Abstract
SARS-Cov-2 infection is frequently associated with Nervous System manifestations. However, it is not clear how SARS-CoV-2 can cause neurological dysfunctions and which molecular processes are affected in the brain. In this work, we examined the frontal cortex tissue of patients who died of COVID-19 for the presence of SARS-CoV-2, comparing qRT-PCR with ddPCR. We also investigated the transcriptomic profile of frontal cortex from COVID-19 patients and matched controls by RNA-seq analysis to characterize the transcriptional signature.
Our data showed that SARS-CoV-2 could be detected by ddPCR in 8 (88%) of 9 examined samples while by qRT-PCR in one case only (11%). Transcriptomic analysis revealed that 11 genes (10 mRNAs and 1 lncRNA) were differential expressed when frontal cortex of COVID-19 patients were compared to controls. These genes fall into categories including hypoxia, hemoglobin-stabilizing protein, hydrogen peroxide processes. This work demonstrated that the quantity of viral RNA in frontal cortex is minimal and it can be detected only with a very sensitive method (ddPCR). Thus, it is likely that SARS-CoV-2 does not actively infect and replicate in the brain; its topography within encephalic structures remains uncertain. Moreover, COVID-19 may have a role on brain gene expression, since we observed an important downregulation of genes associated to hypoxia inducting factor system (HIF) that may inhibit the capacity of defense system during infection and oxigen deprivation, showing that hypoxia, well known multi organ condition associated to COVID-19, also marked the brain.
Keywords: SARS-CoV-2, Frontal cortex, Transcriptome, Hypoxia, Hemoglobin
1. Introduction
COVID-19, a disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection evolved in the last year into a global pandemic (Zou et al., 2020), is primary characterized by respiratory symptoms. The most common symptoms are fever, cough, fatigue, shortness of breath, but some COVID-19 patients evolve to acute respiratory distress syndrome and/or multi organ failure (Huang, 2020, Guan et al., 2020). COVID-19 patients experience moderate to severe hypoxemia that can contribute to multiple organ dysfunction, especially affecting the kidney, heart and central nervous system (CNS). Indeed, neurological, and cardiovascular effects have been described (Helms et al., 2020, Wichmann et al., 2020). COVID-19 is particularly detrimental in patients with multiple comorbidities (e. g. cardiovascular diseases, diabetes, obesity, pulmonary disorders, and cancer). The cytokine storm induced by SARS-CoV-2 may be uncontrolled and might cause a wide range of symptoms, including several neurological manifestations. Neurological phenomena, observed during and after the acute COVID-19 phase, have now been described in several studies reporting both CNS symptoms (psychomotor retardation, dizziness, confusion, delirium, ataxia, encephalitis, stroke, seizures) and vegetative/peripheral manifestations (vomiting, hypotension, severe asthenia, myalgia, neuralgia, headache, olfactory, and gustatory dysfunction, Guillain-Barré syndrome) (Ahmad and Rathore, 2020, Baig et al., 2020, Mao et al., 2020, Poloni et al., 2020a, Poloni et al., 2020b, Zhao et al., 2020, Zito et al., 2020).
Neurological manifestations caused by COVID-19 may be due to immune-mediated mechanisms or direct viral invasion of CNS. Particularly, SARS-CoV-2 may enter the CNS through different routes: 1) the trans-synaptic route, particularly intranasal to the olfactory bulbs and the basal frontal lobes; 2) the endothelial-astrocytic route by crossing the blood brain barrier or through transport by infected leukocytes (Trojan horse mechanism) (Baig et al., 2020). Available autopsy data from brains of COVID-19 patients show unspecific findings of brain congestion, oedema, neuronal damage, and inflammatory infiltrates with features of encephalitis and meningitis. Such findings appear attributable more to hypoxic and immune-mediated phenomena rather than to virus-induced lesions; they do not clearly establish the presence of SARS-CoV-2 in the brain and its role in neuronal damage remains unclear (Matschke et al., 2020, Solomon et al., 2020, von Weyhern et al., 2020). Viral protein detection is very scant in CNS and limited to isolate neurons in the medulla oblongata. On the other hand, few papers have been published about the presence of viral RNA in the human brain and no correlation has been found between severity of neurological clinical signs and the presence of viral protein or RNA in the human brain of COVID-19 patients (Frank, 2020). Nonetheless, in a recent series, RNA was detected in about half of the cases (Matschke et al., 2020), but the consequences of brain infection in terms of molecular alteration have yet to be revealed.
In this paper, we investigated the presence of SARS-CoV-2 in the brain of elderly patients died of COVID-19 and the possible effect of SARS-CoV-2 infection on transcriptomic profile in frontal cortex comparing COVID-19 patients and matched controls by RNA-seq analysis. Fronto-basal cortex was chosen from the assumption that SARS‐CoV‐2 may invade the brain through the olfactory pathway, as recently reported (Politi et al., 2020).
2. Materials and methods
2.1. Tissue collection and preparation
Autoptic human brain samples were provided by Unit of Legal Medicine and Forensic Sciences, Institute of Legal Medicine of University of Pavia and the Abbiategrasso Brain Bank (ABB) of Golgi Cenci Foundation (Milan, Italy). The ABB autopsy and sampling protocol (Poloni et al., 2020a, Poloni et al., 2020b) was approved by the Ethics Committee of the University of Pavia on October 6th, 2009 (Committee report 3/2009). Patients who died for SARS-CoV-2 infection underwent forensic autopsies, ordered by the Prosecutor. The samples were taken according to Italian laws about personal data treatment, the reference law is the authorization n9/2016 of the guarantor of privacy, then replaced by Regulation (EU) 2016/679 of the European Parliament and of the Council. The mean of post mortem-interval was 8 days (ranging 3 to 15 days, Table 1 ) for COVID-19 cases. All the bodies were stored at 4 °C until the time of the autopsy. The preservation of the body was adequate in all 9 COVID-19 cases considered for the study and none were excluded. A reconstruction of the patients' clinical history was carried out through a retrospective evaluation of medical charts by a forensic medical doctor and a neurologist with expertise in neuropathology, who carried out also the neuropathological evaluation. COVID-19 cases (5 women and 4 men; median age 83 years), were clinically defined for the presence or absence of dementia, and for the presence or absence of comorbidities including cardiovascular disease, diabetes, obesity, pulmonary disorders and cancer (Table 1). Frontal lobe samples from 8 non-COVID-19 cases provided by ABB were used for comparison. They were selected with age and clinical characteristics similar to those of the COVID-19 group. The control ABB cases differ from the COVID-19 cases for the post mortem-time which was much shorter (from 2 to 16 h) (Table 1). The PMD was necessarily different because the ABB protocol implies an autopsy performed no later than 30 h after death (Poloni et al., 2020a, Poloni et al., 2020b). Instead, the COVID-19 autopsies followed the course of the forensic roles. After the brain removal, one slice of anterior frontal lobe was fresh cut into about 10 mm thickness and frozen at −80 degreesC. An adjacent slice was fixed in formalin, included in paraffin and cut into 8 μm thick serial sections. Tissue samples for this study included cortical gray matter and subcortical white matter. The sections were stained with Hematoxylin and Eosin and Luxol Fast Blue (LFB) to evaluate vascular, architectural and structural tissue abnormalities, inflammatory infiltrates, and myelin loss. All the cases were studied for the presence of underlying AD pathology. For this purpose, reactions for TAU (AT8 ThermoScientific MN1020) and beta-amyloid (4G8, BioLegend) were performed on additional sections including frontal, temporal and parieto-occipital lobes, and hippocampi. The inflammatory infiltrates were characterized through antibodies against microglia (CD68) and lymphocytes (CD3, CD20). Anti-SARS-CoV-2 antibodies were used to detect viral inclusions.
Table 1.
Summary of the anagraphical and clinical characteristics of COVID-19 and NON-COVID-19 cases. Abbreviations: PMD = Post Mortem Delay; Dem = Dementia including Vascular Dementia (VaD) and Alzheimer's Disease (AD); Com = Comorbidities including cardiovascular disease, diabetes, obesity, pulmonary disorders and cancer.
| CASE N. | AGE | SEX | PMD (hours) | CLINICAL CONDITION | |
|---|---|---|---|---|---|
| COVID-19 CASES | 1 | 74 | F | 168 | Dem (AD) |
| 2 | 87 | M | 168 | Dem (VaD) + Com | |
| 3 | 67 | M | 120 | Com | |
| 4 | 94 | F | 72 | Dem (AD + VaD) + Com | |
| 5 | 80 | F | 360 | Com | |
| 6 | 83 | F | 312 | Dem (AD) + Com | |
| 7 | 92 | M | 144 | Dem (AD) + Com | |
| 8 | 81 | M | 168 | Dem (AD + VaD) + Com | |
| 9 | 90 | F | 264 | Com | |
| NON-COVID-19 CASES | 10 | 104 | F | 6 | Dem (AD) |
| 11 | 80 | M | 15 | Dem (AD + VaD) | |
| 12 | 79 | M | 16 | Com | |
| 13 | 78 | F | 8 | Dem (AD) + Com | |
| 14 | 84 | F | 2 | Dem (AD + VaD) | |
| 15 | 85 | F | 15 | Dem (AD + VaD) + Com | |
| 16 | 84 | M | 10 | Dem (VaD) + Com | |
| 17 | 79 | M | 3 | Com |
2.2. RNA extraction
Frozen slices from frontal cortex were used for isolation of total RNA by Trizol reagent (Life Science Technologies, Monza, Italy) according to the manufacturer’s instructions. Quantification and quality control of RNAs have been done using a Nanodrop ND-100 Spectrophotometer (Nanodrop Technologies, Wilmington, USA) and a 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit, Waldbronn, Germany); RNAs with a 260:280 ratio of ≥ 1.5 and an RNA integrity number of ≥ 8 were selected to deep sequencing.
2.3. Quantitative PCR and droplet digital PCR analysis of SARS-CoV-2 in frontal cortex tissues
2.3.1. Quantitative PCR (qPCR)
Dual labelled TaqMan probes with 5′-6-FAM fluorescent dye and 3′- BHQ-1 quencher for SARS-CoV-2 target sequences N1 was used for the detection of viral RNA. For internal reference control, a pair of primers and TaqMan probe for RNase P (RP), labelled with 5′-HEX fluorescent dye and 3′-BHQ-1 quenchers were used. Primers use have been indicated by US Centers for Disease Control and Prevention [CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel].
For reaction mix 5 µl of extracted RNA, 5 µl of Reliance One-Step RT-qPCR Supermix (BioRad, Richmond, CA), 1 µl of RT enzyme and 4 µl of water have been used for qPCR in CFX96 (BioRad, Richmond, CA). qPCR analysis has been considered valid in all samples in which RP gene has been detected. Positive samples were determinate by Cycle threshold (Ct) of N1 and N2 gene minor of 40.
2.3.2. Droplet digital PCR (ddPCR)
For the detection of SARS-CoV-2, the One-Step RT-ddPCR Advanced Kit for Probes (BioRad, Richmond, CA) was used. For each 20 μl reaction, 5 μl of Supermix, 2 μl of Reverse transcriptase, 1 μl of 300 mM DTT, 1 μl of Probe (1:40), 1 μl of both Forward and Reverse primers (1:10) and 4 μl of H2O were provided and 5 μl of starting RNA was added. The oligonucleotides and probes used in RT‐ddPCR were the same used in RT‐qPCR. For the droplet generation in RT‐ddPCR, 20 μl of reaction volume was transferred to 8‐channel disposable droplet generation cartridge and 70 μl of droplet generation oil for probe was added in adjacent oil wells. The cartridge was placed into a QX200 droplet generator (BioRad, Richmond, CA), which makes the partition of each sample into droplets. The droplets were carefully transferred to a semi‐skirted 96‐well PCR plate (BioRad, Richmond, CA), sealed using the PX1 PCR Plate Sealer (BioRad, Richmond, CA) for PCR in 2720 Thermal Cycler (Applied Biosystems, USA). After PCR of targets presented in the droplets, the QX200 droplet reader (BioRad, Richmond, CA) was used to analyze each droplet individually, which counts positive and negative droplets to establish absolute quantification of samples (concentration).
The QuantaSoft 1.6 software was used to view fluorescence data in 1D amplitude, concentration data, copy number data, events (number of positive, negative or total droplet counts). The multi‐well threshold tool was used in all the wells according to results of specificity assay in negative samples to discriminate between positive and negative droplets. The software automatically reported the copy number of each sample.
2.4. Libraries preparation for RNA-Seq and bioinformatic data analysis
Sequencing libraries of COVID-19 patients and matched controls were prepared with the CORALL Total RNA-Seq Library Prep Kit (Lexogen, Vienna, Austria), using 1 µg of total RNA. Quality of sequencing libraries was assessed by 2100 Bioanalyzer with a DNA1000 assay (Agilent, Waldbronn, Germany) and DNA High Sensitivity assay (Agilent, Waldbronn, Germany).
RNA processing was carried out using Illumina NextSeq 500 Sequencing (Illumina, San Diego, CA). FastQ files were generated via llumina bcl2fastq2 (Version 2.17.1.14 - http://support.illumina.com/downloads/bcl-2fastq-conversion-software-v217.html) starting from raw sequencing reads produced by Illumina NextSeq sequencer. Gene and transcript intensities were computed using STAR/RSEM software using Gencode Release 19 (GRCh37.p13) as a reference, using the “stranded” option. Differential expression analysis for mRNA was performed using R package EBSeq. Differential expression analysis for long non-coding RNAs (lncRNAs) was performed with the R package DESeq.2. Coding and non coding genes were considered differentially expressed and retained for further analysis with |log2(disease sample/healthy control)| ≥ 1 and a FDR ≤ 0.1. We imposed minimum |Log2FC| of 1 and a FDR lower than 0.1 as thresholds to differentially expressed genes. This choice is motivated by the decision to maximize the sensitivity of this analysis, in order to perform a massive screening and identify candidate genes to be validated with real-time analysis (Gagliardi et al., 2018, Zucca et al., 2019).
RNA sequencing data is available in GEO repository (GSE164332).
2.5. Pathway analysis
Gene enrichment analysis was performed on coding genes. We performed a Gene Ontology (GO) analysis for biological processes, cellular components and molecular function and a BioPlanet 2019 (Huang et al., 2019a, Huang et al., 2019b) via enrichR web tool and clusterProfiler (Yu et al., 2012).
2.6. qPCR validation
PCR oligonucleotide for genes pairs were selected spanning introns to optimize amplification from mRNA templates and avoiding nonspecific amplification products, using NCBI’s Primer- BLAST or online Primer 3.0. Moreover, primers were designed in specific regions that do not overlap with Antisense sequences (primers upon request). Total cDNAs were prepared using iScript™ cDNA Synthesis Kit (BioRad, Richmond, CA). qPCR reactions were performed with SYBR Green SuperMix (BioRad, Richmond, CA), using 1 μl of cDNA template (or water control). Cycle threshold (Ct) values normalized against those determined for GAPDH. Fold-expression differences relative to healthy controls were determined using the 2ΔΔCt method. Significance of gene expression changes relative to controls was analysed using one-way ANOVA (Kruskal-Wallis) and the Dunns post-test for all possible test pairings using Prism GraphPad 5.02 software (GraphPad Software, San Diego, CA).
3. Results
3.1. Neuropathology
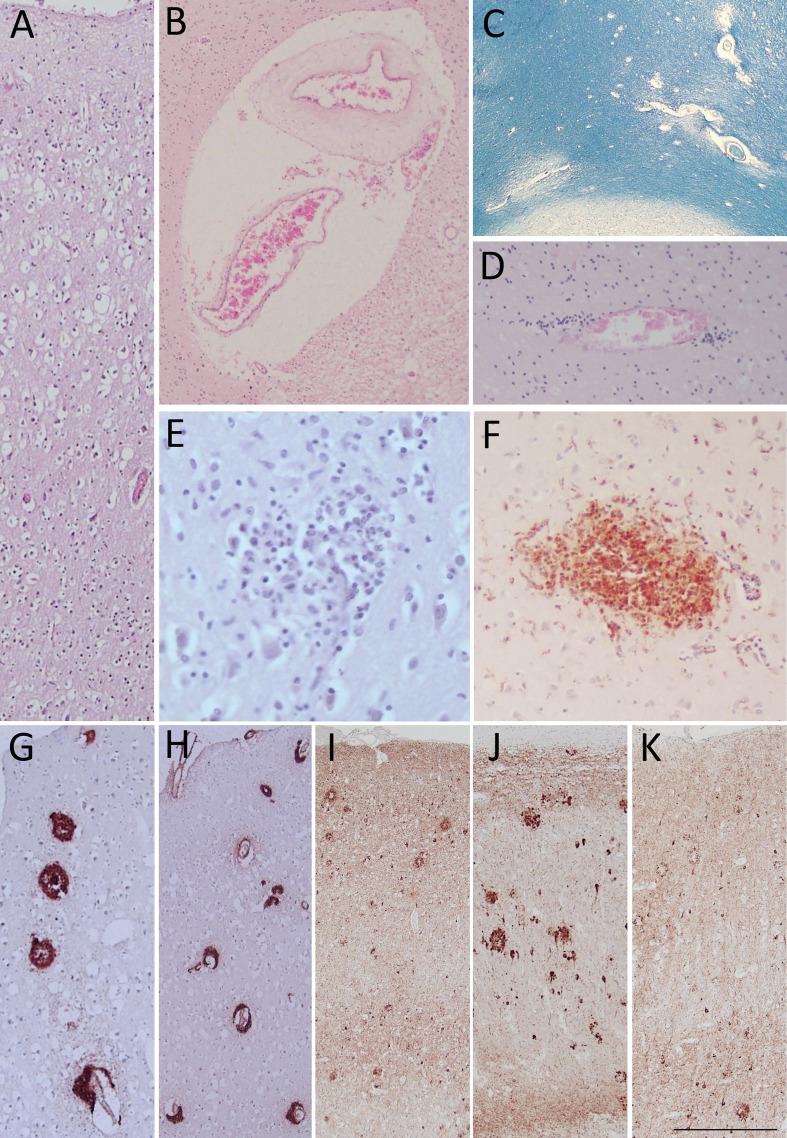
In all COVID-19 cases, the cortical cytoarchitecture was deeply altered, due to severe hypoxia, agonal changes and postmortem artifacts. Hematoxylin-Eosin staining showed widespread edema with marked perineuronal space dilatation and neuronal loss in cortical gray matter, particularly in the supragranular layers (A). No fresh vascular lesions were found but some COVID-19 cases had pre-existing small vessel disease (SVD). Moderate to severe SVD was present in cases 2, 4 and 8, with perivascular space dilatation, arteriolosclerosis (Fig. 1 B), and myelin loss (Fig. 1C). Moderate to severe SVD was also present in non-COVID cases (cases 11, 14, 15 and 16). Nobody showed a clear picture of vasculitis but COVID-19 cases frequently had perivascular infiltrates that represented the main inflammatory feature in subjects without dementia and with a low amount of viral RNA (Fig. 1D). On the other hand, COVID-19 cases with dementia and higher load of viral RNA presented multifocal inflammatory infiltrates and nodules, mainly in cortical gray matter (Fig. 1E). The nodules were composed almost entirely by CD68-positive microglial cells (Fig. 1F), as activation of innate immunity. Lymphocytes were very rare and viral inclusions were detected in very few cells of the lower brainstem (data not shown). Definite AD diagnosis was made in COVID-19 cases 1, 4, 6, 7, 8 (Fig. 1G-K), and in non-COVID cases 10, 11, 13, 14, and 15. Despite the difference in post-mortem delay between the COVID-19 cases and the ABB controls, they appear to be comparable on a histological level and there is adequate immunohistochemical antigenic detection also in COVID-19 brains.
Fig. 1.
Widespread oedema is clearly evident in supragranular layers of gray matter (A) by Hematoxylin and Eosin (HE) staining. Small vessel disease (SVD) is identified by perivascular space dilatation, features of arteriolosclerosis (B), and myelin loss (C) observed respectively by HE and Luxol Fast Blue stainings. In cases with a low amount of viral RNA perivascular inflammatory infiltrates appear as the most prominent inflammatory feature (D; HE staining); while in cases with dementia and higher amount of viral RNA there are more prominent microglial nodule as shown by both, HE (E) and CD68 antibody (F). In COVID-19 cases affected by Alzheimer’s Disease, 4G8 antibody detects amyloid plaques (G) and, possibly, cerebral amyloid angiopathy (H). In the same cases, AT8 immunoreactivity shows neuritic plaques, tangles and threads in frontal cortex (I), hippocampus (J) and occipital cortex (K). Three COVID-19 cases with dementia are shown in panels A-C and E-K, while panel D shows a COVID-19 case without dementia carrying al low viral RNA load. Images include frontal lobes (A-I), hippocampus (J) and occipital lobe (K). Scale bar: 256 µm (A,G); 340 µm (B,H,I,J,K); 807 µm (C); 122 µm (D,F); 73 µm (E). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. SARS-CoV 2 detection in frontal cortex
RNA extracted from frontal cortex of both COVID-19 patients and controls have been analyzed for SARS-CoV-2 nucleocapsid gene 1 (nCoV_N1) and human Ribonuclease P (hRP), as controls of RNA extraction. The analysis of viral RNA detection by qPCR was positive for viral RNA (nCoV_N1) only in one patient while in all samples the expression of hRP gene was present, demonstrating that the RNA was correctly extracted (Table supplementary 1A). Moreover, we repeated the analysis by ddPCR, a more sensitive technique for RNA detection and we quantified the viral RNA in 8 (88%) samples of 9 examined patients (Table supplementary 1B).
3.3. RNA-seq differentially expressed mRNAs and lncRNAs
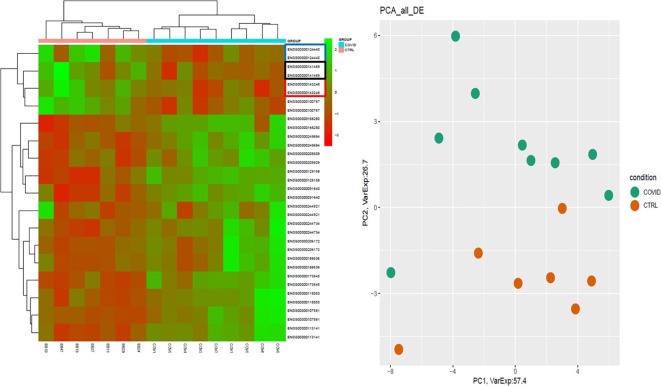
In COVID-19 patients, RNA-seq data reported 11 differentially expressed genes (DEGs), 10 were coding RNAs while 1 was a lncRNA. Concerning coding genes, 4 out of 10 have been found down-regulated while 6 out of 10 up-regulated. Heat-map separately representing the expression levels of all dysregulated mRNAs and lncRNA in COVID-19 and control subjects is represented in Fig. 2 A. Here, different expression profiles in COVID-19 and controls can be visibly distinguished. We also performed a Principal Component Analysis (PCA) shown in Fig. 2B, where COVID-19 patients’ samples are clearly separated from controls cluster.
Fig. 2.
Expression profiles of differentially expressed genes (DEGs) between COVID-19 patients and healthy controls. In panel (A), Heatmap of log-normalized RNA expression counts for patients and controls are represented, while in panel (B) PCA of DEGs is shown.
Moreover, in COVID-19 patients, an interesting data is about the number of deregulated (DE) transcripts in COVID-19 patients compared to controls, in fact, only 10 coding mRNA e 1 lncRNA have been found altered (Table 2 ).
Table 2.
Differentially expressed genes in COVID-19 group respect to controls. Gene name and measured log2FC are reported for each transcript. Only transcripts with |log2(disease sample/healthy control)|≥1. In rtPCR validation column, validation tissue is reported for selected transcripts.
| gene_name | log2FoldChange | gene_biotype | qPCR validation |
|---|---|---|---|
| SLC14A1 | −1,683191231 | protein_coding | |
| HIF3A | −1,496533825 | protein_coding | validated |
| PAPLN | −1,432307747 | protein_coding | |
| RGS5 | −1,295800416 | protein_coding | validated |
| SERGEF | 1,047286497 | protein_coding | |
| NCL | 1,089753339 | protein_coding | |
| ZNF622 | 1,202542764 | protein_coding | |
| HBA2 | 2,369463648 | protein_coding | validated |
| HBB | 2,493031576 | protein_coding | validated |
| HBA1 | 2,715506437 | protein_coding | validated |
| CTB-36O1.7 | 3,439830495 | processed_pseudogene |
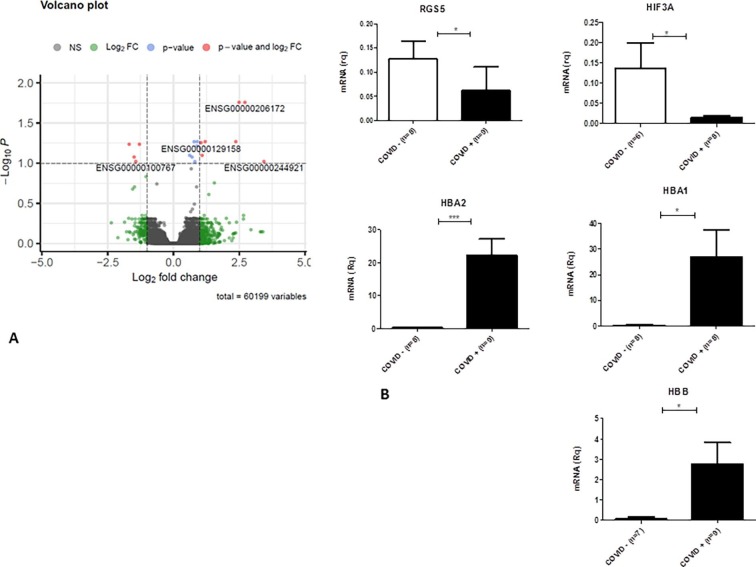
The volcano plot shows the most significant DEGs, validated by qPCR, in COVID-19 patients that confirm the different degree of alteration in the population (Fig. 3 ). We considered the low number of DEGs in COVID-19 patients an interesting datum. The most deregulated genes are up-regulated mRNA coding for hemoglobin subunits (HBA, and HBB) and down-regulated genes associated to hypoxia and infalmmation (SLC14A1, HIF3A and RGS5). This data also correlate with neuropathological findings that showed inflammatory infiltrates in COVID-19 patients’ tissue.
Fig. 3.
Volcano Plot and qPCR validation. Volcano plot of differentially expressed genes (DEGs) between COVID-19 patients and healthy controls (A). Gene ID of the most deregulated transcripts are reported, red dots represent DEGs based on p-value and Fold Change. Most DEGs have been validated by qPCR (B). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Pathway analysis
BioPlanet2019 pathway, GO and KEGG enrichment analyses for DEGs were performed to identify the most important pathways and molecular features associated to DEGs.
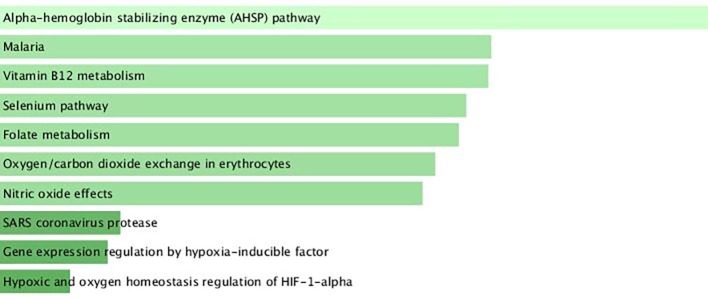
The most relevant data concern pathway analysis that showed that DEGs are associated to SARS coronavirus protease and Alpha hemoglobin stabilizing protein (AHSP) pathway (Fig. 4 ).
Fig. 4.
BioPlanet2019 pathway analysis for DEGs in COVID-19 patients compared to healthy controls. In panel A BioPlanet2019 pathways enriched by deregulated genes are shown. The length of the bar represents the significance of that specific gene-set or term. The brighter the color, the more significant that term is. In panel B is showed the cluster gram of DEGs associated to enriched pathways.
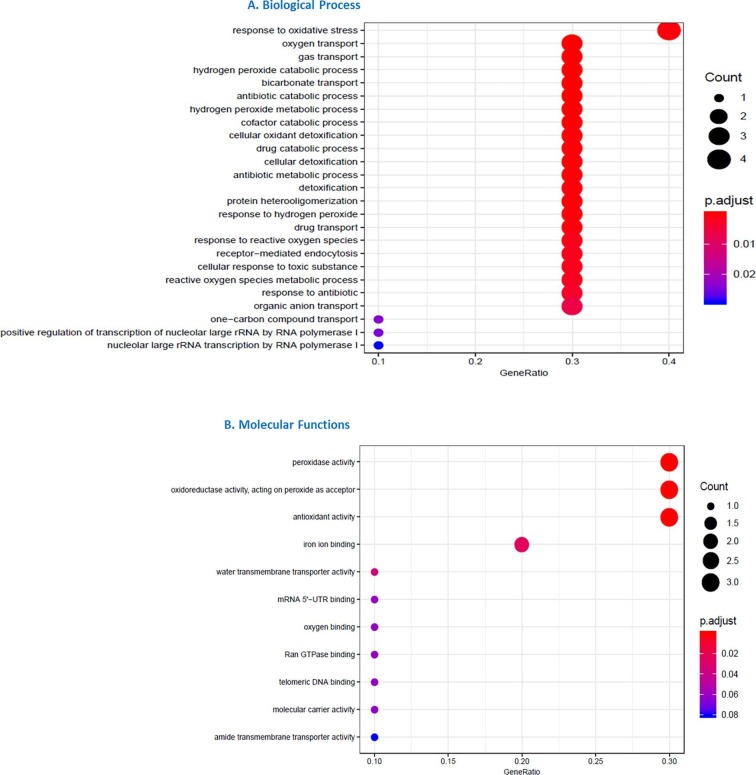
The GO biological processes enriched terms for response to oxidative stress, oxygen and gas transport and hydrogen peroxide catabolic and metabolic process, underline the significant involvement of respiratory aspect in COVID-19 phenotype (Fig. 5 ).
Fig. 5.
GO analysis for DEGs in COVID-19 patients compared to healthy controls by AnnotationHub. DEG enriched GO terms for Biological process (A) and Molecular Function (B) are shown.
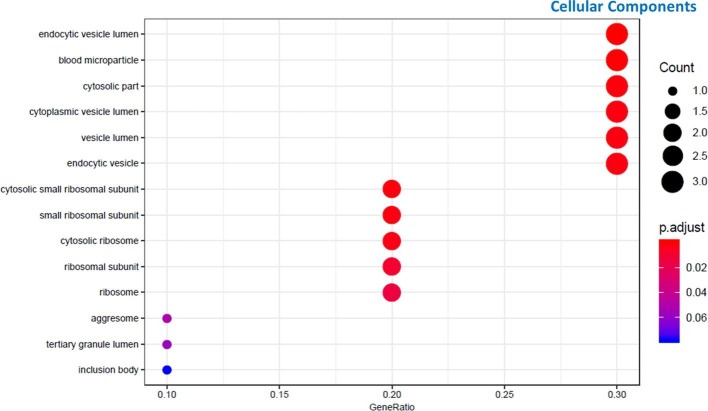
Moreover, also cellular detoxification and response to toxic substance pathways have been found involved. Analysis of molecular functions also confirmed the involvement of peroxide and antioxidant activity. About cellular component, the most representative element was vesicles lumen (Supplementary figure 1).
4. Discussion
SARS-CoV-2 infection can cause severe pulmonary disease and complications with possible multi-organ consequences involving CNS (Renu et al., 2020). The most frequent neurological manifestation of COVID-19 in patients with dementia is a non-specific encephalopathy with behavioral changes, such as psychomotor retardation and delirium, typical of several infective-inflammatory diseases (Poloni et al., 2020a, Poloni et al., 2020b). These clinical manifestations may be due to non-specific pathological phenomena that can be attributed to inflammatory state, hypoxia, and sepsis, possibly superimposed on pre-existing neurodegenerative or vascular pathologies. At gross neuropathological examination, we could not find fresh macroscopic vascular lesions. All COVID-19 brains show congestion, oedema, and neuronal damage. These alterations are not SARS-CoV-2-specific but related to hypoxic-agonic and postmortem phenomena. Also, other pathological findings such as AD pathology and SVD are clearly pre-existent conditions and cannot be considered virus-specific,. On the other hand, we observed perivascular inflammatory infiltrates and nodules that are, probably, attributable to SARS-CoV-2. The inflammatory infiltrates consist essentially of cells of the innate immunity (monocyte-macrophage-microglia series) with very rare lymphocytes. Although the small number of cases does not allow us to draw firm conclusions, it is interesting to note that the inflammatory load appears greater in cases with dementia and with the greatest amount of viral RNA.
For a deeper understanding of the pathogenic processes induced by SARS-CoV-2 in the CNS, the neuropathological data should be supplemented with biochemical data. Therefore, detecting the presence of the virus in CNS, and the biochemical alterations induced by the virus, is crucial for the interpretation of the brain pathophysiology of COVID-19. Viral SARS-CoV-2 infection causes alterations in transcriptome of affected cells and tissues, as lung and peripheral blood mononuclear cells (PBMCs) (Islam and Khan, 2020, Yang et al., 2020, Xiong et al., 2020). Starting from the evidence that transcriptomic profile may be altered by SARS-CoV-2 infection and considering previous data (Asadi-Pooya and Simani, 2020, Baig et al., 2020, Koralnik and Tyler, 2020) about a CNS involvement, we performed RNA-sequencing of frontal basal cortex to characterize the gene expression profile of COVID-19 patients. First, we checked for the presence of viral RNA demonstrating that SARS-CoV-2 RNA was detectable by qRT-PCR only in one sample, while using a more sensitive method, ddPCR, it was appreciable in almost all COVID-19 samples (88%). Interestingly, literature reports that in COVID-19 brain tissue (not specifically in frontal cortex) viral RNA is detectable in about 40–50% of samples or fewer (Matschke et al., 2020, Solomon et al., 2020). In this work, we showed that detection of virus in brain tissue was increasable to 90% through ddPCR (Suo et al., 2020). Indeed, it should be considered that the time elapsed between the acute infection and the death is relevant for virus clearance and, probably, affects the presence of SARS-CoV-2 inside the brain. Thus, it is necessary to use an extremely sensitive method. In any case, the low amount of viral RNA detected does not allow to distinguish whether the virus is located in brain parenchyma or in brain blood vessels. However, these data, together with the absence of a specific neuropathologic picture of encephalitis, testify to the absence of viral replicative activity within the brain.
About transcriptome profiling results, we found that 11 genes (10 mRNAs and 1 lncRNA) were differential expressed when frontal cortex of COVID-19 patients was compared to controls. These genes fall into categories including hypoxia, hemoglobin stabilizing protein, hydrogen peroxide processes, which all play important roles in SARS-CoV-2 infection (Cavezzi et al., 2020, Jahani et al., 2020, Thomas et al., 2020). Interesting genes HIF3A, SLC14A1 and RGS5 have been found down-regulated. HIF3A belongs to hypoxia-inducible factor family genes; they are transcription factors that respond to decrease in cellular environment oxygen or hypoxia (Smith et al., 2008, Wilkins et al., 2016). Inhibition of HIF-1 and dysregulation in Akt/mTOR/HIF-1 has been found in SARS-CoV-2 infected cells (Appelberg et al., 2020) and it has been shown that inhibition of HIF-1 can promote replication of influenza A virus and severe inflammation mediated via promotion of autophagy (Zhao et al., 2020). Also increasing HIF-1α level promotes the defense capacity of macrophages in cases of infection (Zhao et al., 2020). Our data showed a reduction of HIF suggesting a possible inhibition of defense capacity (Zhao et al., 2020). In fact, HIF pathway usually plays a protective role after injury and promotes cell recovery that in COVID-19 patients is not possible due to HIF down-regulation (Corrado and Fontana, 2020). Previous work reported that the HIF response is variable depending upon the type of hypoxic stress (Mandic et al., 2018). Particularly, HIF was induced by chronic or intermittent hypoxia (Powell and Fu, 2008). Our COVID-19 cases had a severe acute hypoxia due pulmonary failure, which was not corrected through mechanical ventilation. As highlighted by the neuropathological picture, this peculiar condition caused an extreme hypoxic suffering of the brain, which, probably, induced a down-regulation of the HIF response, thus creating a vicious circle that might be important in COVID-19 pathogenesis. Moreover, SLC2A1 has been described as part of HIF-1 signaling (Appelberg et al., 2020, Dengler et al., 2014), and also RGS5, involved in endothelial apoptosis, is regulated by HIF (Jin et al., 2009).
About up-regulated genes, the most relevant data concerned hemoglobin subunits (HBB, A1 and A2) increase. Hb up-regulation may be linked to downregulation of mTOR activity, suggesting that the overexpression of Hb may act as response to mTOR downregulation (Codrich et al., 2017). Moreover, Hb overexpression may be a protective system against oxidative stress. In fact, it has been demonstrated that Hb overexpression may be neuroprotective (Amri et al., 2017).
About lncRNA CTB-36O1.7, an uncharacterized gene, it has been found associated to Multiple Sclerosis in brain (Chiricosta et al., 2020). Although its role is still unknown, this finding may confirm a role of lncRNA CTB-36O1.7 in inflammation and in the regulation of microglia. The neuropathological picture observed in our COVID-19 cases is characterized by microglial activation, in the absence of clear signs of encephalitis due to viral replication. Thus, our data corroborates the involvement of lncRNA CTB-36O1.7 in the activation of microglia and innate immunity rather than in adaptive immunity linked to an active infection.
5. Conclusions
In conclusion, our data confirm the presence of viral RNA at very low amount in frontal cortex tissues. As a matter of fact, COVID-19 detection is variable; therefore a sensitive method is required to reveal the viral RNA.
This work demonstrates that the quantity of viral RNA is minimal, thus, it is likely that SARS-CoV-2 does not actively infect and replicate in the brain. Also, this work showed that hypoxia, a well-known condition associated to COVID-19 infection, is a multi-organ feature (lung, kidney, and hearth) that marks even the CNS, and specifically brain cortex. Our data showed an important deregulation of hypoxia inducting factor system that inhibits defense system from infection. Our data also suggested an activation of protective mechanism by Hb in response to oxidative stress and deficit in mTOR system. Furthermore, as demonstrated by the neuropathological picture and the up-regulation of lncRNA CTB-36O1.7, the activation of microglia seems to have an important role in the pathogenesis of the neurological manifestations of COVID-19. Therefore, the topography and intensity of microglial activation should be investigated further, and comparison studies with non-SARS-CoV-2 cases should be conducted.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank the Abbiategrasso Brain Bank (ABB) donors and the COVID-19 patients who donated the noblest organ of their body.
We also would like to thank Dr. Antonio Traversi and Prysmian Group for the support.
Funding
Project was funded by the Italian Ministry of Health (Ricerca Corrente 2020)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbi.2021.05.012.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
References
- Ahmad I., Rathore F.A. Neurological manifestations and complications of COVID-19: A literature review. J Clin Neurosci. 2020;77:8–12. doi: 10.1016/j.jocn.2020.05.017. PMID: 32409215; PMCID: PMC7200361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amri F., Ghouili I., Tonon M., Amri M., Masmoudi-Kouki O. Hemoglobin-Improved Protection in Cultured Cerebral Cortical Astroglial Cells: Inhibition of Oxidative Stress and Caspase Activation. Frontiers in Endocrinology. 2017;8:67. doi: 10.3389/fendo.2017.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- K.S. Appelberg S. Gupta svensson akujsärvi, Sara & Ambikan, Anoop & Mikaeloff, Flora & Saccon, Elisa & Végvári, Akos & Benfeitas, Rui & Sperk, Maike & Ståhlberg, Marie & Krishnan, Shuba & Singh, Kamal & Penninger, Josef & Mirazimi, Ali & Neogi, Ujjwal. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerging Microbes & Infections. 9 2020 1 36 10.1080/22221751.2020.1799723. [DOI] [PMC free article] [PubMed]
- Asadi-Pooya A.A., Simani L. Central nervous system manifestations of COVID-19: A systematic review. J Neurol Sci. 2020;15(413) doi: 10.1016/j.jns.2020.116832. PMID: 32299017; PMCID: PMC7151535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig A.M., Khaleeq A., Ali U., Syeda H. Evidence of the COVID-19 Virus Targeting the CNS: Tissue Distribution, Host-Virus Interaction, and Proposed Neurotropic Mechanisms. ACS Chem Neurosci. 2020;11(7):995–998. doi: 10.1021/acschemneuro.0c00122. PMID: 32167747; PMCID: PMC7094171. [DOI] [PubMed] [Google Scholar]
- Cavezzi A., Troiani E., Corrao S. COVID-19: hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin Pract. 2020;10(2):1271. doi: 10.4081/cp.2020.1271. Published 2020 May 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiricosta L., Gugliandolo A., Bramanti P., Mazzon E. Could the Heat Shock Proteins 70 Family Members Exacerbate the Immune Response in Multiple Sclerosis? An in Silico Study. Genes (Basel). 2020;11(6):615. doi: 10.3390/genes11060615. Published 2020 Jun 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codrich M., Bertuzzi M., Russo R., Francescatto M., Espinoza S., Zentilin L., Giacca M., Cesselli D., Beltrami A.P., Ascenzi P., Zucchelli S., Persichetti F., Leanza G., Gustincich S. Neuronal hemoglobin affects dopaminergic cells' response to stress. Cell Death Dis. 2017;8(1) doi: 10.1038/cddis.2016.458. PMID: 28055011; PMCID: PMC5386368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrado C., Fontana S. Hypoxia and HIF Signaling: One Axis with Divergent Effects. Int. J. Mol. Sci. 2020;21:5611. doi: 10.3390/ijms21165611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler V.L., Galbraith M., Espinosa J.M. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49(1):1–15. doi: 10.3109/10409238.2013.838205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S. Catch me if you can: SARS-CoV-2 detection in brains of deceased patients with COVID-19. Lancet Neurol. 2020;19(11):883–884. doi: 10.1016/S1474-4422(20)30371-9. Epub 2020 Oct 5. PMID: 33031734; PMCID: PMC7535625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi S., Zucca S., Pandini C., Diamanti L., Bordoni M., Sproviero D., Arigoni M., Olivero M., Pansarasa O., Ceroni M., Calogero R., Cereda C. Long non-coding and coding RNAs characterization in Peripheral Blood Mononuclear Cells and Spinal Cord from Amyotrophic Lateral Sclerosis patients. Sci Rep. 2018;8(1):2378. doi: 10.1038/s41598-018-20679-5. PMID: 29402919; PMCID: PMC5799454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. China Medical Treatment Expert Group for Covid-19. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. Epub 2020 Feb 28. PMID: 32109013; PMCID: PMC7092819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C., Collange O., Boulay C., Fafi-Kremer S., Ohana M., Anheim M., Meziani F. Neurologic Features in Severe SARS-CoV-2 Infection. N Engl J Med. 2020;382(23):2268–2270. doi: 10.1056/NEJMc2008597. Epub 2020 Apr 15. PMID: 32294339; PMCID: PMC7179967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497-506. doi: 10.1016/S0140-6736(20)30183-5. Epub 2020 Jan 24. Erratum in: Lancet. 2020 Jan 30; PMID: 31986264; PMCID: PMC7159299. [DOI] [PMC free article] [PubMed]
- Huang R, Grishagin I, Wang Y, et al. The NCATS BioPlanet - An Integrated Platform for Exploring the Universe of Cellular Signaling Pathways for Toxicology, Systems Biology, and Chemical Genomics. Front Pharmacol. 2019;10:445. Published 2019 Apr 26. doi:10.3389/fphar.2019.00445. [DOI] [PMC free article] [PubMed]
- Huang C., et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam ABMMK, Khan MA. Lung transcriptome of a COVID-19 patient and systems biology predictions suggest impaired surfactant production which may be druggable by surfactant therapy. Sci Rep. 2020 Nov 10;10(1):19395. doi: 10.1038/s41598-020-76404-8. [DOI] [PMC free article] [PubMed]
- Jahani M., Dokaneheifard S., Mansouri K. Hypoxia: A key feature of COVID-19 launching activation of HIF-1 and cytokine storm. J Inflamm. 2020;17:33. doi: 10.1186/s12950-020-00263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., An X., Ye Z., Cully B., Wu J., Li J. RGS5, a hypoxia-inducible apoptotic stimulator in endothelial cells. J Biol Chem. 2009;284(35):23436–23443. doi: 10.1074/jbc.M109.032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koralnik I.J., Tyler K.L. COVID-19: A Global Threat to the Nervous System. Ann Neurol. 2020;88(1):1–11. doi: 10.1002/ana.25807. PMID: 32506549; PMCID: PMC7300753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milica Mandic Velislava Tzaneva Vincent Careau Perry, S.f. Hif-1α paralogs play a role in the hypoxic ventilatory response of larval and adult zebrafish (Danio rerio) The Journal of Experimental Biology. 222 2018 jeb.195198 10.1242/jeb.195198. [DOI] [PubMed]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., Miao X., Li Y., Hu B. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan. China. JAMA Neurol. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. PMID: 32275288; PMCID: PMC7149362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschke J, Lütgehetmann M, Hagel C, Sperhake JP, Schröder AS, Edler C, Mushumba H, Fitzek A, Allweiss L, Dandri M, Dottermusch M, Heinemann A, Pfefferle S, Schwabenland M, Sumner Magruder D, Bonn S, Prinz M, Gerloff C, Püschel K, Krasemann S, Aepfelbacher M, Glatzel M. Neuropathology of patients with COVID-19 in Germany: a post-mortem case series. Lancet Neurol. 2020 Nov;19(11):919-929. doi: 10.1016/S1474-4422(20)30308-2. Epub 2020 Oct 5. PMID: 33031735; PMCID: PMC7535629. [DOI] [PMC free article] [PubMed]
- Politi LS, Salsano E, Grimaldi M. Magnetic Resonance Imaging Alteration of the Brain in a Patient with Coronavirus Disease 2019 (COVID-19) and Anosmia. JAMA Neurology; Published Online May 29, 2020; doi:10.1001/jamaneurol.2020.2125. [DOI] [PubMed]
- Poloni TE, Carlos AF, Cairati M, Cutaia C, Medici V, Marelli E, Ferrari D, Galli A, Bognetti P, Davin A, Cirrincione A, Ceretti A, Cereda C, Ceroni M, Tronconi L, Vitali S, Guaita A. Prevalence and prognostic value of Delirium as the initial presentation of COVID-19 in the elderly with dementia: An Italian retrospective study. EClinicalMedicine. 2020 Sep;26:100490. doi: 10.1016/j.eclinm.2020.100490. Epub 2020 Jul 30. PMID: 32838241; PMCID: PMC7392565. [DOI] [PMC free article] [PubMed]
- Poloni TE, Medici V, Carlos AF, Davin A, Ceretti A, Mangieri M, Cassini P, Vaccaro R, Zaccaria D, Abbondanza S, Bordoni M, Fantini V, Fogato E, Cereda C, Ceroni M, Guaita A.Abbiategrasso Brain Bank Protocol for Collecting, Processing and Characterizing Aging Brains. J Vis Exp. 2020 Jun 3;(160). doi: 10.3791/60296. [DOI] [PubMed]
- Powell F.L., Fu Z. HIF-1 and ventilatory acclimatization to chronic hypoxia. Respir Physiol Neurobiol. 2008;164(1–2):282–287. doi: 10.1016/j.resp.2008.07.017. PMID: 18708172; PMCID: PMC2700119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renu K, Prasanna PL, Valsala Gopalakrishnan A. Coronaviruses pathogenesis, comorbidities and multi-organ damage - A review. Life Sci. 2020 Aug 15;255:117839. doi: 10.1016/j.lfs.2020.117839. Epub 2020 May 22. PMID: 32450165; PMCID: PMC7243768. [DOI] [PMC free article] [PubMed]
- Smith T.G., Robbins P.A., Ratcliffe P.J. The human side of hypoxia-inducible factor. Br J Haematol. 2008;141(3):325–334. doi: 10.1111/j.1365-2141.2008.07029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon I.H., Normandin E., Bhattacharyya S., Mukerji S.S., Keller K., Ali A.S., et al. Neuropathological Features of Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMc2019373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Q., Wang X., Sajid M., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Y., Zhang Q., Liu Y., Xiong Y., Chen Y. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerging microbes & infections. 2020;9(1):1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Stefanoni D, Dzieciatkowska M, Issaian A, Nemkov T, Hill RC, Francis RO, Hudson KE, Buehler PW, Zimring JC, Hod EA, Hansen KC, Spitalnik SL, D'Alessandro A. Evidence of Structural Protein Damage and Membrane Lipid Remodeling in Red Blood Cells from COVID-19 Patients. J Proteome Res. 2020 Nov 6;19(11):4455-4469. doi: 10.1021/acs.jproteome.0c00606. Epub 2020 Oct 26. PMID: 33103907; PMCID: PMC7640979. [DOI] [PMC free article] [PubMed]
- von Weyhern C.H., Kaufmann I., Neff F., Kremer M. Early evidence of pronounced brain involvement in fatal COVID-19 outcomes. Lancet. 2020;395(10241) doi: 10.1016/S0140-6736(20)31282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann D., Sperhake J.P., Lutgehetmann M. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann Intern Med. 2020;173:268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins S.E., Abboud M.I., Hancock R.L., Schofield C.J. Targeting Protein-Protein Interactions in the HIF System. ChemMedChem. 2016;11:773. doi: 10.1002/cmdc.201600012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wu S., Yu Z., Huang J., Zhong X., Liu X., Zhu H., Xiao L., Deng Q., Sun W. Transcriptomic analysis reveals novel mechanisms of SARS-CoV-2 infection in human lung cells. Immun Inflamm Dis. 2020;8(4):753–762. doi: 10.1002/iid3.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L., Han Y., He Q. clusterProfiler: an R package for comparing biological themes among gene clusters.OMICS: A Journal of. Integrative Biology. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Shen D., Zhou H., Liu J., Chen S. Guillain-Barré syndrome associated with SARS-CoV-2 infection: causality or coincidence? Lancet Neurol [Internet]. 2020;19(5):383–384. doi: 10.1016/S1474-4422(20)30109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito A., Alfonsi E., Franciotta D., Todisco M., Gastaldi M., Cotta Ramusino M., Ceroni M., Costa A. COVID-19 and Guillain-Barré Syndrome: A Case Report and Review of Literature. Front Neurol. 2020;21(11):909. doi: 10.3389/fneur.2020.00909. PMID: 32973665; PMCID: PMC7471770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucca S., Gagliardi S., Pandini C., Diamanti L., Bordoni M., Sproviero D., Arigoni M., Olivero M., Pansarasa O., Ceroni M., Calogero R., Cereda C. RNA-Seq profiling in peripheral blood mononuclear cells of amyotrophic lateral sclerosis patients and controls. Sci Data. 2019;5(6) doi: 10.1038/sdata.2019.6. PMID: 30720798; PMCID: PMC6362931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.