Abstract
Key points
microRNAs (miRs) are small non‐coding molecules that regulate post‐transcriptional target gene expression.
miRs are involved in regulating cellular activities in response to mechanical loading in all physiological systems, although it is largely unknown whether this response differs with increasing magnitudes of load.
miR‐221, miR‐222, miR‐21‐5p and miR‐27a‐5p were significantly increased in ex vivo cartilage explants subjected to increasing load magnitude and in in vivo joint cartilage exposed to abnormal loading.
TIMP3 and CPEB3 are putative miR targets in chondrocytes
Identification of mechanically regulated miRs that have potential to impact on tissue homeostasis provides a mechanism by which load‐induced tissue behaviour is regulated, in both health and pathology, in all physiological systems.
Abstract
MicroRNAs (miRs) are small non‐coding molecules that regulate post‐transcriptional target gene expression and are involved in mechano‐regulation of cellular activities in all physiological systems. It is unknown whether such epigenetic mechanisms are regulated in response to increasing magnitudes of load. The present study investigated mechano‐regulation of miRs in articular cartilage subjected to ‘physiological’ and ‘non‐physiological’ compressive loads in vitro as a model system and validated findings in an in vivo model of abnormal joint loading. Bovine full‐depth articular cartilage explants were loaded to 2.5 MPa (physiological) or 7 MPa (non‐physiological) (1 Hz, 15 min) and mechanically‐regulated miRs identified using next generation sequencing and verified using a quantitative PCR. Downstream targets were verified using miR‐specific mimics or inhibitors in conjunction with 3′‐UTR luciferase activity assays. A subset of miRs were mechanically‐regulated in ex vivo cartilage explants and in vivo joint cartilage. miR‐221, miR‐222, miR‐21‐5p and miR‐27a‐5p were increased and miR‐483 levels decreased with increasing load magnitude. Tissue inhibitor of metalloproteinase 3 (TIMP3) and cytoplasmic polyadenylation element binding protein 3 (CPEB3) were identified as putative downstream targets. Our data confirm miR‐221 and ‐222 mechano‐regulation and demonstrates novel mechano‐regulation of miR‐21‐5p and miR‐27a‐5p in ex vivo and in vivo cartilage loading models. TIMP3 and CPEB3 are putative miR targets in chondrocytes. Identification of specific miRs that are regulated by increasing load magnitude, as well as their potential to impact on tissue homeostasis, has direct relevance to other mechano‐sensitive physiological systems and provides a mechanism by which load‐induced tissue behaviour is regulated, in both health and pathology.
Keywords: articular cartilage, CPEB3, mechanical load, miRNA, TIMP3
Key points
microRNAs (miRs) are small non‐coding molecules that regulate post‐transcriptional target gene expression.
miRs are involved in regulating cellular activities in response to mechanical loading in all physiological systems, although it is largely unknown whether this response differs with increasing magnitudes of load.
miR‐221, miR‐222, miR‐21‐5p and miR‐27a‐5p were significantly increased in ex vivo cartilage explants subjected to increasing load magnitude and in in vivo joint cartilage exposed to abnormal loading.
TIMP3 and CPEB3 are putative miR targets in chondrocytes
Identification of mechanically regulated miRs that have potential to impact on tissue homeostasis provides a mechanism by which load‐induced tissue behaviour is regulated, in both health and pathology, in all physiological systems.
Introduction
Mechanical loading is essential with respect to regulating the functional capabilities of physiological systems including the musculoskeletal, cardiovascular and nervous system; this is achieved, at the cell and tissue level, by adapting to changes in mechanical load and/or metabolic stress applied. One of the major musculoskeletal tissues, articular cartilage, primarily functions to dissipate mechanical forces across the synovial joint surface and facilitates smooth, low‐friction movement. The biomechanical integrity of articular cartilage is reliant on the biochemical composition of the extracellular matrix (ECM) (Gilbert & Blain, 2018), and maintenance of cartilage tissue homeostasis, effected by the chondrocytes, is similarly dependent on mechanical load (Buckwalter et al. 2005). Joint articular cartilage is predominantly exposed to dynamic compressive forces, although both tensile strain and shear stresses also result from everyday movement (Lee et al. 2005). Application of moderate, physiological mechanical loads is essential for maintaining cartilage homeostasis by promoting anabolic activities such as increased production of ECM molecules, whereas abnormal, non‐physiological joint loading, as characterized by either overload or insufficient load, disrupts the homeostatic balance, favouring catabolism and cartilage degeneration, comprising the hallmark of osteoarthritis (OA) (Felson, 2013).
Mechano‐regulation of cellular activities within physiological systems is known to occur through epigenetic mechanisms (e.g. RNA silencing). Primary contributors to RNA silencing are the microRNAs (miR), which are small (20–2 3bp), non‐coding cytoplasmic RNAs that control the post‐transcriptional regulation of one‐third of all genes and are important in development, homeostasis and degeneration of tissues, including articular cartilage (Goldring & Marcu, 2012). Epigenetic studies have demonstrated that mechanical force has an impact on cellular responses through regulation of miR expression levels in tendon fibroblasts (Mendias et al. 2012), smooth muscle cells (Song et al. 2012), trabecular meshwork cells (Luna et al. 2011) and endothelial cells (Qin et al. 2010; Weber et al. 2010; Zhou et al. 2011). A small number of miRs were also identified as being mechanosensitive in chondrocytes (Dunn et al. 2009; Guan et al. 2011; Jin et al. 2014; Yang et al. 2016; Cheleschi et al. 2017). However, these studies were performed on isolated cells devoid of a substantial ECM, a feature known to be critical for cell–matrix mechano‐communications (Guilak et al. 2006).
Therefore, using articular cartilage as a model system, the present study aimed to identify miRs that respond to ‘physiological’ and ‘non‐physiological’ mechanical loads and to investigate the regulation of their potential downstream target genes.
Materials and methods
Reagents were from Sigma (Poole, UK) unless otherwise specified; molecular biology reagents and plastic ware were certified RNase and DNase‐free. Culture medium consisted of Dulbecco's modified Eagle's medium/Ham's F12‐glutamax (1:1; Life Technologies, Paisley, UK) supplemented with 100 μg mL−1 penicillin, 100 U mL−1 streptomycin, 50 μg mL−1 ascorbate‐2‐phosphate and 1 × insulin‐transferrin‐selenium ethanolamine (1 × ITS‐X; Life Technologies).
Preparation of articular cartilage explants and high‐density primary chondrocytes
Full depth articular cartilage explants were removed (5 mm biopsy punch; Selles Medical Limited, Hull, UK) from the metacarpophalangeal joint of <3‐week old bovine calves within 6 h of slaughter (F. Drury & Sons Abattoir, Swindon, UK); ethical approval was not required. Cartilage explants were equilibrated in culture medium for 3 days prior to mechanical load. Primary chondrocytes were isolated from full depth cartilage utilizing the same tissue source as explants, and enzymatic digestion was performed (Al‐Sabah et al. 2016). All cultures were maintained in 5% CO2 and 20% O2 at 37°C. Each experiment utilized tissue from between two and three animals, and repeat experiments utilized tissue from independent animals.
In vitro application of mechanical load to cartilage explants
Cartilage explants were subjected to either a ‘physiological’ (2.5 MPa, 1 Hz) or a ‘non‐physiological’ load (7 MPa, 1 Hz) for 15 min using the ElectroForce 3200 (TA Instruments, New Castle, DE, USA) (Al‐Sabah et al. 2016) and gene expression analysed at 2, 6 and 24 h post‐load; unloaded explants served as controls. Explants were immediately snap frozen and remained in liquid nitrogen (<48 h) until RNA extraction. Loading regimes were selected based on articular cartilage literature demonstrating that ≤5 MPa is generally accepted as a ‘physiological’ load (Grodzinsky et al. 2000; Fehrenbacher et al. 2003), whereas peak loads >5 MPa are considered degradative (i.e. ‘non‐physiological’) (Fehrenbacher et al. 2003); the frequency was set at 1 Hz, which has been demonstrated to resemble a human fast walking speed (Bader et al. 2011).
In vivo application of mechanical load
Twelve‐week old male C57Bl6 mice (∼25 g; Envigo, Huntington, UK) were randomly assigned to either experimental or control groups and randomly allocated to MB1 cages (960 cm2) in groups of five (12:12 h light/dark photocycles, with food and water available ad libitum). Animal husbandry and procedures were performed in compliance with the Animals (Scientific Procedures) Act 1986 [Home Office licence P287E87DF] according to Home Office and ARRIVE guidelines (Kilkenny et al. 2010). Mice were anaesthetized with isoflurane and custom‐built cups used to hold the right ankle and knee in flexion with a 30o offset prior to the application of a 0.5 N pre‐load (ElectroForce13200; TA Instruments, Elstree, UK). A single 12 N load at a velocity of 1.4 mm s−1 was then applied resulting in anterior cruciate ligament (ACL) rupture as described previously (Gilbert et al. 2018); mechanical loading was always conducted in the morning. Buprenorphine (0.05 mg kg−1) was administered s.c. to mice at the start of the experiment; animals were able to move freely and were monitored for welfare and lameness until termination of the experiment. Mice were culled by cervical dislocation at either day 1 or 7 post‐load and the knee articular cartilage was dissected and processed for histology (toluidine blue staining) as described previously (Gilbert et al. 2018) or immediately snap frozen and remained in liquid nitrogen until RNA extraction. These early time points allowed assessment of mechanically regulated miRs in cartilage prior to overt degenerative changes and ECM loss. Nine animals were utilized for quantification of miR levels and the representative histology depicting the loading model phenotype is derived from experiments published in Gilbert et al. (2018).
RNA extraction and reverse transcription for mRNA analysis
Total RNA was extracted from cartilage explants/chondrocytes using 500 μL of Trizol reagent (Invitrogen, Paisley, UK) (Al‐Sabah et al. 2016). RNA integrity was assessed (2100 Bioanalyzer and associated RNA 6000 Nano kit; Agilent Technologies, Wokingham, UK) and RNA integrity numbers >8.5 were observed. cDNA (total volume of 20 μL) was synthesized from 300 ng of total RNA using Superscript III reverse transcriptase in conjunction with 0.5 μg of random primers (Promega, Southampton, UK) in accordance with the manufacturer's instructions (Invitrogen).
RNA extraction and reverse transcription for miR analysis
Total RNA was extracted from cartilage explants/chondrocytes as described above, except 1 mL of Trizol reagent was used. After ethanol precipitation, total RNA was purified using a mirVana miR Isolation Kit (Ambion, Paisley, UK) in accordance with the manufacturer's instructions. RNA integrity numbers of >8.0 were observed. cDNA of mature miRs was generated separately from total RNA (5 ng) using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Paisley, UK) involving 50 U of MultiScribe Reverse Transcriptase and stem‐looped reverse transcription primers, specific to individual miRs, from TaqMan MicroRNA Assays (Applied Biosystems, Paisley, UK) in accordance with the manufacturer's instructions.
miR next generation sequencing and bioinformatic analysis
Mechanically‐regulated articular cartilage miRs were identified using next generation sequencing (NGS) using >3.5 μg of RNA per sample. Procedures were conducted in accordance with the manufacturers’ instructions. Library preparation was conducted on 450 ng of total RNA using the NEB Next Small RNA Library Prep Set for Illumina (Multiplex Compatible: BioLabs, Hitchin, UK) and amplified cDNA was purified using a QIAquick PCR Purification Kit (Qiagen, Crawley, UK). miR libraries were selected by running purified cDNA samples on 8% (v/v) polyacrylamide gels and excising bands located at ∼140 bp (Crowe et al. 2016). A Multiplex Compatible kit (NEB Next Small RNA Library Prep Set for Illumina) was used to elute and purify the miRs, and the concentration of miR libraries assessed prior to analysis on a HiSeq Sequencing System (The Genome Analysis Centre, Norwich, UK). miR deep sequencing data (raw FASTQ files) were run through FastQC and Cutadapt (Martin 2011), and trimmed FASTQ files were aligned against known bos taurus miR sequences from miRBase (http://www.mirbase.org). Quantification was determined by counting aligned reads against a reference, using a combination of RSamTools and ShortRead (Li et al. 2009) bioconductor packages. Differential expression was assessed using DESeq2 (Love et al. 2014). Global experimental variance was analysed using principal component analysis to assess for outlier samples and statistical significance from differential expression tests was determined by retaining miRs that had an adjusted P < 0.05.
Manipulation of miR expression levels in high‐density chondrocyte cultures
Primary bovine chondrocytes were seeded onto six‐well culture plates (VWR, Lutterworth, UK) at a density of 4 × 106 cells per well in antibiotic‐free culture media and incubated at 37°C for 24 h prior to transfection. Chondrocytes were transfected for 48 h with 50 nm mirVana miR inhibitors (Applied Biosystems) or 50 nm miScript miR mimics (Qiagen) using DharmaFECT1 lipid reagent (Dharmacon, Cambridge, UK) in accordance with the manufacturer's instructions; mirVana miR Inhibitor Negative Control #1 (Applied Biosystems) and AllStars negative control small interfering RNA (siRNA) (Qiagen) were utilized as transfection controls (50 nm).
Quantification of miRNA and mRNA transcripts
Quantification of mRNA or miR in experimental samples was performed using a MxPro3000 QPCR system (Agilent Technologies, Stockport, UK) and measured using either reference gene primers (MWG‐Biotech AG, Ebersberg, Germany) or bovine‐specific TaqMan probes (Applied Biosystems, Paisley, UK) in conjunction with either Brilliant III Ultra‐Fast SYBR Green QPCR Master Mix (Agilent Genomics, Berkshire, UK) or TaqMan Fast Advanced Master Mix (Applied Biosystems, Paisley, UK). Reference gene primers (200 nM final concentration (Al‐Sabah et al. 2016)) including SDHA, YWHAZ, HPRT, 18 s and β‐actin were validated as per MIQE guidelines (Bustin et al. 2009). Cycling conditions were: 95°C‐3 min (1 cycle), 95°C‐15 s followed by 60°C‐30 s (40 cycles) with an additional dissociation cycle of 95°C‐1min, 60°C‐30 s followed by 95°C‐30 s (1 cycle) to confirm primer specificity with SYBR Green detection. Relative quantification was calculated using the 2−ΔΔCT method (Livak & Schmittgen, 2001), with unloaded controls as a reference group to quantify relative changes in transcript expression. Fold change was normalized to the geometric mean of 2–3 reference genes whose expression was identified as stable under the experimental condition using RefFinder software (https://www.heartcure.com.au/for-researchers/).
Luciferase activity assays
The 3′‐UTR of mRNAs, containing the predicted binding site of target miRs, were cloned into pmirGLO dual‐luciferase miRNA target expression vector (Promega, Southampton, UK) by In‐Fusion (Takara Bio Europe, Saint‐Germain‐en‐Laye, France) and construct sequences verified (for primer sequences, see Table 1). SW1353 chondrosarcoma cells (∼20 000 cells cm–2) were co‐transfected with 50 nm miRNA mimics with the reporter plasmids (500 ng mL−1) (Barter et al. 2015); transfection of 50 nm AllStars negative control siRNA with the reporter plasmids was used as control. Following a 24 h transfection, cells were lysed and luciferase levels were determined using a Promega GloMax luminometer and the Dual‐Luciferase reporter assay system (Promega).
Table 1.
Sequences of primers used to clone the 3′‐UTR of mRNAs containing the predicted binding site of target miRs
| Target | 5′‐ to 3′ Oligo sequence | Annealing temperature (°C) |
|---|---|---|
| TIMP3 UTR | F 5′‐GCTCGCTAGCCTCGACTGAGCTTCCCTTGGACACT‐3′ R 5′‐CGACTCTAGACTCGAGCTAAAGGGAAAGGCGGAT‐3′ | 60 |
| CPEB3 UTR | F 5′‐GCTCGCTAGCCTCGAAAGGAGGGAAAAGAGAGGGC‐3′ R 5′‐CGACTCTAGACTCGAAACAGAGCACCGCAAAGTAC‐3′ | 60 |
Statistical analysis
Quantitative PCR (qPCR) data are presented as the mean ± 95% confidence intervals (CIs) after normalization to identified reference genes for explants (SDHA and YWHAZ), transfected cells (HPRT and YWHAZ) or in vivo model (U6, 18s and β‐actin) and further normalized to untreated controls. Experiments were performed on explants (n = 6), transfected cells (n = 3) and in vivo studies (n = 9), with three independent repeats for explant and cell studies. Data were assessed for normality and differences in variances and transformed where required. One‐way ANOVA and Fisher's post hoc test were performed to determine significance of mechanical load or manipulation of miR expression levels on gene expression, respectively; the results were considered statistically significant at P < 0.05 (Minitab, version 17; Minitab, LLC, State College, PA, USA).
Results
Identification and validation of mechanically‐regulated miRs in cartilage explants
NGS was performed on cartilage explants subjected to a 2.5 or 7 MPa load (1 Hz, 15 min) to identify mechano‐sensitive miRs (Table 2); unloaded explants served as controls. NGS analysis was conducted at 2, 6 or 24 h post‐load to investigate temporal differences in miR expression. A small number of annotated miRs were significantly regulated by load at 2 h (8 miRs) (Table 2) and 6 h (5 miRs) (Table 2), with 17 miRs detected at 24 h post‐load (Table 2). A greater number of miRs were only regulated by the non‐physiological 7 MPa load (Table 2). Given the number of significant changes at 24 h post‐load (Table 2), the top five most significantly up‐regulated (i.e. miR‐222, ‐27a‐5p, ‐221, ‐543 and ‐21‐5p) and the two most significantly down‐regulated (i.e. miR‐451 and ‐483) were validated for this time point using TaqMan qPCR on individual RNA samples (Fig. 1). Relative to unloaded, a 7 MPa load increased expression of miR‐21‐5p (two‐fold; P = 0.034) (Fig. 1 A), miR‐27‐5p (2.56‐fold; P = 0.001) (Fig. 1 B), miR‐221 (3.85‐fold; P < 0.001) (Fig. 1 C) and miR‐222 (3.78‐fold; P < 0.001) (Fig. 1 D), and decreased miR‐483 expression (two‐fold; P = 0.047) (Fig. 1 F). A loading magnitude‐dependent regulation of miRs was observed between explants subjected to a 7 MPa vs. 2.5 MPa load: miR‐27‐5p (2.4‐fold, P < 0.001) (Fig. 1 B), miR‐221 (2.55‐fold, P = 0.011) (Fig. 1 C) and miR‐222 (2.83‐fold, P = 0.002) (Fig. 1 D). Although miR‐seq indicated that a 7 MPa load down‐regulated miR‐451 levels (2.1‐fold, P = 0.002) (Table 2) and up‐regulated miR‐543 levels (2.62‐fold, P < 0.001; P = 0.005) (Table 2) relative to unloaded cartilage, this was not verified using qPCR (Fig. 1 E and G).
Table 2.
Mean fold‐change and statistical significance of mechanically‐regulated miRs in articular chondrocytes subjected to loads of 2.5 or 7 MPa (1 Hz, 15 min), to represent a physiological or non‐physiological load respectively, and analysed 2, 6 and 24 h post‐cessation of load (unloaded explants served as controls)
| UL vs. 2.5MPa | UL vs. 7MPa | 2.5 vs. 7MPa | ||||
|---|---|---|---|---|---|---|
| FC | Padj | FC | Padj | FC | Padj | |
| Analysed 2 h post‐cessation of load | ||||||
| miR‐27a‐5p | 4.420 | 2.79 × 10–22 | 7.092 | 3.08 × 10–40 | ||
| miR‐2898 | 0.588 | 0.008 | 0.408 | 3.16 × 10–10 | ||
| miR‐2478 | 0.566 | 0.001 | ||||
| miR‐98 | 1.698 | 0.001 | ||||
| miR‐23b‐3p | 1.539 | 0.004 | ||||
| miR‐1260b | 0.647 | 0.025 | ||||
| miR‐23a | 1.464 | 0.034 | ||||
| miR‐148b | 1.644 | 0.039 | ||||
| Analysed 6 h post‐cessation of load | ||||||
| miR‐486 | 0.521 | 0.002 | ||||
| miR‐677 | 2.149 | 5.50 × 10–5 | 1.933 | 0.001 | ||
| miR‐222 | 1.568 | 0.008 | ||||
| miR‐2889 | 2.393 | 1.54 × 10–6 | ||||
| miR‐1249 | 0.562 | 0.013 | ||||
| Analysed 24 h post‐cessation of load | ||||||
| miR‐222 | 1.814 | 0.003 | 7.409 | 8.81 × 10–51 | 4.085 | 4.00 × 10–25 |
| miR‐27a‐5p | 3.019 | 1.14 × 10–16 | 2.027 | 5.84 × 10–7 | ||
| miR‐221 | 3.393 | 1.63 × 10–13 | 2.593 | 3.58 × 10–8 | ||
| miR‐543 | 2.619 | 1.30 × 10–9 | 1.725 | 0.005 | ||
| miR‐21‐5p | 2.267 | 6.67 × 10–6 | 1.719 | 0.013 | ||
| miR‐495 | 1.775 | 1.45 × 10–4 | ||||
| miR‐451 | 0.481 | 0.002 | ||||
| miR‐425‐5p | 0.626 | 0.010 | 0.672 | 0.037 | ||
| miR‐20a | 1.603 | 0.012 | ||||
| miR‐7 | 1.699 | 0.017 | ||||
| miR‐760‐3p | 1.715 | 0.017 | ||||
| miR‐2318 | 1.8518 | 0.022 | ||||
| miR‐2344 | 1.829 | 0.030 | ||||
| miR‐431 | 1.817 | 0.030 | ||||
| miR‐155 | 1.472 | 0.042 | ||||
| miR‐100 | 1.429 | 0.048 | ||||
| miR‐483 | 0.599 | 0.025 | ||||
Data are representative of three independent experiments (n = 6 explants per individual experiment).
Figure 1. Validation of mechanically‐regulated miRs in cartilage explants using qPCR.
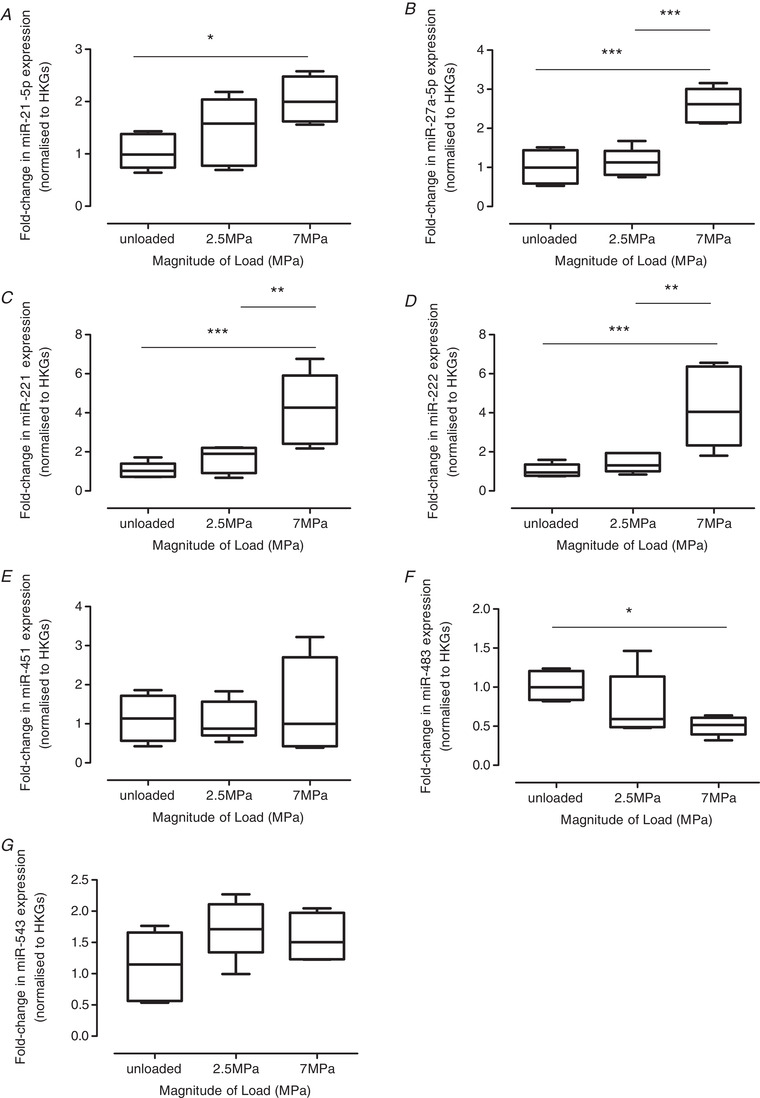
qPCR validation of mechanically‐regulated miRs, identified by NGS, in cartilage explants subjected to loads of 2.5 or 7 MPa (1 Hz, 15 min) and analysed 24 h post‐cessation of load for (A) miR‐21‐5p, (B) miR‐27a‐5p, (C) miR‐221, (D) miR‐222, (E) miR‐451, (F) miR‐483 and (G) miR‐453; unloaded explants served as controls. miR levels were normalized to the geometric mean of two reference genes (SDHA, YWHAZ) and further normalized relative to the unloaded control cDNAs. Data are presented as box plots depicting the mean ± 95% CI (n = 6 explants) and are representative of three independent experiments. Statistical analysis was performed using one‐way ANOVA with Tukey's post hoc test.
In vivo validation of miR‐21‐5p, miR‐27‐5p, miR‐221 and miR‐222 mechano‐regulation
The physiological relevance of identified mechanically‐regulated miRs was determined in a murine in vivo model of post‐traumatic OA, in which ACL rupture induces mechanical instability, by applying an abnormal load to the knee joint (Gilbert et al. 2018). Synovial infiltration occurs rapidly followed by extensive joint degeneration as characterized by articular cartilage loss and bone remodelling by day 21 (Fig. 2 A) (Gilbert et al. 2018). However, at the earlier stages analysed in the present study, the articular cartilage is intact. Of the original miRs identified (Table 1), four that were successfully validated by qPCR (miR‐221, ‐222, ‐21‐5p and ‐27‐5p) (Fig. 1) were subsequently analysed in vivo. No significant effects were detected after 1 day of destabilization; however, after 7 days of mechanical instability, miR‐221 (2.20‐fold, P < 0.001) (Fig. 2 B), miR‐222 (1.56‐fold, P = 0.070) (Fig. 2 C), miR21‐5p (4.75‐fold, P = 0.002) (Fig. 2 D) and miR‐27‐5p (4.21‐fold, P = 0.003) (Fig. 2 E) were up‐regulated compared to naïve mice (control). Furthermore, miR‐221 (2.05‐fold, P = 0.003) (Fig. 2 B), miR‐222 (1.90‐fold, P = 0.030) (Fig. 2 C), miR21‐5p (7.99‐fold, P = 0.001) (Fig. 2 D) and miR‐27‐5p (3.19‐fold, P = 0.013) (Fig. 2 E) were all significantly up‐regulated compared to mice after 1 day of joint instability.
Figure 2. Validation of mechanically regulated miRNAs in a murine in vivo model of load‐induced joint degeneration.
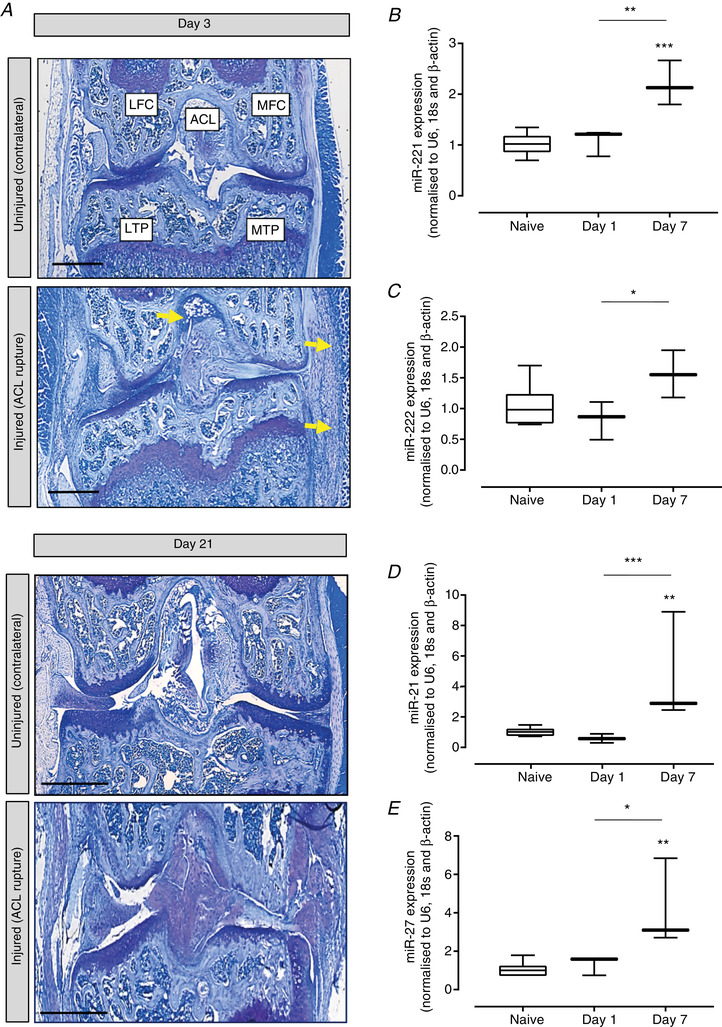
A, toluidine blue staining of a representative mouse knee joint at days 3 and 21 after ACL rupture to induce joint instability/joint degeneration. MTP, medial tibial plateau; MFC, medial femoral condyle; LTP, tibial plateau; LFC, lateral femoral condyle; ACL, anterior cruciate ligament. Yellow indicates inflammatory cell infiltrate. Validation of differential expression of (B) miR‐221, (C) miR‐222, (D) miR‐21‐5p and (E) miR‐27‐5p in articular cartilage after normalization to the geometric mean of the reference genes U6, β‐actin and 18s and further normalization to the uninjured knee cartilage. Data are presented as box plots depicting the mean ± 95% CI (n = 3 animals per experimental time point). Statistical analysis was performed using one‐way ANOVA with Tukey's post hoc test. [Color figure can be viewed at wileyonlinelibrary.com]
miR target gene validation
Potential miR target genes identified by NGS were determined using Targetscan (http://www.targetscan.org) in conjunction with an assessment of their relevance to mechanical load or cartilage homeostasis as determined using the literature; putative target genes were examined by manipulation of expression levels using specific miR mimics or inhibitors (Fig. 3). Three putative miR‐21‐5p targets were selected: cytoplasmic polyadenylation element binding protein 3 (CPEB3), matrix metalloproteinase 13 (MMP13) and tissue inhibitor of metalloproteinase 3 (TIMP3). miR‐221 and miR‐222 seed sites are identical; hence, the selected putative target genes included: CPEB3, leukaemia inhibitory factor receptor (LIFR) and TIMP3. miR‐27a target gene validation was not performed because a consistent reduction in miR‐27a expression was not achieved using specific antagomirs. qPCR analysis confirmed that mimic‐induced elevations in miR‐221 levels resulted in a significant reduction in TIMP3 (P = 0.006) (Fig. 3 A). Conversely, inhibition of miR‐221 expression correlated with a significant increase in TIMP3 transcription (P = 0.003) (Fig. 3 B). Similarly, a mimic‐induced increase in miR‐222 levels led to a significant reduction in TIMP3 (P = 0.006) (Fig. 3 C). Conversely, inhibition of miR‐222 expression correlated with a significant increase in TIMP3 (P = 0.025) (Fig. 3 D). No other putative target genes were robustly regulated by miR‐221 or 222. Mimic‐induced miR‐21 levels resulted in a significant reduction in TIMP3 (P = 0.006) (Fig. 3 E) and CPEB3 transcription (P = 0.015) (Fig. 3 G). Conversely, inhibition of miR‐21 expression correlated with a significant increase in TIMP3 (P = 0.010) (Fig. 3 F) and no significant effect on CPEB3 (P = 0.068) (Fig. 3 H). MMP13 expression was not consistently regulated by miR‐21 (data not shown).
Figure 3. Validation of TIMP3 and CPEB3, putative target genes of miR‐221, miR‐222 or miR‐21, using TaqMan qPCR.
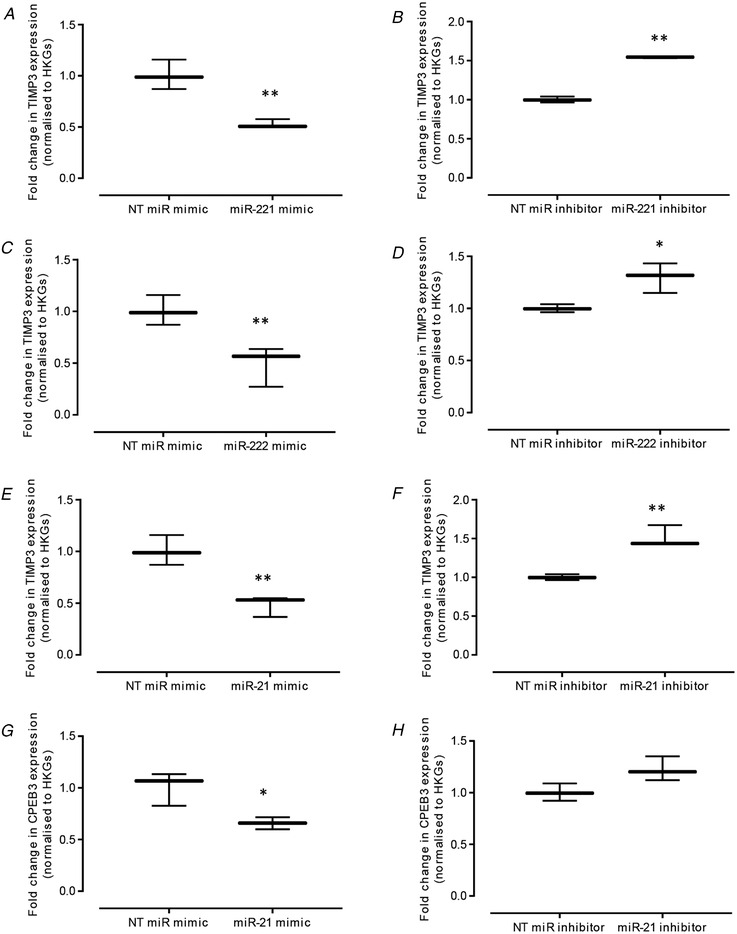
Primary bovine chondrocytes were treated with either 50 nm miR mimic, 50 nm inhibitor or negative control siRNAs for each respective miR for 48 h, prior to analysis of the effect of overexpression and knockdown of miR‐221 on TIMP3 transcription (A and B), miR‐222 on TIMP3 transcription (C and D), and miR‐21 on TIMP3 (E and F) and CPEB3 (G and H) transcript levels after normalization to the geometric mean of the reference genes HPRT and YWHAZ and further normalization to respective negative control siRNAs. Data are presented as the mean ± 95% CI (n = 3 wells) and are representative of three independent experiments. Statistical analysis was performed using one‐way ANOVA with Tukey's post hoc test.
Activation of the 3′‐UTR of target mRNAs containing the predicted seed sites was investigated (Fig. 4). Addition of miR‐21 mimic significantly suppressed luciferase activity regulated by TIMP3 (P = 0.053) (Fig. 4 A) and Cpeb3 3′‐UTRs (P = 0.010) (Fig. 4 B). Furthermore, miR‐222 mimic also significantly inhibited CPEB3 3′‐UTR regulated luciferase activity (P = 0.010) (Fig. 4 B). Surprisingly, and in contrast to qPCR validation, miR‐222 overexpression significantly increased luciferase activity regulated by TIMP3 3′‐UTR (P = 0.030) (Fig. 4 A). Although this contradicts the miR‐222 mimic data demonstrating a significant TIMP3 reduction (Fig. 3 C), it does substantiate the load‐induced TIMP3 observed in the in vitro loading model where TIMP3 transcription was elevated in response to 7 MPa load (4.8‐fold, P < 0.001) (Fig. 4 C).
Figure 4. Verification of 3′‐UTR activation of target mRNAs containing the predicted miR seed sites using a luciferase promoter assay.
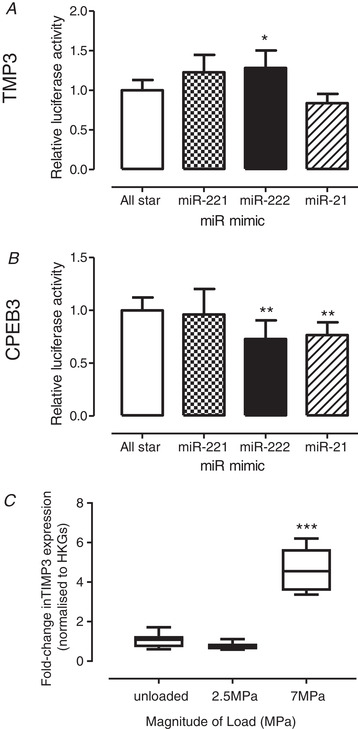
SW1353 chondrosarcoma cells were co‐transfected with reporter plasmids containing either (A) TIMP3 or (B) CPEB3 3′‐UTRs and 50 nm miR‐221, miR‐222 or miR‐21‐5p mimics, or the negative control siRNA, for 24 h and luciferase levels were determined. Data are presented as the mean ± 95% CI (n = 3 wells) and are representative of three independent experiments. C, TIMP3 mRNA levels, as assessed using qPCR, in cartilage explants subjected to loads of 2.5 or 7 MPa (1 Hz, 15 min) and analysed 24 h post‐cessation of load; unloaded explants served as controls. mRNA levels were normalized to the geometric mean of two reference genes (SDHA, YWHAZ) and further normalized relative to the unloaded control cDNAs. Data are presented as box plots depicting the mean ± 95% CI (n = 6 explants) and are representative of three independent experiments. Statistical analysis was performed using one‐way ANOVA with Tukey's post hoc test.
Discussion
Physiological forces are critical for maintaining tissue homeostasis, and the involvement of epigenetic mechanisms such as mechano‐regulation of miR expression occurs in many tissues, including articular cartilage (Dunn et al. 2009; Guan et al. 2011; Jin et al. 2014; Yang et al. 2015; Cheleschi et al. 2017). However, our understanding of miR involvement in response to different magnitudes of mechanical forces and, specifically, its impact on controlling mechanically induced tissue homeostasis is still in its infancy. The present study investigated the mechano‐regulation of miRs in articular cartilage tissue explants subjected to ‘physiological’ and ‘non‐physiological’ loads in vitro and validated regulated miRs in a murine in vivo model of abnormal joint loading. In addition, the study identified downstream miR targets to provide insight on mechanisms of mechanically mediated cartilage homeostasis. Importantly, the seed regions of the miRs of interest analysed in the present study are evolutionarily conserved across bovine, mouse and human species, indicating their potential physiological relevance.
Analysis of the miR‐seq data illustrated that (i) a miR‐mediated response to a 15 min loading episode was most noticeable at 24 h post‐load and (ii) differentially regulated miRs were largely responsive to non‐physiological compressive loads; the small number of miRs that were significantly regulated in response to physiological load probably reflects the loading regimen period. The miRs that were identified and validated to be most robustly altered by non‐physiological load compared to unloaded controls and to physiological load were miR‐221 and miR‐222. This confirms the mechano‐sensitive nature of miR‐221 and miR‐222, previously shown in cardiomyocytes after cardiac overload (El‐Armouche et al. 2010), as well as in tendon fibroblasts (Mendias et al. 2012), engineered cartilage constructs in response to a catabolic loading regimen (Hecht et al. 2019) and anterior weight‐bearing cartilage relative to the posterior non‐weight bearing tissue in bovine stifle joints (Dunn et al. 2009).
Chondrogenic markers COL2A1 and SOX9 have been identified as putative gene targets for miR‐221 and miR‐222 (conserved seed site) that may influence cartilage homeostasis (Lolli et al. 2014); furthermore, miR‐221 silencing strongly enhanced in vivo cartilage repair (Lolli et al. 2016). miR‐221 inhibition also enhanced expression of chondrocyte‐like phenotypic markers in intervertebral disc cells (Penolazzi et al. 2018). Therefore, miR‐221 and miR‐222 induction, observed in the cartilage explants in response to non‐physiological load (Al Sabah A., Duance V. C., Blain E. J., unpublished observations), suggests an attempt to remodel the cartilage tissue to confer a more appropriate biomechanical response.
Analysis of downstream target genes identified robust regulation of TIMP3 only. However, although TIMP3 was clearly regulated via overexpression/inhibition studies in primary chondrocytes, this did not reflect observations in the SW1353 chondrosarcoma cell line for 3′‐UTR activity using the luciferase assay or recapitulate events in tissue demonstrating that other, as yet unidentified, targets are regulated by miR‐221 and miR‐222 to elicit effects. These conflicting findings may be explained by the different experimental systems used in the present study, thus potentially masking the effects of other regulatory contributors with respect to the influence of miR‐221 and miR‐222 on Timp3 expression. Another possibility that might explain the simultaneous elevation of both the tested miRs and TIMP3 is a regulatory loop , such that elevated TIMP3 expression induces higher expression of these miRs to reduce Timp3 transcript levels in cells over time. Analysis at time points beyond 24 h post‐load would provide insight as to whether potential regulatory loops exist.
Two other miRs robustly regulated by a magnitude‐dependent load in our in vitro and in vivo loading models were miR‐21‐5p and miR‐27a‐5p. To the best of our knowledge, this is the first report of the mechano‐regulation of these miRs in articular cartilage. However, miR‐21 mechano‐regulation occurs in other cell types; tensile strain induced miR‐21 expression in human aortic smooth muscle cells (Song et al. 2012), and both laminar (Weber et al. 2010) and oscillatory shear stress (Zhou et al. 2011) elevated miR‐21 levels in human umbilical vein endothelial cells. By contrast, pulsatile shear stress inhibited miR‐21 expression in these endothelial cells (Zhou et al. 2011), revealing the mechano‐sensitive nature of these molecules. In the present study, both TIMP3 and CPEB3 were identified as downstream targets of miR‐21‐5p; however, as noted previously, TIMP3 is not negatively correlated with miR‐21‐5p levels in our model systems. Furthermore, CPEB3 levels were not significantly regulated in the present study, indicating that, although these genes are direct targets of miR‐21‐5p in primary chondrocytes, they are not directly regulated in our models. As a result of the complexities of such signalling mechanisms in the tissue, further experiments are clearly required to determine the interplay of these miRs and their mechanistic activities in cartilage homeostasis, which both remain beyond the scope of the present study.
miR‐27a‐5p was robustly regulated by mechanical load both in vitro and in vivo. Mechano‐regulation of miR‐27a in articular cartilage is a novel finding and corroborates studies demonstrating up‐regulation of both miR‐27a and miR‐27b in endothelial cells subjected to laminar flow (Urbich et al. 2012) and endothelial cells exposed to cyclic tensile strain (Wang et al. 2017). Downstream targets of miR‐27‐5p, which are known to be regulated in in situ cartilage explants in response to non‐physiological load Al‐Sabah et al. (unpublished data), include WNT signalling molecules such as DKK2 (Tao et al. 2015; Wu et al. 2019) and sFRP1 (Wu et al. 2019). Future studies will explore the relationship between mechano‐sensitive miR‐27‐5p and downstream regulation of WNT signalling components in cartilage homeostasis.
A reduction in miR‐483 levels was observed in response to non‐physiological load and is the first report of its mechano‐sensitivity in articular cartilage. Its potential role in cartilage homeostasis is not well defined, with conflicting evidence suggesting anabolic (Yang et al. 2015) and catabolic outcomes (Xu et al. 2017; Wang et al. 2019); hence, its observed reduction in response to abnormal load may reflect an attempt at tissue remodelling.
A limitation of the present study is use of immature articular cartilage removed from underlying subchondral bone, which could influence mechano‐biological outcomes. However, to mitigate this limitation, we validated identified miRs in an in vivo model of abnormal joint loading to confirm their mechano‐regulation; interestingly, many of the miRs regulated by load in our in vitro and in vivo models have also been reported to be differentially expressed in OA (Tardif et al. 2009; Zhang et al. 2014; Song et al. 2015; Wang et al. 2019), lending weight to their relevance in cartilage homeostasis.
In conclusion, the loading magnitude‐dependent regulation of specific miRs identified in the present study, as well as their potential to impact on tissue homeostasis, has direct relevance to other physiological systems that are mechano‐sensitive. Furthermore, it provides a pivotal mechanism by which load‐induced tissue behaviours are regulated, in both health and pathology, and is critical to understand with respect to successful tissue engineering strategies in physiological systems.
Additional information
Competing interests
The authors declare that they have no competing interests.
Author contributions
PS, VCD, DAY and EJB were responsible for study conception and design. PS, SJG, SC, JT, AJS and EJB were responsible for data acquisition and statistical analysis. PS, SJG, SC, MJB, VCD, DAY and EJB were responsible for data analysis and interpretation. EJB was responsible for manuscript preparation. PS, SJG, SC, JT, AJS, MJB, VCD, DAY and EJB were responsible for critical revision of the draft. All authors approved the final article submitted for publication and take full responsibility for integrity of the study.
Funding
The sponsors had no role in the study design, collection, analysis and interpretation of data; in the writing of the article; nor in the decision to submit the article for publication.
Supporting information
Statistical Summary Document
Acknowledgements
This study was funded by The President's Research scholarship (Cardiff University), Versus Arthritis (18461, 19424 and 510390), the Cardiff Institute for Tissue Engineering and Repair research bursary and the JGW Patterson Foundation. Equipment was provided by the Biomechanics and Bioengineering Research Centre Versus Arthritis (18461, 510390).
Biography
Dr Sophie Gilbert has worked in the connective tissue field for 25 years focussing on the relationship between biomechanics and biology of joint tissues. She has extensive research experience in extracellular matrix biology in normal and disease states including its epigenetic regulation under mechanical load. She has been pivotal in developing and characterising cell and animal models to investigate the signalling mechanisms that drive mechanical load and inflammation induced joint pathology. These include a novel, and recently published, anterior cruciate ligament (ACL) rupture model of post‐traumatic osteoarthritis which in 2018 received the award for “Excellence in Basic, Clinical, and Translational Science” bestowed by the Journal of Orthopaedic Research. Her goal is to elucidate potential mechanisms of joint destruction that may be targeted in the treatment or diagnosis of arthritis.

Edited by: Michael Hogan & Alicia D'Souza
Data availability statement
The data that support the findings of this study are being made openly available in GEO (GSE158571).
References
- Al‐Sabah A, Stadnik P, Gilbert SJ, Duance VC & Blain EJ (2016). Importance of reference gene selection for articular cartilage mechanobiology studies. Osteoarthritis Cartilage 24, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader DL, Salter DM & Chowdhury TT (2011). Biomechanical influence of cartilage homeostasis in health and disease. Arthritis 2011, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter MJ, Tselepi M, Gomez R, Woods S, Hui W, Smith GR, Shanley DP, Clark IM & Young DA (2015). Genome‐wide microRNA and gene analysis of mesenchymal stem cell chondrogenesis identifies an essential role and multiple targets for miR‐140‐5p. Stem Cells 33, 3266–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckwalter JA, Mankin HJ & Grodzinsky AJ (2005). Articular cartilage and osteoarthritis. Instr Course Lect 54, 465–480. [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J & Wittwer CT (2009). The MIQE guidelines: minimum information for publication of quantitative real‐time PCR experiments. Clin Chem 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Cheleschi S, De Palma A, Pecorelli A, Pascarelli NA, Valacchi G, Belmonte G, Carta S, Galeazzi M & Fioravanti A (2017). Hydrostatic pressure regulates microRNA expression levels in osteoarthritic chondrocyte cultures via the Wnt/beta‐catenin pathway. Int J Mol Sci 18, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe N, Swingler TE, Le LT, Barter MJ, Wheeler G, Pais H, Donell ST, Young DA, Dalmay T & Clark IM (2016). Detecting new microRNAs in human osteoarthritic chondrocytes identifies miR‐3085 as a human, chondrocyte‐selective, microRNA. Osteoarthritis Cartilage 24, 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W, DuRaine G & Reddi AH (2009). Profiling microRNA expression in bovine articular cartilage and implications for mechanotransduction. Arthritis Rheum 60, 2333–2339. [DOI] [PubMed] [Google Scholar]
- El‐Armouche A, Schwoerer AP, Neuber C, Emons J, Biermann D, Christalla T, Grundhoff A, Eschenhagen T, Zimmermann WH & Ehmke H (2010). Common microRNA signatures in cardiac hypertrophic and atrophic remodeling induced by changes in hemodynamic load. PLoS One 5, e14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher A, Steck E, Rickert M, Roth W & Richter W (2003). Rapid regulation of collagen but not metalloproteinase 1, 3, 13, 14 and tissue inhibitor of metalloproteinase 1, 2, 3 expression in response to mechanical loading of cartilage explants in vitro. Arch Biochem Biophys 410, 39–47. [DOI] [PubMed] [Google Scholar]
- Felson DT (2013). Osteoarthritis as a disease of mechanics. Osteoarthritis Cartilage 21, 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SJ & Blain EJ (2018). Cartilage mechanobiology: how chondrocytes respond to mechanical load. In Mechanobiology in Health and Disease, ed. Verbruggen S, pp. 99–126. Elsevier, London. [Google Scholar]
- Gilbert SJ, Bonnet CS, Stadnik P, Duance VC, Mason DJ & Blain EJ (2018). Inflammatory and degenerative phases resulting from anterior cruciate rupture in a non‐invasive murine model of post‐traumatic osteoarthritis. J Orthop Res 36, 2118‐2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring MB & Marcu KB (2012). Epigenomic and microRNA‐mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol Med 18, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodzinsky AJ, Levenston ME, Jin M & Frank EH (2000). Cartilage tissue remodeling in response to mechanical forces. Annu Rev Biomed Eng 2, 691–713. [DOI] [PubMed] [Google Scholar]
- Guan YJ, Yang X, Wei L & Chen Q (2011). MiR‐365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. FASEB J 25, 4457–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak F, Alexopoulos LG, Upton ML, Youn I, Choi JB, Cao L, Setton LA & Haider MA (2006). The pericellular matrix as a transducer of biomechanical and biochemical signals in articular cartilage. Ann N Y Acad Sci 1068, 498–512. [DOI] [PubMed] [Google Scholar]
- Hecht N, Johnstone B, Angele P, Walker T & Richter W (2019). Mechanosensitive MiRs regulated by anabolic and catabolic loading of human cartilage. Osteoarthritis Cartilage 27, 1208‐1218, [DOI] [PubMed] [Google Scholar]
- Jin L, Zhao J, Jing W, Yan S, Wang X, Xiao C & Ma B (2014). Role of miR‐146a in human chondrocyte apoptosis in response to mechanical pressure injury in vitro. Int J Mol Med 34, 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne WJ, Cuthill IC, Emerson M & Altman DG (2010). Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. J Pharmacol Pharmacother 1, 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Grad S, Wimmer M & Alini M (2005). The influence of mechanical stimuli on articular cartilage tissue engineering. In Topics in Tissue Engineering, ed. Ashammakhi N & Reis RL, pp. 1–32. eBook. https://www.oulu.fi/spareparts/ebook_topics_in_t_e_vol2/abstracts/alini_0102.pdf [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, & 1000 Genome Project Data Processing Subgroup (2009). The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ & Schmittgen TD (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐delta delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lolli A, Lambertini E, Penolazzi L, Angelozzi M, Morganti C, Franceschetti T, Pelucchi S, Gambari R & Piva R (2014). Pro‐chondrogenic effect of miR‐221 and slug depletion in human MSCs. Stem Cell Rev 10, 841–855. [DOI] [PubMed] [Google Scholar]
- Lolli A, Narcisi R, Lambertini E, Penolazzi L, Angelozzi M, Kops N, Gasparini S, van Osch GJ & Piva R (2016). Silencing of antichondrogenic microRNA‐221 in human mesenchymal stem cells promotes cartilage repair in vivo. Stem Cells 34, 1801–1811. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna C, Li G, Qiu J, Epstein DL & Gonzalez P (2011). MicroRNA‐24 regulates the processing of latent TGFbeta1 during cyclic mechanical stress in human trabecular meshwork cells through direct targeting of FURIN. J Cell Physiol 226, 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011). Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet.J 17, 10–12. [Google Scholar]
- Mendias CL, Gumucio JP & Lynch EB (2012). Mechanical loading and TGF‐beta change the expression of multiple miRNAs in tendon fibroblasts. J Appl Physiol 113, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penolazzi L, Lambertini E, Bergamin LS, Roncada T, De Bonis P, Cavallo M & Piva R (2018). MicroRNA‐221 silencing attenuates the degenerated phenotype of intervertebral disc cells. Aging 10, 2001–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Wang X, Wang Y, Tang Z, Cui Q, Xi J, Li YS, Chien S & Wang N (2010). MicroRNA‐19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc Natl Acad Sci U S A 107, 3240–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Hu B, Qu H, Bi C, Huang X & Zhang M (2012). Mechanical stretch modulates microRNA 21 expression, participating in proliferation and apoptosis in cultured human aortic smooth muscle cells. PLoS One 7, e47657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Jin EH, Kim D, Kim KY, Chun CH & Jin EJ (2015). MicroRNA‐222 regulates MMP‐13 via targeting HDAC‐4 during osteoarthritis pathogenesis. BBA Clin 3, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H, Wang L, Zhou J, Pang P, Cai S, Li J, Mei S & Li F (2015). The transcription factor ccaat/enhancer binding protein beta (C/EBPbeta) and miR‐27a regulate the expression of porcine Dickkopf2 (DKK2). Sci Rep 5, 17972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif G, Hum D, Pelletier JP, Duval N & Martel‐Pelletier J (2009). Regulation of the IGFBP‐5 and MMP‐13 genes by the microRNAs miR‐140 and miR‐27a in human osteoarthritic chondrocytes. BMC Musculoskelet Disord 10, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbich C, Kaluza D, Fromel T, Knau A, Bennewitz K, Boon RA, Bonauer A, Doebele C, Boeckel JN, Hergenreider E, Zeiher AM, Kroll J, Fleming I & Dimmeler S (2012). MicroRNA‐27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood 119, 1607–1616. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang H, Sun Q, Yang J, Zeng C, Ding C, Cai D, Liu A & Bai X (2019). Chondrocyte mTORC1 activation stimulates miR‐483‐5p via HDAC4 in osteoarthritis progression. J Cell Physiol 234, 2730–2740. [DOI] [PubMed] [Google Scholar]
- Wang L, Bao H, Wang KX, Zhang P, Yao QP, Chen XH, Huang K, Qi YX & Jiang ZL (2017). Secreted miR‐27a induced by cyclic stretch modulates the proliferation of endothelial cells in hypertension via GRK6. Sci Rep 7, 41058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Baker MB, Moore JP & Searles CD (2010). MiR‐21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem Biophys Res Commun 393, 643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Gu Q, Chen X, Mi W, Wu T & Huang H (2019). MiR‐27a targets DKK2 and SFRP1 to promote reosseointegration in the regenerative treatment of peri‐implantitis. J Bone Miner Res 34, 123–134. [DOI] [PubMed] [Google Scholar]
- Xu R, Li J, Wei B, Huo W & Wang L (2017). MicroRNA‐483‐5p modulates the expression of cartilage‐related genes in human chondrocytes through down‐regulating TGF‐β1 expression. Tohoku J Exp Med 243, 41–48. [DOI] [PubMed] [Google Scholar]
- Yang M, Zhang L & Gibson GJ (2015). Chondrocyte miRNAs 221 and 483–5p respond to loss of matrix interaction by modulating proliferation and matrix synthesis. Connect Tissue Res 56, 236–243. [DOI] [PubMed] [Google Scholar]
- Yang X, Guan Y, Tian S, Wang Y, Sun K & Chen Q (2016). Mechanical and IL‐1beta responsive miR‐365 contributes to osteoarthritis development by targeting histone deacetylase 4. Int J Mol Sci 17, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jia J, Yang S, Liu X, Ye S & Tian H (2014). MicroRNA‐21 controls the development of osteoarthritis by targeting GDF‐5 in chondrocytes. Exp Mol Med 46, e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ, Li JY & Chien S (2011). MicroRNA‐21 targets peroxisome proliferators‐activated receptor‐alpha in an autoregulatory loop to modulate flow‐induced endothelial inflammation. Proc Natl Acad Sci U S A 108, 10355–10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical Summary Document
Data Availability Statement
The data that support the findings of this study are being made openly available in GEO (GSE158571).


