Abstract
Known modifiable risk factors account for a small fraction of premenopausal breast cancers. We investigated associations between pre-diagnostic circulating amino acid and amino acid-related metabolites (N = 207) and risk of breast cancer among predominantly premenopausal women of the Nurses’ Health Study II using conditional logistic regression (1057 cases, 1057 controls) and multivariable analyses evaluating all metabolites jointly. Eleven metabolites were associated with breast cancer risk (q-value < 0.2). Seven metabolites remained associated after adjustment for established risk factors (p-value < 0.05) and were selected by at least one multivariable modeling approach: higher levels of 2-aminohippuric acid, kynurenic acid, piperine (all three with q-value < 0.2), DMGV and phenylacetylglutamine were associated with lower breast cancer risk (e.g., piperine: ORadjusted (95%CI) = 0.84 (0.77–0.92)) while higher levels of creatine and C40:7 phosphatidylethanolamine (PE) plasmalogen were associated with increased breast cancer risk (e.g., C40:7 PE plasmalogen: ORadjusted (95%CI) = 1.11 (1.01–1.22)). Five amino acids and amino acid-related metabolites (2-aminohippuric acid, DMGV, kynurenic acid, phenylacetylglutamine, and piperine) were inversely associated, while one amino acid and a phospholipid (creatine and C40:7 PE plasmalogen) were positively associated with breast cancer risk among predominately premenopausal women, independent of established breast cancer risk factors.
Subject terms: Risk factors, Epidemiology
Introduction
Breast cancer is the most common malignancy among women in the United States, with more than 250,000 cases diagnosed each year1. Known modifiable risk factors are estimated to account for only around one-third of postmenopausal breast cancers2–4, and an even smaller fraction of premenopausal cancers2,5. Thus, new strategies are needed for the identification of modifiable risk factors, especially for premenopausal breast cancers.
Metabolites are small molecules that are produced and consumed by cellular metabolism. The study of the complete collection of metabolites, called metabolomics, provides a direct signature of cellular activity in the body and has emerged as a powerful tool for the diagnosis, characterization, and prediction of disease. Metabolomic methods have uncovered biomarkers for a wide variety of cancers including colorectal, gastric, pancreatic, liver, ovarian, breast, urinary, esophageal, and lung6. In breast cancer, metabolomics has proven useful for tumor biology characterization, predicting treatment response, anticipating recurrence, and estimating prognosis7.
More recently, prospective epidemiological studies have used metabolomics to identify metabolite risk factors for several cancers including pancreatic8–10, prostate11,12, liver13, colorectal14, ovarian15,16, endometrial17, and breast cancer18–22. For breast cancer, studies have used both targeted18,21,23 and untargeted20,22 methods to discover metabolomic risk factors associated with diet20,23, body mass index (BMI)21, microbiota metabolism20, lipid, amino acid, and other metabolic pathways18,20,22. While several studies stratified results by estrogen receptor (ER) status18,21,23, no prospective metabolomic breast cancer studies have investigated differential effects by menopausal status.
In this study, we assessed the association of over 200 prospectively measured circulating amino acid and amino acid-related metabolites with risk of breast cancer among the predominantly premenopausal women (1057 cases and 1057 matched controls) of the Nurses’ Health Study II (NHSII).
Results
Study population
1057 cases and 1057 matched controls were included in this study (Table 1). Women were an average 53 years old and predominantly premenopausal (80%) at the time of blood collection. At diagnosis, 42% of the women were premenopausal and 46% were postmenopausal. The mean time between blood collection and diagnosis was 8 years (SD = 4.4), ranging from 10 months to 17.4 years. (1st quartile: 4.25 years; 3rd quartile: 11.6 years).
Table 1.
Characteristics of breast cancer cases and matched controls at blood collection in the Nurses’ Health Study II, mean (SD) or %.
| Cases (n = 1057) | Controls (n = 1057) | |
|---|---|---|
| Age at blood collectiona (y) | 44.7 (4.5) | 44.8 (4.4) |
| Age at menarche (y) | 12.4 (1.3) | 12.8 (1.4) |
| Time between sample collection and diagnosis (y) | 8.0 (4.4) | |
| Parity and age at first birth (%): | ||
| Nulliparous | 21.1 | 15.9 |
| 1–2 children, ≥25 y | 39.2 | 34.9 |
| 1–2 children, <25 y | 14.7 | 15.9 |
| 3+ children, <25 y | 11.3 | 16.6 |
| 3+ children, ≥25 y | 13.8 | 14.2 |
| Ever breastfed (%) | 63.1 | 65.0 |
| Family history of breast cancer (%) | 17.4 | 10.8 |
| Personal history of benign breast disease (%) | 22.1 | 15.6 |
| BMI at age 18 (kg/m2) | 20.8 (2.9) | 21.1 (3.1) |
| Weight change between age 18 and blood collection (kg) | 11.6 (12.0) | 12.6 (13.2) |
| Physical activity, MET-h/wk | 18.0 (15.3) | 18.1 (15.5) |
| Alcohol consumption (g/day) | 3.8 (6.9) | 3.3 (5.6) |
| Past/current exogenous hormone useb (%) | 86.3 | 86.7 |
| Menopausal status at blood collectiona (%) | ||
| Premenopausal | 80.2 | 79.7 |
| Postmenopausal | 12.7 | 13.1 |
| Unknown | 7.1 | 7.3 |
| Menopausal status at diagnosisa (%) | ||
| Premenopausal | 42.0 | 42.2 |
| Postmenopausal | 46.4 | 47.1 |
| Unknown | 11.6 | 10.7 |
| Caucasiana (%) | 97.2 | 98.4 |
| Fasting (>8 h) at blood collectiona (%) | 68.7 | 74.7 |
aMatching factor
bOral contraceptive or menopausal hormone therapy
Conditional logistic regression (CLR)
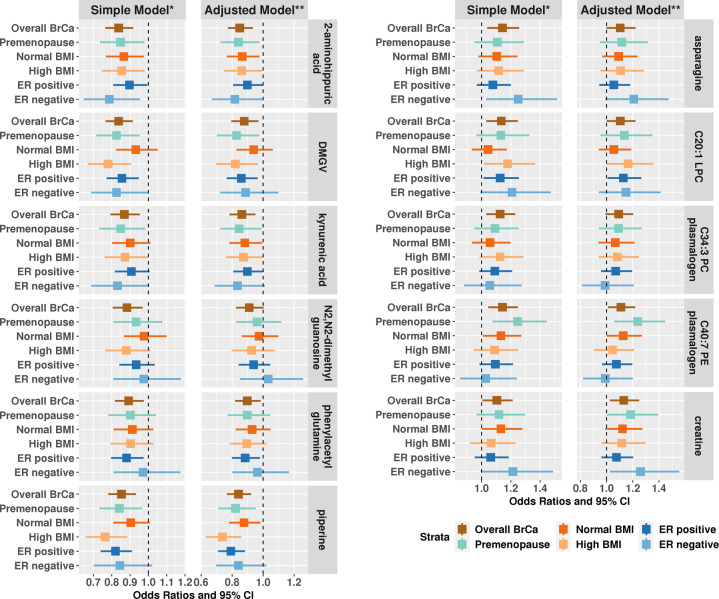
Eleven metabolites were significantly associated with risk of breast cancer based on the simple model (Fig. 1 and Supplementary Table 1). Six metabolites were associated with lower risk while five metabolites were associated with higher risk of overall breast cancer. Dimethylguanidino valeric acid (DMGV; OR per 1-SD increase (95% CI) = 0.84 (0.77–0.92)), 2-aminohippuric acid (OR (95% CI) = 0.84 (0.76–0.92)), and piperine (OR (95% CI) = 0.85 (0.78–0.93)) had the strongest inverse associations. C40:7 phosphatidylethanolamine (PE) plasmalogen (OR (95% CI) = 1.14 (1.05–1.25)) and asparagine (OR (95% CI) = 1.14 (1.04–1.26)) had the strongest positive associations. Creatine was the only metabolite with q-value > 0.2 in the simple model but is included here as q-value < 0.2 in both Lasso models. Results were similar when we included adjustment for breast cancer risk factors (DMGV: 0.88 (0.79–0.97); 2-aminohippuric acid: 0.85 (0.77–0.93); C40:7 PE: plasmalogen: 1.11 (1.01–1.22); asparagine: 1.10 (1.00–1.22)) and when we restricted to premenopausal women (Fig. 1 and Supplementary Table 2) or ER+ tumors (Fig. 1 and Supplementary Table 3).
Fig. 1. Odds ratios and 95% confidence intervals (CI) per 1 SD increase for metabolites significantly associated with risk of overall breast cancer (p-value < 0.05 and q-value < 0.2) in Nurses’ Health Study II, among premenopausal women only, by BMI category (<25, ≥25)***, and by ER status (ER positive, ER negative)****.
Creatine, although not selected by the conditional logistic regression (p-value < 0.05, q-value > 0.2), is shown here for completeness, as is was selected by the multivariable models. *Simple model: adjusts for matching factors including menopause status at blood draw, time of blood draw, date/season of blood draw, luteal day at blood draw, fasting status at blood draw, menopausal status at diagnosis and race. **Adjusted model: in addition to matching factors, this model adjusts for BMI at age 18, weight change between age 18 and time of blood draw, age at menarche, parity and age at first birth, family history of breast cancer, personal history of benign breast disease, physical activity, alcohol consumption, exogenous hormone use, and breast feeding history. ***All p-interaction with BMI category were >0.07 except for DMGV p-interaction = 0.04 (simple and adjusted model). ****All p-heterogeneity by ER status were >0.13 except for asparagine p-heterogeneity = 0.02/0.03 (simple/adjusted model).
Among the 11 selected metabolites, only DMGV showed effect modification by BMI with stronger associations among women with high BMI (adjusted model, high BMI: OR (95% CI) = 0.82 (0.70–0.97); normal BMI: OR (95% CI) = 0.94 (0.83–1.06); p-interaction = 0.04). Significant interactions with BMI were observed for several additional metabolites that were not significant overall; for example C36:4 DAG/TAG fragment (high BMI: OR (95% CI) = 0.92 (0.80–1.06); normal BMI: OR (95% CI) = 1.21 (1.06–1.37); p-interaction = 0.005), serine (high BMI: OR (95% CI) = 1.19 (1.04–1.38); normal BMI: OR (95% CI) = 0.93 (0.83–1.05); p-interaction = 0.008), and proline betaine (high BMI: OR (95% CI) = 0.82 (0.72–0.94); normal BMI: OR (95% CI) = 1.04 (0.92–1.17); p-interaction = 0.017).
Of the 11 selected metabolites, we only observed stronger associations with risk of ER- tumors in the adjusted model (ER−; Supplementary Table 3) for asparagine (ER− tumors: OR (95% CI) = 1.21 (0.99–1.47); ER+ tumors: OR (95% CI) = 1.06 (0.94–1.18); p-heterogeneity = 0.03). Three additional metabolites were suggestively different by ER status: betaine (ER− tumors: OR (95% CI) = 0.89 (0.73–1.09); ER+ tumors: OR (95% CI) = 1.07 (0.96–1.20); p-heterogeneity = 0.02), 4-acetamidobutanate (ER− tumors: OR (95% CI) = 0.89 (0.72–1.09); ER+ tumors: OR (95% CI) = 0.97 (0.87–1.08); p-heterogeneity = 0.03), and histidine (ER− tumors: OR (95% CI) = 1.06 (0.87–1.29); ER+ tumors: OR (95% CI) = 1.01 (0.91–1.13); p-heterogeneity = 0.05).
In an exploratory analysis including metabolites with >10% missing values (N = 19; four metabolites had >90% missingness), metoprolol (45% missing values) was nominally significantly associated with risk of breast cancer (likelihood-ratio test p-value = 0.04; data not shown). Women with detectable metoprolol levels had a 23% higher risk (presence–absence indicator p-value = 0.07) of breast cancer compared to women with undetectable metoprolol. However, among women with measured metoprolol, higher levels were associated with lower risk (OR per one unit increase in log-transformed and standardized metabolite levels = 0.91, p-value = 0.14). The remaining metabolites with high missingness were not associated with risk of breast cancer.
Multivariable models of the joint association of all metabolites
The inverse association of piperine with risk of breast cancer met the threshold for statistical significance (p-value < 0.05 and q-value < 0.20) in all three simple (without adjustment for risk factors) multivariable models, Lasso, Elastic Net, and Random Forests (Tables 2 and 3). In addition, DMGV and N2,N2-dimethylguanosine were detected in the Random Forests model with minimal adjustment, satisfying a q-value threshold of 0.05. Higher levels of C40:7 PE plasmalogen and creatine were associated with increased breast cancer risk in CLR Lasso models (q-value < 0.20). These associations remained significant after further adjustment for risk factors (nominal p < 0.05) with the exception of N2,N2-dimethylguanosine in Random Forests (p = 0.05) and piperine in Elastic Net (nominal p-value < 0.05, q-value > 0.2).
Table 2.
All metabolites that met q-value < 0.2 (marked as √ in the table) in at least one primary model analysis that adjusts for matching factors (*marks p-value < 0.05a).
| Metabolites | Logisticb | Lassob | Elastic netb | Random forestb | Number of √ | Logisticc | Lassoc | Elastic netc | Random forestc |
|---|---|---|---|---|---|---|---|---|---|
| DMGV | √* | * | √* | 2 | * | √* | |||
| 2-aminohippuric acid | √* | * | 1 | √* | * | ||||
| piperine | √* | √* | √* | √* | 4 | √* | √* | * | √* |
| kynurenic acid | √* | * | 1 | √* | √* | ||||
| N2,N2-dimethylguanosine | √* | √* | 2 | ||||||
| Phenylacetyl-glutamine | √* | * | 1 | * | √* | ||||
| C34:3 PC plasmalogen | √* | * | * | 1 | √* | √* | |||
| C20:1 LPC | √* | 1 | |||||||
| C40:7 PE plasmalogen | √* | √* | * | 2 | * | √* | * | ||
| Asparagine | √* | * | * | * | 1 | * | * | ||
| Creatine | * | √* | * | 1 | * | √* |
aFour metabolites that met threshold for statistical significance in Lasso only but had Logistic regression raw p values > 0.3 were excluded (C12:1 carnitine, C22:5 LPC, C46:2 TAG, glycine)
bSimple model adjusts for matching factors including menopause status at blood draw, time of blood draw, date/season of blood draw, luteal day at blood draw, fasting status at blood draw, menopausal status at diagnosis and race
cAdjusted model: in addition to matching factors, this model adjusts for BMI at age 18, weight change between age 18 and time of blood draw, age at menarche, parity and age at first birth, family history of breast cancer, personal history of benign breast disease, physical activity, alcohol consumption, exogenous hormone use, and breast feeding history
Table 3.
All metabolites that met q-value < 0.2 in at least one primary model analysis that adjusts for matching factors. M1 is the simple modela (accounting for matching factors) and M2 is the adjusted modelb (adjusting for matching factors and breast cancer risk factors).
| Metabolite | Model | Logistic odds ratio | Logistic P-value | Lasso odds ratio | Lasso P-value | Elastic net odds ratio | Elastic net P-value | Random forest P-value |
|---|---|---|---|---|---|---|---|---|
| DMGV | M1 | 0.84 | 9.23E−05 | 0.87 | 4.00E−02 | 0.90 | 5.60E−02 | <0.01 |
| M2 | 0.88 | 1.10E−02 | 0.89 | 6.80E−02 | 0.96 | 1.64E−01 | <0.01 | |
| 2-aminohippuric acid | M1 | 0.84 | 1.42E−04 | 0.93 | 2.24E−01 | 0.94 | 2.48E−01 | 0.02 |
| M2 | 0.85 | 7.72E−04 | 0.92 | 1.56E−01 | 0.96 | 9.60E−02 | 0.02 | |
| Piperine | M1 | 0.85 | 4.59E−04 | 0.80 | 0.00E+00 | 0.85 | 0.00E+00 | <0.01 |
| M2 | 0.84 | 2.79E−04 | 0.78 | 0.00E+00 | 0.92 | 1.60E−02 | <0.01 | |
| Kynurenic acid | M1 | 0.87 | 3.00E−03 | 0.87 | 2.80E−02 | 0.90 | 5.60E−02 | 0.1 |
| M2 | 0.86 | 2.92E−03 | 0.86 | 1.20E−02 | 0.96 | 8.80E−02 | 0.11 | |
| N2,N2-dimethyl-guanosine | M1 | 0.88 | 8.47E−03 | 1.00 | 1.00E+00 | 0.97 | 6.04E−01 | <0.01 |
| M2 | 0.91 | 6.01E−02 | 1.00 | 5.04E−01 | 0.98 | 3.04E−01 | 0.05 | |
| Phenylacetyl-glutamine | M1 | 0.89 | 1.15E−02 | 0.88 | 2.80E−02 | 0.90 | 5.20E−02 | 0.13 |
| M2 | 0.90 | 2.13E−02 | 0.89 | 2.40E−02 | 0.96 | 9.20E−02 | 0.19 | |
| C34:3 PC plasmalogen | M1 | 1.13 | 7.64E−03 | 1.24 | 2.80E−02 | 1.09 | 1.28E−01 | 0.01 |
| M2 | 1.09 | 7.44E−02 | 1.29 | 1.60E−02 | 1.03 | 1.64E−01 | <0.01 | |
| C20:1 LPC | M1 | 1.13 | 9.00E−03 | 1.08 | 2.44E−01 | 1.06 | 3.80E−01 | 0.17 |
| M2 | 1.10 | 5.18E−02 | 1.14 | 6.00E−02 | 1.02 | 2.76E−01 | 0.25 | |
| C40:7 PE plasmalogen | M1 | 1.14 | 3.23E−03 | 1.25 | 0.00E+00 | 1.11 | 6.80E−02 | 0.01 |
| M2 | 1.11 | 3.01E−02 | 1.29 | 0.00E+00 | 1.03 | 1.28E−01 | 0.04 | |
| Asparagine | M1 | 1.14 | 6.10E−03 | 1.22 | 2.80E−02 | 1.13 | 3.20E−02 | 0.02 |
| M2 | 1.10 | 5.41E−02 | 1.18 | 4.40E−02 | 1.04 | 1.20E−01 | 0.03 | |
| Creatine | M1 | 1.10 | 3.94E−02 | 1.27 | 0.00E+00 | 1.16 | 1.20E−02 | 0.06 |
| M2 | 1.13 | 1.51E−02 | 1.24 | 0.00E+00 | 1.05 | 5.60E−02 | 0.07 |
aSimple model: adjusts for matching factors including menopausal status at blood draw, time of blood draw, date/season of blood draw, luteal day at blood draw, fasting status at blood draw, menopausal status at diagnosis and race
bAdjusted model: in addition to matching factors, this model adjusts for BMI at age 18, weight change between age 18 and time of blood draw, age at menarche, parity and age at first birth, family history of breast cancer, personal history of benign breast disease, physical activity, alcohol consumption, exogenous hormone use, breast feeding history
When all eleven identified metabolites were assessed together in an adjusted lasso CLR model, ten metabolites remained independently associated with risk of breast cancer. The direction of association in the lasso CLR model was consistent with the previous CLR and multivariable models except for N2,N2-dimethylguanosine who’s coefficient was estimated to be equal to zero, reflecting its high correlation with kynurenic acid and 2-aminohippuric acid (Fig. 2).
Fig. 2. Heatmap of all pairwise correlations among the 11 metabolites associated with breast cancer risk.
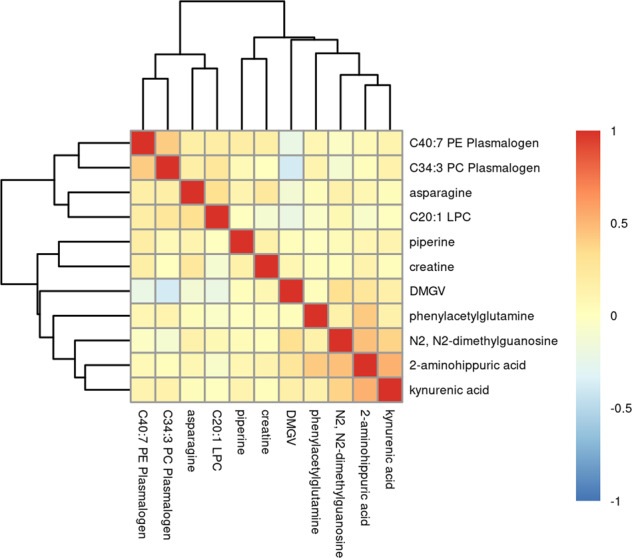
Positive correlations are shown in shades of red while inverse correlations are shown in shades of blue.
Discussion
We conducted a large-scale study of 207 circulating amino acid and amino acid-related metabolites, and risk of breast cancer in a nested case-control study (1057 cases and 1057 matched controls) within NHSII, a cohort of predominantly premenopausal women. Higher levels of six metabolites, 2-aminohippuric acid, DMGV, kynurenic acid, N2, N2-dimethylguanosine, phenylacetyl glutamine, and piperine, were associated with lower breast cancer risk while higher levels of asparagine, creatine, and three lipids, C20:1 LPC, C34:3 PC plasmalogen, C40:7 PE plasmalogen, were associated with increased breast cancer risk. Inverse associations between 2-aminohippuric acid, DMGV, kynurenic acid, phenylacetyl glutamine and piperine, and the positive associations with creatine and C40:7 PE plasmalogen remained statistically significant after adjusting for established risk factors and were selected by at least one multivariable modeling approach (Lasso, Elastic Net, Random Forests). Notably, associations between 2-aminohippuric acid, piperine, and kynurenic acid remained significant even after multiple testing correction. None of the metabolites showed effect modification by BMI, except DMGV. None of the metabolites showed heterogeneity by ER status, except asparagine.
Piperine is a polyphenol responsible for the pungency of black and long pepper and exhibits a wide range of properties: anti-diabetic, anti-inflammatory, immunomodulatory, reduction of insulin resistance, and enhanced drug bioavailability24–26. Piperine also inhibits tumorigenesis, tumor angiogenesis, cancer cell proliferation, cancer cell migration and invasion, and enhances apoptosis and autophagy27. Experimental and cell line studies identified anti-breast cancer specific mechanisms of action, including decreased matrix metalloproteinase 9 (MMP-9) and MMP-13 expression, induced apoptosis through activation of caspase-3 and inhibition of human epidermal growth factor receptor 2 (HER2) gene expression28. Synergetic effects of piperine and chemotherapy drugs (paclitaxel, doxorubicin), hormone therapy drugs (tamoxifen), radiotherapy, TRAIL-based and nano-delivery-based therapy drugs (paclitaxel, rapamycine) were observed28. Notably, piperine inhibited growth and motility29, and enhanced efficacy of TRAIL-based therapy30 in triple-negative breast cancer cells, the most aggressive breast cancer subtype. In a previous study of the associations of diet-related metabolites and breast cancer risk within the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer screening trial (n = 1242, 621 cases), piperine was found to be modestly correlated with liquor consumption (correlation = 0.16) and similar to our study, inversely associated with breast cancer risk (OR comparing 90th versus 10th percentile = 0.74 (0.56–0.99, p = 0.045)), after adjusting for BMI and other potential confounders23. Similarly, piperine was associated with lower risk of breast cancer (OR per 1 SD increase = 0.94 (0.89–0.99), p = 0.01) in the SU.VI.MAX cohort (n = 400, 200 cases)20. Notably, PLCO women were postmenopausal while the SU.VI.MAX cohort included premenopausal and postmenopausal women at the time of blood collection suggesting that the association between piperine and breast cancer may be independent of menopausal status.
Dimethylguanadino valeric acid (DMGV), an organic keto acid, is the product of transamination of asymmetric dimethylarginine (ADMA), which inhibits nitric oxide signaling that is crucial to endothelial function—excess ADMA is associated with increased risk of cardiovascular disease31,32. Plasma DMGV is positively associated with incident coronary artery disease, cardiovascular mortality, nonalcoholic fatty acid liver disease, and type II diabetes33,34. Circulating DMGV is directly correlated with resistance to the metabolic benefits of exercise35. Consumption of vegetables and red wine are associated with lower circulating DMGV, while sugar-sweetened beverage consumption is associated with higher circulating DMGV34,36. DMGV was associated with lower breast cancer risk in our study. The association between DMGV and breast cancer risk has not been previously assessed in prospective cohort studies. However, this metabolite was correlated with liver fat in the offspring cohort of the Framingham Heart Study (β = 0.02, 95% CI: 0.018–0.022, p < 10−23)33. In addition, in the same study, baseline DMGV levels were associated with higher risk of type 2 diabetes, with replication of this association in the Malmo Diet and Cancer study and the Jackson Heart Study33. Large prospective studies are required to validate the association between DMGV and breast cancer risk. If replicated, experimental studies will be needed to understand the complex relationship between DMGV, diet, physical activity, CVD, type II diabetes, and breast cancer. Of note is that this analysis was in predominantly premenopausal women, among whom adiposity is also inversely associated with risk of breast cancer for reasons that are still not fully understood37.
Although the measurement platform used here was optimized to measure amino acids and related metabolites, not lipids, our analysis included a small number of lipids and identified a few significant associations. Plasmalogens are a subclass of phospholipids (components of the cell membrane and involved in cell signaling and cell cycle regulation38) that constitute 15–20% of all phospholipids in cell membranes39. Plasmalogens have head groups that are usually either phosphatidylcholine (PC plasmalogens) or ethanolamine (PE plasmalogens), and are characterized by an ether bond to an alkenyl group in the sn-1 position while the sn-2 position is usually occupied by polyunsaturated fatty acids39. Certain cancers exhibit altered plasmalogen levels: circulating plasmalogens are depressed in pancreatic cancer patients40 and increased in gastric carcinoma patients41 compared to healthy controls. Our study identified two plasmalogens, C34:3 PC plasmalogen and C40:7 PE plasmalogen, associated with increased risk of breast cancer. The fatty acid component of specific lipids may also reflect dietary or metabolic processes; notably C40:7 PE plasmalogen is highly unsaturated but the position of the double bonds cannot be determined in this metabolomics assay. Among 74 women with breast cancer, the levels of the majority of measured phospholipids (LPC, LPE, PC, and PE) were higher in tumor tissue when compared to normal breast tissue samples38. Similar trends were observed in another study comparing tumor to normal tissue in 257 participants with breast cancer, and these lipids were correlated with cancer progression and patient survival42. Contrary to our findings, in the European Prospective Investigation into Cancer (EPIC) cohort, PC plasmalogens, including C34:3 PC plasmalogen, were inversely associated with risk of breast cancer18,19. However, neither study stratified their results by menopausal status, thus making a direct comparison difficult. Additional studies are needed to evaluate how plasmalogens are associated with risk of breast cancer and if this relationship is modulated by menopausal status.
LPCs are derived from phosphatidylcholines after hydrolysis of one of the fatty acid groups. In the liver, LPCs upregulate genes involved in cholesterol biosynthesis, while circulating LPCs activate many inflammatory and oxidative stress signaling pathways, and are associated with inflammatory diseases such as atherosclerosis and multiple sclerosis43. Circulating LPCs have mixed associations with certain cancers. For instance, circulating LPCs are elevated in ovarian cancer patients but depressed in leukemia patients relative to healthy controls44; LPCs showed inverse associations with risk of endometrioid and clear cell ovarian tumors, with stronger inverse associations among premenopausal women, in NHS and NHSII15,16. While most LPCs measured in EPIC were inversely associated with risk, one of our top hits was LPC C20:1 that was positively associated with risk18. In an earlier study nested within the EPIC cohort of 774 participants including 362 breast cancer cases, C18:0 LPC was identified as inversely associated with breast cancer risk, after adjusting for potential risk factors19, though analyses were not stratified by menopausal status.
Our study identified three amino acid derivatives associated with breast cancer risk. High levels of phenylacetylglutamine were associated with decreased breast cancer risk while high levels of asparagine and creatine were associated with increased risk. Phenylacetylglutamine is formed from phenylacetate and glutamine and is found as a normal constituent of human urine45. Phenylacetylglutamine is a host microbiome cometabolite associated with bacterial phenylalanine metabolism46–49. Clostridium difficile, F. prausnitzii, Bifidobacterium, Subdoligranulum, and Lactobacillus are all positively associated with hippuric acid48,50, while Bifidobacterium is positively associated with phenylacetylglutamine and microbes of the Christensellaceae, Ruminococcaceae, and Lachnospiracaea families are negatively associated with phenylacetylglutamine47,51. While not directly linked to breast cancer risk, high serum levels of phenylacetylglutamine is a potential early marker of kidney dysfunction in chronic kidney disease51. Glutamine, a precursor to phenylacetylglutamine, has been associated with breast cancer risk in a nested case-control study within the French SU.VI.MAX cohort (n = 211 cases). High levels of glutamine were associated with increased risk (OR per SD increase =1.33, 95% CI: 1.07–1.66) and this association persisted among the subgroup of premenopausal women (p for interaction = 0.003)52. Notably, elevated expression of asparagine synthetase correlates with lung metastasis in breast cancer patients53. Furthermore, in vitro breast cancer studies show asparagine bioavailability promotes invasion and metastasis and that this effect is prevented by limiting asparagine bioavailability53. In a nested case-control study within the EPIC cohort (n = 1624 cases), asparagine was inversely associated with breast cancer risk (OR = 0.87 per SD increase, 95% CI: 0.80–0.95, FDR p = 0.06), in contrast to the direction of association in our study18. However, the EPIC study participants were overwhelmingly (> 70%) postmenopausal at the time of blood collection, in contrast to our population with 80% premenopausal women. Differences in the menopausal status may partially explain the observed opposite directions of association between asparagine and breast cancer risk.
Creatine is obtained from meat consumption and synthesized endogenously from arginine, glycine, and methionine. Most creatine is found in skeletal muscle, and a significant amount is also found in the brain. Omnivores obtain roughly 50% of their daily creatine from meat and 50% is biosynthesized, while vegetarians biosynthesize most of their creatine54 and have significantly lower muscular creatine levels than meat eaters55. Creatine is broken down to creatinine in a first-order reaction, the rate of which decreases with age and decreased muscle mass56. Creatine/creatinine metabolism plays an important role in energy metabolism in skeletal muscle tissue, and thus disturbances in this pathway are associated with many muscle diseases, whether as a cause or consequence57. To the best of our knowledge, no previous work has reported a link between creatine and breast cancer risk. However, the association we observed in this analysis is consistent with the positive association between red meat and risk of breast cancer among premenopausal women in the NHSII cohort58.
Kynurenic acid and 2-aminohippuric acid are benzenoids inversely associated with breast cancer risk in our study. 2-amminohippuric acid is a glycine conjugate of anthranilic acid and can be synthesized in the liver59, but little is known about its biological function. Both kynurenic acid and 2-aminohippuric acid are part of the kynurenine branch of the tryptophan pathway, an essential amino acid and a precursor to many biologically active metabolites60,61. Studies of the function of kynurenic acid suggest pleiotropic roles in disease. On the one hand, kynurenic acid has been shown to have anti-inflammatory and anti-ulcerative properties in animal models, as well as antioxidative properties in vitro in human cells62. On the other hand, circulating kynurenic acid was associated with increased risk of insulin resistance63. Kynurenic acid acts as both an anti-inflammatory and immunosuppressive factor which in turn allows tumor proliferation64. Tryptophan metabolism plays an important role in tumor progression and malignancy with several cancers expressing tryptophan-degrading enzymes such as IDO165,66. However, the direct role of kynurenic acid remains unclear, with both proliferative and antiproliferative effects on human glioblastoma cells, an antiproliferative effect on human colon adenocarcinoma cells, and decreased DNA synthesis and inhibited migration in both cell types62. Neither kynurenic acid nor 2-aminohippuric acid have been previously associated with breast cancer risk.
N2,N2-dimethylguanosine (DMGU) is a purine nucleoside and a primary degradation product of transport RNA. Elevated circulating DMGU levels may indicate cellular stress and is associated with several diseases, including pulmonary arterial hypertension67, solid tumors68, incident type II diabetes69, and all-cause mortality70. Elevated levels of N2,N2-dimethylguanosine were found in patients with acute leukemia and breast cancer71. Elevated circulating levels of this metabolite are associated with lower risk of breast cancer in our study.
Importantly, there are several studies suggesting lifestyle and dietary changes may impact circulating levels of the here identified metabolites; however, the relationships are complex and warrant further study. For example, vegetables intake was positively associated with piperine (manuscript in preparation) and inversely associated with DMGV34 but both metabolites are inversely associated with risk. The Mediterranean diet was positively associated with C40:7 PE plasmalogen72 though this metabolite was positively associated with risk. Further, physical activity is inversely associated with creatine and positively associated with asparagine73 though we observed positive associations with breast cancer risk for both metabolites. This work supports the fact that these metabolites are potentially modifiable, however the complex relationships with breast cancer risk preclude clear recommendations and more investigation is required.
Our study has several strengths and limitations. Notably, we conducted a prospective analysis of amino acid and amino acid-related metabolomics and risk of breast cancer among a large number of predominantly premenopausal women. We had detailed information on sample collection characteristics and risk factors, which we included in our statistical approaches. Although metabolomics was measured at only one point in time, the identified metabolites are reasonably stable over time74 (ICCs or correlation over 1–2 years ≥0.75 for nine metabolites; no data are available for 2-aminohippuric acid and C20:1 LPC). Our cohort consisted of registered nurses, a group that are not representative of the general population (e.g., social economic status), however there is no evidence suggesting that breast carcinogenesis is different in this group of women. Additionally, our cohort also included predominantly Caucasian women. However, while the prevalence of risk factors often differs across population subgroups, many breast cancer risk factors have been documented to operate similarly across ethnic groups, as would be expected from a common underlying biology75–88. Nonetheless, it is crucial that future studies conduct similar analyses in racially and ethnically diverse cohorts. While we had reasonable power in most of our analyses, we had limited power among ER negative tumors. Lastly, the uniqueness of our data measured among predominantly premenopausal women make replication studies challenging. We conducted this analysis in a hypothesis generating framework and hope that other cohorts will follow and analyze metabolomics data stratifying by menopausal status.
In summary, we identified eleven metabolites associated with risk of breast cancer among premenopausal women. Seven metabolites remained associated after adjustment for established risk factors and were selected by at least one multivariable modeling approach: higher levels of 2-aminohippuric acid, kynurenic acid, piperine, DMGV and phenylacetylglutamine were associated with lower breast cancer risk while higher levels of creatine and C40:7 PE plasmalogen were associated with increased breast cancer risk. Additional prospective cohort studies are needed to assess these associations considering menopausal status. If these findings are validated, experimental studies are warranted to understand the underlying biological mechanisms driving changes in metabolite levels.
Methods
Study population
In 1989, 116,429 female registered nurses aged 25–42 y returned a mailed questionnaire and were enrolled in the NHSII. Participants have been followed biennially since 1989 with questionnaires collecting information on reproductive history, lifestyle factors, diet, medication use, and new disease diagnoses.
In 1996–1999, 29,611 NHSII participants aged 32–54 y contributed blood samples, as previously described89. Of these, 18,521 women who had not used oral contraceptives, been pregnant or breastfed in the previous 6 months provided samples timed within the menstrual cycle, targeting the early follicular (days 3–5 of the cycle) and mid-luteal (7–9 days prior to expected start of next cycle) phases. The remaining women donated a single untimed sample. Follicular plasma was separated and frozen by the participants and returned with the luteal sample; samples were collected and shipped overnight to our laboratory where we processed and archived aliquots of white blood cell, red blood cell, and plasma in liquid nitrogen freezers (≤−130 °C). Follow-up in the blood subcohort is high (96% in 2011).
The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. The return of the self-administered questionnaire and blood sample was considered to imply consent.
Case and control selection
Cases of breast cancer were identified after blood collection among women who had no reported cancer (other than nonmelanoma skin). Thousand and fifty-seven cases (invasive cases n = 780) were diagnosed between 1999 and 2011. Breast cancer cases were reported by the participant, which were confirmed by medical record reviews (n = 1015) or verbally by the nurse (n = 42). Given the high confirmation rate by medical record for breast cancer in this cohort (99%), all cases are included in this analysis.
One control was matched per case by the following factors: age (+/− 2 y), menopausal status and postmenopausal hormone therapy (HT) use at blood collection and diagnosis (premenopausal, postmenopausal and not taking HT, postmenopausal and taking HT, and unknown), and month (+/− 1 month), time of day (+/− 2 h), fasting status at blood collection (≤8h after a meal or unknown; >8h), race/ethnicity (African–American, Asian, Hispanic, Caucasian, other) and luteal day (+/− 1 day; timed samples only).
Covariate information
Data on breast cancer risk factors, including anthropometric measures, reproductive history, and lifestyle factors, were collected from questionnaires administered biennially and at the time of blood collections. Case characteristics, including invasive vs. in situ, histologic grade, estrogen and progesterone receptor (ER, PR), were extracted from pathology reports. As previously described90, immunohistochemical results for ER and PR, read manually by a study pathologist, were included for cases with available tumor tissue included in tissue microarrays.
Laboratory assay
Plasma metabolites were profiled at the Broad Institute of MIT and Harvard (Cambridge, MA) using a liquid chromatography tandem mass spectrometry (LC-MS) method designed to measure polar metabolites such as amino acids, amino acids derivatives, dipeptides, and other cationic metabolites as described previously33,74,91,92. Pooled plasma reference samples were included every 20 samples and results were standardized using the ratio of the value of the sample to the value of the nearest pooled reference multiplied by the median of all reference values for the metabolite. Samples were run together, with matched case-control pairs (as sets) distributed randomly within the batch, and the order of the case and controls within each pair randomly assigned. Therefore, the case and its control were always directly adjacent to each other in the analytic run, thereby limiting variability in platform performance across matched case–control pairs. In addition, 238 quality control (QC) samples, to which the laboratory was blinded, were also profiled. These were randomly distributed among the participants’ samples.
Hydrophilic interaction liquid chromatography (HILIC) analyses of water soluble metabolites in the positive ionization mode were conducted using an LC-MS system comprised of a Shimadzu Nexera X2 U-HPLC (Shimadzu Corp.; Marlborough, MA) coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific; Waltham, MA). Metabolites were extracted from plasma (10 µL) using 90 µL of acetonitrile/methanol/formic acid (74.9:24.9:0.2 v/v/v) containing stable isotope-labeled internal standards (valine-d8, Sigma-Aldrich; St. Louis, MO; and phenylalanine-d8, Cambridge Isotope Laboratories; Andover, MA). The samples were centrifuged (10 min, 9000 × g, 4 °C), and the supernatants were injected directly onto a 150 × 2 mm, 3 µm Atlantis HILIC column (Waters; Milford, MA). The column was eluted isocratically at a flow rate of 250 µL/min with 5% mobile phase A (10 mM ammonium formate and 0.1% formic acid in water) for 0.5 min followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 min. MS analyses were carried out using electrospray ionization in the positive ion mode using full scan analysis over 70–800 m/z at 70,000 resolution and 3 Hz data acquisition rate. Other MS settings were: sheath gas 40, sweep gas 2, spray voltage 3.5 kV, capillary temperature 350 °C, S-lens RF 40, heater temperature 300 °C, microscans 1, automatic gain control target 1e6, and maximum ion time 250 ms. Metabolite identities were confirmed using authentic reference standards or reference samples.
In total, 259 known metabolites were measured in this study. Metabolites not passing our previously conducted processing delay pilot study74 were excluded from this analysis (N = 33). All metabolites (N = 226) included here exhibited good reproducibility within person over 1-2 years74. Two hundred and six metabolites had no missing values among participant samples. One metabolite had <10% missing values and 19 metabolites had ≥10% missing values. Most of the metabolites (N = 191) had a coefficient of variation (CV) < 25% and an intraclass correlation coefficient (ICC) > 0.4 among blinded QC samples. Twenty-five metabolites had CV ≥ 25%, five had ICCs ≤ 0.4, and five metabolites had CV ≥ 25% and ICC ≤ 0.4.
Statistical analysis
Metabolite levels were natural logarithm transformed and standardized prior to statistical analysis. Missing values were imputed by one half the lowest observed value per metabolite, for metabolites with <10% missing values (N = 1). Metabolites with >10% missing values (N = 19) were excluded from the main analysis and evaluated in an exploratory analysis. Association of metabolite levels with breast cancer risk was assessed in metabolite-by-metabolite models and in multivariable analyses that included all metabolites simultaneously.
The association of individual metabolites with breast cancer risk was assessed in conditional logistic regression models (1057 cases and 1057 controls). In a simple model, each metabolite was included without adjustment for other factors. In an adjusted model, the following additional factors were included: BMI at age 18, weight change from age 18 to time of blood draw, age at menarche, parity and age at first birth, family history of breast cancer, diagnosis of benign breast disease, physical activity, alcohol consumption, exogenous hormone use and breastfeeding history. Odds ratios (OR) and 95% confidence intervals (95% CI) were estimated for a one-unit (one standard deviation) increase in the log-transformed and standardized metabolites levels.
We performed analyses restricting to premenopausal women at blood collection (838 cases and 838 controls), and analyses stratified by BMI (<25 kg/m2 [635 cases and 578 controls]; ≥25 kg/m2 [422 cases and 479 controls]) and ER status (ER+ [585 cases and 1057 controls]; ER− [126 cases and 1057 controls]). In a sensitivity analysis, we observed similar results between conditional logistic regression and unconditional logistic regression adjusting for the matching factors. Thus, stratified analyses were conducted using unconditional logistic regression, additionally adjusting for the matching factors. To test for effect modifications by BMI and heterogeneity by ER status, we included cross-product terms of the metabolite with potential effect modifier in conditional logistic models and report the p-value for that interaction. As ER status represents a case characteristic, we assigned each control the ER status of its matched case.
In an exploratory analysis we assessed the association with risk of breast cancer for the 19 metabolites with >10% missing values. We included the continuous metabolite level as well as a presence/absence indicator in the fully adjusted conditional logistic regression model and performed a likelihood-ratio test (full model compared to a model excluding both the metabolite and the presence/absence indicator) to estimate the significance level of the association.
Multivariable analyses evaluating the joint association of the 207 metabolites with breast cancer risk were based on (1) conditional logistic regression with lasso penalty (“Lasso”), (2) conditional logistic regression with an elastic net penalty (“Elastic Net”), and (3) random forests. In the Lasso and Elastic Net analyses, a minimally adjusted model included only the set of 207 metabolites, whereas a fully adjusted model further adjusted for the risk factors noted above. In each analysis, the optimal values of the regularization parameter(s) were estimated as that which minimizes the average deviance in the left-out partitions, in a 10-fold cross validation procedure. A p-value for each metabolite was obtained from a permutation test in which the case/control labels were permuted within each matched stratum. A p-value for each metabolite was calculated as the proportion of permutations (out of 250) in which the magnitude of the coefficient under label permutation was at least as large as the regression coefficient in the observed dataset. Analyses were carried out using the R library clogitL193.
Random forests analyses included a minimally adjusted model that included the 207 metabolites and matching factors. A fully adjusted model also included the additional risk factors noted above. In all analyses, the parameter mtry corresponding to the number of variables randomly sampled as candidates at each split was set to the square root of the total number of covariates in the model. Each classifier was an aggregate of 5000 trees. A p-value for each metabolite was obtained from a permutation test as described above in which the case/control labels were randomly permuted 100 times. Analyses were carried out using the R library random forests94.
To adjust for multiple testing in the conditional logistic regression and the multivariable models we estimated the positive FDR based on the q-value procedure95. Metabolites that satisfied a p-value less than 0.05 and corresponding q-value less than 0.20 in the minimally adjusted model were discussed as primary findings.
Criterion for statistical significance: metabolites that met a p-value < 0.05 and q-value < 0.20 in at least one of the four models (Conditional Logistic Regression, Lasso, Elastic Net, and Random Forests) with minimal adjustment were considered as statistically significant. An exception was made for metabolites that did not meet a p-value threshold < 0.05 in the conditional logistic regression: four metabolites that met the threshold for statistical significance in Lasso only but had high raw p-values (>0.3) in the conditional logistic regression were excluded (C12:1 carnitine, C22:5 LPC, C46:2 TAG, glycine).
A metabolite score was estimated for each participant as a linear combination of all metabolites that met the threshold for statistical significance. The coefficients associated with each metabolite were estimated in a conditional logistic regression model with the Lasso penalty that included all metabolites simultaneously and with full adjustment for all potential confounders.
Analyses were caried out using R96 version 3.6.3 and SAS version 9.4. All statistical tests were two-sided.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This study was funded by the National Cancer Institute (R01 CA050385, UM1 CA186107, P01 CA087969, R01 CA49449, U01 CA176726, and R01 CA67262). We would like to thank the participants and staff of the Nurses’ Health Studies for their valuable contributions as well as the following state cancer registries for their help: A.L., A.Z., A.R., C.A., C.O., C.T., D.E., F.L., G.A., I.D., I.L., I.N., I.A., K.Y., L.A., M.E., M.D., M.A., M.I., N.E., N.H., N.J., N.Y., N.C., N.D., O.H., O.K., O.R., P.A., R.I., S.C., T.N., T.X., V.A., W.A. and W.Y. We would also like to thank Ryan Sheehan for his help with Fig. 2. The authors assume full responsibility for analyses and interpretation of these data.
Author contributions
O.A.Z. and R.B. contributed equally to this study. O.A.Z. and R.B. contributed to data curation, formal analysis, investigation, methodology, manuscript writing, and manuscript review. Y.Z. contributed to formal analysis and manuscript review. L.F. contributed to resources and manuscript review. S.J., J.A.P. and C.B.C. contributed to data curation, resources and manuscript review. S.S.T. contributed to conceptualization, investigation, methodology and manuscript review. A.H.E. contributed to conceptualization, funding acquisition, investigation, methodology, project administration, resources, supervision, manuscript writing, and manuscript review.
Data availability
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1437434997. The demographics and questionnaire data together with the plasma metabolomics profiles are not publicly available for the following reason: data contain information that could compromise research participant privacy. Requests to access these data should be made via http://www.nurseshealthstudy.org/researchers. Analysis results for all metabolites are openly available as part of this figshare metadata record in the files titled “supplementary_tables 1–4.csv”.
Code availability
The code used in this analysis is available upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Oana A. Zeleznik, Raji Balasubramanian
Supplementary information
The online version contains supplementary material available at 10.1038/s41523-021-00262-4.
References
- 1.Harbeck N, et al. Breast cancer. Nat. Rev. Dis. Prim. 2019;5:1–31. doi: 10.1038/s41572-018-0051-2. [DOI] [PubMed] [Google Scholar]
- 2.Dartois L, et al. Proportion of premenopausal and postmenopausal breast cancers attributable to known risk factors: estimates from the E3N-EPIC cohort. Int. J. Cancer. 2016;138:2415–2427. doi: 10.1002/ijc.29987. [DOI] [PubMed] [Google Scholar]
- 3.Sprague BL, et al. Proportion of invasive breast cancer attributable to risk factors modifiable after menopause. Am. J. Epidemiol. 2008;168:404–411. doi: 10.1093/aje/kwn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamimi R, et al. Population attributable risk of modifiable and nonmodifiable breast cancer risk factors in postmenopausal breast cancer. Am. J. Epidemiol. 2016;184:884–893. doi: 10.1093/aje/kww145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maas P, et al. Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States. Jama Oncol. 2016;2:1295–1302. doi: 10.1001/jamaoncol.2016.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armitage EG, Barbas C. Metabolomics in cancer biomarker discovery: current trends and future perspectives. J. Pharm. Biomed. Anal. 2014;87:1–11. doi: 10.1016/j.jpba.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 7.McCartney A, et al. Metabolomics in breast cancer: a decade in review. Cancer Treat. Rev. 2018;67:88–96. doi: 10.1016/j.ctrv.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Jiao L, et al. A prospective targeted serum metabolomics study of pancreatic cancer in postmenopausal women. Cancer Prev. Res. 2019;12:237–246. doi: 10.1158/1940-6207.CAPR-18-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayers JR, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med. 2014;20:1193. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu X, et al. Prospective metabolomics study identifies potential novel blood metabolites associated with pancreatic cancer risk. Int. J. Cancer. 2018;143:2161–2167. doi: 10.1002/ijc.31574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, et al. Prospective serum metabolomic profiling of lethal prostate cancer. Int. J. Cancer. 2019;145:3231–3243. doi: 10.1002/ijc.32218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Jacobs EJ, Carter BD, Gapstur SM, Stevens VL. Plasma metabolomic profiles and risk of advanced and fatal prostate cancer. Eur. Urol. Oncol. 2019;4:56–65. doi: 10.1016/j.euo.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Loftfield E, et al. Prospective investigation of serum metabolites, coffee drinking, liver cancer incidence, and liver disease mortality. J. Natl Cancer Inst. 2019;112:286–294. doi: 10.1093/jnci/djz122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perttula K, et al. Untargeted lipidomic features associated with colorectal cancer in a prospective cohort. BMC Cancer. 2018;18:996. doi: 10.1186/s12885-018-4894-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeleznik OA, et al. Circulating lysophosphatidylcholines, phosphatidylcholines, ceramides, and sphingomyelins and ovarian cancer risk: a 23-year prospective study. J. Natl Cancer Inst. 2019;112:628–636. doi: 10.1093/jnci/djz195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeleznik OA, et al. A prospective analysis of circulating plasma metabolites associated with ovarian cancer risk. Cancer Res. 2020;80:1357–1367. doi: 10.1158/0008-5472.CAN-19-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Troisi J, et al. Metabolomic signature of endometrial cancer. J. Proteome Res. 2018;17:804–812. doi: 10.1021/acs.jproteome.7b00503. [DOI] [PubMed] [Google Scholar]
- 18.His M, et al. Prospective analysis of circulating metabolites and breast cancer in EPIC. BMC Med. 2019;17:178. doi: 10.1186/s12916-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kühn T, et al. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016;14:13. doi: 10.1186/s12916-016-0552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lécuyer L, et al. Diet-related metabolomic signature of long-term breast cancer risk using penalized regression: an exploratory study in the SU.VI.MAX cohort. Cancer Epidemiol. Biomark. Prev. 2019;29:396–405. doi: 10.1158/1055-9965.EPI-19-0900. [DOI] [PubMed] [Google Scholar]
- 21.Moore SC, et al. A metabolomics analysis of body mass index and postmenopausal breast cancer risk. J. Natl Cancer Inst. 2018;110:588–597. doi: 10.1093/jnci/djx244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo HJ, et al. Analysis of metabolites and metabolic pathways in breast cancer in a Korean prospective cohort: the Korean Cancer Prevention Study-II. Metabolomics. 2018;14:85. doi: 10.1007/s11306-018-1382-4. [DOI] [PubMed] [Google Scholar]
- 23.Playdon MC, et al. Nutritional metabolomics and breast cancer risk in a prospective study. Am. J. Clin. Nutr. 2017;106:637–649. doi: 10.3945/ajcn.116.150912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derosa, G., Maffioli, P. & Sahebkar, A. In Anti-inflammatory Nutraceuticals and Chronic Diseases 173–184 (Springer, 2016).
- 25.Meghwal M, Goswami T. Piper nigrum and piperine: an update. Phytother. Res. 2013;27:1121–1130. doi: 10.1002/ptr.4972. [DOI] [PubMed] [Google Scholar]
- 26.Yadav, V., Krishnan, A. & Vohora, D. A systematic review on Piper longum L.: Bridging traditional knowledge and pharmacological evidence for future translational research. J. Ethnopharmacol. 247, 112255 (2019). [DOI] [PubMed]
- 27.Zadorozhna, M., Tataranni, T. & Mangieri, D. Piperine: role in prevention and progression of cancer. Mol. Biol. Rep. 46, 1–13 (2019). [DOI] [PubMed]
- 28.Aumeeruddy MZ, Mahomoodally MF. Combating breast cancer using combination therapy with 3 phytochemicals: piperine, sulforaphane, and thymoquinone. Cancer. 2019;125:1600–1611. doi: 10.1002/cncr.32022. [DOI] [PubMed] [Google Scholar]
- 29.Greenshields AL, et al. Piperine inhibits the growth and motility of triple-negative breast cancer cells. Cancer Lett. 2015;357:129–140. doi: 10.1016/j.canlet.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 30.Abdelhamed S, et al. Piperine enhances the efficacy of TRAIL-based therapy for triple-negative breast cancer cells. Anticancer Res. 2014;34:1893–1899. [PubMed] [Google Scholar]
- 31.Rodionov RN, Murry DJ, Vaulman SF, Stevens JW, Lentz SR. Human alanine-glyoxylate aminotransferase 2 lowers asymmetric dimethylarginine and protects from inhibition of nitric oxide production. J. Biol. Chem. 2010;285:5385–5391. doi: 10.1074/jbc.M109.091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willeit, P. et al. Asymmetric dimethylarginine and cardiovascular risk: systematic review and meta‐analysis of 22 prospective studies. J. Am. Heart Assoc.4, e001833 (2015). [DOI] [PMC free article] [PubMed]
- 33.O’Sullivan JF, et al. Dimethylguanidino valeric acid is a marker of liver fat and predicts diabetes. J. Clin. Investig. 2017;127:4394–4402. doi: 10.1172/JCI95995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ottosson F, et al. Dimethylguanidino valerate: a lifestyle‐related metabolite associated with future coronary artery disease and cardiovascular mortality. J. Am. Heart Assoc. 2019;8:e012846. doi: 10.1161/JAHA.119.012846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robbins JM, et al. Association of dimethylguanidino valeric acid with partial resistance to metabolic health benefits of regular exercise. JAMA Cardiol. 2019;4:636–643. doi: 10.1001/jamacardio.2019.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernández‐Alonso P, et al. Plasma metabolites associated with frequent red wine consumption: a metabolomics approach within the PREDIMED Study. Mol. Nutr. Food Res. 2019;63:1900140. doi: 10.1002/mnfr.201900140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoemaker MJ, et al. Association of body mass index and age with subsequent breast cancer risk in premenopausal women. JAMA Oncol. 2018;4:e181771–e181771. doi: 10.1001/jamaoncol.2018.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita Y, et al. Differences in elongation of very long chain fatty acids and fatty acid metabolism between triple-negative and hormone receptor-positive breast cancer. BMC Cancer. 2017;17:589. doi: 10.1186/s12885-017-3554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Messias MCF, Mecatti GC, Priolli DG, de Oliveira Carvalho P. Plasmalogen lipids: functional mechanism and their involvement in gastrointestinal cancer. Lipids Health Dis. 2018;17:41. doi: 10.1186/s12944-018-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchie SA, et al. Metabolic system alterations in pancreatic cancer patient serum: potential for early detection. BMC Cancer. 2013;13:416. doi: 10.1186/1471-2407-13-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lv J, Lv C-Q, Xu L, Yang H. Plasma content variation and correlation of plasmalogen and GIS, TC, and TPL in gastric carcinoma patients: a comparative study. Med Sci. Monit. Basic Res. 2015;21:157–160. doi: 10.12659/MSMBR.893908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hilvo M, et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71:3236–3245. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- 43.Law, S.-H. et al. An updated review of lysophosphatidylcholine metabolism in human diseases. Int. J. Mol. Sci.20, 1149 (2019). [DOI] [PMC free article] [PubMed]
- 44.Libert, D. M., Nowacki, A. S. & Natowicz, M. R. Metabolomic analysis of obesity, metabolic syndrome, and type 2 diabetes: amino acid and acylcarnitine levels change along a spectrum of metabolic wellness. PeerJ6, e5410 (2018). [DOI] [PMC free article] [PubMed]
- 45.PubChem Database. Vol. Phenylacetylglutamine, CID=92258 (National Center for Biotechnology Information) https://pubchem.ncbi.nlm.nih.gov/compound/Phenylacetylglutamine Accessed 27 March 2020.
- 46.Lees HJ, Swann JR, Wilson ID, Nicholson JK, Holmes E. Hippurate: the natural history of a mammalian–microbial cometabolite. J. Proteome Res. 2013;12:1527–1546. doi: 10.1021/pr300900b. [DOI] [PubMed] [Google Scholar]
- 47.Li M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl Acad. Sci. USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pallister, T. et al. Hippurate as a metabolomic marker of gut microbiome diversity: Modulation by diet and relationship to metabolic syndrome. Sci. Rep.7, 1–9 (2017). [DOI] [PMC free article] [PubMed]
- 49.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl Acad. Sci. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Behr C, et al. Gut microbiome-related metabolic changes in plasma of antibiotic-treated rats. Arch. Toxicol. 2017;91:3439–3454. doi: 10.1007/s00204-017-1949-2. [DOI] [PubMed] [Google Scholar]
- 51.Barrios, C. et al. Gut-microbiota-metabolite axis in early renal function decline. PLoS ONE10, e034311 (2015). [DOI] [PMC free article] [PubMed]
- 52.Lécuyer L, et al. Plasma metabolomic signatures associated with long-term breast cancer risk in the SU. VI. MAX prospective cohort. Cancer Epidemiol. Prev. Biomark. 2019;28:1300–1307. doi: 10.1158/1055-9965.EPI-19-0154. [DOI] [PubMed] [Google Scholar]
- 53.Knott SR, et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature. 2018;554:378–381. doi: 10.1038/nature25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riesberg LA, Weed SA, McDonald TL, Eckerson JM, Drescher KM. Beyond muscles: the untapped potential of creatine. Int. Immunopharmacol. 2016;37:31–42. doi: 10.1016/j.intimp.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delanghe J, et al. Normal reference values for creatine, creatinine, and carnitine are lower in vegetarians. Clin. Chem. 1989;35:1802–1803. doi: 10.1093/clinchem/35.8.1802. [DOI] [PubMed] [Google Scholar]
- 56.Brosnan JT, Brosnan ME. Creatine metabolism and the urea cycle. Mol. Genet. Metab. 2010;100:S49–S52. doi: 10.1016/j.ymgme.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 57.Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol. Rev. 2000;80:1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- 58.Farvid MS, Cho E, Chen WY, Eliassen AH, Willett WC. Dietary protein sources in early adulthood and breast cancer incidence: prospective cohort study. BMJ. 2014;348:g3437. doi: 10.1136/bmj.g3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naito J, Sasaki E, Ohta Y, Shinohara R, Ishiguro I. Anthranilic acid metabolism in the isolated perfused rat liver: detection and determination of anthranilic acid and its related substances using high-performance liquid chromatography with electrochemical detection. Biochem. Pharmacol. 1984;33:3195–3200. doi: 10.1016/0006-2952(84)90076-5. [DOI] [PubMed] [Google Scholar]
- 60.Badawy, A. A. B. Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int. J. Tryptophan Res.10, 1178646917691938 (2017). [DOI] [PMC free article] [PubMed]
- 61.Richard DM, et al. L-Tryptophan: basic metabolic functions, behavioral research and therapeutic indications. Int J. Tryptophan Res. 2009;2:45–60. doi: 10.4137/IJTR.S2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walczak, K., Wnorowski, A., Turski, W. A. & Plech, T. Kynurenic acid and cancer: facts and controversies. Cell. Mol. Life Sci. 77, 1531–1550 (2019). [DOI] [PMC free article] [PubMed]
- 63.Yu E, et al. Association of tryptophan metabolites with incident type 2 diabetes in the PREDIMED trial: a case–cohort study. Clin. Chem. 2018;64:1211–1220. doi: 10.1373/clinchem.2018.288720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wirthgen E, Hoeflich A, Rebl A, Günther J. Kynurenic acid: the Janus-faced role of an immunomodulatory tryptophan metabolite and its link to pathological conditions. Front. Immunol. 2018;8:1957. doi: 10.3389/fimmu.2017.01957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019;18:379–401. doi: 10.1038/s41573-019-0016-5. [DOI] [PubMed] [Google Scholar]
- 66.Labadie BW, Bao R, Luke JJ. Reimagining IDO pathway inhibition in cancer immunotherapy via downstream focus on the Tryptophan–Kynurenine–Aryl hydrocarbon axis. Clin. Cancer Res. 2019;25:1462–1471. doi: 10.1158/1078-0432.CCR-18-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhodes CJ, et al. Plasma metabolomics implicates modified transfer RNAs and altered bioenergetics in the outcomes of pulmonary arterial hypertension. Circulation. 2017;135:460–475. doi: 10.1161/CIRCULATIONAHA.116.024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seidel A, Brunner S, Seidel P, Fritz GI, Herbarth O. Modified nucleosides: an accurate tumour marker for clinical diagnosis of cancer, early detection and therapy control. Br. J. Cancer. 2006;94:1726–1733. doi: 10.1038/sj.bjc.6603164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ottosson F, Smith E, Gallo W, Fernandez C, Melander O. Purine metabolites and carnitine biosynthesis intermediates are biomarkers for incident type 2 diabetes. J. Clin. Endocrinol. Metab. 2019;104:4921–4930. doi: 10.1210/jc.2019-00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Balasubramanian, R. et al. Metabolomic profiles associated with all-cause mortality in the Women’s Health Initiative. Int. J. Epidemiol.49, dyz211 (2019). [DOI] [PMC free article] [PubMed]
- 71.Levine L, Waalkes TP, Stolbach L. Brief communication: serum levels of N 2, N 2-dimethylguanosine and pseudouridine as determined by radioimmunoassay for patients with malignancy. J. Natl Cancer Inst. 1975;54:341–343. [PubMed] [Google Scholar]
- 72.Li, J. et al. The Mediterranean diet, plasma metabolome, and cardiovascular disease risk. Eur. Heart J.41, 2645–2656 (2020). [DOI] [PMC free article] [PubMed]
- 73.Ding M, et al. Metabolome-Wide Association Study of the relationship between habitual physical activity and plasma metabolite levels. Am. J. Epidemiol. 2019;188:1932–1943. doi: 10.1093/aje/kwz171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Townsend MK, et al. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin. Chem. 2013;59:1657–1667. doi: 10.1373/clinchem.2012.199133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eliassen AH, Hankinson SE, Rosner B, Holmes MD, Willett WC. Physical activity and risk of breast cancer among postmenopausal women. Arch. Intern. Med. 2010;170:1758–1764. doi: 10.1001/archinternmed.2010.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adams-Campbell LL, Rosenberg L, Rao RS, Palmer JR. Strenuous physical activity and breast cancer risk in African-American women. J. Natl Med. Assoc. 2001;93:267–275. [PMC free article] [PubMed] [Google Scholar]
- 77.Colditz GA, et al. Family history, age, and risk of breast cancer. J. Am. Med. Assoc. 1993;270:338–343. doi: 10.1001/jama.1993.03510030062035. [DOI] [PubMed] [Google Scholar]
- 78.Palmer JR, Boggs DA, Adams-Campbell LL, Rosenberg L. Family history of cancer and risk of breast cancer in the Black Women’s Health Study. Cancer Causes Control. 2009;20:1733–1737. doi: 10.1007/s10552-009-9425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kilfoy BA, et al. Family history of malignancies and risk of breast cancer: prospective data from the Shanghai women’s health study. Cancer Causes Control. 2008;19:1139–1145. doi: 10.1007/s10552-008-9181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hunter DJ, et al. Oral contraceptive use and breast cancer: a prospective study of young women. Cancer Epidemiol. Biomark. Prev. 2010;19:2496–2502. doi: 10.1158/1055-9965.EPI-10-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rosenberg L, Palmer JR, Wise LA, Adams-Campbell LL. A prospective study of female hormone use and breast cancer among black women. Arch. Intern. Med. 2006;166:760–765. doi: 10.1001/archinte.166.7.760. [DOI] [PubMed] [Google Scholar]
- 82.Colditz GA, et al. The use of estrogens and progestins and the risk of breast cancer in postmenopausal women. N. Engl. J. Med. 1995;332:1589–1593. doi: 10.1056/NEJM199506153322401. [DOI] [PubMed] [Google Scholar]
- 83.Rosenberg L, Boggs DA, Wise LA, Adams-Campbell LL, Palmer JR. Oral contraceptive use and estrogen/progesterone receptor-negative breast cancer among African American women. Cancer Epidemiol. Biomark. Prev. 2010;19:2073–2079. doi: 10.1158/1055-9965.EPI-10-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fung, T. T. et al. Dietary patterns and the risk of postmenopausal breast cancer. Int. J. Cancer116, 116–121 (2005). [DOI] [PubMed]
- 85.Boggs DA, et al. Fruit and vegetable intake in relation to risk of breast cancer in the Black Women’s Health Study. Am. J. Epidemiol. 2010;172:1268–1279. doi: 10.1093/aje/kwq293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Agurs-Collins T, Rosenberg L, Makambi K, Palmer JR, Adams-Campbell L. Dietary patterns and breast cancer risk in women participating in the Black Women’s Health Study. Am. J. Clin. Nutr. 2009;90:621–628. doi: 10.3945/ajcn.2009.27666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boggs DA, et al. Tea and coffee intake in relation to risk of breast cancer in the Black Women’s Health Study. Cancer Causes Control. 2010;21:1941–1948. doi: 10.1007/s10552-010-9622-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ganmaa D, et al. Coffee, tea, caffeine and risk of breast cancer: a 22-year follow-up. Int J. Cancer. 2008;122:2071–2076. doi: 10.1002/ijc.23336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eliassen AH, et al. Endogenous steroid hormone concentrations and risk of breast cancer among premenopausal women. J. Natl Cancer Inst. 2006;98:1406–1415. doi: 10.1093/jnci/djj376. [DOI] [PubMed] [Google Scholar]
- 90.Fortner RT, et al. Parity, breastfeeding, and breast cancer risk by hormone receptor status and molecular phenotype: results from the Nurses’ Health Studies. Breast Cancer Res. 2019;21:40. doi: 10.1186/s13058-019-1119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mascanfroni ID, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat. Med. 2015;21:638. doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paynter NP, et al. Metabolic predictors of incident coronary heart disease in women. Circulation. 2018;137:841–853. doi: 10.1161/CIRCULATIONAHA.117.029468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Avalos M, Pouyes H, Grandvalet Y, Orriols L, Lagarde E. Sparse conditional logistic regression for analyzing large-scale matched data from epidemiological studies: a simple algorithm. BMC Bioinform. 2015;16:S1. doi: 10.1186/1471-2105-16-S6-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 95.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 2003;31:2013–2035. doi: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- 96.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (2020) https://www.R-project.org/.
- 97.Zeleznik, O. A. et al. Metadata record for the article: Circulating amino acids and amino acid-related metabolites and risk of breast cancer among predominantly premenopausal women. (figshare 10.6084/m9.figshare.14374349, 2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1437434997. The demographics and questionnaire data together with the plasma metabolomics profiles are not publicly available for the following reason: data contain information that could compromise research participant privacy. Requests to access these data should be made via http://www.nurseshealthstudy.org/researchers. Analysis results for all metabolites are openly available as part of this figshare metadata record in the files titled “supplementary_tables 1–4.csv”.
The code used in this analysis is available upon request.