% polygalacturonate, 0.5% pectin, 0.5% xylan, and 0.1% total leaf extract from I. pseudacorus.
TABLE 1.
Botrytis strains analyzed by MLST and infection tests.
| No. | Name | Species | Year | Host plant | Location | Source/references | Use |
| 1 | B05.10 | Bcin | 1994 | Grapevine | Italy | Büttner et al., 1994 | IT; fullseq |
| 2 | T4 | Bcin | 2005 | Tomato | France, Vaucluse | Fournier et al., 2005 | IT; fullseq |
| 3 | C12_S_E1_5 | Bcin | 2012 | Strawberry | China, Zhejiang, Shaoxing | Z. Ma | MLST |
| 4 | C12_S_E7_2 | Bcin | 2012 | Strawberry | China, Zhejiang, Shaoxing | Z. Ma | MLST |
| 5 | C12_S_E7_4 | Bcin | 2012 | Strawberry | China, Zhejiang, Shaoxing | Z. Ma | MLST |
| 6 | CH14_ES_11_1 | Bcin | 2014 | Blueberry | Chile | E. Silva | MLST |
| 7 | CH14_ES_11_5 | Bcin | 2014 | Blueberry | Chile | E. Silva | MLST |
| 8 | D06_1_30 | Bcin | 2006 | Grapevine | GER, Neustadt-Weinstr. | Leroch et al., 2013 | MLST |
| 9 | D08_H_8_3 | Bcin | 2008 | Strawberry | GER, Vechta | R. Weber | IT; MLST |
| 10 | D08_H_8_4 | Bcin “S”* | 2008 | Strawberry | GER, Vechta | Leroch et al., 2013 | IT; fullseq |
| 11 | D08_H_8_6 | Bcin B | 2008 | Strawberry | GER, Vechta | Leroch et al., 2013 | MLST |
| 12 | D09_Bc11 | Bcin | 2009 | Strawberry | GER | van Kan et al., 2017 | MLST |
| 13 | D09_K_2_3 | Bcin | 2009 | Strawberry | GER, Grafschaft-Oeverich | Leroch et al., 2013 | MLST |
| 14 | D09_K_4_1 | Bcin “S”* | 2009 | Strawberry | GER, Grafschaft-Oeverich | Leroch et al., 2013 | MLST |
| 15 | D09_K_4_2 | Bcin B | 2009 | Strawberry | GER, Grafschaft-Oeverich | This work | IT; MLST |
| 16 | D10_B_F1_6 | Bcin “S”* | 2010 | Strawberry | GER, Breisach, Buchholz | Leroch et al., 2013 | MLST |
| 17 | D10_B_F3_5 | Bcin “S”* | 2010 | Strawberry | GER, Breisach, Buchholz | Leroch et al., 2013 | MLST |
| 18 | D10_B_S3_16 | Bcin B | 2010 | Strawberry | GER, Breisach, Rombach | Leroch et al., 2013 | IT; fullseq |
| 19 | D10_B_S6_1 | Bcin B | 2010 | Strawberry | GER, Breisach, Rombach | This work | MLST |
| 20 | D10_K_S11_6 | Bcin B | 2010 | Strawberry | GER, Grafschaft-Oeverich | This work | IT; MLST |
| 21 | D10_MR_S19 | Bcin | 2010 | Strawberry | GER, Grafschaft-Oeverich | Rupp et al., 2017 | MLST |
| 22 | D10_K_S12_13 | Bcin | 2010 | Strawberry | GER, Koblenz, Oeverich | This work | IT |
| 23 | D11_H_R3_7 | Bcin B | 2011 | Strawberry | GER, Hamburg | R. Weber | MLST |
| 24 | D11_KL_tax4 | Bcin | 2011 | Taxus spp. | GER, Kaiserslautern | This work | MLST |
| 25 | D11_M_E01 | Bcin | 2011 | Strawberry | GER, Meckenheim | This work | IT; MLST |
| 26 | D11_M_E07 | Bcin | 2011 | Strawberry | GER, Meckenheim | This work | IT; MLST |
| 27 | D11_M_W04 | Bcin | 2011 | Grapevine | GER, Meckenheim | This work | IT; MLST |
| 28 | D11_T_B09 | Bcin | 2011 | Broad bean | GER, Trier | This work | IT; MLST |
| 29 | D11_T_B14 | Bcin | 2011 | Broad bean | GER, Trier | This work | IT; MLST |
| 30 | D11_T_B25 | Bcin B | 2011 | Broad bean | GER, Trier | This work | IT; MLST |
| 31 | D11_T_B45 | Bcin | 2011 | Strawberry | GER, Trier | This work | IT; MLST |
| 32 | D11_T_E12 | Bcin B | 2011 | Strawberry | GER, Trier | This work | MLST |
| 33 | D11_T_E15 | Bcin B | 2011 | Strawberry | GER, Trier | This work | IT; MLST |
| 34 | D11_T_E18 | Bcin B | 2011 | Strawberry | GER, Trier | This work | MLST |
| 35 | D11_T_WF43 | Bcin | 2011 | Grapevine | GER, Trier | This work | MLST |
| 36 | D11_T_WH08 | Bcin | 2011 | Grapevine | GER, Trier | This work | IT; MLST |
| 37 | D12_E_cal13 | Bcin | 2012 | Caltha palustris | GER, Kaiserslautern | This work | MLST |
| 38 | D12_BH20_4 | Bcin | 2012 | Raspberry | GER, Buxtehude | R. Weber | MLST |
| 39 | D12_H_BioH1 | Bcin | 2012 | Raspberry | GER, Buxtehude | R. Weber | MLST |
| 40 | D12_Pepper1 | Bcin | 2012 | Pepper | GER | J. Schumacher | MLST |
| 41 | D13_MR_S9 | Bcin | 2013 | Strawberry | GER, Bruchsal, Lichtenau | Rupp et al., 2017 | MLST |
| 42 | D13_B_KF1_25 | Bcin | 2013 | Strawberry | GER, Bruchsal, Sanweier | This work | MLST |
| 43 | D13_MR_S11 | Bcin | 2013 | Strawberry | GER, Bruchsal, Hirschberg | Rupp et al., 2017 | MLST |
| 44 | D13_E_IL12 | Bcin I | 2013 | Iris pseudacorus | GER, Kaiserslautern | This work | MLST |
| 45 | D13_E_IL4 | Bcin I | 2013 | Iris pseudacorus | GER, Kaiserslautern | This work | MLST |
| 46 | D13_E_IL7 | Bcin I | 2013 | Iris pseudacorus | GER, Kaiserslautern | This work | MLST |
| 47 | D13_MR_S2 | Bcin | 2013 | Strawberry | GER, Koblenz-Herold | Rupp et al., 2017 | MLST |
| 48 | D13_MR_S1 | Bcin | 2013 | Strawberry | GER, Bruchsal, Lichtenau | Rupp et al., 2017 | MLST |
| 49 | D13_MR_S4 | Bcin | 2013 | Strawberry | GER, Bruchsal, Lichtenau | Rupp et al., 2017 | MLST |
| 50 | D13_MR_S14 | Bcin | 2013 | Strawberry | GER, Grafschaft-Eckendorf | Rupp et al., 2017 | MLST |
| 51 | D13_MR_S29 | Bcin | 2013 | Strawberry | GER, Klein Altendorf | Rupp et al., 2017 | MLST |
| 52 | D14_Heid15 | Bcin B | 2014 | Blueberry | GER, Grethem | This work | MLST |
| 53 | D14_Ki11 | Bcin | 2014 | Cherry | GER, Jork | R. Weber | MLST |
| 54 | D14_Ki12 | Bcin B | 2014 | Cherry | GER, Jork | R. Weber | MLST |
| 55 | G09_S04 | Bcin | 2009 | Strawberry | Greece | Leroch et al., 2013 | MLST |
| 56 | G09_S33 | Bcin “S”* | 2009 | Strawberry | Greece | Leroch et al., 2013 | IT; fullseq |
| 57 | G11_MG1_E5 | Bcin | 2011 | Strawberry | Greece, Manolada | G. Karaoglanidis | MLST |
| 58 | G11_MG1_E7 | Bcin | 2011 | Strawberry | Greece, Manolada | G. Karaoglanidis | MLST |
| 59 | G11_MG1_E22 | Bcin | 2011 | Strawberry | Greece, Manolada | G. Karaoglanidis | MLST |
| 60 | G13_EBio4 | Bcin | 2013 | Strawberry | Greece | G. Karaoglanidis | MLST |
| 61 | N11_K_W02 | Bcin | 2011 | Grapevine | Norway, Kvelland | This work | MLST |
| 62 | N11_K_W03 | Bcin B | 2011 | Grapevine | Norway, Kvelland | This work | MLST |
| 63 | N11_K_W06 | Bcin B | 2011 | Grapevine | Norway, Kvelland | This work | IT; MLST |
| 64 | N11_K_W08 | Bcin | 2011 | Grapevine | Norway, Kvelland | Leroch et al., 2013 | MLST |
| 65 | N11_K_W11 | Bcin “S”* | 2011 | Grapevine | Norway, Kvelland | Leroch et al., 2013 | IT; MLST |
| 66 | N11_K_W14 | Bcin | 2011 | Grapevine | Norway, Kvelland | This work | IT; MLST |
| 67 | N11_S_E08 | Bcin “S”* | 2011 | Strawberry | Norway, Søgne | Leroch et al., 2013 | IT; MLST |
| 68 | N11_S_E09 | Bcin B | 2011 | Strawberry | Norway, Søgne | This work | IT; MLST |
| 69 | N11_S_E10 | Bcin B | 2011 | Strawberry | Norway, Søgne | This work | IT; MLST |
| 70 | N11_S_E15 | Bcin | 2011 | Strawberry | Norway, Søgne | Leroch et al., 2013 | IT; MLST |
| 71 | SA12_Ro | Bcin | 2012 | Rooibush | South Africa | L. Mostert | MLST |
| 72 | SAS405 | Bcin | <1990 | Grapevine | Italy | Faretra et al., 1988 | IT; MLST |
| 73 | U10_SC_BRO1 | Bcin | 2010 | Strawberry | United States, South Carolina | G. Schnabel | MLST |
| 74 | U11_M_E1 | Bcin B | 2011 | Strawberry | United States, North Carolina | G. Schnabel | MLST |
| 75 | U11_SC_BR02 | Bcin | 2011 | Strawberry | United States, South Carolina | G. Schnabel | MLST |
| 76 | V1750 | Bcin | ? | Cucumber | Japan | Schumacher et al., 2013 | IT |
| 77 | 2230 | B. fabae | ? | Broad bean | France | A.S. Walker, INRA | IT; MLST |
| 78 | 2235 | B. fabae | ? | Broad bean | France | A.S. Walker, INRA | IT; MLST |
| 79 | 2240 | B. fabae | ? | Broad bean | France | A.S. Walker, INRA | IT; MLST |
| 80 | 11001 | B. fabae | ? | Broad bean | France | A.S. Walker, INRA | IT |
| 81 | 11002 | B. fabae | ? | Broad bean | France | Leroch et al., 2013 | IT; fullseq |
| 82 | D12_B_B02 | B. fabae | 2012 | Broad bean | GER, Bonn | M. Heupel | MLST |
| 83 | D12_B_B28 | B. fabae | 2012 | Broad bean | GER, Bonn | M. Heupel | MLST |
| 84 | G12 | B. fabae | 2012 | Broad bean | Greece | G. Karaoglanidis | IT; fullseq |
| 85 | G12_B03B | B. fabae | 2012 | Broad bean | Greece | G. Karaoglanidis | MLST |
| 86 | D08_H_8_15 | B. pseud | 2008 | Strawberry | GER, Vechta | Leroch et al., 2013 | MLST |
| 87 | D11_KL_cal2 | B. pseud | 2011 | Caltha palustris | GER, Kaiserslautern | This work | MLST |
| 88 | D11_M_E27 | B. pseud | 2011 | Strawberry | GER, Meckenheim | This work | IT; MLST |
| 89 | D11_T_B18 | B. pseud | 2011 | Broad bean | GER, Trier | This work | IT; MLST |
| 90 | D11_T_E01 | B. pseud | 2011 | Strawberry | GER, Trier | This work | MLST |
| 91 | D12_E_cal10 | B. pseud | 2012 | Caltha palustris | GER, Kaiserslautern | This work | MLST |
| 92 | D13_E_IF4 | B. pseud | 2013 | Iris pseudacorus | GER, Kaiserslautern | This work | MLST |
| 93 | N11_K_W15 | B. pseud | 2011 | Grapevine | Norway, Kvelland | This work | IT; MLST |
| 94 | N11_S_E06 | B. pseud | 2011 | Strawberry | Norway, Søgne | Leroch et al., 2013 | IT; MLST |
| 95 | VD110 | B. pseud | 2007 | Grapevine | France, Courteron | Walker et al., 2011 | IT; fullseq |
| 96 | VD184 | B. pseud | 2007 | Blackberry | France, Courteron | Walker et al., 2011 | IT |
| 97 | VD256 | B. pseud | 2007 | Grapevine | France, Courteron | Walker et al., 2011 | MLST |
| 98 | MUCL2830 | B. calthae | 1961 | Caltha palustris | Canada, Quebec | Leroch et al., 2013 | MLST |
| 99 | Pae14 | B. paeoniae | 2013 | Peony | GER, Wonsheim | This work | MLST |
| 100 | GBc_5 | B. sinovitic. | 2010 | Grapevine | China, Xinjiang | Zhou et al., 2014 | MLST |
*Strains marked as Bcin “S” have been described to belong to “Botrytis group S,” based on sequencing of mrr1, hsp60, fg1020, nep2, and ms547 (Leroch et al., 2013). ? means unknown.
Extraction of genomic DNA was performed using conidia from sporulating agar plates. Conidia were washed from a sporulating malt agar extract plate with sterile water, filtered through glass wool, centrifuged, and suspended in water at a concentration of 1–5 × 107 conidia ml–1; 400 μl of the spore suspension was mixed with 200 μl CTAB solution (per 100 ml: 2.5 g sorbitol; 1 g N-laurylsarcosine; 0.8 g N-cetyl-trimethylammonium bromide; 4.7 g NaCl; 1 g Na-EDTA; and 1 g polyvinylpyrrolidone) in a 2 ml screw-capped microfuge tube and homogenized with glass beads (1 mm diameter) with FastPrep (MP Biomedicals Germany GmbH, Eschwege, Germany; 45 and 30 s at level 6, interrupted by cooling on ice). After centrifugation (14,000 rpm, 2 min), the clear supernatant was transferred into a fresh tube, and 100 μl chloroform was added. After vortexing for 5 s and centrifugation (14,000 rpm, 3 min), the upper phase was transferred into a new tube, and the DNA precipitated by the addition of an equal volume of 2-propanol. After centrifugation (14,000 rpm, 5 min), the DNA pellet was washed with 70% ethanol, air-dried, and dissolved in 15–30 μl TE buffer. Concentration and integrity of the DNA were estimated by agarose gel electrophoresis.
PCR Assays
For genetic analysis of Botrytis strains, ca. 10 ng of total Botrytis DNA was used for PCR assays with diagnostic primers, using 20 μl reactions containing 0.2 mM dNTPs, 0.25 μM primers, 40 mM Tris–HCl, 0.02% Tween 20, 2 mM MgCl2, pH 8.3, and 0.5 units Taq polymerase. Amplification was performed in different thermocyclers set at 5 min 94°C, followed by 30–35 cycles with 30 s 94°C, 30 s 52–60°C, and 1 min per kb 72°C. Electrophoresis was performed with 1 × TAE buffer in 1–3% agarose gels. Annealing temperatures were adjusted to ca. 5°C below the melting temperatures of the primers. Primers used in this study are listed in Supplementary Table 1.
Selection of Genes for Multilocus Sequence Typing
To identify genes suitable for MLST of B. cinerea and related Botrytis species, we first searched for genes in FUNYBASE, a database containing 246 families of single-copy orthologs obtained from 21 fungal genomes (Marthey, 20081). Two of the most variable FUNYBASE genes, ms547 (Bcin12g03020) and fg1020 (Bcin04g02090), were selected. These have been used previously for differentiation of B. pseudocinerea and B. cinerea (Walker et al., 2011), and between B. cinerea strains (Leroch et al., 2013). No other FUNYBASE gene was found to be suitable for MLST, because of low diversity between the four tested Botrytis spp. and/or between B. cinerea strains. For identification of further genes useful for differentiation, genome sequences from six B. cinerea strains and one strain each of B. fabae, B. pseudocinerea, and B. calthae (unpublished data) were reference aligned to the sequenced genome of B. cinerea strain B05.102, and searched for complete homologous gene models. Genes yielding the highest diversity scores between the four Botrytis species were tested for high resolution among the sequenced B. cinerea strains, conservation in other ascomycetes (preferred), predicted function (preferred), and location on different chromosomes in the B. cinerea B05.10 reference genome. Based on these criteria, a final list of 10 genes, including the previously used nep2, ms547, and fg1020, was selected for MLST (Table 2). Primers for these genes were designed to match the sequences of B. cinerea, B. fabae, B. pseudocinerea, and B. calthae; therefore they were partly degenerated, containing mixtures of variable nucleotides. For B. calthae, sequence comparisons of several strains (Plesken et al., 2015b) confirmed species-specific similarities for nep2 and ms547, and the suitability of the primers. Primer positions were chosen to obtain genomic PCR fragments with heterogeneous size, to facilitate their differentiation on the gels.
TABLE 2.
Genes used for MLST of Botrytis isolates.
| Code | Gene ID (name)* | Predicted function | CDS size (bp) | Fragment sizes (bp) |
|
| First PCR | Second PCR | ||||
| MLST1 | Bcin01g07220 | ATP pheromone transporter | 4551 | 1258 | 1350 |
| MLST2 | Bcin05g07690 | Glycoside hydrolase | 1769 | 1100 | 1192 |
| MLST3 | Bcin06g01710 | Aromatic amino acid decarboxylase | 1695 | 1019 | 1111 |
| MLST4* | Bcin09g03030 (Bcdpb2) | DNA polymerase ε subunit B | 3347 | 1011 | 1103 |
| MLST5 | Bcin11g01310 | Protease | 1701 | 921 | 1013 |
| MLST6 | Bcin15g03910 | Palmitoyl protein thioesterase | 1148 | 971 | 1063 |
| MLST7 | Bcin16g03460 | 2OG-Fe(II) oxygenase | 968 | 833 | 925 |
| MLST8* | Bcin12g03020 (ms547/Bcdbp7) | ATP-dependent RNA helicase | 2634 | 935 | 1027 |
| MLST9* | Bcin02g07770 (Bcnep2) | Necrosis-inducing protein 2 | 845 | 842 | 934 |
| MLST10* | Bcin04g02090 (fg1020/Bcufd2) | Ubiquitin fusion degradation protein | 3788 | 910 | 1002 |
*Gene names were taken from http://fungi.ensembl.org/Botrytis_cinerea/, and from Marthey (2008).
Indel-PCR has been used for preliminary identification of Botrytis species: A 122 bp deletion upstream of the coding region in the B. fabae homolog of B. cinerea strain B05.10 gene Bcin13g02260 (Rigotti et al., 2002), a 24 bp deletion in the B. pseudocinerea homolog of Bcin09p02270 (Plesken et al., 2015a), and a 9 bp deletion in the B. calthae homolog of Bcin16g02210 (Plesken et al., 2015b). Furthermore, an 18 bpand a 21 bp indel in the mrr1 coding region showing variable configuration in different B. cinerea isolates (Leroch et al., 2013) allowed differentiation within B. cinerea (cf. Table 4).
TABLE 4.
List of primers used for PCR-based differentiation of Botrytis species and genotypes of B. cinerea, depending on the insertion (ins) or deletion (del) state of diagnostic indels.
| Primers | Bcal9-fw | Bps24-fw | Bfab122-fw | mrr1-18-fw | BcB15-fw | mrr1-21-fw | BcIris6-fw |
| Bcal9-rv | Bps24-rv | Bfab122-rv | mrr1-18-rv | BcB15-rv | mrr1-21-rv | BcIris6-rv | |
| Gene ID | Bcin16g02210 | Bcin09g02270 | Bcin13g02260 | Bcin05g01790 | Bcin11g00620 | Bcin05g01790 | Bcin01g05500 |
| Indel size | 9 bp | 24 bp | 122 bp | 18 b | 15 bp | 21 bp | 6 bp |
| PCR size | 64/55 bp | 136/112 bp | 355/233 bp | 200/182 bp | 99/84 bp | 149/128 bp | 85/79 bp |
| B. cin | del | ins | ins | ins/del | ins | ins/del | ins |
| B. cin B | del | ins | ins | ins | del (39)* | ins/del | ins |
| B. cin Iris | del | ins | ins | ins | ins | ins | del (21)* |
| B. fabae | del | ins | del (31)* | ins | ins | ins | ins |
| B. pseud | del | del (69)* | ins | ins | ins | del | ins |
| B. calthae | ins (17)* | ins | ins | ins | ins | ins | ins |
Text in bold, indicating the diagnostic state of the Botrytis species or B. cinerea group to be identified. In parentheses: Number of isolates tested.
Preparation of DNA for Mass Sequencing
A two-step PCR was performed, following the protocol “Procedure & Checklist -Preparing SMRTbellTM Libraries using PacBio® Barcoded Universal Primers for Multiplex SMRT® Sequencing,” with modifications. Instead of Phusion HF polymerase, a self-prepared Taq polymerase was used. PCR conditions were as follows: The first-round PCR with gene-specific primers attached to universal forward and reverse primer sequences (30 bp each) was performed in 20 μl reactions, using ca. 1 ng of Botrytis DNA, 0.2 mM dNTPs, 0.25 μM primers, 1 μl Taq polymerase, and 1 × Taq buffer (20 mM Tris–HCl, pH 8.5, 16 mM ammonium sulfate, 0.01% Tween 20, and 2 mM MgCl2). PCR was performed with 5 min initial denaturation at 94°C, 22 cycles of 30 s at 94°C, 30 s at 55°C, and 80 s at 72°C, and a final 5 min at 72°C. Products from the first-round PCR were checked for their sizes and amounts on an agarose gel, diluted 1:200, and used as templates for the second-round PCR using barcoding primers (for each strain, a unique pair of 16 bp barcodes 5’-attached to universal forward and reverse primers) in 25 μl with 0.25 mM dNTPs, 0.3 μM primers, 1 μl Taq polymerase, 1 × Taq buffer, and 1 μl first-round PCR product, with the same thermocycling conditions.
Concentrations of the final PCR products were estimated by their gel staining intensities, and from 100 strains, 10 PCR fragments per strain (representing MLST1-10 amplicons) mixed to yield a final concentration of 1 μg ml–1 for each fragment. A total of 1000 fragments were sequenced in one PacBio RSII SMRT cell according to the supplier’s protocol, yielding an average sequence depth of 32x. Reads of inserts were extracted and aligned to obtain the consensus sequences.
Sequence data are found in Supplementary File 1 (Botrytis mrr1 sequence data) and Supplementary File 2 (Botrytis MLST1-10 sequence data).
Infection Tests
Infection tests were performed with detached leaves of tomato (Lycopersicon esculentum), faba bean (Vicia faba), Phaseolus bean (Phaseolus vulgaris), salad (Lactuca sativa), barley (Hordeum vulgaris), yellow iris (I. pseudacorus), placed on wet filter paper, and with apple fruit (cv. Golden Delicious). Inoculations were performed with 20 μl conidial suspensions containing 2 × 105 conidia ml–1 in Gamborg GB5 medium supplemented with 25 mM glucose. Inoculation of Iris leaves was performed either with agar plugs of 1 mm thickness and 2 mm diameter inoculated 1 day before with 2000 conidia, or with conidial suspensions as described above, after wiping away cuticular waxes of the Iris leaves with wet gloved fingers to allow more consistent infection. Nevertheless, infection by the B. cinerea strains varied to some extent with the age and size of the Iris leaves used. Apple fruits were inoculated by pipetting 20 μl conidial suspensions into wounds created by a cork borer with 2 mm diameter. After incubation of inoculated leaves and fruits in humid chambers at ambient light and temperature conditions for the indicated times, lesion diameters were measured with a digital caliper. Mean values of at least replicates are shown in the figures, with standard deviations.
Tree Building and Statistical Data Evaluation
Sequence alignments and tree building were performed by PhyML for maximum-likelihood phylogenetic tree reconstruction, using SeaView version 4 software (Gouy et al., 2010). Distance-based tree reconstruction was performed by BioNJ or by maximum-parsimony, which gave similar results for all trees generated. Statistical analyses were performed using the GraphPad Prism software. The detailed analysis method is depicted in the individual figure legends. Infection and growth experiments were carried out at least three times, with two or three technical replicates per sample.
Results
Use of Indels in mrr1 for Preliminary Classification of B. cinerea Strains
Previously, genetic analysis of B. cinerea strains from fungicide-treated strawberry fields has revealed a high polymorphism of the mrr1 gene, which showed up to 5% sequence divergence between the sequenced B. cinerea reference strains and several strains resistant to multiple fungicides from German strawberry fields. Further sequencing of the genes fg1020, ms547, and nep2 supported a genetic separation between this strawberry population originally referred to as “group S” and isolates similar to reference strains B05.10 and T4 (Leroch et al., 2013). Because of the high variability of mrr1, a comparative sequence analysis was performed. An alignment of ca. 2030 bp mrr1 segment covering the coding region was done with 26 B. cinerea strains, two B. fabae strains, four B. pseudocinerea strains, and one B. calthae strain. Several insertions-deletions (indels) are present in mrr1, including an 18 bp indel and a 21 bp indel in the coding region, which are useful for preliminary characterization of Botrytis isolates (Leroch et al., 2013). The dendrogram for mrr1 revealed a division of strains of B. cinerea and other Botrytis species into several distinct groups, but did not show their correct phylogenetic relationships, as several B. cinerea strains grouped closer to other Botrytis species than to other B. cinerea strains (Supplementary Figure 1). It was therefore evident that the high diversity of mrr1 is useful for preliminary assessment of genetic variability in Botrytis, but not for estimation of phylogenetic relationships.
Identification of Genes Suitable for MLST-Based Identification and Phylogenetic Placement of Botrytis Strains
To establish a reliable scheme for multilocus sequencing (MLST) of B. cinerea and closely related (clade 1) Botrytis species, 10 genes were selected which provided high-resolution differentiation and phylogenetic information of the strains analyzed (see section “Materials and Methods”; Table 2). Inter-strain variability of the MLST genes was three- to fourfold higher compared to hsp60, and a total of 10.5% of their sequences contained variable sites (Supplementary Table 2).
The 10 concatenated MLST sequences were aligned and used for tree building. This procedure was used for differentiation of 95 Botrytis strains (74 B. cinerea, eight B. fabae, 10 B. pseudocinerea, and one each of B. calthae, B. sinoviticola, and Botrytis paeoniae). The B. cinerea strains were obtained from 12 plant species, 10 countries, and 11 different years of sampling (Table 1). Eight B. cinerea strains from German strawberry fields showing multiple resistance to seven major anti-Botrytis fungicide classes (Rupp et al., 2017) were included into the analysis. For comparison, sequences were included from eight strains of B. cinerea and one strain each of B. fabae, B. pseudocinerea, and B. calthae for which genome sequences were available.
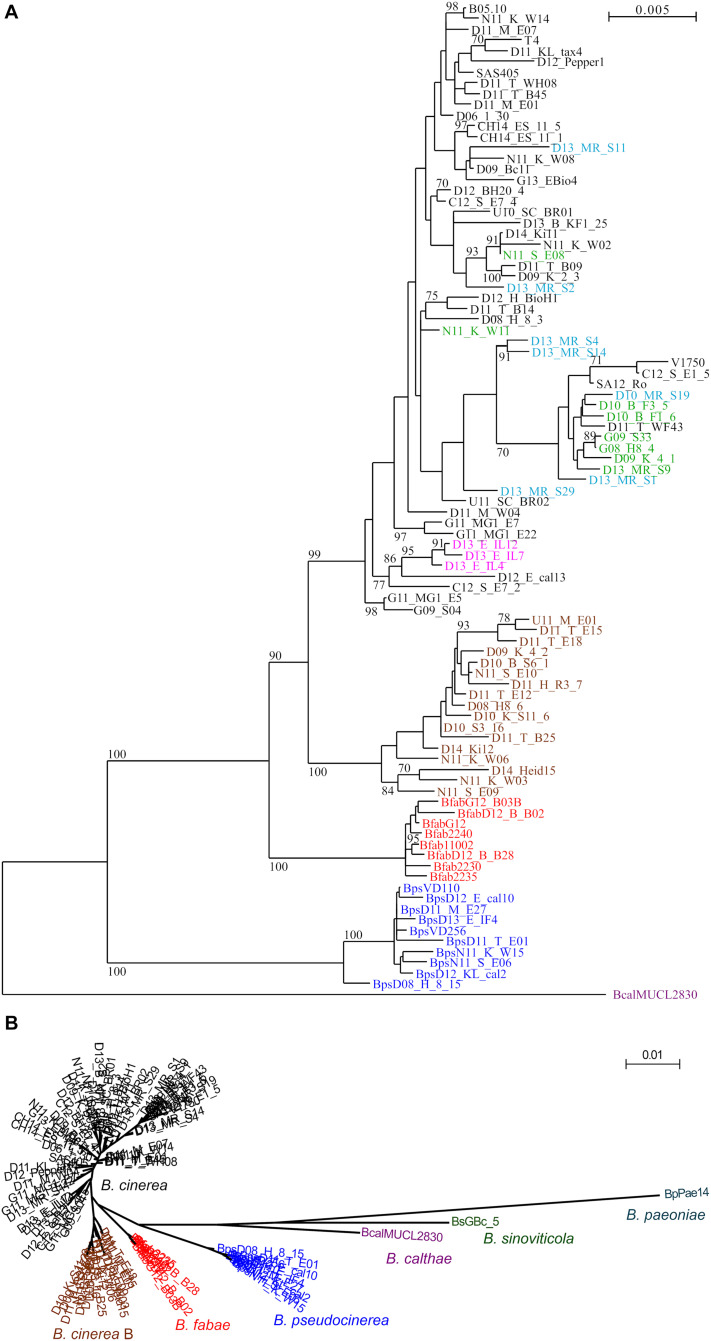
PhyML trees generated with all strains clearly supported the expected separation of the clade 1 species B. sinoviticola, B. calthae, B. pseudocinerea, B. fabae, and B. cinerea, and the larger phylogenetic distance of the clade 2 species B. paeoniae (Figures 1A,B). B. cinerea strains were found to form more or less distinct clusters. Most interestingly, strains in one cluster, in the following referred to as B. cinerea (group) B, were clearly separated from all other B. cinerea strains. The separation of B. cinerea B strains from the remaining B. cinerea strains was also observed with the trees generated with most of the individual MLST sequences (Supplementary Figure 2). Strains previously identified as B. cinerea group S, which have been found to be dominating in German strawberry fields (Leroch et al., 2013), were found to be partly clustered within the B. cinerea branches (Figure 1). They were found to include five or six of the seven strawberry strains with resistance to multiple fungicides, but also strains isolated in Greece, South Africa, United States, China, and Japan, several of them being collected from other host species. The two sequenced strains B05.10 and T4, which have been widely used for phenotypic and genetic studies, clustered relatively close to each other, despite their known sequence diversity and different contents of transposable elements (Amselem et al., 2011).
FIGURE 1.
(A) PhyML-generated tree of 93 Botrytis strains, based on concatenated sequences of genes MLST1–MLST10, with 9524 nucleotides compared. Bootstrap values > 70 based on 1000 replicates are shown. Bcal (purple): B. calthae; Bps (blue): B. pseudocinerea; Bfab (red): B. fabae. B. cinerea group B (brown); B. cinerea Iris isolates (pink). B. cinerea strawberry isolates previously described as “group S” (Leroch et al., 2013) are shown in green, and B. cinerea strawberry isolates with multiple fungicide resistances are shown in light blue. (B) Circular PhyML tree including B. paeoniae and B. sinoviticola, illustrating genetic separation of all species, and of B. cinerea group B from other B. cinerea strains.
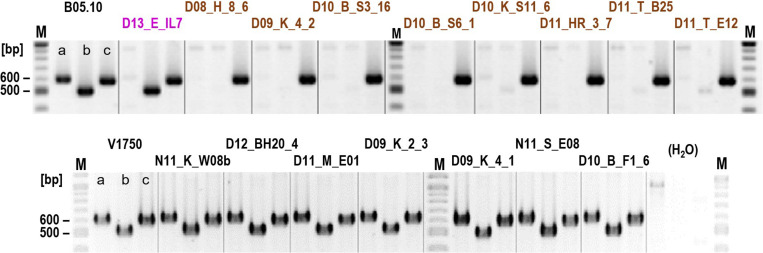
B. cinerea B Strains Lack Biosynthesis Gene Cluster for the Phytotoxin Botcinic Acid
Botrytis cinerea produces two phytotoxins, the sesquiterpenoid botrydial (BOT) and the polyketide botcinic acid (BOA), which have been shown to be required together for full virulence (Dalmais et al., 2011). We tested the strains for the presence of bot and boa genes by PCR. Whereas the bot2 gene was detected in all strains, both boa6 (encoding a polyketide synthase) and boa17 (the last gene of the boa cluster) were missing in all 16 tested B. cinerea B strains (Figure 2). In contrast, all other tested B. cinerea strains contained boa6, and most strains also contained boa17, which might not to be required for botcinic acid biosynthesis (Porquier et al., 2019). These data were consistent with genome sequence data from strain D10_S3_16, which revealed the complete absence of the boa cluster (not shown). Therefore, B. cinerea B strains apparently have lost the ability to produce botcinic acid.
FIGURE 2.
Detection of genes involved in the synthesis of botrydial and botcinic acid in B. cinerea strains. Agarose gel-separated PCR products for boa17 (lanes a), boa6 (lanes b), and bot2 (lanes c) are shown. B. cinerea B strains are marked in brown and the Iris isolate in pink.
Infection Tests Reveal Different Degrees of Host Specificity of Botrytis Clade I Species
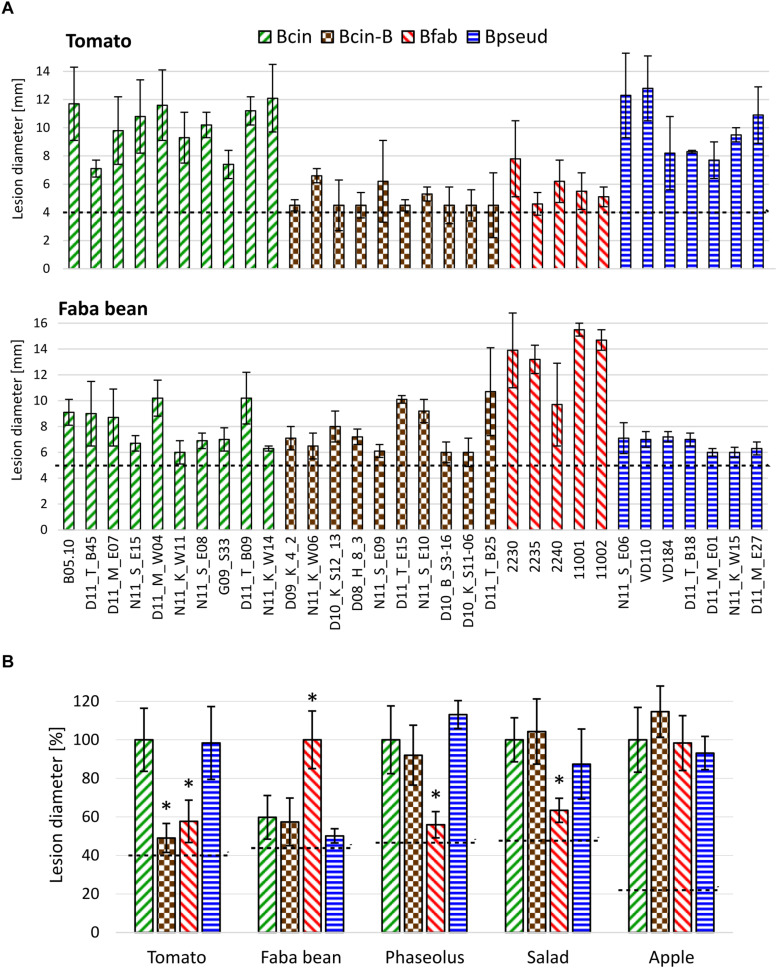
To compare virulence and host specificity of B. cinerea, B. cinerea B, B. fabae, and B. pseudocinerea, infection tests were performed with strains of each of these taxa on tissues of five plant species. All strains were able to form primary lesions under the inoculation droplets, but differences were observed in the size of expanding lesions (Figure 3 and Supplementary Figure 3). Overall, B. cinerea and B. pseudocinerea strains showed similar infection behavior, except for smaller lesions caused by B. pseudocinerea on faba bean. As expected, B. fabae was most aggressive on its host faba bean, but induced only small expanding lesions on leaves of tomato, Phaseolus bean, and salad. B. cinerea B strains formed significantly smaller lesions on tomato leaves compared to common B. cinerea strains. No significant differences were observed between B. cinerea and B. cinerea B in lesion formation on apple fruit.
FIGURE 3.
Lesion formation by strains of B. cinerea, B. cinerea B, B. fabae, and B. pseudocinerea on tissues of different plants. (A) Lesions caused by individual isolates on tomato and faba bean leaves. (B) Mean lesion sizes of strains (shown in A and Supplementary Figure 3) from different Botrytis taxa on five different plant tissues, indicated in percent lesion size produced by B. cinerea (for faba bean: B. fabae). The dotted lines indicate the average sizes of the inoculation droplets. The p-values by one-way ANOVA followed by Dunnett’s multiple comparisons post hoc test (control: B. cinerea) are indicated. *p < 0.001.
Discovery of Pink B. cinerea Strains From the Monocot Iris pseudacorus
In 2012 and 2013, five and 19 Botrytis strains, respectively, were recovered from flowers and leaves of yellow iris (I. pseudacorus) plants grown at two adjacent sites along a creek in the Palatine Forest in Germany. The strains showed growth characteristics and sporulation typical for B. cinerea. B. convoluta, a pathogen specific to Iris spp. (Whetzel and Drayton, 1932), has been reported to cause mainly rhizome infections. We did not observe infections on the rhizomes of the Iris plants, and were unable to obtain Botrytis isolates from them. DNA analysis confirmed the identity of the strains as B. cinerea or B. pseudocinerea. All isolates from 2012, and 17 isolates from 2013 were found to contain a 21 bp insertion, consistent with their initial classification as “group S” according to Leroch et al. (2013). Two isolates, obtained from flowers, were identified as B. pseudocinerea, based on a 24 bp deletion in Bcin09p02270 (Plesken et al., 2015a), which was confirmed for strain Bps_D12_E_IF4 by sequencing (Figure 1). With mrr1 and concatenated MLST1-10 genes, four B. cinerea Iris strains were found to form a cluster within B. cinerea (Supplementary Figure 1). Based on the mrr1 sequence of these strains, a primer-induced restriction analysis (PIRA-PCR) was designed (Haliassos et al., 1989), which resulted in a 72 bp PCR fragment which could be cleaved with the restriction enzyme AccI with DNA of all of the B. cinerea Iris strains, but not with DNA from any other B. cinerea strains tested. Furthermore, a 6 bp indel in Bcin01g05500 was identified (see below), which was in the deleted state only in the B. cinerea Iris population (Table 3).
TABLE 3.
Genetic and phenotypic differentiation of B. cinerea Iris strains.
| Isolate | Mycel. color | mrr1 indel | mrr1 PIRA PCR* | Bcin01g05500 indel | Bikaverin genes |
Mating type | Haplotype** | |||
| bcbik1 | bcbik2 | bcbik3 | bcbik5 | |||||||
| PCR size (bp) | – | 149/128 | 76 | 85/79 | 679 | 711 | 1278 | 797 | – | – |
| D12_E_IL1 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-1 | 1 |
| D12_E_IL2 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-1 | 1 |
| D12_E_IL4& | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-2 | 5 |
| D12_E_IL6& | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-1 | 1 |
| D12_E_IL9 | Pink | ins | 62 + 14 bp | del | yes | yes | yes | yes | MAT1-2 | 2 |
| D13_E_IF1 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-1 | 1 |
| D13_E_IF5 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-2 | 2 |
| D13_E_IF8 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-1 | 1 |
| D13_E_IL1 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-2 | 2 |
| D13_E_IL2 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-1 | 4 |
| D13_E_IL3 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-2 | 2 |
| D13_E_IL4$ | Gray | ins | 62 + 14 bp | del | no | no | no | no | MAT1-2 | 3 |
| D13_E_IL5 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-2 | 2 |
| D13_E_IL6 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-2 | 2 |
| D13_E_IL7$ | Pink | ins | 62 + 14 bp | del | yes | yes | yes | yes | MAT1-2 | 2 |
| D13_E_IL8 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-2 | 2 |
| D13_E_IL9 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-2 | 2 |
| D13_E_IL1 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-2 | 2 |
| D13_E_IL11 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-1 | 1 |
| D13_E_IL12$,& | Pink | ins | 62 + 14 bp | del | yes | yes | yes | yes | MAT1-2 | 5 |
| D13_E_IL13 | Pink | ins | 62 + 14 bp | del | yes | n.a. | n.a. | n.a. | MAT1-2 | 2 |
| BpsD13_E_IF2 | Gray | del | No product | ins | n.a. | n.a. | n.a. | n.a. | n.a. | – |
| BpsD13_E_IF4 | Gray | del | No product | ins | n.a. | n.a. | n.a. | n.a. | n.a. | – |
| V1750 | Pink | ins | 76 bp | ins | yes | yes | yes | yes | n.a. | – |
| B05.10 | Gray | del | 76 bp | ins | no | yes | yes | yes | MAT1-1 | – |
| B. cinerea® | Gray | ins/del | 76 bp | ins | no | n.a. | n.a. | n.a. | n.a. | – |
| B. fabae G12 | Gray | del | 76 bp | ins | no | n.a. | n.a. | n.a. | n.a. | – |
| B. pseud VD110 | Gray | del | No product | ins | no | n.a. | n.a. | n.a. | n.a. | – |
*Products after digestion with AccI. **Based on data in the table and MLST or mrr1 sequencing data. § Randomly chosen B. cinerea non-Iris isolates (n = 20). $Used for MLST sequencing. &Used for mrr1 sequencing. n.a.: not analyzed.
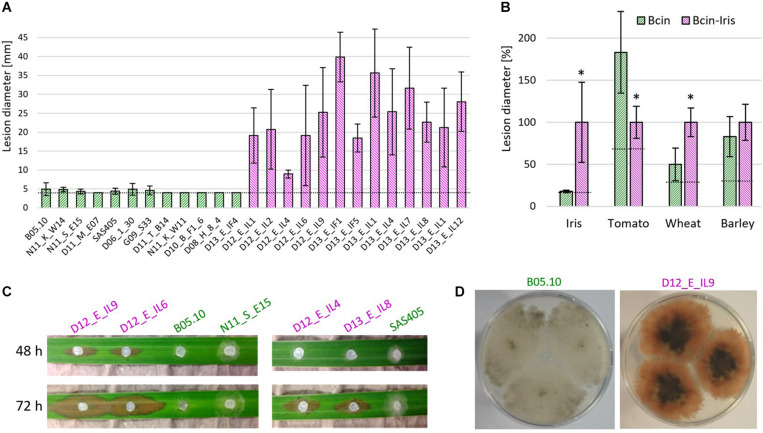
Remarkably, all but one of the 22 B. cinerea Iris strains revealed a pink color when cultured on ME agar (Figure 4D). The pink color has been observed previously in rare B. cinerea strains, and is caused by the production of the red pigment bikaverin. Bikaverin is produced by several Fusarium spp. and encoded by a cluster containing six genes including a polyketide synthase. Bikaverin producing B. cinerea strains have gained a complete, functional ortholog of this cluster by horizontal gene transfer (Campbell et al., 2013; Schumacher et al., 2013). Gray B. cinerea strains contain a degenerated gene cluster lacking Bcbik1, the gene for the polyketide synthase. The presence of bcbik1 in the B. cinerea Iris population was confirmed by PCR with all 20 pink strains, but not for the two gray Iris strains. For three pink Iris strains and the pink control strain, V1750 (Schumacher et al., 2013), the presence of bcbik2, bcbik3, and bcbik5 was confirmed by PCR. Consistent with the genome sequence, B05.10 DNA also revealed the presence of bcbik2, bcbik3, and bcbik5, whereas in the gray Iris strain D13_E_IL04, the four bcbik genes appeared to be missing (Table 3). Analysis of the mating type genes revealed the presence of both MAT1-1 and MAT1-2 genotypes, indicating that even in this small population, sexual reproduction might still occur (Table 3).
FIGURE 4.
B. cinerea Iris strains are highly aggressive on Iris pseudacorus leaves and produce the red pigment bikaverin. (A) Lesion development on detached leaves, 5 days after inoculation with 12 B. cinerea and 13 B. cinerea Iris strains. (B) Mean lesion sizes induced by B. cinerea (green) and B. cinerea Iris (pink; shown in A and Supplementary Figure 4) on leaves of different plant species. The dotted lines indicate the average sizes of the inoculation droplets. Values for B. cinerea and B. cinerea Iris were compared by unpaired t-test. *p < 0.001. n.s.: not significant. (C) Symptoms on I. pseudacorus leaves induced by different B. cinerea strains. (D) Pictures of colonies of gray B. cinerea (left) and pink B. cinerea Iris (right) strains.
Taken together, these data demonstrate that the pink Iris strains, and the gray strain D13_E_IL4, represent a genetically distinct B. cinerea population, in the following referred to as B. cinerea Iris. Based on the ability to produce bikaverin, mating type, and MLST sequences, five haplotypes were distinguished among the Iris strains.
B. cinerea Iris Strains Are Highly Aggressive on I. pseudacorus Leaves
Infection tests on detached leaves of different plants were performed. All of the Iris strains were highly aggressive on I. pseudacorus leaves, causing expanding lesions 5 days after inoculation. In contrast, strain B05.10 and several other B. cinerea strains usually developed only primary lesions, which sometimes expanded to a small extent (Figures 4A–C). Compared to common B. cinerea strains, the Iris strains showed significantly reduced lesion formation on tomato leaves. In contrast, virulence on wheat leaves was higher for the Iris strains than for the other B. cinerea strains, while no significant differences in lesion formation were observed on barley (Figure 4B and Supplementary Figure 4). Because monocot and dicot plants differ in the pectin content of their cell walls, we reasoned that the difference in virulence could be related to their cell wall degrading ability. When the growth of the isolates was tested on media containing different carbon sources, however, no significant differences were observed between two B. cinerea reference strains and four Iris strains (Supplementary Figure 5).
Identification of New Indels for PCR-Based Identification of B. cinerea Iris and Group B Strains
Indel-PCR has been used for preliminary identification of Botrytis species, e.g., B. fabae and B. pseudocinerea (Rigotti et al., 2002; Plesken et al., 2015a). By performing blastn searches with codon sequences of predicted genes in published and self-generated genome assemblies of B. cinerea, B. fabae, B. pseudocinerea, and B. calthae, two more useful indels were identified, and flanking primers were designed which amplified the DNA from all tested clade 1 Botrytis species. A 6 bp indel in Bcin01g05500 was suitable for identification of B. cinerea Iris strains, and a 15 bp indel in Bcin11g00620 allowed identification of B. cinerea group B strains. The location of the indels, their specificity, and the primers used are summarized in Table 4.
Discussion
In this study, the genetic diversity of a large number of B. cinerea strains and smaller collections of B. pseudocinerea and B. fabae was investigated. Single PCR-based genotyping of Botrytis strains is useful for a preliminary classification, using polymorphic sequences and indels of selected genes, as demonstrated in this and previous studies (Plesken et al., 2015a). Indels of various sizes are frequently found in fungal genomes, but most of them are located outside of the coding sequences. Because of their rare occurrence and likely stability, indels in protein coding sequences have been used as high potential phylogenetic markers (Ajawatanawong and Baldauf, 2013). In our study, all preliminary assignments of species based on indel markers that were further analyzed by sequencing could be confirmed. For robust identification of isolates with a high sensitivity for differentiation, an MLST scheme has been established, using 10 genes which were selected for high variability and phylogenetic reliability. Using PacBio sequencing, 10 PCR products each of ca. 100 isolates can be sequenced in a single run, using appropriate barcodes. The genes selected for MLST provide more sequence information and show a considerably higher diversity than hsp60, g3pdh, and rbp2, which have been used for species differentiation of Botrytis species (Staats et al., 2005). Previous studies of B. cinerea populations, using PCR-based markers such as RAPD, ALFP, and microsatellite or simple sequence repeats (SSR; Walker, 2016), are efficient and provide valuable informations about population structures, but no sequence data for further strain comparisons. The MLST scheme developed in this study allows differentiation of B. cinerea and other Botrytis strains with high resolution. The design of the MLST primers also allowed amplification and sequencing of genes of two clade 2 species, B. paeoniae and Botrytis mali (not shown). They are therefore likely to be useful for genotyping of strains from all Botrytis species, but might require sequence adjustments for optimal performance.
The MLST analysis revealed a clear separation and phylogenetic placement of all Botrytis species. Intraspecific diversity of B. pseudocinerea and B. fabae strains appeared to be lower than that of B. cinerea. This might be partially explained by their smaller sample sizes, and remains to be confirmed with more strains from similarly diverse locations as those from B. cinerea. The phylogenetic tree reconstructed from the MLST sequences revealed several clusters with different bootstrap support. A previously identified B. cinerea population, B. cinerea “group S” (Leroch et al., 2013), was found to be much less evident on the basis of the MLST data (Figure 2), which does not justify to maintain group S as a separate taxonomic unit. Studies on B. cinerea group S strains in New Zealand and France (Johnston et al., 2014; Walker, 2016) and sequencing of additional genes, as shown below, did not support such a clear separation of group S strains.
A major discovery was the identification of a population, referred to as B. cinerea B, which was clearly separated from all other B. cinerea strains. Confirmation for genetic separation of the B. cinerea B population came from the observation that all of them lacked probably the whole botcinic acid gene (boa) cluster, which is present in all other B. cinerea strains and the majority of sequenced Botrytis species (Valero-Jiménez et al., 2020). Interestingly, the boa cluster is located in B. cinerea at the left terminus of chromosome 1, directly adjacent to the telomere (van Kan et al., 2017). It seems therefore likely that parts of the left arm of chromosome 1 have been deleted in the B. cinerea B population. Single mutants defective in botcinic acid synthesis have been shown to be unaffected in their virulence toward bean leaves, whereas double mutants defective in both botcinic acid and botrydial formation showed reduced infection (Dalmais et al., 2011). In a recent study, however, bot2 boa6 double mutants were not found to be significantly affected in their virulence (Leisen et al., 2020). The existence of a B. cinerea group which lacks botcinic acid synthesis capacity also indicates that botcinic acid has no major role for plant infection. B. cinerea B strains were found on a variety of host plants (strawberry, grapevine, broad bean, cherry, and blueberry) in Germany and Norway, which indicates that they are widely distributed in Europe and maybe worldwide. Similar to B. pseudocinerea, which exists as a minor sister species in sympatry with B. cinerea, B. cinerea B was always found as minor populations together with common B. cinerea isolates. The occurrence of B. cinerea B remains to be analyzed systematically. For this, the PCR primers that detect the 15 bp deletion in Bcin11g00620, and the loss of boa6 will facilitate a rapid screening for B. cinerea B genotypes.
The genetic separation between B. cinerea and B. cinerea group B indicates that genetic exchange between these groups has been strongly reduced or even stopped. Lack of genetic exchange by sexual recombination is an essential requirement for the definition of species and subspecies (Taylor et al., 2000). However, other concepts for species definition based on differences in morphology and ecology are much less clear. Similar to B. pseudocinerea, B. cinerea B isolates are morphologically indistinghuishable from B. cinerea regarding mycelium growth, sporulation, and sclerotium formation. Furthermore, they have been isolated from the same hosts as B. cinerea, indicating that they occupy a similar ecological niche (Walker et al., 2011). Nevertheless, B. pseudocinerea has been found to be more abundant in the spring, and is preferentially found on dead tissue parts from grapevine rather than on living berries (Walker et al., 2011). In commercial fields and orchards, B. pseudocinerea disappeared rapidly after fungicide treatments, and were almost never found to acquire fungicide resistance (Plesken et al., 2015a). B. cinerea B strains were found to be less virulent on tomato leaves than B. cinerea strains, which also indicates diverse ecological adaptions. Further studies are necessary to clarify these differences between B. cinerea B and B. cinerea. While our data indicate that a sexual barrier has already been formed between them, the loss sexual compatibility should be confirmed experimentally, as reported for B. pseudocinerea and B. cinerea (Walker et al., 2011). Further studies with B. cinerea group B populations are required to confirm whether or not they might represent a minor subspecies of B. cinerea.
The isolation of a genetically homogeneous B. cinerea population from the wild monocot plant I. pseudacorus was unexpected. B. cinerea and other clade 1 Botrytis species are known to infect almost only dicotyledonous hosts (Staats et al., 2005; van Kan, 2006), although B. cinerea can also attack some monocot plants. Iris isolates were considerably more aggressive on Iris than common B. cinerea strains. In contrast, they showed lower virulence on tomato leaves. Infection assays were performed with detached leaves, which might not yield the same results as leaves attached to intact plants. Nevertheless, differences in the infection behavior of Iris and other B. cinerea strains were obvious, in particular on Iris leaves. Several lines of evidence confirmed that the Iris strains are genetically distinct. While pink variants of B. cinerea are very rare, all but one of the Iris strains were pink and contained bcbik1 encoding the polyketide synthase for bikaverin biosynthesis. Further genetic evidences, including MLST analysis of three Iris strains, a single nucleotide polymorphism in mrr1 detected by PIRA-PCR, and a 6 bp deletion in Bcin01g05500 detected by PCR, confirmed that B. cinerea Iris represents a distinct, possibly local population. Even this small population was found to contain different haplotypes. Both mating type loci MAT1-1 and MAT1-2 were detected in similar numbers among the Iris strains, indicating that sexual exchange might still occur. Surprisingly, isolate D13_E_IL4, which clustered with the other Iris strains and was aggressive on Iris, did not show a pink color. PCR analysis revealed that this isolate not only lacked bcbik1 but also bcbik2, bcbik3, and bcbik5, which are retained in B05.10 and all other B. cinerea isolates tested. Therefore, the loss of the whole bik gene cluster from D13_E_IL4 must have occurred in recent history. Bikaverin has antibiotic, anticancer, and antioomycete activity (Son et al., 2008) and has been shown to play a role in antagonistic interactions with other microbes (Spraker et al., 2016). Whether the pigment provides a selective advantage for B. cinerea remains to be investigated.
In summary, we have shown that MLST based on 10 suitable genes is a powerful tool for differentiation and classification of Botrytis clade 1 and possibly also for clade 2 strains, which makes it possible to recognize populations with different degrees of genetic separation. This work might stimulate further research into the intraspecific diversity of B. cinerea and other Botrytis species. It confirms previous evidences that genetic adaptation of B. cinerea toward certain hosts can occur (Mercier et al., 2019). Modern genome sequencing technologies and advanced editing tools such as CRISPR/Cas (Leisen et al., 2020) will enable a comparative molecular and phenotypic analysis of B. cinerea isolates with different host specificity, to uncover the mechanisms determining host range and host adaptation of the gray mold fungus.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
CP did the sampling and genetic characterization of the isolates. PP prepared the PCR fragments for PacBio sequencing. BR, ZN, LZ, and PP performed phenotypic and PCR studies with the isolates. BH performed PacBio sequencing. KK evaluated sequencing data. MH did the conception and design of the study, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Fransziska Schlesinger, Jaqueline Altensell, and Sibylle Rumsey for their help with PCR and infection assays. We are grateful to our colleagues Roland Weber, George Karaoglanidis, Anne-Sophie Walker, Zhonghua Ma, Guoqing Li, Evelyn Silva, Guido Schnabel, and Lizel Mostert for providing us with strains from their countries.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.663027/full#supplementary-material
References
- Ajawatanawong P., Baldauf S. (2013). Evolution of protein indels in plants, animals and fungi. BMC Evol. Biol. 2013:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanio A., Desnos-Ollivier M., Garcia-Hermoso D., Bretagne S. (2017). Investigating clinical issues by genotyping of medically important fungi: why and how? Clin. Microbiol. Rev. 30 671–707. 10.1128/CMR.00043-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem J., Cuomo C. A., van Kan J. A. L., Viaud M., Benito E. P., Couloux A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. 10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner P., Koch F., Voigt K., Quidde T., Risch S., Blaich R., et al. (1994). Variations in ploidy among isolates of Botrytis cinerea: implications for genetic and molecular analyses. Curr. Genet. 25 445–450. 10.1007/BF00351784 [DOI] [PubMed] [Google Scholar]
- Campbell M. A., Staats M., van Kan J. A. L., Rokas A., Slot J. C. (2013). Repeated loss of an anciently horizontally transferred gene cluster in Botrytis. Mycologia 105 1126–1134. 10.3852/12-390 [DOI] [PubMed] [Google Scholar]
- Chen Y., Frazzitta A. E., Litvintseva A. P., Fang C., Mitchell T. G., Springer D. J., et al. (2015). Next generation multilocus sequence typing (NGMLST) and the analytical software program MLSTEZ enable efficient, cost-effective, high-throughput, multilocus sequencing typing. Fungal. Genet. Biol. 75 64–71. 10.1016/j.fgb.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais B., Schumacher J., Moraga J., Le Pêcheur P., Tudzynski B., Collado I. G., et al. (2011). The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol. Plant Pathol. 12 564–579. 10.1111/j.1364-3703.2010.00692.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R., van Kan J. A. L., Pretorius Z. A., Hammond-Kosack K. E., Di Pietro A., Spanu P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13 414–430. 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diolez A., Marches F., Fortini D., Brygoo Y. (1995). Boty, a long-terminal-repeat retroelement in the phytopathogenic fungus Botrytis cinerea. Appl. Environ. Microbiol. 61 103–108. 10.1128/AEM.61.1.103-108.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elad Y., Pertot I., Prado A., Stewart A. (2016). “Plant hosts of Botrytis spp,” in Botrytis – the Fungus, the Pathogen and its Management in Agricultural Systems, eds Fillinger S., Elad Y. (Cham: Springer; ), 413–486. [Google Scholar]
- Faretra F., Antonacci E., Pollastro S. (1988). Sexual behaviour and mating system of Botryotinia fuckeliana, teleomorph of Botrytis cinerea. J. Gen. Microbiol. 134 2543–2550. [Google Scholar]
- Fournier E., Giraud T., Albertini C., Brygoo Y. (2005). Partition of the Botrytis cinerea complex in France using multiple gene genealogies. Mycologia 97 1251–1267. 10.3852/mycologia.97.6.1251 [DOI] [PubMed] [Google Scholar]
- Fournier E., Levis C., Fortini D., Leroux P., Giraud T., Brygoo Y. (2003). Characterization of Bc-hch, the Botrytis cinerea homolog of the Neurospora crassa het-c vegetative incompatibility locus, and its use as a population marker. Mycologia 95 251–261. [PubMed] [Google Scholar]
- Giraud T., Fortini D., Levis C., Lamarque C., Leroux P., Lobuglio K., et al. (1999). Two sibling species of the Botrytis cinerea complex, transposa and vacuma, are found in sympatry on numerous host plants. Phytopathology 89 967–973. 10.1094/PHYTO.1999.89.10.967 [DOI] [PubMed] [Google Scholar]
- Giraud T., Fortini D., Levis C., Leroux P., Brygoo Y. (1997). RFLP markers show genetic recombination in Botryotinia fuckeliana (Botrytis cinerea) and transposable elements reveal two sympatric species. Mol. Biol. Evol. 14 1177–1185. 10.1093/oxfordjournals.molbev.a025727 [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27 221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Haliassos A., Chomel J. C., Grandjouan S., Kruh J., Kaplan J. C., Kitzis A. (1989). Detection of minority point mutations by modified PCR technique: a new approach for a sensitive diagnosis of tumor-progression markers. Nucleic Acids. Res. 17 8093–8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde K. D., Nilsson R. H., Alias S. A., Ariyawansa H. A., Blair J. E., Cai L., et al. (2014). One stop shop: backbones trees for important phytopathogenic genera: I: Botrytis. Fungal Divers. 67 21–125. [Google Scholar]
- Johnston P. R., Hoksbergen K., Park D., Beever R. E. (2014). Genetic diversity of Botrytis in New Zealand vineyards and the significance of its seasonal and regional variation. Plant Pathol. 63 888–898. [Google Scholar]
- Kerssies A. (1990). A selective medium for Botrytis cinerea to be used in a spore-trap. Neth. J. Pl. Path. 96 247–250. [Google Scholar]
- Kretschmer M., Leroch M., Mosbach A., Walker A.-S., Fillinger S., Mernke D., et al. (2009). Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog. 5:e1000696. 10.1371/journal.ppat.1000696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisen T., Bietz F., Werner J., Wegner A., Schaffrath U., Scheuring D., et al. (2020). CRISPR/Cas with ribonucleoprotein complexes and transiently selected telomere vectors allows highly efficient marker-free and multiple genome editing in Botrytis cinerea. PLoS Pathog. 16:e1008326. 10.1371/journal.ppat.1008326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroch M., Plesken C., Weber R. W. S., Kauff F., Scalliet G., Hahn M. (2013). Grey mould populations in German strawberry fields are resistant to multiple fungicides and dominated by a novel clade Closely related to Botrytis cinerea. Appl. Environ. Microbiol. 79 159–167. 10.1128/AEM.02655-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis C., Fortini D., Brygoo Y. (1997). Flipper, a mobile Fot1-like transposable element in Botrytis cinerea. Mol. Gen. Genet. 254 674–680. [DOI] [PubMed] [Google Scholar]
- Liu Q., Li G., Li J., Chen S. (2016). Botrytis eucalypti, a novel species isolated from diseased Eucalyptus seedlings in South China. Mycol. Progress 15 1057–1079. 10.1007/s11557-016-1229-1 [DOI] [Google Scholar]
- Maiden M. C. J. (2006). Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60 561–588. 10.1146/annurev.micro.59.030804.121325 [DOI] [PubMed] [Google Scholar]
- Marthey S. (2008). FUNYBASE: a FUNgal phYlogenomic dataBASE. BMC Bioinformatics 9:456. 10.1186/1471-2105-9-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F., Blancard D., Lecomte P., Levis C., Dubos B., Fermaud M. (2003). Phenotypic differences between vacuma and transposa subpopulations of Botrytis cinerea. Eur. J. Plant Pathol. 109 479–488. [Google Scholar]
- Martinez F., Dubos B., Fermaud M. (2005). The Role of saprotrophy and virulence in the population dynamics of Botrytis cinerea in vineyards. Phytopathology 95 692–700. 10.1094/PHYTO-95-0692 [DOI] [PubMed] [Google Scholar]
- Mercier A., Carpentier F., Duplaix C., Auger A., Pradier J. M., Viaud M., et al. (2019). The polyphagous plant pathogenic fungus Botrytis cinerea encompasses host-specialized and generalist populations. Environ. Microbiol. 21 4808–4821. 10.1111/1462-2920.14829 [DOI] [PubMed] [Google Scholar]
- Odds F. C., Jacobsen M. D. (2008). Multilocus sequence typing of pathogenic Candida species. Eukaryotic Cell 7 1075–1084. 10.1128/EC.00062-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesken C., Weber R. W. S., Rupp S., Leroch M., Hahn M. (2015a). Botrytis pseudocinerea is a significant pathogen of several crop plants but susceptible to displacement by fungicide-resistant B. cinerea strains. Appl. Environ. Microbiol. 81 7048–7056. 10.1128/AEM.01719-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesken C., Westrich L. D., Hahn M. (2015b). Genetic and phenotypic characterization of Botrytis calthae. Plant Pathol. 64 128–136. [Google Scholar]
- Porquier A., Moraga J., Morgant G., Dalmais B., Simon A., Sghyer H., et al. (2019). Botcinic acid biosynthesis in Botrytis cinerea relies on a subtelomeric gene cluster surrounded by relics of transposons and is regulated by the Zn2Cys6 transcription factor BcBoa13. Curr. Genet. 65 965–980. 10.1007/s00294-019-00952-4 [DOI] [PubMed] [Google Scholar]
- Richards J., Xiao C. L., Jurick Ii W. (2020). Botrytis spp.: a contemporary perspective and synthesis of recent scientific developments of a widespread genus that threatens global food security. Phytopathology 111 432–436. 10.1094/PHYTO-10-20-0475-IA [DOI] [PubMed] [Google Scholar]
- Rigotti S., Gindro K., Richter H., Viret O. (2002). Characterization of molecular markers for specific and sensitive detection of Botrytis cinerea Pers. Fr. in strawberry (Fragaria xananassa Duch.) using PCR. FEMS Microbiol. Lett. 209 169–174. 10.1111/j.1574-6968.2002.tb11127.x [DOI] [PubMed] [Google Scholar]
- Rowe H. C., Kliebenstein D. J. (2007). Elevated genetic variation within virulence-associated Botrytis cinerea polygalacturonase loci. Mol. Plant Microbe Interact. 20 1126–1137. 10.1094/MPMI-20-9-1126 [DOI] [PubMed] [Google Scholar]
- Rupp S., Weber R. W. S., Rieger D., Detzel P., Hahn M. (2017). Spread of Botrytis cinerea strains with multiple fungicide resistance in German horticulture. Front. Microbiol. 7:2075. 10.3389/fmicb.2016.02075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J., Gautier A., Morgant G., Studt L., Ducrot P.-H., Le Pêcheur P., et al. (2013). A functional bikaverin biosynthesis gene cluster in rare strains of Botrytis cinerea is positively controlled by VELVET. PLoS One 8:e53729. 10.1371/journal.pone.0053729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S. W., Kim H. Y., Choi G. J., Lim H. K., Jang K. S., Lee S. O., et al. (2008). Bikaverin and fusaric acid from Fusarium oxysporum show antioomycete activity against Phytophthora infestans. J. Appl. Microbiol. 104 692–698. 10.1111/j.1365-2672.2007.03581.x [DOI] [PubMed] [Google Scholar]
- Spraker J. E., Sanchez L. M., Lowe T. M., Dorrestein P. C., Keller N. P. (2016). Ralstonia solanacearum lipopeptide induces chlamydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J. 10 2317–2330. 10.1038/ismej.2016.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats M., van Baarlen P., Schouten A., van Kan J. A. L., Bakker F. T. (2007). Positive selection in phytotoxic protein-encoding genes of Botrytis species. Fungal Genet. Biol. 44 52–63. 10.1016/j.fgb.2006.07.003 [DOI] [PubMed] [Google Scholar]
- Staats M., van Baarlen P., van Kan J. A. L. (2005). Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Mol. Biol. Evol. 22 333–346. 10.1093/molbev/msi020 [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Jacobson D. J., Kroken S., Kasuga T., Geiser D. M., Hibbett D. S., et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31 21–32. 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- Valero-Jiménez C. A., Steentjes M. B. F., Slot J. C., Shi-Kunne X., Scholten O. E., van Kan J. A. L. (2020). Dynamics in secondary metabolite gene clusters in otherwise highly syntenic and stable genomes in the fungal genus Botrytis. Genome Biol. Evol. 12 2491–2507. 10.1093/gbe/evaa218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kan J. A. L. (2006). Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11 247–253. 10.1016/j.tplants.2006.03.005 [DOI] [PubMed] [Google Scholar]
- van Kan J. A. L., Stassen J. H. M., Mosbach A., van der Lee T. A. J., Faino L., Farmer A. D., et al. (2017). A gapless genome sequence of the fungus Botrytis cinerea. Mol. Plant Pathol. 18 75–89. 10.1111/mpp.12384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A.-S. (2016). “Diversity within and between species of Botrytis,” in Botrytis – the Fungus, the Pathogen and its Management in Agricultural Systems, Chap. 6, eds Fillinger S., Elad Y. (Cham: Springer; ), 95–124. [Google Scholar]
- Walker A.-S., Gautier A. L., Confais J., Martinho D., Viaud M., Le P., et al. (2011). Botrytis pseudocinerea, a new cryptic species causing grey mould in French vineyards in sympatry with Botrytis cinerea. Phytopathology 101 1433–1445. 10.1094/PHYTO-04-11-0104 [DOI] [PubMed] [Google Scholar]
- Walker A.-S., Gladieux P., Decognet V., Fermaud M., Confais J., Roudet J., et al. (2015). Population structure and temporal maintenance of the multihost fungal pathogen Botrytis cinerea: causes and implications for disease management. Environ. Microbiol. 17 1261–1274. 10.1111/1462-2920.12563 [DOI] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F.-M., Zhao H., Zhang Z., Kaloshian I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342 118–123. 10.1126/science.1239705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetzel H. H., Drayton F. L. (1932). A new species of Botrytis on rhizomatous iris. Mycologia 24:469. [Google Scholar]
- Zhang J., Yang H., Yu Q. Y., Wu M. D., Yang L., Zhuang W. Y., et al. (2016). Botrytis pyriformis sp. nov., a novel and likely saprophytic species of Botrytis. Mycologia 108 682–696. 10.3852/15-340 [DOI] [PubMed] [Google Scholar]
- Zhou Y. J., Zhang J., Wang X. D., Yang L., Jiang D. H., Li G. Q., et al. (2014). Morphological and phylogenetic identification of Botrytis sinoviticola, a novel cryptic species causing grey mould disease of table grapes (Vitis vinifera) in China. Mycologia 106 43–56. 10.3852/13-032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.