Abstract
Environmental factors such as nutrition, stress, and toxicants can influence epigenetic programming and phenotypes of a wide variety of species from plants to humans. The current study was designed to investigate the impacts of hatchery spawning and rearing on steelhead trout (Oncorhynchus mykiss) vs the wild fish on a molecular level. Additionally, epigenetic differences between feeding practices that allow slow growth (2 years) and fast growth (1 year) hatchery trout were investigated. The sperm and red blood cells (RBC) from adult male slow growth/maturation hatchery steelhead, fast growth/maturation hatchery steelhead, and wild (natural-origin) steelhead were collected for DNA preparation to investigate potential alterations in differential DNA methylation regions (DMRs) and genetic mutations, involving copy number variations (CNVs). The sperm and RBC DNA both had a large number of DMRs when comparing the hatchery vs wild steelhead trout populations. The DMRs were cell type specific with negligible overlap. Slow growth/maturation compared to fast growth/maturation steelhead also had a larger number of DMRs in the RBC samples. A number of the DMRs had associated genes that were correlated to various biological processes and pathologies. Observations demonstrate a major epigenetic programming difference between the hatchery and wild natural-origin fish populations, but negligible genetic differences. Therefore, hatchery conditions and growth/maturation rate can alter the epigenetic developmental programming of the steelhead trout. Interestingly, epigenetic alterations in the sperm allow for potential epigenetic transgenerational inheritance of phenotypic variation to future generations. The impacts of hatchery exposures are not only important to consider on the fish exposed, but also on future generations and evolutionary trajectory of fish in the river populations.
Keywords: steelhead, hatchery, rearing conditions, epigenetic inheritance, fish, genomics, phenotypic variation, sperm, generational
Introduction
Epigenetics is a complementary mechanism with genetics for the molecular control of biology [1, 2]. A range of environmental factors including toxicants, stress, and nutrition can developmentally alter a variety of phenotypes in species from plants to humans through epigenetics [3]. Epigenetics is defined as “molecular factors and processes around DNA that regulate genome activity independent of DNA sequence, and are stable mitotically” [3, 4]. The ability of environmental factors to alter epigenetic programming, while not changing DNA sequence, provides a molecular mechanism for the environment to directly impact phenotypic variation and evolution [1, 3]. Although direct exposures of somatic cells to environmental factors can influence the individual exposed, epigenetic change in the germline (sperm or egg) can be transmitted to the next generation, and is termed “epigenetic inheritance” [2, 3]. In the event, the germline epigenetic alterations are transmitted to subsequent generations, in the absence of continued direct exposure, this is defined as “epigenetic transgenerational inheritance” [3, 5]. Therefore, environmental factors have the ability to developmentally impact epigenetic programming to influence the phenotype of the individual exposed. In addition, if the germline (e.g. sperm) is affected the potential for generational effects on phenotypic variation develop. The current study is designed to investigate the molecular effects of hatchery rearing on steelhead trout (Oncorhynchus mykiss).
Hatcheries involve both aquaculture facilities for food supply, and operations for sustaining sufficient numbers of endangered fish such as the salmon populations in the Pacific Northwest, USA [6, 7]. Research shows that hatchery-reared fish differ from wild fish both in phenotype and in having reduced reproductive success in the wild [8–11]. In steelhead trout (Oncorhynchus mykiss), it has been shown that the offspring of fish experiencing even a single generation of hatchery rearing show marked phenotypic changes [8–22] (Table 1). This has also been observed in a variety of different hatchery salmon and trout species [12, 23–45] (Table 1). Pathologies observed include decreased fitness of hatchery-reared fish and offspring in the wild, and changes in age at spawning, morphology, growth rate, brain morphology, anti-predator behavior, and migration [8–45].
Table 1:
Summary hatchery impacts on fish phenotypes (literature review data summary)
| Species | Phenotypic alteration | References |
|---|---|---|
| Steelhead | Decreased fitness of hatchery reared | [8–10, 12–16, 21, 22] |
| Brook Trout | Individuals or their offspring in the wild | [23] |
| Atlantic Salmon | [12, 24–26] | |
| Chinook Salmon | [12, 27] | |
| Coho Salmon | [12, 28, 45] | |
| Rainbow Trout | [29] | |
| Steelhead | Change in age at spawning | [19] |
| Sockeye Salmon | [30] | |
| Steelhead | Change in lateral line morphology and | [11, 18] |
| Coho Salmon | Otoliths | [31] |
| Steelhead | Increased growth rate and size | [15] |
| Steelhead | Change in brain morphology | [20] |
| Chinook Salmon | [32] | |
| Coho Salmon | [33] | |
| Steelhead | Gene expression differences | [17] |
| Atlantic Salmon | Change in body shape | [34] |
| Rainbow Trout | [35] | |
| Atlantic salmon | Reduced antipredator behavior | [36, 37] |
| Atlantic salmon | Increased growth rate and size | [26, 38–40] |
| Atlantic salmon | Delayed hatch | [41] |
| Atlantic salmon | Altered migration | [26, 40, 42] |
| Atlantic salmon | Change in gut microbiome | [43] |
| Chinook Salmon | Reduced egg size | [44] |
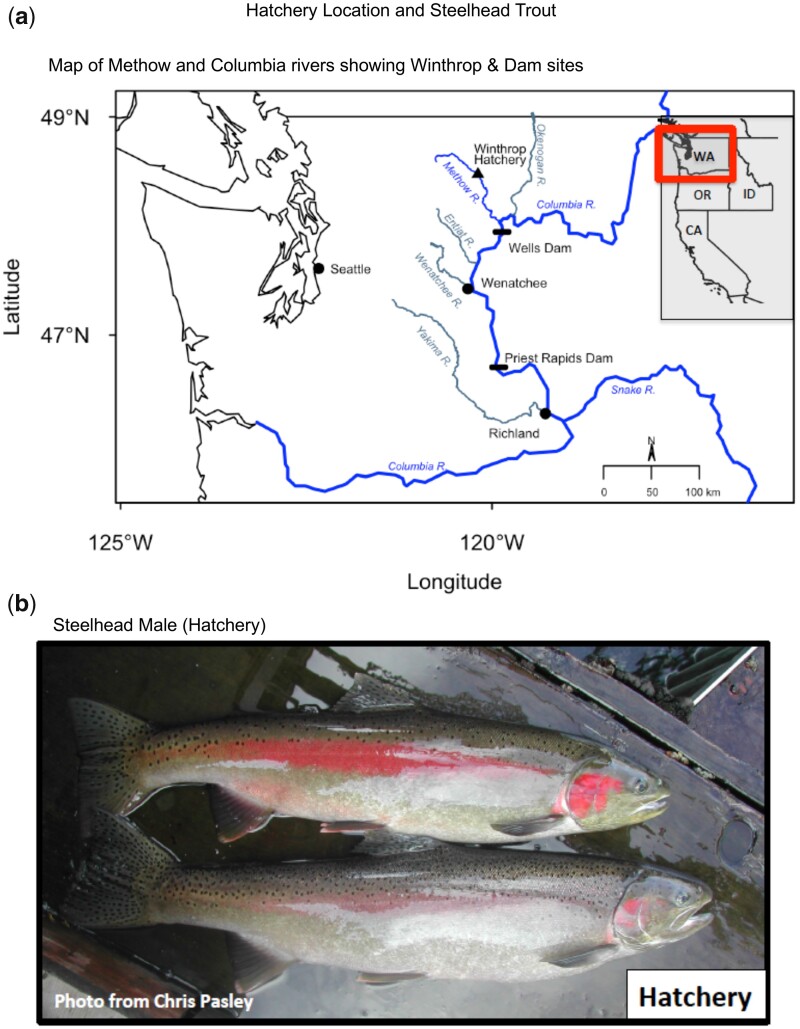
A model anadromous migratory salmonid fish species selected for the current study is the steelhead trout. A brood-stock hatchery operation (Winthrop National Fish Hatchery, WNFH, US Fish and Wildlife Service) on the Methow River in Washington State, USA was selected to compare wild natural-origin populations and hatchery populations of steelhead trout (Fig. 1). This hatchery used hatchery-reared steelhead crossed with natural-origin fish from the same river as broodstock to produce offspring that were raised in hatcheries until the time of release as smolts, at which point they can migrate downstream to the ocean. Some steelhead were placed on a high level (plane) of nutrition and reached smolt stage within 1 year (fast growth/maturation), at which time they were released. Others were placed on a lower level (plane) of nutrition, and took 2 years to reach smolt stage (slow growth/maturation) prior to release. Natural-origin steelhead trout generally take 2 years to reach smolt stage in this river system. After returning from the ocean, fast growth/maturation (S1), slow growth/maturation (S2), and natural-origin (N) fish were collected and sampled.
Figure 1:
Hatchery locations, rivers, adjacent dams, and steelhead trout. (a) Map of Methow River and Columbia River confluence, and Winthrop hatchery. (b) Steelhead males (hatchery-origin)
The Methow River summer-run steelhead is part of an upper Columbia River evolutionary significant unit currently listed as threatened under the Endangered Species Act. For the past 60 years, the large number of hatchery fish released that breed with wild fish has had a significant influence on the wild steelhead population. Therefore, there is no distinct wild Methow River steelhead population without a molecular influence from hatchery impacts. The term natural-origin is used to refer to fish spawned in the wild and live their entire lives in the wild. Hatchery fish refers to fish generated through artificial rebreeding and crosses in the juvenile stages in the hatchery before being released into the wild with an adipose fin clip. In the current study, the wild (natural-origin) and hatchery (slow 2 years and fast 1-year juvenile growth/maturation) are compared. Natural and hatchery-origin adult steelhead returning in the Methow river in 2013 and 2014 and captured in winter and spring of 2014 and 2015 were used in this study (Supplementary Table S1).
The molecular effects of hatchery rearing and growth rate on somatic cells (red blood cells, RBC) and germline cells (sperm) were investigated. Genome-wide molecular analyses of differential DNA methylation regions (DMR) and genetic mutations (copy number variation, CNV) were performed. The hypothesis tested was that the hatchery spawning and rearing conditions alter the epigenetic programming of the steelhead somatic cells (RBC) and germline (sperm), such that later in life following return the adult steelhead will have altered epigenetics, with potential generational impacts through the sperm. This will result in an altered phenotypic variation, fitness, and evolutionary trajectory of the wild population.
Results
The steelhead trout was selected as a model salmonid fish species for the current study due to the migratory nature of the fish and availability of the trout genome sequence to facilitate molecular studies. The adult phenotypes and color after migration and return from the Pacific Ocean are generally similar between the hatchery and wild steelhead populations, as shown for the hatchery fish in Fig. 1. The steelhead was collected on the Methow River near Winthrop Washington, USA, during spring following the previous summer migration (Fig. 1a). A steelhead hatchery is located in Winthrop Washington that uses a brood-stock population of adult river caught fish each year to spawn and rear the steelhead. An alternate fish hatchery also used was at Wells Dam below the mouth of the Methow River on the Columbia River (Fig. 1 and Supplementary Table S1). During collection, both hatchery (determined by a fin clip) and wild (natural-origin) steelhead were obtained. Since each cell type in the body has a unique epigenome (e.g. DNA methylation patterns), purified cell types are required for epigenetic analysis to allow unambiguous data interpretation. For the current study, both sperm and purified RBC (that contain nuclei in fish) samples were collected from the males. The DNA was isolated from the cell types obtained from adult male hatchery and wild steelhead (Fig. 1b). The isolated DNA from the RBC and sperm samples were then processed for the molecular studies.
The DMRs between the hatchery and wild male steelhead populations were identified in both the sperm and RBC separately. For each treatment group, equal amounts of DNA from three to five different males were pooled and three pools created for n = 9–15 animals per group. The treatment groups comprised fish from the Winthrop National Hatchery-fast growth/maturation (WNFH-S1; i.e. S1), Winthrop National Hatchery-slow growth/maturation (WNFH-S2; i.e. S2), Winthrop National Hatchery standard hatchery (WNFH-H; i.e. H), and non-hatchery reared natural-origin wild fish (N). Information on the individual fish collections, characteristics, and labeling are presented in Supplementary Table S1. The information includes sample group, identification number, floy tag number, sex (males), date collection, natural or hatchery type, hatchery location, approximate age at collection, rearing (growth) strategy, and sample pool for each individual (Supplementary Table S1). Individual males used for the study had no difference in the length or age between the groups (Supplementary Table S1). Equal amounts of DNA from each individual upon collection was used in the hatchery or wild population pools, and then the DNA was fragmented through sonication and the methylated DNA immunoprecipitated (MeDIP) with an antibody to methylcytosine, as described [46] in the Methods. The MeDIP DNA was used to prepare libraries for next-generation sequencing (Seq) for an MeDIP-Seq analysis to identify the DMRs, see Methods. This genome-wide epigenetic analysis assesses >90% of the genome compared to other procedures such as reduced representation bisulfite sequencing (RRBS) that assesses <10% of the genome [47]. The sperm and RBC DNA samples were analyzed separately with MeDIP-Seq.
A comparison of the standard hatchery WNFH-H (H), fast 1-year growth/maturation WNFH-S1 (S1), slow 2-year growth/maturation WNFH-S2 (S2), and wild (natural-origin) (N) populations involved 2 years of collection with N1 in the first year collection for standard hatchery H vs N1 and a distinct collection of N2 in the second year collection for the S1 vs N2 and the S2 vs N2 comparisons. Therefore, these analyses were done separately. Summary of the individual fish and characteristics is presented in Supplementary Table S1. The MeDIP-Seq procedure was used to identify the DMRs between the group comparisons (Table 2). The analyses at several different statistically significant P values are shown for single sites (1000 bp window), and for multiple adjacent window sites (≥2 1000 bp windows) with each being statistically significant with edgeR and false discovery rate (FDR), as described in the Methods. As the statistical threshold decreases (P-value) the number of identified DMRs decreases, as expected, and the edgeR P < 1e−05 significance level was selected for subsequent use and data presentation. This generally correlated to an FDR of P < 0.05. Although the data analysis focused on the most stringently selected DMRs, the other DMRs at a lower statistical threshold are anticipated to also be important, but more variable between individuals. The DMR numbers for the sperm and RBC are presented in Table 2. For DMRs with multiple adjacent windows, the most predominant number of adjacent sites is 2 (1000 bp each) with the highest number of adjacent sites being 8. Therefore, the more stringently identified DMRs (all sites at P < 1e−05) provide a reasonable set of DMRs (i.e. signature) to be used for further analysis. The lists of DMRs and genomic features comparing hatchery vs wild steelhead for the sperm are presented in Supplementary Table S2, and for the RBC in Supplementary Table S3.
Table 2:
DMR number, variation, and statistics. (a) Sperm wild natural (N1) vs hatchery (H) DMR number and statistics. (b) Sperm wild natural (N2) vs hatchery S1 DMR number and statistics. (c) Sperm wild natural (N2) vs hatchery S2 DMR number and statistics. (d) Sperm hatchery S1 vs S2 number and statistics. (e) RBC wild natural (N1) vs hatchery (H) DMR number and statistics. (f) RBC wild natural (N2) vs hatchery S1 DMR number and statistics. (g) RBC wild natural (N2) vs Hatchery S2 number and statistics. (h) RBC hatchery S1 vs S2 number and statistics. The All Sites represent all DMRs with at least one 100 bp site and multiple windows represent ≥2100 bp adjacent sites. The number of single and multiple sites for DMRs at P < 1e−05 are presented. The hatchery S1 is the 1-year fast growth/maturation to smolt, and the S2 is the 2-year slow growth/maturation to smolt origin of fish
| (a) Sperm wild (N1) vs Hatchery DMRs | |||||
|---|---|---|---|---|---|
| P-value | All sites | Multiple windows | |||
| 0.001 | 4352 | 217 | |||
| 1e−04 | 1328 | 83 | |||
| 1e−05 | 577 | 34 | |||
| 1e−06 | 296 | 14 | |||
| 1e−07 | 167 | 8 | |||
| Number of significant sites | 1 | 2 | 3 | ||
| Number of DMR | 543 | 32 | 2 | ||
| (b) Sperm Wild (N2) vs Hatchery S1 DMRs | |||||
|---|---|---|---|---|---|
| P-value | All sites | Multiple windows | |||
| 0.001 | 7332 | 323 | |||
| 1e−04 | 1689 | 71 | |||
| 1e−05 | 454 | 24 | |||
| 1e−06 | 149 | 13 | |||
| 1e−07 | 64 | 6 | |||
| Number of significant sites | 1 | 2 | ≥4 | ||
| Number of DMR | 430 | 21 | 3 | ||
| (c) Sperm Wild (N2) vs Hatchery S2 DMRs | |||||
|---|---|---|---|---|---|
| P-value | All sites | Multiple windows | |||
| 0.001 | 6253 | 283 | |||
| 1e−04 | 1811 | 109 | |||
| 1e−05 | 693 | 56 | |||
| 1e−06 | 313 | 35 | |||
| 1e−07 | 167 | 22 | |||
| Number of significant sites | 1 | 2 | ≥3 | ||
| Number of DMR | 637 | 46 | 10 | ||
| (d) Sperm Hatchery S1 vs S2 DMRs | |||||
|---|---|---|---|---|---|
| P-value | All sites | Multiple windows | |||
| 0.001 | 2560 | 56 | |||
| 1e−04 | 573 | 26 | |||
| 1e−05 | 194 | 11 | |||
| 1e−06 | 102 | 4 | |||
| 1e−07 | 63 | 3 | |||
| Number of significant sites | 1 | 2 | 5 | ||
| Number of DMR | 183 | 10 | 1 | ||
| (e) RBC Wild (N1) vs Hatchery DMRs | |||||
|---|---|---|---|---|---|
| P-value | All sites | Multiple windows | |||
| 0.001 | 6795 | 609 | |||
| 1e−04 | 1754 | 113 | |||
| 1e−05 | 509 | 27 | |||
| 1e−06 | 182 | 8 | |||
| 1e−07 | 79 | 3 | |||
| Number of significant sites | 1 | 2 | 3 | ||
| Number of DMR | 482 | 22 | 5 | ||
| (f) RBC Wild (N2) vs Hatchery S1 DMRs | |||||
|---|---|---|---|---|---|
| P-value | All sites | Multiple windows | |||
| 0.001 | 5596 | 206 | |||
| 1e−04 | 1268 | 47 | |||
| 1e−05 | 389 | 14 | |||
| 1e−06 | 175 | 6 | |||
| 1e−07 | 73 | 2 | |||
| Number of significant sites | 1 | 2 | 4 | ||
| Number of DMR | 375 | 12 | 2 | ||
| (g) RBC Wild (N2) vs Hatchery S2 DMRs | |||||
|---|---|---|---|---|---|
| P-value | All sites | Multiple windows | |||
| 0.001 | 4978 | 129 | |||
| 1e−04 | 1191 | 46 | |||
| 1e−05 | 398 | 26 | |||
| 1e−06 | 170 | 17 | |||
| 1e−07 | 77 | 9 | |||
| Number of significant sites | 1 | 2 | ≥3 | ||
| Number of DMR | 372 | 23 | 3 | ||
| (h) RBC Hatchery S1 vs S2 DMRs | |||||
|---|---|---|---|---|---|
| P-value | All sites | Multiple windows | |||
| 0.001 | 11611 | 665 | |||
| 1e−04 | 3556 | 188 | |||
| 1e−05 | 1097 | 61 | |||
| 1e−06 | 438 | 18 | |||
| 1e−07 | 193 | 9 | |||
| Number of significant sites | 1 | 2 | ≥3 | ||
| Number of DMR | 1036 | 54 | 7 | ||
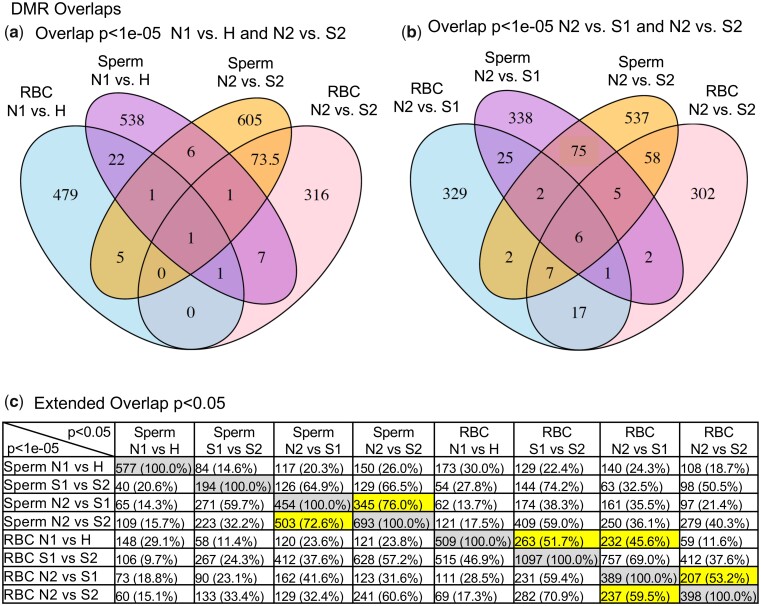
A major hatchery condition that can be altered is the nutrition and amount of food intake that can promote a fast growth 1-year juvenile maturation or a more normal slow growth 2-year juvenile maturation. A group of WNFH fast (S1) and slow (S2) growth/maturation steelhead were compared with the wild natural-origin (N) groups (i.e. N1 and N2). Both the N2 vs S1 or S2 comparisons are presented in Table 2 for the identification of DMRs. Using a P < 1e−05 statistical threshold, similar numbers of DMRs were obtained for the wild natural-origin (N2), hatchery-S1, or hatchery-S2 comparisons. An overlap of these DMR sets is shown in Fig. 2b for sperm and Fig. 3b for RBC. A comparison of the hatchery S1 vs hatchery S2 is also provided with a higher number of DMRs for RBC and lower number for sperm (Table 2). The Venn diagram overlaps of all the comparisons (Figs 2b and 3b) suggests each group comparison was primarily distinct with the largest overlaps between the N2 vs S1 and N2 vs S2, as well as the S1 vs S2 and N1 vs H. Therefore, at P < 1e−05, the DMRs are primarily distinct for each comparison.
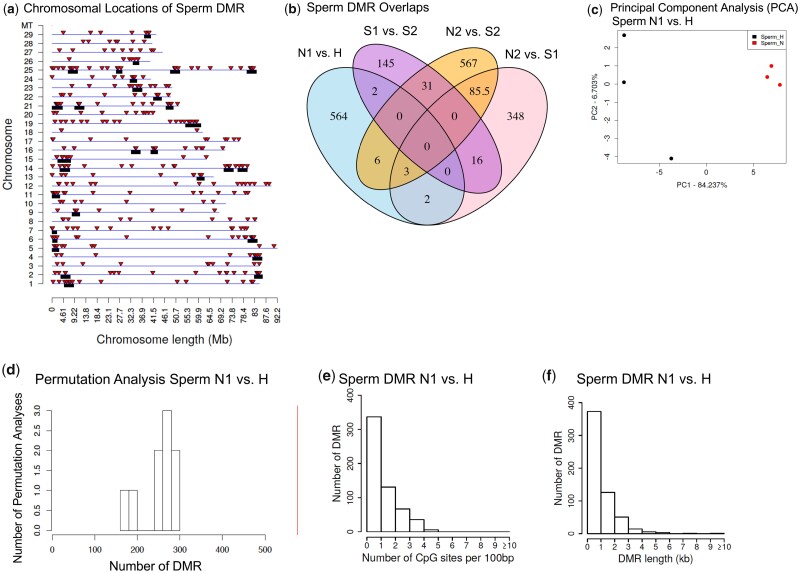
Figure 2:
sperm DMR N1 vs H chromosomal locations and analysis. (a) DMR chromosomal locations on the individual chromosomes vs size of chromosomes. All DMRs at a P-value threshold of P < 1e−05 are shown with red arrowheads and clusters DMRs with black boxes. In addition, 212 DMRs were located on the unplaced concatenated scaffolds not shown. (b) Sperm comparisons DMR overlaps, P < 1e−05. (c) Principal component analysis (PCA) of sperm DMRs N1 vs H. (d) Permutation analysis of sperm DMRs N1 vs H. (e) The number of sperm DMRs at different CpG densities. All DMRs at a P-value threshold of 1e−05 are shown. (f) The DMR lengths (kb) for sperm DMRs. All DMRs at a P-value threshold of 1e−05 are shown
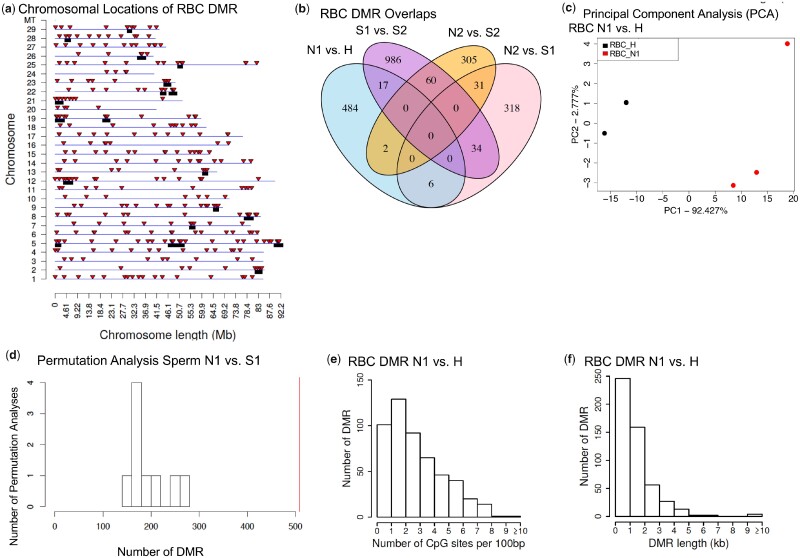
Figure 3:
RBC DMR N1 vs H chromosomal locations and analysis. (a) DMR chromosomal locations on the individual chromosomes vs size of chromosomes. All DMRs at a P-value threshold of P < 1e−05 are shown with red arrowheads and DMR clusters with black boxes. In addition, 175 DMRs were located on the unplaced concatenated scaffolds not shown. (b) RBC comparisons DMR overlaps, P < 1e−05. (c) Principal component analysis (PCA) of RBC DMRs N1 vs H. (d) Permutation analysis of RBC DMRs N1 vs H. (e) The number of RBC DMRs at different CpG densities. All DMRs at a P-value threshold of 1e−05 are shown. (f) The DMR lengths (kb) for RBC DMRs. All DMRs at a P-value threshold of 1e−05 are shown
The chromosomal locations of the DMRs for the sperm are presented in Fig. 2a and for the RBC in Fig. 3a. The DMRs are present on all of the annotated steelhead trout genome chromosomes for both sperm and RBC DMRs. The red arrowheads indicate the locations of the DMRs. Previous studies have demonstrated that DMRs can cluster on the genome to give statistically over-represented groups of DMRs [48]. These clusters of DMRs are shown in Figs 2a and 3a as black boxes on the chromosomes, and may function as epigenetic control regions [49]. Therefore, the DMRs are present throughout the genome and not isolated to specific regions or chromosomes. The RefSeq version of the Omyk 1.0 steelhead trout genome was used and obtained from NCBI. The read alignment rate was generally 76–83% for the steelhead sequencing with ∼55–100 million reads per pool for comparison. The steelhead trout genome is now nearly fully assembled, but some of the sequence is still in unplaced contigs not localized to a specific chromosome. In Figs 2 and 3, chromosomes marked as #1–#29 and mitochondrial DNA (MT) all have DMRs mapped to chromosomes. The number of DMRs associated with unplaced contigs is listed in the figure legends. Therefore, the lack of a complete genome annotation provides an underestimate of the genome map findings, but is sufficient to allow interpretation of the majority of the data. The chromosomal locations of the wild (N2) vs hatchery S1 (i.e. 1-year fast growth/maturation) or S2 (i.e. 2-year slow growth/maturation), and the hatchery S1 vs S2 are shown in Fig. 4a–d and in Supplementary Fig. S1. The lists of DMRs are presented in Supplementary Tables S2–S9 with genome location, statistical edgeR P values, log-fold change [(+) indicating an increase in DNA methylation, and (−) indicating a decrease in DNA methylation] between the comparisons, length (kb), CpG density, and gene associations. Although a mixture of DNA methylation increases and decreases at various DMRs were observed for all analyses, the sperm generally had a higher number of increases in DNA methylation at DMR (Supplementary Tables S2–S12).
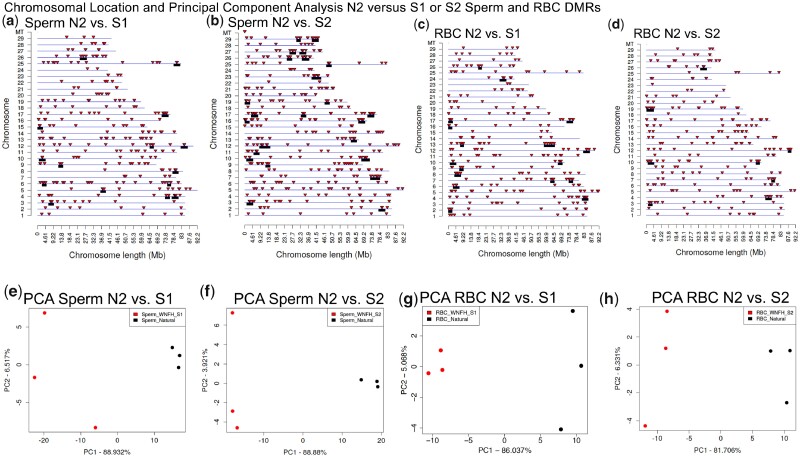
Figure 4:
DMR chromosomal locations on the individual chromosomes with red arrowheads indicating DMR and black boxes clusters of DMRs. All DMRs at a P-value threshold of P < 1e−05 are shown. (a) Sperm N2 vs S1 DMRs: 174 DMRs were located on the unplaced concatenated scaffold. (b) Sperm N2 vs S2 DMRs: 300 DMRs were located on the unplaced concatenated scaffold. (c) RBC N2 vs S1 DMRs: 110 DMRs were located on the unplaced concatenated scaffold. (d) RBC N2 vs S2 DMRs: 130 DMRs were located on the unplaced concatenated scaffold. Principal component analysis (PCA) using only DMR sites. (e) Sperm DMRs N2 vs S1 (f) Sperm DMRs N2 vs S2. (g) RBC DMRs N2 vs S1. (h) RBC DMRs N2 vs S2. Legend inserts with color N2 or S1 or S2
Analysis of potential similarities in the natural (N1) vs hatchery (H) DMRs (WNFH-H vs N1) at P < 1e−05 between the sperm (577 DMRs) and RBC (509 DMRs) demonstrated only 25 DMRs with overlap. The list of these overlapped DMRs and associated genes is presented (Supplementary Table S10). The most common associated gene sites for both sperm and RBC were uncharacterized LOC, but genes involved in signaling and epigenetics were present. Although the DMR was primarily cell specific at the threshold of P < 1e−05, an expanded sperm and RBC overlaps are presented in Fig. 5 to determine if greater overlap was present at a reduced statistical threshold. Interestingly, an extended overlap comparing the P < 1e−05 DMRs with DMRs at a lower stringency threshold P < 0.05 identified a greater degree of overlap among all the comparisons (Fig. 5c). Generally, a 15–30% overlap was observed unless the S1 and S2 populations were involved, which often had a 45–65% overlap. Therefore, using a lower statistical threshold allowed higher overlap of the DMRs between the comparisons. The altered S1 and S2 hatchery growth had an increased variation and allowed greater overlaps in DMRs. The N2 vs S1 and N2 vs S2 had an over 70% overlap in this comparison (Fig. 5c). The N1 vs H sperm had less overlap, but the RBC N1 vs H had ∼50% overlap with the S1 and S2 RBC. Therefore, the hatchery growth/maturation comparisons indicated DMR had good overlap for the RBC, but were more distinct for the sperm DMRs.
Figure 5:
DMR overlaps (a) DMR overlap between sperm and RBC DMR (N1 vs H and N2 vs S2). (b) Sperm and RBC overlap P < 1e−05 N2 vs S1 and N2 vs S2. (c) Extended DMR overlap P < 1e−05 vs P < 0.05. The horizontal line indicating the number of DMR overlap and percentage at P < 0.05. Gray highlight is anticipated 100% overlap and yellow highlight overlaps mentioned in the text
Previous studies have demonstrated that MeDIP-Seq identified DMRs generally have low-density CpG content, and exist in CpG deserts [50]. Analysis of the CpG density of the DMRs identified in the current study also demonstrated the DMRs to have a low CpG density (Figs 2e and 3e and Supplementary Fig. S2). The predominant CpG density in the data sets was one or two CpG per 100 bp for both the sperm and RBC. In the CpG deserts of a few thousand bases, the CpG can cluster to presumably act as regulatory sites [50]. The size of the DMRs was found to be predominantly one or two thousand bases Figs 2f and 3f and Supplementary Fig. S2. A few DMRs were between 5 and 10 kb, but the majority were smaller. Similar observations were made with the other comparison of slow and fast growth (N2 vs S1, N2 vs S2, S1 vs S2) for (P < 1e−05) DMRs, as presented in Supplementary Fig. S2. Therefore, the DMR genomic features identified were similar to DMRs previously characterized [3, 50, 51]. This is significant since the MeDIP-Seq procedure used in the current study optimally assessed lower density CpG deserts, while other procedures based on bisulfite (i.e. RRBS) assesses only a small percentage of the genome (<10%) and is optimal for high-density CpG sites.
A permutation analysis was performed to help verify the significance of the DMRs identified. This analysis involves randomly shuffling the samples between groups to obtain a null distribution for the number of DMRs expected due to random chance. Internal population epigenetic variation is generally associated with hypervariable or metastable DMRs, as previously described [52, 53]. There was generally a higher number of DMRs in the full analysis when compared to the sperm and RBC DMRs from the permutation comparisons of the hatchery and wild populations (Figs 2d and 3d and Supplementary Fig. S3). Two of the comparisons, sperm N2 vs S1 and sperm S1 vs S2, did not show a higher number of DMRs with the full analysis (Supplementary Fig. S3C and G). Although only a six-pool sub-comparison is not ideal for a thorough permutation analysis, the sperm with the S1 and S2 populations do show a higher degree of variation. The DMR analysis was also supported with a principal component analysis (PCA) of the DMRs, which demonstrated good separation of the various groups for all comparisons (Figs 2c, 3c, and 4e–h, and Supplementary Fig. S4. Since pools of animals were used, it is difficult to determine if the Fig. 1 parameters such as age, size, hatchery, or collection data create variability in the data, however, the PCA and permutation analyses indicate the DMR identified for the group comparisons is not a major factor. Within the specific groups, the variation shown for each group in the PCA may be associated with the variables shown in Supplementary Table S1.
The potential genetic variation between the hatchery and wild steelhead populations was also investigated. A type of genetic mutation previously shown to be one of the most common and stable is CNVs of repeat elements [51, 54, 55]. Previously, we have shown the ability to use CNV as a measure of genetic variation in wild populations [56]. Sequencing read depth data from the WNFH-H and N (i.e. N1) groups was used for a CNV analysis (CNV-Seq). The CNV for the hatchery and natural-origin wild populations for both the sperm and RBC are presented in Supplementary Fig. S5. CNV analysis did not show either hatchery or natural-origin samples to have more CNVs than the other. The sperm had higher numbers of CNVs than the RBC. None of the CNVs were common between the different animal pools. Therefore, in contrast to the epigenetic changes observed, negligible genetic changes between the hatchery and wild populations were observed. A limitation in this analysis is the read depth only allowed the identification of large size CNV as presented. The negligible effect of hatchery conditions on genetics has been previously reported [57, 58].
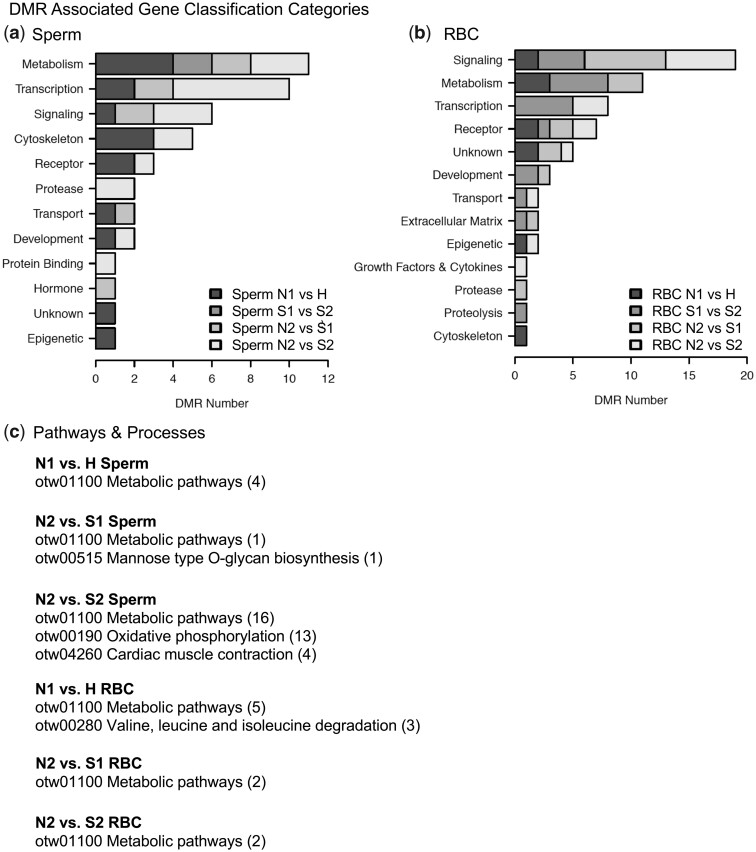
The potential gene associations with the DMRs were investigated using the steelhead trout genome annotations included in the RefSeq genome. The DMR associated genes are presented in Supplementary Tables S2–S9 for all the comparisons. The majority of DMRs did not have known gene associations. The cell-specific DMR associated gene functional categories are presented in Fig. 6a for sperm and Fig. 6b for RBC. The most predominant gene classification categories associated with all the group comparisons are transcription, metabolism, signaling, transport, development, protease, and epigenetics (Fig. 6). These gene categories are anticipated as they are the most abundant in most species and cell-type genomes. When the hatchery vs natural genes are put into a KEGG pathway analysis, the metabolic pathways (DCXR, ITPA, and PLCB3) and cytokine-cytokine receptor interactions (IL15, IL21) pathways were identified for the sperm, and proteasome (psme9a, psme1) for the RBC DMR associated genes (Fig. 6b). Limited numbers of DMR associated genes were present in the pathways, except for the S1 vs S2 comparison. Therefore, further analyses used a combination of all the comparison DMR associated genes.
Figure 6:
DMR associated gene categories. The gene classification is listed and correlated to the number of DMR associated genes within the specific classification category for (a) sperm DMR associated gene categories and (b) RBC DMR associated gene categories. (c) Pathways and processes with multiple genes
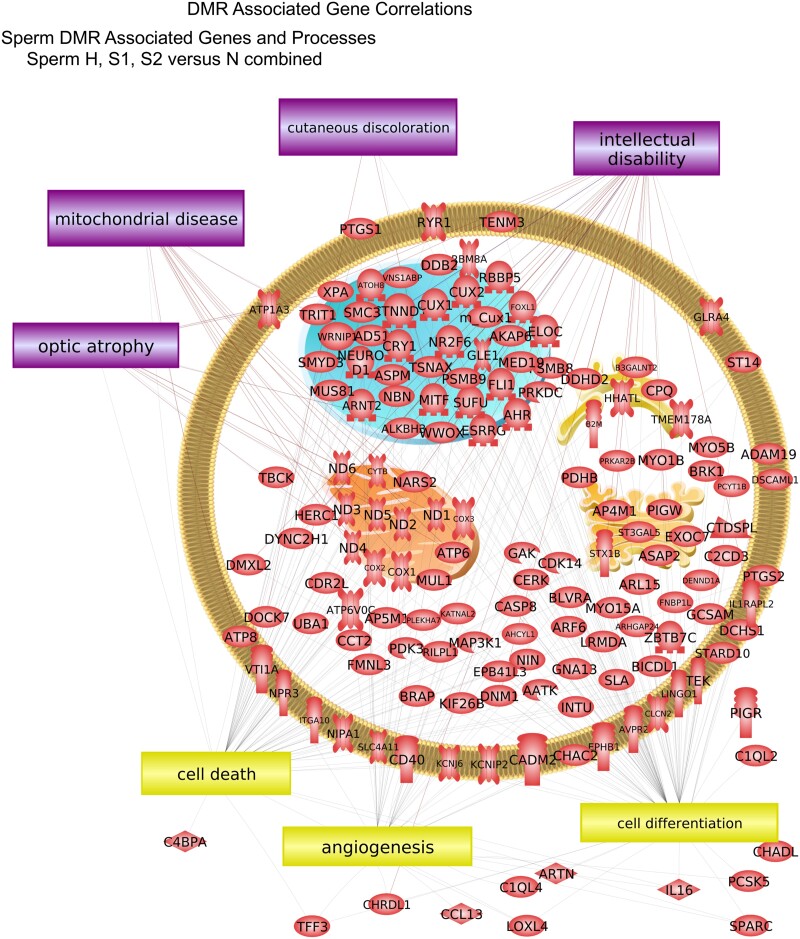
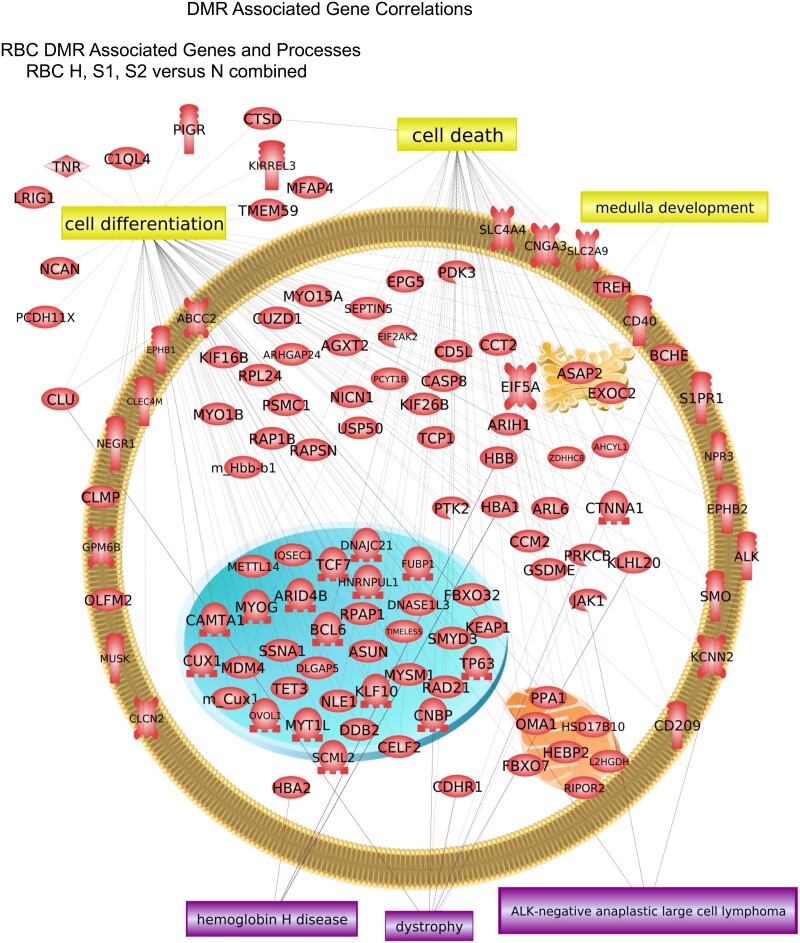
The DMR associated genes were analyzed for physiological processes and pathology correlations using the steelhead annotations provided by NCBI. The predominant physiological processes were cell death, angiogenesis, and cell differentiation for sperm DMR associated genes (Fig. 7). Cell differentiation, cell death, and medulla development are predominant for RBC DMR associated genes (Fig. 8). The predominant sperm DMR gene-associated pathologies were intellectual disability, cutaneous discoloration, mitochondrial disease, and optic atrophy in sperm DMR associated genes (Fig. 7). Pathologies of hemoglobin disease, dystrophy, and anaplastic large cell lymphoma in RBC DMR associated genes were predominant (Fig. 8). Additional cellular processes and pathway correlations with statistical significance are presented in Supplementary Table S11 for the sperm DMR associated genes and in Supplementary Table S12 for the RBC DMR associated genes. The processes or pathologies are listed with the total number of genes, the overlap number of genes, and the P-value significance of the correlation is presented. Each of the different comparisons’ DMR associated genes are combined to identify the potential associated processes and pathologies correlated with the hatchery and rearing conditions.
Figure 7:
Sperm DMR associated gene correlations. Cellular localization of associated genes with processes and pathologies in box
Figure 8:
RBC DMR associated gene correlations. Cellular localization of associated genes with processes and pathologies in box
Discussion
Observations indicate hatchery spawning and rearing induces epigenetic alterations when compared to a wild (natural-origin) populations of steelhead trout. In addition, feeding and growth rate (1-year fast growth S1 vs 2-year slow growth S2) of juveniles results in altered epigenetic programming in adulthood. The hatchery conditions impose nutritional, behavioral, or other types of stressors on the fish that can developmentally promote altered epigenetic programming and phenotypic variation. The exposure of somatic cells leading to epigenetic (e.g. DNA methylation) change may impact the exposed individuals’ health and phenotypes. Therefore, the hatchery populations can have reduced fertility, abnormal health, and survival when compared to the wild populations (Table 1). Since environmental exposures generally cannot directly alter genetic mutations, epigenetics provides a molecular mechanism for the physiological changes observed. In the event, the hatchery induced epigenetic alterations (i.e. epimutations) that appear in the germline (egg or sperm), then the impacts may persist across generations. Previously, the transmission of epigenetic information through the germline to alter a variety of phenotypes in a number of different species, including fish, have been observed [3]. This epigenetic inheritance can influence the next generation’s phenotypes and health. If the germline epigenetic changes persist between generations in the absence of environmental exposures, then this is termed epigenetic transgenerational inheritance [3, 5]. Therefore, the hatchery impacts may not only be on the exposed individuals, but on subsequent generations. This has the potential to dramatically impact the wild natural-origin populations and their evolutionary trajectory. The current study demonstrates the ability of hatchery conditions to promote epigenetic alterations in both somatic cells (RBC) and germ cells (sperm), so both directly exposed fish populations and future generations need to be considered.
Previous studies have demonstrated hatchery impacts on fish phenotypic alterations and health (Table 1). Hatchery rearing has been shown to decrease reproductive success in steelhead trout by about 40% per captive-reared generation in the first two generations when fish are moved back to natural environments [8, 13]. Hatchery rearing of salmonids affects lateral line and auditory structures [11]. Phenotypic changes and decreases in fitness can occur even when wild fish are incorporated as broodstock in hatchery operations [9]. Although the relationships of the molecular epigenetic alterations and DMRs found in the current study cannot currently be causally related directly to the phenotypes, hatchery-induced epigenetic alterations (i.e. epimutations) provide a potential molecular mechanism for the phenotypic variation and health effects previously observed.
The hatchery fish are exposed to hatchery conditions and reared through development to the smolt stage, so all early development periods are impacted. Primordial germ cells (PGCs) in fish species, including salmonids, are specified by the localization of germplasm components into certain cells early in embryonic development [59]. These germplasm components include the products of the vasa and nanos genes [59]. PGCs migrate to the gonadal anlagen, then undergo mitotic proliferation and differentiate to become either spermatogonia or oogonia [59]. The PGC is a critical stage for the male germline when dramatic epigenetic reprogramming occurs. Puberty in male salmonids is associated with growth and size, photoperiod, and hormones including 11-ketotestosterone [60]. Once spermatogenesis is initiated, spermatogonia are surrounded by Sertoli cells to form cyst-like structures (spermatocysts) inside which the germ cells undergo mitotic divisions, produce spermatocytes that undergo miotic divisions to form spermatids, which then undergo spermiogenesis to produce spermatozoa. The spermatozoa are released from the spermatocysts into the testis tubules [61]. The earlier stages of spermatogenesis are regulated by follicle-stimulating hormone (FSH) from the pituitary, while the later stages are regulated by luteinizing hormone (LH) [62]. Any of these developmental stages may be impacted by environmental factors that result in epigenetic changes to the germ cells.
There is evidence that epigenetic shifts during early life play a significant role in directing the life history and phenotypes of fish during adulthood. This is clearly demonstrated in cases of temperature-dependent sex determination, as seen with sea bass [63]. In Atlantic salmon, the temperature of embryonic development affects growth rate of the smolt stage, and is associated with larval myogenin expression and DNA methylation levels [64]. Similarly, stressors during embryonic and larval stages for salmon induce changes in the immune response at 4 months of age that are associated with changes in DNA methylation in gill tissue [65]. These early developmental impacts on later-life health and disease involve the developmental origin of health and disease (DOHAD) mechanisms observed in many species, including humans. Epigenetics is one of the primary molecular mechanisms involved in this phenomenon.
In the current study, the epigenetic alterations observed were DMRs identified in the hatchery steelhead somatic and germline cells, compared to natural-origin populations. The presence of a reproducible DMR is termed an epimutation [1, 3, 4]. The molecular procedure used to identify the DMRs was methylated DNA immunoprecipitation (MeDIP) followed by Seq for an MeDIP-Seq analysis [66]. The MeDIP procedure is biased to low-density CpG analysis (<10% CpG) [67], which constitutes ∼95% of the genome [68]. In contrast, due to the informatics and alignment issues, a bisulfite procedure (RRBS) can be biased to higher density CpG analysis (>10%), which constitutes ∼5% of the genome [47]. Therefore, the current study used MeDIP-Seq to capture the majority of the genome, while previous studies have used bisulfite (i.e. RRBS) procedures [58, 69]. The DMRs identified in the current study had low CpG content, and exist primarily in CpG deserts [50]. Evolutionarily, CpG methylation has been shown to increase susceptibility for C to T conversions, and leads to regions of the genome with low CpG content. Small clusters of CpG in these CpG deserts were likely conserved due to regulatory roles for these CpG clusters [50]. The genomic locations of the DMRs also demonstrate a genome-wide distribution on most chromosomes. Although the assembly of the steelhead trout genome is not complete, it has been significantly improved recently and the most recent reference genome (Oncorhynchus mykiss, Omyk_1.0, accession GCF_002163495.1, https://www.ncbi.nlm.nih.gov/assembly/GCF_002163495.1), was used in the current study. The comparison of the hatchery and wild (natural-origin) steelhead populations identified a large number of single-site DMRs and a smaller set of DMRs using a more stringent selection of adjacent site DMRs. Therefore, the epimutations identified in this study appear to be due to alterations in epigenetic programming between the hatchery and wild natural-origin steelhead populations. Although variables such as age, size, and hatchery conditions will impact an individual’s epigenome, these variables were controlled for in this study to the extent possible. The permutation analyses and PCA plots did not identify any obvious sources of unexplained variability.
In the current study, comparisons were also made between hatchery-reared steelhead fed at a high plane of nutrition and reaching smolt stage within 1 year (fast-growth; WNFH-S1), and hatchery-reared steelhead fed less and reaching smolt stage within 2 years (slow-growth; WNFS-S2), or non-hatchery reared wild natural-origin steelhead that typically take 2 years to mature to the smolt stage (N). Similar to what was found when comparing hatchery-reared to wild steelhead trout, the DMRs identified occurred primarily in CpG deserts, and a large proportion of them was present in intergenic regions. A comparison of the slow and fast growth/maturation hatchery conditions also demonstrated similar genomic features in the DMRs identified. Therefore, the epimutation characteristics found in the current study are similar to those previously identified in other species following environmental exposures [1, 3, 70].
Genetic analysis in the current study used an evaluation of CNVs to estimate gene sequence similarity between treatment groups. Observations indicate there was negligible genetic variation between these populations. Negligible genetic mutations have been previously observed between the hatchery and wild populations compared [57, 58, 71]. This is to be expected in the Methow River fisheries system examined, as hatchery-reared broodstock are crossed with stream-raised “wild” natural-origin steelhead at each generation. In addition, hatchery-reared fish can spawn with wild fish in the river. This blends the genetic backgrounds of the populations examined. These observations suggest hatchery or feeding rate-induced epigenetic alterations may have a significant role in the phenotype and health differences between the populations. However, as with any biological system, the integration of genetics and epigenetics will be required to fully realize the phenotypic variation and environmentally induced health effects. The initial environmentally induced epigenetic alterations are anticipated to generationally lead to genetic changes, as previously described [72], such that an integration of both molecular mechanisms is involved.
The DMR associated genes indicate the epigenetic alterations observed may have major effects on genome activity, potentially correlating to the phenotypic alterations known to occur (Table 1). The majority of DMRs did not have gene associations, suggesting distal regulation of genomic activity may be involved, as previously suggested for other species [48]. For those DMRs with associated genes, a large number of gene classification categories were implicated in the hatchery vs natural-origin steelhead studies including transcription, development, protease, transport, immune, epigenetic, metabolism, and hormone (Fig. 6). Specific cellular pathways were also identified, but only 2–3 genes were represented in each pathway. A large number of cellular processes and pathologies were correlated with the DMR associated genes for both the sperm and RBC (Supplementary Tables S11 and S12 and Figs 6–8). Observations suggest a number of different physiological processes and pathologies may potentially be affected by the epigenetic DNA methylation alterations observed. These potential alterations in genome activity are speculated to correlate to the phenotypic variation and health impacts of the hatchery steelhead populations, but future research is needed to identify causal relationships.
Accelerated growth and maturation rates in the hatchery promoted the highest level of epigenetic changes in the mature fish. The 1-year S1 matured smolt as an adult had the highest level of epigenetic change compared to the 2-year S2 matured smolt as an adult which was similar to the wild natural-origin population. However, the S2 matured population also had a high level of epigenetic change compared to the wild natural-origin population. This suggests other hatchery environmental factors than growth in the S2 population also exist and contribute to the hatchery impacts on the epigenetic programming. Clearly, moving to a more normal hatchery condition to mimic the natural-origin wild population [73] will help improve the physiology and health of the steelhead trout population and reduce the abnormal phenotypes observed in Table 1. In addition to the early developmental rearing and nutrition parameters, a large number of other parameters can impact the migration and adult fish development [7, 74]. This can include the impacts of fin clips [75], migration impacts of Dams [76], behavioral impacts from predators [77], water temperature and quality [78], and ocean conditions [79]. Therefore, the epigenetic analysis of the adult fish in the current study will incorporate not only the hatchery rearing impacts, but these other variables as well. This needs to be considered in the data interpretation.
The current study supports a potential role for hatchery and growth rate-induced epigenetic change in steelhead trout. The observations correlate with the phenotypic variation between hatchery and wild natural-origin fish populations. Although the programming and signatures of DMRs were found to be associated with specific cell types, the functional impacts of these DMRs need to be further investigated. A recent study has also examined the genetic and epigenetic differences between hatchery-reared and natural steelhead trout from the Methow River system [58]. In this study, Restriction Site Associated DNA Sequencing (RAD-Seq) was used to assess genetic similarity by identifying single nucleotide polymorphisms (SNPs). Similar to the current study, negligible genetic differences were found. Gavery et al. [58] used RBBS to evaluate DNA methylation, and again similar to the current study, found epigenetic differences between hatchery-raised and natural fish. The current study provides a more genome-wide analysis due to the MeDIP-Seq focus on lower density genome regions, and a CNV analysis that also reinforces the previously published genetic results. Therefore, the current hatchery conditions used can impact phenotypic variation through environmentally induced epigenetic transgenerational inheritance.
Conclusions
Clearly, the current study supports a critical role for epigenetics when considering the molecular source for hatchery impacts, however, it will be an integration of epigenetics and genetics that will influence the molecular control of phenotypic variation. Since the sperm were found to have epigenetic alterations, the generational impacts through epigenetic inheritance also need to be considered. The potential that these epigenetic germline effects can be transmitted in the absence of direct exposure through epigenetic transgenerational inheritance now needs to be assessed. In hatchery operations involving food production, such as aquaculture, or when the hatchery fish are not allowed to breed with the wild fish population, impacts on the wild population will be reduced. However, in the event a hatchery population can breed with a wild fish population, the long-term impacts on the wild population and the environment could be dramatic and alter the future trajectory of the wild population. Further research into environmental epigenetic impacts on hatchery and wild fish populations is now needed. Interestingly, the epigenetic alterations observed could be used as biomarkers to further identify hatchery impacts on the fish, as well as correlate with phenotypic alterations observed in the future.
Methods
Animal Studies
As described in the Supplementary Methods [58], adult steelhead trout, Oncorhynchus mykiss, were collected from the Methow River and Winthrop National Fish Hatchery in Winthrop, WA, USA. The purified sperm and RBCs were collected for DNA preparations and epigenetic analysis. The sample collections were obtained by the WNFH staff and the staff of Dr Penny Swanson’s laboratory at the Northwest Fisheries Science Center, and the National Marine Fisheries Service (NMFS, NOAA) in Seattle WA, who had the appropriate vertebrate animal approvals for the study. All fish were reared and sampled according to the WNFH and University of Washington Institutional Animal Care and Use Committee (Dr Penny Swanson, Protocol #2313-90). Samples were shipped on dry ice and stored at −80°C for analysis. The initial natural and hatchery origin adult steelhead returning to the Methow river in summer 2013 and captured spring 2014 were used as designated N1 and H. Subsequent years were collected in a similar manner for the N2 and S1 or S2 populations (Supplementary Table S1).
Epigenetic Analysis, Statistics, and Bioinformatics
DNA was isolated from sperm and RBCs as described in the Supplementary Methods [66]. Methylated DNA immunoprecipitation (MeDIP), followed by Seq (MeDIP-Seq) was performed. MeDIP-Seq, sequencing libraries, Seq, and bioinformatics analysis were performed as described in the Supplementary Methods [66]. All molecular data has been deposited into the public database at NCBI (GEO # GSE145887), and R code computational tools are available at GitHub (https://github.com/skinnerlab/MeDIP-seq) and www.skinner.wsu.edu.
Supplementary Material
Acknowledgements
We acknowledge the advice and critical technical assistance of the staff with the USFWS, WNFH, and the Dr Penny Swanson laboratory at NMFS, NOAA at NWFSC in Seattle, WA. We acknowledge the contributions of Dr Penny Swanson who assisted in the experimental design, sample collection, and facilitating funding from NOAA. We thank Ms Jayleana Barton for technical assistance, Ms Amanda Quilty for editing, and Ms Heather Johnson for assistance in preparation of the manuscript.
Data Availability
All sequence data have been deposited to NCBI, GEO # GSE145887.
Funding
The research was supported by the National Oceanic and Atmospheric Administration (NOAA; grant numbers RA133F15SE1307 and RA133F14SE3610) and National Institutes of Health (NIH; grant number ES012974) grant to M.K.S.
Conflict of interest statement
The authors have declared that no competing interests exist.
Supplementary data
Supplementary data are available at EnvEpig online.
References
- 1. Jirtle RL, Skinner MK.. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007;8:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bonasio R. The expanding epigenetic landscape of non-model organisms. J Exp Biol 2015;218:114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Skinner MK. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol Cell Endocrinol 2014;398:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics 2011;6:838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anway MD, Cupp AS, Uzumcu M, Skinner MK.. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005;308:1466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McClure MM, Utter FM, Baldwin C, Carmichael RW, Hassemer PF, Howell PJ, Spruell P, Cooney TD, Schaller HA, Petrosky CE.. Evolutionary effects of alternative artificial propagation programs: implications for viability of endangered anadromous salmonids. Evol Appl 2008;1:356–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naish KA, Taylor JE 3rd, Levin PS, Quinn TP, Winton JR, Huppert D, Hilborn R.. An evaluation of the effects of conservation and fishery enhancement hatcheries on wild populations of salmon. Adv Mar Biol 2007;53:61–194. [DOI] [PubMed] [Google Scholar]
- 8. Araki H, Cooper B, Blouin MS.. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science 2007;318:100–3. [DOI] [PubMed] [Google Scholar]
- 9. Fraser DJ. How well can captive breeding programs conserve biodiversity? A review of salmonids. Evol Appl 2008;1:535–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frankham R. Genetic adaptation to captivity in species conservation programs. Mol Ecol 2008;17:325–33. [DOI] [PubMed] [Google Scholar]
- 11. Brown AD, Sisneros JA, Jurasin T, Coffin AB.. Effects of hatchery rearing on the structure and function of salmonid mechanosensory systems. Adv Exp Med Biol 2016;875:117–24. [DOI] [PubMed] [Google Scholar]
- 12. Christie MR, Ford MJ, Blouin MS.. On the reproductive success of early-generation hatchery fish in the wild. Evol Appl 2014;7:883–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ford MJ, Murdoch AR, Hughes MS, Seamons TR, LaHood ES.. Broodstock history strongly influences natural spawning success in hatchery steelhead (Oncorhynchus mykiss). PLoS One 2016;11:e0164801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Araki H, Ardren WR, Olsen E, Cooper B, Blouin MS.. Reproductive success of captive-bred steelhead trout in the wild: evaluation of three hatchery programs in the Hood river. Conserv Biol 2007;21:181–90. [DOI] [PubMed] [Google Scholar]
- 15. Reisenbichler R, Rubin S, Wetzel L, Phelps S.. Natural selection after release from a hatchery leads to domestication in steelhead, Oncorhynchus mykiss, in Stock Enhancement and Sea Ranching: Developments, Pitfalls and Opportunities, 2nd edn, Leber KM, Kitada S, Blankenship HL, Svåsand T, eds. 2004, Blackwell Publishing Ltd., Hoboken, NJ, p. 371–84. [Google Scholar]
- 16. Christie MR, Marine ML, French RA, Blouin MS.. Genetic adaptation to captivity can occur in a single generation. Proc Natl Acad Sci U S A 2012;109:238–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Christie MR, Marine ML, Fox SE, French RA, Blouin MS.. A single generation of domestication heritably alters the expression of hundreds of genes. Nat Commun 2016;7:10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brown AD, Sisneros JA, Jurasin T, Nguyen C, Coffin AB.. Differences in lateral line morphology between hatchery- and wild-origin steelhead. PLoS One 2013;8: e59162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abadia-Cardoso A, Anderson EC, Pearse DE, Garza JC.. Large-scale parentage analysis reveals reproductive patterns and heritability of spawn timing in a hatchery population of steelhead (Oncorhynchus mykiss). Mol Ecol 2013;22: 4733–46. [DOI] [PubMed] [Google Scholar]
- 20. Kihslinger RL, Nevitt GA.. Early rearing environment impacts cerebellar growth in juvenile salmon. J Exp Biol 2006;209: 504–9. [DOI] [PubMed] [Google Scholar]
- 21. Araki H, Cooper B, Blouin MS.. Carry-over effect of captive breeding reduces reproductive fitness of wild-born descendants in the wild. Biol Lett 2009;5: 621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moore M, Berejikian BA, Tezak EP.. Variation in the early marine survival and behavior of natural and hatchery-reared Hood Canal steelhead. PLoS One 2012;7: e49645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fraser DJ, Walker L, Yates MC, Marin K, Wood JLA, Bernos TA, Zastavniouk C.. Population correlates of rapid captive-induced maladaptation in a wild fish. Evol Appl 2019;12: 1305–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reed TE, Prodohl P, Hynes R, Cross T, Ferguson A, McGinnity P.. Quantifying heritable variation in fitness-related traits of wild, farmed and hybrid Atlantic salmon families in a wild river environment. Heredity (Edinb)), 2015;115:173–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Milot E, Perrier C, Papillon L, Dodson JJ, Bernatchez L.. Reduced fitness of Atlantic salmon released in the wild after one generation of captive breeding. Evol Appl 2013;6:472–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skaala O, Besnier F, Borgstrom R, Barlaup B, Sorvik AG, Normann E, Ostebo BI, Hansen MM, Glover KA.. An extensive common-garden study with domesticated and wild Atlantic salmon in the wild reveals impact on smolt production and shifts in fitness traits. Evol Appl 2019;12:1001–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janowitz-Koch I, Rabe C, Kinzer R, Nelson D, Hess MA, Narum SR.. Long-term evaluation of fitness and demographic effects of a Chinook Salmon supplementation program. Evol Appl 2019;12:456–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Theriault V, Moyer GR, Jackson LS, Blouin MS, Banks MA.. Reduced reproductive success of hatchery coho salmon in the wild: insights into most likely mechanisms. Mol Ecol 2011;20:1860–9. [DOI] [PubMed] [Google Scholar]
- 29. Miller LM, Close T, Kapuscinski AR.. Lower fitness of hatchery and hybrid rainbow trout compared to naturalized populations in Lake Superior tributaries. Mol Ecol 2004;13:3379–88. [DOI] [PubMed] [Google Scholar]
- 30. Tillotson MD, Barnett HK, Bhuthimethee M, Koehler ME, Quinn TP.. Artificial selection on reproductive timing in hatchery salmon drives a phenological shift and potential maladaptation to climate change. Evol Appl 2019;12:1344–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sweeting RM, Beamish RJ, Neville CM.. Crystalline otoliths in teleosts: comparisons between hatchery and wild coho slamon (Oncorrhynchus kisutch) in the Straight of Georgia. Rev Fish Biol Fisheries 2004;14:361–9. [Google Scholar]
- 32. Kihslinger RL, Lema SC, Nevitt GA.. Environmental rearing conditions produce forebrain differences in wild Chinook salmon Oncorhynchus tshawytscha. Comp Biochem Physiol A Mol Integr Physiol 2006;145: 145–51. [DOI] [PubMed] [Google Scholar]
- 33. Kotrschal A, Sundstrom LF, Brelin D, Devlin RH, Kolm N.. Inside the heads of David and Goliath: environmental effects on brain morphology among wild and growth-enhanced coho salmon Oncorhynchus kisutch. J Fish Biol 2012;81: 987–1002. [DOI] [PubMed] [Google Scholar]
- 34. Stringwell R, Lock A, Stutchbury CJ, Baggett E, Taylor J, Gough PJ, Garcia de Leaniz C.. Maladaptation and phenotypic mismatch in hatchery-reared Atlantic salmon Salmo salar released in the wild. J Fish Biol 2014;85: 1927–45. [DOI] [PubMed] [Google Scholar]
- 35. Pulcini D, Wheeler PA, Cataudella S, Russo T, Thorgaard GH.. Domestication shapes morphology in rainbow trout Oncorhynchus mykiss. J Fish Biol 2013;82: 390–407. [DOI] [PubMed] [Google Scholar]
- 36. de Mestral LG, Herbinger CM.. Reduction in antipredator response detected between first and second generations of endangered juvenile Atlantic salmon Salmo salar in a captive breeding and rearing programme. J Fish Biol 2013;83:1268–86. [DOI] [PubMed] [Google Scholar]
- 37. Salvanes AGV. Are antipredator behaviours of hatchery Salmo salar juveniles similar to wild juveniles? J Fish Biol 2017;90:1785–96. [DOI] [PubMed] [Google Scholar]
- 38. Harvey AC, Solberg MF, Troianou E, Carvalho GR, Taylor MI, Creer S, Dyrhovden L, Matre IH, Glover KA.. Plasticity in growth of farmed and wild Atlantic salmon: is the increased growth rate of farmed salmon caused by evolutionary adaptations to the commercial diet? BMC Evol Biol 2016;16:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vainikka A, Kallio-Nyberg I, Heino M, Koljonen ML.. Divergent trends in life-history traits between Atlantic salmon Salmo salar of wild and hatchery origin in the Baltic Sea. J Fish Biol 2010;76: 622–40. [DOI] [PubMed] [Google Scholar]
- 40. Horreo JL, Valiente AG, Ardura A, Blanco A, Garcia-Gonzalez C, Garcia-Vazquez E.. Nature versus nurture? Consequences of short captivity in early stages. Ecol Evol 2018;8: 521–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. De Paoli T, Fuller-Tyszkiewicz M, Krug I.. Insecure attachment and maladaptive schema in disordered eating: the mediating role of rejection sensitivity. Clin Psychol Psychother 2017;24:1273–84. [DOI] [PubMed] [Google Scholar]
- 42. Barlaup BT, Rund H, Normann ES, Stranzl S, Mahlum S, Vollset KW.. Out of sync: monitoring the time of sea entry of wild and hatchery salmon Salmo salar smolt using floating passive-integrated transponder antennae. J Fish Biol 2018;93:455–64. [DOI] [PubMed] [Google Scholar]
- 43. Lavoie C, Courcelle M, Redivo B, Derome N.. Structural and compositional mismatch between captive and wild Atlantic salmon (Salmo salar) parrs' gut microbiota highlights the relevance of integrating molecular ecology for management and conservation methods. Evol Appl 2018;11: 1671–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heath DD, Heath JW, Bryden CA, Johnson RM, Fox CW.. Rapid evolution of egg size in captive salmon. Science 2003;299: 1738–40. [DOI] [PubMed] [Google Scholar]
- 45. Chittenden CM, Biagi CA, Davidsen JG, Davidsen AG, Kondo H, McKnight A, Pedersen OP, Raven PA, Rikardsen AH, Shrimpton JM, Zuehlke B, McKinley RS, Devlin RH.. Genetic versus rearing-environment effects on phenotype: hatchery and natural rearing effects on hatchery- and wild-born coho salmon. PLoS One 2010;5: e12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guerrero-Bosagna C, Settles M, Lucker B, Skinner M.. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One 2010;5:e13100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg KJ, Gu J, Echipare L, O'Geen H, Lister R, Pelizzola M, Xi Y, Epstein CB, Bernstein BE, Hawkins RD, Ren B, Chung WY, Gu H, Bock C, Gnirke A, Zhang MQ, Haussler D, Ecker JR, Li W, Farnham PJ, Waterland RA, Meissner A, Marra MA, Hirst M, Milosavljevic A, Costello JF.. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 2010;28:1097–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Skinner MK, Manikkam M, Haque MM, Zhang B, Savenkova M.. Epigenetic transgenerational inheritance of somatic transcriptomes and epigenetic control regions. Genome Biol 2012;13:R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haque MM, Nilsson EE, Holder LB, Skinner MK.. Genomic clustering of differential DNA methylated regions (epimutations) associated with the epigenetic transgenerational inheritance of disease and phenotypic variation. BMC Genomics 2016;17:418.1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Skinner MK, Guerrero-Bosagna C.. Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genomics 2014;15:692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nozawa M, Kawahara Y, Nei M.. Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci USA 2007;104:20421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E.. Metastable epialleles in mammals. Trends Genet 2002;18: 348–51. [DOI] [PubMed] [Google Scholar]
- 53. Dolinoy DC, Das R, Weidman JR, Jirtle RL.. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res 2007;61:30R–7R. [DOI] [PubMed] [Google Scholar]
- 54. Gazave E, Darré F, Morcillo-Suarez C, Petit-Marty N, Carreño A, Marigorta UM, Ryder OA, Blancher A, Rocchi M, Bosch E, Baker C, Marquès-Bonet T, Eichler EE, Navarro A.. Copy number variation analysis in the great apes reveals species-specific patterns of structural variation. Genome Res 2011;21:1626–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Poptsova M, Banerjee S, Gokcumen O, Rubin MA, Demichelis F.. Impact of constitutional copy number variants on biological pathway evolution. BMC Evol Biol 2013;13:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Skinner MK, Gurerrero-Bosagna C, Haque MM, Nilsson EE, Koop JAH, Knutie SA, Clayton DH.. Epigenetics and the evolution of Darwin's Finches. Genome Biol Evol 2014;6:1972–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Le Luyer J, Laporte M, Beacham TD, Kaukinen KH, Withler RE, Leong JS, Rondeau EB, Koop BF, Bernatchez L.. Parallel epigenetic modifications induced by hatchery rearing in a Pacific salmon. Proc Natl Acad Sci USA 2017;114:12964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gavery MR, Nichols KM, Goetz GW, Middleton MA, Swanson P.. Characterization of genetic and epigenetic variation in sperm and red blood cells from adult hatchery and natural-origin steelhead, Oncorhynchus mykiss. G3 (Bethesda) 2018;8:3723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoshizaki G, Takeuchi Y, Kobayashi T, Ihara S, Takeuchi T.. Primordial germ cells: the blueprint for a piscine life. Fish Physiol Biochem 2002;26:3–12. [Google Scholar]
- 60. Campbell B, Dickey JT, Swanson P.. Endocrine changes during onset of puberty in male spring Chinook salmon, Oncorhynchus tshawytscha. Biol Reprod 2003;69:2109–17. [DOI] [PubMed] [Google Scholar]
- 61. Uribe MC, Grier HJ, Mejia-Roa V.. Comparative testicular structure and spermatogenesis in bony fishes. Spermatogenesis 2014;4:e983400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Knapp R, Carlisle SL.. Testicular function and hormonal regulation in fishes, in Fishes, Hormones and Reproduction in Vertebrates, Norris DO, Lopez KH, eds. 2011, Academic Press, New York, NY. p. 43–63. [Google Scholar]
- 63. Navarro-Martin L, Vinas J, Ribas L, Diaz N, Gutierrez A, Di Croce L, Piferrer F.. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet 2011;7:e1002447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Burgerhout E, Mommens M, Johnsen H, Aunsmo A, Santi N, Andersen O.. Genetic background and embryonic temperature affect DNA methylation and expression of myogenin and muscle development in Atlantic salmon (Salmo salar). PLoS One 2017;12:e0179918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Uren Webster TM, Rodriguez-Barreto D, Martin SAM, Van Oosterhout C, Orozco-terWengel P, Cable J, Hamilton A, Garcia De Leaniz C, Consuegra S.. Contrasting effects of acute and chronic stress on the transcriptome, epigenome, and immune response of Atlantic salmon. Epigenetics 2018;13:1191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ben Maamar M, Nilsson E, Sadler-Riggleman I, Beck D, McCarrey JR, Skinner MK.. Developmental origins of transgenerational sperm DNA methylation epimutations following ancestral DDT exposure. Dev Biol 2019;445:280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, Strbenac D, Robinson MD, Clark SJ.. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 2011;6:34–44. [DOI] [PubMed] [Google Scholar]
- 68. Ben Maamar M, Sadler-Riggleman I, Beck D, Skinner MK.. Genome-wide mapping of DNA methylation 5mC by Methylated DNA immunoprecipitation (MeDIP)-sequencing. DNA Modif Methods Mol Biol 2021;2198:301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou L, Ng HK, Drautz-Moses DI, Schuster SC, Beck S, Kim C, Chambers JC, Loh M.. Systematic evaluation of library preparation methods and sequencing platforms for high-throughput whole genome bisulfite sequencing. Sci Rep 2019;9:10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nilsson E, Sadler-Riggleman I, Skinner MK.. Environmentally induced epigenetic transgenerational inheritance of disease. Environ Epigenet 2018;4:dvy016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Blankenship SM, Campbell MR, Hess JE, Hess MA, Kassler TW, Kozfkay CC, Matala AP, Narum SR, Paquin MM, Small MP, Stephenson JJ, Warheit KI, Moran P.. Major lineages and metapopulations in Columbia River Oncorhynchus mykiss are structured by dynamic landscape features and environments. Trans Am Fish Soc 2011;140:665–84. [Google Scholar]
- 72. Skinner MK, Guerrero-Bosagna C, Haque MM.. Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics 2015;10:762–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gavery MR, Nichols KM, Berejikian BA, Tatara CP, Goetz GW, Dickey JT, Van Doornik DM, Swanson P.. Temporal dynamics of DNA methylation patterns in response to rearing juvenile steelhead (Oncorhynchus mykiss) in a hatchery versus simulated stream environment. Genes (Basel) 2019;10:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Johnsson JI, Brockmark S, Naslund J.. Environmental effects on behavioural development consequences for fitness of captive-reared fishes in the wild. J Fish Biol 2014;85:1946–71. [DOI] [PubMed] [Google Scholar]
- 75. Bert B, Chmielewska J, Bergmann S, Busch M, Driever W, Finger-Baier K, Hößler J, Köhler A, Leich N, Misgeld T, Nöldner T, Reiher A, Schartl M, Seebach-Sproedt A, Thumberger T, Schönfelder G, Grune B.. Considerations for a European animal welfare standard to evaluate adverse phenotypes in teleost fish. EMBO J 2016;35:1151–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Arkoosh MR, Kagley AN, Anulacion BF, Boylen DA, Sandford BP, Loge FJ, Johnson LL, Collier TK.. Disease susceptibility of hatchery snake river spring-summer Chinook salmon with different juvenile migration histories in the Columbia River. J Aquat Anim Health 2006;18:223–31. [DOI] [PubMed] [Google Scholar]
- 77. Furey NB, Armstrong JB, Beauchamp DA, Hinch SG.. Migratory coupling between predators and prey. Nat Ecol Evol 2018;2:1846–53. [DOI] [PubMed] [Google Scholar]
- 78. Fullerton AH, Torgersen CE, Lawler JJ, Steel EA, Ebersole JL, Lee SY.. Longitudinal thermal heterogeneity in rivers and refugia for coldwater species: effects of scale and climate change. Aquat Sci 2018;80:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hall ER, Wickes L, Burnett LE, Scott GI, Hernandez D, Yates KK, Barbero L, Reimer JJ, Baalousha M, Mintz J, Cai WJ, Craig JK, DeVoe MR, Fisher WS, Hathaway TK, Jewett EB, Johnson Z, Keener P, Mordecai RS, Noakes S, Phillips C, Sandifer PA, Schnetzer A, Styron J.. Acidification in the U.S. Southeast: causes, Potential Consequences and the Role of the Southeast Ocean and Coastal Acidification Network. Front Mar Sci 2020;7:1–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data have been deposited to NCBI, GEO # GSE145887.