Abstract
Antimicrobial resistance seriously threatened human health, and new antimicrobial agents are desperately needed. As one of the largest classes of plant secondary metabolite, flavonoids can be widely found in various parts of the plant, and their antibacterial activities have been increasingly paid attention to. Based on the physicochemical parameters and antibacterial activities of sixty-six flavonoids reported, two regression equations between their ACD/LogP or LogD7.40 and their minimum inhibitory concentrations (MICs) to gram-positive bacteria were established with the correlation coefficients above 0.93, and then were verified by another sixty-eight flavonoids reported. From these two equations, the MICs of most flavonoids against gram-positive bacteria could be roughly calculated from their ACD/LogP or LogD7.40, and the minimum MIC was predicted as approximately 10.2 or 4.8 μM, more likely falls into the range from 2.6 to 10.2 μM, or from 1.2 to 4.8 μM. Simultaneously, both tendentiously concave regression curves indicated that the lipophilicity is a key factor for flavonoids against gram-positive bacteria. Combined with the literature analyses, the results also suggested that the cell membrane is the main site of flavonoids acting on gram-positive bacteria, and which likely involves the damage of phospholipid bilayers, the inhibition of the respiratory chain or the ATP synthesis, or some others.
Subject terms: Secondary metabolism, Natural products, Antimicrobials, Natural products
Introduction
Antimicrobial resistance (AMR) has been seriously threatened human public health and global economic development, and new antimicrobial agents are desperately needed1,2. Antibiotics, as the secondary metabolites produced by many bacteria, actinomycetes and fungi, showed remarkably antimicrobial activities, while they also bring some toxic side effects to human body, and are unavoidable to lead to the resistance3. Many plant ingredients present weaker antimicrobial activities, while some of them can reverse the resistance of antimicrobial agents4. Simultaneously, most of them are considered nontoxic to human body because of their ubiquity in all sorts of plant derived foods and beverages.
As one of the largest classes of plant secondary metabolite, flavonoids can be widely found in various parts of the plants, such as fruit, vegetables, nuts and tea4. These compounds have a wide range of pharmacological activities including antibiosis, antioxidation, and coronary heart disease prevention, etc. It is worth noting that some flavonoids can enhance the sensitivity of bacteria to antibiotics, and even reverse the AMR4,5. Thereout, the antibacterial activities of flavonoids have been paid more and more attention to. Recently, several investigations were performed for the antimicrobial activities of flavonoids, and the probable relationships between their chemical structures and antimicrobial activities were also summarized4–6. However, the regularity conclusions on the structure–activity relationships of flavonoids against bacteria still need to be further explored.
During our researches on antimicrobial agents7–9, it is vaguely found that the antimicrobial activities of flavonoids are not related to their special structure, while may be related to their polarities or lipid-water partition coefficients. Many data of plant flavonoids, involving their chemical structures and antibacterial activities reported in previous papers, were searched and analyzed for proving it. The inhibitory activities of plant flavonoids against gram-positive bacteria especially Staphylococcus aureus can be widely searched, while those against gram-negative ones and fungi were reported too few to carry out statistical analyses4,6. Thereby, the former was our focus in this research. As the inhibitory activities of a compound against different pathogenic bacteria are varied, this paper will pay more attention to the inhibitory activities of these flavonoids against Staphylococcus aureus, a species most reported in the literature.
Results
Structure, antibacterial activity, and physicochemical parameter
Sixty-six flavonoids (1 to 66) shown on Figs. 1, 2, 3, 4, 5 and 6, reported in six papers10–15, were selected for the preliminary structure-physicochemical parameter-activity analyses of plant flavonoids against gram-positive bacteria, especially Staphylococcus aureus. These flavonoids include three subclasses as flavonols, dihydroflavones and dihydroflavonols. Regression analyses indicated that no universal correlation between the antimicrobial activity (expressed as minimum inhibitory concentration, MIC) and the physicochemical parameter Gibbs energy, LogP (Partition coefficient), MR (Molar Refractivity), CMR (Calculated Molar Refractivity), tPSA (Topological Polar Surface Area), or solubility (SolDB) could be established for these flavonoids. However, probable correlations between the antimicrobial activities (MIC, or MIC90 which expressed as the MIC of a compound to 90% test isolates of a specific pathogen) and the physicochemical parameter CLogP (Calculated Partition coefficient), ACD/LogP, or LogD7.40 (Log10 of distribution coefficient at pH 7.40) were respectively discovered, and the physicochemical parameters and antimicrobial activities of these compounds were listed in Tables 1, 2, 3, 4, 5 and 6 for further analyses10–15.
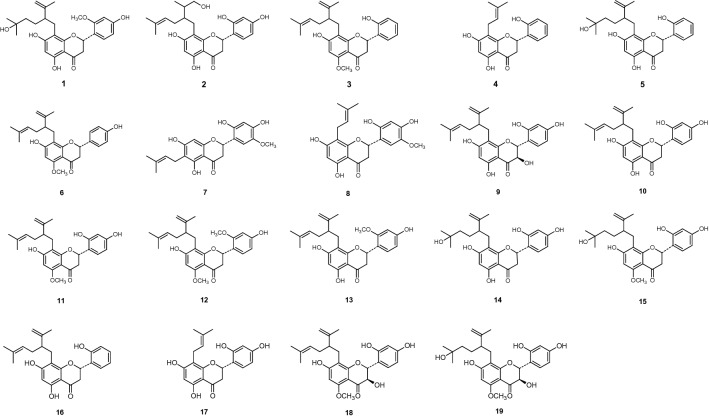
Figure 1.
Chemical structures of compounds 1 to 1910.
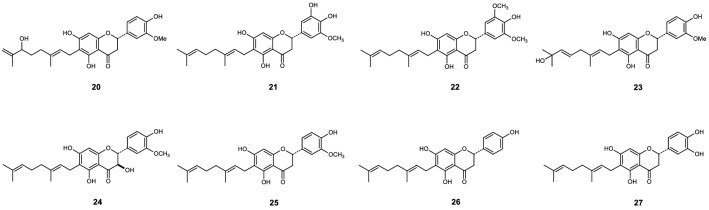
Figure 2.
Chemical structures of compounds 20 to 2711.
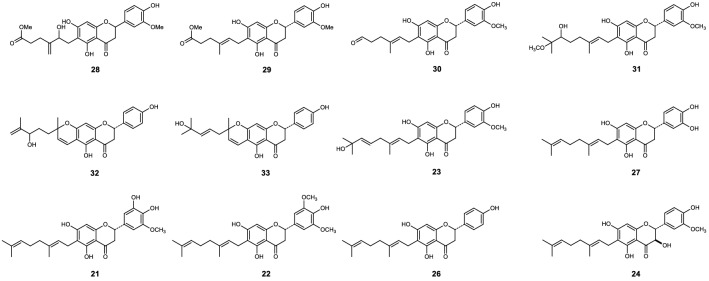
Figure 3.
Chemical structures of compounds 21 to 24, and 26 to 3312.
Figure 4.
Chemical structures of compounds 34 to 4413.
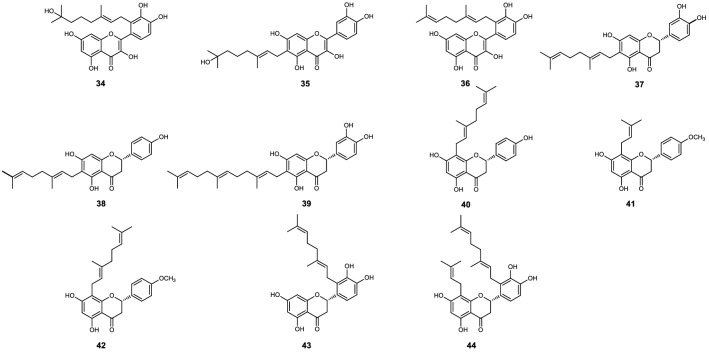
Figure 5.
Chemical structures of compounds 17, and 45 to 5414.
Figure 6.
Chemical structures of compounds 55 to 6615.
Table 1.
Physico-chemical parameters and antimicrobial activities of compounds 1 to 1910.
| Compounds | CLogPa | ACD/LogPb | LogD7.40b | MIC (μM)c | |
|---|---|---|---|---|---|
| S. aureus | B. subtilis | ||||
| 1 | 4.67 | 5.55 | 5.38 | L | L |
| 2 | 4.08 | 5.09 | 4.92 | 11.3 | 11.3 |
| 3 | 6.31 | 7.02 | 6.80 | 11.8 | 5.9 |
| 4 | 4.35 | 5.29 | 5.09 | 14.7 | 14.7 |
| 5 | 4.52 | 5.52 | 5.35 | – | – |
| 6 | 6.36 | 7.02 | 6.81 | 23.7 | 23.7 |
| 7 | 3.53 | 4.18 | 4.09 | 25.9 | 25.9 |
| 8 | 3.58 | 4.18 | 3.98 | 25.9 | 25.9 |
| 9 | 4.50 | 5.74 | 5.50 | 22.7 | 22.7 |
| 10 | 5.58 | 6.52 | 6.33 | 5.9 | 5.9 |
| 11 | 5.64 | 6.30 | 6.08 | 5.7 | 5.7 |
| 12 | 6.46 | 7.05 | 6.83 | 5.5 | 5.5 |
| 13 | 6.40 | 7.27 | 7.09 | 5.7 | 5.7 |
| 14 | 3.86 | 4.80 | 4.63 | L | L |
| 15 | 3.92 | 4.58 | 4.37 | L | L |
| 16 | 6.25 | 7.24 | 7.06 | 12.2 | 6.1 |
| 17 | 3.68 | 4.56 | 4.37 | 14.0 | 7.0 |
| 18 | 4.57 | 5.53 | 5.26 | – | – |
| 19 | 2.84 | 3.81 | 3.56 | – | – |
aThe CLogP values were calculated using software ChemBioDraw Ultra 12.0.
bThe ACD/Log P and LogD7.40 values were calculated using software ACD/Labs 6.0.
cMIC, minimum inhibitory concentration; S. aureus, Staphylococcus aureus; B. subtilis, Bacillus subtilis; L, lower activity than other compounds while no data was given; –, no data was given.
Table 2.
Physico-chemical parameters and antimicrobial activities of compounds 20 to 2711.
| Compounds | CLogPa | ACD/LogPb | LogD7.40b | MIC (μM)c | |
|---|---|---|---|---|---|
| S. aureus | S. epidermidis | ||||
| 20 | 4.22 | 5.56 | 5.34 | 35.2 | 70.4 |
| 21 | 5.68 | 6.54 | 6.32 | 4.4 | 8.8 |
| 22 | 6.01 | 6.61 | 6.39 | 8.5 | 8.5 |
| 23 | 4.22 | 5.18 | 4.96 | 140.8d | 140.8d |
| 24 | 5.15 | 6.25 | 5.97 | 4.4 | 4.4 |
| 25 | 6.23 | 7.02 | 6.81 | 18.2 | 9.1 |
| 26 | 6.38 | 7.32 | 7.12 | 19.6 | 9.8 |
| 27 | 5.78 | 6.72 | 6.51 | 9.4 | 9.4 |
aThe CLogP values were calculated using software ChemBioDraw Ultra 12.0.
bThe ACD/Log P and LogD7.40 values were calculated using software ACD/Labs 6.0.
cMIC, minimum inhibitory concentration; S. aureus, Staphylococcus aureus ATCC 25923; S. epidermidis, Staphylococcus epidermidis ATCC 12228.
dBoth MICs of compound 23 against S. aureus ATCC 25923 and S. epidermidis ATCC 12228 were more than 32 μg/mL (70.4 μM). As microdilution broth method was used to test MIC, we set 64 μg/mL (140.8 μM) as their MICs.
Table 3.
Physico-chemical parameters and antimicrobial activities of compounds 21 to 24, and 26 to 3312.
| Compounds | CLogPa | ACD/LogPb | LogD7.40b | MIC (μM)c | ||
|---|---|---|---|---|---|---|
| MRSA 6975 | MRSA 630 | MRSA 6205 | ||||
| 28 | 1.83 | 3.27 | 3.04 | 140.2 | 280.4d | 280.4d |
| 29 | 3.68 | 4.60 | 4.38 | 36.2 | 144.6 | 72.3 |
| 30 | 3.28 | 4.27 | 4.05 | 19.4 | 155.2 | 77.6 |
| 31 | 3.91 | 4.67 | 4.46 | 32.9 | 263.2 | 263.2 |
| 32 | 4.47 | 6.10 | 5.76 | 18.9 | 151.5 | 151.5 |
| 33 | 4.47 | 5.63 | 5.29 | 37.9 | 151.5 | 151.5 |
| 23 | 4.22 | 5.18 | 4.96 | 17.6 | 35.2 | 35.2 |
| 27 | 5.78 | 6.72 | 6.51 | 18.8 | 37.7 | 37.7 |
| 21 | 5.68 | 6.54 | 6.32 | 17.6 | 8.8 | 8.8 |
| 22 | 6.01 | 6.61 | 6.39 | 17.1 | 17.1 | 8.5 |
| 26 | 6.38 | 7.32 | 7.12 | 4.9 | 9.8 | 4.9 |
| 24 | 5.15 | 6.25 | 5.97 | 8.8 | 8.8 | 17.6 |
aThe CLogP values were calculated using software ChemBioDraw Ultra 12.0.
bThe ACD/Log P and LogD7.40 values were calculated using software ACD/Labs 6.0.
cMIC, minimum inhibitory concentration; MRSA 6975, methicillin-resistant Staphylococcus aureus 6975; MRSA 630, methicillin-resistant Staphylococcus aureus 630; MRSA 6205, methicillin-resistant Staphylococcus aureus 6205.
dThe MICs of compound 28 against MRSA 630 and 6205 were more than 64 μg/mL (140.2 μM). As microdilution broth method was used to test MIC and the three physico-chemical parameters were small, we set 128 μg/mL (280.4 μM) as their MICs.
Table 4.
Physico-chemical parameters and antimicrobial activities of compounds 34 to 4413.
| Compounds | CLogPa | ACD/LogPb | LogD7.40b | MIC (μM)c | |
|---|---|---|---|---|---|
| S. aureus | B. subtilis | ||||
| 34 | 3.46 | 4.52 | 3.84 | 140.2 | 140.2 |
| 35 | 3.71 | 4.52 | 3.93 | 140.2 | 140.2 |
| 36 | 5.18 | 6.20 | 5.53 | 73.0 | 73.0 |
| 37 | 5.78 | 6.72 | 6.51 | 9.5 | 9.5 |
| 38 | 6.38 | 7.32 | 7.12 | 19.7 | 9.8 |
| 39 | 7.81 | 8.75 | 8.54 | 32.6 | 16.3 |
| 40 | 6.43 | 7.32 | 7.13 | 9.8 | 39.4 |
| 41 | 4.98 | 5.94 | 5.75 | 90.8 | 90.8 |
| 42 | 7.01 | 7.97 | 7.78 | 19.0 | 19.0 |
| 43 | 5.78 | 6.74 | 6.5 | 37.9 | 37.9 |
| 44 | 7.73 | 8.84 | 8.64 | 8.2 | 16.3 |
aThe CLogP values were calculated using software ChemBioDraw Ultra 12.0.
bThe ACD/Log P and LogD7.40 values were calculated using software ACD/Labs 6.0.
cMIC, minimum inhibitory concentration; S. aureus, Staphylococcus aureus 209P; B. subtilis, Bacillus subtilis NBRC 3134.
Table 5.
Physico-chemical parameters and antimicrobial activities of compounds 17, and 45 to 5414.
| Compounds | CLogPa | ACD/LogPb | LogD7.40b | MIC90 (μM)c | |
|---|---|---|---|---|---|
| MRSA (22) | MSSA (7) | ||||
| 45 | 2.55 | 3.79 | 3.67 | 167.8 | 335.7 |
| 46 | 2.60 | 3.79 | 3.53 | 167.8 | 167.8 |
| 47 | 2.65 | 3.92 | 3.59 | 42.1 | 42.1 |
| 48 | 3.42 | 4.67 | 4.35 | 81.4 | 40.6 |
| 49 | 2.65 | 4.11 | 3.67 | 84.5 | 84.5 |
| 17 | 3.68 | 4.56 | 4.37 | 175.4 | 350.8 |
| 50 | 4.40 | 5.29 | 5.10 | 183.6 | 183.6 |
| 51 | 2.44 | 3.19 | 2.96 | > 918.3 | > 918.3 |
| 52 | 3.32 | 4.51 | 4.27 | 87.8 | 87.8 |
| 53 | 1.37 | 2.42 | 2.11 | 1734.6d | 1734.6d |
| 54 | 3.37 | 4.64 | 4.34 | 88.3 | 88.3 |
aThe CLogP values were calculated using software ChemBioDraw Ultra 12.0.
bThe ACD/Log P and LogD7.40 values were calculated using software ACD/Labs 6.0.
cMIC90, minimum inhibitory concentration to 90% test isolates; MRSA (22), twenty-two isolates of methicillin-resistant Staphylococcus aureus; MSSA (7), seven isolates of methicillin-susceptible Staphylococcus aureus.
dBoth MIC90s of compound 53 against MRSA and MSSA were more than 250 μg/mL (867.3 μM). As microdilution broth method was used to test MIC, we set 500 μg/mL (1734.6 μM) as their MIC90s.
Table 6.
Physico-chemical parameters and antimicrobial activities of compounds 55 to 6615.
| Compounds | CLogPa | ACD/LogPb | LogD7.40b | MIC (μM)c | |
|---|---|---|---|---|---|
| MRSA G31 | MRSA G47 | ||||
| 55 | 5.58 | 6.52 | 6.33 | 14.7 | 7.4 |
| 56 | 7.58 | 8.76 | 8.70 | 12.0 | 6.0 |
| 57 | 3.78 | 4.72 | 4.51 | 16.2 | 32.3 |
| 58 | 5.53 | 6.52 | 6.33 | 14.7 | 14.7 |
| 59 | 4.94 | 5.89 | 5.67 | 28.4 | 7.1 |
| 60 | 4.99 | 5.89 | 5.68 | 28.4 | 14.2 |
| 61 | 5.71 | 6.60 | 6.35 | 29.4 | 14.7 |
| 62 | 4.86 | 5.81 | 5.62 | 28.4 | 28.4 |
| 63 | 4.81 | 5.81 | 5.62 | 28.4 | 28.4 |
| 64 | 3.68 | 4.56 | 4.37 | 35.1 | 35.1 |
| 65 | 6.25 | 7.24 | 7.06 | 122.4d | 122.4d |
| 66 | 2.44 | 3.19 | 2.96 | 1469.2 | 734.6 |
aThe CLogP values were calculated using software ChemBioDraw Ultra 12.0.
bThe ACD/Log P and LogD7.40 values were calculated using software ACD/Labs 6.0.
cMIC, minimum inhibitory concentration; MRSA G31 and G47, methicillin-resistant Staphylococcus aureus G31 and G47.
dBoth MICs of compound 65 against MRSA G31 and G47 were more than 25 μg/mL (61.2 μM). As microdilution broth method was used to test MIC, we set 50 μg/mL (122.4 μM) as their MICs.
Data analysis and correlation establishment
The regression analyses for the physicochemical parameters CLogP, ACD/LogP, or LogD7.40 and the antimicrobial activities (MIC or MIC90) of these flavonoids to a certain pathogenic bacterium were respectively performed, and their regression curves were showed on Fig. S1 to S6 in Supplementary Information. From these figures, nearly all regression curves indicate that the antibacterial activities of these flavonoids present similar change characteristics along with the increase of their LogP or LogD7.40. First, the antibacterial activities will dramatically increase when the LogP or LogD7.40 increase up to a specific value. Along with the further increase of LogP or LogD7.40, the antibacterial activities will first increase tendentiously and then decrease. Simultaneously, their regression equations between the physicochemical parameter (x) and the MIC (y), together with the correlation coefficients (r), were respectively presented on Fig. S1 to S6, and summarily listed in Table 7. Most correlation coefficients (r) were more than 0.90 (Table 7). This indicated that there is a good correlation between the physicochemical parameter CLogP, ACD/LogP, or LogD7.40 and the antimicrobial activities (MIC), of these flavonoids to a certain pathogenic bacterium.
Table 7.
Regression equations between the physicochemical parameter (x) and the antimicrobial activity (y) to a certain pathogenic microorganism.
| Compounds | Parametersb | Pathogenic bacteriaa | Regression equation (rc) |
|---|---|---|---|
| 1 to 19 | CLogP | S. aureus | y = −14.562x5 + 368.41x4 − 3689.3x3 + 18274x2 − 44755x + 43,369 (0.8514) |
| ACD/LogP | y = −6.1684x5 + 180.3x4 − 2090x3 + 12006x2 − 34172x + 38,560 (0.7592) | ||
| LogD7.40 | y = −5.3777x5 + 151.88x4 − 1700.4x3 + 9430.9x2 − 25910x + 28,225 (0.7331) | ||
| CLogP | B. subtilis | y = −13.392x5 + 345.58x4 − 3527.6x3 + 17792x2 − 44315x + 43,606 (0.8093) | |
| ACD/LogP | y = −8.1245x5 + 238.87x4 − 2785.6x3 + 16098x2 − 46081x + 52,265 (0.8168) | ||
| LogD7.40 | y = −6.8012x5 + 193.67x4 − 2186.6x3 + 12230x2 − 33875x + 37,175 (0.7660) | ||
| 20 to 27 | CLogP | S. aureus | y = 8.524x4 − 204.37x3 + 1846.7x2 − 7432.5x + 11,221 (0.7878) |
| ACD/LogP | y = −35.117x5 + 1129.5x4 − 14526x3 + 93407x2 − 300407x + 386,677 (0.9998) | ||
| LogD7.40 | y = −32.854x5 + 1023.1x4 − 12742x3 + 79351x2 − 247194x + 308,250 (0.9998) | ||
| CLogP | S. epidermidis | y = 27.806x4 − 635.4x3 + 5428.8x2 − 20549x + 29,078 (0.9228) | |
| ACD/LogP | y = 49.336x5 − 1560.6x4 + 19638x3 − 122806x2 + 381390x − 470,183 (0.9999) | ||
| LogD7.40 | y = 50.433x5 − 1541.1x4 + 18726x3 − 113020x2 + 338580x − 402,381 (0.9999) | ||
| 21 to 24, and 26 to 33 | CLogP | MRSA 6975 | y = 1.8495x4 − 35.986x3 + 255.94x2 − 793.24x + 933.27 (0.9727) |
| ACD/LogP | y = 4.3462x4 − 100.91x3 + 865.04x2 − 3250.5x + 4550 (0.9737) | ||
| LogD7.40 | y = 3.9824x4 − 89.302x3 + 738.93x2 − 2679.6x + 3624.8 (0.9724) | ||
| CLogP | MRSA 630 | y = 6.6617x4 − 108.73x3 + 631.7x2 − 1592.5x + 1668.4 (0.8594) | |
| ACD/LogP | y = 6.7373x4 − 144.65x3 + 1138.9x2 − 3954.4x + 5320.3 (0.8317) | ||
| LogD7.40 | y = 0.7482x5 − 12.599x4 + 58.825x3 + 49.926x2 − 1007.8x + 2110.7 (0.8409) | ||
| CLogP | MRSA 6205 | y = 13.675x4 − 236.81x3 + 1465.8x2 − 3839.6x + 3692.3 (0.8157) | |
| ACD/LogP | y = −1.087x5 + 45.877x4 − 674.86x3 + 4547.7x2 − 14384x + 17,445 (0.7738) | ||
| LogD7.40 | y = −5.6301x5 + 162.46x4 − 1831.6x3 + 10059x2 − 26887x + 28,095 (0.7847) | ||
| 34 to 44 | CLogP | S. aureus | y = 5.0053x3 − 76.039x2 + 329.45x − 293.29 (0.9720) |
| ACD/LogP | y = 4.1508x3 − 74.07x2 + 387.42x − 479.73 (0.9651) | ||
| LogD7.40 | y = 3.2426x3 − 54.386x2 + 256.74x − 225.91 (0.9643) | ||
| CLogP | B. subtilis | y = 3.2197x3 − 46.825x2 + 177.07x − 40.407 (0.9622) | |
| ACD/LogP | y = 3.0215x3 − 52.494x2 + 254.63x − 216.12 (0.9619) | ||
| LogD7.40 | y = 2.3606x3 − 38.598x2 + 166.87x − 63.384 (0.9607) | ||
| 17, 45 to 54 | CLogP | MRSA | y = −187.27x3 + 2012.3x2 − 7014.5x + 8045.6 (0.9982) |
| ACD/LogP | y = −141.22x3 + 2038.8x2 − 9673x + 15,205 (0.9972) | ||
| LogD7.40 | y = −138.02x3 + 1870.5x2 − 8315.2x + 12,249 (0.9964) | ||
| CLogP | MSSA | y = −186.51x3 + 1996x2 − 6936.4x + 7973.6 (0.9830) | |
| ACD/LogP | y = −129.02x3 + 1872.9x2 − 8962.9x + 14,287 (0.9789) | ||
| LogD7.40 | y = −146.22x3 + 1942.5x2 − 8495.7x + 12,386 (0.9760) | ||
| 55 to 66 | CLogP | MRSA G31 | y = −17.547x5 + 454.87x4 − 4632.7x3 + 23200x2 − 57189x + 55,596 (0.9999) |
| ACD/LogP | y = −12.103x5 + 371.48x4 − 4493.1x3 + 26798x2 − 78893x + 91,819 (0.9999) | ||
| LogD7.40 | y = −10.79x5 + 322.43x4 − 3789.8x3 + 21934x2 − 62584x + 70,524 (0.9999) | ||
| CLogP | MRSA G47 | y = −14.185x5 + 357.83x4 − 3530.1x3 + 17044x2 − 40327x + 37,493 (0.9997) | |
| ACD/LogP | y = −9.8483x5 + 294.98x4 − 3468.7x3 + 20038x2 − 56931x + 63,738 (0.9997) | ||
| LogD7.40 | y = −8.6059x5 + 250.58x4 − 2858.5x3 + 15993x2 − 43945x + 47,542 (0.9996) |
aThe antimicrobial activity (y) was expressed as MIC or MIC90 to a certain pathogenic microorganism. S. aureus, Staphylococcus aureus; S. epidermidis, Staphylococcus epidermidis; B. subtilis, Bacillus subtilis; MRSA, methicillin-resistant Staphylococcus aureus; MSSA methicillin-susceptible Staphylococcus aureus.
bCLogP was calculated using software ChemBioDraw Ultra 12.0, and ACD/LogP and LogD7.40 were calculated using software ACD/Labs 6.0.
cr, correlation coefficients.
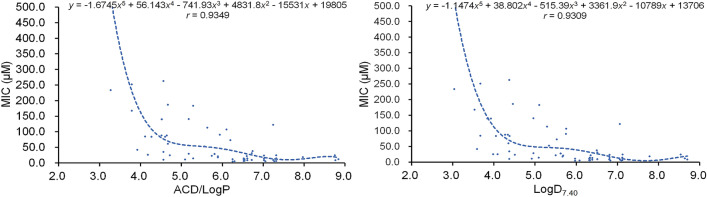
As we pointed out above, the antimicrobial activities of a compound against different pathogenic bacteria were varied, and even against the same one in different determination conditions. Thereby, the regression analyses were respectively performed for these flavonoids reported in different papers. Considering that the pathogenic bacteria used for antibacterial experiments mainly involved S. aureus, S. epidermidis, and B. subtilis, the same compound should present similar inhibitory activities and identical antibacterial mechanism to these gram-positive bacteria. Thereby, we put the physicochemical parameters and the average MICs to S. aureus, S. epidermidis, or/and B. subtilis (Tables 1, 2, 3, 4, 5 and 6), of these flavonoids together for further regression analyses. The results indicated that the correlation between CLogP and antibacterial activities (MICs) is weak with a correlation coefficient of 0.8412, while that between ACD/LogP or LogD7.40 (x) and MICs (y) is more reliable (Fig. 7). The regression equations were respectively expressed as y = − 1.6745x5 + 56.143x4 − 741.93x3 + 4831.8x2 − 15531x + 19,805 and y = − 1.1474x5 + 38.802x4 − 515.39x3 + 3361.9x2 − 10789x + 13,706, with the correlation coefficients of 0.9349 and 0.9309, respectively. These further proved, by a larger sample, that the inhibitory activities of these flavonoids to gram-positive bacteria will nonlinearly increase as the ACD/LogP or LogD7.40 increase to approximately 7.0, and then decrease along with the further increase of ACD/LogP or LogD7.40.
Figure 7.
Polynomial regression analyses for the physicochemical parameters ACD/LogP or LogD7.40 (x) and the average MICs (y) to gram-positive bacteria including S. aureus, S. epidermidis, or/and B. subtilis, of compounds 1 to 66.
Verification
To verify the above correlations, other sixty-eight flavonoids (Fig. 8) including flavone, isoflavone, flavonol, flavanonol, dihydroflavone, dihydroisoflavone, flavane, and chalcone subclasses etc., reported in seven papers4,16–21, were selected for the comparison of theoretical and reported MICs. Using above two regression equations y = − 1.6745x5 + 56.143x4 − 741.93x3 + 4831.8x2 − 15531x + 19,805 and y = − 1.1474x5 + 38.802x4 − 515.39x3 + 3361.9x2 − 10789x + 13,706 (x is the ACD/LogP or LogD7.40, and y is the antimicrobial activities (MICs)), the theoretical MICs of these flavonoids can be calculated. Considering that many factors, such as determination method, concentration of bacterial suspension, and test medium used, may influence on the determination of MIC5, the results reported would fluctuate within a reasonable range of the actual values. Thereout, the predicted MICs ranged from 1/4 × to 4 × the determined one were acceptable (marked as A), especially those ranged from 1/2 × to 2 × the determined one, were considered as complete coincidence (marked as C) since the MICs were generally determined by double dilution method22. Simultaneously, those more than or equal to the minimum value when the determined MICs were no upper limit were also regarded as complete coincidence (marked as C). Otherwise, those were unacceptable (marked as U). The results (Table 8) indicated that the predicted MICs were in acceptable or complete coincidence with the measured ones for approximate 85.3% flavonoids. Although the antibacterial activities of ten flavonoids (14.7%) are unsatisfactorily predicted, there are six compounds with the predicted MICs falling into the range of 1/8 × to 8 × determined ones. This together indicated that the MICs of most flavonoids against gram-positive bacteria can be roughly calculated from their ACD/LogP or LogD7.40 although the predicted values are not in accordance with their tested ones for a few flavonoids. At least, these indicated that the ACD/LogP or LogD7.40 is a key factor for the inhibitory activities of plant flavonoids against gram-positive bacteria.
Figure 8.
Table 8.
Comparison of predicted and reported antibacterial activities of some flavonoids.
| Compounds | Molecular weight | Predicted antibacterial activitiesa | Measured antibacterial activities | Coincidenceb | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| By LogP | By LogD7.40 | ||||||||
| LogP | MIC (μM) | LogD7.40 | MIC (μM) | MIC (μg/mL) | MIC (μM) | ||||
| 67 | 338.35 | 4.20 | 121.21 | 3.77 | 162.30 | 62.5 | 184.7 | C | 16 |
| 68 | 336.34 | 5.55 | 54.21 | 4.82 | 52.69 | 62.5 | 185.8 | A | 16 |
| 69 | 314.29 | 2.16 | 2467.72 | 1.72 | 2794.56 | ≥ 200 | ≥ 636.4 | C | 17 |
| 70 | 298.29 | 3.10 | 695.19 | 2.32 | 1381.83 | ≥ 200 | ≥ 670.5 | C | 17 |
| 71 | 240.25 | 3.93 | 183.65 | 3.62 | 206.48 | 50–200 | 208.1–832.5 | C | 17 |
| 72 | 406.47 | 7.33 | 14.22 | 6.89 | 19.22 | 1.56–3.13 | 3.8–7.7 | C | 4 |
| 73 | 270.24 | 2.83 | 1030.88 | 2.16 | 1683.78 | > 125–240 | > 462.6–888.1 | C | 4 |
| 74 | 432.38 | 1.70 | 4166.24 | 0.76 | 7234.60 | 1–2 | 2.3–4.6 | U | 4 |
| 75 | 542.62 | 8.63 | 23.00 | 8.17 | 13.80 | 2.3–37.5 | 4.4–69.1 | C | 4 |
| 76 | 422.47 | 6.59 | 31.55 | 6.40 | 32.27 | 0.5–4 | 1.2–9.5 | A | 4 |
| 77 | 424.49 | 6.60 | 31.25 | 6.42 | 31.74 | 2.9 | 6.8 | U | 4 |
| 78 | 324.37 | 5.49 | 54.80 | 5.44 | 47.91 | 0.3–0.6 | 0.9–1.9 | U | 4 |
| 79 | 408.49 | 5.95 | 48.36 | 5.70 | 45.79 | 0.6–1.22 | 1.5–3.0 | U | 4 |
| 80 | 406.51 | 8.35 | 21.21 | 7.93 | 10.00 | 0.125–2 | 0.3–4.9 | A | 4 |
| 81 | 338.4 | 4.95 | 61.35 | 4.82 | 52.69 | 3.13–16 | 9.3–47.3 | C | 4 |
| 82 | 338.4 | 4.95 | 61.35 | 4.82 | 52.69 | 3.13–6.25 | 9.4–18.9 | A | 18,19 |
| 83 | 286.28 | 2.57 | 1471.41 | 2.44 | 1185.40 | > 25–100 | > 87.3–349.3 | C | 18,19 |
| 84 | 256.25 | 2.76 | 1137.07 | 2.61 | 946.52 | > 100 | > 390.2 | C | 18,19 |
| 85 | 418.39 | 0.61 | 11,968.2 | 0.46 | 9405.99 | > 50 | > 119.5 | C | 18,19 |
| 86 | 352.34 | 5.67 | 52.85 | 5.07 | 49.83 | 12.5 | 35.5 | C | 18,19 |
| 87 | 268.26 | 3.15,16 | 644.51 | 2.91 | 621.60 | > 25–100 | > 93.2–372.8 | C | 18,19 |
| 88 | 382.41 | 5.38 | 55.79 | 5.12 | 49.51 | > 25–50 | > 65.4–130.8 | C | 18,19 |
| 89 | 354.35 | 4.15,16 | 130.39 | 3.48 | 258.70 | 12.5–25 | 35.3–70.6 | C | 18,19 |
| 90 | 382.41 | 5.75 | 51.77 | 5.75 | 45.21 | > 50 | > 130.8 | U | 18,19 |
| 91 | 424.53 | 6.32 | 39.42 | 6.32 | 34.31 | 3.13–6.25 | 7.4–14.7 | A | 18,19 |
| 92 | 370.44 | 4.41 | 93.10 | 4.40 | 68.92 | 6.25–12.5 | 16.9–33.7 | A | 18,19 |
| 93 | 324.37 | 4.18 | 124.77 | 4.18 | 88.60 | 6.25–12.5 | 19.3–38.5 | A | 18,19 |
| 94 | 322.35 | 6.64 | 30.07 | 6.63 | 26.09 | 12.5–25 | 38.8–77.6 | C | 18,19 |
| 95 | 302.24 | 2.07 | 2746.05 | 1.40 | 3919.38 | > 125 | > 413.6 | C | 20 |
| 96 | 254.24 | 2.88 | 960.17 | 2.33 | 1364.52 | > 125 | > 491.7 | C | 20 |
| 97 | 302.24 | 2.62 | 1376.51 | 1.95 | 2158.21 | 500 | 1654.3 | C | 20 |
| 98 | 318.24 | 2.11 | 2619.40 | 1.42 | 3839.98 | 62.5–125 | 196.4–392.8 | U | 20 |
| 99 | 238.24 | 3.76 | 242.43 | 3.74 | 170.26 | > 125 | > 524.7 | U | 20 |
| 100 | 270.24 | 2.1 | 2650.61 | 1.57 | 3284.32 | ≥ 125 | ≥ 462.6 | C | 20 |
| 101 | 286.24 | 2.52 | 1571.57 | 2.22 | 1564.90 | > 125 | > 436.7 | C | 20 |
| 102 | 284.26 | 3.04 | 760.45 | 2.50 | 1096.06 | > 125 | > 439.7 | C | 20 |
| 103 | 300.26 | 3.00 | 806.77 | 2.33 | 1364.52 | > 125 | > 416.3 | C | 20 |
| 104 | 302.24 | 2.54 | 1530.86 | 2.14 | 1724.98 | 31.3–62.5 | 103.4–206.8 | U | 20 |
| 105 | 360.31 | 3.02 | 783.32 | 2.32 | 1381.83 | > 125 | > 346.9 | C | 20 |
| 106 | 284.26 | 3.22 | 578.89 | 2.45 | 1170.12 | > 125 | > 439.7 | C | 20 |
| 107 | 254.24 | 2.51 | 1592.25 | 2.19 | 1623.47 | 31.3–62.5 | 122.9–245.8 | U | 20 |
| 108 | 418.39 | 0.61 | 11,968.2 | 0.46 | 9405.99 | > 128 | > 305.9 | C | 21 |
| 109 | 460.43 | 2.46 | 1699.00 | 2.31 | 1399.32 | > 128 | > 278.0 | C | 21 |
| 110 | 550.51 | 2.24 | 2239.77 | 2.06 | 1897.96 | > 128 | > 232.5 | C | 21 |
| 111 | 418.39 | 1.04 | 8107.91 | 0.87 | 6546.46 | > 128 | > 305.9 | C | 21 |
| 112 | 302.28 | 2.36 | 1929.91 | 2.19 | 1623.47 | > 128 | > 423.5 | C | 21 |
| 113 | 270.28 | 3.23 | 570.00 | 3.10 | 468.93 | 64–128 | 236.8–473.6 | C | 21 |
| 114 | 256.25 | 3.40 | 436.17 | 3.26 | 366.65 | 128 | 499.5 | C | 21 |
| 115 | 354.35 | 5.03 | 59.77 | 4.48 | 64.08 | 32–64 | 90.3–180.6 | C | 21 |
| 116 | 352.34 | 4.63 | 73.82 | 4.07 | 102.86 | 32–64 | 90.8–181.6 | C | 21 |
| 117 | 368.38 | 5.33 | 56.22 | 5.08 | 49.76 | 64 | 173.73 | A | 21 |
| 118 | 352.38 | 5.69 | 52.59 | 5.40 | 48.13 | 16 | 45.41 | C | 21 |
| 119 | 406.47 | 7.33 | 14.22 | 7.16 | 13.23 | 8 | 19.68 | C | 21 |
| 120 | 338.35 | 5.24 | 57.06 | 4.69 | 55.65 | 16–32 | 47.3–94.6 | C | 21 |
| 121 | 368.38 | 4.70 | 69.97 | 4.27 | 79.27 | 64 | 173.7 | A | 21 |
| 122 | 422.47 | 7.13 | 17.41 | 6.89 | 19.22 | 16 | 37.9 | C | 21 |
| 123 | 368.38 | 4.56 | 78.51 | 4.27 | 79.27 | 32–64 | 86.9–173.7 | C | 21 |
| 124 | 354.35 | 5.47 | 54.98 | 5.21 | 49.04 | 32–64 | 90.3–180.6 | C | 21 |
| 125 | 354.35 | 5.47 | 54.98 | 5.21 | 49.04 | 32 | 90.3 | C | 21 |
| 126 | 384.42 | 4.83 | 64.65 | 4.67 | 56.23 | 32–64 | 83.2–166.5 | C | 21 |
| 127 | 424.49 | 6.69 | 28.60 | 6.50 | 29.62 | 8 | 18.9 | C | 21 |
| 128 | 368.38 | 5.99 | 47.55 | 5.98 | 41.74 | 16 | 43.4 | C | 21 |
| 129 | 384.38 | 3.76 | 242.43 | 3.76 | 164.91 | ≥ 128 | ≥ 333.0 | C | 21 |
| 130 | 368.38 | 5.61 | 53.56 | 5.59 | 46.85 | 16–32 | 43.4–86.9 | C | 21 |
| 131 | 384.38 | 3.86 | 205.75 | 3.86 | 140.83 | > 128 | > 333.0 | C | 21 |
| 132 | 410.46 | 5.79 | 51.17 | 5.79 | 44.71 | > 128 | > 311.9 | U | 21 |
| 133 | 340.37 | 4.10 | 140.56 | 4.10 | 98.63 | 16 | 47.0 | A | 21 |
| 134 | 382.41 | 5.75 | 51.77 | 5.75 | 45.21 | > 50–128 | > 130.8–334.7 | A | 21 |
Antibacterial activities were expressed as MICs of flavonoids to gram-positive bacteria which include S. aureus, S. epidermidis and B. subtilis.
aLogP and LogD7.40 were calculated using software ACD/Labs 6.0.
bC, Complete coincidence; A, Acceptable; U, Unacceptable.
Discussion and conclusion
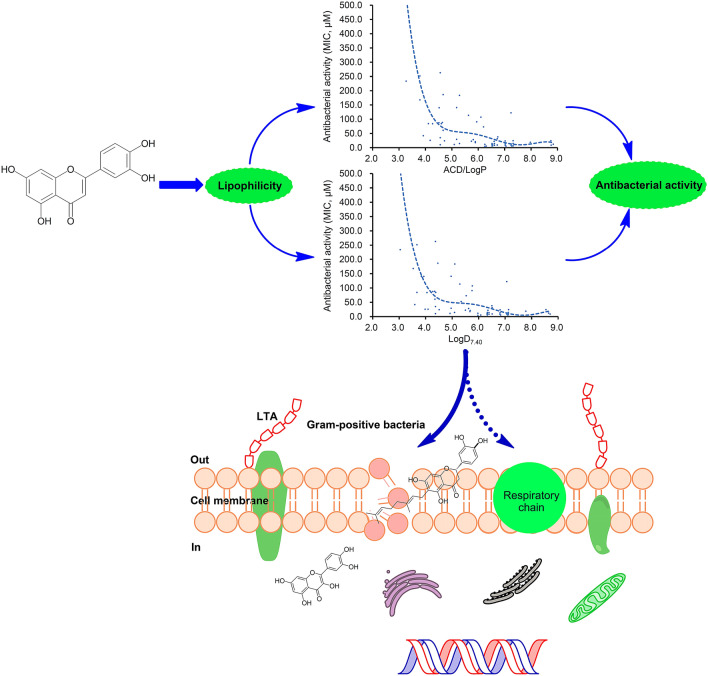
Flavonoids can be widely found in various parts of the plant, and their antibacterial activities have been paid more and more attention to, especially after some of them were discovered to have the potency to enhance the susceptibility of some antibiotics to bacteria4,5. Based on the related data of plant flavonoids reported, many related physicochemical parameters were calculated, using software ChemBioDraw Ultra 12.0 and ACD/Labs 6.0, for the discovery of the correlations between the physicochemical parameters and the MICs of flavonoids against gram-positive bacteria. Two regression equations between the ACD/LogP or LogD7.40 (x) and the antimicrobial activities (MICs) (y) were established as y = − 1.6745x5 + 56.143x4 − 741.93x3 + 4831.8x2 − 15531x + 19,805 and y = − 1.1474x5 + 38.802x4 − 515.39x3 + 3361.9x2 − 10789x + 13,706. From these two equations, the MICs of most flavonoids against gram-positive bacteria (mainly Staphylococcus and Bacillus) could be roughly calculated from their ACD/LogP or LogD7.40, and their minimum value was predicted as approximately 10.2 or 4.8 μM. Considering that the experimental MICs would fluctuate within a reasonable range5, the minimum MIC of plant flavonoids will likely fall into the range from 2.6 to 10.2 μM, or from 1.2 to 4.8 μM, predicted from their ACD/LogP or LogD7.40.
After all, the antibacterial activities of a compound to different pathogens are varied, and so these two regression equations, mainly valuable for Staphylococcus and Bacillus, may not always be suitable for flavonoids to other gram-positive bacteria. However, the acceptable range from 1/4 × to 4 × the determined MICs will increase the applicability of these two equations used for the prediction of plant flavonoids to other gram-positive bacteria. To say the least, if necessary, similar regression equations can be also established from the physicochemical parameters and the MICs to other gram-positive bacteria, of flavonoids. Thereby, we concluded that the MICs of most flavonoids against gram-positive bacteria can be roughly calculated from their physicochemical parameters ACD/LogP or LogD7.40.
Lipophilicity is a very important descriptor indicating membrane permeation23, and generally expressed as LogP which is valid only for a single electrical species. For ionizable drugs, LogD that refers to a pH-dependent mixture of all electrical species presented at any given pH was regarded as a better descriptor reflecting the actual partitioning and lipophilicity24,25. Generally, most flavonoids contain two or more phenolic hydroxyl groups4–6, and present similar weak acidity with the pKa of 7.0 to 10.0. Thereby, their LogD will correspondingly decrease along with the increase of environmental pH from about 5.0. Considering the pH in human blood or in the media of MIC determination was approximately 7.40, their LogD at pH 7.40 were selected. These together above indicate that the lipophilicity of plant flavonoids is a key factor for their inhibitory activities to gram-positive bacteria. As the lipophilicity is closely related to membrane permeability26, the tendentiously concave regression curves between the antibacterial activity and the LogP or LogD7.40 also indicate that the cell membrane is probably an important site of flavonoids acting on gram-positive bacteria.
Different antibacterial mechanisms of plant flavonoids were reported4–6, such as causing cell-membrane damage, inhibition on various synthase involving the nucleic acid synthesis, the bacterial respiratory chain, or the cell envelope synthesis. However, the results above suggested that the antibacterial activities of these plant flavonoids had no obvious relationship with the specific fragments of their structures, while presented great relationship with their lipophilicities. Simultaneously, the antibacterial activities of plant flavonoids will dramatically increase as the LogP or LogD increases from 2.5 to 4.0 which range the membrane permeability remarkably decrease while the affinity to lipid bilayer greatly increase27–29. According to this, plant flavonoids may not target specific synthases, but more likely to nonspecifically act on the cell-membrane bilayer or the respiratory chain to kill bacteria. This deduction was indirectly supported by many researches which were reviewed in three paper4–6, such as follows: (1) two mechanisms may be involved the interactions of flavonoids with lipid bilayers, which include the interactions at the membrane interface between the polar heads of phospholipids and the more hydrophilic flavonoids, and the partition of the more hydrophobic flavonoids in the interior of the lipid bilayer30; (2) nonspecific interactions of flavonoids with phospholipids can lead to the changes of the membrane properties31; (3) The increased activities of more lipophilic flavonoids are due to the enhanced membrane affinity of their long acyl chains32; (4) Some lipophilic flavonoids can decrease the fluidity and integrity of cellular membrane to inhibit gram-positive bacteria33,34, such as sophoraflavanone G and 3-arylideneflavanones.
Although many other antibacterial mechanisms acting on various synthase for the nucleic acid or cell envelope syntheses were mentioned in these reviews4,6, two facts found from the researches of the cited literature are worth further discussing. First, most flavonoids used for mechanism exploration have the cLogP ranged from about 2.0 to 4.0, and are easy to infiltrate into the bacterial cell, while they present very weak antibacterial activities with the MICs more than 250 μg/mL. Second, most experiments were achieved by the determination of enzyme activities in vitro35,36, the molecular docking of flavonoids with various synthases37, the proteomics technology without the combination of related experiments and the consideration of first the chicken or the egg38. Another thing should be considered is whether some molecules can pass through the cell membrane and infiltrate into the bacterial cell or not. Moreover, previous works indicated the antibacterial activity to gram-positive bacteria was observed only four of fourteen flavonoids, while only four of seven flavonoids with DNA gyrase inhibition showed weak inhibitory activity to gram-positive bacteria20. Simultaneously, the authors pointed out that mechanisms other than DNA gyrase inhibition may also play a role in the antibacterial activity. Thereby, the conclusion that some of these flavonoids studied are potent inhibitors of DNA gyrase is worth reconsidering20. In fact, this work just right indicated that the inhibitory activity of flavonoids against gram-positive bacteria did not correlate with their in vitro DNA gyrase inhibition to a large extent. This was also supported by previous publication39. These together further confirmed that the cell-membrane should be the main region of plant flavonoids acting on Gram-positive bacteria, and which likely involving the disruption or damage of phospholipid bilayers, the inhibition of the respiratory chain or ATP synthesis, or some others.
According to the regression equations and above conclusions, the inhibitory activities of flavonoids to gram-positive bacteria will increase when the alkyl especially isopentyl were introduced into the structures of flavonoids no matter carbon position it is introduced into. This can be interpreted that the introduction of alkyl would increase the lipophilicity of flavonoids or the LogP, and thereout increase their interactions with phospholipids of cell membrane. However, the introduction of too many alkyls will overmuch increase the LogP of these flavonoids, and which will lead their lipophilicities too large to pass through the hydrophilic region of phospholipid bilayers. This was proved by previous similar work26,32,40. On the contrary, the inhibitory activities of flavonoids to gram-positive bacteria will decrease when polar groups, such as hydroxyl and glycosyl, were introduced into their structures. This can be interpreted as that the excessive hydrophilicity of flavonoids will hinder its infiltration into phospholipid bilayers and interaction with hydrophobic region of cell membrane.
Based on the physicochemical parameters and MICs of various flavonoids, the regression equations and above conclusions were achieved. For a certain subclass of flavonoids, the regression equations with larger correlation coefficient can be established for their more accurate MIC predictions, and then can be further used for the structural design and optimization to obtain more efficient antibacterial activity.
As the inhibitory activities of plant flavonoids against gram-negative bacteria were reported less, it is difficult to draw a statistical conclusion. Considering that the cell envelope of gram-negative bacteria was different from that of gram-positive ones, it is worth further exploring whether the above regression equations and above conclusions are suitable for plant flavonoids against gram-negative bacteria. However, these can provide good references for their related researches. Referring to the above conclusions, the anti-MRSA activities of trimethylhydroquinone, vitamin K3 and carnosic acid were successfully predicted and verified by our laboratory9,41.
In conclusion, the MICs of most flavonoids against gram-positive bacteria can be roughly calculated from their physicochemical parameters ACD/LogP or LogD7.40, and the lipophilicity is a key factor of plant flavonoids against gram-positive bacteria. Combined with the analyses of previous publications, the results also suggest that the cell membrane may be the main site of plant flavonoids acting on gram-positive bacteria, and which likely involves the damage of phospholipid bilayers, the inhibition of the respiratory chain or ATP synthesis, or some others. Base on this, the inhibitory activities and mechanisms of plant flavonoids to gram-positive bacteria were diagrammatically presented as Fig. 9.
Figure 9.
Diagrammatic presentation for the inhibitory activities and mechanisms of plant flavonoids to gram-positive bacteria.
Methods
Information and data
The structures, antimicrobial activities and other related information of plant flavonoids were unsystematically searched from Google academic search engine, and several databases SciFinder, Medline, Elsevier, ACS, ScienceDirect, Wiley Online Library, Springer-Link, and RSC, using keywords flavonoid and antimicrobial, or and antibacterial, and or and anti-MRSA. Furthermore, the relevant references in the obtained literature were also tracked. The structures, antibacterial activities, and other related information of flavonoids were collected from the obtained literature that can provide more than five or more flavonoids. As the antimicrobial activities of a certain compound against different pathogenic strains were varied, compounds reported in different papers were independently collected for the following analyses. Finally, the structures of selected compounds were drawn using software ChemBioDraw Ultra 12.0.
Simulation calculation of physicochemical parameters
The physicochemical parameters Gibbs energy, LogP, CLogP, MR, CMR and tPSA were calculated using software ChemBioDraw Ultra 12.0. Moreover, another software ACD/Labs 6.0 was also used for the calculations of physicochemical parameters LogP, LogD7.40 and solubility (SolDB).
Data analysis and correlation establishment
The physicochemical parameters and antibacterial activities of flavonoids reported in the same paper were respectively listed in a table, even those of the same compound. The regression analyses between the calculated values of each parameter and the antimicrobial activities (expressed as MICs) of all compounds in a table were respectively performed using Microsoft Excel software. It is noting that compounds without related antimicrobial information were not considered for the regression analyses, while they can be used for the following discussion. The physicochemical parameters significantly correlating with the antimicrobial activities were selected for the further analyses of correlations between the physicochemical parameters and antimicrobial activities of flavonoids.
Verification
Some other flavonoids were searched from above several databases, and the chemical structures of various flavonoids presented in previous publications were also drawn using software ChemBioDraw Ultra 12.0. The physicochemical parameters LogP and LogD7.40 of these flavonoids were respectively calculated by software ACD/Labs 6.0, and then their antimicrobial activities (MICs) were respectively predicted using the above regression equations. Comparing with the predicted MICs with the determined one, the regression equations can be verified.
Supplementary Information
Acknowledgements
This work was financially supported by grants from the National Natural Science Foundation of China (No. 82073745, 81960636 and 81660578).
Author contributions
G.Y. initiated the project, hypothesis and design, analyzed the data and obtained regression equations, wrote the manuscript text, took part in the search and collection of flavonoids data, and prepared the figures and tables; Y.G. performed the search and collection of flavonoids data, and took part in the preparation of figures and tables; H.Y., S.L., Y.S. and S.C. took part in the data processing and analysis. All authors reviewed the manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-90035-7.
References
- 1.Laxminarayan R, Sridhar D, Blaser M, Wang M, Woolhouse M. Achieving global targets for antimicrobial resistance. Science. 2016;353:874–875. doi: 10.1126/science.aaf9286. [DOI] [PubMed] [Google Scholar]
- 2.Kurosu M, Siricilla S, Mitachi K. Advances in MRSA drug discovery: where are we and where do we need to be? Exp. Opin. Drug Discov. 2013;8:1095–1116. doi: 10.1517/17460441.2013.807246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu X, Xu L, Yuan G, Wang Y, Qu Y, Zhou M. Synergistic combination of two antimicrobial agents closing each other's mutant selection windows to prevent antimicrobial resistance. Sci. Rep. 2018;8:7237. doi: 10.1038/s41598-018-25714-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Górniak I, Bartoszewski R, Króliczewski J. Comprehensive review of antimicrobial activities of plant flavonoids. Phytochem. Rev. 2019;18:241–272. doi: 10.1007/s11101-018-9591-z. [DOI] [Google Scholar]
- 5.Cushnie TPT, Lamb AJ. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents. 2005;26:343–356. doi: 10.1016/j.ijantimicag.2005.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Y, Yang W, Tang F, Chen X, Ren L. Antibacterial activities of flavonoids: structure-activity relationship and mechanism. Curr. Med. Chem. 2015;22:132–149. doi: 10.2174/0929867321666140916113443. [DOI] [PubMed] [Google Scholar]
- 7.Xu X, et al. Synergistic combination of two antimicrobial agents closing each other's mutant selection windows to prevent antimicrobial resistance. Sci. Rep. 2018;8:7237. doi: 10.1038/s41598-018-25714-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan G, et al. Azalomycin F5a, a polyhydroxy macrolide binding to the polar head of phospholipid and targeting to lipoteichoic acid to kill methicillin-resistant Staphylococcus aureus. Biomed. Pharmacother. 2019;109:1940–1950. doi: 10.1016/j.biopha.2018.11.067. [DOI] [PubMed] [Google Scholar]
- 9.Yuan G, Zhu X, Li P, Zhang Q, Cao J. New activity for old drug: In vitro activities of vitamin K3 and menadione sodium bisulfite against methicillin-resistant Staphylococcus aureus. Afr. J. Pharm. Pharmacol. 2014;8:364–371. [Google Scholar]
- 10.Kuroyanagi M, Arakawa T, Hirayama Y, Hayashi T. Antibacterial and antiandrogen flavonoids from Sophora flavescens. J. Nat. Prod. 1999;62:1595–1599. doi: 10.1021/np990051d. [DOI] [PubMed] [Google Scholar]
- 11.Šmejkal K, et al. Antibacterial C-geranylflavonoids from Paulownia tomentosa fruits. J. Nat. Prod. 2008;71:706–709. doi: 10.1021/np070446u. [DOI] [PubMed] [Google Scholar]
- 12.Navrátilová A, et al. Minor C-geranylated flavanones from Paulownia tomentosa fruits with MRSA antibacterial activity. Phytochemistry. 2013;89:104–113. doi: 10.1016/j.phytochem.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Inui S, et al. Solophenols B−D and solomonin: New prenylated polyphenols isolated from propolis collected from the solomon islands and their antibacterial activity. J. Agric. Food Chem. 2012;60:11765–11770. doi: 10.1021/jf303516w. [DOI] [PubMed] [Google Scholar]
- 14.Sasaki H, Kashiwada Y, Shibata H, Takaishi Y. Prenylated flavonoids from Desmodium caudatum and evaluation of their anti-MRSA activity. Phytochemistry. 2012;82:136–142. doi: 10.1016/j.phytochem.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Tsuchiya H, Sato M, et al. Comparative study on the antibacterial activity of phytochemical flavanones against methicillin-resistant Staphylococcus aureus. J. Ethnopharmacol. 1996;50:27–34. doi: 10.1016/0378-8741(96)85514-0. [DOI] [PubMed] [Google Scholar]
- 16.Edziri H, et al. Antibacterial, antifungal and cytotoxic activities of two flavonoids from Retama raetam flowers. Molecules. 2012;17:7284–7293. doi: 10.3390/molecules17067284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sufian AS, Ramasamy K, Ahmat N, Zakaria ZA, Yusof MIM. Isolation and identification of antibacterial and cytotoxic compounds from the leaves of Muntingia calabura L. J. Ethnopharmacol. 2013;146:198–204. doi: 10.1016/j.jep.2012.12.032. [DOI] [PubMed] [Google Scholar]
- 18.Fukai T, et al. Antimicrobial activity of licorice flavonoids against methicillin-resistant Staphylococcus aureus. Fitoterapia. 2002;73:536–539. doi: 10.1016/S0367-326X(02)00168-5. [DOI] [PubMed] [Google Scholar]
- 19.Fukai T, Marumo A, Kaitou K, Kanda T, Terada S, Nomura T. Anti-Helicobacter pylori flavonoids from licorice extract. Life Sci. 2002;71:1449–1463. doi: 10.1016/S0024-3205(02)01864-7. [DOI] [PubMed] [Google Scholar]
- 20.Ohemeng KA, Schwender CF, Fu KP, Barrett JF. DNA gyrase inhibitory and antibacterial activity of some flavones(l) Bioorg. Med. Chem. Lett. 1993;3:225–230. doi: 10.1016/S0960-894X(01)80881-7. [DOI] [Google Scholar]
- 21.Hatano T., Shintani Y., Aga Y., Shiota S., Tsuchiya T., Yoshida T. Phenolic constituents of Licorice. VIII.1) structures of glicophenone and glicoisoflavanone, and effects of Licorice phenolics on methicillin-resistant Staphylococcus aureus. Chem. Pharm. Bull. 2000;48(9):1286–1292. doi: 10.1248/cpb.48.1286. [DOI] [PubMed] [Google Scholar]
- 22.Clinicaland Laboratory Standards Institute . CLSI Document M07-A9. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard. 19. Pennsylvania: Wayne; 2012. [Google Scholar]
- 23.Malkia A, Murtomaki L, Urtti A, Kontturi K. Drug permeation in biomembranes: In vitro and in silico prediction and influence of physicochemical properties. Eur. J. Pharm. Sci. 2004;23:13–47. doi: 10.1016/j.ejps.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 24.Bhal SK, Kassam K, Peirson IG, Pearl GM. The rule of five revisited: Applying log d in place of log p in drug-likeness filters. Mol. Pharm. 2007;4:556–560. doi: 10.1021/mp0700209. [DOI] [PubMed] [Google Scholar]
- 25.Hou T. J., Zhang W., Xia K., Qiao X. B., Xu X. J. ADME evaluation in drug discovery. 5. Correlation of Caco-2 permeation with simple molecular properties. J. Chem. Inf. Comput. Sci. 2004;44(5):1585–1600. doi: 10.1021/ci049884m. [DOI] [PubMed] [Google Scholar]
- 26.Testa B, Crivori P, Reist M, Carrupt PA. The influence of lipophilicity on the pharmacokinetic behavior of drugs: Concepts and examples. Perspect. Drug Discov. 2000;19:179–211. doi: 10.1023/A:1008741731244. [DOI] [Google Scholar]
- 27.Jaehde U, Goto T, de Boer AG, Breimer DDB. Blood-brain barrier transport rate of quinoline antibacterials evaluated in cerebrovascular endothelial cell cultures. Eur. J. Pharm. Sci. 1993;1:49–55. doi: 10.1016/0928-0987(93)90017-5. [DOI] [Google Scholar]
- 28.Le Brun PPH, Fox PLA, de Vries ME, Bodde HE. In vitro penetration of some b-adrenoreceptor blocking drugs through porcine buccal mucosa. Int. J. Pharm. 1989;49:141–145. doi: 10.1016/0378-5173(89)90113-0. [DOI] [Google Scholar]
- 29.Tsuchiya H. Membrane interactions of phytochemicals as their molecular mechanism applicable to the discovery of drug leads from plants. Molecules. 2015;20:18923–18966. doi: 10.3390/molecules201018923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arora A, Byrem TM, Nair MG, Strasburg GM. Modulation of liposomal membrane fluidity by flavonoids and isoflavonoids. Arch. Biochem. Biophys. 2000;373:102–109. doi: 10.1006/abbi.1999.1525. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto Y, et al. Antibacterial and antifungal activities of new acylated derivatives of epigallocatechin gallate. Front. Microbiol. 2012;3:53. doi: 10.3389/fmicb.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuchiya H, Iinuma M. Reduction of membrane fluidity by antibacterial sophoraflavanone G isolated from Sophora exigua. Phytomedicine. 2000;7:161–165. doi: 10.1016/S0944-7113(00)80089-6. [DOI] [PubMed] [Google Scholar]
- 33.Budzynska A, et al. Synthetic 3-arylideneflavanones as inhibitors of the initial stages of biofilm formation by Staphylococcus aureus and Enterococcus faecalis. Z. Naturforsch. C. 2011;66:104–114. doi: 10.1515/znc-2011-3-403. [DOI] [PubMed] [Google Scholar]
- 34.Singh SP, Konwarh R, Konwar BK, Karak N. Molecular docking studies on analogues of quercetin with D-alanine: D-alanine ligase of Helicobacter pylori. Med. Chem. Res. 2013;22:2139–2150. doi: 10.1007/s00044-012-0207-7. [DOI] [Google Scholar]
- 35.Wu D, et al. DAlanine: D-alanine ligase as a new target for the flavonoids quercetin and apigenin. Int. J. Antimicrob. Agents. 2008;32:421–426. doi: 10.1016/j.ijantimicag.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Jeong KW, et al. Screening of flavonoids as candidate antibiotics against Enterococcus faecalis. J. Nat. Prod. 2009;72:719–724. doi: 10.1021/np800698d. [DOI] [PubMed] [Google Scholar]
- 37.Elmasri WA, et al. Multitargeted flavonoid inhibition of the pathogenic bacterium Staphylococcus aureus: A proteomic characterization. J. Proteome Res. 2017;16:2579–2586. doi: 10.1021/acs.jproteome.7b00137. [DOI] [PubMed] [Google Scholar]
- 38.Havsteen B. Flavonoids, a class of natural products of high pharmacological potency. Biochem. Pharm. 1983;32:1141–1148. doi: 10.1016/0006-2952(83)90262-9. [DOI] [PubMed] [Google Scholar]
- 39.Hansch C., Fujita T. ρ–σ–π analysis. A method for the correlation of biological activity and chemical structure. J. Am. Chem. Soc. 1964;86:1616–1626. doi: 10.1021/ja01062a035. [DOI] [Google Scholar]
- 40.Baláž Š. Lipophilicity in trans-bilayer transport and subcellular pharmacokinetics. Perspect. Drug Discov. 2000;19:179–211. doi: 10.1023/A:1008775707749. [DOI] [Google Scholar]
- 41.Yuan G, Li P, Yang H. Anti-MRSA activity of carnosic acid in rosemary. Chin. J. Mod. Appl. Pharm. 2012;29:571–574. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.