Abstract
Recent years witnessed the discovery of ubiquitous and diverse 5′-end RNA cap-like modifications in prokaryotes as well as in eukaryotes. These non-canonical caps include metabolic cofactors, such as NAD+/NADH, FAD, cell wall precursors UDP-GlcNAc, alarmones, e.g. dinucleotides polyphosphates, ADP-ribose and potentially other nucleoside derivatives. They are installed at the 5′ position of RNA via template-dependent incorporation of nucleotide analogues as an initiation substrate by RNA polymerases. However, the discovery of NAD-capped processed RNAs in human cells suggests the existence of alternative post-transcriptional NC capping pathways. In this review, we compiled growing evidence for a number of these other mechanisms which produce various non-canonically capped RNAs and a growing repertoire of capping small molecules. Enzymes shown to be involved are ADP-ribose polymerases, glycohydrolases and tRNA synthetases, and may potentially include RNA 3′-phosphate cyclases, tRNA guanylyl transferases, RNA ligases and ribozymes. An emerging rich variety of capping molecules and enzymes suggests an unrecognized level of complexity of RNA metabolism.
Keywords: capping, non-canonical capping, RNA processing
1. Introduction
Cell fitness is highly dependent on fast and adequate changes of gene expression to cope with changing conditions. Gene expression is mainly regulated by adjusting RNA levels by production or degradation, but also by the differential utilization of RNA as defined by its sequence, structure or epitranscriptomic modifications. RNA capping is an important mechanism, affecting the overall fate of the respective RNA. Canonical eukaryotic 5′ RNA caps are attached to the nascent RNA early during its synthesis by RNA polymerase (RNAP) and can subsequently affect almost all its cellular roles. They enable initiation of protein synthesis, serve as an identifier for recruiting protein factors for pre-mRNA splicing, polyadenylation and nuclear export, and affect stability and susceptibility of RNA to nucleases [1].
Until recently, it was assumed that just eukaryotic cells are endowed with a 5′ RNA caps, a 5′-5′ linked N7-methyl guanosine (m7G)1 added co-transcriptionally to the 5′ ends of transcripts producing m7G-RNA [2,3]. 5′ ends of prokaryotic RNAs were considered less heterogeneous. 5′ triphosphorylated RNA (ppp-RNA) results from the incorporation of nucleoside triphosphate as the initiating substance of the nascent RNA by RNAP. Prokaryotic ppp-RNA was supposed to be diversified only by removing some of its phosphates (becoming diphosphorylated, monophosphorylated or hydroxylated). The phosphorylation state of RNA 5′ end defines RNA longevity and susceptibility to degradation, therefore affecting gene regulation and cell survival. Monophosphorylated transcripts are preferred substrates of the main degradation endonucleases (e.g. RNase E in Escherichia coli or the 5′ exonuclease RNase J in Bacillus subtilis), which makes them vulnerable to rapid attack by either of them [4,5].
Recent years revealed several surprising facts about bacterial 5′ RNA ends, some of them with consequential findings in eukaryotes. Besides the high percentage of diphosphorylated RNA (pp-RNA; 35%–50% mRNA) [6], a surprising number of different cap-like structures, including dinucleotide analogues, metabolic cofactors and cell wall precursors, were revealed at the 5′ ends of bacterial and eukaryotic RNAs [7–10]. The discovery of bacterial non-canonical (NC) caps challenged the perception of eukaryotic RNA 5′ end uniqueness [8,9,11–14] blasting off the new research field of RNA 5′ end epitranscriptomics.
Table 1.
List of abbreviations used in the text.
| list of abbreviations | |||
|---|---|---|---|
| ADP | adenosine diphosphate | NC | non-canonical |
| ADPR | adenosine diphosphate ribose | NGD | nicotinamide guanine dinucleotide |
| ATP | adenosine triphosphate | NMN | nicotinamide mononucleotide |
| CoA | coenzyme A | NMNAT | nicotinamide mononucleotide adenylyltransferase |
| FAD | flavin adenine dinucleotide | NPnNs | dinucleotide polyphosphates |
| GDP | guanosine diphosphatase | PARP | poly (ADP-ribose) polymerase |
| GMP | guanosine monophosphatase | ppp-RNA | triphosphorylated ribonucleic acid |
| GTP | guanosine triphosphatase | pp-RNA | diphosphorylated ribonucleic acid |
| GTase | RNA guanylyltransferase | p-RNA | monophosphorylated ribonucleic acid |
| GTPase | guanosine 5′-triphosphatase | PRNTase | polyribonucleotidyltransferase |
| m7G | N7-methyl guanosine | RdRp | RNA-dependent RNA polymerase |
| MTase | methyltransferase | RNAP | RNA polymerase |
| NAAD | nicotinic acid adenine dinucleotide | TPase | RNA triphosphatase |
| NAD+ | nicotinamide adenine dinucleotide | UDP-Glc | uridine diphosphate glucose |
| NADH | reduced nicotinamide adenine dinucleotide | UDP-GlcNAc | uridine diphosphate N-acetylglucosamine |
| NAM | nicotinamide | VPg | viral protein genome-linked (VPg) |
| NaMN | nicotinic acid mononucleotide | VSV | vesicular stomatitis virus |
Table 2.
Summary of the known and potential canonical and non-canonical 5′ RNA capping enzymes described in the text and their 5′ RNA cap products (for details see the text).
| non-canonical 5' RNA capping enzyme | cap |
|---|---|
| multi-subunit and single-subunit RNA polymerase, primase | variety of dinucleotides and nucleotide analogues (e.g. NAD+, NADH, FAD, UDP-Glc, UDP-GlcNAc, NPnN, CoA) |
| GTase | m7G |
| RdRp L protein of VSV | m7G |
| RNA ligase | ADP |
| tRNA synthetase (LysU) | Ap4N |
| RNA phosphate cyclase (RtcA) | ADP |
| PARPs | ADPR |
| NMNATs | NAD+; capping of RNA not proved |
| CD38 | ADPR |
| ribozymes | NAD+, FAD, CoA |
2. Canonical 5′ RNA caps and capping mechanism
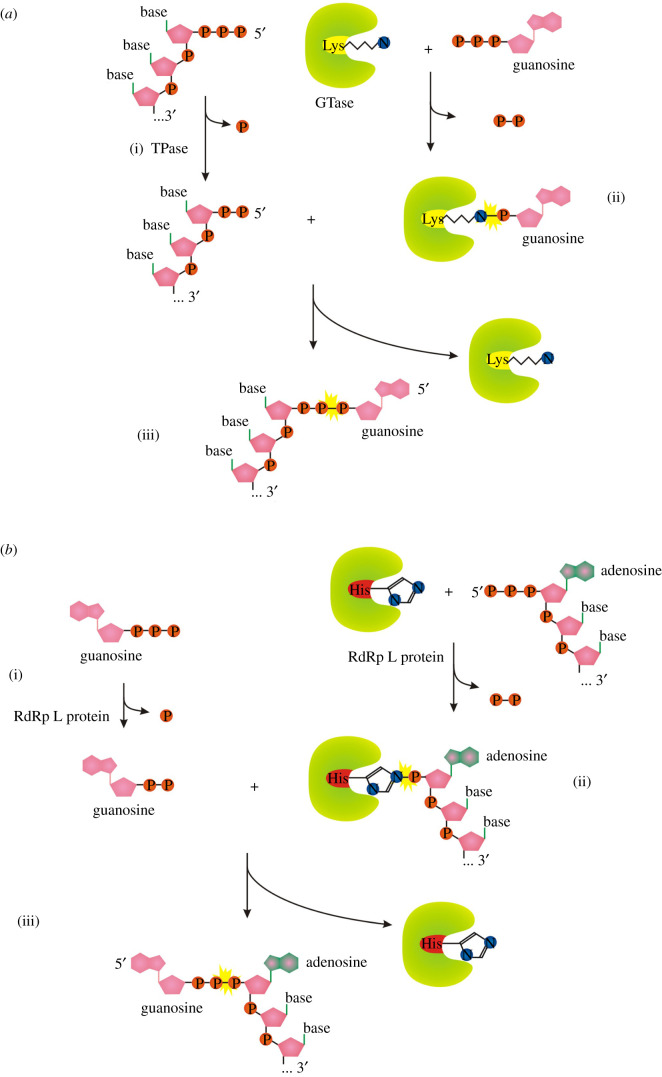
All known eukaryotes synthesize m7G-RNA by the enzymatic activities of three enzymes: RNA triphosphatase (TPase), RNA guanylyltransferase (GTase) and guanine-N7 methyltransferase (guanine-N7 MTase) [1]. RNA TPase removes the γ-phosphate from the 5′ triphosphate to generate 5′ diphosphate RNA (figure 1a(i)). GTase transfers a GMP group from GTP to the 5′ diphosphate via a lysine-GMP covalent intermediate (figures 1a(ii), (iii) and 2a). The guanine-N7 MTase then adds a methyl group to the N7 amine of the guanine cap to form the cap 0 structure. +1 nucleotide of the resulting m7G-RNA can be further methylated at the 2′-O-ribose of the first (cap 1) or first and second nucleotides (cap 2) [1]. Hypermethylation of 5′ m7G cap (cap 4 structure) can be found in trypanosomes and other Kinetoplastida [15]. A different methylation level further diversifies the function and localization of capped RNAs [1].
Figure 1.
(a) Canonical mechanisms of m7G cap formation in eukaryotes. (b) Alternative pathway in vesicular stomatitis virus. Orange ‘P’ symbolizes the phosphorus group, adenosine nucleoside is in green and pink, guanosine nucleoside is in pink-only, pink pentagon represents ribose. Yellow asterisks highlight the new emerging linkage.
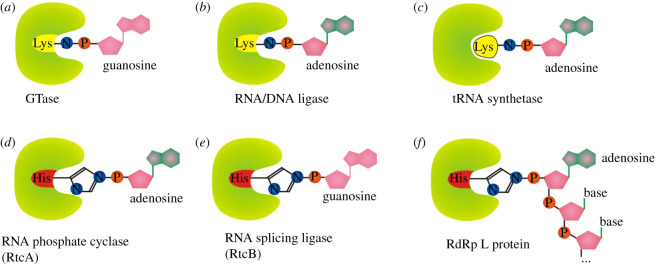
Figure 2.
Nucleotide-enzyme, RNA-enzyme or nucleotide-amino acid covalently bound intermediates used during 5′ aminoacylation of RNA by different enzymes (3′ aminoacylation by RtcB). (a) RNA guanylyl transferase (GTase), (b) RNA/DNA ligase, (c) tRNA synthetase (LysU). (d) RNA phosphate cyclase (RtcA), (e) RNA splicing ligase (RtcB), (f) RNA-dependent RNA polymerase L protein from VSV.
The described canonical capping mechanism is common for eukaryotic organisms as well as some DNA viruses (e.g. vaccinia virus) and double-stranded RNA viruses (e.g. reoviruses). By contrast, vesicular stomatitis virus (VSV), and probably other non-segmented negative-strand RNA viruses (Mononegavirales), use a different approach to cap RNA [16]. Its multifunctional RNA-dependent RNA polymerase (RdRp) L protein carries out an unconventional mechanism that involves the stepwise action of guanosine 5′-triphosphatase (GTPase) (figure 1b(i)), followed by RNA: GDP polyribonucleotidyl transferase (PRNTase) (figure 1b(ii), (iii)). The main difference is RdRp L protein-mediated transfer of the 5′-monophosphorylated pre-mRNA (figure 1b(ii)) to GDP through a covalent enzyme-phosphorylated RNA intermediate (figures 1b(ii,iii) and 2f). Interestingly the transfer mechanism involves the formation of a phosphoramide bond to histidine instead of lysine as occurs in the conventional system (compare figure 2a versus f). The sequence of methylation events in conventional versus VSV systems has a reversed order [16].
3. A great variety of non-canonical 5′ RNA caps was discovered in bacteria, eukaryotes and their mitochondria
First indications of a vast spectrum of 5′ RNA modifications were published in two mass spectroscopy-based papers from 2009, which screened for small-molecule conjugates of RNA in E. coli and Streptomyces venezuelae [8,9]. Besides NAD- and CoA-linked RNA, they identified succinyl-, acetyl- and methylmalonyl-thioester derivatives of CoA. All these metabolites were proved to localize at the 5′ end of cellular RNAs and can be therefore classified as 5′ NC caps. Authors of these studies also identified 3′-aminoacyl adenosine monophosphates (originated most likely from aminoacylated tRNAs, but their existence on other RNA species is not excluded) and 17 [9] or 24 [8] additional unknown species attached to unspecified sites of RNA, suggesting an unprecedented range of bacterial RNA modifications in vivo. The existence of NAD-RNA was confirmed in bacteria, yeast, human and plant cells, as well as in mitochondria [11,12,17–20]. Other NC caps were later identified in vivo—flavin adenine dinucleotide (FAD), uridine diphosphate glucose (UDP-Glc), uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) and dinucleotide polyphosphates (NPnNs) [7,10,14,21].
4. Unrelated RNA polymerases are the main, but not the only enzymes installing NC caps
Most of the discovered NC capping is believed to originate from the incorporation of abundant cell metabolites (nucleotide analogues or dinucleotides) to RNA during the initiation step of template-dependent transcription by DNA dependent RNAPs. The nucleotide part of the metabolite pairs with the transcription start site of template DNA forming the 5′ end of nascent RNA.
The first argument for NC ab initio incorporation by RNAP was the simple observation that NAD+ incorporated only in RNA, which typically initiates with ATP. Additionally, there is a high correlation between the promoter sequence and the efficiency of NAD+ incorporation [17,22–24], supporting the template dependency of this process. Importantly, NC caps were found only on non-processed RNAs initially—in yeast NAD-RNA on pre-mRNAs, and on mitochondrial transcripts that are not 5′ end processed [25]. Another argument for RNAP II-dependent NAD+ capping is the utilization of different transcription start sites for NAD versus m7G-capped RNAs of the same gene observed in Arabidopsis thaliana and Saccharomyces cerevisiae [22,26].
Initiation with NC nucleosides seems to be a common feature of structurally and evolutionary unrelated families of transcriptases: (i) multi-subunit RNAPs—bacterial RNAPs and eukaryotic RNAP II, (ii) single-subunit RNAPs—mitochondrial and bacteriophage T7 RNAP [12,13,27–29]. Recently, a replicative enzyme, primase, which makes short RNA primers to be extended by DNA polymerase was shown to cap RNA with NAD+ and FAD in vitro [29]. Capping seems ubiquitous, yet there is a conspicuous outlier—a chloroplast. Up until now, only two chloroplast RNAs were found to be NAD-capped [18,26]. One of them was rRNA, which suggests that the capping could be done by one of the phage-type single-subunit polymerase transcribing rrn operon of Arabidopsis chloroplast [30]. This leaves the question whether the main plastid RNAP of bacterial type is capable of NAD+/NADH incorporation. We tested cyanobacterial RNAPs, the closest bacterial relatives of chloroplast RNAPs (of Synechococcus elongatus PCC 7942 and Synechocystis sp. PCC 6803) and found that they can efficiently incorporate NAD+/NADH into an RNA (C.J. 2020, unpublished data). We used conditions similar to those published for E. coli RNAP [13] for assessing formation of the two nucleotides-long RNA (0.5 mM cofactor and 50 µM CTP as next nucleotide, linear DNA fragment containing RNA I promoter in 20 mM Tris–HCl (pH 7.9), 40 mM KCl, 10 mM MgCl2). Whether the relatively low in vivo NAD+/NADH concentration in chloroplast [31] prevents its incorporation as initiating NC cap, or the capped RNA is processed very fast remains to be answered in prospective research.
Despite all these arguments for ab initio capping by RNAP, there are indications for other post-transcriptional mechanisms to obtain 5′-NC-capped RNA. The most convincing one is in mammalian cells, where intronic small nucleolar RNAs (snoRNAs) and small Cajal body RNAs (scaRNAs) were reported to be NAD-capped [32]. While the processed intronic RNAs are inherently monophosphorylated [33–35] it can be deduced that 5′ NAD-caps can be added independently of RNAP action. This review focuses on known and potential alternative pathways of NC RNA capping with abundant AMP analogues.
5. RNA polymerases cap RNA with NAD+/NADH: could enzymes of NAD+ biosynthesis pathway cap RNA post-transcriptionally?
Nicotinamide dinucleotide (NAD) is a cofactor required for many cellular oxidases and reductases. As a redox coenzyme, it shuttles between the oxidized form (NAD+) and the reduced form (NADH). However, it has been uncovered in the last few years that the role of NAD+ in cells is much broader. Besides being a redox coenzyme, NAD+ is consumed and used as a co-substrate in the enzymatic reactions by several types of enzymes (e.g. NAD-dependent DNA ligases, poly (ADP-ribose) polymerases (PARPs), ADP-ribosyl cyclases, ADP-ribosyltransferases that modify proteins and small molecules) [36] and excitingly it was found as a 5′ cap of RNAs from bacteria as well as eukaryotes [8,11–13].
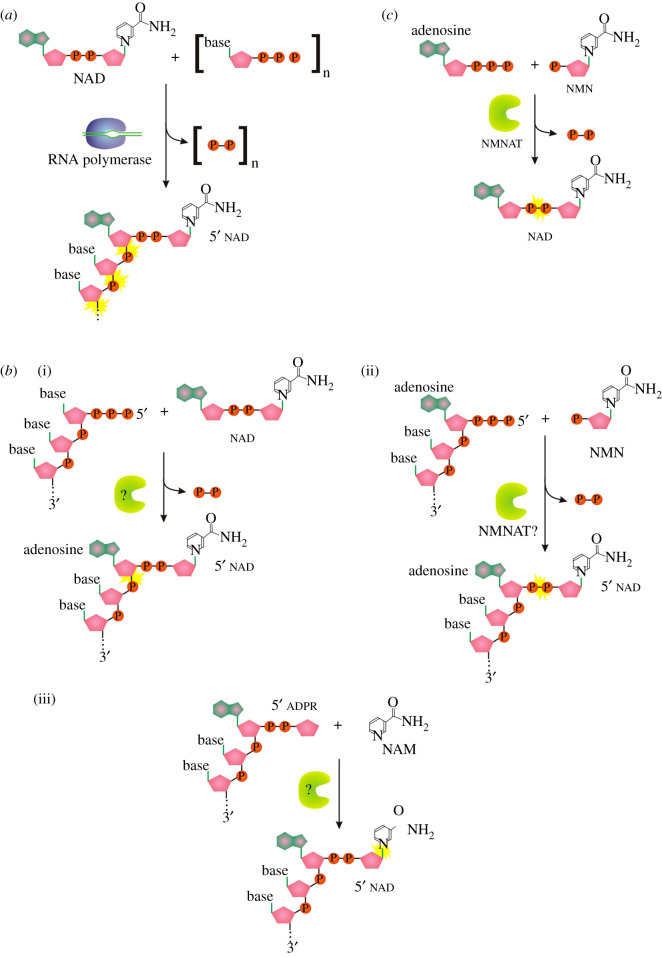
NADylated RNA (NAD-RNA) was one of the first discovered modifications of the 5′ end of bacterial RNA. Although expected to originate mainly from transcription initiation with NAD+ (figure 3a) [12], it was also demonstrated to appear on mammalian processed RNA species, providing the proof of the existence of post-transcriptional NAD-capping process [32]. Post-transcriptional NAD+ capping may result from at least three different events (figure 3b): (i) covalent binding of the whole NAD+ to any nucleoside triphosphate on the 5′ end of RNA, (ii) transfer of nicotinamide ribonucleotide to 5′ ATP-terminated RNA, or (iii) transfer of nicotinamide to ADP ribosylated RNA (ADPR-RNA). So far, no post-transcriptionally NAD-capping enzyme has been described, and identification of such an enzyme would provide a useful tool for studying NAD capping function and regulation.
Figure 3.
(a) Mechanism of ab initio NAD capping by RNAP. (b) Three possible pathways to produce NAD-RNA post-transcriptionally. (c) Known function of NMNATs in NAD synthesis. Symbols as described in figure 1.
A suitable post-transcriptional NAD-capping candidate may be recruited from nicotinamide mononucleotide adenylyltransferases (NMNATs) or other enzymes of the NAD+ biosynthesis pathway. NMNATs catalyse the transfer of the AMP moiety of ATP to nicotinamide mononucleotide (NMN) to form NAD+ (figure 3c), and they also catalyse the reverse reaction (the generation of ATP and NMN from NAD+ or alternative dinucleotides and pyrophosphate [37]). We speculate that the analogous activity of NMNAT might be useful for capping of 5′ATP-terminated RNA with a nicotinamide ribonucleotide/nicotinic acid mononucleotide moiety (figure 3b(ii)) and potentially also for decapping of NADylated RNA.
It was previously shown that the enzymatic substrate-binding pocket of some NMNATs is not strictly specific to ATP, but it can also adopt phosphorylated proteins [38] or different nucleotides and dinucleotides (e.g. reduced or phosphorylated forms of NAD (NADH, NADP+, respectively) [39], nicotinic acid adenine dinucleotide (NAAD), nicotinamide guanine dinucleotide (NGD) [37]). These findings indicate plasticity of the active site of at least some NMNATs, permitting speculations about possible RNA capping abilities of enzymes from this family.
Cells usually code for more isoforms of NMNATs (two in bacteria [40,41], two in yeast [42], three in human cells [37]). Due to different subcellular localization, tissue specificity of particular isoforms and their co-expression, they are expected to carry on defined, non-redundant functions. However, their specific functions are still enigmatic. Human NMNAT1 is a nuclear protein, NMNAT2 and NMNAT3 are localized to the Golgi complex and the mitochondria, respectively. Mitochondrial localization of NMNAT3 makes it an interesting target for further testing of its potential capping activity since 40–70% of NAD+ in cells resides in the mitochondria [43,44] and up to 15% of human mitochondrial RNA and up to 60% of yeast mitochondrial RNA is NAD-capped (although in yeast mitochondria, strictly unprocessed transcripts were found to be NAD+-capped [25]). Moreover, kinetic properties showed that particularly NMNAT3 exhibits a high tolerance towards substrate modifications and it can convert NAAD, NGD and NADH to the same extent as NAD+ [37].
Two NMNATs with different substrate specificities were identified in E. coli: NadD (YbeN) which preferentially uses nicotinic acid mononucleotide (NaMN; 20 times more efficiently than NMN) [41,45,46] and NadR, which prefers NMN (170 times more efficiently than NaMN) [40]. Cyanobacterial NMNAT (slr0787 or NadM_SYNY3) also harbours a highly conserved sequence that classifies it as NUDIX hydrolase (hydrolases cleaving nucleoside diphosphates linked to moiety X) specific for ADP-ribose and 2′-phospho-ADP-ribose [47].
To test a hypothesis of NMNATs' ability to cap RNA with NAD+ we overproduced and purified recombinant NadD and NadR proteins from E. coli. We tested their potential capping and decapping activities in vitro by incubating them with radiolabelled ppp-RNA or NAD-RNA in conditions suitable for NAD synthesis as specified by Rafaelli et al. for NadR (50 mM Hepes, pH 8.6, 10 mM MgCl2, 1 mM NMN) [40] and by Mehl et al. for NadD [41] (100 mM Tris–HCl, pH 8.0, with 2 mM MgCl2, 5 mM NMN) but did not observe either capping or decapping activity (J.W. 2020, unpublished data). The absence of activity of NadR and NadD towards RNA may reflect the absence of such a mechanism in bacteria, or alternatively the need for condition optimization or auxiliary factors. NAD+ post-transcriptional capping can also be performed by another unknown enzyme. All things considered, a test of the involvement of eukaryotic NMNAT in NC RNA capping is still worthwhile.
6. Capping RNA with dinucleotide polyphosphates (NpnNs) by RNA polymerase and aminoacyl tRNA synthetase
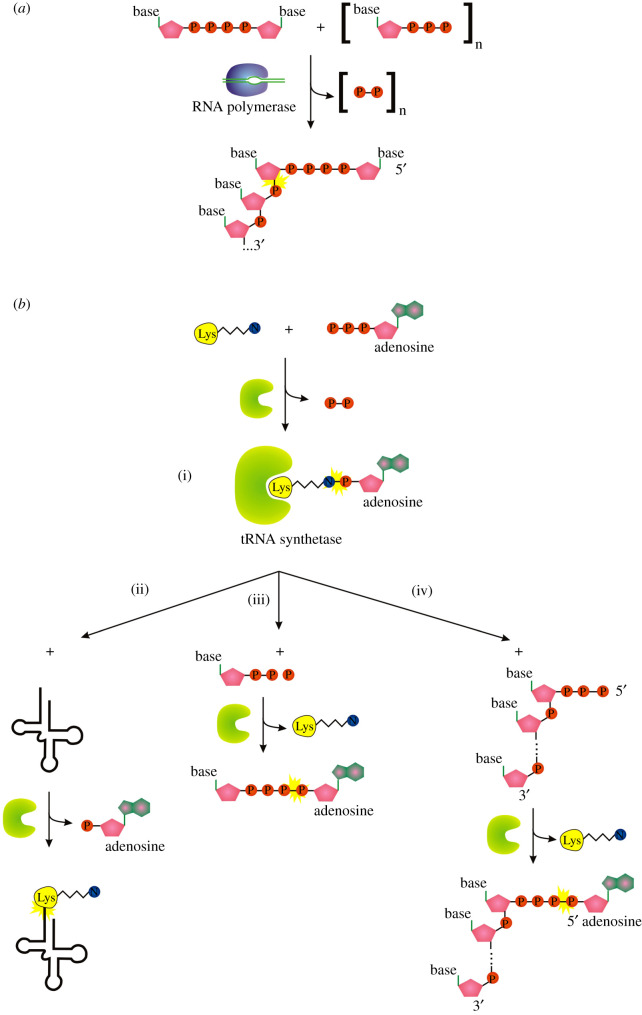
During the last 2 years, the 5′ RNA cap family was expanded by a number of new caps formed by dinucleotide polyphosphates [10,14,21,48]. For more than five decades, dinucleoside polyphosphates (NpnNs) were known as signalling molecules in all life domains, but their target and physiological function were elusive [49,50]. Various Np3Ns and Np4Ns are being synthesized by a side reaction during aminoacyl-tRNA synthesis by lysyl-tRNA synthetase (LysU, figure 4b, reactions (i) and (ii) versus (i) and (iii)) [50–52]. But the range of producers is possibly wider while overproduction of other aminoacyl -tRNA synthetases (methionyl- phenylalanyl- or valyl-) affects the NpnN levels in E. coli as well [51,53]. In bacteria under stress conditions like oxidative stress or heat shock, the concentration of NpnNs can increase from the μM to the mM range [54–56], which is why they are thought to act as alarmones [53]. In eukaryotes, they play a role in neuronal signalling, immune response and cardiovascular function [57].
Figure 4.
Pathways capping RNA with Np4N cap. (a) RNAP ab initio capping. (b) Aminoacyl tRNA synthetase pathway mediating nucleotidyl transfer dependent on amino acid-AMP covalent intermediate. (i) Formation of an amino acid-AMP intermediate, (ii) charging of tRNA, (iii) production of Np4A alarmone, (iv) Ap4N capping of RNA. Symbols as described in figure 1.
Papers from two independent groups demonstrated that RNA can be capped in vivo by a number of different NpnNs and their methylated forms: Ap3A, m6Ap3A, Ap3G, m7Gp4Gm, Ap5A, m6Ap5G, m6Ap4G, m6Ap5A and 2mAp5G [10,14]. The type of capping is specific for the growth phase and can be induced by disulphide stress. All presented NpnN studies are consistent with the hypothesis that NpnN capping may be the signal-transfer pathway that mediates transmission of NpnN levels to bacterial gene expression by changing the lifetime of capped RNAs. Lifetime is affected by decapping enzymes (primarily ApaH), which are specifically inhibited by inducers of disulfide stress, stabilizing its target RNAs [10,14,21]. This statement is consistent with findings of previous work that ApaH mutation causes Ap4A accumulation in E. coli [58]. The NpnN-RNA is further stabilized by methylation of the cap, inhibiting RppH (RNA pyrophosphohydrolase)-dependent degradation. This is probably caused by introducing the positive charge to the purine ring by the methylation followed by loss of the interaction with two RppH arginines responsible for the purines binding [10]. The process of in vivo methylation of NpnN caps needs to be deciphered in future studies.
The Belasco group demonstrated that RNAP is expected to be the enzyme responsible for NpnN capping of RNA in vivo (figure 4a). They manipulated the promoter sequence upstream of the site of transcription initiation and showed that it influences both Np4A incorporation into nascent transcripts in vitro and levels of Np4 capping in vivo [14]. The ninefold preference of RNAP to initiate transcription with NpnN rather than ATP also supports the RNAP-dependent origin of most NpnN-RNA in the cell [21]; however, it was not observed by the Cahová group [10] (the authors observed at least two times better incorporation of ATP compared with Ap6A or Ap4G, similarly to NAD+ incorporation).
Interestingly, Ap4N-RNA can be produced in vitro by lysyl-tRNA synthetase LysU in the presence of triphosphorylated RNA, ATP and lysine (figure 4b, reactions (i) and (iv)). A supplementary role for aminoacyl-tRNA synthetases or other cellular enzymes in capping ppp-RNA by nucleotidyl transfer to the RNA 5′ end cannot be excluded; however, the contribution is likely to be relatively small [14]. All enzymatic reactions carried by LysU (and probably also by other aminoacyl tRNA synthetases [53]) require the amino acid-NTP covalent intermediate (figure 2c and 4b) which draws a parallel to other nucleotidyl transferases known to cap RNA in vitro or in vivo. In contrast to other discussed mechanisms in this review, the amino acid is not covalently bound to the capping enzyme but is tightly bound in the active centre of the enzyme.
Concurrently with Np4N level rise during heat shock and oxidative stress, relevant RNA capping [14] is expected to rise. In connection with this notion, it is interesting that heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40 in E. coli bind Ap4A [59]. It would be interesting to test whether Ap4A-RNA can interact with these enzymes and localize them to the site of RNA translation. Particularly chaperone activity of DnaK/GroEL localized at the site of emerging nascent protein during heat shock could be beneficial for its correct folding which can be compromised by high temperature.
Decapping of RNA by ApaH may be a more widespread phenomenon than previously expected. ApaH-like phosphatase (TbALPH1) is the major mRNA decapping enzyme of trypanosomes where no clear orthologues to Dcp2 (the main decapping enzyme in most studied eukaryotes) or its associated proteins are present. While the genome of trypanosomes is transcribed as long, polycistronic pre-mRNAs of up to 100 protein-coding genes and lacks conventional promoters, the missing transcriptional regulation in these parasites must be supplemented by post-transcriptional mechanisms, including the regulation of mRNA decay. RNAs of trypanosomes (and other Kinetoplastida species) have a unique, heavily methylated cap4 structure which can be removed by TbALPH [60]. Similarly, methylated Np4Ns of E. coli are removed by ApaH [10], whereas bacterial RppH decapping is fully inhibited by the cap methylation. This parallel may be useful for studying ApaH-like recognition and decapping mechanisms of methylated cap structures in higher organisms and may shed light on the evolution of similar structures.
7. ADP-ribose is a potential new major non-canonical RNA 5′ cap with at least four possible mechanisms for capping
ADP-ribosylation is a reversible chemical modification catalysed by ADP-ribosyltransferases called poly (ADP-ribose) polymerases (PARPs). The essence of the process is a transfer of monomers or polymers of ADP-ribose nucleotide from NAD+ cofactor onto macromolecular targets such as proteins and nucleic acids (see the ADPR transfer to RNA, figure 5b). ADP-ribosylation affects a wide range of key biological processes: DNA-damage repair, DNA replication, transcription, cell division, signal transduction, stress and infection responses, microbial pathogenicity and ageing [61].
Figure 5.
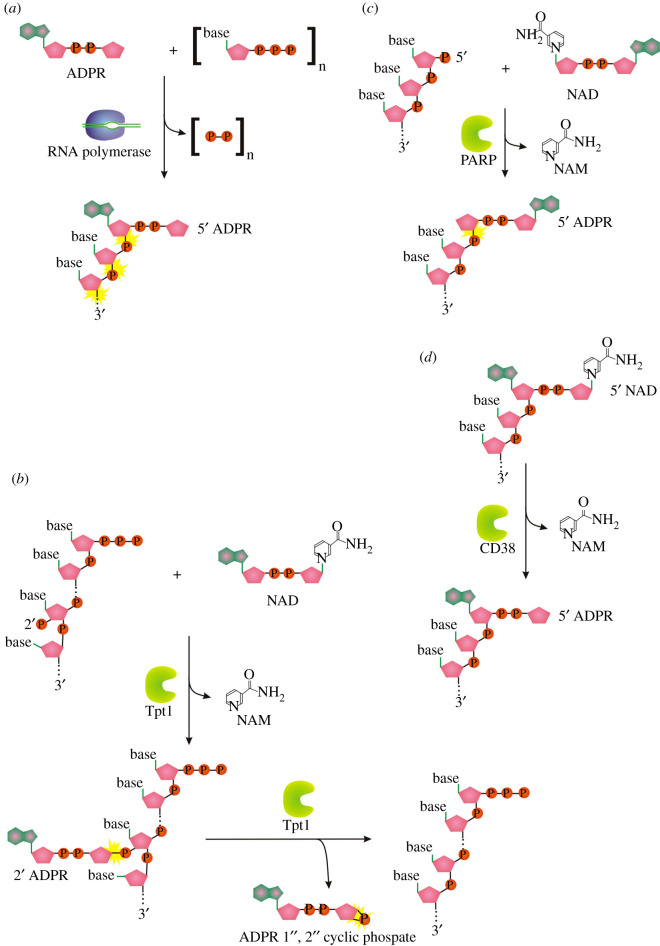
(a) Proposed mechanism for 5′ RNA capping with ADPR by transcription initiation by RNA polymerase. (b) Tpt1-mediated transfer of an internal RNA 2′-monophosphate (2′ p) to NAD+ to form a 2′- OH RNA, ADP-ribose 1″,2″ cyclic phosphate, and nicotinamide [36]. (c) Proposed reaction mechanism for Tpt1-dependent 5′ RNA capping with ADPR. (d) Mechanism of decapping of NADylated RNA by ADP ribosyl cyclase (CD38) producing ADPR-capped RNA. Symbols as described in figure 1.
The canonical function of 2′-phosphotransferase (Tpt1), a member of PARP family, is the NAD+-dependent conversion of an internal RNA 2′-phosphate to a 2′-OH using ADPR-RNA intermediate (figure 5b) [36]. Intriguingly, despite Tpt1 homologs being found in all domains of life, 2′-phosphate tRNAs are normally produced during tRNA splicing only in fungi. Other organisms use different splicing mechanisms [36]. Therefore, substrates and the biological functions of the Tpt1 homologs in these species were enigmatic. Recently, a subset of Tpt1 enzymes (bacterial and archaeal Tpt1, and other members from PARP family from eukaryotes: human PARP10, PARP11, PARP15, TRPT1) were demonstrated to catalyse NAD+-dependent ADP-ribosylation of an RNA or DNA 5′-monophosphate terminus in vitro, producing ADPR-RNA (figure 5c) [62,63].
Although ADPR-RNAs are not yet found in vivo, the conditions used during in vitro experiments strongly suggest that the same reactions may occur in cells [62,63]. Another hint is the existence of ADP-ribosylhydrolases, able to decap ADPR-RNA [62]. Moreover, the existence of analogous 5′ ribosylated DNA was already predicted by experiments in cell-free extracts and preliminary in vivo data [64–66]. ADPR-RNA can be alternatively attached to the RNA 5′ end during transcription initiation by RNAP (figure 5a) but ADPR concentration in the cell is three orders of magnitude lower than ATP or NAD+—4.3 × 10−6 versus 9.6 × 10−3 and 2.6 × 10−3 M, respectively [67], which makes the process of ab initio capping by RNAP less plausible but does not exclude its usage by RNAP as initiating substrates in a specific situation.
As recently discovered, ADPR-RNA can be also produced by hydrolysis of N-glycosidic bond of NAD-RNA by eukaryotic glycohydrolase CD38 in vitro (figure 5d) [68].
If found on RNA in vivo, ADP-ribose would be a new NC RNA cap, possibly present in all domains of life. It could be part of anti-viral immunity as ADP-ribosylation-dependent response of a host, which could be counteracted by virus-induced decapping of ADP-ribosylated RNAs [62]. This hypothesis is supported by a recent discovery that a conserved macrodomain-containing nsP3 protein of many viruses (including SARS-Cov2, the cause of COVID-19) hydrolyses ADP-ribosylated RNAs [69].
8. Potential capping enzymes producing A-pp-RNA: RNA ligases, RNA 3′ phosphate cyclases and tRNA guanylyl transferases
8.1. RNA ligases
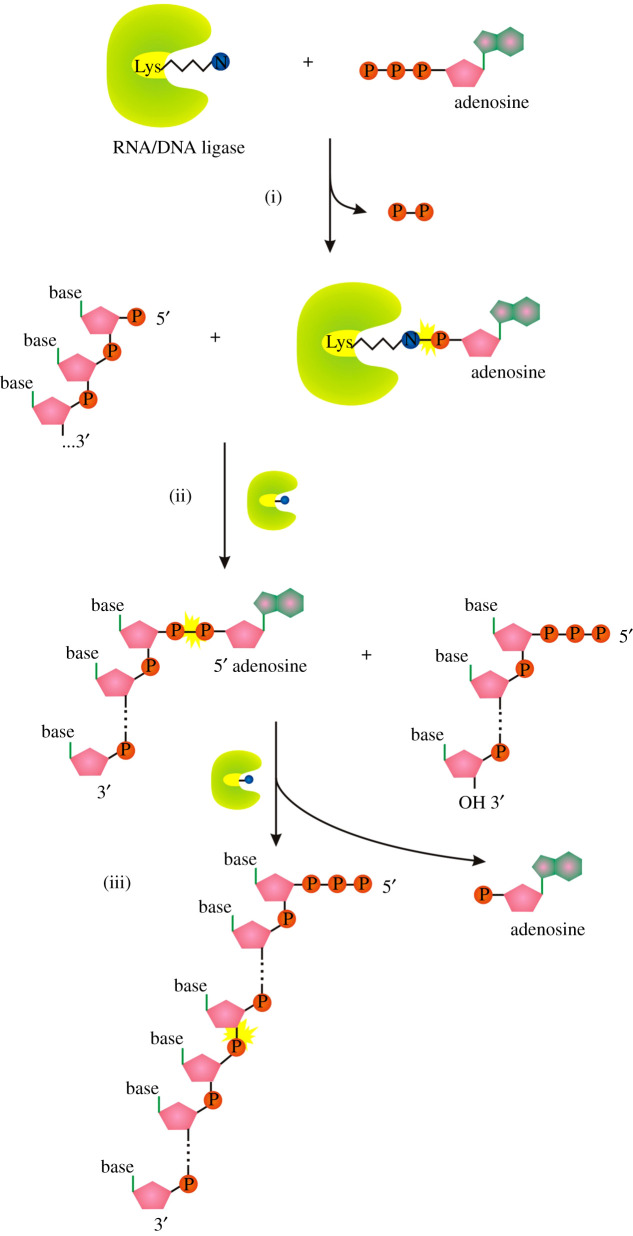
ATP- and NAD-dependent DNA ligases and ATP-dependent RNA ligases comprise a superfamily together with the above-mentioned GTP-dependent mRNA capping enzymes (guanylyl transferases, figures 1a and 2a). Similarly to GTase, DNA/RNA ligases catalyse 3-step nucleotidyl transfer to polynucleotide 5′ ends via covalent enzyme-(lysyl-N)-AMP intermediates (figure 2b) [70]. In the first step, attack on the α-phosphorus of ATP or NAD+ by the enzyme results in the release of pyrophosphate or nicotinamide mononucleotide (NMN), respectively, and formation of a covalent enzyme-(lysyl-N)–NMP intermediate (figure 6a reaction (i)). In the next step, the AMP (p-A) is transferred to the 5′ end of p-DNA/p-RNA forming A-pp-DNA/A-pp-RNA (ii). In the last step, DNA/RNA ligases catalyse attack by the 3′-OH of the nick on the A-pp-DNA/RNA to join two polynucleotides and release AMP (iii). At least in the case of DNA ligases, the final ligation step has variable efficiency, therefore releasing high levels of adenylated nucleic acid intermediate [71], the efficiency of RNA ligases potentially releasing adenylated RNA is unknown.
Figure 6.
Mechanism of RNA ligation by ATP-dependent RNA ligases. Symbols as described in figure 1.
8.2. RNA 3′-phosphate cyclase RtcA
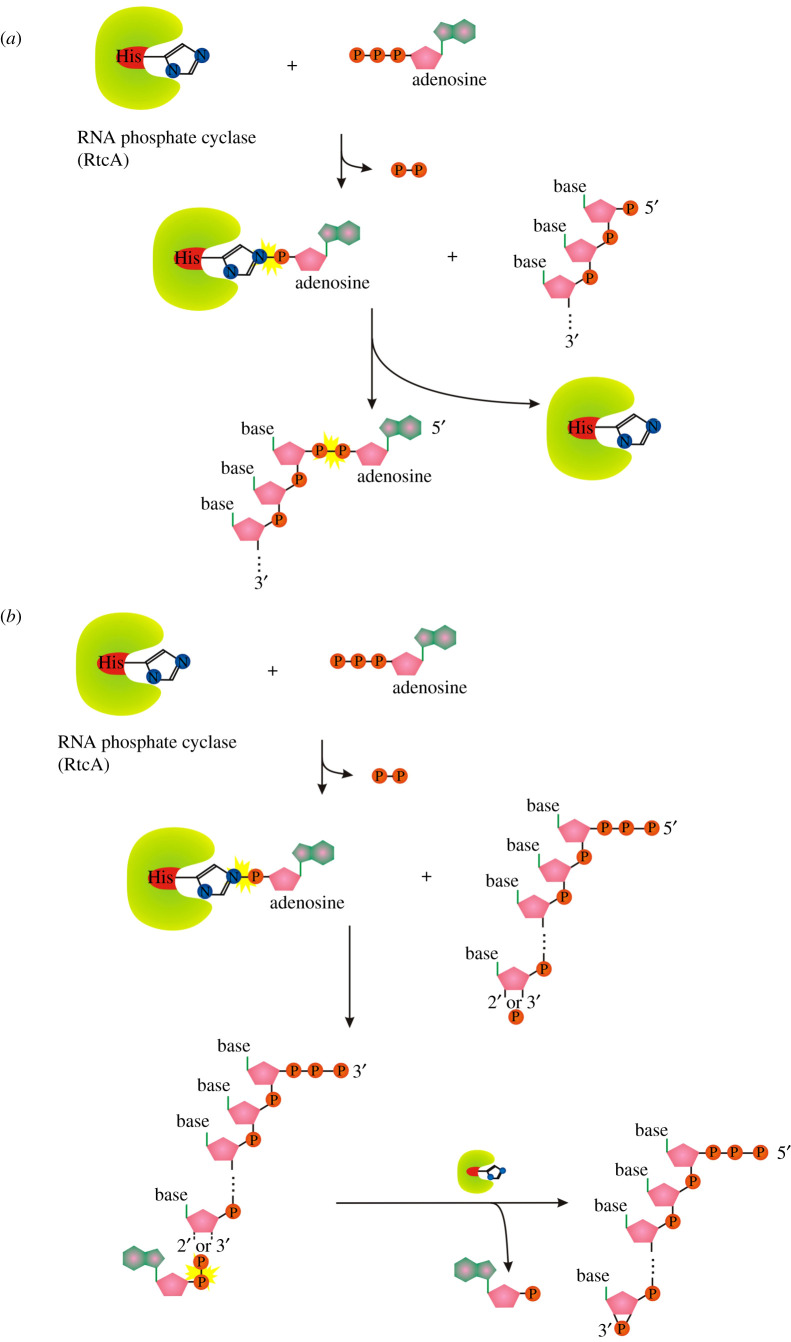
A similar ligase-like mechanism of 5′-DNA/RNA adenylylation is catalysed by the RNA 3′-phosphate cyclase (RtcA) [72]. Escherichia coli RtcA catalyses adenylylation of 5′-p ends of DNA or RNA strands using RtcA-histidinyl-N-AMP intermediate to form A-pp-DNA and A-pp-RNA products (figure 7a). The canonical function of RtcA is a catalysis of the ATP-dependent conversion of a 3′-p-RNA or 2′-p-RNA end to a 2′,3′-cyclic phosphate via covalent enzyme-histidinyl-N-AMP and RNA-3′pp-A intermediates (figure 7b) [73]. RtcA is coregulated in an operon with an RNA ligase, RtcB, that splices RNA 5′-OH ends to either 3′-phosphate or 2′,3′-cyclic phosphate ends (figure 8) [74]. RtcA might serve an end healing function in an RNA repair pathway, by converting RNA 2′-phosphates, which cannot be spliced by RtcB, to 2′,3′-cyclic phosphates that can be sealed.
Figure 7.
(a) Mechanism of 5′ RNA/DNA adenylylation by RtcA. (b) Mechanism of RtcA 2′,3′ phosphate cyclization. Symbols as described in figure 1.
Figure 8.
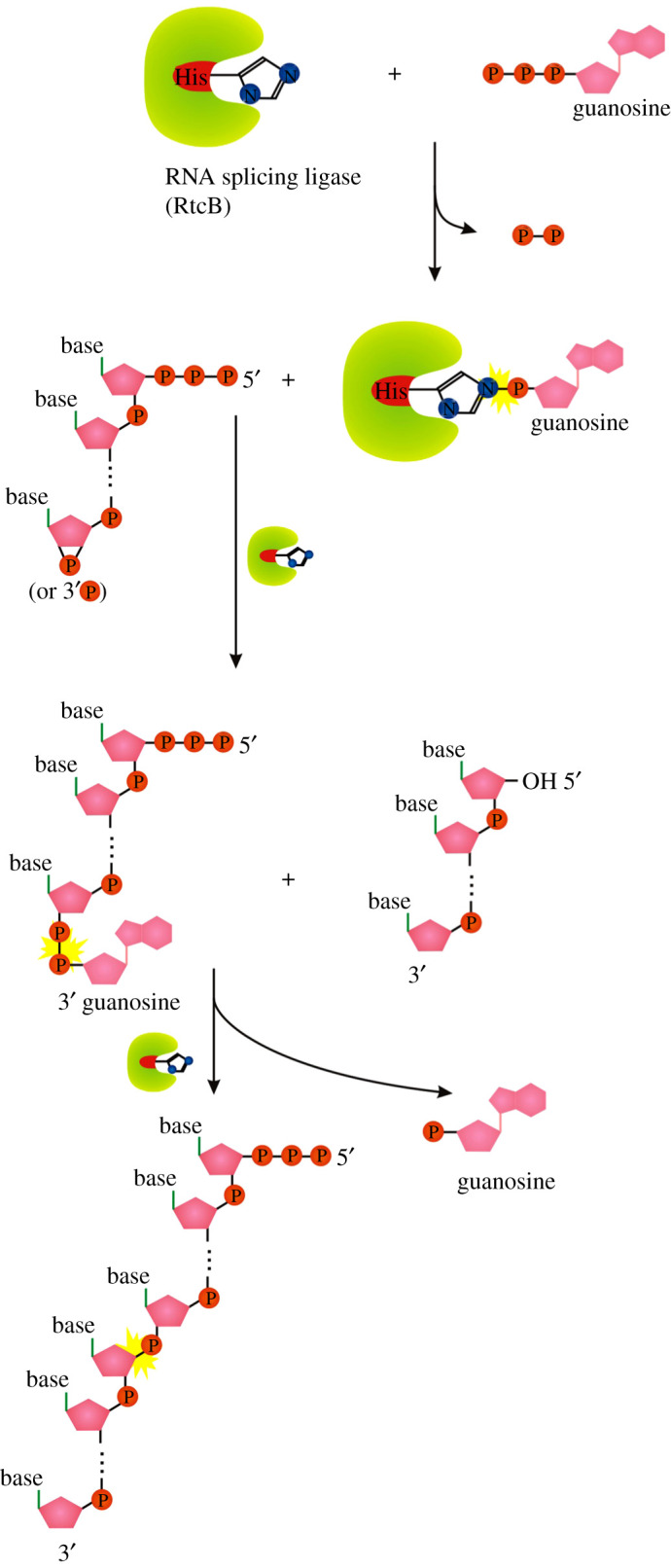
Splicing mechanism of RtcB. Symbols as described in figure 1.
The main difference among the conventional DNA/RNA ligases, RtcA and RtcB is the amino acid used to form the reaction intermediate (RtcA with ATP/RtcB with GTP form a covalent RtcA-histidinyl-N-AMP/RtcB-histidinyl-N-GTP intermediate, ligases use enzyme-lysyl-N-AMP intermediate; figure 2b,d,e). RtcB and RtcA are structurally unrelated proteins with no similarities in their active sites [75–77]. They also lack structural similarity with RNA/DNA ligases, either globally or with respect to their active site architectures [75,78].
The ability of E. coli RtcA to catalyse adenylylation of 5′p ends of DNA or RNA strands to form A-pp-DNA and A-pp-RNA products opens the possibility of new cellular functions in RNA capping and therefore affecting the properties of capped RNAs. Such possibilities need to be tested.
8.3. tRNA guanylyl transferases
5′-5′ mRNA GTases, and therefore also canonical caps, are unique for eukaryotes and missing in prokaryotes and archaea. Interestingly, a similar enzyme (tRNA guanylyl transferases; tRNA GTase), is present in all three domains of life (THG1 in human, Thg1 in yeast and Thg1-like proteins (TLPs) in archaea and bacteria) [79,80]. tRNA GTase is the only known example of an enzyme that catalyses templated nucleotide addition in the 3′-5′ direction, opposite to that of all known DNA and RNAPs [81] resulting in ppp-G-RNA (compare with G-ppp-RNA produced by mRNA GTase). In the first step, it uses ATP to activate the monophosphorylated tRNAHis (p-tRNAHis generated by ribonuclease P) producing adenylated tRNAHis (A-pp-tRNAHis) intermediate (figure 9(i)). In the second step, the hydroxyl of a GTP nucleotide attacks the activated intermediate, yielding the triphosphorylated form of tRNAHis (ppp-G-tRNAHis) and AMP (figure 9(ii)) [80–82].
Figure 9.
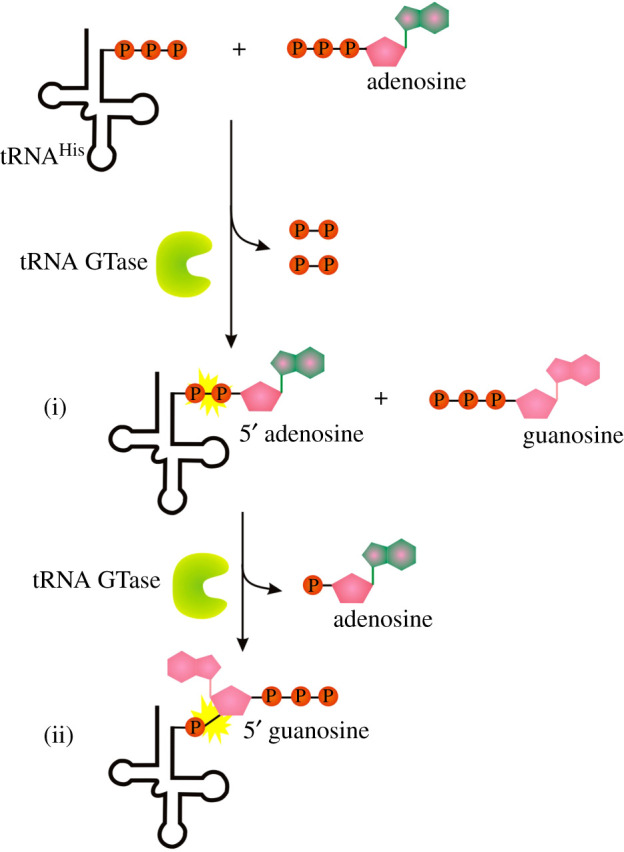
Mechanism of 3′-5′ GTP (ppp-G) addition by tRNA guanylyl transferases. Symbols as described in figure 1.
The adenylylated intermediate product of the pathway (A-pp-tRNA) can be classified as NC-capped RNA but while the process of adenylylation and subsequent guanylyl transfer is specifically bound to recognition of anticodon of tRNAHis [79], its wider use in RNA NC capping is questionable.
9. FAD capping by RNA polymerase and potentially flavin biosynthesis enzymes
Flavin adenine dinucleotide is one of the cellular metabolites reported as covalently bound to the RNA in prokaryotes, eukaryotes and viruses [7]. It is readily incorporated by cellular and mitochondrial RNAPs to 5′ end of the nascent transcript [13,27]. FAD as well as NADH serves as NC initiating substrates for a primase and affect replication primer processing in vitro [29].
Whether there are other enzymes producing FAD-RNA remains an unanswered question. We speculate that enzymes from the flavin mononucleotide adenylyl transferase (FMNAT) family could be a possible candidate, analogically to NMNATs as candidates for NAD+ capping.
10. Non-canonical RNA capping by ribozymes
Intriguingly, not only proteins can cap RNA with metabolite cofactors. Ribozymes able to make NAD+-, FAD- and CoA-capped RNA by attaching small-molecule precursors to a 5′-terminal ATP were evolved in vitro [83]. Ribozymes are generally believed to be functional fossils from RNA world [84]. Hypothetically, the ribozyme reactions might once have been the main pathway, and could also today still be working unrecognized in in vivo synthesis of capped RNA [85].
11. Protein caps of the viral genome
Some positive sense single-stranded RNA viruses have viral protein genome-linked (VPgs) covalently attached to the 5′ end of their genome. In caliciviruses, VPgs were confirmed to interact with the cap-binding protein (eIF4E) and to be essential for translation [86]. These viral cap-like structures are not small molecules but proteins, and thus can be classified as NC 5′ RNA caps.
12. The extent of NC capping and cellular roles of capped RNAs are still unclear
A number of potential physiological roles of NC capping are suggested in two recent reviews, including RNA folding and stability, cellular localization, etc. [85,87]. Yet only a few actual examples are published, e.g. increased NADylation of RNAIII, a central quorum-sensing regulator of Staphylococcus aureus repressed production of toxins, thus decreasing cytotoxicity of the bacterium [88]. Therefore, so far, the extent, impact or functions of NC-capped RNAs remain largely unknown. To demonstrate this point, we would like to highlight a few enigmatic and still controversial aspects of NAD capping, the most studied NC capping to date, thanks to the number of NAD-RNA isolation and quantification techniques [11,24,26].
It is unclear if a majority of the capped transcript are full-sized or truncated species. In bacteria (E. coli, B. subtilis and Staphylococcus aureus), the abundant NAD-capped species are 5'-terminal fragments of certain mRNAs [11]. Fragmented NAD-RNAs were also found in B. subtilis dormant spores [89]. Nevertheless, validation of some mRNA species detected also full-length transcripts in B. subtilis and S. aureus [17,88]. These discrepancies may be caused by the bias of the original method, where only short RNAs (less than 200 nt) were used for NGS sequencing library preparation [11]. Indeed in yeast, mammals and plants, the observed NAD-RNAs were mostly of the full length [18,25,26,32]. However, recent preprint publication from yeast reports yet again a large number of short mRNA 5′-terminal fragments [22], echoing findings in bacteria. The important question to answer is how these short NAD+-capped RNAs are produced, whether the presence of NAD+ at the 5′-end affects either transcription initiation, termination or degradation of full-length RNAs to make species of shorter yet defined length, stable enough to be detected, or whether the short length is just an artefact of NAD-RNA identification process.
The impact of capping on transcription and post-transcriptional processing of RNA is known only for a few examples. In S. cerevisiae and A. thaliana, RNAP II uses different transcription start sites (TSS) for NAD-RNA and for m7G-capped RNAs of the same gene, but the biological function of alternative TSSs remains unsolved. It was shown for E. coli transcription that 5′-NAD can stabilize short transcript and prevent their release improving chances for RNAP to escape into a productive elongation [13]. Mammalian and plant NAD-RNAs can be spliced and polyadenylated [18,32], suggesting they are treated at least by some of the cellular machinery as canonical mRNAs.
The translatability of NC-capped mRNA is a conundrum. Reports on the translatability of NADylated mRNAs are conflicting. While in human and yeast cells NAD-RNAs do not support translation but instead promote mRNA decay [22,32], NAD-RNA species of plants are enriched in the polysomal fraction and associate with translating ribosomes, suggesting that they are translated [18]. Reports from bacteria about NAD-RNA translation are scarce. In S. aureus 5′-NAD cap on RNAIII might impair translation of a small open reading frame inside this regulatory RNA in vivo, but the impact of additional factors cannot be ruled out. Both NAD+-RNAIII and ppp-RNAIII supported the formation of stable translational initiation complexes in vitro [88]. In summary, utilization of NAD-mRNAs for its main purpose (translation) is probably ceased either by the truncation of the resulting NAD-RNA by an unknown mechanism (mostly in bacteria) or by its inability to be translated (in human and yeast cells). The translatability is feasible in mitochondria, where NAD+ capping seems to be higher than in other cell compartments or prokaryotic cells (up to 15% in human and 60% in yeast [19]). It is hard to imagine that so high number of mitochondrial transcripts would be made just for being degraded, so NAD+ capping of mitochondrial RNAs seems to be at least tolerated if not required for efficient translation.
13. Summary and perspectives
In this review, we compiled information about a number of enzymes beyond RNAPs capable of in vitro or in vivo addition of NC caps to RNA. All the reviewed enzymes with proven capping activity decorate RNA by 5′-5′ addition of nucleoside connected by a variable number of phosphates or phosphate-ribose-phosphate bridges. Interestingly, most of these enzymes share the principal mechanism of cap addition with canonical 5′ RNA capping: they use amino acid—nucleotide covalent linkage as an intermediate for nucleotidyl transfer to RNA/nucleotide (figure 2). This common feature can serve as a useful clue for searching other undiscovered capping mechanisms. For example, adenylylation enzymes (AMPylators) using intermediates with covalent phosphodiester bonds between amino acid (serine, tyrosine or threonine) side chain of the enzyme and adenosine monophosphate [90] could be a possible source of capping enzymes. Despite NAD+ seeming to be one of the most ubiquitous NC caps, no post-transcriptional capping enzyme was identified for it so far. We tested a possibility of NAD-RNA capping by enzymes of NAD+ synthesis pathway (E. coli NMNATs NadD and NadR) but did not observe any capping or decapping activity in tested conditions.
Initially, NC capping was perceived as RNAP-dependent stoichiometric event affected by the ratio of concentrations of NTPs to other NTP-analogues. This review focuses on the facts which challenge this perception, showing that cells are probably packed with enzymes capping and decapping RNA with NC caps. Whether all NC capping events are stochastic side reactions of moonlighting enzymes with established primary function, or there exist dedicated machinery for NC capping, remains to be shown. Expanding repertoire of caps and capping enzymes suggests if not a physiological function then at least a major physiological impact of these caps.
Supplementary Material
Footnotes
Data accessibility
This article has no additional data.
Authors' contributions
This is a review paper with unpublished experimental results discussed. J.W. wrote the manuscript and performed experiments, C.J. performed experiments, Y.Y. wrote and edited the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the Royal Society URF to Y.Y. and the Leverhulme Trust [J.W., Y.Y. grant no. RPG-2018-437].
References
- 1.Ramanathan A, Robb GB, Chan SH. 2016. mRNA capping: biological functions and applications. Nucleic Acids Res. 44, 7511–7526. ( 10.1093/nar/gkw551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei CM, Gershowitz A, Moss B. 1975. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4, 379–386. ( 10.1016/0092-8674(75)90158-0) [DOI] [PubMed] [Google Scholar]
- 3.Schaller H, Gray C, Herrmann K, Furuichi Y, Morgan M, Muthukrishnan S, Shatkin AJ. 1975. Reovirus messenger RNA contains a methylated, blocked 5′-terminal structure: m7G(5′)ppp(5′)GmpCp-. Proc. Natl Acad. Sci. USA 72, 2468–2468. ( 10.1073/pnas.72.6.2468-a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C. 2007. 5′-to-3′ Exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129, 681–692. ( 10.1016/j.cell.2007.02.051) [DOI] [PubMed] [Google Scholar]
- 5.Mackie GA. 1998. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395, 720–723. ( 10.1038/27246) [DOI] [PubMed] [Google Scholar]
- 6.Luciano DJ, Vasilyev N, Richards J, Serganov A, Belasco JG. 2017. A novel RNA phosphorylation state enables 5′ end-dependent degradation in Escherichia coli. Mol. Cell 67, 44–54. ( 10.1016/j.molcel.2017.05.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, et al. 2019. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 47, e130. ( 10.1093/nar/gkz751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR. 2009. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 5, 879–881. ( 10.1038/nchembio.235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowtoniuk WE, Shen Y, Heemstra JM, Agarwal I, Liu DR. 2009. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc. Natl Acad. Sci. USA 106, 7768–7773. ( 10.1073/pnas.0900528106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudeček O, et al. 2020. Dinucleoside polyphosphates act as 5′-RNA caps in bacteria. Nat. Commun. 11, 1052. ( 10.1038/s41467-020-14896-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cahová H, Winz M-L, Höfer K, Nübel G, Jäschke A. 2015. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519, 374–377. ( 10.1038/nature14020) [DOI] [PubMed] [Google Scholar]
- 12.Bird JG, et al. 2016. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535, 444–447. ( 10.1038/nature18622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julius C, Yuzenkova Y. 2017. Bacterial RNA polymerase caps RNA with various cofactors and cell wall precursors. Nucleic Acids Res. 45, 8282–8290. ( 10.1093/nar/gkx452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luciano DJ, Levenson-Palmer R, Belasco JG. 2019. Stresses that raise Np4A levels induce protective nucleoside tetraphosphate capping of bacterial RNA. Mol. Cell 75, 957–966. ( 10.1016/j.molcel.2019.05.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267, 9805–9815. ( 10.1016/S0021-9258(19)50165-X) [DOI] [PubMed] [Google Scholar]
- 16.Ogino T, Yadav SP, Banerjee AK. 2010. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl Acad. Sci. USA 107, 3463–3468. ( 10.1073/pnas.0913083107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frindert J, Zhang Y, Burhenne U, Haefeli WE, Jä A. 2018. Identification, biosynthesis, and decapping of NAD-capped RNAs in B. subtilis. Cell Rep. 24, 47. ( 10.1016/j.celrep.2018.07.047) [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Li S, Zhao Y, You C, Le B, Gong Z, Mo B, Xia Y, Chen X. 2019. NAD+-capped RNAs are widespread in the Arabidopsis transcriptome and can probably be translated. Proc. Natl Acad. Sci. USA 116,12 094–12 102. ( 10.1073/pnas.1903682116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bird JG, et al. 2018. Highly efficient 5′ capping of mitochondrial RNA with nad+and NADH by yeast and human mitochondrial RNA polymerase. Elife 7, e42179. ( 10.7554/eLife.42179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grudzien-Nogalska E, Bird JG, Nickels BE, Kiledjian M. 2018. ‘NAD-capQ’ detection and quantitation of NAD caps. RNA 24, 1418–1425. ( 10.1261/rna.067686.118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luciano DJ, Belasco JG. 2020. Np4A alarmones function in bacteria as precursors to RNA caps. Proc. Natl Acad. Sci. USA 117, 3560–3567. ( 10.1073/pnas.1914229117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, et al.2020Extensive 5′-surveillance guards against non-cnanonical NAD-caps of nuclear mRNAs in yeast correspondence. bioRxiv ( 10.1101/2020.04.28.065920) [DOI]
- 23.Bird J, Nickels B, Ebright R. 2017. RNA capping by transcription initiation with non-canonical initiating nucleotides (NCINs): determination of relative efficiencies of transcription initiation with NCINs and NTPs. Bio-protocol 7, e2336. ( 10.21769/bioprotoc.2336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vvedenskaya IO, et al. 2018. CapZyme-Seq comprehensively defines promoter-sequence determinants for RNA 5′ capping with NAD+. Mol. Cell 70, 553–564. ( 10.1016/j.molcel.2018.03.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walters RW, Matheny T, Mizoue LS, Rao BS, Muhlrad D, Parker R. 2017. Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 114, 480–485. ( 10.1073/pnas.1619369114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Zhong H, Zhang S, Shao X, Ni M, Cai Z, Chen X, Xia Y. 2019. NAD tagSeq reveals that NAD+-capped RNAs are mostly produced from a large number of protein-coding genes in Arabidopsis. Proc. Natl Acad. Sci. USA 116, 12 072–12 077. ( 10.1073/pnas.1903683116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Julius C, Riaz-Bradley A, Yuzenkova Y. 2018. RNA capping by mitochondrial and multi-subunit RNA polymerases. Transcription 9, 292–297. ( 10.1080/21541264.2018.1456258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malygin AG, Shemyakin MF. 1979. Adenosine, NAD and FAD can initiate template-dependent RNA a synthesis catalyzed by Escherichia coli RNA polymerase. FEBS Lett. 102, 51–54. ( 10.1016/0014-5793(79)80926-6) [DOI] [PubMed] [Google Scholar]
- 29.Julius C, Salgado PS, Yuzenkova Y. 2020. Metabolic cofactors NADH and FAD act as non-canonical initiating substrates for a primase and affect replication primer processing in vitro. Nucleic Acids Res. 48, 7298–7306. ( 10.1093/nar/gkaa447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courtois F, Merendino L, Demarsy E, Mache R, Lerbs-Mache S. 2007. Phage-type RNA polymerase RPOTmp transcribes the RRN operon from the PC promoter at early developmental stages in Arabidopsis. Plant Physiol. 145, 712–721. ( 10.1104/pp.107.103846) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gakière B, Hao J, de Bont L, Pétriacq P, Nunes-Nesi A, Fernie AR.. 2018. NAD+ biosynthesis and signaling in plants. CRC Crit. Rev. Plant Sci. 37, 259–307. ( 10.1080/07352689.2018.1505591) [DOI] [Google Scholar]
- 32.Jiao X, Doamekpor SK, Bird JG, Nickels BE, Tong L, Hart RP, Kiledjian M. 2017. 5′ End nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell 168, 1015–1027. ( 10.1016/j.cell.2017.02.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nag MK, Thai TT, Ruff EA, Selvamurugan N, Kunnimalaiyaan M, Eliceiri GL. 1993. Genes for E1, E2, and E3 small nucleolar RNAs. Proc. Natl Acad. Sci. USA 90, 9001–9005. ( 10.1073/pnas.90.19.9001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiss T, Filipowicz W. 1993. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 12, 2913–2920. ( 10.1002/j.1460-2075.1993.tb05953.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tycowski KT, Shu MD, Steitz JA. 1993. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 7, 1176–1190. ( 10.1101/gad.7.7a.1176) [DOI] [PubMed] [Google Scholar]
- 36.Lin H. 2007. Nicotinamide adenine dinucleotide: beyond a redox coenzyme. Org. Biomol. Chem. 5, 2541–2554. ( 10.1039/b706887e) [DOI] [PubMed] [Google Scholar]
- 37.Berger F, Lau C, Dahlmann M, Ziegler M. 2005. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 280, 36 334–36 341. ( 10.1074/jbc.M508660200) [DOI] [PubMed] [Google Scholar]
- 38.Ma X, et al. 2020. Nicotinamide mononucleotide adenylyl transferase uses its NAD+ substrate-binding site to chaperone phosphorylated TAU. Elife 9, e51859. ( 10.7554/eLife.51859) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfoh R, Pai EF, Saridakis V. 2015. Nicotinamide mononucleotide adenylyltransferase displays alternate binding modes for nicotinamide nucleotides. Acta Crystallogr. Sect. D Biol. Crystallogr. 71, 2032–2039. ( 10.1107/S1399004715015497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raffaelli N, Lorenzi T, Mariani PL, Emanuelli M, Amici A, Ruggieri S, Magni G. 1999. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 181, 5509–5511. ( 10.1128/jb.181.17.5509-5511.1999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehl RA, Kinsland C, Begley TP. 2000. Identification of the Escherichia coli nicotinic acid mononucleotide adenylyltransferase gene. J. Bacteriol. 182, 4372–4374. ( 10.1128/JB.182.15.4372-4374.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emanuelli M, Amici A, Carnevali F, Pierella F, Raffaelli N, Magni G. 2003. Identification and characterization of a second NMN adenylyltransferase gene in Saccharomyces cerevisiae. Protein Expr. Purif. 27, 357–364. ( 10.1016/S1046-5928(02)00645-9) [DOI] [PubMed] [Google Scholar]
- 43.Di Lisa F, Ziegler M.. 2001. Pathophysiological relevance of mitochondria in NAD+ metabolism. FEBS Lett. 492, 4–8. ( 10.1016/S0014-5793(01)02198-6) [DOI] [PubMed] [Google Scholar]
- 44.Nikiforov A, Dölle C, Niere M, Ziegler M. 2011. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: From entry of extracellular precursors to mitochondrial NAD generation. J. Biol. Chem. 286, 21 767–21 778. ( 10.1074/jbc.M110.213298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, Zhou T, Kurnasov O, Cheek S, Grishin NV, Osterman A. 2002. Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD. Structure 10, 69–79. ( 10.1016/S0969-2126(01)00693-1) [DOI] [PubMed] [Google Scholar]
- 46.Dahmen W, Webb B, Preiss J. 1967. The deamido-diphosphopyridine nucleotide and diphosphopyridine nucleotide pyrophosphorylases of Escherichia coli and yeast. Arch. Biochem. Biophys. 120, 440–450. ( 10.1016/0003-9861(67)90262-7) [DOI] [PubMed] [Google Scholar]
- 47.Raffaelli N, Lorenzi T, Amici A, Emanuelli M, Ruggieri S, Magni G. 1999. Synechocystis sp. slr0787 protein is a novel bifunctional enzyme endowed with both nicotinamide mononucleotide adenylyltransferase and ‘Nudix’’ hydrolase activities'. FEBS Lett. 444, 222–226. ( 10.1016/S0014-5793(99)00068-X) [DOI] [PubMed] [Google Scholar]
- 48.Jagodnik J, Gourse RL. 2020. Deciphering the RNA capping process in bacteria. Proc. Natl Acad. Sci. USA 117, 4445–4446. ( 10.1073/pnas.2000341117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finamore F, Warner A. 1963. The occurrence of P1, P4-diguanosine 5′-tetraphosphate in brine shrimp eggs. J. Biol. Chem. 238, 344–348. [PubMed] [Google Scholar]
- 50.Zamecnik PG, Stephenson ML, Janeway CM, Randerath K. 1966. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 24, 91–97. ( 10.1016/0006-291X(66)90415-3) [DOI] [PubMed] [Google Scholar]
- 51.Brevet A, Chen J, Leveque F, Plateau P, Blanquet S. 1989. In vivo synthesis of adenylylated bis(5′-nucleosidyl) tetraphosphates (Ap4N) by Escherichia coli aminoacyl-tRNA synthetases. Proc. Natl Acad. Sci. USA 86, 8275–8279. ( 10.1073/pnas.86.21.8275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plateau P, Blanquet S. 1982. Zinc-dependent synthesis of various dinucleoside 5′,5′″-P1,P3-tri- or 5′,5′″-P1,P4-tetraphosphates by Escherichia coli lysyl-tRNA synthetase. Biochemistry 21, 5273–5279. ( 10.1021/bi00264a024) [DOI] [PubMed] [Google Scholar]
- 53.Varshavsky A. 1983. Diadenosine 5′, 5′′′-P1, P4-tetraphosphate: a pleiotropically acting alarmone? Cell 34, 711–712. ( 10.1016/0092-8674(83)90526-3) [DOI] [PubMed] [Google Scholar]
- 54.VanBogelen RA, Kelley PM, Neidhardt FC. 1987. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J. Bacteriol. 169, 26–32. ( 10.1128/jb.169.1.26-32.1987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee PC, Bochner BR, Ames BN. 1983. AppppA, heat-shock stress, and cell oxidation. Proc. Natl Acad. Sci. USA 80, 7496–7500. ( 10.1073/pnas.80.24.7496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bochner BR, Lee PC, Wilson SW, Cutler CW, Ames BN. 1984. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 37, 225–232. ( 10.1016/0092-8674(84)90318-0) [DOI] [PubMed] [Google Scholar]
- 57.Boulos S, Razin E, Nechushtan H, Rachmin I. 2016. Diadenosine tetraphosphate (Ap4A) in health and disease, pp. 207–219. Cham, Switzerland: Springer. [Google Scholar]
- 58.Farr SB, Arnosti DN, Chamberlin MJ, Ames BN. 1989. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc. Natl Acad. Sci. USA 86, 5010–5014. ( 10.1073/pnas.86.13.5010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnstone DB, Farr SB. 1991. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 10, 3897–3904. ( 10.1002/j.1460-2075.1991.tb04959.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kramer S, McLennan AG. 2019. The complex enzymology of mRNA decapping: enzymes of four classes cleave pyrophosphate bonds. Wiley Interdiscip. Rev. RNA 10, e1511. ( 10.1002/wrna.1511) [DOI] [PubMed] [Google Scholar]
- 61.Palazzo L, Mikoč A, Ahel I. 2017. ADP-ribosylation: new facets of an ancient modification. FEBS J. 284, 2932–2946. ( 10.1111/febs.14078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Munnur D, et al. 2019. Reversible ADP-ribosylation of RNA. Nucleic Acids Res. 47, 5658–5669. ( 10.1093/nar/gkz305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Munir A, Banerjee A, Shuman S. 2018. NAD+-dependent synthesis of a 5-phospho-ADP-ribosylated RNA/DNA cap by RNA 2-phosphotransferase Tpt1. Nucleic Acids Res. 46, 9617–9624. ( 10.1093/nar/gky792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zarkovic G, et al. 2018. Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: New insights into DNA ADP-ribosylation. Nucleic Acids Res. 46, 2417–2431. ( 10.1093/nar/gkx1318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matta E, Kiribayeva A, Khassenov B, Matkarimov BT, Ishchenko AA. 2020. Insight into DNA substrate specificity of PARP1-catalysed DNA poly(ADP-ribosyl)ation. Sci. Rep. 10, 1–11. ( 10.1038/s41598-020-60631-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Belousova EA, Ishchenko AA, Lavrik OI. 2018. DNA is a new target of Parp3. Sci. Rep. 8, 4176. ( 10.1038/s41598-018-22673-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD.. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599. ( 10.1038/nchembio.186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abele F, Höfer K, Bernhard P, Grawenhoff J, Seidel M, Krause A, Kopf S, Schröter M, Jäschke A. 2020. A novel NAD-RNA decapping pathway discovered by synthetic light-up NAD-RNAs. Biomolecules 10, 513. ( 10.3390/biom10040513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rack JGM, Palazzo L, Ahel I. 2020. (ADP-ribosyl)hydrolases: structure, function, and biology. Genes Dev. 34, 263–284. ( 10.1101/gad.334631.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shuman S, Lima CD. 2004. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 14, 757–764. ( 10.1016/j.sbi.2004.10.006) [DOI] [PubMed] [Google Scholar]
- 71.Gong C, Martins A, Bongiorno P, Glickman M, Shuman S. 2004. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 279, 20 594–20 606. ( 10.1074/jbc.M401841200) [DOI] [PubMed] [Google Scholar]
- 72.Chakravarty AK, Shuman S. 2011. RNA 3′-phosphate cyclase (RtcA) catalyzes ligase-like adenylylation of DNA and RNA 5′-monophosphate ends. J. Biol. Chem. 286, 4117–4122. ( 10.1074/jbc.M110.196766) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Das U, Shuman S. 2013. 2′-Phosphate cyclase activity of RtcA: a potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA 19, 1355–1362. ( 10.1261/rna.039917.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Das U, Chakravarty AK, Remus BS, Shuman S. 2013. Rewriting the rules for end joining via enzymatic splicing of DNA 3′-PO4 and 5′-OH ends. Proc. Natl Acad. Sci. USA 110, 20 437–20 442. ( 10.1073/pnas.1314289110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanaka N, Smith P, Shuman S. 2010. Structure of the RNA 3′-phosphate cyclase-adenylate intermediate illuminates nucleotide specificity and covalent nucleotidyl transfer. Structure 18, 449–457. ( 10.1016/j.str.2010.01.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chakravarty AK, Smith P, Shuman S. 2011. Structures of RNA 3′-phosphate cyclase bound to ATP reveal the mechanism of nucleotidyl transfer and metal-assisted catalysis. Proc. Natl Acad. Sci. USA 108, 21 034–21 039. ( 10.1073/pnas.1115560108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Englert M, et al. 2012. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc. Natl Acad. Sci. USA 109, 15 235–15 240. ( 10.1073/pnas.1213795109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shuman S. 2009. DNA ligases: progress and prospects. J. Biol. Chem. 284, 17 365–17 369. ( 10.1074/jbc.R900017200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hyde SJ, Rao BS, Eckenroth BE, Jackman JE, Doublié S. 2013. Structural studies of a bacterial tRNAHIS guanylyltransferase (Thg1)-like protein, with nucleotide in the activation and nucleotidyl transfer sites. PLoS ONE 8, e67465. ( 10.1371/journal.pone.0067465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heinemann IU, Randau L, Tomko RJ, Söll D. 2010. 3′-5′ tRNAHis guanylyltransferase in bacteria. FEBS Lett. 584, 3567–3572. ( 10.1016/j.febslet.2010.07.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublié S. 2010. tRNAHis guanylyltransferase (THG1), a unique 3′-5′ nucleotidyl transferase, shares unexpected structural homology with canonical 5′-3′ DNA polymerases. Proc. Natl Acad. Sci. USA 107,20 305–20 310. ( 10.1073/PNAS.1010436107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jackman JE, Phizicky EM. 2006. tRNAHis guanylyltransferase adds G-1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA 12, 1007–1014. ( 10.1261/rna.54706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang F, Bugg CW, Yarus M. 2000. RNA-catalyzed CoA, NAD, and FAD synthesis from phosphopantetheine, NMN, and FMN. Biochemistry 39, 15 548–15 555. ( 10.1021/bi002061f) [DOI] [PubMed] [Google Scholar]
- 84.Gilbert W. 1986. Origin of life: the RNA world. Nature 319, 618. ( 10.1038/319618a0) [DOI] [Google Scholar]
- 85.Julius C, Yuzenkova Y. 2019. Noncanonical RNA-capping: discovery, mechanism, and physiological role debate. Wiley Interdiscip. Rev. RNA. 10, 1512. ( 10.1002/wrna.1512) [DOI] [PubMed] [Google Scholar]
- 86.Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberté JF, Roberts L. 2005. Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Rep. 6, 968–972. ( 10.1038/sj.embor.7400510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vasilyev N, Gao A, Serganov A. 2019. Noncanonical features and modifications on the 5′-end of bacterial sRNAs and mRNAs. Wiley Interdiscip. Rev. RNA. 10, e1509. ( 10.1002/wrna.1509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morales-Filloy HG, Zhang Y, Nübel G, George SE, Korn N, Wolz C, Jäschke A. 2020. The 5′-NAD cap of RNAIII modulates toxin production in staphylococcus aureus isolates. J. Bacteriol. 202, e00591-19. ( 10.1128/JB.00591-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Craft DL, Korza G, Zhang Y, Frindert J, Jschke A, Caimano MJ, Setlow P. 2020. Analysis of 5′-NAD capping of mRNAs in dormant spores of Bacillus subtilis. FEMS Microbiol. Lett. 367, fnaa143. ( 10.1093/femsle/fnaa143) [DOI] [PubMed] [Google Scholar]
- 90.Itzen A, Blankenfeldt W, Goody RS. 2011. Adenylylation: renaissance of a forgotten post-translational modification. Trends Biochem. Sci. 36, 221–228. ( 10.1016/j.tibs.2010.12.004) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.