Highlights
-
•
Archaea-specific primers in high-throughput sequencing found archaea reads.
-
•
Archaea community with DNA-based analysis was different from RNA-based analysis.
-
•
Archaea are a natural part of microbiomes in drinking water distribution systems.
-
•
Active archaea reads were high in non-disinfected and cold distribution systems.
Keywords: Archaea diversity, Drinking water distribution system, Chlorination, High-throughput sequencing, 16S rRNA gene, 16S rRNA gene transcript
Graphical abstract
Abstract
The knowledge about the members of active archaea communities in DWDS is limited. The current understanding is based on high-throughput 16S ribosomal RNA gene (DNA-based) amplicon sequencing that reveals the diversity of active, dormant, and dead members of the prokaryote (bacteria, archaea) communities. The sequencing primers optimized for bacteria community analysis may underestimate the share of the archaea community. This study characterized archaea communities at five full-scale drinking water distribution systems (DWDS), representing a variety of drinking water production units (A-E); A&B use artificially recharged non-disinfected groundwater (ARG), the other DWDS's supplied water disinfected by using ultraviolet (UV) light and chlorine compounds, C&D were surface waterworks and E was a ground waterworks. For the first time for archaea community analyses, this study employed the archaea-specific high-throughput sequencing primers for 16S ribosomal RNA (rRNA) as a target (reverse-transcribed cDNA; an RNA-based approach) in addition to the previously used 16S rRNA gene target (rDNA; a DNA-based approach) to reveal the active fraction of the archaea present in DWDS. The archaea community structure in varying environmental conditions in the water and biofilm of the five DWDSs were investigated by taking into consideration the system properties (cold or hot water system) and water age (distance from the treatment plants) in samples from each season of one year.
The RNA-based archaea amplicon reads were obtained mostly from cold water samples from DWDSs (A–B) distributing water without disinfection where the DNA-based and RNA-based analysis created separate clusters in a weighted beta-diversity analysis. The season and location in DWDS A further affected the diversity of these archaea communities as was seen by different clusters in beta-diversity plots. The recovery of archaea reads was not adequate for analysis in any of the disinfected samples in DWDSs C–E or non-disinfected hot water in DWDSs A–B when utilizing RNA-based template. The metabolically active archaea community of DWDSs thus seemed to be effectively controlled by disinfection of water and in the hot water systems by the temperature. All biofilms regardless of DWDS showed lower species richness values (mainly Nitrososphaeria class) than non-disinfected water from DWDSs A–B where several archaea classes occurred (e.g. Woesearchaeia, Nitrososphaeria, Micrarchaeia, Methanomicrobia, Iairchaeia, Bathyarchaeia) indicating only part of the archaea members were able to survive in biofilms. Thus, Archaea has been shown as a significant part of normal DWDS biota, and their role especially in non-disinfected DWDS may be more important than previously considered.
1. Introduction
Drinking water distribution systems (DWDS) have diverse biotic communities from wide taxonomic ranges such as archaea, bacteria, eukaryotes, and viruses (Wielen et al., 2009; Prest et al., 2016; Inkinen et al., 2016, 2019). Although archaea are an important domain of microbes, most of these communities have uncultured nature and thus they are not fully understood in the discussion of DWDS microbiomes (Prest et al., 2016; Steen et al., 2019). Therefore, research reports related to archaea in DWDS are rare (Wielen et al., 2009; Roeselers et al., 2015; Bautista-de los Santos et al., 2016; Bradley et al., 2020). In drinking water treatment plants, ammonia-oxidizing archaea (AOA) have shown high abundance and are responsible for the nitrification activities in the water treatment filtration processes (Wielen et al., 2009; Kasuga et al., 2010). Ammonium can be present in DWDS of ground waterworks and at surface waterworks too when chloramination is used for disinfection, and thus in these systems, the proportion of AOA communities could be high (Wielen et al., 2009; Waak et al., 2019; Bradley et al., 2020).
Until recently, it was thought that only a few types of bacteria in the genera Nitrosomonas, Nitrosospira, and Nitrosococcus have a key role in ammonia oxidation. However, Koenneke et al. (2005) discovered the important role of archaea in ammonia oxidation. Since their discovery, AOA has often been found to outnumber the ammonia-oxidizing Bacteria in many habitats. The occurrence of archaea and their metabolic functions in the Earth's natural habitats (e.g. aquatic environments, soils) and man-made systems (e.g. wastewater treatment plants) have been reported (Wielen et al., 2009; Auguet et al., 2010; Gonzalez-Martinez et al., 2018). Archaea can survive in nutrient-poor oligotrophic settings, such as subsurface environments, in a viable state with an extremely low metabolic rate (Hoehler and Jørgensen, 2013). Archaea have even been suggested to be crucial in the development of microbial co-occurrences and to play a vital role in maintaining microbial ecosystems in the soil environment (Shi et al., 2019).
The microbial growth in DWDS occurs within the biofilms at the inner surfaces of the pipelines and may change the physical, chemical, and biological characteristics of drinking water (Lie et al., 2016). Therefore, two approaches; disinfection (such as chlorination, UV) and intensive removal of nutrients (biologically available organic carbon and phosphorus) are commonly practiced for keeping DWDSs biologically stable (Ikonen et al., 2017; Zhang et al., 2017; Bautista-de los Santos, et al. 2019; Dai et al., 2020). Further, the availability of micro-nutrients such as iron, manganese, and zinc in DWDS increases the autotrophic communities (Liu et al., 2016). The grow-die cycle of biofilm communities in the DWDS might increase the availability of nutrients for further biological growth (Fish et al., 2015; Lie et al., 2016).
In ecological microbiome studies including the drinking water environment, the major focus has been to study the Bacteria domain. For Bacteria, widely utilized universal primer pairs detect also Archaea, but their Archaea coverage might be poor (Klindworth et al., 2013; Roeselers et al., 2015; Thijs et al., 2017; Zhang et al., 2017). New archaea primers have been designed and utilized for better coverage and specificity (Stahl and Amann, 1991; Gantner et al., 2011). In recent years, varying archaea-specific primers have been successfully utilized to better describe archaea diversity in varying habitats, including DWDS biofilms (Fish et al., 2015), wastewater, and soil (Shi et al., 2019). The aforementioned microbial community studies have targeted the 16S ribosomal RNA genes (DNA-based method) and thus detect active, dormant, and dead community members. In previous studies of bacteria and eukaryotes in DWDSs, ribosomal RNA (RNA-based method) has been utilized for reverse-transcribed RNA prior to sequencing as complementary DNA (cDNA) to capture active community members (Inkinen et al., 2016, 2019). By using high-throughput techniques, an RNA-based template has recently been used to study archaea communities in the bovine rumen (Kang et al., 2013), and anaerobic digestion (Vrieze et al., 2018). In one earlier study, an RNA-based template was also used as a target for archaea in a fluorescent oligonucleotide probe (Manz et al., 1993).
This study comprehensively investigated archaea communities existing in full-scale DWDSs by using an archaea-specific primer pair for high-throughput sequencing utilizing DNA- and RNA-based templates. The study was extended to assess the impact of DWDS properties with a strong focus on evaluating both the overall and active/dormant archaea communities. To capture the archaea diversity, varying environmental conditions were included: non-disinfected systems and systems with post-treatment disinfection (UV, chlorination, or chloramination), surface water, artificial recharge groundwater or groundwater as source waters for drinking water production, as well as samples from the various DWDS habitats (water or biofilm), system properties (cold or hot water system), location in the DWDS (water age) and seasons of the year (winter, spring, summer, and autumn).
2. Material and methods
2.1. DWDS sample and water quality data collection
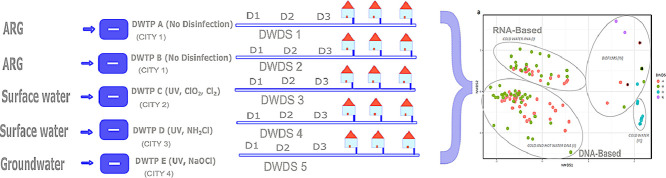
Water samples were collected from five different DWDSs (A–E) of Finland. Each DWDS served waterworks with varying raw water sources, purification processes, and varying disinfection treatments as follows: artificial groundwater production without any disinfection (DWDSs A–B, in the same city), surface waterworks with chlorine dioxide (ClO2), and chlorine (Cl2) disinfection (DWDS C), surface waterworks with chloramine (NH2Cl) disinfection (DWDS D) and groundwater distributed with sodium hypochlorite (NaOCl) (DWDS E). All disinfection treatments (DWDSs C–E) included UV disinfection before chlorination.
A total of 180 samples; including cold drinking water (N = 119), hot water (N = 40), biofilm, i.e. pipeline biofilm or water meter soft deposits (N = 16), and water meter related water (N = 5) samples were analysed. A detailed description of the sampling, sample processing as well as specific details of drinking water distribution system (DWDS) characteristics, sampling details, and water quality measurement results can be found in our previous research papers focusing on eukaryotic communities (Inkinen et al., 2019) and physico-chemical water quality (Ikonen et al., 2017).
In each DWDS A–E, large volume water samples (100 L) from taps were collected and filtered using a dead-end ultrafiltration method (DEUF) (Smith and Hill 2009; Inkinen et al., 2019). A hollow-fiber polysulfone filter (ASAHI Rexeed-25A, Asahi Kasei Medical Co., Ltd., Tokyo, Japan) was attached to a tap for sample collection and eluted later in the laboratory (Smith and Hill 2009). The average flow of water during sample collection was 3 L/min. The samples were collected in two consecutive weeks in each of the four seasons: winter (January–February), spring (March-May), summer (August–September), and autumn (October–December), in the year 2015. Water sampling in all seasons and within each DWDS included three different locations at increasing distances (cumulative pipe length; 1–9 km, 3–26 km, and 11–36 km) from the waterworks (Inkinen et al., 2019). Besides, hot water samples were collected merely from the second location (3–26 km). One to three biofilm samples were collected from each DWDS (A–E) as pipeline biofilm or water meter soft deposits. Biofilm samples from the late autumn season included three pipeline samples from the second location in waterworks D–E using 15 cm detachable pieces of pipe as pipe collectors. Water meter biofilms included samples from varying locations within DWDSs (two each in DWDS A–C, one in DWDS D, and three in DWDS E). Biofilm of the pipe collector was removed by shaking 1350 rpm for 3 × 5 min (Heidolph Vibramax, Schwabach, Germany) with sterile 2 mm glass beads (Karl Hecht GmbH & Co. KG, Germany) followed by rinsing with 5-ml sample water (Inkinen et al., 2019). Biofilm from water meters was detached by brushing and collected using a pipette. All biofilm samples were sonicated for 1 min in 40 kHz (Branson Ultrasonics, Danbury, USA). Two water meter related water samples were collected from each DWDSs A–B and one in DWDS C. Filtration through a polycarbonate membrane (Nuclepore Polycarbonate, Whatman, Kent, UK) with a pore size of 0.4 µm was conducted on the water samples (ca. 75 ml DEUF eluate, range 65–145 ml) and biofilms (10 ml suspension) for a secondary concentration of bacteria and archaea biomass. The final DEUF eluate volumes for analysis corresponded to water volumes of 8–25 L (mean 14 L). Negative controls (N = 35) were taken at different sample processing steps as earlier described (Inkinen et al., 2019). Positive controls for archaea were not available. Membranes were kept frozen at −75 °C before extraction of nucleic acids.
2.2. DNA and RNA extraction, and PCR amplification
DNA and RNA were extracted from the membranes as described earlier (Inkinen et al., 2019). In brief, total nucleic acids were extracted to a final elution volume of 100 µl using a Chemagic DNA Plant kit (Perkin Elmer, Waltham, MA, USA) following the manufacturer's instructions without removing the RNA. The RNA was purified from a 30 µl subsample of the extracted nucleic acids using an Ambion TURBO DNA-free™ kit (Life Technologies, Carlsbad, CA, USA) followed by reverse transcription to complementary DNA (cDNA) using the Invitrogen™ Superscript III First-Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA, USA).
Barcoded primers A340F (5′-CCCTAYGGGGYGCASCAG-3′) (Gantner et al., 2011) and 915R (5′-TGTGCTCCCCCGCCAATTCCT-3′) (Stahl and Amann, 1991), designed specifically for archaea, were used in a tagged amplicon PCR before the Illumina MiSeq sequencing process. The PCR reaction was conducted as described earlier (Inkinen et al., 2019) and included ca. 1–10 ng of nucleic acids. Illumina libraries were constructed using a ca. 100 ng purified amplicon pool, and gel electrophoresis was used for pooling and size selection at LGC Genomics GmbH (Berlin, Germany).
2.3. Next-generation sequencing and bioinformatics
Sequencing was performed on an Illumina MiSeq platform that produced 300 base pair (bp) paired-end reads (Illumina, Inc., San Diego, CA, US). The data were denoised by using the DADA2 protocol (software version 1.8) to produce amplicon sequence variants (ASVs) (Callahan et al., 2016). Since the overlap was just 25 reads with the archaea primers used (A340F–915R), the initial pre-processing of reads for low-quality removal reduced the whole overlap region in the upstream analysis. However, the information from both the R1 and R2 reads together is more informative for taxonomy identification with higher accuracy. The R1 and R2 reads were stitched with ambiguous "N" as done in an earlier study (Jeraldo et al., 2014). Before stitching, all reads were checked for missing overlap regions. The code for stitching Read 1 (file1.R1.preprocessed.cleaned.fastq) and Read 2 (file1.R2.preprocessed.cleaned.fastq) was as follows: paste file.R1.preprocessed.cleaned.fastq -d " " file.R2.preprocessed.cleaned.fastq | sed 's/ /NNNNNNNNNN/g' > stitched_reads_file1.R1_R2.fastq.
During the insertion of ambiguous “N”, the R1 and R2 processed reads were joined as a single read (R1-N-R2) by mapping and aligning to the full-length 16S region before proceeding further in the analysis pipeline. The sequence table was constructed, and chimeras were removed using a “per-sample” method (Callahan et al., 2016). The final ASV table was constructed and the taxonomy was assigned using the SILVA database version 128 release (Quast et al., 2013; Ylimaz et al., 2014). Then, the downstream processing of the ASV table was performed using the QIIME software package (version 1.9.1) (Caporaso et al., 2010) and visualized in the MicrobiomeAnalyst software package (Dhariwal et al., 2017; Chong et al., 2020).
2.4. Data analyses and statistics
The initial alpha-rarefaction limit (402 total sequences) for the entire data including all three domains (Archaea, Bacteria, Eukaryota) was determined based on sample versus negative controls present in the same beta-diversity plots produced in the QIIME software and visualized in the EMPeror software (Vázquez-Baeza et al., 2013), according to the total read counts. A new ASV table containing only archaea (rarefraction 375) was filtered from the entire table (Archaea, Bacteria, Eukaryota) for further analysis in MicrobiomeAnalyst. Samples that produced less than 375 archaea reads were excluded from the analysis afterward based on beta-diversities, alpha-diversity values, and taxa profile comparisons. During analysing each subsample type in MicrobiomeAnalyst, the data was rarefied into the lowest number of reads of the particular subsample type for avoiding the biased of the high variation of read numbers (Dhariwal et al., 2017; Chong et al., 2020). The rarefraction limit for each subsample type is described in each table and figure. Only sample sets (e.g. DWDS, sample type, location, DNA/RNA) that produced on average enough archaea reads (archaea reads >30% relative abundance) for analysis were chosen to produce representative archaea community characterization results for the given environment.
Alpha-diversity values were calculated from the ASV table using standard metrics such as observed ASVs, Chao1, ACE, Simpson, and Shannon. Alpha-diversity indices were compared between different sample water types with non-parametric Kruskal-Wallis and Mann-Whitney tests in IBM SPSS (version 27).
Beta-diversity values (weighted Bray-Curtis and unweighted Binary Jaccard distance matrices) were calculated from the ASV table and differences between the samples were displayed using an analysis of similarity (ANOSIM) for non-metric multidimensional scaling (NMDS) plots. Unweighted beta-diversity (Jaccard) was reported if the visual analysis showed different main observations compared to the weighted beta-diversity (Bray-Curtis). Taxa summaries by relative abundance (%) were calculated for representative taxa (class and genus level). The absolute abundance of ASVs for the entire dataset was visualized as a heatmap, hierarchically clustered on a taxon scale (archaeal class) based on the default Euclidean distance and average clustering method. The taxa summaries, heatmap clustering analysis as well as the alpha- and beta-diversities of the entire archaea data were calculated and visualized using the MicrobiomeAnalyst software.
The majority of the statistical analyses were performed on the water data from DWDSs A–B as these contained enough reads to capture the characteristics of the archaea community. Only the RNA of the hot water samples failed to yield enough archaea reads in DWDSs A–B. Three main subsets: 1) the entire dataset of 119 archaea samples (main drivers i.e. clusters of the archaea communities), 2) cold water from non-disinfected DWDSs A–B consisting of 84 samples excluding water meter samples (DNA/RNA, DWDS, season, location) and 3) second sampling locations of DWDSs A–B including 29 samples (cold versus hot water, only DNA) were used for statistical analysis. The statistical significance level of P < 0.05 was used in all statistical analyses.
2.5. Availability of data and material
Primer clipped reads were deposited in the Sequence Read Archive (SRA) of NCBI under existing BioSample accession numbers from SAMN10653499 to SAMN10653948 of the BioProject PRJNA509718 (Inkinen et al., 2019) with a new submission (SUB6956053) describing archaea sequences. All samples and controls in this project are listed in a metadata file (Supplementary file 6, Table S3).
3. Results
3.1. Archaea reads coverage and data description of all domains
From the 360 samples, 335 samples produced reads in high-throughput sequencing analyses. A rarefaction value of 402 for the total reads based on controls was used to filter out low total read samples which yielded 289 samples for further analysis. Overall, 768,600 DNA-based reads (Table 1) and 3722,244 RNA-based reads (Table 2) were obtained while accounting for Bacteria, Archaea, and Eukaryota reads together.
Table 1.
Summary of all DNA-based library reads (rarefraction 402). The archaea read numbers are marked bold if the sampling location was considered to contain enough archaea reads for analysis. Only location 3 was included for analysis in DWDS D. Others include Eukaryota or unassigned domain reads.
| DWDS | Source | Total reads (100%) | Archaea reads (83.8%) | Bacteria reads (10.0%) | Other (6.2%) | |||||||||||
| Avg. | Min | Max | N | Avg. | Min | Max | % | Avg. | Min | Max | % | Avg. | Min | Max | ||
| A | Cold water | 3810 | 610 | 10,450 | 25 | 3750 | 610 | 10,450 | 98.4 | <5 | 0 | 30 | 0.0 | 60 | 60 | 60 |
| Hot water | 2410 | 500 | 4570 | 6 | 2150 | 440 | 4290 | 89.6 | 120 | 40 | 200 | 4.9 | 130 | 20 | 340 | |
| Biofilm | 62,710 | 37,140 | 88,280 | 2 | 62,420 | 36,930 | 87,910 | 99.5 | 30 | 30 | 40 | 0.1 | 260 | 180 | 340 | |
| B | Cold water | 6950 | 610 | 52,890 | 26 | 6880 | 590 | 52,750 | 99.0 | 0 | 0 | <5 | 0.0 | 70 | 0 | 390 |
| Hot water | 4290 | 1190 | 8900 | 7 | 3630 | 1110 | 8830 | 84.8 | 440 | <5 | 1220 | 10.3 | 210 | 30 | 690 | |
| Biofilm | 15,570 | 14,480 | 16,670 | 2 | 15,330 | 14,280 | 16,370 | 98.4 | <5 | 0 | 1 | 0.0 | 240 | 190 | 290 | |
| C | Cold water | 1520 | 400 | 6440 | 20 | <5 | 0 | 20 | 0.2 | 870 | 40 | 5920 | 57.0 | 650 | 10 | 1580 |
| Hot water | 1360 | 490 | 2720 | 5 | 0 | 0 | 0 | 0.0 | 700 | 70 | 1700 | 51.7 | 660 | 360 | 1670 | |
| Biofilm | 3460 | 3460 | 3460 | 1 | 0 | 0 | 0 | 0.0 | 2190 | 2190 | 2190 | 63.3 | 1270 | 1270 | 1270 | |
| D | Cold water | 5740 | 500 | 45,640 | 15 | 4660 | 0 | 44,860 | 81.0 | 140 | 0 | 350 | 2.4 | 950 | 40 | 1990 |
| Hot water | 2570 | 740 | 3980 | 6 | 10 | 0 | 60 | 0.4 | 180 | 0 | 550 | 7.0 | 2380 | 740 | 3830 | |
| Biofilm | 8040 | 1830 | 14,630 | 4 | 2790 | 0 | 11,140 | 34.6 | 2880 | 50 | 10,730 | 35.8 | 2380 | 1340 | 3900 | |
| E | Cold water | 1410 | 440 | 3570 | 7 | 160 | 20 | 530 | 11.0 | 850 | 0 | 3520 | 60.0 | 410 | 0 | 1350 |
| Hot water | 7440 | 1300 | 20,280 | 6 | <5 | 0 | 30 | 0.1 | 7370 | 1300 | 20,170 | 99.0 | 70 | 0 | 180 | |
| Biofilm | 31,440 | 27,280 | 35,590 | 2 | 31,230 | 26,930 | 35,530 | 99.3 | 50 | 0 | 100 | 0.2 | 160 | 60 | 260 | |
Table 2.
Summary of all the RNA-based library reads (rarefraction 402). Only cold water and the biofilm from DWDSs A–B contain sufficient archaea read counts for further analysis (bold numbers). Others include Eukaryota or unassigned domain reads.
| DWDS | Source | Total reads (100%) | Archaea reads (12.7%) | Bacteria reads (87.3%) | Other (0.004%) | |||||||||||
| Avg. | Min | Max | N | Avg. | Min | Max | % | Avg. | Min | Max | % | Avg. | Min | Max | ||
| A | Cold water | 4600 | 470 | 9310 | 21 | 1990 | 70 | 5700 | 43.2 | 2610 | 400 | 6300 | 56.8 | <5 | 0 | 10 |
| Hot water | 12,820 | 410 | 30,340 | 8 | 100 | 0 | 310 | 0.8 | 12,710 | 410 | 30,310 | 99.2 | 0 | 0 | 0 | |
| Biofilm | 17,770 | 15,090 | 20,450 | 2 | 13,460 | 12,990 | 13,940 | 75.8 | 4310 | 2100 | 6510 | 24.2 | 0 | 0 | 0 | |
| B | Cold water | 6210 | 410 | 17,270 | 22 | 4180 | 50 | 16,510 | 67.2 | 2030 | 220 | 6330 | 32.7 | <5 | 0 | 10 |
| Hot water | 18,120 | 1170 | 48,430 | 7 | 170 | 0 | 400 | 0.9 | 17,950 | 1170 | 48,380 | 99.1 | 0 | 0 | 0 | |
| Biofilm | 17,860 | 2260 | 33,460 | 2 | 7660 | 90 | 15,220 | 42.9 | 10,200 | 2170 | 18,230 | 57.1 | 0 | 0 | 0 | |
| C | Cold water | 22,080 | 1760 | 84,160 | 22 | 0 | 0 | <5 | 0.0 | 22,080 | 1760 | 84,160 | 100.0 | 0 | 0 | 0 |
| Hot water | 32,160 | 420 | 53,210 | 7 | <5 | 0 | 10 | 0.0 | 32,160 | 420 | 53,200 | 100.0 | <5 | 0 | 10 | |
| Biofilm | 26,300 | 2670 | 49,940 | 2 | 10 | 0 | 20 | 0.0 | 26,300 | 2660 | 49,940 | 100.0 | 0 | 0 | 0 | |
| D | Cold water | 14,190 | 640 | 35,090 | 23 | 10 | 0 | 110 | 0.0 | 14,190 | 640 | 35,090 | 100.0 | 0 | 0 | 0 |
| Hot water | 3840 | 2160 | 5610 | 6 | 0 | 0 | 0 | 0.0 | 3840 | 2160 | 5600 | 100.0 | <5 | 0 | <5 | |
| Biofilm | 23,150 | 10,560 | 44,240 | 4 | <5 | 0 | 20 | 0.0 | 23,150 | 10,560 | 44,220 | 100.0 | 0 | 0 | 0 | |
| E | Cold water | 7490 | 480 | 31,850 | 18 | 170 | 0 | 1560 | 2.3 | 7320 | 480 | 31,850 | 97.7 | <5 | 0 | 20 |
| Hot water | 8890 | 870 | 14,170 | 6 | <5 | 0 | <5 | 0.0 | 8880 | 870 | 14,170 | 100.0 | <5 | 0 | 10 | |
| Biofilm | 4790 | 730 | 16,780 | 5 | 220 | 0 | 830 | 4.6 | 4570 | 730 | 16,600 | 95.4 | 0 | 0 | <5 | |
Not all samples produced archaea reads. More archaea reads were obtained in the DNA-based library than the RNA-based library, which contained more bacterial reads (Tables 1 and 2). Only certain locations were chosen for further analysis of the archaea. In the DNA-based library, all samples from waterworks A and B, cold water and water meter biofilm samples from waterworks D (location 3), as well as biofilm samples from waterworks E were selected for further archaea analyses (Table 1). In the RNA-based library, only cold water and biofilms from the non-disinfected DWDSs A and B contained enough archaea reads for analysis (Table 2).
3.2. Archaea communities in water and biofilms
A total of 119 samples were analysed for their archaea communities and produced ≥375 reads classified as archaea (rarefaction value of archaea sub-sample) from the selected sampling locations (Table 1, Table 2). The final dataset consisted of cold water (55 DNA-based and 37 RNA-based samples), hot water (17 DNA-based samples), and water meter biofilms (7 DNA-based and 3 RNA-based samples). Overall, the data contained 7847 different amplicon sequence variants (ASVs) that resulted in the further analyses of 1384 ASVs after removal of features that occurred only in one sample. From these 1384 ASVs (Supplementary file 1: Table S1), only 36 ASVs (2.6%) could be identified at the genus level and 138 ASVs at the order level (10%). Most ASVs achieved identification only at the class level (1327 ASVs, 96%). On average, 6483 archaea reads were obtained per sample (minimum 375 to maximum 87,908 reads).
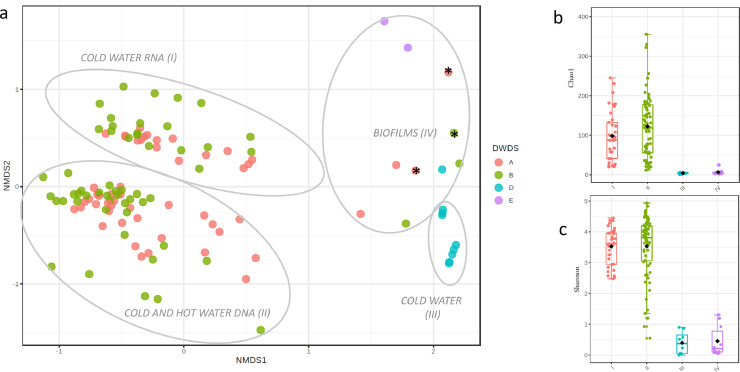
The weighted beta-diversity of the archaea communities formed clear clusters (R = 0.69, P < 0.001, ANOSIM): RNA-based (cluster I) and DNA-based (cluster II) ASV in the non-disinfected DWDS A–B water samples, reads from disinfected DWDS D water samples (cluster III) and water meter biofilms at all the studied DWDSs (cluster IV) (Fig. 1a). The alpha-diversities, species richness according to the Chao1 index, and species richness and evenness according to Shannon diversity estimate, indicated higher archaea diversity in the water from DWDSs A–B compared to the water from DWDS D and the water meter biofilms (Fig. 1b–c).
Fig. 1.
Archaea community separated in clusters from RNA-based reads libraries from DWDSs A–B cold water samples(cluster I), DNA-based reads libraries from DWDSs A–B cold and hot water (cluster II), and RNA and DNA -based reads libraries from the cold water of disinfected DWDS D (cluster III) and water meter biofilms (cluster IV) presented as a) non-metric multidimensional scaling (NMDS) plots of weighted Bray-Curtis beta-diversities and b) Chao1 and c) Shannon alpha-diversity indexes. RNA-based sequencing reads libraries from biofilm samples are marked with an asterisk(*).
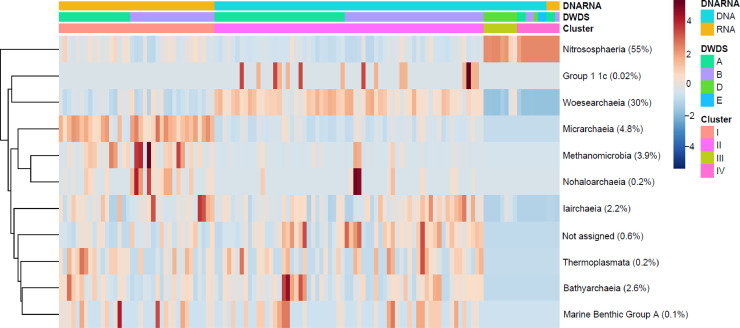
A heatmap clustering analysis revealed that non-disinfected water from DWDSs A–B contained the highest abundance of archaea reads from the classes Micrarchaeia, Methanomicrobia, and Nanohaloarchaeia in the RNA-based library (cluster I) and classes Woesearchaeia and Group 11c in the DNA-based library (cluster II) (Fig. 2). A relative abundance table (% per sample) further revealed that most of the archaeal classes in this study were present in the DWDSs A–B water samples (clusters I–II) in contrast to the DWDS D water samples (cluster III) and biofilms (cluster IV) that consisted of only two or three archaeal classes (Supplementary file 2: Table S2). At the genera level, only Canditatus Nitrosoarchaeum, Candidatus Methanoperedens, and Candidatus Nitrosotalea were abundant in both water sample RNA-based libraries (average 10–14%, Cluster 1) and DNA-based libraries (average 2–8%, cluster II).
Fig. 2.
A heatmap clustering analysis of the archaeal classes presented by representative clusters (Fig. 1). The average relative abundance (%) for each archaea class is presented in brackets. Red indicates a high abundance and blue a low abundance of the archaeal class in the sampling location. Clusters: DWDSs A–B cold water, RNA-based (cluster I), DWDSs A–B cold and hot water, DNA-based (cluster II), cold water from disinfected DWDS D (cluster III), and water meter biofilms (cluster IV). Archaea represent the following phyla (the main classes in brackets): Thaumarchaeota (Nitrososphaeria, Group 1 1c, Marine Benthic Group A), Nanoarchaeaeota (Woesearchaeia, Nanohaloarchaeia), Euryarchaeota (Methanomicrobia, Thermoplasmata), Diapherotrites (Micrarchaeia, Iairchaeia) and Crerchaeota (Bathyarchaeia).
Moreover, water from DWDS D (cluster III) and the biofilms (cluster IV) contained a notably high abundance of reads from one class: Nitrososphaeria (Fig. 2), forming on average 84% and 99% relative abundance in these samples, respectively (Supplementary file 2). Of this class, the genus Canditatus Nitrosoarchaeum was highly abundant (90–100%) in disinfected systems i.e. DWDS D water samples taken in the winter, spring, and summer season as well as the DWDS D–E biofilm samples (Supplementary file 2). Another genus, Candidatus Nitrosotenuis, was notably abundant (40–99%) in the non-disinfected systems DWDSs A–B water meter biofilms of both RNA-based and DNA-based sequence read libraries excluding one biofilm (DNA) that consisted mainly of Canditatus Nitrosoarchaeum (Supplementary file 2).
3.3. Archaea communities in cold non-disinfected water
3.3.1. The effect of the season
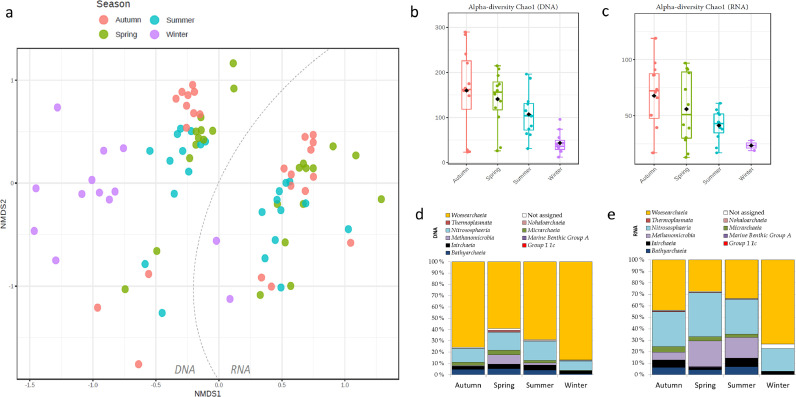
The beta-diversity of cold water from DWDSs A–B (N = 84) indicated separate RNA-based and DNA-based clusters (R = 0.54, P < 0.001, ANOSIM) followed by a minor effect of the season (R = 0.18, P < 0.001, ANOSIM) (Fig. 3a). The species richness (Chao1) of the total archaea community (N = 47, DNA-based) was significantly affected by the season (P < 0.01, Kruskal-Wallis) (Fig. 3b). The same phenomenon was seen in the active community (N = 37, RNA-based) but was not statistically significant (Fig. 3c). In both the total and active communities, the number of estimated species reached the lowest values in winter and grouped apart from samples from other seasons (Fig. 3a–c). Moreover, the average relative abundance of the archaeal class Woesearchaeia seemed higher in winter compared to other seasons, while the occurrence of a few other classes (Methanomicrobia, Nitrosphaeria, Bathyarchaeia) seemed less abundant in winter in both the DNA-based and RNA-based samples (Fig. 3d–e). Moreover, the relative abundance of the taxa profiles suggested the Nitrososphaeria class was particularly active in the autumn, spring, and summer seasons and Methanomicrobia to be active in all seasons based on their higher abundance of RNA-based compared to DNA-based (Fig. 3d–e).
Fig. 3.
Archaea community characteristics of DWDSs A–B cold water samples coloured according to the season. a) The beta-diversity in a weighted Bray-Curtis matrix, DNA-based and RNA-based, alpha-diversity species richness Chao1 from b) DNA-based and c) RNA-based and the relative abundance of archaea classes (%) from d) DNA-based and e) RNAbased sequencing reads libraries.
3.3.2. The effect of the location
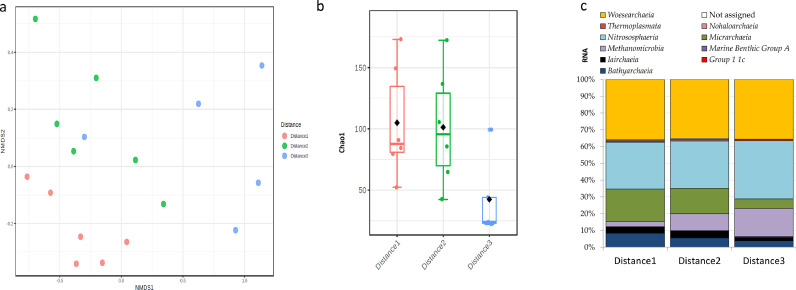
The effect of the distance from the waterworks, i.e., the sampling location reflecting the water age, was notable in DWDS A for both the active (N = 17, Fig. 4, Supplementary file 3: Fig. S1) and total (N = 23, Supplementary file 3: Fig. S2–S4) archaea communities. Differences in the RNA-based sequence libraries according to the sampling location for DWDS A are visible in the weighted beta-diversity plot (R = 0.64, P < 0.001, ANOSIM) (Fig 4a). The location also affected the alpha-diversities and the lowest alpha-diversity values were noted in location 3 according to the Chao1 richness index (Fig. 4b) and Shannon evenness index (Supplementary file 3: Fig. S1) although these were not statistically significant (P = 0.06–0.07). The beta-diversity analysis by location revealed that the total (DNA-based) archaea community showed similar characteristics to the active (RNA–based) archaea community (R = 0.57, P < 0.001, ANOSIM). Additionally, the alpha-diversity values were significantly different between the locations (P < 0.05, Kruskal-Wallis) (Supplementary file 3: Fig. S2–S3). However, the taxa abundance profiles of the DNA-based sequences appeared similar via visual inspection between the locations (Supplementary file 3: Fig. S4). Instead, the RNA-based sequence libraries seemed to display some minor changes in the relative abundances of a few archaeal classes between the locations (Fig. 4c). The relative abundance of Methanomicrobia and Nitrososphaeria increased whereas the relative abundance of Bathyarchaeia and Micrarchaea decreased in location three compared to the first location. The same effect was not evident in DWDS B as beta-diversity (RNA-based, N = 20; DNA-based, N = 24) and alpha-diversities were not different between the locations (P > 0.05) (Supplementary file 4: Fig. S5–S6).
Fig. 4.
Characteristics of RNA-based archaea community in cold water of DWDS A according to the increasing distance from the waterworks, a) the beta-diversity in a weighted Bray-Curtis matrix, b) a boxplot of the alpha-diversity according to the Chao1 richness index and c) the taxa abundance by class level (%). The archaeal subset rarefaction value 517 is used for alpha-diversity. Distance 1 = 1–9 km, Distance 2 = 3–26 km, Distance 3 = 11–36 km from drinking water production plant.
3.3.3. Archaea communities between cold and hot water systems
The groups of cold and hot water samples from the sampling location 2 in DWDSs A–B (DNA-based, N = 29) created distinct clusters in a weighted beta-diversity Bray-Curtis analysis (R = 0.37, P < 0.001, ANOSIM), in which also the winter samples (three cold water and one hot water) created separate clusters regardless of the water system (Supplementary file 5: Fig. S7). The archaea community characteristics in the cold and hot water systems within comparable time points (spring, summer, and autumn seasons) are presented in Table 3. The alpha-diversities measured as Chao1 and Shannon indexes were similar for the groups (P > 0.05) (Table 3). The archaeal class abundance (%) profiles were similar for the cold and hot water samples except for a few classes that seemed slightly more abundant in hot water (Bathyarchaea) or cold water (Methanomicrobia) (Table 3).
Table 3.
Average (N = 12) alpha-diversity values and relative abundance estimates of the archaeal classes. Cold and hot water from DWDSs A–B sampling location 2 (spring, summer, and autumn seasons), alpha-diversity rarefaction value of subset 443.
| Community estimator | Analysis/taxa | Cold water | Hot water |
| Alpha-diversity | Richness, Chao 1 | 186 | 128 |
| Evenness, Shannon | 4.1 | 4.0 | |
| Relative abundance (%), class | Bathyarchaeia | 5.4% | 14.0% |
| Group 1 1c | 0.1% | 0.2% | |
| Iairchaeia | 3.6% | 4.5% | |
| Marine Benthic Group A | 0.4% | 0.5% | |
| Methanomicrobia | 3.9% | 0.8% | |
| Micrarchaeia | 3.6% | 5.4% | |
| Nitrososphaeria | 19.3% | 18.9% | |
| Nanohaloarchaeia | 0.2% | 0.0% | |
| Thermoplasmata | 0.6% | 0.8% | |
| Not assigned | 1.4% | 2.6% | |
| Woesearchaeia | 61.4% | 52.3% |
4. Discussion
4.1. Active or dormant (RNA-based) archaea communities exist in DWDSs and display different characteristics to the total (DNA-based) archaea community
To our knowledge, our work is the first to combine both approaches—using cDNA derived from ribosomal RNA to describe the active or dormant fraction of an archaea community and using the rRNA gene (DNA) to describe the total community—in order to obtain the most realistic picture of the archaea community inhabiting full-scale DWDSs. Despite the successful recovery in the archaea with the DNA-based method with 85–100% reads of the total reads in the DWDSs A–B samples on average, the same recovery rate was not achieved by using the RNA-based method. In this study, the maximum bacteria recovery (sequence reads identified as bacteria) with RNA-based method in DWDSs A–B was 57% of cold water and biofilm samples due to successful archaea recovery (Table 2). This deviated from bacteria recovery in all samples from DWDSs C–E as well as in the hot water samples from DWDSs A–B, which was on average ≥95%. Interestingly, in those samples containing enough RNA-based sequence reads for comparison, the DNA-based and RNA-based archaea communities were crucially different, as shown by the separate clusters in the beta-diversity analysis as earlier reported in the aerobic digestion process (Vrieze et al., 2018). The lower archaea recovery with the RNA-based method could be due to the lower abundance of active or dormant archaea compared to the corresponding bacteria due to an average higher growth rate of bacteria than archaea (Vrieze et al., 2018). The phenomenon may be dependent on the environmental conditions, for example, higher optimum temperature and lower pH in the soil for nitrification potential is usually preferred for ammonia-oxidizing archaea than for ammonia-oxidizing bacteria (Gubry-Rangin et al., 2010; Zeng et al., 2011; Mukhtar et al., 2019). Regardless of the methodology, archaea are considered to be less abundant than bacteria in drinking water distribution systems and related habitats such as bottled groundwater, with archaea being detected at very low levels (Manz et al., 1993; França et al., 2015; Bautista-de los Santos et al., 2016). In one earlier DWDS study, specific probes were used to target rRNA, but archaea were not detected above the estimated detection limit of 103 to 104 ribosomes per cell (Manz et al., 1993). The low abundance of RNA of some species might play a greater role in metabolism than previously thought, and has been reported for novel methanogen species in the rumen (Kang et al., 2013).
4.2. Archaea communities show more diversity in non-disinfected water compared to biofilms or disinfected water
Of the potential ecological disturbances, active or dormant archaea seemed especially sensitive to disinfection based on the lack of archaea reads for analysis recovered from any of the samples collected from the disinfected systems DWDSs C–E. To support this, in an extensive meta-study utilizing DNA, archaea sequences were detected in less than 20% in chlorinated systems and almost 90% in disinfectant residual-free systems (Bautista-de los Santos et al., 2016). In the present study, water samples taken from non-disinfected DWDSs A–B were more diverse with higher species richness than the biofilms or disinfected DWDS D water and were grouped apart from them according to their beta-diversity. Lower diversity in biofilms to water suggests that only part of the archaea members present in water were able to attach in biofilms and survive there, a finding similar to our earlier observations when DWDS bacteria were studied (Inkinen et al., 2016). Earlier observations by others report a generally lower abundance of microbial communities in disinfected water systems and sensitivity of some important archaea to chlorination, such as ammonia-oxidizing archaea in water treatment processes and DWDSs (Kasuga et al., 2010; Bautista-de los Santos et al., 2016; Dai et al., 2020). Archaea, similarly to other microorganisms, may be protectively sheltered in biofilms against chlorination (Schwering et al., 2013; Fish and Boxall, 2018).
Biofilms showed different characteristics to water with a lower ASV richness and a high abundance of only a few taxa, mainly Nitrosophaeria. Previous research has similarly reported archaea as part of the biofilm community in DWDSs with low archaea diversity which may involve different interactions with the environment than more diverse bacteria (Fish et al., 2015). Overall, archaeal phyla Euryarchaeota, Nanoarchaeaeota, Diapherotrites, Thaumarchaeota, and Crenarchaeota have been found in both DNA-based and RNA-based templates, and these have been also detected earlier in DWDSs or drinking water-related environments (França et al., 2015; Roeselers et al., 2015; Bautista-de los Santos et al., 2016; Dai et al., 2020). Similar to the abovementioned studies, we could not reach beyond the phylum or class level. Only <3% of the ASVs were identified at the genus level (Supplementary file 1) which suggests that typical archaea in DWDSs are not fully covered in the current SILVA reference database.
4.3. Water temperature, season, and location in DWDS affect archaea communities
Archaea were not recovered from the RNA-based library of the hot water samples suggesting the high water temperature, on average 53.8–57.7 °C in the DWDSs A–B water samples (Inkinen et al., 2019) effectively controlled the active and dormant fraction of the archaea communities. The selective pressure of hot water has been shown in earlier studies to affect also bacterial and eukaryotic communities (Dai et al., 2018; Inkinen et al., 2019). Slightly lower species richness was observed in hot water compared to the archaea in cold water, however, this difference was not found to be statistically significant. Our earlier work from the same system found a reduced number of eukaryotic diversity in hot water as compared to the cold water systems (Inkinen et al., 2019). In DNA-based libraries of the present work, the same archaeal classes occurred in cold and hot water with an increased relative abundance of Bathyarchaiea suggesting this class may have at some point had a competitive advantage due to the ecological disturbance of the high temperature.
The season had an impact on the archaea communities with the lowest species richness in the winter season in DWDSs A–B in both DNA-based and RNA-based sequencing read libraries. This may be in part due to changes in the water temperature and also absorbance varies between the seasons in the studied DWDSs (Ikonen et al., 2017). Previous studies have highlighted this phenomenon for total cell counts and bacterial communities in DWDSs where the seasonal variation is typical and correlates especially with temperature (Pinto et al., 2014; Prest et al., 2016; Schleich et al., 2019). Some archaea members, e.g. Methanomicrobia, were less abundant in the winter than other seasons. This class has earlier been reported to flourish in the summer season in littoral sediment in the sea (Yan et al., 2018). Moreover, changes in raw water quality could also explain the changes in the DWDSs because structures of archaea communities in freshwater are known to change along with the seasons of the year (Li et al., 2015).
The location within the DWDSs might affect the archaea communities too. In this study, the sampling locations in DWDS A created separate clusters in terms of their beta-diversity, and differences in the ASV richness values were observed. In an earlier study of the same five DWDSs targeting eukaryote communities, the effect of location on the species richness was even more pronounced covering all DWDSs (Inkinen et al., 2019). In the same system also heterotrophic bacteria plate counts varied between locations in DWDSs A and B (Ikonen et al., 2017). Moreover, earlier studies have reported changes in bacterial community structures between different locations in full-scale DWDSs and even within a single building (Pinto et al., 2014; Inkinen et al., 2016; Schleich et al., 2019). Possible reasons for spatial differences could include changes in environmental factors or archaea species regrowth, interactions with biofilms, or the differences in survival. The relative abundance of the class Nitrososphaeria—the most abundant class in the biofilm samples—was increased in RNA-based sequencing read libraries at DWDS A location 3 suggesting the detachment of biofilms in the water (Wingender and Flemming, 2004). Moreover, the archaea that were found solely in the disinfected DWDS D water at location 3 (the farthest sampling location) may have originated from biofilms, as the water samples were clustered close to the biofilm in a beta-diversity plot and contained mainly the archaeal genus Candidatus Nitrosoarcheaum (Nitrososphaeria class), which is a genus found especially rich in the biofilms of disinfected systems (DWDSs D–E).
Our study has indicated not only Bacteria but also a wide set of Archaea can grow in DWDS, especially in unchlorinated systems. The domain Archaea and its functions are not well understood yet, and more work is required to develop the taxonomic database to enable the assignment of the sequencing reads to the genus level. Without such information, the exact implications of the findings to the water systems management are difficult to make. Of the identified Archaea in our study, DWDS D distributing water disinfected by using chloramination implicated that the farthest location from the waterworks (19 km's distance) was inhabited with AOA. This might tentatively affect the efficiency of the post-chlorination and/or cause disinfection byproducts (Waak et al., 2019). In the future, more phenomena could be explained by using a holistic approach studying all members of the microbiome. For example, in addition to 16S and 18S rRNA gene studies, also metagenomics and virus communities could be investigated. Eventually, one future aspect could be even to engineer the DWDS microbiome in a way that the microbes causing adverse health effects could be suspended (Neu and Hammes, 2020). This would require further understanding of the relationships between different domains of lives occurring in the DWDS.
5. Conclusions
-
•
Archaea communities including active or dormant archaea were revealed in the drinking water system at detectable levels in certain environmental conditions, especially DWDS supplying for non-disinfected waters.
-
•
Cold non-disinfected water from the two separate waterworks from the same city and both using artificial groundwater sources contained an especially diverse archaea community.
-
•
In non-disinfected water, up to ten archaeal classes were present as opposed to less diverse biofilms, and in disinfected water that accounted only for a few highly abundant taxa.
-
•
Disinfection seemed to control both total and active archaea occurrence in DWDS water and hot water temperature seemed to control the active archaea occurrence in DWDS water.
-
•
The season and distance from the waterworks both affected the archaea communities.
-
•
The functional role of archaea in DWDS may be more important than previously thought and should be further studied.
Authors’ contribution
J Inkinen analysed the original data and drafted the initial version of the manuscript. TP, ITM, ET, MK, and JP participated in conceptualization, funding acquisition, supervision, investigation, and allocation of resources. BJ denoised the sequence data. SS, A-MH, AP, J Ikonen, and AK participated in the investigation and AT on manuscript preparation. All the authors participated in editing the initial draft and commented and approved the final submission version.
Funding
This work was supported by the Academy of Finland (DWDSOME, project numbers 275549) for conducting the project; and Maa- ja vesitekniikan tuki ry (Grant number 41713) for preparing the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
All personnel of the waterworks participated in the sampling campaign consecution as well as at Water Microbiology Laboratory in the Finnish Institute for Health and Welfare, Kuopio, Finland are gratefully acknowledged. Gareth Attwood is acknowledged for grammatical advice on the article.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.wroa.2021.100101.
Appendix. Supplementary materials
References
- Auguet J., Barberan A., Casamayor E.O. Global ecological patterns in uncultured Archaea. ISME J. 2010;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- Bautista-de los Santos Q.M., Schroeder J., Sevillano-Rivera M.C., Sungthong R., Ijaz U.Z., Sloan W.T., Pinto A.J. Emerging investigators series: microbial communities in full-scale drinking water distribution systems – a meta-analysis. Environ. Sci. Water Res. Technol. 2016;2:631–644. doi: 10.1039/C6EW00030D. [DOI] [Google Scholar]
- Bradley T.C., Haas C.N., Sales C.M. Nitrification in premise plumbing: a review. Water (Basel) 2020;12(3):830. doi: 10.3390/w12030830. [DOI] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Liu P., Zhou G., Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020;15:799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- Dai, D., Rhoads, W.J., Edwards, M.A., Pruden, A., 2018. Shotgun metagenomics reveals taxonomic and functional shifts in hot water microbiome due to temperature setting and stagnation. Front. Microbiol.9, 1–17. https://doi.org/10.3389/fmicb.2018.02695. [DOI] [PMC free article] [PubMed]
- Dai Z., Sevillano-Rivera M.C., Calus S.T., Bautista-de los Santos Q.M., Eren A.M., van der Wielen P.W.J.J. 2020. Disinfection Exhibits Systematic Impacts on the Drinking Water microbiome. Microbiome 8, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A., Chong J., Habib S., King I.L., Agellon L.B., Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucl. Acids Res. 2017;45:W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish K.E., Boxall J.B. Biofilm microbiome (re)growth dynamics in drinking water distribution systems are impacted by chlorine concentration. Front. Microbiol. 2018;9:1–21. doi: 10.3389/fmicb.2018.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish K.E., Collins R., Green N.H., Sharpe R.L., Douterelo I., Osborn A.M., Boxall J.B. Characterisation of the physical composition and microbial community structure of biofilms within a model full-scale drinking water distribution system. PLoS ONE. 2015:1–22. doi: 10.1371/journal.pone.0115824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- França L., Lopéz-Lopéz A., Rosselló-Móra R., Costa M.S. Microbial diversity and dynamics of a groundwater and a still bottled natural mineral water. Environ. Microbiol. 2015;17:577–593. doi: 10.1111/1462-2920.12430. [DOI] [PubMed] [Google Scholar]
- Gantner S., Andersson A.F., Alonso-Sáez L., Bertilsson S. Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods. 2011;84:12–18. doi: 10.1016/j.mimet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Martinez A., Sihvonen M., Muñoz-Palazon B., Mikola A., Vahala R. Microbial ecology of full-scale wastewater treatment systems in the Polar Arctic Circle: archaea, bacteria and fungi. Sci. Rep. 2018:1–11. doi: 10.1038/s41598-018-20633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubry-Rangin C., Nicol G.W., Prosser J.I. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol. Ecol. 2010;74:566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- Hoehler T.M., Jørgensen B.B. Microbial life under extreme. Nat. Rev. Microbiol. 2013;11:83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- Ikonen J.M., Hokajärvi A.-.M., Heikkinen J., Pitkänen T., Ciszek R., Kolehmainen M. Drinking water quality in distribution systems of surface and ground waterworks in Finland. J .Water Secur. 2017;3:1–10. doi: 10.15544/jws.2017.004. [DOI] [Google Scholar]
- Inkinen J., Jayaprakash B., Santo Domingo J.W., Keinänen-Toivola M.M., Ryu H., Pitkänen T. Diversity of ribosomal 16S DNA- and RNA-based bacterial community in an office building drinking water system. J. Appl. Microbiol. 2016;120:1723–1738. doi: 10.1111/jam.13144. [DOI] [PubMed] [Google Scholar]
- Inkinen J., Jayaprakash B., Siponen S., Hokajärvi A., Pursiainen A., Ikonen J. Active eukaryotes in drinking water distribution systems of ground and surface waterworks. Microbiome. 2019;7:1–17. doi: 10.1186/s40168-019-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeraldo P., Kalari K., Chen X., Bhavsar J., Mangalam A., White B. IM-TORNADO: a Tool for Comparison of 16S Reads from Paired-End Libraries. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0114804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.H., Evans P., Morrison M., Mcsweeney C. Identification of metabolically active proteobacterial and archaeal communities in the rumen by DNA- and RNA-derived 16S rRNA gene. J. Appl. Microbiol. 2013;115:644–653. doi: 10.1111/jam.12270. [DOI] [PubMed] [Google Scholar]
- Kasuga I., Nakagaki H., Kurisu F., Furumai H. Predominance of ammonia-oxidizing archaea on granular activated carbon used in a full-scale advanced drinking water treatment plant. Water Res. 2010;44:5039–5049. doi: 10.1016/j.watres.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucl. Acids Res. 2013;41:1–11. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M., Bernhard A., de la Torre J., Walker C.B., Waterbury J.B., Stahl D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Li J., Zhang J., Liu L., Fan Y., Li L., Yang Y. Annual periodicity in planktonic bacterial and archaeal community composition of eutrophic Lake Taihu. Sci. Rep. 2015;5:1–14. doi: 10.1038/srep15488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Gunawan C., Barraud N., Rice S.A., Harry E.J., Amal R. Understanding, monitoring, and controlling biofilm growth in drinking water distribution systems. Environ. Sci. Technol. 2016;2016(17):8954–8976. doi: 10.1021/acs.est.6b00835. 50. [DOI] [PubMed] [Google Scholar]
- Manz W., Szewzyk U., Ericsson P., Amann R., Schleifer K.H., Stenström T.A. In situ identification of bacteria in drinking water and adjoining biofilms by hybridization with 16S and 23S rRNA-directed fluorescent oligonucleotide probes. Appl. Environ. Microbiol. 1993;59(7):2293–2298. doi: 10.1128/AEM.59.7.2293-2298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar H., Lin Y.P., Lin C.M., Lin Y.R. Relative abundance of ammonia oxidizing archaea and bacteria influences soil nitrification responses to temperature. Microorganisms. 2019;7(11):526. doi: 10.3390/microorganisms7110526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu L., Hammes F. Feeding the Building Plumbing Microbiome: the Importance of Synthetic Polymeric Materials for Biofilm Formation and Management. Water (Basel) 2020;12 doi: 10.3390/w12061774. 1774. [DOI] [Google Scholar]
- Pinto A., Schroeder J., Lunn M., Sloan W., Raskin L. Spatial-temporal survey and occupancy-abundance modeling to predict bacterial community dynamics in the drinking water microbiome. MBio. 2014;5:1–13. doi: 10.1128/mBio.01135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prest E.I., Weissbrodt D.G., Hammes F., Van Loosdrecht M.C.M., Vrouwenvelder J.S. Long-term bacterial dynamics in a full-scale drinking water distribution system. PLoS ONE. 2016;11:1–20. doi: 10.1371/journal.pone.0164445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Glo F.O., Yarza P. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeselers G., Coolen J., van der Wielen P.W.J.J, Jaspers M.C., Atsma A., de Graaf B., Schuren F. Microbial biogeography of drinking water: patterns in phylogenetic diversity across space and time. Environ. Microbiol. 2015;17:2505–2514. doi: 10.1111/1462-2920.12739. [DOI] [PubMed] [Google Scholar]
- Schleich C., Chan S., Pullerits K., Besmer M.D., Paul C.J., Rådström P., Keucken A. Mapping dynamics of bacterial communities in a full-scale drinking water distribution system using flow cytometry. Water (Switzerland) 2019;11:1–14. doi: 10.3390/w11102137. [DOI] [Google Scholar]
- Schwering M., Song J., Louie M., Turner R.J., Ceri H. Multi-species biofilms defined from drinking water microorganisms provide increased protection against chlorine disinfection. Biofouling. 2013;29:917–928. doi: 10.1080/08927014.2013.816298. [DOI] [PubMed] [Google Scholar]
- Shi Y., Fan K., Li Y., Yang T. Archaea enhance the robustness of microbial co-occurrence networks in Tibetan Plateau soils. Soil Sci. Soc. Am. J. 2019;83:1093–1099. doi: 10.2136/sssaj2018.11.0426. [DOI] [Google Scholar]
- Smith C.M., Hill V.R. Dead-end hollow-fiber ultrafiltration for recovery of diverse microbes from water. Appl. Environ. Microbiol. 2009;75(16):5284–5289. doi: 10.1128/AEM.00456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D.A., Amann R. Development and application of nucleic acid probes in bacterial systematics. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons Ltd.; Chichester, UK: 1991. pp. 205–248. [DOI] [Google Scholar]
- Steen A.D., Crits-Christoph A., Carini P., DeAngelis K.M., Fierer N., Lloyd K.G., Cameron Thrash J. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 2019;13:3126–3130. doi: 10.1038/s41396-019-0484-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs S., De Beeck M.O., Beckers B., Truyens S., Stevens V., Van Hamme J.D. Comparative evaluation of four bacteria-specific primer pairs for 16S rRNA gene surveys. Front. Microbiol. 2017;8:1–15. doi: 10.3389/fmicb.2017.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y., Pirrung M., Gonzalez A., Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2:2–5. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze J.De, Pinto A.J., Sloan W.T., Boon N., Ijaz U.Z. The active microbial community more accurately reflects the anaerobic digestion process: 16S rRNA (gene) sequencing as a predictive tool. Microbiome. 2018;6:1–13. doi: 10.1186/s40168-018-0449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waak M.B., Hozalski R.M., Hallé C. Comparison of the microbiomes of two drinking water distribution systems—with and without residual chloramine disinfection. Microbiome. 2019;7:87. doi: 10.1186/s40168-019-0707-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielen P.W.J.J.Van Der, Voost S., Kooij D.Van Der. Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl. Environ. Microbiol. 2009;75:4687–4695. doi: 10.1128/AEM.00387-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender J., Flemming H. Contamination potential of drinking water distribution network biofilms. Water Sci. Technol. 2004;49:277–286. doi: 10.2166/wst.2004.0861. [DOI] [PubMed] [Google Scholar]
- Yan L., Yu D., Hui N., Naanuri E., Viggor S., Gafarov A. Distribution of archaeal communities along the coast of the Gulf of Finland and their response to oil contamination. Front. Microbiol. 2018;9:15. doi: 10.3389/fmicb.2018.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucl. Acids Res. 2014;2014(42):D643–D648. doi: 10.1093/nar/gkt1209. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G., Zhang J., Chen Y., Yu Z., Yu M., Li H. Relative contributions of archaea and bacteria to microbial ammonia oxidation differ under different conditions during agricultural waste composting. Bioresour. Technol. 2011;102:9026–9032. doi: 10.1016/j.biortech.2011.07.076. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Oh S., Liu W. Impact of drinking water treatment and distribution on the microbiome continuum: an ecological disturbance’s perspective. Environ. Microbiol. 2017;19:3163–3174. doi: 10.1111/1462-2920.13800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primer clipped reads were deposited in the Sequence Read Archive (SRA) of NCBI under existing BioSample accession numbers from SAMN10653499 to SAMN10653948 of the BioProject PRJNA509718 (Inkinen et al., 2019) with a new submission (SUB6956053) describing archaea sequences. All samples and controls in this project are listed in a metadata file (Supplementary file 6, Table S3).