Abstract
It is now becoming clear that neutrophils and eosinophils are heterogeneous cells with potentially multiple subsets in health and disease. With greater marker coverage by multi-color flow cytometry and single-cell level sequencing of granulocyte populations, novel phenotypes of these cells began to emerge. Intriguingly, many newly described subsets blend distinctions between classical myeloid lineage phenotypes, which are especially true for tissue resident or recruited cells in contexts of inflammation and disease. This includes reports of neutrophils with features of eosinophils, monocytes and dendritic cells, and eosinophil subsets expressing neutrophil markers. Moreover, novel studies show the ability of immature neutrophils to transdifferentiate into mature cells belonging to other myeloid lineages (eosinophils, monocytes/macrophages). In this review, we summarize novel findings in this exciting research frontier and shed light on potential processes driving the plasticity and heterogeneity of granulocyte subsets. Specifically, we discuss the hematopoietic flexibility of granulocyte precursors in bone marrow and the adaptation of myeloid cells to local tissue microenvironments. The understanding of such intermediate and developmental phenotypes is very important, as it can teach us about origins of functionally distinct myeloid cells during inflammation, and explain reasons for successes and failures of biologics targeting terminally differentiated granulocytes.
Keywords: Eosinophils, Neutrophils, Monocytes, Macrophages, Myeloid cells, Subsets, Heterogeneity, Plasticity, Hematopoiesis, Single-cell sequencing, Bone marrow
Introduction
Both neutrophil and eosinophil research fields are currently in the state of renaissance, which is fueled by the advent of next-generation sequencing and multi-panel flow cytometry approaches. In particular, new evidence is coming to light that both neutrophils and eosinophils are highly heterogeneous and do not exist only in singularly differentiated states (1–3). It is becoming apparent that there are multiple granulocyte subsets with different roles in health and disease (2–5). Classical characterization of neutrophils and eosinophils and their precursors is based on density gradient and histological examination of shape, size, nuclear morphology, and granular content. Limited flow cytometry marker sets were identified over time to accurately match the morphological description of different granulocytes in blood and other tissues. However, morphological and histochemical analysis of neutral versus red-staining cells can be quite subjective and does not reflect developmental trajectories and functional properties of different varieties of neutrophils and eosinophils. With greater marker coverage by multi-color flow cytometry and single-cell level sequencing of granulocyte populations, novel phenotypes began to emerge. Intriguingly, there are many cases when this new data started blending the distinctions between classical neutrophil and eosinophil phenotypes. It is especially true for tissue dwelling cells in contexts of inflammation and subset emergence in disease. Although neutrophils and eosinophils express distinct sets of granule proteins and play distinct functional roles in innate immunity (phagocytosis and intracellular killing of microorganisms by neutrophils; helminth defense, contribution to hypersensitivity reactions, release of lipid mediators by eosinophils), there are also substantial overlaps in their functions. Both cell types are able to produce ROS bursts and release extracellular DNA traps, as well as contribute pro-inflammatory, tissue remodeling, and pro-resolving mediators via degranulation or de novo synthesis. Potentially, two different processes are responsible for such phenotypic and functional plasticity of granulocyte lineages: hematopoietic flexibility of granulocyte precursors in bone marrow and adaptation of cells to local tissue microenvironments in the face of different inflammatory stimuli (Fig. 1). In this review, we would like to briefly summarize findings in this interesting emerging area and discuss potential drivers of granulocyte plasticity and heterogeneity in the following three chapters by (1) bringing attention to the increased reporting of mixed myeloid phenotypes in different microenvironmental contexts and (2) discussing novel evidence for granulocyte hematopoiesis as a significant source of subset heterogeneity.
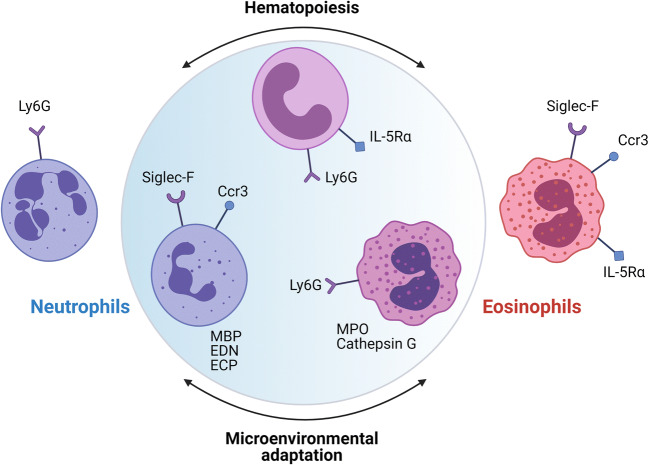
Fig. 1.
Aside from terminally differentiated mature eosinophils and neutrophils defined by conventional flow cytometry and granular protein markers, intermediate phenotypes with mixed neutrophil-eosinophil characteristics are coming to light. Two different processes may be responsible for such plasticity of granulocyte lineages: hematopoietic flexibility of granulocyte precursors and adaptation of different subsets to local tissue and cytokine microenvironments
Myeloid plasticity of granulocytes
Neutrophils with features of other myeloid cells
Classical neutrophils
Neutrophils are the most abundant type of circulating myeloid cells, constituting 50–70% of peripheral blood in humans and 10–25% in mice (6). They are also a surprisingly dominant granulocyte population in normal bone marrow. Neutrophils exhibit ring-like or segmented nuclear morphology and neutral granular staining. Neutrophil granule contents include myeloid peroxidase, lysozyme, alkaline phosphatase, neutrophil elastase, and β-glucuronidase. Conventional flow cytometry markers for mice are Gr-1 and Ly6G, high expression of which is used to sufficiently capture all neutrophil populations (these markers are absent in human neutrophils). CD15, CD66b, and CD16 are used in flow cytometry identification of circulating human neutrophils. Neutrophils are emerging as highly heterogeneous cells, especially in the contexts of disease and inflammation. CD101, CXCR2, CD33, CD10, and CD49d are additional markers used to describe specific subsets of neutrophils in conjunction with the classically defined markers listed above. Neutrophils are short-lived cells, with a classically cited half-life of 7–9 h for human circulating neutrophils (7). Newer studies suggest that the half-life of human circulating neutrophils could actually be more than 5 days (8). Hypoxia and inflammatory cytokines, as well as growth factors (e.g., GM-CSF, TNF), can significantly extend neutrophil survival in tissues (9, 10). For decades the dogma has been that blood neutrophils as phagocytes migrate rapidly into tissues in response to infection or injury, forming the first line of host defense. New evidence shows that there are tissue resident neutrophils that act as the first responders to pathogens and injury and have multiple immunosuppressive and immunoregulatory roles in eradicating pathogens, minimizing tissue damage, and contributing to tissue repair (11). Altogether, neutrophils persist long enough in blood and tissues to be responsive to differential stimuli regulatory for their survival, phenotype, and function. The following examples of neutrophil phenotypic plasticity may represent their hematopoietic flexibility or specification in response to different microenvironmental cues.
Neutrophils expressing eosinophil markers
Interestingly, there are multiple reports of neutrophils expressing markers more typical for an eosinophil lineage. Gorski et al. (12) report expression of functional IL-5 receptor alpha (IL-5Rα) by murine neutrophils infiltrating lungs in response to influenza A viral infection and on neutrophils from the inflamed lungs of children with treatment-refractory asthma. The authors only examined asthmatic individuals, not healthy controls, and did not provide functional characterization of IL-5Rα(+) neutrophils. However, they reported IL-5Rα expression in the human differentiated neutrophil-like HL-60 cell line under normal culture conditions. They also showed in mice that IL-5 suppressed neutrophil respiratory burst, a classical neutrophil effector function (12). The same Ly6G(+)IL-5Rα(+) mouse lung neutrophil population was also reported in a study by Mesnil et al. (13). Our group found that the majority of murine bone marrow Ly6G(high) neutrophils also express IL-5Rα (14). This is a surprising finding, as IL-5Rα is a marker typically expressed on cells of eosinophil lineage. In the same report, we also pursued the idea that according to this receptor expression, IL-5 would unlock a hidden hematopoietic plasticity of bone marrow neutrophils. Indeed, normal murine bone marrow neutrophils were able to transdifferentiate to red-staining Siglec-F(+) eosinophils with mature granules and expression of eosinophil peroxidase when treated with IL-5 (14). Altogether, this shows that although IL-5 is important for the development of eosinophils, IL-5Rα as a marker is not specific to eosinophil lineage.
A glycan-binding protein Siglec-F is another marker typically attributable to eosinophil lineage (although also expressed by alveolar macrophages). Pfirschke et al. (15) report that neutrophils infiltrating murine lung adenocarcinomas are of CD11b(+)Ly6G(+)Siglec-F(high) phenotype and are distinct from Siglec-F(high) eosinophils, Siglec-F(high) macrophages, and CD11b(+)Ly6G(+) myeloid-derived suppressor cells. They further show that Siglec-F(high) neutrophils can live in tumors for several days and significantly associate with the lung tumor burden; however, they do not provide specific functional characterization of this neutrophil subset (15). The half-life of tumor-infiltrating Siglec-F(high) neutrophils was significantly longer than that of blood or lung Siglec-F(low) neutrophils from the same mice. The authors discuss that these cells are likely tumor promoting in the context of cancer (by association of the phenotype with lung tumor burden) but could be beneficial in the context of homeostasis and normal tissue repair. A single-cell transcriptomic analysis of neutrophil diversification in mouse models of myocardial infarction found a similar long-lived Siglec-F(high) neutrophil subset that was characteristic for late stage inflammation of the infarcted heart (16). Atypical Ly6G(+)Siglec-F(+) neutrophils were found in olfactory neuroepithelium of mice at steady state and during inflammation and had functions supporting neurogenesis (17). A similar Siglec-F(+) neutrophil subset was reported in the mouse nasal mucosa in an allergic rhinitis model (18). Moreover, flow cytometry analysis of neutrophils from models of influenza infection in mice detected induced expression of a novel chemokine receptor repertoire, which included expression of chemokine receptors CCR3 and CCR5, which are more typical for eosinophils responsive to chemokines eotaxin and RANTES (19). Stimulation of these receptors with corresponding ligands increased neutrophil migratory capacity, phagocytic activity, and NETosis. The authors suggest that this enhanced chemokine receptor repertoire represents a mechanism to fine tune effector functions of neutrophils infiltrating tissues. Similarly, a study by Hartl et al. (20) reported that infiltrated neutrophils from patients with chronic inflammatory lung diseases and rheumatoid arthritis highly expressed CCR3 and CCR5, which are absent on human circulating neutrophils. Another study found that asthma-related inflammation promotes lung metastasis of breast cancer cells through the CCL11-CCR3 pathway, which directly promoted neutrophil recruitment (21). Yet another study showed that along with eosinophil recruitment, eotaxin CCL11 and CCR3 are important in the pulmonary recruitment of neutrophils with potentially pathogenic roles in murine bleomycin-induced lung fibrosis models (22). In humans, eotaxin CCL11 and eotaxin-2 (CCL24) induced recruitment not only of eosinophils but also neutrophils and macrophages following cutaneous injection of these chemokines in atopic and nonatopic volunteers (23).
Such context-dependent phenotypic confusion goes beyond surface marker expression by flow cytometry. A study by Abu-Ghazaleh et al. (24) quantified expression of typical eosinophil granule proteins in different populations of peripheral blood granulocytes. This study found measurable quantities of eosinophil major basic protein (MBP), eosinophil-derived neurotoxin (EDN), and eosinophil cationic protein (ECP) in highly purified blood neutrophils. A study by Monteseirin et al. identified human neutrophils from allergic patients as a novel source of eosinophil cationic protein (ECP) in IgE-mediated processes (25). Another study found expression of ECP in human neutrophils from the inflamed nasal tissue of patients with allergic rhinitis and chronic rhinosinusitis by immunohistochemical staining (26).
Neutrophils with features of other myeloid lineages
Additional literature evidence shows that neutrophils are capable of incredible myeloid plasticity. Matsushima et al. (27) published that both immature and mature neutrophils isolated from murine bone marrow can differentiate into a previously unrecognized “hybrid” population showing dual properties of both neutrophils and dendritic cells (DCs) when cultured with GM-CSF. The resulting cells express markers of both neutrophils (Ly6G, CXCR2) and DCs (CD11c, MHCII, CD80, and CD86). They also have dendritic morphology, probing motion, podosome formation, and the ability to present protein antigens to naïve CD4 T cells. However, they also retain intrinsic abilities of neutrophils to capture exogenous material, extrude neutrophil extracellular traps, and kill bacteria. Therefore, bone marrow neutrophils retain sufficient plasticity to differentiate into neutrophil-DC hybrids that can participate in both innate and adaptive immune responses (27). Neutrophil-DC hybrids were also reported in experimentally induced inflammatory lesions in mice (28) and murine models of invasive fungal infections (29).
According to descriptive immunophenotypic and cell culture studies, neutrophils also have the ability to directly “transdifferentiate” into monocytes/macrophages (30). Koffel et al. provided direct experimental evidence that inflammatory signals relayed by the MKK6-p38MAPK cascade induce monocytic cell differentiation from band-stage neutrophils (31). Another study by Ding et al. (32) showed that the monocytes that repopulate in mice after chemotherapy (cyclophosphamide treatment) acquire a neutrophil precursor gene signature, including expression of primary and secondary granules, and assume immunosuppressive functions. Our own data shows that Ly6G(+)IL-5Rα(+) neutrophils purified from normal murine bone marrow transdifferentiate not only to Siglec-F(+) eosinophils with IL-5 treatment but also into Ly6C(+) monocytes/macrophages when cultured with M-CSF (14). Additional abundant evidence shows that different neutrophil subsets may acquire characteristics of myeloid-derived suppressor cells (MDSCs) and assume immunosuppressive functions in human cancer and mouse models of tumorigenesis (33–36).
By far, this is not an exhaustive list of examples of unexpected neutrophil plasticity and heterogeneity in different tissue and activation contexts. Overall, it challenges the long-standing dogma that neutrophils are a homogeneous population of terminally differentiated short-lived cells with strictly first responder functions in host defense. For further examples and reading on the subject, I refer the reader to excellent reviews by Silvestre-Roig et al. (37), Ng et al. (38) and Ballesteros et al. (39).
Eosinophils with atypical phenotypes
Classical eosinophils
Compared to neutrophils, mature eosinophils are far less frequent in peripheral blood circulation and as residents in homeostatic tissues (as low as 1% of all hematopoietic cells), although their numbers increase dramatically during tissue repair and remodeling, helminth defense, and allergic inflammation. Eosinophils exhibit ring-like or segmented nuclear morphology and red eosin-positive granular staining. Eosinophil granule contents include major basic protein (MBP), eosinophil-derived neurotoxin (EDN), eosinophil cationic protein (ECP), and eosinophil peroxidase (EPO). Human eosinophils uniquely contain Charcot-Leyden crystals (CLC or galectin-10). For further reading on the differences between eosinophil and neutrophil granule proteins and their functional significance, we refer our reader to an excellent review by Gigon et al. (40). Conventional flow cytometry markers for mice are Siglec-F, IL-5Rα, and Ccr3. Siglec-F is frequently used alone to capture eosinophil populations. SIGLEC8 and CCR3 are conventionally used in flow cytometry identification of human neutrophils. Similar to neutrophils, new evidence shows that there are tissue resident eosinophils in multiple organ systems, as well as heterogeneous populations of these cells with different roles in health and disease (3, 5, 41–43). Despite heterogeneity of mature eosinophils, there is no evidence that immature or mature eosinophils can transdifferentiate to other myeloid lineages. However, at the precursor level, differentiating eosinophils can have neutrophilic features, which is reviewed below.
Eosinophils expressing neutrophil markers
Recent studies demonstrated that Ly6G/Gr1, a marker typical for neutrophil lineage, makes an unexpected cameo appearance in eosinophils (44). A study by Percopo et al. (45) first described Siglec-F(+)Gr1(high) eosinophils as a distinct subpopulation within the lungs of allergen-challenged mice. This eosinophil population persisted in eosinophil granule protein-deficient (EPX-/- and MBP-1-/-) mice but was absent in an eosinophil-deficient ΔdblGATA strain, which suggests it could be in an eosinophil precursor state prior to the mature granule formation checkpoint. Functionally, Siglec-F(+)Gr1(high) eosinophils had a distinct cytokine profile (CXCL13, IL-27, IL-13 identified only in this population), which points to their distinct immunomodulatory potential. Ly6G was also detected in 50% of Siglec-F(+) bone marrow eosinophils (45). A subsequent study by Limkar et al. determined that IL-5 drives Ly6G/Gr1 surface antigen expression in conventional bone marrow-derived murine eosinophil cultures (46). Noteworthy, this study also found that Ly6G-expressing eosinophils were less responsive to eotaxin-1. Moreover, our recent study (14) showed that Siglec-F(+)Ly6G(+) eosinophils can be directly derived from Siglec-F(-)Ly6G(high)IL-5Rα(+) neutrophil populations when treated with IL-5, which can potentially explain the existence of such mixed neutrophil-eosinophil phenotypes in mice. Interestingly, we found that G-CSF treatment of bone marrow neutrophils was sufficient to induce a Siglec-F(+)Ly6G(+) phenotype in the absence of IL-5 (14). Collectively, all evidence points to a mixed Siglec-F(+)Ly6G(+) neutrophil-eosinophil phenotype as a transient developmental stage of eosinophils originating either from earlier precursors or neutrophils with myeloid plasticity potential. Importantly, it also demonstrates that Ly6G alone (even at high levels) is not a sufficient neutrophil marker. Reliance on Ly6G expression alone without the characterization of additional markers, or the developmental state of the cells of interest, can lead to serious confusion between neutrophil and eosinophil populations. As an unwanted consequence, this can lead to erroneous interpretation of data.
In support of developmental origins of a mixed neutrophil-eosinophil phenotype, a study by Kim et al. (47) showed that terminally differentiating eosinophils co-express neutrophil primary granule proteins (cathepsin G, neutrophil elastase), as well as eosinophil-specific granule proteins (MBP), in a temporal manner. Interestingly, eosinophils from patients with type 1 diabetes mellitus express high levels of neutrophil granule markers: myeloid alpha-defensins and myeloperoxidase (48). On a mixed functional level, a study by Esnault et al. (49) showed that human eosinophils can produce matrix metalloproteinase-9 (MMP9), which leads to release of IL-1β, which potentially bridges eosinophilic and neutrophilic responses associated with treatment-refractory asthma.
Hematopoietic origins of granulocyte heterogeneity
The examples illustrated above suggest that the flexibility of granulocytes in different developmental states has significant potential to contribute to myeloid plasticity and heterogeneity. Single-cell sequencing approaches are perfectly suited to reexamine hematopoiesis of different cells in bone marrow and other tissues and now shed a new light on the developmental trajectories of myeloid cells. Such analyses began to reveal the complexity of hematopoietic differentiation (50). One of the most exciting insights was made in regard to the modeling of discrete vs. continuous hematopoiesis trajectories (51, 52). Under the discrete differentiation dogma, which is based on studies of flow-sorted populations, hematopoietic stem cells (HSCs) differentiate to mature lineages following a tree-like hierarchy of oligo-, bi-, and unipotent progenitors. New evidence, based on next-generation sequencing, supports the continuous differentiation model with no obvious boundaries in the hierarchy (53). Under the new model, individual HSCs gradually acquire lineage characteristics along multiple directions without passing hierarchically organized progenitor populations (51, 54). This continuous hematopoietic model fits well with the plasticity potential of myeloid cells during lineage differentiation. In this section, we summarize the plasticity and heterogeneity of neutrophil and eosinophil granulocytes according to their developmental trajectories (Fig. 2).
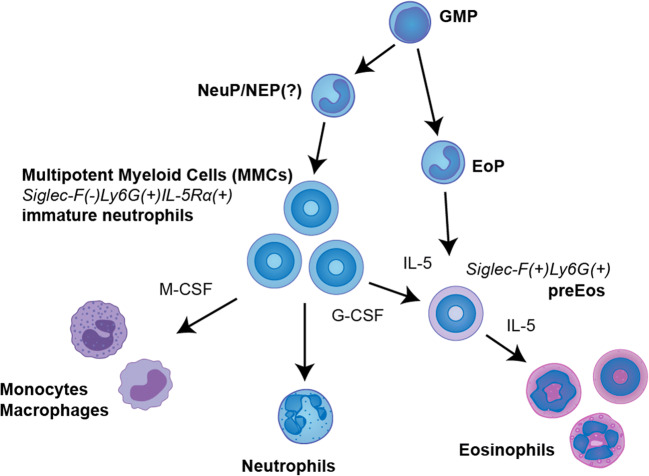
Fig. 2.
Diversity of granulocyte subsets may be fueled by developmental flexibility of myeloid lineages. In particular, immature band-stage Ly6G(+)IL-5Rα(+) neutrophils (also termed multipotent myeloid cells (MMCs) to reflect their plasticity) have been shown to transdifferentiate to eosinophils and monocytes when treated with appropriate inflammatory stimuli. Accordingly, eosinophils can be generated not only from previously defined unipotent eosinophil progenitors but also from MMCs outside of eosinophil lineage, which results in generation of intermediate phenotypes. GMP, granulocyte-monocyte progenitors; NeuP/NEP, neutrophil progenitors/precursors (myelocytes?); EoP, eosinophil progenitors (myelocytes); MMCs, immature neutrophils acting as multipotent myeloid cells; preEos, eosinophil precursors (metamyelocytes)
Neutrophil hematopoiesis
Neutrophils are the most abundant cells in normal bone marrow. In healthy human subjects, neutrophil lineage cells represent approximately 46–74% of the whole bone marrow cellularity (55, 56). Our bone marrow Ly6G(high) neutrophil estimates in naïve wild-type mice are 15–50% of all CD45(+) hematopoietic cells, which varies according to sex and age; neutrophils significantly increase in aged mice (14). Neutrophils develop from granulocyte-monocyte progenitors (GMPs), which in turn develop from common myeloid progenitors (CMPs). The current model of subsequent neutrophil development defines two major phases: (1) the proliferative stage, where GMPs develop into myeloblasts, promyelocytes, and myelocytes (all with mononuclear morphology) and (2) the non-proliferative stage where myelocytes develop into non-proliferating metamyelocytes, band-stage immature neutrophils, and mature neutrophils with characteristic ring-shaped and segmented nuclear morphology. Each developmental stage is characterized by distinct surface antigen marker changes (57–59), galectin expression (60), and changes in transcription factor expression (61). Different neutrophil differentiation stages also correspond to distinct stages of granulopoiesis and are characterized by expression of different granule proteins (61, 62). However, the defined differentiation pathways from GMP to terminally mature neutrophils are poorly understood (6, 63). In particular, the common progenitor specific to neutrophil lineage is not well defined (38, 59, 61, 64). Interestingly, there is also no specific inducer of neutrophil differentiation. Only granulocyte colony-stimulating factor (G-CSF) is used to culture bone marrow-derived neutrophils, which is a pan-granulocytic inducer (65).
Recent single-cell RNA-seq studies of neutrophil lineage differentiation report up to eight distinct populations of neutrophils in the bone marrow and blood of mice and humans, which acquire distinct expansion, trafficking, and effector functions as they mature (63, 66). Four other studies report identification of a neutrophil progenitor (preNeu, NeuP, or hNEP/eNEP), which seem to be committed and unipotent early- and late-stage neutrophil progenitors as shown by adoptive transfer experiments and IdU labeling (63, 64, 67, 68). However, despite differences in nomenclature, these studies seem to describe overlapping populations (38) and do not cross-reference these cells with the known neutrophil promyelocyte and myelocyte stages described earlier. Importantly, all recent studies agree that bone marrow medullary neutrophil subsets can be uniquely defined by specific phenotypic, proliferative, transcriptional, and functional characteristics. The consensus model of bone marrow neutrophil heterogeneity (which summarizes all single-cell studies) describes at least three critical pools at steady state: (1) a pre-neutrophil promyelocyte/myelocyte population as a proliferative pool that can rapidly amplify neutrophil numbers on demand; (2) non-proliferative metamyelocyte immature/band-stage neutrophils that represent a reservoir of neutrophils and can be rapidly deployed to circulation; and (3) mature neutrophils that can have effector functions (38). In the steady state, only mature neutrophils are detected in peripheral blood circulation, although immature neutrophils are also released in circulation during inflammation (63).
Interestingly, examples of murine neutrophils acquiring characteristics of other myeloid lineages (summarized in the section above) typically describe a Ly6G(+) cell with well-defined ring-like nuclear morphology, which suggests that these cells are non-proliferative metamyelocytes that belong to band-stage/immature pools of neutrophils. For example, a study describing the transdifferentiation of neutrophils to monocytes/macrophages specifies that these are band-stage neutrophils (31). Our study also shows that Siglec-F(-)Ly6G(+)IL-5Rα(+) murine bone marrow neutrophils that can transdifferentiate to eosinophils and monocytes are band-stage cells based on ring-like nuclear morphology (14). We confirmed that, despite their plasticity potential, these cells were not proliferative in culture. Based on their non-proliferative nature and ability to contribute to more than lineage, we called this neutrophil phenotype “multipotent myeloid cells” (MMCs). A significant implication of these findings is that immature neutrophils that can be rapidly deployed into circulation represent not only an emergency reservoir of neutrophils but also other myeloid cells as well. Such cells can quickly adapt to local microenvironmental or cytokine milieus in target tissues and differentially contribute to inflammation based on their new functional specialization. It remains to be seen whether or not proliferative cells in earlier stages of neutrophil differentiation exhibit similar developmental plasticity and can potentially contribute to inflammatory processes outside of bone marrow.
Eosinophil hematopoiesis
Unlike neutrophils, single-cell RNA-seq studies of eosinophil developmental trajectories are currently lacking. This is likely complicated by technical hurdles, since eosinophils are less numerous than neutrophils, have high RNAse content, and are hard to maintain outside of tissue environments. However, the currently known eosinophil developmental scheme closely follows stages of differentiation typical for neutrophils. Eosinophil progenitors (EoPs) are derived from GMP/CMP common myeloid progenitors (Lin(-)Il7rα(-)c-Kit(+)Sca-1(+/-)CD34(+)CD16/32(+/-)). EoPs are described as Lin(-)CD34(+)IL-5Rα(+) myelocytes with a mononuclear appearance and are the earliest eosinophil committed cell expressing IL-5Rα described in literature in mice and humans (69–71). In studies of in situ hematopoiesis, EoPs are the only developmental subset during allergic inflammation considered to traffic from bone marrow and further differentiate in lung tissue (72–74). A recent study (75) identified protein kinase Tribbles homolog 1 (Trib1) as a key factor regulating eosinophil identity in the post-EoP stage. Trib1 deletion resulted in Ly6G(+) eosinophils with neutrophilic characteristics and functions, pointing to a very late divergence between eosinophils and neutrophils. Such examples of mixed neutrophil-eosinophil phenotypes with mixed markers and granular content are abundant in literature (25, 45–47, 76, 77), which shows the insufficiency of our knowledge of neutrophil and eosinophil divergence. Eosinophil precursors (PreEos, EoPre) are metamyelocytes that express both Siglec-F and Ly6G; they are also developing mature eosinophil granular content. A Siglec-F(+)Ly6G(+) eosinophil subset was described by recent studies (45, 46, 78) and is especially prevalent during differentiation and inflammation due to its transient nature. Our recent study (14) demonstrated that Siglec-F(+)Ly6G(+) eosinophils can be differentiated from Ly6G(+)IL-5Rα(+) neutrophils, which shows that subsets of eosinophils may have developmental origins outside of eosinophil lineage. Interestingly, G-CSF is also sufficient to generate this eosinophil phenotype from neutrophils, which bypasses the need for IL-5 stimulation (14). At the tissue level, we showed that extracellular matrix constituents, such as Tenascin C, are also sufficient to promote eosinophil developmental phenotypes not dependent on IL-5 (79). At the granular formation checkpoint, PreEos express markers Epx, Mbp (Prg2), Ikzf3, and Xbp1 and further undergo full granular maturation to become mature red-staining eosinophils (Eos) with a Siglec-F(+)Ccr3(+)Ly6G(-) phenotype (80, 81). Mature eosinophils can assume multiple phenotypes due to their differential activation by cytokines and localization in different tissue compartments (41). We summarized all emerging markers and nomenclature of mature eosinophil heterogeneity in our recent review (5). At least three different eosinophil subsets can be found in lung tissue (5). It is plausible that tissue eosinophil subsets in homeostasis and inflammation exist on the spectrum of lineage commitment and differentiation, with potentially distinct myeloid origins. Currently, there is a significant gap in our understanding of developmental trajectories, tissue origins, proliferative capacities, and functional differences between different eosinophil subsets. Development of single-cell sequencing approaches, lineage tracing in vivo, and manipulation in mouse models will be necessary to carefully map the heterogeneity of eosinophil granulocytes.
Conclusions
Myeloid cell subsets are shaped by and uniquely adapt to their immediate tissue environments. Both microenvironmental plasticity and hematopoietic flexibility may be prerequisites to the rapid functional adaptation of neutrophil and eosinophil subsets to diverse biological stimuli in health and disease. From an evolutionary standpoint, the plasticity and ability of immature neutrophils to contribute to mature pools of eosinophils, monocytes, and dendritic cells may represent a winning emergency response strategy to quickly mobilize very large numbers of functionally flexible cells when and where they are needed most. Such myeloid plasticity also bears significant implications for the development and application of biologics (mepolizumab, benralizumab, reslizumab) targeting the IL-5 axis of eosinophil development and activation. For example, targeting mature eosinophils may potentiate “neutrophil-like” phenotypes which do not necessitate IL-5 stimulation or promote flexibility of developmental trajectories originating from outside of eosinophil lineage. Currently, we know very little about which myeloid developmental trajectories are suppressed or promoted by biologic therapies. This is even more actual today in light of studies showing that severe COVID-19 infection is characterized by a dysregulated myeloid cell compartment (82). Both on the basic science and clinical fronts, new next-generation approaches in conjunction with careful phenotyping are necessary to elucidate the biology of myeloid diversification and unravel the true face of inflammation.
Author contributions
Corresponding author outlined and structured the concepts in this review, wrote and edited the manuscript, and designed and prepared figures. Final version of the manuscript was approved by S.B.
Funding
This work was supported by the National Institutes of Health (NIH/NIAID) grant R01AI127783 to Dr. Berdnikovs. Additionally, this study was supported by the Ernest S. Bazley Foundation.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Summary sentence
Both developmental flexibility of immature myeloid populations and adaptations to different tissue microenvironments drive heterogeneity and plasticity of neutrophil and eosinophil subsets.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Giese MA, Hind LE, Huttenlocher A. Neutrophil plasticity in the tumor microenvironment. Blood. 2019;133:2159–2167. doi: 10.1182/blood-2018-11-844548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong CW. Current Understanding in neutrophil differentiation and heterogeneity. Immune Netw. 2017;17:298–306. doi: 10.4110/in.2017.17.5.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JJ, Jacobsen EA, McGarry MP, Schleimer RP, Lee NA. Eosinophils in health and disease: the LIAR hypothesis. Clin Exp Allergy. 2010;40:563–575. doi: 10.1111/j.1365-2222.2010.03484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bugl S, Wirths S, Muller MR, Radsak MP, Kopp HG. Current insights into neutrophil homeostasis. Ann N Y Acad Sci. 2012;1266:171–178. doi: 10.1111/j.1749-6632.2012.06607.x. [DOI] [PubMed] [Google Scholar]
- 5.Abdala-Valencia H, Coden ME, Chiarella SE, Jacobsen EA, Bochner BS, Lee JJ, Berdnikovs S. Shaping eosinophil identity in the tissue contexts of development, homeostasis, and disease. J Leukoc Biol. 2018;104:95–108. doi: 10.1002/JLB.1MR1117-442RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hidalgo A, Chilvers ER, Summers C, Koenderman L. The neutrophil life cycle. Trends Immunol. 2019;40:584–597. doi: 10.1016/j.it.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Dancey JT, Deubelbeiss KA, Harker LA, Finch CA. Neutrophil kinetics in man. J Clin Invest. 1976;58:705–715. doi: 10.1172/JCI108517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillay J, den Braber I, Vrisekoop N, Kwast LM, de Boer RJ, Borghans JA, Tesselaar K, Koenderman L. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 2010;116:625–627. doi: 10.1182/blood-2010-01-259028. [DOI] [PubMed] [Google Scholar]
- 9.Walmsley SR, Print C, Farahi N, Peyssonnaux C, Johnson RS, Cramer T, Sobolewski A, Condliffe AM, Cowburn AS, Johnson N, Chilvers ER. Hypoxia-induced neutrophil survival is mediated by HIF-1alpha-dependent NF-kappaB activity. J Exp Med. 2005;201:105–115. doi: 10.1084/jem.20040624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox S, Leitch AE, Duffin R, Haslett C, Rossi AG. Neutrophil apoptosis: relevance to the innate immune response and inflammatory disease. J Innate Immun. 2010;2:216–227. doi: 10.1159/000284367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Filippo K, Rankin SM. The secretive life of neutrophils revealed by intravital microscopy. Front Cell Dev Biol. 2020;8:603230. doi: 10.3389/fcell.2020.603230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorski SA, Lawrence MG, Hinkelman A, Spano MM, Steinke JW, Borish L, Teague WG, Braciale TJ. Expression of IL-5 receptor alpha by murine and human lung neutrophils. PLoS One. 2019;14:e0221113. doi: 10.1371/journal.pone.0221113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mesnil C, Raulier S, Paulissen G, Xiao X, Birrell MA, Pirottin D, Janss T, Starkl P, Ramery E, Henket M, Schleich FN, Radermecker M, Thielemans K, Gillet L, Thiry M, Belvisi MG, Louis R, Desmet C, Marichal T, Bureau F. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest. 2016;126:3279–3295. doi: 10.1172/JCI85664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong BM, Walker MT, Rodriguez R, Coden ME, Nagasaka R, Doan TC, Politanska Y, Abdala-Valencia H, Berdnikovs S. 2021. More than neutrophils: Lin(+)Ly6G(+)IL-5Ralpha(+) multipotent myeloid cells (MMCs) are dominant in normal murine bone marrow and retain capacity to differentiate into eosinophils and monocytes. J Leukoc Biol [DOI] [PMC free article] [PubMed]
- 15.Pfirschke C, Engblom C, Gungabeesoon J, Lin Y, Rickelt S, Zilionis R, Messemaker M, Siwicki M, Gerhard GM, Kohl A, Meylan E, Weissleder R, Klein AM, Pittet MJ. Tumor-promoting Ly-6G(+) SiglecF(high) cells are mature and long-lived neutrophils. Cell Rep. 2020;32:108164. doi: 10.1016/j.celrep.2020.108164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calcagno DM, Zhang C, Toomu A, Huang K, Ninh VK, Miyamoto S, Aguirre AD, Fu Z, Heller Brown J, King KR. SiglecF(HI) Marks late-stage neutrophils of the infarcted heart: a single-cell transcriptomic analysis of neutrophil diversification. J Am Heart Assoc. 2021;10:e019019. doi: 10.1161/JAHA.120.019019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa K, Asano K, Yotsumoto S, Yamane T, Arita M, Hayashi Y, Harada H, Makino-Okamura C, Fukuyama H, Kondo K, Yamasoba T, Tanaka M. Frontline science: conversion of neutrophils into atypical Ly6G(+) SiglecF(+) immune cells with neurosupportive potential in olfactory neuroepithelium. J Leukoc Biol. 2021;109:481–496. doi: 10.1002/JLB.1HI0620-190RR. [DOI] [PubMed] [Google Scholar]
- 18.Matsui M, Nagakubo D, Satooka H, Hirata T. A novel Siglec-F(+) neutrophil subset in the mouse nasal mucosa exhibits an activated phenotype and is increased in an allergic rhinitis model. Biochem Biophys Res Commun. 2020;526:599–606. doi: 10.1016/j.bbrc.2020.03.122. [DOI] [PubMed] [Google Scholar]
- 19.Rudd JM, Pulavendran S, Ashar HK, Ritchey JW, Snider TA, Malayer JR, Marie M, Chow VTK, Narasaraju T. Neutrophils induce a novel chemokine receptors repertoire during influenza pneumonia. Front Cell Infect Microbiol. 2019;9:108. doi: 10.3389/fcimb.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartl D, Krauss-Etschmann S, Koller B, Hordijk PL, Kuijpers TW, Hoffmann F, Hector A, Eber E, Marcos V, Bittmann I, Eickelberg O, Griese M, Roos D. Infiltrated neutrophils acquire novel chemokine receptor expression and chemokine responsiveness in chronic inflammatory lung diseases. J Immunol. 2008;181:8053–8067. doi: 10.4049/jimmunol.181.11.8053. [DOI] [PubMed] [Google Scholar]
- 21.Bekaert S, Rocks N, Vanwinge C, Noel A, Cataldo D. Asthma-related inflammation promotes lung metastasis of breast cancer cells through CCL11-CCR3 pathway. Respir Res. 2021;22:61. doi: 10.1186/s12931-021-01652-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huaux F, Gharaee-Kermani M, Liu T, Morel V, McGarry B, Ullenbruch M, Kunkel SL, Wang J, Xing Z, Phan SH. Role of Eotaxin-1 (CCL11) and CC chemokine receptor 3 (CCR3) in bleomycin-induced lung injury and fibrosis. Am J Pathol. 2005;167:1485–1496. doi: 10.1016/S0002-9440(10)61235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menzies-Gow A, Ying S, Sabroe I, Stubbs VL, Soler D, Williams TJ, Kay AB. Eotaxin (CCL11) and eotaxin-2 (CCL24) induce recruitment of eosinophils, basophils, neutrophils, and macrophages as well as features of early- and late-phase allergic reactions following cutaneous injection in human atopic and nonatopic volunteers. J Immunol. 2002;169:2712–2718. doi: 10.4049/jimmunol.169.5.2712. [DOI] [PubMed] [Google Scholar]
- 24.Abu-Ghazaleh RI, Dunnette SL, Loegering DA, Checkel JL, Kita H, Thomas LL, Gleich GJ. Eosinophil granule proteins in peripheral blood granulocytes. J Leukoc Biol. 1992;52:611–618. doi: 10.1002/jlb.52.6.611. [DOI] [PubMed] [Google Scholar]
- 25.Monteseirin J, Vega A, Chacon P, Camacho MJ, El Bekay R, Asturias JA, Martinez A, Guardia P, Perez-Cano R, Conde J. Neutrophils as a novel source of eosinophil cationic protein in IgE-mediated processes. J Immunol. 2007;179:2634–2641. doi: 10.4049/jimmunol.179.4.2634. [DOI] [PubMed] [Google Scholar]
- 26.Yu WT, Wang Y, Zhou Y, Zhu Y, Chen S, Wang YJ, Kong WJ, Chen JJ. Expression of eosinophilic cationic protein in neutrophils and its effect on objective evaluation of nasal inflammation. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2019;33:581–585. doi: 10.13201/j.issn.1001-1781.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Matsushima H, Geng S, Lu R, Okamoto T, Yao Y, Mayuzumi N, Kotol PF, Chojnacki BJ, Miyazaki T, Gallo RL, Takashima A. Neutrophil differentiation into a unique hybrid population exhibiting dual phenotype and functionality of neutrophils and dendritic cells. Blood. 2013;121:1677–1689. doi: 10.1182/blood-2012-07-445189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geng S, Matsushima H, Okamoto T, Yao Y, Lu R, Page K, Blumenthal RM, Ward NL, Miyazaki T, Takashima A. Emergence, origin, and function of neutrophil-dendritic cell hybrids in experimentally induced inflammatory lesions in mice. Blood. 2013;121:1690–1700. doi: 10.1182/blood-2012-07-445197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fites JS, Gui M, Kernien JF, Negoro P, Dagher Z, Sykes DB, Nett JE, Mansour MK, Klein BS. An unappreciated role for neutrophil-DC hybrids in immunity to invasive fungal infections. PLoS Pathog. 2018;14:e1007073. doi: 10.1371/journal.ppat.1007073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guilliams M, Mildner A, Yona S. Developmental and functional heterogeneity of monocytes. Immunity. 2018;49:595–613. doi: 10.1016/j.immuni.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Koffel R, Meshcheryakova A, Warszawska J, Hennig A, Wagner K, Jorgl A, Gubi D, Moser D, Hladik A, Hoffmann U, Fischer MB, van den Berg W, Koenders M, Scheinecker C, Gesslbauer B, Knapp S, Strobl H. Monocytic cell differentiation from band-stage neutrophils under inflammatory conditions via MKK6 activation. Blood. 2014;124:2713–2724. doi: 10.1182/blood-2014-07-588178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding ZC, Aboelella NS, Bryan L, Shi H, Zhou G. The Monocytes That Repopulate in Mice After Cyclophosphamide Treatment Acquire a Neutrophil Precursor Gene Signature and Immunosuppressive Activity. Front Immunol. 2020;11:594540. doi: 10.3389/fimmu.2020.594540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veglia F, Hashimoto A, Dweep H, Sanseviero E, De Leo A, Tcyganov E, Kossenkov A, Mulligan C, Nam B, Masters G, Patel J, Bhargava V, Wilkinson P, Smirnov D, Sepulveda MA, Singhal S, Eruslanov EB, Cristescu R, Loboda A, Nefedova Y, Gabrilovich DI 2021 Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. J Exp Med 218 [DOI] [PMC free article] [PubMed]
- 34.Pillay J, Tak T, Kamp VM, Koenderman L. Immune suppression by neutrophils and granulocytic myeloid-derived suppressor cells: similarities and differences. Cell Mol Life Sci. 2013;70:3813–3827. doi: 10.1007/s00018-013-1286-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millrud CR, Bergenfelz C, Leandersson K. On the origin of myeloid-derived suppressor cells. Oncotarget. 2017;8:3649–3665. doi: 10.18632/oncotarget.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Negorev D, Beier UH, Zhang T, Quatromoni JG, Bhojnagarwala P, Albelda SM, Singhal S, Eruslanov E, Lohoff FW, Levine MH, Diamond JM, Christie JD, Hancock WW, Akimova T. Human neutrophils can mimic myeloid-derived suppressor cells (PMN-MDSC) and suppress microbead or lectin-induced T cell proliferation through artefactual mechanisms. Sci Rep. 2018;8:3135. doi: 10.1038/s41598-018-21450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silvestre-Roig C, Fridlender ZG, Glogauer M, Scapini P. Neutrophil diversity in health and disease. Trends Immunol. 2019;40:565–583. doi: 10.1016/j.it.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng LG, Ostuni R, Hidalgo A. Heterogeneity of neutrophils. Nat Rev Immunol. 2019;19:255–265. doi: 10.1038/s41577-019-0141-8. [DOI] [PubMed] [Google Scholar]
- 39.Ballesteros I, Rubio-Ponce A, Genua M, Lusito E, Kwok I, Fernandez-Calvo G, Khoyratty TE, van Grinsven E, Gonzalez-Hernandez S, Nicolas-Avila JA, Vicanolo T, Maccataio A, Benguria A, Li JL, Adrover JM, Aroca-Crevillen A, Quintana JA, Martin-Salamanca S, Mayo F, Ascher S, Barbiera G, Soehnlein O, Gunzer M, Ginhoux F, Sanchez-Cabo F, Nistal-Villan E, Schulz C, Dopazo A, Reinhardt C, Udalova IA, Ng LG, Ostuni R, Hidalgo A. Co-option of Neutrophil Fates by Tissue Environments. Cell. 2020;183:1282–97 e18. doi: 10.1016/j.cell.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Gigon L, Yousefi S, Karaulov A, Simon HU. Mechanisms of toxicity mediated by neutrophil and eosinophil granule proteins. Allergol Int. 2021;70:30–38. doi: 10.1016/j.alit.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Abdala Valencia H, Loffredo LF, Misharin AV, Berdnikovs S. Phenotypic plasticity and targeting of Siglec-F(high) CD11c(low) eosinophils to the airway in a murine model of asthma. Allergy. 2016;71:267–271. doi: 10.1111/all.12776. [DOI] [PubMed] [Google Scholar]
- 42.Rothenberg ME. A hidden residential cell in the lung. J Clin Invest. 2016;126:3185–3187. doi: 10.1172/JCI89768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreev D, Liu M, Kachler K, Llerins Perez M, Kirchner P, Kolle J, Giessl A, Rauber S, Song R, Aust O, Gruneboom A, Kleyer A, Canete JD, Ekici A, Ramming A, Finotto S, Schett G, Bozec A 2020 Regulatory eosinophils induce the resolution of experimental arthritis and appear in remission state of human rheumatoid arthritis. Ann Rheum Dis [DOI] [PubMed]
- 44.Lacy P. Gr1 makes an unexpected cameo appearance in eosinophils. J Leukoc Biol. 2020;107:363–365. doi: 10.1002/JLB.2CE1119-473R. [DOI] [PubMed] [Google Scholar]
- 45.Percopo CM, Brenner TA, Ma M, Kraemer LS, Hakeem RM, Lee JJ, Rosenberg HF. SiglecF+Gr1hi eosinophils are a distinct subpopulation within the lungs of allergen-challenged mice. J Leukoc Biol. 2017;101:321–328. doi: 10.1189/jlb.3A0416-166R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Limkar AR, Mai E, Sek AC, Percopo CM, Rosenberg HF. Frontline Science: Cytokine-mediated developmental phenotype of mouse eosinophils: IL-5-associated expression of the Ly6G/Gr1 surface Ag. J Leukoc Biol. 2020;107:367–377. doi: 10.1002/JLB.1HI1019-116RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim K, Hwang SM, Kim SM, Park SW, Jung Y, Chung IY. Terminally differentiating eosinophils express neutrophil primary granule proteins as well as eosinophil-specific granule proteins in a temporal manner. Immune Netw. 2017;17:410–423. doi: 10.4110/in.2017.17.6.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuwirth A, Dobes J, Oujezdska J, Ballek O, Benesova M, Sumnik Z, Vcelakova J, Kolouskova S, Obermannova B, Kolar M, Stechova K, Filipp D. Eosinophils from patients with type 1 diabetes mellitus express high level of myeloid alpha-defensins and myeloperoxidase. Cell Immunol. 2012;273:158–163. doi: 10.1016/j.cellimm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Esnault S, Kelly EA, Johnson SH, DeLain LP, Haedt MJ, Noll AL, Sandbo N, Jarjour NN. Matrix metalloproteinase-9-dependent release of IL-1beta by human eosinophils. Mediat Inflamm. 2019;2019:7479107. doi: 10.1155/2019/7479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orfao A, Matarraz S, Perez-Andres M, Almeida J, Teodosio C, Berkowska MA, van Dongen JJM, EuroFlow Immunophenotypic dissection of normal hematopoiesis. J Immunol Methods. 2019;475:112684. doi: 10.1016/j.jim.2019.112684. [DOI] [PubMed] [Google Scholar]
- 51.Cheng H, Zheng Z, Cheng T. New paradigms on hematopoietic stem cell differentiation. Protein Cell. 2020;11:34–44. doi: 10.1007/s13238-019-0633-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karamitros D, Stoilova B, Aboukhalil Z, Hamey F, Reinisch A, Samitsch M, Quek L, Otto G, Repapi E, Doondeea J, Usukhbayar B, Calvo J, Taylor S, Goardon N, Six E, Pflumio F, Porcher C, Majeti R, Gottgens B, Vyas P. Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells. Nat Immunol. 2018;19:85–97. doi: 10.1038/s41590-017-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, Hirche C, Lutz C, Buss EC, Nowak D, Boch T, Hofmann WK, Ho AD, Huber W, Trumpp A, Essers MA, Steinmetz LM. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol. 2017;19:271–281. doi: 10.1038/ncb3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Macaulay IC, Svensson V, Labalette C, Ferreira L, Hamey F, Voet T, Teichmann SA, Cvejic A. Single-cell RNA-sequencing reveals a continuous spectrum of differentiation in hematopoietic cells. Cell Rep. 2016;14:966–977. doi: 10.1016/j.celrep.2015.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matarraz S, Lopez A, Barrena S, Fernandez C, Jensen E, Flores J, Barcena P, Rasillo A, Sayagues JM, Sanchez ML, Hernandez-Campo P, Hernandez Rivas JM, Salvador C, Fernandez-Mosteirin N, Giralt M, Perdiguer L, Orfao A. The immunophenotype of different immature, myeloid and B-cell lineage-committed CD34+ hematopoietic cells allows discrimination between normal/reactive and myelodysplastic syndrome precursors. Leukemia. 2008;22:1175–1183. doi: 10.1038/leu.2008.49. [DOI] [PubMed] [Google Scholar]
- 56.Matarraz S, Lopez A, Barrena S, Fernandez C, Jensen E, Flores-Montero J, Rasillo A, Sayagues JM, Sanchez ML, Barcena P, Hernandez-Rivas JM, Salvador C, Fernandez-Mosteirin N, Giralt M, Perdiguer L, Laranjeira P, Paiva A, Orfao A. Bone marrow cells from myelodysplastic syndromes show altered immunophenotypic profiles that may contribute to the diagnosis and prognostic stratification of the disease: a pilot study on a series of 56 patients. Cytometry B Clin Cytom. 2010;78:154–168. doi: 10.1002/cyto.b.20513. [DOI] [PubMed] [Google Scholar]
- 57.Elghetany MT. Surface antigen changes during normal neutrophilic development: a critical review. Blood Cells Mol Dis. 2002;28:260–274. doi: 10.1006/bcmd.2002.0513. [DOI] [PubMed] [Google Scholar]
- 58.Elghetany MT, Ge Y, Patel J, Martinez J, Uhrova H. Flow cytometric study of neutrophilic granulopoiesis in normal bone marrow using an expanded panel of antibodies: correlation with morphologic assessments. J Clin Lab Anal. 2004;18:36–41. doi: 10.1002/jcla.20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yvan-Charvet L, Ng LG. Granulopoiesis and neutrophil homeostasis: a metabolic, daily balancing act. Trends Immunol. 2019;40:598–612. doi: 10.1016/j.it.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 60.Abedin MJ, Kashio Y, Seki M, Nakamura K, Hirashima M. Potential roles of galectins in myeloid differentiation into three different lineages. J Leukoc Biol. 2003;73:650–656. doi: 10.1189/jlb.0402163. [DOI] [PubMed] [Google Scholar]
- 61.Lawrence SM, Corriden R, Nizet V (2018) The ontogeny of a neutrophil: mechanisms of granulopoiesis and homeostasis. Microbiol Mol Biol Rev 82 [DOI] [PMC free article] [PubMed]
- 62.Benarafa C, Simon HU. Role of granule proteases in the life and death of neutrophils. Biochem Biophys Res Commun. 2017;482:473–481. doi: 10.1016/j.bbrc.2016.11.086. [DOI] [PubMed] [Google Scholar]
- 63.Evrard M, Kwok IWH, Chong SZ, Teng KWW, Becht E, Chen J, Sieow JL, Penny HL, Ching GC, Devi S, Adrover JM, Li JLY, Liong KH, Tan L, Poon Z, Foo S, Chua JW, Su IH, Balabanian K, Bachelerie F, Biswas SK, Larbi A, Hwang WYK, Madan V, Koeffler HP, Wong SC, Newell EW, Hidalgo A, Ginhoux F, Ng LG. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity. 2018;48:364–79 e8. doi: 10.1016/j.immuni.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 64.Zhu YP, Padgett L, Dinh HQ, Marcovecchio P, Blatchley A, Wu R, Ehinger E, Kim C, Mikulski Z, Seumois G, Madrigal A, Vijayanand P, Hedrick CC. Identification of an early unipotent neutrophil progenitor with pro-tumoral activity in mouse and human bone marrow. Cell Rep. 2018;24:2329–41 e8. doi: 10.1016/j.celrep.2018.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Basu S, Hodgson G, Katz M, Dunn AR. Evaluation of role of G-CSF in the production, survival, and release of neutrophils from bone marrow into circulation. Blood. 2002;100:854–861. doi: 10.1182/blood.V100.3.854. [DOI] [PubMed] [Google Scholar]
- 66.Xie X, Shi Q, Wu P, Zhang X, Kambara H, Su J, Yu H, Park SY, Guo R, Ren Q, Zhang S, Xu Y, Silberstein LE, Cheng T, Ma F, Li C, Luo HR. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat Immunol. 2020;21:1119–1133. doi: 10.1038/s41590-020-0736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim MH, Yang D, Kim M, Kim SY, Kim D, Kang SJ. A late-lineage murine neutrophil precursor population exhibits dynamic changes during demand-adapted granulopoiesis. Sci Rep. 2017;7:39804. doi: 10.1038/srep39804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dinh HQ, Eggert T, Meyer MA, Zhu YP, Olingy CE, Llewellyn R, Wu R, Hedrick CC. Coexpression of CD71 and CD117 Identifies an early unipotent neutrophil progenitor population in human bone marrow. Immunity. 2020;53:319–34 e6. doi: 10.1016/j.immuni.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnston LK, Bryce PJ. Understanding interleukin 33 and its roles in eosinophil development. Front Med (Lausanne) 2017;4:51. doi: 10.3389/fmed.2017.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, Gurish MF, Takatsu K, Akashi K. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med. 2005;201:1891–1897. doi: 10.1084/jem.20050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mori Y, Iwasaki H, Kohno K, Yoshimoto G, Kikushige Y, Okeda A, Uike N, Niiro H, Takenaka K, Nagafuji K, Miyamoto T, Harada M, Takatsu K, Akashi K. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. J Exp Med. 2009;206:183–193. doi: 10.1084/jem.20081756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sehmi R, Smith SG, Kjarsgaard M, Radford K, Boulet LP, Lemiere C, Prazma CM, Ortega H, Martin JG, Nair P. Role of local eosinophilopoietic processes in the development of airway eosinophilia in prednisone-dependent severe asthma. Clin Exp Allergy. 2016;46:793–802. doi: 10.1111/cea.12695. [DOI] [PubMed] [Google Scholar]
- 73.Radinger M, Bossios A, Sjostrand M, Lu Y, Malmhall C, Dahlborn AK, Lee JJ, Lotvall J. Local proliferation and mobilization of CCR3(+) CD34(+) eosinophil-lineage-committed cells in the lung. Immunology. 2011;132:144–154. doi: 10.1111/j.1365-2567.2010.03349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Southam DS, Widmer N, Ellis R, Hirota JA, Inman MD, Sehmi R. Increased eosinophil-lineage committed progenitors in the lung of allergen-challenged mice. J Allergy Clin Immunol. 2005;115:95–102. doi: 10.1016/j.jaci.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 75.Mack EA, Stein SJ, Rome KS, Xu L, Wertheim GB, Melo RCN, Pear WS. Trib1 regulates eosinophil lineage commitment and identity by restraining the neutrophil program. Blood. 2019;133:2413–2426. doi: 10.1182/blood.2018872218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Engblom C, Pfirschke C, Zilionis R, Da Silva MJ, Bos SA, Courties G, Rickelt S, Severe N, Baryawno N, Faget J, Savova V, Zemmour D, Kline J, Siwicki M, Garris C, Pucci F, Liao HW, Lin YJ, Newton A, Yaghi OK, Iwamoto Y, Tricot B, Wojtkiewicz GR, Nahrendorf M, Cortez-Retamozo V, Meylan E, Hynes RO, Demay M, Klein A, Bredella MA, Scadden DT, Weissleder R, Pittet MJ. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecF(high) neutrophils. Science. 2017;358:eaal5081. doi: 10.1126/science.aal5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takashima A, Yao Y. Neutrophil plasticity: acquisition of phenotype and functionality of antigen-presenting cell. J Leukoc Biol. 2015;98:489–496. doi: 10.1189/jlb.1MR1014-502R. [DOI] [PubMed] [Google Scholar]
- 78.Johnston LK, Hsu CL, Krier-Burris RA, Chhiba KD, Chien KB, McKenzie A, Berdnikovs S, Bryce PJ. IL-33 Precedes IL-5 in regulating eosinophil commitment and is required for eosinophil homeostasis. J Immunol. 2016;197:3445–3453. doi: 10.4049/jimmunol.1600611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doan TC, Jeong BM, Coden ME, Loffredo LF, Bhattacharyya S, Chiarella SE, Varga J, Abdala-Valencia H, Berdnikovs S. Matrix protein tenascin-C expands and reversibly blocks maturation of murine eosinophil progenitors. J Allergy Clin Immunol. 2018;142:695–8 e4. doi: 10.1016/j.jaci.2018.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fulkerson PC, Rothenberg ME. Eosinophil development, disease involvement, and therapeutic suppression. Adv Immunol. 2018;138:1–34. doi: 10.1016/bs.ai.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 81.Bouffi C, Kartashov AV, Schollaert KL, Chen X, Bacon WC, Weirauch MT, Barski A, Fulkerson PC. Transcription factor repertoire of homeostatic eosinophilopoiesis. J Immunol. 2015;195:2683–2695. doi: 10.4049/jimmunol.1500510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schulte-Schrepping J, Reusch N, Paclik D, Bassler K, Schlickeiser S, Zhang B, Kramer B, Krammer T, Brumhard S, Bonaguro L, De Domenico E, Wendisch D, Grasshoff M, Kapellos TS, Beckstette M, Pecht T, Saglam A, Dietrich O, Mei HE, Schulz AR, Conrad C, Kunkel D, Vafadarnejad E, Xu CJ, Horne A, Herbert M, Drews A, Thibeault C, Pfeiffer M, Hippenstiel S, Hocke A, Muller-Redetzky H, Heim KM, Machleidt F, Uhrig A, Bosquillon de Jarcy L, Jurgens L, Stegemann M, Glosenkamp CR, Volk HD, Goffinet C, Landthaler M, Wyler E, Georg P, Schneider M, Dang-Heine C, Neuwinger N, Kappert K, Tauber R, Corman V, Raabe J, Kaiser KM, Vinh MT, Rieke G, Meisel C, Ulas T, Becker M, Geffers R, Witzenrath M, Drosten C, Suttorp N, von Kalle C, Kurth F, Handler K, Schultze JL, Aschenbrenner AC, Li Y, Nattermann J, Sawitzki B, Saliba AE, Sander LE, Deutsche C-OI. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–40 e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]