Abstract
Survival analysis models the distribution of time until an event of interest, such as discharge from the hospital or admission to the ICU. When a model’s predicted number of events within any time interval is similar to the observed number, it is called well-calibrated. A survival model’s calibration can be measured using, for instance, distributional calibration (D-CALIBRATION) [Haider et al., 2020] which computes the squared difference between the observed and predicted number of events within different time intervals. Classically, calibration is addressed in post-training analysis. We develop explicit calibration (X-CAL), which turns D-CALIBRATION into a differentiable objective that can be used in survival modeling alongside maximum likelihood estimation and other objectives. X-CAL allows practitioners to directly optimize calibration and strike a desired balance between predictive power and calibration. In our experiments, we fit a variety of shallow and deep models on simulated data, a survival dataset based on MNIST, on length-of-stay prediction using MIMIC-III data, and on brain cancer data from The Cancer Genome Atlas. We show that the models we study can be miscalibrated. We give experimental evidence on these datasets that X-CAL improves D-CALIBRATION without a large decrease in concordance or likelihood.
1. Introduction
A core challenge in healthcare is to assess the risk of events such as onset of disease or death. Given a patient’s vitals and lab values, physicians should know whether the patient is at risk for transfer to a higher level of care. Accurate estimates of the time-until-event help physicians assess risk and accordingly prescribe treatment strategies: doctors match aggressiveness of treatment against severity of illness. These predictions are important to the health of the individual patient and to the allocation of resources in the healthcare system, affecting all patients.
Survival Analysis formalizes this risk assessment by estimating the conditional distribution of the time-until-event for an outcome of interest, called the failure time. Unlike supervised learning, survival analysis must handle datapoints that are censored: their failure time is not observed, but bounds on the failure time are. For example, in a 10 year cardiac health study [Wilson et al., 1998, Vasan et al., 2008], some individuals will remain healthy over the study duration. Censored points are informative, as we can learn that someone’s physiology indicates they are healthy-enough to avoid onset of cardiac issues within the next 10 years.
A well-calibrated survival model is one where the predicted number of events within any time interval is similar to the observed number [Pepe and Janes, 2013]. When this is the case, event probabilities can be interpreted as risk and can be used for downstream tasks, treatment strategy, and human-computable risk score development [Sullivan et al., 2004, Demler et al., 2015, Haider et al., 2020]. Calibrated conditional models enable accurate, individualized prognosis and may help prevent giving patients misinformed limits on their survival, such as 6 months when they would survive years. Poorly calibrated predictions of time-to-event can misinform decisions about a patient’s future.
Calibration is a concern in today’s deep models. Classical neural networks that were not wide or deep by modern standards were found to be as calibrated as other models after the latter were calibrated (boosted trees, random forests, and SVMs calibrated using Platt scaling and isotonic regression) [Niculescu-Mizil and Caruana, 2005]. However, deeper and wider models using batchnorm and dropout have been found to be overconfident or otherwise miscalibrated [Guo et al., 2017]. Common shallow survival models such as the Weibull Accelerated Failure Times (AFT) model may also be miscalibrated [Haider et al., 2020]. We explore shallow and deep models in this work.
Calibration checks are usually performed post-training. This approach decouples the search for a good predictive model and a well-calibrated one [Song et al., 2019, Platt, 1999, Zadrozny and Elkan, 2002]. Recent approaches tackle calibration in-training via alternate loss functions. However, these may not, even implicitly, optimize a well-defined calibration measure, nor do they allow for explicit balance between prediction and calibration [Avati et al., 2019]. Calibration during training has been explored recently for binary classification [Kumar et al., 2018]. Limited evaluations of calibration in survival models can be done by considering only particular time points: this model is well-calibrated for half-year predictions. Recent work considers D-CALIBRATION [Haider et al., 2020], a holistic measure of calibration of time-until-event that measures calibration of distributions.
In this work, we propose to improve calibration by augmenting traditional objectives for survival modeling with a differentiable approximation of D-CALIBRATION, which we call explicit calibration (X-CAL). X-CAL is a plug-in objective that reduces obtaining good calibration to an optimization problem amenable to data sub-sampling. X-CAL helps build well-calibrated versions of many existing models and controls calibration during training. In our experiments 2, we fit a variety of shallow and deep models on simulated data, a survival dataset based on MNIST, on length-of-stay prediction using MIMIC-III data, and on brain cancer data from The Cancer Genome Atlas. We show that the models we study can be miscalibrated. We give experimental evidence on these datasets that X-CAL improves D-CALIBRATION without a large decrease in concordance or likelihood.
2. Defining and Evaluating Calibration in Survival Analysis
Survival analysis models the time t > 0 until an event, called the failure time. t is often assumed to be conditionally distributed given covariates x. Unlike typical regression problems, there may also be censoring times c that determine whether t is observed. We focus on right-censoring in this work, with observations (u, δ, x) where u = min(t, c) and . If δ = 1 then u is a failure time. Otherwise u is a censoring time and the datapoint is called censored. Censoring times may be constant or random. We assume censoring-at-random: t ⫫ c | x.
We denote the joint distribution of (t, x) by P and the conditional cumulative distribution function (CDF) of t | x by F (sometimes denoting the marginal CDF by F when clear). Whenever distributions or CDFs have no subscript parameters, they are taken to be true data-generating distributions and when they have parameters θ they denote a model. We give more review of key concepts, definitions, and common survival analysis models in Appendix A.
2.1. Defining Calibration
We first establish a common definition of calibration for binary outcome. Let x be covariates and let d be a binary outcome distributed conditional on x. Let them have joint distribution P(d, x). Define riskθ(x) as the modeled probability Pθ(d = 1 | x), a deterministic function of x. Pepe and Janes [2013] define calibration as the condition that
| (1) |
That is, the frequency of events is r among subjects whose modeled risks are equal to r. For a survival problem with joint distribution P(t, x), we can define risk to depend on an observed failure time instead of the binary outcome d = 1. With Fθ as the model CDF, the definition of risk for survival analysis becomes riskθ(t, x) = Fθ(t | x), a deterministic function of (t, x). Then perfect calibration is the condition that, for all sub-intervals I = [a, b] of [0, 1],
| (2) |
This is because, for continuous F (an assumption we keep for the remainder of the text), CDFs transform samples of their own distribution to Unif(0, 1) variates. Thus, when model predictions are perfect and Fθ = F, the probability that Fθ(t | x) takes a value in interval I is equal to |I|. Since the expectation is taken over x, the same holds when Fθ(t | x) = F(t), the true marginal CDF.
2.2. Evaluating Calibration
Classical tests and their recent modifications assess calibration of survival models for a particular time of interest t∗ by comparing observed versus modeled event frequencies [Lemeshow and Hosmer Jr, 1982, Grønnesby and Borgan, 1996, D’agostino and Nam, 2003, Royston and Altman, 2013, Demler et al., 2015, Yadlowsky et al., 2019]. They apply the condition in Equation (1) for the classification task t < t∗ | x. These tests are limited in two ways 1) it is not clear how to combine calibration assessments over the entire range of possible time predictions [Haider et al., 2020] and 2) they answer calibration in a rigid yes/no fashion with hypothesis testing. We briefly review these tests in Appendix A.
D-calibration
Haider et al. [2020] develop distributional calibration (D-CALIBRATION) to test the calibration of conditional survival distributions across all times. D-CALIBRATION uses the condition in Equation (2) and checks the extent to which it holds by evaluating the model conditional CDF on times in the data and checking that these CDF evaluations are uniform over [0, 1]. This uniformity ensures that observed and predicted numbers of events within each time interval match.
To set this up formally, recall that F denotes the unknown true CDF. For each individual x, let Fθ(t | x) denote the modeled CDF of time-until-failure. To measure overall calibration error, D-CALIBRATION accumulates the squared errors of the equality condition in Equation (2) over sets that cover [0, 1]:
| (3) |
The collection is chosen to contain disjoint contiguous intervals I ⊆ [0, 1], that cover the whole interval [0, 1]. Haider et al. [2020] perform a χ2-test to determine whether a model is well-calibrated, replacing the expectation in Equation (3) with a Monte Carlo estimate.
Properties
Setting aside the hypothesis testing step, we highlight two key properties of D-CALIBRATION. First, D-CALIBRATION is zero for the correct conditional model. This ensures that the correct model is not wrongly mischaracterized as miscalibrated. Second, for a given model class and dataset, smaller D-CALIBRATION means a model is more calibrated. This means that it makes sense to minimize D-CALIBRATION. Next, we make use of these properties and turn D-CALIBRATION into a differentiable objective.
3. X-cal: A Differentiable Calibration Objective
We measure calibration error with D-CALIBRATION (Equation (3)) and propose to incorporate it into our training and minimize it directly. However, the indicator function poses a challenge for optimization. Instead, we derive a soft version of D-CALIBRATION using a soft set membership function. We then develop an upper-bound to soft D-CALIBRATION that we call X-CAL that supports subsampling for stochastic optimization with batch data.
3.1. Soft Membership D-CALIBRATION
We replace the membership indicator for a set I with a differentiable function. Let γ > 0 be a temperature parameter. Let σ(x) = (1 + exp[−x])−1. For point u and the set I = [a, b], define soft membership ζγ as
| (4) |
where γ → ∞ makes membership exact. This is visualized in Figure 2 in Appendix G. We propose the following differentiable approximation to Equation (3), which we call soft D-CALIBRATION, for use in a calibration objective:
| (5) |
We find that γ = 104 allows for close-enough approximation to optimize exact D-CALIBRATION.
3.2. Stochastic Optimization via Jensen’s Inequality
Soft D-CALIBRATION squares an expectation over the data, meaning that its gradient includes a product of two expectations over the same data. Due to this, it is hard to obtain a low-variance, unbiased gradient estimate with batches of data, which is important for models that rely on stochastic optimization. To remedy this, we develop an upper-bound on soft D-CALIBRATION, which we call X-CAL, whose gradient has an easier unbiased estimator.
Let Rγ,θ(t, x, I) denote the contribution to soft D-CALIBRATION error due to one set I and a single sample (t, x) in Equation (5): Rγ,θ(t, x, I) := ζγ (Fθ(t | x); I) − |I|. Then soft D-CALIBRATION can be written as:
For each term in the sum over sets I, we proceed by in two steps. First, replace the expectation over data with an expectation over sets of samples of the mean of Rγ,θ where S is a set of size M. Second, use Jensen’s inequality to switch the expectation and square.
| (6) |
We call this upper-bound X-CAL and denote it by . To summarize, by soft indicator approximation and by Jensen’s inequality. As M → ∞, the slack introduced due to Jensen’s inequality vanishes (in practice we are constrained by the size of the dataset). We now derive the gradient with respect to θ, using :
| (7) |
We estimate Equation (7) by sampling batches S of size M from the empirical data.
Analyzing this gradient demonstrates how X-CAL works. If the fraction of points in bin I is larger than |I|, X-CAL pushes points out of I. The gradient of ζγ pushes points in the first half of the bin to have smaller CDF values and similarly points in the second half are pushed upwards.
While this works well for intervals not at the boundary of [0, 1], some care must be taken at the boundaries. CDF values in the last bin may be pushed to one and unable to leave the bin. Since the maximum CDF value is one, for any b > 1. Making use of this property, X-CAL extends the right endpoint of the last bin so that all CDF values are in the first half of the bin and therefore are pushed to be smaller. The boundary condition near zero is similar. We provide further analysis in Appendix I.
X-CAL can be added to loss functions such as negative log likelihood (NLL) and other survival modeling objectives such as Survival-CRPS (CRPS) [Avati et al., 2019]. For example, the full X-CALIBRATED maximum likelihood objective for a model Pθ and λ > 0 is:
| (8) |
Choosing γ
For small γ, soft D-CALIBRATION is a poor approximation to D-CALIBRATION. For large γ, gradients vanish, making it hard to optimize D-CALIBRATION. We find that setting γ = 10000 worked in all experiments. We evaluate the choice of γ in Appendix G.
Bound Tightness
The slack in Jensen’s inequality does not adversely affect our experiments in practice. We successfully use small batches, e.g. < 1000, for datasets such as MNIST. We always report exact D-CALIBRATION in the results. We evaluate the tightness of this bound and show that models ordered by the upper-bound are ordered in D-CALIBRATION the same way in Appendix H.
3.3. Handling Censored Data
In presence of right-censoring, failure times are censored more often than earlier times. So, applying the true CDF to only uncensored failure times results in a non-uniform distribution skewed to smaller values in [0, 1]. Censoring must be taken into account.
Let x be a censored point with observed censoring time u and unobserved failure time t. Recall that . In this case c = u = u and δ = 0. Let Ft = F(t | x), Fc = F(c | x), and Fu = F(u | x). We first state the fact that, under t ⫫ c | x, a datapoint observed to be censored at time u has Ft ∼ Unif(Fu, 1) for true CDF F (proof in Appendix C). This means that we can compute the probabilty that t falls in each bin I = [a, b]:
| (9) |
Haider et al. [2020] make this observation and suggest a method for handling censoring points: they contribute in place of the unobserved :
| (10) |
This estimator does not change the expectation defining D-CALIBRATION, thereby preserving the property that D-CALIBRATION is 0 for a calibrated model. We soften Equation (9) with:
where we have used a one-sided soft indicator for in the right-hand term. We use ζγ,cens in place of ζγ for censored points in soft D-CALIBRATION. This gives the following estimator for soft D-CALIBRATION with censoring:
| (11) |
The upper-bound of Equation (11) and its corresponding gradient can be derived analogously to the uncensored case. We use these in ours experiments on censored data.
4. Experiments
We study how X-CAL allows the modeler to optimize for a specified balance between prediction and calibration. We augment maximum likelihood estimation with X-CAL for various settings of coefficient λ, where λ = 0 corresponds to vanilla maximum likelihood. Maximum likelihood for survival analysis is described in Appendix A (Equation (12)). For the log-normal experiments, we also use Survival-CRPS (CRPS) [Avati et al., 2019] with X-CAL since S-CRPS enjoys a closed-form for log-normal. S-CRPS was developed to produce calibrated survival models but it optimizes neither a calibration measure nor a traditional likelihood. See Appendix B for a description of S-CRPS.
Models, Optimization, and Evaluation
We use log-normal, Weibull, Categorical and Multi-Task Logistic Regression (MTLR) models with various linear or deep parameterizations. For the discrete models, we optionally interpolate their CDF (denoted in the tables by NI for not-interpolated and I for interpolated). See Appendix E for general model descriptions. Experiment-specific model details may be found in Appendix F. We use γ = 10000. We use 20 D-CALIBRATION bins disjoint over [0, 1] for all experiments except for the cancer data, where we use 10 bins as in Haider et al. [2020]. For all experiments, we measure the loss on a validation set at each training epoch to chose a model to report test set metrics with. We report the test set NLL, test set D-CALIBRATION and Harrell’s Concordance Index [Harrell Jr et al., 1996] (abbreviated CONC) on the test set for several settings of λ. We compute concordance using the Lifelines package [Davidson-Pilon et al., 2017]. All reported results are an average of three seeds.
Data
We discuss differences in performance on simulated gamma data, semi-synthetic survival data where times are conditional on the MNIST classes, length of stay prediction in the Medical Information Mart for Intensive Care (MIMIC-III) dataset [Johnson et al., 2016], and glioma brain cancer data from The Cancer Genome Atlas (TCGA). Additional data details may be found in Appendix D.
4.1. Experiment 1: Simulated Gamma Times with Log-Linear Mean
Data
We design a simulation study to show that a conditional distribution may achieve good concordance and likelihood but will have poor D-CALIBRATION. After adding X-CAL, we are able to improve the exact D-CALIBRATION. We sample from a multivariate normal with σ2 = 10.0. We sample times t conditionally from a gamma with mean μ that is log-linear in x and constant variance 1e-3. The censoring times c are drawn like the event times, except with a different coefficient for the log-linear function. We experiment with censored and uncensored simulations, where we discard c and always observe t for uncensored. We sample a train/validation/test sets with 100k/50k/50k datapoints, respectively.
Results
Due to high variance in x and low conditional variance, this simulation has low noise. With large, clean data, this experiment validates the basic method on continuous and discrete models in the presence of censoring. Table 1 demonstrates how increasing λ gracefully balances D-CALIBRATION with NLL and concordance for different models and objectives: log-normal trained via NLL and with S-CRPS, and the categorical model trained via NLL, without CDF interpolation. For results on more models and choices of λ see Table 9 for uncensored results and Table 10 for censored in Appendix J.
Table 1:
Gamma simulation, censored
| λ | 0 | 1 | 10 | 100 | 500 | 1000 | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Log-Norm nll | nll | −0.059 | −0.049 | 0.004 | 0.138 | 0.191 | 0.215 |
| d-cal | 0.029 | 0.020 | 0.005 | 2e-4 | 6e-5 | 7e-5 | |
| conc | 0.981 | 0.969 | 0.942 | 0.916 | 0.914 | 0.897 | |
|
| |||||||
| Log-Norm s-crps | nll | 0.038 | 0.084 | 0.143 | 0.201 | 0.343 | 0.436 |
| d-cal | 0.017 | 0.007 | 0.001 | 1e-4 | 5e-5 | 8e-5 | |
| conc | 0.982 | 0.978 | 0.963 | 0.950 | 0.850 | 0.855 | |
|
| |||||||
| Cat-ni | nll | 0.797 | 0.799 | 0.822 | 1.149 | 1.665 | 1.920 |
| d-cal | 0.009 | 0.006 | 0.002 | 2e-4 | 6e-5 | 6e-5 | |
| conc | 0.987 | 0.987 | 0.987 | 0.976 | 0.922 | 0.861 | |
4.2. Experiment 2: Semi-Synthetic Experiment: Survival MNIST
Data
Following Pölsterl [2019], we simulate a survival dataset conditionally on the MNIST dataset [LeCun et al., 2010]. Each MNIST label gets a deterministic risk score, with labels loosely grouped together by risk groups (Table 5 in Appendix D.2). Datapoint image xi with label yi has time ti drawn from a Gamma with mean equal to risk(yi) and constant variance 1e-3. Therefore ti ⫫ xi | yi and times for datapoints that share an MNIST class are identically drawn. We draw censoring times c uniformly between the minimum failure time and the 90th percentile time, which resulted in about 50% censoring. We use PyTorch’s MNIST with test split into validation/test. The model does not see the MNIST class and learns a distribution over times given pixels xi. We experiment with censored and uncensored simulations, where we discard c and always observe t for uncensored.
Results
This semi-synthetic experiment tests the ability to tune calibration in presence of a high-dimensional conditioning set (MNIST images) and through a typical convolutional architecture. Table 2 demonstrates that the deep log-normal models started off miscalibrated relative to the categorical model for λ = 0 and that all models were able to significantly improve in calibration. See Table 11 and Table 12 for more uncensored and censored survival-MNIST results.
Table 2:
Survival-mnist, censored
| λ | 0 | 1 | 10 | 100 | 500 | 1000 | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Log-Norm nll | nll | 4.337 | 4.377 | 4.483 | 4.682 | 4.914 | 5.151 |
| d-cal | 0.392 | 0.074 | 0.020 | 0.005 | 0.005 | 0.007 | |
| conc | 0.902 | 0.873 | 0.794 | 0.696 | 0.628 | 0.573 | |
|
| |||||||
| Log-Norm s-crps | nll | 4.950 | 4.929 | 4.859 | 4.749 | 4.786 | 4.877 |
| d-cal | 0.215 | 0.122 | 0.051 | 0.010 | 0.002 | 9e-4 | |
| conc | 0.891 | 0.881 | 0.874 | 0.868 | 0.839 | 0.815 | |
|
| |||||||
| Cat-ni | nll | 1.733 | 1.734 | 1.765 | 1.861 | 2.074 | 3.030 |
| d-cal | 0.018 | 0.014 | 0.004 | 5e-4 | 5e-4 | 4e-4 | |
| conc | 0.945 | 0.945 | 0.927 | 0.919 | 0.862 | 0.713 | |
4.3. Experiment 3: Length of Stay Prediction in MIMIC-III
Data
We predict the length of stay (in number of hours) in the ICU, using data from the MIMIC-III dataset. Such predictions are important both for individual risk predictions and prognoses and for hospital-wide resource management. We follow the preprocessing in Harutyunyan et al. [2017], a popular MIMIC-III benchmarking paper and repository 3. The covariates are a time series of 17 physiological variables (Table 6 in Appendix D.3) including respiratory rate and glascow coma scale information. There is no censoring in this task. We skip imputation and instead use missingness masks as features. There are 2, 925, 434 and 525, 912 instances in the training and test sets. We split the training set in half for train and validation.
Results
Harutyunyan et al. [2017] discuss the difficult of this task when predicting fine-grained lengths-of-stay, as opposed to simpler classification tasks like more/less one week stay. The true conditionals are high in entropy given the chosen covariates Table 3 demonstrates this difficulty, as can be seen in the concordances. We report the categorical model with and without CDF interpolation and the log-normal trained with S-CRPS. NLL for the log-normal is not reported because S-CRPS does not optimize NLL and did poorly on this metric. The log-normal trained with NLL was not able to fit this task on any of the three metrics. All three models reported are able to reduce D-CALIBRATION. Results for all models and more choices of λ may be found in Table 13. The categorical models with and without CDF interpolation match in concordance for λ = 0 and λ = 1000. However, the interpolated model achieves better D-CALIBRATION. This may be due to the lower-bound ℓ > 0 on a discrete model’s D-CALIBRATION (Appendix E).
Table 3:
mimic-iii length of stay
| λ | 0 | 1 | 10 | 100 | 500 | 1000 | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Log-Norm s-crps | d-cal | 0.859 | 0.639 | 0.155 | 0.046 | 0.009 | 0.005 |
| conc | 0.625 | 0.639 | 0.575 | 0.555 | 0.528 | 0.506 | |
|
| |||||||
| Cat-ni | Test nll | 3.142 | 3.177 | 3.167 | 3.088 | 3.448 | 3.665 |
| d-cal | 0.002 | 0.002 | 0.001 | 2e-4 | 1e-4 | 1e-4 | |
| conc | 0.702 | 0.700 | 0.699 | 0.690 | 0.642 | 0.627 | |
|
| |||||||
| Cat-i | nll | 3.142 | 3.075 | 3.073 | 3.073 | 3.364 | 3.708 |
| d-cal | 4e-4 | 2e-4 | 2e-4 | 1e-4 | 5e-5 | 4e-5 | |
| conc | 0.702 | 0.702 | 0.702 | 0.695 | 0.638 | 0.627 | |
4.4. Experiment 4: Glioma data from The Cancer Genome Atlas
We use the glioma (a type of brain cancer) dataset 4 collected as part of the TCGA program and studied in [Network, 2015]. We focus on predicting time until death from the clinical data, which includes tumor tissue location, time of pathological diagnosis, Karnofsky performance score, radiation therapy, demographic information, and more. Censoring means they did not pass away. The train/validation/test sets are made of 552/276/277 datapoints respectively, of which 235/129/126 are censored, respectively.
Results
For this task, we study the Weibull AFT model, reduce the deep log-normal model from three to two hidden layers, and study a linear MTLR model (with CDF interpolation) in place of the deep categorical due to the small data size. MTLR is more constrained than linear categorical due to shared parameters. Table 4 demonstrates these three models’ ability to improve D-CALIBRATION. MTLR is able to fit well and does not give up much concordance. Results for all models and more choices of λ may be found in Table 14.
Table 4:
The Cancer Genome Atlas, glioma
| λ | 0 | 1 | 10 | 100 | 500 | 1000 | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Log-Norm nll | nll | 14.187 | 6.585 | 4.639 | 4.181 | 4.403 | 4.510 |
| d-cal | 0.059 | 0.024 | 0.010 | 0.003 | 0.002 | 0.004 | |
| conc | 0.657 | 0.632 | 0.703 | 0.805 | 0.474 | 0.387 | |
|
| |||||||
| Weibull | nll | 4.436 | 4.390 | 4.292 | 4.498 | 4.475 | 4.528 |
| d-cal | 0.035 | 0.028 | 0.009 | 0.003 | 0.004 | 0.007 | |
| conc | 0.788 | 0.785 | 0.777 | 0.702 | 0.608 | 0.575 | |
|
| |||||||
| mtlr-ni | nll | 1.624 | 1.620 | 1.636 | 1.658 | 1.748 | 1.758 |
| d-cal | 0.009 | 0.007 | 0.005 | 0.003 | 0.002 | 0.002 | |
| conc | 0.828 | 0.829 | 0.824 | 0.818 | 0.788 | 0.763 | |
5. Related Work
Deep Survival Analysis
Recent approaches to survival analysis parameterize the failure distribution as a deep neural network function of the [Ranganath et al., 2016, Alaa and van der Schaar, 2017, Katzman et al., 2018]. Miscouridou et al. [2018] and Lee et al. [2018] use a discrete categorical distribution over times interpreted ordinally, which can approximate any smooth density with sufficient data. The categorical approach has also been used when the conditional is parameterized by a recurrent neural network of sequential covariates [Giunchiglia et al., 2018, Ren et al., 2019]. Miscouridou et al. [2018] extend deep survival analysis to deal with missingness in x.
Post-training calibration methods
Practitioners have used two calibration methods for binary classifiers, which modify model predictions maximize likelihood on a held-out dataset. Platt scaling [Platt, 1999] works by using a scalar logistic regression built on top of predicted probabilities. Isotonic regression [Zadrozny and Elkan, 2002] uses a nonparametric piecewise linear transformation instead of the logistic regression. These methods do not reveal an explicit balance between prediction quality and calibration during model training. X-CAL allows practitioners to explore this balance while searching in the full model space.
Objectives
When an unbounded loss function (e.g. NLL) is used and the gradients are a function of x, the model may put undue focus on explaining a given outlier x*, worsening calibration during training. For this reason, robust objectives have been explored. Avati et al. [2019] consider continuous ranked probability score (CRPS) [Matheson and Winkler, 1976], a robust proper scoring rule for continuous outcomes, and adapt it to S-CRPS for survival analysis by accounting for censoring. However, S-CRPS does not provide a clear way to balance predictive power and calibration. Kumar et al. [2018] develop a trainable kernel-based calibration measure for binary classification but they do not discuss an optimizable calibration metric for survival analysis.
Brier Score
The Brier Score [Brier and Allen, 1951] decomposes into a calibration metric (numerator of Hosmer-Lemeshow) and a discrimination term encouraging patients with the same failure status at t* to have the same failure probability at t*. To capture entire distributions over time, the Integrated Brier Score is used. The Inverse Probability of Censoring Weighting Brier Score [Graf et al., 1999] handles censoring but requires estimation of the censoring distribution, a whole survival analysis problem (with censoring due to the failures) on its own [Gerds and Schumacher, 2006, Kvamme and Borgan, 2019]. X-CAL can balance discrimination and calibration without estimation of the censoring distribution.
6. Discussion
Model calibration is an important consideration in many clinical problems, especially when treatment decisions require risk estimates across all times in the future. We tackle the problem of building models that are calibrated over individual failure distributions. To this end, we provide a new technique that explicitly targets calibration during model training. We achieve this by constructing a differentiable approximation of D-CALIBRATION, and using it as an add-on objective to maximum likelihood and S-CRPS. As we show in our experiments, X-CAL allows for explicit and direct control of calibration on both simulated and real data. Further, we showed that searching over the X-CAL λ parameter can strike the practitioner-specified balance between predictive power and calibration.
Marginal versus Conditional Calibration
D-CALIBRATION is 0 for the true conditional and marginal distributions of failure times. This is because D-CALIBRATION measures marginal calibration, i.e. x is integrated out. Conditional calibration is the stronger condition that Fθ(t | x) is calibrated for all x. This is in general infeasible even to measure (let alone optimize) [Vovk et al., 2005, Pepe and Janes, 2013, Barber et al., 2019] without strong assumptions since for continuous x we usually observe just one sample. However, among the distributions that have 0 D-CALIBRATION, the true conditional distribution has the smallest NLL. Therefore, X-CALIBRATED objectives with proper scoring rules (like NLL) have an optimum only for the true conditional model in the limit of infinite data.
D-Calibration and Censoring
Equation (10) in Section 3.3 provides a censored version of D-CALIBRATION that is 0 for a calibrated model, like the original D-CALIBRATION (Equation (3)). However, this censored calibration measure is not equal to D-CALIBRATION in general for miscalibrated models. For a distribution Fθ with non-zero D-CALIBRATION, for any censoring distribution G, estimates of the censored version will assess Fθ to be more uniform than if exact D-CALIBRATION were able to be computed using all true observed failure times. This happens especially in the case of heavy and early censoring because a lot of uniform weight is assigned [Haider et al., 2020, Avati et al., 2019]. This means that the censored objective can be close to 0 for a miscalibrated model on a highly censored dataset.
An alternative strategy that avoids this issue is to use inverse weighting methods (e.g. Inverse Propensity Estimator of outcome under treatment [Horvitz and Thompson, 1952], Inverse Probability of Censoring-Weighted Brier Score [Graf et al., 1999, Gerds and Schumacher, 2006] and Inverse Probability of Censoring-Weighted binary calibration for survival analysis [Yadlowsky et al., 2019]). Inverse weighting would preserve the expectation that defines D-CALIBRATION for any censoring distribution. One option is to adjust with p(c | x). This requires c ⊥ t | x and solving an additional censored survival problem p(c | x). Nevertheless, if a censoring estimate is provided, the methodology in this work could then be applied to an inverse-weighted D-CALIBRATION. There is then a trade-off between the censored estimator proposed by Haider et al. [2020] that we use (no modeling G) and inverse-weighted estimators (which preserve D-CALIBRATION for miscalibrated models).
Broader Impact
In this paper, we study calibration of survival analysis models and suggest an objective for improving calibration during model training. Since calibration means that modeled probabilities correspond to the actual observed risk of an event, practitioners may feel more confident about using model outputs directly for decision making e.g. to decide how many emergency room staff members qualified for performing a given procedure should be present tomorrow given all current ER patients. But if the distribution of event times in these patients differs from validation data, because say the population has different demographics, calibration should not provide the practitioner with more confidence to directly use such model outputs.
Acknowledgments
This work was supported by:
• NIH/NHLBI Award R01HL148248
• NSF Award 1922658 NRT-HDR: FUTURE Foundations, Translation, and Responsibility for Data Science.
• NSF Award 1514422 TWC: Medium: Scaling proof-based verifiable computation
• NSF Award 1815633 SHF
We thank Humza Haider for sharing the original D-calibration experimental data, Avati et al. [2019] for publishing their code and the Cancer Genome Atlas Research Network for making the glioma data public. We thank all the reviewers for thoughtful feedback.
A. Background on Survival Analysis and Related Work
Survival analysis models the probability distribution of a time-until-event. The event is often called a failure time. For example, we may model time until onset of coronary heart disease given a patient’s current health status [Wilson et al., 1998, Vasan et al., 2008].
Survival analysis differs from standard probabilistic regression problems in that data may be censored. For example, a patient may leave a study before developing the studied condition, or may not develop the condition before the study ends. In these cases, the time that a patient leaves or the study ends is called the censoring time. These are cases of right-censoring, where it is only known that the failure time is greater than the observed censoring time.
We review key definitions in survival analysis. See George et al. [2014] for a review. For textbooks, see Andersen et al. [2012], Kalbfleisch and Prentice [2002], and Lawless [2011].
A.1. Notation
Let t be a continuous random variable denoting the failure time with CDF F and density f. The survival function is defined as 1 minus the CDF: . Censoring times are considered random variables c with CDF G, survival function , and density g. In general these distributions may be conditional on covariates x.
For datapoints i, let ti be failure times and ci be censoring times. Let us focus on right-censoring where and the observed data consists of (xi, ui, δi). In general we cannot throw away censored points, since p(t | x, t < c) ≠ p(t | x) and we would therefore biasedly estimate the failure distribution F.
A.2. Assumptions About Censoring
It may seem that we need to model c to estimate the parameters of f, but under certain assumptions, we can write the likelihood (with respect to f’s parameters) for a dataset with censoring without estimating the censoring distribution. In this work, we assume:
Assumption.
Censoring-at-random. t is distributed marginally or conditionally on x. c is either a constant, distributed marginally, or distributed conditionally on x. In any case, it must hold that t ⫫ c | x.
Assumption.
Non-informative Censoring. The censoring time c’s distribution parameters θc are distinct from parameters θt of t’s distribution.
A.3. Likelihoods
Under the two censoring assumptions, the log-likelihood can be derived to be
| (12) |
and can be maximized to learn parameters θt of f without an estimate of G. This can be interpreted as follows: an uncensored individual has δi = 1, meaning ui = ti. This point contributes through the failure density f(ui) = f(ti), as in standard regression likelihoods. Censored points contribute through failure survival function because there failure time is known to be greater than ui. Full discussions of survival likelihoods can be found in Kalbfleisch and Prentice [2002], Lawless [2011], Andersen et al. [2012].
A.4. Testing Calibration
Classical goodness-of-fit tests [Lemeshow and Hosmer Jr, 1982, Grønnesby and Borgan, 1996, D’agostino and Nam, 2003] and their recent modifications [Demler et al., 2015] assess calibration of survival analysis models for a particular time of interest t∗. These take the following steps:
pick a time t* at which to measure calibration
evaluate model probability pi = pθ(t < t* | xi) of failing by time t*
sort pi into K groups gk defined by quantiles (e.g. K = 2 corresponds to partitioning the data into a low-risk group and high-risk group)
compute the observed # of events using e.g. (1−KMk[t∗])|gk| where KMk the Kaplan-Meier estimate [Kaplan and Meier, 1958] of the survival function just on data in gk’s
compute the expected #,
let
gives a χ2 test statistic
small p-value → model not calibrated
Figure 1:
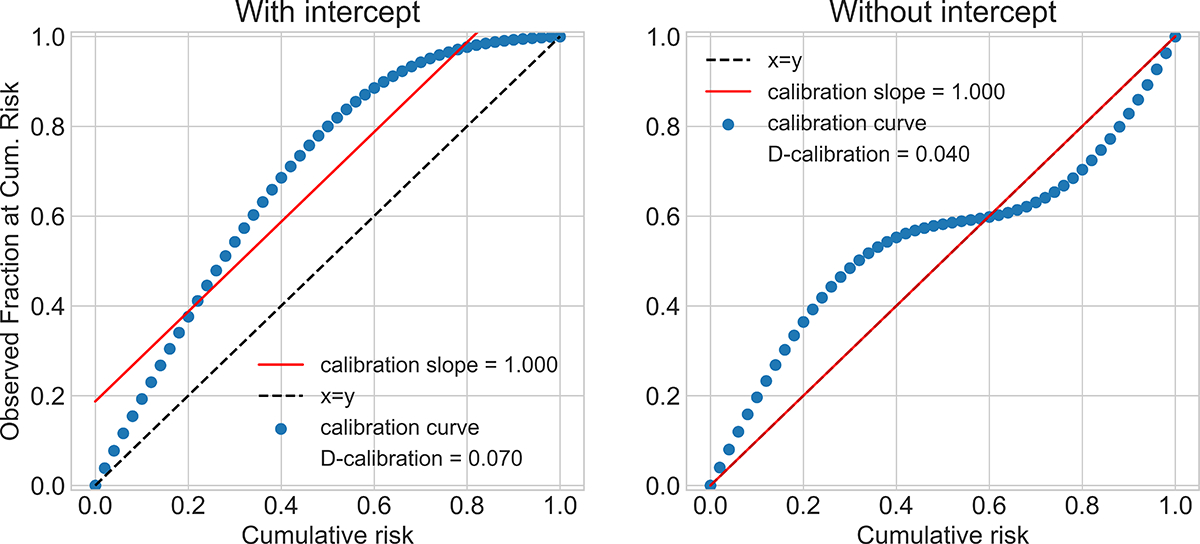
Sub-optimal calibration curves that result in optimal calibration slope.
Demler et al. [2015] review these tests and propose some modifications when there are not enough individuals assigned to each bin. These tests are limited in two ways: they answer calibration in a rigid yes/no fashion with hypothesis testing, and it is not clear how to combine calibration assessments over the entire range of possible time predictions.
A.5. Calibration Slope
Calibration Slope
Recent publications in machine learning [Avati et al., 2019] and in medicine [Besseling et al., 2017] use the calibration slope to evaluate calibration [Stevens and Poppe, 2020]. First, a calibration curve is computed by plotting, for each quantile ρ ∈ [0, 1], the fraction of observed samples with a failure time smaller than that quantile’s time . Then, report the slope of the best-fit line to this curve. When a model is well-calibrated, the true and predicted densities are close and the best fit line has slope 1.0. However, slope can be 1.0 (with intercept 0.0) even when the model is not well-calibrated.
Here, we construct two possible calibration curves that cannot result from well-calibrated models. However, the resulting calibration slope is close to 1.0. Avati et al. [2019] use a line of best fit with non-zero intercept. We plot hypothetical calibration curves in Figure 1 such that the corresponding best fit line has slope 1.0, with and without intercept terms. Stevens and Poppe [2020] make a related observation about calibration slope: a near-zero intercept of the line of best fit, or other evidence of calibration, should always be reported alongside near-1 slope when claiming a model is calibrated. However, we demonstrate here that even slope 1 and intercept 0 can result from poorly calibrated models. The interested reader should see Stevens and Poppe [2020] for an assessment of recent publications in medicine that report only slope and for the history of slope-only as a “measure of spread” [Cox, 1958].
B. Survival CRPS
S-CRPS is proposed by Avati et al. [2019]:
where y is the event time, c is an indicator for censorship and is the CDF from the model. See Avati et al. [2019] Appendix B for a detailed derivation of S-CRPS objective for a log-normal model.
C. CDF of Survival Time is Uniform for Censored Patient
Consider the data distribution P(t, c | x) and using the conditional P(t | x) of this distribution to evaluate D-CALIBRATION on this data. For a point that is censored at time c, P(t | x) would simply condition on the event t > c for constant c, yielding P(t | t > c, x). However, the true failure distribution for such a point is P(t | t > c, c = c, x). Under censoring-at-random,
| (13) |
Let F be the failure CDF. Let pt be the density of t | x. Apply transformation z = F(t|x). To compute z’s density, we need:
Applying change of variable to compute z’s density:
Therefore, z is uniform distributed over [0, 1]. So conditioning on set (t > c, x) = (z > F(c|x), x) gives the result:
The CDF value of the unobserved time for a censored datapoint is uniform above the failure CDF applied to the censoring time. Haider et al. [2020] (Appendix B) give an alternate proof in terms of expected counts.
D. Extra Data Details
D.1. Data Details for Simulation Study
For the gamma simulation, we draw x from a D = 32 multivariate Normal with 0 mean and diagonal covariance with σ2 = 10.0. We draw failure times t conditionally on x from a gamma distribution with mean μ log-linear in x. The weights of the linear function are drawn uniformly. The gamma distribution has constant variance 1e-3. This is achieved by setting and β = μi/1e-3.
Censoring times are drawn like failure times but with a different set of weights for the linear function. This means t ⫫ c | x.
D.2. Data Details for MNIST
As described in the main text, we follow Pölsterl [2019] to simulate a survival dataset conditionally on the MNIST dataset [LeCun et al., 2010]. Each MNIST label gets a deterministic risk score, with labels loosely grouped together by risk groups. See Table 5 for an example of the risk groups and risk scores for the MNIST classes.
Datapoint image xi with label yi has time ti drawn from a Gamma whose mean is the risk score and whose variance is constant 1e-3. Therefore ti is independent of xi given yi and times for datapoints that share an MNIST class are identically drawn.
For each split of the data (e.g. training set), we draw censoring times uniformly between the minimum failure time in that split and the 90th percentile time, which, due to the particular failure distributions, resulted in about 50% censoring.
Table 5:
Risk scores for digit classes.
| Digit | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|---|
| Risk Group | most | least | lower | lower | lower | higher | least | most | least | most |
| Risk Score | 11.25 | 2.25 | 5.25 | 5.0 | 4.75 | 8.0 | 2.0 | 11.0 | 1.75 | 10.75 |
Table 6:
The 17 selected clinical variables. The second column shows the source table(s) of a variable from mimic-iii database. The third column lists the “normal” values used in the imputation step. Table reproduced from Harutyunyan et al. [2017].
| Variable table | Impute value | Modeled as |
|---|---|---|
| Capillary refill rate | 0.0 | categorical |
| Diastolic blood pressure | 59.0 | continuous |
| Fraction inspired oxygen | 0.21 | continuous |
| Glascow coma scale eye opening | 4 spontaneously | categorical |
| Glascow coma scale motor response | 6 obeys commands | categorical |
| Glascow coma scale total | 15 | categorical |
| Glascow coma scale verbal response | 5 oriented | categorical |
| Glucose | 128.0 | continuous |
| Heart Rate | 86 | continuous |
| Height | 170.0 | continuous |
| Mean blood pressure | 77.0 | continuous |
| Oxygen saturation | 98.0 | continuous |
| Respiratory rate | 19 | continuous |
| Systolic blood pressure | 118.0 | continuous |
| Temperature | 36.6 | continuous |
| Weight | 81.0 | continuous |
| pH | 7.4 | continuous |
D.3. Data Details for MIMIC-III
We show the 17 physiological variables we use in Table 6. The table is reproduced from Harutyunyan et al. [2017]. This dataset differs from other MIMIC-III length of stay datasets because one stay in the ICU of a single patient produces many datapoints: remaining time at each hour after admission. After excluding ICU transfers and patients under 18, there are 2, 925, 434 and 525, 912 instances in the training and test sets. We split the training set in half for train and validation.
D.4. Data Details for The Cancer Genome Atlas Glioma Data
We use the glioma (a type of brain cancer) data5 collected as part of the TCGA program and studied in [Network, 2015]. TCGA comprises clinical data and molecular from 11,000 patients being treated for a diverse set of cancer types. We focus on predicting time until death from the clinical data, which includes:
tumor tissue site
time of initial pathologic diagnosis
radiation therapy
Karnofsky performance score
histological type
demographic information
Censoring means they did not pass away. The train/validation/test sets are made of 552/276/277 datapoints respectively, of which 235/129/126 are censored, respectively.
To download this data, use the firebrowse. tool, select the Glioma (GBMLGG) cohort, and then click the blue clinical features bar on the right hand side. Select the “Clinical Pick Tier 1” file.
We standardized the features and then clamped their maximum absolute value at 5.0. This is in part because we were working with the Weibull AFT model, which is very sensitive to large variance in covariates.
E. Model Descriptions
We describe the models we use in the experiments. For all models, the parameterization as a function of x varies in complexity (e.g. linear or deep) depending on task.
Log-normal model
When logT is Normal with mean μ and variance σ2, we say that T is log-normal with location μ and scale σ. We parameterize μ and σ as functions of x (small ReLU networks with 1 to 3 hidden layers, depending on experiment).
Weibull Model
The Weibull Accelerated Failure Times (AFT) model sets logT = β0+β⊤X+σW where σ is a scale parameter and W is Gumbel. It follows that T ∼ Weibull(λ, k) with scale λ = exp[β⊤X] and concentration k = σ−1 [Liu, 2018]. We constrain k ∈ (1, 2).
Interpolation for Discrete Models
The next two models predict for a finite set of times and therefore have a discontiuous CDF. These models have a lower-bound ℓ > 0 on D-CALIBRATION because the CDF values will not be Unif(0, 1) distributed. However, ℓ decreases to 0 as the number of discrete times increases. For any fixed number of times, minimizing D-CALIBRATION will still improve calibration, which we observe in our experiments.
We optionally use linear interpolation to calculate the CDF. Suppose a time t falls into bin k which covers time interval (ta, tb). If we do not use interpolation, then the CDF value P(T ≤ t) we calculate is the sum of the probabilities of bins whose indices are smaller than or equal to k. If we use linear interpolation, we replace the probability of bin k, P(k), in the summation by:
Categorical Model
We parameterize a categorical distribution over discrete times by using a neural network function of x with a size B output. Interpreted ordinally, this can approximate continuous survival distributions as B → ∞ [Lee et al., 2018, Miscouridou et al., 2018]. The time for each bin is set to training data percentiles so that each next bin captures the range of times for the next (100/B)th percentile of training data, using only uncensored times.
Multi-Task Logistic Regression (mtlr)
MTLR differs from the Categorical Model because there is some relationship between the probability of the bins. Assume we have K bins. In the linear case Yu et al. [2011], suppose our input is x and parameters Θ = (θ1, . . . , θK−1). The probability for bin k < K is:
and the probability for bin K is :
F. Experimental Details
F.1. Gamma Simulation
We use a 4-layer neural network of hidden-layer sizes 128, 64, 64 units, with ReLU activations to parameterize the categorical and log-normal distributions. For categorical we use another linear transformation to map to 50 output dimensions. For the log-normal model, two copies of the above neural network are used, one to output the location and the other to output the log of the log-normal scale parameter. For MTLR, we use a linear transformation from covariates to 50 dimensions and use a softmax layer to output the probability for the 50 bins. We use 0 dropout, 0 weight decay, learning rate 1e-3 and batch size 1000 for 100 epochs in this experiment.
F.2. Survial MNIST
The model does not see the MNIST class and learns a distribution over times given pixels xi. We use a convolutional neural network. We use several layers of 2D convolutions with a kernel of size 2 and stride of size 1. The sequence of channel numbers is 32, 64, 128, 256 with the last layer containing scalars. After each convolution, we use ReLU, then dropout, then size 2 max pooling.
For categorical and log-normal models, this CNN output is mapped through a three-hidden-layer ReLU neural network with hidden sizes 512, 1024, 1024. Between the fully connected layers, we use ReLU then dropout. Again, with the log-normal, separate networks are used to output the location and log-scale. For MTLR, the CNN output is linearly mapped to the 50 bins. For categorical, we use 0.2 dropout for uncensored and 0.1 for censored. In MTLR, we use dropout 0.2. In lognormal, we use dropout 0.1. We use weight decay 1e-4, learning rate 1e-3, and batch size 5000 for 200 epochs.
F.3. MIMIC-III
The input is high-dimensional (about 1400) because it is a concatenated time series and because missingness masks are used. We use a 4-layer neural network of hidden-layer sizes 2048, 1024, 1024 units with ReLU activations. For the categorical model, we use B = 20 categorical output bins. For the log-normal model, we use one three-hidden neural network of hidden-layer sizes 128, 64, 64 units and an independent copy to output the location and log-scale parameters. We use dropout 0.15, learning rate 1e-3 and weight decay 1e-4 for 200 epochs at batch size 5000.
F.4. The Cancer Genome Atlas, Glioma
The Weibull model has parameters scale and concentration. The scale is set to exp[β⊤x] for regression parameters β, plus a constant 1.0 for numerical stability. We optimize the concentration parameter in (1, 2). The log-normal model is as described in the simulated gamma experiment, except that it has two instead of three hidden layers, due to small data sample size. The categorical and MTLR models are also as described in the simulated gamma experiment, except that they have 20 instead of 50 bins, and are linear, again due to small data sample size.
We standardize this data and then clamp all covariates at absolute value 5.0. For all models, we train for 10,000 epochs at learning rate 1e-2 with full data batch size 1201. We use 10 D-CALIBRATION bins for this experiment as studied in Haider et al. [2020], rather than the 20 bins used in all other experiments.
G. Exploring Choice of γ soft-indicator parameter
There is a trade-off in setting the soft membership parameter γ. Larger values approximate the indicator function better, but can have worse gradients because the values lie in the flat region of the sigmoid. See Figure 2 for an example of how gamma changes the soft indicator for a given set I = [0.45, 0.55]. We choose γ = 10000 in all of the experiments and find that it allows us to minimize exact d-cal (D-CAL). We explore other choices in Table 7. We see the expected improvement in approximation as γ increases. Then, as γ gets too large, exact D-CAL stops improving as a function of λ.
H. Exploring Slack due to Jensen’s Inequality
We trained the Categorical model on the gamma simulation data with γ = 10, 000 and batch size 10, 000 for all λ. The trained models are evaluated on the training set (size 100, 000) with two different test batch sizes, 500 and 1000. Table 8 demonstrates that the upper-bounds for both batch sizes preserve model ordering with respect to exact D-CALIBRATION. The bound for batch size 10, 000 is quite close to the exact D-CALIBRATION.
Figure 2:
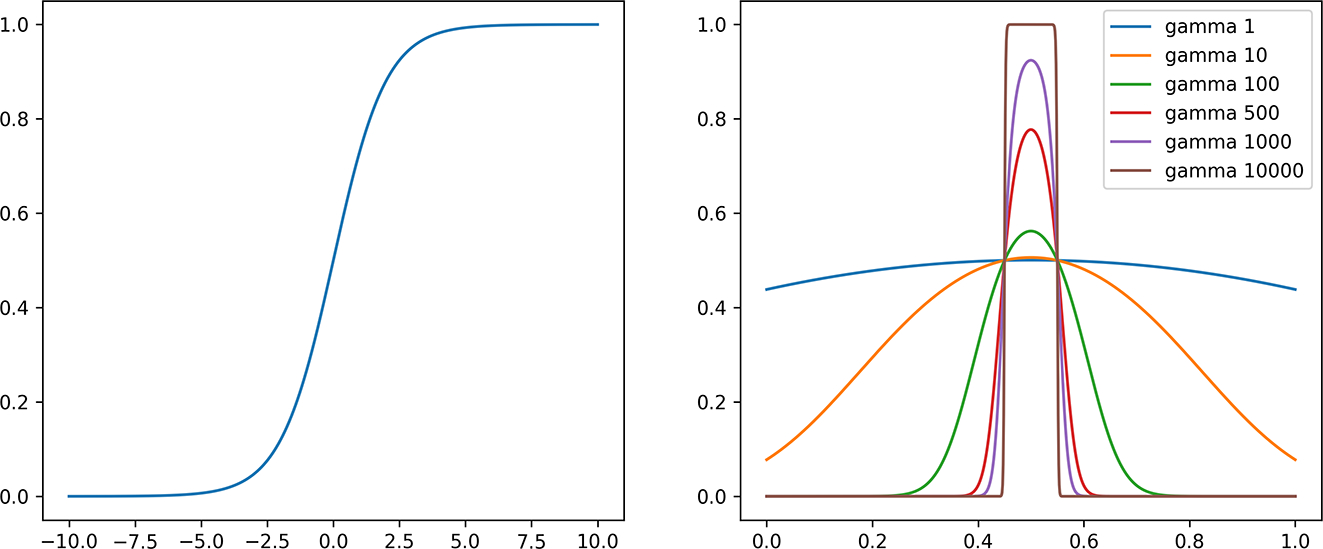
Left: the sigmoid function. Right: choice of hyper-parameter gamma in soft indicator function for set I = [0.45, 0.55].
Table 7:
Exact D-Cal, Soft-Dcal, and NLL at end of training, evaluated on training data for models trained with λ = 10 and batch size 1,000. Approximation improves as γ increases. Gradients vanish when γ gets too large. All experiments are better in calibration than the λ = 0 MLE model, which has exact D-cal 0.09.
| γ | 10 | 102 | 103 | 104 | 105 | 106 | 107 | 5 × 107 |
|---|---|---|---|---|---|---|---|---|
| Exact D-Cal | 0.2337 | 0.0095 | 0.0079 | 0.0039 | 0.0025 | 0.0014 | 0.0015 | 0.0048 |
| Soft D-Cal | 0.4599 | 0.0604 | 0.0074 | 0.0039 | 0.0025 | 0.0014 | 0.0015 | 0.0048 |
| NLL | 2.1180 | 1.1362 | 1.0793 | 1.2508 | 1.6993 | 2.3873 | 2.6940 | 3.4377 |
I. Modification of soft indicator for the first and the last interval
In our soft indicator,
is a differentiable approximation for . When b is the upper boundary of all the u values, for example, 1 for CDF values, the b in the soft indicator can be replaced by any value that is greater than b. We use 2 to replace 1 for the upper boundary when b = 1 in our experiments. Similarly we use a = −1 to replace a = 0 for the lower boundary when a = 0.
Table 8:
Slack in the upper-bound preserves modeling ordering with respect to exact d-calibration
| λ | Batch Size | Exact D-Cal | Upper-bound |
|---|---|---|---|
| 0 | 500 | 0.05883 | 0.0605 |
| 10000 | ” | 0.0589 | |
| 1 | 500 | 0.02204 | 0.0238 |
| 10000 | ” | 0.0221 | |
| 5 | 500 | 0.00963 | 0.0114 |
| 10000 | ” | 0.0097 | |
| 10 | 500 | 0.00482 | 0.0066 |
| 10000 | ” | 0.0048 | |
| 50 | 500 | 0.00040 | 0.0021 |
| 10000 | ” | 0.0004 | |
| 100 | 500 | 0.00022 | 0.0021 |
| 10000 | ” | 0.0003 | |
| 500 | 500 | 0.00015 | 0.0020 |
| 10000 | ” | 0.0002 | |
| 1000 | 500 | 0.00006 | 0.0019 |
| 10000 | ” | 0.0001 | |
Consider the term in our upper-bound (eq. (6)) for the last interval I = [a, b], where b = 1, . The gradient of this term with respect to one CDF value ui is:
If
then the fraction of points in the interval is larger than the size of the interval. We want to move the points out of the interval. In the last interval, in order to move points out of the interval, we can only make the values smaller, which means we want the gradient with respect to u to be positive. (recall that we are moving in the direction of the negative gradient to minimize the objective). However, for points that are greater than (a + b)/2, the above gradient will be negative because term (a + b − 2ui) is negative. This is not ideal. Changing the value b from 1 to 2 can resolve the issue. Since CDF values are all smaller than 1, (a + b)/2 will always be greater than u if we use b = 2 for the last interval. The above optimization issue only applies on the first and last interval because for intervals in the middle, we can move the points either to left or right to lower the fraction of points in the interval.
J. Full Results: More Models and Choices of Lambda
Table 9:
Gamma simulation, uncensored (full results)
| λ | 0 | 1 | 5 | 10 | 50 | 100 | 500 | 1000 | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Log-Norm nll | nll | 0.381 | 0.423 | 0.507 | 0.580 | 0.763 | 0.809 | 0.870 | 0.882 |
| d-cal | 0.271 | 0.060 | 0.021 | 0.011 | 0.001 | 4e-4 | 1e-4 | 7e-5 | |
| CONC | 0.982 | 0.955 | 0.931 | 0.908 | 0.841 | 0.835 | 0.809 | 0.802 | |
|
| |||||||||
| Log-Norm s-crps | nll | 0.455 | 0.614 | 0.730 | 0.781 | 0.837 | 0.848 | 0.869 | 0.965 |
| d-cal | 0.055 | 0.014 | 0.004 | 0.002 | 2e-4 | 1e-4 | 1e-4 | 1e-4 | |
| conc | 0.979 | 0.975 | 0.968 | 0.959 | 0.940 | 0.931 | 0.864 | 0.811 | |
|
| |||||||||
| Cat-ni | nll | 0.998 | 1.042 | 1.129 | 1.197 | 1.788 | 2.098 | 3.148 | 3.688 |
| d-cal | 0.074 | 0.023 | 0.008 | 0.005 | 4e-4 | 4e-4 | 2e-4 | 1e-4 | |
| conc | 0.986 | 0.986 | 0.985 | 0.985 | 0.973 | 0.960 | 0.877 | 0.748 | |
|
| |||||||||
| Cat-i | nll | 0.997 | 1.001 | 1.029 | 1.083 | 1.763 | 2.083 | 3.167 | 3.788 |
| d-cal | 0.002 | 0.002 | 0.001 | 0.002 | 5e-4 | 5e-4 | 1e-4 | 1e-4 | |
| conc | 0.986 | 0.986 | 0.986 | 0.985 | 0.972 | 0.960 | 0.874 | 0.699 | |
|
| |||||||||
| mtlr-NI | nll | 1.287 | 1.409 | 1.589 | 1.612 | 2.356 | 2.590 | 3.267 | 3.509 |
| d-cal | 0.027 | 0.027 | 0.015 | 0.008 | 5e-4 | 2e-4 | 2e-4 | 2e-4 | |
| conc | 0.986 | 0.986 | 0.983 | 0.981 | 0.952 | 0.940 | 0.909 | 0.899 | |
|
| |||||||||
| mtlr-I | nll | 1.392 | 1.419 | 1.616 | 1.823 | 2.165 | 2.612 | 2.982 | 3.184 |
| d-cal | 0.048 | 0.034 | 0.017 | 0.009 | 7e-4 | 2e-4 | 1e-4 | 1e-4 | |
| conc | 0.986 | 0.986 | 0.982 | 0.980 | 0.958 | 0.934 | 0.918 | 0.917 | |
Table 10:
Gamma simulation, censored (full results). For categorical model with interpolation, the d-cal is already very low at λ = 0 so it is hard to optimize this one further.
| λ | 0 | 1 | 5 | 10 | 50 | 100 | 500 | 1000 | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Log-Norm nll | nll | −0.059 | −0.049 | −0.022 | 0.004 | 0.099 | 0.138 | 0.191 | 0.215 |
| d-cal | 0.029 | 0.020 | 0.008 | 0.005 | 7e-4 | 2e-4 | 6e-5 | 7e-5 | |
| conc | 0.981 | 0.969 | 0.950 | 0.942 | 0.927 | 0.916 | 0.914 | 0.897 | |
|
| |||||||||
| Log-Norm s-crps | nll | 0.038 | 0.084 | 0.119 | 0.143 | 0.185 | 0.201 | 0.343 | 0.436 |
| d-cal | 0.017 | 0.007 | 0.003 | 0.001 | 1e-4 | 1e-4 | 5e-5 | 8e-5 | |
| conc | 0.982 | 0.978 | 0.971 | 0.963 | 0.952 | 0.950 | 0.850 | 0.855 | |
|
| |||||||||
| Cat-ni | nll | 0.797 | 0.799 | 0.805 | 0.822 | 1.023 | 1.149 | 1.665 | 1.920 |
| d-cal | 0.009 | 0.006 | 0.003 | 0.002 | 3e-4 | 2e-4 | 6e-5 | 6e-5 | |
| conc | 0.987 | 0.987 | 0.987 | 0.987 | 0.982 | 0.976 | 0.922 | 0.861 | |
|
| |||||||||
| Cat-i | nll | 0.783 | 0.782 | 0.788 | 0.795 | 0.948 | 1.124 | 1.686 | 1.994 |
| d-cal | 7e-5 | 1e-4 | 6e-5 | 8e-5 | 2e-4 | 2e-4 | 4e-5 | 6e-5 | |
| conc | 0.987 | 0.987 | 0.987 | 0.987 | 0.983 | 0.976 | 0.933 | 0.847 | |
|
| |||||||||
| mtlr-NI | nll | 0.873 | 0.875 | 0.875 | 0.977 | 1.271 | 1.412 | 1.747 | 1.900 |
| d-cal | 0.004 | 0.004 | 0.003 | 0.004 | 4e-4 | 2e-4 | 2e-4 | 2e-4 | |
| conc | 0.987 | 0.987 | 0.987 | 0.985 | 0.973 | 0.965 | 0.951 | 0.943 | |
|
| |||||||||
| mtlr-I | nll | 0.829 | 0.830 | 0.866 | 0.981 | 1.266 | 1.414 | 1.762 | 1.912 |
| d-cal | 0.004 | 0.004 | 0.004 | 0.004 | 5e-4 | 1e-4 | 6e-5 | 7e-5 | |
| conc | 0.988 | 0.988 | 0.987 | 0.985 | 0.971 | 0.963 | 0.947 | 0.939 | |
Table 11:
Survival-mnist, uncensored (full results)
| λ | 0 | 1 | 5 | 10 | 50 | 100 | 500 | 1000 | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Log-Norm nll | nll | 4.344 | 4.407 | 4.530 | 4.508 | 4.549 | 4.571 | 5.265 | 5.417 |
| d-cal | 0.328 | 0.104 | 0.018 | 0.020 | 0.011 | 0.010 | 0.005 | 0.005 | |
| conc | 0.886 | 0.867 | 0.754 | 0.759 | 0.725 | 0.713 | 0.541 | 0.509 | |
|
| |||||||||
| Log-Norm s-crps | nll | 4.983 | 4.940 | 4.853 | 4.759 | 4.714 | 4.673 | 4.852 | 5.118 |
| d-cal | 0.212 | 0.132 | 0.081 | 0.059 | 0.020 | 0.007 | 0.003 | 0.003 | |
| conc | 0.889 | 0.878 | 0.866 | 0.861 | 0.873 | 0.873 | 0.820 | 0.798 | |
|
| |||||||||
| Cat-ni | nll | 1.726 | 1.730 | 1.737 | 1.755 | 1.824 | 1.860 | 2.076 | 3.073 |
| d-cal | 0.019 | 0.013 | 0.008 | 0.005 | 9e-4 | 9e-4 | 6e-4 | 3e-4 | |
| conc | 0.945 | 0.945 | 0.945 | 0.937 | 0.921 | 0.916 | 0.854 | 0.690 | |
|
| |||||||||
| Cat-i | nll | 1.726 | 1.731 | 1.735 | 1.741 | 1.782 | 1.809 | 1.953 | 2.157 |
| d-cal | 0.007 | 0.005 | 0.003 | 0.002 | 6e-4 | 3e-4 | 4e-4 | 3e-4 | |
| conc | 0.945 | 0.945 | 0.945 | 0.945 | 0.940 | 0.937 | 0.897 | 0.830 | |
|
| |||||||||
| mtlr-ni | nll | 1.747 | 1.745 | 1.749 | 1.772 | 1.832 | 1.850 | 2.075 | 2.419 |
| d-cal | 0.018 | 0.014 | 0.008 | 0.004 | 0.001 | 0.001 | 8e-4 | 0.002 | |
| conc | 0.944 | 0.945 | 0.945 | 0.944 | 0.934 | 0.934 | 0.870 | 0.808 | |
|
| |||||||||
| mtlr-i | nll | 1.746 | 1.746 | 1.752 | 1.756 | 1.779 | 1.802 | 1.975 | 2.560 |
| d-cal | 0.005 | 0.004 | 0.003 | 0.002 | 5e-4 | 4e-4 | 8e-4 | 0.001 | |
| conc | 0.944 | 0.944 | 0.945 | 0.944 | 0.941 | 0.936 | 0.886 | 0.806 | |
Table 12:
Survival-mnist, censored (full results)
| λ | 0 | 1 | 5 | 10 | 50 | 100 | 500 | 1000 | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Log-Norm nll | nll | 4.337 | 4.377 | 4.433 | 4.483 | 4.602 | 4.682 | 4.914 | 5.151 |
| d-cal | 0.392 | 0.074 | 0.033 | 0.020 | 0.008 | 0.005 | 0.005 | 0.007 | |
| conc | 0.902 | 0.873 | 0.829 | 0.794 | 0.728 | 0.696 | 0.628 | 0.573 | |
|
| |||||||||
| Log-Norm s-crps | nll | 4.950 | 4.929 | 4.873 | 4.859 | 4.672 | 4.749 | 4.786 | 4.877 |
| d-cal | 0.215 | 0.122 | 0.071 | 0.051 | 0.018 | 0.010 | 0.002 | 9e-4 | |
| conc | 0.891 | 0.881 | 0.871 | 0.874 | 0.866 | 0.868 | 0.839 | 0.815 | |
|
| |||||||||
| Cat-ni | nll | 1.733 | 1.734 | 1.738 | 1.765 | 1.827 | 1.861 | 2.074 | 3.030 |
| d-cal | 0.018 | 0.014 | 0.008 | 0.004 | 8e-4 | 5e-4 | 5e-4 | 4e-4 | |
| conc | 0.945 | 0.945 | 0.944 | 0.927 | 0.920 | 0.919 | 0.862 | 0.713 | |
|
| |||||||||
| Cat-i | nll | 1.731 | 1.731 | 1.741 | 1.750 | 1.779 | 1.805 | 1.955 | 2.113 |
| d-cal | 0.007 | 0.006 | 0.003 | 0.002 | 3e-4 | 4e-4 | 4e-4 | 3e-4 | |
| conc | 0.945 | 0.944 | 0.945 | 0.945 | 0.942 | 0.938 | 0.901 | 0.843 | |
|
| |||||||||
| mtlr-NI | nll | 1.126 | 1.118 | 1.125 | 1.136 | 1.174 | 1.193 | 1.350 | 1.482 |
| d-cal | 0.021 | 0.017 | 0.012 | 0.009 | 0.006 | 0.006 | 0.006 | 0.007 | |
| conc | 0.958 | 0.960 | 0.961 | 0.960 | 0.949 | 0.943 | 0.897 | 0.880 | |
|
| |||||||||
| mtlr-I | nll | 1.126 | 1.118 | 1.125 | 1.136 | 1.174 | 1.193 | 1.350 | 1.482 |
| d-cal | 0.021 | 0.017 | 0.012 | 0.009 | 0.006 | 0.006 | 0.006 | 0.007 | |
| conc | 0.958 | 0.960 | 0.961 | 0.960 | 0.949 | 0.943 | 0.897 | 0.880 | |
Table 13:
mimic-iii length of stay (full results)
| λ | 0 | 1 | 5 | 10 | 50 | 100 | 500 | 1000 | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Log-Norm s-crps | d-cal | 0.860 | 0.639 | 0.210 | 0.155 | 0.066 | 0.046 | 0.009 | 0.005 |
| conc | 0.625 | 0.639 | 0.577 | 0.575 | 0.558 | 0.555 | 0.528 | 0.506 | |
|
| |||||||||
| Cat-ni | nll | 3.142 | 3.177 | 3.101 | 3.167 | 3.086 | 3.088 | 3.448 | 3.665 |
| d-cal | 0.002 | 0.002 | 0.002 | 0.001 | 3e-4 | 2e-4 | 1e-4 | 1e-4 | |
| conc | 0.702 | 0.700 | 0.701 | 0.699 | 0.695 | 0.690 | 0.642 | 0.627 | |
|
| |||||||||
| Cat-i | nll | 3.142 | 3.075 | 3.157 | 3.073 | 3.002 | 3.073 | 3.364 | 3.708 |
| d-cal | 4-e4 | 3e-4 | 3e-4 | 3e-4 | 4e-4 | 1e-4 | 5e-5 | 4e-5 | |
| conc | 0.702 | 0.702 | 0.701 | 0.702 | 0.698 | 0.695 | 0.638 | 0.627 | |
Table 14:
The Cancer Genome Atlas, glioma (full results)
| λ | 0 | 1 | 5 | 10 | 50 | 100 | 500 | 1000 | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Weibull | nll | 4.436 | 4.390 | 4.313 | 4.292 | 4.441 | 4.498 | 4.475 | 4.528 |
| d-cal | 0.035 | 0.028 | 0.014 | 0.009 | 0.003 | 0.003 | 0.004 | 0.007 | |
| conc | 0.788 | 0.785 | 0.781 | 0.777 | 0.731 | 0.702 | 0.608 | 0.575 | |
|
| |||||||||
| Log-Norm nll | nll | 14.187 | 6.585 | 4.841 | 4.639 | 4.181 | 4.181 | 4.403 | 4.510 |
| d-cal | 0.059 | 0.024 | 0.012 | 0.010 | 0.003 | 0.003 | 0.002 | 0.004 | |
| conc | 0.657 | 0.632 | 0.673 | 0.703 | 0.778 | 0.805 | 0.474 | 0.387 | |
|
| |||||||||
| Log-Norm s-crps | nll | 5.784 | 5.801 | 5.731 | 5.698 | 5.047 | 4.892 | 4.750 | 4.712 |
| d-cal | 0.258 | 0.2585 | 0.257 | 0.252 | 0.100 | 0.0702 | 0.044 | 0.025 | |
| conc | 0.798 | 0.798 | 0.798 | 0.810 | 0.568 | 0.507 | 0.420 | 0.363 | |
|
| |||||||||
| Cat-ni | nll | 1.718 | 1.742 | 1.746 | 1.758 | 1.800 | 1.799 | 1.810 | 1.826 |
| d-cal | 0.008 | 0.003 | 0.002 | 0.002 | 0.003 | 0.003 | 0.003 | 0.002 | |
| conc | 0.781 | 0.771 | 0.775 | 0.775 | 0.765 | 0.765 | 0.758 | 0.748 | |
|
| |||||||||
| Cat-i | nll | 1.711 | 1.718 | 1.733 | 1.726 | 1.743 | 1.787 | 1.781 | 1.789 |
| d-cal | 0.003 | 0.001 | 8e-4 | 0.001 | 0.002 | 0.002 | 0.002 | 0.002 | |
| conc | 0.778 | 0.779 | 0.780 | 0.798 | 0.804 | 0.803 | 0.806 | 0.802 | |
|
| |||||||||
| mtlr-NI | nll | 1.624 | 1.620 | 1.636 | 1.636 | 1.666 | 1.658 | 1.748 | 1.758 |
| d-cal | 0.009 | 0.007 | 0.007 | 0.005 | 0.003 | 0.003 | 0.002 | 0.002 | |
| conc | 0.828 | 0.829 | 0.822 | 0.824 | 0.814 | 0.818 | 0.788 | 0.763 | |
|
| |||||||||
| mtlr-I | nll | 1.616 | 1.626 | 1.612 | 1.612 | 1.632 | 1.640 | 1.636 | 1.753 |
| d-cal | 0.003 | 0.003 | 0.002 | 0.001 | 0.001 | 0.001 | 9e-4 | 0.001 | |
| conc | 0.827 | 0.825 | 0.831 | 0.829 | 0.824 | 0.823 | 0.825 | 0.783 | |
Footnotes
Code is available at https://github.com/rajesh-lab/X-CAL
Contributor Information
Mark Goldstein, New York University.
Xintian Han, New York University.
Aahlad Puli, New York University.
Adler J. Perotte, Columbia University
Rajesh Ranganath, New York University.
References
- Alaa AM and van der Schaar M Deep multi-task gaussian processes for survival analysis with competing risks. In Proceedings of the 31st International Conference on Neural Information Processing Systems, pages 2326–2334. Curran Associates Inc., 2017. [Google Scholar]
- Andersen PK, Borgan O, Gill RD, and Keiding N Statistical models based on counting processes. Springer Science & Business Media, 2012. [Google Scholar]
- Avati A, Duan T, Zhou S, Jung K, Shah NH, and Ng AY Countdown regression: Sharp and calibrated survival predictions. In Globerson A and Silva R, editors, Proceedings of the Thirty-Fifth Conference on Uncertainty in Artificial Intelligence, UAI 2019, Tel Aviv, Israel, July 22–25, 2019, page 28. AUAI Press, 2019. URL http://auai.org/uai2019/proceedings/papers/28.pdf. [Google Scholar]
- Barber RF, Candes EJ, Ramdas A, and Tibshirani RJ The limits of distribution-free conditional predictive inference. arXiv preprint arXiv:1903.04684, 2019. [Google Scholar]
- Besseling J, Reitsma JB, Gaudet D, Brisson D, Kastelein JJ, Hovingh GK, and Hutten BA Selection of individuals for genetic testing for familial hypercholesterolaemia: development and external validation of a prediction model for the presence of a mutation causing familial hypercholesterolaemia. European heart journal, 38(8):565–573, 2017. [DOI] [PubMed] [Google Scholar]
- Brier GW and Allen RA Verification of weather forecasts. In Compendium of meteorology, pages 841–848. Springer, 1951. [Google Scholar]
- Cox DR Two further applications of a model for binary regression. Biometrika, 45(3/4):562–565, 1958. [Google Scholar]
- D’agostino R and Nam B-H Evaluation of the performance of survival analysis models: discrimination and calibration measures. Handbook of statistics, 23:1–25, 2003. [Google Scholar]
- Davidson-Pilon C, Kalderstam J, Jacobson N, Zivich P, Kuhn B, Williamson M, Moncada-Torres A, Stark K, Anton S, Noorbakhsh J, et al. Camdavidsonpilon/lifelines: v0.24.0. Context, 604(40F), 2017. [Google Scholar]
- Demler OV, Paynter NP, and Cook NR Tests of calibration and goodness-of-fit in the survival setting. Statistics in medicine, 34(10):1659–1680, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George B, Seals S, and Aban I Survival analysis and regression models. Journal of nuclear cardiology, 21(4):686–694, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerds TA and Schumacher M Consistent estimation of the expected brier score in general survival models with right-censored event times. Biometrical Journal, 48(6):1029–1040, 2006. [DOI] [PubMed] [Google Scholar]
- Giunchiglia E, Nemchenko A, and van der Schaar M Rnn-surv: A deep recurrent model for survival analysis. In International Conference on Artificial Neural Networks, pages 23–32. Springer, 2018. [Google Scholar]
- Graf E, Schmoor C, Sauerbrei W, and Schumacher M Assessment and comparison of prognostic classification schemes for survival data. Statistics in medicine, 18(17–18):2529–2545, 1999. [DOI] [PubMed] [Google Scholar]
- Grønnesby JK and Borgan Ø A method for checking regression models in survival analysis based on the risk score. Lifetime data analysis, 2(4):315–328, 1996. [DOI] [PubMed] [Google Scholar]
- Guo C, Pleiss G, Sun Y, and Weinberger KQ On calibration of modern neural networks. In Proceedings of the 34th International Conference on Machine Learning-Volume 70, pages 1321–1330. JMLR.org, 2017. [Google Scholar]
- Haider H, Hoehn B, Davis S, and Greiner R Effective ways to build and evaluate individual survival distributions. Journal of Machine Learning Research, 21(85):1–63, 2020.34305477 [Google Scholar]
- Harrell FE Jr, Lee KL, and Mark DB Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics in medicine, 15(4):361–387, 1996. [DOI] [PubMed] [Google Scholar]
- Harutyunyan H, Khachatrian H, Kale DC, Steeg GV, and Galstyan A Multitask learning and benchmarking with clinical time series data. arXiv preprint arXiv:1703.07771, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz DG and Thompson DJ A generalization of sampling without replacement from a finite universe. Journal of the American statistical Association, 47(260):663–685, 1952. [Google Scholar]
- Johnson AE, Pollard TJ, Shen L, Li-wei HL, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, and Mark RG Mimic-iii, a freely accessible critical care database. Scientific data, 3: 160035, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbfleisch JD and Prentice RL The Statistical Analysis of Failure Time Data. Wiley Series in Probability and Statistics. John Wiley & Sons, Inc., 2 edition, 2002. [Google Scholar]
- Kaplan EL and Meier P Nonparametric estimation from incomplete observations. Journal of the American statistical association, 53(282):457–481, 1958. [Google Scholar]
- Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, and Kluger Y Deepsurv: personalized treatment recommender system using a cox proportional hazards deep neural network. BMC medical research methodology, 18(1):24, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Sarawagi S, and Jain U Trainable calibration measures for neural networks from kernel mean embeddings. In International Conference on Machine Learning, pages 2805–2814, 2018. [Google Scholar]
- Kvamme H and Borgan Ø The brier score under administrative censoring: Problems and solutions. arXiv preprint arXiv:1912.08581, 2019. [Google Scholar]
- Lawless JF Statistical Models and Methods for Lifetime Data. Wiley Series in Probability and Statistics. John Wiley & Sons, Inc., 2 edition, 2011. [Google Scholar]
- LeCun Y, Cortes C, and Burges C Mnist handwritten digit database. ATT Labs; [Online]. Available: http://yann.lecun.com/exdb/mnist, 2, 2010. [Google Scholar]
- Lee C, Zame WR, Yoon J, and van der Schaar M Deephit: A deep learning approach to survival analysis with competing risks. In Thirty-Second AAAI Conference on Artificial Intelligence, 2018. [Google Scholar]
- Lemeshow S and Hosmer DW Jr. A review of goodness of fit statistics for use in the development of logistic regression models. American journal of epidemiology, 115(1):92–106, 1982. [DOI] [PubMed] [Google Scholar]
- Liu E Using weibull accelerated failure time regression model to predict survival time and life expectancy. BioRxiv, page 362186, 2018. [Google Scholar]
- Matheson JE and Winkler RL Scoring rules for continuous probability distributions. Management science, 22(10):1087–1096, 1976. [Google Scholar]
- Miscouridou X, Perotte A, Elhadad N, and Ranganath R Deep survival analysis: Nonparametrics and missingness. In Machine Learning for Healthcare Conference, pages 244–256, 2018. [Google Scholar]
- Network CGAR Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. New England Journal of Medicine, 372(26):2481–2498, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu-Mizil A and Caruana R Predicting good probabilities with supervised learning. In Proceedings of the 22nd international conference on Machine learning, pages 625–632, 2005. [Google Scholar]
- Pepe M and Janes H Methods for evaluating prediction performance of biomarkers and tests. In Risk assessment and evaluation of predictions, pages 107–142. Springer, 2013. [Google Scholar]
- Platt JC Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. In ADVANCES IN LARGE MARGIN CLASSIFIERS, pages 61–74. MIT Press, 1999. [Google Scholar]
- Pölsterl S Sebastian pölsterl, Jul 2019. URL https://k-d-w.org/blog/2019/07/survival-analysis-for-deep-learning/. [Google Scholar]
- Ranganath R, Perotte A, Elhadad N, and Blei D Deep survival analysis. arXiv preprint arXiv:1608.02158, 2016. [Google Scholar]
- Ren K, Qin J, Zheng L, Yang Z, Zhang W, Qiu L, and Yu Y Deep recurrent survival analysis. In Proceedings of the AAAI Conference on Artificial Intelligence, volume 33, pages 4798–4805, 2019. [Google Scholar]
- Royston P and Altman DG External validation of a cox prognostic model: principles and methods. BMC medical research methodology, 13(1):33, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Diethe T, Kull M, and Flach P Distribution calibration for regression. In International Conference on Machine Learning, pages 5897–5906, 2019. [Google Scholar]
- Stevens RJ and Poppe KK Validation of clinical prediction models: what does the “calibration slope” really measure? Journal of clinical epidemiology, 118:93–99, 2020. [DOI] [PubMed] [Google Scholar]
- Sullivan LM, Massaro JM, and D’Agostino RB Sr. Presentation of multivariate data for clinical use: The framingham study risk score functions. Statistics in medicine, 23(10):1631–1660, 2004. [DOI] [PubMed] [Google Scholar]
- Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, and Kannel WB General cardiovascular risk profile for use in primary care. 2008. [DOI] [PubMed] [Google Scholar]
- Vovk V, Gammerman A, and Shafer G Algorithmic learning in a random world. Springer Science & Business Media, 2005. [Google Scholar]
- Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, and Kannel WB Prediction of coronary heart disease using risk factor categories. Circulation, 97(18):1837–1847, 1998. [DOI] [PubMed] [Google Scholar]
- Yadlowsky S, Basu S, and Tian L A calibration metric for risk scores with survival data. In Machine Learning for Healthcare Conference, pages 424–450, 2019. [Google Scholar]
- Yu C-N, Greiner R, Lin H-C, and Baracos V Learning patient-specific cancer survival distributions as a sequence of dependent regressors. In Advances in Neural Information Processing Systems, pages 1845–1853, 2011. [Google Scholar]
- Zadrozny B and Elkan C Transforming classifier scores into accurate multiclass probability estimates. In Proceedings of the eighth ACM SIGKDD international conference on Knowledge discovery and data mining, pages 694–699, 2002. [Google Scholar]


