Abstract
More than 12,000 plant species (ca. 10% of flowering plants) exude latex when their tissues are injured. Latex is produced and stored in specialized cells named “laticifers”. Laticifers form a tubing system composed of rows of elongated cells that branch and create an internal network encompassing the entire plant. Laticifers constitute a recent evolutionary achievement in ecophysiological adaptation to specific natural environments; however, their fitness benefit to the plant still remains to be proven. The identification of Euphorbia lathyris mutants (pil mutants) deficient in laticifer cells or latex metabolism, and therefore compromised in latex production, allowed us to test the importance of laticifers in pest resistance. We provided genetic evidence indicating that laticifers represent a cellular adaptation for an essential defense strategy to fend off arthropod herbivores with different feeding habits, such as Spodoptera exigua and Tetranychus urticae. In marked contrast, we also discovered that a lack of laticifer cells causes complete resistance to the fungal pathogen Botrytis cinerea. Thereafter, a latex-derived factor required for conidia germination on the leaf surface was identified. This factor promoted disease susceptibility enhancement even in the non-latex-bearing plant Arabidopsis. We speculate on the role of laticifers in the co-evolutionary arms race between plants and their enemies.
Key words: latex, Euphorbia lathyris, disease resistance, disease susceptibility, plant defense
Laticifers are a specialized plant cell type responsible for latex biosynthesis and accumulation. Through the analysis of Euphorbia lathyris mutants deficient in laticifer cells and latex production, this study demonstrates that laticifer cells are essential for resistance to insect herbivores but, at the same time, constitute a source of disease susceptibility to fungal infection.
Introduction
Laticifers are highly specialized plant cells that grow continuously by elongation, becoming the longest cell type in plants and assembling into a branched tubing system within the plant body. Although it is common and extensively developed in thousands of different vascular plant species (Lewinsohn, 1991), the organization and function of this cellular system remain poorly understood. Laticifers produce and accumulate latex, a fluid that comprises the cytosol of the cell and contains a huge variety of substances in solution and colloidal suspension, including carbohydrates, organic acids, fats, proteins, mucilages, sterols, rubbers, and essential oils (Hagel et al., 2008). Latex is stored within the laticifers, generating turgor pressure in these cells. Because the laticiferous system defines a continuous cellular space along the plant body, latex oozes copiously in response to pre-existing turgor pressure whenever the plant is injured. Metabolites that originate in latex encompass valuable bioproducts such as natural rubber from the rubber tree (Hevea brasiliensis), opiates from the opium poppy (Papaver somniferum), papain (peptidase) from the papaya tree (Carica papaya), cardiac glycosides from milkweeds (Asclepias spp.), and phenolic glucosides from hemp (Cannabis sativa) (Esau, 1965). Each species is enriched in different latex molecules following species-specific patterns. Laticifer cell structures originate early in embryogenesis (Mahlberg, 1993; Castelblanque et al., 2016); they are widely distributed in almost all plant tissues and are especially abundant in leaves (Castelblanque et al., 2016). Laticifers are usually associated with vascular tissues, particularly the phloem, and this proximity may allow a direct transfer of transported nutrients to the laticifer, supplying its intense biosynthetic demands (Prado and Demarco, 2018). In an evolutionary context, laticifers are regarded as a recently evolved cell type. They have only been found in the angiosperm fossil record since the Eocene epoch, 50 million years ago (Mya), whereas angiosperm fossils originate in the Cretaceous period, 140 Mya (Mahlberg and Sabharwal, 1968; Lange, 2015). It is also generally admitted that laticifers have polyphyletic origins within vascular plants, having appeared several times in different clades by convergent evolution (Mahlberg and Sabharwal, 1968; Hagel et al., 2008; Ramos et al., 2019). Nevertheless, other studies have claimed that laticifer features are also consistent with divergent evolution in which laticifers existed in the last common ancestor of laticiferous clades but were lost in some species (Lange, 2015).
Regarding the function of laticifers, latex contains a wide array of secondary metabolites, but none of them enter the primary metabolism of the cell; for instance, latex starch does not function as an exploitable carbohydrate in the laticifers (Spilatro and Mahlberg, 1986). The main constituents of latex are specialized metabolites and defense proteins, and laticifers provide an adaptation for the storage and isolation of such compounds from the rest of the plant. Stored under pressure, only upon the physical rupture of laticifer tubular structures is latex released to the environment. Evidence for a defensive function of latex and its effect on insects abounds (Agrawal and Konno, 2009; Konno, 2011; Dussourd, 2017), but much less is known about its effects on pathogens. In the first instance, latex can function as a physical defense against insect herbivores because of entrapment in latex. The sticky rubber-like precursors contained in latex hamper insect performance by gluing their mouthparts or trapping their body parts while, at the same time, latex compounds facilitate rapid wound closure to prevent infection by pathogens (Konno, 2011). This machinery has been referred to as a “squirt gun” defense (Becerra and Venable, 1990), whereby latex travels quickly through the canal system of laticifers to provide a physical barrier to an herbivore. In the second instance, latex can function as a chemical defense because of the potential antibiotic effects of secondary metabolites stored in latex and the presence of other latex constituents such as hydrolytic enzymes and defense proteins, which may play defensive roles against intruders. Transcriptomic and proteomic analysis of latex from different plant species has revealed an abundance of defense proteins, including peptidases, chitinases, lectins, pathogenesis-related proteins, and protease inhibitors (Kitajima et al., 2012, 2016, 2018). However, information on latex proteins is lacking, and it is difficult to have a broad concept of their diversity and function, particularly if certain proteins are targeted at specific enemies or if they hold complementary activities against diverse attackers (Konno, 2011; Souza et al., 2011; Ramos et al., 2015; Freitas et al., 2016). In addition, recent evidence indicates that a latex secondary metabolite (i.e., the sesquiterpene lactone taraxinic acid β-D-glucopyranosyl ester) produced by dandelion (Taraxacum officinale agg.) decreases the performance of its major native insect root herbivore, Melolontha melolontha (Huber et al., 2016). Also, chemical profiling of Euphorbia peplus latex revealed an important array of diterpenoids and some triterpenoids. Interestingly, a high number of those diterpenoids showed potent antifeedant activity against a generalist plant-feeding insect, the cotton bollworm (Helicoverpa armigera), whereas the major E. peplus triterpenoid (peplusol) was not active against this insect. Conversely, this acyclic triterpene presented antifungal activity against pathogenic fungi (Rhizoctonia solani, Colletotrichum litchi, and Fusarium oxysporum), whereas E. peplus diterpenoids did not inhibit fungal growth (Hua et al., 2017). An extensively studied example is the production of cardenolides in latex from milkweed plants (Asclepias spp.). Cardenolides are a group of cardiac-active steroids, and up to 200 different structures have been reported, all sharing the same mode of action (inhibition of the common Na+/K+ ATPase in animal cells), making them toxic to a wide array of animals, including insects. This case has been studied especially in the context of chemical ecology and plant–herbivore co-evolution because cardenolide production has a genetic basis and is subjected to natural selection by herbivores (Agrawal, 2005; Rasmann et al., 2009; Zuest et al., 2019).
Although the importance of latex in plant defense against herbivores is recognized, the principles that orchestrate the integrated activity of different latex compounds against different primary consumers still require better description (Ramos et al., 2019). Also, it remains unclear whether the appearance of laticifers, as a recently evolved defense strategy, exerts positive (synergistic) or negative (trade-off) effects on previously evolved defenses. In fact, classical works in the field have reported various evidence for trade-offs between pathogen and herbivore resistance (Felton and Korth, 2000), although at the same time it has been recognized that a clear dichotomy between pathogen- and herbivore-specific defense pathways does not always exist, as multiple pathways are elicited during the attack by both types of organisms.
Conclusions regarding the role of laticifers in plant defense are based mostly on observations of how latex and its products affect the growth and performance of herbivores and pathogens, but no genetic evidence for such a definitive biological role is available. The Euphorbiaceae (spurge) family is one of the largest flowering plant families; it contains about 300 genera and nearly 8000 species, of which 4500 are latex-producing species (Lewinsohn, 1991). Recently, we characterized the laticiferous system of Euphorbia lathyris (caper spurge) and identified a laticifer-associated gene expression pattern (Castelblanque et al., 2016). A survey for E. lathyris mutants compromised in laticifer differentiation and latex production enabled the identification of pil (poor in latex) mutants (Castelblanque et al., 2016). These mutants revealed that laticifers are not essential for plant development and fitness, at least when plants are grown under controlled conditions. The availability of pil mutants offers a unique opportunity to acquire new information on the role of laticifers, as a complex plant trait, in host defense. Here we provide evidence for the importance of laticifers in mediating resistance to insect herbivores and, concurrently, disease susceptibility to fungal infection.
Results
Severe susceptibility of pil mutants to spodoptera exigua
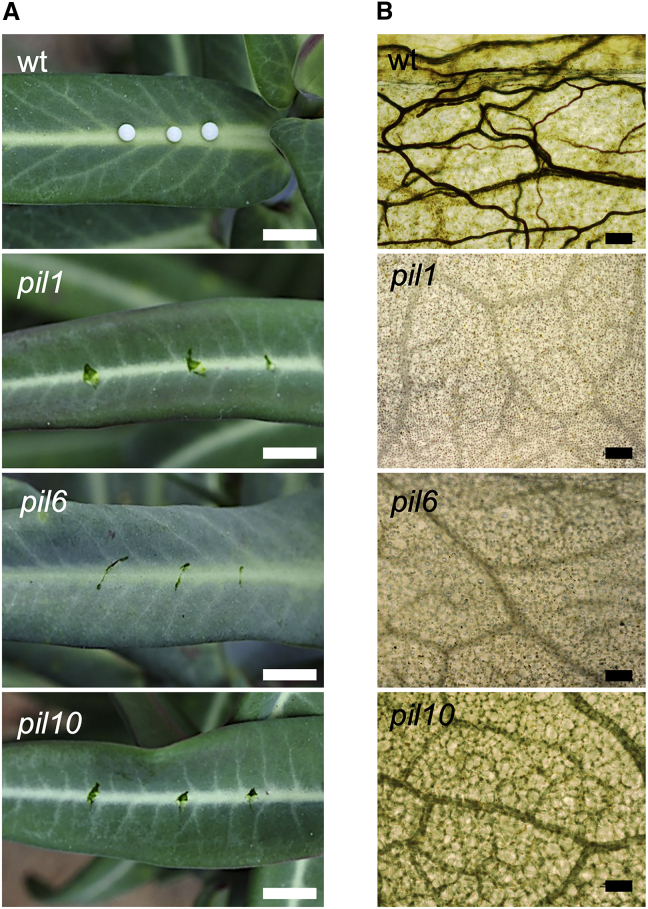
Three non-allelic recessive E. lathyris pil mutants impaired in latex production, pil1, pil6, and pil10 (Figure 1A), were used throughout this study. pil1 and pil6 mutants are defective in laticifer cell elongation (Castelblanque et al., 2016) and lack the characteristic cellular network of tubular laticifer cell structures observed in wild-type leaves upon whole-mount staining with Sudan black (Figure 1B), a colorant that selectively detects latex constituents (Castelblanque et al., 2016). pil10 is a metabolic mutant in which the differentiation and growth of laticifers is not affected but latex biosynthesis and accumulation are impaired. Therefore, pil10 plants do not stain with Sudan black (Figure 1B) despite the presence of latex-devoid laticifer cells and a laticifer cell population density similar to that observed in wild-type plants (Castelblanque et al., 2016).
Figure 1.
E. lathyris mutants used in this study.
(A) Comparison of latex oozing after pricking leaves from wild-type, pil1, pil6, and pil10 plants. Scale bars represent 1 cm.
(B) Whole-mount Sudan black B staining of leaf blade sectors showing the staining of laticifer cells in wild-type plants and its absence in pil1, pil6, and pil10 mutants. Scale bars represent 100 μm.
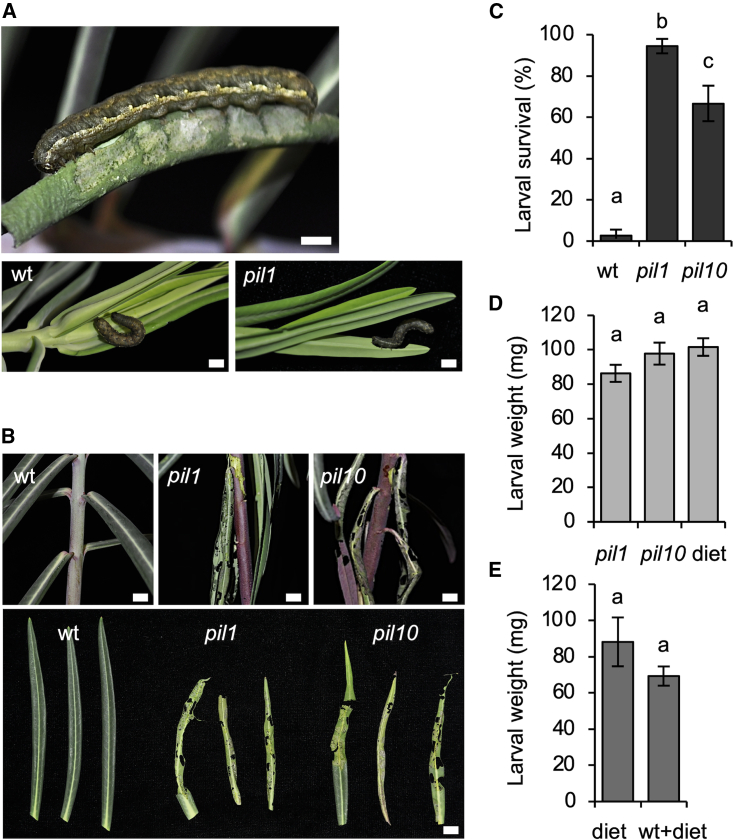
To address the importance of laticifer cells and latex production to defense against arthropod herbivores, we first studied the performance of larvae of Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae), a polyphagous insect with typical chewing mouthparts (Figure 2A). E. lathyris wild-type plants and the latex-deficient pil1 and pil10 mutants were infested with a fixed number of fourth instar larvae (LIV) in climatic chambers, and growth and performance of the larvae, as well as consequences for the host plant, were monitored. Results (Figure 2B) indicated that wild-type plants completely resisted S. exigua, and the plants showed no symptoms of insect feeding. Moreover, none of the S. exigua LIV larvae survived to the end of the experiments carried out with wild-type plants (Figure 2C). Notably, when S. exigua was assayed on the E. lathyris pil1 and pil10 mutants, both of which lack latex production, plants were severely damaged and vastly consumed by S. exigua LIV (Figure 2B). Therefore, the two latex-defective mutants proved to be an excellent food source for S. exigua larvae: most larvae survived to the end of the experiments, in marked contrast to observations on wild-type plants (Figure 2C). In addition, larval weight at the end of the experiment was the same as that of larvae grown on artificial diet (Figure 2D), indicating that the two pil mutants do not contain secondary metabolites with antibiotic properties. Moreover, the larval weights of specimens fed on E. lathyris wild-type plants supplemented with artificial diet or on artificial diet alone did not differ (Figure 2E). This suggests that resistance of wild-type plants against this herbivore should be attributed to antixenosis (Kogan and Ortman, 1978) rather than antibiosis.
Figure 2.
Response of latex- and laticifer-defective E. lathyris plants to infestation by S. exigua.
(A) LIV larvae of S. exigua on E. lathyris wild-type and pil1 plants. Scale bars represent 1 cm.
(B) Feeding injury caused by S. exigua LIV on E. lathyris pil1 and pil10 mutants and lack of damage on wild-type plants. Scale bars represent 1 cm.
(C) Survival of LIV larvae of S. exigua on E. lathyris wild-type, pil1, and pil10 mutants. Error bars represent mean ± SE (one-way ANOVA, p < 0.0001).
(D) Weight of LV larvae of S. exigua fed on E. lathyris pil1 and pil10 mutants and on artificial diet. Error bars represent mean ± SE (one-way ANOVA, p = 0.1438).
(E) Weight of LV larvae of S. exigua fed on artificial diet alone or on E. lathyris wild-type plants supplemented with artificial diet. Error bars represent mean ± SE (one-way ANOVA, p = 0.1287).
Severe susceptibility of pil mutants to Tetranychus urticae
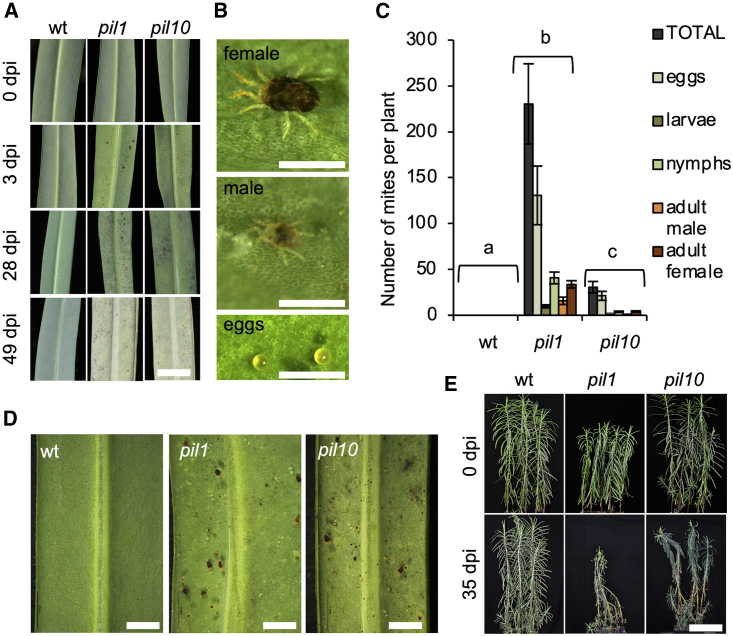
We next wondered whether other arthropod species with different feeding habits/mouthparts could take advantage of the lack of latex to infest E. lathyris. The spider mite Tetranychus urticae Koch (Acari: Prostigmata) is a highly polyphagous and cosmopolitan herbivore with short stylets that enable it to suck cell contents from epidermal and subepidermal plant cells. Wild-type, pil1, and pil10 plants were artificially infested with T. urticae adult females, and plants were regularly inspected over a period of 49 days post infestation (dpi) to assess plant damage. Some plants were subjected to a destructive sampling at 14 dpi to assess mite population density. Chlorosis appeared progressively on the leaves of both pil1 and pil10 plants but not on those of wild-type plants (Figure 3A), and symptoms were consistent with the greater number of mites (including adults [both males and females], nymphs, larvae, and eggs) identified on pil1 and pil10 plants (Figures 3B and 3C). Mites occupied the abaxial side of the leaves in the susceptible mutants (e.g., pil1), but there was no sign of infestation in wild-type plants (Figure 3D). At later stages of infestation, mites colonized entire mutant plants (Supplemental Figure 1A) and aggregated in young apical regions where plants showed a characteristic stress response (e.g., leaf curling and retracted growth from the shoot apical meristem) (Supplemental Figure 1B) typical of heavily infested plants and related to the ballooning behavior of T. urticae. The acute infestations caused pil1 and pil10 plants to collapse 4–5 weeks after inoculation, whereas the wild-type plants remained intact and free of infestation by T. urticae (Figure 3E). However, T. urticae infestations were more severe on pil1 than on pil10 plants. pil1 plants are defective in laticifer differentiation and growth and therefore produce no latex, whereas pil10 plants appear to exhibit normal laticifer differentiation and growth but are metabolically affected in latex production (Castelblanque et al., 2016). It seems very likely that pil10 laticifer cells can still produce a residual amount of latex, albeit not in sufficient amounts to confer the complete resistance to T. urticae observed in the wild-type parental plants (Figure 2C).
Figure 3.
Response of latex- and laticifer-defective E. lathyris plants to infestation by the spider mite T. urticae.
(A) Artificial infestation of wild-type, pil1, and pil10 plants with T. urticae; infested plants were inspected visually for 49 days post infestation (dpi) to monitor chlorosis and plant damage. Scale bars represent 1 cm.
(B) Detail of a female, a male, and egg deposition of T. urticae on the leaf surface of pil1 plants. Scale bars represent 500 μm.
(C) Quantification of the number of mites identified at different stages, including adults (both male and female), nymphs, larvae, and eggs at 14 dpi. Error bars represent mean ± SE (one-way ANOVA, p < 0.0001).
(D) Strong proliferation of T. urticae in pil1 and pil10 plants occurs most prominently on the abaxial side of the leaf, whereas no mites are observed in wild-type plants. Scale bars represent 2 mm.
(E) Severe infestation was observed in pil1 and pil10 mutants 35 dpi while wild-type plants, growing side by side, showed no signs of infestation. Scale bars represent 25 cm.
Susceptibility to the whitefly Aleyrodes proletella remains intact in pil1 and pil10 plants
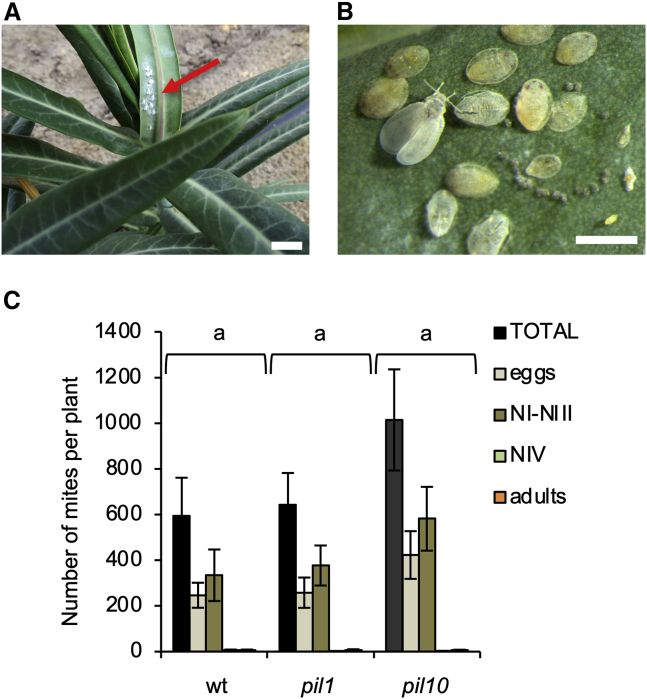
Next, we wondered whether E. lathyris would also show resistance to the whitefly Aleyrodes proletella L. (Hemiptera: Aleyrodidae), another arthropod with a different type of feeding habit. Unlike previous arthropods, A. proletella is a phloem-sucking insect that carries long, flexible stylets, which are pushed through the leaf surface of host plants and maneuvered to reach phloem vessels and gain access to phloem sap. In contrast to arthropods with chewing mouthparts (e.g., S. exigua) or short stylets (e.g., T. urticae), phloem-feeding insects with long stylets have evolved to suppress or weaken the jasmonic acid (JA)-inducible plant defense pathway by stimulating the salicylic acid-inducible pathway (Pieterse and Dicke, 2007; Walling, 2008). Therefore, we performed comparative quantitative experiments on the performance of this whitefly on pil1, pil10, and wild-type plants of E. lathyris under controlled climatic conditions. Upon infestation with the same number of adult whiteflies, wild-type, pil1, and pil10 plants were similarly infested by A. proletella (Figure 4A and 4B), as similar numbers of eggs, nymphs, and adults were recorded on both genotypes (Figure 4C) by the end of the experiments. These results indicate that latex appears to play no role in the susceptibility of E. lathyris to the phloem-feeding whitefly A. proletella, but instead represents an essential defense strategy that mediates survival when E. lathyris plants confront either a chewing insect (S. exigua) or a short-stylet piercing-sucking mite (T. urticae). These findings help to explain why pil mutant plants progressively succumbed and collapsed when grown in the open field (Supplemental Figure 2A–2D) because of heavy infestation by surrounding herbivores (i.e., T. urticae; Supplemental Figure 2E), whereas nearby wild-type plants remained healthy and completed their life cycle.
Figure 4.
Response of latex-defective E. lathyris plants to infestation by A. proletella.
(A) Natural infestation of E. lathyris wild-type plants by A. proletella. Scale bars represent 1 mm.
(B) Different life stages of A. proletella: winged adult whiteflies, eggs, and nymphs (NI to NIV or puparia). Scale bars represent 1 mm.
(C) Composition of A. proletella populations on E. lathyris wild-type and pil1 mutant plants at the end of the experiment showed neither quantitative nor qualitative differences, and both plant types were suitable hosts for this herbivore. Error bars represent mean ± SE (one-way ANOVA, p = 0.2461).
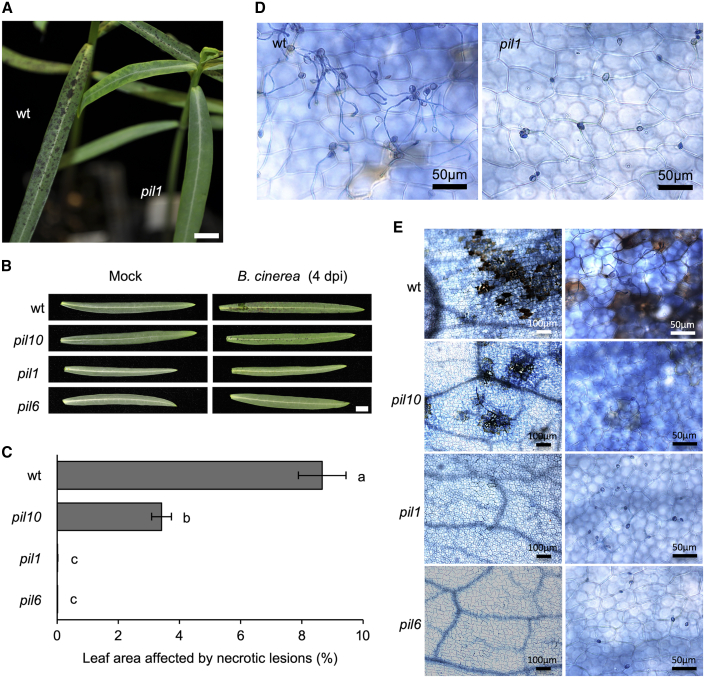
pil mutants show full resistance to the necrotrophic fungal pathogen Botrytis cinerea
Latex production in laticifers is controlled by the plant hormone JA (Laosombut et al., 2016; Castelblanque et al., 2018), and applications of JA commonly activate resistance to pests and fungal pathogens (Mengiste, 2012). We therefore asked whether the resistance of E. lathyris to a fungal pathogen was altered in pil mutants. Toward this end, we evaluated how plants responded to the gray mold fungus Botrytis cinerea. Spraying plants with a suspension of B. cinerea spores revealed that wild-type plants supported fungal growth and responded with the rapid development of severe necrosis (Figure 5A and 5B). Macroscopic evaluation of disease symptoms by measuring the percentage of leaf area affected by necrotic lesions at 4 dpi revealed that E. lathyris was highly susceptible to B. cinerea (Figure 5C). In marked contrast, the parallel inoculation of pil1 and pil6 plants revealed that these mutants did not support fungal growth and consequently developed little or no necrosis on inoculated leaves (Figure 5A and 5B). The percentage of leaf area affected by necrotic lesions reflected little or almost no infection (Figure 5C). Meanwhile, although the severity of B. cinerea infection in pil10 plants was far less than that in wild-type plants, its necrotic lesion area was higher than that of the pil1 and pil6 mutants (Figure 5C). This strongly indicates that the disappearance of disease susceptibility (or the acquired resistance) to B. cinerea, common to the three pil mutants, probably derives from a lack of latex production. This result revealed that latex has opposing roles in resistance: it generally confers resistance to herbivores and concurrently promotes susceptibility to fungi.
Figure 5.
Response of latex- and laticifer-defective E. lathyris plants to inoculation with the fungal pathogen B. cinerea.
(A) Disease symptoms in wild-type plants and lack of them in the pil1 mutant at 4 dpi. Scale bars represent 1 cm.
(B) Phenotype of leaves from inoculated wild-type, pil1, pil6, and pil10 plants at 4 dpi. Scale bars represent 1 cm.
(C) Percentage of leaf area affected by necrotic lesions at 4 dpi. Error bars represent mean ± SE (one-way ANOVA, p < 0.0001).
(D) Germinated conidia in wild-type surface leaves and absence of germination in pil1 mutants, as revealed by trypan blue staining at 4 dpi.
(E) Severe cellular lesions in wild-type plants, minor lesions in the pil10 mutant, and absence of lesions in pil1 and pil6 mutants, as revealed by trypan blue staining at 4 dpi.
Lack of conidia germination on pil mutants
We next examined the growth of the fungus and the progression of cell death disease symptoms by staining the inoculated leaves with trypan blue (TB). TB staining of wild-type plants revealed that the majority of conidia germinated on the leaf surface, and the plant supported the vegetative growth of hyphae. This was followed by the appearance of localized plant cell death at sites where enlarging hyphae penetrated the tissues (Figure 5D and 5E). By contrast, TB staining of pil1- and pil6-inoculated leaves revealed neither conidial germination on the leaf surface nor signs of plant cell death (Figure 5D and 5E). For pil10, germination of some conidia occurred, eventually giving rise to small localized spots of cell death that did not spread as widely as those of wild-type plants (Figure 5E). As discussed above for T. urticae, a residual production of latex in pil10 plants may explain their differences from pil1 and pil6 mutants, as the latter were devoid of laticifer cells. These observations suggested that active laticifers are the source of a host susceptibility factor required for the germination of fungal conidia. To the best of our knowledge, these results reveal the existence of a genetically controlled pre-penetration mechanism of resistance in E. lathyris.
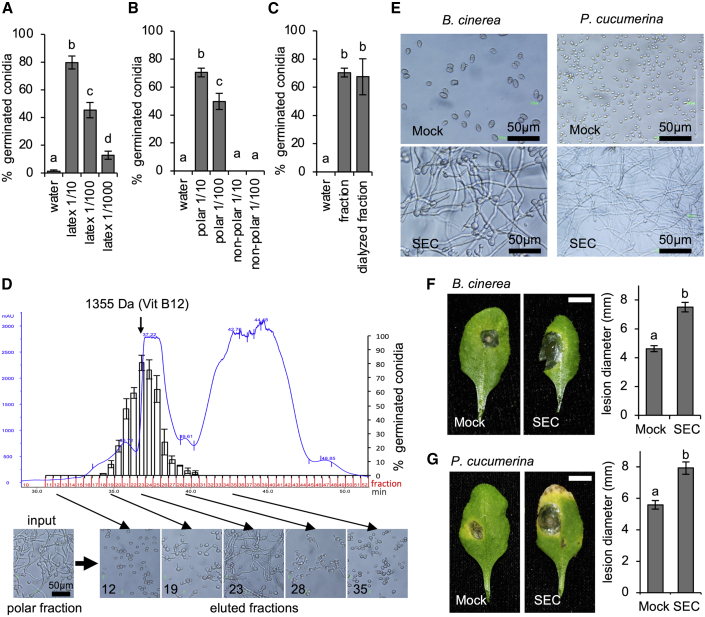
Latex promotes germination of fungal spores
Next, we reasoned that latex might be enriched in a susceptibility factor required for conidia germination. We tested this hypothesis by studying the effect of latex on in vitro germination of B. cinerea conidia. When suspended in water, conidia did not germinate in vitro (Figure 6A) and remained in a dormant state. Conversely, the addition of an aliquot of the latex that oozed after pricking an E. lathyris leaf provoked germination of more than 80% of the suspended conidia, which then initiated vegetative growth and formed long, branching filamentous hyphae. This pro-germination effect was progressively reduced upon serial dilutions of the latex (Figure 6A). Moreover, separation of the latex content into a polar and a non-polar fraction by extraction with an organic solvent revealed that the full pro-germination activity was partitioned into the polar fraction (Figure 6B). Additionally, the polar fraction was stable and supported 24 h of dialysis through a dialysis membrane with a cutoff of 1000 Da (Figure 6C), suggesting that the factor consisted of molecule(s) larger than 1 kDa. Further fractionation of the polar fraction by size-exclusion chromatography (SEC) through a fast protein liquid chromatography (FPLC)–SEC column revealed that the pro-germination activity was recovered in the column fractions that co-eluted with a 1.3-kDa molecular marker (Figure 6D).
Figure 6.
A factor isolated from E. lathyris latex promotes in vitro conidial germination in B. cinerea and P. cucumerina and enhances Arabidopsis susceptibility to both pathogens.
(A)B. cinerea conidial germination after 24 h in water supplemented with latex supernatant at different dilutions (1/10, 1/100, and 1/1000). Error bars represent mean ± SE (one-way ANOVA, p < 0.0001).
(B)B. cinerea conidial germination in water supplemented with latex polar and non-polar fractions after heptane partition. Error bars represent mean ± SE (one-way ANOVA, p < 0.0001).
(C)B. cinerea conidial germination after 24 h in water supplemented with the polar fraction before and after dialysis with a 1000 molecular weight cutoff membrane. Error bars represent mean ± SE (one-way ANOVA, p < 0.0001).
(D) FPLC–SEC chromatogram of the polar fraction and B. cinerea conidial germination after 24 h in water supplemented with the eluted fractions.
(E) Conidial germination in B. cinerea and P. cucumerina after 24 h in water (Mock) or water supplemented with the FPLC–SEC fraction (SEC).
(F)Arabidopsis Col-0 plants inoculated with a drop of B. cinerea conidia suspension (Mock) or conidia supplemented with the FPLC–SEC fraction (SEC), and lesion diameters at 3 dpi. Scale bars represent 1 cm. Error bars represent mean ± SE (one-way ANOVA, p < 0.0002).
(G)Arabidopsis Col-0 plants inoculated with a drop of P. cucumerina conidia suspension (Mock) or conidia supplemented with the FPLC–SEC fraction (SEC), and lesion diameters at 11 dpi. Scale bars represent 1 cm. Error bars represent mean ± SE (one-way ANOVA, p < 0.0001).
A latex-derived factor fosters disease susceptibility of Arabidopsis plants to B. cinerea and P. cucumerina
We next asked whether the factor present in E. lathyris latex was specific for B. cinerea or could promote germination of other plant pathogenic fungi (i.e., Plectosphaerella cucumerina). Comparative in vitro germination assays of spores from B. cinerea and P. cucumerina in the presence and absence of the 1.3-kDa FPLC–SEC purified factor (Figure 6E) revealed that spores from both fungal species rapidly germinated and initiated vegetative growth when the purified factor was present. Therefore, the pro-germination activity of the latex factor does not appear to be specific to B. cinerea. We then wondered whether the latex-derived factor could increase the pathogenicity of the fungi. If so, one would expect that its external application to a non-latex-bearing plant would cause the host to support increased pathogen growth, which would in turn generate disease enhancement. Arabidopsis thaliana is a natural host for B. cinerea and P. cucumerina. Therefore, we inoculated the leaves of Arabidopsis Columbia-0 (Col-0) plants with a drop of spores from either B. cinerea or P. cucumerina without (mock) or with the purified FPLC–SEC fractions. We followed the progression of disease and recorded the necrotic lesion diameter at 3 dpi for B. cinerea and at 11 dpi for P. cucumerina. As expected, mock-treated Arabidopsis plants responded to both fungi (Figure 6F and 6G) with the development of necrotic lesions. Remarkably, the presence of the latex-derived factor promoted a dramatic enlargement of the necrotic lesions generated by both fungi at the inoculation site (Figure 6F and 6G). The disease-promoting effect of the latex-derived factor was particularly striking in the case of B. cinerea: the necrotizing area was nearly double that of mock-treated plants. Thus, the latex-derived factor from E. lathyris further promoted the pathogenicity of both fungi on susceptible Arabidopsis plants. Whether this is due to an additive effect over the normal mechanism that controls plant susceptibility or results from complementation of a pre-existing mechanism remains unknown. If the former holds, it may be that Arabidopsis plants can synthesize a molecule that resembles the one present in latex and that is recognized by the fungi to facilitate conidia germination.
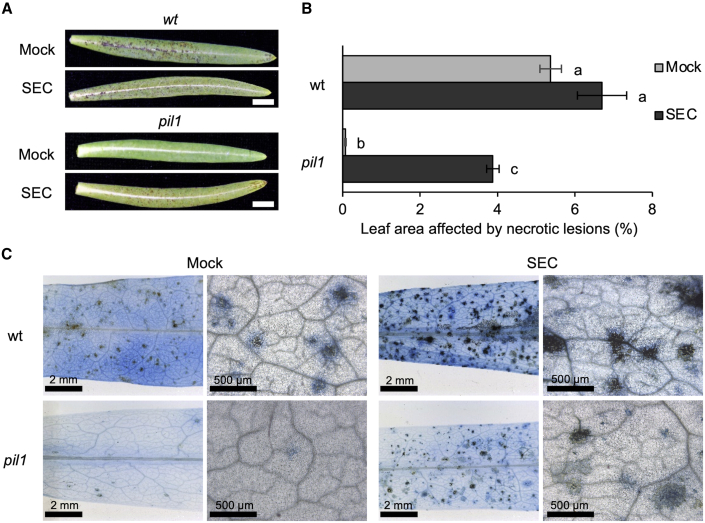
A latex-derived factor restores the susceptibility of pil1 plants to B. cinerea
Following the previous observations, we tested whether the exogenous application of the susceptibility factor from latex could reverse the observed notable resistance of pil1 plants to B. cinerea. The selected FPLC–SEC fraction was exogenously applied to E. lathyris wild-type and pil1 plants at the moment of B. cinerea inoculation, and the response to infection was recorded at 4 dpi. Figure 7A and 7B show that the leaf area affected by necrotic lesions was practically zero in pil1, but this was reversed upon application of the FPLC–SEC fraction, leading to a dramatic increase in the leaf area affected by necrosis. Also, the susceptibility of the wild-type plants showed partial enhancement, albeit not statistically significant (Figure 7B), upon supplementation with the FPLC–SEC fraction, similar to the results shown above for Arabidopsis (Figure 6F). TB staining confirmed that application of the FPLC–SEC fraction promoted the appearance of cellular lesions after fungal inoculation of pil1 plants (Figure 7C). Therefore, latex is enriched in a compound that is able to foster fungal growth, and that complements the disease resistance phenotype of pil1 plants to B. cinerea.
Figure 7.
Response of E. lathyris plants to inoculation with the fungal pathogen B. cinerea complemented with the FPLC–SEC fraction.
(A) Disease symptoms in wild-type plants and the pil1 mutant inoculated with a B. cinerea conidia suspension (Mock) or conidia supplemented with the FPLC–SEC fraction (SEC) at 4 dpi. Scale bars represent 1 cm.
(B) Percentage of leaf area affected by necrotic lesions at 4 dpi. Error bars represent mean ± SE (one-way ANOVA, p = 0.0024).
(C) Cellular lesions in wild-type and pil1 plants revealed by trypan blue staining at 4 dpi.
Discussion
The bulk of evidence suggests that the production of latex by laticifers has a defensive role, particularly against herbivores (Agrawal and Konno, 2009; Konno, 2011; Dussourd, 2017; Zuest et al., 2019). However, this perspective has been derived primarily from cause-and-effect experiments whereby latex-bearing plants deprived of latex by physical methods (cutting and washing of leaves) promoted better performance of the herbivore (Agrawal and Konno, 2009; Dussourd, 2017; Konno et al., 2004). However, genetic evidence is still pending to accurately evaluate the hypothesis that laticifers and latex production represent a specific ecophysiological adaptation related to host defense and to address the fitness benefits conferred by plant latex production. The availability of pil mutants of the latex-bearing plant E. lathyris has been instrumental toward reaching this goal.
The observation that the strong natural resistance of wild-type E. lathyris plants against S. exigua (a mandibulate chewing insect) and T. urticae (a stylet-based cell feeder arthropod) is blocked in pil mutants, and the susceptibility that ensues when latex production is impeded in pil mutants, provide a strong indication that wild-type E. lathyris plants are not hosts for these herbivores and that latex is essential for their defense. Our results provide the first genetic evidence supporting the importance of latex production as a critical trait to prevent herbivory. The findings for S. exigua suggest that latex indeed provides antixenosis against this herbivore to wild-type E. lathyris plants. Interestingly, the observation that the whitefly A. proletella (a sap-sucking insect) infested pil1, pil10, and wild-type plants to a similar degree suggests that A. proletella may have developed a surveillance strategy whereby the stylet detects the presence of laticifer cells, which profusely expand throughout the leaf lamina, and avoids touching them while finding its route toward the phloem vessels, thus permitting E. lathyris to serve as a host for this insect. Therefore, latex-mediated antixenosis against one herbivore (e.g., S. exigua, T. urticae) cannot be generalized against other herbivores (e.g., A. proletella).
Interestingly, the finding that the strong disease susceptibility of E. lathyris plants toward the necrotrophic fungal pathogen B. cinerea is blocked when latex production is genetically compromised in pil mutants led to the identification of a 1.3-kDa latex-enriched molecule. This latex-derived compound functions as a conidial germination-promoting factor and appears to be key for the progression of fungal infection. Early recognition of this 1.3-kDa plant susceptibility factor on the leaf surface dictates the germination of conidia and the emergence of hyphae, leading to the formation of fungal infective structures that penetrate the plant tissues. This finding thus reveals a pre-penetration phase in the fungal infection process that is ultimately controlled by the activity of a latex-derived factor. This latex-derived factor, when applied exogenously, is able to enhance the normal susceptibility of Arabidopsis plants to two different fungal necrotrophs (i.e., B. cinerea and P. cucumerina). Moreover, the latex-derived factor is also able to reverse the well-known disease resistance phenotype of pil1 plants to B. cinerea, causing them to regain susceptibility similar to that of wild-type plants. During B. cinerea infection, the conidia that land on the surface must undergo attachment, germination, hyphal growth, and appressoria formation during the so-called pre-penetration processes (van Kan, 2006). This process must be initiated by sensing plant cues that signal the fungal pathogen to germinate and initiate the infection process; otherwise, the fungus remains latent and infection does not proceed. Some of the molecules involved in this kind of inter-kingdom signaling have been identified (e.g., strigolactones, cutin monomers, and chitin-related compounds), and they ultimately condition the interaction of the fungus with the plant (Bonfante and Genre, 2015). Examples include the maize wax mutant glossy11, which is devoid of cuticular very-long-chain aldehydes and in which germination and appressorial differentiation of the fungus Blumeria graminis become impeded (Hansjakob et al., 2011). The Arabidopsis mutant botrytis resistant1 (bre1) displays strong resistance to B. cinerea infection and was found to be defective in its cuticular membrane (Bessire et al., 2007). Although a more comprehensive characterization is needed, the susceptibility factor isolated from E. lathyris latex appears to function as a cue for B. cinerea to initiate conidial germination on the leaf surface and hence provides a good system for the study of early stages in the infection process. Our future goals are the deep characterization and purification of the 1.3-kDa factor and the investigation of its mode of action, including how it is transported from the laticifer cell to the leaf surface and what type of recognition and signaling takes place in the conidia to promote fungal growth.
In summary, latex production in E. lathyris is required for resistance to herbivores but is a source of disease susceptibility to fungal pathogens. The opposing roles of laticifer cells in mediating herbivore resistance and fungal susceptibility, together with the polyphyletic origin of laticifer cells during angiosperm evolution, represent a paradigmatic example of ecosystem adaptation mediated by cell speciation. According to evolutionary ecology, particular traits are constrained because gains in their expression come at the expense of other important traits (Felton and Korth, 2000; Cipollini et al., 2014; Hahn, et al., 2019). We speculate that in the co-evolutionary arms race between plants and their enemies in natural environments, selection pressures exerted by prevailing pests favor the development of new host adaptive traits for survival, such as the acquisition of laticifers and latex to fend off herbivores. However, at the expense of this new adaptive trait, other primary consumers seemed favored (i.e., fungi), reducing the capacity of the plant to withstand disease.
Methods
Plant material and plant growth conditions
E. lathyris wild-type plants and pil mutants used here have been described previously (Castelblanque et al., 2016). Plants were grown in a growth chamber (19°C–23°C, 85% relative humidity, 120–150 μmol m−2 s−1 fluorescent illumination, 16-h light photoperiod). Arabidopsis Col-0 plants were grown in a growth chamber (19°C–23°C, 85% relative humidity, 100 μmol m−2 s−1 fluorescent illumination, 10-h light photoperiod).
Laticifer staining
Entire plants were immersed in fixative (formaldehyde/acetic acid/ethanol, 3.5:10:50) overnight at 4°C. Plant sectors were washed with 70% ethanol and stained with Sudan black B (0.1% [w/v] in 70% ethanol) for 3–4 h at room temperature, washed with 70% ethanol and then with water, and placed in 2.5 M NaOH until the leaves were cleared. Tissues were observed under an Eclipse E600 (Nikon) light microscope.
Stock colonies and feeding experiments for arthropods
Early larval instars (LI and LII) of S. exigua were obtained from existing colonies at the Department of Genetics of Universitat de València. S. exigua larvae were reared on artificial diet as described by Bell and Joachim (1976) in a climatic chamber with a temperature of 25°C and a 12-h light photoperiod. Initial specimens (all stages) of the two-spotted spider mite, T. urticae, and the cabbage whitefly, A. proletella, were obtained from infested E. lathyris plants collected in field experiments. Mites were subsequently reared in a climatic chamber (25°C, 12-h light photoperiod) on fully expanded detached leaves of Phaseolus vulgaris L. (Fabaceae). Leaves were placed upside down on top of sponges covered with cotton in water-containing trays that served both as a water source for leaves and mites and as a barrier against mite dispersal. This population was reared for about 3 months before the onset of the experiments. Whiteflies were reared in a greenhouse (25°C ± 10°C, natural photoperiod) on E. lathyris plants for 4 weeks before the onset of the experiments.
Infestation experiments for arthropods
The performance of S. exigua larvae on wild-type plants and pil1 and pil10 mutants was assessed under controlled environmental conditions (the same conditions used for rearing). Two-month-old plants in groups of six were enclosed in mesh cages and infested with six larvae each. Cohorts of larvae of the same age (LIV) were directly obtained from the stock colonies, individually weighed, and transferred to plants. Larval survival and larval weight were measured again 6 days later. Two controls were used: one in which larvae were reared on artificial diet only and another in which artificial diet was added to cages where larvae had been placed on wild-type plants. The performance of T. urticae was tested on E. lathyris wild-type plants and pil1 and pil10 mutants. In the case of T. urticae, a few days prior to the start of the experiments, several hundred ovipositing females were haphazardly taken from the rearing units, transferred to detached bean leaves for 24 h, and removed thereafter. Leaves with eggs less than 24 h old were maintained separately for 12 days in a climatic chamber under the same environmental conditions used for rearing. Adult females less than 48 h old were then selected to infest 6-week-old plants (20 females per plant). Two weeks later, a destructive sampling was performed to assess the numbers of juvenile (eggs, larvae, and nymphs) and adult spider mites on each plant. Additional plants were followed for up to five extra weeks to assess mite damage. Experiments were conducted in a climatic chamber using the same environmental conditions as before. Twelve plants (= replicates per plant genotype) were used: six to assess mite dynamics and six to assess mite damage. Similarly, the performance of A. proletella whiteflies was tested on wild-type and pil1 plants. These experiments were performed in a greenhouse under the same environmental conditions used for rearing. Two-month-old plants in groups of four were enclosed in mesh cages and infested with adult whiteflies taken directly from the stock colony (30 individuals per plant). The numbers of juvenile (NI to NIV or puparia) and adult whiteflies on each plant were recorded 2 weeks after infestation.
B. cinerea and P. cucumerina fungal spore preparations
The B. cinerea strain used was CECT2100 (Spanish Type Culture Collection, Universitat de València). It was routinely cultured on potato dextrose agar (PDA) supplemented with lyophilized tomato leaves at 24°C. Conidia were collected from 4-week-old cultures by scraping surface plates and washing with sterile water, then filtering through cotton to remove debris. Conidia were washed with sterile water twice (5000 rpm, 10 min, 25°C), quantified with a hemacytometer, resuspended in water, and adjusted to the final concentration. When conidia were used to inoculate plants (E. lathyris or Arabidopsis), they were resuspended in Gamborg's B5 medium (Duchefa, the Netherlands) supplemented with 10 mM sucrose and 10 mM potassium phosphate at pH 6 (Benito et al., 1998). P. cucumerina (Ton and Mauch-Mani, 2004) was grown on half-strength PDA at 24°C. Conidia were collected from 4-week-old cultures following the same protocol used for B. cinerea conidia, resuspended in water, and adjusted to the final concentration.
E. lathyris plant inoculation with B. cinerea
Conidia suspension was prepared as described above at a concentration of 1.0 × 106 conidia ml−1, supplemented with 0.02% (v/v) Silwet, and incubated for 2–3 h at room temperature. E. lathyris 3-week-old wild-type and pil plants were then inoculated by spraying the conidia suspension. The experiments were conducted in inoculation chambers, and each experiment used at least 12 plants per genotype and three different chambers. Each inoculation chamber contained all the genotypes, and 10 ml of conidia suspension was used for each chamber. Mock plants (controls) were sprayed with suspension buffer. All plants were maintained in the inoculation chambers at 100% relative humidity, and sampling was performed at 4 dpi to evaluate the disease symptoms by means of the percentage of leaf area affected by necrotic lesions. Additionally, leaf samples were stained with lactophenol–TB (Koch and Slusarenko, 1990) and examined under a light microscope (Leica DM5000). Each assay was repeated at least three times.
Percentage of leaf area affected by necrotic lesions
To estimate the severity of infection caused by B. cinerea in different plant genotypes, we photographed all leaves from each treated plant with the adaxial side up using a non-reflective background. The resulting digital images were later analyzed with ImageJ software (National Institutes of Health, Bethesda, MD, USA; https://imagej.nih.gov/ij/) to determine the area of necrotic lesions and the total leaf area using the color thresholding method.
E. lathyris latex collection, in vitro conidial germination, and susceptibility factor isolation
Fresh latex was harvested from 2-month-old E. lathyris plants by cutting shoots with a scalpel blade. Latex was collected in tubes and stored immediately at −80°C. It was then centrifuged (15 000 rpm, 5 min, 4°C), the precipitates were discarded, and the supernatant was filtered through a 45-μm membrane and used for in vitro conidial germination assays and susceptibility factor isolation. For in vitro conidial germination assays, B. cinerea and P. cucumerina conidia were isolated as previously described and resuspended in sterile water at final concentrations of 1.0 × 106 and 5.0 × 106 conidia ml−1, respectively. Each assay consisted of 250 μl of liquid medium and 25 μl of conidia. After 24 h of incubation at 23°C and a 16-h photoperiod, conidia germination rate was recorded under a microscope using at least 100 conidia for each sample. Negative (sterile water or phosphate buffer as the liquid medium) and positive controls (Gamborg's B5 medium supplemented with 10 mM sucrose and 10 mM potassium phosphate at pH 6) were included in all experiments.
Latex supernatant was partitioned into polar and non-polar fractions. Two volumes of heptane and 0.25 volumes of phosphate buffer (pH 7) were added to 1 volume of latex supernatant, and the mixture was homogenized at 60°C for 1 h. After soft centrifugation (5000 rpm, 2 min, 25°C), two phases were obtained: an upper phase with the heptane extract (non-polar fraction) and a lower phase with the aqueous phase (polar fraction). They were concentrated, resuspended in water (polar fraction) or isopropanol (non-polar fraction), filtered through a 45-μm membrane, and tested in the in vitro conidial germination assay.
The polar fraction was further purified by gel-filtration chromatography/SEC in an FPLC system equipped with a Superdex 75 10/300 GL column (GE Healthcare). The separation in this case is made according to molecular size, first eluting molecules with higher molecular weights. The sample volume was 500 μl, the eluent was 0.05 M phosphate buffer with 0.15 M NaCl at pH 7.0, and the flow rate was 0.5 ml min−1. Standard molecules were tested in the FPLC–SEC system with the same conditions to determine the approximate molecular weight (MW) of the SEC fractions: M1 (aprotinin, MW = 6512), M2 (vitamin B12, MW = 1355), and M3 (folic acid, MW = 441). All FPLC–SEC eluted fractions were collected, concentrated, resuspended in water, filtered through a 45-μm membrane, and tested in the in vitro conidial germination assay.
In parallel, the polar fraction was thoroughly dialyzed against distilled water using a membrane with a 1000 molecular weight cutoff (Pur-A-Lyzer, Sigma-Aldrich) at room temperature for 24 h with three changes of water. The dialyzed fraction was concentrated to the initial volume, filtered through a 45-μm membrane, and tested in the in vitro conidial germination assay.
B. cinerea and P. cucumerina inoculations in Arabidopsis
Five-week-old Arabidopsis plants were inoculated by applying 6-μl droplets of spore suspension of B. cinerea and P. cucumerina at 2.5 × 104 and 5.0 × 106 conidia ml−1, respectively. The challenged plants were maintained at 100% relative humidity. Disease symptoms were evaluated by determining the lesion diameter of at least 50 lesions at 3 dpi for B. cinerea and 11 dpi for P. cucumerina.
Statistical analysis
Each experiment was performed at least three times. Unless stated, data represent means ± SD (n = 3 biological replicates). Significant differences were assessed with one-way ANOVA, and means were compared using Duncan’s post hoc test with a p < 0.05 level of significance; the different letters above the bars in figures indicate different homogeneous groups with statistically significant differences.
Funding
This work was supported by a grant from the Agencia Estatal de Investigación (AEI), Spain (RTI2018-098501-B-I00).
Acknowledgments
We thank Begoña Balaguer and Miguel Ozáez for their assistance in the experiments. The authors declare no competing interests.
Author contributions
P.V., L.C., and J.A.J. designed the experiments; J.A.J. and E.A.-F. performed the arthropod experiments; J.G.-A., C.M.-A., F.J.E., and L.C. performed the fungal pathogen experiments; J.J.R. and L.C. performed the open field tests; P.V. and L.C. wrote the article.
Published: September 11, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Agrawal A.A. Natural selection on common milkweed (Asclepias syriaca) by a community of specialized insect herbivores. Evol. Ecol. Res. 2005;7:651–667. [Google Scholar]
- Agrawal A.A., Konno K. Latex: a model for understanding mechanisms, ecology, and evolution of plant defense against herbivory. Annu. Rev. Ecol. Evol. Syst. 2009;40:311–331. [Google Scholar]
- Becerra J.X., Venable D.L. Rapid-terpene-bath and squirt-gun defense in Bursera schlechtendalii and the counterploy of chrysomelid beetles. Biotropica. 1990;22:320–323. [Google Scholar]
- Bell R.A., Joachim F.G. Techniques for rearing laboratory colonies of tobacco hornworms and pink bollworms Lepidoptera-Sphingidae-Gelechiidae. Ann. Entomol. Soc. America. 1976;69:365–373. [Google Scholar]
- Benito E.P., ten Have A., van't Klooster J.W., van Kan J.A.L. Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea. Eur. J. Plant Pathol. 1998;104:207–220. [Google Scholar]
- Bessire M., Chassot C., Jacquat A.C., Humphry M., Borel S., Petetot J.M.C., Metraux J.P., Nawrath C. A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J. 2007;26(8):2158–2168. doi: 10.1038/sj.emboj.7601658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P., Genre A. Arbuscular mycorrhizal dialogues: do you speak 'plantish' or 'fungish? Trends Plant Sci. 2015;20(3):150–154. doi: 10.1016/j.tplants.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Castelblanque L., Balaguer B., Marti C., Rodriguez J.J., Orozco M., Vera P. Novel insights into the organization of laticifer cells: a cell comprising a unified whole system. Plant Physiol. 2016;172:1032–1044. doi: 10.1104/pp.16.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelblanque L., Balaguer B., Marti C., Orozco M., Vera P. LOL2 and LOL5 loci control latex production by laticifer cells in Euphorbia lathyris. New Phytol. 2018;219:1467–1479. doi: 10.1111/nph.15253. [DOI] [PubMed] [Google Scholar]
- Cipollini D., Walters D., Voelckel C. Costs of resistance in plants: from theory to evidence. Insect-Plant Interactions. 2014;47:263–307. [Google Scholar]
- Dussourd D.E. Behavioral sabotage of plant defenses by insect folivores. Annu. Rev. Entomol. 2017;62:15–34. doi: 10.1146/annurev-ento-031616-035030. [DOI] [PubMed] [Google Scholar]
- Esau K. Wiley; New York: 1965. Plant Anatomy. [Google Scholar]
- Felton G.W., Korth K.L. Trade-offs between pathogen and herbivore resistance. Curr. Opin. Plant Biol. 2000;3:309–314. doi: 10.1016/s1369-5266(00)00086-8. [DOI] [PubMed] [Google Scholar]
- Freitas C.D.T., Viana C.A., Vasconcelos I.M., Moreno F.B.B., Lima J.V., Oliveira H.D., Moreira R.A., Monteiro-Moreira A.C.O., Ramos M.V. First insights into the diversity and functional properties of chitinases of the latex of Calotropis procera. Plant Physiol. Biochem. 2016;108:361–371. doi: 10.1016/j.plaphy.2016.07.028. [DOI] [PubMed] [Google Scholar]
- Hagel J.M., Yeung E.C., Facchini P.J. Got milk? The secret life of laticifers. Trends Plant Sci. 2008;13:631–639. doi: 10.1016/j.tplants.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Hahn P.G., Agrawal A.A., Sussman K.I., Maron J.L. Population variation, environmental gradients, and the evolutionary ecology of plant defense against herbivory. Am. Naturalist. 2019;193:20–34. doi: 10.1086/700838. [DOI] [PubMed] [Google Scholar]
- Hansjakob A., Riederer M., Hildebrandt U. Wax matters: absence of very-long-chain aldehydes from the leaf cuticular wax of the glossy11 mutant of maize compromises the prepenetration processes of Blumeria graminis. Plant Pathol. 2011;60(6):1151–1161. [Google Scholar]
- Hua J., Liu Y., Xiao C.-J., Jin S.-X., Lou S.-H., Li S.-H. Chemical profile and defensive function of the latex of Euphorbia peplus. Phytochemistry. 2017;136:56–64. doi: 10.1016/j.phytochem.2016.12.021. [DOI] [PubMed] [Google Scholar]
- Huber M., Epping J., Gronover C.S., Fricke J., Aziz Z., Brillatz T., Swyers M., Köllner T.G., Vogel H., Hammerbacher A. A latex metabolite benefits plant fitness under root herbivore attack. PLoS Biol. 2016;14 doi: 10.1371/journal.pbio.1002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S., Taira T., Oda K., Yamato K.T., Inukai Y., Hori Y. Comparative study of gene expression and major proteins' function of laticifers in lignified and unlignified organs of mulberry. Planta. 2012;235:589–601. doi: 10.1007/s00425-011-1533-6. [DOI] [PubMed] [Google Scholar]
- Kitajima S., Miura K., Aoki W., Yamato K.T., Taira T., Murakami R., Aburaya S. Transcriptome and proteome analyses provide insight into laticifer's defense of Euphorbia tirucalli against pests. Plant Physiol. Biochem. 2016;108:434–446. doi: 10.1016/j.plaphy.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Kitajima S., Aoki W., Shibata D., Nakajima D., Sakurai N., Yazaki K., Munakata R., Taira T., Kobayashi M., Aburaya S. Comparative multi-omics analysis reveals diverse latex-based defense strategies against pests among latex-producing organs of the fig tree (Ficus carica) Planta. 2018;247:1423–1438. doi: 10.1007/s00425-018-2880-3. [DOI] [PubMed] [Google Scholar]
- Koch E., Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan M., Ortman E.F. Antixenosis—a new term proposed to define painter's “nonpreference” modality of resistance. Bull. Entomol. Soc. America. 1978;24:175–176. [Google Scholar]
- Konno K. Plant latex and other exudates as plant defense systems: roles of various defense chemicals and proteins contained therein. Phytochemistry. 2011;72:1510–1530. doi: 10.1016/j.phytochem.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Konno K., Hirayama C., Nakamura M., tateishi K., Tamura Y., Hattori M., Kohno K. Papain protects papaya trees from herbivorous insects: role of cysteine proteases in latex. Plant J. 2004;37:370–378. doi: 10.1046/j.1365-313x.2003.01968.x. [DOI] [PubMed] [Google Scholar]
- Lange B.M. The evolution of plant secretory structures and emergence of terpenoid chemical diversity. Annu. Rev. Plant Biol. 2015;66:139–159. doi: 10.1146/annurev-arplant-043014-114639. [DOI] [PubMed] [Google Scholar]
- Laosombut T., Arreewichit P., Nirapathpongporn K., Traipern P., Kongsawadworakul P., Viboonjun U., Narangajavana J. Differential expression of methyl jasmonate-responsive genes correlates with laticifer vessel proliferation in phloem tissue of rubber tree (Hevea brasiliensis) J. Plant Growth Regul. 2016;35:1049–1063. [Google Scholar]
- Lewinsohn T. The geographical distribution of plant latex. Chemoecology. 1991;2:64–68. [Google Scholar]
- Mahlberg P.G. Laticifers—an historical-perspective. Bot. Rev. 1993;59(1):1–23. [Google Scholar]
- Mahlberg P.G., Sabharwal P.S. Origin and early development of nonarticulated laticifers in embryos of Euphorbia marginata. Am. J. Bot. 1968;55:375–381. [Google Scholar]
- Mengiste T. Plant immunity to necrotrophs. Annu. Rev. Phytopathology. 2012;50:267–294. doi: 10.1146/annurev-phyto-081211-172955. [DOI] [PubMed] [Google Scholar]
- Pieterse C.M.J., Dicke M. Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends Plant Sci. 2007;12:564–569. doi: 10.1016/j.tplants.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Prado E., Demarco D. Laticifers and secretory ducts: similarities and differences. In: Hufnagel L., editor. Ecosystem Services and Global Ecology. IntechOpen; 2018. pp. 103–123. [Google Scholar]
- Ramos M.V., Demarco D., da Costa Souza I.C., Teixeira de Freitas C.D. Laticifers, latex, and their role in plant defense. Trends Plant Sci. 2019;24:553–567. doi: 10.1016/j.tplants.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Ramos M.V., Pereira D.A., Souza D.P., Silva M.L.S., Alencar L.M.R., Sousa J.S., Queiroz J.F.N., Freitas C.D.T. Peptidases and peptidase inhibitors in gut of caterpillars and in the latex of their host plants. Planta. 2015;241(1):167–178. doi: 10.1007/s00425-014-2174-3. [DOI] [PubMed] [Google Scholar]
- Rasmann S., Johnson M.D., Agrawal A.A. Induced responses to herbivory and jasmonate in three milkweed species. J. Chem. Ecol. 2009;35:1326–1334. doi: 10.1007/s10886-009-9719-0. [DOI] [PubMed] [Google Scholar]
- Souza D.P., Freitas C.D.T., Pereira D.A., Nogueira F.C., Silva F.D.A., Salas C.E., Ramos M.V. Laticifer proteins play a defensive role against hemibiotrophic and necrotrophic phytopathogens. Planta. 2011;234(1):183–193. doi: 10.1007/s00425-011-1392-1. [DOI] [PubMed] [Google Scholar]
- Spilatro S.R., Mahlberg P.G. Latex and laticifer starch content of developing leaves of Euphorbia pulcherrima. Am. J. Bot. 1986;73:1312–1318. doi: 10.1002/j.1537-2197.1990.tb13542.x. [DOI] [PubMed] [Google Scholar]
- Ton J., Mauch-Mani B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 2004;38:119–130. doi: 10.1111/j.1365-313X.2004.02028.x. [DOI] [PubMed] [Google Scholar]
- van Kan J.A.L. Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 2006;11(5):247–253. doi: 10.1016/j.tplants.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Walling L.L. Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 2008;146:859–866. doi: 10.1104/pp.107.113142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuest T., Petschenka G., Hastings A.P., Agrawal A.A. Toxicity of milkweed leaves and latex: chromatographic quantification versus biological activity of cardenolides in 16 asclepias species. J. Chem. Ecol. 2019;45:50–60. doi: 10.1007/s10886-018-1040-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.