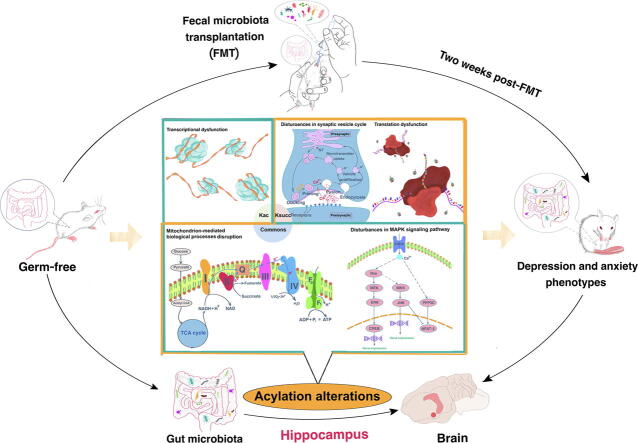
Graphical abstract
Keywords: Posttranslational modifications, Acetylation, Succinylation, Gut microbiota, Depression, Hippocampus
Abstract
Introduction
Major depressive disorder is caused by gene–environment interactions, and the host microbiome has been recognized as an important environmental factor. However, the underlying mechanisms of the host–microbiota interactions that lead to depression are complex and remain poorly understood.
Objectives
The present study aimed to explore the possible mechanisms underlying gut microbiota dysbiosis-induced depressive-like behaviors.
Methods
We used high-performance liquid chromatography-tandem mass spectrometry to analyze alterations in the hippocampal lysine acetylome and succinylome in male mice that had received gut microbiota from fecal samples of either patients with major depressive disorder or healthy controls. This was followed by bioinformatic analyses.
Results
A total of 315 acetylation sites on 223 proteins and 624 succinylation sites on 494 proteins were differentially expressed in the gut microbiota-dysbiosis mice. The significantly acetylated proteins were primarily associated with carbon metabolism disruption and gene transcription suppression, while the synaptic vesicle cycle and protein translation were the most significantly altered functions for succinylated proteins. Additionally, our findings suggest that gut microbiota dysbiosis disturbs mitochondria-mediated biological processes and the MAPK signaling pathway through crosstalk between acetylation and succinylation on relevant proteins.
Conclusions
This is the first study to demonstrate modifications in acetylation and succinylation in gut microbiota-dysbiosis mice. Our findings provide new avenues for exploring the pathogenesis of gut microbiota dysbiosis-related depression, and highlight potential targets for depression treatment.
Introduction
Human diseases are caused by gene–environment interactions, and the human microbiome has been recognized as an important environmental factor in disease [1]. The Human Microbiome Project (HMP) [2], [3] has generated abundant evidence that promotes our understanding of microbiota characterization and how the microbiome influences human health and disease. Findings from multi-omics studies of the microbiome, including the HMP, have elucidated the mechanisms of host–microbiota interactions during pregnancy and in preterm birth [4], type 2 diabetes [5], and inflammatory bowel disease [6]. They have also demonstrated the effects of the gut–brain axis [7] on brain function and behavior in psychiatric disorders, such as schizophrenia, anxiety, and depression.
Previously, we found significant alterations in the gut microbiomes of patients with major depressive disorder (MDD) [8], [9], and showed that there are sex-specific differences in these changes [10]. When fecal samples were transplanted from individuals with MDD to germ-free (GF) mice, the gut microbiome-remodeled mice exhibited significant depressive- and anxiety-like behaviors [9], [11], [12], which suggests a causal role of gut microbiota dysbiosis in the development of depression. To further understand the mechanisms of host–microbiota interactions that occur during depression, we have previously used -omics techniques to detect molecular alterations in gut microbiota-dysbiosis mice at the gene, protein, and metabolite levels. We found that these biological changes are associated with various cellular processes, including metabolism, axon guidance, glucocorticoid receptor pathway signaling, and PKC–CREB signaling [9], [11], [12], [13], [14], [15], [16]. However, the underlying mechanisms of the effects of gut microbiota on host function and behavior remain incompletely understood.
Proteins carry out most activities that are essential for life, and their functions are regulated by a variety of posttranslational modifications (PTMs), which play a pivotal role in regulating biological processes [17]. In recent decades, advances in high-throughput mass spectrometry have enabled us to identify thousands of PTM sites. Particularly, lysine (Lys) residues are a target of many PTMs, and acetylation is one of the most abundant and evolutionarily conserved PTMs [18]. Histone acetylation may represent the mechanism by which the gut microbiota induces depressive-like behaviors [19]. Acetylation also modifies thousands of non-histone proteins located in various cellular compartments [20] and regulates a range of biological processes related to gene regulation, signal transduction, and metabolism [21], [22]. Moreover, it has been reported that acetyl-lysine reader bromodomain-containing proteins can occur acetylation crosstalk with other PTMs, such as succinylation [23]. Thus, Lys acylation may be a functional modification that impacts host–microbiota interactions.
In the present study, we generated a comprehensive map of Lys-acetylation (Kac) and Lys-succinylation (Ksucc) dynamics in the hippocampus in response to gut microbiota dysbiosis. We also analyzed the correlated pathophysiological changes of gut microbiota dysbiosis-induced depressive-like behaviors, using proteomic analyses of Kac- and Ksucc-modified proteins in hippocampal tissue from mice that received either ‘depression microbiota’ (microbiota from fecal samples of MDD patients) or ‘healthy microbiota’ (microbiota from fecal samples of healthy controls). For this analysis, we used tandem-mass-tag (TMT) labeling, and high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) analysis.
Materials and methods
Fecal microbiota transplantation (FMT)
The FMT procedure was performed as described previously [9]. Briefly, fecal samples derived from either patients with MDD or healthy controls were colonized to adult GF Kunming mice to generate ‘depression microbiota’ and ‘healthy microbiota’ recipient mice, respectively. Prior to the fecal sample collection, written informed consent was obtained from all MDD patients and healthy controls. This research was approved by the Ethics Committee of Chongqing Medical University (Chongqing, China; 2017013) and was performed in accordance with the National Institutes of Health Guidelines. More details are given in the Supplemental Information.
Behavioral tests
Behavioral tests were carried out 2 weeks after the FMT, which is typically used as the benchmark time point for assessing disease-related phenotypes [24], [25]. Depressive-like behavior was assessed by quantifying immobility time during the last 5 min of a forced swimming test (FST), as previously described [9]. Additionally, anxiety-like behavior was assessed by measuring the distance traveled within the central 25% of an experimental field during the last 5 min of an open-field test (OFT). The results were analyzed with the Mann–Whitney U nonparametric test using SPSS v21.0 (IBM Corp., Armonk, NY, USA).
Sample collection and preparation
After the behavioral tests were performed, the hippocampus—the key brain area for the generation and regulation of emotion [26]—was obtained from the six ‘depression microbiota’ recipient mice with the most significant behavioral phenotypes, as well as from six randomly selected ‘healthy microbiota’ recipient mice. A 2-D Quant Kit (GE Healthcare, Chicago, IL, USA) was used to determine the total protein concentration of the pooled samples, followed by trypsin digestion. Peptides were reconstituted and labelled with a 6-plex TMT kit (ThermoFisher Scientific, Waltham, MA, USA). More details are given in the Supplemental Information.
HPLC fractionation and Kac/Ksucc peptide enrichment
Each sample was fractionated using a high-pH reverse-phase HPLC system within an Agilent 300Extend C18 column (Agilent, Folsom, CA, USA). The Kac/Ksucc peptides were enriched by incubating the fractions with pre-washed antibody beads (PTM Biolabs, Hangzhou, China). More details are given in the Supplemental Information.
Liquid chromatography-mass spectrometry identification
Peptides were then dissolved using 0.1% formic acid and loaded to a reverse-phase pre-column inserted into an EASY-nLC 1000 UPLC system (Thermo Fisher Scientific). The resulting peptides were then analyzed using a Q Exactive™ Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific) in triplicate, and the MS/MS data were processed by using MaxQuant software. The false discovery rate (FDR) thresholds were set at 1% for proteins, peptides, and modification sites. The probability of site localization was set at > 0.75. More details are given in the Supplemental Information.
Conserved sequence and PTM correlation analysis
Conserved sequences, constituted by amino acid residues in specific positions of modifier 21-mers, were explored using motif-X [27]. Lys was set as the central residue, with a window width of 21. The minimal occurrence was set to 20, and significance was set to 0.000001. Additionally, to perform the PTM correlation analysis, the present data were compared with known PTMs in the PhosphoSitePlus public database [28] and with phosphorylation sites [29] that we had previously identified in hippocampal tissue from gut microbiome-remodeled mice.
Secondary structure prediction
To predict the local secondary structures of proteins from primary sequences, we analyzed the secondary structures surrounding the acetylated and succinylated Lys, as well as those surrounding all Lys, using NetSurfP v1.0 software [30] with default parameters. The Wilcoxon test was used to calculate p-values, and significance was set at p < 0.05.
Cellular localization of Kac and Ksucc proteomes
The subcellular localizations of Kac and Ksucc proteins were predicted based on the annotations in Wolfpsort software [31], Ingenuity Pathway Analysis (IPA) software, and the public UniProt database [32]. Additionally, we compared our data with the mitochondrial proteome, as annotated in the MitoCarta database [33].
Functional annotation analysis
Gene Ontology (GO) annotations and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment pathways were analyzed using the OmicsBean system (http://www.omicsbean.cn). The p-values were calculated using Fisher’s exact test with a hypergeometric algorithm, and significance was set as p < 0.05. Additionally, functional protein domains were analyzed based on the PFAM database [34], and terms with FDR < 0.05 were considered significant.
Protein–protein interaction (PPI) network analysis
PPI analysis was performed using the STRING database (https://string-db.org/), and an interaction with a confidence score > 0.7 was selected. The PPI networks were visualized in Cytoscape [35], and the MCODE plug-in was used to identify highly connected clusters [36].
Protein complex analysis
We used the annotated CORUM database [37] for protein complex enrichment analysis. Overrepresented complexes were identified using Fisher’s exact test, and p < 0.05 was considered significant.
Results
Depressive- and anxiety-like behaviors in gut microbiome-remodeled mice
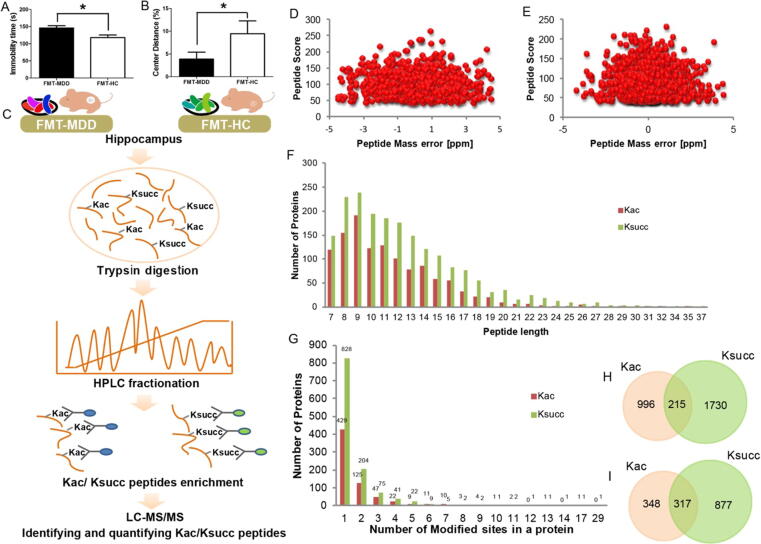
Eleven ‘depression microbiota’ recipient mice and 10 ‘healthy microbiota’ recipient mice were initially used for the behavioral tests. Using the concepts of stress susceptibility and resiliency [38], the ‘depression microbiota’ recipient mice were divided into susceptible and resilient subgroups based on their behavioral results. As a result, only six susceptible ‘depression microbiota’ recipient mice and six randomly selected ‘healthy microbiota’ recipient mice were used for further analysis. There was an increase in immobility during the FST (Fig. 1A) and a decrease in the center distance proportion during the OFT (Fig. 1B) in the susceptible ‘depression microbiota’ recipient mice compared with ‘healthy microbiota’ recipient mice, indicating increased depressive- and anxiety-like behaviors in gut microbiota-dysbiosis mice.
Fig. 1.
Profiling the Lys acetylation and succinylation proteome in hippocampal samples from gut microbiome-remodeled mice. (A) There was a significant increase in immobility time in the forced swimming test (FST) in ‘depression microbiota’ recipient mice (n = 6) compared with ‘healthy microbiota’ recipient mice (n = 6), indicating significant depressive-like behavior. *p < 0.05. (B) There was a decrease in the center distance proportion in the open field test (OFT) in ‘depression microbiota’ recipient mice (n = 6) compared with ‘healthy microbiota’ recipient mice (n = 6), indicating significant anxiety-like behavior. *p < 0.05. (C) Workflow for the identification and quantification of lysine acetylation and succinylation in hippocampal tissue from gut microbiome-remodeled mice. (D, E) Distribution of mass error of all identified acetylated and succinylated peptides, respectively. (F) Distribution of peptide length of all identified acetylated and succinylated peptides. (G) The number of acetylation and succinylation sites within each modified protein. (H) Venn diagram showing the overlapped results between identified Lys acetylation and succinylation sites in hippocampal tissue from gut microbiome-remodeled mice. (I) Venn diagram showing the overlapped results between identified acetylated and succinylated proteins in hippocampal tissue from gut microbiome-remodeled mice.
Microbial regulation of Kac and Ksucc proteomes in the hippocampus
To identify global changes in Kac and Ksucc in response to gut microbiota dysbiosis, we assessed Kac and Ksucc in hippocampal samples from ‘depression microbiota’ and ‘healthy microbiota’ recipient mice using HPLC-MS/MS (Fig. 1C). The mass error distribution was near zero, and most of them were < 5 parts per million (PPM) (Fig. 1D and E). The length of most of the peptides ranged from 7 to 19 amino acids (Fig. 1F), which indicates that the sample preparation reached the standards. Altogether, 1211 Kac sites on 665 proteins and 1945 Ksucc sites on 1194 proteins were identified, with an FDR < 1% at the protein, peptide, and site levels. Different numbers of Kac and Ksucc sites existed in the proteins, from 1 to 29 (Fig. 1G). Gut microbiota dysbiosis induced high levels of acylation in some proteins, including SPTAN1 with 17 Kac sites and 29 Ksucc sites, SPTBN1 with 14 Ksucc sites, and GLUD1 with 13 Kac sites. To investigate the overlaps (and the extent of such overlaps) between the Kac and Ksucc sites, we compared all identified Kac and Ksucc sites in the hippocampus samples, and revealed that 215 sites were both acetylated and succinylated (Fig. 1H). Concurrently, both acetylation and succinylation occurred in 317 proteins (Fig. 1I). Taken together, these results suggest that gut microbiota dysbiosis can result in global changes in Kac and Ksucc.
Conserved sequence and PTM correlation analysis
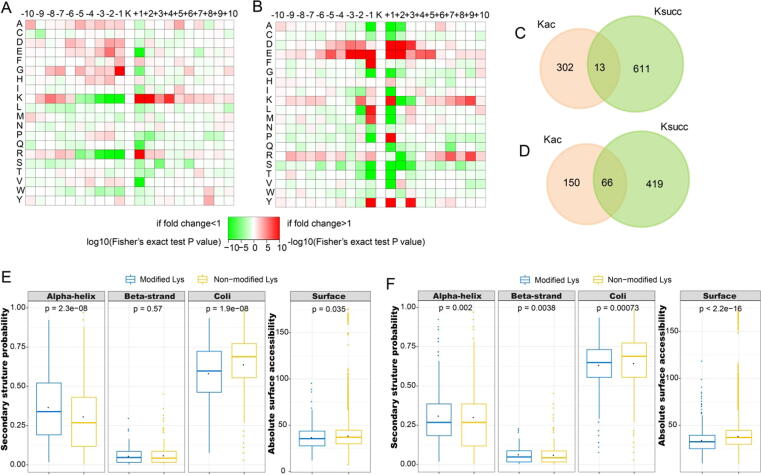
To obtain a more detailed understanding of the Kac and Ksucc sites that were influenced by gut microbiota dysbiosis, we evaluated the conserved sequences surrounding the identified Kac and Ksucc sites. The results suggested a preference for Lys at the +1 or +2 position and Arg at the +1 position relative to the Kac sites (Fig. 2A), while there was a preference for Asp at the +1 or +2 position and Glu at the −3 to +2 position relative to the Ksucc sites (Fig. 2B). Accordingly, motif analysis identified KacK and Ksucc*E motifs as the most conserved sequences for Kac and Ksucc sites, respectively (Table S1). These results demonstrate that Kac sites that are influenced by gut microbiota dysbiosis tend to be near basic amino acids, while Ksucc sites tend to be near acidic amino acids. In addition, PTM correlation analysis indicated that 238 of the Kac sites that we identified had been reported previously (Table S2), and that 347 Kac sites and 485 Ksucc sites overlapped with ubiquitylation (Table S3). Furthermore, we identified 133 Kac sites and 197 Ksucc sites located within six residues upstream or downstream of a range of known PTMs, including monomethylation, dimethylation, N-glycosylation, O-linked β-N-acetylglucosamine, and ubiquitylation sites (Table S4). In hippocampal tissue from the gut microbiota-dysbiosis mice, 72% and 65% of the Kac and Ksucc sites, respectively, that colocalized with phosphorylation sites were located near phosphoserine residues (Table S5).
Fig. 2.
Properties of the acetylation and succinylation proteome. (A, B) Heat maps showing features of the flanking sequences for all Lys acetylation and succinylation sites, respectively. Multiple hypothesis tests were performed for the p-values. (C) Venn diagram showing the overlapped results between significant Lys acetylation and succinylation sites in hippocampal tissue from gut microbiota-dysbiosis mice. (D) Venn diagram showing the overlapped results between significant acetylated and succinylated proteins in hippocampal tissue from gut microbiota-dysbiosis mice. (E) Secondary structure distribution and surface accessibility prediction of significantly acetylated sites. (F) Secondary structure distribution and surface accessibility prediction of significantly succinylated sites.
Screening for Kac and Ksucc sites that were significantly modified in response to gut microbiota dysbiosis
To select the significant molecules, we quantified all of the identified Kac and Ksucc sites in gut microbiome-remodeled mice. Of these sites, 920 Kac sites on 512 proteins and 1455 Ksucc sites on 937 proteins were quantified. Differential site modifications were selected using the thresholds of p < 0.05 and fold-change ≥ 1.2 or ≤ 0.83. In total, 177 Kac sites on 137 proteins were up-regulated, and 138 Kac sites on 86 proteins were down-regulated (Table S6) in ‘depression microbiota’ recipient mice compared with ‘healthy microbiota’ recipient mice. Importantly, eight Kac sites with > 10-fold change in abundance were identified, one site was increased and seven sites were decreased. Additionally, 380 Ksucc sites on 287 proteins were up-regulated, and 244 Ksucc sites on 207 proteins were down-regulated (Table S7). However, only three Ksucc sites with > 10-fold decrease in abundance were identified. In addition, only 13 acetyl-Lys sites were found to be succinylated (Fig. 2C), and only 66 proteins were both acetylated and succinylated (Fig. 2D and Table S8).
Prediction of the secondary structures of significant Kac and Ksucc proteins
To determine how gut microbiota dysbiosis affects protein structure and function, we predicted the secondary structures of proteins that exhibited significant changes in Kac and Ksucc. Approximately 41.98% of the Kac sites were located in ordered regions—36.51% in α-helices and 5.47% in β-strands—and the remaining 58.01% were located in disordered regions (Fig. 2E and Table S9). Most of the Ksucc sites were located in coils (63.04%), followed by α-helices (30.76%) and β-strands (6.18%) (Fig. 2F and Table S10). These results indicate that gut microbiota dysbiosis may influence protein function via Kac and Ksucc. Next, we evaluated the surface accessibility of Kac and Ksucc sites, and found that 36.53% of Kac sites and 33.76% of Ksucc sites were surface-exposed, compared with 38.22% and 37.87% of non-modified Lys residues, respectively. Therefore, Kac and Ksucc induced by gut microbiota dysbiosis can affect the surface properties of proteins.
Cellular localization of significant Kac and Ksucc proteins
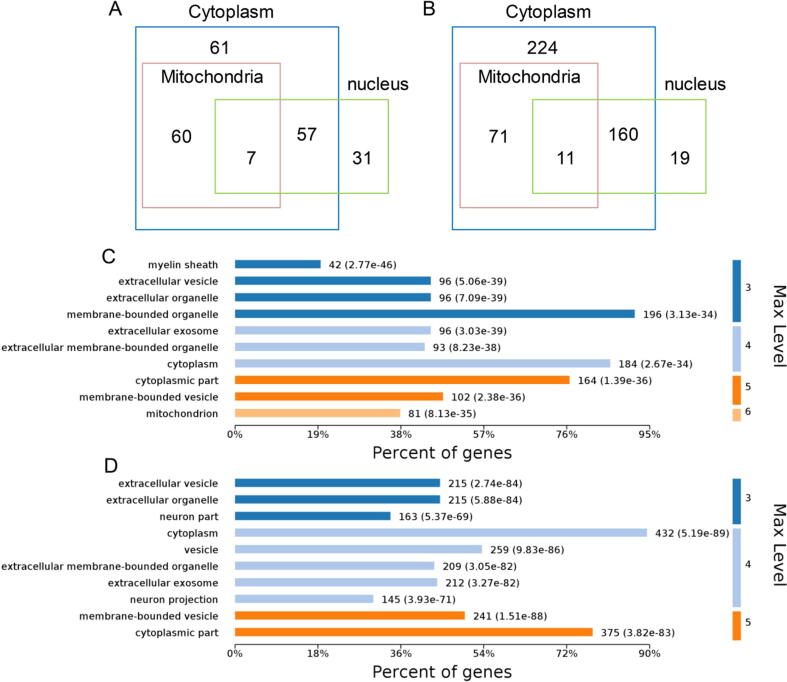
To determine whether gut microbiota dysbiosis regulates Kac and Ksucc of proteins in various cellular compartments, we predicted the cellular localization of all significantly acetylated and succinylated proteins. As expected, the Kac modification was abundant on nuclear proteins, while 36% of cytoplasmic acetylated proteins were localized partially or exclusively to the mitochondria (Fig. 3A). These results indicate that the gut microbiota may regulate intra-nuclear processes and mitochondrial protein functions through the acetylation of relevant proteins. Moreover, a comparable number of succinylated proteins were annotated as mitochondrial, while only 19 Ksucc proteins were localized exclusively to the nucleus (Fig. 3B). Approximately half of the succinylated proteins were cytoplasmic, which suggests that Ksucc has a critical role in regulating extensive cytosolic processes. To determine the likelihood of mitochondrial localization, we compared our MS/MS data with the MitoCarta database and found that 5% of mitochondrial proteins underwent Kac changes, whereas 5.6% of mitochondrial proteins exhibited Ksucc changes (Table S11).
Fig. 3.
Annotations of the acetylation and succinylation proteome. (A, B) Venn diagrams showing the cellular localization of significantly acetylated and succinylated proteins, respectively. (C, D) Bar graphs showing the annotated cellular components enriched in significant acetylated and succinylated proteins, respectively. Max Level means the maximal annotated level of this term in the GO graph (tree), and the number indicates the depth of the GO term level.
Functional annotation of significant Kac and Ksucc proteins
To further reveal the biological functions of the acetylated and succinylated proteins, we assigned GO annotations to these significantly altered proteins (Fig. S1 and Fig. S2). Acetylated proteins were primarily annotated in the mitochondria in the cellular component category (Fig. 3C), whereas succinylated proteins were annotated in the cytoplasm (Fig. 3D). Additionally, carboxylic acid metabolism and protein complex subunit organization were significantly enriched in Kac and Ksucc proteins, respectively, in the biological process category. Poly(A) RNA binding was significantly annotated for both acetylated and succinylated proteins in molecular function category. Domain analysis revealed that acetylated proteins were enriched for core histone H2A/H2B/H3/H4, 14-3-3 protein, and aldehyde dehydrogenase (Table S12), while succinylated proteins were enriched for HSP70 protein and cation-transporting ATPase (Table S13).
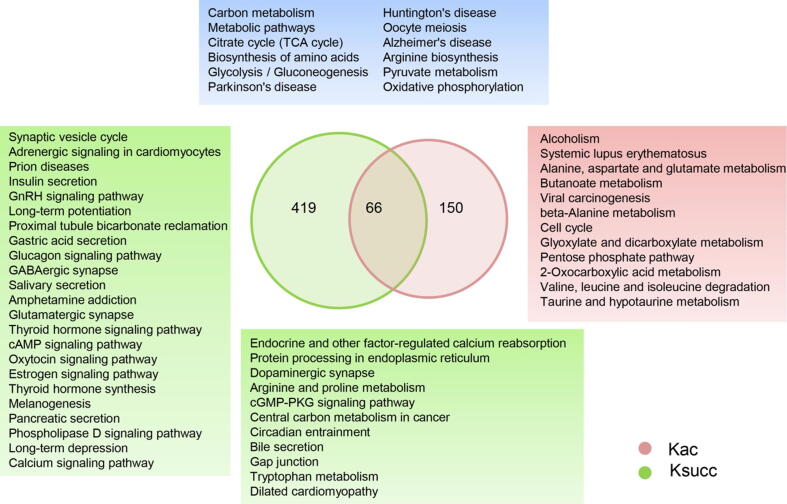
We also performed KEGG pathway enrichment analysis to obtain a better understanding of the biological functions of the acetylated and succinylated proteins that were regulated by gut microbiota. In the gut microbiota-dysbiosis mice, carbon metabolism was the most significantly altered function according to the acetylated protein enrichment analysis (Fig. S3), while involvement in the synaptic vesicle cycle was unique to succinylated proteins (Fig. S4 and Fig. S5). However, 58 and 82 proteins in metabolic pathways were acetylated and succinylated, respectively. These results suggest that the gut microbiota may regulate host functions through Kac and Ksucc, and that these two PTMs have both unique and shared features. We therefore performed an overlap analysis to identify common pathways that were significantly enriched (Benjamini–Hochberg-adjusted p < 0.01) in both acetylated and succinylated proteins (Fig. 4). Notably, nearly all of the overlapping pathways were partly or exclusively localized to the mitochondria and involved with acetyl-CoA and succinyl-CoA, including glycolysis/gluconeogenesis, the citrate cycle (TCA cycle), biosynthesis of amino acids, pyruvate metabolism, and oxidative phosphorylation. Most of the proteins in these pathways were modified by both acetylation and succinylation. These results indicate that gut microbiota dysbiosis may affect mitochondria-mediated biological processes by altering the levels of acetylation and succinylation of relevant proteins, which then leads to the development of depression.
Fig. 4.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways overlap between significantly acetylated and succinylated proteins in gut microbiota-dysbiosis mice. Venn diagram displaying the number of common and unique proteins between both post-translational modifications. The blue area displays the common KEGG pathways enriched in both succinylated and acetylated proteins, the green area shows the unique KEGG pathways enriched in succinylated proteins, and the red area shows the unique KEGG pathways enriched in acetylated proteins.
PPI networks of significant Kac and Ksucc proteins
To obtain a further understanding of the complicated biological processes regulated by gut microbiota dysbiosis, we created PPI networks for the acetylated and succinylated proteins. Our datasets showed two networks with highly connected clusters of nodes that extended out to form parts of pathways and protein complexes (Fig. 5A and B). Using the MCODE plug-in, we identified two highly interconnected clusters from the acetylated PPI network as being related to systemic lupus erythematosus (Fig. 5C) and the cell cycle (Fig. 5D). Three clusters were identified among the succinylated proteins, including proteins related to the ribosome (Fig. 5E), endocytosis (Fig. 5F), and oxidative phosphorylation (Fig. 5G). Taken together, the decreased acetylation of histone proteins and succinylation of ribosomal proteins in gut microbiota-dysbiosis mice suggest disturbances in transcription and translation.
Fig. 5.
Protein–protein interaction (PPI) network analysis for the significantly acetylated and succinylated proteins. (A) PPI network for the acetylated proteins. (B) PPI network for the succinylated proteins. (C) Subnetwork for the acetylated proteins involved in systemic lupus erythematosus. (D) Subnetwork for the acetylated proteins related to the cell cycle. (E) Subnetwork for the succinylated proteins related to the ribosome. (F) Subnetwork for the succinylated proteins involved in endocytosis. (G) Subnetwork for the succinylated proteins involved in oxidative phosphorylation.
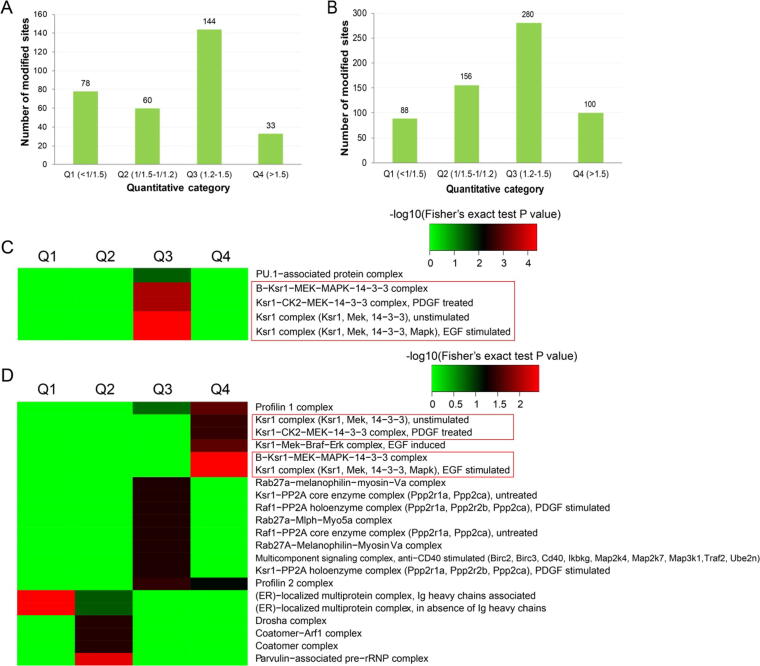
Protein complex analysis
Protein complex enrichment analysis was performed to identify complexes that were regulated by Kac and Ksucc. Because proteins within the same complex have similar functions, we analyzed the up-regulated and down-regulated proteins separately. Furthermore, to uncover the degree of protein complex acylation, we divided the proteins that exhibited significant changes in Kac and Ksucc into four quantiles (Q1–Q4) based on fold-change values (Fig. 6A and B). Based on this analysis, we identified five protein complexes enriched in Kac proteins (Fig. 6C) and 21 enriched in Ksucc proteins (Fig. 6D). Four protein complexes were significantly modified by both acetylation and succinylation, including the unstimulated KSR1 complex (KSR1, MEK, 14-3-3), the B–KSR1–MEK–MAPK–14-3-3 complex, the PDGF-treated KSR1–CK2–MEK–14-3-3 complex, and the EGF-stimulated KSR1 complex (KSR1, MEK, 14-3-3, MAPK). This finding suggests that the gut microbiota regulates the physiological functions of these protein complexes through Lys acylation. These protein complexes are primarily involved in the MAPK signaling pathway. Therefore, we speculate that gut microbiota dysbiosis-induced depression is associated with alterations in the acetylation and succinylation of relevant proteins in the MAPK signaling pathway.
Fig. 6.
Protein complex analysis for the significantly acetylated and succinylated proteins. (A, B) Degree of modification of significantly acetylated and succinylated proteins, respectively. (C, D) Heat maps generated by protein complex enrichment-based cluster analysis for acetylated and succinylated proteins, respectively.
Discussion
PTMs can modify proteins at any time during their lifecycle, and significantly increase the diversity and complexity of the proteome by altering the activity, localization, and PPI of target proteins [39]. To date, more than 300 PTMs have been documented [40]. Kac is among the most abundant and evolutionarily conserved PTMs, and is involved in regulating various biological processes. A previous study has reported that hippocampal acetylation may improve prenatal stress-induced depressive-like behaviors [41], and the histone deacetylase inhibitor can also alleviate depressive-like symptoms [42]. In addition, a significant decrease in the acetylation of membrane-associated tubulin has been reported in the postmortem prefrontal cortex of MDD patients relative to controls [43]. There is some crosstalk between Kac and other PTMs, such as succinylation. However, little is known about Kac and Ksucc in gut microbiota dysbiosis-induced depression. Increasing evidence indicates that both surface factors (e.g., polysaccharides, peptidoglycans, and other antigens) on microbiota and gut microbial metabolites (e.g., bacteriocins, neuromodulators, choline, bile acids, and short-chain fatty acids) are important mediators that modulate brain function [7]. This suggests, at least in part, that these microbial factors may act as potential mediators that modulate the PMTs of hippocampal proteins through the microbiota–gut–brain axis.
In the present study, Kac and Ksucc sites that were influenced by gut microbiota dysbiosis tended to be near basic and acidic amino acids, respectively. There was some correlation between changes in these two PTMs and those of other modifications, including monomethylation, dimethylation, N-glycosylation, O-linked β-N-acetylglucosamine, and ubiquitylation. Moreover, the limited overlap between Kac and Ksucc modifications suggests that they participate in different regulatory mechanisms. Numerous proteins carrying the Kac modification localized to the nucleus and mitochondria. In certain cases, site-specific acetylation is sufficient to alter nucleosome function and chromatin folding [44], [45], which causes changes in gene expression. Acetyl-Lys residues on histones can act as an “epitope” to recruit protein complexes containing acetyl-Lys bromodomain domains, such as histone acetyltransferase, methyltransferase, and transcription activating factor [46]. Thus, we speculate that gut microbiota dysbiosis may disrupt intra-nuclear processes by acetylating relevant nucleoproteins. Additionally, mitochondria are key centers for metabolism and energy production, and Kac is the main mechanism that regulates mitochondrial function [47]. Altogether, 63% of mitochondrial proteins are Lys acetylated [47], which indicates that gut microbiota dysbiosis may cause mitochondrial dysfunction through the acetylation of relevant proteins. However, consistent with previous findings, Ksucc proteins in the gut microbiota-dysbiosis mice were widely localized to the cytoplasm, mitochondria, and nucleus [48], [49]. Therefore, we infer that gut microbiota dysbiosis may affect various cellular processes by altering the succinylation of relevant proteins.
Proteins with altered Kac in gut microbiota-dysbiosis mice were primarily involved in carbon metabolism, which can be divided into one-carbon and central carbon metabolisms. Abundant Kac and Ksucc sites on proteins related to carbon metabolism have been identified in Aeromonas hydrophila [50]. Carbon metabolism plays an important role in regulating health and disease [51], [52], [53]. Previous studies have confirmed that perturbations in carbon metabolism are involved in the pathogenesis of depression [54], [55], [56], [57], and that the absence of gut microbiota leads to similar changes in GF mice [58]. Thus, we suggest that, in depression, gut microbiota dysbiosis may influence host brain function by altering the acetylation levels of proteins related to carbon metabolism. Additionally, down-regulation of histone protein (e.g., H2AFZ, HIST1H2BM, and H2AFV) acetylation was observed in gut microbiota-dysbiosis mice, indicating a potential role for the gut microbiota in regulating transcription.
Enrichment analysis of Ksucc proteins found disturbances in various biological processes, especially the synaptic vesicle cycle. Synaptic vesicles, located at axon terminals, are the core elements in the physiology of presynaptic terminals and are primarily involved in the secretion of neurotransmitters, such as glutamate and γ-aminobutyric acid (GABA). A number of preclinical studies have reported that perturbances in glutamatergic, GABAergic, and cholinergic neurotransmitters are related to the development of depression [59], [60], [61]. Previous findings also indicated that the expression levels of proteins related to the synaptic vesicle cycle and vesicle organization, such as synaptotagmins, synaptophysin, Rab, and SNAP proteins, are altered by the absence of gut microbiota [62]. Compared with healthy controls, significant alterations in the gut microbiome were found in fecal samples from patients with MDD [8], [9], [63], [64], with increased levels of Actinobacteria, Bacteroidetes, and Proteobacteria, but reduced levels of Firmicutes. The gut microbiota has a potential role in regulating the expression of postsynaptic GABA receptors [65] as well as N-methyl-d-aspartate receptors [66]. Taken together, these findings suggest that gut microbiota dysbiosis may affect host brain functions by disrupting neurotransmitter release and function. Additionally, the gut microbiota may affect protein translation [67]. Consistent with this finding, we detected a down-regulation of succinylation on many ribosomal proteins, suggesting a potential role for translational dysfunction in the pathogenesis of gut microbiota dysbiosis-related depression.
Overlap analysis suggested that the gut microbiota may regulate host functions by altering Kac and Ksucc; these two PTMs have unique and shared features. Some pivotal metabolic pathways, including glycolysis/gluconeogenesis, the citrate cycle, biosynthesis of amino acids, pyruvate metabolism, and oxidative phosphorylation, were identified in both the Kac and Ksucc protein enrichment analyses. Significantly, many key enzymes in these pathways (e.g., PGM2, ALDOA, GOT2, MDH1, GLUD1, ABAT, ALDH1B1, and DLAT) were regulated by both acetylation and succinylation. These common metabolic pathways are partly or exclusively localized to mitochondria. We also found that many proteins in the respiratory chain were regulated by both acetylation and succinylation, such as NDUFA10, CYCS, SLC25A5, and ATP5C1. Previous studies have found that mitochondrial dysfunction is associated with depression [68], [69], and that PTMs act as important regulators of mitochondrial function [70]. Thus, gut microbiota dysbiosis-induced depression may be associated with mitochondrial dysfunction, characterized by alterations in the levels of Kac and Ksucc of relevant proteins. In addition, many protein complexes involved in MAPK signaling were regulated by both acetylation and succinylation, such as the B–KSR1–MEK–MAPK–14-3-3 complex. Numerous studies have confirmed the potential role of disrupted MAPK signaling in depression [71], [72]. Therefore, we speculate that gut microbiota dysbiosis-induced depression is related to changes in the acetylation and succinylation of proteins in the MAPK signaling pathway.
There were some limitations to the present study. First, only hippocampal tissue from male mice was analyzed to characterize the Lys acetylome and succinylome. Thus, other emotion-related brain areas should be taken into consideration in further studies to obtain a more comprehensive understanding of the underlying mechanisms of host–microbiota interactions. Furthermore, sex differences in FMT-based dysbiosis-induced mice, as well as sex-specific alterations in PMT patterns, require further investigation. Second, the main alterations identified in the present study were not experimentally verified because of a lack of commercially available antibodies for the specific Kac and Ksucc sites addressed here; however, the mass spectrometry data were highly reliable, with average peptide scores as high as 100, as determined by Mascot software. We will also perform functional studies in our future research in this field. Third, the fecal sample collection may also have been a limitation, as previously described [9], because the pooled fecal samples used in this study were from subjects recruited from the same clinical site. Therefore, further studies using individual fecal samples from ethnically diverse donors are required, to identify the specific gut microbiota strains that contribute to the onset of depression.
Conclusions
This study aimed to elucidate the underlying mechanisms by which gut microbiota dysbiosis induces depressive-like behaviors, by analyzing alterations in the hippocampal Lys acetylome and succinylome in gut microbiota-dysbiosis mice. A total of 315 Kac sites on 223 proteins and 624 Ksucc sites on 494 proteins were identified as being significantly expressed in the gut microbiota-dysbiosis mice. The significantly acetylated proteins were primarily associated with carbon metabolism disruption and gene transcription suppression, while the synaptic vesicle cycle and protein translation were the most significantly altered functions for the succinylated proteins. In addition, gut microbiota dysbiosis may disturb mitochondria-mediated biological processes and the MAPK signaling pathway through crosstalk between acetylation and succinylation on relevant proteins. Our study provides new avenues for exploring the pathogenesis of gut microbiota dysbiosis-related depression, and highlights potential targets for treating depression.
Compliance with Ethics Requirements
This study was approved by the Ethics Committee of Chongqing Medical University (Chongqing, China; 2017013) and was performed in accordance with NIH Guidelines.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the Non-Profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (Grant no. 2019PT320002), the National Key R&D Program of China (Grant no. 2017YFA0505700), the Natural Science Foundation Project of China (Grant no. 81820108015), the Chongqing Science & Technology Commission (cstc2018jcyjAX0341), and the Yongchuan District Science & Technology Committee (Ycstc2017cb5001). We thank the Experimental Animal Research Center at Third Military Medical University (Chongqing, China) for providing the germ-free mice.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.12.002.
Contributor Information
Hong Wei, Email: weihong63528@163.com.
Peng Xie, Email: xiepeng@cqmu.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Backhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101(44):15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Integrative H.M.P.R.N.C. The integrative human microbiome project. Nature 2019; 569(7758): 641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed]
- 3.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fettweis J.M., Serrano M.G., Brooks J.P., Edwards D.J., Girerd P.H., Parikh H.I. The vaginal microbiome and preterm birth. Nat Med. 2019;25(6):1012–1021. doi: 10.1038/s41591-019-0450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou W., Sailani M.R., Contrepois K., Zhou Y., Ahadi S., Leopold S.R. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature. 2019;569(7758):663–671. doi: 10.1038/s41586-019-1236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd-Price J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., Poon T.W. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cryan J.F., O'Riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M. The microbiota-gut-brain axis. Physiol Rev. 2019;99(4):1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z., Li J., Gui S., Zhou C., Chen J., Yang C. Comparative metaproteomics analysis shows altered fecal microbiota signatures in patients with major depressive disorder. NeuroReport. 2018;29(5):417–425. doi: 10.1097/wnr.0000000000000985. [DOI] [PubMed] [Google Scholar]
- 9.Zheng P., Zeng B., Zhou C., Liu M., Fang Z., Xu X. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol Psychiatry. 2016;21(6):786–796. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 10.Chen J.J., Zheng P., Liu Y.Y., Zhong X.G., Wang H.Y., Guo Y.J. Sex differences in gut microbiota in patients with major depressive disorder. Neuropsychiatr Dis Treat. 2018;14:647–655. doi: 10.2147/ndt.S159322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C., Yang X., Zeng B., Zeng L., Gong X., Zhou C. Proteomic analysis of olfactory bulb suggests CACNA1E as a promoter of CREB signaling in microbiota-induced depression. J Proteomics. 2019;194:132–147. doi: 10.1016/j.jprot.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 12.Li B., Guo K., Zeng L., Zeng B., Huo R., Luo Y. Metabolite identification in fecal microbiota transplantation mouse livers and combined proteomics with chronic unpredictive mild stress mouse livers. Transl Psychiatry. 2018;8(1):34. doi: 10.1038/s41398-017-0078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J.J., Zeng B.H., Li W.W., Zhou C.J., Fan S.H., Cheng K. Effects of gut microbiota on the microRNA and mRNA expression in the hippocampus of mice. Behav Brain Res. 2017;322(Pt A):34–41. doi: 10.1016/j.bbr.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Huo R., Zeng B., Zeng L., Cheng K., Li B., Luo Y. Microbiota modulate anxiety-like behavior and endocrine abnormalities in hypothalamic-pituitary-adrenal axis. Front Cell Infect Microbiol. 2017;7:489. doi: 10.3389/fcimb.2017.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Y., Zeng B., Zeng L., Du X., Li B., Huo R. Gut microbiota regulates mouse behaviors through glucocorticoid receptor pathway genes in the hippocampus. Transl Psychiatry. 2018;8(1):187. doi: 10.1038/s41398-018-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng L., Zeng B., Wang H., Li B., Huo R., Zheng P. Microbiota modulates behavior and protein kinase C mediated cAMP response element-binding protein signaling. Sci Rep. 2016;6:29998. doi: 10.1038/srep29998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salomon D., Orth K. What pathogens have taught us about posttranslational modifications. Cell Host Microbe. 2013;14(3):269–279. doi: 10.1016/j.chom.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayasu E.S., Burnet M.C., Walukiewicz H.E., Wilkins C.S., Shukla A.K., Brooks S. Ancient regulatory role of lysine acetylation in central metabolism. mBio. 2017;8(6):e01894–17. doi: 10.1128/mBio.01894-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caspani G., Kennedy S., Foster J.A., Swann J. Gut microbial metabolites in depression: understanding the biochemical mechanisms. Microb Cell. 2019;6(10):454–481. doi: 10.15698/mic2019.10.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhary C., Weinert B.T., Nishida Y., Verdin E., Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15(8):536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 21.Ali I., Conrad R.J., Verdin E., Ott M. Lysine acetylation goes global: from epigenetics to metabolism and therapeutics. Chem Rev. 2018;118(3):1216–1252. doi: 10.1021/acs.chemrev.7b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narita T., Weinert B.T., Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019;20(3):156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 23.Weinert B.T., Scholz C., Wagner S.A., Iesmantavicius V., Su D., Daniel J.A. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4(4):842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 24.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 25.Koren O., Goodrich J.K., Cullender T.C., Spor A., Laitinen K., Bäckhed H.K. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150(3):470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell S., Macqueen G. The role of the hippocampus in the pathophysiology of major depression. J Psychiatry Neuroscience: JPN. 2004;29(6):417–426. [PMC free article] [PubMed] [Google Scholar]
- 27.Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. Curr Protoc Bioinformatics. 2011; Chapter 13: Unit 13.15-24. doi: 10.1002/0471250953.bi1315s35. [DOI] [PubMed]
- 28.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 2015; 43(Database issue): D512-520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed]
- 29.Wang H., Liu L., Rao X., Zeng B., Yu Y., Zhou C. Integrated phosphoproteomic and metabolomic profiling reveals perturbed pathways in the hippocampus of gut microbiota dysbiosis mice. Transl Psychiatry. 2020;10(1):346. doi: 10.1038/s41398-020-01024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen B., Petersen T.N., Andersen P., Nielsen M., Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res 2007; 35(Web Server issue): W585-587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed]
- 32.The U.C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–d432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bader G.D., Hogue C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giurgiu M., Reinhard J., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019;47(D1):D559–d563. doi: 10.1093/nar/gky973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charney D.S. Psychobiological mechanisms of resilience and vulnerability: implications for successful adaptation to extreme stress. Am J Psychiatry. 2004;161(2):195–216. doi: 10.1176/appi.ajp.161.2.195. [DOI] [PubMed] [Google Scholar]
- 39.Yakubu R.R., Weiss L.M., Silmon de Monerri N.C. Post-translational modifications as key regulators of apicomplexan biology: insights from proteome-wide studies. Mol Microbiol. 2018;107(1):1–23. doi: 10.1111/mmi.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witze E.S., Old W.M., Resing K.A., Ahn N.G. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4(10):798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y., Zhang J., Zhang L., Dang S., Su Q., Zhang H. Hippocampal acetylation may improve prenatal-stress-induced depression-like behavior of male offspring rats through regulating AMPARs expression. Neurochem Res. 2017;42(12):3456–3464. doi: 10.1007/s11064-017-2393-7. [DOI] [PubMed] [Google Scholar]
- 42.Chen W.-Y., Zhang H., Gatta E., Glover E.J., Pandey S.C., Lasek A.W. The histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) alleviates depression-like behavior and normalizes epigenetic changes in the hippocampus during ethanol withdrawal. Alcohol (Fayetteville, N.Y.) 2019;78:79–87. doi: 10.1016/j.alcohol.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh H., Chmura J., Bhaumik R., Pandey G.N., Rasenick M.M. Membrane-associated α-tubulin is less acetylated in postmortem prefrontal cortex from depressed subjects relative to controls: cytoskeletal dynamics, HDAC6, and depression. J Neurosci: Off J Soc Neurosci. 2020;40(20):4033–4041. doi: 10.1523/JNEUROSCI.3033-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann H., Hancock S.M., Buning R., Routh A., Chapman L., Somers J. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36(1):153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shogren-Knaak M., Ishii H., Sun J.-M., Pazin M.J., Davie J.R., Peterson C.L. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science (New York, N.Y.). 2006;311(5762):844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 46.Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J.-P., Barsyte-Lovejoy D. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149(1):214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baeza J., Smallegan M.J., Denu J.M. Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem Sci. 2016;41(3):231–244. doi: 10.1016/j.tibs.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie L., Li J., Deng W., Yu Z., Fang W., Chen M. Proteomic analysis of lysine succinylation of the human pathogen Histoplasma capsulatum. J Proteomics. 2017;154:109–117. doi: 10.1016/j.jprot.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., Wang G., Song L., Mu P., Wang S., Liang W. Global analysis of protein lysine succinylation profiles in common wheat. BMC Genom. 2017;18(1):309. doi: 10.1186/s12864-017-3698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun L., Yao Z., Guo Z., Zhang L., Wang Y., Mao R. Comprehensive analysis of the lysine acetylome in Aeromonas hydrophila reveals cross-talk between lysine acetylation and succinylation in LuxS. Emerg Microbes Infect. 2019;8(1):1229–1239. doi: 10.1080/22221751.2019.1656549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anandhan A., Jacome M.S., Lei S., Hernandez-Franco P., Pappa A., Panayiotidis M.I. Metabolic dysfunction in Parkinson's disease: bioenergetics, redox homeostasis and central carbon metabolism. Brain Res Bull. 2017;133:12–30. doi: 10.1016/j.brainresbull.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ducker G.S., Rabinowitz J.D. One-carbon metabolism in health and disease. Cell Metab. 2017;25(1):27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y., Chen Y., Fang J. Post-transcriptional and post-translational regulation of central carbon metabolic enzymes in cancer. Anti-Cancer Agents Med Chem. 2017;17(11):1456–1465. doi: 10.2174/1871520617666170327110712. [DOI] [PubMed] [Google Scholar]
- 54.Kim J.-M., Stewart R., Kim S.-W., Yang S.-J., Shin I.-S., Yoon J.-S. Predictive value of folate, vitamin B12 and homocysteine levels in late-life depression. Br J Psychiatry: J Mental Sci. 2008;192(4):268–274. doi: 10.1192/bjp.bp.107.039511. [DOI] [PubMed] [Google Scholar]
- 55.Liu L., Zhou X., Zhang Y., Liu Y., Yang L., Pu J. The identification of metabolic disturbances in the prefrontal cortex of the chronic restraint stress rat model of depression. Behav Brain Res. 2016;305:148–156. doi: 10.1016/j.bbr.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Liu L., Zhou X., Zhang Y., Pu J., Yang L., Yuan S. Hippocampal metabolic differences implicate distinctions between physical and psychological stress in four rat models of depression. Transl Psychiatry. 2018;8(1):4. doi: 10.1038/s41398-017-0018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou X., Liu L., Zhang Y., Pu J., Yang L., Zhou C. Metabolomics identifies perturbations in amino acid metabolism in the prefrontal cortex of the learned helplessness rat model of depression. Neuroscience. 2017;343:1–9. doi: 10.1016/j.neuroscience.2016.11.038. [DOI] [PubMed] [Google Scholar]
- 58.Wang H., Liu L., Rao X., Chai T., Zeng B., Zhang X. Commensal microbiota regulation of metabolic networks during olfactory dysfunction in mice. Neuropsychiatr Dis Treat. 2020;16:761–769. doi: 10.2147/NDT.S236541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duman R.S., Sanacora G., Krystal J.H. Altered connectivity in depression: GABA and glutamate neurotransmitter deficits and reversal by novel treatments. Neuron. 2019;102(1):75–90. doi: 10.1016/j.neuron.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma K., Xu A., Cui S., Sun M.R., Xue Y.C., Wang J.H. Impaired GABA synthesis, uptake and release are associated with depression-like behaviors induced by chronic mild stress. Transl Psychiatry. 2016;6(10) doi: 10.1038/tp.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pytka K., Dziubina A., Młyniec K., Dziedziczak A., Żmudzka E., Furgała A. The role of glutamatergic, GABA-ergic, and cholinergic receptors in depression and antidepressant-like effect. Pharmacol Rep. 2016;68(2):443–450. doi: 10.1016/j.pharep.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 62.Yu Y., Raka F., Adeli K. The role of the gut microbiota in lipid and lipoprotein metabolism. J Clin Med. 2019;8(12) doi: 10.3390/jcm8122227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang H., Ling Z., Zhang Y., Mao H., Ma Z., Yin Y. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 64.Naseribafrouei A., Hestad K., Avershina E., Sekelja M., Linløkken A., Wilson R. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motility: Off J Eur Gastrointestinal Motility Soc. 2014;26(8):1155–1162. doi: 10.1111/nmo.12378. [DOI] [PubMed] [Google Scholar]
- 65.Bravo J.A., Forsythe P., Chew M.V., Escaravage E., Savignac H.M., Dinan T.G. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. PNAS. 2011;108(38):16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fröhlich E.E., Farzi A., Mayerhofer R., Reichmann F., Jačan A., Wagner B. Cognitive impairment by antibiotic-induced gut dysbiosis: Analysis of gut microbiota-brain communication. Brain Behav Immun. 2016;56:140–155. doi: 10.1016/j.bbi.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwartz M.H., Wang H., Pan J.N., Clark W.C., Cui S., Eckwahl M.J. Microbiome characterization by high-throughput transfer RNA sequencing and modification analysis. Nat Commun. 2018;9(1):5353. doi: 10.1038/s41467-018-07675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bansal Y., Kuhad A. Mitochondrial Dysfunction in Depression. Curr Neuropharmacol. 2016;14(6):610–618. doi: 10.2174/1570159X14666160229114755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karabatsiakis A., Böck C., Salinas-Manrique J., Kolassa S., Calzia E., Dietrich D.E. Mitochondrial respiration in peripheral blood mononuclear cells correlates with depressive subsymptoms and severity of major depression. Transl Psychiatry. 2014;4 doi: 10.1038/tp.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stram A.R., Payne R.M. Post-translational modifications in mitochondria: protein signaling in the powerhouse. Cell Mol life Sci : CMLS. 2016;73(21):4063–4073. doi: 10.1007/s00018-016-2280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Humo M., Ayazgök B., Becker L.J., Waltisperger E., Rantamäki T., Yalcin I. Ketamine induces rapid and sustained antidepressant-like effects in chronic pain induced depression: Role of MAPK signaling pathway. Prog Neuro-Psychopharmacol Biol Psychiatry. 2020;100 doi: 10.1016/j.pnpbp.2020.109898. [DOI] [PubMed] [Google Scholar]
- 72.Wang J.Q., Mao L. The ERK pathway: molecular mechanisms and treatment of depression. Mol Neurobiol. 2019;56(9):6197–6205. doi: 10.1007/s12035-019-1524-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.