Abstract
Machine learning (ML) has been widely used in microbiome research for biomarker selection and disease prediction. By training microbial profiles of samples from patients and healthy controls, ML classifiers constructs data models by community features that highly correlated with the target diseases, so as to determine the status of new samples. To clearly understand the host-microbe interaction of specific diseases, previous studies always focused on well-designed cohorts, in which each sample was exactly labeled by a single status type. However, in fact an individual may be associated with multiple diseases simultaneously, which introduce additional variations on microbial patterns that interferes the status detection. More importantly, comorbidities or complications can be missed by regular ML models, limiting the practical application of microbiome techniques. In this review, we summarize the typical ML approaches of single-label classification for microbiome research, and demonstrate their limitations in multi-label disease detection using a real dataset. Then we prospect a further step of ML towards multi-label classification that potentially solves the aforementioned problem, including a series of promising strategies and key technical issues for applying multi-label classification in microbiome-based studies.
Keywords: Machine learning, Microbiome, Single-label classification, Multi-label classification
1. Introduction
Microbiome analysis characterizes the dynamics of complex microbial communities, thus provides opportunities to investigate the associations between microbial profiles and human diseases [1], [2], [3]. Recently years, the scale of publicly-available microbiome data is increasing intensively due to high-throughput sequencing. Usually, microbiome features can be surveyed on shallow taxonomy by clustering amplicon-based DNA reads into OTUs (Operational Taxonomic Units) [4], [5], or species/strain level taxonomy and metabolic functions by decoding shotgun metagenomic sequences [6], [7]. Such features (e.g. species, OTU, functions, etc.) are quantified by either sequence count, or normalized into relative abundance by sequence proportion. Then machine learning (ML) algorithms uncover unique patterns of microbiome features under different statuses, thus promote microbiome-based disease detection and treatment [8], [9], [10]. As an important technique of machine learning, supervised classification has been widely used in prediction of inflammatory bowel disease (IBD) [11], [12], cancer [13], [14], diabetes [15], gingivitis [16], [17] and other diseases based on human microbiome profiles [18], [19]. By constructing classifiers and models using taxonomical or functional profiles from patients and their healthy control as training data, ML classifiers determine the status of new samples. In addition, some ML approaches such as support vector machines (SVM) [20] and random forest (RF) [21] can further measure the importance of each feature during the model training, which can identify microbial biomarkers that highly contribute to the classification [2], [22], [23].
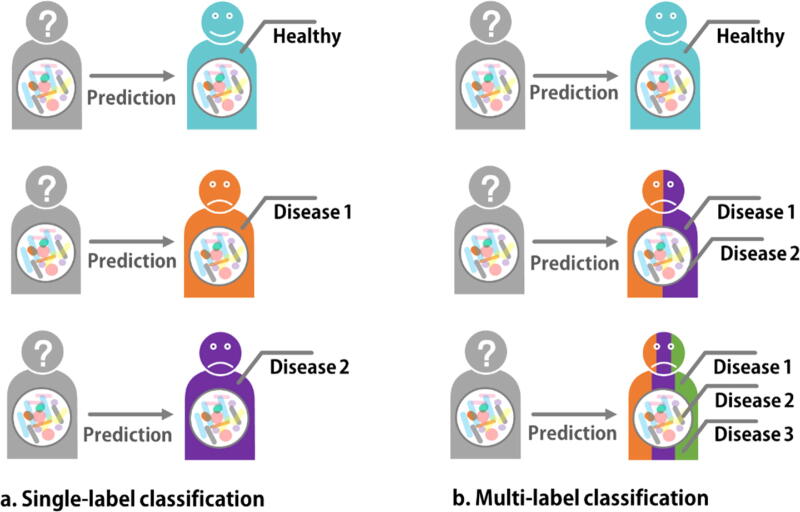
To clearly understand the interaction between microbes and healthy status, previously research cohorts are always well designed, in which a sample has only one exact label that describes its healthy status, e.g. a sample is either confident healthy, or associated with a definite disease (Fig. 1a). Nevertheless, such strategy exhibits its limitations in practical and clinical applications, since a patient may have more than one label (multiple diseases, also denoted as complications or comorbidities; Fig. 1b). For example, in American Gut Project [24] cohort, 8297 of 13,545 patients were marked with at least 2 diseases. In this case, regular classifiers do not work well for the prediction could be significantly interfered by the co-effect and interactions of multiple diseases. More importantly, since only a single label (e.g. a specific disease) was presented in the prediction result, comorbidities or complications were always missed or omitted by single-label ML models [2], [22].
Fig. 1.
Comparison of single-label classification and multi-label classification. a. Single-label classification requires a sample has one label (status). b. Multi-label classification can detect more than one status for each sample.
In this work, we summarize the typical and classical machine learning methods for microbiome research, and demonstrate their limitations in disease recognition using American Gut Project dataset. Then we prospect a further step to solve the aforementioned problems by a series of promising strategies on multi-label classification [25], [26], [27], [28]. Finally, we also raise and discuss some key technical issues in applying multi-label classification into microbiome-based disease detection.
2. Single-label classification in microbiome studies
Microbiome-based disease detection can be considered as a classification problem using microbial profiles, which are parsed from DNA sequences by bioinformatics tools such as UPARSE [5], QIIME/QIIME2 [29], [30], Parallel-Meta3 [31], MetaPhlAn2 [6], HUMANn2 [7], Kraken [32], according to the sequencing method and type [3]. Given microbiome profiles for n samples ( is the microbial profile of a sample that can be represented by normalized richness of features like species, OTU, function, etc.) and their corresponding status meta-data (label) , ML classifier solves a function that maps the profiles to their meta-data, thus predict the status of a new subject based on its profile. Usually, the classifier requires each subject has a single label (status), which is known as single-label classification. Here label yi in meta-data Y is a discrete variable that , here cj is a status (e.g. a specific disease). Specifically, when (e.g., is healthy and is IBD), the ML works as binary classifier that only differentiates IBD samples from healthy ones; When , it becomes multiple-category classifier that can determine the disease type of a new sample from multiple disease categories (Fig. 1a). Here we review the commonly used single-label classification approaches, including logistic regression, support vector machine, k nearest neighbors, random forest, gradient boosting tree and neural networks (Table 1).
Table 1.
Characteristics of machine learning methods widely used for microbiome-based disease detection.
| ML approach | Feature importance measurement | Interpretability | Package and applicable programming language |
|---|---|---|---|
| LR | Y | Excellent | Scikit-learn (Python) [33] |
| SVM | Y | Good | Scikit-learn (Python), LibSVM (Python/R/Java) [34] |
| k-NN | N | Weak | Scikit-learn (Python) |
| RF | Y | Good | Scikit-learn (Python) randomForest (R) [35] |
| GBDT | Y | Good | Xgboost (Python/R/C++) [36], [37] Lightgbm (Python/R/C++) [38] Catboost (Python/R/C++) [39] |
| Neural Networks | N | Weak | Tensorflow (Python/Java) [40] PyTorch (Python) [41]Keras (Python) [42] |
Logistic regression (LR) is a typical linear model for binary classification that utilizes a logistic function to model a binary dependent variable [43]. Basically, it calculates the probability for the occurrence of a specified event, e.g., a microbiome sample is healthy or disease. Due to the advantages in efficiency and interpretability, it is commonly used as a benchmark in microbiome-based disease detection [9], [44], although the performance is not as well as other methods. Different from LR, support vector machine (SVM) captures non-linear associations of microbiome profiles and host status to maximize the margin between healthy and disease samples [20], which achieves much better performance than LR. Noteworthy, as binary classifiers, LR and SVM can also be extended as multi-category classifier, by assigning a respective classifier for each disease. Another method is k-nearest neighbors (k-NN), which directly label a new sample by its k nearest neighbors [45]. One crucial problem of k-NN is how to appropriately measure the neighborship among microbiomes [46] by geometry-based distance metrices such as Bray-Curtis, JSD, JCCARD [47], or phylogeny-based algorithms like UniFrac [48] or Meta-Storms [49]. Recently, A search-based strategy employed microbiome search engine (MSE) [50] to separate unhealthy microbiomes from health ones by outlier novelty score, and then recognize their detailed disease type via a phylogeny-distance based k-NN, which outperforms traditional ML implementations in sensitivity, robustness and speed [51].
To further improve the performance for microbiome disease detection, ensemble classification approaches are developed by integrating individual ML methods [52], [53]. As an ensemble classifier, random forest (RF) constructs a multitude of decision trees through random selection of samples and features in training data, and then combines the predicted status of new samples by voting [2], [8], [9], [21]. Different from RF, gradient boosting decision tree (GBDT) assigns a weight to each microbiome sample, builds the tree-like model in a stage-wise fashion [54], [55] and then update parameters iteratively to minimize estimation errors [56]. Both RF and GBDT are not only superior to individual ML methods in precision, but can also evaluate the elucidate the contribution of each microbial feature for classification [22], [23].
In traditional ML, feature extraction from input data is fundamental for accuracy and sensitivity, e.g. select out biomarker species that play as signatures during the development of a disease, while such process always requires artificial efforts [57]. Deep learning performs feature extraction automatically and trains deep neural networks in an end-to-end way [58], which can alleviate the high dimensionality introduced by the complexity of microbial communities. Neural networks (such as deep neural networks (DNNs) [59], recurrent neural networks (RNNs) [60], convolutional neural networks (CNNs) [61], etc.) have been successfully transited from image analysis to microbiome research. In computer vision, CNNs make convolution operation for neighboring pixels to generate new variables. However, neighborship relations between microbes are not well-defined in a community. Therefore, Sharma et al. [62] developed a novel method based on CNNs by incorporating a stratified approach to group OTUs into phylum clusters. Lo et al. [63] also modeled microbiome profiles with a negative binomial distribution and solved over fitting problem by data augmentation technique in CNNs.
3. Limitations of single-label classification on real microbiome dataset
To measure the feasibility of single-label classifiers in handling microbiomes with multiple labels, we performed the disease detection using a subset of American Gut Project [24] cohort (refer to Materials and Methods for details). 16S rRNA amplicon microbiomes were collected from 3433 healthy hosts as control and 10,826 patients recorded with five target diseases, including Irritable bowel syndrome (IBS), Autoimmune, Lung disease, Migraine and Thyroid (Table 2). For each target disease, microbiome samples were divided into two groups: i) Single Disease group (SD) that contains controls and samples only with this target disease; ii) Multiple Disease group (MD) that contains controls and samples with this target disease and other comorbidities. Controls in each group were randomly selected from the healthy samples, and the sample number was set as equal to disease samples. We implemented two ensemble single-label classifiers of RF and GBDT to detect the target disease using OTU level profiles in each group, respectively. Performance was evaluated by AUC (Area Under the receiver operating characteristic Curve) using 5-fold cross-validation (refer to Materials and Methods for detailed configurations and parameters).
Table 2.
Brief summary of samples labeled with target diseases.
| Target disease | Total number of disease samples | Number of single-disease samples | Number of comorbidities samples |
|---|---|---|---|
| IBS | 2351 | 1064 | 1287 |
| Autoimmune | 2301 | 487 | 1814 |
| Lung disease | 2251 | 1248 | 1003 |
| Migraine | 2109 | 938 | 1171 |
| Thyroid | 1814 | 559 | 1255 |
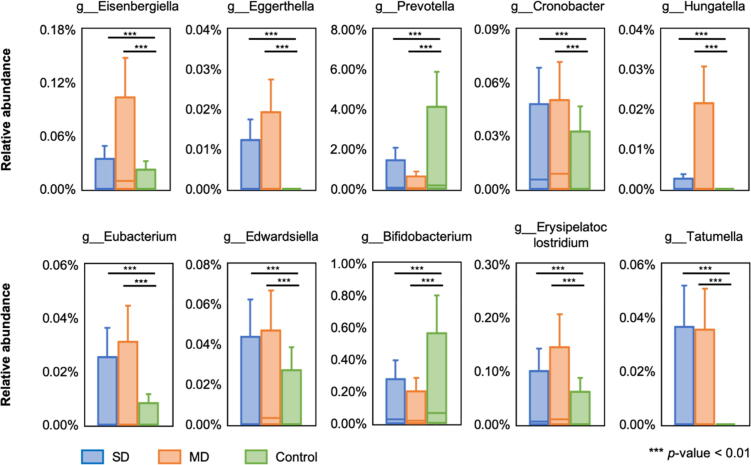
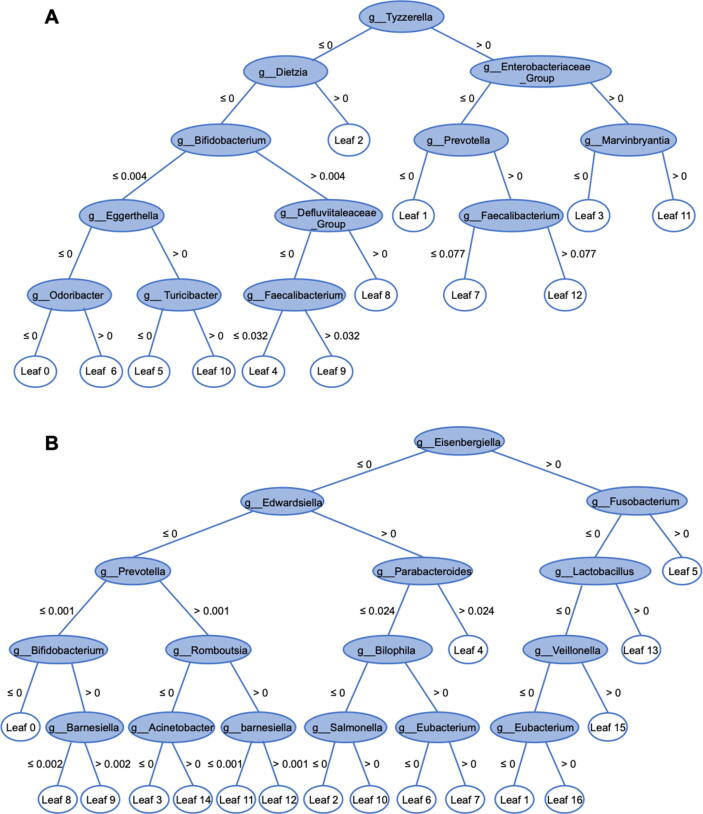
As results shown in Table 3, to detect the target disease in single-disease group, classifiers trained by SD outperformed those by MD, mainly due to eliminating additional variations on microbiota patterns of comorbidities. On the other side, classifiers trained by MD was superior to those by SD on multi-disease samples. Then we further dissected the microbial biomarkers and ML models between SD and MD that led to such results. A distribution-free test [64], [65] on autoimmune samples showed that biomarkers selected from SD were shared with MD (Fig. 2; taxonomy was annotated on genus level; refer to Materials and Methods for details). However, the decision tree constructed by GBDT binary classifier from SD was quite different from that from MD (Fig. 3; e.g. the structure and interactions between nodes in the MD tree were more complicated), implying the variation of microbial interactions between single disease and multiple disease. Therefore, influences of comorbidities on microbiota should be considered for ML model design and construction in practical cases. Notably, although the precision on target disease detection can be optimized, neither of the single-label ML classifier is able to detect the comorbidities or complications beyond the target disease.
Table 3.
Results of single-label classifiers on target diseases detection.
| a. Performance (AUC) on IBS | ||||
|---|---|---|---|---|
| Testing set | SD |
MD |
||
| Training set | SD | MD | SD | MD |
| RF | 0.681 ± 0.039 | 0.661 ± 0.032 | 0.718 ± 0.025 | 0.757 ± 0.018 |
| GBDT | 0.713 ± 0.025 | 0.689 ± 0.036 | 0.731 ± 0.022 | 0.787 ± 0.015 |
| b. Performance (AUC) on Lung disease | ||||
|---|---|---|---|---|
| Testing set | SD |
MD |
||
| Training set | SD | MD | SD | MD |
| RF | 0.671 ±0.28 | 0.538 ±0.023 | 0.618 ±0.024 | 0.727 ±0.025 |
| GBDT | 0.659 ±0.017 | 0.571 ±0.021 | 0.614 ±0.006 | 0.754 ±0.019 |
| c. Performance (AUC) on Migraine | ||||
|---|---|---|---|---|
| Testing set | SD |
MD |
||
| Training set | SD | MD | SD | MD |
| RF | 0.686 ±0.018 | 0.619 ± 0.021 | 0.670 ± 0.021 | 0.749 ± 0.017 |
| GBDT | 0.682 ± 0.019 | 0.642 ± 0.015 | 0.656 ± 0.018 | 0.764 ± 0.017 |
| d. Performance (AUC) on Thyroid | ||||
|---|---|---|---|---|
| Testing set | SD |
MD |
||
| Training set | SD | MD | SD | MD |
| RF | 0.714 ± 0.021 | 0.683 ± 0.017 | 0.755 ± 0.032 | 0.769 ± 0.032 |
| GBDT | 0.728 ± 0.026 | 0.700 ± 0.025 | 0.764 ± 0.024 | 0.794 ± 0.024 |
| e. Performance (AUC) on Autoimmune | ||||
|---|---|---|---|---|
| Testing set | SD |
MD |
||
| Training set | SD | MD | SD | MD |
| RF | 0.664 ± 0.05 | 0.650 ± 0.036 | 0.715 ± 0.039 | 0.776 ± 0.035 |
| GBDT | 0.689 ± 0.041 | 0.665 ± 0.031 | 0.741 ± 0.022 | 0.790 ± 0.041 |
Fig. 2.
Microbial biomarkers of autoimmune selected from SD and MD by distribution-free independence test.
Fig. 3.
Decision tree of GBDT binary classifier constructed from SD (A) was less complicated than that from MD (B). In each tree internal nodes represent taxa on genus-level, leaf nodes represent labels, and branch weights represent criteria for decision.
4. Multi-label classification: one step forward of machine learning for microbiome
Different from single-label ML classifiers (Fig. 1a), multi-label classification allows each sample to have more than one status (label; Fig. 1b). It is natural to introduce multi-label classification into microbiome-based disease detection for a sample (patient) may have multiple labels (comorbidities or complications). Here we introduce two schemes for multi-label classification: algorithm adaption and problem transformation [27].
Algorithm adaptation processes multi-label data by directly modifying single-label classifiers. For example, ML-kNN (multi-label k-nearest neighbors) combines the k-NN and Bayesian rule to determine the label set of a new sample [66]. Another example is a decision tree algorithm named C4.5 [67] that makes leaves represent a set of labels and modifying entropy-like function [68] for multi-label classification. Recently, a new ML-DT (Multi-Label Decision Tree) algorithm has been developed based on the non-parametric predictive inference model on multinomial data, which achieves a robust performance using precise probabilities [69].
Problem transformation, as the name suggests, transforms the multi-label problem into single-label ones by binary relevance, calibrated label ranking or class chains. Binary relevance bases on a one-against-all strategy that converts m (m > 1) labels into separate m binary classification problems, and determines each label by a binary classifier. Although it provides a simple and efficient solution, binary relevance ignores the possible correlations between labels thus leads to erroneous results [70]. To tackle such disadvantage, calibrated label ranking transforms m-label classification into label ranking problem [71] by considering the relevance in pairwise labels and constructs m * (m − 1)/2 binary classifiers. Hence, each label is voted by m-1 binary classifiers. Additionally, voting probabilities of m-1 binary classifiers can be utilized as features to train a new binary classifier to further improve the performance. Furthermore, one label may depend on some other labels, e.g. diagnosis and treatment of cardiovascular disease has been linked with those of IBD [72]. In this condition, class chains [73] that treats dependent labels as features of binary classifiers will be an ideal option.
5. Key technical issues of multi-label classification for microbiome-based disease detection
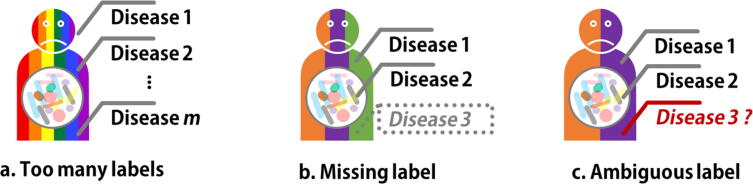
Well-established multi-label classification methods also exhibit shortages in processing microbiome datasets due to the high data complexity, data heterogeneity and microbe-disease interaction. In the past years, hundreds of microbiome-diseases interactions have been studied and reported, e.g. Disbiome database [74] collected 10,934 experimentally verified microbe-disease associations between 372 diseases and 1622 microbes. A general challenge is that such a large number of labels can lead to unexpected high computational cost (Fig. 4a). For example, to train a 100-label classification model (a sample has more than one disease from a total number of 100 diseases), binary relevance approach needs 100 binary classifiers, and calibrated label ranking requires up to 4950 classifiers. Recently, embedding methods such as SLEEC (Sparse Local Embeddings for Extreme Classification) algorithm [75] are proposed for many-label challenge. It projects labels into lower dimension-space vectors, constructs a regression for each label, and decodes the predicted labels via compressed techniques. To fit large-scale datasets, SLEEC uses unsupervised k-means algorithm to partition training data into several smaller subsets before the projection step. However, due to omitting the label information, the pre-partition may affect the quality of afterward projection. Therefore, embedding methods are further improved by incorporating feature vectors and label information using graph embedding algorithm [76] and an adaptive feature agglomeration technique like DEFRAG (aDaptive Extreme FeatuRe AGglomeration) [77].
Fig. 4.
Three key technical issues in multi-label classification. a. Too many labels in training data leads to unexpected high computational cost. b. Missed label reduces the detection sensitivity. c. Ambiguous label introduces false positive results.
Label missing is another common problem in multi-label classification (Fig. 4b). It is possible that in the American Gut Project cohort, some multi-disease samples were incorrectly grouped in SD for inadequate clinical examinations, making a ‘Negative’ or ‘Not provided’ record for some diseases in meta-data. Such label missing may also occur in multi-label classification result due to the low sensitivity when detecting multiple statuses at the same time. Here we introduce two alternatives including graph-based method and low-rank method to improve the sensitivity. The former one, graph-based method, estimates the comprehensive labels derived from label-specific graph [78] or label vectors [79]. The later one, low rank method, formulates multi-label learning as a matrix completion problem that contains side information [80], which can be estimated by empirical risk minimization framework [81] to avoid label missing.
In real-world scenarios, it is possible that the diseases meta-data is based on hosts’ personal experience or other unreliable conclusions without clinical diagnosis or confirmation from medical professionals. Such ambiguous labels in training data (Fig. 4c) may introduce false positive results. Partial multi-label approaches can eliminate errors caused by ambiguous or erroneous labels, mainly by maintaining a confidence value for each candidate label [82]. Based on how to calculate the confidence value, partial multi-label approaches are generally divided into two types, two-stage method and end-to-end learning method. Two-stage method estimates the confidence of candidate labels for each sample by iterative label propagation, and then train multi-label classifiers using credible labels with high confidence [83]. This straightforward concept however can be error-prone due to insufficient disambiguation. Different from separating confidence estimation and classifier construction as two stages, the end-to-end method treats confidence values as weights of model training functions [82], [84], [85] and enhance label disambiguation by combining two stages into a unified framework.
6. Conclusion and discussion
In this work, we reviewed typical single-label machine learning methods in microbiome research. While such ML approaches can help in interpreting the pattern of microbiome-disease linkages and predicting the status for newly sequenced samples, a significant limitation is raised, mainly in handling multi-label problems that a single microbiome can be associated with several different healthy conditions. Hence, we prospect one step forward of ML in microbiome filed towards multi-label classification that provides promising opportunities to tackle such limitation in research and application.
Another concern is that interactions among microbes has not been effectively considered by ML classifiers. Although biomarker fractions from single disease and comorbidities were similar (Fig. 1), their hierarchies in the GBDT decision tree are highly diverse (Fig. 2), probably directed by different interactions between microbes. Recently, co-occurrence or correlation among microbes have been widely studied in various ecosystems [86], [87], [88], [89], which survey microbe-microbe interactions from biological aspect. Nevertheless, how to efficiently and effectively integrate such biological information into ML classifiers is still an opening problem for further work [22], [90].
Few studies concentrated on the interpretability of ML model in microbiome studies, however it is meaningful to explain the disease prediction results. Among single-label classification methods, logistic regression has the best interpretability and the lowest performance, while NNs are on the opposite side. Although RF and GBDT also output feature importance, the calculation are too rough for further causal interpretation. Advanced statistical methods such as single index model that combines flexibility of modeling with interpretability of (linear) coefficients [91], [92] may provide a potential solution for balancing the interpretability and performance. Meanwhile, host heterogeneity on age, gender, diet, life style and other factors [93], as well as the sparsity, variance, and high-dimensionality [94] of microbiome data can also confound the disease detection and interpretation, which should be evaluated and considered in experiment design and ML analysis.
7. Materials and methods
7.1. Experiment design and datasets
The American Gut Project cohort contains 29,344 subjects including 15,799 healthy controls and 13,545 patients. The disease statuses of each subject were obtained from the original questionnaire -based meta-data that consists of information in diet, health status and hygiene. 16S rRNA OTU profiles of gut microbiomes (by close-OTU-picking) were download from Qiita [95], and taxonomy annotation on genus level was parsed by GreenGenes 13-8 database [96] using Parallel-META 3 [31]. The relative abundance on OTU and genus level was directly calculated by sequence count, and then normalized by 16S rRNA gene copy number from PICRUSt 2 [97]. We also drop subjects without microbiome samples.
A subject was treated either as a patient if recorded as ‘Diagnosed by a medical professional (doctor, physician assistant)’ for a specified disease in the meta-data, or as healthy if marked as ‘I do not have this condition’ for all diseases. Finally, we collected data of 3433 healthy samples and 10,826 patients. For each target disease, microbiome samples were selected and divided into two groups: Single Disease group (SD) contains controls and samples only with the target disease; Multiple Disease group (MD) contains controls and samples with the target disease and other comorbidities. Controls samples in each group were randomly selected from the 3,433 healthy samples, and the sample number was set as equal to disease samples.
For each target disease we performed two experiments. First, we assessed the ML classifiers in distinguishing disease samples and healthy controls in SD group. Classifier models were constructed by SD group and MD group. Specifically, 5-fold cross-validation was employed when detecting SD samples by models trained from SD group (in which 80% of the samples were randomly selected as the training set for model construction and the remaining 20% were the testing set for validation). Meanwhile, in each of the 5 folds we also randomly select the same number of samples from MD group to train another model for target disease detection in the identical SD testing set. AUCs of the SD-trained model and MD-trained model were recorded for comparison. Secondly, we then assessed the ML classifiers in detecting MD group, and models were also constructed by SD group and MD group in the previous procedure.
7.2. Machine learning methods and biomarker selection
Two popular ensemble single-label classification methods, random forest and GBDT were employed to construct single-label classifiers. Random forest was implemented by ‘scikit-learn’ package in python, the ‘number of trees’ is set as 500, while other parameters were kept as default configuration. GBDT was implemented by ‘lightgbm’ package in python with parameters of ‘learning rate’ = 0.02, ‘maximum tree depth’=6, ‘number of boosted trees’ = 1000, ‘maximum tree leaves’ = 64, ‘subsample ratio’=0.8 and ‘colsample_bytree’=0.8. Biomarkers analysis was performed by distribution-free test (‘mvtpy’ package in python) on genus-level abundance between disease and control samples, and the top 10 taxa on the test statistic with p-value < 0.01were selected out as biomarkers.
7.3. Code and data availability
All datasets and code in this work are available at https://github.com/BruceQD/Microbiome-based-disease-detection. All other relevant data is available upon request.
8. Author statement
S.W and Y.C. contributed to the description and summary of algorithms, and performed the analysis. Z.L., J.L. and F.Z. reviewed and edited the manuscript before submission. X.S. conceived the idea and wrote the manuscript. Author order was determined by mutual agreement.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
XS acknowledges support of grants 31771463 and 32070086 from National Natural Science Foundation of China. SW acknowledges support of grant ZR2019PF012 from Shandong Provincial Natural Science Foundation of China.
References
- 1.Knight R. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 2.LaPierre N. MetaPheno: a critical evaluation of deep learning and machine learning in metagenome-based disease prediction. Methods. 2019;166:74–82. doi: 10.1016/j.ymeth.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su X. Method development for cross-study microbiome data mining: challenges and opportunities. Computational and Structural. Biotechnol J. 2020 doi: 10.1016/j.csbj.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 5.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 6.Truong D.T. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12(10):902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 7.Franzosa E.A. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15(11):962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Namkung J. Machine learning methods for microbiome studies. J Microbiol. 2020;58(3):206–216. doi: 10.1007/s12275-020-0066-8. [DOI] [PubMed] [Google Scholar]
- 9.Topçuoğlu B.D. A framework for effective application of machine learning to microbiome-based classification problems. Mbio. 2020;11(3) doi: 10.1128/mBio.00434-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cammarota G. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat Rev Gastroenterol Hepatol. 2020 doi: 10.1038/s41575-020-0327-3. [DOI] [PubMed] [Google Scholar]
- 11.Gevers D. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halfvarson J. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. doi: 10.1038/nmicrobiol.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wirbel J. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poore G.D. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579(7800):567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Bajaj J.S. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302(1):G168–G175. doi: 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang S. Predictive modeling of gingivitis severity and susceptibility via oral microbiota. ISME J. 2014;8(9):1768–1780. doi: 10.1038/ismej.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang S. Longitudinal multi-omics and microbiome meta-analysis identify an asymptomatic gingival state that links gingivitis, periodontitis, and aging. mBio. 2021;12(2) doi: 10.1128/mBio.03281-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duvallet C. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun. 2017;8(1):1784. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vangay P., Hillmann B.M., Knights D. Microbiome Learning Repo (ML Repo): a public repository of microbiome regression and classification tasks. GigaScience. 2019;8(5) doi: 10.1093/gigascience/giz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortes C., Vapnik V. Support-vector networks. Machine Learn. 1995;20(3):273–297. [Google Scholar]
- 21.Breiman L. Random forests. Machine Learn. 2001;45(1):5–32. [Google Scholar]
- 22.Duvallet C. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun. 2017;8(1):1–10. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasolli E. Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput Biol. 2016;12(7) doi: 10.1371/journal.pcbi.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald D. American Gut: an open platform for citizen science microbiome research. Msystems. 2018;3(3):e00031–e118. doi: 10.1128/mSystems.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, W., et al., The Emerging Trends of Multi-Label Learning. arXiv preprint arXiv:2011.11197; 2020.
- 26.Tsoumakas G., Katakis I. Multi-label classification: an overview. Int J Data Warehous Min (IJDWM) 2007;3(3):1–13. [Google Scholar]
- 27.Zhang M.-L., Zhou Z.-H. A review on multi-label learning algorithms. IEEE Trans Knowl Data Eng. 2013;26(8):1819–1837. [Google Scholar]
- 28.Gibaja E., Ventura S. Multi-label learning: a review of the state of the art and ongoing research. Wiley Interdiscip Rev: Data Min Knowledge Disc. 2014;4(6):411–444. [Google Scholar]
- 29.Caporaso J.G. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolyen E. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing G. Parallel-META 3: comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities. Sci Rep. 2017;7(1):1–11. doi: 10.1038/srep40371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedregosa F. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 34.Chang C.-C., Lin C.-J. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol (TIST) 2011;2(3):1–27. [Google Scholar]
- 35.RColorBrewer S., Liaw M.A. University of California, Berkeley; Berkeley, CA, USA: 2018. Package ‘randomForest’. [Google Scholar]
- 36.Chen T., Guestrin C. Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining. 2016. Xgboost: A scalable tree boosting system. [Google Scholar]
- 37.Chen, T., et al., Xgboost: extreme gradient boosting. R package version 0.4-2, 2015: p. 1–4.
- 38.Ke G, et al. Lightgbm: A highly efficient gradient boosting decision tree. in Advances in neural information processing systems; 2017.
- 39.Prokhorenkova L, et al. CatBoost: unbiased boosting with categorical features. in Advances in neural information processing systems. 2018.
- 40.Abadi M, et al. Tensorflow: A system for large-scale machine learning. in 12th {USENIX} symposium on operating systems design and implementation ({OSDI} 16); 2016.
- 41.Paszke A, et al., Pytorch: An imperative style, high-performance deep learning library. arXiv preprint arXiv:1912.01703, 2019.
- 42.Ketkar N. Deep learning with Python. Springer; 2017. Introduction to keras; pp. 97–111. [Google Scholar]
- 43.Kleinbaum DG, et al., Logistic regression. 2002: Springer.
- 44.Song K., Wright F., Zhou Y.-H. Systematic comparisons for composition profiles, taxonomic levels, and machine learning methods for microbiome-based disease prediction. Front Mol Biosci. 2020;7:423. doi: 10.3389/fmolb.2020.610845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson L.E. K-nearest neighbor. Scholarpedia. 2009;4(2):1883. [Google Scholar]
- 46.Comin M. Comparison of microbiome samples: methods and computational challenges. Brief Bioinform. 2020 doi: 10.1093/bib/bbaa121. [DOI] [PubMed] [Google Scholar]
- 47.Ricotta C., Podani J. On some properties of the Bray-Curtis dissimilarity and their ecological meaning. Ecol Complexity. 2017;31:201–205. [Google Scholar]
- 48.McDonald D. Striped UniFrac: enabling microbiome analysis at unprecedented scale. Nat Methods. 2018;15(11):847–848. doi: 10.1038/s41592-018-0187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jing G. Dynamic Meta-Storms enables comprehensive taxonomic and phylogenetic comparison of shotgun metagenomes at the species level. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz910. [DOI] [PubMed] [Google Scholar]
- 50.Jing G. Microbiome search engine 2: a Platform for taxonomic and functional search of global microbiomes on the whole-microbiome level. mSystems. 2021;6(1) doi: 10.1128/mSystems.00943-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su X. Multiple-disease detection and classification across cohorts via microbiome search. Msystems. 2020;5(2) doi: 10.1128/mSystems.00150-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, Z.-H., Ensemble Learning. Encyclopedia of biometrics, 2009. 1: p. 270–3.
- 53.Polikar R. Ensemble machine learning. Springer; 2012. Ensemble learning; pp. 1–34. [Google Scholar]
- 54.Friedman J.H. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001:1189–1232. [Google Scholar]
- 55.Friedman J.H. Stochastic gradient boosting. Comput Stat Data Anal. 2002;38(4):367–378. [Google Scholar]
- 56.Ruder, S., An overview of gradient descent optimization algorithms. arXiv preprint arXiv:1609.04747, 2016.
- 57.Pouyanfar S. A survey on deep learning: algorithms, techniques, and applications. ACM Comput Surveys (CSUR) 2018;51(5):1–36. [Google Scholar]
- 58.Glasmachers T. Limits of End-to-End Learning, in Proceedings of the Ninth Asian Conference on Machine Learning, Z. Min-Ling and N. Yung-Kyun, Editors. 2017, PMLR: Proceedings of Machine Learning Research. p. 17--32.
- 59.Deng Y. A hierarchical fused fuzzy deep neural network for data classification. IEEE Trans Fuzzy Syst. 2016;25(4):1006–1012. [Google Scholar]
- 60.Mou L., Ghamisi P., Zhu X.X. Deep recurrent neural networks for hyperspectral image classification. IEEE Trans Geosci Remote Sens. 2017;55(7):3639–3655. [Google Scholar]
- 61.Gu J. Recent advances in convolutional neural networks. Pattern Recogn. 2018;77:354–377. [Google Scholar]
- 62.Sharma D., Paterson A.D., Xu W. TaxoNN: ensemble of neural networks on stratified microbiome data for disease prediction. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lo C., Marculescu R. MetaNN: accurate classification of host phenotypes from metagenomic data using neural networks. BMC Bioinf. 2019;20(12):314. doi: 10.1186/s12859-019-2833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui H., Zhong W. A distribution-free test of independence based on mean variance index. Comput Stat Data Anal. 2019;139:117–133. [Google Scholar]
- 65.Cui H., Li R., Zhong W. Model-free feature screening for ultrahigh dimensional discriminant analysis. J Am Stat Assoc. 2015;110(510):630–641. doi: 10.1080/01621459.2014.920256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang M.-L., Zhou Z.-H. ML-KNN: A lazy learning approach to multi-label learning. Pattern Recogn. 2007;40(7):2038–2048. [Google Scholar]
- 67.Quinlan JR. C4. 5: programs for machine learning. 2014: Elsevier.
- 68.Clare A., King R.D. European conference on principles of data mining and knowledge discovery. Springer; 2001. Knowledge discovery in multi-label phenotype data. [Google Scholar]
- 69.Moral-García S. Non-parametric predictive inference for solving multi-label classification. Appl Soft Comput. 2020;88 [Google Scholar]
- 70.Zhang M.-L. Binary relevance for multi-label learning: an overview. Front Comp Sci. 2018;12(2):191–202. [Google Scholar]
- 71.Dery, L., Multi-label Ranking: Mining Multi-label and Label Ranking Data. arXiv preprint arXiv:2101.00583, 2021.
- 72.Argollo M. Comorbidities in inflammatory bowel disease: a call for action. Lancet Gastroenterol Hepatol. 2019;4(8):643–654. doi: 10.1016/S2468-1253(19)30173-6. [DOI] [PubMed] [Google Scholar]
- 73.Read J. Classifier chains for multi-label classification. Machine Learn. 2011;85(3):333. [Google Scholar]
- 74.Janssens Y. Disbiome database: linking the microbiome to disease. BMC Microbiol. 2018;18(1):1–6. doi: 10.1186/s12866-018-1197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bhatia, K., et al. Sparse Local Embeddings for Extreme Multi-label Classification. in NIPS. 2015.
- 76.Tagami, Annexml Y. Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining. 2017. Approximate nearest neighbor search for extreme multi-label classification. [Google Scholar]
- 77.Jalan A, Kar P. Accelerating extreme classification via adaptive feature agglomeration. arXiv preprint arXiv:1905.11769; 2019.
- 78.Sun Y.-Y., Zhang Y., Zhou Z.-H. Proceedings of the AAAI Conference on Artificial Intelligence. 2010. Multi-label learning with weak label. [Google Scholar]
- 79.Wu B. 22nd International Conference on Pattern Recognition. IEEE; 2014. Multi-label learning with missing labels. [Google Scholar]
- 80.Xu M, Jin R, Zhou Z-H. Speedup matrix completion with side information: Application to multi-label learning. In: Advances in neural information processing systems. 2013.
- 81.Yu H-F, et al. Large-scale multi-label learning with missing labels. in International conference on machine learning; 2014. PMLR.
- 82.Xie M.-K., Huang S.-J. Proceedings of the AAAI Conference on Artificial Intelligence. 2018. Partial multi-label learning. [Google Scholar]
- 83.Fang J.-P., Zhang M.-L. Proceedings of the AAAI Conference on Artificial Intelligence. 2019. Partial multi-label learning via credible label elicitation. [DOI] [PubMed] [Google Scholar]
- 84.He S. IEEE International Conference on Data Mining (ICDM) IEEE; 2019. Discriminatively relabel for partial multi-label learning. [Google Scholar]
- 85.Yu G. 2018 IEEE International Conference on Data Mining (ICDM) IEEE; 2018. Feature-induced partial multi-label learning. [Google Scholar]
- 86.Friedman J., Alm E.J. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8(9) doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Faust K. Microbial co-occurrence relationships in the human microbiome. PLoS comput biol. 2012;8(7) doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kurtz Z.D. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. 2015;11(5) doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu G. Guild-based analysis for understanding gut microbiome in human health and diseases. Genome Med. 2021;13(1):22. doi: 10.1186/s13073-021-00840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jackson M.A. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun. 2018;9(1):1–8. doi: 10.1038/s41467-018-05184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liang H. Estimation and testing for partially linear single-index models. Ann Stat. 2010;38(6):3811. doi: 10.1214/10-AOS835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Y., Tong T., Li G. SIMEX estimation for single-index model with covariate measurement error. AStA Adv Statist Anal. 2019;103(1):137–161. [Google Scholar]
- 93.Vujkovic-Cvijin I. Host variables confound gut microbiota studies of human disease. Nature. 2020;587(7834):448–454. doi: 10.1038/s41586-020-2881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu L.Z. Assessment and selection of competing models for zero-inflated microbiome data. PLoS ONE. 2015;10(7) doi: 10.1371/journal.pone.0129606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gonzalez A. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods. 2018;15(10):796–798. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McDonald D. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Douglas G.M. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38(6):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets and code in this work are available at https://github.com/BruceQD/Microbiome-based-disease-detection. All other relevant data is available upon request.