Abstract
The recent rise in the prevalence of chronic allergic diseases among children has increased disease burden and reduced quality of life, especially for children with comorbid allergic diseases. Predicting the occurrence of allergic diseases can help prevent its onset for those in high risk groups. Herein, we aimed to construct prediction models for asthma, atopic dermatitis (AD), and asthma-AD comorbidity (also known as atopic march) using a genome-wide association study (GWAS) and family history data from patients of Korean heritage. Among 973 patients and 481 healthy controls, we evaluated single nucleotide polymorphism (SNP) heritability for each disease using genome-based restricted maximum likelihood (GREML) analysis. We then compared the performance of prediction models constructed using Least Absolute Shrinkage and Selection Operator (LASSO) and penalized ridge regression methods. Our results indicate that the addition of family history risk scores to the prediction model greatly increase the predictability of asthma and asthma-AD comorbidity. However, prediction of AD was mostly attributable to GWAS SNPs.
Keywords: Asthma, Atopic dermatitis, Prediction model, Family history, Genome-wide association study
To the Editor:
The prevalence of chronic allergic diseases in children has risen recently, increasing disease burden and leading to lower quality of life, especially in children who have developed comorbid allergic diseases, known as atopic march.1 Predicting allergic disease occurrence can help prevent those in high risk groups from developing diseases. Unfortunately, the predictability and validity of constructed models differ greatly, and most have not proven clinically useful.2 Genome-wide prediction of childhood asthma based on highest-ranked single nucleotide polymorphisms (SNPs) has suggested that the genetic origins of asthma are diverse.3 Herein, we aimed to construct prediction models for asthma, atopic dermatitis (AD), and asthma-AD comorbidity using a genome-wide association study (GWAS) and family history data from patients of Korean heritage. We evaluated SNP heritability for each disease and compared the performance of our prediction models built upon distinct factors.
A total of 973 patients with both allergic disease and sensitization were included in the study, along with 481 healthy controls (Supplementary Table 1). Among those with allergic disease, there were 498 patients with asthma, 632 patients with AD, and 157 patients with asthma-AD comorbidity. All allergic disease subjects had confirmed allergic sensitization, defined by allergen-specific IgE levels >0.7 kUA/L to at least 1 of the 9 common food or airborne allergens. AD was diagnosed by pediatric allergists for patients with a SCORAD (SCORing Atopic Dermatitis) index of over > 30. Asthma was confirmed based on consistent respiratory symptoms verified by a physician, the presence of either ≥12% increase in forced expiratory volume in 1 second (FEV1) in response to a bronchodilator, or bronchial hyperresponsiveness defined as ≥ 20% decrease in FEV1 with inhalation of <16 mg/mL methacholine. DNA was extracted from whole blood samples, and genome-wide SNPs and genetic variant information were identified using the Illumina HumanCoreExome-24 v1.0 BeadChip kit (Illumina Inc., San Diego, CA, USA). Method specifications including confirmation standards for allergic sensitization, quality control, and imputation methods for genotype data are provided in the Supplementary Methods and Supplementary Table 2.
To investigate the overall influence of genotype on each disease, we calculated the proportion of phenotypic variance explained by SNPs using genome-based restricted maximum likelihood (GREML) analysis4 with Genome-wide Complex Trait Analysis (GCTA) 64 version 1.92.0 beta 2 software (http://cnsgenomics.com/software/gcta/). For this analysis, SNPs were pruned using a sliding window protocol with a maximum of 500 kbp and linkage disequilibrium (LD) composite measure threshold of 0.2, and the genetic relationship matrix was calculated. Sex and 10 genotype principal component (PC) scores were included as covariates. We then used Least Absolute Shrinkage and Selection Operator (LASSO) and penalized ridge regression to develop prediction models for each disease. Risk scores of each disease were calculated as described by Gim et al5 using the disease status of relatives and their kinship coefficients. A positive family history was considered if family members, including patient parents and siblings only, had asthma, AD, allergic rhinitis, or food allergy; otherwise, the family history was considered negative. SNPs were selected using absolute best linear unbiased prediction (BLUP) values. BLUPs were calculated by GCTA64 after adjustment for sex and 10 genotype PC scores, and the top 100, 500, 1000, 5000, and 10 000 SNPs with the largest absolute BLUPs were selected. The list of SNPs used for the models with best performance for each disease is shown in Supplementary Table 3. Prediction model performance was evaluated by comparing the area under the curve (AUC) of receiver operating characteristic (ROC) curves obtained by 10-fold cross validation. The detailed procedures for both analyses are provided in the Supplementary Methods.
Table 1 shows the estimated SNP heritability by GREML analysis. Genotyped SNPs accounted for 54.88% (p = 0.0714), 4.64% (p > 0.4), and 12.75% (p > 0.4) of the phenotypic variance of AD, asthma, and asthma-AD comorbidity, respectively. Our SNP heritability estimates might be smaller than the previous studies and SNP heritability tends to be smaller than heritability estimated by the family-based samples.6 The difference may also be related with the difference in subject ethnicity and the genotyping chip designed mainly for use in Caucasians.
Table 1.
Estimated SNP heritability for each disease calculated with GREML analysis.
| Phenotype | Hsnp2 | SE | P-value |
|---|---|---|---|
| Asthma | 0.0464 | 0.3805 | 0.4411 |
| Atopic dermatitis | 0.5488 | 0.3812 | 0.0714 |
| Asthma-AD comorbidity | 0.1275 | 0.4456 | 0.4212 |
Hsnp2, SNP heritability at the liability scale. SE, standard error. AD, Atopic Dermatits, P-values were calculated by likelihood ratio tests
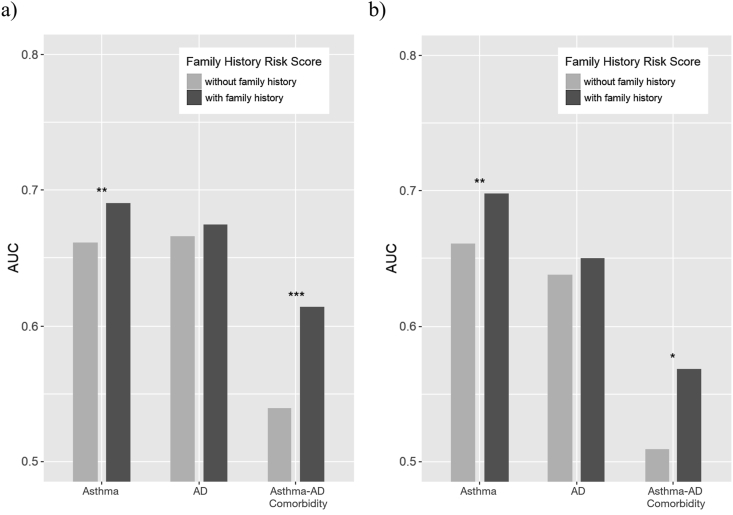
Addition of the family history risk scores significantly increased prediction model performance for asthma and asthma-AD comorbidity, as shown in Fig. 1. For asthma, AUC increased from 0.661 to 0.691 (DeLong test p = 7.14 × 10−3) for the LASSO prediction model, and from 0.661 to 0.698 (p = 8.48 × 10−3) for the ridge regression model. Similarly, the prediction model performance for asthma-AD comorbidity increased significantly with addition of the family history risk scores (LASSO: 0.539 to 0.614, p = 9.01 × 10−4; ridge regression: 0.509 to 0.568, p = 0.0424). Prediction model performance also increased for AD, but not as significantly (LASSO: 0.666 to 0.675, p = 0.360; ridge regression: 0.638 to 0.650, p = 0.408). The number of SNPs used for each model and the sensitivity and specificity estimates at the inflection points are presented in Supplementary Table 4.
Fig. 1.
Vertical bar plots of AUCs of the (a) LASSO and (b) ridge regression prediction models with or without the addition of family history risk scores. The top 100, 500, 1,000, 5,000, and 10,000 SNPs selected with best linear unbiased prediction values (BLUPs) were separately used for prediction model building, and the best AUCs are shown. ∗ paired DeLong test p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. AD, Atopic Dermatits
Applying the LASSO prediction model of asthma to UK Biobank data, we achieved an AUC of 0.567 (p < 2.2 × 10−16). The odds ratio (OR) of the polygenic risk score (PRS) estimated from the logistic regression was 10.129 (p = 0.02). This analysis is described in the Supplementary Methods. However, we could not validate the prediction models for AD and asthma-AD comorbidity, and those combined with the family history risk scores using UK Biobank data, because it contained insufficient family history information.
Our results show that the addition of family history risk scores increased the overall prediction model performance for asthma, suggesting that family factors other than genotype contribute to the predictive power of the model. Asthma heritability is only partly explained by genetic variants, with the range of heritability estimates varying from 35% to 95% in previous studies.7 Similarly, the predictive value of positive family history alone has been reported to range from 11% to 37%.8 Although sole use of family history may be unreliable for asthma predictability, its addition to GWAS may improve prediction model performance. Further, the addition of family history risk scores greatly increased the predictability of asthma-AD comorbidity, despite the initial AUC being around 0.50. On the other hand, prediction of AD may be mostly attributable to GWAS SNPs.
Family history risk scores can explain additional family factors such as shared environmental effects or genetic effects that cannot be detected by GWAS. Several limitations are present in our study, including the retrospective collection of family history from medical records and the relatively small sample size of the GWAS cohort. Additionally, an age factor might have contributed to bias, since the AD group was younger than the other groups; the effects of GWAS SNP heritability are the most influential at a young age while the effects of environmental factors increase with time. In conclusion, a prediction model using both GWAS data and family history can help predict the development of allergic diseases. Consideration of family history may enhance predictive performance when accompanied with genetic risk of allergic diseases.
Abbreviations
AD, atopic dermatitis. SNP, single nucleotide polymorphism. GWAS, genome-wide association study. PRS, polygenic risk score. LASSO, least absolute shrinkage and selection operator. PC, principal component. BLUP, best linear unbiased predictor. AUC, area under curve. ROC, receiver operating characteristic curve.
Consent for publication
All authors agree with the publication of this manuscript.
Author contributions
Jaehyun Park performed the analytical computations. Haerin Jang developed the theory and wrote the manuscript. Mina Kim and Jung Yeon Hong contributed to sample preparation used for GWAS. Yoon Hee Kim and Myung Hyun Sohn contributed to the interpretation of the results. Sungho Won and Kyung Won Kim conceived the study and were in charge of overall direction and planning. All authors discussed the results and contributed to the final manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Ethics approval
The study was approved by the institutional review board of Severance Hospital (Seoul, Korea; IRB no. 4–2004–0036). Written consent was provided from all participants prior to the study.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government(MIST) (No. 2019R1F1A1058910).
Footnotes
Full list of author information is available at the end of the article
Supplementary data to this article can be found online at https://doi.org/10.1016/j.waojou.2021.100539.
Contributor Information
Sungho Won, Email: won1@snu.ac.kr.
Kyung Won Kim, Email: kwkim@yuhs.ac.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Dharmage S.C., Lowe A.J., Matheson M.C., Burgess J.A., Allen K.J., Abramson M.J. Atopic dermatitis and the atopic march revisited. Allergy. 2014;69:17–27. doi: 10.1111/all.12268. [DOI] [PubMed] [Google Scholar]
- 2.Colicino S., Munblit D., Minelli C., Custovic A., Cullinan P. Validation of childhood asthma predictive tools: a systematic review. Clin Exp Allergy. 2019;49:410–418. doi: 10.1111/cea.13336. [DOI] [PubMed] [Google Scholar]
- 3.Spycher B.D., Henderson J., Granell R. Genome-wide prediction of childhood asthma and related phenotypes in a longitudinal birth cohort. J Allergy Clin Immunol. 2012;130:503–509. doi: 10.1016/j.jaci.2012.06.002. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S.H., Wray N.R., Goddard M.E., Visscher P.M. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gim J., Kim W., Kwak S.H. Improving disease prediction by incorporating family disease history in risk prediction models with large-scale genetic data. Genetics. 2017;207:1147–1155. doi: 10.1534/genetics.117.300283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J., Zeng J., Goddard M. Concepts, estimation and interpretation of SNP-based heritability. Nat Genet. 2017;49:1304–1310. doi: 10.1038/ng.3941. [DOI] [PubMed] [Google Scholar]
- 7.Kim K.W., Ober C. Lessons learned from GWAS of asthma. Allergy Asthma Immunol Res. 2019;11:170–187. doi: 10.4168/aair.2019.11.2.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke W., Fesinmeyer M., Reed K., Hampson L., Carlsten C. Family history as a predictor of asthma risk. Am J Prev Med. 2003;24:160–169. doi: 10.1016/s0749-3797(02)00589-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.