Abstract
Objective:
Investigate the gut microbiome in progressive multiple sclerosis (MS) and how it relates to clinical disease.
Methods:
We sequenced the microbiota from healthy controls, relapsing remitting MS (RRMS), and progressive MS patients and correlated the levels of bacteria with clinical features of disease, including EDSS, quality of life, and brain MRI lesions/atrophy. We colonized mice with MS-derived Akkermansia and induced experimental autoimmune encephalomyelitis.
Results:
Microbiota β-diversity differed between MS patients vs. controls but did not differ between RRMS vs. progressive MS or differ based on disease modifying therapies. Disease status had the greatest effect on the microbiome β-diversity, followed by BMI, race, and sex. In both progressive and RRMS, we found increased Clostridium bolteae, Ruthenibacterium lactatiformans and Akkermansia and decreased Blautia wexlerae, Dorea formicigenerans, and Erysipelotrichaceae CCM. Unique to progressive MS, we found elevated Enterobacteriaceae and Clostridium g24 FCEY and decreased Blautia and Agathobaculum. Several Clostridium species were associated with higher EDSS and fatigue scores. Contrary to the view that elevated Akkermansia in MS has a detrimental role, we found that Akkermansia was linked to lower disability, suggesting a beneficial role. Consistent with this, we found that Akkermansia isolated from MS patients ameliorated EAE, which was linked to a reduction in RORγt+ and IL-17 producing γδ T cells.
Interpretation:
While some microbiota alterations are shared in relapsing and progressive MS, we identified unique bacteria associated with progressive MS and clinical measures of disease. Furthermore, elevated Akkermansia in MS may be a compensatory beneficial response in the MS microbiome.
INTRODUCTION
Multiple sclerosis (MS) is an autoimmune disease triggered by environmental factors that act on a genetically susceptible host. One of the most poorly understood aspects of MS is the biology associated with the transition from the relapsing remitting (RR) to the progressive form (1). Progressive MS is more refractory to therapy, is associated with greater physical disability, and with greater impairment of quality of life including fatigue and depression. Thus, there is a critical need to understand factors associated with progressive MS and how to modify them.
The microbiome encompasses trillions of organisms that can affect neurologic disease by modulating immune cells that traffic from the gut to the brain, secreting neuroactive metabolites, altering endocrine signaling pathways, and triggering afferent neurons (2). Studies in animal models demonstrate that the gut microbiota can affect neuroinflammation. Germ-free and antibiotic treated mice are resistant to both induced and spontaneous experimental autoimmune encephalomyelitis (EAE), the animal model for multiple sclerosis (3-6). Administering bacteria that promote T regulatory cells, including polysaccharide A positive Bacteroides fragilis, can ameliorate EAE (7), whereas administering bacteria that induce Th17 cells can worsen EAE. In addition, bacteria may act together through biomimicry and inflammation to induce spinal cord inflammation (8). Colonizing spontaneous EAE mice with microbiota from RRMS patients worsens disease (9, 10) and is linked with decreased IL-10 producing T regs (10). Incubating MS microbiota with human PBMCs induced proinflammatory responses in vitro (9), suggesting that the MS microbiota both lack beneficial microbes that regulate autoimmunity and have an overabundance of proinflammatory bacteria.
Several laboratories, including ours, have reported microbiome alterations in RRMS (9-19), including increases in Akkermansia and decreases in butyrate producing bacteria. Microbiota alterations in MS may be primary drivers of the disease or may instead reflect alterations secondary to the disease process. We found that the host can shape the microbiome through the secretion of microRNAs (20). Furthermore, we found that MS patients and mice at peak EAE have elevated intestinal miR-30d, which increases the levels of Akkermansia and ameliorates EAE (20). This suggests that elevated Akkermansia may result from positive host selection, rather than contributing to MS. Studies have found low Prevotella in RRMS patients and administering human-derived Prevotella histicola improved EAE (21), suggesting that increasing depleted bacteria can lessen neuroinflammation. We found that administering a probiotic to RRMS patients reversed the inflammatory phenotype of peripheral monocytes, and that immune changes were durable months after the probiotic was stopped (15). Taken together, these studies demonstrate that the microbiota plays an important role in MS and that modulating the microbiota has therapeutic potential.
To date there have been few studies of the microbiome in progressive MS. A Russian cohort of 15 primary progressive (PPMS) subjects had elevated Gemmiger and Rumincoccus and an increase in the family Verrocomicrobiaceae, which contains Akkermansia (22). A Belgian cohort of 28 PPMS patients had lower Gemmiger and Butyricicoccus and higher Methanobrevibacter, Sporobacter, and Clostridium cluster IV in PPMS (23). A Japanese cohort of 15 SPMS subjects had elevated Clostridium and decreased butyrate producers (24). We investigated the microbiome in progressive MS, compared it to subjects with relapsing disease, and identified microbial taxa that correlated with disability, quality of life, and brain magnetic resonance imaging (MRI) measures (lesions and atrophy).
METHODS
Study subjects and clinical metadata.
Microbiome study subjects were recruited from the CLIMB study conducted at the MS center at the Brigham & Women’s Hospital. Subjects were selected based on a diagnosis of RRMS or progressive MS, and willingness to participate in a microbiome study, and meeting the inclusion/exclusion criteria (below). The protocols for this study received prior approvals from all institutional review boards, and informed consent was obtained from each subject. Study subjects collected a stool sample at home, then shipped samples overnight on icepacks to the laboratory, and samples were frozen at −80°C upon receipt. Inclusion criteria for subjects with multiple sclerosis included a diagnosis of MS according to the latest McDonald’s Criteria. Disease subtypes were further classified as relapsing remitting MS or progressive MS which included both primary and secondary progressive. Exclusion criteria included pregnancy, history of gastrointestinal surgery, intestinal bowel disease, and antibiotics within the past 3 months. Quality of life was assessed in 95 RRMS and 27 progressive MS patients with the validated Neuro-QOL questionnaire (25). Patients in our CLIMB study undergo yearly 3T MRI scans using a consistent high-resolution protocol (26). We employed automated pipelines to quantify brain T2 lesion volume (26), and normalized whole brain (27) and deep gray matter (28) volumes. Healthy subjects from the PhenoGenetic Project (29), a resource of individuals who are self-reported to be free of chronic infectious and inflammatory diseases and recallable by demographic or genotypic feature for biosampling, were approached to provide a stool sample. Collection and processing procedures were identical to the ones used for the MS patients.
Microbiome Analysis.
DNA was extracted using the DNAeasy PowerLyzer Microbiome DNA extraction kit (Qiagen), and the V4 region of the 16S rRNA gene was amplified using barcoded primers developed by the Earth Microbiome Project and as previously described (15). Paired-end reads were sequenced on the MiSeq at Harvard Biopolymers facility, and sequence analysis was performed in QIIME2. Samples were sequenced on two MiSeq runs, with 173 samples sequenced on both runs, 57 samples sequenced only on the first run, and 52 samples only sequenced on the second run. Denoising and quality filtering of data was performed using DADA2 for each run, then paired samples were merged. Any sample with less than 1000 reads was then removed from analysis. For taxonomic assignment, sequences from EZ-biocloud database formatted for QIIME and released May 2018 was used as a reference set (30). The RDP classifier was trained against the V4 region of this database, then was used to identify sequences. Significant differences in alpha-diversity was performed by the non-parametric Kruskal Wallis test, differences in beta-diversity was performed by PERMANOVA, with correction for multiple comparison testing, and assessment of contributors to microbiome variation was determined by the ADONIS test. After relative abundance was calculated, species that had less than 10% prevalence in any group (HC, RRMS, progressive) were removed from differential testing and correlation analysis. Compositional differences were determined using linear discriminant analysis effect size (LEfSe) with the alpha set at 0.05, and the effect size set at greater than 2 (31). To adjust these findings for other factors that may affect the microbiome, the microbiome multivariable associations with linear models (MaAsLin) tool (32) was used to identify compositional differences while adjusting for age, BMI, sex, race, and ethnicity. To identify bacteria linked with EDSS, MRI measurements, and quality of life, Spearman correlations were performed in R and were adjusted for age using R package “ppcor”.
Isolation and Identification of MS-derived Akkermansia.
Stool samples from 6 individuals with detectable Akkermansia via 16S V4 rRNA sequencing underwent 7 10-fold serial dilution in pre-reduced anaerobically sterilized (PRAS) saline and 100 μL of the 10−4 through 10−7 dilutions were plated on minimal mucin media (Anaerobe Systems). 8-10 colonies were isolated in pure culture per microbiota donor after incubation of 3-7 days. Isolates were frozen in Brucella broth (Difco) plus 15% glycerol, and the near full length 16S rRNA gene was amplified using the 8F and 1510R primers according to previous methods (33). After PCR, primers were removed by ExoSapIT, then sequenced by Sanger Sequencing at the Dana Farber Sequencing Core. Forward and reverse sequences were then quality trimmed and joined using UGene Software. Identification and percent identity were then performed using batch BLAST, National Centers for Biotechnology Information (NCBI). Finally, phylogenetic tree of isolates and references sequences was constructed using Phylogeny.fr.
Effect of MS-derived Akkermansia on EAE.
MS-derived Akkermansia strains BWH-J5, BWH-I7, and BWH-H3, as well as the Type strain of Akkermansia muciniphila, were grown in BHI+mucin broth as previously described (34). Bacteroides cellulosilyticus strain BWH-E5, which is not altered in RR or progressive MS, was administered as a control bacterium. Live bacteria (OD600 0.32, 200 μL) or vehicle control of BHI+mucin broth were delivered to 9-week-old female C57BL6J mice (n = 10-14/group) by oral gavage 3 times a week beginning 2.2 weeks prior to disease induction and receiving bacteria treatment up to 2 weeks post disease induction. Experimental autoimmune encephalomyelitis was induced by injecting 150 μg of myelin oligodendrocyte glycoprotein (MOG) and Complete Freund’s Adjuvant (CFA) subcutaneously and administering a peritoneal injection of 200 ng of pertussis toxin on the same day, and 48 hours later, and disability scores were determined by standard scoring criteria as previously described (34). Differences between the groups were determined by Friedman’s test and Dunn’s correction for multiple comparisons. Animals were housed in a biosafety level 2 facility (BSL2) using autoclaved cages and aseptic handling procedures and kept under a 12-hour light/dark cycle. All animal experiments were conducted according to an IACUC approved protocol.
Effect of MS-derived Akkermansia on T cells in EAE.
For immunologic analysis, a second cohort of 9-week-old female C57BL6J mice (n = 5 per group) received the same microbiota strains, dosing, and immunization as described above. 10 days post disease induction, splenocytes were isolated for flow cytometric analyses. Red blood cells were lysed with ACK, and dead cells were stained with fixable viability dye Aqua Zombie (1:1000 diluted in PBS; Biolegend). Surface markers were stained for 25 min at 4°C in FACs buffer (Mg2+ and Ca2+ free HBSS with 2% FCS, 0.4% EDTA [0.5 M] and 2.5% HEPES [1M]), cells were fixed in Cytoperm/Cytofix (eBioscience), permeabilized with Perm/Wash Buffer (eBiosciences), and then stained for intracellular markers. Extracellular antibodies used were: PerCp-Cy5.5-anti-TCRβ (H57-597; 1:800; Biolegend), BV785-anti-CD4 (RM4-5; 1:400; Biolegend), BV711-anti-CD8a (53-6.7; 1:400; BD), and APC-anti-TCDγδ (GL-3,1:100, ThermoFisher). To measure T cell transcription factors, intracellular antibodies used were FITC-anti-Foxp3 (FJK-16s; 1:100; ThermoFisher), and BV421-anti-RORγT (Q31-378, 1:100, BD). For intracellular cytokine staining, cells were first stimulated for 4 h with PMA (phorbol 12-myristate 13-aceate; 50 ng ml−1; Sigma-Aldrich) and ionomycin (1 μM; Sigma-Aldrich) and a protein-transport inhibitor containing monensin (1 μg ml−1 GolgiStop; BD Biosciences) before detection by staining with antibodies. Intracellular antibodies used were BV421-anti-IFN-γ (XMG1.2; 1:400; Biolegend), PE-Cy7-anti-IL-17A (eBio17B7; 1:400; ThermoFisher), FITC-anti-IL-10 (Jes5-16E3; 1:100; BioLegend), and PE-anti-GM-CSF (MP1-22E9, 1:100, ThermoFisher). Flow-cytometric acquisition was performed on a Fortessa (BD Biosciences) by using DIVA software (BD Biosciences) and data were analyzed with FlowJo software versions 10.1 (TreeStar Inc). Cells were gated on lymphocytes, single cells, live cells, then T cell subsets were divided into TCRβ+CD4+, TCRβ+CD8+, or TCRβ-TCRγδ+ cell populations, and then transcription factors (FoxP3 and RORγT) and cytokines (IL-10, GM-CSF, IL-17, and INFγ) were measured.
RESULTS
Patients with progressive MS have alterations in intestinal microbiota composition.
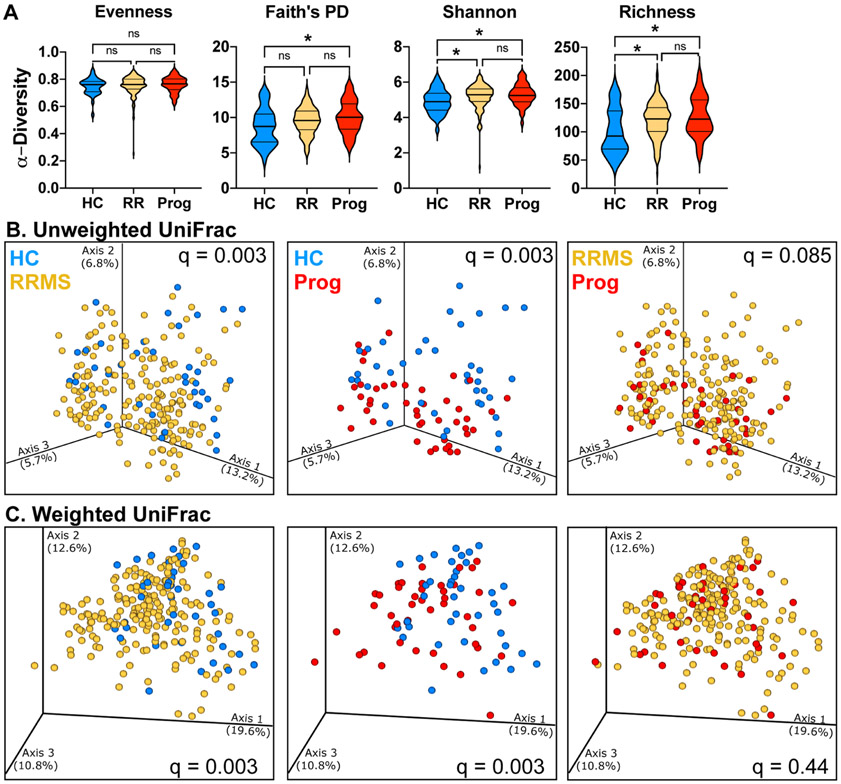
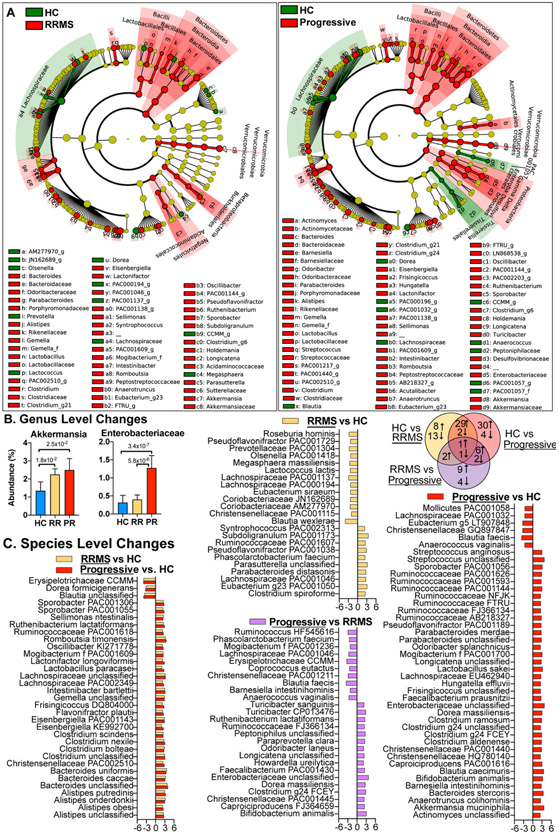
We analyzed the microbiota in 40 healthy controls, 199 RRMS patients, and 44 progressive patients by sequencing the V4 region of the microbial 16S rRNA gene. Examining alpha-diversity, we found no changes in evenness, but found slightly elevated phylogenetic diversity, Shannon diversity, and richness in both RRMS and progressive MS compared to healthy controls (Fig 1A). Examining β-diversity, we found that overall microbiota community structure differed between both progressive and relapsing patients vs. healthy controls but did not differ between progressive and RRMS (Fig 1B,C). At the phylum through genus (Fig 2AB) and at species levels (Fig 2C), we found that progressive patients had unique changes compared to RRMS and healthy controls including an increase in Enterobacteriaceae, Ruminococcaceae FJ366134 and Clostridaceae g24 FCEY and a decrease in Dorea longicatena, Anaerococcus vaginalis, and Blautia faecis. Progressive MS patients shared microbiota alterations with RRMS patients compared to healthy controls, including an increase in Akkermansia at the genus level and Clostridium bolteae at the species level and a decrease in Dorea formicigenerans and unclassified Blautia at the species level. C. bolteae is associated with the induction of Th17 cells (35) and was originally isolated from an autistic patient (36) and has been reported to be elevated in neuromyelitis optica (37). In addition to increased abundance, C. bolteae had a higher prevalence in progressive MS (50% of subjects) compared to healthy controls (12% of subjects). There were two bacteria that were altered in all three comparisons, including a reduction in the not yet cultured Erysipelotrichaceae CCMM and an increase in recently discovered Ruthenibacterium lactatiformans, which was one of the most significantly elevated bacteria in both RRMS and progressive MS.
Figure 1. Microbiota α- and β-diversity in relapsing and progressive MS.
A) Alpha diversity metrics for Evenness, Faith’s phylogenetic diversity, Shannon diversity, and richness (number of features) were calculated at an average sampling depth of 5,000 reads per sample in healthy controls (n = 40), RRMS (n = 199), and progressive MS (n = 44). * p < 0.05 Kruskal-Wallis. B-C) Principal coordinate analysis of intestinal microbiota samples based on unweighted (B) and weighted (C) UniFrac distances show significantly different clustering between HC and RRMS, between HC and progressive MS, but not between RRMS and progressive MS, q = PERMANOVA p-values adjusted for false discovery rate. Each dot represents the microbiota from one individual. HC, healthy control (blue), RRMS (yellow), or progressive MS (red).
Figure 2. Compositional differences in the microbiota of progressive and relapsing MS.
Microbiota was sequenced in healthy controls (HC, n=40), RRMS (n = 199), and Progressive MS (n = 44) subjects. A) Differences are visualized on a cladogram, which shows all changes at the phylum level (inner dots, outer wedge label) through genus level (outer dots, labeled with small letters for abbreviation). Red (MS) or green (HC) circles indicate increased levels in corresponding groups, yellow circles indicates a taxon present but not differentially abundant. Size of the dot corresponds to the overall abundance of that taxon in the microbiome. B) The relative abundance of selected microbiota altered in progressive MS. C) LDA effect size of significantly altered bacteria at the lowest classifiable levels and Venn diagram showing the number of bacteria increased or decreased in each comparison. Positive LDA effect size = up in the underlined group.
Microbiota changes adjusted for host factors.
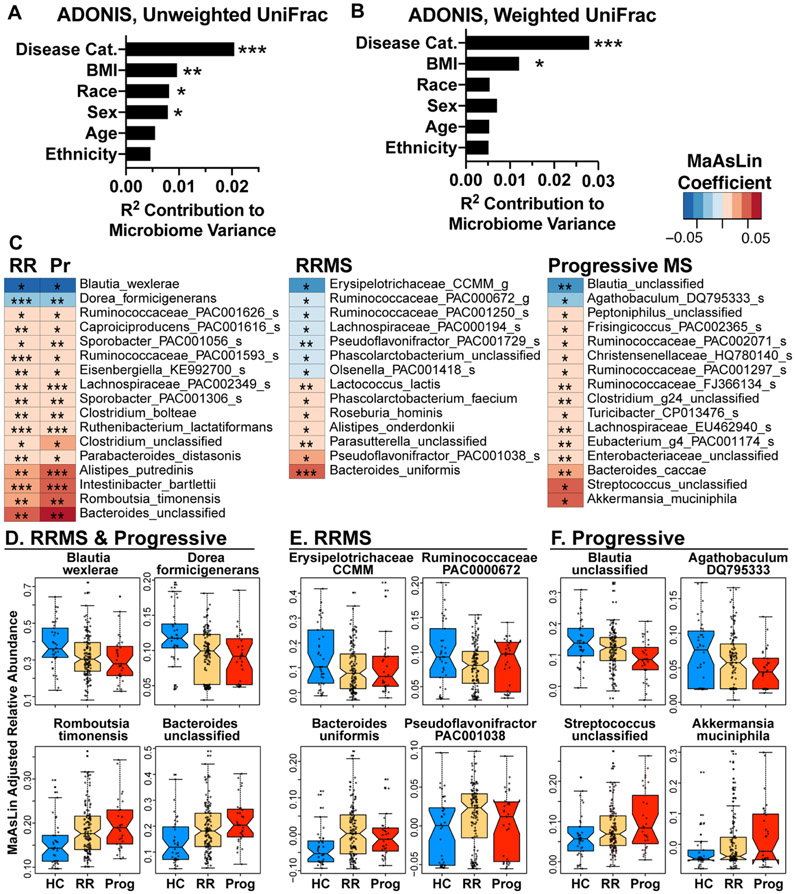
Because host factors can affect the microbiome, we measured the effect disease status, age, BMI, sex, race, and ethnicity on microbiome β-diversity. For this analysis, we restricted the dataset to subjects with a reported BMI, race, and ethnicity (HC = 38, RRMS = 135, progressive MS = 31 subjects). We found that disease status had the largest effect on microbiome composition (p = 0.001), followed by BMI, race, and sex, whereas age and ethnicity did not have a significant effect. We then used the MaAsLin test to identify bacteria in MS vs. HC while adjusting for these variables. As shown in Figure 3C, we found that 14 bacteria were increased and 2 decreased in both progressive and RRMS vs. healthy controls, with the largest increases in Romboutsia timonensis and unclassified Bacteroides, and the largest decreases in Blautia wexlerae and Dorea formicigenerans (Fig 3D). RRMS had unique differences vs. HC in our adjusted model, including decreased Erysipelotrichaceae CCMM and Ruminococcaceae PAC000672 and increased Bacteroides uniformis and Pseudoflavonifractor PAC001038 (Fig 3E). Progressive MS had unique changes vs. HC with the largest increases in Akkermansia muciniphila and Streptococcus and the largest decreases in Blautia unclassified and Agathobaculum DQ795333. Many of the bacteria we identified in our adjusted model (Fig 3) were similar to the bacteria we identified with LEfSe (Fig 2). This is consistent with our finding that disease status had the greatest effect on microbiota composition compared to the other demographic variables we examined (Fig 3A-B).
Figure 3. Microbiota differences adjusted for host variables.
A-B) Effect of host factors on microbiome beta-diversity was measured using the ADONIS test of unweighted and weighted UniFrac distances. Analysis was restricted to subjects with complete demographic information and a recorded BMI, (n = 38 HC, n = 135 RRMS, n = 31 progressive MS) C) Microbiota altered in RR or progressive MS vs. healthy control, MaAsLin, adjusted for age, BMI, sex, race, and ethnicity. D-F) Abundance of the two most decreased taxa and two most increased taxa in both RRMS and progressive MS vs. HC (D), unique to RRMS (E), or unique to progressive MS (F). * p < 0.05, ** p < 0.01, ***p < 0.001.
The effect of disease modifying therapy on the microbiota.
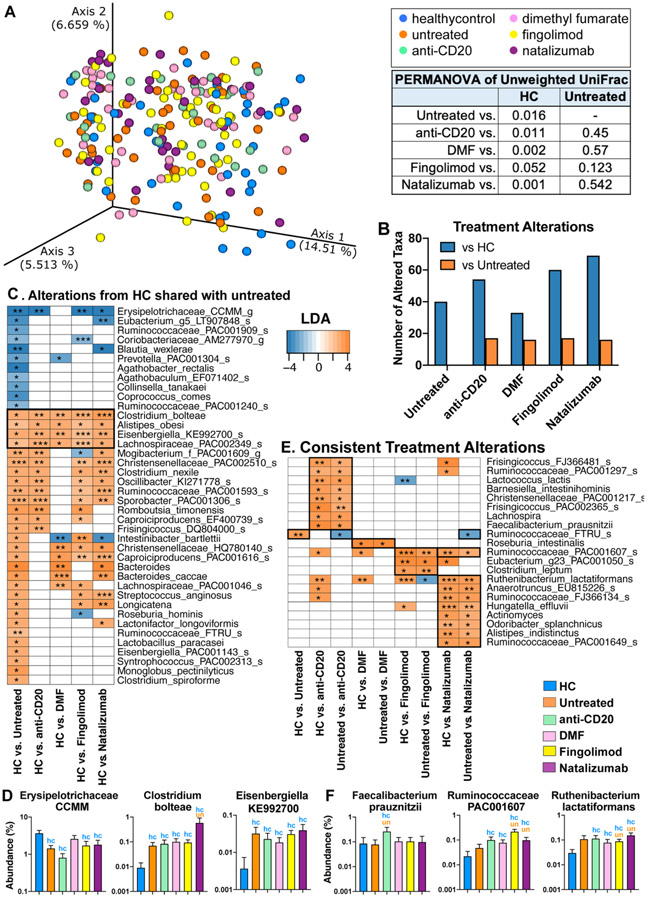
To determine the extent to which disease modifying therapy (DMT) affected the microbiome, we examined changes in beta-diversity in patients receiving four commonly used treatments at our center including anti-CD20 (rituximab and ocrelizumab), dimethyl fumarate, fingolimod, and natalizumab. We found that the overall microbiota composition in subjects on DMTs did not differ from untreated MS patients in beta-diversity, whereas all MS treatment subgroups differed from healthy controls (p < 0.05, PERMANOVA) (Fig 4A-B). These data suggest that disease status has a greater effect on the microbiota than treatment. While there were no changes in beta-diversity, we found changes in specific bacteria that were linked to therapy. Although for each DMT, we found fewer bacteria that differed vs. untreated MS (orange bars) than differed vs. HC (blue bars bars) (Fig 4B). As shown in Fig 4C-D, many of the bacteria that differed between untreated-MS vs. HC also differed between treated-MS vs. HC including an increase in Clostridium bolteae, Eisenbergiella KE992700, Alistipes obesi, and Lachnospiraceae PAC002349 and a decrease in Erysipelotrichaceae CCMM. For each treatment, we found unique bacteria modulated by the DMT vs. both untreated MS and HC (Fig 4E-F). Specifically, we found that anti-CD20 increased Faecalibacterium prauznitzii, and DMF increased Roseburia intestinalis, two butyrate producers reported to be reduced in MS (13, 17, 18). In addition, we found that fingolimod and natalizumab increased Ruminococcaceae PAC001607. Finally, we found increased Ruthenibacterium lactatiformans in all DMTs vs. HC, suggesting that treatment may partially contribute to the increased Ruthenibacterium lactatiformans we identified in Fig 2-3.
Figure 4. The effect of treatment on the MS microbiota.
A) PCoA of unweighted UniFrac distances of RRMS and progressive subjects not on treatment (n = 33) or treated with anti-CD20 (n = 25), dimethyl fumarate (n = 33), fingolimod (n = 57), or natalizumab (n = 36), or healthy controls (n = 40). PERMANOVA test for clustering reveals differences between healthy controls and MS patients on treatment, but not between untreated MS patients and those on therapy. B) Number of taxa altered comparing DMT group vs. healthy controls (blue bar) or vs. untreated MS (orange bar). C) Linear discriminant analysis (LDA) effect size of bacteria altered in untreated MS vs. HC, and whether those bacteria are similarly altered in each DMT group. D) Representative bacteria consistently altered in MS, regardless of treatment status. blue hc = significantly different from healthy controls, orange un = significantly different from untreated MS patients, p < 0.05, LDA >2 LEfSe. E) Bacteria consistently altered by DMT vs. HC and DMT vs. untreated MS. F) Representative bacteria consistently altered by treatment compared to both healthy control and untreated MS patients.
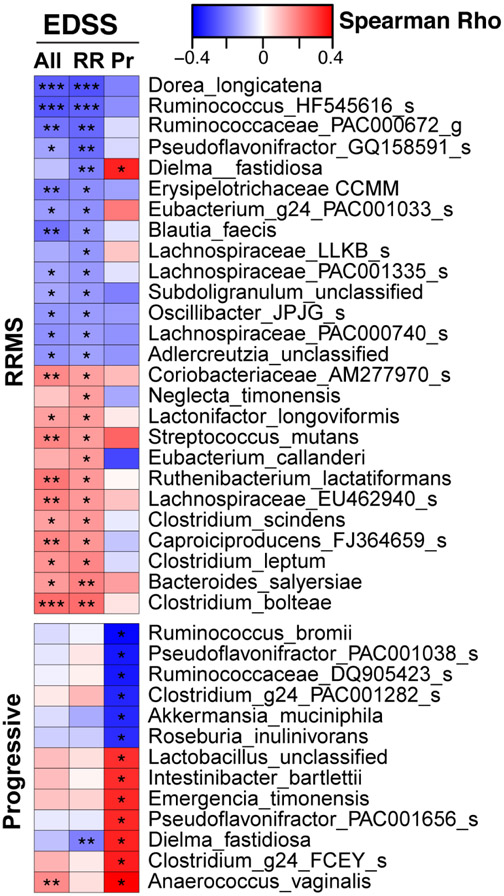
Identification of bacteria associated with disability.
To determine whether bacteria we identified in progressive and RRMS were associated with disability, we examined the relationship between the microbiota and the expanded disability status score (EDSS), adjusting for age (Fig 5). Because progressive MS patients have greater disability than RRMS patients, we examined the relationship between the microbiome and EDSS for each disease category separately as well as for the two categories together. In both progressive and relapsing MS, we found several Clostridium species that were associated with worse EDSS, including Clostridium g24 FCEY (closely related to C. bolteae) in progressive MS and C. bolteae, C. leptum, and C. scindens in RRMS. Butyrate producers had negative correlations with EDSS including Ruminococcaceae HF545616 in RRMS, and Ruminococcus bromii and Roseburia inulinovorans in progressive MS. This is consistent with a potential beneficial role for butyrate producing bacteria in MS (38). Unexpectedly, we found that Akkermansia was negatively correlated with EDSS in progressive patients. Thus, contrary to the view that Akkermansia has a detrimental role in MS, our findings raise the possibility that elevated Akkermansia could represent a compensatory microbiome response to the disease.
Figure 5. Microbiota associated with disability.
Microbiota correlations with EDSS scores show unique relationships in RRMS (n = 198) and progressive MS (n = 43), Spearman correlations, adjusted for age. * p < 0.05, ** p < 0.01, *** p < 0.001.
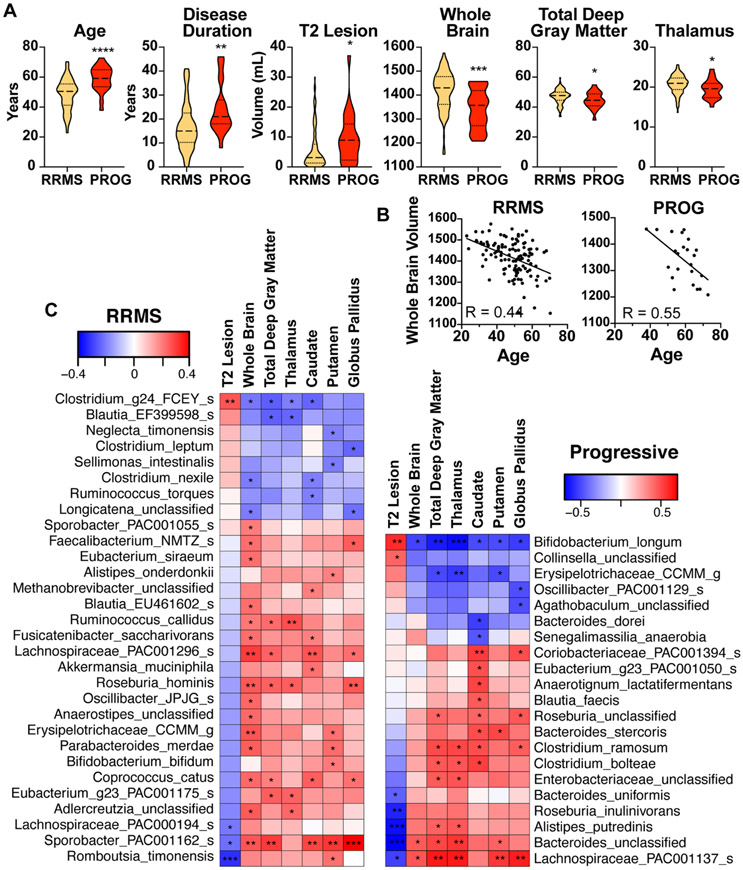
Identification of bacteria associated with MRI measures of disease.
Atrophy is more severe in progressive vs. RRMS (39, 40), particularly in gray matter areas (39, 41). Furthermore, brain volume loss in the first year of the disease is a strong predictor of future neurologic impairment (42, 43). The microbiome can alter neurogenesis (44), myelination (45) and influence inflammation in the brain (46), but its relationship to MRI metrics in MS is unknown. To determine whether there were associations between the gut microbiome and MRI measures of disease severity, we identified a cohort of progressive and RRMS patients in our CLIMB longitudinal cohort study for which we obtained quantitative measures of brain T2 lesions and both whole brain and regional deep gray matter volumes from 3T MRI. Progressive patients had higher T2 lesion volume and lower normalized whole brain, total deep gray, and thalamic volumes (Fig 6A). Because brain volume negatively correlated with age (Fig 6B), we adjusted our MRI-microbiota analysis for age. In RRMS, we found several Clostridium species associated with increased T2 lesion volume and decreased brain volume, including C. leptum, C. nexile, and Clostridium g24 FCEY (Fig 6C), consistent with our EDSS data (Fig 5), suggesting a detrimental role. In RRMS, several bacteria had a negative correlation with T2 lesion volume and positive correlation with brain volume, including Sporobacter PAC00162, Akkermansia muciniphila, Erysipelotrichaceae CCMM (Fig 6C), consistent with our observation that some of these bacteria were associated with lower disability (Fig 5). However, in progressive MS, we found opposite relationships between the microbiota and MRI measures. For example, Erysipelotrichaceae CCMM had a negative correlation with brain volume and several Clostridium species positively correlated with increased brain volume. This may reflect different biologic processes in RRMS vs progressive MS.
Figure 6. Associations between the microbiota and MRI brain measurements in progressive MS.
A) Age, disease duration, and brain 3T MRI measurements in progressive MS (n = 23) and RRMS (n = 116) patients, t-test. B) Brain volume negatively correlated with age, linear regression, p <0.001, R = 0.44 and 0.55 for RRMS and progressive MS, respectively. C) Bacteria that correlate with lesion volume (upper section) and brain volume (lower section) in RRMS and progressive MS. Spearman correlation adjusted for age. * p < 0.05, ** p < 0.01, *** p < 0.001.
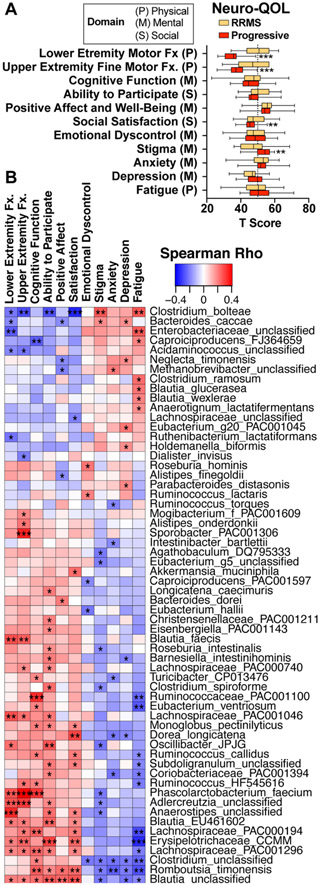
Identification of bacteria associated with quality of life.
Quality of life is an important global indicator of MS disease status and is routinely measured as part of our CLIMB longitudinal cohort study (47, 48). We asked whether there was an association between the gut microbiota and quality of life using the validated NeuroQOL questionnaire, which measures 3 major domains of quality of life: physical, mental, and social. For the first six metrics (“lower extremity motor function” through “satisfaction”), higher scores indicate higher quality of life, and for the last five metrics (“emotional dyscontrol” through “fatigue”), higher scores indicate lower quality of life (Fig 7A). Because there were no differences in fatigue, anxiety and depression in our progressive vs. RRMS subjects (Fig 7A), we analyzed a combined cohort of progressive and RRMS patients. We found that Enterobacteriaceae, C. bolteae and other Clostridia positively correlated with fatigue suggesting a potential detrimental role whereas Erysipelotrichaceae CCMM, Romboutsia timonensis, and Lachnospiraceae PAC00194 negatively correlated with fatigue, depression, and anxiety, suggesting a potential beneficial role (Fig 7C).
Figure 7. Microbiota associations with quality of life.
A) Quality of life measurements in 95 relapsing (RRMS) and 27 progressive patients were assessed using the NeuroQOL questionnaire across three domains: physical, mental, and social. Departure from the population norm (T score = 50) in RRMS and progressive patients. B) Microbiota correlations with quality of life, Spearman correlation adjusted for age. * p<0.05, **<0.01, ***< 0.001.
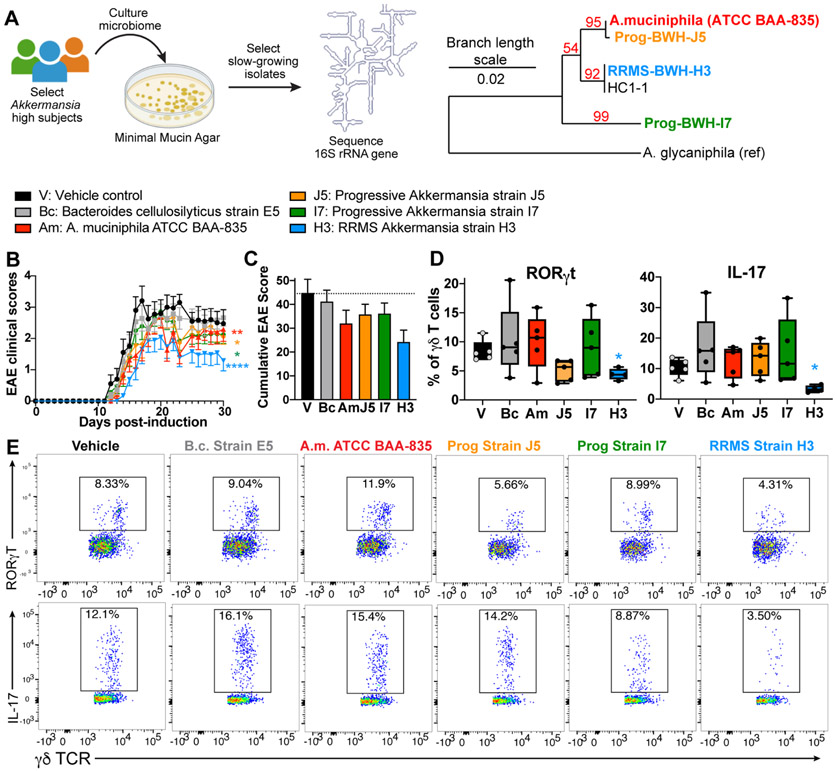
MS-associated Akkermansia ameliorates EAE.
As discussed above, we found that Akkermansia was negatively correlated with EDSS and MRI burden of disease (Fig 5, 6) suggesting that Akkermansia has a beneficial role in MS. Thus, we investigated whether MS-associated Akkermansia had a beneficial effect in the MS model of EAE. We identified progressive and RRMS patients with high Akkermansia and were able to distinguish three subtypes based on 16S rRNA V4 sequences (Fig 8A). We then isolated bacteria on minimal mucin media from 6 subjects, identified bacterial strains using Sanger sequencing, and recovered Akkermansia isolates from 4 of our subjects (HC1, RRMS1, Prog1, Prog2). The strain Prog2-BWH-J5 was nearly identical (99.7%) to the Type strain of Akkermansia muciniphila and strain RRMS1- BWH-H3 was 98.7% identical to the Type strain, indicating a new sub-strain of A. muciniphila. Strain Prog1- BWH-I7 was only 96.5% identical, indicating a new species of Akkermansia. To test the role MS-derived Akkermansia in disease, we colonized C57/BL6 mice with Akkermansia strains BWH-J5, BWH-H3, and BWH-I7 and found that they all lowered disease in the MOG/C57 model of EAE, with the strongest protective effect from strain H3 (Fig 8B). As a control, we used Bacteroides cellulosilyticus, a commensal gut microbe that was not altered in MS. In addition, we measured immune responses in an independent experiment 10 days post-immunization and found that Akkermansia strain BWH-H3 reduced RORγT positive γδ T cells and IL-17 producing γδ T cells. No effect was observed in FoxP3, RORγt, IL-10, IFNγ, or IL-17 expression in CD4 or CD8 T cells (not shown). These findings support a beneficial role for Akkermansia in MS and suggest that there may be strain-specific effects on CNS autoimmunity.
Figure 8. MS-associated Akkermansia ameliorates EAE.
A) Stool samples from MS patients with high levels of Akkermansia, corresponding to 3 different V4 16S rRNA sequences, were plated on minimal mucin agar, and slow-growing strains were isolated and identified by 16S rRNA Sanger sequencing. Phylogenetic tree constructed from near full length 16S rRNA sequences shows three phylotypes of Akkermansia isolated from HC and MS patients. B) Akkermansia isolated from RRMS and progressive MS subjects reduce EAE score in the C57/MOG model, whereas the control bacteria Bacteroides cellulosilyticus (Bc) does not, n = 10-14 mice per group. * p < 0.05, ** p < 0.01, **** p < 0.0001 Freidman’s test with Dunn’s correction for multiple comparisons. C) Cumulative EAE scores. D-E) Mice were colonized with B. cellulosilyticus, Akkermansia muciniphila Type strain, and three MS-derived Akkermansia strains, n = 5 per group. EAE was induced, and immunologic responses were measured 10 days later. D) Levels of RORγT+ γδ T cells in unstimulated splenocytes and levels of IL-17 production in PMA/ionomycin stimulated splenocytes. * p < 0.05, one-way ANOVA. E) Representative FACs plots of RORγT and IL-17 production from splenic γδ-T cells.
DISCUSSION
While the microbiome plays a clear role in relapsing-remitting MS, there are few studies that have characterized the microbiome in progressive MS and connected changes in the progressive microbiome to clinical disease. Approximately half of the microbiota changes we found were unique in progressive MS vs. HC compared to RRMS vs. HC. We identified two bacteria that were altered in both types of MS, but more prominent in progressive MS. Erysipelotrichaceae CCMM was decreased in RRMS vs. HC and decreased even further in progressive MS vs. both RRMS and HC. Consistent with a potentially beneficial role, Erysipelotrichaceae CCMM was associated and increased motor and cognitive function and decreased EDSS and fatigue. The sequence of the V4 region of the 16S rRNA from Erysipelotrichaceae CCMM in our study was identical to two recently described bacteria, Faecalibacillus intestinalis and Faecalibacillus faecis (49). Little is known about the functions of these bacteria in neurologic or autoimmune disease. Ruthenibacterium lactatiformans was increased in RRMS vs. HC and increased even further in progressive MS vs. both RRMS and HC. R. lactatiformans was associated with increased EDSS and decreased lower extremity motor function, consistent with a potentially detrimental role. R. lactatiformans is a lactate producing member of the Ruminococcaceae family (50) and lactate is hypothesized to contribute to disease progression in MS by contributing to mitochondria dysfunction (51, 52). Serum lactate has been reported to be higher in MS vs. HC, higher in progressive vs. RRMS, and positively correlated with EDSS (51, 52). Elevated CSF lactate has been found in RRMS patients and was associated with long-term disease progression (53).
We identified six bacteria that were specifically elevated in progressive MS vs. both HC and RRMS including Enterobacteriaceae, Bifidobacterium animalis, Clostridium g24 FCEY, Dorea massiliensis, Longicatena, and Ruminococcaceae FJ366134. Two bacteria were uniquely decreased in progressive MS, including Lachnospiraceae PAC001046 and Phascolarctobacterium faecium. Adjusting for age, sex, race, ethnicity, and BMI, we confirmed that Enterobacteriaceae, Clostridium g24 FCEY, and Ruminococcaceae FJ366134 were uniquely elevated in progressive MS. We also found that Clostridium g24 FCEY was associated with greater disability, and Enterobacteriaceae was associated with fatigue. Clostridium g24 FCEY is a not-yet-cultivated member of the Lachnospiraceae family, and is closely related to Clostridium bolteae, which we found to be elevated in both RR and progressive MS and associated with disability and fatigue. Enterobacteriaceae is a family of Gram-negative facultative anaerobes which encompasses many important gut bacteria, which include E. coli, Shigella and Salmonella. The Enterobacteriaceae family is difficult to speciate based on 16S sequencing alone because E. coli and Shigella have identical 16S rRNA sequences. Some species may be commensal while others are pathogenic (54, 55). Several Enterobacteriaceae strains can attach to the intestinal mucosa and stimulate immune responses (56). Thus, additional studies using shotgun metagenomics are needed to identify these bacteria at the species level and to determine whether progressive MS patients have an enrichment in other bacteria with the property of adhering to the intestinal mucosa.
Adjusting for age, we identified two bacteria that were uniquely decreased in progressive MS, Agathobaculum DQ795333 and Blautia unclassified. The sequence from Agathobaculum DQ795333 was 99.6% similar to Agathobaculum butyriciproducens, a recently discovered butyrate-producing bacteria in the Ruminococcaceae family (57). The gut microbiota can lessen inflammatory disease by producing butyrate and inducing T regulatory cells, and several studies have reported decreased butyrate producers in MS (58, 59). The sequence from unclassified Blautia was 99.6% similar to Blautia luti, an acetate and succinate producing member of the Lachnospiraceae family. Blautia play important roles in carbohydrate fermentation which supports cross-feeding networks in the microbiome, and Blautia species have been proposed as bacteria with high potential for use as next-generation probiotics (60). Of note, Blautia luti and Blautia wexlerae (which we found decreased in both RR and progressive MS), were reported to be depleted in children with obesity and insulin resistance and had negative correlations with markers of inflammation in the stool (61). Furthermore, secreted products from B. luti and B. wexlerae can exert an anti-inflammatory effect on peripheral blood mononuclear cells (61). In our study, Blautia unclassified had the strongest association with 9 out of 11 quality of life parameters, including a positive correlation with motor function, cognitive function, and affect, and a negative correlation with fatigue and depression.
We identified alterations in the microbiota that were similar in both progressive and RRMS patients vs. healthy controls, including depletion of Erysipelotrichaceae CCMM and Blautia wexlerae (discussed above), and Dorea formicigenerans. D. formicigenerans is a member of the Lachnospiraceae family and produces abundant amounts of formic acid (62). Studies have shown that administering formic acid to pigs increases levels of beneficial microbes and suppresses pathogenic members of the Enterobacteriaceae family (enterotoxigenic E. coli and Salmonella) (63, 64). We found several Clostridium species were elevated in both progressive and RRMS patients, including Clostridium bolteae, C. nexile, and C. scindens, which have all been reported to be elevated in new-onset, treatment naive MS patients (17). Adjusting for confounders including BMI, age, race, ethnicity, and sex, we confirmed that C. bolteae was elevated in both groups. C. bolteae was originally isolated from an autistic child and may induce Th17 cells by direct attachment to the mucosa (35). It has been shown that gut microorganisms may act together via molecular mimicry and induction of Th17 cells to worsen spinal cord inflammation in the EAE model (8), and the bacteria that we identified may contribute to disease through multiple mechanisms. We have also found that C. bolteae is elevated in patients with neuromyelitis optica spectrum disorders and shares protein sequence homology with aquaporin 4 (37), suggesting that it may also act by molecular mimicry.
We observed elevated Akkermansia at the genus level in both progressive and RRMS patients. At the species level, Akkermansia muciniphila was significantly increased in progressive MS (p = 0.02 MaAsLin), while there was only a trend of increased A. muciniphila in our RRMS subjects (p = 0.10 MaAsLin). This could reflect additional strain and species level variation in Akkermansia in RRMS, which we were able to identify through use of a new taxonomic reference database from EzBioCloud (30). We and others have previously observed elevated levels of Akkermansia in RRMS (9-11, 18), and elevated Akkermansia in MS is also observed in a recent meta-analysis of microbiota alterations in autoimmunity (65). Of note, Akkermansia has been reported to have a beneficial role in multiple diseases (66, 67). Akkermansia improves metabolism in obese and diabetic mice (66, 68), improves cancer check point immunotherapy (69), is associated with the anti-seizure effects of a ketogenic diet (70) and improves disease in an animal model of ALS (67). Furthermore, the anti-inflammatory properties of Akkermansia can be strain-specific (71), warranting further study of Akkermansia strains in neurologic and inflammatory diseases.
We found that Akkermansia negatively correlated with disability and T2 lesion volume, and positively correlated with brain volume. This was unexpected given that it has been assumed that elevated Akkermansia in MS is detrimental (9, 11). To directly test the in vivo properties of Akkermansia, we isolated Akkermansia from progressive and RRMS patients, colonized animals with these strains prior to EAE induction and found that MS-derived Akkermansia ameliorated EAE. Akkermansia muciniphila strain BWH-H3 had the strongest protective effect, which was associated with decrease in RORγT+ and IL-17 producing γδ T cells. There are large populations of γδ T cells in the intestinal mucosa which respond rapidly to the microbiota (72). This microbiota-γδ T cell interaction could be relevant in MS as γδ T cells traffic to the CNS and produce high levels of IL-17 in EAE, and IL-17 producing γδ T are elevated in the blood of MS patients (73). Consistent with our findings in the C57 model, investigators found that mice with higher levels of Akkermansia had less progression in the NOD progressive EAE model (74). Humans co-evolved with the gut microbiota and developed the production of microRNAs to selectively enhance the growth of specific bacteria (20). We previously found that MS patients and mice at peak EAE had increased levels of the microRNA miR-30d in the gut, which increases Akkermansia levels (34). Taken together, our findings suggest that elevated Akkermansia may be a beneficial compensatory microbiome response in MS.
Alterations in the microbiota may reflect differences in patient populations, rather than be drivers of disease. In our study, healthy controls, RRMS, and progressive MS subjects were well-matched for BMI, sex, gender, race, and ethnicity. MS disproportionately affects women and whites of European descent (75, 76), which is reflected in the study population that we recruited from our center (Table 1). Progressive MS patients were on average older than RRMS patients, consistent with the observation that age is one of the greatest risk factors for progressive MS (43). The microbiome composition in adults is relatively stable and similar between young adults (20-40 years of age) and middle aged (40-60 years) adults, but can differ in adults over 60 years of age (77). In order to address this, we adjusted microbiota correlations with EDSS, MRI, and quality of life for age. We determined the contribution of demographic factors to microbiome variation, and found that after disease status, BMI had the largest contribution to variation in the microbiome. Race and sex affected β diversity based on unweighted UniFrac distances but did not affect β diversity based on weighted UniFrac distances. We found no contribution of age or ethnicity to microbiome variation in our cohort.
Table 1:
Study Subject Demographics
| HC | RRMS | Prog | |
|---|---|---|---|
| Number of Subjects | 40 | 199 | 44 |
| Number Female Subjects (%) | 28 (70.0%) | 152 (76.4%) | 31 (70.4%) |
| Age (years) ± St. dev. | 45.4 ± 9.2 | 49.3 ± 9.5 | 57.8 ± 7.6 |
| Body Mass Index ± St. dev. | 28 ± 7.9 | 27.1 ± 6.3 | 28.4 ± 6.5 |
| Race | |||
| White | 38 | 188 | 42 |
| Black | 1 | 7 | 1 |
| More than one race | 1 | 3 | 1 |
| Unknown, not reported | 0 | 1 | 0 |
| Ethnicity | |||
| Not Hispanic or Latino | 39 | 193 | 43 |
| Hispanic or Latino | 1 | 6 | 1 |
| Disease Duration ± St. dev. | NA | 16.3 ± 10 | 24 ± 10.9 |
| EDSS ± St. dev. | NA | 1.8 ± 1.2 | 5 ± 1.8 |
| Treatment | |||
| Untreated | NA | 40 | 5 |
| Fingolimod | NA | 47 | 2 |
| Dimethyl fumarate | NA | 29 | 5 |
| Natalizumab | NA | 28 | 3 |
| anti-CD20 | NA | 19 | 6 |
| Interferon-beta | NA | 14 | 5 |
| Glatiramer acetate | NA | 12 | 3 |
| Mycophenolate mofetil | NA | 1 | 4 |
| Methylprednisolone | NA | 3 | 1 |
| teriflunomide | NA | 0 | 3 |
| Methotrexate | NA | 1 | 0 |
| Other | NA | 0 | 2 |
| More than one DMT | NA | 1 | 3 |
| Recently Off Treatment | NA | 4 | 2 |
Several of our findings were found in other MS studies of progressive MS (17). A Russian cohort of 15 PPMS subjects also reported elevated Verrocomicrobiaceae (the family that contains Akkermansia) (22), Consistent with our observation of decreased butyrate producing bacteria in progressive MS (Agathobaculum), a Belgian cohort of 28 PPMS patients reported lower Butyricicoccus (23) and Japanese cohort of 15 SPMS subjects reported decreased butyrate producers (24). Furthermore, the Japanese cohort reported elevated Clostridium species, similar to our results in progressive MS (24). Our cohort of 44 progressive MS subjects is the largest progressive MS population studied to date. Future longitudinal investigations will further clarify the relationship between the microbiome and disease progression over time.
We observed a correlation between the microbiome and quality of life. The mechanism for this association is not clear, but it may be related to the microbial production of neurotransmitters (78, 79). While there is debate on whether bacterial neurotransmitters cross the blood brain barrier, it has been shown that administering gamma amino butyric acid (GABA) producing Lactobacillus reduces anxiety and depression in animal models (78), and GABA producing Bacteroides correlates with functional MRI connectivity in patients with major depressive disorder (79). Self-reported fatigue is a risk factor for the transition to progressive MS (80). Our study identifies bacteria that are associated with fatigue and other measurements of quality of life, which may provide an avenue to affect this through the microbiome.
We had previously found that disease modifying therapies, including glatiramer acetate and interferon, normalized some of the MS-associated changes in specific bacteria (11), and others have reported on MS treatment effects in MS (81). However, in this larger study, we found that disease status had a much greater effect on the microbiome than therapy. This suggests that currently used disease modifying therapies primarily act on immune mechanisms of the disease rather than through the gut. A limitation of our study is that we examined changes in microbiota in a cross-sectional study. Thus, interindividual variability may mask treatment-induced changes. Further work comparing the microbiota before and after therapy could help better define the effect of DMTs on the gut microbiota. In addition, there may be a differential effect of therapy on the microbiome in RRMS vs progressive MS. In our cohort, we had 19-47 RRMS subjects per treatment group and only 2-6 progressive MS subjects per treatment group. In a separate analysis of only RRMS subjects, the majority of taxonomic changes were similar (not shown). Because we had small numbers of progressive subjects in each subject group, we could not analyze them separately.
In summary, we have shown that there are unique changes in the microbiome in progressive MS, and that other features of the MS microbiota were observed in both relapsing and progressive disease. Importantly, we found correlations between the microbiome and both clinical and MRI measures of disease supporting a role for the microbiome in the disease process. We experimentally validated our finding of a correlation between Akkermansia and less disease in MS by transferring MS-derived Akkermansia into EAE mice and demonstrating a beneficial effect. This finding can serve as a framework to test additional candidates that we identified in our study. Furthermore, comparative genomic analysis may be an approach that could identify strain-specific factors in Akkermansia that confer protection in MS. A major question is whether microbiota manipulation is a viable therapeutic avenue to treat MS. We previously reported that a probiotic containing Lactobacillus, Bifidobacterium and Streptococcus strains promoted anti-inflammatory immunity in RRMS patients and healthy controls and some of these immune changes persisted after discontinuation of the probiotic (15). We have now identified new bacterial species that may find usefulness for the treatment of progressive MS including Akkermansia. Although it is now accepted that the microbiome plays an important biologic role in MS, it must be emphasized that the mechanisms by which the microbiome affects MS have not been well-defined and many confounding factors exist. Nonetheless, our findings support the possibility that microbiota manipulation may one day be used as a treatment for MS and identify unique microbiota changes in progressive disease.
ACKNOWLEDGEMENT
This work was supported by NIH Grant 5R01NS087226, The NextGen Collaborative Grant from the Brigham Research Institute and the Water Cove Charitable Foundation. LMC was supported by the Nancy Davis Race to Erase MS Young Investigator Award. AHM was supported by a clinician-scientist development award from National MS Society.
Footnotes
POTENTIAL CONFLICTS OF INTEREST
Nothing to report.
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1002/ana.26084
DATA AVAILABILITY
The microbiota 16S rRNA sequence data has been submitted to the National Center for Biotechnology Information (NCBI) Short Read Archives (SRA) under Bioproject accession number PRJNA721421, which is publicly available. The deidentified metadata, including diagnosis, age, treatment, race, ethnicity, BMI, sex, and EDSS have been included along with the sequencing data. EDSS is recorded under the column heading host_phenotype.
REFERENCES
- 1.Baecher-Allan C, Kaskow BJ, Weiner HL. Multiple Sclerosis: Mechanisms and Immunotherapy. Neuron. 2018;97(4):742–68. Epub 2018/02/23. doi: 10.1016/j.neuron.2018.01.021. PubMed PMID: 29470968. [DOI] [PubMed] [Google Scholar]
- 2.Cox LM, Weiner HL. Microbiota Signaling Pathways that Influence Neurologic Disease. Neurotherapeutics. 2018;15(1):135–45. Epub 2018/01/18. doi: 10.1007/s13311-017-0598-8. PubMed PMID: 29340928; PMCID: PMC5794708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479(7374):538–41. Epub 2011/10/28. doi: 10.1038/nature10554. PubMed PMID: 22031325. [DOI] [PubMed] [Google Scholar]
- 4.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108 Suppl 1 (Supplement 1):4615–22. Epub 2010/07/28. doi: 10.1073/pnas.1000082107. PubMed PMID: 20660719; PMCID: PMC3063590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, Kasper LH. Role of Gut Commensal Microflora in the Development of Experimental Autoimmune Encephalomyelitis. The Journal of Immunology. 2009;183(10):6041–50. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- 6.Yokote H, Miyake S, Croxford JL, Oki S, Mizusawa H, Yamamura T. NKT Cell-Dependent Amelioration of a Mouse Model of Multiple Sclerosis by Altering Gut Flora. AJPA. 2010;173(6):1714–23. doi: 10.2353/ajpath.2008.080622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochoa-Reparaz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 2010;3(5):487–95. Epub 2010/06/10. doi: 10.1038/mi.2010.29. PubMed PMID: 20531465. [DOI] [PubMed] [Google Scholar]
- 8.Miyauchi E, Kim SW, Suda W, Kawasumi M, Onawa S, Taguchi-Atarashi N, Morita H, Taylor TD, Hattori M, Ohno H. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature. 2020;585(7823):102–6. Epub 2020/08/28. doi: 10.1038/s41586-020-2634-9. PubMed PMID: 32848245. [DOI] [PubMed] [Google Scholar]
- 9.Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK, Hauser SL, Crabtree-Hartman E, Sand IK, Gacias M, Zhu Y, Casaccia P, Cree BAC, Knight R, Mazmanian SK, Baranzini SE. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A. 2017;114(40):10713–8. Epub 2017/09/13. doi: 10.1073/pnas.1711235114. PubMed PMID: 28893978; PMCID: PMC5635915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, Liu C, Klotz L, Stauffer U, Baranzini SE, Kumpfel T, Hohlfeld R, Krishnamoorthy G, Wekerle H. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A. 2017;114(40):10719–24. Epub 2017/09/13. doi: 10.1073/pnas.1711233114. PubMed PMID: 28893994; PMCID: PMC5635914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, Patel B, Mazzola MA, Liu S, Glanz BL, Cook S, Tankou S, Stuart F, Melo K, Nejad P, Smith K, Topcuolu BD, Holden J, Kivisakk P, Chitnis T, De Jager PL, Quintana FJ, Gerber GK, Bry L, Weiner HL. Alterations of the human gut microbiome in multiple sclerosis. Nature communications. 2016;7(1):12015. Epub 2016/06/29. doi: 10.1038/ncomms12015. PubMed PMID: 27352007; PMCID: PMC4931233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tremlett H, Fadrosh DW, Faruqi AA, Hart J, Roalstad S, Graves J, Spencer CM, Lynch SV, Zamvil SS, Waubant E, Centers USNoPM. Associations between the gut microbiota and host immune markers in pediatric multiple sclerosis and controls. BMC Neurol. 2016;16(1):182. Epub 2016/09/23. doi: 10.1186/s12883-016-0703-3. PubMed PMID: 27652609; PMCID: PMC5031272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, Chihara N, Tomita A, Sato W, Kim SW, Morita H, Hattori M, Yamamura T. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS One. 2015;10(9):e0137429. Epub 2015/09/15. doi: 10.1371/journal.pone.0137429. PubMed PMID: 26367776; PMCID: PMC4569432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Soldan MMP, Luckey DH, Marietta EV, Jeraldo PR, Chen X. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Scientific Reports. 2016;6. PubMed PMID: 27346372; PMCID: PMC4921909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tankou SK, Regev K, Healy BC, Tjon E, Laghi L, Cox LM, Kivisakk P, Pierre IV, Hrishikesh L, Gandhi R, Cook S, Glanz B, Stankiewicz J, Weiner HL. A probiotic modulates the microbiome and immunity in multiple sclerosis. Ann Neurol. 2018;83(6):1147–61. Epub 2018/04/22. doi: 10.1002/ana.25244. PubMed PMID: 29679417; PMCID: PMC6181139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cosorich I, Dalla-Costa G, Sorini C, Ferrarese R, Messina MJ, Dolpady J, Radice E, Mariani A, Testoni PA, Canducci F, Comi G, Martinelli V, Falcone M. High frequency of intestinal TH17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci Adv. 2017;3(7):e1700492. Epub 2017/07/15. doi: 10.1126/sciadv.1700492. PubMed PMID: 28706993; PMCID: PMC5507635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ventura RE, Iizumi T, Battaglia T, Liu M, Perez-Perez GI, Herbert J, Blaser MJ. Gut microbiome of treatment-naive MS patients of different ethnicities early in disease course. Sci Rep. 2019;9(1):16396. Epub 2019/11/11. doi: 10.1038/s41598-019-52894-z. PubMed PMID: 31705027; PMCID: PMC6841666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantarel BL, Waubant E, Chehoud C, Kuczynski J, DeSantis TZ, Warrington J, Venkatesan A, Fraser CM, Mowry EM. Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Investig Med. 2015;63(5):729–34. Epub 2015/03/17. doi: 10.1097/JIM.0000000000000192. PubMed PMID: 25775034; PMCID: PMC4439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tremlett H, Fadrosh D, Faruqi A, Zhu F, Hart J, Roalstad S, Graves J, Lynch S, Waubant E. Gut microbiota in early pediatric multiple sclerosis: a case– control study. European journal of neurology. 2016;23(8):1308–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, Comstock LE, Gandhi R, Weiner HL. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe. 2016;19(1):32–43. Epub 2016/01/15. doi: 10.1016/j.chom.2015.12.005. PubMed PMID: 26764595; PMCID: PMC4847146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shahi SK, Freedman S, Luckey D, Karau M, Marietta E, Gibson-Corley K, Patel R, Rodriguez M, David CS, Taneja V. Human gut-derived commensal Prevotella histicola suppress experimental autoimmune encephalomyelitis in humanized mice. Journal of immunology. 2017. [Google Scholar]
- 22.Kozhieva M, Naumova N, Alikina T, Boyko A, Vlassov V, Kabilov MR. Primary progressive multiple sclerosis in a Russian cohort: relationship with gut bacterial diversity. BMC Microbiol. 2019;19(1):309. Epub 2020/01/01. doi: 10.1186/s12866-019-1685-2. PubMed PMID: 31888483; PMCID: PMC6937728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynders T, Devolder L, Valles-Colomer M, Van Remoortel A, Joossens M, De Keyser J, Nagels G, D’hooghe M, Raes J. Gut microbiome variation is associated to Multiple Sclerosis phenotypic subtypes. Annals of Clinical and Translational Neurology. 2020. PubMed PMID: 32162850; PMCID: PMC7187717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takewaki D, Suda W, Sato W, Takayasu L, Kumar N, Kimura K, Kaga N, Mizuno T, Miyake S, Hattori M, Yamamura T. Alterations of the gut ecological and functional microenvironment in different stages of multiple sclerosis. Proc Natl Acad Sci U S A. 2020;117(36):22402–12. Epub 2020/08/26. doi: 10.1073/pnas.2011703117. PubMed PMID: 32839304; PMCID: PMC7486801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cella D, Lai JS, Nowinski CJ, Victorson D, Peterman A, Miller D, Bethoux F, Heinemann A, Rubin S, Cavazos JE, Reder AT, Sufit R, Simuni T, Holmes GL, Siderowf A, Wojna V, Bode R, McKinney N, Podrabsky T, Wortman K, Choi S, Gershon R, Rothrock N, Moy C. Neuro-QOL: brief measures of health-related quality of life for clinical research in neurology. Neurology. 2012;78(23):1860–7. Epub 2012/05/11. doi: 10.1212/WNL.0b013e318258f744. PubMed PMID: 22573626; PMCID: PMC3369516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier DS, Guttmann CRG, Tummala S, Moscufo N, Cavallari M, Tauhid S, Bakshi R, Weiner HL. Dual-Sensitivity Multiple Sclerosis Lesion and CSF Segmentation for Multichannel 3T Brain MRI. J Neuroimaging. 2018;28(1):36–47. Epub 2017/12/14. doi: 10.1111/jon.12491. PubMed PMID: 29235194; PMCID: PMC5814929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu R, Tauhid S, Glanz BI, Healy BC, Kim G, Oommen VV, Khalid F, Neema M, Bakshi R. Whole Brain Volume Measured from 1.5T versus 3T MRI in Healthy Subjects and Patients with Multiple Sclerosis. J Neuroimaging. 2016;26(1):62–7. Epub 2015/06/30. doi: 10.1111/jon.12271. PubMed PMID: 26118637; PMCID: PMC4755143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu R, Hurwitz S, Tauhid S, Bakshi R. Automated segmentation of cerebral deep gray matter from MRI scans: effect of field strength on sensitivity and reliability. BMC Neurol. 2017;17(1):172. Epub 2017/09/07. doi: 10.1186/s12883-017-0949-4. PubMed PMID: 28874119; PMCID: PMC5584325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, Feng T, Lee M, Asinovski N, Frohlich I, Imboywa S, Von Korff A, Okada Y, Patsopoulos NA, Davis S, McCabe C, Paik HI, Srivastava GP, Raychaudhuri S, Hafler DA, Koller D, Regev A, Hacohen N, Mathis D, Benoist C, Stranger BE, De Jager PL. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344(6183):519–23. Epub 2014/05/03. doi: 10.1126/science.1249547. PubMed PMID: 24786080; PMCID: PMC4910825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67(5):1613–7. Epub 2016/12/23. doi: 10.1099/ijsem.0.001755. PubMed PMID: 28005526; PMCID: PMC5563544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. Epub 2011/06/28. doi: 10.1186/gb-2011-12-6-r60. PubMed PMID: 21702898; PMCID: PMC3218848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tickle T, Huttenhower CM. Multivariate statistical framework that finds associations between clinical metadata and microbial community abundance or function. 2014. [Google Scholar]
- 33.Cox LM, Sohn J, Tyrrell KL, Citron DM, Lawson PA, Patel NB, Iizumi T, Perez-Perez GI, Goldstein EJC, Blaser MJ. Description of two novel members of the family Erysipelotrichaceae: Ileibacterium valens gen. nov., sp. nov. and Dubosiella newyorkensis, gen. nov., sp. nov., from the murine intestine, and emendation to the description of Faecalibaculum rodentium. Int J Syst Evol Microbiol. 2017;67(5):1247–54. Epub 2017/01/20. doi: 10.1099/ijsem.0.001793. PubMed PMID: 28100298; PMCID: PMC5817276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, Rezende RM, Moreira TG, Tankou SK, Cox LM, Wu M, Song A, Dhang FH, Wei Z, Costamagna G, Weiner HL. Oral Administration of miR-30d from Feces of MS Patients Suppresses MS-like Symptoms in Mice by Expanding Akkermansia muciniphila. Cell Host Microbe. 2019;26(6):779–94 e8. Epub 2019/12/01. doi: 10.1016/j.chom.2019.10.008. PubMed PMID: 31784260; PMCID: PMC6948921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S, Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nunez G, Camp JG, Hattori M, Umesaki Y, Honda K. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163(2):367–80. Epub 2015/09/29. doi: 10.1016/j.cell.2015.08.058. PubMed PMID: 26411289; PMCID: PMC4765954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y, Liu C, Molitoris DR, Tomzynski TJ, Lawson PA, Collins MD, Finegold SM. Clostridium bolteae sp. nov., isolated from human sources. Syst Appl Microbiol. 2003;26(1):84–9. Epub 2003/05/16. doi: 10.1078/072320203322337353. PubMed PMID: 12747414. [DOI] [PubMed] [Google Scholar]
- 37.Pandit L, Cox LM, Malli C, D'Cunha A, Rooney T, Lokhande H, Willocq V, Saxena S, Chitnis T. Clostridium bolteae is elevated in neuromyelitis optica spectrum disorder in India and shares sequence similarity with AQP4. Neurol Neuroimmunol Neuroinflamm. 2021;8(1):e907. Epub 2020/11/06. doi: 10.1212/NXI.0000000000000907. PubMed PMID: 33148687; PMCID: PMC7643530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294(1):1–8. Epub 2009/02/19. doi: 10.1111/j.1574-6968.2009.01514.x. PubMed PMID: 19222573. [DOI] [PubMed] [Google Scholar]
- 39.Bakshi R, Thompson AJ, Rocca MA, Pelletier D, Dousset V, Barkhof F, Inglese M, Guttmann CR, Horsfield MA, Filippi M. MRI in multiple sclerosis: current status and future prospects. Lancet Neurol. 2008;7(7):615–25. Epub 2008/06/21. doi: 10.1016/S1474-4422(08)70137-6. PubMed PMID: 18565455; PMCID: PMC2586926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holland CM, Charil A, Csapo I, Liptak Z, Ichise M, Khoury SJ, Bakshi R, Weiner HL, Guttmann CR. The relationship between normal cerebral perfusion patterns and white matter lesion distribution in 1,249 patients with multiple sclerosis. J Neuroimaging. 2012;22(2):129–36. Epub 2011/03/31. doi: 10.1111/j.1552-6569.2011.00585.x. PubMed PMID: 21447022. [DOI] [PubMed] [Google Scholar]
- 41.Pirko I, Lucchinetti CF, Sriram S, Bakshi R. Gray matter involvement in multiple sclerosis. Neurology. 2007;68(9):634–42. Epub 2007/02/28. doi: 10.1212/01.wnl.0000250267.85698.7a. PubMed PMID: 17325269. [DOI] [PubMed] [Google Scholar]
- 42.Andravizou A, Dardiotis E, Artemiadis A, Sokratous M, Siokas V, Tsouris Z, Aloizou AM, Nikolaidis I, Bakirtzis C, Tsivgoulis G, Deretzi G, Grigoriadis N, Bogdanos DP, Hadjigeorgiou GM. Brain atrophy in multiple sclerosis: mechanisms, clinical relevance and treatment options. Auto Immun Highlights. 2019;10(1):7. Epub 2020/04/08. doi: 10.1186/s13317-019-0117-5. PubMed PMID: 32257063; PMCID: PMC7065319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller DH, Lublin FD, Sormani MP, Kappos L, Yaldizli O, Freedman MS, Cree BAC, Weiner HL, Lubetzki C, Hartung HP, Montalban X, Uitdehaag BMJ, MacManus DG, Yousry TA, Gandini Wheeler-Kingshott CAM, Li B, Putzki N, Merschhemke M, Haring DA, Wolinsky JS. Brain atrophy and disability worsening in primary progressive multiple sclerosis: insights from the INFORMS study. Ann Clin Transl Neurol. 2018;5(3):346–56. Epub 2018/03/22. doi: 10.1002/acn3.534. PubMed PMID: 29560379; PMCID: PMC5846448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohle L, Mattei D, Heimesaat MM, Bereswill S, Fischer A, Alutis M, French T, Hambardzumyan D, Matzinger P, Dunay IR, Wolf SA. Ly6C(hi) Monocytes Provide a Link between Antibiotic-Induced Changes in Gut Microbiota and Adult Hippocampal Neurogenesis. Cell Rep. 2016;15(9):1945–56. Epub 2016/05/24. doi: 10.1016/j.celrep.2016.04.074. PubMed PMID: 27210745. [DOI] [PubMed] [Google Scholar]
- 45.Hoban AE, Stilling RM, Ryan FJ, Shanahan F, Dinan TG, Claesson MJ, Clarke G, Cryan JF. Regulation of prefrontal cortex myelination by the microbiota. Transl Psychiatry. 2016;6(4):e774. Epub 2016/04/06. doi: 10.1038/tp.2016.42. PubMed PMID: 27045844; PMCID: PMC4872400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao CC, Patel B, Yan R, Blain M, Alvarez JI, Kebir H, Anandasabapathy N, Izquierdo G, Jung S, Obholzer N, Pochet N, Clish CB, Prinz M, Prat A, Antel J, Quintana FJ. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22(6):586–97. Epub 2016/05/10. doi: 10.1038/nm.4106. PubMed PMID: 27158906; PMCID: PMC4899206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glanz BI, Healy BC, Rintell DJ, Jaffin SK, Bakshi R, Weiner HL. The association between cognitive impairment and quality of life in patients with early multiple sclerosis. J Neurol Sci. 2010;290(1-2):75–9. Epub 2009/12/01. doi: 10.1016/j.jns.2009.11.004. PubMed PMID: 19944429. [DOI] [PubMed] [Google Scholar]
- 48.Glanz BI, Degano IR, Rintell DJ, Chitnis T, Weiner HL, Healy BC. Work productivity in relapsing multiple sclerosis: associations with disability, depression, fatigue, anxiety, cognition, and health-related quality of life. Value Health. 2012;15(8): 1029–35. Epub 2012/12/19. doi: 10.1016/j.jval.2012.07.010. PubMed PMID: 23244804. [DOI] [PubMed] [Google Scholar]
- 49.Seo B, Jeon K, Baek I, Lee YM, Baek K, Ko G. Faecalibacillus intestinalis gen. nov., sp. nov. and Faecalibacillus faecis sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2019;69(7):2120–8. Epub 2019/05/18. doi: 10.1099/ijsem.0.003443. PubMed PMID: 31099736. [DOI] [PubMed] [Google Scholar]
- 50.Shkoporov AN, Chaplin AV, Shcherbakova VA, Suzina NE, Kafarskaia LI, Bozhenko VK, Efimov BA. Ruthenibacterium lactatiformans gen. nov., sp. nov., an anaerobic, lactate-producing member of the family Ruminococcaceae isolated from human faeces. Int J Syst Evol Microbiol. 2016;66(8):3041–9. Epub 2016/05/08. doi: 10.1099/ijsem.0.001143. PubMed PMID: 27154556. [DOI] [PubMed] [Google Scholar]
- 51.Amorini AM, Nociti V, Petzold A, Gasperini C, Quartuccio E, Lazzarino G, Di Pietro V, Belli A, Signoretti S, Vagnozzi R, Lazzarino G, Tavazzi B. Serum lactate as a novel potential biomarker in multiple sclerosis. Biochim Biophys Acta. 2014;1842(7):1137–43. Epub 2014/04/15. doi: 10.1016/j.bbadis.2014.04.005. PubMed PMID: 24726946. [DOI] [PubMed] [Google Scholar]
- 52.Esmael A, Talaat M, Egila H, Eltoukhy K. Mitochondrial dysfunction and serum lactate as a biomarker for the progression and disability in MS and its correlation with the radiological findings. Neurol Res. 2021:1–9. Epub 2021/03/05. doi: 10.1080/01616412.2021.1893567. PubMed PMID: 33657991. [DOI] [PubMed] [Google Scholar]
- 53.Albanese M, Zagaglia S, Landi D, Boffa L, Nicoletti CG, Marciani MG, Mandolesi G, Marfia GA, Buttari F, Mori F, Centonze D. Cerebrospinal fluid lactate is associated with multiple sclerosis disease progression. J Neuroinflammation. 2016;13:36. Epub 2016/02/13. doi: 10.1186/s12974-016-0502-1. PubMed PMID: 26863878; PMCID: PMC4750170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parsot C, Sansonetti PJ. The virulence plasmid of Shigellae: an archipelago of pathogenicity islands? Pathogenicity islands and other mobile virulence elements: American Society of Microbiology; 1999. p. 151–65. [Google Scholar]
- 55.Kaper JB, Mellies JL, Nataro JP. Pathogenicity islands and other mobile genetic elements of diarrheagenic Escherichia coli. Pathogenicity islands and other mobile virulence elements: American Society of Microbiology; 1999. p. 33–58. [Google Scholar]
- 56.Kim M, Galan C, Hill AA, Wu WJ, Fehlner-Peach H, Song HW, Schady D, Bettini ML, Simpson KW, Longman RS, Littman DR, Diehl GE. Critical Role for the Microbiota in CX(3)CR1(+) Intestinal Mononuclear Phagocyte Regulation of Intestinal T Cell Responses. Immunity. 2018;49(1):151–63.e5. Epub 2018/07/08. doi: 10.1016/j.immuni.2018.05.009. PubMed PMID: 29980437; PMCID: PMC6051886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahn S, Jin T-E, Chang D-H, Rhee M-S, Kim HJ, Lee SJ, Park D-S, Kim B-C. Agathobaculum butyriciproducens gen. nov. & sp. nov., a strict anaerobic, butyrate-producing gut bacterium isolated from human faeces and reclassification of Eubacterium desmolans as Agathobaculum desmolans comb. nov. International Journal of Systematic and Evolutionary Microbiology. 2016;66(9):3656–61. Epub 2016/06/24. doi: 10.1099/ijsem.0.001195. PubMed PMID: 27334534. [DOI] [PubMed] [Google Scholar]
- 58.Mirza A, Forbes JD, Zhu F, Bernstein CN, Van Domselaar G, Graham M, Waubant E, Tremlett H. The multiple sclerosis gut microbiota: A systematic review. Mult Scler Relat Disord. 2020;37:101427. Epub 2020/03/17. doi: 10.1016/j.msard.2019.101427. PubMed PMID: 32172998. [DOI] [PubMed] [Google Scholar]
- 59.Noto D, Miyake S. Gut dysbiosis and multiple sclerosis. Clin Immunol. 2020:108380. Epub 2020/03/15. doi: 10.1016/j.clim.2020.108380. PubMed PMID: 32169440. [DOI] [PubMed] [Google Scholar]
- 60.Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, Zhao J, Zhang H, Chen W. Blautia-a new functional genus with potential probiotic properties? Gut Microbes. 2021;13(1):1–21. Epub 2021/02/03. doi: 10.1080/19490976.2021.1875796. PubMed PMID: 33525961; PMCID: PMC7872077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benitez-Paez A, Gomez Del Pugar EM, Lopez-Almela I, Moya-Perez A, Codoner-Franch P, Sanz Y. Depletion of Blautia Species in the Microbiota of Obese Children Relates to Intestinal Inflammation and Metabolic Phenotype Worsening. mSystems. 2020;5(2). Epub 2020/03/27. doi: 10.1128/mSystems.00857-19. PubMed PMID: 32209719; PMCID: PMC7093825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taras D, Simmering R, Collins MD, Lawson PA, Blaut M. Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2002;52(Pt 2):423–8. Epub 2002/04/05. doi: 10.1099/00207713-52-2-423. PubMed PMID: 11931151. [DOI] [PubMed] [Google Scholar]
- 63.Ren C, Wang Y, Lin X, Song H, Zhou Q, Xu W, Shi K, Chen J, Song J, Chen F, Zhang S, Guan W. A Combination of Formic Acid and Monolaurin Attenuates Enterotoxigenic Escherichia coli Induced Intestinal Inflammation in Piglets by Inhibiting the NF-kappaB/MAPK Pathways with Modulation of Gut Microbiota. J Agric Food Chem. 2020;68(14):4155–65. Epub 2020/03/24. doi: 10.1021/acs.jafc.0c01414. PubMed PMID: 32202779. [DOI] [PubMed] [Google Scholar]
- 64.Luise D, Correa F, Bosi P, Trevisi P. A Review of the Effect of Formic Acid and Its Salts on the Gastrointestinal Microbiota and Performance of Pigs. Animals (Basel). 2020;10(5):887. doi: 10.3390/ani10050887. PubMed PMID: 32438743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volkova A, Ruggles KV. Predictive Metagenomic Analysis of Autoimmune Disease Identifies Robust Autoimmunity and Disease Specific Microbial Signatures. Frontiers in microbiology. 2021;12(418):621310. Epub 2021/03/23. doi: 10.3389/fmicb.2021.621310. PubMed PMID: 33746917; PMCID: PMC7969817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cani PD, de Vos WM. Next-Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Frontiers in microbiology. 2017;8(1765):1765. Epub 2017/10/12. doi: 10.3389/fmicb.2017.01765. PubMed PMID: 29018410; PMCID: PMC5614963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, Kleimeyer C, Moresi C, Harnik Y, Zur M, Zabari M, Brik RB, Kviatcovsky D, Zmora N, Cohen Y, Bar N, Levi I, Amar N, Mehlman T, Brandis A, Biton I, Kuperman Y, Tsoory M, Alfahel L, Harmelin A, Schwartz M, Israelson A, Arike L, Johansson MEV, Hansson GC, Gotkine M, Segal E, Elinav E. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019;572(7770):474–80. Epub 2019/07/23. doi: 10.1038/s41586-019-1443-5. PubMed PMID: 31330533. [DOI] [PubMed] [Google Scholar]
- 68.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KC, Aalvink S, Martinez LO, Dumas ME, Maiter D, Loumaye A, Hermans MP, Thissen JP, Belzer C, de Vos WM, Cani PD. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23(1):107–13. Epub 2016/11/29. doi: 10.1038/nm.4236. PubMed PMID: 27892954. [DOI] [PubMed] [Google Scholar]
- 69.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragon L, Jacquelot N, Qu B, Ferrere G, Clemenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–7. Epub 2017/11/04. doi: 10.1126/science.aan3706. PubMed PMID: 29097494. [DOI] [PubMed] [Google Scholar]
- 70.Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell. 2018;174(2):497. Epub 2018/07/17. doi: 10.1016/j.cell.2018.06.051. PubMed PMID: 30007420; PMCID: PMC6062008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhai R, Xue X, Zhang L, Yang X, Zhao L, Zhang C. Strain-Specific Anti-inflammatory Properties of Two Akkermansia muciniphila Strains on Chronic Colitis in Mice. Front Cell Infect Microbiol. 2019;9:239. Epub 2019/07/25. doi: 10.3389/fcimb.2019.00239. PubMed PMID: 31334133; PMCID: PMC6624636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duan J, Chung H, Troy E, Kasper DL. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe. 2010;7(2):140–50. Epub 2010/02/18. doi: 10.1016/j.chom.2010.01.005. PubMed PMID: 20159619; PMCID: PMC4048034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wo J, Zhang F, Li Z, Sun C, Zhang W, Sun G. The Role of Gamma-Delta T Cells in Diseases of the Central Nervous System. Frontiers in immunology. 2020;11:580304. Epub 2020/11/17. doi: 10.3389/fimmu.2020.580304. PubMed PMID: 33193380; PMCID: PMC7644879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colpitts SL, Kasper EJ, Keever A, Liljenberg C, Kirby T, Magori K, Kasper LH, Ochoa-Repáraz J. A bidirectional association between the gut microbiota and CNS disease in a biphasic murine model of multiple sclerosis. Gut Microbes. 2017;137(5):00-. PubMed PMID: 28708466; PMCID: PMC5730387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amezcua L, McCauley JL. Race and ethnicity on MS presentation and disease course. Mult Scler. 2020;26(5):561–7. Epub 2020/01/23. doi: 10.1177/1352458519887328. PubMed PMID: 31965878; PMCID: PMC7160002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harbo HF, Gold R, Tintore M. Sex and gender issues in multiple sclerosis. Ther Adv Neurol Disord. 2013;6(4):237–48. Epub 2013/07/17. doi: 10.1177/1756285613488434. PubMed PMID: 23858327; PMCID: PMC3707353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghosh TS, Das M, Jeffery IB, O'Toole PW. Adjusting for age improves identification of gut microbiome alterations in multiple diseases. Elife. 2020;9. Epub 2020/03/12. doi: 10.7554/eLife.50240. PubMed PMID: 32159510; PMCID: PMC7065848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108(38):16050–5. Epub 2011/08/31. doi: 10.1073/pnas.1102999108. PubMed PMID: 21876150; PMCID: PMC3179073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, McDonald D, Dietrich D, Ramadhar TR, Lekbua A, Mroue N, Liston C, Stewart EJ, Dubin MJ, Zengler K, Knight R, Gilbert JA, Clardy J, Lewis K. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol. 2019;4(3):396–403. Epub 2018/12/12. doi: 10.1038/s41564-018-0307-3. PubMed PMID: 30531975; PMCID: PMC6384127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vaughn C, Kavak K, Bushra A, Noyes E, Edwards K, Goodman A, Coyle P, Krupp L, Jubelt B, Gottesman M, Benedict R, Weinstock-Guttman B. Self-reported fatigue and lower limb problems predictive of conversion to secondary progressive multiple sclerosis in an aging sample of patients (S10.004). Neurology. 2017;88(16 Supplement):S10.004. [Google Scholar]
- 81.Sand IK, Zhu Y, Ntranos A, Clemente JC, Cekanaviciute E, Brandstadter R, Crabtree-Hartman E, Singh S, Bencosme Y, Debelius J. Disease-modifying therapies alter gut microbial composition in MS. Neurology-Neuroimmunology Neuroinflammation. 2019;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The microbiota 16S rRNA sequence data has been submitted to the National Center for Biotechnology Information (NCBI) Short Read Archives (SRA) under Bioproject accession number PRJNA721421, which is publicly available. The deidentified metadata, including diagnosis, age, treatment, race, ethnicity, BMI, sex, and EDSS have been included along with the sequencing data. EDSS is recorded under the column heading host_phenotype.