Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is a devastating pathogen in the swine industry worldwide. miRNAs are reported to be involved in virus–host interaction. Here, we used high-throughput sequencing and miRNA inhibitors to screen possible miRNAs that can inhibit PRRSV infection on its target cell, porcine alveolar macrophages. We observed that miR-218 was downregulated upon virus infection, and knockdown of miR-218 significantly enhanced PRRSV replication. Overexpression of miR-218 resulted in a decrease in PRRSV replication, and this overexpression did not alter viral genomic RNA levels, but rather increased antiviral interferon signaling. Further analysis revealed that miR-218 regulated PRRSV replication by directly targeting porcine suppressor of cytokine signaling 3 (SOCS3), a JAK2 kinase inhibitor. Knockdown of the endogenous SOCS3 expression led to augmentation of type I interferon genes and resulted in decreased PRRSV replication, and vice versa. During PRRSV infection in vivo and in vitro, cellular miR-218 expression was downregulated and SOCS3 expression was upregulated, further supporting the inverse correlation between miR-218 and SOCS3 expression. The data on SOCS3 depletion in combination with miR-218 inhibition suggested that the antiviral activity of miR-218 required the SOCS3-mediated signaling pathway. Similarly, miR-218 negatively regulated PRRSV replication in Marc-145 cells, as well as the replication of porcine epidemic diarrhea virus and transmissible gastroenteritis virus in Vero and ST cells respectively. Taken together, these results demonstrate that PRRSV-induced miR-218 downregulation serves to inhibit the type I interferon response and may provide a novel therapeutic target for treatment of PRRSV and other viral infections.
Keywords: porcine reproductive and respiratory syndrome virus, host–virus interaction, miR-218, type I interferon response, suppressor of cytokine signaling 3 (SOCS3)
Abbreviations: 3'UTR, 3'-untranslated region; ANOVA, analysis of variance; GBP1, interferon-induced guanylate-binding protein 1; hpi, hours postinfection; HP-PRRSV, high-pathogenic PRRSV; IFA, immunofluorescence assay; IFIT1, interferon-induced protein with tetratricopeptide repeats 1; IFN, interferon; ISG, IFN-stimulated gene; ISG15, IFN-stimulated gene 15; MOI, multiplicity of infection; OASL, 2'-5'-oligoadenylate synthetase-like protein; PAM, porcine alveolar macrophage; PEDV, porcine epidemic diarrhea virus; PRRSV, porcine reproductive and respiratory syndrome virus; qPCR, quantitative real-time PCR; SOCS3, suppressor of cytokine signaling 3; TCID50, 50% tissue culture infective dose; TGEV, transmissible gastroenteritis virus
Porcine reproductive and respiratory syndrome virus (PRRSV) is an enveloped, single-stranded positive RNA virus, belonging to the family Arteriviridae of the order Nidovirales (1, 2). PRRSV is the etiological agent of porcine reproductive and respiratory syndrome (PRRS), which is characterized by reproductive failure in sows and severe respiratory symptoms in piglets and growing pigs. PRRS was first described in the United States in 1987 and in Europe in 1990 (3, 4), and since then this disease has spread around most pig-producing countries and has become an economically devastating disease in the swine industry worldwide. To control this disease, researchers have developed different vaccines. However, due to the high antigenic heterogeneity of PRRSV, the use of current vaccines has some limitations (5, 6). Therefore, it is worthwhile to explore the immune regulatory molecules against PRRSV infection from the host’s perspective.
miRNAs, a class of endogenous noncoding RNAs of ∼22 nucleotides, play important roles in the regulation of gene expression at the posttranscriptional level. miRNAs are initially transcribed from the genome as primary miRNAs and processed into the final single-stranded mature miRNAs through a series of intermediates by biogenesis machinery. Mature miRNAs are then incorporated into RNA-induced silencing complex where miRNAs bind to their target mRNAs and result in mRNA destabilization and/or translational repression (7, 8). In animals, the 5'-proximal seed region (at nucleotides 2–8) of miRNAs binds to complementary sequences within the 3'-untranslated region (3'UTR) of the target mRNA (9). It is estimated that more than half of the protein coding genes in mammals can be regulated by miRNAs (10), thus miRNAs can participate in a series of cellular processes including DNA replication and reparation, cell proliferation and differentiation, and ontogenesis (11, 12, 13). In addition, miRNAs are also involved in the repertoire of virus–host interactions and affect viral replication (14, 15, 16).
During virus infection, type Ⅰ interferons (IFNs) with antiviral activity, such as IFN-α and IFN-β, are produced by fibroblasts and monocytes (17). Once released, type I IFNs bind to their cognate receptors on target cells, which activate the Jak-STAT signaling pathway to induce transcription of IFN-stimulated genes (ISGs) (18, 19, 20). Recent evidence reveals that miRNAs can regulate the replication of several viruses through managing the production of IFNs and ISGs (21, 22). Likewise, a few miRNAs, such as miRNA-23, miR-26a, miR-30c, and miRNA-373, attribute to modulate PRRSV replication by targeting IFN or its signaling pathways (23, 24, 25). Since the details of miRNA-mediated regulation of viral replication have just begun to emerge, a comprehensive investigation of their roles in PRRSV pathogenesis will contribute to a better understanding of host–pathogen interactions.
In the present study, we obtained the differently expressed miRNA profiles by deep sequencing of HP-PRRSV-infected alveolar macrophages. Based on the screening data, we have investigated miR-218 induction during PRRSV infection in vitro and in vivo. We found that miR-218 negatively regulates PRRSV replication by targeting SOCS3, thus clarifying one of the molecular mechanisms underlying PRRSV pathogenesis.
Results
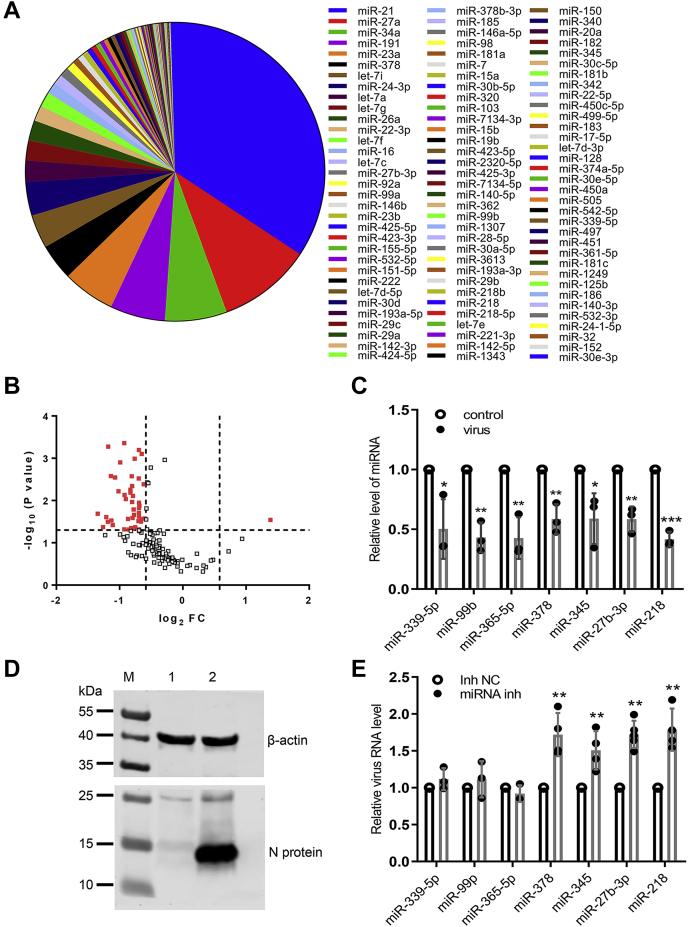
PRRSV infection alters miRNA profiles of PAMs
To investigate the miRNAs involved in the host response against PRRSV infection, we utilized a high-throughput deep sequencing to profile miRNA in porcine alveolar macrophages (PAMs) at 24 h postinfection (hpi) of a HP-PRRSV strain HuN4. Mock-treated PAMs were used as control. The top 100 expressed miRNAs obtained from mock-treated PAMs are shown in Figure 1A. Comparing with previously reported miRNAs related to PRRSV infection, most of them are present in this figure, such as miR-23, miR-378, miR-26a, miR-22, let-7f, miR-29a, miR-181, miR-30c, miR-505, and miR-125b (26), suggesting that our deep sequencing data are reliable. After virus treatment, most miRNAs were downregulated, as shown in the volcano plot (Fig. 1B). Among them, seven miRNAs including miR-339-5p, miR-99b, miR-365-5p, miR-378, miR-345, miR-27b-3p, and miR-218 all ranked inside the top 100 and were significantly downregulated (Fig. 1C). Like our findings, Zhen et al. reported that miR-378 was downregulated upon PRRSV infection as well (27). The PRRSV replication in PAMs was confirmed by western blot analysis (Fig. 1D). Next, the effect of these seven miRNAs on PRRSV infection was explored by blocking their endogenous expression using specific inhibitors. PAMs were transfected with each of seven miRNA inhibitors or negative control for 24 h, and cells were then inoculated with PRRSV at an MOI of 0.1 for 24 h. Relative quantitative PCR (qPCR) results showed that inhibition of miR-378, miR-345, miR-27b-3p, or miR-218 had significantly increased the expression levels of viral RNA (Fig. 1E), indicating that these four miRNAs negatively regulate PRRSV replication. Consistent with our data, miR-378 has been reported to be an effective suppressor against PRRSV replication (27).
Figure 1.
Expression profiles of miRNA in PAMs upon PRRSV infection.A, top 100 most abundant miRNAs in PAMs. B, Volcano plot of differential expression patterns of miRNA in PAMs after HP-PRRSV HuN4 infection. The vertical lines correspond to 1.5-fold up and down, respectively, and the horizontal line represents a p value of 0.05. The red dots in the plot represent the differently expressed miRNA with statistical significance. C, relative expression of seven differently expressed miRNAs. D, Western blot analysis confirmed PRRSV HuN4 infection in PAMs. PAMs were infected with HuN4 at a MOI of 0.1 for 24 h. Cell lysates were subjected to western blot assay with indicated antibodies. M, marker; Lane 1, control-treated PAMs; Lane 2, HuN4-treated PAMs. E, PAMs were transfected with the inhibitor of each of miR-339-5p, miR-99b, miR-365-5p, miR-378, miR-345, miR-27b-3p, miR-218, or negative control. At 24 h posttransfection, cells were infected with PRRSV HuN4 at a MOI of 0.1 for 24 h. The levels of viral RNA were determined with qPCR. Data are presented as the mean ± SD. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. The p value was calculated using Student’s t-test (two-tailed).
PRRSV replication-mediated miR-218 downregulation facilitates PRRSV replication in PAMs
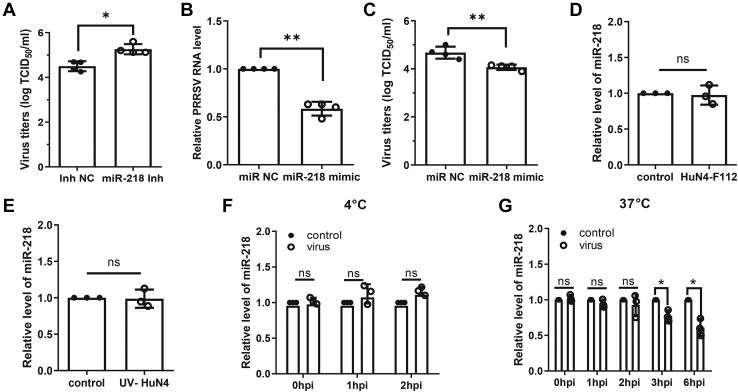
Since the miR-218 was significantly downregulated upon virus infection and the miR-218 inhibitor resulted in an increase in viral RNA, we further investigated the regulatory function of miR-218 inhibitor on PRRSV replication. The virus titers of collected cell supernatants were measured by TCID50 assay, and the result showed that treatment with miR-218 inhibitor resulted in a marked increase in PRRSV titers in contrast to treatment with miRNA negative control (Fig. 2A). Meanwhile, the commercial miRNA mimic was used to overexpress miR-218 in PAMs. At 24 h post miR-218 mimic transfection, PAMs were infected with PRRSV for an additional 24 h. The qPCR results showed that overexpression of miR-218 significantly reduced viral RNA load compared with the negative control (Fig. 2B). The results of TCID50 assay also displayed that miR-218 overexpression led to a significant decrease in viral titers (Fig. 2C). No toxicity of miR-218 mimic at concentrations of <150 nM was evident for PAMs (Fig. S1). These findings indicate that miR-218 has the potential to inhibit PRRSV replication in PAMs.
Figure 2.
PRRSV-mediated miR-218 downregulation facilitates PRRSV replication in PAMs.A, miR-218 inhibitor increases PRRSV titers. PAMs were transfected with miR-218 inhibitor or inhibitor negative control (inh NC) for 24 h, and cells were infected with HP-PRRSV HuN4 strain at an MOI of 0.1 for 24 h. Viral titers of supernatants were evaluated with TCID50. B and C, miR-218 mimic represses PRRSV replication. PAMs were transfected with miR-218 mimic or miRNA negative control (miR NC) for 24 h, and cells were then inoculated with HuN4 at an MOI of 0.1 for 24 h. Viral RNA levels were determined by qPCR (B) and virus titers were determined by TCID50 (C). D, PRRSV low-virulent strain HuN4-F112 does not induce the downregulation of miR-218. PAMs were inoculated with virus for 24 h. Total RNA was extracted, and the miR-218 level was determined by qPCR. E, miR-218 is not reduced upon inactivated PRRSV HuN4 incubation. PAMs were inoculated with UV-inactivated HuN4 for 24 h. The miR-218 level was determined by qPCR. F, virus attachment does not affect miR-218 expression. PAMs were incubated with virus at 4 °C for indicated time points. The miR-218 level was determined by qPCR. G, viral replication is required for miR-218 downregulation. PAMs were incubated with virus at 37 °C for indicated time points. The miR-218 level was determined by qPCR. Data are presented as the mean ± SD. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. The p value was calculated using two-way ANOVA with Bonferroni’s posttest.
To determine whether low-virulent PRRSV has a similar effect on miR-218 expression, PAMs were treated with attenuated live PRRSV vaccine strain HuN4-F112, which is a Marc-145-adaptive strain and cannot efficiently replicate in PAMs (28). The result showed that the expression level of miR-218 was not reduced in HuN4-F112-treated PAMs compared with the control group (Fig. 2D). Similarly, UV-inactivated HuN4 did not result in the decrease of miR-218 expression (Fig. 2E), indicating that virus replication is required for the downregulation of miR-218. Furthermore, we identified the dynamic changes of miR-218 expression at different stages of virus infection. As shown in Figure 2F, the virus attachment did not affect miR-218 expression; as shown in Figure 2G, the downregulation of miR-218 occurred after the virus entered the cell and during its biosynthesis (after two hpi). Altogether, our findings indicate that virus replication-mediated miR-218 downregulation facilitates PRRSV replication.
miR-218 positively links to the antiviral responses
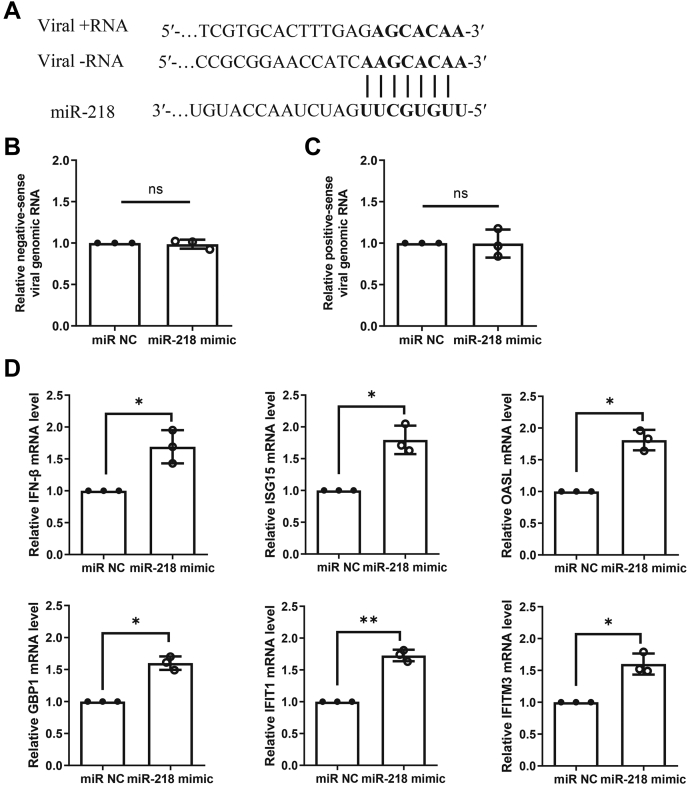
Previous studies have demonstrated that miRNA-mediated viral regulation can happen by directly targeting the viral genome or by targeting host genes involved in viral replication (29, 30). During the replication cycle of PRRSV, the positive-sense RNA genome is transcribed into an intermediate, negative-sense RNA molecule, which functions as templates for the synthesis of positive-sense genomic RNA (31). Thus, to uncover the potential function of miR-218 during PRRSV replication, we compared the miR-218 seed sequence with PRRSV genomic RNA and found that there were at least two paired regions between miR-218 and negative/positive-sense viral RNA (Fig. 3A). To test whether miR-218 directly targets viral RNA and acts as a direct antiviral factor, we infected PAMs with PRRSV for 2 h and then transfected miR-218 mimic for 8 h and followed by detecting its effect on viral RNAs. As shown in Figure 3, B and C, overexpression of miR-218 did not affect the expression levels of negative-sense genomic RNA and positive-sense genomic RNA, suggesting that miR-218 regulates PRRSV replication not through directly targeting viral genomic RNA.
Figure 3.
miR-218 does not directly interact with viral genomic RNA but alters type I IFN signaling pathway.A, diagram of the predicted target site for miR-218 in PRRSV genomic RNA. In this model, we hypothesized that miR-218 could inhibit PRRSV replication by either targeting nine nucleotides in viral negative-sense genomic RNA and or targeting seven nucleotides in viral positive-sense genomic RNA as indicated. B and C, miR-218 mimic does not affect the levels of viral genomic RNA. PAMs were incubated with PRRSV for 2 h, and miR-218 mimic or negative control was then transfected for 8 h. The relative levels of viral negative-sense and positive-sense genomic RNA were determined by qPCR. D, overexpression of miR-218 enhances type I IFN signaling pathway. PAMs were transfected with miR-218 mimic or negative control for 24 h. Total RNA was extracted, and the mRNA levels of IFN-β, ISG15, OASL, GBP1, IFIT1, and IFITM3 were determined by qPCR. Data are presented as the mean ± SD. ∗, p < 0.05; ∗∗, p < 0.01. The p value was calculated using two-way ANOVA with Bonferroni’s posttest. OASL, 2′-5′-oligoadenylate synthetase-like protein.
Given that miR-218 can regulate PRRSV replication in PAMs, it is worthwhile to further investigate the molecular mechanisms. Type I IFN is the key innate immune cytokine produced in large quantities by cells to trigger antiviral function (32), we next determined whether miR-218 could modify innate immune response pathways of type I IFN. PAMs were treated with miR-218 mimic for 24 h, and cells were then collected for detecting the transcriptional levels of IFN-β and several ISGs. The results showed that the mRNA levels of IFN-β and several ISGs including ISG15, 2′-5′-oligoadenylate synthetase-like protein (OASL), GBP1, IFIT1, and IFITM3 were increased in the presence of miR-218 mimic compared with the negative control (Fig. 3D). These findings suggest that miR-218 regulates PRRSV replication through modulating type I IFN response.
miR-218 negatively regulates the expression of porcine SOCS3
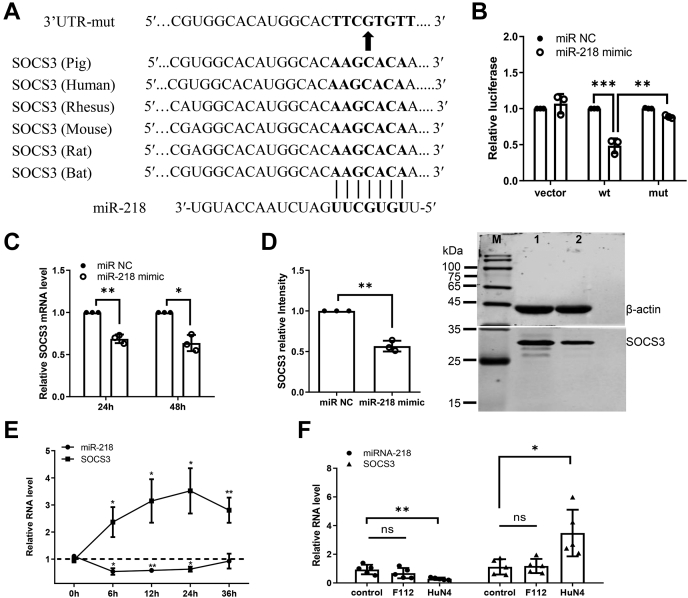
To explore the molecular mechanism by which miR-218 regulates type I IFN response, TargetScan and miRDB were used to determine the potential target gene of miR-218. One putative binding site for miR-218 was identified in the 3’UTR of porcine suppressor of cytokine signaling 3 (SOCS3), a critical regulator of infection and inflammation (33). The pairing region between the miR-218 seed sequence and SOCS3 3'UTR seed binding site was shown in Figure 4A. To validate the direct association between SOCS3 and miR-218, a region of SOCS3 3'UTR containing the predicted target site of miR-218 was cloned into a dual-luciferase reporter vector pmirGLO, and the same 3'UTR region with mutations in the seed binding site was also constructed into the same vector to obtain the mutant plasmid (Supplementary material S1). HEK-293 T cells were cotransfected with each of an empty vector, wild-type SOCS3 3'UTR plasmid or mutant plasmid in the presence of miR-218 mimic or miRNA negative control for 48 h. The results showed that miR-218 mimic significantly inhibited the luciferase activity of wild-type SOCS3 3'UTR plasmid-transfected cells relative to miRNA control. In contrast, transfection with mutant SOCS3 3'UTR abolished the inhibition effect of miR-218 mimic on the luciferase activity (Fig. 4B), suggesting that miR-218 can directly interact with SOCS3 3’UTR. To further verify the regulatory effect of miR-218 on SOCS3, PAMs were transfected with miR-218 mimic for 24 h and 48 h, and the mRNA and protein expression levels of SOCS3 were determined. The qPCR results showed that SOCS3 mRNA level was significantly reduced following the transfection with miR-218 mimic (Fig. 4C). Western blot analysis also revealed that treatment with miR-218 mimic markedly reduced SOCS3 protein expression (Fig. 4D). These data demonstrate that miR-218 negatively regulates SOCS3 expression at the transcriptional level by targeting SOCS3 3’UTR.
Figure 4.
miR-218 negatively regulates the expression of porcine SOCS3.A, diagram of the predicted miR-218 binding sites in SOCS3 3'UTR of different species. miR-218 seed regions and mutated miR-218 are shown in bold black. B, miR-218 directly targets SOCS3 3'UTR. HEK-293T cells were cotransfected with either miR-218 mimic or negative control, along with empty vector, SOCS3 3'UTR wild-type, or mutant-type reporter plasmids. After 48 h, transfected cells were collected and lysed for dual-luciferase reporter assay. C, miR-218 mimic decreases the levels of SOCS3 mRNA. PAMs were transfected with miR-218 mimic for 24 h and 48 h respectively, and the SOCS3 mRNA levels were determined by qPCR. D, miR-218 mimic results in decreased levels of SOCS3 protein. At 48 h posttransfection with miR-218 mimic or negative control, PAMs were lysed and subjected to western blot. M, marker; Lane 1, negative control siRNA-transfected PAMs; Lane 2, SOCS3 siRNA-transfected PAMs. The densitometric analysis was performed using Image J. E, the negative correlation between miR-218 and SOCS3 upon PRRSV infection in vitro. PAMs were infected with PRRSV strain HuN4 and were sampled at different time points as indicated. Total RNA was extracted to detect the expression levels of miR-218 and SOCS3 mRNA using qPCR. F, the correlation between miR-218 and SOCS3 upon PRRSV inoculation in vivo. qPCR analysis was used to determine the miR-218 and SOCS3 RNA level in lung samples from pigs inoculated with mock, vaccine strain HuN4-F112, or HP-PRRSV strain HuN4 for 21 days. Data are presented as the mean ± SD. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. The p value was calculated using two-way ANOVA with Bonferroni’s posttest.
Next, we determined whether there is a correlation between miR-218 and SOCS3 during PRRSV infection in vitro and in vivo. After incubating PAMs with mock or PRRSV for different time points, the expression levels of miR-218 and SOCS3 were assessed by qPCR. As shown in Figure 4E, the expression levels of miR-218 decreased within 24 hpi. Whereas the expression levels of SOCS3 significantly increased at 6 hpi and reached a peak at 24 hpi. These results suggest that the expression of SOCS3 is negatively correlated with miR-218 during PRRSV infection in vitro. To confirm the in vitro data, in vivo samples were also examined. Total RNA was extracted from the lung of pigs that were inoculated with mock, vaccine strain HuN4-F112, and HP-PRRSV strain HuN4 for 21 days as previously described (28). The expression levels of miR-218 and SOCS3 were determined by qPCR. The levels of miR-218 were significantly lower in HuN4-inoculated pigs compared with mock controls and HuN4-F112 pigs. On the contrary, infection by HuN4 resulted in an increase in SOCS3 expression levels in comparison with the control group or HuN4-F112 group (Fig. 4F). Taken together, these results demonstrate that virus-induced miR-218 downregulation leads to an increase of SOCS3 expression, which might impair the antiviral response to facilitate PRRSV replication.
SOCS3 augments PRRSV replication by blocking type I IFN signaling pathway
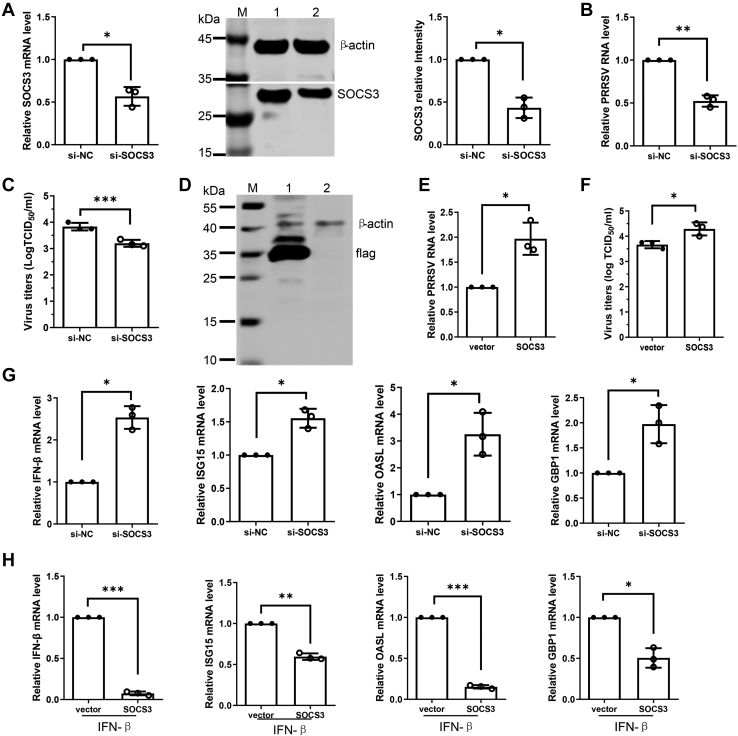
Moreover, the effect of SOCS3 on PRRSV replication was investigated using genetic modification methods. The SOCS3 mRNA level in PAMs transfected with SOCS3-specific siRNA was significantly decreased relative to control siRNA. Western blot analysis of detergent lysates collected from cells transfected with SOCS3 siRNA also revealed a clear reduction in the level of SOCS3 protein (Fig. 5A). At 24 h post siRNA transfection, PAMs were inoculated with PRRSV for an additional 24 h. We observed that the viral RNA levels were greatly decreased in the SOCS3-specific transfection group compared with the control siRNA group (Fig. 5B). Knockdown of endogenous SOCS3 with siRNA also reduced virus titer as measured by TCID50 assay (Fig. 5C). To confirm siRNA data, another SOCS3-specific siRNA was introduced to repeat the above experiments. The results showed that the SOCS3 siRNA resulted in the decrease of virus RNA level and titers (Fig. S2). To further confirm the effect of SOCS3 on PRRSV replication, we tried to overexpress SOCS3 in primary PAMs, but the expression level of SOCS3 was not detectable (data not shown), which might be due to low transfection efficiency. Therefore, the immortalized PAMs were used to overexpress SOCS3, and the cells were then infected with PRRSV. Western blot analysis confirmed that SOCS3 was overexpressed successfully in the immortalized PAMs (Fig. 5D). As shown in Figure 5E, SOCS3 overexpression increased the levels of viral RNA in contrast to the vector control. TCID50 assay results confirmed the positive effect of SOCS3 overexpression on virus titer in comparison with the vector control (Fig. 5F). These data indicate that SOCS3 negatively regulates PRRSV replication.
Figure 5.
SOCS3 promotes PRRSV replication by blocking type I IFN signaling pathway.A, verification of SOCS3 knockdown efficiency. Primary PAMs were transfected with SOCS3-specific siRNA (si-SOCS3) or negative control (si-NC) for 24 h, and the knockdown efficiency of SOCS3 was determined by qPCR and western blot. M, marker; Lane 1, negative control siRNA-treated PAMs; Lane 2, SOCS3 siRNA-treated PAMs. B and C, depletion of endogenous SOCS3 decreases PRRSV replication. After siRNA transfection for 24 h, PAMs were infected with PRRSV for 24 h. The effect of SOCS3 knockdown on PRRSV replication was determined by qPCR (B) and TCID50 assay (C). D, verification of SOCS3 overexpression. Immortalized PAMs were transfected with pCAGGS-SOCS3 or vector control for 24 h, and cell lysates were subjected to western blot to detect SOCS3 expression. M, marker; Lane 1, pCAGGS-SOCS3-transfected PAMs; Lane 2, vector control-transfected PAMs. E and F, overexpression of SOCS3 increases PRRSV replication. After pCAGGS-SOCS3 transfection for 24 h, the immortalized PAMs were exposed to PRRSV for 24 h. The levels of viral RNA were determined by qPCR (E), and virus titers were detected by TCID50 (F). G, knockdown SOCS3 expression increases the expression of several ISGs. Primary PAMs were transfected with control or SOCS3 specific siRNA for 24 h. Total RNA was extracted, and the mRNA levels of IFN-β, ISG15, OASL, and GBP1 were determined by qPCR. H, SOCS3 overexpression results in the decreased expression of several ISGs. Immortalized PAMs were transfected with vector control or pCAGGS-SOCS3 for 24 h and then treated with IFN-β (20 ng/ml) for 24 h. Total RNA was extracted, and the mRNA levels of IFN-β, ISG15, OASL, and GBP1 were determined by qPCR. Data are presented as the mean ± SD. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. The p value was calculated using two-way ANOVA with Bonferroni’s posttest. OASL, 2′-5′-oligoadenylate synthetase-like protein.
Since miR-218 negatively regulates type I IFN response, we assessed the effect of SOCS3 on the signaling pathway of type I IFN by modifying SOCS3 expression in PAMs. Here, the mRNA levels of IFN-β and several ISGs including ISG15, OASL, and GBP1 were analyzed by qPCR. The results showed that depletion of endogenous SOCS3 expression significantly enhanced the mRNA levels of IFN-β and three ISGs, ISG15, OASL, and GBP1, in contrast to the control siRNA treatment (Figs. 5G and S3). To confirm the effect of SOCS3 on type I IFN signaling pathway, the immortalized PAMs were transfected with a eukaryotic expression plasmid containing SOCS3 and followed by IFN-β stimulation. We observed that the mRNA levels of IFN-β, ISG15, OASL, and GBP1 were significantly decreased in SOCS3 transfection group compared with the control group (Fig. 5H). Overall, these findings suggest that PRRSV induced-miR-218 downregulation upregulates SOCS3 expression, which may serve as a negative regulator of type I IFN response to promote PRRSV replication.
Antiviral function of miR-218 is mainly through targeting SOCS3
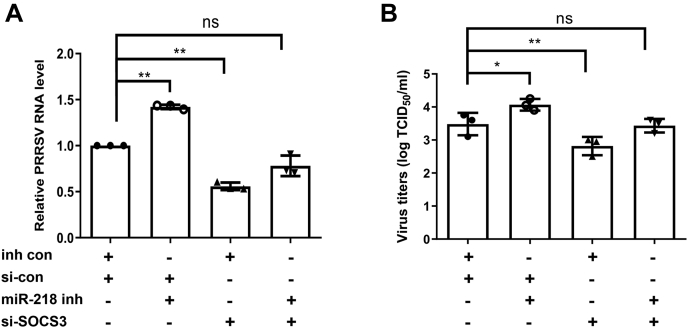
To elucidate the role of SOCS3 in the antiviral function of miR-218, we carried out miR-218 and SOCS3 RNA interference knockdown experiments. We confirmed that inhibition of miR-218 effectively promoted PRRSV replication in PAMs, as shown by viral RNA and virus titer. Whereas depletion of SOCS3 by specific siRNA remarkably suppressed PRRSV replication (Fig. 6, A and B). When the miR-218 was inhibited by its inhibitor in SOCS3-depleted cells, the increase of both viral RNA and virus titer by miR-218 inhibitor was blocked by the depletion of SOCS3 expression (Fig. 6, A and B). These results indicate that SOCS3 is beneficial for PRRSV replication, and the virus-induced miR-218 downregulation impairs the host antiviral response through the SOCS3-mediated signaling pathway.
Figure 6.
PRRSV-induced miR-218 downregulation facilitates PRRSV replication via SOCS3-mediated signaling. PAMs were transfected with control siRNA (si-con) or SOCS3 siRNA (si-SOCS3) for 24 h and followed by transfection with miR-218 inhibitor (miR-218 inh) or inhibitor control (inh con) for 24 h. The cells were then infected with PRRSV for an additional 24 h. The levels of viral RNA were determined by qPCR (A), and virus titers were detected by TCID50 (B). Data are presented as the mean ± SD. ∗, p < 0.05; ∗∗, p < 0.01. The p value was calculated using two-way ANOVA with Bonferroni’s posttest.
miR-218 negatively regulates PRRSV replication in Marc-145 cells
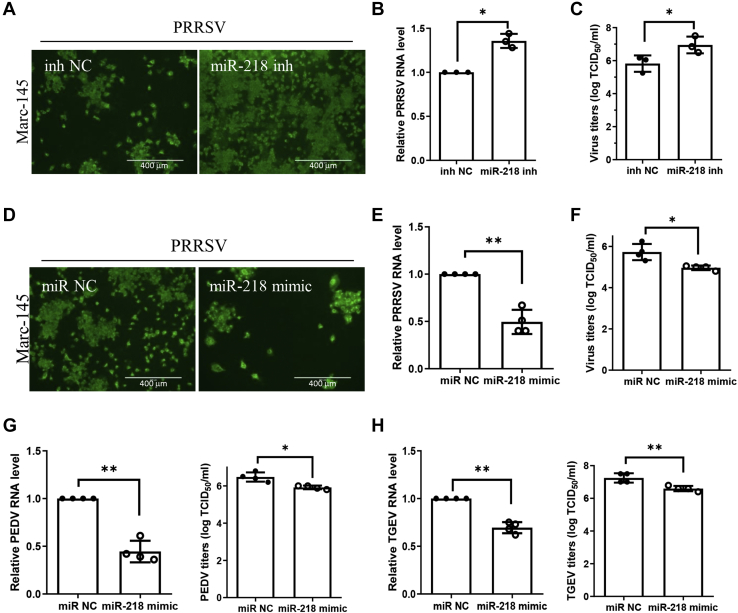
To verify the negative regulation of PRRSV replication by miR-218, we examined the antiviral effect of miR-218 in Marc-145 cells. Marc-145 cells were transfected with miR-218 inhibitor for 24 h, and cells were then infected with PRRSV at an MOI of 0.1 for 24 h. Cell monolayers were fixed for IFA analysis, and we observed notably more PRRSV-positive cells with miR-218 inhibitor than the control group (Fig. 7A). The levels of viral RNA were increased in miR-218 inhibitor-treated group compared with the control group (Fig. 7B). Transfection with miR-218 inhibitor consequently augmented viral titers as measured by TCID50 (Fig. 7C). Similarly, when Marc-145 cells were transfected with miR-218 mimic, the number of PRRSV-positive cells was lower in miR-218 mimic-treated cells than in control-treated cells (Fig. 7D). The viral RNA levels were significantly decreased with miR-218 mimic treatment (Fig. 7E), and the reduced titers of virus in infected cell cultures containing miR-218 mimic were confirmed by measuring TCID50 (Fig. 7F). These data indicate that suppression of PRRSV replication by miR-218 in Marc-145 cells is consistent with that in PAMs, suggesting that the evolutionarily conserved miR-218 might have a similar function in antiviral effect in different cell lines.
Figure 7.
miR-218 may have a broad-spectrum antiviral property.A–C, knockdown endogenous miR-218 facilitates PRRSV replication in Marc-145 cells. Marc-145 cells were transfected with miR-218 inhibitor or control for 24 h before they were inoculated with PRRSV HuN4. At 24 h after inoculation, the cell monolayers were fixed and examined for virus infection by IFA (scale bar = 400 μm) (A); the levels of viral RNA were determined by qPCR (B); and virus titers were evaluated with TCID50 (C). D–F, miR-218 mimic inhibits PRRSV replication in Marc-145 cells. Marc-145 cells were transfected with miR-218 mimic or negative control for 24 h, and cells were then inoculated with PRRSV for 24 h. PRRSV infection was examined by IFA (scale bar = 400 μm) (D), and viral RNA levels were determined by qPCR (E), and virus titers were evaluated with TCID50 (F). G, miR-218 overexpression decreases PEDV replication. Vero E6 cells were transfected with miR-218 mimic for 24 h, and cells were then inoculated with PEDV for 24 h. The viral RNA levels were determined by qPCR and virus titers were evaluated with TCID50. H, miR-218 overexpression decreases TGEV replication. ST cells were transfected with miR-218 mimic for 24 h, and cells were then infected with TGEV for 24 h. The levels of viral RNA were determined by qPCR and virus titers were evaluated with TCID50. Data are presented as the mean ± SD. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. The p value was calculated using two-way ANOVA with Bonferroni’s posttest.
miR-218 may have a broad-spectrum antiviral property
Next, we evaluated the antiviral effect of the evolutionarily conserved miR-218 in two other members of the order Nidovirales, PEDV (porcine epidemic diarrhea virus) and TGEV (transmissible gastroenteritis virus). Vero E6 cells were transfected with miR-218 mimic for 24 h, and cells were then inoculated with PEDV at an MOI of 0.1 for another 24 h. Cells and supernatant were harvested for detecting viral RNA levels by qPCR and viral titers by TCID50 respectively. The results showed that overexpression of miR-218 significantly inhibited PEDV replication as indicated by RNA levels and viral titers (Fig. 7G). ST cells were also treated with miR-218 mimic as above, and cells were infected with TGEV at an MOI of 0.1 for 24 h. As shown in Figure 7H, miR-218 overexpression also reduced TGEV loads as measured by qPCR and TCID50. These results indicate that the evolutionarily conserved miR-218 may have a broad-spectrum antiviral property.
Discussion
The first line of host defense against viruses is the innate immune system, in which IFNs are glycoproteins with strong antiviral and immunomodulatory activities. The published data suggest that PRRSV is susceptible to IFNs in vivo and in vitro (34, 35). To establish effective infection, viruses must circumvent the powerful IFN responses by modulating or evading these defenses (36). PRRSV, like other viruses, has evolved numerous strategies to interfere with the different steps of initial antiviral host responses and establish persistent infection (37, 38, 39). Emerging evidence has shown that miRNAs play key roles in regulating the inflammatory process and immune responses, see reviews (40, 41, 42). Here, our deep sequencing data indicate that the abundant miRNAs such as miR-21, miR-27a, miR-34, miR-191, and miR-23a are highly associated with innate immune responses in porcine macrophages (43, 44). During PRRSV infection, we observed that the expression levels of numerous miRNAs were modulated. Analysis with miRNA inhibitors revealed that four out of seven downregulated miRNAs including miR-378, miR-345, miR-27b-3p, and miR-218, would augment PRRSV replication, suggesting that viruses have evolved to modify host miRNAs to evade antiviral immune responses, leading to enhanced viral replication and pathogenesis. Among them, miR-378 had been reported to be a negative regulator for PRRSV replication (23).
miR-218 was first described in prostate cancer and gastric cancer (45, 46) and has been shown to serve as a tumor suppressor by inhibiting glioblastoma invasion, migration, and proliferation in different types of cancer (47). Recently, miR-218 has been shown to contribute to other processes, including apoptosis and inflammation response (48, 49). Last year, Li et. al revealed the critical regulatory role of cellular miR-218 in the pathogenesis of Kaposi’s sarcoma-associated herpesvirus infection (50). Therefore, we further examined the regulatory role of miR-218 on PRRSV replication by using either miRNA mimic or inhibitor in PAMs. Interestingly, like miR-378, miR-218 was also found as an effective PRRSV replication suppressor (Fig. 1). Additionally, we observed that PRRSV vaccine strain HuN4-F112 and UV-inactivated HP-PRRSV HuN4 could not significantly reduce miR-218 expression, and the decreased expression of miR-218 only occurred after virus entered into the cells, indicating that PRRSV replication is required for miR-218 downregulation. All these data suggest that PRRSV replication induces the downregulation of miR-218 to facilitate virus replication.
Next, we explored the mechanisms underlying the effects of miR-218 on PRRSV replication. The regulatory role of miRNA within a cell is to manipulate the protein levels via direct binding to mRNA and influencing translation efficiency or mRNA abundance. Meanwhile, miRNAs can also affect virus replication and pathogenesis by either direct targeting of viral RNA genome or targeting to host transcriptome. Thus, we investigated the effect of miR-218 on the expression of PRRSV genomic RNA including positive-sense RNA and negative-sense RNA (51). Even though PRRSV genomic RNA harbored at least two miR-218 binding sites, overexpression of miR-218 did not alter viral RNA abundance or stability (Fig. 3). The possible reason that miR-218 is unable to bind and inhibit viral replication could be (1) viral genome is not functionally sensitive to miR-218-guided repression (2); miR-218-viral genome interaction is not a positive balance of minimum free energy (3); the other factors might affect the thermodynamic state of the miR-218–viral genome interaction. These findings indicate that the mechanism by which miR-218 affects PRRSV replication might be through targeting the host target gene.
Based on the bioinformatic analysis results, we found SOCS3 as the putative target gene of miR-218. This may explain the probable actions of SOCS3 in the regulation of immunity and inflammation. Therefore, we hypothesized that miR-218 binds to SOCS3 mRNA in PAMs to repress SOCS expression and, thus, indirectly regulate PRRSV replication. In this study, we found that miR-218 can directly bind to porcine SCOS3 3’UTR and thus decrease its expression in PAMs (Fig. 4). Additionally, the expression of cellular miR-218 in PRRSV-infected PAMs was decreased, while the SOCS3 expression was increased. Further, to establish in vivo correlation between miR-218 and SOCS3, we evaluated the expression of miR-218 and SOCS3 in PRRSV-infected lung samples by qPCR. Consistent with the in vitro results, PRRSV infection induced significantly lower expression of miR-218 and higher expression of SOCS3 in porcine lungs. Based on our data, we demonstrated that the increased expression of SOCS3 following PRRSV infection is mediated by the downregulated miR-218.
In human, SOCS3 was associated with inflammation and Jak/STAT-mediated signaling pathway (52). Thus, we examined the regulatory function of SOCS3 in anti-PRRSV activities in porcine cells. Our results demonstrate that virus-mediated upregulation of SOCS3 inhibits type I IFN signaling pathway, facilitating PRRSV replication in PAMs (Fig. 5). Although our data have shown that the downregulation of miR-218 upon PRRSV infection can mediate the upregulation of its target gene SOCS3, we needed direct evidence to confirm our hypothesis. We found that the augmentation effect of the miR-218 inhibitor on virus replication was inhibited by SOCS3 knockdown treatment (Fig. 6). These data suggest that SOCS3 is the target effector of miR-218 that negatively regulates the antiviral activity of type I IFN, indicating that SOCS3 may allow PRRSV to evade the protective innate immune response within host cells.
Since the regulatory role of miR-218 on PRRSV replication in Marc-145 cells was also confirmed, we proposed that miR-218 might have a broad-spectrum antiviral property. Two porcine coronaviruses, PEDV and TGEV, were selected since these viruses belong to the same order Nidovirales as PRRSV. As expected, miR-218 overexpression inhibited both PEDV and TGEV replication (Fig. 7), suggesting that miR-218 has a broad-spectrum antiviral activity. However, further studies are required to test this hypothesis. Similar to our findings, other miRNAs such as miR-124, miR-24, and miR-744 have also been reported to have a broad-spectrum antiviral target by regulating p38 MAPK signaling (53).
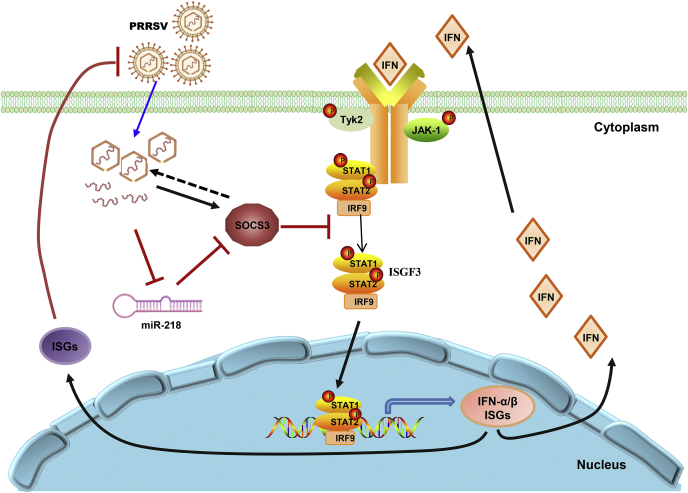
In summary, we used a high-throughput deep sequencing and inhibitors to screen miRNAs that have an inhibitory effect on the replication of PRRSV. The identification of the miR-218 as a potent antiviral miRNA with some inhibitory effects that extend beyond the Arteriviridae led us to investigate the role of this miRNA in influencing the host–pathogen interface. Our findings have shown that the downregulation of miR-218 augments PRRSV replication by upregulating the SOCS3 signaling pathway, which suppresses the antiviral activity of type I IFN (Fig. 8). Although the signaling intermediates between PRRSV and miR-218 remain to be elucidated, we have demonstrated that PRRSV infection results in the downregulation of miR-218 in vitro and in vivo. In conclusion, we have uncovered a novel mechanism in which PRRSV uses miR-218 to suppress cellular antiviral defenses, which may present a potential therapeutic target against PRRSV replication.
Figure 8.
A schematic representation of the proposed model for the mechanism by which miR-218 assists PRRSV in evading the host innate immune response. PRRSV infection results in a decrease of miR-218 that negatively regulates SOCS3 expression. The upregulated SOCS3 can block the interferon signaling pathway. This establishes a model by which PRRSV-induced miR-218 downregulation augments virus replication via SOCS3-mediated suppression of ISGs expression.
Experimental procedures
Cells and viruses
Primary PAMs were obtained from 5-week-old specific pathogen-free pigs. The immortalized PAM cell line was a gift from Dr Yan-dong Tang (54). Both primary and immortalized PAMs were cultured in RPMI-1640 medium (Gibco, USA), supplemented with 10% fetal bovine serum (FBS, Hyclone, USA). Marc-145, Vero-E6, ST, and HEK-293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco, USA) supplemented with 10% FBS. The HP-PRRSV strain HuN4 (GenBank no. EF635006) and low-virulent strain HuN4-F112 (GenBank No. MG972944) were grown and titrated in Marc-145 cells. The PEDV strain CV777 (GenBank no. KT323979) was propagated and titrated in Vero E6 cells. The TGEV strain H165 (GenBank no. EU074218) was titrated in ST cells.
miRNA deep sequencing and analysis
Primary PAMs freshly isolated from pigs were inoculated with PRRSV strain HuN4 at an MOI of 0.1 or mock control for 24 h. Total RNA was extracted from cells by using Trizol (Sigma-Aldrich, Germany) and digested with DNase I (TaKaRa, Japan) according to the manufacturer's instructions. Sequencing libraries were generated using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (number E7300 L, New England Biolabs). Sequencing was performed using Illumina HiSeq XTen platform. Clean reads were obtained by removing low-quality reads (Q-value≤20), with 5-primer contaminants, without the 3’-primer, without the insert, with poly(A), and shorter than 18 nucleotides from the raw data reads. Clean reads were then aligned with the reference genome, Rfam database 13.0, and RepBase database to discard the degraded fragments of mRNA or other noncoding RNAs, such as rRNA, scRNA, snoRNA, snRNA, tRNA, repeat sequences using Bowtie tools software. Clean reads were searched against miRBase 22.0 to identify known miRNAs in Sus scrofa. Novel miRNAs were predicted to find step-loops or hairpin structures using the miRDeep2 software. The miRNA expression level was calculated and normalized to the total number of reads in each library, respectively. We identified differentially expressed miRNAs with a false discovery rate <0.05 and fold change ≥1.5 threshold. Pairwise differential expression comparisons were made between mock and PRRSV-infected PAMs.
RNA oligo transfection and virus incubation
RNA oligos including miRNA inhibitor, miRNA mimic, and siRNA were synthesized by Genepharma (Suzhou, China). The RNA oligo sequences are listed in Table S1. The miRNA mimic (at a concentration of 50 nM), miRNA inhibitor (50 nM), siRNA (100 nM), and the corresponding negative control were transfected into target cells for 24 h using lipofectamine RNAiMAX reagent (Invitrogen, USA) according to the manufacturer’s instructions.
After all the different indicated treatments, cell monolayers were inoculated with HP-PRRSV HuN4 at an MOI of 0.1 unless otherwise stated. After 60 min of incubation, cell monolayers were washed and then further incubated in fresh culture media for an additional 24 h or indicated time points. The cells and supernatants were then collected for qPCR and virus titer determination separately.
RNA extraction and quantitative PCR
Total RNA was extracted with Trizol according to the manufacturer’s instruction. Reverse transcription was performed with 1 μg total RNA to obtain cDNA via primescript RT reagent with gDNA eraser kit (Cat #RR047 A, TaKaRa, Japan), which contains DNase treatment to eliminate genomic DNA contamination. And stem-loop primers were used for reverse transcription of miRNAs. Quantitative PCR (qPCR) was carried out as previously described (55) by specific primers listed in Table S2. The relative miRNA expression levels were normalized to small nuclear RNA U6, and the relative mRNA levels of other genes were normalized to β-actin. Finally, data were analyzed with the cycle threshold (ΔΔCt) method relative to the negative control group (56).
TCID50 assay
Virus titration was performed as previously described with a slight modification (57). Virus samples were collected and clarified by centrifugation at 8000×g for 10 min prior to virus titration. TCID50 assays were performed on Marc-145 cells for PRRSV, on Vero E6 cells for PEDV, and on ST cells for TGEV following the method of Reed & Muench as previously described (58). Briefly, cell monolayers (104 cells per well) in 96-well plates (Corning, USA) were inoculated with 100 μl tenfold serial dilutions of each virus stock and incubated for 4–5 days prior to observation of the presence of cytopathic effect.
Dual-luciferase reporter assay
A partial sequence of SOCS3 3’UTR, about 100 bp containing the predicated seeding sequence of miR-218, was synthesized and cloned into a dual-luciferase reporter vector, pmirGLO (Promega, USA). This constructed plasmid was named a wild-type reporter plasmid. The same 3'UTR region with mutations in the seed binding site listed in Figure 4 and Supplementary material S1 was synthesized and cloned into the same vector, and this recombinant plasmid was named as mutant-type reporter plasmid. HEK-293 T cells were transfected with either miR-218 mimic or negative control, along with pmirGLO-SOCS3 3′UTR wild-type or mutant-type reporter plasmid using Lipofectamine 2000 (Invitrogen, USA). After cotransfection for 48 h, the cells were lysed with passive lysis buffer, and luciferase reporter gene assays were performed with a dual-luciferase reporter assay kit (Promega, USA) according to the manufacturer’s instructions. Firefly luciferase enzyme activities were normalized to the renilla luciferase activities. Data represent the means ± standard deviations (SD) from three independent experiments.
Plasmids construction and transfection
Porcine SOCS3 protein entire coding sequence was amplified from PAMs cDNA using the primers listed in Table S2 and then cloned into the eukaryotic expression vector pCAGGS with C-terminal-flag. The nucleotide sequence of SOCS3 was determined to ensure that the correct clone was used in this study. The plasmid was extracted using the Endotoxin-Free Plasmid DNA Miniprep Kit (Tiangen, China) according to the manufacturer's instructions. The plasmid carrying SOCS3 at a final concentration of 1 μg/ml was transfected into cells with X-tremeGENE HP DNA transfection reagent (Roche, Switzerland) as recommended by the manufacturer. The vector pCAGGS was used as the negative control. At 24 h posttransfection, the cells were infected with PRRSV, followed by the indicated analysis.
Animal infection model
Fifteen 5-week-old SPF pigs were randomly divided into three groups, five piglets for each group. Piglets in group 1 were the infection control. Piglets in group 2 were inoculated intramuscularly each with 105.0 TCID50 PRRSV vaccine strain (HuN4-F112). Piglets in group 3 were infected with PRRSV (104.0 TCID50 per pig) intramuscularly (2 ml) and intranasally (2 ml). The animals were kept in separate rooms. Clinical signs and fever were recorded daily. All the animals were euthanized 21 days postchallenge.
Western blot
Western blot assays were performed as previously described (59). In brief, cells were lysed by RIPA Lysis Buffer (Haigene, China) containing nuclease and protease inhibitors. Cell lysates (50 μg per well) were separated by 12% SDS-PAGE gel under reducing conditions and transferred to a PVDF membrane (Merck Millipore, USA). Membranes were incubated with the indicated primary antibodies and appropriate secondary antibodies. The mouse anti-SOCS3 mAb was purchased from Santa Cruz (sc-73045) and used at 1:200. The mouse anti-FLAG mAb (F-1804, Sigma) was diluted at 1:1000, and the mouse anti-β-actin mAb (A2228, Sigma) was used at 1:3000. The mouse anti-PRRSV nucleocapsid (N) protein mAb was produced and purified in our laboratory and was diluted at 1:10,000. The IRDye 680 conjugated goat anti mouse IgG was from Li-Cor Biosciences and used at 1:10,000. Finally, the membranes were scanned with Odyssey infrared imaging system (Li-Cor Biosciences, USA). The densitometric analysis was performed by Image J. The expression levels of indicated proteins were normalized by comparison with β-actin.
Immunofluorescence assay
IFA was carried out as described previously with slight modification (60). Marc-145 cells were plated into 12-well plates at 5✕105 cells/well and cultured overnight. Once the treatment was done as indicated, Marc-145 cells were fixed with 4% paraformaldehyde for 10 min at 4 °C. After being dried up, the cells were penetrated with 0.1% Triton-X100 for 30 min at room temperature. Cells were then washed twice with PBS and 2% bovine serum albumin was used as a blocking reagent. The cell monolayers were incubated with mAb SDOW17 for PRRSV N protein at 1:500 (Rural Technologies, USA) for 2 h at 37 °C. After being washed three times with PBS, cells were then incubated with goat anti mouse IgG conjugated with FITC at 1:500, (Thermo Fisher Scientific, USA) for 40 min at 37 °C. After three times of washing, the cell images were captured with an inverted fluorescence microscope equipped with a camera (Evos FL, USA).
CCK-8 assay
The cytotoxicity effect of miR-218 mimic was determined by CCK-8 assay kit (Dojindo, Japan). Briefly, the PAM monolayers in 96-well plates were transfected with either miR-218 mimic or negative control at concentrations of 30, 50, 100, and 150 nM. At 48 h posttransfection, 10 μl CCK-8 solution was added to each well of the plate and incubated for another 3 h at 37 °C. Then, the microplates were measured by absorbance photometer at 450 nm.
Statistical analysis
All statistical analyses were carried out using Prism 8.0.1 (GraphPad Software, Inc) and MS-Excel. We performed at least three independent reproducible results for most experiments. Differences between the experimental and control groups were tested by using two-way ANOVA with Bonferroni’s posttest. Data are presented as the mean ± standard deviations (SD) from three or more independent experiments. A p value of <0.05 was considered statistically significant.
Ethical approval
The SPF pigs were owned by Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, Heilongjiang Province, China. The entire procedure was carried out in strict accordance with the protocol approved by the Animal Care and Use Committee of Harbin Veterinary Research Institute of Chinese Academy of Agricultural Sciences (Approval ID: 200720–01), and experiments were performed according to the regulations and guidelines established by this committee.
Data availability
All data for these studies is contained within this article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Gebremeskel Mamu Werid for careful reading of this article and thoughtful feedback. We are grateful to Changwen Li and Xiaoye Lu for providing veterinary and animal care support.
Author contributions
Y. W., Lin Zhang, and Lu Zhang conceived the project and carried out the experiments; Lin Zhang, Lu Zhang, Y. P., J. G., Y. X., and Y. W. analyzed the data; X. L., Z. T., and H. C. contributed materials/analysis tools and analyzed the data; Y. W. Lin Zhang, and Lu Zhang wrote and reviewed the manuscript.
Funding and additional information
This research was supported by National Natural Science Foundation of China Grants 31572497 & 31372416, and Heilongjiang province matching grant for national Key R&D Program (GX18B052).
Edited by Ronald Wek
Supporting information
References
- 1.Cavanagh D. Nidovirales: A new order comprising Coronaviridae and arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 2.Meulenberg J.J. PRRSV, the virus. Vet. Res. 2000;31:11–21. doi: 10.1051/vetres:2000103. [DOI] [PubMed] [Google Scholar]
- 3.Collins J.E., Benfield D.A., Christianson W.T., Harris L., Hennings J.C., Shaw D.P., Goyal S.M., McCullough S., Morrison R.B., Joo H.S., Gorcyca D.C. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Invest. 1992;4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- 4.Meulenberg J.J., Hulst M.M., de Meijer E.J., Moonen P.L., den Besten A., de Kluyver E.P., Wensvoort G., Moormann R.J. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rappe J.C., García-Nicolás O., Flückiger F., Thür B., Hofmann M.A., Summerfield A., Ruggli N. Heterogeneous antigenic properties of the porcine reproductive and respiratory syndrome virus nucleocapsid. Vet. Res. 2016;47:117. doi: 10.1186/s13567-016-0399-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vu H.L.X., Pattnaik A.K., Osorio F.A. Strategies to broaden the cross-protective efficacy of vaccines against porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 2017;206:29–34. doi: 10.1016/j.vetmic.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Dong H., Lei J., Ding L., Wen Y., Ju H., Zhang X. MicroRNA: Function, detection, and bioanalysis. Chem. Rev. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 8.Mohr A.M., Mott J.L. Overview of microRNA biology. Semin. Liver Dis. 2015;35:3–11. doi: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 10.Chekulaeva M., Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 12.Bartel D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidigal J.A., Ventura A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015;25:137–147. doi: 10.1016/j.tcb.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbu M.G., Condrat C.E., Thompson D.C., Bugnar O.L., Cretoiu D., Toader O.D., Suciu N., Voinea S.C. MicroRNA Involvement in signaling pathways during viral infection. Front Cell Dev Biol. 2020;8:143. doi: 10.3389/fcell.2020.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan X., Wang L., Sun G., Yan W., Yang Y. Understanding the cross-talk between host and virus in poultry from the perspectives of microRNA. Poult. Sci. 2020;99:1838–1846. doi: 10.1016/j.psj.2019.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trobaugh D.W., Klimstra W.B. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017;23:80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J., Zinkernagel R.M., Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 18.Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forster S.C., Tate M.D., Hertzog P.J. MicroRNA as type I interferon-regulated transcripts and modulators of the innate immune response. Front Immunol. 2015;6:334. doi: 10.3389/fimmu.2015.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong R.R., Abd-Aziz N., Affendi S., Poh C.L. Role of microRNAs in antiviral responses to dengue infection. J. Biomed. Sci. 2020;27:4. doi: 10.1186/s12929-019-0614-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q., Huang C., Yang Q., Gao L., Liu H.C., Tang J., Feng W.H. MicroRNA-30c modulates type I IFN responses to facilitate porcine reproductive and respiratory syndrome virus infection by targeting JAK1. J. Immunol. 2016;196:2272–2282. doi: 10.4049/jimmunol.1502006. [DOI] [PubMed] [Google Scholar]
- 24.Li L., Wei Z., Zhou Y., Gao F., Jiang Y., Yu L., Zheng H., Tong W., Yang S., Zheng H., Shan T., Liu F., Xia T., Tong G. Host miR-26a suppresses replication of porcine reproductive and respiratory syndrome virus by upregulating type I interferons. Virus Res. 2015;195:86–94. doi: 10.1016/j.virusres.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J., Shi X., Zhang X., Wang A., Wang L., Yang Y., Deng R., Zhang G.P. MicroRNA 373 facilitates the replication of porcine reproductive and respiratory syndrome virus by its negative regulation of type I interferon induction. J. Virol. 2017;91 doi: 10.1128/JVI.01311-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du T., Nan Y., Xiao S., Zhao Q., Zhou E.M. Antiviral strategies against PRRSV infection. Trends Microbiol. 2017;25:968–979. doi: 10.1016/j.tim.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Zhen Y., Wang F., Liang W., Liu J., Gao G., Wang Y., Xu X., Su Q., Zhang Q., Liu B. Identification of differentially expressed non-coding RNA in porcine alveolar macrophages from Tongcheng and large white pigs Responded to PRRSV. Sci. Rep. 2018;8:15621. doi: 10.1038/s41598-018-33891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian Z.J., An T.Q., Zhou Y.J., Peng J.M., Hu S.P., Wei T.C., Jiang Y.F., Xiao Y., Tong G.Z. An attenuated live vaccine based on highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) protects piglets against HP-PRRS. Vet. Microbiol. 2009;138:34–40. doi: 10.1016/j.vetmic.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Bernier A., Sagan S.M. The Diverse roles of microRNAs at the Host⁻Virus Interface. Viruses. 2018;10:440–465. doi: 10.3390/v10080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skalsky R.L., Cullen B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang Y., Snijder E.J. The PRRSV replicase: Exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 2010;154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mesev E.V., LeDesma R.A., Ploss A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019;4:914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carow B., Rottenberg M.E. SOCS3, a Major regulator of infection and inflammation. Front Immunol. 2014;5:58. doi: 10.3389/fimmu.2014.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brockmeier S.L., Loving C.L., Eberle K.C., Hau S.J., Buckley A., Van Geelen A., Montiel N.A., Nicholson T., Lager K.M. Interferon alpha inhibits replication of a live-attenuated porcine reproductive and respiratory syndrome virus vaccine preventing development of an adaptive immune response in swine. Vet. Microbiol. 2017;212:48–51. doi: 10.1016/j.vetmic.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Luo R., Fang L., Jin H., Jiang Y., Wang D., Chen H., Xiao S. Antiviral activity of type I and type III interferons against porcine reproductive and respiratory syndrome virus (PRRSV) Antiviral Res. 2011;91:99–101. doi: 10.1016/j.antiviral.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 36.Finlay B.B., McFadden G. Anti-immunology: Evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 37.Huang C., Zhang Q., Feng W.H. Regulation and evasion of antiviral immune responses by porcine reproductive and respiratory syndrome virus. Virus Res. 2015;202:101–111. doi: 10.1016/j.virusres.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ke H., Yoo D. The viral innate immune antagonism and an alternative vaccine design for PRRS virus. Vet. Microbiol. 2017;209:75–89. doi: 10.1016/j.vetmic.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nan Y., Wu C., Gu G., Sun W., Zhang Y.J., Zhou E.M. Improved vaccine against PRRSV: Current Progress and future perspective. Front Microbiol. 2017;8:1635. doi: 10.3389/fmicb.2017.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bronevetsky Y., Ansel K.M. Regulation of miRNA biogenesis and turnover in the immune system. Immunol. Rev. 2013;253:304–316. doi: 10.1111/imr.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holla S., Balaji K.N. Epigenetics and miRNA during bacteria-induced host immune responses. Epigenomics. 2015;7:1197–1212. doi: 10.2217/epi.15.75. [DOI] [PubMed] [Google Scholar]
- 42.Roy S., Sen C.K. MiRNA in innate immune responses: Novel players in wound inflammation. Physiol. Genomics. 2011;43:557–565. doi: 10.1152/physiolgenomics.00160.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy S. miRNA in macrophage development and function. Antioxid. Redox Signal. 2016;25:795–804. doi: 10.1089/ars.2016.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Essandoh K., Li Y., Huo J., Fan G.C. MiRNA-mediated macrophage Polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46:122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tie J., Pan Y., Zhao L., Wu K., Liu J., Sun S., Guo X., Wang B., Gang Y., Zhang Y., Li Q., Qiao T., Zhao Q., Nie Y., Fan D. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. Plos Genet. 2010;6:e1000879. doi: 10.1371/journal.pgen.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leite K.R., Sousa-Canavez J.M., Reis S.T., Tomiyama A.H., Camara-Lopes L.H., Sañudo A., Antunes A.A., Srougi M. Change in expression of miR-let7c, miR-100, and miR-218 from high grade localized prostate cancer to metastasis. Urol. Oncol. 2011;29:265–269. doi: 10.1016/j.urolonc.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Duan F., Wang K., Dai L., Zhao X., Feng Y., Song C., Cui S., Wang C. Prognostic significance of low microRNA-218 expression in patients with different types of cancer: Evidence from published studies. Medicine (Baltimore) 2016;95:e4773. doi: 10.1097/MD.0000000000004773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X., Yao X., Xie T., Chang Z., Guo Y., Ni H. Exosome-derived uterine miR-218 isolated from cows with endometritis regulates the release of cytokines and chemokines. Microb. Biotechnol. 2020;13:1103–1117. doi: 10.1111/1751-7915.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M., Guo Q., Cai H., Wang H., Ma Z., Zhang X. miR-218 regulates diabetic nephropathy via targeting IKK-β and modulating NK-κB-mediated inflammation. J. Cell Physiol. 2020;235:3362–3371. doi: 10.1002/jcp.29224. [DOI] [PubMed] [Google Scholar]
- 50.Li W., Wang Q., Feng Q., Wang F., Yan Q., Gao S.J., Lu C. Oncogenic KSHV-encoded interferon regulatory factor upregulates HMGB2 and CMPK1 expression to promote cell invasion by disrupting a complex lncRNA-OIP5-AS1/miR-218-5p network. Plos Pathog. 2019;15:e1007578. doi: 10.1371/journal.ppat.1007578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasternak A.O., Spaan W.J., Snijder E.J. Nidovirus transcription: How to make sense…? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 52.Pedroso J.A.B., Ramos-Lobo A.M., Donato J., Jr. SOCS3 as a future target to treat metabolic disorders. Hormones (Athens) 2019;18:127–136. doi: 10.1007/s42000-018-0078-5. [DOI] [PubMed] [Google Scholar]
- 53.McCaskill J.L., Ressel S., Alber A., Redford J., Power U.F., Schwarze J., Dutia B.M., Buck A.H. Broad-spectrum inhibition of respiratory virus infection by MicroRNA mimics targeting p38 MAPK signaling. Mol. Ther. Nucleic Acids. 2017;7:256–266. doi: 10.1016/j.omtn.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang T.Y., Liu Y.G., Li L., Wang G., Wang H.M., Zhang H.L., Zhao S.F., Gao J.C., An T.Q., Tian Z.J., Tang Y.D., Cai X.H. Porcine alveolar macrophage CD163 abundance is a pivotal switch for porcine reproductive and respiratory syndrome virus infection. Oncotarget. 2018;9:12174–12185. doi: 10.18632/oncotarget.24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J., Guo L., Yang L., Xu J., Zhang L., Feng L., Chen H., Wang Y. Metalloprotease ADAM17 regulates porcine epidemic diarrhea virus infection by modifying aminopeptidase N. Virology. 2018;517:24–29. doi: 10.1016/j.virol.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Xu J., Zhang L., Xu Y., Zhang H., Gao J., Wang Q., Tian Z., Xuan L., Chen H., Wang Y. PP2A facilitates porcine reproductive and respiratory syndrome virus replication by Deactivating irf3 and limiting type I interferon production. Viruses. 2019;11:948–965. doi: 10.3390/v11100948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chua B.H., Phuektes P., Sanders S.A., Nicholls P.K., McMinn P.C. The molecular basis of mouse adaptation by human enterovirus 71. J. Gen. Virol. 2008;89:1622–1632. doi: 10.1099/vir.0.83676-0. [DOI] [PubMed] [Google Scholar]
- 59.Luo X., Guo L., Zhang J., Xu Y., Gu W., Feng L., Wang Y. Tight Junction protein Occludin is a porcine epidemic diarrhea virus Entry factor. J. Virol. 2017;91 doi: 10.1128/JVI.00202-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo L., Luo X., Li R., Xu Y., Zhang J., Ge J., Bu Z., Feng L., Wang Y. Porcine epidemic diarrhea virus infection inhibits interferon signaling by targeted Degradation of STAT1. J. Virol. 2016;90:8281–8292. doi: 10.1128/JVI.01091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for these studies is contained within this article.