Abstract
Aquaporin-4 (AQP4) is a predominant water channel expressed in astrocytes in the mammalian brain. AQP4 is crucial for the regulation of homeostatic water movement across the blood–brain barrier (BBB). Although the molecular mechanisms regulating AQP4 levels in the cerebral cortex under pathological conditions have been intensively investigated, those under normal physiological conditions are not fully understood. Here we demonstrate that AQP4 is selectively expressed in astrocytes in the mouse cerebral cortex during development. BMP signaling was preferentially activated in AQP4-positive astrocytes. Furthermore, activation of BMP signaling by in utero electroporation markedly increased AQP4 levels in the cerebral cortex, and inhibition of BMP signaling strongly suppressed them. These results indicate that BMP signaling alters AQP4 levels in the mouse cerebral cortex during development.
Subject terms: Development of the nervous system, Molecular neuroscience
Introduction
Aquaporin-4 (AQP4) is a predominant water channel in the mammalian brain and is mainly expressed at the endfeet of astrocytes1–3. In the cerebral cortex, AQP4 plays crucial roles in controlling water homeostasis. AQP4 regulates water movement across the blood–brain barrier (BBB), which restricts large polar substances and potentially neurotoxic compounds in circulation from passively diffusing into the brain4,5. Changes in AQP4 expression are often associated with various diseases such as epilepsy, edema and glioblastoma6–10. Thus, uncovering the molecular mechanisms regulating the expression of AQP4 is an important research topic. However, our understanding of the mechanisms of regulating AQP4 expression in the cerebral cortex is still rudimentary.
Previous reports have investigated the regulation of AQP4 expression in the context of inflammation in the mouse brain11. The induction of oxidative stress activates the p38 mitogen-activated protein kinase (MAPK) pathway, inducing transcriptional activation of AQP412. Activation of the cysteinyl leukotriene receptor 2 induces AQP4 upregulation through the p38 and ERK signaling pathways in astrocytes13. In contrast to the regulation of AQP4 expression under pathological conditions, the mechanisms regulating AQP4 expression under normal physiological conditions in the cerebral cortex are not fully understood.
Bone morphogenetic proteins (BMPs), members of the TGF-β family of signaling molecules, have numerous functions in the brain14–18. Here, we show that BMP signaling was activated in astrocytes in the cerebral cortex. Activation of BMP signaling increased AQP4 levels in astrocytes in the mouse cerebral cortex in vivo. Consistently, inhibition of BMP signaling decreased AQP4 levels in astrocytes. Taken together with a previous study showing that activation of BMP signaling increased AQP4 expression in cultured astrocytes19, these results clearly indicate that BMP signaling changes AQP4 levels in astrocytes under normal physiological conditions.
Results
Expression patterns of AQP4 in the developing mouse cerebral cortex
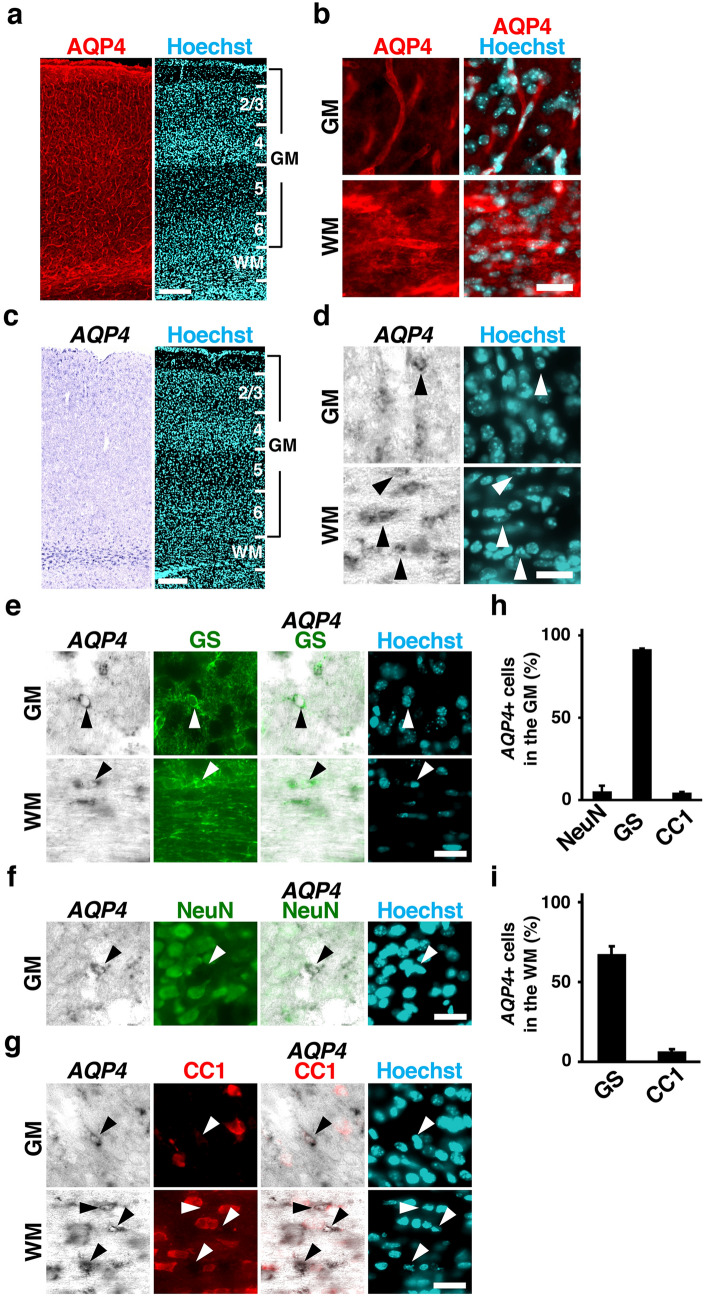
To investigate the expression patterns of AQP4, we performed immunohistochemistry for AQP4 using the mouse cerebral cortex at postnatal day 16 (P16) (Fig. 1a,b). AQP4 was expressed throughout the cortex, though the expression levels of AQP4 were higher in the white matter (WM) than that in the gray matter (GM). Consistent with a previous study showing that AQP4 is distributed at the endfeet of astrocytes attached to blood vessels20, vascular structures were labeled with AQP4 immunostaining (Fig. 1b). Next, we investigated the distribution patterns of AQP4-positive cells using in situ hybridization for AQP4. Consistent with the results of AQP4 immunostaining, AQP4 mRNA was preferentially distributed in the WM but was also observed in the GM (Fig. 1c,d).
Figure 1.
Expression patterns of AQP4 in the developing mouse cerebral cortex. (a,b) Sections of the mouse cerebral cortex at P16 were subjected to AQP4 immunohistochemistry and Hoechst 33342 staining. Low magnification images (a) and high magnification images (b) corresponding to the gray matter (GM) and the white matter (WM) are shown. AQP4 was expressed throughout the developing mouse cerebral cortex. The expression level of AQP4 was higher in the WM than in the GM. (c,d) Sections of the mouse cerebral cortex at P16 were subjected to in situ hybridization for AQP4 and Hoechst 33342 staining. Low magnification images (c) and high magnification images (d) corresponding to the GM and the WM are shown. AQP4 mRNA was preferentially distributed in the WM but was also observed in the GM. (e–g) Sections of the mouse cerebral cortex at P16 were subjected to in situ hybridization for AQP4 and immunohistochemistry for either GS (e), NeuN (f) or CC1 (g). High-magnification images of the GM and the WM are shown. AQP4 was preferentially expressed in GS-positive cells (e, arrowheads), while AQP4 signals were not colocalized with NeuN-positive cells or CC1-positive cells (f and g, arrowheads). (h,i) The percentages of AQP4-positive cells co-expressing NeuN, GS, and CC1 in the GM (h) and the WM (i). n = 3 animals for each condition. Bars represent mean ± SD. Numbers indicate the corresponding layers in the cerebral cortex. GM, gray matter; WM, white matter. Scale bars 200 µm (a,c) and 25 µm (b,d–g).
Although it has been reported that AQP4 is expressed only in astrocytes in the cerebral cortex of adult mice1,2, it remained unknown which cell types express AQP4 during development. To examine which cell types express AQP4 in the mouse cerebral cortex during development, we combined in situ hybridization for AQP4 and immunohistochemistry for either the astrocyte marker glutamine synthetase (GS), the neuron marker NeuN or the oligodendrocyte marker CC1 (Fig. 1e–g). Consistent with previous reports using the adult brain1,2, AQP4 was preferentially expressed in GS-positive cells in the mouse cerebral cortex at P16, while AQP4 signals were not co-localized with NeuN-positive cells or CC1-positive cells (Fig. 1h,i) (GM; NeuN, 5 ± 3%; GS, 92 ± 0.5%; CC1, 5 ± 0.3%) (WM; NeuN, 0%; GS, 67 ± 5%; CC1, 7 ± 1%). These results suggest that AQP4 is mainly expressed in astrocytes in the developing mouse cerebral cortex.
BMP signaling is activated in AQP4-positive astrocytes in the developing mouse cerebral cortex
We next investigated the molecular mechanisms regulating AQP4 levels during cortical development. We performed immunohistochemistry for phospho-Smad1/5/8 (pSmad) using cortical sections of mice at P16. Consistent with a previous report21, pSmad, which is induced by the activation of BMP signaling, was observed in the mouse cerebral cortex at P16, more predominantly in the WM (Fig. 2a).
Figure 2.
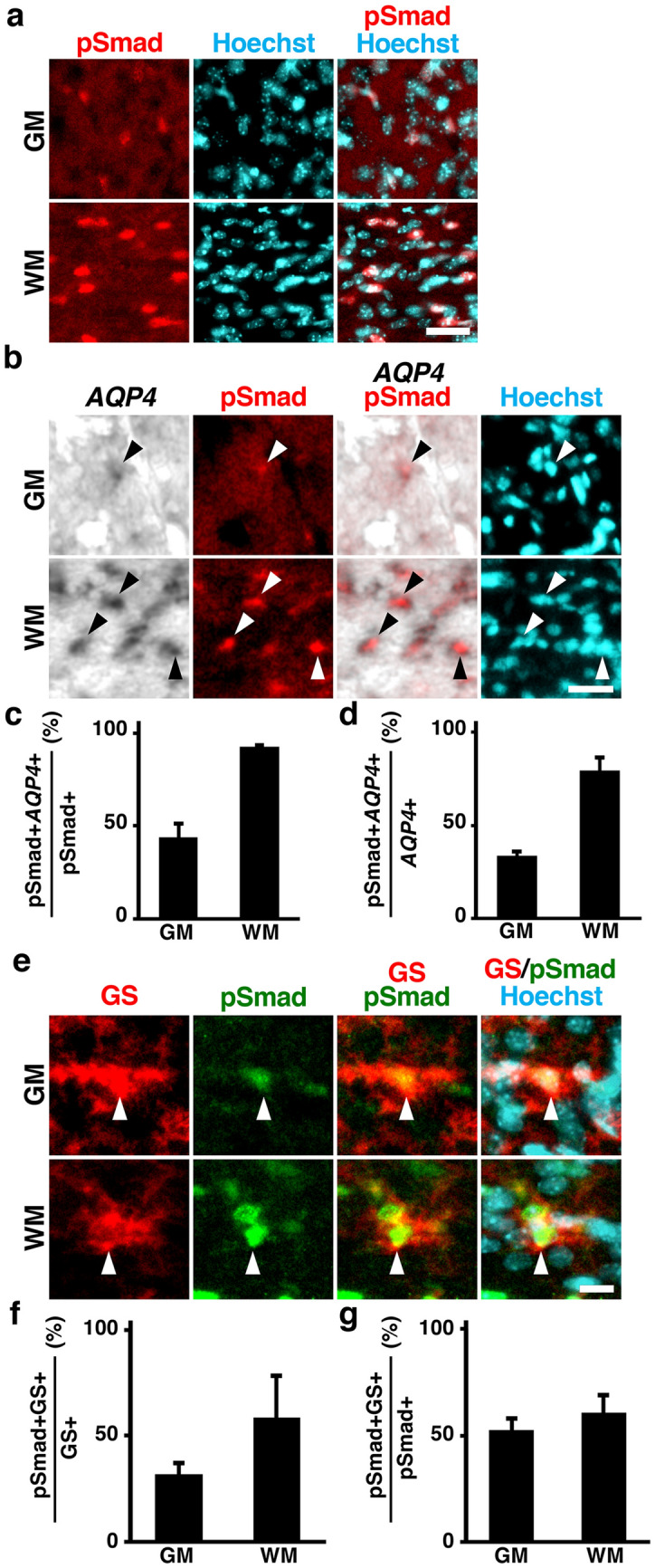
BMP signaling is activated in AQP4-positive astrocytes in the developing mouse cerebral cortex. (a) Sections of the mouse cerebral cortex at P16 were subjected to immunohistochemistry for pSmad and Hoechst 33342 staining. pSmad signals were more predominant in the WM compared with the GM. (b) Sections were subjected to immunohistochemistry for pSmad and in situ hybridization for AQP4. High magnification images are shown. Many AQP4-positive astrocytes were also labeled with pSmad (arrowheads). (c) The percentages of pSmad-positive cells co-expressing AQP4 in the GM and the WM. (d) The percentages of AQP4-positive cells co-labeled with pSmad in the GM and the WM. (e) Sections of the mouse cerebral cortex at P16 were subjected to immunohistochemistry for pSmad and GS. High magnification images are shown. pSmad signals were observed in GS-positive cells (arrowheads). (f) The percentages of GS-positive cells co-labeled with pSmad in the GM and the WM. (g) The percentages of pSmad-positive cells co-expressing GS in the GM and the WM. GM, gray matter; WM, white matter. n = 3 animals for each condition. Bars represent mean ± SD. Scale bars 25 μm (a,b) and 10 µm (e).
Because both AQP4 and pSmad were predominantly distributed in the WM, we hypothesized that BMP signaling affects AQP4 levels. To test this, we performed immunohistochemistry for pSmad and in situ hybridization for AQP4 and found that most pSmad-positive cells in the WM expressed AQP4 (92 ± 1%) (Fig. 2b,c). Furthermore, most of the AQP4-positive cells in the WM were positive for pSmad (79 ± 6%) (Fig. 2d). Consistent with our observation, the percentage of pSmad-positive cells co-expressing AQP4 and that of AQP4-positive cells co-labeled with pSmad were smaller in the GM (Fig. 2c,d).
To examine whether pSmad is distributed in astrocytes, we performed immunohistochemistry for pSmad and GS. We found that pSmad was mainly observed in GS-positive cells in the WM (59 ± 20%) (Fig. 2e,f). The percentage of GS-positive cells co-labeled with pSmad was smaller in the GM (Fig. 2e,f). More than half of pSmad-positive cells expressed GS in the GM and the WM (GM, 53 ± 6%; WM, 61 ± 8%) (Fig. 2g). These results suggest that BMP signaling is activated in AQP4-positive astrocytes, especially in the WM, during development. Consistent with our observations, a previous study demonstrated that BMP receptors were preferentially expressed in astrocytes in the developing mouse cerebral cortex22.
Activation of BMP signaling increases AQP4 levels in the mouse cerebral cortex
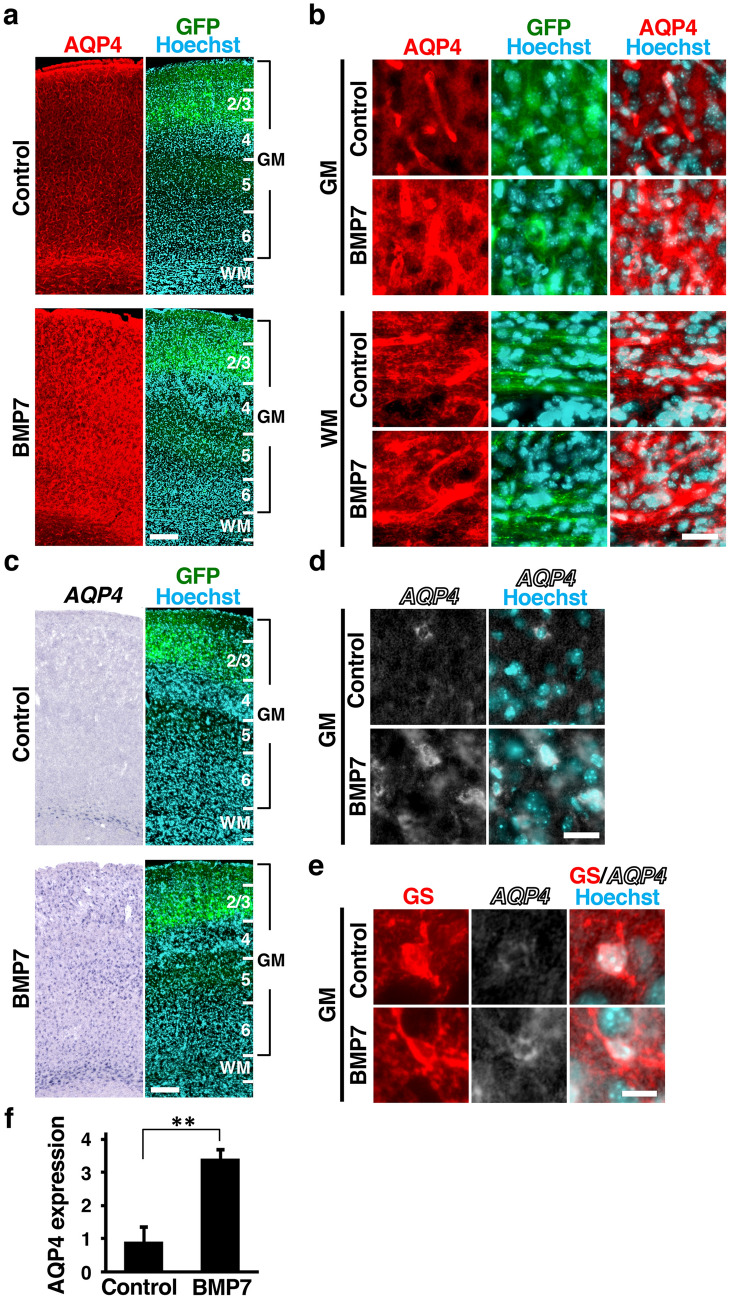
To directly test whether BMP signaling alters AQP4 levels, we stimulated BMP signaling by introducing BMP7 into the mouse cerebral cortex using in utero electroporation (IUE) at embryonic day 14 (E14) and examined AQP4 expression at P16. Previous studies and the Allen Brain Atlas (https://portal.brain-map.org/) have shown that BMP family members such as BMP3 and BMP7 are expressed in the cerebral cortex during development23,24. Because it was reported that BMP3 does not have the ability to activate Smad1/5/8 or to induce bone and cartilage tissues in vivo, and because most BMP ligands share Smad1/5/8 as downstream mediators25, here we utilized BMP7 to activate BMP signaling. In situ hybridization for BMP7 confirmed that BMP7 mRNA was indeed produced as a result of introducing BMP7-expressing plasmids into the mouse cerebral cortex (Supplementary Fig. S1a). Furthermore, immunostaining for pSmad demonstrated that BMP signaling was strongly activated by the introduction of BMP7-expressing plasmids (Supplementary Fig. S1b). In addition, it should be noted that the layer structure of the cerebral cortex was not affected by BMP7 electroporation (Supplementary Fig. S2).
We next performed immunohistochemistry and in situ hybridization for AQP4 and found that the activation of BMP signaling markedly increased both AQP4 protein and AQP4 mRNA in the cerebral cortex (Fig. 3a–d). The increase of AQP4 was clearer in the GM than in the WM, presumably because AQP4 expression in the GM of the control brain were lower than those in the WM (Fig. 3a,c). We performed in situ hybridization for AQP4 and immunohistochemistry for GS and found that AQP4 mRNA was increased in GS-positive cells (Fig. 3e). Our quantification of the intensity of AQP4 immunoreactivity showed that activation of BMP signaling significantly increased AQP4 protein in the cerebral cortex (control, 0.92 ± 0.43; BMP7, 3.41 ± 0.26; p < 0.01, Student’s t-test) (Fig. 3f). Taken together with a previous study showing that activation of BMP signaling increased AQP4 expression in cultured astrocytes19, these results indicate that activation of BMP signaling is sufficient for increasing AQP4 levels in astrocytes in the cerebral cortex.
Figure 3.
Activation of BMP signaling increases AQP4 levels in the mouse cerebral cortex. CAG-EGFP plus either pCAG-BMP7 or pCAG control vector was electroporated at E14, and the brains were dissected at P16. (a,b) Coronal sections stained with anti-AQP4 antibody, anti-GFP antibody and Hoechst 33342. Low magnification images (a) and high magnification images of the GM and the WM (b) are shown. The immunoreactivity of AQP4 was markedly increased by activation of BMP signaling. (c,d) Sections were subjected to in situ hybridization for AQP4 and Hoechst 33342 staining. Low magnification images (c) and high magnification images (d) are shown. Activation of BMP signaling significantly increased mRNA expression levels of AQP4. (e) Sections were subjected to in situ hybridization for AQP4 and immunohistochemistry for GS. High magnification images are shown. Activation of BMP signaling increased mRNA expression levels of AQP4 in GS-positive cells. (f) Quantification of AQP4 immunoreactivity in the cerebral cortex. Activation of BMP signaling significantly increased AQP4 immunoreactivity. n = 3 animals for each condition. Bars represent mean ± SD. **p < 0.01, Student’s t-test. Numbers indicate the corresponding layers in the cerebral cortex. GM, gray matter; WM, white matter. Scale bars 200 µm (a,c), 25 µm (b,d) and 10 µm (e).
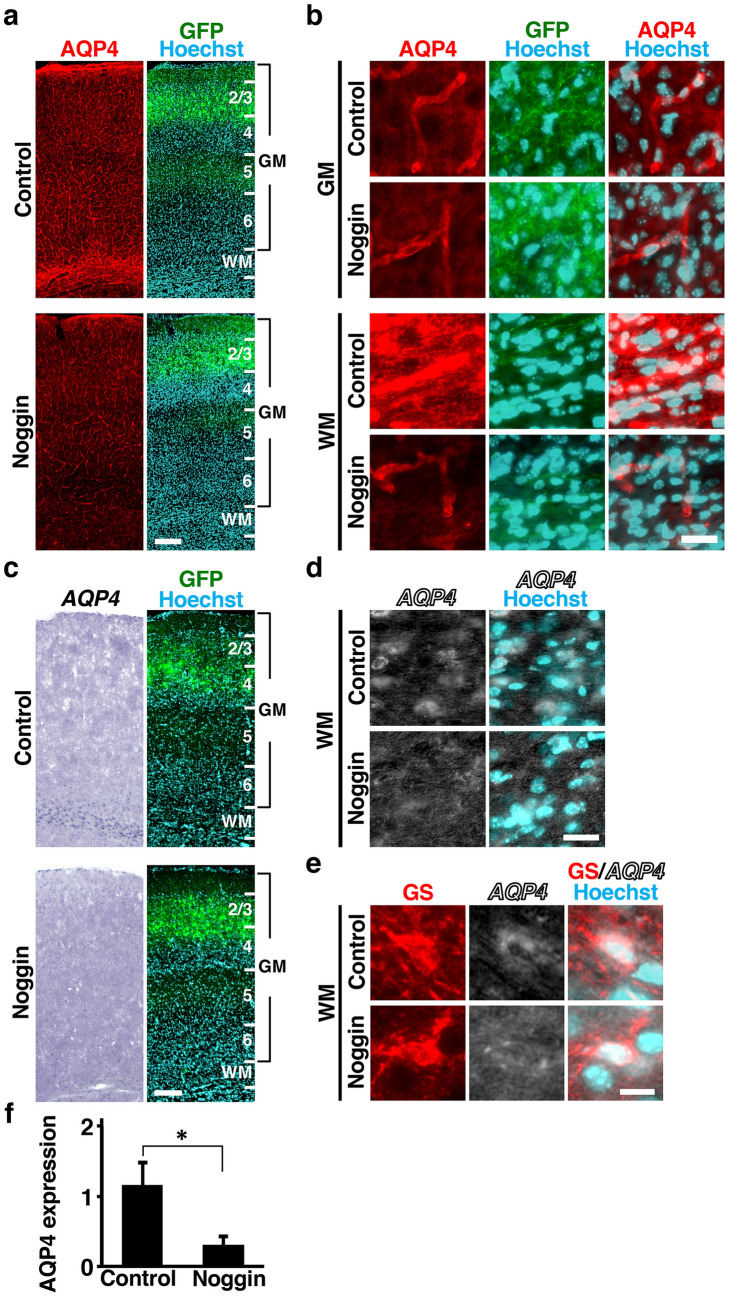
Inhibition of BMP signaling reduces AQP4 levels in the mouse cerebral cortex
We next examined whether endogenous BMP signaling is required for inducing AQP4 levels. To test this, we used noggin, which binds specifically to BMPs and antagonizes BMP signaling by blocking the interaction of BMPs with their receptors26. Noggin was introduced into the mouse cerebral cortex using IUE at E14, and AQP4 expression was examined at P16. Immunostaining showed that pSmad signals were markedly reduced by noggin (Supplementary Fig. S1c), suggesting that BMP signaling was inhibited by the introduction of noggin-expressing plasmids using IUE. In addition, the layer structure of the cerebral cortex was not affected by noggin electroporation (Supplementary Fig. S2). Immunohistochemistry and in situ hybridization consistently showed that noggin markedly reduced AQP4 levels (Fig. 4a–d). The reduction of AQP4 signals in the WM was obvious, whereas that in the GM was not clearly observable (Fig. 4a,c). This seems to be because AQP4 levels in the GM of the control cerebral cortex were lower than those in the WM. We performed in situ hybridization for AQP4 and immunohistochemistry for GS and found that noggin significantly decreased AQP4 mRNA in GS-positive cells in the WM (Fig. 4e). Our quantification for the intensity of AQP4 immunoreactivity showed that the inhibition of BMP signaling significantly decreased AQP4 protein levels in the WM (control, 1.16 ± 0.33; noggin, 0.30 ± 0.13; p < 0.05, Student’s t-test) (Fig. 4f).
Figure 4.
Inhibition of BMP signaling reduces AQP4 levels in the mouse cerebral cortex. pCAG-EGFP plus either pCAG-noggin or pCAG control vector was electroporated at E14, and the brains were dissected at P16. (a,b) Coronal sections stained with anti-AQP4 antibody, anti-GFP antibody and Hoechst 33342. Low magnification images (a) and high magnification images (b) of the GM and the WM are shown. The immunoreactivity of AQP4 in the WM was markedly decreased by noggin. (c,d) Sections were subjected to in situ hybridization for AQP4 and Hoechst 33342 staining. Low magnification images (c) and high magnification images of the WM (d) are shown. Noggin significantly decreased mRNA expression levels of AQP4 in the WM. (e) Sections are subjected to in situ hybridization for AQP4 and immunohistochemistry for GS. High magnification images of the WM are shown. (f) Quantification of AQP4 immunoreactivity in the WM. Inhibition of BMP signaling by noggin significantly decreased AQP4 immunoreactivity in the WM. n = 3 animals for each condition. Bars represent mean ± SD. *p < 0.05, Student’s t-test. Numbers indicate the corresponding layers in the cerebral cortex. GM, gray matter; WM, white matter. Scale bars 200 µm (a,c), 25 µm (b,d) and 10 µm (e).
To further confirm the importance of endogenous BMP signaling, we utilized a soluble dominant-negative BMP receptor (sBMPR) which lacks both transmembrane and serine/threonine kinase domains27. sBMPR was introduced into the mouse cerebral cortex using IUE at E14, and the cerebral cortex was examined at P16. Consistent with the results obtained using noggin, immunohistochemistry showed that sBMPR reduced AQP4 levels (Supplementary Fig. S3a,b), Our quantification showed that sBMPR significantly decreased AQP4 immunoreactivity in the WM (control, 0.85 ± 0.28; sBMPR, 0.4 ± 0.13; p < 0.05, Student’s t-test) (Supplementary Fig. S3c). These results demonstrate that endogenous BMP signaling is essential for the AQP4 levels found in astrocytes. Taken together, our findings indicate that BMP signaling increases AQP4 levels in the mouse cerebral cortex during development.
Discussion
Our experiments demonstrated that BMP signaling was activated in astrocytes which expressed AQP4. Activation of BMP signaling increased AQP4 levels in astrocytes. Consistently, inhibition of BMP signaling decreased AQP4 levels in astrocytes in the cerebral cortex. Our findings indicate that BMP signaling increases AQP4 levels in the mouse cerebral cortex during development.
The physiological regulation of AQP4 levels
Although previous studies have examined the regulatory mechanisms of AQP4 levels, most of the studies investigated these mechanisms under pathological conditions such as inflammatory responses, hepatic encephalopathy, epilepsy, brain edema and glioblastoma6–11. For example, the exposure of astrocytes to ammonium chloride, which is an in vitro model of hepatic encephalopathy, activates the p38 MAP kinase pathway, and as a result, AQP4 expression is upregulated12. Oxygen–glucose deprivation induces AQP4 upregulation in astrocytes through the p38 MAP kinase and ERK signaling pathways13. However, little was known about the regulatory mechanisms of AQP4 levels under normal physiological conditions. A previous pioneering study using cultured astrocytes reported that BMP activation increased the expression levels of AQP4 protein in astrocytes in vitro19. Our gain- and loss-of-function experiments clearly demonstrated that endogenous BMP signaling increases AQP4 levels in the mouse cerebral cortex in vivo during development. Because a previous study reported that BMP signaling promoted astrocyte maturation, increased AQP4 expression and downregulated EGF receptor expression19, it seems possible that activation of BMP signaling induced astrocyte maturation and, as a result, AQP4 expression was increased in the cerebral cortex.
BMP can activate both canonical and non-canonical intracellular signaling pathways. In the canonical BMP signaling pathway, activation of BMP receptors phosphorylates Smad1/5/8, which makes a complex with Smad4. Then, the complex translocates into the nucleus and regulates gene expression. In the non-canonical BMP signaling pathway, BMP activates several molecules, which are distinct from Smad, such as p38 MAP kinase, JNK, ERK, PI3 kinase and Cdc42. Because previous studies demonstrated that p38 MAP kinase regulates AQP4 expression downstream of ammonium chloride and oxygen–glucose deprivation, it seems plausible that p38 MAP kinase mediates the effect of BMP on AQP4 levels in the cerebral cortex. In addition, because our experiments showed that Smad1/5/8 was activated in AQP4-positive astrocytes, it remains possible that the canonical BMP signaling pathway is involved in AQP4 expression. Further investigation would be necessary in order to uncover the entire picture of the mechanisms underlying the physiological regulation of AQP4 levels.
Previous studies using IUE reported that radial migration of cortical neurons was inhibited by introducing either BMP or dominant-negative BMP receptors at E17.5 or P0, respectively21,28. In contrast, our experiments using BMP and noggin did not show obvious effects on the cortical layers at P16 (Supplementary Fig. S2). It seemed plausible that neuronal migration was delayed by activation or inhibition of BMP signaling but finished by P16.
Possible roles of AQP4 in the brain
The importance of AQP4 in the brain has been investigated intensively. It was reported that AQP4 has an important role in water and waste clearance in the brain. The deletion of AQP4 expression resulted in the reduction of interstitial solute clearance, and in the accumulation of amyloid β, a peptide thought to be pathogenic in Alzheimer’s disease29. Other studies reported that AQP4 is involved in sleep architecture. A cohort study of adult humans showed that genetic variants in the AQP4 genome are related to sleep quality30. Another study reported an AQP4-haplotype associated with distinct modulations of non-rapid eye movement (NREM) sleep31. It seems plausible that the regulation of AQP4 expression by BMP signaling, which we uncovered in this study, is involved in the regulation of water and waste clearance and sleep quality. It would be intriguing to examine whether BMP signaling alters water and solute clearance and sleep quality through regulating AQP4 expression.
The BBB is important for brain homeostasis because the BBB restricts large polar substances and potentially neurotoxic compounds in the circulation from passively diffusing into the brain4,5. Therefore, it is important to elucidate the mechanism underlying the structural and functional development of the BBB. AQP4 is one of the major functional components of the BBB and is crucial for water transport at the blood–brain interface1–3,5. Our data showed that BMP signaling is necessary and sufficient for the expression of AQP4 in vivo, raising the possibility that BMP signaling regulates the function of the BBB by modulating AQP4 expression. Interestingly, it was reported that the structure of the BBB is also regulated by BMP signaling32. Taken together, BMP signaling may regulate the BBB not only structurally but also functionally through regulating AQP4 expression.
Methods
Animals
ICR mice were purchased from SLC (Hamamatsu, Japan) and reared on a normal 12 h light/dark schedule. The day of conception and that of birth were counted as embryonic day 0 (E0) and postnatal day 0 (P0), respectively. All procedures were performed in accordance with protocols approved by the Animal Care Committee of Kanazawa University and with the ARRIVE guidelines.
In utero electroporation (IUE) procedure
IUE using mice was performed as described previously with slight modifications33,34. Briefly, pregnant ICR mice were anesthetized, and the uterine horns were exposed. Approximately 1–2 μl of DNA solution was injected into the lateral ventricle of embryos using a pulled glass micropipette. Each embryo within its uterus was placed between a tweezer-type electrode (CUY650 P3, NEPA Gene). Square electric pulses (45 V, 50 ms) were passed 5 times at 1 s intervals using an electroporator (ECM 830, BTX). Care was taken to quickly place embryos back into the abdominal cavity to avoid temperature loss. The wall and skin of the abdominal cavity were sutured, and embryos were allowed to develop normally.
Plasmids
Noggin was kindly provided by Dr. Daisuke Saito (Kyushu University) and was inserted into pCAG vector, yielding pCAG-noggin. sBMPR was kindly provided by Dr. Naoto Ueno (National Institute for Basic Biology, Okazaki, Japan) and was inserted into pCAG vector, yielding pCAG-sBMPR. The cDNA for mouse BMP7 was amplified using PCR with the following primers: 5′-ATTCTCGAGGCCACCATGCACGTGCGCTCGCTGC-3′ and 5′-AAGCGGCCGCCTAGTGGCAGCCACAGG-3′. The amplified fragment was digested with XhoI and NotI, and then subcloned into pCAG vector. pCAG-EGFP was described previously35. Plasmids were purified using the EndoFree Plasmid Maxi Kit (QIAGEN). To activate BMP signaling, a mixture of pCAG-EGFP (0.5 mg/ml) plus either pCAG-BMP7 or pCAG control plasmid (5 mg/ml) in PBS was used. To inhibit BMP signaling, a mixture of pCAG-EGFP (0.5 mg/ml) plus either pCAG-noggin, pCAG-sBMPR or pCAG control plasmid (5 mg/ml) was used. Fast Green solution was added at a final concentration of 0.3% to monitor the injection.
Tissue preparation
Tissue sections were prepared as described previously36,37. Mice were deeply anesthetized and transcardially perfused with 4% paraformaldehyde (PFA)/PBS. Brains were dissected and post-fixed with an overnight immersion in 4% PFA/PBS, cryoprotected with 2-day immersion in 30% sucrose/PBS, and embedded in OCT compound. Coronal sections of 20–30 μm or 50 μm thickness were made using a cryostat.
Immunohistochemistry
Immunohistochemistry was performed as described previously with slight modifications38,39. Briefly, free-floating sections were permeabilized with 0.3% Triton X-100/PBS, incubated overnight with primary antibodies in 2% bovine serum albumin (BSA)/PBS, and then washed with PBS 3 times for 10 min each. The sections were incubated with secondary antibodies and Hoechst 33342 in 2% BSA/PBS for 2 h, and then washed with PBS 3 times for 10 min each. The sections were mounted on slides with Mowiol (Sigma-Aldrich). Antibodies used were as follows: rabbit anti-aquaporin-4 (AQP4) antibody (Millipore), rabbit anti-glutamine synthetase (GS) antibody (Sigma-Aldrich), mouse anti-GS antibody (Roche), rabbit anti-NeuN antibody (Cell Signaling Technology), mouse CC1 antibody (Calbiochem), rabbit anti-phospho-Smad1/5/8 antibody (Cell Signaling Technology), rabbit anti-Cux1 antibody (Santa Cruz), rat anti-Ctip2 antibody (Abcam), donkey secondary antibodies conjugated with Cy3 (Jackson ImmunoResearch), donkey secondary antibodies conjugated with Alexa Fluor 647 (Molecular Probes) and donkey secondary antibodies conjugated with Alexa Fluor 488 (Molecular Probes).
In situ hybridization
In situ hybridization was performed as described previously with slight modifications37,40,41. Briefly, sections of 20–30 μm thickness were incubated overnight with digoxigenin-labeled RNA probes in hybridization buffer (50% formamide, 5 × SSC, 5 × Denhardt's solution, 0.3 mg/ml yeast RNA, 0.1 mg/ml herring sperm DNA, and 1 mM dithiothreitol). The sections were then incubated with an alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche) and were visualized using NBT/BCIP as substrates. In some experiments, the sections were then subjected to immunohistochemistry and Hoechst 33342 staining.
To make an RNA probe for mouse AQP4, the cDNA fragment for mouse AQP4 was amplified with RT-PCR using the following primers: 5'-AAAGAATTCTTTTGCTCGGTGTGGGAT-3' and 5'-AAAGAATTCGAAGCAAGAAACCCGCAA-3'. The amplified fragment was digested with EcoRI and then subcloned into pCRII-TOPO vector, and digoxigenin-labeled RNA probes were made. To make an RNA probe for mouse BMP7, digoxigenin-labeled RNA probes were made using pCAG-BMP7 plasmid.
Microscopy
Epifluorescence microscopy was performed with BZ-9000 and BZ-X710 microscopes (KEYENCE).
Quantification and statistical analysis
Coronal sections were stained with Hoechst 33342 plus either anti-GS antibody, anti-NeuN antibody, CC1 antibody, anti-AQP4 antibody or anti-phospho-Smad1/5/8 antibody. In some experiments, the sections were subjected to in situ hybridization for AQP4. Images captured using a BZ-9000 microscope were used for quantification. The background signal was removed by subtracting the average signal intensity of negative cells.
For cell counting, the numbers of immunopositive cells in columns with 200 μm width were manually counted using the “Cell Counter” tool of Image J software.
To measure the average intensities of AQP4 immunoreactivity, the regions in columns with 200 μm width corresponding to the GM or the WM were selected using the “Rectangle Selection” function of ImageJ. Average signal intensities in the regions were measured using the “ROI Manager” tool of ImageJ. To minimize variation in signal intensity depending on the position of each section, the average intensity of AQP4 on the electroporated side and that on the contralateral non-electroporated side of the cortex in the same brain section were measured, and the former was divided by the latter. Values in graphs are mean ± SD. An unpaired two-tailed Student's t-test was used to determine statistical significance. P values<0.05 were considered statistically significant.
Supplementary Information
Acknowledgements
We thank Dr. Naoto Ueno (National Institute for Basic Biology) and Dr. Daisuke Saito (Kyushu University) for plasmids. We are grateful to Zachary Blalock and the Kawasaki lab members for their helpful support. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan Agency for Medical Research and Development (AMED), Takeda Science Foundation, the Uehara Memorial Foundation, the Naito Foundation, the Kanazawa University SAKIGAKE project 2018 and the Kanazawa University CHOZEN project.
Author contributions
K.M., Y.S. and H.K. designed the research. K.M., N.M., K.S., T.H.-H., K.M. and Y.S. performed experiments. K.M., Y.S. and H.K. wrote the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-89997-5.
References
- 1.Verkman AS, Binder DK, Bloch O, Auguste K, Papadopoulos MC. Three distinct roles of aquaporin-4 in brain function revealed by knockout mice. Biochim. Biophys. Acta. 2006;1758:1085–1093. doi: 10.1016/j.bbamem.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen S, et al. Specialized membrane domains for water transport in glial cells: High-resolution immunogold cytochemistry of aquaporin-4 in rat brain. J. Neurosci. 1997;17:171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rash JE, Yasumura T, Hudson CS, Agre P, Nielsen S. Direct immunogold labeling of aquaporin-4 in square arrays of astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Proc. Natl. Acad. Sci. USA. 1998;95:11981–11986. doi: 10.1073/pnas.95.20.11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manley GT, et al. Aquaporin-4 deletion in mice reduces brain edema after acute water intoxication and ischemic stroke. Nat. Med. 2000;6:159–163. doi: 10.1038/72256. [DOI] [PubMed] [Google Scholar]
- 5.Haj-Yasein NN, et al. Glial-conditional deletion of aquaporin-4 (Aqp4) reduces blood-brain water uptake and confers barrier function on perivascular astrocyte endfeet. Proc. Natl. Acad. Sci. USA. 2011;108:17815–17820. doi: 10.1073/pnas.1110655108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binder DK, Nagelhus EA, Ottersen OP. Aquaporin-4 and epilepsy. Glia. 2012;60:1203–1214. doi: 10.1002/glia.22317. [DOI] [PubMed] [Google Scholar]
- 7.Salman MM, et al. Transcriptome analysis suggests a role for the differential expression of cerebral aquaporins and the MAPK signalling pathway in human temporal lobe epilepsy. Eur. J. Neurosci. 2017;46:2121–2132. doi: 10.1111/ejn.13652. [DOI] [PubMed] [Google Scholar]
- 8.Papadopoulos MC, Manley GT, Krishna S, Verkman AS. Aquaporin-4 facilitates reabsorption of excess fluid in vasogenic brain edema. FASEB J. 2004;18:1291–1293. doi: 10.1096/fj.04-1723fje. [DOI] [PubMed] [Google Scholar]
- 9.Saadoun S, Papadopoulos MC, Davies DC, Krishna S, Bell BA. Aquaporin-4 expression is increased in oedematous human brain tumours. J. Neurol. Neurosurg. Psychiatry. 2002;72:262–265. doi: 10.1136/jnnp.72.2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warth A, Mittelbronn M, Wolburg H. Redistribution of the water channel protein aquaporin-4 and the K+ channel protein Kir4.1 differs in low- and high-grade human brain tumors. Acta Neuropathol. 2005;109:418–426. doi: 10.1007/s00401-005-0984-x. [DOI] [PubMed] [Google Scholar]
- 11.Hsu Y, Tran M, Linninger AA. Dynamic regulation of aquaporin-4 water channels in neurological disorders. Croat. Med. J. 2015;56:401–421. doi: 10.3325/cmj.2015.56.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan CF, Zhu SM, Zheng YY. Ammonia induces upregulation of aquaporin-4 in neocortical astrocytes of rats through the p38 mitogen-activated protein kinase pathway. Chin. Med. J. (Engl) 2010;123:1888–1892. doi: 10.3760/cma.j.issn.0366-6999.2010.14.011. [DOI] [PubMed] [Google Scholar]
- 13.Qi LL, et al. CysLT2 receptor-mediated AQP4 up-regulation is involved in ischemic-like injury through activation of ERK and p38 MAPK in rat astrocytes. Life Sci. 2011;88:50–56. doi: 10.1016/j.lfs.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 14.Furuta Y, Piston DW, Hogan BL. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development. 1997;124:2203–2212. doi: 10.1242/dev.124.11.2203. [DOI] [PubMed] [Google Scholar]
- 15.Gross RE, et al. Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron. 1996;17:595–606. doi: 10.1016/s0896-6273(00)80193-2. [DOI] [PubMed] [Google Scholar]
- 16.Shimogori T, Banuchi V, Ng HY, Strauss JB, Grove EA. Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development. 2004;131:5639–5647. doi: 10.1242/dev.01428. [DOI] [PubMed] [Google Scholar]
- 17.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda S, Taga T. Cell fate determination regulated by a transcriptional signal network in the developing mouse brain. Anat. Sci. Int. 2005;80:12–18. doi: 10.1111/j.1447-073x.2005.00097.x. [DOI] [PubMed] [Google Scholar]
- 19.Scholze AR, Foo LC, Mulinyawe S, Barres BA. BMP signaling in astrocytes downregulates EGFR to modulate survival and maturation. PLoS One. 2014;9:e110668. doi: 10.1371/journal.pone.0110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munk AS, et al. PDGF-B is required for development of the glymphatic system. Cell Rep. 2019;26(11):2955-2969.e3. doi: 10.1016/j.celrep.2019.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena M, Agnihotri N, Sen J. Perturbation of canonical and non-canonical BMP signaling affects migration, polarity and dendritogenesis of mouse cortical neurons. Development. 2018;145:dev147157. doi: 10.1242/dev.147157. [DOI] [PubMed] [Google Scholar]
- 22.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segklia A, et al. Bmp7 regulates the survival, proliferation, and neurogenic properties of neural progenitor cells during corticogenesis in the mouse. PLoS One. 2012;7:e34088. doi: 10.1371/journal.pone.0034088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choe Y, Kozlova A, Graf D, Pleasure SJ. Bone morphogenic protein signaling is a major determinant of dentate development. J. Neurosci. 2013;33:6766–6775. doi: 10.1523/JNEUROSCI.0128-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazono K, Kamiya Y, Morikawa M. Bone morphogenetic protein receptors and signal transduction. J. Biochem. 2010;147:35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- 26.Zimmerman LB, De Jesus-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 27.Natsume T, et al. Interaction between soluble type I receptor for bone morphogenetic protein and bone morphogenetic protein-4. J. Biol. Chem. 1997;272:11535–11540. doi: 10.1074/jbc.272.17.11535. [DOI] [PubMed] [Google Scholar]
- 28.Choe Y, Pleasure SJ. Meningeal Bmps regulate cortical layer formation. Brain Plast. 2018;4:169–183. doi: 10.3233/BPL-170048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iliff JJ, et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci. Transl. Med. 2012;4:147ra111. doi: 10.1126/scitranslmed.3003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rainey-Smith SR, et al. Genetic variation in Aquaporin-4 moderates the relationship between sleep and brain Abeta-amyloid burden. Transl. Psychiatry. 2018;8:47. doi: 10.1038/s41398-018-0094-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulv Larsen SM, et al. Haplotype of the astrocytic water channel AQP4 is associated with slow wave energy regulation in human NREM sleep. PLoS Biol. 2020;18:e3000623. doi: 10.1371/journal.pbio.3000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Araya R, et al. BMP signaling through BMPRIA in astrocytes is essential for proper cerebral angiogenesis and formation of the blood-brain-barrier. Mol. Cell. Neurosci. 2008;38:417–430. doi: 10.1016/j.mcn.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakimoto M, et al. Classic cadherins mediate selective intracortical circuit formation in the mouse neocortex. Cereb. Cortex. 2015;25:3535–3546. doi: 10.1093/cercor/bhu197. [DOI] [PubMed] [Google Scholar]
- 34.Hoshiba Y, et al. Sox11 balances dendritic morphogenesis with neuronal migration in the developing cerebral cortex. J. Neurosci. 2016;36:5775–5784. doi: 10.1523/JNEUROSCI.3250-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sehara K, et al. Whisker-related axonal patterns and plasticity of layer 2/3 neurons in the mouse barrel cortex. J. Neurosci. 2010;30:3082–3092. doi: 10.1523/JNEUROSCI.6096-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayakawa I, Kawasaki H. Rearrangement of retinogeniculate projection patterns after eye-specific segregation in mice. PLoS One. 2010;5:e11001. doi: 10.1371/journal.pone.0011001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwai L, et al. FoxP2 is a parvocellular-specific transcription factor in the visual thalamus of monkeys and ferrets. Cereb. Cortex. 2013;23:2204–2212. doi: 10.1093/cercor/bhs207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinh Duong TA, et al. FGF signaling directs the cell fate switch from neurons to astrocytes in the developing mouse cerebral cortex. J. Neurosci. 2019;39:6081–6094. doi: 10.1523/JNEUROSCI.2195-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toda T, et al. Birth regulates the initiation of sensory map formation through serotonin signaling. Dev. Cell. 2013;27:32–46. doi: 10.1016/j.devcel.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki H, Crowley JC, Livesey FJ, Katz LC. Molecular organization of the ferret visual thalamus. J. Neurosci. 2004;24:9962–9970. doi: 10.1523/JNEUROSCI.2165-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebisu H, Iwai-Takekoshi L, Fujita-Jimbo E, Momoi T, Kawasaki H. Foxp2 regulates identities and projection patterns of thalamic nuclei during development. Cereb. Cortex. 2017;27:3648–3659. doi: 10.1093/cercor/bhw187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.