Abstract
Purpose
To identify global gene expression changes in the corneal epithelium of keratoconus (KC) patients compared to non-KC myopic controls.
Methods
RNA-sequencing was performed on corneal epithelium samples of five progressive KC and five myopic control patients. Selected results were validated using TaqMan quantitative PCR (qPCR) on 31 additional independent samples, and protein level validation was conducted using western blot analysis on a subset. Immunohistochemistry was performed on tissue microarrays containing cores from over 100 KC and control cases. WNT10A transcript levels in corneal epithelium were correlated with tomographic indicators of KC disease severity in 15 eyes. Additionally, WNT10A was overexpressed in vitro in immortalized corneal epithelial cells.
Results
WNT10A was found to be underexpressed in KC epithelium at the transcript (ratio KC/control = 0.59, P = 0.02 per RNA-sequencing study; ratio = 0.66, P = 0.03 per qPCR) and protein (ratio = 0.07, P = 0.06) levels. Immunohistochemical analysis also indicated WNT10A protein was decreased in Bowman's layer of KC patients. In contrast, WNT10A transcript level positively correlated with increased keratometry (Kmax ρ = 0.57, P = 0.02). Finally, WNT10A positively regulated COL1A1 expression in corneal epithelial cells.
Conclusions
A specific Wnt ligand, WNT10A, is reduced at the mRNA and protein level in KC epithelium and Bowman's layer. This ligand positively regulates collagen type I expression in corneal epithelial cells. The results suggest that WNT10A expression in the corneal epithelium may play a role in progressive KC.
Keywords: keratoconus, corneal epithelium, Wnt signaling, WNT10A
Keratoconus is a common ectasia of the cornea with a reported prevalence as high as 265 per 100,000.1–3 The hallmark of keratoconus is biomechanical failure of the cornea, leading to characteristic corneal protrusion and blurred vision due to myopia and irregular astigmatism. Keratoconus does not manifest abruptly; it is a progressive disease that starts in adolescence, and these younger patients are especially prone to gradual steepening of the cornea.4,5 In contrast, most older keratoconus patients tend to have stable disease without any apparent progression. Despite decades of investigation, the underlying mechanisms, gene expression, and cytokine changes that lead to keratoconus remain elusive.6–12 Although considered a predominantly stromal disease, alterations in epithelial biology have been demonstrated in keratoconus and have been suggested to promote disease pathogenesis. However, no distinct biological pathway linking aberrant epithelium to loss of mechanical strength in keratoconus has been established.6,7,13–18 As keratoconus often leads to progressive vision loss, identifying the molecular mechanisms regulating keratoconus development and progression may provide new therapeutic avenues to explore.
To identify key keratoconus regulators, transcriptomic studies of keratoconus corneas have been performed7–10,19–21; however, most were conducted using whole corneas containing all layers, rather than focusing on epithelium.8,9,19–21 One study did compare epithelial gene expression in keratoconus and control cases but used cases for which disease severity and progression status were not clearly defined.7 Using samples from progressing cases is important as end-stage keratoconus corneas may show secondary changes, which mask early molecular drivers of disease activity. The advent of corneal collagen crosslinking therapies designed to arrest disease progression, in which epithelium is removed as part of the standard procedure, provides the opportunity to examine samples relatively early in the disease course. Here we performed RNA-sequencing (RNA-seq) of corneal epithelium obtained from mild to moderate, progressive keratoconus patients during corneal collagen crosslinking as well as controls to investigate progressive disease in greater detail.
Methods
Ethics
This study was approved by the Johns Hopkins University School of Medicine Institutional Review Board (IRB00112675). All study procedures adhered to the tenets of the Declaration of Helsinki, and all study participants provided written informed consent.
RNA-Sequencing and PCR Validation
Mild to moderate, progressive keratoconus patients undergoing epithelium-off corneal collagen crosslinking or healthy mild to moderate myopic control patients undergoing photorefractive keratectomy were selected based on clinical examination and Scheimpflug tomography (Pentacam, Oculus, Germany; see Supplementary Table S1 for keratoconus patients’ clinical characteristics). Specifically, in order to treat progressive cases as early as possible, eyes were defined as having progressive keratoconus if there was a documented increase in steepest central keratometry or maximal keratometry on corneal tomography of 0.4 diopters over 3 to 4 months or more than 1 diopter a year. Patients 25 years or younger with evidence of keratoconus on tomography and distance spectacle-corrected vision 20/25 or worse were also defined as having progressive disease. Importantly, keratoconus was ruled out in all control patients prior to study enrollment.
For the purpose of the RNA and western blot studies described herein, corneal epithelial samples were acquired during corneal collagen crosslinking or photorefractive keratectomy. These samples were obtained in a sterile fashion after topical application of proparacaine HCl 0.5% ophthalmic drops (Bausch & Lomb, Rochester, NY, USA) and 5% betadine ophthalmic solution (Alcon, Fort Worth, TX, USA). A 9-mm metal well was placed on the central cornea avoiding the limbus, and 20% alcohol was applied for 30 seconds to facilitate epithelial removal. The alcohol was removed with cellulose sponges followed by thorough irrigation with balanced salt solution. The central epithelia were promptly debrided with sterile cellulose sponges and immediately placed in the operating room into individual RNAlater tubes (Thermo Fisher, Waltham, MA, USA). Samples were kept refrigerated (4°C) in RNAlater for 48 hours and then frozen at −80°C. The samples were homogenized with sterile individual tips (Tissueruptur II; Qiagen, Hilden, Germany). RNA was extracted using the RNAeasy Plus kit (Qiagen). RNA quality was checked with a Nanodrop (Thermo Fisher) and a bioanalyzer nano RNA chip (Agilent, Santa Clara, CA, USA). Samples with a RNA Integrity Number (RIN) score over 8 were used for RNA-seq. RNA-seq was performed on corneal epithelium derived from five keratoconus and five myopic controls. Library preparation was performed according to the manufacturer's instructions with the TruSeq Stranded Library prep protocol (Illumina, San Diego, CA, USA) at the Johns Hopkins School of Medicine sequencing core. Sequencing was paired-end 75 nucleotides with a total target of 400 million reads on the Illumina NextSeq 500 High 150 Output sequencing instrument (40 million reads per sample). The libraries were sequenced to provide FASTA raw data.
RNA-Seq Statistical Analyses
Using CLCGenomicsServer 8.5.4, the FASTA reads were aligned to the NCBI transcriptome GRCm38 with 53,853 transcripts. The transcript name identifiers from the fragments per kilobase of transcript per million mapped reads (FPKM) files were updated to current HGNC/NCBI nomenclature. FPKM values of raw data files equivalent to 0.0 were treated as nulls; the remaining actual values were transformed into log2 notation. Quality control examination of the raw log2 signals in box plot and histogram showed technically consistent results across the samples. These log2 values were quantile normalized together for further analysis. Differential expression analysis was conducted with a two-tailed, two-way t-test ANOVA in which samples of similar demographics were paired as the second degree of freedom using the Partek GS 6.6 analytic platform (Partek, Inc., St. Louis, MO, USA). The results are reported as differential expression of the transcripts in ratio, linear fold change and their statistical significance in uncorrected P-values. A standard deviation analysis was performed for each class-class comparison using only those transcripts that had NCBI Entrez gene annotation and a mean FPKM log2 value >−0.2 in the lower-expressed class.
Ingenuity
The QIAGEN Ingenuity Pathway Analysis (IPA) platform was used to evaluate the biological significance of the RNA-seq gene expression results. Genes with adequately high expression (average log2 FPKM >−0.2) and >2 SD differential expression between keratoconus and normal control samples were deemed to show significant differential expression and were compared to those genes that were not, looking for enrichment in IPA's literature-based categories. These broad categories were their Canonical Pathways, Upstream Analysis, Diseases and Functions, and Regulator Effects. These categories’ degrees of enrichment were scored by Fisher's exact test P values, activation z scores for the first three, and consistency scores for the last.
Quantitative PCR
A total of 500 ng RNA was reverse transcribed to cDNA using the iScript cDNA synthesis kit (Thermo Fisher) and a thermal cycler (T100; Bio-Rad, Hercules, CA, USA). All quantitative PCR (qPCR) studies were performed in duplicate using TaqMan assays (Thermo Fisher; see Supplementary Table S2). The t-test analysis identified RPL13A as an appropriate housekeeping gene with stable expression in keratoconus samples and controls per t-test (Supplementary Fig. S1). The transcriptomic results in 10 sequenced samples were validated by quantitative RT-PCR (qRT-PCR). A larger-scale qRT-PCR study was conducted with additional 15 keratoconus samples and 16 controls. The ΔΔCt method was used to calculate relative gene expression compared to the reference gene (RPL13A).22 Statistical analysis was performed in Excel (Microsoft, Redmond, WA, USA) using a two-tailed t-test. A P value <0.05 was considered statistically significant.
Western Blotting Analysis
Standard western blotting techniques were used. Tissue samples from four keratoconus patients and three myopic controls kept in RNAlater and frozen in −80°C were used for this assay.23 The samples were homogenized as described above in RIPA buffer (Boston BioProducts, Ashland, MA, USA). As for the cell culture assay described below, cells were lysed with RIPA buffer and a combination of protease and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO, USA). Protein quantification was performed using a BCA assay (Thermo Fisher). Gel electrophoresis was performed on Tris-Glycine 4%–12% gels (Thermo Fisher) with 10 µg protein per lane. The proteins were then transferred to a polyvinylidene fluoride membrane using a semidry transfer apparatus, blocked in 5% bovine serum albumin/0.1% Tween 20–Tris-buffered saline (TBS) for 1 hour at room temperature. The membrane was then suspended in a blocking buffer with the appropriate primary antibodies (either 1:1000 diluted primary anti-WNT10A rabbit polyclonal antibody [GTX111191; GeneTex, Irvine, CA, USA] or 1:2000 diluted primary anti-COL1A1 polyclonal sheep antibody [AF6220; R&D Systems, Minneapolis, MN, USA]) and incubated at 4°C overnight. The membrane was then washed three times in 0.1% Tween 20/TBS solution and incubated in blocking buffer containing 1:5000 dilution of horseradish peroxidase–conjugated anti-rabbit or anti-sheep secondary antibody. A chemiluminescent substrate was then applied to the membrane (ECL, Thermo Fisher) and the bands were imaged on an x-ray film. Image analysis was performed with ImageJ version 1.52h (National Institutes of Health, Bethesda, MD, USA), and WNT10A/COL1A1 relative intensity was recorded compared to glyceraldehyde 3-phosphate dehydrogenase.24 A Mann-Whitney test was used to compare keratoconus band intensity to controls using Prism 8 (GraphPad Software, La Jolla, CA, USA).
Immunohistochemistry Studies
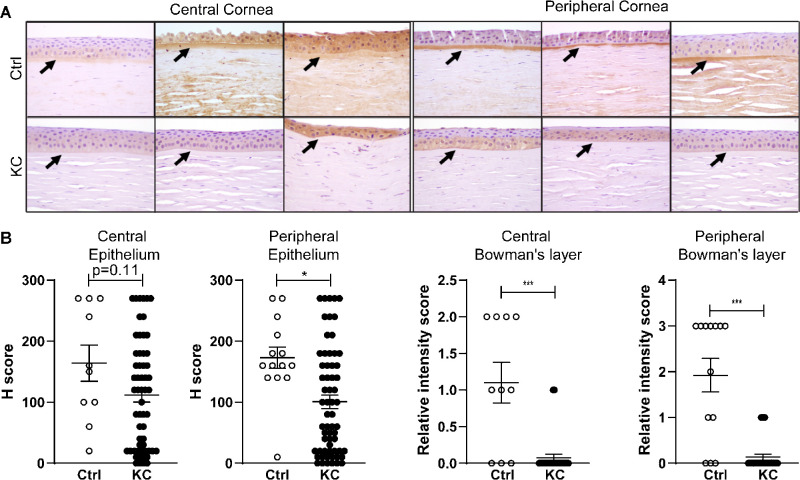
Full-thickness corneal tissues acquired from keratoconus cases during keratoplasty as well as autopsy eyes were used.25 Full-thickness punches of 1 mm diameter of representative central and peripheral corneal cores were then embedded in a 16-by-13 array, generating a tissue microarray measuring 15.6 × 19.5 × 5 mm. Five-micron sections were cut from these blocks and placed on slides for staining. Briefly, the slides were deparaffinized and rehydrated on a heating plate with xylene and graded ethanol series, washed and incubated with hydrogen peroxide, blocked in normal goat serum and 0.1% Tween 20/TBS, and then incubated overnight at room temperature in blocking buffer containing WNT10A antibody at a 1:200 dilution (SAB3500393; Sigma-Aldrich). The rest of the procedure was carried out using Vectastatin Elite ABC Kit Peroxidase HRP (PK-6101; Vector Laboratories, Burlingame, CA, USA): the slides were washed in TBS and incubated with blocking buffer and rabbit biotinylated secondary antibody supplied in the kit and according to the manufacturer's instructions, as well as incubated with ABC reagent and then peroxidase substrate solution. The slides were then counterstained with hematoxylin and dehydrated in ethanol gradient and xylene. The samples were independently examined by three skilled observers (CGE, RNP, USS), and staining scores were reached by consensus. Acellular Bowman's layer staining scores were evaluated separately for peripheral and central cornea with a staining score including 0 (none), 1 (weak), 2 (moderate), and 3 (strong). Epithelial staining was evaluated for both extent of cellular cytoplasmic staining (in percentages by decile; i.e., 0%, 10%, 20%) and intensity (0–3). A combined epithelial staining “H-score” was obtained by multiplying extent by intensity with the final score between 0 and 300.26 Light microscopy was used to capture images. A Mann-Whitney test was used to compare keratoconus staining scores to controls using Prism 8 (GraphPad Software).
Gene Expression/Disease Phenotype Correlation
Preoperative tomographic data (Pentacam; Oculus, Wetzlar, Germany) were collected for keratoconus eyes that were included in the PCR validation study (n = 15). Specifically, data on preoperative steep central anterior corneal keratometry (K2), maximal anterior corneal keratometry (Kmax), corneal thickness (pachymetry) at the thinnest point, and corneal volume were retrieved from the medical records. No linear correlation was found, and therefore Spearman's rank correlation coefficients were calculated to test whether monotonic relationships exist between WNT10A relative corneal epithelial transcript level per qPCR and tomographic disease severity markers. A P value <0.05 was considered statistically significant. This analysis was performed using STATA statistical software (v16.0) (StataCorp LLC, College Station, TX, USA).
Overexpression Plasmid Assay
A WNT10A overexpression plasmid was gifted from Marian Waterman (Addgene plasmid #35920; http://n2t.net/addgene:35920; RRID:Addgene_35920) and transformed using NEB 10-β competent Escherichia coli C3019H (New England Biolabs, MA, USA).27 A naked control plasmid sharing a similar backbone (pcDNA3.1) was kindly gifted from the Don Zack laboratory and transformed similarly. Different passages of immortalized corneal epithelial cells (hTCEpi) that were produced by Dr. Danielle M. Robertson at the James V. Jester laboratory, given to Dr. Shukti Chakravarti and then kindly gifted to our laboratory, were used and treated as single biological specimens.28 A total of 100,000 cells (each) of six different passages were seeded in 9.5-cm2 tissue culture–treated plates (Nest Biotechnology, Wuxi, China) and grown to 80% confluence in four identically configured six-well plates. Each well containing a different passage of hTCEpi cells was individually transfected in serum-free media (Opti-Mem; Thermo Fisher) using Lipofectamine STEM reagent (Thermo Fisher) with 500 ng of either a WNT10A overexpression plasmid or an empty, control plasmid. Forty-eight hours after transfection, all cells achieved similar confluence and the cells were either lysed with RLT Plus buffer (Thermo Fisher) or with RIPA buffer and a cocktail of protease and phosphatase inhibitor (Sigma-Aldrich).
Results
Global Transcriptomic Results and PCR Validation
The RNA-seq study was conducted in 10 samples (five keratoconus, five controls), with affected patients showing a mean steep keratometry value of 55.6 diopters (see Supplementary Table S1 for clinical characteristics). We identified 53,853 transcripts, of which 11,072 were differentially expressed between the two groups with an average FPKM >−0.2 (Fig. 1A). Of these, the reads from 551 genes were two or more standard deviations from the mean. This included 310 upregulated and 241 downregulated genes in the keratoconus samples as compared to controls. The top dysregulated genes are presented in Supplementary Table S3 and in Figure 1B.
Figure 1.
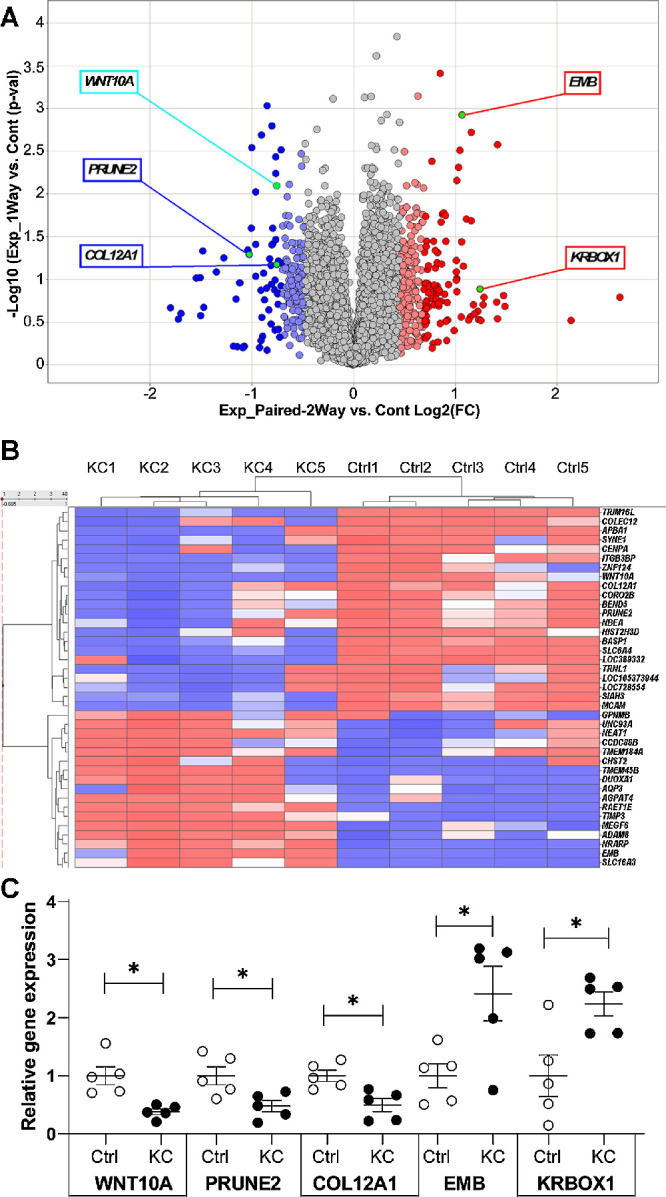
(A) Volcano plot depicting the results of the RNA-seq study. Five qPCR validated genes are highlighted. (B) Heat map representation of the genes with highest differential expression. Ctrl, control; KC, keratoconus. (C) qPCR validation of five genes shown to be dysregulated in the RNA-seq study. WNT10A is underexpressed in the corneal epithelium of keratoconus eyes.
IPA revealed that the top canonical pathways involved in this RNA-seq study included antigen presentation, communication between innate and adaptive immune cells, interferon signaling, graft-versus-host disease signaling, and allograft rejection signaling (Supplementary Table S4). Top activated upstream regulators included IFNA2, IFNG, IFNL1, and TNF, while NKX2-3 was inhibited, potentially consistent with the known association between atopic disease and keratoconus (Supplementary Table S4). Additionally, the transcriptomic analysis was associated with skin disorders, organismal injury, endocrinopathies, gastrointestinal disease, and metabolic diseases. High consistency scores were found for top regulator effect networks with an emphasis on antiviral response and antigen presentation. These high consistency scores are indicators of node expression that is consistent with the predicted direction based on available literature and suggest that these effectors are of potential biological significance based on the results of the sequencing study. Molecular and cellular functions found to be affected by the current transcriptomic study included protein trafficking, protein synthesis, cell death and survival, and cellular assembly, function, and maintenance.
In order to validate the RNA sequencing data, five dysregulated genes from Supplementary Table S3 (WNT10A, PRUNE2, COL12A1, EMB, KRBOX1) were examined with TaqMan qPCR. These qPCR studies confirmed the results of initial RNA-seq analyses, with statistically significant changes noted for both RNA-seq and qPCR (Fig. 1B). WNT10A transcripts were found to be underexpressed in keratoconus epithelium in both the RNA-seq study (ratio: 0.59, P = 0.02) and the qPCR analyses (ratio: 0.38, P = 0.01). PRUNE2 and COL12A1 were similarly validated as downregulated (respectively, ratio 0.48, P < 0.03; ratio 0.49, P < 0.01) while both EMB and KRBOX1 were validated as being overexpressed (Fig. 1B, respectively, ratio 2.42, P < 0.04; ratio = 2.246, P = 0.02).
WNT10A Alterations in Keratoconus Epithelium
Given the previously reported relationship between aberrant Wnt signaling and keratoconus, as well as recent genome-wide association study (GWAS) evidence implicating a WNT10A exonic variant with reduced corneal thickness, we focused on altered WNT10A expression in keratoconus.6–10
Additionally, other members of the Wnt signaling family were found to be dysregulated in our RNA-seq data set. Specifically, analysis of genes in the canonical pathway defined by Kyoto Encyclopedia of Genes and Genomes (KEGG) revealed reductions in TCF15 (1.8-fold, P = 0.15) and LEF1 (2.1-fold, P = 0.12). Only minimal, nonsignificant changes were found in planar cell polarity pathway genes, which are involved in cellular apical-basal orientation.29 The lack of involvement of the planar cell polarity pathway is of interest as a WNT10A mutation leads to differentiation defects that simulate KLF4 deficiency, and KLF4 has been shown to regulate corneal epithelial polarity.30,31 However, calcium pathway components associated with Wnt showed either an increase in CAMK2A (2.1-fold, P = 0.07) and PLCH1 (1.9-fold, P = 0.06) or a decrease in PLCB1 (−2.2-fold, P = 0.05). In summary, we observed not only changes in WNT10A transcription but also other dysregulations in canonical Wnt signaling and the Wnt calcium pathway.
To further validate the findings of the preliminary study, we used qPCR to examine WNT10A mRNA levels in 31 additional samples, including 15 progressive keratoconus patients undergoing crosslinking and 16 myopic controls. While some variation was seen in control cases, analysis of this independent validation cohort also showed significantly reduced expression of WNT10A transcripts in keratoconus epithelium (ratio 0.66, P = 0.03; Fig. 2A).
Figure 2.
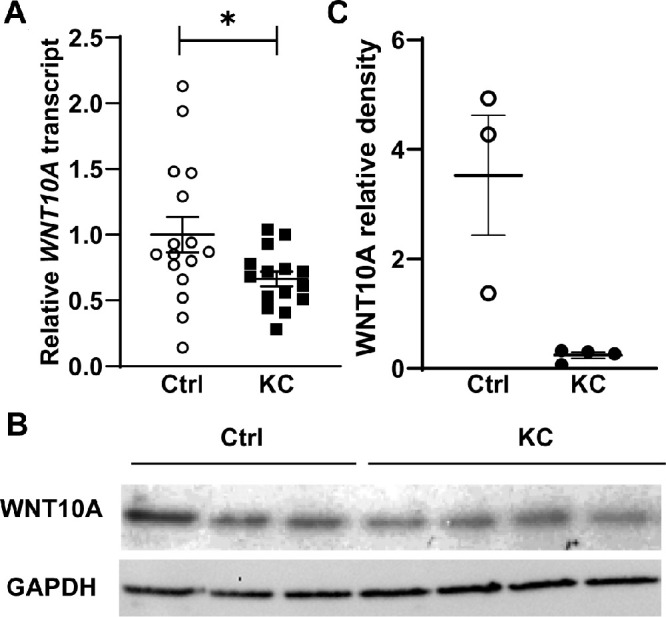
(A) qPCR validation of WNT10A underexpression in keratoconus corneal epithelium using 31 independent samples. (B) WNT10A is underexpressed at the protein level in keratoconus corneal epithelium per western blot (band size 46 kDa). (C) Relative densitometry of the blot shown in (B) shows WNT10A underexpression compared to glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
We then examined whether WNT10A protein levels were also lower in keratoconus epithelium. While sufficient material was available in only a small number of cases, western blot analysis of protein extracts from affected (n = 4) and control cases (n = 3) showed a trend toward reduced WNT10A expression in keratoconus patients (ratio: 0.07, P = 0.06; Figs. 2B, 2C).
In Keratoconus, WNT10A Is Largely Absent From Bowman's Layer
In order to examine expression of WNT10A protein in both corneal epithelium and underlying layers, we used tissue microarrays with representative cores from both peripheral and central cornea of keratoconus and healthy controls. For assessments of Bowman's layer, 58 central corneal and 62 peripheral keratoconus specimens, as well as 11 central and 13 peripheral controls, contained sufficient tissue for analysis on stained slides. For epithelial staining studies, 58 central corneal and 62 peripheral keratoconus specimens, as well as 10 and 14 control specimens, could be scored. Immunohistochemical analysis of control tissues showed WNT10A protein in the epithelium, Bowman's layer, and corneal stroma (Fig. 3A). Epithelial staining was generally weak to moderate in controls, with more variation seen centrally. These intensity values were multiplied by the percentage of cells staining in order to derive a combined “H score.” While median central epithelial staining was somewhat lower in keratoconus cases than controls (120 vs. 155, respectively) this difference was not statistically significant (P = 0.11). The differences in peripheral staining scores of keratoconus epithelium versus controls were more prominent (median score 80 vs. 160, respectively; P < 0.01). These data support our western analysis and suggest that in keratoconus epithelium, WNT10A protein levels are reduced, most prominently in the periphery of the cornea.
Figure 3.
(A) Immunohistochemical staining of WNT10A in control and keratoconus central and peripheral corneas. Epithelium, Bowman's layer (arrows), and anterior stroma are shown. Notable WNT10A staining of Bowman's layer in control corneas (arrows aim at Bowman's) and absent staining in keratoconus Bowman's layer. (B) Relative intensity scores demonstrate little to no WNT10A staining in keratoconus Bowman's layer: central score 0 [0,0] (median [interquartile range]) vs. 2 [1,2] in controls and peripherally 0 [0,0] vs. 3 [3,3] in controls. WNT10A staining in the central corneal epithelium of keratoconus and controls corneas is similar: keratoconus epithelium score 120 [20,180] vs. 155 [90,270] in controls. Peripheral keratoconus epithelium has lower staining score for WNT10A: 80 [20,165] vs. 160 [140, 217.5] in controls.
Differences in WNT10A protein were even more pronounced in Bowman's layer. Moderate to strong expression was seen in this acellular collagen layer below the epithelium in most control cases, but the protein was undetectable in most keratoconus cases (Fig. 3A). The median intensity keratoconus center score was 0 vs. 2 in controls (P << 0.01), while the median keratoconus peripheral score was 0 vs. 3 in controls (P << 0.01; Fig. 3B). Our immunohistochemical analysis of WNT10A expression thus suggests that changes in keratoconus epithelium are associated with even more prominently reduced protein levels in the underlying Bowman's layer.
WNT10A Relative Gene Expression in the Corneal Epithelium Positively Correlates With Keratoconus Severity
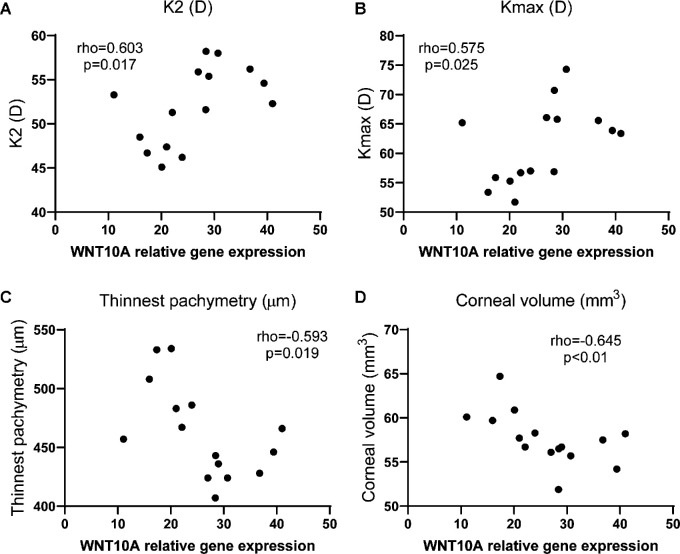
In order to further validate the association between WNT10A corneal epithelial transcript levels and keratoconus, we sought to evaluate whether relative gene expression levels correlate with disease severity. Corneal tomography is considered the gold standard in diagnosis and follow-up of keratoconus, and corneas with more severe disease have higher keratometry values, lower thickness (pachymetry), and lower volume. In the 15 eyes from our independent validation cohort used in this analysis, WNT10A relative transcript level correlated positively with pre-crosslinking keratoconus tomographic severity indices: K2 – steep central anterior corneal keratometry (Spearman's rank correlation [ρ] = 0.6; P = 0.02); Kmax – maximal anterior corneal keratometry (ρ = 0.57; P = 0.02); pachymetry (corneal thickness) at the thinnest point (ρ = −0.59, P = 0.02); and corneal volume (ρ = −0.64, P < 0.01) as shown in Figure 4.
Figure 4.
WNT10A relative gene expression positively correlates with increased corneal curvature, a marker of keratoconus severity: (A) K2 and (B) Kmax. WNT10A negatively correlates with pachymetry at the thinnest point (C) and corneal volume (D), both decreased in more severe keratoconus.
WNT10A Promotes COL1A1 Expression in Immortalized Corneal Epithelial Cells
We next assessed whether direct stimulation with WNT10A can regulate COL1A1 expression in immortalized corneal epithelial cells. These experiments were performed at equivalent confluence levels in order to avoid differences in collagen expression due to cell density. AXIN2 was chosen as a readout because it is a marker of canonical Wnt signaling.32,33 COL1A1 was chosen since Wnt signaling and WNT10A in particular have been shown to regulate collagen type I production.34,35 As expected, increased expression of WNT10A mRNA (ratio: 72.63, P << 0.01) and protein (ratio: 6.47, P = 0.02) was noted in corneal epithelial cells after a construct encoding WNT10A was introduced (Fig. 5). In contrast, Axin2 transcript levels remained relatively stable (ratio: 1.13, P = 0.59), suggesting no significant induction of canonical Wnt signaling. COL1A1 transcript levels were found to be significantly increased (ratio: 1.7, P = 0.02), and a more prominent elevation in COL1A1 protein level was also noted (ratio: 11, P = 0.02).
Figure 5.
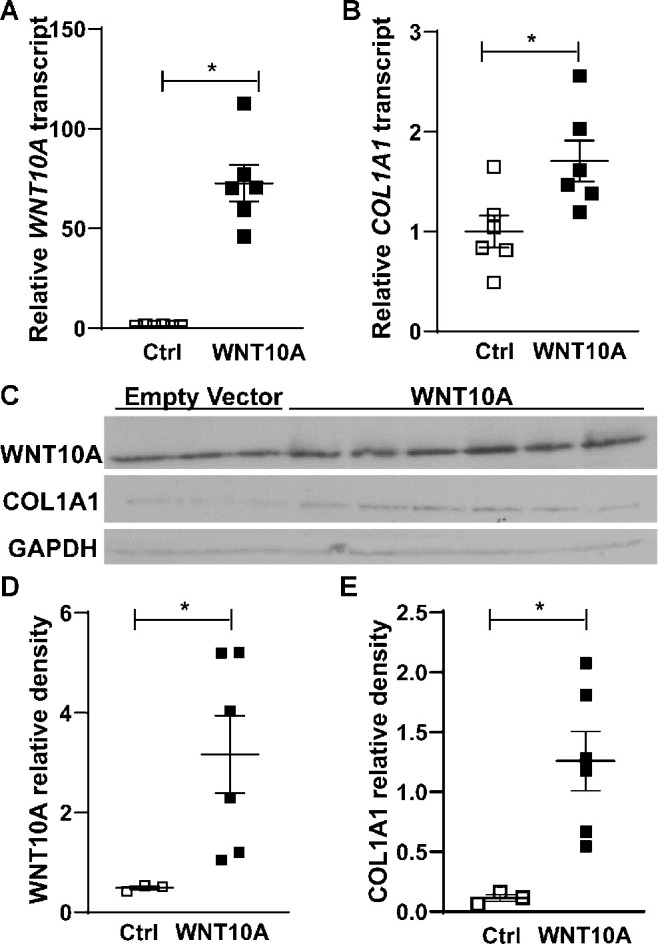
WNT10A overexpression in hTCEpi cells. Both WNT10A transcript (A) and protein levels (C, D) are elevated in the WNT10A overexpression plasmid wells (band size for WNT10A, 46 kDA; COL1A1, ∼120 kDA). COL1A1 transcript (B) and protein (C, D) are also increased in wells overexpressing WNT10A.
Discussion
The concept that epithelial alterations may play a role in the development of keratoconus has been suggested in the past, but molecular understanding of how this might occur has been limited.6,7,13–18 The results of this study provide a potential mechanism through which the keratoconus epithelium may affect corneal strength and disease progression. The main findings include underexpression of WNT10A transcript and protein in patient samples, absence of WNT10A protein in keratoconus from Bowman's layer, and a correlation between WNT10A transcript level and disease severity. We also found that WNT10A positively regulates COL1A1 expression in corneal epithelial cells in vitro.
Collagen type I is a major component of the corneal extracellular matrix, and we hypothesize that when WNT10A levels are low, reduced production of collagen I in the epithelium may affect the tensile strength of the epithelium itself. Moreover, collagen I is also localized to Bowman's layer, and Wnt dysregulation at the level of the epithelium may affect collagen I density in the underlying Bowman's layer, thus compromising Bowman's biomechanical strength.36,37 In this context, reduced production of collagen I may also potentiate Bowman's layer breaks typical of keratoconus and lead to disease progression. We also speculate that reduced WNT10A signaling in the epithelium may lead to additional alterations in epithelial factors that crosstalk with the stroma and alter stromal cells that affect overall integrity of the cornea.
Wnt signaling has been shown to be dysregulated in keratoconus. Genome-wide association studies identified an exonic variant in WNT10A as a risk factor for reduced central corneal thickness, a hallmark of keratoconus; another intronic variant in WNT7B was associated with central corneal thickness in Latinos.38,39 However, WNT10A expression in the corneal epithelium was not characterized in these studies. Full-thickness corneal specimens obtained during keratoplasty for keratoconus were sequenced and showed Wnt dysregulation, specifically in Wnt receptors FZD1 and FZD7 but also in AXIN2 and other regulatory genes.8 A Wnt ligand—WNT5A—was initially shown to be dysregulated in this study of full-thickness corneal tissue, but a follow-up study showed similar expression in keratoconus patients and controls.19 Nevertheless, DNA methylation profiles were shown to be different for WNT3 and WNT5A in keratoconus full-thickness corneal samples.40 Of note, the control group used in these studies comprised full-thickness abnormal corneas that underwent surgery for reasons other than keratoconus (e.g., ulcers and bullous keratopathy), so the results may have been skewed by comparison to abnormal as opposed to healthy controls. A recent study reported RNA sequencing of full-thickness corneas from keratoconus patients of African American and Middle Eastern ancestry who had decreased WNT5A and WNT9A expression, as well as the Wnt signaling intermediate secreted frizzled-related protein 1 (SFRP1).21
In another study of keratoconus, an RNA array was used to compare the corneal epithelium of keratoconus patients undergoing corneal transplant surgery; this study demonstrated Wnt signaling dysregulations, specifically in LEF1, PITX2, and SFRP1.6 SFRP1 is a Wnt signaling modulator, and in this study, SFRP expression was associated with patient age, and the protein localized mostly in the basal epithelium. The same group later showed that tear SFRP1 protein concentration was lower in keratoconus patients despite an overall increase in total tear protein.41 In another study, the group showed that direct stimulation of corneal epithelial cells with SFRP1 led to increased metabolic activity of cultured corneal epithelial cells in vitro.17 Finally, the group published an RNA-seq study done in the corneal epithelium only, in a similar manner to our study, which showed downregulation of genes associated with Wnt signaling but also Notch1 and Hedgehog.7 Neither WNT10A nor the progression status mentioned in that study. A different RNA-seq study performed on full-thickness corneal tissue examined coding and long noncoding RNAs and also found elevated SFRP1; additionally, it showed that a long noncoding member of Wnt (WNT4-2:1) was elevated in keratoconus.9 Of note, the keratoconus group comprised tissue obtained from patients aged 24 to 64 years, but disease progression status was not clearly defined. Additionally, the control group comprised older patients (61–77 years old), so the lack of age match may have affected the results.
However, to our knowledge, no Wnt ligand had previously been implicated specifically in keratoconus alterations in corneal epithelium at the mRNA or protein level.6–10 Our results on the dysregulation of a specific Wnt signaling ligand—WNT10A—are complementary to previous reports on SFRP upregulation in keratoconus, given that SFRP is a known modulator of Wnt signaling and can inhibit the pathway in many contexts.42 Overall, these results are suggestive of reduced Wnt signaling in keratoconus epithelium.6,41
WNT10A, a Wnt signaling pathway ligand, is known to be expressed in the central regions of mature corneal epithelium.43 In addition to evidence that Wnt signaling is dysfunctional in keratoconus, a WNT10A exonic variant has been linked to reduced corneal thickness and an increased risk of keratoconus.38 Moreover, WNT10A depletion in mouse skin fibroblasts is associated with multiple phenotypes similar to those seen in keratoconus, including delayed wound healing and reduced stromagenesis, fibroblast cellularity and activation, and collagen production.38,44 Recently, a genome-wide association study identified two WNT10A missense mutations associated with corneal hysteresis and corneal resistance factor, both known to be reduced in keratoconus.45 With this in mind, we focused on WNT10A and examined its pattern of dysregulation in keratoconus epithelium.
The keratoconus corneal epithelium can be perceived as an optical buffer to the abnormally shaped underlying cornea in keratoconus. In a dual-Schiempflug combined with placido-disk tomography study of moderate to severe keratoconus, maximal anterior keratometry was found to be more than 2 diopters higher after removal of the epithelium, but no changes were found in the posterior corneal surface to account for this difference in keratometry.46 Shear stress and mechanotransduction are likely to play a role in modeling the corneal epithelium in keratoconus, and Wnt signaling specifically is known to participate in mechanotransduction.47 Therefore, our observation of WNT10A dysregulation in the corneal epithelium could potentially be the result of an attempt of the corneal epithelial cells to mitigate or respond to the ultrastructural abnormality of the cornea characteristic of keratoconus, as suggested recently.48
Another notable finding in this study is the absence of WNT10A from Bowman's layer, in contrast to the diffuse expression seen in controls; this is of particular interest as this anatomic structure is often fragmented in keratoconus.49,50 With the highest tissue elasticity within the cornea, Bowman's layer is thought to support the normal architecture of the cornea by providing mechanical strength.51 In fact, Bowman's layer transplantation has been used to treat advanced keratoconus.52 Therefore, the integrity of Bowman's layer may play a role in maintaining the biomechanical properties of the cornea, which are known to be markedly reduced in keratoconus. The corneal stroma extracellular matrix is made up primarily of collagen type I, and this type of collagen also localizes to Bowman's layer.36,37 Others have suggested that during development, corneal epithelial cells produce some of Bowman's layer components, including collagen type I.53–55 However, our demonstration that WNT10A positively regulates collagen type I expression in corneal epithelial cells links Wnt more directly to structural proteins critical to corneal stability. We hypothesize that WNT10A underexpression in the corneal epithelium leads to reduced collagen type I deposition in the underlying Bowman's layer. Furthermore, the corneal epithelium itself appears to contribute to the mechanical strength of the cornea.56 Reduced collagen I in the epithelium may therefore also alter corneal stiffness and decrease overall biomechanical stability. We therefore hypothesize that reduced WNT10A expression in the corneal epithelium may negatively affect collagen type I production within this layer of the cornea and reduce its internal biomechanical strength, thus promoting disease progression in keratoconus.
The observation that WNT10A relative transcript level in the corneal epithelium correlates with tomographic disease severity markers further supports the hypothesis that WNT10A is involved in the disease process. The positive monotonic correlation between WNT10A transcript levels and disease severity may be perceived as at odds with the finding that WNT10A is underexpressed in earlier progressive keratoconus compared to controls. However, we hypothesize that although WNT10A transcript levels are generally lower in keratoconus in comparison to controls, a compensatory mechanism increases WNT10A expression in advanced keratoconus, characterized by increased corneal curvature and thinner pachymetry measurements.
A unique strength of our study is the use of a large number of verified patient samples. All corneal epithelial patient samples used in the RNA-seq study originated from patients with mild to moderate, progressive disease, whereas some previous studies used full-thickness corneas tissue obtained during keratoplasty, which is typically reserved for end-stage disease.8,9,19,20 Therefore, these samples may provide a more precise representation of the transcriptomic profile of cells with early yet active keratoconus. In addition, a large number of keratoconus samples were obtained for both the large qPCR validation cohort and immunohistochemistry studies. Together, the high sample numbers from over 100 individual keratoconus patients and controls strongly support our findings regarding WNT10A expression in keratoconus. Overall, the transcriptomic data obtained from this clinically well characterized cohort of early progressive keratoconus patients’ corneal epithelium have generated a transcriptional data set that will hopefully be useful for other researchers in the field. For instance, our work has focused on Wnt signaling, but the pathway analysis of the transcriptomic results has suggested interferon dysregulation and activation of immune response. These data, together with the typical geographical clustering seen in keratoconus and disease onset in early adolescence, are also suggestive of altered immune responses and viral etiology, which could provide additional prospects for research.
Overall, the results of the current study provide additional evidence of Wnt signaling pathway dysregulation in keratoconus, specifically in the corneal epithelium and Bowman's layer. We have also shown that WNT10A positively regulates collagen I production in corneal epithelial cells. These studies also suggest that the Wnt signaling pathway represents a possible therapeutic target, as its manipulation in the corneal epithelium may induce collagen I production and thus increase tensile strength in keratoconus. However, additional studies are required to verify this hypothesis.
Supplementary Material
Acknowledgments
The authors thank Regina “Kay” Morgan, Tammy Kephart, Rebecca Scarborough, Cailyn Sonneborn, and Antionette Price.
Supported by National Eye Institute Grant K08EY027474, Wilmer Biostatistics Core Grant P30EY001765, an unrestricted departmental grant to Wilmer Eye Institute from Research to Prevent Blindness, and philanthropic grants from Debbie Colson and Jeffrey Williams, Ellen A. Cherniavsky, Hymowitz Family Foundation, Tyrone and Jennifer Throop, the Kahn Foundation, and Donald Jump.
Disclosure: J.W. Foster, None; R.N. Parikh, None; J. Wang, None; K.S. Bower, None; M. Matthaei, None; S. Chakravarti, None; A.S. Jun, None; C.G. Eberhart, None; U.S. Soiberman, None
References
- 1. Godefrooij DA, de Wit GA, Uiterwaal CS, Imhof SM, Wisse RP.. Age-specific incidence and prevalence of keratoconus: a nationwide registration study. Am J Ophthalmol. 2017; 175: 169–172. [DOI] [PubMed] [Google Scholar]
- 2. Klintworth GK, Damms T.. Corneal dystrophies and keratoconus. Curr Opin Ophthalmol. 1995; 6(4): 44–56. [DOI] [PubMed] [Google Scholar]
- 3. Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998; 42(4): 297–319. [DOI] [PubMed] [Google Scholar]
- 4. Choi JA, Kim MS.. Progression of keratoconus by longitudinal assessment with corneal topography. Invest Ophthalmol Vis Sci. 2012; 53(2): 927–35. [DOI] [PubMed] [Google Scholar]
- 5. McMahon TT, Edrington TB, Szczotka-Flynn L, Olafsson HE, Davis LJ, Schechtman KB.. Longitudinal changes in corneal curvature in keratoconus. Cornea. 2006; 25(3): 296–305. [DOI] [PubMed] [Google Scholar]
- 6. Sutton G, Madigan M, Roufas A, McAvoy J.. Secreted frizzled-related protein 1 (SFRP1) is highly upregulated in keratoconus epithelium: a novel finding highlighting a new potential focus for keratoconus research and treatment. Clin Exp Ophthalmol. 2010; 38(1): 43–8. [DOI] [PubMed] [Google Scholar]
- 7. You J, Corley SM, Wen L, et al.. RNA-seq analysis and comparison of corneal epithelium in keratoconus and myopia patients. Sci Rep. 2018; 8(1): 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kabza M, Karolak JA, Rydzanicz M, et al.. Collagen synthesis disruption and downregulation of core elements of TGF-beta, Hippo, and Wnt pathways in keratoconus corneas. Eur J Hum Genet. 2017; 25(5): 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khaled ML, Bykhovskaya Y, Yablonski SER, et al.. Differential expression of coding and long noncoding RNAs in keratoconus-affected corneas. Invest Ophthalmol Vis Sci. 2018; 59(7): 2717–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kabza M, Karolak JA, Rydzanicz M, et al.. Multiple differentially methylated regions specific to keratoconus explain known keratoconus linkage loci. Invest Ophthalmol Vis Sci. 2019; 60(5): 1501–1509. [DOI] [PubMed] [Google Scholar]
- 11. Wisse RP, Kuiper JJ, Gans R, Imhof S, Radstake TR, Van der Lelij A.. Cytokine expression in keratoconus and its corneal microenvironment: a systematic review. Ocul Surf. 2015; 13(4): 272–283. [DOI] [PubMed] [Google Scholar]
- 12. Wisse RPL, Kuiper JJW, Radstake TRD, Broen JCA.. Quantification of double stranded DNA breaks and telomere length as proxies for corneal damage and replicative stress in human keratoconus corneas. Transl Vis Sci Technol. 2019; 8(4): 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chaerkady R, Shao H, Scott SG, Pandey A, Jun AS, Chakravarti S.. The keratoconus corneal proteome: loss of epithelial integrity and stromal degeneration. J Proteomics. 2013; 87: 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Joseph R, Srivastava OP, Pfister RR.. Differential epithelial and stromal protein profiles in keratoconus and normal human corneas. Exp Eye Res. 2011; 92(4): 282–298. [DOI] [PubMed] [Google Scholar]
- 15. Srivastava OP, Chandrasekaran D, Pfister RR.. Molecular changes in selected epithelial proteins in human keratoconus corneas compared to normal corneas. Mol Vis. 2006; 12: 1615–1625. [PubMed] [Google Scholar]
- 16. Nielsen K, Vorum H, Fagerholm P, et al.. Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study. Exp Eye Res. 2006; 82(2): 201–209. [DOI] [PubMed] [Google Scholar]
- 17. You J, Munoz-Erazo L, Wen L, Hodge C, Madigan MC, Sutton G.. In-vitro effects of secreted frizzled-related protein 1 (SFRP1) on human corneal epithelial cells. Curr Eye Res. 2018; 43(4): 455–459. [DOI] [PubMed] [Google Scholar]
- 18. You J, Wen L, Roufas A, Madigan MC, Sutton G.. Expression of SFRP family proteins in human keratoconus corneas. PLoS One. 2013; 8(6): e66770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karolak JA, Ginter-Matuszewska B, Tomela K, et al.. Further evaluation of differential expression of keratoconus candidate genes in human corneas. PeerJ. 2020; 8: e9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Macé M, Galiacy SD, Erraud A, et al.. Comparative transcriptome and network biology analyses demonstrate antiproliferative and hyperapoptotic phenotypes in human keratoconus corneas. Invest Ophthalmol Vis Sci. 2011; 52(9): 6181–6191. [DOI] [PubMed] [Google Scholar]
- 21. Shinde V, Hu N, Mahale A, et al.. RNA sequencing of corneas from two keratoconus patient groups identifies potential biomarkers and decreased NRF2-antioxidant responses. Sci Rep. 2020; 10(1): 9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001; 25(4): 402–408. [DOI] [PubMed] [Google Scholar]
- 23. Bennike TB, Kastaniegaard K, Padurariu S, et al.. Comparing the proteome of snap frozen, RNAlater preserved, and formalin-fixed paraffin-embedded human tissue samples. EuPA Open Proteom. 2016; 10: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schneider CA, Rasband WS, Eliceiri KW.. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012; 9(7): 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lackner EM, Matthaei M, Meng H, Ardjomand N, Eberhart CG, Jun AS.. Design and analysis of keratoconus tissue microarrays. Cornea. 2014; 33(1): 49–55. [DOI] [PubMed] [Google Scholar]
- 26. Hirsch FR, Varella-Garcia M, Bunn PA Jr, et al.. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003; 21(20): 3798–3807. [DOI] [PubMed] [Google Scholar]
- 27. Najdi R, Proffitt K, Sprowl S, et al.. A uniform human Wnt expression library reveals a shared secretory pathway and unique signaling activities. Differentiation. 2012; 84(2): 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robertson DM, Li L, Fisher S, et al.. Characterization of growth and differentiation in a telomerase-immortalized human corneal epithelial cell line. Invest Ophthalmol Vis Sci. 2005; 46(2): 470–478. [DOI] [PubMed] [Google Scholar]
- 29. Butler MT, Wallingford JB.. Planar cell polarity in development and disease. Nat Rev Mol Cell Biol. 2017; 18(6): 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu M, Horrell J, Snitow M, et al.. WNT10A mutation causes ectodermal dysplasia by impairing progenitor cell proliferation and KLF4-mediated differentiation. Nat Commun. 2017; 8: 15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tiwari A, Swamynathan S, Jhanji V, Swamynathan SK.. KLF4 coordinates corneal epithelial apical-basal polarity and plane of cell division and is downregulated in ocular surface squamous neoplasia. Invest Ophthalmol Vis Sci. 2020; 61(5): 15–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lim X, Tan SH, Yu KL, Lim SB, Nusse R.. Axin2 marks quiescent hair follicle bulge stem cells that are maintained by autocrine Wnt/β-catenin signaling. Proc Natl Acad Sci USA. 2016; 113(11): E1498–E1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F.. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002; 22(4): 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang KY, Yamada S, Izumi H, et al.. Critical in vivo roles of WNT10A in wound healing by regulating collagen expression/synthesis in WNT10A-deficient mice. PLoS One. 2018; 13(3): e0195156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Z, Guo Q, Tian H, Lv P, Zhou C, Gao X.. Effects of WNT10A on proliferation and differentiation of human dental pulp cells. J Endod. 2014; 40(10): 1593–1599. [DOI] [PubMed] [Google Scholar]
- 36. Newsome DA, Foidart JM, Hassell JR, Krachmer JH, Rodrigues MM, Katz SI.. Detection of specific collagen types in normal and keratoconus corneas. Invest Ophthalmol Vis Sci. 1981; 20(6): 738–750. [PubMed] [Google Scholar]
- 37. Marshall GE, Konstas AG, Lee WR.. Immunogold fine structural localization of extracellular matrix components in aged human cornea. I. Types I-IV collagen and laminin. Graefes Arch Clin Exp Ophthalmol. 1991; 229(2): 157–163. [DOI] [PubMed] [Google Scholar]
- 38. Cuellar-Partida G, Springelkamp H, Lucas SE, et al.. WNT10A exonic variant increases the risk of keratoconus by decreasing corneal thickness. Hum Mol Genet. 2015; 24(17): 5060–5068. [DOI] [PubMed] [Google Scholar]
- 39. Gao X, Nannini DR, Corrao K, et al.. Genome-wide association study identifies WNT7B as a novel locus for central corneal thickness in Latinos. Hum Mol Genet. 2016; 25(22): 5035–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kabza M, Karolak JA, Rydzanicz M, et al.. Multiple differentially methylated regions specific to keratoconus explain known keratoconus linkage loci. Invest Ophthalmol Vis Sci. 2019; 60(5): 1501–1509. [DOI] [PubMed] [Google Scholar]
- 41. You J, Hodge C, Wen L, McAvoy JW, Madigan MC, Sutton G.. Tear levels of SFRP1 are significantly reduced in keratoconus patients. Mol Vis. 2013; 19: 509. [PMC free article] [PubMed] [Google Scholar]
- 42. Surana R, Sikka S, Cai W, et al.. Secreted frizzled related proteins: Implications in cancers. Biochim Biophys Acta. 2014; 1845(1): 53–65. [DOI] [PubMed] [Google Scholar]
- 43. Nakatsu MN, Ding Z, Ng MY, Truong TT, Yu F, Deng SX.. Wnt/beta-catenin signaling regulates proliferation of human cornea epithelial stem/progenitor cells. Invest Ophthalmol Vis Sci. 2011; 52(7): 4734–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang K-Y, Yamada S, Izumi H, et al.. Critical in vivo roles of WNT10A in wound healing by regulating collagen expression/synthesis in WNT10A-deficient mice. PLoS ONE. 2018; 13(3): e0195156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simcoe MJ, Khwaja AP, Eye UKB, Consortium V, Hysi PG, Hammond CJ.. Genome-wide association study of corneal biomechanical properties identifies over 200 loci providing insight into the genetic aetiology of ocular diseases. Hum Mol Genet. 2020; 29(18): 3154–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ziaei M, Meyer J, Gokul A, Vellara H, McGhee CNJ.. Direct measurement of anterior corneal curvature changes attributable to epithelial removal in keratoconus. J Cataract Refract Surg. 2018; 44(1): 71–77. [DOI] [PubMed] [Google Scholar]
- 47. Abuammah A, Maimari N, Towhidi L, et al.. New developments in mechanotransduction: cross talk of the Wnt, TGF-β and Notch signalling pathways in reaction to shear stress. Curr Opin Biomed Eng. 2018; 5: 96–104. [Google Scholar]
- 48. Amit C, Padmanabhan P, Narayanan J.. Deciphering the mechanoresponsive role of β-catenin in keratoconus epithelium. Sci Rep. 2020; 10(1): 21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sykakis E, Carley F, Irion L, Denton J, Hillarby MC.. An in depth analysis of histopathological characteristics found in keratoconus. Pathology. 2012; 44(3): 234–239. [DOI] [PubMed] [Google Scholar]
- 50. Sawaguchi S, Fukuchi T, Abe H, Kaiya T, Sugar J, Yue BY.. Three-dimensional scanning electron microscopic study of keratoconus corneas. Arch Ophthalmol. 1998; 116(1): 62–68. [DOI] [PubMed] [Google Scholar]
- 51. Last JA, Thomasy SM, Croasdale CR, Russell P, Murphy CJ.. Compliance profile of the human cornea as measured by atomic force microscopy. Micron. 2012; 43(12): 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van Dijk K, Liarakos VS, Parker J, et al.. Bowman layer transplantation to reduce and stabilize progressive, advanced keratoconus. Ophthalmology. 2015; 122(5): 909–917. [DOI] [PubMed] [Google Scholar]
- 53. Tisdale AS, Spurr-Michaud SJ, Rodrigues M, Hackett J, Krachmer J, Gipson IK.. Development of the anchoring structures of the epithelium in rabbit and human fetal corneas. Invest Ophthalmol Vis Sci. 1988; 29(5): 727–736. [PubMed] [Google Scholar]
- 54. Gordon MK, Foley JW, Birk DE, Fitch JM, Linsenmayer TF.. Type V collagen and Bowman's membrane: quantitation of mRNA in corneal epithelium and stroma. J Biol Chem. 1994; 269(40): 24959–24966. [PubMed] [Google Scholar]
- 55. Linsenmayer TF, Smith GN Jr, Hay ED. Synthesis of two collagen types by embryonic chick corneal epithelium in vitro. Proc Natl Acad Sci USA. 1977; 74(1): 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ziaei M, Gokul A, Vellara H, Lu LM, Patel DV, McGhee CNJ.. Measurement of in vivo biomechanical changes attributable to epithelial removal in keratoconus using a noncontact tonometer. Cornea. 2020; 39(8): 946–951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.